Abstract
The budding yeast Saccharomyces cerevisiae has been used as a model organism for the basic mechanism of aging, which provides useful assay systems for measuring both replicative and chronological lifespans. In the course of our screening program for substances that extend replicative lifespan, cucurbitacin B (CuB) was found as a hit compound from a compound library, which contains cerebrosides, phenols, sesquiterpenoid, triterpenoids, and sterols isolated from natural products by our research group. Importantly, it prolonged not only the replicative lifespan but also the chronological lifespan in yeast. CuB increased ATG32 gene expression, suggesting that CuB induces autophagy. Indeed, the GFP signal generated from the cleavage of GFP-Atg8, which is a signature of autophagy, was increased upon CuB treatment. On the other hand, CuB failed to increase the chronological lifespans when either ATG2 or ATG32, essential autophagy genes, was deleted, indicating that the lifespan extension by CuB depends on autophagy induction. Furthermore, CuB significantly increased superoxide dismutase (Sod) activity and the survival rate of yeast under oxidative stress, while it decreased the amount of reactive oxygen species (ROS) and malondialdehyde (MDA) production, indicating that CuB has activity to antagonize oxidative stress. Additionally, CuB did not affect replicative lifespans of sod1, sod2, uth1, and skn7 mutants with the K6001 background, indicating that aging-related genes including SOD1, SOD2, UTH1, and SKN7 participate in the antiaging effect of CuB. These results suggest that CuB exerts antiaging activity by regulating autophagy, ROS, antioxidative ability, and aging-related genes. Finally, we discuss the possible intracellular targets of CuB based on the phenotypic comparison between the CuB and global gene deletion databases.
1. Introduction
Yeast has the replicative lifespan and chronological lifespan. The replicative lifespan measures the number of daughters of a single mother cell, which can asexually produce prior to senescence. The chronological lifespan is defined as the length of time a yeast cell can survive in the nondividing G0 state [1]. Both of them are regulated by many environmental and genetic factors [2]. In our study, we used K6001 yeast as an aging model, which is a mutant yeast strain derived from W303. A unique character of K6001, in which only mother cells can produce daughter cells in glucose medium, but not in galactose medium [3], can be used for measuring replicative lifespan. Under the guidance of K6001 replicative lifespan assay, we have isolated several compounds with antiaging activity from natural products [4, 5].
Autophagy is a highly conserved pathway in organisms from yeast to human, which involves degradation of damaged organelles and proteins and circulation of amino acids and other metabolites [6]. It regulates the genomic integrity via suppression of cell division in yeast under starvation [7]. Besides, decreased autophagy leads to downregulation of proteostasis, which is one of the hallmarks for aging. Dysfunction or decreased expression of autophagy genes leads to shorter lifespan in yeast and fruit fly. Conversely, enhanced autophagy promotes the longevity in aging models and protects against aging and age-related disorders [8]. Specially, the lifespan-extending effects of rapamycin in C. elegans, fruit fly, and mice are abolished by knocking out or knocking down of ATG genes [9]. In addition, caloric restriction and resveratrol (RES) induced autophagy dependent on Sir2 function in yeast and C. elegans, and the deleted and silenced ATG genes avoid the antiaging effect of caloric restriction, RES, and overexpression of SIR2 [10]. Therefore, autophagy has a close link to aging.
Oxidative stress is another cause of aging. Free radicals produced through aerobic metabolism accumulate over time and accelerate aging [11]. In yeast, the accumulation of ROS promotes replicative and chronological aging. The upregulation of antioxidant removes excess ROS and maintains redox balance, eventually delaying the progression of aging in yeast [12]. In our previous studies, we showed that antioxidative activity plays an important role in the antiaging effects of cholesterol and parishin [4, 5].
CuB has been shown as a promising compound for anticancer treatment in recent years, and it also shows antioxidative and anti-inflammatory activities [13, 14]. The regulation of STAT3 and Raf/MEK/ERK signaling pathways by CuB is likely to be involved in its antitumor activity such as induction of apoptosis in tumor cells [15]. Moreover, CuB was found to induce autophagy in cells [16]. In the present study, we identified CuB as a compound with antiaging activity in yeast, resulting from our screening for antiaging compounds from a compound library using yeast replicative and chronological lifespan assays. Here, we report a unique activity of CuB, which exerts antiaging effects in yeast through regulating autophagy and oxidative stress.
2. Materials and Methods
2.1. CuB, Yeast Strains, and Medium
CuB was isolated from Pedicellus melo in accordance with a methodology that was previously used by our group. The chemical structure of CuB (Figure 1(a)) was identified by comparing HR ESI-MS, 1H NMR, and 13C NMR data with reported data [17]. 1H NMR (500 MHz, acetone-d 6): δ 0.91 (3H, s), 1.02 (3H, s), 1.11 (1H, q, J = 13.0 Hz), 1.28 (3H, s), 1.32 (3H, s), 1.39 (3H, s), 1.40 (1H, m), 1.44 (3H, s), 1.51 (3H, s), 1.55 (3H, s), 1.84 (1H, m), 1.96 (3H, s), 1.97 (2H, m), 2.11 (1H, m), 2.40 (1H, dd, J = 5.5, 19.0 Hz), 2.51 (1H, d, J = 14.5 Hz), 2.66 (1H, d, J = 6.5 Hz), 3.02 (1H, d, J = 12.5 Hz), 3.40 (1H, d, J = 14.5 Hz), 4.45 (1H, m), 4.51 (1H, s), 4.56 (1H, m), 5.82 (1H, d, J = 3.0 Hz), 6.81 (1H, d, J = 16.0 Hz), and 6.98 (1H, d, J = 16.0 Hz); 13C NMR (125 MHz, acetone-d 6): δ 19.2, 20.2, 20.6, 21.8, 22.0, 24.7, 25.1, 26.4, 27.1, 29.9, 34.2, 37.1, 43.6, 46.6, 49.0, 49.1, 49.4, 51.1, 51.4, 59.1, 71.4, 72.3, 79.6, 80.1, 120.8, 122.3, 142.0, 151.0, 170.3, 203.5, 212.7, and 213.7; and high-resolution ESI-TOF-MS m/z 581.3085, calcd for C32H46NaO8 (M+Na)+ 581.3090. All the yeast strains used in the present study are listed in Table 1.
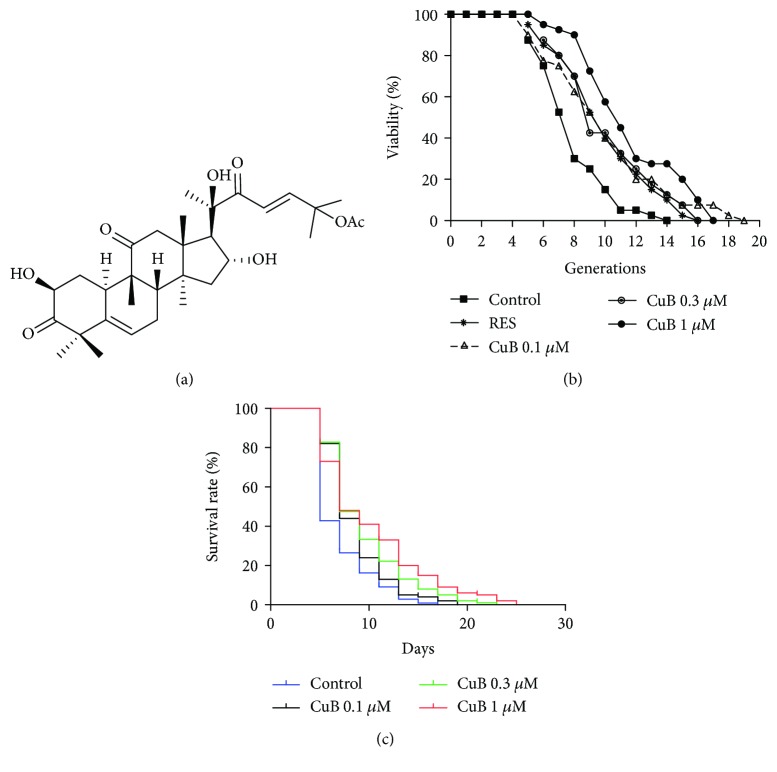
Figure 1.
Chemical structure and antiaging activity of cucurbitacin B (CuB). (a) Chemical structure of CuB. (b) Effect of CuB on the replicative lifespan of K6001 yeast. RES (10 μM) was used as the positive control. (c) Effect of CuB on the chronological lifespan of YOM36 yeast.
