Abstract
Due to the heterogenous lipid environment in which integral membrane proteins are embedded, they should follow a set of assembly rules, which govern transmembrane protein folding and topogenesis accordingly to a given lipid profile. Recombinant strains of bacteria have been engineered to have different membrane phospholipid compositions by molecular genetic manipulation of endogenous and foreign genes encoding lipid biosynthetic enzymes. Such strains provide a means to investigate the in vivo role of lipids in many different aspects of membrane function, folding and biogenesis. In vitro and in vivo studies established a function of lipids as molecular chaperones and topological determinants specifically assisting folding and topogenesis of membrane proteins. These results led to the extension of the Positive Inside Rule to Charge Balance Rule, which incorporates a role for lipid-protein interactions in determining membrane protein topological organization at the time of initial membrane insertion and dynamically after initial assembly. Membrane protein topogenesis appears to be a thermodynamically driven process in which lipid-protein interactions affect the potency of charged amino acid residues as topological signals. Dual topology for a membrane protein can be established during initial assembly where folding intermediates in multiple topological conformations are in rapid equilibrium (thus separated by a low activation energy), which is determined by the lipid environment. Post-assembly changes in lipid composition or post-translational modifications can trigger a reorganization of protein topology by inducing destabilization and refolding of a membrane protein. The lipid-dependent dynamic nature of membrane protein organization provides a novel means of regulating protein function.
Keywords: membrane protein, phospholipid, topogenesis, Charge Balance Rule, protein folding
1.0. Introduction
The work of Günter Blobel firmly established the basic principles governing the targeting and initial insertion of proteins into the membrane utilizing the translocon machinery [1]. Extensive studies have detailed the finer points of membrane protein assembly to give a picture of the initial steps of insertion of proteins into the membrane as well as their translocation across the membrane [2,3]. These studies have progressed in parallel with detailing the driving forces and interactions responsible for final folding of proteins in the lipid bilayer. Most of the latter studies have focused on membrane protein sequence determinants which are required for proper and uniform insertion of hydrophobic transmembrane domains (TMDs) into and exclusion of hydrophilic extramembrane domains (EMDs) from the hydrophobic core of the lipid bilayer [4]. This primarily protein centric focus has resulted in less attention paid to the role of the diverse nature and dynamic properties of the cellular lipidome, which may exceed the complexity of the proteome of any given cell [5].
TMD insertion into and EMD exclusion from the lipid bilayer can be easily modeled based on physical principles governing the partitioning of these domains (depending on their length and hydrophobicity) between an aqueous and an organic phase [6]. Less defined is the molecular basis for the orientation of TMDs and EMDs with respect to the plane of the lipid bilayer. The Positive Inside Rule [7] was formulated originally by the statistically based observation that over 80% of membrane protein EMDs exposed to the cytoplasm carry a net positive charge. Experimental manipulation of EMD charges further supports this general rule [7,8] and the functioning of positively charged amino acids as retention signals. However, the Positive Inside Rule does not explain the cytoplasmic orientation of the remaining 20% of EMDs that are net negative or neutral. It is also not clear why positively charged residues are retained in the cytosol and appear to be more potent topological determinants than negatively charged residues under physiological conditions. The strong inward negative membrane potential across many membranes has been suggested as the orientation force favoring cytoplasmic orientation of positively charged EMDs [7,9,10]. However, acidophiles have a positive inward membrane potential and still obey the Positive Inside Rule at least statistically by orienting net positively charged EMDs toward the cytoplasm [11]. An early study of the effect of progressively increasing the membrane content of negatively charged phospholipids suggested that interaction of positively charged amino acids in EMDs with negatively charged lipid headgroups favored orientation of net positively charged EMDs toward the cytoplasm during initial translocon-dependent membrane insertion of proteins [12]. As will be discussed, this appears not to be the basis for the Positive Inside Rule. Furthermore, neither the current understanding of translocon-dependent protein insertion into membranes nor the Positive Inside Rule can explain co-existence of membrane proteins with dual or multiple topologies in the same or different cell membranes, changes in topological organization of membrane proteins post-assembly in response to changes in the membrane lipid composition and posttranslational modification, or the positive inside negative outside bias in positioning EMDs. Although the translocon/insertion machinery may perform some of the initial co-translational catalysis, the presence of equal or different amounts of oppositely oriented protein conformers within the same membrane is well beyond the control of the translocon.
In order to fully understand the process of membrane protein synthesis and assembly, studies must also be focused on what happens to protein chains after they leave the translocon and distribute themselves across the lipid bilayer. The lipid bilayer is a diverse and dynamic mixture of amphipathic molecules held together by non-covalent hydrophobic forces [5,13]. Due to the dynamic nature of the lipid bilayer during the cell cycle with respect to overall composition, distribution of lipid species laterally and between the monolayers, and between multiple organelle membranes, the temporal effect of neighboring lipids on organization of membrane proteins must be considered. Finally, the potential for changes in topology induced by post-assembly modification of membrane proteins within a changing lipid environment must be addressed. Based on extensive studies in whole cells, isolated membranes and reconstituted proteoliposomes, we have postulated the Charge Balance Rule [14,15] as an extension of the Positive Inside Rule to incorporate the role of lipid-protein interactions in dynamic organization of membrane proteins.
2.0. Complexity of the lipidome
Three major phospholipids (phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL)) comprise about 95% of the lipids found in the inner membrane of a simple organism such as Escherichia coli [13]. The remaining 5% is made up of precursors to the major phospholipids as well as some modified phospholipids. The inner leaflet of the outer membrane is almost exclusively composed of PE while the outer leaflet is a monolayer of the lipid A component of lipopolysaccharide. The overall phospholipid composition of E. coli is about 70–80% of zwitterionic PE with the remainder being mostly anionic PG plus CL. Unrecognized in many in vitro studies is that the inner membrane content is roughly 50–60% PE given the high PE content of the outer membrane. Eukaryotic membranes are considerably more complex since they contain sterols, sphingolipids, complex glycolipids and the additional phospholipid headgroups of choline and inositol. Additional complexity comes from variation in fatty acid composition within each lipid class ranging from 12–26 carbons with different degrees of saturation and asymmetric distribution of fatty acids esterified to the glycerol backbone, which could be physiologically important [16]. Therefore, the diversity of lipid species in a simple organism such as E. coli is in the 1000’s while diversity in higher organisms is considerably higher. Furthermore, lipid composition in all cell types is not uniform either laterally along the membrane, between the two leaflets of the lipid bilayer, between different membranes, or temporally during the cell cycle.
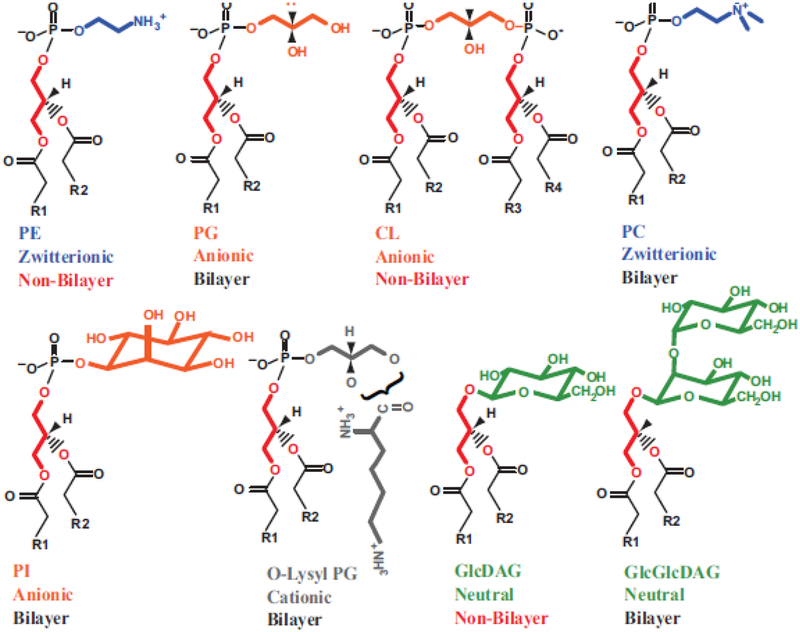
The properties of a membrane bilayer are a complex sum of the properties of its individual species, which ultimately affects the structure and function of embedded and peripheral membrane proteins at the time of initial membrane insertion as well as after proteins traffic to their final location [5]. Although the physical and chemical properties of single lipids and lipid mixtures have been extensively studied (see Fig. 1), it is not clear how these properties translate into physiological function. Therefore, the use of simple lipid mixtures in studies carried out solely in vitro has the potential to result in inaccurate or incomplete information about a specific lipid’s role. However, given the complexity of lipid compositions, in vivo manipulation of membrane lipid composition results in complex pleiotropic effects that are difficult to sort out.
Figure 1.
