Abstract
The neurotrophic tyrosine receptor kinase (NTRK) gene family encodes three tropomyosin receptor kinases (TRKA, TRKB, TRKC) that contribute to central and peripheral nervous system development and function. NTRK gene fusions are oncogenic drivers of various adult and paediatric tumours. Several methods have been used to detect NTRK gene fusions including immunohistochemistry, fluorescence in situ hybridisation, reverse transcriptase polymerase chain reaction, and DNA- or RNA-based next-generation sequencing. For patients with TRK fusion cancer, TRK inhibition is an important therapeutic target. Following the FDA approval of the selective TRK inhibitor, larotrectinib, as well as the ongoing development of multi-kinase inhibitors with activity in TRK fusion cancer, testing for NTRK gene fusions should become part of the standard diagnostic process. In this review we discuss the biology of NTRK gene fusions, and we present a testing algorithm to aid detection of these gene fusions in clinical practice and guide treatment decisions.
Keywords: ntrk gene fusions, TRKA, TRKB, TRKC, trk fusion cancer, cancer screening, tyrosine kinase inhibitor, tumour-agnostic biomarker
Introduction
Fusions involving neurotrophic tyrosine receptor kinases (NTRK) were among the first gene translocations described in cancer.1 Selective inhibition of the resulting tropomyosin receptor kinase (TRK) fusion proteins offers a precision medicine approach to the treatment of a range of tumour types.2
NTRK structure and function
Tropomyosin receptor kinase A, B and C (TRKA, TRKB and TRKC) encoded by the NTRK1, NTRK2 and NTRK3 genes located on human chromosomes 1q23.1, 9q21.33 and 15q25.3, respectively, are receptor tyrosine kinases expressed in human neuronal tissue.3–5
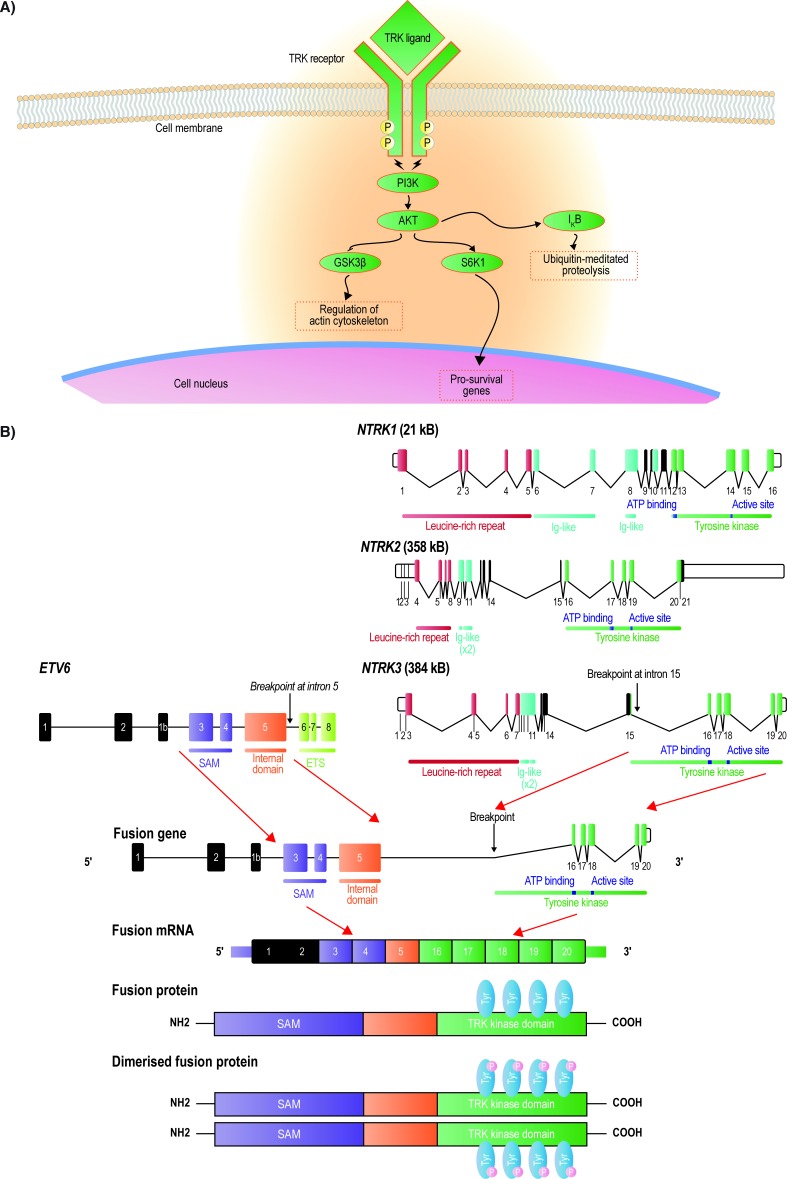
All three TRK receptors comprise an extracellular ligand-binding domain, a transmembrane region and an intracellular adenosine triphosphate-binding domain.2 6 TRK receptors are activated when neurotrophin ligands bind to the extracellular domain of the receptor (figure 1A). The neurotrophins are specific to each receptor: nerve growth factor (NGF) activates TRKA, brain-derived neurotrophic growth factor (BDNF) and neurotrophin 4/5 activate TRKB and neurotrophin 3 activates TRKC.2 Ligand–receptor interaction triggers receptor homodimerisation, phosphorylation of the kinase domain and activation of downstream signalling pathways that play pivotal roles in the development and function of the central and peripheral nervous systems.2
Figure 1.
