Abstract
BACKGROUND
In the context of kidney transplantation, genomic incompatibilities between donor and recipient may lead to allosensitization against new antigens. We hypothesized that recessive inheritance of gene-disrupting variants may represent a risk factor for allograft rejection.
METHODS
We performed a two-stage genetic association study of kidney allograft rejection. In the first stage, we performed a recessive association screen of 50 common gene-intersecting deletion polymorphisms in a cohort of kidney transplant recipients. In the second stage, we replicated our findings in three independent cohorts of donor–recipient pairs. We defined genomic collision as a specific donor–recipient genotype combination in which a recipient who was homozygous for a gene-intersecting deletion received a transplant from a nonhomozygous donor. Identification of alloantibodies was performed with the use of protein arrays, enzyme-linked immunosorbent assays, and Western blot analyses.
RESULTS
In the discovery cohort, which included 705 recipients, we found a significant association with allograft rejection at the LIMS1 locus represented by rs893403 (hazard ratio with the risk genotype vs. nonrisk genotypes, 1.84; 95% confidence interval [CI], 1.35 to 2.50; P= 9.8×10−5). This effect was replicated under the genomic-collision model in three independent cohorts involving a total of 2004 donor–recipient pairs (hazard ratio, 1.55; 95% CI, 1.25 to 1.93; P = 6.5×10−5). In the combined analysis (discovery cohort plus replication cohorts), the risk genotype was associated with a higher risk of rejection than the nonrisk genotype (hazard ratio, 1.63; 95% CI, 1.37 to 1.95; P = 4.7×10−8). We identified a specific antibody response against LIMS1, a kidney-expressed protein encoded within the collision locus. The response involved predominantly IgG2 and IgG3 antibody subclasses.
CONCLUSIONS
We found that the LIMS1 locus appeared to encode a minor histocompatibility antigen. Genomic collision at this locus was associated with rejection of the kidney allograft and with production of anti-LIMS1 IgG2 and IgG3. (Funded by the Columbia University Transplant Center and others.)
APPROXIMATELY 20% OF THE KIDNEY waiting list in the United States consists of candidates whose allografts have failed.1 Acute rejection is one of the strongest predictors of decreased allograft survival. Although human genetic variation and donor–recipient genetic interactions are likely to be involved in the determination of allograft outcomes, there have been few well-designed and adequately powered genetic investigations in this area.
Even though the dramatic improvements in short-term outcomes after transplantation have not resulted in similar improvements in long-term graft survival, new genetic strategies for organ matching may offer practical means by which rejection can be avoided and graft survival can be prolonged.2,3 HLA matching has been studied extensively, with an emphasis on donor-specific HLA alloantibodies.2,4–6 However, the incidence of donor-specific HLA alloantibodies among all allograft recipients is relatively low (15 to 25%).7–10 Increasing evidence suggests an important role for minor histocompatibility anti-gens that result in immune activation without HLA-directed response. The clinical significance of minor histocompatibility antigens is high-lighted by the observation that recipients of allografts from HLA-identical siblings require immunosuppression despite perfect HLA matching, and there are survival differences when analyses are stratified according to panel-reactive anti-body titers.3,11 It is estimated that approximately 56% of allograft failures can be attributed to immunologic reactions, with 38% of the reactions being against non-HLA factors and only 18% being due to HLA mismatch.12 In the past decade, several minor histocompatibility antigens have been successfully identified.13–18 Angiotensin II type 1 (AT1) receptor is perhaps the best-studied example, with preexisting and acquired anti–AT1 receptor antibodies predicting allograft rejection and failure.15,19–21 Nevertheless, the over-all incidence of known alloantibodies to minor histocompatibility antigens is low, which suggests that there are contributions from additional antigens.14
We hypothesized that the recipient’s inheritance of variants that disrupt kidney genes predisposes the recipient to allosensitization and rejection. In this study, we tested common copy-number polymorphisms that intersect genes, since such variants have a profound effect on gene function and expression.22 Using a genetic association screen of high-priority deletions, we sought to identify specific loci associated with kidney allograft rejection.
Methods
STUDY DESIGN AND CLINICAL OUTCOME
The study design had two stages. The first stage (the discovery phase) involved a screen of 50 high-priority copy-number polymorphisms (see below) in kidney allograft recipients who had undergone transplantation at the Columbia University Irving Medical Center in New York. Signals that reached a P value of less than 0.05 were advanced to the second stage (the replication phase). The replication phase involved genotyping of the top signals from the discovery phase in additional cohorts involving donor–recipient pairs. The primary outcome in the genetic association study was the first rejection in a time-to-event analysis, defined as a rejection event (anti-body-mediated rejection or T-cell–mediated rejection) that occurred between the date of transplantation and the date of allograft biopsy showing such an event.
DISCOVERY PHASE
We used a publicly available catalogue of known copy-number variants generated with the use of 2.1 M NimbleGen comparative genome hybridization arrays.23,24 From this data set, a total of 3266 copy-number variants were mapped to the human reference genome hg18 (accessed July 2010). To optimize the power of this study, we selected only copy-number polymorphisms that had a global minor allele frequency of more than 10%, corresponding to the expected homozygosity rates of more than 1% (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org).
Next, we intersected this set with a transcribed segment of the genome to identify potential gene-disrupting variants. A total of 180 gene-intersecting copy-number polymorphisms met our criteria, of which 87 (48%) were deletions. On the basis of the HapMap3 data, we found a single-nucleotide polymorphism (SNP) that was an informative tag (r2>0.8) for 50 (57%) of the identified deletions. These were prioritized for targeted genotyping in the discovery cohort, which involved 705 kidney recipients who had undergone transplantation at Columbia University and had been followed for a mean of 8.6 years (Table S2 in the Supplementary Appendix).
