Abstract
Fungal highly reducing polyketide synthases (HRPKSs) biosynthesize polyketides using a single set of domains iteratively. Product release is a critical step in HRPKS function to ensure timely termination and enzyme turnover. Nearly all of the HRPKSs characterized to date employ a separate thioesterase (TE) or acyltransferase enzyme for product release. In this study, we characterized two fungal HRPKSs that have fused C-terminal TE domains, a new domain architecture for fungal HRPKSs. We showed that both HRPKS-TEs synthesize aminoacylated polyketides in an ATP-independent fashion. The KU42 TE domain selects cysteine and homocysteine, and catalyzes transthioesterification using the side chain thiol group as the nucleophile. In contrast, the KU43 TE domain selects leucine methyl ester and performs a direct amidation of the polyketide, a reaction typically catalyzed by nonribosomal peptide synthetase (NRPS) domains. The characterization of these HRPKS-TE enzymes showcase the functional diversity of HRPKS enzymes, and provides potential TE domains as biocatalytic tools to diversify HRPKS structures.
Keywords: Natural product, cysteine, aminoacylation, templated biosynthesis
Graphical Abstract

INTRODUCTION
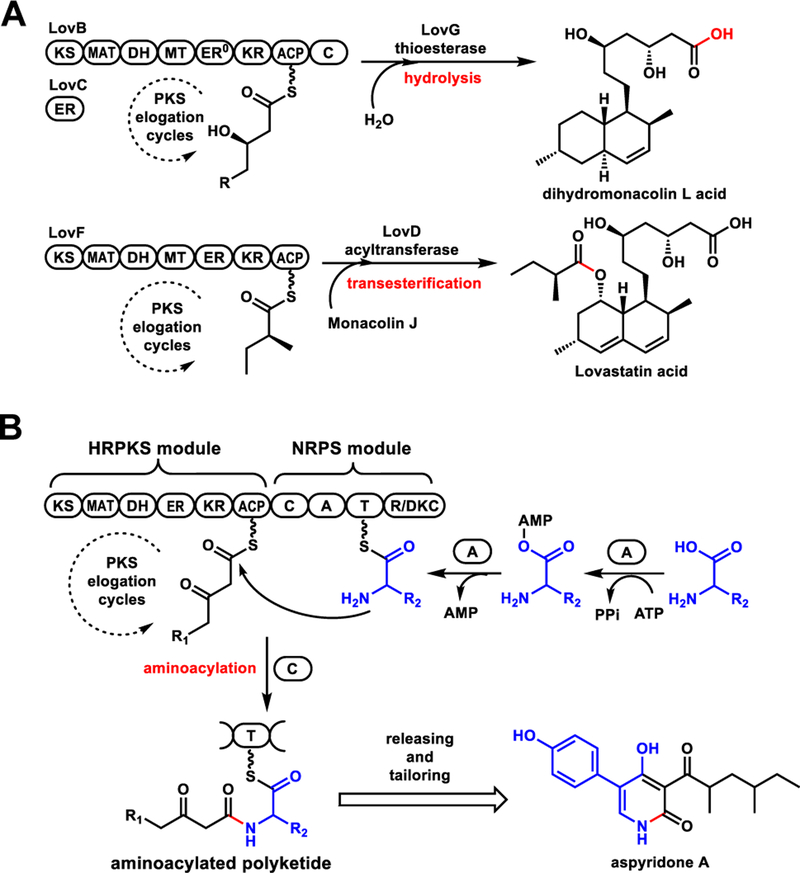
In thiol-templated biosynthesis catalyzed by polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs), product release is an essential step to ensure enzyme turnover. In bacterial PKSs where multidomain modules are arranged in an assembly line like fashion, C-terminal thioesterase (TE) domains are used to release the polyketide product via hydrolysis or macrocyclization.1 Fungal highly reducing PKSs (HRPKSs)2, on the other hand, consist of a single set of catalytic domains that is used iteratively. The domain architecture of fungal HRPKSs resembles a single module of bacterial type I modular PKSs. In contrast to bacterial product release strategies, many fungal HRPKSs studied to date use a separate, dissociated TE enzyme or acyltransferase to release the polyketide chain through hydrolysis or transesterification (Figure 1A).3 Both examples are represented in the lovastatin biosynthetic pathway:4 the TE LovG hydrolyzes dihydromonacolin L acid from LovB; while the acyltransferase LovD transfers α-methylbutyrate from LovF to monacolin L acid and afford lovastatin acid(Figure 1A). In tandem PKS systems, product of the HRPKS can be offloaded by the N-terminal starter unit–ACP transacylase (SAT) domain of a nonreducing PKS (NRPKS).5 Other modes of product release from HRPKS not catalyzed by TE have been reported in several recent studies.6
Figure 1.
Known product releasing mechanisms of fungal HRPKSs. (A) Product release is catalyzed by a separate TE or acyltransferase through hydrolysis or transesterification; (B) C domain serves as the releasing domain in the biosynthesis of fungal aminoacylated polyketide products. Domain abbreviations: KS, ketosynthase; MAT, malonyl-CoA:ACP transacylase; DH, dehydratase; MT, methyltransferase; ER, enoyl reductase; KR, ketoreductase; ACP, acyl carrier protein; C, condensation; A, adenylation; T, thiolation; R, reductase.
Many fungal polyketides are aminoacylated and further modified to have significant biological activities, exemplified by cytochalasin E (antiangiogensis),7 pseurotin A (immunosuppressant)8 and UCS1025A (anticancer).9 These natural products are synthesized by HRPKS-NRPS hybrid enzymes.10 The adenylation domain in the C-terminal NRPS module uses one ATP molecule to activate a specific amino acid followed by acyl transfer to the thiolation (T) domain. The condensation domain (C) then serves as a HRPKS releasing domain in principle, by transferring the completed polyketide acyl chain from the ACP domain of HRPKS module to the T domain in the NRPS module as an aminoacylated polyketide, and forming an amide bond (Figure 1B).10
The iterative nature of fungal HRPKSs dictates that a single set of domains is used repeatedly during elongation and tailoring of the polyketide chain. Hence, the releasing domain/enzyme is necessary to ensure that the correct polyketide product, once formed, is released promptly. Previous studies showed that in the absence of releasing domains, the HRPKS comes to a halt due to occupancy of the ACP domain,11 or functions aberrantly to yield products of incorrect sizes.12 Therefore, understanding the different mechanisms of product release is a key aspect of decoding HRPKS programming rules. Genome mining of fungal HRPKSs with an emphasis on finding unusual product release domains can lead to discovery of new products. For example, we previously identified a clade of HRPKSs that are terminated with a C-terminal domain with sequence homology to carnitine O-acyltransferase (cAT).13 The reversible nature of the cAT was a key feature that enabled complete execution of Tv6–931 programming steps.