Schematic figure showing the TRK receptor tyrosine kinases, activating neurotrophins and the major signal transduction pathways (A) and the genomic structures of NTRK1, NTRK2, and NTRK3, with the size of each gene in parentheses (B). The ETV6 and NTRK3 gene fusion and the resultant constitutively active TRK fusion protein is a typical example. GSK3ß, glycogen synthase kinase 3 beta; Ig, immunoglobulin; mRNA, messenger ribonucleic acid; NTRK, neurotrophic tyrosine receptor kinase; PI3K, phosphoinositide-3-kinase; SAM, sterile alpha motif; TRK, tropomyosin receptor kinase.
Binding of NGF to TRKA results in activation of the RAS mitogen-activated protein kinase pathway leading to increased proliferation and cellular growth mediated by extracellular signal-related kinase (ERK) signalling.2 Phospholipase C-γ (PLCγ) and phosphoinositide-3-kinase (PI3K) are also activated.2 BDNF binding to TRKB activates the RAS-ERK, PI3K and PLC-γ pathways resulting in neuronal differentiation and survival, and NT3 binding to TRKC preferentially activates the PI3K/AKT pathway, which prevents apoptosis and increases cell survival.2 The proper regulation of TRK receptor levels and their activation is critical to normal cell function. Upregulation of TRK receptors has been reported in a number of central nervous system-related disorders, for example, TRKB in epilepsy, neuropathic pain or depression.2
NTRK gene fusions
NTRK gene fusions result from intra-chromosomal or inter-chromosomal rearrangements that juxtapose the 3’ region of the NTRK gene with the 5’ sequence of a fusion partner gene expressed by the tumour cell progenitor (figure 1B).2 The NTRK gene fusion transcript encodes a protein composed of the N-terminus of the fusion partner with the TRK partner tyrosine kinase domain.2 In most characterised fusions, the 5’ partner gene sequence encodes one or more dimerisation domains,7 resulting in a constitutively active fusion protein.7 This constitutive activation results in uninterrupted downstream signalling messages,7 8 thereby acting as a true oncogenic driver. Although fusions may occur in any of the three NTRK genes,9 most of those identified to date involve either NTRK3 or NTRK1.7 9 10
TRK fusion cancer
Fusions involving the NTRK1, 2 and 3 genes have been identified as oncogenic drivers and diagnostic markers in various cancer types (table 1).7 9–38 TRK fusion proteins are often mutually exclusive of other known fusion proteins involving kinases.39 Specific NTRK gene fusions are associated with certain tumours,9 for example, the ETV6-NTRK3 gene fusion is exhibited by 90%–100% of mammary analogue secretory carcinomas,11 >90% of secretory breast cancers,12 and is present in most cases of infantile fibrosarcoma34 and congenital mesoblastic nephroma.40 In contrast some cancers have many different fusion partners.7 In lung cancer, seven different gene fusions involving the NTRK1 gene leading to constitutive TRKA tyrosine kinase domain activation have been described (table 1), for example, rearrangement of the 5’ portion of the myosin phosphatase Rho-interacting protein (MPRIP) gene fused to the 3’ portion of NTRK1 or rearrangement between CD74 and NTRK1.7 9
Table 1.