Summary of glycerolipid physical and chemical properties [5]. Glycerolipid headgroups range from net charged (anionic or cationic) to neutral (either uncharged or zwitterionic). R1 and R2 denote acyl chains of fatty acids esterified to diacylglycerol (DAG). Depending on the shape of the molecular when considering the ionized headgroup and the fatty acid composition, these lipids can either be bilayer (cylindrical shaped) or non-bilayer (prism shaped) prone. PE can assume either a cylindrical shape when both fatty acids are fully saturated or a prism shape when at least one fatty acid is unsaturated. CL is non-bilayer in the presence of divalent cations, which is the physiological state. Temperature and fatty acid composition affect both the fluidity (lower with saturated fatty acids and at lower temperatures) and the bilayer to non-bilayer transition, which occurs as temperature is raised. Although cellular membranes are bilayer to maintain barrier function, the presence of non-bilayer prone lipids introduces lateral stress and local disorder within the lipid bilayer.
The combined results from in vivo, in situ and reconstituted systems can result in definitive understanding of the role of specific lipid-protein interactions in determining protein structure and function. Our approach to understand the role of specific lipids in various cellular processes has been to alter membrane phospholipid through generation of null mutations in genes encoding enzymes responsible for the committed steps to synthesis of the major phospholipids of E. coli (Fig. 2). Surprisingly, cells completely lacking PE, PG plus CL or CL are viable under set growth conditions but display several phenotypes that are related to the absence of the respective lipids [17]. The molecular basis for selected phenotypes were further investigated in whole cells [18], isolated membranes [19] and proteoliposomes composed of defined lipid compositions and purified proteins [20]. Further analysis of the phenotypes of these null mutants have uncovered roles for specific phospholipids in membrane protein folding and topological organization, DNA replication, cell division, protein translocation across membranes, solute transport, energy transduction, and organization of phospholipids into domains [13]. Most significant for this review is the role of membrane lipid composition in the organization of membrane proteins after exit from the translocon and insertion into the lipid bilayer.
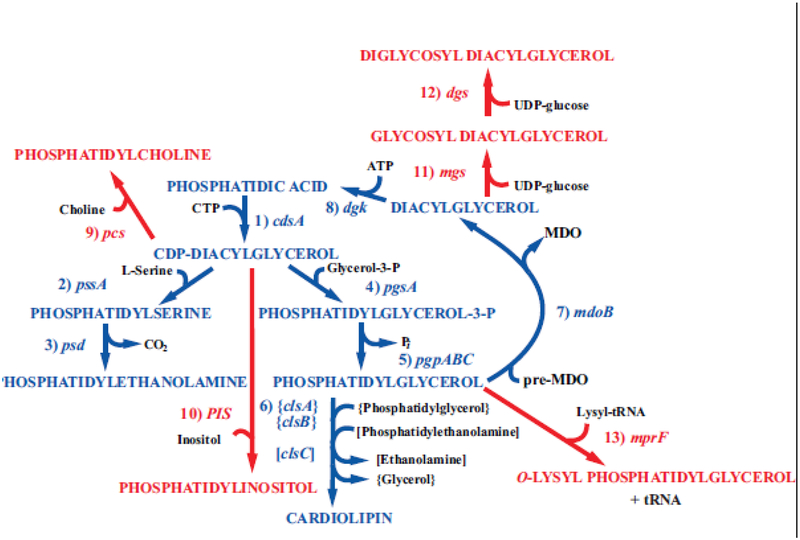
Figure 2.
Synthesis of native and foreign lipids in E. coli. The enzymes with their respective genes named catalyze the following steps for synthesis of native phospholipids noted in blue: 1. CDP-diacylglycerol synthase; 2. phosphatidylserine synthase; 3. phosphatidylserine decarboxylase; 4. phosphatidylglycerophosphate synthase; 5. phosphatidylglycerophosphate phosphatases encoded by three genes; 6. cardiolipin synthases encoded by 3 genes; 7. phosphatidylglycerol:pre-membrane derived oligosaccharide (MDO) sn-glycerol-1-P transferase; 8. diacylglycerol kinase. The enzymes with their respective genes named and their source catalyze the following steps for synthesis of phospholipids foreign to E. coli noted in red: 9. phosphatidylcholine synthase (Legionella pneumophila [21,22]); 10. phosphatidylinositol synthase (Saccharomyces cerevisiae [23]); 11. glucosyl diacylglycerol synthase (Acholeplasma laidlawii [24]); 12. diglucosyl diacylglycerol synthase (Acholeplasma laidlawii [25]); 13. lysyl t-RNA:phosphatidylglycerol lysine transferase (Staphylococcus aureus [26]). Figure (modified) and legend reprinted by permission from Springer Nature: [27] Copyright 2018.
3.0. Requirement of PE for secondary active transporter function
Null mutants of the pssA gene of E. coli lack phosphatidylserine (PS) synthase, which catalyzes the committed step (Fig. 2) to PE biosynthesis [13,28]. The mutant lacks all amino-containing phospholipids (i.e. PS and PE). The phospholipid to protein ratio is unchanged with an increase in the remaining anionic phospholipids PG and CL and their anionic precursors. The mutant requires medium supplemented with all the amino acids. Lack of growth on μmolar but growth on mmolar lactose as a carbon source is consistent with the earlier observation that the lactose permease (LacY) [29], when reconstituted in proteoliposomes, required PE to support energy-dependent uphill transport of lactose but not energy-independent downhill transport [30,31]. A similar requirement for uptake of phenylalanine (PheP) [32], γ-aminobutyric acid (GabP) [33], melibiose (MelB) [34] and sucrose (CscB) [35,36] suggested a general PE requirement for secondary active transporters. Although there are some effects on energy metabolism in PE-lacking cells [37,38] (especially when grown in a chemically defined, less rich medium [39]), subsequent studies ruled out altered energy metabolism as the basis for the lack of uphill transport [20,40].
4.0. PE as a lipochaperone
Lack of recognition of LacY synthesized in PE-lacking cells [19,41,42] by a monoclonal antibody (mAb4B1) specific for a conformational epitope (domain P7, Fig. 3) missing in mutants of LacY not competent for active transport suggested a structural defect in LacY assembled in cells lacking PE [43,44]. These initial studies led to detailed investigation of how lipid-protein interactions determine protein structure at the time of initiation folding as well as dynamically post-assembly.
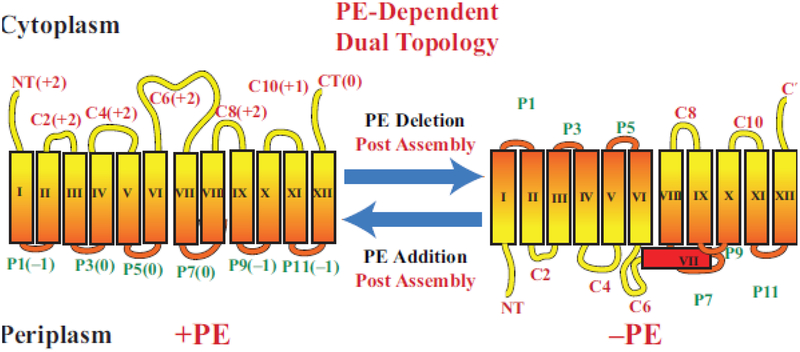
Figure 3.
Topological organization of LacY as a function of membrane lipid composition. TMDs (Roman numerals) and EMDs (Arabic numerals) are sequentially numbered from the N-terminus to C-terminus with EMDs exposed to the periplasm (P) or cytoplasm (C) as in wild type cells. Net charge of EMDs is shown. Topology of LacY is shown after initial assembly in PE-containing cells (+PE) or after initial assembly in PE-lacking (–PE). The interconversion of topological conformers and the ratio of native to inverted conformer are reversible in both directions depending on the dynamic level of PE in membranes. Figure (modified) and legend reprinted by permission from Springer Nature: [27] Copyright 2018.
LacY, although completely delipidated when extracted from membranes using sodium dodecyl sulfate, retains significant amounts of secondary and tertiary structure. LacY from wild type cells when subjected to sodium dodecyl sulfate-polyacryamide gel electrophoresis and Western blotting retains sufficient conformational memory to be recognized by the conformationally specific mAb4B1. LacY from PE-lacking cells is not recognized by mAb4B1. However, transfer of unreactive LacY from a sodium dodecyl sulfate-polyacryamide gel to a solid support pre-blotted with PE restores recognition by mAb4B1. This method, coined an Eastern-Western blot, was used to screen the properties of phospholipids that restore native conformation of the epitope within domain P7 of LacY synthesized in PE-lacking cells. Anionic PG and CL alone were ineffective, as were PEs containing only unsaturated fatty acids (such as dioleyl PE) in the absence of PG and CL. These unsaturated fatty acid derivatives of PE are prone to non-bilayer hexagonal II phase formation at room temperature unless they are mixed with PG and CL to form a lamellar bilayer phase, which supports refolding of domain P7. Phosphatidlycholine (PC) species containing only unsaturated fatty acids with or without the presence of anionic phospholipids do not support refolding of domain P7. This result is consistent with previous findings that similar PC containing mixtures used to reconstitute purified LacY in proteoliposomes only supported downhill but not uphill transport of substrates. The P7 domain epitope was also restored in membranes isolated from PE-lacking cells by in situ synthesis of PE [19]. Taken together the lamellar state of PE is necessary during initial folding of LacY, which retains conformational memory in at least EMD P7 after complete delipidation and can support refolding of misfolded LacY. Moreover, solubilization of PE-deficient membranes in the presence of added PE followed by Western blot analysis did not result in restoration of mAb4B1 recognition, indicating that renaturation in the presence of PE rather than exposure of denatured LacY to PE is required to reform the native epitope. Since the conformation of epitope P7 is dependent on PE during folding but not after proper folding (i.e. native LacY delipidated by SDS), these experiments established PE as a lipochaperone.