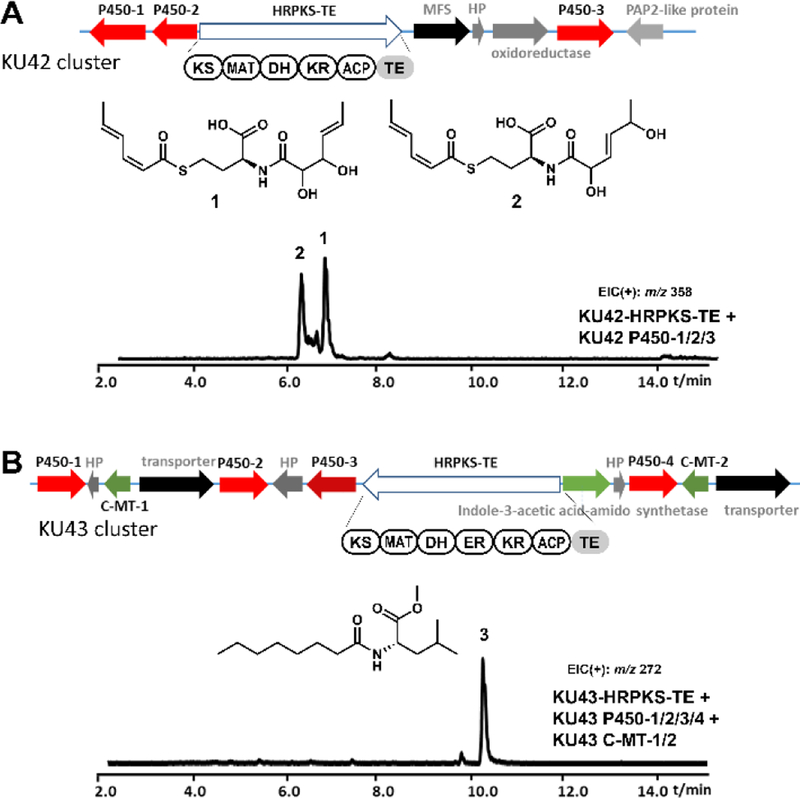
Recently, our genome mining efforts identified a group of fungal HRPKSs with fused terminal TE domains that differ from all characterized HRPKSs to date. These architecturally distinct HRPKSs had no known associated products.14 Two such gene clusters, KU42 and KU43, mined from the genomes of basidiomycete fungal species, Punctularia strigosozonata and Hydnomerulius pinastri, respectively, contain genes encoding HRPKS-TE belonging to this family (Figure 2).14 Bioinformatic analysis indicated that within the KU42 cluster, genes encoding an HRPKS-TE, three P450s, an oxidoreductase, a PAP2-like protein, a hypothetical protein, and a major facilitator superfamily transporter (Figure 2A) are found. When KU42 HRPKS-TE and the three P450s were introduced together into engineered yeast strain RC01, two aminoacylated products (1 and 2, as shown in Figure 2A) were identified.14 In parallel, genes from KU43 clusters encode an HRPKS-TE, four P450s, two carboxyl methyltransferases (C-MTs), and others (Figure 2B). The expression of the KU43 HRPKS-TE together with the four P450s and two C-MTs in engineered yeast strains resulted in the production of an aminoacylated polyketide 3 (Figure 2B).14 The structures of aminoacylated polyketides were surprising based on known fungal HRPKS programming rules, since enzymes responsible for amino acid activation and incorporation, such as NRPS or CoA-ligase, are absent in the two gene clusters.
Figure 2.
Genome mining of fungal secondary metabolite biosynthetic gene clusters containing a gene encoding HRPKS-TE megasynthase. (A) KU42 cluster mined from P. strigosozonata and the production of 1 and 2 in yeast. (B) KU43 cluster mined from H. pinastri and the production of 3 in yeast. HP, hypothetical protein; PAP2, type 2 phosphatidic acid phosphatase; MFS, major facilitator superfamily; C-MT, carboxyl methyltransferase. Shown are the traces of extracted-ion chromatogram. In control traces from strains transformed with vector only, these compounds were not found.
Here we studied the biosynthetic mechanisms of these HRPKS-TE enzymes and showed the amino acids are directly incorporated by the fused TE domains. In the case of KU42, we demonstrate the TE domain can catalyze a transthioesterification with thiol containing amino acids followed by a S-N shift to form the amide bond.
RESULTS AND DISCUSSIONS
Sequence analysis of the KU42 TE domain.
1 and 2 are closely related compounds containing a S, N-diacylated homocysteine. The acyl groups appear to derive from the same dienoyl triketide precursor. In the absence of any apparent amino acid activation domain, we hypothesized that 1 and 2 were formed by the KU42 HRPKS-TE domain, and homocysteine is used to release the polyketide chains. The amino and the thiol groups of homocysteine serve as nucleophiles to perform aminoacylation and thiolation, respectively, to form the diacylated adduct. We propose the amino acid specificity is likely controlled by the TE domain. Sequence analysis revealed that the TE domain, like many other PKS TE domains, is a member of the serine α/β hydrolase domain superfamily.15 The TE domain contains a cysteine residue (C2040) instead of serine as the conserved active site nucleophile, while the other two residues of the conserved catalytic triad, D2056 and H2225, have the usual identity (Figure S1).1b, 16
Homology searches against predicted fungal protein sequences revealed that similar TE domains are encoded on a diverse set of fungal genomes. Of 500 BLAST hits retrieved (with E values between 2e-41 and 0.127), thirty-seven coded for cysteine as the active site nucleophile, while 452 had serine (seven coded for alanine at this position; three other sequences had other residues or alignment gaps in the conserved position). The occurrence of cysteine or serine in this key active site position was independent of phylogeny (Figure S2), suggesting that mutation to cysteine has occurred multiple times in the evolution of this domain. Cysteine active site nucleophiles have been observed in some starter-unit:ACP transacylase domains in fungal nonreducing PKSs, although the biochemical reason for this substitution over serine is unclear.17 A recent report from the Wencewicz group reported a cysteine nucleophile in a TE domain is required for cyclization of the β-lactone product obafluorin.18 It was proposed that the thioester intermediate is at a higher ground state energy compared to the oxyester, and thus is favorable in driving β-lactone formation.18
Functional characterization of KU42-HRPKS-TE.
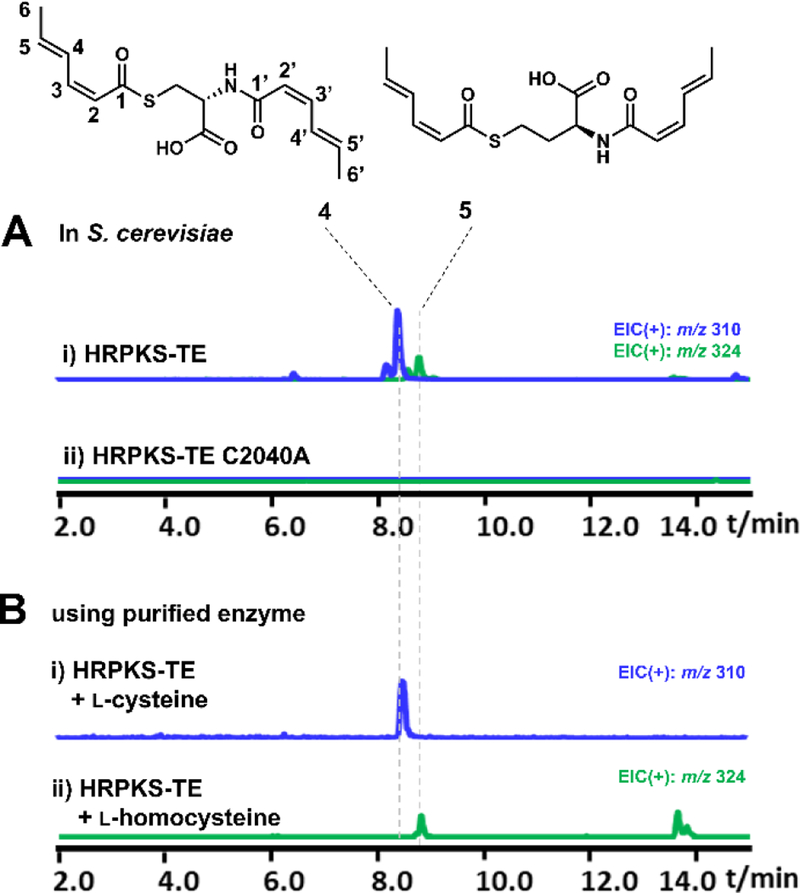
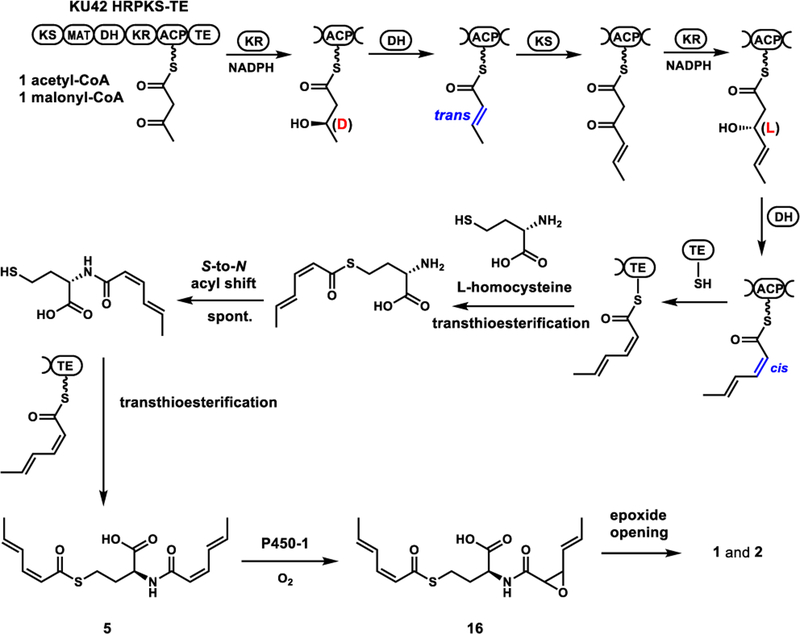
To identify the product synthesized by KU42-HRPKS-TE alone, the gene was cloned under ADH2 promoter on a 2μ plasmid and transformed into the engineered S. cerevisiae strain BJ5464-NpgA.11 After four days of culturing followed by extraction with ethyl acetate (containing 1% acetic acid), we observed accumulation of several new metabolites (Figure 3A). The major product is 4 with molecular weight (MW) of 309, and the minor product is 5 with MW of 323. These compounds were isolated and characterized by NMR (Table S3 and S4, Figure S12–S21). Compound 4 was elucidated to be disorbyl cysteine, in which both of the thiol group and the amine group of cysteine were acylated with sorbic acid (Figure 3A). Based on the value of proton coupling constants obtained from 1H NMR spectrum (J=11.2 Hz for H2-H3 and H2’-H3’, 14.0 Hz for H4-H5 and H4’-H5’) and published literature values for sorbate with differently olefin stereochemistry,19 the double bond configurations in 4 were assigned to be 2-cis-4-trans-sorbyl. Similar to 4, the structure of 5 was also elucidated in which the l-cysteine unit in compound 4 was replaced by l-homocysteine (Figure 3A). 5 is therefore the likely oxidized to 1 and 2 when the P450 enzymes were coexpressed in yeast (Figure 2A).