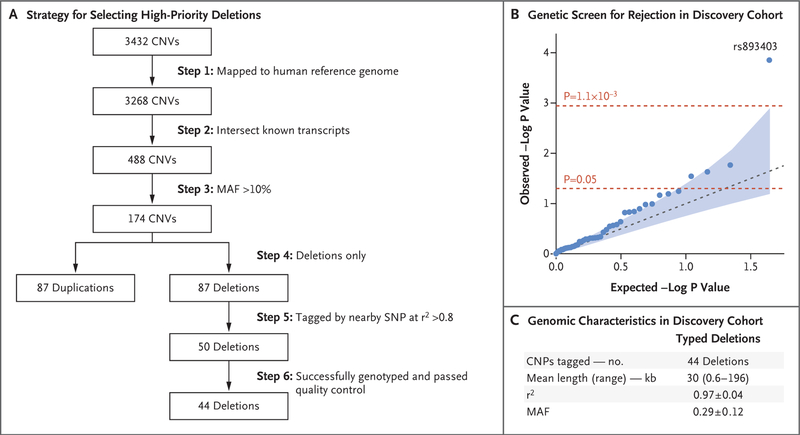
For the 50 genotyped SNPs, we performed strict quality-control analysis of genotypes that included per-SNP and per-recipient genotyping rates of more than 95%, elimination of monomorphic SNPs, and elimination of markers that significantly deviated from the Hardy–Weinberg equilibrium within each ethnic group. In total, 44 SNPs passed all the quality-control filters (Fig. 1, and Table S3 in the Supplementary Appendix).
Figure 1. Discovery Phase.
Panel A shows our strategy for selecting high-priority deletions for tagging and typing in the discovery cohort. A total of 44 of 50 deletions were successfully tagged and genotyped in the discovery cohort; 6 of 50 deletion-tagging single-nucleotide polymorphisms (SNPs) were either monomorphic or failed our genotype quality-control analysis. Annotations were based on the human reference genome hg18 (accessed in July 2010). Copy-number polymorphisms (CNPs) were common copy-number variants (CNVs with an allele frequency of >1%). MAF denotes minor allele frequency. Panel B shows the probability–probability plot for the genetic screen for rejection in the discovery cohort of 705 recipients under a recessive model. The blue dots represent P values for 44 successfully typed common deletions; the red dotted lines represent significance thresholds of 0.05 (unadjusted analysis) and 0.0011 (Bonferroni-corrected for 44 independent tests). The blue dotted line indicates the expectation under the null hypothesis, and the shaded area corresponds to a 95% confidence interval for the null hypothesis of no association. The top SNP (rs893403) represents a near-perfect tag (r2 = 0.98) for a common 1.5-kb deletion (CNVR915.1) on chromosome 2q12.3. Panel C shows the genomic characteristics of the 44 CNP-tagging SNPs that were tested in the discovery phase. Plus–minus values are means ±SD. Details are provided in Table S3 in the Supplementary Appendix.
REPLICATION PHASE
The replication cohorts (Table S4 in the Supplementary Appendix) included the Belfast cohort (387 donor–recipient pairs; mean follow-up, 9.2 years; overall rejection rate, 24%), the TransplantLines Genetics cohort (833 donor–recipient pairs; mean follow-up, 6.6 years; overall rejection rate, 36%), and the Torino cohort (784 donor–recipient pairs; mean follow-up, 9.6 years; overall rejection rate, 22%), providing a total of 2004 donor–recipient pairs for analysis. The genotype quality-control and analytical procedures that we used for replication were similar to those used in the discovery phase (see the Supplemental Methods section in the Supplementary Appendix).
MOLECULAR ANALYSES
Detailed methods regarding the deletion break-point mapping, functional genomic annotations, sequence motif analysis, tissue immunohisto-chemical and in situ hybridization studies, detection of anti-LIMS1 antibodies, and cell-culture experiments are provided in the Supplemental Methods section in the Supplementary Appendix. Testing for expression quantitative trait loci (eQTL) was conducted with the use of Genotype-Tissue Expression (GTEx) and the Nephrotic Syndrome Study Network (NEPTUNE) data sets. Detection of anti-LIMS1 antibodies was performed by protein arrays (ProtoArray, Invitrogen) and confirmed by means of enzyme-linked immunosorbent assay and Western blots.
STATISTICAL ANALYSIS
The association of genetic predictors and base-line covariates with the primary outcome was tested with the use of the cause-specific hazards models by treating death and allograft loss as censoring events. In recipient-only analyses, the deletion-tagging alleles were coded under a recessive model in which a risk genotype was defined according to a recipient’s homozygosity for a deletion-tagging allele. The full models were additionally adjusted for age, sex, ethnic group, donation type (living or cadaveric donor), HLA mismatch, and sensitization factors. We prespecified a Bonferroni-adjusted significance threshold for statistical significance in the discovery phase (alpha level of 0.05 ÷ 44, or 1.1×10−3). On the basis of the observed distribution of P values, we estimated a positive false discovery rate (Q value) for each of the tested polymorphisms. In the combined donor–recipient analyses, we defined genomic collision as a specific donor–recipient genotype combination in which a recipient who was homozygous for a deletion-tagging allele received a transplant from a nonhomozygous donor. We defined genomewide significance as an alpha level of 5.0×10−8, as generally accepted for genetic association studies.
RESULTS
DISCOVERY PHASE
To test for the effect of deletion homozygosity on the risk of rejection, we used time-to-event survival analysis under a recessive model for the deletion-tagging alleles. In the Kaplan–Meier analysis, we observed that a single SNP (rs893403) surpassed a Bonferroni-corrected significance threshold and reached a false discovery rate of 0.3%. Kidney transplant recipients who were homozygous for the deletion-tagging allele had an approximately 84% higher risk of rejection than those who did not have this genotype (hazard ratio, 1.84; 95% confidence interval [CI], 1.35 to 2.50; P = 9.8×10−5) (Fig. 2A). This effect was robust to adjustments for ethnic group (adjusted hazard ratio, 1.80; 95% CI, 1.32 to 2.45; P = 2.0×10−4) as well as for age, sex, donation type, HLA-mismatch status, and sensitization risk factors, including history of transplantation, transfusion, and pregnancy (adjusted hazard ratio, 1.83; 95% CI, 1.31 to 2.53; P = 2.9×10−4). The top SNP, rs893403, represented a near-perfect tag for a 1.5-kb deletion on chromosome 2q12.3 (CNVR915.1, r2 = 0.98).