NTRK gene fusions identified in adult and paediatric cancers by relative frequency of NTRK gene fusions
| Fusion partner | |||
| Tumour | NTRK1 | NTRK2 | NTRK3 |
| Adult cancers | |||
| High frequency (>80%) | |||
| Mammary analogue secretory carcinomas | ETV6 11 | ||
| Secretory breast carcinoma | ETV6 12 | ||
| Intermediate frequency (5%–25%) | |||
| Papillary thyroid cancer | TFG, 13 SSBP2, 9 SQSTM1, 9 TPR, 7 PPL 7 | ETV6, 9 43 RBPMS 9 | |
| Low frequency (<5%) | |||
| Appendiceal cancer | LMNA 18 | ||
| Glioma/glioblastoma | ARHGEF2, 19 BCAN, 20 21 CHTOP, 19 NFASC 20 | BCR, 18 AFAP1, 9 SQSTM1 9 | AFAP1, 18 ZNF710, 18 EML4 18 |
| Astrocytoma | QK1, 7 NACC2 7 | ||
| Gastrointestinal stromal tumour | ETV6 15 | ||
| Head and neck cancer | PAN3 9 | LYN 9 | |
| Lung cancer | CD74, 7 GRPAP1, 23 IRF2BP2, 18 MPRIP, 7 P2RY8, 18 SQSTM1, 24 TPM3 18 | TRIM24 9 | |
| Sarcoma | TPM3, 9 LMNA 18 | TPM4 10 | |
| Breast cancer | CGN, 25 GATAD2B, 25 LMNA, 25 MDM4, 25 PEAR1, 25 TPM3, 10 25 | ETV6 25 | |
| Acute lymphoblastic leukaemia, acute myeloid leukaemia, histiocytosis, multiple myeloma, dendritic cell neoplasms | ETV6 26 | ||
| Uterine sarcoma | LMNA, 27 TPM3, 27 TPR 27 | RBPMS 27 | |
| Cholangiocarcinoma | LMNA, 10 RABGAP1L 28 | ||
| Pancreatic cancer | CTRC 10 | ||
| Melanoma | DDR2, 29 GON4L, 29 TRIM63 29 | TRAF2 29 | ETV6 9 |
| Colorectal cancer | LMNA, 10 TPM3, 10 SCYL3 30 | ETV6 18 | |
| Paediatric cancers | |||
| High frequency (>80%) | |||
| Secretory breast carcinoma | ETV6 12 | ||
| Infantile fibrosarcoma and other mesenchymal tumours | SQSTM1, 31 TPM3, 41 LMNA 41 | EML4, 32 41 ETV6 34 63 | |
| Cellular and mixed congenital mesoblastic nephroma | TPR, 40 LMNA 40 | EML4,32 40 ETV633 40 | |
| Intermediate frequency (5%–25%) | |||
| Papillary thyroid cancer | TPR, 35 IRF2BP2, 10 TPM3 14 | ETV6 35 | |
| Spitz tumours | TP53, 16 LMNA 16 | ETV6, 17 MYH9, 17 MYO5A 17 | |
| Paediatric high-grade gliomas | TPM3 36 | AGBL4, 36 VCL 36 | ETV6, 36 BTB1 36 |
| Low frequency (<5%) | |||
| Ganglioglioma | TLE 38 | ||
| Astrocytoma | NACC2, 37 QK1 37 | ||
Epidemiology of TRK fusion cancer
NTRK gene fusions may occur in as many as 1% of all solid tumours.7 10 They are found in numerous tumour types in both adult and paediatric patients2 7 10 (table 1). Two main categories of tumours are identified: rare cancers with a high frequency (>80%) of NTRK gene fusions and more common cancers with a lower frequency of NTRK gene fusions (either 5%–25% or <5%; table 1). A high frequency of NTRK gene fusions have been identified in mammary analogue secretory carcinomas (90%–100%)11 and secretory breast carcinomas (>90%)12 in adult patients, and in infantile fibrosarcomas (91%–100%),34 other mesenchymal tumours (100%)41 and congenital mesoblastic nephromas (83%)42 in paediatric patients. NTRK gene fusions are found at a lower frequency in radiation-associated papillary thyroid cancer (14.5%)43 in adult patients and papillary thyroid cancer (26%)35 and Spitzoid tumours (16%)16 in paediatric or adolescent patients. The reported frequency of NTRK gene fusions in common cancer types is generally <5%, including head and neck cancer (0.2%),9 lung cancer (0.2%–3.3%),7 9 colorectal cancer (0.7%–1.5%),9 44 skin cutaneous melanoma (0.3%),9 and sarcoma (1%).9
Treatments targeting NTRK gene fusions
A number of TRK inhibitors are emerging which can be subdivided into those that are selective inhibitors for TRK and those that are multi-kinase inhibitors active against a range of targets including TRK.45 Larotrectinib is currently the only selective TRK inhibitor and was approved by the Food and Drug Administration (FDA) in November 2018.46 Data on 55 larotrectinib-treated paediatric and adult patients with TRK fusion-positive advanced solid tumours, representing 17 unique cancer types, have been evaluated.10 Objective tumour responses, based on independent radiologic review, were seen in 75% of patients.10 At 1 year, 71% of the responses were ongoing and 55% of patients remained progression-free.10 The median duration of response had not been reached after a median follow-up of 8.3 months.10 The same was true for median progression-free survival after a median follow-up of 9.9 months.10 Larotrectinib was well tolerated. Adverse events were predominantly of grade 1 and no patient discontinued larotrectinib due to drug-related adverse events.10 Furthermore, no adverse event of grade 3 or 4 that was considered by the investigators to be related to larotrectinib occurred in more than 5% of patients.10 Among infants, children and adolescents (n=24), larotrectinib was well tolerated and showed a high response rate in those with advanced, TRK fusion-positive solid tumours (n=17).47 Five of these children (median age, 2 years; range, 0.4–12 years) with locally advanced soft tissue tumours achieved a partial response to larotrectinib (RECIST v1.1) and underwent surgical resection after a median of six cycles (range, 4–9 cycles) of treatment.48 Similar findings were reported by Drilon et al10 for two children with locally advanced infantile fibrosarcoma. Larotrectinib treatment resulted in sufficient tumour shrinkage to allow for limb-sparing surgery with pathologic assessment confirming negative margins (R0 surgery). Both patients were progression-free without larotrectinib treatment after 4.8 months and 6.0 months of follow-up.