5.0. Experimental basis for the Charge Balance Rule
5.1. Generation of mutants in phospholipid metabolism
The experiments using mAb4B1 recognition of the P7 domain as function of lipid environment for LacY strongly suggested a lipid requirement for native protein folding. In order to investigate the influence of membrane lipid composition on protein structure and function, a set of fully viable E. coli “lipid mutants” was developed in which native and introduced foreign lipid content can be systematically controlled at steady state, titrated in a dose-dependent manner or varied temporally during the cell growth [13,14,45]. The pssA null mutant when transformed by a plasmid expressing PS synthase constitutively or under regulation of the tet promoter (controlled by anhydrotetracycline in the growth medium) displays wild type phospholipid composition or a dose-dependent level of PE (from 2–70%), respectively. Introducing foreign genes (Fig. 2) in the null pssA mutant results in full replacement of PE by PC and about 30–40% mono- and diglucosyl diacylglycerol or O-lysyl-PG of total glycerophospholipids. The only common property of these lipids is the ability to buffer the high negative charge density of the membrane surface due to PG and CL, rather than any common physical or structural property.
5.2. Development of methods to measure membrane protein topological organization
We first focused on LacY because of its reported dependence on PE for full function, the availability of a large array of genetic and biochemical tools [46], and eventually a detailed crystal structure [47,48]. To probe organization of LacY in the cytoplasmic membrane of whole E. coli cells, we utilized the substituted cysteine accessibility method applied to TMD orientation (SCAM™) [49] to determine the orientation of LacY with respect to the plane of the lipid bilayer (Fig. 4). This method is based on the accessibility to a membrane impermeable sulfhydryl reagent of single cysteine replacements in EMDs of a cysteine-less derivative of LacY. Fig. 3 displays the expected topological organization of LacY when expressed in cells containing wild type levels of PE [14,15,45]. However, in the pssA null mutant, LacY assumes a vastly different organization with the N-terminal 6-TMD bundle completely inverted with respect to the plane of the membrane bilayer and the last 5-TMD bundle. The low hydrophobicity TMD VII (contains two asparate residues) acts as a hinge point for this inverted structure and is now exposed to the periplasm.
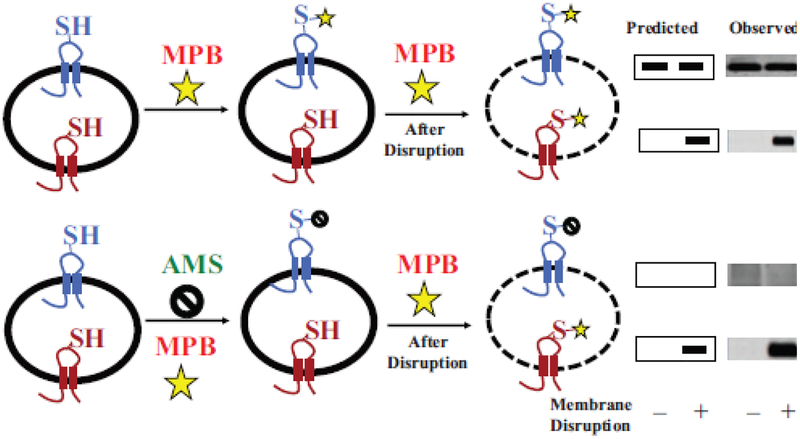
Figure 4.
General strategy for SCAM™ using impermeable thiol reagents. A target membrane protein containing a single cysteine replacement exposed either to the exterior (blue) or interior (red) side of a cell membrane, membrane vesicle or proteoliposome is shown. Half of the sample is treated with the detectable thiol reagent 3-(N-maleimidylpropionyl) biocytin (MPB) to specifically label the externally exposed cysteine (top panel), and the other half is treated with the non-detectable thiol reagent 4-acetamido-4’-maleimidylstilbene-2,2’-disulfonic acid (AMS) (bottom panel) to protect external cysteines during subsequent MPB lableing. Both halves are either kept intact (–) or disrupted (+) by sonication or detergent treatment to expose the interior cysteine followed by treatment with MPB to specifically label the previously inaccessible internal cysteine residue. Labeling by MPB that can be blocked completely by pretreatment with AMS is an independent verification of an outside facing residue (bottom panel). Labeling by MPB that cannot be blocked by such AMS treatment is an independent verification of a residue that is facing the cytoplasm. The target protein is immunoprecipitated and resolved by sodium dodecyl sulfate-polyacrylamine gel electrophoresis and biotinylated protein is detected using avidin-horse radish peroxidase with the predicted and observed results shown on the right. A protein with dual topology would display an increase in the amount of labeled protein after disruption in the upper panel and reduced amount of labeling detected after disruption in the lower panel. The modified figure and legend were reprinted by permission from Springer Nature: [49] Copyright 2019.
5.3. The Positive Inside Rule appers to be violated in PE-lacking cells
Native LacY assembled in wild type E. coli strictly follows the Positive Inside Rule (Fig. 3). All cytoplasmically exposed EMDs have a net positive charge even though they contain a mixture of basic and acidic amino acids. The periplasmically exposed EMDs are either net negative or neutral and contain no positive residues. Nevertheless, the Positive Inside Rule is violated in cells lacking PE by positioning net positively charged EMDs facing the periplasm. Changes in the charge distribution of residues within the cytoplasmically exposed EMDs led to a clearer understanding of how protein charged residues determine TMD orientation as a function of membrane lipid composition [15]. Changing the net positive charge (either adding a positive or removing a negative residue) within the cytoplasmic EMD surface (EMD NT, C2, C4 or C6) of the N-terminal 6-TMD bundle in a position independent manner prevented inversion of LacY in the absence of PE. Inversion of LacY in the presence of PE required an addition of six negative charges distributed along the cytoplasmic surface of the N-terminal bundle. Increasing the hydrophobicity of TMD VII by replacing one aspartic residue by isoleucine prevented inversion in PE-lacking cells. However, the thermodynamic barrier to exposing a now more hydrophobic TMD to an aqueous environment could be overcome by increasing the negative charge of the N-terminal bundle cytoplasmic surface in PE-containing cells [15]. The more hydrophobic TMD VII does not pose a barrier to exposure to aqueous exposure [35] presumably due to the higher translocation potential of negative residues in the absence of PE.
At intermediate levels of PE (using tet promoter regulated pssA), the ratio of native to inverted LacY was proportional to the membrane PE content [50]. Therefore, not only altering the net charge of EMDs [15,36], but also membrane lipid composition can result in a mixture of topological conformers for a membrane protein. These results provide a mechanistic basis for the existence of proteins that assume multiple topological organizations in the same or different membranes.
These results strongly suggest that the presence of PE, which dilutes the high negative membrane surface charge density contributed by PG and CL, suppresses the membrane translocation potential of acidic residues. Also, generation of a mixture of topological conformers of a membrane protein is dependent on lipid-protein interactions and most likely not dependent on the protein insertion machinery because precise timing of these events on both sides of the membrane is well beyond the control of the membrane insertion and folding machineries. How this is accomplished is not clear, but a possibility is the presence of PE increases the effective pKa of acidic residues rendering them more neutral. In the absence of PE, acidic residues express their full potential as translocation signals.
5.4. Generalizing the role of PE to net neutral lipids
The effect of lipid composition is not structurally specific for PE. Expression of the foreign zwitterionic (but net neutral) lipid PC [21], neutral mono- [24] or diglucosyl diacylglycerol [25] or cationic O-lysyl-PG (unpublished result, M. Bogdanov and W. Dowhan) in PE-lacking cells fully substitutes for PE in maintaining native LacY topological organization. The effectiveness of uncharged lipids in maintaining native topology rules out a direct interaction between acidic amino acid residues and PE as the mechanism by which PE suppresses the translocation potential of acidic residues. The inversion of LacY induced by increasing the anionic lipid content of the membrane surface rules out interaction of positively charged protein residues with acidic lipid headgroups as the basis for the Positive Inside Rule [12].