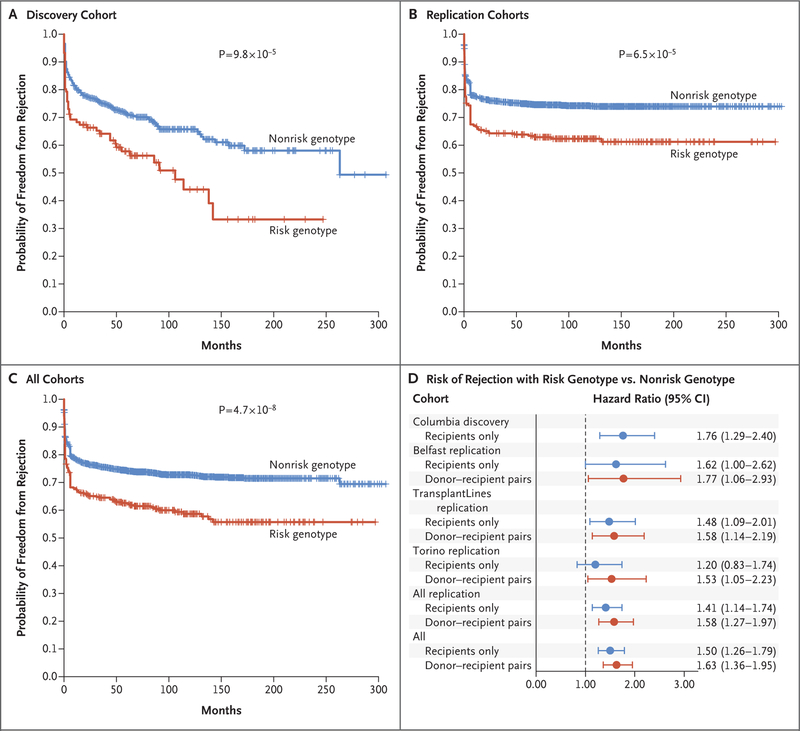
Figure 2. Effects of rs893403 on Rejection-free Allograft Survival in Study Cohorts.
Panel A shows the results in the discovery phase (involving 705 kidney transplant recipients [the Columbia cohort] who had either a non-risk genotype [blue] or a risk genotype [red]). Tick marks indicate censored data. Panel B shows the results in the replication phase, which involved a stratified analysis of three other cohorts (Belfast, TransplantLines, and Torino) that included a total of 2004 donor–recipient pairs. The P values correspond to the minimally adjusted model, with adjustment for cohort only (if applicable). Panel C shows the results in all the cohorts combined, which involved a stratified analysis of the four cohorts (i.e., 2709 kidney transplants [in 705 recipients from the discovery cohort and 2004 donor–recipient pairs from the replication cohorts]). Panel D shows the estimated hazard ratios (with 95% confidence intervals) of rejection in each of the four cohorts individually, in all the replication cohorts, and in all the cohorts combined. The effects were estimated before (blue [recipient only]) and after (red [donor–recipient pairs]) accounting for donor compatibility in order to show that the inclusion of genetic information from the donors resulted in consistently improved hazard ratio estimates.
REPLICATION PHASE
We genotyped rs893403 in kidney transplant recipients and their matched donors from three international kidney transplant cohorts (the Belfast, TransplantLines, and Torino cohorts). After standard genotype quality control was assessed, there were 2004 donor–recipient pairs available across the three replication cohorts. For each replication cohort, we first applied the same approach as in the discovery phase, using the recipient’s homozygosity for the deletion-tagging allele as a predictor in the model of rejection. Despite the diversity of our replication cohorts, we found a direction-consistent effect of the risk genotype in all three cohorts (Table S5 in the Supplementary Appendix), and a combined meta-analysis confirmed that recipients with the risk genotype had a higher risk of acute rejection than those with a nonrisk genotype independently of age, sex, donation type, HLA-mismatch status, and transplantation center (adjusted hazard ratio, 1.41; 95% CI, 1.14 to 1.74; P = 1.6×10−3). In the combined stratified analysis of the discovery and replication cohorts, a high-risk genotype conveyed a risk of rejection that was 50% higher than that observed with a nonrisk genotype (adjusted hazard ratio, 1.50; 95% CI, 1.26 to 1.79; P = 5.4×10−6). The significance of this association surpassed our prespecified Bonferroni-adjusted threshold (alpha level of 1.1×10−3).
Subsequently, we used the genetic information from the donor–recipient pairs to test the genomic-collision hypothesis. The risk of genomic collision was defined according to recipient homozygosity in the absence of donor homozygosity. In findings consistent with our hypothesis, the effect estimates became larger after the analysis accounted for donor genotypes within each cohort as directly compared with recipient-only analyses (Table 1). The effect estimates were direction-consistent and similar in magnitude in each of the three replication cohorts, with adjusted hazard ratios of 1.77 (95% CI, 1.06 to 2.93; P = 2.8×10−2) in the Belfast cohort, 1.58 (95% CI, 1.14 to 2.19; P = 5.7×10−3) in the TransplantLines cohort, and 1.53 (95% CI, 1.05 to 2.23; P = 2.6×10−2) in the Torino cohort. In the combined stratified analysis of all the replication data sets, the donor–recipient pairs with the collision genotype had a risk of allograft rejection that was 58% higher than the risk among pairs without the collision genotype, and this effect was significant (adjusted hazard ratio, 1.58; 95% CI, 1.27 to 1.97; P = 5.1×10−5) (Fig. 2B).
Table 1.
