Abstract
Mammalian tissue development is an intricate, spatiotemporal process of self-organization that emerges from gene regulatory networks of differentiating stem cells. A major goal in stem cell biology is to gain a sufficient understanding of gene regulatory networks and cell-cell interactions to enable the reliable and robust engineering of morphogenesis. Here, we review advances in synthetic biology, single cell genomics, and multiscale modeling, which, when synthesized, provide a framework to achieve the ambitious goal of programming morphogenesis in complex tissues and organoids.
Keywords: Synthetic biology, Systems biology, Organoids, Stem cell self-organization, Multicellular systems, Single cell genomics
Self-organizing Multicellular Systems
The need for novel human-based cellular systems is vital due to shortages of donor organs, ineffective drug development pipelines, and a scarcity of predictive individualized human models. Animal models have thus far been the tools of choice to investigate developmental processes. However, fundamental differences in the biology between humans and, for example, mice [1], limit the translation of lessons learned from animal models to humans. The discovery of a method to generate a complex tissue akin to the optic cup from pluripotent stem cells in vitro [2], prompted a surge of studies involving organoids (see Glossary), gastruloids, and other forms of complex self-organizing tissues [3–5]. Self-organizing systems (reviewed by Simunovic and Brivanlou [3]) provide a new platform to understand developmental biology by emulating embryological process ex-vivo (a.k.a. “building to understand”) [6, 7].
Organoids, a prominent example of self-organizing cellular systems, have been grown in 3-D from human adult or pluripotent stem cells. Several examples of human organoids have been developed that can emulate structures and functions associated with human organs such as gut, kidney, liver, lung, and brain [8]. Organoids display the exciting potential to model key aspects of human development and disease processes, as well as advance efforts towards precision medicine and human disease modeling. However, stem cell-derived organoids often lack subsets of stromal cells, immune components, or a vascular system, and often fail to differentiate fully into mature phenotypes [9]. Current in vitro morphogenesis models frequently lack spatiotemporal control, robustness, and reproducibility that are exhibited in vivo by natural biological systems such as developing organs [10]. For example, soluble factors added to culture media affect the entire tissue with minimum spatial control. These limitations impede the development of organoids and negatively impact their utility in biology and medicine. Hence, the rapidly evolving fields of stem cell engineering and organoid technology face key challenges to systematically understand, control, and direct local and global morphogenetic events towards desired fates.
The collective properties of multicellular systems including final cellular composition, tissue identity and patterning arise from individual cell behavior. Recent technological advances in single cell analysis provide the chance to examine and link cell state to models of self-organization, morphogenesis, and tissue-level behaviors. Additionally, using engineering approaches can offer unprecedented capacities to exert biological control, probe the underlying design principles of multicellular systems, and generate miniature organs with phenotypes closer to native tissues. An integrative approach which combines cellular engineering with high resolution tissue analyses and in silico models provide an opportunity to rationally program morphogenesis and advance generation of human-based multicellular systems.
Genetically guided morphogenesis by transcription factors
A substantial portion of macromolecular players in developmental processes in complex animals are genomically encoded in the form of genetic regulatory codes [11]. These developmental gene regulatory networks (GRNs) control system-wide spatial positioning of specific cellular functions, progressive pattern formation, and emergence of organ forms and functions [12]. The ability to map and to manipulate GRNs can be exploited to program and guide morphogenesis in vitro in a predictable fashion. In fact, several past studies have used transcription factors to generate a homogenous cell population from stem cells or somatic cells [13–17]. More recently, transcription factor-based engineering has successfully been applied to generate complex human tissues (Box 1). Transcription factor triggered-morphogenesis can elicit spontaneous production of signaling cues that are required for tissue development and spontaneous morphogenesis. For instance, engineering heterogeneous levels of GATA6 transcription factor induced self-vascularizing fetal liver tissue from human induced pluripotent stem cells (Box 1) [18]. Additionally, cell fates triggered by transcription factors could override the effect of medium [18, 19]. Therefore, while traditional tissue engineering relies on engineering extrinsic cellular microenvironment, engineering tissues through manipulating cells’ intrinsic genetic programs can provide a new complementary approach for advanced organogenesis in a dish.
Box 1. Transcription Factor-triggered Complex Tissues.
The most straightforward approach towards genetically guided morphogenesis is to activate key transcription factors that have been identified as integral for the development of a given tissue. In fact, a few past biological studies have used this simple form of genetic circuit for engineering complex tissues as outlined below:
Thyroid:
Endocrine cell types of the thyroid are generated from uniquely double positive NKX2.1+/PAX8+ cells during the development of the gut tube. Antonica et al. directly expressed NKX2.1 and PAX8 using a doxycycline inducible switch for the target genes in mouse embryonic stem cells. After growing embryoid bodies for 4 days, the cultures were induced with 3 days of doxycycline which yielded NKX2.1+/PAX8+ cells capable of developing 3D thyroid follicle-like structures with the addition of thyroid stimulating hormone (TSH) into the medium [77].
Fetal liver:
During embryogenesis, tissues are developed through co-differentiation of multiple progenitor cells via reciprocal cell-cell interactions. Taking advantage of this design principle, human induced pluripotent stem cells (hiPSCs) were engineered with different copy number of an inducible GATA6-expressing circuit [18]. By transient and heterogeneous induction of GATA6 in hiPSCs, formation of all three human germ layers, symmetry breaking and co-differentiation of endoderm and mesoderm tissues were triggered. The cultures were kept in pluripotency medium for the first 5 days and subsequently transferred to basal medium without addition of growth factors. After 14 days, cultures formed complex 3D self-vascularized fetal liver-like tissue with different subsets of stromal niches. During the course of study, Gata6-engineered cells progressively self-organized into intricate patterns and self-produced signaling cues necessary for tissue morphogenesis. This study proposes genetic heterogeneity and co-development can act as two design principles for engineering multicellular self-organizing systems.
Thymus:
Bredenkamp et al. used forced expression of Foxn1 in mouse embryonic fibroblasts to generate Foxn1-induced thymic epithelial cells (iTECs) and tested their ability to support T-cell development. Transplantation of aggregates formed from iTECs mixed with PDGFR-αβ+ mesenchymal cells and CD45+Lin− immature thymocytes into nude Foxn1−/− mice generated functional thymus tissue capable of supporting generation of CD4+ and CD8+ T cells [78].
Pancreas:
Chen et al. demonstrated the potential for renewable sources of pancreatic β cells from conversion of intestinal crypts by transiently expressing β cell enriched transcription factors Pdx1, MafA, and Ngn3. This approach resulted in rapid conversion of intestinal organoids into a tissue with glucose responsive, insulin producing “neoislets” [79].