LacY reconstituted into proteoliposomes composed of native E. coli phospholipids displays full energy-dependent active transport of substrate against a concentration gradient [30,40]. Active transport does not occur when PE is replaced by dioleoyl PC [30], a favorite phospholipid used to study membrane proteins reconstituted into proteoliposomes. The molecular basis for this defect was thought to be due to need for the ionizable ethanolamine headgroup of PE. However, PC supports uphill transport when expressed as a foreign lipid in vivo [51] as did palmitoyl oleoyl PC, PC derived from E. coli and neutral glycolipids in vitro [40]. Therefore, zwitterionic phospholipids and net neutral lipids support uphill transport dependent on their fatty acid composition and the common property of the apparent dilution of the high negative charge due to PG and CL.
5.5. Extension of the Positive Inside Rule to the Charge Balance Rule
The above studies form the basis for the Charge Balance Rule that incorporates the influence of lipid environment on the topological organization and function of membrane proteins [15,52–54]. The effect of net neutral lipids on topology explains why positive residues are dominant over negative residues as topological determinants and why some net negative EMDs (containing a mixture of negative and positive residues) are exposed to the cytoplasm. Also, final topological arrangement is a thermodynamic balance between short-range lipid-protein charge effects and long-range protein properties, as evidenced by the effect of changes in the hydrophobicity of TMD VII on final topological organization. LacY is not unique in its cooperative response to changes in EMD charges and the lipid environment. Very similar results were obtained in studies of PheP, GabP and CscB [32,33,35,36]. Recent studies with CscB [36] further established that there is little dependence on the position of charged residues within EMDs as a factor in determining topological organization. Both lipid- and protein charge-dependent topological dynamics of SecG [55,56] as well as large-scale conformational reversible transmembrane reorientations of the colicin Ia [57], a proteorhodopsin [58], transcription factor Nrf1 [59] and scramblase 1 [60] suggest a mechanism driven by the Charge Balance Rule as discussed in detail in [54]. Table 1 summarizes studies that support the Positive Inside Rule and the Charge Balance Rule. Finally, as further documented below, membrane protein topological organization is not fixed at the time of initial assembly but is highly dynamic and independent of factors other than the properties of a protein and its lipid environment.
TABLE 1.
Summary of studies supporting the Positive Inside Rule and the Charge Balance Rule
| Testing membrane protein assembly rules | Positive Inside Rule | Charge Balance Rule |
|---|---|---|
| Statistically | Genome wide [61–63] | Bacterial small multidrug resistance (SMR) proteins [64,65] |
| In vivo | Chimeric construct [66] SMR proteins [67–70], LacY [36] Hybrid chimeric and mutant constructs [71–75] Preprolactin [76] Asialoglycoprotein receptor subunit HI [75,77–80] P-glycoprotein [81–83] Chloroplastic Toc34 [84] |
LacY [18,24,52] GabP [33] CscB [35] PheP [32,35] |
| Sequence position and lipid specificity | Chimeric constructs [9,12,85] | LacY [15] PheP [35] CscB [35,36] |
| In vitro (liposomes) | None that we know of | LacY [20,86] OEP7 [87] Toc34 [88] GltPh Pyrococcus horikoshii [89] Proteorhodopsin [58] Sperm ATPase [90] |
| Single-molecule force microscopy | None that we know of | LacY [91,92] |
| In real-time | None that we know of | LacY [86,93] |
| Crystallographically | Database [94] | None |
5.6. Dynamic organization of membrane proteins
Due to the assumed high energy barrier to post-assembly topological reorganization of membrane proteins, prevailing dogma is that topological organization is fixed during initial translocon-directed bilayer insertion of TMDs. Availability of the tet promoter regulated pssA gene provided a means of testing this assumption [50,51]. Induction of new LacY synthesis was terminated in cells either containing (plus inducer) or lacking (minus inducer) PE to establish an assembled pool of native or inverted LacY, respectively (Fig. 3). Inducer was then removed from the former cells or added to the latter cells. As PE levels declined or increased, the amount of native LacY conformer proportionally decreased or increased. PheP undergoes a similar lipid-dependent post-assembly topological rearrangement [32].
These results indicate that at least for some proteins there is a low energy barrier for large in vitro post-assembly topological rearrangements, which further supports the importance of lipid-protein interactions in determining membrane protein organization. Such large transmembrane rearrangements of hydrophilic EMDs can be facilitated by charged amino acid residues that stabilize water defects within the lipid bilayer [95] thus lowering the free energy barrier for flipping [96].
6.0. In vitro verification of the Charge Balance Rule
Topological organization may be influenced by other cellular factors such as the translocon at the time of initial assembly or protein chaperones once proteins are initially folded. However, in vitro studies strongly indicate that such additional factors are of secondary importance in establishing and maintaining membrane protein topology. The ratio of native to inverted conforms of LacY, when initially reconstituted into liposomes, is directly proportional to the PE content with the remaining phospholipid being a mixture of PG and CL [86] exactly mimicking in vivo results [50]. The native orientation under the reconstitution conditions employed is inverted with respect to that in cells with the cytoplasmic domains exposed on the surface of proteoliposomes. Energy-dependent uphill transport and proper folding of LacY is dependent on the presence of PE, PC (dependent on fatty acid composition) or monoglucosyl diacylglycerol [40].
6.1. Lipid-dependent topological rearrangement in vitro
In order to determine whether post-assembly changes in topology can be demonstrated in vitro, a novel method of changing liposome lipid composition [97] was adapted to change the lipid composition of proteoliposomes (termed fliposomes) after initial assembly. ß-Methyl cyclodextrin (ßMCD) catalyzes the rapid exchange of phospholipids between the outer leaflets of liposomes and multilamellar vesicles (MLV) without fusion or disruption of liposome integrity. Initially, steady state experiments were conducted in which LacY resided in PG/CL proteoliposomes either containing or lacking PE [86]. The proteoliposomes were mixed in the presence of ßMCD and MLVs containing either PG/CL or PE, respectively. Topological organization was measured before and after mixing using SCAM™, which established the same topological reorganization, dependent solely on a change in lipid composition, as observed in vivo.
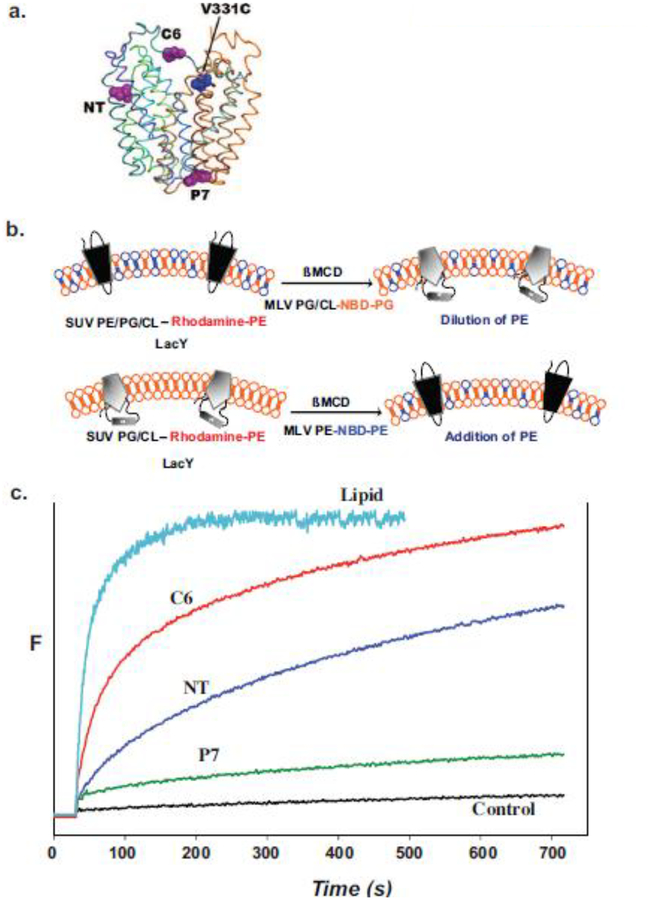
To establish whether such topological inversion can occur on a physiologically significant time scale, the rate of lipid induced topological changes was measured using Förster resonance energy transfer (FRET) [98] (Fig. 5). An acceptor chromophore (IAEDANS, 5-({2-[(iodoacetyl)amino]ethyl}amino)naphthalene-1-sulfonic acid) was placed in a cytoplasmic EMD within the C-terminal 6-TMD helical bundle of LacY, whose topology is insensitive to the lipid environment (Fig. 5a). A FRET donor (tryptophan) was engineered in either the NT, C6 or P7 EMD. Only orientation of EMDs NT and C6 are sensitive to lipid environment with these EMDs being close enough to the chromophore to elicit energy transfer only in the native conformation. The rate of lipid transfer between proteoliposomes and MLVs was monitored with FRET between trace amounts of fluorescent phospholipids in the vesicles (Fig. 5b). As shown in Fig. 5c, transfer of PE to PE-lacking proteoliposomes containing LacY is rapid at room temperature followed by a slower but still rapid increase in FRET when assessing EMD NT or C6. As expected, there is little change in FRET from EMD P7. A similar result was obtained with a rapid decrease in FRET transfer when PE was diluted in proteoliposomes by addition of PG. Rates were easily analyzed for experiments carried out at room temperature but were too fast to analyze at 37°C. Therefore, such interconversions of topological proceed at a physiologically significant rate, independent of other cellular factors.