Cox Proportional-Hazards Association Analysis of LIMS1 Collision Genotype with Allograft Rejection in a Time-to-Event Analysis, According to Cohort.
| Cohort | Population | Minimally Adjusted Model* | Fully Adjusted Model† | ||
|---|---|---|---|---|---|
| Hazard Ratio (95% CI) | P Value | Hazard Ratio (95% CI) | P Value | ||
| Columbia discovery cohort | 705 Recipients | 1.84 (1.35–2.50) | 9.8×10−5 | 1.76 (1.29–2.40) | 3.8×10−4 |
| Belfast replication cohort | 387 Donor–recipient pairs | 1.70 (1.03–2.82) | 4.0×10−2 | 1.77 (1.06–2.93) | 2.8×10−2 |
| TransplantLines replication cohort | 833 Donor–recipient pairs | 1.53 (1.13–2.09) | 6.6×10−3 | 1.58 (1.14–2.19) | 5.7×10−3 |
| Torino replication cohort | 784 Donor–recipient pairs | 1.49 (1.02–2.16) | 3.9×10−2 | 1.53 (1.05–2.23) | 2.6×10−2 |
| All replication cohorts | 2004 Donor–recipient pairs | 1.55 (1.25–1.93) | 6.5×10−5 | 1.58 (1.27–1.97) | 5.1×10−5 |
| All cohorts | 2709 Transplants | 1.63 (1.37–1.95) | 4.7×10−8 | 1.63 (1.36–1.95) | 9.4×10−8 |
The analysis was adjusted for cohort only (if applicable).
The analysis was adjusted for recipient’s age, sex, race, ethnic group, HLA-mismatch status, and cohort (if applicable).
Next, we performed a pooled analysis of all four cohorts, which involved 2709 transplants (in 705 unmatched recipients from the discovery phase and in 2004 donor–recipient pairs from the replication phase). We note that the inclusion in this analysis of the Columbia cohort, which lacked donor genotype data, resulted in an expected predictor misclassification frequency of 1.7% in the combined data set, biasing the result slightly toward the null. Despite this limitation, the collision genotype was associated with the rejection risk, reaching genomewide significance (hazard ratio, 1.63; 95% CI, 1.37 to 1.95; P = 4.7×10−8). The genomic-collision model was superior to recipient-only recessive or additive models (Table S6 and Figs. S1 through S3 in the Supplementary Appendix).
The effect of the collision genotype was robust to multivariate adjustment for age, race, ethnic group, and HLA-mismatch status (adjusted hazard ratio, 1.63; 95% CI, 1.36 to 1.95; P = 9.4×10−8) (Fig. 2C and 2D) and was consistent under alternative statistical models (Table S7 in the Supplementary Appendix). The association was driven predominantly by T-cell–mediated rejection, the most common type of rejection, but other rejection types also contributed (Fig. S4 in the Supplementary Appendix). This effect was three times as high as the risk due to per-allele HLA mismatch in the same model (adjusted hazard ratio, 1.21; 95% CI, 1.16 to 1.28; P = 1.9×10−14). Although our study was not adequately powered to test for the association with allograft failure, we observed a nonsignificantly higher risk of failure in the collision genotype group than in the noncollision genotype group (adjusted hazard ratio, 1.12; 95% CI, 0.90 to 1.39; P = 0.32) (Fig. S5 in the Supplementary Appendix).
FUNCTIONAL ANNOTATION OF THE 2Q12.3 LOCUS
The top SNP, rs893403, resides on chromosome 2q12.3 in the intronic portion of LIMS1. This gene encodes a protein that is involved in cell adhesion and integrin signaling found in focal adhesion plaques. The risk allele, rs893403-G, is frequent in persons of European and African ancestry but absent in persons of East Asian ancestry and tags a common 1.5-kb deletion (CNVR915.1) that is downstream of LIMS1 (r2 = 0.98 in the HapMap European population). CNVR915.1 was originally annotated to intersect LOC100288532, a gene in the human reference genome hg18 that was removed in subsequent releases of the human genome. Our deletion breakpoint mapping and detailed functional annotations of the region indicated that the rs893403-G risk allele was associated with lower messenger RNA (mRNA) expression of LIMS1 and GCC2, the neighboring gene, across multiple GTEx project tissues than was the alternative allele. Furthermore, rs893403 has a direction-consistent cis-eQTL effect on the LIMS1 mRNA level in the kidney tubulointerstitium. (Details are provided in the Supplemental Results section, Table S8, and Figs. S6 through S13 in the Supplementary Appendix.)
Using immunohistochemical studies, we confirmed that LIMS1 was strongly expressed in human kidneys and other commonly transplanted organ tissues, such as heart and lung. Within the kidney, LIMS1 staining is strongest in the distal nephron, including the basolateral surface of distal tubules in the cortex and medulla, the medullary thick ascending limb of the loop of Henle, and medullary collecting ducts. The proximal-to-distal gradient of LIMS1 expression was consistent with human kidney single nuclear RNA sequencing data and was confirmed by means of RNAscope in situ hybridization. Moreover, cell-surface LIMS1 was induced by hypoxia in HEK293 cell lines, a finding consistent with previously reported hypoxia-induced LIMS1 gene expression in cultured endothelial cells.25 In contrast, GCC2 was detected predominantly in the cytoplasmic compartment of proximal tubules (most strongly in S3) and in kidney vascular smooth muscle. (Details are provided in Table S10 and Figs. S14 through S19 in the Supplementary Appendix.)