Genetically-guided morphogenesis via integration of systems and synthetic biology
Design principles of how collective morphogenetic behaviors arise from cells’ genetically encoded GRNs are inherently complex and incompletely understood. Advances in systems biology such as spatiotemporal single cell analysis and computational modeling provide the possibility to reverse engineer in vivo developmental processes to delineate the choice of transcription factors or other candidate pathways to interrogate in vitro. Subsequently, in vitro morphogenetic engineering can be carried out by overexpressing target genes or devising more sophisticated designer gene circuits that are interfaced with the candidate cellular GRNs. For instance, layered gene circuits and cascades can be exploited for stepwise controlled cellular differentiation. Synthetic biology is an emerging field that offers a compendium of toolsets germane to engineering of such genetic circuits. The designer gene circuits can provide spatiotemporal control over cell differentiations, migration, and cell-cell communications, and eventually in vitro organogenesis. Hence, they provide tools capable of steering in vitro morphogenetic processes (refer to Box 2 for explanations). Here, we will review advances in single cell genomics, multiscale modeling, and mammalian synthetic biology. We believe the integration of these areas can establish a synergistic pipeline for coherent design and engineering of human tissue morphogenesis in vitro (Box 2).
Box 2. Morphogenetic Engineering 101.
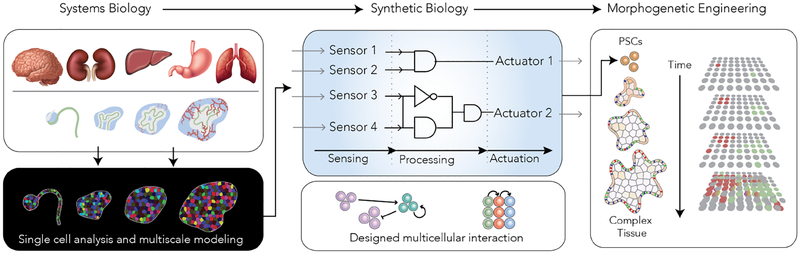
A simple transcription factor (TF)-based switch is an open loop genetic circuit without any feedback control. This simplest form of genetic circuit, is either constitutively active or can be inducible by a small molecule such as doxycycline. Engineering these simple circuits have already shown exciting biological potentials with generation of tissue patterns, functions and developmentally-relevant states [18, 77–79]. A single TF-induced cellular system made through overexpression of one TF may be thought as minimal morphogenetic units that, if characterized properly, will advance our biological understanding of morphogenesis and can improve our synthetic potentials. These units can be assembled in a plug-and-play format for generation of advanced tissues or organoids. For instance, TF-angiogenic or TF-innervating unit can be incorporated in current organoids to achieve additional tissue complexity. While open loop circuits will prove to be invaluable, in natural biological systems feedback loops and sensors provide robustness and maintain dynamic function within a fuzzy cellular context. In this line, Del Vecchio et al. have argued that engineering such feedback control mechanisms can augment cell fate programming (a.k.a. synthetic genetic feedback controller) [80]. Additionally, when building multicellular system engineering feedback loops can control size and improve cellular composition. Additionally, feedback-driven morphogenesis has been suggested before [81] and enables development of artificial tissue homeostasis [82]. Synthetic biology can provide designer genetic circuits with features such feedback regulations, temporal controls and cascades. Its contribution to morphogenesis of complex cellular systems can be two folds: First, it can help to decode molecular algorithms of cells to understand how they cooperate to encode a collective cell behavior (e.g. by using gene circuits to record cellular memory or to rewire existing GRNs). Second, it utilizes designer synthetic gene circuits to provide ability to spatiotemporally control and program multicellular systems. In both cases, modern systems biology approaches such as single cell genomics and multiscale modeling can generate working models that can identify set of key GRNs or display behavior of molecular networks. The model can guide the synthetic biology efforts while the synthetic biology toolbox alters the parameters and connections to experiment/improve the model’s accuracy. Hence, the integration of both parts remains pivotal to understand and engineer multicellular self-organization and morphogenetic functions (Figure I).
Figure I.
Integrating Systems and Synthetic Biology for Morphogenetic Engineering.
Single-cell analysis: A blueprint for decoding genetic regulatory codes
Single-cell RNA sequencing (scRNA-seq) has garnered much interest in recent years. Unlike bulk sequencing, which is prone to masking differences in cells due to averaging, single-cell approaches can capture the inherent heterogeneity between cells [20]. Data generated from these methods, when coupled with spatial and temporal information, can serve as a substrate for a bottom-up understanding of self-organizing processes such as morphogenesis. In this section, we describe several early examples of how single cell analytics can yield quantitative information on cell state, cell type, and localization that will be required for understanding self-organizing morphogenesis. The theoretical utility of the combination of data was explored by Pettit and colleagues when they developed a clustering method capable of incorporating both expression and spatial information to identify known and novel cell types. Inspired by image segmentation techniques, their method, which relies on a Hidden Markov Random Field model, clustered cells in Platynereis dumerilii brain more in a spatially coherent, and seemingly more biologically accurate manner as compared to traditional clustering algorithms that do not consider spatial information [21].
A barrier to the broader application of techniques to integrate spatial and transcriptional data is that readily-accessible methods to isolate and extract RNA from thousands of single cells requires tissue dissociation and subsequent loss of spatial information. Low throughput methods that involve manual isolation, such as laser capture microdissection, can maintain some spatial information, but these methods do not generate data of sufficient scale for systems biology studies [22–24]. Likewise, in situ hybridization and in situ sequencing methods currently do not scale well either in terms of the number of cells, or the number of genes assayed (Box 3). However, it is possible to computationally infer the localization of single cells by integrating scRNA-seq with data from in situ methods (Figure 1).
Box 3: In situ Molecular Profiling.
The development of fluorescence in situ hybridization (FISH) was a first step in deriving spatial information of RNA data [83]. Since then, advances in image detection and the commercialization of fluorescent probes have paved the way for techniques such as single-molecule FISH (smFISH), which utilizes the hybridization of fluorescent probes to target single mRNA molecules. Variants of this approach have focused on improvements to sensitivity and taken advantage of high throughput DNA synthesizers to create large numbers of singly labeled probes to cut down on false positives and negatives from nonspecific binding [84], allowing for a cleaner and more accurate signal.
Other smFISH variants have focused on methods to multiplex different mRNAs. Lubeck et al. developed seqFISH, in which transcripts are fixed in cells and subjected to rounds of hybridization of the same probe sequences with different fluorophores. Samples are imaged and probe stripped after each round, creating a sequential barcode of fluorescent spots that is theoretically capable of identifying F^N unique mRNAs, where F is the number of available fluorophores and N is the number of rounds of hybridization [85]. SeqFISH has since been used to localize cell types in the hippocampus [86].
To achieve the resolution required to map RNA to specific cellular compartments, Chen et al. developed Expansion FISH (ExFISH). This method relies on expansion microscopy (ExM), in which RNA molecules are linked to an expandable gel. The gel is expanded, and RNAs undergo traditional FISH, allowing for super-resolution imaging [87].
Recently developed sequencing-based approaches that capture spatial information have involved the use of in situ sequencing without RNA extraction. Ke et al. demonstrated the feasibility of sequencing chemistry within fixed cells and tissues with gap-targeted and barcode-targeted sequencing approaches that utilize padlock probing, rolling-circle amplification (RCA), and sequencing-by-ligation chemistry to analyze up to four-base-pair fragments in mRNA. Specificity was demonstrated through the gap-targeted approach by sequencing a different motif of beta-actin transcript (ACTB) in human and mouse fibroblasts that differ by a SNV and finding that 208 of 215 reads mapped to the correct cell type. Using the same approach, sensitivity was demonstrated by studying codon 12 of KRAS in KRAS mutant A549 and wild-type ONCO-DG1 and finding that mutations of a single cell could be detected at a 1:1000 spike-in ratio. Additionally, the barcode-targeted approach was utilized for multiplexed in situ profiling of 39 transcripts in breast cancer tissue [88].