Figure 3.
Characterization of KU42 HRPKS-TE in vivo and in vitro. Shown are the extracted-ion chromatogram. (A) Expression of HRPKS-TE and C2040A mutant in yeast and the structures of the compounds produced; (B) LC-MS analysis of the in vitro assays of purified HRPKS-TE in the presence of malonyl-CoA, NADPH and l-cysteine or l-homocysteine.
In order to confirm that KU42 HRPKS-TE alone is sufficient for the biosynthesis of 4 and 5, recombinant enzyme (246.2 kDa) was purified from yeast to homogeneity by anti-FLAG resin column chromatography (Figure S3). Assay of the enzyme in the presence of malonyl-CoA, NADPH, and l-cysteine yielded 4 as expected (Figure 3B). When l-cysteine was replaced by l-homocysteine, the production of 5 was confirmed (Figure 3B). When the enzyme was assayed in the presence of equimolar amount of l-cysteine and l-homocysteine (1 mM), the production of both 4 and 5 was detected with 4 as the predominant product (Figure S4). ATP was not necessary in these assays to activate the amino acids, as expected since there is no adenylation domain in the enzyme. The collective in vivo and in vitro assays confirm the HRPKS-TE alone is able to add two molecules of the polyketide product onto either cysteine (preferred substrate) or homocysteine, forming the S, N-diacylated products without the use of a condensation or adenylation domain.
KU42 HRPKS-TE substrate scope and proposed reaction mechanism.
We then investigated the function of TE domain in KU42 HRPKS-TE. We first constructed the TE active site mutant C2040A, in which the putative cysteine nucleophile was mutated into alanine. When transformed into yeast, the mutant lost the ability to biosynthesize 4 and 5 (Figure 3A), indicating that both polyketide chain release and amino acid incorporation require the cysteine nucleophile. Mutation of the putative general base H2225 to either Ala or Asn similarly abolished the function of the enzyme. When the active site nucleophile was changed from cysteine to serine (C2040S) and expressed in yeast, we also did not observe formation of either 4 or 5. We further subjected these culture supernatants to untargeted metabolite analysis using XCMS (Figure S5).20 This analysis revealed that the C2040S mutant, but not C2040A or H2225A, produced an unique compound not produced by wild type HRPKS-TE, and has (ion) mass of 113.0597 Da, corresponding to the molecular formula of sorbic acid (C6H8O2) plus a proton. This peak indicates the C2040S mutant retained the ability to synthesize the polyketide product, but is unable to perform aminoacylation or thiolation. Instead, the product appears to be prone to hydrolysis, analogous to the cysteine to serine mutation observed in obafluorin TE domain.18 Although it is not clear whether such hydrolysis occurs as a S2040-sorbyl oxyster, or direct hydrolysis from the ACP, this result confirms the necessity of the cysteine as an active site nucleophile.
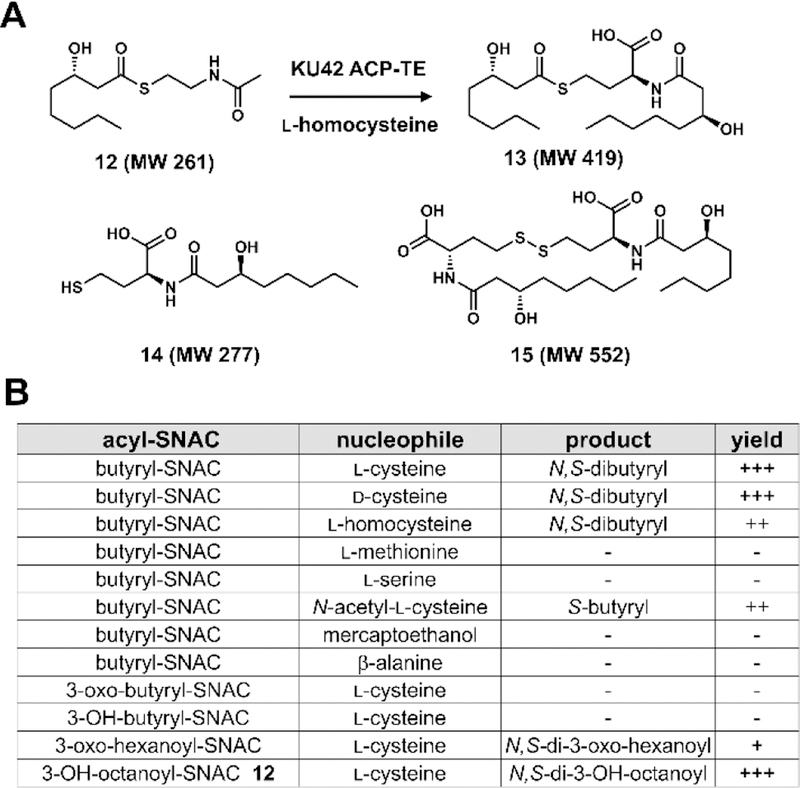
We attempted to express and purify the standalone TE domain from E. coli BL21(DE3), but failed to obtain soluble protein. We were successful in expression of the TE domain together with upstream ACP domain, ACP-TE, as N-terminal His6 tagged fusion protein (Figure S3). To test the function of ACP-TE, acyl-S-N-acetyl cysteamine (SNAC) compounds were added to the enzyme and analyzed by LC-MS to determine whether one or two acyl groups are transferred to the amino acid nucleophile. Due to instability of sorbyl-SNAC, we used an available acyl-SNAC, (S)-3-hydroxyoctanoyl-SNAC (12)21, as a substrate analog (Figure 4A). Compared to the negative control, we were able to detect formation of three different products (Figure S6): one has the MW of 419, which is expected for the diacylated product 13; another product has the MW of 277, which corresponds to the MW of the monoacylated product 14; the third compound has the MW of 552, which corresponds to that of 15, a dimer of 14 formed though oxidative coupling of free thiol groups to form a disulfide (Figure 4A). When tris-(2-carboxyethyl)phosphine (TCEP) was included in the assay, formation of 15 was no longer detected and only 13 and 14 were formed. This supports the proposed relationship of 14 and 15. Together with the mutagenesis results, these results confirmed that the TE domain is indeed responsible for the acylation reactions.
Figure 4.
Assaying KU42 TE domain with different acyl and amino acid substrates reveal biochemical mechanism. (A) Product analysis of the in vitro assays of KU42 ACP-TE in the presence of l-homocysteine and 12. In the negative control, boiled enzyme was used. (B) Summary of products formed by using different amino acids and acyl-SNAC substrates.