Figure 5.
Monitoring lipid-dependent topological changes in fliposomes. a. Schematic native structure of LacY showing the position of an engineered tryptophan residue in either EMD NT (residue 14), C6 (residue 205) or P7 (residue 250) relative to the chromophore at position V331C. b. LacY engineered to display high FRET intensity in the native conformation (upper left) or low FRET intensity in the inverted conformation (lower left) was reconstituted into small unilamellar vesicles (SUV) with or without PE, respectively. The SUVs contained trace amounts rhodamine labeled PE. Multilamellar vesicles (MLV) containing PG and CL with a trace amount of 6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl-PG (NBD-PG) or PE with a trace amount of NBD-PE were added to the SUVs containing native or inverted LacY, respectively. Transfer of lipids between SUVs and MLVs was initiated by addition of ß-methyl cyclodextrin (ßMCD)-loaded MLVs to the SUV suspension. The rate of lipid transfer was monitored by FRET between rhodamine- and NBD-labeled lipids. The rate of LacY topological change was monitored by FRET between a tryptophan residue and a chromophore in the C-terminal six TMD bundle of LacY. c. Time scale for change in FRET upon addition of PE to proteoliposomes containing LacY in the absence of PE. Control indicates lack of changes in FRET when SUVs and MLVs both lack PE. Figure 5b was reproduced from[98] by permission of the National Academy of Sciences, USA. Figure 5c was constructed based on data presented in [98].
6.2. Effect of post-translational modification on membrane protein topology
According to the Charge Balance Rule, a change in the net charge of an EMD post-assembly should also induce a change in topological organization. To test this hypothesis, LacY was engineered to contain a protein kinase-dependent phosphorylation site in EMD C6 [93]. Multiple negative charges were introduced into the EMDs of the N-terminal helical bundle so that the native topology predominated in the presence of PE but was one negative charge short of inducing the inverted topology [15]. Rapid phosphorylation of this LacY derivative in PE-containing proteoliposomes induced rapid topological rearrangement of LacY on the same time scale as lipid-induced rearrangements.
Studies of phosphorylation-induced changes in protein topology in model proteoliposomes that reflect the lipid composition of various eukaryotic membranes confirmed the potential of such changes in these membranes but also revealed a more complex dependence on lipid composition [93]. The diversity of lipid species in eukaryote membranes was accompanied by an increased complexity of lipid effects on the rate and extent of protein flipping. Protein flipping rates (native to inverted conformer) and extent of flipping increased along the secretory pathway in proteoliposomes mimicking the lipid composition of the endoplasmic reticulum to the Golgi and finally plasma membrane. Flipping rates (including mitochondria) were in the same range as in E. coli phospholipids except for a significantly lower rate in endosome lipids and endoplasmic reticulum lipids unless the latter was supplemented with cholesterol. The extent of flipping was 40–60% except for only 20% in endosome lipids versus 85% in E. coli lipids. The effect of cholesterol was complex in that it exhibited some properties of a neutral lipid mimicking PE but also increased lipid order in the presence of sphingomyelin while decreasing order in its absence. Differences in lipid order did not correlate with any effects on rate or extent of flipping. Given that LacY may not reflect the properties of many eukaryotic proteins yet undergoes significant topological changes in mixtures of eukaryotic lipids strongly indicates that such transitions can occur in eukaryotic membranes and represent an additional mode of cellular regulation.
There are other post-translational modifications that change the net charge of extramembrane domains and therefore have the potential to alter the topology of membrane proteins. Acylation of lysine, deamidation of glutamine or asparagine, adenylylation of hydroxyl residues, succinylation of lysine and sulfation of tyrosine decrease the net positive charge of EMDs. Glycosylation of membrane proteins has been proposed to prevent topological changes post-assembly due to the increased thermodynamic barrier to moving a hydrated carbohydrate through the bilayer. However, the ability of half of LacY to undergo such a transition suggests that many N-glycosylated membrane proteins could undergo topological changes as was demonstrated for transcription factor Nrf1 [59].
There are at least two examples where phosphorylation of a membrane protein appears to alter its topology. CD38, which exists in multiple topology states, synthesizes cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate, which are messengers for Ca+2-mobilization. CD38 contains a single TMD and exists in two orientations with its catalytic site in one of its EMDs [99,100]. Phosphomimic changing the serine residues to aspartate in the cytoplasmic-facing N-terminal domain results in flipping of this domain to the opposite side of the membrane. Scamblase 1, which transfers aminophospholipids between leaflets of the lipid bilayer, also exists in two dynamic topologically different conformers during differentiation of primary monocytes to macrophages, which could coincides with the change in the PS asymmetric distribution between the two leaflets of the plasma membrane also observed during monocyte-to-macrophage differentiation [60]. The protein is also subject to phosphorylation so that a combination of changes in membrane lipid composition and phosphorylation may be responsible for its dynamic organization. Table 2 summarizes reports of proteins that undergo topological rearrangements either during initial synthesis or after final assembly.
TABLE 2.
Experimental evidences for co- and post-translational and post-insertional membrane protein topological dynamics and instability
| Co-translational/Co-insertional | Po st-translational | Post-insertional |
|---|---|---|
| In vivo | ||
| Triggered by change of lipid composition | Governed by molecular chaperones: | Triggered by change of lipid composition: |
| LacY [15,18] PheP [32] GabP [33] CscB [35,36] Hepatitis B vims envelope protein [101] Triggered by changing of positive inside bias: Hybrid chimeric constructs [73,77] |
Hepatitis C protein NS4B [102–104] Triggered by change of positive inside bias: Lep constructs [69] Governed by topogenic sequences Aquaporin 1 [105,106] Erythrocyte Band 3 [107,108] |
LacY [15,18] PheP[32] Triggered by change of positive inside bias: EmrE [70] |
| In vitro | ||
| Triggered by change of lipid composition | None that we know of | Triggered by change of lipid composition: |
| GltPh Pyrococcus horikoshii [89] Proteorhodopsin [58] Sperm ATPase [90] LacY [20] |
LacY [20,40,92,98] Colicin la channel [57] Triggered by membrane depolarization Colicin la channel [109,110] Colicin A channel [111] Governed by insertion-deinsertion cycle of SecA SecG [56,112] |
|
7.0. Discussion
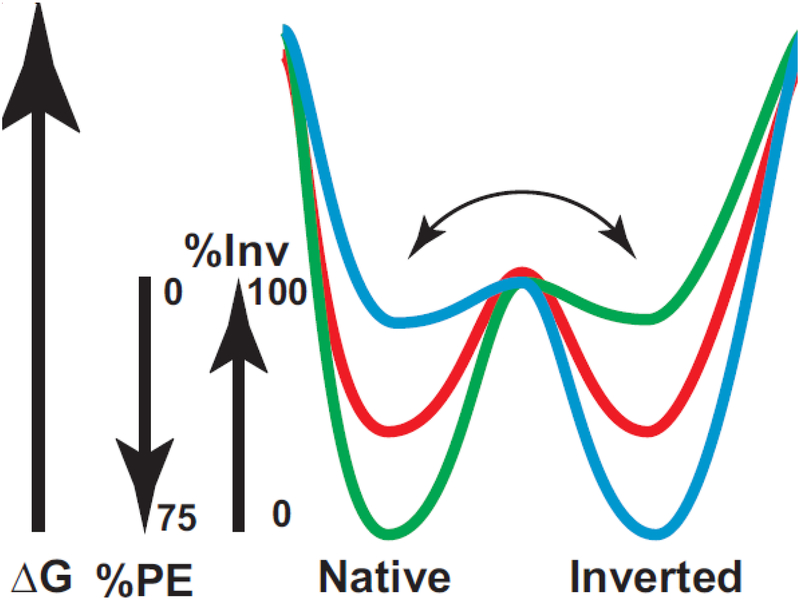
It is now clear that topological organization of membrane proteins is governed by both the protein sequence and its lipid environment. Membrane proteins can undergo topological rearrangements either at the time of initial translocon-directed membrane insertion or dynamically post-assembly in response to changes in the local lipid environment and posttranslational modification. The generation of membrane proteins with dual topology or the determination of TMD orientation need not involve the translocon machinery or other cellular factors other than the protein sequence and its lipid environment. We postulate that during folding of a membrane protein the distribution among multiple topological conformers, which are in rapid equilibrium with each other, is determined by the lipid composition (Fig. 6). Co-translational folding in the membrane would follow the energy landscape resulting in higher energy minima for the inverted conformation at high PE and the native conformation at low PE. At intermediate PE levels both conformers would co-exist dependent on the equilibrium established during folding. Further folding of conformers populated during early folding would result in a high energy barrier between conformer states preventing interconversion among the final conformers. Post-assembly changes in the lipid environment or protein modification may destabilize the conformer mixture allowing partial unfolding and rapid redistribution to the new mixture of stable conformer states. The Charge Balance Rule provides a mechanistic basis for the stable existence of multiple topological conformers for homodimers such as EmrE [113] or monomeric proteins that reside in either the same of different membranes [99,100]. The Charge Balance Rule explains why positive charge is dominant over negative charge as a topological determinant and provides a means for inclusion of acidic residues on the cytoplasmic face of membrane proteins for functional or structural purposes without affecting topology governed by the Positive Inside Rule.