The in situ sequencing and hybridization techniques described above have since progressed from barcode and gap-targeted approaches. Fluorescent in situ RNA sequencing (FISSEQ) begins with the reverse transcription of RNA in fixed cells and the incorporation of aminoallyl dUTP. The amine-modified cDNA amplicons are fixed to the cellular protein matrix and circularized before undergoing RCA and crosslinking in the presence of aminoallyl-dUTP. The resulting RCPs are single-stranded DNA nanoballs consisting of tandem copies of the cDNA sequence that is then sequenced via SOLiD sequencing by ligation. In human primary fibroblasts, FISSEQ was capable of demonstrating differential RNA expression and localization using 30 base pair reads in a wound healing assay [89, 90].
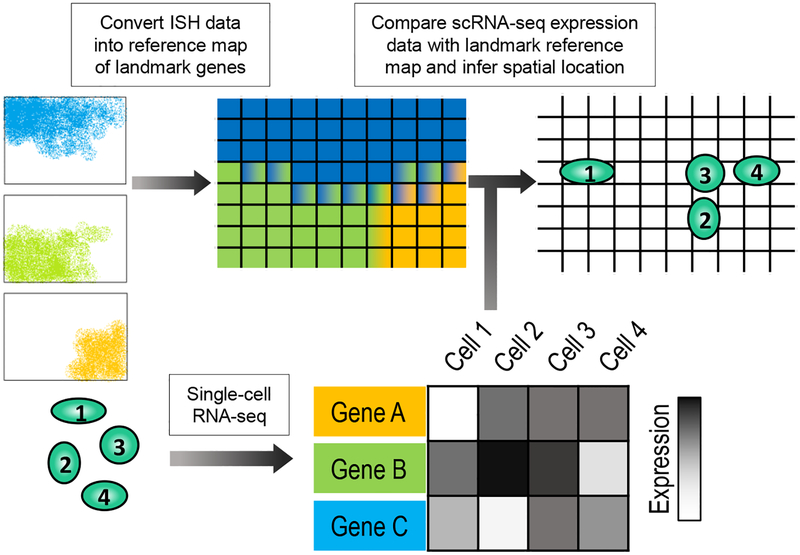
Figure 1. Inferring Location from scRNA-seq.
A generalization of the spatial inference strategies presented by Satija and colleagues [25], Achim and colleagues [26], and Halpern and colleagues [27]. In situ hybridization data is used to create a spatial reference map of landmark genes. In parallel, single-cell RNA-seq is performed on dissociated cells. Landmark gene expression from scRNA-seq data is compared to the reference map, and location of each dissociated cell is inferred.
One such approach is Seurat [25], which combines gene expression profiles in the form of scRNA-seq data and in situ hybridization data from a small set of landmark genes to infer spatial localization. In this method, the tissue of interest is spatially discretized into domains, or bins, that hold a binary value for each landmark gene, representing if it is expressed in that domain or not. Using this binary spatial map, statistical models are built for gene expression in each bin. Finally, each cell’s landmark gene expression is compared to the statistical models, and Seurat assigns a probability of the cell originating from each bin. Seurat successfully mapped 851 single cells in a developing zebrafish embryo using a discretized reference map of 47 genes. Additionally, Seurat has been used with unsupervised analysis of single-cell RNA-seq data, identifying rare sub-populations of cells, such as spatially characterizing prechordal plate progenitors, endodermal progenitors, and primordial germ cells [25]. In a separate but analogous approach, Achim and colleagues utilized FISH expression atlases and single-cell RNA-seq data to spatially characterize cells in the brain of the annelid Platynereis dumerilii [26].
Recently, Halpern and colleagues applied an equivalent approach in the mouse liver, combining smFISH of just six highly expressed landmark genes, diverse in expression along the lobule axis (“zonation”), with massively parallel single-cell RNA-seq. This resulted in the spatial expression profiles, or zonation profiles, of all liver genes, 50% of which were found to be significantly zonated, many of which peak at intermediate lobules [27]. Traditionally, hepatocytes are classified into one of two roles, periportal or pericentral. This study provided evidence for a non-binary spatial heterogeneity in the liver, revealing different roles for hepatocytes located between the two extremes. A more accurate representation of the division of labor will give rise to a more biological realistic metabolic network model. This method has also demonstrated the feasibility of applying such inference techniques in mammalian organs.
Paramount to understanding the underpinnings of self-organization will be to uncover cell-cell interactions. Camp and colleagues turned to scRNA-seq and established receptor-ligand pairs to determine the extent of cell crosstalk during liver bud development. In their screening method, a potential cell-cell interaction network was created based on an expression cutoff of receptor-ligand pairings between each cell-cell combination. Predictions from the screening were validated by knocking down receptor-ligand pairs and determining the effect on differentiation, as well as by utilizing chemical inhibitors to test for inhibition on liver bud development. These experiments demonstrated the importance of endothelial cell and hepatic endoderm crosstalk in differentiation and implicating a role for VEGF signaling in liver bud development. The cell-cell interactions predicted by this type of receptor-ligand analysis represent one method of uncovering potential crosstalk between cells during the self-organization process [28].
In addition to yielding quantitative information on cell state, cell type, and localization, single cell analytics can also be used to reconstruct or map developmental GRNs, which will be necessary to model and perform dynamic simulations of cell state transitions, and to design synthetic circuits for improved organoid engineering. Li and Luo and coworkers developed Sinova, a bioinformatics pipeline utilizing single cell RNA-seq expression data that was able to spatiotemporally reconstruct growth plate development, identifying and reconstructing previously unknown molecular cascades. This included the prediction of previously unknown transcription factors, such as the Klf and Fos families, and the reconstruction of GRNs across different developmental stages [29]. Buganim and colleagues utilized single-cell technologies to profile 48 genes at different time points during reprogramming. This work validated key gene-gene relationships (including a role of Sox2 in activating Oct4 through preservation of high levels of genes such as Nr5a2), demonstrated that pluripotency activation is possible with a combination of factors distinct from Oct4, Sox2, Nanog, Klf4, and c-Myc (such as Lin28, Sall4, Esrrb, and Dppa2), and thus revealed novel reprogramming factors [30]. Camp and colleagues utilized single cell RNA-seq to compare the cellular compositions of human cerebral organoids with fetal neocortex and reconstruct the gene regulatory networks and cell lineage underlying the differentiation process, ultimately finding that the organoid system represents a fairly accurate model of human corticogenesis [31]. Similarly, Gouti and colleagues mapped the GRN underpinning in vivo and in vitro neural-mesodermal differentiation and described how retinoic acid and Wnt signaling interact with the GRNs to balance neural and mesodermal differentiation [32]. Many other studies that map developmental GRNs are reviewed by Kumar and colleagues [33]. As a collective, these studies demonstrate the promise and ability of utilizing single-cell profiling to aide fate engineering.