The substrate scope of the TE domain was further assayed using different acyl-SNAC and amino acid substrates (Figure 4B). When l-cysteine was used as the nucleophile, several acyl-SNAC substrates with different acyl chain length (from C4 to C8) could be used as the acyl donors by TE domain. Among the C4 substrates, only butyryl-SNAC was accepted by the TE domain (Figure S6). Using butyryl-SNAC as the acyl donor, different amino acid nucleophiles were tested for formation of mono- and dibutyryl adducts. One key trend that emerged, which is consistent with amino acid incorporated into 4 and 5, is that amino acids without a free thiol group cannot be used as nucleophiles, such as l-serine and l-methionine. Interestingly, d-cysteine can also be accepted by the TE domain as nucleophile to form the dibutyryl-d-cysteine product. Importantly, when N-acetyl-l-cysteine was used as the nucleophile, we observed formation of a monobutyryl adduct by LC-MS. This product, unlike 14, does not undergo air oxidation to form a dimer, suggesting that the free thiol group is acylated by the butyryl unit.
From these in vitro assay results of TE domain with different substrates, a possible catalytic mechanism for the formation of diacylated products 4 and 5 can be proposed. We propose the TE domain is a bona fide acyltransferase that can catalyze the transthioesterification between the side chain thiol nucleophile and the sorbyl-S-ACP thioester to release the product. When l-homocysteine is used as the releasing nucleophile, TE domain would catalyze the transthioesterification reaction to yield S-sorbyl homocysteine. This is followed by a spontaneous intramolecular S-to-N acyl shift to form the N-sorbyl homocysteine to regenerate a free thiol group. This type of uncatalyzed rearrangement reaction is widely used in native chemical ligation to synthesize peptides and small proteins.22 With the thiol group now free to react, the TE domain catalyzes the second transthioesterification reaction of N-sorbyl homocysteine to form the disorbyl homocysteine 5 (Figure 6). Therefore, the TE domain does not catalyze direct aminoacylation, but instead catalyzes the unprecedented release of polyketide products using the side chain thiol of cysteine or homocysteine.
Figure 6.
The proposed biosynthetic pathway of 5.
Delineating the stereoselectivity of KR and DH domains of KU42 HRPKS-TE in the synthesis of 2-cis-4-trans-sorbyl polyketide chain.
A surprise in the structures of 4 and 5 is that they contain both cis- and trans-double bonds. Several HRPKSs have been proposed to biosynthesize polyketides with both cis and trans olefins, such as in the sordarin pathway.23 Cox and coworkers recently reported StPKS1 from Strobilurus tenacellus can produce a polyketide containing both cis- and trans-olefins.24 However, the programming rules that govern the formation of mixed olefins from a single set of KR and DH domains are unknown. A working hypothesis is that the KR domain reduces the β-keto functionality with opposite stereochemistry after the two chain elongation steps. The two oppositely configured alcohol substrates binds in the DH active site differently for syn elimination, leading to either cis or trans products. We previously showed in the hypothemycin HRPKS, the stereochemistry (l or d) of KR-catalyzed β-ketoreduction is chain-length dependent.25 It is also well documented that stereochemistry of the reduced alcohol can affect the stereoselectivity of the DH-catalyzed dehydration reaction.26
To test this hypothesis, we first verified the HRPKS product is indeed the 2-cis-4-trans sorbyl chain, and the TE domain is not involved in trans-cis isomerization of the second olefin. We constructed a truncated version of KU42 HRPKS-TE lacking the TE domain (HRPKS ΔTE). Assay of the truncated enzyme in the presence of only malonyl-CoA yielded triacetic acid lactone (Figure S7), confirming the elongation domains are active. Addition of NADPH led to no detectable polyketide products, since there is no releasing domain (Figure S7). To enable release, we added a heterologous TE domain from an unrelated PKS13, which was previously demonstrated to have promiscuous substrate specificity in ACP-thioester hydrolysis,27 into the reaction mixture. Under this condition, we observed turnover of a new product that had the similar UV spectrum with 2-trans-4-trans-sorbic acid, but different retention time (Figure S7). We synthesized an authentic standard of 2-cis-4-trans-sorbic acid (Figure S28), which matched that released from the enzymatic assay. This confirmed the polyketide produced by KU42 HRPKS is indeed the 2-cis-4-trans-sorbate, and no post-PKS isomerization of a trans-trans product is required.
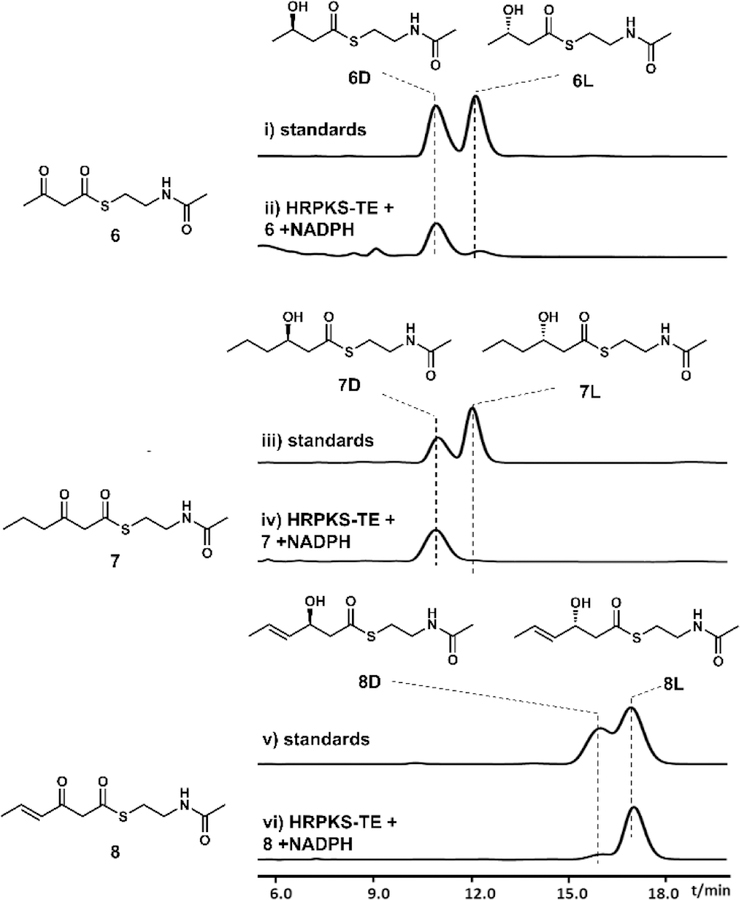
We next examined the stereoselectivity of the KR domain towards β-keto substrates of different sizes. We assayed the HRPKS-TE with three different β-ketoacyl-SNAC compounds in the presence of NADPH, and analyzed the β-keto reduced products using chiral HPLC and synthetic standards (Figure 5). The diketide acetoacetyl-SNAC (6)25 and triketide 3-oxohexanoyl-SNAC (7)25 were both reduced to the corresponding β-hydroxyacyl-SNAC, and the retention time matched to that of the 6d and 7d (the d configuration of the hydroxyl products),25 respectively (Figure 5). Hence the KR specificity does not vary with chain length. We then assayed the product stereochemistry using 3-oxo-4-trans-hexenoyl-SNAC (8)28, of which the acyl chain is the natural triketide of the KR domain. Surprisingly, the product elution time matched that of 8l. To further verify the stereochemistry of this product, we scaled up the reaction and purified sufficient amounts of 8l by HPLC. The structure was verified by NMR analysis (Table S5, Figure S22 and S23) and the optical rotation ([α]D27 +13, c =0.2, CHCl3) matched to the reported value of 8l ([α]D25 +17, c =0.42, CH2Cl2).28 Our studies therefore revealed an unexpected layer of fungal HRPKS programming complexity, in that the stereoselectivity of the KR domain depends on the degree of unsaturation of the substrate. In this case, changing from a single bond in the substrate (in 7) to the double bond (in 8) completely reversed the stereochemical outcome. The stereochemical switch from diketide 6 (in d configuration) to triketide 8 (in l configuration) is most likely the cause for the subsequent different dehydration stereochemical outcomes.
Figure 5.
Biochemical characterization of the function of KU42-HRPKS-TE KR domain in vitro. Shown are the HPLC traces at 240 nm.