Table 1.
Yeast strains used in the present study.
| Strains | Genotype | Source |
|---|---|---|
| K6001 | MATa, ade2-1, trp1-1, can1-100, leu2-3,112, his3-11,15, GAL, psi+, ho::HO::CDC6 (at HO), cdc6::hisG, ura3::URA3 GAL-ubiR-CDC6 (at URA3) | Gifted by Professor Michael Breitenbach |
| Δuth1 of K6001, Δskn7 of K6001, Δsod1 of K6001, Δsod2 of K6001, Δatg2 of K6001, Δatg32 of K6001 | Replace the UTH1 gene, SKN7 gene, SOD1 gene, SOD2 gene, ATG2 gene, and ATG32 gene in K6001 with kanamycin gene, respectively | Constructed by Professor Akira Matsuura |
| BY4741 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Gifted by Professor Akira Matsuura |
| YOM36 | Prototrophic derivative of BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) | Gifted by Professor Akira Matsuura |
| Δatg2 of YOM36, Δatg32 of YOM36 | Replace the ATG2 gene and ATG32 gene in YOM36 with kanamycin gene, respectively | Constructed by Professor Akira Matsuura |
| YOM38 containing plasmid pR316-GFP-ATG8 | Prototrophic derivative of BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0) containing plasmid pR316-GFP-ATG8 | Constructed by Professor Akira Matsuura |
| YO2458 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YOR202w::KanMX4 | Provided by Professor Yoshikazu Ohya |
2.2. Replicative and Chronological Lifespan Assays
The replicative lifespan assay was performed following a previously described methodology [5]. In brief, the K6001 strain was inoculated in galactose medium and cultured for 24–28 h in a shaking incubator at 160 rpm and 28°C. After washing with phosphate-buffered saline (PBS) for three times, approximately 4000 cells were spread onto glucose agar plates (2% glucose, 2% hipolypeptone, 1% yeast extract, and 2% agar) supplemented with 0, 0.1, 0.3, or 1 μM CuB or 10 μΜ RES. Afterward, the yeast cells were cultured at 28°C for 2 days. Forty microcolonies from each group were observed under microscopy, and the number of daughter cells in each microcolony was counted. The replicative lifespan assays of K6001 mutants (Δuth1, Δskn7, Δsod1, Δsod2, Δatg2, and Δatg32 of K6001) were conducted similarly to those of the wild-type K6001. RES was procured from J&K Scientific Ltd. (Beijing, China) and was used as positive control.
The chronological lifespan assay was conducted in accordance with a previous methodology [18]. Briefly, YOM36 yeast cells were cultured in synthetic defined (SD) medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate (BD Difco), 0.5% ammonium sulfate, and 0.2% glucose) for 24 h and then inoculated into the SD medium containing 0, 0.1, 0.3, or 1 μM CuB with the initial OD600 value of 0.01. Cultures were grown in a shaker at 180 rpm and 30°C. Growth kinetics was recorded by measuring the OD600 value every 2 or 4 h until the stationary phase was reached. In addition, the survival rate was measured by counting colony-forming units (CFUs) every 2 days. The CFUs on day 3 was denoted as 100% survival.
2.3. Real-Time Polymerase Chain Reaction Analysis
Wild-type BY4741 were incubated with the negative control or 1 μM CuB in glucose medium overnight. RNA was extracted through a hot phenol method. cDNA was synthesized through the reverse transcription method using the HiFi-MMLV cDNA Kit (CoWin Biotech, Beijing, China) and 5 μg of RNA. Real-time polymerase chain reaction (RT-PCR) was performed in reference to a previous study [5] by using CFX96 Touch (Bio-Rad, Hercules, USA) and SYBR Premix Ex Taq (Takara, Otsu, Japan). The thermal cycling parameters were as follows: for ATG2 and ATG32, 40 cycles, 94°C for 15 s, 51.6°C for 15 s, and 68°C for 20 s. The primers used for RT-PCR are as follows: for ATG2, sense 5′-GCT CCT GTC AGA TCG TTT AT-3′ and antisense 5′-TTC AGA CTC CTT CCC AAA TG-3′; for ATG32, sense 5′-ACC GTC TCA TCC CTT TAA AC-3′ and antisense 5′-CTT CCT CAA AAG CCT CAT CT-3′; and for TUB1, sense 5′-CCA AGG GCT ATT TAC GTG GA-3′ and antisense 5′-GGT GTA ATG GCC TCT TGC AT-3′. The 2−ΔΔCt method was used to analyze relative gene expression data. The mRNA levels of ATG2 and ATG32 were normalized to those of TUB1.
2.4. GFP–ATG8 of Yeast Analyzed with Fluorescent Microscopy
YOM38 yeast cells containing pR316-GFP-ATG8 or YOM36 yeast cells were cultured in the SD medium with the initial OD600 value of 0.1 and treated with 0, 0.1, 0.3, or 1 μM CuB or 300 μM RES for 22 h. Subsequently, yeast cells were stained with 20 μg/mL DAPI in dark for 10 min and washed with the PBS for three times. Yeast cells were observed using a two-photon confocal fluorescence microscope (Olympus FV1000BX-51, Tokyo, Japan) or fluorescent microscope (Leica DMI3000 B, Wetzlar, Germany). Pictures were acquired using image acquisition and analysis software.
2.5. Western Blot Analysis
YOM38 yeast cells containing the pR316-GFP-ATG8 plasmid were cultured as described in the previous section. At first, yeast cells were incubated with 300 μM RES or CuB at doses of 0, 0.1, 0.3, or 1 μM, and the yeast cells were collected at the specified time point and washed three times with PBS. After that, the samples were ultrasonicated for five times (1 min for each time), freeze-thawed for five times, and sonicated for five times again. The cell lysates were centrifuged, and the protein concentrations of the supernatant were measured with BCA Protein Assay Kit (CoWin Biotech, Beijing, China). Approximately 20 μg protein was separated with SDS-PAGE and transferred to PVDF membranes. The membranes were incubated with primary antibodies followed by secondary antibodies. Antigens were visualized using ECL Western Blot Kit (CoWin Biotech, Beijing, China). The primary antibodies used are as follows: anti-GFP antibody (Medical & Biological Laboratories, Nagoya, Japan) and anti-β-actin antibody (CoWin Biotech, Beijing, China). The secondary antibodies used are as follows: horseradish peroxidase-linked anti-rabbit and anti-mouse IgGs (CoWin Biotech, Beijing, China).
2.6. Antioxidative Assay
BY4741 yeast cells with the initial OD600 value of 0.1 were cultured in liquid glucose medium and treated with 0, 0.1, 0.3, or 1 μM CuB or 10 μM RES for 24 h. Afterward, 5 μL of yeast culture in each group with the same OD600 value was dropped onto glucose agar plates containing 9 mM H2O2. After 3 days, the growth of yeast was observed and photographed. Besides, further spots of 1/10, 1/100, and 1/1000 dilutions in each group were also dropped onto glucose agar plates containing 9 mM H2O2, and the growth was observed after 5 days.
Another method was performed to quantify the effect of CuB on the oxidative stress response of yeast. Similar to that in the former method, BY4741 yeast cells were treated with 0, 0.1, 0.3, or 1 μM CuB or 10 μM RES. 200 yeast cells from each group were spread onto glucose agar plates supplemented with or without 5 mM H2O2. The growth of yeast was observed after 2 days, and the number of microcolonies in each plate was counted. The survival rate was calculated as the ratio of the number of microcolonies in the presence of 5 mM H2O2 divided by the number of microcolonies in the absence of 5 mM H2O2.
2.7. ROS, MDA, and Sod Enzyme Assays
CuB- or RES-treated BY4741 yeast cells were cultured for 23 h in liquid glucose medium at 28°C. DCFH-DA (2′,7′-dichlorodihydrofluorescein diacetate, 1 μL, 10 mM) was added to 1 mL of the cells. The mixture was then incubated at 28°C in the dark with shaking for 1 h. The cells were quickly washed with PBS thrice in the dark, and the DCF (2′,7′-dichlorofluorescein) fluorescence intensity of 1 × 107 cells was recorded using the SpectraMax M3 multimode microplate reader (Molecular Devices Corporation, California, USA) under the excitation wavelength of 488 nm and emission wavelength of 525 nm.