To date, a spatially aware, cell type composition tool has yet to be developed, and utilizing spatial and composition information to improve cell fate engineering remains a goal in the field. Such approaches will be limited by data collection methods. In the landmark gene expression maps discussed above, for example, biologists are limited to systems in which a subset of genes can generate a highly resolved map. Ultimately, however, spatial, temporal, and regulatory information gleaned from these single cell analyses will not only elucidate specific developmental design elements, but will also be incorporated into multiscale models and simulations that will unravel suitable design paths for in vitro engineering of organogenesis.
Multiscale Modeling
Multiscale computational approaches provide a method of faithfully describing self-organization. Because cell-cell and cell-environment interactions are critical determinants of cell fate decisions and because the these interactions are highly heterogeneous in nature, multiscale approaches that model systems from the molecular and cellular level are particularly apt. These models evolve through time based on a set of biophysically-inspired rules, ultimately capturing interactions between cells and their environment that generate emergent phenomena. For example, Uzkudun and colleagues have used multiscale models and simulations to map GRNs that explain proximodistal patterning of the limb bud [34]. In general, there are many diverse approaches to performing multiscale modeling; below we discuss agent-based modeling and its application to complex cell population phenomena.
Agent-based models discretize the system being modeled into autonomous entities, each of which act according to a set of rules at each discrete time step throughout a simulation [35]. In such a framework, each agent is fully characterized by a set of inputs, outputs, possible states, and variables stored in its memory. Additionally, an agent must have a set of transition functions that bring the agent from one state to the next, a set of functions that map an input and memory variable to an output and updated memory variable, as well as an initial state and initial memory. Briefly, at each time step, each agent receives an input signal, and depending on its current memory, it may change states, releasing an output and updating its memory. In a biological setting, this could be paralleled to a cell that receives a chemical or spatial cue and transitions phenotypes based on a set of biological rules producing some form of output. Of course, an agent in biology is not limited to the cell, and for a complex system, a single model often includes many distinct types of agents.
Because this family of models is easily adaptable to biological systems, several groups have implemented them in various settings. Agent-based models have been used to simulate the initiation frequency and location of sprouting angiogenesis [36], leukocyte trafficking in the microvasculature [37], and Nanog and Rex1 heterogeneity in mouse embryonic stem cell (mESC) cultures [38] among others.
In particular, to simulate sprouting initiation, Walpole and coworkers spatially divided each cell into eight membrane nodes and a single centroid node, which were connected by a series of links. Adjacent cells were connected by intercellular links. Implementing rules for VEGF and DLL4 signaling, the model was able to accurately predict tip cell location and sprouting frequency [36]. To simulate differences between fluctuations in Nanog and Rex1 expression levels in mESC cultures due to noise and due to deterministic oscillations, Herberg and colleagues built a model of self-renewal, considering cell signaling between individual cells which stored information on transcription factor dynamics. Combining this method with experimentally observed results, it was found that the bistability of Nanog levels is due to deterministic regulation sustained by Fgf4/Erk signaling, and changes between the two stable states are helped by random noise [38].
Cell-based models, exemplified by cellular automata (CA) and cellular Potts (PC) models, can be considered as a sub-category of agent based models in which there is only a single agent type (Box 4). Cell-based models have proven useful to explore complexity in several developmental contexts. For example, Krupinski and colleagues built a cell-based model to describe early morphogenesis of the mammalian embryo. In their model, blastomeres are elastic spheres that interact mechanically with each other, with each blastomere containing a specific genetic network that defines its mechanical properties. Cell division is modeled as a discrete event that conserves total cell volume during that time step. Cells divide either randomly, with content split symmetrically, or along the direction of a polarized vector, with content split asymmetrically. Their multiscale model was subsequently described by equations pertaining to mechanical motion and to genetic network dynamics [39]. In another application of cell-based models, Leung et al. developed an agent-based CP model simulating the development of the genital tubercle. Their model restricted the cells to a 2D lattice with dynamics governed by signaling networks encompassing SHH, FGF10, and androgen [40].
Box 4: Cell-Based Models.
As agent-based models, cell-based models discretize the system to be modeled into individual cells or subcellular components. These models come in two types – on-lattice and off-lattice. In the first, cells or cellular components are constrained to the coordinates of a grid. In the second, any cell or cellular component’s position is represented by any real number.
Two popular on-lattice or lattice-based models are cellular automata (CA) and cellular Potts (CP) models (Figure I). In CA, cells are assigned to a single site on the lattice and each site holds at most one cell. In CP models, each cell spans several sites on the grid. The simplicity of CA allows for fast simulation of large numbers of cells, whereas the CP model offers a more realistic representation of cell shape. The lattice-structure of these models offer ease in determining which cells are neighboring, which are just any pairs in adjacent lattice sites. However, the lattice results in a low degree of spatial flexibility, and processes requiring cell movement or cell division require shifting the rest of the tissue [91]. Unlike agent-based models, in which multiple types of agents can interact with each other and the environment directly, cells in on-lattice cell-based models are thus typically static and homogeneous, while the environment moves through them.
Off-lattice models can offer more flexibility in cell shape and location, and thus these models are often used in mechanical studies of cell populations. Vertex models are one specific category of off-lattice cell-based approaches for characterizing aspects of self-organization in the context of cell mechanics. These models represent the shape of the tissue of interest by a set of vertices that are typically defined as the points of contact between three or more cells. In defining vertices in such a way, cellular interfaces and volumes can be determined based on the location of the vertices. Models can be two or three dimensional, and either apical or lateral. The motion of each of the vertices is determined by equations describing internal and external forces acting on the vertex. Vertex models are easily adapted to biological systems influenced by cellular mechanics. As such, they have been used to study a variety of processes in developmental biology, most notably epithelial morphogenesis [92–94]. Recently, a modified vertex model was used to study the formation of respiratory appendages in Drosophila oogenesis [94].
The benefits of implementing a vertex model include the ability to describe the shape of a tissue based on changes in cell shape and rearrangements. However, as with any discrete approach, a vertex model could become cumbersome with a large number of parameters. In this case, continuous approaches may offer an easier solution, despite generalizing the characteristics and behavior of the tissue and losing details of mechanics at the cellular level.
Figure I.
Examples of How Cell-based Modeling Can be Used to Represent Self-organizing Processes. Phenotypic changes, as well as mechanical changes, can be modeled by taking discrete steps through time. At each step, changes in spatial or chemical cues drive changes in the system based on a set of pre-defined rules.
The history of agent-based (and cell-based) models can be traced back to the 1940s when von Neumann, Ulam, and Conway first introduced Cellular Automata. Regardless of terminology, all of these multiscale models represent powerful yet underutilized approaches capable of simulating self-organizing processes. The goal of simulation will be to gain insight on the effect of manipulation on system parameters, not only to help elucidate the complexities of self-organization, but also to generate more information that can be used in further experimental and computational studies.