PKS DH domains generally catalyze the syn dehydration of their substrates.29 Cane and coworkers showed that in the biosynthesis of fostriecin, the double bond configurations produced by DH domains were determined by the stereo-configuration of the β-hydroxy group that controlled by KR domains.26b We therefore propose that because of the different stereochemistry of the β-hydroxyl group in 6d and 8l, the substrates will bind differently in the active site of the DH domain, leading to trans and cis geometry of the dehydrated olefin, respectively. When diketide substrate 6d was incubated with KU42 HRPKS-TE, we detected the formation of a new product 9 (MW 187) (Figure S8). The compound was purified from scaled-up in vitro assays and shown to be trans-crotonoyl-SNAC (Table S6, Figure S24 and S25). No dehydration product was detected when 6l was used as the substrate (Figure S8), indicating strict substrate specificity of the DH domain at the diketide stage. When 8l was tested as the substrate, we also detected the formation of a dehydration product 10 (MW 213) (Figure S8). However after purification from scaled-up in vitro assays, the product was shown to be 2-trans-4-trans-sorbyl-SNAC (Table S7, Figure S26 and S27), not the expected 2-cis-4-trans-sorbyl-SNAC. Assaying DH domains that produce cis-configured olefin has documented difficulties due to isomerization of the double bond that is α,β to the thioester.26d, 30 Similarly, we proposed that the formation of 2-trans-4-trans-sorbyl-SNAC is due to the rapid cis-to-trans isomerization of 2-cis-4-trans sorbyl product, which could be catalyzed by nucleophiles such as N-acetylcysteamine through a Michael addition and a retro-Michael addition (see Figure S9).31 N-acetylcysteamine itself could form through the decomposition of acyl-SNAC substrates. Although we were not able to prove directly the dehydration specificity of the DH domain in these experiments, our demonstration of unexpected KR programming rules and comparison to knowledge accumulated for the bacterial DH domains, can rationalize the formation of cis and trans olefin in the same polyketide product by a single fungal HRPKS (Figure 6).
Characterization of the roles of the three P450s from KU42 cluster.
Our results showed that the major product of KU42 HRPKS-TE when expressed in yeast or assayed in vitro is 4, the disorbyl-l-cysteine. However, we also showed that when KU42 HRPKS-TE was co-expressed with three P450s enzymes from KU42 cluster in yeast, the products were 1 and 2, which are derived from the homocysteine containing 5 (Figure 2A).13 The two hydroxyl groups in 1 and 2 are presumably formed from water-mediated opening of an epoxide precursor. To explore the roles of these P450s in the biosynthesis of 1 and 2, co-expression experiments of the HRPKS-TE and individual P450 in S. cerevisiae RC0132 were performed (Figure S10). Coexpression with either P450–2 or P450–3 did not lead to the formation of 1 and 2. When KU42 HRPKS was co-expressed with P450–1, both 1 and 2 could be detected (Figure S10). Therefore, we assign P450–1 to be an epoxidase that catalyzes the selective epoxidation of one of the cis olefin in 5 to form an unstable product 16, which can be readily hydrolyzed to form 1 and 2 under culturing condition (Figure 6). P450–1 appears to be selective for the homocysteine adduct 5, as no cysteine derived analogs of 1 and 2 can be detected in the culture.
The overall biosynthetic pathway to 1 and 2 from KU42 HRPKS-TE is shown in Figure 6. Formation of the 2-cis-4-trans-sorbyl polyketide chain is proposed to involve the programmed stereoselectivities of the KR domain, which in turn influences the dehydration stereochemistry. The TE domain then catalyzes two transthioesterification steps using either cysteine or homocysteine. The second transthioesterification follows an S-N rearrangement, to afford 4 or 5, respectively. While 4 does not appear to be further processed, 5 is selectively epoxidized by P450–1 to 16, which can be attacked by water to afford both 1 and 2. The exact natural product of the KU42 pathway is unknown and is subject to future studies.
Factors that influence cysteine to homocysteine incorporation levels.
Our in vitro experiments clearly indicated HRPKS-TE in fact prefers cysteine over homocysteine, yet based on in vivo reconstitution results with P450–1, homocysteine is the preferred substrate to yield the natural product of the pathway. Therefore, the in vivo ratio of cysteine to homocysteine should impact product distributions, and the roles of other enzymes in the cluster. To test this idea, we utilized CRISPRi-mediated gene repression, coupled to a targeted metabolomics readout. We integrated the KU42 HRPKS-TE gene into a yeast haploid strain, and mated this strain to 41 CRISPRi strains33 expressing different guide RNAs directed against one of 17 genes involved in homocysteine and cysteine biosynthetic pathways (Figure S11). The resulting diploid strains were then cultured in 96-well plates in the presence or absence of anhydrotetracycline (ATc; which induces guide RNA expression). The relative titers of cysteine-derived 4 and homocysteine-derived 5 in each well were measured via LC/MS. Among the genes tested, repression of CYS4, which encodes for cystathionine β-synthase that converts homocysteine to cystathionine (a cysteine precursor), was found to result in the greatest shift in relative abundance of each product. We observed a ~ 5x increase in the titer of 5 and a ~ 2x decrease in titer of 4 (Figure S11), for an overall ~10 fold change in the relative amounts of 5 and 4.
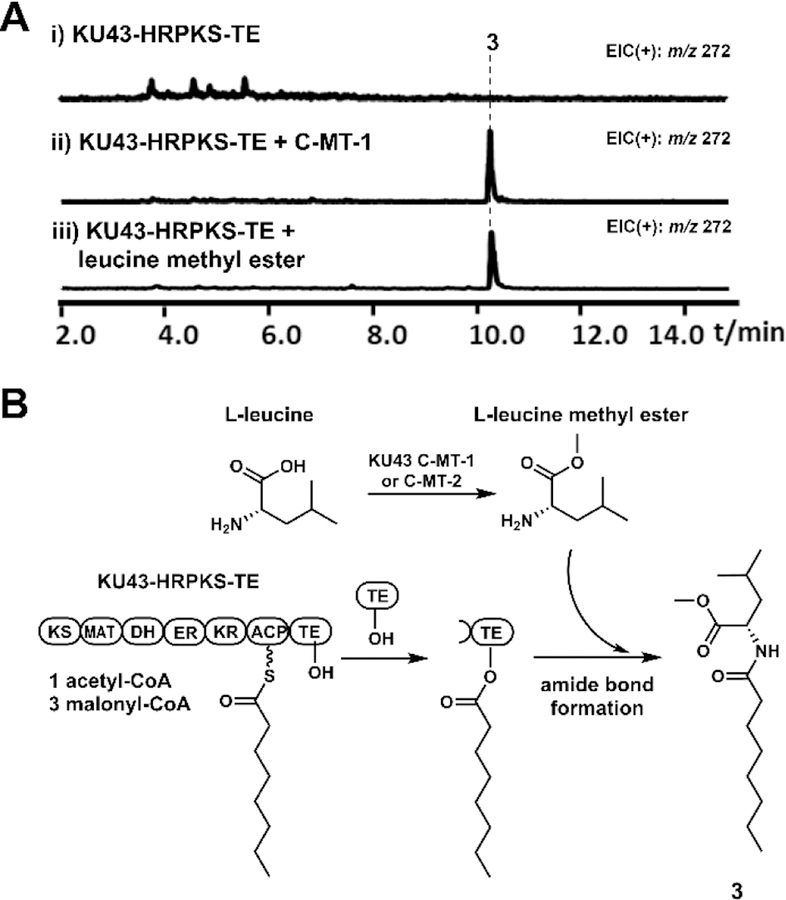
KU43-HRPKS-TE is responsible for the biosynthesis of 3.
Our studies above showed how the KU42 HRPKS-TE can synthesize disorbyl homocysteine products, using TE to catalyze transthioesterification and release the HRPKS product. In contrast, the product of KU43 HRPKS-TE, 3, is formed by a direct amidation between octanoate and leucine methyl ester (Figure 7). Analysis of KU43-HRPKS-TE indicated that it contains the domain organization of KS-MAT-DH-ER-KR-ACP-TE. We reasoned the TE domain could be responsible for the amidation reaction since no enzyme responsible for amino acid activation and incorporation is present in the KU43 gene cluster. Different from the KU42-HRPKS TE, sequence alignment results indicated that the active site of KU43-HRPKS TE is serine, the same as the canonical TEs (Figure S1).
Figure 7.