BY4741 yeast cells were treated with CuB or RES in liquid glucose medium for 24 or 48 h at 28°C with the initial OD of 0.1. Yeast cells were collected through centrifugation and five cycles of ultrasonication (1 min for each time). Then, the cells were frozen for 5 min in liquid nitrogen, subsequently thawed for 2 min in a 37°C water bath, and sonicated for five cycles. The cell lysates were centrifuged, and the supernatant was removed for MDA quantification. MDA levels were quantified by using the MDA assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) following the manufacturer's instructions.
BY4741 yeast cells were treated in a manner similar to that in MDA assay. Briefly, cells were treated with CuB or RES in the liquid glucose medium for 24 or 48 h at 28°C. Then, yeast cells were collected and subjected to five 1 min cycles of ultrasonication. Subsequently, the total super oxide dismutase (T-Sod) and Sod1 enzyme activities of the supernatant were tested using SOD Assay Kits (A001-1, A001-2) (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) following the manufacturer's instructions.
2.8. Growth Curves of Yeast Cells with CuB
To test growth inhibition in liquid culture, the YPD medium containing 0, 1.56, 6.25, 25, or 50 μM CuB was used. Cultures of Y02458, his3∆ of BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, and YOR202w::KanMX4) were treated with CuB and grown at 25°C for 25 h. Optical density from 0 to 25 hours was measured at 10 min intervals using an absorption spectrometer at 660 nm. The assay was repeated at least 2 times.
2.9. High-Dimensional Phenotypic Analysis
High-dimensional phenotypic analysis was conducted in accordance with a previous report [19]. Yeast cells were cultured in YPD medium with or without CuB, and cells in the logarithmic phase were fixed with 3.7% formaldehyde. Yeast cells, nuclear DNA, and actin were stained with 20 mg/mL fluorescein isothiocyanate-Con A, 4,6-diamidino-2-phenylindole, and rhodamine phalloidin, respectively. Morphological changes were observed through fluorescence microscopy. CalMorph was used to characterize each yeast cell through the quantification of 501 morphological parameters.
2.10. Fitness
To estimate fitness, we employed a previously published dataset of logarithmic strain growth rate coefficients for haploid nonessential gene deletion mutants grown on a basal medium (LSC basal) [20]. We used the p value calculated as the significance of lower fitness from the wild type of each strain based on one tail of the estimated probability distribution [20, 21] and calculated FDR as described previously [21].
2.11. Data Processing
Coefficient of variation values were highly dependent on the mean values in a nonlinear manner [22] and therefore were not suited for normalization. Instead, we defined noise values as the residuals between observed and predicted values, as described previously [21, 23].
2.12. Detection of Specific Morphologically Abnormal Mutants
The probability distribution of the wild-type replicates for each trait of the 501 parameters was estimated using maximum likelihood estimation with one of four probability density functions (gamma, beta, Gaussian, and beta-binomial distribution), as described previously [24]. We calculated the p value of every nonessential deletion mutant as morphological abnormality from the wild type for each trait (two-sided one-sample test) and identified the lowest p value among 501 traits as the “specific morphological abnormality,” as described previously [21]. Maximum likelihood estimation and calculation of the p value were performed using the gamlss function in R software's (http://www.r-project.org) gamlss package [25]. The FDR, a rate of type I errors in the rejected null hypothesis due to multiple comparisons, was calculated using the qvalue R function in the qvalue package [26]. We plotted specific morphologically abnormal mutants as circles filled with green.
2.13. Plot of Single-Gene Deletion Strains with Increased Lifespan
To plot single-gene deletion strains with increased lifespan, we employed a previously published dataset of replicative lifespan for nonessential gene deletion mutants [27].
2.14. Statistical Analysis
All statistical analyses were performed using GraphPad Prism 5 (GraphPad Software Inc.). Survival analysis was used for chronological lifespan assay. Analysis of variance was used to determine the significant differences among groups in all experiments, followed by two-tailed multiple t-tests with Bonferroni correction. A p value of less than 0.05 was considered statistically significant.
3. Results
3.1. CuB Has Significant Antiaging Effects
We have previously isolated a number of antiaging substances, such as cholesterol and parishin, from natural products on the basis of the K6001 lifespan assay [4, 5]. In the present study, we again used the K6001 lifespan assay to screen for antiaging compounds from a compound library and identified the active compound CuB (Figure 1(a)) as a potential antiaging compound. Subsequently, we performed chronological lifespan assay to confirm antiaging effects of CuB. As shown in Figure 1(b) and Table 2, the average replicative lifespan of each treatment group was as follows: 6.95 ± 0.34 generations for the control group, 9.03 ± 0.47 generations for the group treated with 10 μM RES, 8.90 ± 0.55 generations for the group treated with 0.1 μM CuB, 8.95 ± 0.51 generations for the group treated with 0.3 μM CuB, and 9.60 ± 0.49 generations for the group treated with 1 μM CuB. These results indicate that treatment with 0.1, 0.3, and 1 μM CuB significantly prolongs the replicative lifespan of K6001 (p < 0.05, p < 0.05, and p < 0.01, respectively). As shown in Figure 1(c), CuB increased the survival rate of yeast (p < 0.001, p < 0.001, and p < 0.001, respectively). CuB significantly increased the days at which viability is equal to 0 of yeast from 15 ± 1.15 days to 17 ± 1.15, 21 ± 1.15, and 23.67 ± 0.67 days, respectively. These results suggest that CuB has significant antiaging effects on yeast cells.
Table 2.
Replicative lifespan of K6001 and its mutants.
| Figures | Yeast strains | Treatment (μM) | Replicative lifespan (generations) |
|---|---|---|---|
| Figure 1(b) | K6001 | Control | 6.95 ± 0.34 |
| RES-10 | 9.03 ± 0.47∗ | ||
| CuB-0.1 | 8.90 ± 0.55∗ | ||
| CuB-0.3 | 8.95 ± 0.51∗ | ||
| CuB-1.0 | 9.60±0.49∗∗ | ||
|
| |||
| Figures 2(b) and 2(c) | K6001 | Control | 7.05 ± 0.35 |
| RES-10 | 9.05 ± 0.55∗ | ||
| CuB-1.0 | 9.65±0.61∗∗ | ||
| atg2 mutant of K6001 | Control | 6.95 ± 0.43 | |
| RES-10 | 7.30 ± 0.40 | ||
| CuB-1.0 | 7.18 ± 0.45 | ||
| atg32 mutant of K6001 | Control | 6.73 ± 0.42 | |
| RES-10 | 7.40 ± 0.44 | ||
| CuB-1.0 | 7.10 ± 0.47 | ||
|
| |||
| Figure 4 | K6001 | Control | 6.95 ± 0.34 |
| RES-10 | 9.03 ± 0.47∗ | ||
| CuB-1.0 | 9.60±0.49∗∗ | ||
| sod1 mutant of K6001 | Control | 6.93 ± 0.43 | |
| RES-10 | 7.73 ± 0.39 | ||
| CuB-1.0 | 7.45 ± 0.46 | ||
| sod2 mutant of K6001 | Control | 7.35 ± 0.37 | |
| RES-10 | 6.85 ± 0.47 | ||
| CuB-1.0 | 7.10 ± 0.33 | ||
| uth1 mutant of K6001 | Control | 9.95 ± 0.63 | |
| RES-10 | 9.30 ± 0.59 | ||
| CuB-1.0 | 9.90 ± 0.69 | ||
| skn7 mutant of K6001 | Control | 8.43 ± 0.58 | |
| RES-10 | 9.13 ± 0.52 | ||
| CuB-1.0 | 9.30 ± 0.60 | ||
Replicative lifespan was shown as average ± SEM; ∗ and ∗∗ represent significant differences compared to the corresponding control groups (p < 0.05, p < 0.01.).