In the context of self-organization, the agent-based modeling is appealing for several reasons. First it is a discrete model which unlike a continuous approach, is able to account for the heterogeneity in a system. Because the system is composed of autonomous agents, individual agents can serve different functions, follow distinct sets of rules, and have different properties, implying that individual agents have unique responses to system inputs. As an example, this type of model can account for spatial considerations, in which a cell located closer to a cue will respond differently, and not just to a different degree, than a cell located further away. Second, the agent-based model is an interaction-based model in that interactions between agents and the environment drive the simulation at each time step. This allows for the consideration of lower-level detail, giving a more biologically faithful representation of the system as long as the set of rules and parameters does not become cumbersome. Finally, multiple agent-based programming languages exist that can be adapted to model biological systems including NetLogo [41] and AgentSpeak [42].
Synthetic Biology for Morphogenetic Engineering
Understanding how individual signaling elements assemble into a morphogenetic complex behavior in time and space remains as a challenge in decoding tissue development. A synthetic biology approach provides a “build to understand” methodology to answer such questions. It additionally provides a toolbox to guide tissue self-organization towards final desired states. In fact, pattern formation in non-mammalian systems has been engineered using synthetic biology-based strategies [43].
A simple genetic circuit in synthetic biology is composed of sensor(s), processor(s), and actuator(s). Sensors recognize cellular or environmental cues (inputs). Processors determine the type of response. Actuators produce a defined response (output). These elements can shape genetic circuits that can be interfaced with cellular states or micro-environmental contexts and initiate cell fate programming, cellular communications and program collective tissue behaviors such as morphogenesis. Ultimately, the goal is to define a set of rules and morphogenetic modules in a plug-and-play format to create defined morphogenetic behaviors. A limited set of such modules was described before including ones for cell proliferation or death, adhesion, migration and sorting [44].
An ability to couple molecular inputs relevant to a cell state or an environmental context (cell state sensor) to a user-defined output is vital for spatiotemporal control in multicellular systems. Several sensors have been developed to identify biological (e.g., a protein) or non-biological (e.g., light) signals [45–48]. For example, through promoter engineering and signal amplification using a positive feedback loop, an integrative framework for selective and robust sensing of transcription factors were achieved [49]. These synthetic promoters can act as a valuable tool to spatially limit the function of genetic circuits in cell type of interest. MicroRNA (miRNA) levels can differentiate between a wide range of cell types and have been employed as classifiers for cell types of interest [50]. By incorporation of miRNA target sites in a transcript of interest, a library of miRNA-responsive sensors (Figure 2a) has been constructed [51]. For instance, tissue specific miRNA sensor encoding Bim apoptotic protein were engineered to enrich for cell type of interest such as cardiomyocytes after stem cell differentiation [52]. To achieve programmable integration of multiple inputs, Xie and coworkers [53] generated a “classifier” gene circuit that integrates information from six endogenous miRNA profiles to differentiate two cell lines with ON:OFF ratio of ~ 8- to 10-folds. In fact, during mammalian development cell death plays a key role in sculpting the organs and select for fittest cell population [54]. Therefore, miRNA-based cell death circuit can be used to guide in vitro tissue formation or reduce aberrant cell fates within developing organoids.
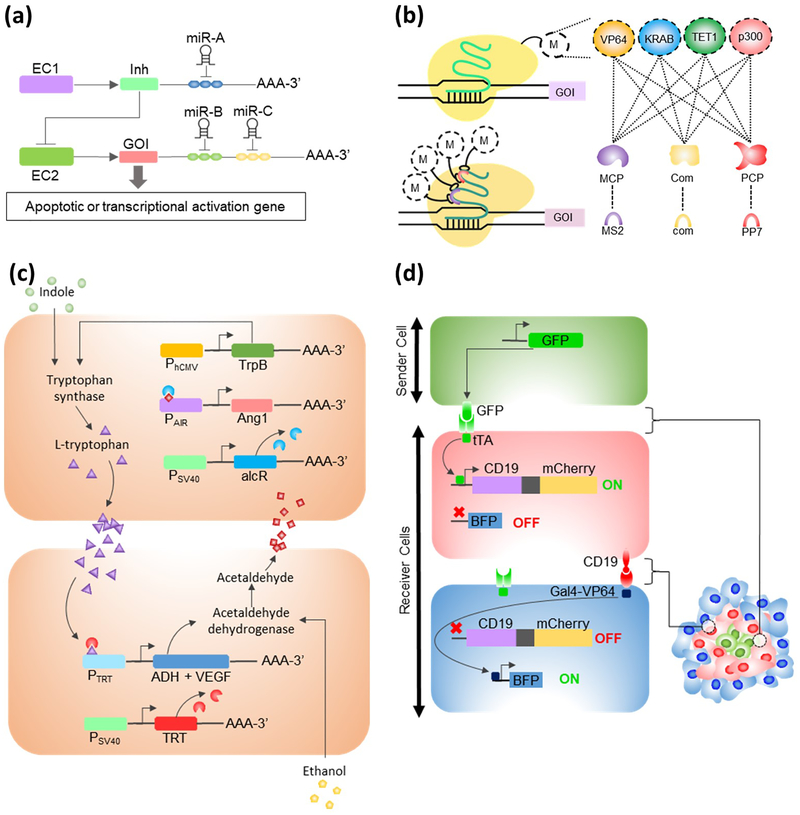
Figure 2. Synthetic Biology Tools for Programming Cell and Tissue.
(A) Complex layered microRNA (miR) classification circuits incorporating gene of interest (GOI) can be engineered via cell specific, high and/or low microRNA sensors. The circuit requires high levels of miR-A and low levels of miR-B and C for transcription of the GOI. EC, Engineered DNA Circuit (B) The dCas9 system can be used to implement multiplexed activation and repression of GOI’s by various effector proteins, including transcription activators (VP64) and repressors (KRAB), DNA demethylases (TET1), and chromatin remodelers (p300), either (i.) tethered directly to the Cas9 protein, or (ii.) fused to an RNA binding domain such as MCP, Com, or PCP. M, modular effector domain [66]. (C) Bacchus and colleagues engineered “sender/receiver” and “processor” cells [71]. The genetic circuit interactions shown were successfully utilized to generate a two-way communication system that controls the permeability of an endothelial cell layer by VEGF and Ang1 expression. TRT, l-tryptophan-dependent transactivator; AlcR, acetaldehyde-dependent transactivator; VEGF, vascular endothelial growth factor; Ang1, Angiopoietin-1. (D) Morsut and colleagues developed “synNotch” system [73] which incorporates modified Notch signaling pathway components into cells to create multi-layered self-organizing ring patterns. Sender cell displays GFP on plasma membrane that interacts with anti-GFP synNotch on receiver cell. SynNotch releases tTA and expresses both CD19 and red fluorescent reporter proteins. CD19 triggers anti-CD19 synNotch receptor on the adjacent cell and induces BFP. Figures 2B, 2C, and 2D are redrawings based on Shalem et al., Bacchus et al., and Morsut et al. respectively.
Artificial RNA switches such as RNA aptamers and aptazymes, are another class of genetic sensors that binds molecules of interest and control gene expression in mammalian cells [48, 55]. To this end, Wnt- and NF-κB-responsive RNA aptamers were produced which could link activation of underlying pathways to target gene induction. However, engineering aptamers with high affinity to a molecule of interest is not always feasible, which hamper the modularity of this approach. Other seminal studies have developed sensors that can interface an external environmental cue to cell’s transcriptional program. A modular extracellular sensor architecture (MESA) is a customizable example that acts via ligand-induced receptor dimerization, a protease action and release of a cognate transcriptional modulator [45]. Combinatorial usage of MESAs can provide a platform for multi-parametric environmental sensing [56].