Favourable preliminary results were seen with entrectinib in two Phase I clinical trials of paediatric and adult patients with NTRK, ROS1 or ALK fusions21 leading to further investigations in patients with NTRK gene fusions. TRK inhibitors developed to overcome acquired resistance to first-generation TRK inhibitors are already in development.45 LOXO-195 (BAY 2731954) has demonstrated efficacy against treatment-resistant alleles of NTRK gene fusions in patients with TRK fusion-positive cancers.49 Repotrectinib, a TRK, ROS1 and ALK inhibitor, has demonstrated confirmed responses in patients with ROS1 or NTRK3 fusion-positive cancers who had relapsed on earlier-generation inhibitors.50
Testing methods for TRK fusion cancers
For optimal clinical efficacy of TRK inhibitors, an effective diagnostic strategy to detect NTRK gene fusions in tumour samples is essential to guide treatment selection. Approaches that may be used to directly or indirectly detect the presence of a gene fusion in clinical tissue samples51 include immunohistochemistry (IHC), fluorescence in situ hybridisation (FISH), reverse transcriptase polymerase chain reaction (RT-PCR) and next-generation sequencing (NGS) using DNA or RNA (table 2).10 18 32 34 52–58
Table 2.
Overview of testing methods currently available for NTRK gene fusions
| Assay | Advantages | Disadvantages |
| IHC | Low cost52 53
Readily available34 Detects TRKA, B and C18 Turnaround time 1–2 days53 |
May not be specific for NTRK gene fusion as it detects both wild-type and fusion proteins18
Possible false positives34 Possible false negatives for fusions involving TRKC60 There is no standardisation of scoring algorithms52 |
| FISH | The location of the target within the cell is visible54 55
Several targets can be detected in one sample using several fluorophores54 Requires knowledge of only one of the two fusion partners when using break-apart probes NTRK gene fusions with unknown partners can be detected using break-apart FISH FISH is readily available in most laboratories and institutes |
The target sequence must be known for conventional FISH otherwise three separate tests are required for NTRK1, NTRK2 and NTRK3
56
Complex chromosomal translocations can result in false positive signals56 False negative results may be above 30%63 |
| RT-PCR | High sensitivity and specificity34
Low cost per assay52 |
Target sequences must be known (i.e., cannot readily detect novel fusion partners)32 52
A comprehensive multiplex RT-PCR assay might be challenging because of the potentially large number of possible 5’ fusion partners52 57 |
| NGS | May detect novel fusion partners (depending on the assay used)32
Can be used to evaluate multiple actionable targets simultaneously while preserving limited tissue32 Currently used for NTRK testing10 RNA-NGS is preferred over DNA-NGS as sequencing for RNA-based testing is focused on coding sequences not introns56 |
Commercially available DNA-based NGS platforms may not be capable of identifying all NTRK gene fusions, especially those involving NTRK2 and NTRK3, which have large intronic regions58
DNA-NGS is limited by intron size56 RNA-NGS is limited by RNA quality56 |
Immunohistochemistry
IHC enables detection of TRK overexpression as a surrogate for the presence of an NTRK gene fusion and provides a time-efficient and tissue-efficient technique that may be used for routine screening18 (figure 2A). Studies employing pan-TRK monoclonal antibody cocktails have demonstrated positive TRK expression in tumour samples.18 59–61 However, some studies indicate that interpretation of IHC data may be more challenging than initially ascertained.62 In an analysis of 11,502 formalin-fixed paraffin-embedded (FFPE) tumour samples of various cancer types for the presence of gene fusions, 31 cases (0.27%) with NTRK gene fusions were identified62 by NGS. Of the 28 cases that were assessed by pan-TRK IHC, 21 scored positive (≥1% of tumour cells staining at any intensity above background), giving a sensitivity of only 75%, and 45% of tumours with NTRK3 fusions scored negative by IHC. False negative cases could be related to sample preparation, for example, fixation. Therefore, it is important to check if internal controls such as endothelial cells are positive, or to use external controls such as positive cell lines. Similarly, positive IHC results must be followed with confirmatory testing using a molecular method to verify the presence of a fusion, as overexpression of wildtype TRK proteins may also be detected.
Figure 2.
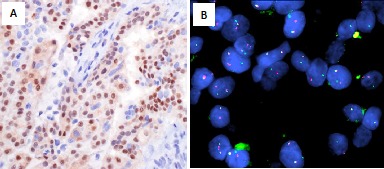
Secretory carcinoma of the breast aka juvenile carcinoma: low-grade basal tumour. (A) Immunohistochemistry. Nuclear staining of TRK detected by pan-TRK IHC. (B) FISH. t(12:15) ETV6-NTRK3 fusion using an ETV6 break-apart probe. Due to the prevalence of ETV6-NTRK3 gene fusions, an ETV6 break-apart probe is typically used. FISH image provided by courtesy of Dr Hanina Hibshoosh, Columbia University. FISH, fluorescence in situ hybridisation; IHC, immunohistochemistry; TRK, tropomyosin receptor kinase.