3.2. CuB Regulates Autophagy in Yeast
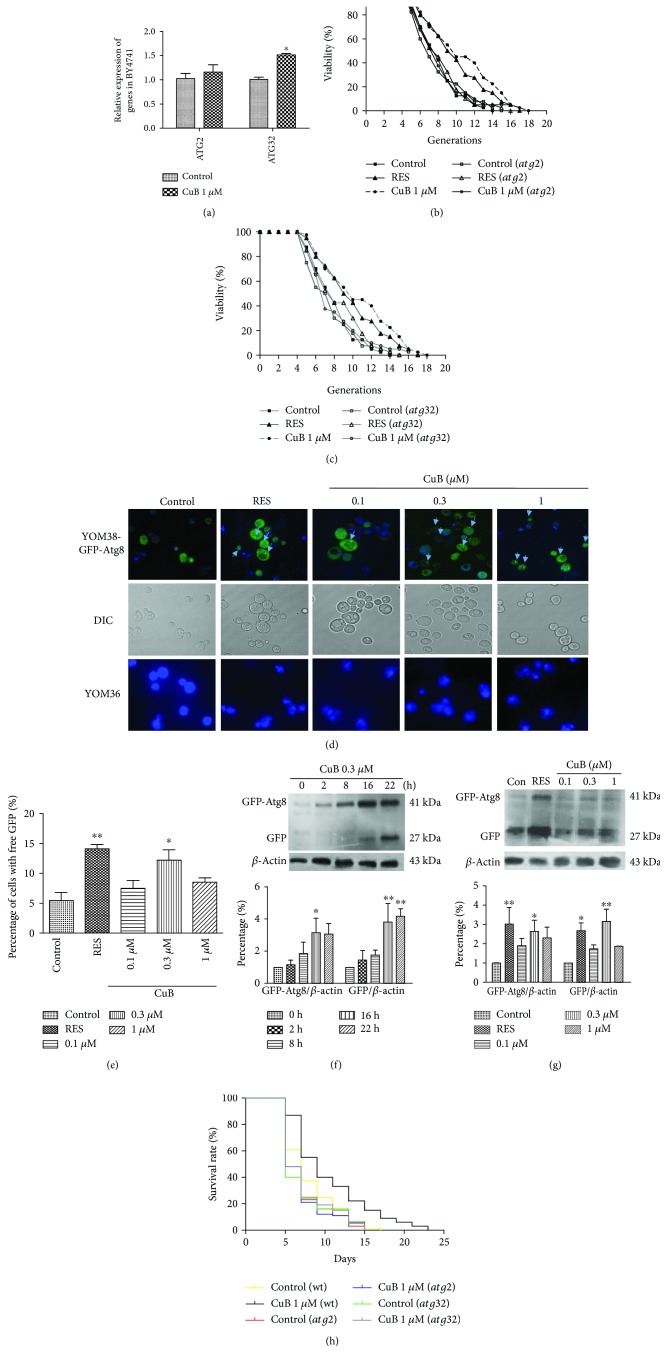
Autophagy is a degenerative process that degrades cellular components for recycling into amino acids and other metabolites. It has essential roles in cell growth, differentiation, development, and aging [8]. Hence, we investigated the effect of CuB on autophagy. ATG2 and ATG32 are two of the most important genes that mediate autophagy in yeast; we first explored ATG2 and ATG32 gene expression in CuB-treated yeast. As shown in Figure 2(a), CuB significantly enhanced ATG32 gene expression but did not significantly affect ATG2 gene expression.
Figure 2.
Effect of CuB on autophagy in yeast. (a) Effect of CuB on ATG2 and ATG32 gene expression after 12 h of treatment. (b, c) Effect of CuB on the replicative lifespan of K6001 and atg2 (b) and atg32 (c) mutants with K6001 background. (d) Fluorescent images of YOM38 yeast contained plasmid pR316-GFP-ATG8 and YOM36 yeast after treatment of RES or different doses of CuB in the SD medium and DAPI staining observed with a two-photon confocal fluorescent microscope. The upper line and middle line showed merged and DIC images of YOM38 yeast cells containing plasmid pR316-GFP-ATG8, respectively. The line below showed fluorescent images of YOM36. (e) Effect of CuB on the percentage of YOM38 cells containing plasmid pR316-GFP-ATG8 with free GFP. Three pictures containing more than 60 cells in each group were used for statistical analysis. (f) Western blot analysis of GFP-Atg8 and free GFP in yeast after shifting to the SD medium containing 0.3 μM CuB for different times. (g) Western blot analysis of GFP-Atg8 and free GFP in yeast after treatment with RES or CuB for 22 h in the SD medium. (h) Effect of CuB on the chronological lifespan of wild-type YOM36 and atg2 and atg32 mutants with YOM36 background. ∗ and ∗∗ indicate significant difference between treatment groups and the corresponding control group (p < 0.05, p < 0.01).
Furthermore, we constructed atg2 and atg32 mutants with K6001 background to do replicative lifespan assay. The changes of the lifespan are shown in Figures 2(b) and 2(c) and Table 2. The significant decrease in replicative lifespan of atg2 and atg32 mutants was not observed compared with that of K6001. After the treatment of CuB, the replicative lifespan of atg2 and atg32 mutants was also not affected. These results suggested that Atg2 and Atg32 did not take an important role in the replicative lifespan of yeast. However, they were required in the replicative lifespan extension of CuB.
To test whether CuB induces autophagy, we used the YOM38 strain, which expresses GFP-Atg8 at a physiological level and monitored the level of GFP upon treatment with CuB under fluorescent microscopy. The fluorescent images are displayed in Figure 2(d), and the statistic result is shown in Figure 2(e). CuB significantly enhanced the percentage of cells with green fluorescence at a dose of 0.3 μM as does RES. In order to investigate whether the GFP signal came from autophagy or autofluorescence of yeast, the YOM36 yeast strain was also treated with RES or CuB. As shown in Figure 2(d), YOM36 expressed no green fluorescence. Besides, western blot analysis showed generation of free GFP, which is released into the vacuole during the autophagy flux. CuB increased the amount of released GFP in yeast in a time course manner, and 0.3 μM CuB significantly increased the amount of free GFP (Figures 2(f) and 2(g)). These results confirm that CuB induces autophagy.
Furthermore, we performed chronological lifespan measurements using atg2 and atg32 mutants derived from a phototrophic derivative of BY4741 (YOM36) (Figure 2(h)). The results showed that the survival rate of atg2 and atg32 mutants significantly decreased (p < 0.05, p < 0.01) compared to YOM36 yeast strain. In addition, CuB at 1 μM increased the survival rate of YOM36 yeast during the whole assay (p < 0.001), but failed to prolong the chronological lifespans of atg2 and atg32 mutants. The days at which viability is equal to 0 of atg2 and atg32 mutants were 14 ± 1 and 16 ± 1 days, respectively, and were shorter than those of the wild-type yeast (17 ± 0 days). In addition, CuB extended the days at which viability is equal to 0 of the wild-type yeast from 17 ± 0 days to 19 ± 2 days. However, the days at which viability is equal to 0 of the atg2 and atg32 mutants in CuB-treated groups were 13 ± 2 and 16 ± 1 days, respectively, indicating that CuB failed to prolong the chronological lifespans of atg2 and atg32 mutants. These results indicate that CuB exerts its activity through regulating autophagy, which requires ATG2 and ATG32.
3.3. CuB Improves the Survival Rate of Yeast under Oxidative Conditions
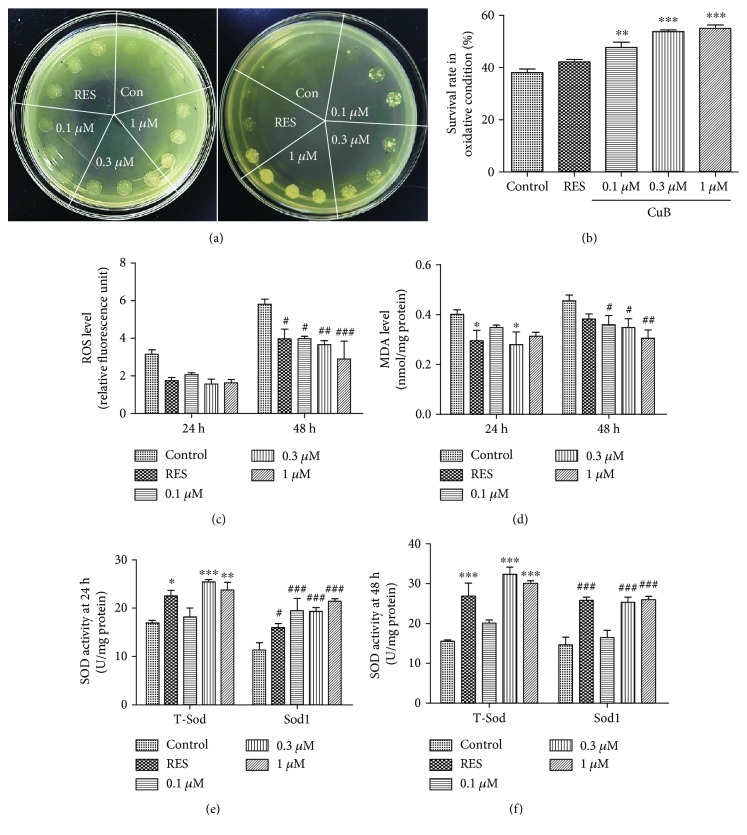
Oxidative stress is a major factor of aging and age-related diseases, and high levels of oxidative stress result in DNA damage, lipid peroxidation, and protein oxidation [28]. Therefore, we performed antioxidative experiments to determine whether antioxidative activity is involved in the antiaging effect of CuB. We used two methods to investigate the effect of CuB on the growth of yeast with 5 μL culture or 1/10, 1/100, and 1/1000 dilutions of each group under oxidative stress at 9 mM H2O2. The growth of yeast in both conditions (Figure 3(a)) was significantly improved, respectively. We also examined the survival rate of yeast under oxidative stress and found that the survival rate was 38.09% ± 1.39 in the control group, 42.19% ± 0.87 in the RES group, 47.74% ± 2.03 in the 0.1 μM CuB group (p < 0.01), 53.73% ± 0.73 in the 0.3 μM CuB group (p < 0.001), and 55.01% ± 1.35 in the 1 μM CuB group (p < 0.001) (Figure 3(b)). These results indicate that the regulation of antioxidative activity has an important role in the antiaging effect of CuB.