SEROREACTIVITY AGAINST LIMS1 IN RECIPIENTS WITH A HIGH-RISK GENOTYPE
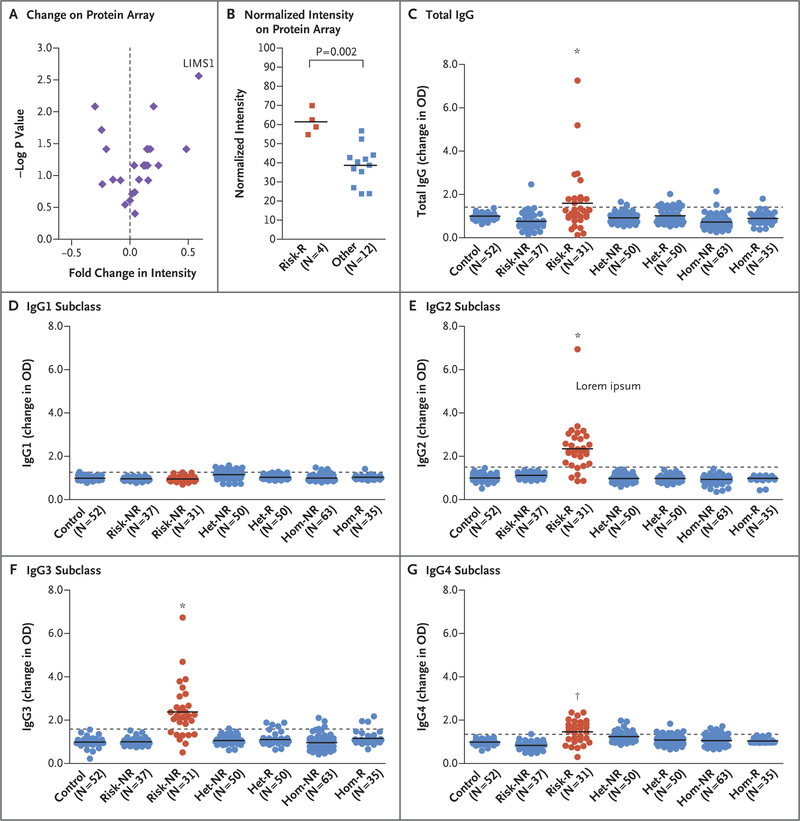
To obtain an unbiased characterization of allo-antibody response in kidney recipients with a high-risk genotype, we used ProtoArray protein arrays to screen serum specimens that had been obtained from recipients. These protein arrays capture the human proteome with 9375 immobilized recombinant human proteins. We tested serum specimens that had been obtained from 16 recipients, including 8 persons with rejection (4 recipients with a high-risk genotype and 4 with a low-risk genotype) and 8 controls who had not had rejection. The seroreactivity to proteins was detected with antihuman IgG as an increased intensity normalized to control protein gradients printed on each array. Despite the small sample size in this experiment, the LIMS1 protein ranked 14th among the 9375 proteins (0.15th percentile) on the array according to the mean intensity in the group of recipients with a high-risk genotype who had allograft rejection. Among the 14 top intensity signals, LIMS1 seroreactivity was most specific to the high-risk rejection group (P = 0.002 for high-risk genotype with rejection vs. all other groups) (Fig. 3A and 3B). No other proteins that were encoded within a 1-Mb window of rs893403 showed significant seroreactivity, although the GCC2 protein was not captured on protein arrays. Finally, we detected a lower-intensity signal for the LIMS2 protein, which shares 92% sequence identity with LIMS1,26 thus suggesting potential cross-reactivity (P=0.007) (Fig. S20 in the Supplementary Appendix).
Figure 3. Detection of Anti-LIMS1 Antibodies in Kidney Transplant Recipients at Genetic Risk for Rejection.
Panel A shows the change in intensity (x axis) as compared with the −log P value (y axis) for the top-ranking proteins on the basis of the mean signal intensity in a protein array; the change is calculated as a ratio of the mean normalized intensity in the high-risk rejection group to the mean normalized intensity of all other groups (termed “fold change”). The findings suggest the presence of anti-LIMS1 reactivity in high-risk recipients with rejection. Panel B shows the normalized intensity levels for LIMS1 on the protein array for the comparison between the high-risk rejection group and all other groups (P = 0.002); the horizontal lines represent the group means. Panel C shows the results of anti-LIMS1 total IgG seroreactivity studies with the use of an enzyme-linked immunosorbent assay that were performed in 318 persons across seven genotype- and phenotype-discordant groups. The results are shown as the change in the optical density (OD), defined as a ratio of the measured OD for each sample to the mean OD of the same 5 normalization controls (serum samples obtained from healthy persons) that were used on each plate. These studies included 52 controls who had not undergone transplantation (Control), 37 recipients who were homozygous for the risk allele and did not have rejection (Risk-NR), 31 recipients who were homozygous for the risk allele and had rejection (Risk-R; in red), 50 recipients who were heterozygous for the risk allele and did not have rejection (Het-NR), 50 recipients who were heterozygous for the risk allele and had rejection (Het-R), 63 recipients who were homozygous for the non-risk–associated allele and did not have rejection (Hom-NR), and 35 recipients who were homozygous for the non-risk–associated allele and had rejection (Hom-R). Total IgG seroreactivity was detected only in recipients with a high-risk genotype who had rejection. Horizontal lines represent group means, and the dotted line represents 3 SD above the mean for the control group. Panels D through G show the anti-LIMS1 reactivity of IgG1, IgG2, IgG3, and IgG4 subclasses, respectively; the results show predominant IgG2 and IgG3 responses. An asterisk indicates a P value of less than 0.001 and a dagger a P value of less than 0.01 for the comparisons of the group of recipients with a high-risk genotype who had rejection as compared with all other groups.
To confirm the protein array findings, we obtained serum specimens from 318 transplant recipients across seven genotype- and phenotype-discordant groups. We detected highly specific IgG reactivity toward LIMS1 in the kidney recipients with a high-risk genotype who had allograft rejection but not in any other control group (Fig. 3C). Next, we investigated whether an anti-body targeting LIMS1 could be injurious. Treatment of cultured human kidney cortical epithelial cells expressing the LIMS1 protein with mouse antihuman LIMS1 antibody disrupted the normal organization of F-actin filaments and showed significant cytotoxicity on lactate dehydrogenase assay as compared with a nonspecific control antibody (Figs. S22 and S23 in the Supplementary Appendix).