Fluorescence in situ hybridisation
Break-apart FISH is a well-established method for detecting clinically relevant gene fusion events52 and is of value in tumours with a high prevalence of NTRK gene fusions involving recurrent fusions24 (figure 2B). The ETV6-NTRK3 gene fusion was one of the first NTRK gene fusions reported and has been identified in numerous cancer types:7 it is amenable for detection using break-apart FISH (figure 2B). As FISH is largely limited to the detection of a single gene fusion, a separate break-apart FISH probe is required for each of the three NTRK genes.56 Furthermore, the 5’ gene fusion partner will not be identified.56 False negatives may result if the deletion is small enough to leave enough of the complementary regions for hybridisation of both FISH probes or if there is a complex FISH pattern with numerous nuclei showing atypical doublet fusion signals and only a few nuclei with split signals.56 Indeed, in one study ETV6 FISH was associated with a 36% false negative rate.63
Reverse transcriptase polymerase chain reaction
RT-PCR provides an alternative or complementary approach to FISH, detecting NTRK gene fusions using primers in the coding sequence of the 5’ fusion partner and the NTRK kinase domain.56 57 A disadvantage of RT-PCR is that the large number of possible 5’ fusion partners may make a comprehensive multiplex RT-PCR assay challenging.57 An alternative approach could be to assess the ratio of 5’ and 3’ amplicons of each of the NTRK genes by multiplex RT-PCR reactions, with an imbalance in the ratio for a specific gene suggesting a possible fusion event.57
Next-generation sequencing
NGS provides a precise method to detect NTRK gene fusions, with high sensitivity and specificity compared with other testing methods.57 An advantage of NGS is that multiple oncogenic events in addition to NTRK gene fusions can be identified from a single tumour sample.57 A wide variety of NGS-based approaches are available for fusion testing with the primary distinguishing factor being whether they are RNA- or DNA-based.56 Access to NGS in a clinical setting may be limited as availability of this technique varies between regions and countries.
DNA-based next-generation sequencing
Although DNA-based NGS panels may detect multiple oncogenic genomic events from one sample, not all DNA-based NGS platforms can identify all NTRK gene fusions, especially those involving NTRK2 and NTRK3 where detection of gene fusions is complicated by the presence of large introns that are typically inadequately sequenced and difficult to analyse56 58 (figure 1B).
RNA-based next-generation sequencing
The advantage of RNA-based NGS over DNA-based NGS is that sequencing is focused on the mature mRNA hence is not affected by intron size.56 A disadvantage is the high reliance on RNA quality, which can be poor if obtained from FFPE samples.56 Many NGS assays now include RNA fusions in their gene panels, and it is likely that NGS diagnostics that depend on RNA for fusion detection will increasingly be used in clinical practice to test for NTRK gene fusions.
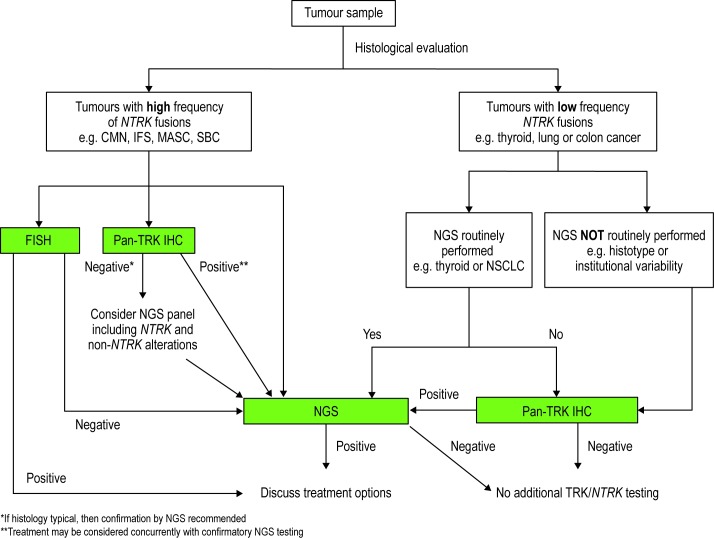
NTRK gene fusion testing algorithm
A proposed screening algorithm for identifying patients with TRK fusion cancer is presented (figure 3). The algorithm incorporates the strengths and availability of each diagnostic technique. The algorithm is based on the categorisation of tumours into two groups based on the incidence of NTRK gene fusion.
Figure 3.
Testing algorithm for TRK fusion cancer. CMN, congenital mesoblastic nephroma; FISH, fluorescence in situ hybridisation; IFS, infantile fibrosarcoma; IHC, immunohistochemistry; MASC, mammary analogue secretory carcinoma; NGS, next-generation sequencing; NSCLC, non-small cell lung cancer; NTRK, neurotrophic tyrosine receptor kinase; SBC, secretory breast carcinoma; TRK, tropomyosin receptor kinase.