Figure 3.
Effect of CuB on oxidative stress in yeast. (a) Effect of CuB on yeast growth with 5 μL yeast culture and 1/10, 1/100, and 1/1000 dilutions under oxidative conditions simulated with 9 mM H2O2. (b) Effect of CuB on the survival rate of yeast under oxidative conditions simulated with 5 mM H2O2. ∗∗ and ∗∗∗ represent significant differences compared to the control group (p < 0.01 and p < 0.001, respectively). (c, d, e, f) Effect of CuB on ROS level, MDA accumulation, T-Sod, and Sod1 enzyme activity at 24 or 48 h. ∗, ∗∗, ∗∗∗ and #, ##, ### indicate significant differences from the corresponding control (p < 0.05, p < 0.01, and p < 0.001, respectively).
3.4. CuB Decreases ROS Accumulation and MDA Production and Increases Sod Activity
ROS are by-products of aerobic respiration and various metabolic processes. They cause secondary reactions, such as lipid peroxidation and protein oxidation [29]. MDA, a biomarker of lipid peroxidation in living cells, has cytotoxic effects [30]. Thus, we tested the change in ROS accumulation and MDA levels in CuB-treated yeast. ROS accumulation in yeast notably decreased after 48 h of incubation with 0.1, 0.3, and 1 μM CuB (Figure 3(c)). Similarly, significant decreases were observed in MDA production by yeast. MDA production decreased after 24 h of incubation with 0.3 μM CuB and 48 h of incubation with 0.1, 0.3, and 1 μM CuB (Figure 3(d)). These results suggested that CuB could significantly inhibit the production of ROS and MDA in yeast.
Sod is a component of antioxidative defense systems that can scavenge ROS and other free radicals in cells. Yeast has two kinds of Sod, Sod1 and Sod2. Sod1 is a stable Sod that localizes in the cytoplasm, and mitochondrial Sod2 is more sensitive to environmental factors. Therefore, we evaluated total-Sod and Sod1 activity in yeast incubated with CuB for 24 or 48 h. As shown in Figures 3(e) and 3(f), T-Sod and Sod1 enzyme activity significantly improved after 24 and 48 h of treatment with 0.3 and 1 μM CuB. Besides, Sod1 activity also improved after 24 h of treatment with 0.1 μM CuB. The results show that CuB can significantly decrease ROS and MDA levels and increase T-Sod and Sod1 enzyme activity in yeast at 0.3 and 1 μM CuB. These results suggest that CuB exerts its antiaging effect by regulating antioxidative activity.
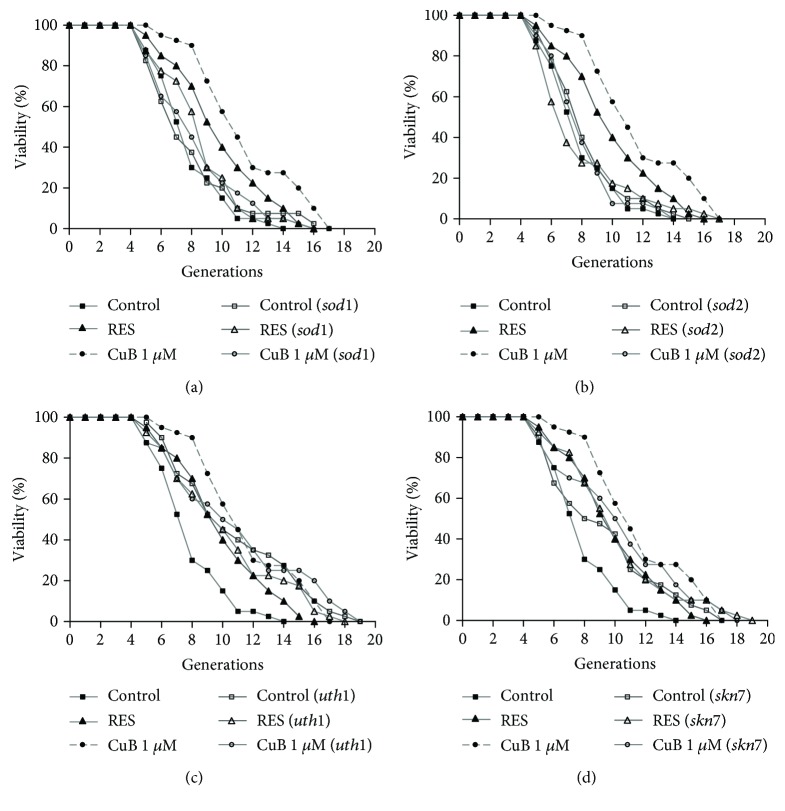
3.5. CuB Does Not Affect the Replicative Lifespans of the Sod1, Sod2, Uth1, and Skn7 Mutants of K6001 Yeast
The results shown in Figure 3 suggest that antioxidative activity is involved in the antiaging activity of CuB and that SOD1 and SOD2 participate in oxidative stress responses. Therefore, we used sod1 and sod2 mutants with the K6001 background to investigate the involvement of SOD1 and SOD2 genes in the antiaging effect of CuB. As shown in Figures 4(a) and 4(b) and Table 2, the average replicative lifespan of the wild-type K6001 yeast was 6.95 ± 0.34 generations under the control treatment, 9.03 ± 0.47 generations under the RES treatment, and 9.60 ± 0.49 generations under the 1 μM CuB treatment. The average replicative lifespan of the sod1 mutant was 6.93 ± 0.43 generations under the control treatment, 7.73 ± 0.39 generations under the RES treatment, and 7.45 ± 0.46 generations under the 1 μM CuB treatment. The average replicative lifespan of the sod2 mutant was 7.35 ± 0.37 generations under the control treatment, 6.85 ± 0.47 generations under the RES treatment, and 7.10 ± 0.33 generations under the 1 μM CuB treatment. These results show that the replicative lifespans of the sod1 and sod2 mutants are unaffected by RES or CuB treatment.
Figure 4.
Effect of CuB on the replicative lifespans of sod1 (a), sod2 (b), uth1 (c), and skn7 (d) mutants with K6001 background. The procedure for the replicative lifespan assay was the same as that for the K6001 lifespan assay.
UTH1 is an aging gene that is involved in the regulation of programmed cell death in yeast [31]. Skn7 is the transcriptional activator of UTH1 and is associated with protection against oxidative stress [32]. In our previous study, UTH1 gene expression in skn7 mutant was significantly decreased [33]. To explore whether UTH1 and SKN7 genes are involved in the lifespan-extending effect of CuB, we measured the replicative lifespans of uth1 and skn7 mutants after CuB treatment. The average replicative lifespan of the uth1 mutant was 9.95 ± 0.63 generations under the control treatment, 9.30 ± 0.59 generations under the RES treatment, and 9.90 ± 0.69 generations under the 1 μM CuB treatment (Figure 4(c) and Table 2). The average replicative lifespan of the skn7 mutant was 8.43 ± 0.58 generations under the control treatment, 9.13 ± 0.52 generations under the RES treatment, and 9.30 ± 0.60 generations under the 1 μM CuB treatment (Figure 4(d) and Table 2). The replicative lifespans of the two mutants after CuB treatment did not significantly change. As the average replicative lifespan of the wild-type K6001 yeast was 6.95 ± 0.34 generations under the control treatment, the replicative lifespan of the uth1 mutant was longer than that of the wild-type K6001. This result was consistent with other reports that deletion of UTH1 increased yeast lifespan [34, 35]. These results indicate that SOD1, SOD2, UTH1, and SKN7 are involved in the antiaging activity of CuB.