Biosynthesis of 3. (A) Product profiles of S. cerevisiae BJ5464-NpgA transformed with KU43 cluster genes. Shown are the extracted MS spectra. (B) The proposed biosynthetic pathway of 3.
Expression of HRPKS-TE alone in BJ5464-NpgA did not yield any new products (Figure 7A). When either C-MT-1 or C-MT-2 (Figure 2B), which are highly identical to each other (96% protein sequence identity), from the cluster was coexpressed with KU43 HRPKS-TE, we detected the production of 3 (Figure 7A). Based on these results, we proposed that C-MT-1 or C-MT-2 is responsible for the biosynthesis of l-leucine methyl ester, which could be used as the nucleophile by KU43 HRPKS-TE to release the octanoate chain biosynthesized by KU43-HRPKS (Figure 7B). To support this hypothesis, we repeated the heterologous expression experiment of KU43 HRPKS-TE with the addition of l-leucine methyl ester as the releasing substrate. As expected, the production of 3 was detected (Figure 7A). Based on these results, we proposed the TE domain of KU43 HRPKS-TE catalyzes polyketide chain release by using l-leucine methyl ester to yield 3 (Figure 7B). Different from the KU42 HRPKS TE, which uses cysteine as the active site nucleophile to catalyze the transthioesterification reaction to yield thiolated polyketide, the KU43 HRPKS-TE uses serine as the active site nucleophile to capture the polyketide product as an oxyester, followed by amide bond formation to yield the aminoacylated polyketide. This is in sharp contrast to the ATP-dependent aminoacylation catalyzed by NRPS domains. Requirement of the serine nucleophile in catalysis was verified by site-directed mutagenesis. Mutation of the active site serine of KU43-HRPKS TE to alanine resulted in no detectable production of 3 when the mutant HRPKS-TE was co-expressed with C-MT-1 in yeast (Figure S29).
Summary.
In this study, we characterized two fungal HRPKSs with fused C-terminal TE domains, a new domain architecture among fungal PKSs. Both KU42 and KU43 HRPKS-TEs produced natural products that have not been reported previously, showing the potential of studying novel HRPKS enzymes for natural product mining. We demonstrated that both TE domains are involved in product release to synthesize aminoacylated polyketides in an ATP-independent fashion. The reactions catalyzed by the two TE domains, however, are very different. The KU42 TE recognizes thiol containing amino acids, and uses the side chain thiol as nucleophile to perform chain release via transthioesterfication. The product can then undergo S-N acyl shift, followed by a second transthioesterification to produce the diacylated amino acid. Although the transthioesterification reactions are known for TE enzymes, the use of cysteine or homocysteine side chain thiol for polyketide product release is reported for the first time. In contrast, the KU43 TE recognizes leucine methyl ester and uses its amino group as nucleophile to release the polyketide chain produced by the HRPKS.
In conclusion, our results show that novel programming rules are present in HRPKS-TEs to biosynthesize different natural products. These TE domains, along with others that can be mined from uncharacterized biosynthetic pathways, can serve as potential biocatalytic tools for diversifying polyketide structures.
EXPERIMENTAL METHODS
Strains and General DNA Manipulation Techniques.
E. coli TOP 10 and E. coli DH10b were used for cloning, following standard recombinant DNA techniques. E. coli BL21(DE3) was used for protein expression. Saccharomyces cerevisiae strain BJ5464-NpgA11 (MATα ura3−52 his3-Δ200 leu2- Δ1 trp1 pep4::HIS3prb1 Δ1.6R can1 GAL) and RC0132 were used as the yeast expression hosts. Polymerase chain reactions were performed using Platinum Pfx DNA polymerase (Invitrogen). DNA restriction enzymes were used as recommended by the manufacturer (New England Biolabs). DNA primers used in this study were synthesized by Integrated DNA Technologies and are listed in Supporting Information, Table S1. PCR products were confirmed by DNA sequencing by Laragen, California, USA.
Compound Isolation and Purification.
To isolate compound 4 and 5, a seed culture of the BJ5464-NpgA strain harboring the expression plasmid of KU42-HRPKS-TE was grown in 40 mL SDCt (uracil dropout) media at 28 °C. After 2 days, this seed culture was then used to inoculate 4 L of YPD medium and cultured for 3 days at 28 °C. The cell pellets was removed by centrifugation and the supernatant was collected and combined. The pH value of the supernatant was adjusted to 3 by adding concentrated HCl. Then the supernatant was extracted with EtOAc twice and the organic layer was combined and concentrated. The crude residue was injected to ISCO-CombiFlash® Rf 200 (Teledyne Isco, Inc) with silico 40g column chromatography for separation. Fractions containing the target compounds were combined, and the target compounds were further purified by semi-preparative HPLC using reverse-phase column (Phenomenex Kinetics, C18, 5 μm, 100 Å, 10 × 250 mm). The titers of compound 4 and 5 are ~1 mg/mL and ~0.3 mg/mL, respectively.
Expression and Purification of KU42-HRPKS-TE and Its Mutants.
For protein expression of KU42-HRPKS-TE and its mutants in yeast strain BJ5464-NpgA, the corresponding genes were cloned into pXW55 vector by yeast in vivo homologous recombination using Frozen-EZ Yeast Transformation II Kit™ (Zymo Research). Single colonies of S. cerevisiae BJ5464-NpgA cells that had been transformed with the expression plasmids were inoculated into 20 mL SDCt (uracil dropout) media and incubated at 28 °C for 2 days. Then, the starter culture was inoculated into 2 L YPD media and incubated at 28 °C. After 2 days, the cell pellets were harvested and suspended in lysis buffer (50 mM NaH2PO4, 150 mM NaCl, 10 mM imidazole, pH 8.0). The cells were lysed by sonication and the cellular debris was removed by high-speed centrifugation (16000 rpm, 1 hour). The enzyme was purified from the supernatant by using anti-FLAG resin column. The fractions containing the target protein were pooled, buffer-exchanged, and concentrated to final buffer (50 mM HEPES, 50 mM NaCl, 10% glycerol, pH 7.0) using an Amicon Ultra-15 (100 kDa) centrifugal filter. Aliquots of the purified proteins were stored at −80 °C. Protein concentration was determined by the Bradford method34 using a BSA calibration curve. The yield of purified KU42-HRPKS-TE is ~2 mg/mL.
Expression and purification of KU42 ACP-TE.
The gene fragment encoding for KU42 ACP-TE was cloned into pET28a between the NdeI and XhoI restriction sites. Single colonies of E. coli BL21(DE3) cells that had been transformed with the expression plasmid was inoculated into 3 mL LB media containing 50 mg/L kanamycin and incubated overnight at 37 °C. Then, the starter culture was inoculated into 500 mL LB media containing 50 mg/L kanamycin, and grew at 37 °C. When the OD600 reached 0.6, the culture was cooled to 16 °C and induced with 0.1 mM IPTG. Cells were harvested after 24 hours by centrifugation. The purification was then carried out at 4 °C. After re-suspended in lysis buffer (50 mM NaH2PO4, 150 mM NaCl, 10 mM imidazole, pH 8.0), the cells were lysed by sonication. And the cellular debris was removed by high-speed centrifugation (16000 rpm, 30 min). Then, the enzyme was purified from the supernatant by using Ni-NTA agarose affinity chromatography to near homogeneity. The purified protein was buffer-exchanged and concentrated to final buffer (50 mM HEPES, 50 mM NaCl, 10% glycerol, pH 7.0). Aliquots of the purified protein were stored at −80 °C. Protein concentration was determined by the Bradford method34 using a BSA calibration curve. The yield of purified KU42 ACP-TE is ~37 mg/mL.
Activity Assay for KU42-HRPKS-TE and Its Mutants.
For in vitro synthesis of 4, 10 μM KU42-HRPKS-TE was incubated with 1 mM l-cysteine, 2 mM malonyl-CoA and 4 mM NADPH in 100 mM HEPES (pH 7.0) buffer. The reaction mixture was incubated at 30 °C overnight. The reaction was quenched with equal volume of acetonitrile. After centrifugation, the supernatant was subjected for LC-MS analysis. For in vitro synthesis of 5, the assay was performed using the same condition except the l-cysteine was replaced by l-homocysteine. All the experiments were done in triplicate. In the manuscript, data collected from one set of experiments are shown.