Engineering cellular computation and signal processing enables development of synthetic gene circuits that integrate and convey multiple inputs towards desired cellular outputs. Often, Boolean logic gates have been employed to illustrate multi-input-multi output network topologies in cells [57, 58]. Saxena and coworkers developed transcription factor-based gene switches and cascades with differential sensitivity to vanillic acid as an input signal. They could recreate temporal dynamics of the major transcription factors Pdx1, Ngn3, and MafA for pancreatic lineage differentiation. Such stepwise genetic activation in progenitor cells could produce glucose-sensitive insulin-secreting beta-like cells [59]. Similar stepwise transcription factor triggered cascades can be exploited to encode co-development of cells and structures in a complex tissue. Through composing genetic circuits, complex network topology can also be achieved and may include auto feedback regulatory loops, genetic cascades, bistable expression networks, time-delay circuit as well as oscillators (Box 2) [60].
The advent of precision genome targeting tools provides us with additional set of transcriptional modulators that enable orthogonal gene activation and repression [61, 62]. These systems include zinc finger proteins, transcription activator-like effectors (TALEs), and the clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 system [62]. Deactivated Cas9 (dCas9) that cannot cleave DNA but retains DNA binding function has been engineered with potent transcriptional modulator domains and chromatin remodelers to achieve effective epigenetic and transcriptional programming tool [62] (Figure 2b). Layered dCas9 synthetic circuits can be built to generate a functional cascade [63]. Additionally, guide RNA (gRNA) engineering using different types of aptamers provided an orthogonal platform for recruitment of different proteins such as activator or repressor domains at defined genomic loci [64–66] (Figure 2b). By extending gRNAs to include modified riboswitches that recognize specific signals in Wnt, NFkB, and oncogenic P53, CRISPR–Cas9-based “signal conductors” have been generated [67]. This circuit enables transcriptional regulation of endogenous genes in response to inflammatory (NFkB) or developmental (β-catenin) and oncogenic cues. Programmability of Cas9 and multiplexing of several orthogonal gRNAs offer possibility for combinatorial activation and repression of different GRN targets.
Cell-cell communication and migration are key steps to encode ordered patterns during morphogenesis. Engineering tunable cell-cell communication with short or long range function can program collective cell behavior and induce coordinated functions in mammalian systems. Recent studies have created both soluble and insoluble synthetic intercellular communications and developed strategies to control mammalian cell motility [68, 69]. Engineering G protein-coupled receptor resulted in directing mammalian cell motility to a bio-inert drug-like small molecule, clozapine-N-oxide (CNO) [70]. A synthetic multicellular two-way communication system was developed using L-tryptophan and acetaldehyde as communicating soluble signals (Figure 2c) [71]. The sender-processor-receiver format can be used to emulate a number of soluble signaling interactions of interest and enables tissue level computation. Cachat and colleagues used the differential adhesive strengths between homotypic and heterotypic E-cadherin and P-cadherin interactions to drive cell sorting when expressed in mammalian cells [72]. Through cadherin-mediated phase separation they could promote mammalian cells to generate intricate 2- and 3-D patterns de novo. In an elegant study, Notch signaling was exploited as a platform to link recognition of an arbitrary extrinsic cellular input to intrinsic transcriptional outputs in an orthogonal manner [73]. Morsut and colleagues designed this platform by replacing extracellular sensor module and intracellular transcriptional module of Notch signaling with diverse ligand binding or transcription factor domains (Figure 2d). This synthetic Notch system (synNotch) was used to generate cellular patterns around GFP expressing “sender cells”. SynNotch is a novel toolset to engineer user-defined tissue level computing, however it lacks amplification signal typically observed in intracellular signaling events. This means one ligand molecule (input) can presumably activate one intracellular signaling molecule (output). Additionally, other modes of cell-cell communication such as exosomes can be engineered to establish exchange of protein or RNA cargos between sender and receiver cells [74].The diverse exosomal inputs signals (e.g. miRNAs) opens myriad of possibilities for transcriptional regulation of morphogenesis or broadcasting cellular fitness or tissue composition during in vitro tissue development.
Concluding Remarks and Future Perspectives: Reading and Writing of Human Tissue Complexity
Organoids and other self-organizing tissues have shown great promises to replicate fundamental facets of human tissue development. At least for therapeutic human translation or disease modeling it is critical to engineer human tissues that can closely mimic their natural counterparts. With this biological design objective in mind, different strategies have been proposed to advance our current approaches relevant to “organogenesis in a dish” [75]. Organ-on-a-chip engineering approaches have focused on probing cell and tissue microenvironment using microfluidic devices and tissue scaffolds with smart biomaterials and controlled mechanical properties [76]. The approach relies on presentation of defined physio-chemical and mechanical cues from outside the cells. Hence, it necessitates knowledge and tools to recapitulate composition and spatiotemporal dynamics of developmental cues presented to the cells, at a given time in morphogenesis.
Synthetic biology-based tissue engineering focuses on engineering cells from inside via genetic circuits with defined functions. It provides a forward engineering approach to generating tissues and organs via genetically programing cell behavior and subsequently tissue morphogenesis. However, with a large space of parameters, finding a best set of candidate genes and pathways for forward engineering might become a challenge (see Outstanding Questions). Yet, the developmental program of mammalian tissues already offer an unmatched library of GRNs that evolved through many years of evolution. If reverse engineered properly, the information can aid synthetic biology for rational engineering of morphogenesis. Additionally, via assembly or rewiring of genetic networks in one or multiple cell population(s), a synthetic biology practice can help in multiscale decoding of organizational principles central for the final integrated cellular behaviors (i.e. self-organization and morphogenesis). By integration of systems and synthetic biology one can envision new generation of programmable tissues and designer organoids that can closely recapitulate human development, shed light to our own engineering design principles or deliver user-defined organ functions. Combined with other engineering approaches such as organ-on-a-chip engineering they can shape the future of modern tissue engineering.
Trends Box.
Stem cell-derived multicellular systems and organoids have opened new opportunities to emulate and understand human development in vitro and provide novel patient specific tissue surrogates for disease modeling.
Single cell sequencing technologies and spatial tissue analysis provide a wealth of information on cellular fate and function and offer invaluable opportunities to decipher developmental processes.
Mammalian synthetic biology utilizes designer synthetic gene circuits to program cell fate and functions towards a desired outcome.
Engineering morphogenesis is an emerging area of science that integrates engineering principles with developmental biology to control and guide collective cell behaviors.
Outstanding Questions box.
What is the best strategy to deliver large genetic circuits to mammalian stem cells without encountering gene silencing or perturbed function over time?
The predictability of synthetic gene networks within the mammalian cells remains poor. How can unintended interactions with host cells and limitations of cellular resources among engineered gene circuits be identified and eliminated?
What computational strategies can be used to identify a core set of tissue parameters and cellular GRNs that are amenable for engineering a given morphogenetic behavior?
How can the balance between engineering defined function and emergence of new behavior be maintained? Can we exploit emergent behaviors to improve morphogenetic engineering?