3.6. CuB Does Not Affect the Growth Nor Morphology of Yeast Cells
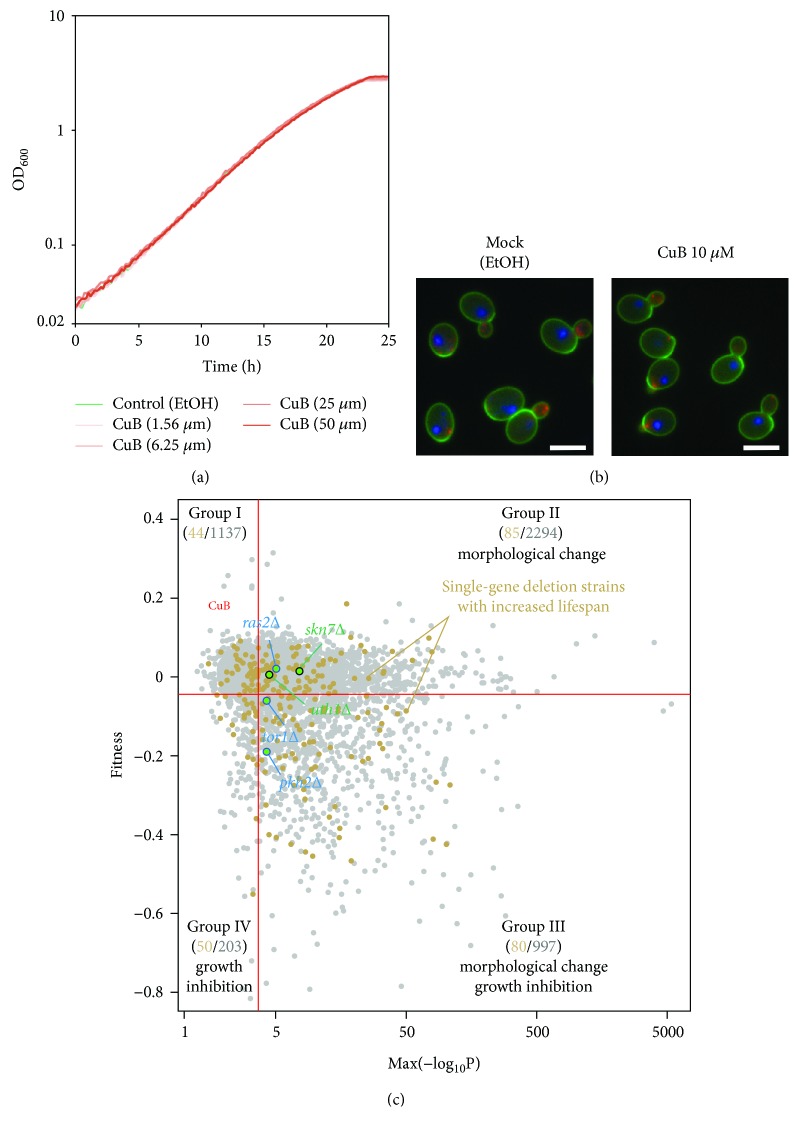
Many active compounds have been investigated in yeast with growth assay and morphological examination to understand their bioactivity. This is because both cell growth and morphology reflect numerous essential cellular processes, such as DNA replication, transcription, translation, vesicular transport, organelle assembly, and cell cycle regulation. Compared with the cells treated with solvent only (1% EtOH), treatment with 1.56 μM to 50 μM CuB failed to inhibit cell growth (Figure 5(a)). Since treatment with 0.1, 0.3, or 1 μM CuB prolongs lifespan, cell growth is less affected by this drug. Examination of yeast morphology after fluorescent staining of the cell wall, actin, and nuclear DNA revealed that treatment with 10 μM CuB did not cause obvious cell morphological changes (Figure 5(b)). After the fluorescent image pictures were quantified with a high-throughput, processing system CalMorph [19], we also statistically analyzed dose-dependent morphological changes with the Jonckheere-Terpstra test [36]. None of the 501 morphological parameters measured by CalMorph exhibited significant dose-dependent changes with FDR of 5%, confirming that CuB-treated cells did not change morphology significantly. These results suggested that CuB neither inhibits cell growth nor affects yeast morphology.
Figure 5.
CuB neither inhibits cell growth nor causes morphological changes. (a) The growth curve of Saccharomyces cerevisiae in the presence and absence of CuB was plotted. The assay was repeated at least twice. (b) Images of cells treated with control (1% EtOH) or CuB (10 μM). The fluorescent dyes fluorescein isothiocyanate-Con A, 4,6-diamidino-2-phenylindole, and rhodamine-phalloidin were used to stain the cell wall, nuclear DNA, and actin, respectively. Scale bar, 5 μm. (c) Scatter plot of mutants with nonessential gene deletions in terms of specific morphological abnormality (x-axis) and growth rate (y-axis). The horizontal and vertical red lines indicate the false discovery rate of 0.01 for growth rate and morphology, respectively. The number shown in each area indicates the number of mutants categorized on the basis of cell growth and morphology (single-gene deletion strains with increased lifespan/mutants categorized on the basis of cell growth and/or morphology).
3.7. Potential CuB Targets
Characterization of nonessential gene deletion mutants in terms of cell growth and morphology can be used to classify genes into four groups: genes not responsible for growth and morphology (group I, 1137 genes), genes only important for morphology (group II, 2294 genes), genes required for both growth and morphology (group III, 997 genes), and genes only required for growth (group IV, 203 genes) (Figure 5(c)) [21]. We attempted to use this information for prediction of the drug target by assuming that inhibition of the gene product by the drug is equivalent to the functional defect by gene deletion. Since CuB neither affects growth nor morphology, CuB likely inhibits the gene function belonging to group I (Figure 5(c)). Our genetic evidence suggested that the aging-related genes, UTH1 and SKN7, participate in antiaging effect of CuB. However, its interaction is likely indirect, because UTH1 and SKN7 belong to group II (Figure 5(c)). We also plotted gene deletion mutants with increased lifespan in Figure 5(c) [27]. Because group I contains 44 aging-related genes, we propose that CuB could target one of these 44 genes listed in Table 3. However, it cannot be excluded that the inhibition of the target by CuB is only partial, and that other genes, including essential ones, could be the main target of the drug.
Table 3.
The targets list of CuB predicted by CalMorph.