IgG subclass analysis showed that the anti-LIMS1 response was predominantly of IgG2 and IgG3 subtype, but weaker IgG4 reactivity was also detected (Fig. 3D through 3G, and Fig. S21 in the Supplementary Appendix). Of 31 recipients with a high-risk genotype who had rejection, 26 (84%) and 23 (74%) tested positive for anti-LIMS1 reactivity by IgG2 and IgG3, respectively. Overall, 29 recipients (94%) tested positive by either IgG2 or IgG3 subtype. IgG2 and IgG3 comprise only a small percentage of total IgG (up to 30% and 8%, respectively), and there were no detectable differences between groups in levels of IgG1, the main IgG subclass, which explains why total IgG had overall lower reactivity than these subtypes.
DISCUSSION
In this study, we examined a genomic-collision scenario in which an allograft recipient was homozygous for a deletion polymorphism and received a kidney allograft from a donor who had at least one normal allele. In the analysis of four large kidney transplant cohorts, we found that the genomic collision at chromosome 2q12.3 led to a risk of rejection that was nearly 60% higher than the risk among donor–recipient pairs with noncollision genotypes. The risk associated with the collision genotype is equivalent to a mismatch of three of six HLA alleles, which is both clinically significant and potentially modifiable by genetic testing and matching. The genomic collision at chromosome 2q12.3 would be expected to occur in approximately 12 to 15% of transplants from unrelated donors among persons of European and African ancestry but would be very rare among persons of East Asian ancestry.
In our study, the collision genotype was associated with the presence of anti-LIMS1 antibodies. We also found that the risk genotype was associated with a lower kidney mRNA level of LIMS1 and that LIMS1 protein was induced on the cell surface under hypoxic conditions. The recessive model potentially supports a loss-of-function effect, and our data point to LIMS1 as the most likely culprit gene, but the precise causal variant underlying this locus is still unclear. We note that rs893403 also regulates mRNA expression of GCC2, encoding a protein of unclear function. We found that the GCC2 protein is expressed in proximal tubule cells, but because recombinant GCC2 is not available and this protein was not captured on protein arrays, we were unable to test for seroreactivity against GCC2 in this study. Further genotype-specific analysis of expression patterns of genes within the LIMS1 locus may be useful, especially in the context of hypoxic injury and other forms of injury.
Several other lines of evidence suggest that genomic incompatibilities beyond the traditional ABO and HLA loci are predictive of allograft rejection. For example, female recipients of organs from male donors are at greater risk for poor graft outcomes, probably because of sensitization to minor histocompatibility antigens encoded on the Y chromosome.27–31 The occurrence of aggressive post-transplantation antiglomerular basement membrane disease in persons affected by the Alport syndrome owing to collagen IV mutations, including loss-of-function variants, exemplifies another proof of concept for this phenomenon.32,33
Taken together, our results provide support for genomic collision at chromosome 2q12.3 contributing to the risk of allograft rejection and point to LIMS1 as a potential minor histocompatibility antigen encoded by this locus. In addition, we found that the LIMS1 protein was expressed in other commonly transplanted tissues, such as the heart and lung, but follow-up studies will be useful in determining whether our findings are generalizable to other organs. The reverse of our hypothesis has previously been tested in the context of bone marrow transplantation: the immune system of a donor who was homozygous for a gene-disrupting deletion may recognize epitopes that are encoded by that gene in the tissues of a recipient, leading to graft-versus-host disease.22 A similar mechanism may also apply to other types of variants that were not examined in this study, such as loss-of-function variants, variants altering the expression of immunogenic proteins, or missense variants that create new immunogenic epitopes. A population-based sequencing study has shown that a large proportion of persons are natural “human gene knockouts” (i.e., have two copies of loss-of-function variants in the same gene) for a number of nonessential genes.34 Given our hypothesis, we speculate that such persons may be at risk for rejection if they receive an allograft expressing an intact protein.
To our knowledge, the LIMS1 locus has not been detected in previous genomewide association studies of kidney transplant rejection. We suspect that this is probably due to the limited sample size of earlier studies and to the fact that limited research has been done in testing the genomic-collision model. Previous studies have involved recipient-only analyses or a standard additive genotype coding scheme.35–37 On the basis of the observed minor allele frequency and the pooled additive effect estimate for the rs893403-G allele in our cohorts, we estimated that our study would have no more than 3.5% power to detect this locus at a Bonferroni-corrected alpha level under additive coding in our discovery cohort. We also estimated that in a recipient-only genomewide association study under an additive model, a minimum of 13,000 kidney transplant recipients would need to be enrolled for the study to have 80% power for detection of this locus at a genomewide significant alpha level of 5×10−8 (assuming a minor allele frequency of 0.50 and a rejection rate of 33%). Although large-scale efforts in genomewide association studies of kidney transplantation are under way,38,39 the largest discovery study of allograft rejection that we are aware of involved only 2094 kidney transplants.37 It remains to be seen whether the combination of genomic profiling with proteome-wide antibody screens can be used effectively to uncover new histocompatibility antigens and potentially improve the precision of organ matching.
Supplementary Material
Acknowledgments
Supported by the Columbia University Transplant Center, a New York Presbyterian Hospital Translational Pilot Grant funded by the New York State Empire Clinical Research Investigator Program, a Columbia University Precision Medicine Pilot Award funded by the Dean of the College of Physicians and Surgeons, and the Columbia Clinical and Translational Science Award Precision Medicine Pilot Program funded by a grant (UL1TR001873) from the NIH National Center for Advancing Translational Sciences and by grants (T35-DK093430, to Ms. Fischman; and T32-DK108741 Supplement, to Ms. Neugut) from the National Institute of Diabetes and Digestive and Kidney Diseases. Replication studies in the Torino cohort were supported by a Department of Excellence Grant 2018–2022 funded by the Italian Ministry of Education to the Department of Medical Sciences of the University of Turin. The Columbia Center for Translational Immunology Flow Cytometry Core is supported in part by a grant (S10RR027050) from the NIH Office of the Director, and the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University is supported by a grant (P30CA013696) from the NIH National Cancer Institute.