Figure 6.
Dual minima energy folding funnel for LacY as a function of membrane lipid composition. The folding of LacY to its lowest free energy state (ΔG) proceeds via a funnel-shaped energy landscape whose shape is defined by the physicochemical properties of the lipid environment (green, 75% PE; red, intermediate % PE; blue, 0% PE). The conformational space available to the population of folding protein conformers at a given lipid composition is defined by the funnel circumference (x-axis) and the internal ΔG (y-axis) of each folding intermediate. As LacY folds to lower energy conformations, it populates thermodynamic traps whose depth and shape determine the percent of the final native or inverted conformation at steady state. Membrane lipid composition affects a late folding event, which is postulated here to define a rapid equilibrium (horizontal arrow) between subsequent pathways leading to either the native, inverted or mixed conformation separated by a high thermodynamic barrier. Figure and legend reproduced from [50] by permission of American Society for Biochemistry and Molecular Biology
Post-assembly changes in topology have important regulatory implications since such changes can alter or modulate protein function either along the internal protein trafficking pathway or within a specific cellular location. Under normal conditions such topological changes provide an unrecognized means of cellular regulation that includes attenuating exposure of epitopes on either side of a membrane. Mutations that eliminate or introduce a new topogenic signals or protein modification sites could result in an alternate topology and function for a protein at its final cellular location. The aberrant lipid-protein interactions during polytopic protein biogenesis can contribute to inherited topological disorders [114], which could arise at the co-translational level of assembly, as recently demonstrated [115]. The generation of dual topology proteins or the determination of TMD orientation need not involve the translocon machinery or other cellular factors other than the protein sequence and the lipid environment.
There remain several unanswered questions. What is the precise mechanism by which lipid composition affects the effective net charge of EMDs? Are the rules for membrane protein assembly different or the same between different organisms? Experiments in proteoliposomes suggest that the membrane potential is not the driving force for the Positive Inside Rule. So what forces determine this charge-dependent orientation of EMDs? Does asymmetric distribution of lipids across the membrane bilayer also affect protein topological organization? Do topological differences originate co-translationally during membrane insertion or are they induced by changes in lipid composition as eukaryotic proteins move through different organelles to their final destination? Answers to these questions will further establish the mechanism underlying the extension of the Positive Inside Rule to the Charge Balance Rule.
Acknowledgements:
This work was supported in whole or in part by National Institutes of Health Grant GM R01 121493 and the John Dunn Research Foundation both to W. D. and the European Union Marie Skłodowska-Curie Grant H2020-MSCA-RISE-2015–690853, NATO Science for Peace and Security Programme-SPS 985291 and Program of Competitive Growth of Kazan Federal University to M. B.
Abbreviations:
- TMD
transmembrane domain
- EMD
extramembrane domain
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- CL
cardiolipin
- PS
phosphatidylserine
- PC
phosphatidlycholine
- LacY
lactose permease
- PheP
phenylalanine permease
- GabP
γ-aminobutyric acid permease
- MelB
melibiose permease
- CscB
sucrose permease
- mAb4B1
monoclonal antibody 4B1
- SCAM™
substituted cysteine accessibility method applied to TMD orientation
- ßMCD
ß-methyl cyclodextrin
- MLV
multilamellar vesicle
- FRET
Förster resonance energy transfer
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Compliance with Ethical Standards: Authors declare no conflicts of interest. No human subjects or animials were used by the authors.
References
- 1.Blobel G (1980) Intracellular protein topogenesis. Proc Natl Acad Sci U S A 77:1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn A, Koch HG, Dalbey RE (2017) Targeting and Insertion of Membrane Proteins. EcoSal Plus 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapoport TA (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450:663–669. [DOI] [PubMed] [Google Scholar]
- 4.Cymer F, von Heijne G, White SH (2015) Mechanisms of integral membrane protein insertion and folding. J Mol Biol 427:999–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowhan W (1997) Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem 66:199–232. [DOI] [PubMed] [Google Scholar]
- 6.Ulmschneider MB, Ulmschneider JP, Schiller N, Wallace BA, von Heijne G, White SH (2014) Spontaneous transmembrane helix insertion thermodynamically mimics translocon-guided insertion. Nat Commun 5:4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Heijne G (2006) Membrane-protein topology. Nat Rev Mol Cell Biol 7:909–918. [DOI] [PubMed] [Google Scholar]
- 8.Andersson H, von Heijne G (1993) Position-specific Asp-Lys pairing can affect signal sequence function and membrane protein topology. J Biol Chem 268:21389–21393. [PubMed] [Google Scholar]
- 9.Andersson H, von Heijne G (1994) Membrane protein topology: effects of Δμ H+ on the translocation of charged residues explain the ‘positive inside’ rule. EMBO J 13:2267–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delgado-Partin VM, Dalbey RE (1998) The proton motive force, acting on acidic residues, promotes translocation of amino-terminal domains of membrane proteins when the hydrophobicity of the translocation signal is low. J Biol Chem 273:9927–9934. [DOI] [PubMed] [Google Scholar]
- 11.van de Vossenberg JL, Albers SV, van der Does C, Driessen AJ, van Klompenburg W (1998) The positive inside rule is not determined by the polarity of the delta psi (transmembrane electrical potential). Mol Microbiol 29:1125–1127. [DOI] [PubMed] [Google Scholar]
- 12.van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B (1997) Anionic phospholipids are determinants of membrane protein topology. EMBO J 16:4261–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowhan W (2013) A retrospective: use of Escherichia coli as a vehicle to study phospholipid synthesis and function. Biochim Biophys Acta 1831:471–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogdanov M, Dowhan W, Vitrac H (2014) Lipids and topological rules governing membrane protein assembly. Biochim Biophys Acta 1843:1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdanov M, Xie J, Heacock P, Dowhan W (2008) To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J Cell Biol 182:925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manni MM, Tiberti ML, Pagnotta S, Barelli H, Gautier R, Antonny B (2018) Acyl chain asymmetry and polyunsaturation of brain phospholipids facilitate membrane vesiculation without leakage. Elife 7: e34394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowhan W (2009) Molecular genetic approaches to defining lipid function. J Lipid Res 50 Suppl:S305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogdanov M, Heacock PN, Dowhan W (2002) A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J 21:2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogdanov M, Dowhan W (1998) Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. EMBO J 17:5255–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Bogdanov M, Dowhan W (2002) Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J 21:5673–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogdanov M, Heacock P, Guan Z, Dowhan W (2010) Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc Natl Acad Sci U S A 107:15057–15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conover GM, Martinez-Morales F, Heidtman MI, Luo ZQ, Tang M, Chen C, Geiger O, Isberg RR (2008) Phosphatidylcholine synthesis is required for optimal function of Legionella pneumophila virulence determinants. Cell Microbiol 10:514–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia W, Dowhan W (1995) Phosphatidylinositol cannot substitute for phosphatidylglycerol in supporting cell growth of Escherichia coli. J Bacteriol 177:2926–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie J, Bogdanov M, Heacock P, Dowhan W (2006) Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J Biol Chem 281:19172–19178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wikström M, Kelly AA, Georgiev A, Eriksson HM, Klement MR, Bogdanov M, Dowhan W, Wieslander Ä (2009) Lipid-engineered Escherichia coli membranes reveal critical lipid headgroup size for protein function. J Biol Chem 284:954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oku Y, Kurokawa K, Ichihashi N, Sekimizu K (2004) Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 150:45–51. [DOI] [PubMed] [Google Scholar]
- 27.Dowhan W, Bogdanov M, Mileykovskaya E, Vitrac H (2017) Functional Roles of Individual Membrane Phospholipids in Escherichia coli and Saccharomyces cerevisiae In: Geiger O (ed) Biogenesis of Fatty Acids, Lipids and Membranes Handbook of Hydrocarbon and Lipid Microbiology. Springer, Cham. [Google Scholar]