In tumours with a high frequency of NTRK gene fusion events, FISH is recommended, with pan-TRK IHC as an alternative if FISH is unavailable. Confirmation by targeted NGS in those cases with positive pan-TRK IHC can be conducted concurrently with treatment considerations. The pattern of TRK staining by IHC may also inform selection of a confirmatory test, as tumours harbouring NTRK1 rearrangements typically show strong, diffuse cytoplasmic staining. In contrast, tumours harbouring NTRK3 rearrangements may have weaker expression but often have at least focal nuclear staining. Negative results from FISH or pan-TRK IHC should be confirmed by NGS, although selection of a broader panel including other receptor tyrosine kinases is warranted as these tumours have a high likelihood of harbouring other diagnostic and/or therapeutic alterations.
In solid tumours where gene fusions are common, but the frequency of NTRK gene fusions is lower (5%–25%), an NGS panel that includes NTRK fusions is recommended as the preferred test for patients. For tumours with a very low frequency of NTRK gene fusions (<5%), but where molecular screening is common, inclusion of NTRK genes in routine NGS analysis is recommended. For tumours with a low frequency of NTRK fusions, where NGS is not available or is not routinely performed for a histotype, pan-TRK IHC should be performed for screening with NGS confirmation of positive IHC results.
In all cases where NGS is recommended, and particularly for those cases in which an NTRK3 rearrangement is favoured by IHC, RNA-based NGS is the ideal testing assay for NTRK gene fusions. Note that this algorithm is not intended to replace the independent medical judgement of the physician in the context of individual clinical circumstances to determine a patient's care.
Conclusions and future directions
NTRK gene fusions have been identified across a range of tumour types and occur at a high frequency in certain rare cancers.2 7 9 20 34 36 42 More common cancers have a low but significant frequency of NTRK gene fusions2 7 9 20 34 36 42 and thus represent a sizeable at-risk patient population. With the recent FDA approval of the selective TRK inhibitor, larotrectinib (Vitrakvi), along with the continued development of multi-kinase inhibitors with activity in TRK fusion cancer, testing for NTRK gene fusions should become part of the standard diagnostic process. Marked differences in the prevalence of NTRK gene fusions across tumour types mean that clinical diagnostic strategies will vary accordingly but will rely on IHC, FISH and NGS assays. The suggested testing algorithm for TRK fusion cancer considers the aetiology of tumours as well as the availability of testing methods to guide detection of these fusions in the clinic. The optimal use of tumour tissue, especially from small biopsies or cytology specimens, and optimisation of multiplexed approaches, remains an area of active research and development.
Take home messages.
The NTRK genes (NTRK1, NTRK2 and NTRK3) encode for TRKA, TRKB and TRKC receptors, three transmembrane proteins, and are normally expressed in neuronal tissue during development.
Fusions involving NTRK genes are oncogenic drivers across a wide range of tumour types and are either highly enriched in select tumour types or infrequently found in other cancers, including common tumours.
NTRK gene fusions should be treated as tumour-agnostic biomarkers.
Specific TRK inhibitors have shown histology-agnostic activity in adult and paediatric patients harbouring NTRK gene fusions providing high durable response rates with a low incidence of adverse events.
IHC, FISH, RT-PCR and NGS are effective screening techniques for identification of TRK fusion cancer. Implementation of these methods can be tailored to individual patients based on histological and clinical presentation.
Acknowledgments
The Submitting Author accepts and understands that any supply made under these terms is made by BMJ to the Submitting Author unless you are acting as an employee on behalf of your employer or a postgraduate student of an affiliated institution that is paying any applicable article publishing charge ('APC') for Open Access articles. Where the Submitting Author wishes to make the Work available on an Open Access basis (and intends to pay the relevant APC), the terms of reuse of such Open Access shall be governed by a Creative Commons licence – details of these licences and which Creative Commons licence will apply to this Work are set out in our licence referred to below.
Footnotes
Handling editor: Newton ACS Wong.
Contributors: All authors contributed equally to the preparation, drafting and reviewing of the manuscript. Medical writing support, including assisting authors with the development of the outline and initial draft, incorporation of comments and preparation of tables and figures, was provided by Penny Butcher, PhD and Alison Scott, PhD; editorial support, including fact-checking, referencing, figure preparation, formatting, proofreading and submission was provided by Annabel Ola, MSc, all of Scion (London, UK), supported by Bayer Healthcare Pharmaceuticals and Loxo Oncology according to Good Publication Practice guidelines (http://annals.org/aim/article/2424869/good-publication-practice-communicating-company-sponsored-medical-research-gpp3).
Competing interests: FP-L has participated in advisory boards for Bayer, Roche, Illumina and Nanostring, and been involved in studies sponsored by Bayer. ERR has had a role as an expert consultant, participated in a meeting and participated in an advisory board for Bayer Healthcare Pharmaceuticals. ARS has had a role as an expert consultant for Merck US, Bristol Meyers Squibb and Bayer Healthcare Pharmaceuticals; participated in meetings for Merck US, Bristol Meyers Squibb and Bayer Healthcare Pharmaceuticals; participated in advisory boards for Merck US, Bristol Meyers Squibb and Bayer Healthcare Pharmaceuticals; and received honoraria from Amgen.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Pulciani S, Santos E, Lauver AV, et al. Oncogenes in solid human tumours. Nature 1982;300:539–42. 10.1038/300539a0 [DOI] [PubMed] [Google Scholar]
- 2. Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 2016;1:e000023 10.1136/esmoopen-2015-000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hugo Gene Nomenclature Committee (HGNC). Symbol report: NTRK1. Available: https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:8031 [Accessed 12 Sep 2018].