Footnotes
The views expressed in this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health (NIH), New York State, or Columbia University.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank all the participating kidney transplant recipients and their donors for contributing biologic samples and clinical data for this study.
Contributor Information
Nicholas J. Steers, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Yifu Li, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Zahida Drace, Immunogenetics and Biology of Transplantation, Città della Salute e della Scienza, University Hospital of Turin, and Medical Genetics, Department of Medical Sciences, University of Turin, Turin
Justin A. D’Addario, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Clara Fischman, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Lili Liu, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Katherine Xu, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Young-Ji Na, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Y. Dana Neugut, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Jun Y. Zhang, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Roel Sterken, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Olivia Balderes, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Drew Bradbury, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Nilgun Ozturk, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Fatih Ozay, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Sanya Goswami, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Karla Mehl, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Jaclyn Wold, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Fatima Z. Jelloul, Department of Pathology, M.D. Anderson Cancer Center, Houston
Mersedeh Rohanizadegan, Division of Genetics and Genomics, Boston Children’s Hospital, and the Department of Medical Oncology, Dana–Farber Cancer Institute, Boston
Christopher E. Gillies, Department of Pediatrics, Division of Pediatric Nephrology, University of Michigan School of Medicine, Ann Arbor
Elena-Rodica M. Vasilescu, Department of Pathology and Cell Biology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
George Vlad, Department of Pathology and Cell Biology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Yi-An Ko, Department of Medicine, Renal, Electrolyte, and Hypertension Division, University of Pennsylvania, Philadelphia
Sumit Mohan, Department of Medicine, Division of Nephrology, Departments of Epidemiology, Mailman School of Public Health, Columbia University, New York
Jai Radhakrishnan, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
David J. Cohen, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Lloyd E. Ratner, Department of Surgery, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Francesco Scolari, Division of Nephrology, Azienda Ospedaliera Spedali Civili of Brescia, Montichiari Hospital, University of Brescia, Brescia
Katalin Susztak, Department of Medicine, Renal, Electrolyte, and Hypertension Division, University of Pennsylvania, Philadelphia
Matthew G. Sampson, Department of Pediatrics, Division of Pediatric Nephrology, University of Michigan School of Medicine, Ann Arbor
Silvia Deaglio, Immunogenetics and Biology of Transplantation, Città della Salute e della Scienza, University Hospital of Turin, and Medical Genetics, Department of Medical Sciences, University of Turin, Turin
Yasar Caliskan, Division of Nephrology, Istanbul Faculty of Medicine, Istanbul University, Istanbul, Turkey
Jonathan Barasch, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Aisling E. Courtney, Nephrology Research Group, Queen’s University of Belfast, Belfast, United Kingdom
Alexander P. Maxwell, Nephrology Research Group, Queen’s University of Belfast, Belfast, United Kingdom
Amy J. McKnight, Nephrology Research Group, Queen’s University of Belfast, Belfast, United Kingdom
Iuliana Ionita-Laza, Biostatistics, Mailman School of Public Health, Columbia University, New York
Stephan J.L. Bakker, Department of Internal Medicine, Division of Nephrology, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands.
Harold Snieder, Department of Epidemiology, Unit of Genetic Epidemiology and Bioinformatics, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands.
Martin H. de Borst, Department of Internal Medicine, Division of Nephrology, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands.
Vivette D’Agati, Department of Pathology and Cell Biology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Antonio Amoroso, Immunogenetics and Biology of Transplantation, Città della Salute e della Scienza, University Hospital of Turin, and Medical Genetics, Department of Medical Sciences, University of Turin, Turin
Ali G. Gharavi, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
Krzysztof Kiryluk, Department of Medicine, Division of Nephrology, Vagelos College of Physicians and Surgeons, Mailman School of Public Health, Columbia University, New York
REFERENCES
- 1.Magee JC, Barr ML, Basadonna GP, et al. Repeat organ transplantation in the United States, 1996–2005. Am J Transplant 2007;7:1424–33. [DOI] [PubMed] [Google Scholar]
- 2.Sellarés J, de Freitas DG, Mengel M, et al. Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant 2012;12:388–99. [DOI] [PubMed] [Google Scholar]
- 3.Opelz G Non-HLA transplantation immunity revealed by lymphocytotoxic anti-bodies. Lancet 2005;365:1570–6. [DOI] [PubMed] [Google Scholar]
- 4.Lachmann N, Terasaki PI, Budde K, et al. Anti-human leukocyte antigen and donor-specific antibodies detected by luminex posttransplant serve as biomarkers for chronic rejection of renal allografts. Transplantation 2009;87:1505–13. [DOI] [PubMed] [Google Scholar]
- 5.Eng HS, Bennett G, Chang SH, et al. Donor human leukocyte antigen specific antibodies predict development and define prognosis in transplant glomerulopathy. Hum Immunol 2011;72:386–91. [DOI] [PubMed] [Google Scholar]
- 6.Willicombe M, Brookes P, Sergeant R, et al. De novo DQ donor-specific antibodies are associated with a significant risk of antibody-mediated rejection and transplant glomerulopathy. Transplantation 2012;94:172–7. [DOI] [PubMed] [Google Scholar]
- 7.Everly MJ, Rebellato LM, Haisch CE, et al. Incidence and impact of de novo donor-specific alloantibody in primary renal allografts. Transplantation 2013;95: 410–7. [DOI] [PubMed] [Google Scholar]