- 28.DeChavigny A, Heacock PN, Dowhan W (1991) Phosphatidylethanolamine may not be essential for the viability of Escherichia coli. J Biol Chem 266:5323–5332. [PubMed] [Google Scholar]
- 29.Wikstrom M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander A, Dowhan W (2004) Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem 279:10484–10493. [DOI] [PubMed] [Google Scholar]
- 30.Chen CC, Wilson TH (1984) The phospholipid requirement for activity of the lactose carrier of Escherichia coli. J Biol Chem 259:10150–10158. [PubMed] [Google Scholar]
- 31.Seto-Young D, Chen CC, Wilson TH (1985) Effect of different phospholipids on the reconstitution of two functions of the lactose carrier of Escherichia coli. J Membr Biol 84:259–267. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W (2003) Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem 278:50128–50135. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Campbell HA, King SC, Dowhan W (2005) Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the gamma-aminobutyric acid permease (GabP) of Escherichia coli. J Biol Chem 280:26032–26038. [DOI] [PubMed] [Google Scholar]
- 34.Hariharan P, Tikhonova E, Medeiros-Silva J, Jeucken A, Bogdanov MV, Dowhan W, Brouwers JF, Weingarth M, Guan L (2018) Structural and functional characterization of protein-lipid interactions of the Salmonella typhimurium melibiose transporter MelB. BMC Biol 16:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitrac H, Bogdanov M, Heacock P, Dowhan W (2011) Lipids and topological rules of membrane protein assembly: balance between long- and short-range lipid-protein interactions. J Biol Chem 286:15182–15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vitrac H, Dowhan W, Bogdanov M (2017) Effects of mixed proximal and distal topogenic signals on the topological sensitivity of a membrane protein to the lipid environment. Biochim Biophys Acta Biomembr 1859:1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogdanov M, Dowhan W (1995) Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J Biol Chem 270:732–739. [DOI] [PubMed] [Google Scholar]
- 38.Mileykovskaya EI, Dowhan W (1993) Alterations in the electron transfer chain in mutant strains of Escherichia coli lacking phosphatidylethanolamine. J Biol Chem 268:24824–24831. [PubMed] [Google Scholar]
- 39.Rowlett VW, Mallampalli V, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, Vitrac H (2017) Impact of Membrane Phospholipid Alterations in Escherichia coli on Cellular Function and Bacterial Stress Adaptation. J Bacteriol 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vitrac H, Bogdanov M, Dowhan W (2013) Proper fatty acid composition rather than an ionizable lipid amine is required for full transport function of lactose permease from Escherichia coli. J Biol Chem 288:5873–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogdanov M, Sun J, Kaback HR, Dowhan W (1996) A phospholipid acts as a chaperone in assembly of a membrane transport protein. J Biol Chem 271:11615–11618. [DOI] [PubMed] [Google Scholar]
- 42.Bogdanov M, Umeda M, Dowhan W (1999) Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J Biol Chem 274:12339–12345. [DOI] [PubMed] [Google Scholar]
- 43.Frillingos S, Kaback HR (1996) Monoclonal antibody 4B1 alters the pKa of a carboxylic acid at position 325 (helix X) of the lactose permease of Escherichia coli. Biochemistry 35:10166–10171. [DOI] [PubMed] [Google Scholar]
- 44.Frillingos S, Wu J, Venkatesan P, Kaback HR (1997) Binding of ligand or monoclonal antibody 4B1 induces discrete structural changes in the lactose permease of Escherichia coli. Biochemistry 36:6408–6414. [DOI] [PubMed] [Google Scholar]
- 45.Dowhan W, Bogdanov M (2012) Molecular genetic and biochemical approaches for defining lipid-dependent membrane protein folding. Biochim Biophys Acta 1818:1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaback HR, Sahin-Toth M, Weinglass AB (2001) The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol 2:610–620. [DOI] [PubMed] [Google Scholar]
- 47.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610–615. [DOI] [PubMed] [Google Scholar]
- 48.Guan L, Mirza O, Verner G, Iwata S, Kaback HR (2007) Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A 104:15294–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogdanov M (2017) Mapping of Membrane Protein Topology by Substituted Cysteine Accessibility Method (SCAM). Methods Mol Biol 1615:105–128. [DOI] [PubMed] [Google Scholar]
- 50.Bogdanov M, Dowhan W (2012) Lipid-dependent generation of dual topology for a membrane protein. J Biol Chem 287:37939–37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bogdanov M, Heacock PN, Dowhan W (2010) Study of polytopic membrane protein topological organization as a function of membrane lipid composition. Methods Mol Biol 619:79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bogdanov M, Xie J, Dowhan W (2009) Lipid-protein interactions drive membrane protein topogenesis in accordance with the positive inside rule. J Biol Chem 284:9637–9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dowhan W, Bogdanov M (2009) Lipid-dependent membrane protein topogenesis. Annu Rev Biochem 78:515–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogdanov M, Vitrac H, Dowhan W (2018) Flip-flopping Membrane Proteins: How the Charge Balance Rule Governs Dynamic Membrane Protein Topology Biogenesis of Fatty Acids, Lipids and Membranes Handbook of Hydrocarbon and Lipid Microbiology. Springer, Cham. [Google Scholar]
- 55.Moser M, Nagamori S, Huber M, Tokuda H, Nishiyama K (2013) Glycolipozyme MPIase is essential for topology inversion of SecG during preprotein translocation. Proc Natl Acad Sci U S A 110:9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishiyama K, Suzuki T, Tokuda H (1996) Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell 85:71–81. [DOI] [PubMed] [Google Scholar]
- 57.Yao XL, Hong M (2006) Effects of anionic lipid and ion concentrations on the topology and segmental mobility of colicin Ia channel domain from solid-state NMR. Biochemistry 45:289–295. [DOI] [PubMed] [Google Scholar]
- 58.Tunuguntla R, Bangar M, Kim K, Stroeve P, Ajo-Franklin CM, Noy A (2013) Lipid bilayer composition can influence the orientation of proteorhodopsin in artificial membranes. Biophys J 105:1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Ren Y, Li S, Hayes JD (2014) Transcription factor Nrf1 is topologically repartitioned across membranes to enable target gene transactivation through its acidic glucose-responsive domains. PLoS One 9:e93458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herate C, Ramdani G, Grant NJ, Marion S, Gasman S, Niedergang F, Benichou S, Bouchet J (2016) Phospholipid Scramblase 1 Modulates FcR-Mediated Phagocytosis in Differentiated Macrophages. PLoS One 11:e0145617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baker JA, Wong WC, Eisenhaber B, Warwicker J, Eisenhaber F (2017) Charged residues next to transmembrane regions revisited: “Positive-inside rule” is complemented by the “negative inside depletion/outside enrichment rule”. BMC Biol 15:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nilsson J, Persson B, von Heijne G (2005) Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins 60:606–616. [DOI] [PubMed] [Google Scholar]
- 63.Wallin E, von Heijne G (1998) Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci 7:1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bay DC, Turner RJ (2013) Membrane composition influences the topology bias of bacterial integral membrane proteins. Biochim Biophys Acta 1828:260–270. [DOI] [PubMed] [Google Scholar]
- 65.Newport TD, Sansom MSP, Stansfeld PJ (2018) The MemProtMD database: a resource for membrane-embedded protein structures and their lipid interactions. Nucleic Acids Res 47:D390–D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Heijne G (1989) Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341:456–458. [DOI] [PubMed] [Google Scholar]
- 67.Kolbusz MA, ter Horst R, Slotboom DJ, Lolkema JS (2010) Orientation of small multidrug resistance transporter subunits in the membrane: correlation with the positive-inside rule. J Mol Biol 402:127–138. [DOI] [PubMed] [Google Scholar]
- 68.Rapp M, Granseth E, Seppälä S, von Heijne G (2006) Identification and evolution of dual-topology membrane proteins. Nat Struct Mol Biol 13:112–116. [DOI] [PubMed] [Google Scholar]
- 69.Seppälä S, Slusky JS, Lloris-Garcera P, Rapp M, von Heijne G (2010) Control of membrane protein topology by a single C-terminal residue. Science 328:1698–1700. [DOI] [PubMed] [Google Scholar]
- 70.Woodall NB, Hadley S, Yin Y, Bowie JU (2017) Complete topology inversion can be part of normal membrane protein biogenesis. Protein Sci 26:824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujita H, Yamagishi M, Kida Y, Sakaguchi M (2011) Positive charges on the translocating polypeptide chain arrest movement through the translocon. J Cell Sci 124:4184–4193. [DOI] [PubMed] [Google Scholar]
- 72.Harley CA, Holt JA, Turner R, Tipper DJ (1998) Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J Biol Chem 273:24963–24971. [DOI] [PubMed] [Google Scholar]