For in vitro synthesis of 2-cis-4-trans-sorbic acid, 10 μM KU42 HRPKS ΔTE was incubated with 2 mM malonyl-CoA, 4 mM NADPH, and 5 μM PKS13-TE in 100 mM HEPES (pH 7.0) buffer. The reaction mixture was incubated at 30 °C overnight. The reaction was quenched with equal volume of acetonitrile. After centrifugation, the supernatant was subjected for LC-MS analysis.
Assay for KU42 ACP-TE Activity Using Different Amino Acid Substrates and Acyl-SNAC Substrates.
For in vitro activity test of KU42 ACP-TE, the following condition was used: the reaction mixture contains 50 μM KU42 ACP-TE, 1 mM amino acid substrate, and 2 mM acyl-SNAC substrate, and incubated at 30 °C for 6 hours. The reaction was quenched by adding equal volume of acetonitrile. After centrifugation, the supernatant was subjected for LC-MS analysis. When tested with different amino acid substrates, butyryl-SNAC was used as the acyl-SNAC substrate. When tested with different acyl-SNAC substrate, l-cysteine was used as the amino acid substrate. All the experiments were done in triplicate. In the manuscript, data collected from one set of experiments are shown.
Assay for the Functions of KU42 HRPKS KR Domain and DH Domain.
To assay the activity of KU42 HRPKS KR domain, 10 μM KU42-HRPKS-TE was incubated with 1 mM β-ketoacyl-SNAC and 2 mM NADPH at 30 °C for 4 hours. The reaction was quenched with equal volume of acetonitrile. After centrifugation, the supernatant was subjected for LC-MS analysis.
To assay the activity of KU42 HRPKS DH domain, 10 μM KU42-HRPKS-TE was incubated with 1 mM β-hydroxyacyl-SNAC at 30 °C for overnight. The reaction was quenched with equal volume of acetonitrile. After centrifugation, the supernatant was subjected for LC-MS analysis.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH 1 U01 GM110706-01 Genome to Natural Products Initiative to Stanford Genome Technology Center, of which YT is a subward recipient. The work in the lab of YT is also supported by 1R35GM118056. The work at Stanford University was also supported by R01GM121932-01A1 to RPS. Chemical characterization studies were supported by shared instrumentation grants from the NSF (CHE-1048804) and the NIH NCRR (S10RR025631). The authors acknowledge Maureen Hillenmeyer, Colin Harvey, Marnix Medema, Mohammad Alanjary, and Elizabeth Sattely for valuable technical discussions.
Footnotes
ASSOCIATED CONTENT
Experimental details, spectroscopic and computational data. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.(a) Du L; Lou L PKS and NRPS release mechanisms. Nat. Prod. Rep 2010, 27, 255–278. [DOI] [PubMed] [Google Scholar]; (b) Horsman ME; Hari TP; Boddy CN Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: logic gate or a victim of fate? Nat. Prod. Rep 2016, 33, 183–202. [DOI] [PubMed] [Google Scholar]
- 2.Cox RJ Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem 2007, 5, 2010–2026. [DOI] [PubMed] [Google Scholar]
- 3.Chooi Y-H; Tang Y Navigating the Fungal Polyketide Chemical Space: From Genes to Molecules. J. Org. Chem 2012, 77, 9933–9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Xie X; Watanabe K; Wojcicki WA; Wang CC; Tang Y Biosynthesis of Lovastatin Analogs with a Broadly Specific Acyltransferase. Chem. Biol 2006, 13, 1161–1169. [DOI] [PubMed] [Google Scholar]; (b) Xu W; Chooi Y-H; Choi JW; Li S; Vederas JC; Da Silva NA; Tang Y LovG: the Thioesterase Required for Dihydromonacolin L Release and Lovastatin Nonaketide Synthase Turnover in Lovastatin Biosynthesis. Angew. Chem. Int. Ed 2013, 52, 6472–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford JM, Dancy BCR, Hill EA, Udwary DW, Townsend CA Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc. Acad. Natl. Sci. U. S. A 2006, 103, 16728–16733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Fujii R; Matsu Y; Minami A; Nagamine S; Takeuchi I; Gomi K; Oikawa H Biosynthetic Study on Antihypercholesterolemic Agent Phomoidride: General Biogenesis of Fungal Dimeric Anhydrides. Org. Lett 2015, 17, 5658–5661. [DOI] [PubMed] [Google Scholar]; (b) Bonsch B; Belt V; Bartel C; Duensing N; Koziol M; Lazarus CM; Bailey AM; Simpson TJ; Cox RJ Identification of genes encoding squalestatin S1 biosynthesis and in vitro production of new squalestatin analogues. Chem. Commun 2016, 52, 6777–6780. [DOI] [PubMed] [Google Scholar]; (c) Liu N; Hung Y-S; Gao S-S; Hang L; Zou Y; Chooi Y-H; Tang Y Identification and Heterologous Production of a Benzoyl-Primed Tricarboxylic Acid Polyketide Intermediate from the Zaragozic Acid A Biosynthetic Pathway. Org. Lett 2017, 19, 3560–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bai J; Yan D; Zhang T; Guo Y; Liu Y; Zou Y; Tang M; Liu B; Wu Q; Yu S; Tang Y; Hu Y A Cascade of Redox Reactions Generates Complexity in the Biosynthesis of the Protein Phosphatase-2 Inhibitor Rubratoxin A. Angew. Chem. Int. Ed 2017, 56, 4782–4786. [DOI] [PubMed] [Google Scholar]; (e) Ugai T, Minami A, Fujii R, Tanaka M, Oguri H, Gomi K, Oikawa H Heterologous expression of highly reducing polyketide synthase involved in betaenone biosynthesis. Chem. Commun 2015, 51, 1878–1881. [DOI] [PubMed] [Google Scholar]
- 7.Büchi G; Kitaura Y; Yuan S-S; Wright HE; Clardy J; Demain AL; Ginsukon T; Hunt N; Wogan GN Structure of Cytochalasin E, a Toxic Metabolite of Aspergillus clavatus. J. Am. Chem. Soc 1973, 95, 5423–5425. [DOI] [PubMed] [Google Scholar]
- 8.Bloch P; Tamm C; Bollinger P; Petcher TJ; Weber HP Pseurotin, a New Metabolite of Pseudeurotium ovalis STOLK Having an Unusual Hetero-Spirocyclic System. Helv. Chim. Acta 1976, 59, 133–137. [DOI] [PubMed] [Google Scholar]
- 9.(a) Nakai R; Ogawa H; Asai A; Ando K; Agatsuma T; Matsumiya S; Yamashita Y; Mizukami T UCS1025A, a Novel Antibiotic Produced by Acremonium sp.. J. Antibiot. (Tokyo) 2000, 53, 294–296. [DOI] [PubMed] [Google Scholar]; (b) Agatsuma T; Akama T; Nara S; Matsumiya S; Nakai R; Ogawa H; Otaki S; Ikeda S; Saitoh Y; Kanda Y UCS1025A and B, New Antitumor Antibiotics from the Fungus Acremonium species. Org. Lett 2002, 4, 4387–4390. [DOI] [PubMed] [Google Scholar]
- 10.(a) Song Z; Cox RJ; Lazarus CM; Simpson TJ Fusarin C Biosynthesis in Fusarium moniliforme and Fusarium venenatum. Chembiochem 2004, 5, 1196–1203. [DOI] [PubMed] [Google Scholar]; (b) Sims JW; Fillmore JP; Warner DD; Schmidt EW Equisetin biosynthesis in Fusarium heterosporum. Chem. Commun 2005, 186–188. [DOI] [PubMed]; (c) Eley KL; Halo LM; Song Z; Powles H; Cox RJ; Bailey AM; Lazarus CM; Simpson TJ Biosynthesis of the 2-Pyridone Tenellin in the Insect Pathogenic Fungus Beauveria bassiana. Chembiochem 2007, 8, 289–297. [DOI] [PubMed] [Google Scholar]; (d) Boettger D; Hertweck C Molecular Diversity Sculpted by Fungal PKS-NRPS Hybrids. Chembiochem 2013, 14, 28–42. [DOI] [PubMed] [Google Scholar]