How can synthetic morphogenetic circuits be developed with the robustness of naturally occurring genetic circuits, which have the advantage of multiple feedback systems and redundant parts?
Acknowledgements
We would like to thank Dr. Samira Kiani for the insightful comments during the preparation of this work as well as Ms. Ashley Lear for helping in the figures. We apologize to anyone whose work could not be cited due to limitations in space.
Glossary
- Aptamer
a nucleic acid or peptide molecule that binds a particular molecular target with high affinity and specificity
- Aptazyme
An aptamer domain fused to a ribozyme domain. Upon the binding of the aptamer, catalytic action of the ribozyme region is activated
- FISH
Fluorescent in situ hybridization. A technique used for visualizing specific sequences of nucleic acids within cells based on complementary base pairing of a florescent or radioactively tagged DNA probe with a target sequence
- Forward engineering
Starting with high-level model which establishes the goals and functions of a system before its physical design or construction
- Gastruloid
A stem cell-based model which emulates spatiotemporal aspects organization and differentiation of gastrulation
- Gene regulatory network (GRN)
A network of interacting genes that collectively controls particular cellular functions
- Genetic circuit
Partly analogous to electronic circuits, it is used in synthetic biology to define an integrated assembly of genetic parts that produces a genetic function
- Morphogenesis
It defines broadly here as events that collectively encode tissue pattern and shapes that are controlled by cell fate decisions, migration, cell-cell interaction and physical cues
- Morphogenetic Engineering
Here it refers to engineering morphogenesis using genetic techniques
- Organoid
A self-organizing, 3-dimentional, multicellular system grown from stem cells in vitro, that contains several organ-specific cell types with associated tissue structures and functions
- Organogenesis in a dish
In vitro approaches directed towards generation of complex tissues and organ-like structures via emulating aspects of developmental processes
- Reverse engineering
decomposition of a system into its functional parts in order to guide understanding of performance objectives and functions of individual components
- Ribozyme
An RNA molecule that can function as an enzyme
- Self-organization
Loosely defined as spontaneous formation of ordered structures via local interactions of less organized parts with no centralized ordering or external template. For more details see Simunovic and Brivanlou [3]
- Synthetic biology
An interdisciplinary field of study for engineering biology that entails (re)design and construction of existing or novel biological systems using well-characterized standardized genetic parts
- Systems biology
An interdisciplinary field of study that aims to develop an integrated understanding of biological systems using quantitative measures of individual components, their interaction dynamics and computational modeling
References
- 1.Seok J et al. (2013) Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110 (9), 3507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eiraku M et al. (2011) Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472 (7341), 51–6. [DOI] [PubMed] [Google Scholar]
- 3.Simunovic M and Brivanlou AH (2017) Embryoids, organoids and gastruloids: new approaches to understanding embryogenesis. Development 144 (6), 976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Brink SC et al. (2014) Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 141 (22), 4231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatehullah A et al. (2016) Organoids as an in vitro model of human development and disease. Nat Cell Biol 18 (3), 246–54. [DOI] [PubMed] [Google Scholar]
- 6.Davies J (2017) Using synthetic biology to explore principles of development. Development 144 (7), 1146–1158. [DOI] [PubMed] [Google Scholar]
- 7.Kicheva A and Rivron NC (2017) Creating to understand - developmental biology meets engineering in Paris. Development 144 (5), 733–736. [DOI] [PubMed] [Google Scholar]
- 8.Clevers H (2016) Modeling Development and Disease with Organoids. Cell 165 (7), 1586–97. [DOI] [PubMed] [Google Scholar]
- 9.Lancaster MA and Knoblich JA (2014) Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345 (6194), 1247125. [DOI] [PubMed] [Google Scholar]
- 10.Huch M et al. (2017) The hope and the hype of organoid research. Development 144 (6), 938–941. [DOI] [PubMed] [Google Scholar]
- 11.Bolouri H and Davidson EH (2002) Modeling transcriptional regulatory networks. Bioessays 24 (12), 1118–29. [DOI] [PubMed] [Google Scholar]
- 12.Peter IS and Davidson EH (2011) A gene regulatory network controlling the embryonic specification of endoderm. Nature 474 (7353), 635–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Y et al. (2014) Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 14 (3), 394–403. [DOI] [PubMed] [Google Scholar]
- 14.Iwafuchi-Doi M and Zaret KS (2014) Pioneer transcription factors in cell reprogramming. Genes Dev 28 (24), 2679–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lis R et al. (2017) Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545 (7655), 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panciera T et al. (2016) Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell Stem Cell 19 (6), 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugimura R et al. (2017) Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545 (7655), 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guye P et al. (2016) Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat Commun 7, 10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busskamp V et al. (2014) Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol 10, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C (2015) Defining cell types and states with single-cell genomics. Genome Res 25 (10), 1491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettit JB et al. (2014) Identifying cell types from spatially referenced single-cell expression datasets. PLoS Comput Biol 10 (9), e1003824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberwine J et al. (1992) Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A 89 (7), 3010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagrath S et al. (2007) Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450 (7173), 1235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang F et al. (2010) RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc 5 (3), 516–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satija R et al. (2015) Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33 (5), 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Achim K et al. (2015) High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol 33 (5), 503–9. [DOI] [PubMed] [Google Scholar]
- 27.Halpern KB et al. (2017) Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542 (7641), 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camp JG et al. (2017) Multilineage communication regulates human liver bud development from pluripotency. Nature 546 (7659), 533–538. [DOI] [PubMed] [Google Scholar]
- 29.Li J et al. (2016) Systematic Reconstruction of Molecular Cascades Regulating GP Development Using Single-Cell RNA-Seq. Cell Rep 15 (7), 1467–1480. [DOI] [PubMed] [Google Scholar]
- 30.Buganim Y et al. (2012) Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150 (6), 1209–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camp JG et al. (2015) Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A 112 (51), 15672–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouti M et al. (2017) A Gene Regulatory Network Balances Neural and Mesoderm Specification during Vertebrate Trunk Development. Dev Cell 41 (3), 243–261 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar P et al. (2017) Understanding development and stem cells using single cell-based analyses of gene expression. Development 144 (1), 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uzkudun M et al. (2015) Data-driven modelling of a gene regulatory network for cell fate decisions in the growing limb bud. Mol Syst Biol 11 (7), 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaul H and Ventikos Y (2015) Investigating biocomplexity through the agent-based paradigm. Brief Bioinform 16 (1), 137–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walpole J et al. (2015) Agent-based model of angiogenesis simulates capillary sprout initiation in multicellular networks. Integr Biol (Camb) 7 (9), 987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey AM et al. (2007) Multi-cell agent-based simulation of the microvasculature to study the dynamics of circulating inflammatory cell trafficking. Ann Biomed Eng 35 (6), 916–36. [DOI] [PubMed] [Google Scholar]