- 4. Hugo Gene Nomenclature Committee (HGNC). Symbol report: NTRK2. Available: https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:8032 [Accessed 12 Sep 2018].
- 5. Hugo Gene Nomenclature Committee (HGNC). Symbol report: NTRK3. Available: https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:8033 [Accessed 12 Sep 2018].
- 6. Klein R, Jing SQ, Nanduri V, et al. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell 1991;65:189–97. 10.1016/0092-8674(91)90419-Y [DOI] [PubMed] [Google Scholar]
- 7. Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov 2015;5:25–34. 10.1158/2159-8290.CD-14-0765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rubin JB, Segal RA. Growth, survival and migration: the Trk to cancer. Cancer Treat Res 2003;115:1–18. [DOI] [PubMed] [Google Scholar]
- 9. Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846 10.1038/ncomms5846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 2018;378:731–9. 10.1056/NEJMoa1714448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skálová A, Vanecek T, Sima R, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol 2010;34:599–608. 10.1097/PAS.0b013e3181d9efcc [DOI] [PubMed] [Google Scholar]
- 12. Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2002;2:367–76. 10.1016/S1535-6108(02)00180-0 [DOI] [PubMed] [Google Scholar]
- 13. Greco A, Mariani C, Miranda C, et al. The DNA rearrangement that generates the TRK-T3 oncogene involves a novel gene on chromosome 3 whose product has a potential coiled-coil domain. Mol Cell Biol 1995;15:6118–27. 10.1128/MCB.15.11.6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ronsley R, Rassekh SR, Shen Y, et al. Application of genomics to identify therapeutic targets in recurrent pediatric papillary thyroid carcinoma. Cold Spring Harb Mol Case Stud 2018;4:pii:a002568 10.1101/mcs.a002568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi E, Chmielecki J, Tang C-M, et al. FGFR1 and NTRK3 actionable alterations in “Wild-Type” gastrointestinal stromal tumors. J Transl Med 2016;14:339 10.1186/s12967-016-1075-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiesner T, He J, Yelensky R, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun 2014;5:3116 10.1038/ncomms4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yeh I, Tee MK, Botton T, et al. NTRK3 kinase fusions in Spitz tumours. J Pathol 2016;240:282–90. 10.1002/path.4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hechtman JF, Benayed R, Hyman DM, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol 2017;41:1547–51. 10.1097/PAS.0000000000000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479–84. 10.1038/nm.3729 [DOI] [PubMed] [Google Scholar]
- 20. Kim J, Lee Y, Cho HJ, et al. NTRK1 fusion in glioblastoma multiforme. PLoS One 2014;9:e91940 10.1371/journal.pone.0091940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drilon A, Siena S, Ou S-HI, et al. Safety and antitumor activity of the multitargeted pan-Trk, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov 2017;7:400–9. 10.1158/2159-8290.CD-16-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshihara K, Wang Q, Torres-Garcia W, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 2015;34:4845–54. 10.1038/onc.2014.406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hartmaier RJ, Albacker LA, Chmielecki J, et al. High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res 2017;77:2464–75. 10.1158/0008-5472.CAN-16-2479 [DOI] [PubMed] [Google Scholar]
- 24. Farago AF, Le LP, Zheng Z, et al. Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol 2015;10:1670–4. 10.1097/01.JTO.0000473485.38553.f0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ross J, Chung J, Elvin J, et al. NTRK fusions in breast cancer: clinical, pathologic and genomic findings. Cancer Res 2018;78:P2-09–15. [Google Scholar]
- 26. Eguchi M, Eguchi-Ishimae M, Tojo A, et al. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25). Blood 1999;93:1355–63. [PubMed] [Google Scholar]
- 27. Chiang S, Cotzia P, Hyman DM, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol 2018;42:791–8. 10.1097/PAS.0000000000001055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ross JS, Wang K, Gay L, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist 2014;19:235–42. 10.1634/theoncologist.2013-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lezcano C, Shoushtari AN, Ariyan C, et al. Primary and metastatic melanoma with NTRK fusions. Am J Surg Pathol 2018;42:1052–8. 10.1097/PAS.0000000000001070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Milione M, Ardini E, Christiansen J, et al. Identification and characterization of a novel SCYL3-NTRK1 rearrangement in a colorectal cancer patient. Oncotarget 2017;8:55353–60. 10.18632/oncotarget.19512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chmielecki J, Bailey M, He J, et al. Genomic profiling of a large set of diverse pediatric cancers identifies known and novel mutations across tumor spectra. Cancer Res 2017;77:509–19. 10.1158/0008-5472.CAN-16-1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Church AJ, Calicchio ML, Nardi V, et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod Pathol 2018;31:463–73. 10.1038/modpathol.2017.127 [DOI] [PubMed] [Google Scholar]