- 11.Ma SM; Li JW; Choi JW; Zhou H; Lee KK; Moorthie VA; Xie X; Kealey JT; Da Silva NA; Vederas JC; Tang Y Complete Reconstitution of a Highly Reducing Iterative Polyketide Synthase. Science 2009, 326, 589–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zabala AO; Chooi Y-H; Choi MS; Lin H-C; Tang Y Fungal Polyketide Synthase Product Chain-Length Control by Partnering Thiohydrolase. ACS Chem. Biol 2014, 9, 1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hang L; Tang M-C; Harvey CJB; Page CG; Li J; Hung Y-S; Liu N; Hillenmeyer MH; Tang Y Reversible Product Release and Recapture by a Fungal Polyketide Synthase Using a Carnitine Acyltransferase Domain. Angew. Chem. Int. Ed 2017, 56, 9556–9560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey CJB; Tang M; Schlecht U; Horecka J; Fischer CR; Lin H-C; Li J; Naughton B; Cherry J; Miranda M; Li YF; Chu AM; Hennessy JR; Vandova GA; Inglis D; Aiyar R; Steinmetz LM; Davis RW; Medema MH; Sattely E; Khosla C; Onge RP St.; Tang Y; Hillenmeyer MH HEx: A heterologous expression platform for the discovery of fungal natural products. Sci. Adv 2018, 4: eaar5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantu DC; Chen Y; Reilly PJ Thioesterases: A new perspective based on their primary and tertiary structures. Protein Sci 2010, 19, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Holmquist M Alpha/Beta-Hydrolase Fold Enzymes: Structures, Functions and Mechanisms. Curr. Protein Pept. Sci 2000, 1, 209–235. [DOI] [PubMed] [Google Scholar]; (b) Rauwerdink A; Kazlauskas RJ How the Same Core Catalytic Machinery Catalyzes 17 Different Reactions: the Serine-Histidine-Aspartate Catalytic Triad of α/β-Hydrolase Fold Enzymes. ACS Catal 2015, 5, 6153–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter JM; Cascio D; Dietrich D; Sato M; Watanabe K; Sawaya MR; Vederas JC; Tang Y Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis. J. Am. Chem. Soc 2015, 137, 9885–9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaffer JE; Reck MR; Prasad NK; Wencewicz TA beta-Lactone formation during product release from a nonribosomal peptide synthetase. Nat. Chem. Biol 2017, 13, 737–744. [DOI] [PubMed] [Google Scholar]
- 19.(a) Crombie L; Crombie WML Stereochemistry of thermolytic base-catalysed decarboxylation to form conjugated diene–acids: synthesis using ethylidenemalonic ester condensation. J. Chem. Soc., Perkin Trans 1 1994, 1267–1274. [Google Scholar]; (b) Schmidt B; Kunz O One-Flask Tethered Ring Closing Metathesis–Electrocyclic Ring Opening for the Highly Stereoselective Synthesis of Conjugated Z/E-Dienes. Eur. J. Org. Chem 2012, 1008–1018.
- 20.Smith CA; Want EJ; O’Maille G; Abagyan R; Siuzdak G XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem 2006, 78, 779–787. [DOI] [PubMed] [Google Scholar]
- 21.Cochrane RV; Sanichar R; Lambkin GR; Reiz B; Xu W; Tang Y; Vederas JC Production of New Cladosporin Analogues by Reconstitution of the Polyketide Synthases Responsible for the Biosynthesis of this Antimalarial Agent. Angew. Chem. Int. Ed 2016, 55, 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Dawson PE; Muir TW; Clark-Lewis I; Kent SB Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [DOI] [PubMed] [Google Scholar]; (b) Conibear AC; Watson EE; Payne RJ; Becker CFW Native chemical ligation in protein synthesis and semi-synthesis. Chem. Soc. Rev 2018, 47, 9046–9068. [DOI] [PubMed] [Google Scholar]
- 23.Kudo F; Matsuura Y; Hayashi T; Fukushima M; Eguchi T Genome mining of the sordarin biosynthetic gene cluster from Sordaria araneosa Cain ATCC 36386: characterization of cycloaraneosene synthase and GDP-6-deoxyaltrose transferase. J. Antibiot (Tokyo) 2016, 69, 541–548. [DOI] [PubMed] [Google Scholar]
- 24.Nofiani R; de Mattos-Shipley K; Lebe KE; Han LC; Iqbal Z; Bailey AM; Willis CL; Simpson TJ; Cox RJ Strobilurin biosynthesis in Basidiomycete fungi. Nat. Commun 2018, 9, 3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H; Gao Z; Qiao K; Wang J; Vederas JC; Tang Y A fungal ketoreductase domain that displays substrate-dependent stereospecificity. Nat. Chem. Biol 2012, 8, 331–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.(a) Xie X; Cane DE Stereospecific Formation of Z-Trisubstituted Double Bonds by the Successive Action of Ketoreductase and Dehydratase Domains from trans-AT Polyketide Synthases. Biochemistry 2018, 57, 3126–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shah DD; You Y-O; Cane DE Stereospecific Formation of E- and Z-Disubstituted Double Bonds by Dehydratase Domains from Modules 1 and 2 of the Fostriecin Polyketide Synthase. J. Am. Chem. Soc 2017, 139, 14322–14330. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liddle E; Scott A; Han LC; Ivison D; Simpson TJ; Willis CL; Cox RJ In vitro kinetic study of the squalestatin tetraketide synthase dehydratase reveals the stereochemical course of a fungal highly reducing polyketide synthase. Chem. Commun 2017, 53, 1727–1730. [DOI] [PubMed] [Google Scholar]; (d) Gay D; You Y-O; Keatinge-Clay A; Cane DE Structure and Stereospecificity of the Dehydratase Domain from the Terminal Module of the Rifamycin Polyketide Synthase. Biochemistry 2013, 52, 8916–8928. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Valenzano CR; You Y-O; Garg A; Keatinge-Clay A; Khosla C; Cane DE Stereospecificity of the Dehydratase Domain of the Erythromycin Polyketide Synthase. J. Am. Chem. Soc 2010, 132, 14697–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M; Zhou H; Wirz M; Tang Y; Boddy CN A Thioesterase from an Iterative Fungal Polyketide Synthase Shows Macrocyclization and Cross Coupling Activity and May Play a Role in Controlling Iterative Cycling through Product Offloading. Biochemistry 2009, 48, 6288–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge H-M; Huang T; Rudolf JD; Lohman JR; Huang S-X; Guo X; Shen B Enediyne Polyketide Synthases Stereoselectively Reduce the β-Ketoacyl Intermediates to β-d-Hydroxyacyl Intermediates in Enediyne Core Biosynthesis. Org. Lett 2014, 16, 3958–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labonte JW; Townsend CA Active Site Comparisons and Catalytic Mechanisms of the Hot Dog Superfamily. Chem. Rev 2013, 113, 2182–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) Vergnolle O; Hahn F; Baerga-Ortiz A; Leadlay PF; Andexer JN Stereoselectivity of Isolated Dehydratase Domains of the Borrelidin Polyketide Synthase: Implications for cis Double Bond Formation. ChemBioChem 2011, 12, 1011–1014. [DOI] [PubMed] [Google Scholar]; (b) Hahn F; Kandziora N; Friedrich S; Leadlay PF Synthesis of complex intermediates for the study of a dehydratase from borrelidin biosynthesis. Beilstein J. Org. Chem 2014, 10, 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeves CD; Hu Z; Reid R; Kealey JT Genes for the Biosynthesis of the Fungal Polyketides Hypothemycin from Hypomyces subiculosus and Radicicol from Pochonia chlamydosporia. Appl. Environ. Microbiol 2008, 74, 5121–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang M-C; Lin H-C; Li D; Zou Y; Li J; Xu W; Cacho RA; Hillenmeyer ME; Garg NK; Tang Y Discovery of Unclustered Fungal Indole Diterpene Biosynthetic Pathways through Combinatorial Pathway Reassembly in Engineered Yeast. J. Am. Chem. Soc 2015, 137, 13724–13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith JD; Schlecht U; Xu W; Suresh S; Horecka J; Proctor MJ; Aiyar RS; Bennett RA; Chu A; Li YF; Roy K; Davis RW; Steinmetz LM; Hyman RW; Levy SF; Onge RP St. A method for high-throughput production of sequence-verified DNA libraries and strain collections. Mol. Syst. Biol 2017, 13: 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradford MM A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.