- 8.Hourmant M, Cesbron-Gautier A, Terasaki PI, et al. Frequency and clinical implications of development of donor-specific and non-donor-specific HLA antibodies after kidney transplantation. J Am Soc Nephrol 2005;16:2804–12. [DOI] [PubMed] [Google Scholar]
- 9.Mohan S, Palanisamy A, Tsapepas D, et al. Donor-specific antibodies adversely affect kidney allograft outcomes. J Am Soc Nephrol 2012;23:2061–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loupy A, Jordan SC. Transplantation: donor-specific HLA antibodies and renal allograft failure. Nat Rev Nephrol 2013;9: 130–1. [DOI] [PubMed] [Google Scholar]
- 11.Joosten SA, van Kooten C. Non-HLA humoral immunity and chronic kidney-graft loss. Lancet 2005;365:1522–3. [DOI] [PubMed] [Google Scholar]
- 12.Terasaki PI. Deduction of the fraction of immunologic and non-immunologic failure in cadaver donor transplants. Clin Transpl 2003;449–52. [PubMed] [Google Scholar]
- 13.Joosten SA, Sijpkens YW, van Ham V, et al. Antibody response against the glomerular basement membrane protein agrin in patients with transplant glomerulopathy. Am J Transplant 2005;5:383–93. [DOI] [PubMed] [Google Scholar]
- 14.Dragun D, Catar R, Philippe A. Non-HLA antibodies against endothelial targets bridging allo- and autoimmunity. Kidney Int 2016;90:280–8. [DOI] [PubMed] [Google Scholar]
- 15.Amico P, Hönger G, Bielmann D, et al. Incidence and prediction of early antibody-mediated rejection due to non-human leukocyte antigen-antibodies. Transplantation 2008;85:1557–63. [DOI] [PubMed] [Google Scholar]
- 16.Sumitran-Holgersson S, Wilczek HE, Holgersson J, Söderström K. Identification of the nonclassical HLA molecules, mica, as targets for humoral immunity associated with irreversible rejection of kidney allografts. Transplantation 2002;74:268–77. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Sigdel T, Vitalone M, Lee SH, Sarwal M. Differential immunogenicity and clinical relevance of kidney compartment specific antigens after renal transplantation. J Proteome Res 2010;9:6715–21. [DOI] [PubMed] [Google Scholar]
- 18.Hess SM, Young EF, Miller KR, et al. Deletion of naïve T cells recognizing the minor histocompatibility antigen HY with toxin-coupled peptide-MHC class I tetramers inhibits cognate CTL responses and alters immunodominance. Transpl Immunol 2013;29:138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dragun D, Müller DN, Hinrich Bräsen J, et al. Angiotensin II type 1–receptor activating antibodies in renal-allograft rejection. N Engl J Med 2005;352:558–69. [DOI] [PubMed] [Google Scholar]
- 20.Banasik M, Boratyńska M, Kościelska-Kasprzak K, et al. Non-HLA antibodies: angiotensin II type 1 receptor (anti-AT1R) and endothelin-1 type A receptor (anti-ETAR) are associated with renal allograft injury and graft loss. Transplant Proc 2014;46:2618–21. [DOI] [PubMed] [Google Scholar]
- 21.Jackson AM, Sigdel TK, Delville M, et al. Endothelial cell antibodies associated with novel targets and increased rejection. J Am Soc Nephrol 2015;26:1161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarroll SA, Bradner JE, Turpeinen H, et al. Donor-recipient mismatch for common gene deletion polymorphisms in graft-versus-host disease. Nat Genet 2009;41: 1341–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature 2010;464:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craddock N, Hurles ME, Cardin N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 2010;464:713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manalo DJ, Rowan A, Lavoie T, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 2005;105:659–69. [DOI] [PubMed] [Google Scholar]
- 26.Braun A, Bordoy R, Stanchi F, et al. PINCH2 is a new five LIM domain protein, homologous to PINCH and localized to focal adhesions. Exp Cell Res 2003; 284:239–50. [DOI] [PubMed] [Google Scholar]
- 27.Candinas D, Gunson BK, Nightingale P, Hubscher S, McMaster P, Neuberger JM. Sex mismatch as a risk factor for chronic rejection of liver allografts. Lancet 1995; 346:1117–21. [DOI] [PubMed] [Google Scholar]
- 28.Prendergast TW, Furukawa S, Beyer AJ III, Browne BJ, Eisen HJ, Jeevanandam V. The role of gender in heart transplantation. Ann Thorac Surg 1998;65:88–94. [DOI] [PubMed] [Google Scholar]
- 29.Roberts DH, Wain JC, Chang Y, Ginns LC. Donor-recipient gender mismatch in lung transplantation: impact on obliterative bronchiolitis and survival. J Heart Lung Transplant 2004;23:1252–9. [DOI] [PubMed] [Google Scholar]
- 30.Gratwohl A, Döhler B, Stern M, Opelz G. H-Y as a minor histocompatibility anti-gen in kidney transplantation: a retrospective cohort study. Lancet 2008;372: 49–53. [DOI] [PubMed] [Google Scholar]
- 31.Kim SJ, Gill JS. H-Y incompatibility predicts short-term outcomes for kidney transplant recipients. J Am Soc Nephrol 2009;20:2025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashtan CE. Renal transplantation in patients with Alport syndrome. Pediatr Transplant 2006;10:651–7. [DOI] [PubMed] [Google Scholar]
- 33.Pedchenko V, Bondar O, Fogo AB, et al. Molecular architecture of the Good-pasture autoantigen in anti-GBM nephritis. N Engl J Med 2010;363:343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulem P, Helgason H, Oddson A, et al. Identification of a large set of rare complete human knockouts. Nat Genet 2015; 47:448–52. [DOI] [PubMed] [Google Scholar]
- 35.Ghisdal L, Baron C, Lebranchu Y, et al. Genome-wide association study of acute renal graft rejection. Am J Transplant 2017;17:201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Brien RP, Phelan PJ, Conroy J, et al. A genome-wide association study of recipient genotype and medium-term kidney allograft function. Clin Transplant 2013; 27:379–87. [DOI] [PubMed] [Google Scholar]
- 37.Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I, et al. Long- and short-term outcomes in renal allografts with deceased donors: a large recipient and donor genome-wide association study. Am J Transplant 2018;18:1370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International Genetics & Translational Research in Transplantation Network (iGeneTRAiN). Design and implementation of the International Genetics and Translational Research in Transplantation Network. Transplantation 2015;99:2401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li YR, van Setten J, Verma SS, et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med 2015;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.