- 73.Kim H, Paul S, Gennity J, Jennity J, Inouye M (1994) Reversible topology of a bifunctional transmembrane protein depends upon the charge balance around its transmembrane domain. Mol Microbiol 11:819–831. [DOI] [PubMed] [Google Scholar]
- 74.Sato M, Hresko R, Mueckler M (1998) Testing the charge difference hypothesis for the assembly of a eucaryotic multispanning membrane protein. J Biol Chem 273:25203–25208. [DOI] [PubMed] [Google Scholar]
- 75.Yamagishi M, Onishi Y, Yoshimura S, Fujita H, Imai K, Kida Y, Sakaguchi M (2014) A few positively charged residues slow movement of a polypeptide chain across the endoplasmic reticulum membrane. Biochemistry 53:5375–5383. [DOI] [PubMed] [Google Scholar]
- 76.Andrews DW, Young JC, Mirels LF, Czarnota GJ (1992) The role of the N region in signal sequence and signal-anchor function. J Biol Chem 267:7761–7769. [PubMed] [Google Scholar]
- 77.Beltzer JP, Fiedler K, Fuhrer C, Geffen I, Handschin C, Wessels HP, Spiess M (1991) Charged residues are major determinants of the transmembrane orientation of a signal-anchor sequence. J Biol Chem 266:973–978. [PubMed] [Google Scholar]
- 78.Goder V, Bieri C, Spiess M (1999) Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J Cell Biol 147:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goder V, Junne T, Spiess M (2004) Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol Biol Cell 15:1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sommer N, Junne T, Kalies KU, Spiess M, Hartmann E (2013) TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim Biophys Acta 1833:3104–3111. [DOI] [PubMed] [Google Scholar]
- 81.Enquist K, Fransson M, Boekel C, Bengtsson I, Geiger K, Lang L, Pettersson A, Johansson S, von Heijne G, Nilsson I (2009) Membrane-integration characteristics of two ABC transporters, CFTR and P-glycoprotein. J Mol Biol 387:1153–1164. [DOI] [PubMed] [Google Scholar]
- 82.Zhang JT, Lee CH, Duthie M, Ling V (1995) Topological determinants of internal transmembrane segments in P-glycoprotein sequences. J Biol Chem 270:1742–1746. [DOI] [PubMed] [Google Scholar]
- 83.Zhang JT, Ling V (1993) Membrane orientation of transmembrane segments 11 and 12 of MDR- and non-MDR-associated P-glycoproteins. Biochim Biophys Acta 1153:191–202. [DOI] [PubMed] [Google Scholar]
- 84.May T, Soll J (1998) Positive charges determine the topology and functionality of the transmembrane domain in the chloroplastic outer envelope protein Toc34. J Cell Biol 141:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andersson H, Bakker E, von Heijne G (1992) Different positively charged amino acids have similar effects on the topology of a polytopic transmembrane protein in Escherichia coli. J Biol Chem 267:1491–1495. [PubMed] [Google Scholar]
- 86.Vitrac H, Bogdanov M, Dowhan W (2013) In vitro reconstitution of lipid-dependent dual topology and postassembly topological switching of a membrane protein. Proc Natl Acad Sci U S A 110:9338–9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schleiff E, Tien R, Salomon M, Soll J (2001) Lipid composition of outer leaflet of chloroplast outer envelope determines topology of OEP7. Mol Biol Cell 12:4090–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qbadou S, Tien R, Soll J, Schleiff E (2003) Membrane insertion of the chloroplast outer envelope protein, Toc34: constrains for insertion and topology. J Cell Sci 116:837–846. [DOI] [PubMed] [Google Scholar]
- 89.McIlwain BC, Vandenberg RJ, Ryan RM (2015) Transport rates of a glutamate transporter homologue are influenced by the lipid bilayer. J Biol Chem 290:9780–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hickey KD, Buhr MM (2011) Lipid bilayer composition affects transmembrane protein orientation and function. J Lipids 2011:208457–208466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dowhan W, Vitrac H, Bogdanov M (2015) May the Force Be With You: Unfolding Lipid-Protein Interactions By Single-Molecule Force Spectroscopy. Structure 23:612–614. [DOI] [PubMed] [Google Scholar]
- 92.Serdiuk T, Sugihara J, Mari SA, Kaback HR, Müller DJ (2015) Observing a Lipid-Dependent Alteration in Single Lactose Permeases. Structure 23:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vitrac H, MacLean DM, Karlstaedt A, Taegtmeyer H, Jayaraman V, Bogdanov M, Dowhan W (2017) Dynamic lipid-dependent modulation of protein topology by post-translational phosphorylation. J Biol Chem:1613–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ikeda M, Arai M, Okuno T, Shimizu T (2003) TMPDB: a database of experimentally-characterized transmembrane topologies. Nucleic Acids Res 31:406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Patel SJ, Van Lehn RC (2018) Characterizing the Molecular Mechanisms for Flipping Charged Peptide Flanking Loops across a Lipid Bilayer. J Phys Chem B 122:10337–10348. [DOI] [PubMed] [Google Scholar]
- 96.Van Lehn RC, Alexander-Katz A (2017) Grafting Charged Species to Membrane-Embedded Scaffolds Dramatically Increases the Rate of Bilayer Flipping. ACS Cent Sci 3:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng HT, Megha, London E (2009) Preparation and properties of asymmetric vesicles that mimic cell membranes: effect upon lipid raft formation and transmembrane helix orientation. J Biol Chem 284:6079–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vitrac H, MacLean DM, Jayaraman V, Bogdanov M, Dowhan W (2015) Dynamic membrane protein topological switching upon changes in phospholipid environment. Proc Natl Acad Sci U S A 112:13874–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao YJ, Lam CM, Lee HC (2012) The membrane-bound enzyme CD38 exists in two opposing orientations. Sci Signal 5:ra67. [DOI] [PubMed] [Google Scholar]
- 100.Zhao YJ, Zhu WJ, Wang XW, Zhang LH, Lee HC (2014) Determinants of the membrane orientation of a calcium signaling enzyme CD38. Biochim Biophys Acta 1853:2095–2103. [DOI] [PubMed] [Google Scholar]
- 101.Dorobantu C, Macovei A, Lazar C, Dwek RA, Zitzmann N, Branza-Nichita N (2011) Cholesterol depletion of hepatoma cells impairs hepatitis B virus envelopment by altering the topology of the large envelope protein. J Virol 85:13373–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Awe K, Lambert C, Prange R (2008) Mammalian BiP controls posttranslational ER translocation of the hepatitis B virus large envelope protein. FEBS Lett 582:3179–3184. [DOI] [PubMed] [Google Scholar]
- 103.Lambert C, Prange R (2003) Chaperone action in the posttranslational topological reorientation of the hepatitis B virus large envelope protein: Implications for translocational regulation. Proc Natl Acad Sci U S A 100:5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lundin M, Monne M, Widell A, Von Heijne G, Persson MA (2003) Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol 77:5428–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR (2000) Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol Biol Cell 11:2973–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Virkki MT, Agrawal N, Edsbacker E, Cristobal S, Elofsson A, Kauko A (2014) Folding of Aquaporin 1: multiple evidence that helix 3 can shift out of the membrane core. Protein Sci 23:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kanki T, Young MT, Sakaguchi M, Hamasaki N, Tanner MJ (2003) The N-terminal region of the transmembrane domain of human erythrocyte band 3. Residues critical for membrane insertion and transport activity. J Biol Chem 278:5564–5573. [DOI] [PubMed] [Google Scholar]
- 108.Ota K, Sakaguchi M, Hamasaki N, Mihara K (1998) Assessment of topogenic functions of anticipated transmembrane segments of human band 3. J Biol Chem 273:28286–28291. [DOI] [PubMed] [Google Scholar]
- 109.Jakes KS, Kienker PK, Slatin SL, Finkelstein A (1998) Translocation of inserted foreign epitopes by a channel-forming protein. Proc Natl Acad Sci U S A 95:4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kienker PK, Qiu X, Slatin SL, Finkelstein A, Jakes KS (1997) Transmembrane insertion of the colicin Ia hydrophobic hairpin. J Membr Biol 157:27–37. [DOI] [PubMed] [Google Scholar]
- 111.Slatin SL, Duche D, Kienker PK, Baty D (2004) Gating movements of colicin A and colicin Ia are different. J Membr Biol 202:73–83. [DOI] [PubMed] [Google Scholar]
- 112.Nagamori S, Nishiyama K, Tokuda H (2002) Membrane topology inversion of SecG detected by labeling with a membrane-impermeable sulfhydryl reagent that causes a close association of SecG with SecA. J Biochem 132:629–634. [DOI] [PubMed] [Google Scholar]
- 113.Fluman N, Tobiasson V, von Heijne G (2017) Stable membrane orientations of small dual-topology membrane proteins. Proc Natl Acad Sci U S A 114:7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schlebach JP, Sanders CR (2015) Influence of Pathogenic Mutations on the Energetics of Translocon-Mediated Bilayer Integration of Transmembrane Helices. J Membr Biol 248:371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roushar FJ, Gruenhagen TC, Penn WD, Li B, Meiler J, Jastrzebska B, Schlebach JP (2018) Contribution of Cotranslational Folding Defects to Membrane Protein Homeostasis. J Am Chem Soc 141:204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]