| ORF | Gene | Description |
|---|---|---|
| YBL052C | SAS3 | Something about silencing |
| YBR007C | DSF2 | Deletion suppressor of mptfive/puffive mutation |
| YBR034C | HMT1 | HnRNP methyl transferase |
| YBR042C | CST26 | Chromosome stability |
| YDL093W | PMT5 | Protein O-mannosyl transferase |
| YDL095W | PMT1 | Protein O-mannosyl transferase |
| YDR006C | SOK1 | Suppressor of kinase |
| YDR099W | BMH2 | Brain modulosignalin homolog |
| YDR110W | FOB1 | Fork blocking less |
| YDR313C | PIB1 | Phosphatidylinositol(3)-phosphate binding |
| YDR486C | VPS60 | Vacuolar protein sorting |
| YEL020C | PXP1 | Peroxisomal protein |
| YER164W | CHD1 | Chromatin organization modifier helicase and DNA-binding domains |
| YFR015C | GSY1 | Glycogen synthase |
| YFR040W | SAP155 | Sit4-associated protein |
| YGL006W | PMC1 | Plasma membrane calcium |
| YGL079W | KXD1 | KxDL homolog |
| YGL200C | EMP24 | Endomembrane protein |
| YGR254W | ENO1 | Enolase |
| YHL002W | HSE1 | Has symptoms of class E mutants; resembles Hbp, STAM, and EAST |
| YIL002C | INP51 | Inositol polyphosphate 5-phosphatase |
| YJL013C | MAD3 | Mitotic arrest-deficient |
| YJL098W | SAP185 | Sit4-associated protein |
| YKL098W | MTC2 | Maintenance of telomere capping |
| YLR176C | RFX1 | Regulatory factor X |
| YMR058W | FET3 | Ferrous transport |
| YMR126C | DLT1 | Defect at low temperature |
| YMR127C | SAS2 | Something about silencing |
| YMR221C | FMP42 | Found in mitochondrial proteome |
| YMR251W | GTO3 | Glutathione transferase omega-like |
| YMR251W-A | HOR7 | Hyperosmolarity-responsive |
| YNL142W | MEP2 | Mourning's ends part II |
| YOL071W | SDH5 | Succinate dehydrogenase |
| YOR311C | DGK1 | Diacylglycerol kinase |
| YPR111W | DBF20 | Dumbbell forming |
| YPL177C | CUP9 | Homeodomain-containing transcriptional repressor |
| YBR054W | YRO2 | Protein with a putative role in response to acid stress |
| YBR238C | YBR238C | Mitochondrial membrane protein |
| YIL089W | YIL089W | Protein of unknown function found in the ER and vacuole lumen |
| YCR101C | YCR101C | Putative protein of unknown function |
| YNL034W | YNL034W | Putative protein of unknown function |
| YDL172C | YDL172C | Dubious open reading frame |
| YDR048C | YDR048C | Dubious open reading frame |
| YGL165C | YGL165C | Dubious open reading frame |
4. Discussion
In China, CuB has been used to treat hepatitis for many years [37]. CuB also has potent anticancer, antioxidative, and anti-inflammatory activities [13, 14]. In the present study, we performed replicative lifespan assays to screen for antiaging substances from compounds library and used chronological lifespan assay to confirm the activity. We identified CuB as a potential antiaging compound. The chronological and replicative lifespan results shown in Figures 1(b) and 1(c) imply that CuB exerts significant antiaging effects in a dose-dependent manner.
We previously applied the replicative lifespan assay to evaluate the antiaging activities of other compounds. However, we found that this assay exhibits deficiencies that may affect the accuracy of its results. For example, random selection and subjective factors affect the quantification of daughter cells in the replicative lifespan assay. Therefore, to increase the accuracy of the bioassay results in the present study, we utilized the chronological lifespan assay to evaluate the antiaging activity of CuB. Yeasts have replicative and chronological lifespans. Replicative aging in yeast resembles the aging of mitotic cells in higher organisms, and chronological aging is similar to the aging of nondividing cells in higher eukaryotes [38]. The results of replicative and chronological lifespan assays suggest that CuB affects both the replicative and chronological lifespans of yeast.
In the present study, we focused on autophagy, oxidative stress, and longevity-related genes to investigate the mechanism of action of CuB. Autophagy levels decrease with age, and autophagy activation ameliorates age-related symptoms and has potential functions against aging [8]. Autophagy-related (ATG) or vacuolar protein-sorting genes encode some of the proteins that mediate autophagy [6]. Autophagy consists of nonselective and selective autophagy. Atg2 participates both nonselective and selective pathways of autophagy, including macroautophagy and pexophagy [39]; Atg32 is only essential for mitophagy [40]. Thus, we analyzed ATG2 and ATG32 expressions and performed replicative lifespan assays with atg2 and atg32 mutants with K6001 background and K6001 yeast and chronological lifespan assays with atg2 and atg32 mutants and YOM36 yeast to investigate the effect of autophagy on the antiaging activity of CuB. The results of gene expression, replicative lifespan, and chronological lifespan shown in Figures 2(a)–2(c) and 2(h) suggest that ATG2 and ATG32 genes have essential roles in the antiaging activity of CuB. Besides, enhanced mitophagy contributes to the extension of longevity by CuB.
Atg8 is an important component for autophagic machinery and participates in the whole process of autophagy, and it is a biomarker of autophagy in yeast [41]. Therefore, we investigated the effect of CuB on autophagy through detection of Atg8 with GFP-ATG8 fluorescent imaging and western blot analysis. Because RES can induce autophagy at 100 μM in mammalian cultured cells [10], we used it as a positive control in experiments for detection of autophagy in the present study. The changes of GFP in yeast in Figures 2(d) and 2(e) and western blot results in Figures 2(f) and 2(g) suggest that CuB significantly induces the autophagy of yeast.
We also investigated the effect of antioxidative activity on the antiaging function of CuB. The changes in the survival rate and Sod, ROS, and MDA levels of yeast shown in Figure 3 demonstrate that CuB exerts its antiaging effect in yeast by regulating antioxidative activity.
Longevity-related and aging-related genes have important roles in the regulation of aging and longevity. UTH1 is a yeast aging gene and takes part in oxidative stress [42]. SKN7 is the transcriptional activator of UTH1 and is related with protection against oxidative stress [32]. SOD1 and SOD2 genes encode Sod1 and Sod2 enzymes, respectively, which are important for redox homeostasis in cells. The replicative lifespan assay results of yeast mutants are shown in Figure 4 and Table 2 and reveal that UTH1, SKN7, SOD1, and SOD2 genes are required in the lifespan-extension activity of CuB. In addition, the replicative lifespan of the uth1 mutant is longer than that of the wild type. This result is consistent with the result of a previous study [35].
To predict the target of CuB, we used CalMorph to analyze the morphological changes exhibited by CuB-treated yeast cells. CalMorph is the image-processing software that has been used to analyze the morphology of 4718 nonessential gene mutants in more than 200 yeast strains. It can effectively identify compound targets [19]. The growth curve and morphological changes of cells treated with CuB are shown in Figure 5 and provide an important clue for the identity of the target genes of CuB. CuB does not cause growth inhibition nor cell morphological changes, and therefore, it could target one of the genes that are not involved in cell growth or morphology. After plotting mutations with long lifespan, we would pinpoint the possible targets of CuB among the 44 genes (Table 3). The results shown in Figure 5(c) suggest that CuB does not directly inhibit the genes mentioned in this study and that CuB might extend the lifespan of yeast by inhibiting upstream of aging-related genes mentioned in this study or by even activating some genes, such as SIR2, SOD1, and SOD2. If CuB activates some genes; activation of the CuB target would not cause growth inhibition or cell morphological changes. In this sense, the possible targets of CuB are not associated with cell growth or cell morphology. Nevertheless, additional efforts are needed to definitively identify the targets of CuB.
5. Conclusions
Overall, we found that CuB significantly prolongs the replicative and chronological lifespans of yeast in a dose-dependent manner. CuB exerts its antiaging effect by regulating autophagy and antioxidative activity. Moreover, the target identification of CuB suggests that 44 proteins that are not associated with cell growth or morphology are potential targets of CuB. However, the specific target of CuB requires further study, such as bioassay on these 44 proteins. Furthermore, given that CuB is already used to treat hepatitis cases in China, it may be developed as an antiaging drug.
Acknowledgments
This work was financially supported by the National Key R&D Program of China (Grant No. 2017YFE0117200), and the National Natural Science Foundation of China (Grant Nos. 21661140001, 21877098, and 21572204). We greatly thank Michael Breitenbach (Salzburg University, Austria) for gifts of the K6001 yeast strain.
Contributor Information
Lan Xiang, Email: lxiang@zju.edu.cn.
Jianhua Qi, Email: qijianhua@zju.edu.cn.
Data Availability
All the figures and table used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
Supplementary Fig. 1. The western blot analysis of GFP-ATG8 and free GFP of yeast after administrating CuB at different times and doses. Each experiment was independently repeated three times.