- 38.Herberg M et al. (2016) Dissecting mechanisms of mouse embryonic stem cells heterogeneity through a model-based analysis of transcription factor dynamics. J R Soc Interface 13 (117). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krupinski P et al. (2011) Simulating the mammalian blastocyst--molecular and mechanical interactions pattern the embryo. PLoS Comput Biol 7 (5), e1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung MC et al. (2016) Computational modeling and simulation of genital tubercle development. Reprod Toxicol 64, 151–61. [DOI] [PubMed] [Google Scholar]
- 41.NetLogo itself: Wilensky U. 1999. NetLogo http://ccl.northwestern.edu/netlogo/. Center for Connected Learning and Computer-Based Modeling, Northwestern University; Evanston, IL. [Google Scholar]
- 42.Bordini RH, Hubner JF, Wooldridge M. Programming Multi-Agent Systems in AgentSpeak Using Jason. John Wiley & Sons, 2007 [Google Scholar]
- 43.Basu S et al. (2005) A synthetic multicellular system for programmed pattern formation. Nature 434 (7037), 1130–4. [DOI] [PubMed] [Google Scholar]
- 44.Cachat E et al. (2014) A library of mammalian effector modules for synthetic morphology. J Biol Eng 8 (1), 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daringer NM et al. (2014) Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth Biol 3 (12), 892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slomovic S and Collins JJ (2015) DNA sense-and-respond protein modules for mammalian cells. Nat Methods 12 (11), 1085–90. [DOI] [PubMed] [Google Scholar]
- 47.Toettcher JE et al. (2011) Light-based feedback for controlling intracellular signaling dynamics. Nat Methods 8 (10), 837–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wieland M et al. (2012) Engineering of ribozyme-based riboswitches for mammalian cells. Methods 56 (3), 351–7. [DOI] [PubMed] [Google Scholar]
- 49.Angelici B et al. (2016) Synthetic Biology Platform for Sensing and Integrating Endogenous Transcriptional Inputs in Mammalian Cells. Cell Rep 16 (9), 2525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown BD et al. (2007) Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol 25 (12), 1457–67. [DOI] [PubMed] [Google Scholar]
- 51.Mullokandov G et al. (2012) High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 9 (8), 840–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miki K et al. (2015) Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell 16 (6), 699–711. [DOI] [PubMed] [Google Scholar]
- 53.Xie Z et al. (2011) Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333 (6047), 1307–11. [DOI] [PubMed] [Google Scholar]
- 54.Penaloza C et al. (2006) Cell death in development: shaping the embryo. Histochem Cell Biol 126 (2), 149–58. [DOI] [PubMed] [Google Scholar]
- 55.Culler SJ et al. (2010) Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330 (6008), 1251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartfield RM et al. (2017) Multiplexing engineered receptors for multiparametric evaluation of environmental ligands. ACS Synth Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benenson Y (2012) Biomolecular computing systems: principles, progress and potential. Nat Rev Genet 13 (7), 455–68. [DOI] [PubMed] [Google Scholar]
- 58.Purcell O and Lu TK (2014) Synthetic analog and digital circuits for cellular computation and memory. Curr Opin Biotechnol 29, 146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saxena P et al. (2016) A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat Commun 7, 11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greber D and Fussenegger M (2007) Mammalian synthetic biology: engineering of sophisticated gene networks. J Biotechnol 130 (4), 329–45. [DOI] [PubMed] [Google Scholar]
- 61.Gao Y et al. (2016) Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods 13 (12), 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thakore PI et al. (2016) Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods 13 (2), 127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiani S et al. (2014) CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat Methods 11 (7), 723–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiani S et al. (2015) Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods 12 (11), 1051–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma H et al. (2016) Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34 (5), 528–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shalem O et al. (2015) High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16 (5), 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y et al. (2016) Directing cellular information flow via CRISPR signal conductors. Nat Methods 13 (11), 938–944. [DOI] [PubMed] [Google Scholar]
- 68.Carvalho A et al. (2014) Genetically encoded sender-receiver system in 3D mammalian cell culture. ACS Synth Biol 3 (5), 264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang WD et al. (2008) Construction of an artificial intercellular communication network using the nitric oxide signaling elements in mammalian cells. Exp Cell Res 314 (4), 699–706. [DOI] [PubMed] [Google Scholar]
- 70.Park JS et al. (2014) Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc Natl Acad Sci U S A 111 (16), 5896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bacchus W et al. (2012) Synthetic two-way communication between mammalian cells. Nat Biotechnol 30 (10), 991–6. [DOI] [PubMed] [Google Scholar]
- 72.Cachat E et al. (2016) 2- and 3-dimensional synthetic large-scale de novo patterning by mammalian cells through phase separation. Sci Rep 6, 20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morsut L et al. (2016) Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell 164 (4), 780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marcus ME and Leonard JN (2013) FedExosomes: Engineering Therapeutic Biological Nanoparticles that Truly Deliver. Pharmaceuticals (Basel) 6 (5), 659–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takebe T et al. (2017) Synergistic Engineering: Organoids Meet Organs-on-a-Chip. Cell Stem Cell 21 (3), 297–300. [DOI] [PubMed] [Google Scholar]
- 76.Bhatia SN and Ingber DE (2014) Microfluidic organs-on-chips. Nat Biotechnol 32 (8), 760–72. [DOI] [PubMed] [Google Scholar]
- 77.Antonica F et al. (2012) Generation of functional thyroid from embryonic stem cells. Nature 491 (7422), 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bredenkamp N et al. (2014) An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts. Nat Cell Biol 16 (9), 902–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen YJ et al. (2014) De novo formation of insulin-producing “neo-beta cell islets” from intestinal crypts. Cell Rep 6 (6), 1046–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Del Vecchio D et al. (2017) A Blueprint for a Synthetic Genetic Feedback Controller to Reprogram Cell Fate. Cell Syst 4 (1), 109–120 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kunche S et al. (2016) Feedback, Lineages and Self-Organizing Morphogenesis. PLoS Comput Biol 12 (3), e1004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miller M et al. (2012) Modular design of artificial tissue homeostasis: robust control through synthetic cellular heterogeneity. PLoS Comput Biol 8 (7), e1002579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singer RH and Ward DC (1982) Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc Natl Acad Sci U S A 79 (23), 7331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raj A et al. (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5 (10), 877–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lubeck E et al. (2014) Single-cell in situ RNA profiling by sequential hybridization. Nat Methods 11 (4), 360–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shah S et al. (2016) In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron 92 (2), 342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen F et al. (2016) Nanoscale imaging of RNA with expansion microscopy. Nat Methods 13 (8), 679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ke R et al. (2013) In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods 10 (9), 857–60. [DOI] [PubMed] [Google Scholar]
- 89.Lee JH et al. (2015) Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 10 (3), 442–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee JH et al. (2014) Highly multiplexed subcellular RNA sequencing in situ. Science 343 (6177), 1360–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Merks Roeland (2015) Cell-based modeling In: Engquist Björn (Ed.) Encyclopedia of Applied and Computational Mathematics, pp. 195–201. [Google Scholar]
- 92.Alt S et al. (2017) Vertex models: from cell mechanics to tissue morphogenesis. Philos Trans R Soc Lond B Biol Sci 372 (1720). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fletcher AG et al. (2017) Mechanocellular models of epithelial morphogenesis. Philos Trans R Soc Lond B Biol Sci 372 (1720). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Misra M et al. (2017) Complex structures from patterned cell sheets. Philos Trans R Soc Lond B Biol Sci 372 (1720). [DOI] [PMC free article] [PubMed] [Google Scholar]