References
- 1.Deprez M. A., Eskes E., Winderickx J., Wilms T. The TORC1-Sch9 pathway as a crucial mediator of chronological lifespan in the yeast Saccharomyces cerevisiae . FEMS Yeast Research. 2018;18(5, article foy048) doi: 10.1093/femsyr/foy048. [DOI] [PubMed] [Google Scholar]
- 2.Polymenis M., Kennedy B. K. Chronological and replicative lifespan in yeast: do they meet in the middle? Cell Cycle. 2012;11(19):3531–3532. doi: 10.4161/cc.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarolim S., Millen J., Heeren G., Laun P., Goldfarb D., Breitenbach M. A novel assay for replicative lifespan in Saccharomyces cerevisiae . FEMS Yeast Research. 2004;5(2):169–177. doi: 10.1016/j.femsyr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y., Lin Y., Cao X., Xiang L., Qi J. Sterols from Mytilidae show anti-aging and neuroprotective effects via anti-oxidative activity. International Journal of Molecular Sciences. 2014;15(12):21660–21673. doi: 10.3390/ijms151221660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y., Sun Y., Weng Y., Matsuura A., Xiang L., Qi J. Parishin from Gastrodia elata extends the lifespan of yeast via regulation of Sir2/Uth1/TOR signaling pathway. Oxidative Medicine and Cellular Longevity. 2016;2016:11. doi: 10.1155/2016/4074690.4074690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyler J. K., Johnson J. E. The role of autophagy in the regulation of yeast life span. Annals of the New York Academy of Sciences. 2018;1418(1):31–43. doi: 10.1111/nyas.13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsui A., Kamada Y., Matsuura A. The role of autophagy in genome stability through suppression of abnormal mitosis under starvation. PLOS Genetics. 2013;9(1, article e1003245) doi: 10.1371/journal.pgen.1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubinsztein D. C., Mariño G., Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Bjedov I., Toivonen J. M., Kerr F., et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster . Cell Metabolism. 2010;11(1):35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morselli E., Mariño G., Bennetzen M. V., et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. Journal of Cell Biology. 2011;192(4):615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harman D. Aging: a theory based on free radical and radiation chemistry. Journal of Gerontology. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 12.Farrugia G., Balzan R. Oxidative stress and programmed cell death in yeast. Frontiers in Oncology. 2012;2:p. 64. doi: 10.3389/fonc.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Y., Fang X., He C., et al. Cucurbitacins: a systematic review of the phytochemistry and anticancer activity. The American Journal of Chinese Medicine. 2015;43(7):1331–1350. doi: 10.1142/S0192415X15500755. [DOI] [PubMed] [Google Scholar]
- 14.Tannin-Spitz T., Bergman M., Grossman S. Cucurbitacin glucosides: Antioxidant and free-radical scavenging activities. Biochemical and Biophysical Research Communications. 2007;364(1):181–186. doi: 10.1016/j.bbrc.2007.09.075. [DOI] [PubMed] [Google Scholar]
- 15.Chan K. T., Li K., Liu S. L., Chu K. H., Toh M., Xie W. D. Cucurbitacin B inhibits STAT3 and the RAF/MEK/ERK pathway in leukemia cell line K562. Cancer Letters. 2010;289(1):46–52. doi: 10.1016/j.canlet.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Zhang T., Li Y., Park K. A., et al. Cucurbitacin induces autophagy through mitochondrial ROS production which counteracts to limit caspase-dependent apoptosis. Autophagy. 2012;8(4):559–576. doi: 10.4161/auto.18867. [DOI] [PubMed] [Google Scholar]
- 17.Wu P. L., Lin F. W., Wu T. S., Kuoh C. S., Lee K. H., Lee S. J. Cytotoxic and anti-HIV principles from the rhizomes of Begonia nantoensis . Chemical and Pharmaceutical Bulletin. 2004;52(3):345–349. doi: 10.1248/cpb.52.345. [DOI] [PubMed] [Google Scholar]
- 18.Maruyama Y., Ito T., Kodama H., Matsuura A. Availability of amino acids extends chronological lifespan by suppressing hyper-acidification of the environment in Saccharomyces cerevisiae . PLoS One. 2016;11(3, article e0151894) doi: 10.1371/journal.pone.0151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohya Y., Sese J., Yukawa M., et al. High-dimensional and large-scale phenotyping of yeast mutants. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):19015–19020. doi: 10.1073/pnas.0509436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warringer J., Ericson E., Fernandez L., Nerman O., Blomberg A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki G., Wang Y., Kubo K., Hirata E., Ohnuki S., Ohya Y. Global study of holistic morphological effectors in the budding yeast Saccharomyces cerevisiae . BMC Genomics. 2018;19(1):p. 149. doi: 10.1186/s12864-018-4526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy S. F., Siegal M. L. Network hubs buffer environmental variation in Saccharomyces cerevisiae . PLoS Biology. 2008;6(11, article e264) doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yvert G., Ohnuki S., Nogami S., et al. Single-cell phenomics reveals intra-species variation of phenotypic noise in yeast. BMC Systems Biology. 2013;7(1):p. 54. doi: 10.1186/1752-0509-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang M., Ohnuki S., Ohya Y. Unveiling nonessential gene deletions that confer significant morphological phenotypes beyond natural yeast strains. BMC Genomics. 2014;15(1):p. 932. doi: 10.1186/1471-2164-15-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stasinopoulos D. M., Rigby R. A. Generalized additive models for location scale and shape (GAMLSS) in R. Journal of Statistical Software. 2007;23(7) doi: 10.18637/jss.v023.i07. [DOI] [Google Scholar]
- 26.Storey J. D. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B. 2002;64(3):479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 27.McCormick M. A., Delaney J. R., Tsuchiya M., et al. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metabolism. 2015;22(5):895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw P. X., Werstuck G., Chen Y. Oxidative stress and aging diseases. Oxidative Medicine and Cellular Longevity. 2014;2014:2. doi: 10.1155/2014/569146.569146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui H., Kong Y., Zhang H. Oxidative stress, mitochondrial dysfunction, and aging. Journal of Signal Transduction. 2012;2012:13. doi: 10.1155/2012/646354.646354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaweł S., Wardas M., Niedworok E., Wardas P. Malondialdehyde (MDA) as a lipid peroxidation marker. Wiadomosci Lekarskie. 2004;57(9-10):453–455. [PubMed] [Google Scholar]
- 31.Ritch J. J., Davidson S. M., Sheehan J. J., Austriaco N. O. P. The Saccharomyces SUN gene, UTH1, is involved in cell wall biogenesis. FEMS Yeast Research. 2010;10(2):168–176. doi: 10.1111/j.1567-1364.2009.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fassler J. S., West A. H. Fungal skn7 stress responses and their relationship to virulence. Eukaryotic Cell. 2011;10(2):156–167. doi: 10.1128/EC.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun K., Xiang L., Ishihara S., Matsuura A., Sakagami Y., Qi J. Anti-aging effects of hesperidin on Saccharomyces cerevisiae via inhibition of reactive oxygen species and UTH1 gene expression. Bioscience Biotechnology and Biochemistry. 2012;76(4):640–645. doi: 10.1271/bbb.110535. [DOI] [PubMed] [Google Scholar]
- 34.Austriaco N. R., Jr. Review: to bud until death: the genetics of ageing in the yeast, Saccharomyces . Yeast. 1996;12(7):623–630. doi: 10.1002/(sici)1097-0061(19960615)12:7<623::aid-yea968>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy B. K., Austriaco N. R., Jr., Zhang J., Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae . Cell. 1995;80(3):485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 36.Jonckheere A. R. A test of significance for the relation between m rankings and k ranked categories. British Journal of Statistical Psychology. 1954;7(2):93–100. doi: 10.1111/j.2044-8317.1954.tb00148.x. [DOI] [Google Scholar]
- 37.Wang Z., Zhu W., Gao M., et al. Simultaneous determination of cucurbitacin B and cucurbitacin E in rat plasma by UHPLC-MS/MS: a pharmacokinetics study after oral administration of cucurbitacin tablets. Journal of Chromatography B Analytical Technologies in the Biomedical and Life Sciences. 2017;1065-1066:63–69. doi: 10.1016/j.jchromb.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 38.Longo V. D., Shadel G. S., Kaeberlein M., Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae . Cell Metabolism. 2012;16(1):18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C. W., Kim J., Huang W. P., et al. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. Journal of Biological Chemistry. 2001;276(32):30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Developmental Cell. 2009;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amar N., Lustig G., Ichimura Y., Ohsumi Y., Elazar Z. Two newly identified sites in the ubiquitin‐like protein Atg8 are essential for autophagy. EMBO reports. 2006;7(6):635–642. doi: 10.1038/sj.embor.7400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandara P. D. S., Flattery-O'Brien J. A., Grant C. M., Dawes I. W. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Current Genetics. 1998;34(4):259–268. doi: 10.1007/s002940050395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. The western blot analysis of GFP-ATG8 and free GFP of yeast after administrating CuB at different times and doses. Each experiment was independently repeated three times.
Data Availability Statement
All the figures and table used to support the findings of this study are included within the article.