- 33. Knezevich SR, Garnett MJ, Pysher TJ, et al. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res 1998;58:5046–8. [PubMed] [Google Scholar]
- 34. Bourgeois JM, Knezevich SR, Mathers JA, et al. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol 2000;24:937–46. 10.1097/00000478-200007000-00005 [DOI] [PubMed] [Google Scholar]
- 35. Prasad ML, Vyas M, Horne MJ, et al. NTRK fusion oncogenes in pediatric papillary thyroid carcinoma in northeast United States. Cancer 2016;122:1097–107. 10.1002/cncr.29887 [DOI] [PubMed] [Google Scholar]
- 36. Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 2014;46:444–50. 10.1038/ng.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones DTW, Hutter B, Jäger N, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 2013;45:927–32. 10.1038/ng.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prabhakaran N, Guzman MA, Navalkele P, et al. Novel TLE4-NTRK2 fusion in a ganglioglioma identified by array-CGH and confirmed by NGS: potential for a gene targeted therapy. Neuropathology 2018;38:380–6. 10.1111/neup.12458 [DOI] [PubMed] [Google Scholar]
- 39. Gross S, Rahal R, Stransky N, et al. Targeting cancer with kinase inhibitors. J Clin Invest 2015;125:1780–9. 10.1172/JCI76094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wegert J, Vokuhl C, Collord G, et al. Recurrent intragenic rearrangements of EGFR and BRAF in soft tissue tumors of infants. Nat Commun 2018;9:2378 10.1038/s41467-018-04650-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davis JL, Lockwood CM, Albert CM, et al. Infantile NTRK-associated mesenchymal tumors. Pediatr Dev Pathol 2018;21:68–78. 10.1177/1093526617712639 [DOI] [PubMed] [Google Scholar]
- 42. Rubin BP, Chen CJ, Morgan TW, et al. Congenital mesoblastic nephroma t(12;15) is associated with ETV6-NTRK3 gene fusion: cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am J Pathol 1998;153:1451–8. 10.1016/S0002-9440(10)65732-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leeman-Neill RJ, Kelly LM, Liu P, et al. ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer 2014;120:799–807. 10.1002/cncr.28484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ardini E, Bosotti R, Borgia AL, et al. The TPM3-NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TrkA kinase inhibition. Mol Oncol 2014;8:1495–507. 10.1016/j.molonc.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kummar S, Lassen UN. Trk inhibition: a new tumor-agnostic treatment strategy. Target Oncol 2018;13:545–56. 10.1007/s11523-018-0590-1 [DOI] [PubMed] [Google Scholar]
- 46. US Food and Drug Adminstration Vitrakvi, 2018. Available: https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=210861 [Accessed 19 Dec 2018].
- 47. Laetsch TW, DuBois SG, Mascarenhas L, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol 2018;19:705–14. 10.1016/S1470-2045(18)30119-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DuBois SG, Laetsch TW, Federman N, et al. The use of neoadjuvant larotrectinib in the management of children with locally advanced TRK fusion sarcomas. Cancer 2018;124:4241–7. 10.1002/cncr.31701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drilon A, Nagasubramanian R, Blake JF, et al. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov 2017;7:963–72. 10.1158/2159-8290.CD-17-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drilon A, Ou S-HI, Cho BC, et al. Repotrectinib (TPX-0005) is a next-generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent-front mutations. Cancer Discov 2018;8:1227–36. 10.1158/2159-8290.CD-18-0484 [DOI] [PubMed] [Google Scholar]
- 51. Pal P, Khan Z. ROS1 [corrected]. J Clin Pathol 2017;70:1001–9. 10.1136/jclinpath-2016-204244 [DOI] [PubMed] [Google Scholar]
- 52. Bubendorf L, Büttner R, Al-Dayel F, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch 2016;469:489–503. 10.1007/s00428-016-2000-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doshi S, Ray D, Stein K, et al. Economic analysis of alternative strategies for detection of ALK rearrangements in non small cell lung cancer. Diagnostics 2016;6:pii:E4. 10.3390/diagnostics6010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kerr KM, López-Ríos F. Precision medicine in NSCLC and pathology: how does ALK fit in the pathway? Ann Oncol 2016;27:iii16–24. 10.1093/annonc/mdw302 [DOI] [PubMed] [Google Scholar]
- 55. Cui C, Shu W, Li P. Fluorescence in situ hybridization: cell-based genetic diagnostic and research applications. Front Cell Dev Biol 2016;4:89 10.3389/fcell.2016.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Davies KD, Le AT, Sheren J, et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J Thorac Oncol 2018;13:1474–82. 10.1016/j.jtho.2018.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beadling C, Wald AI, Warrick A, et al. A multiplexed amplicon approach for detecting gene fusions by next-generation sequencing. J Mol Diagn 2016;18:165–75. 10.1016/j.jmoldx.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 58. Sheikine Y, Kuo FC, Lindeman NI. Clinical and technical aspects of genomic diagnostics for precision oncology. J Clin Oncol 2017;35:929–33. 10.1200/JCO.2016.70.7539 [DOI] [PubMed] [Google Scholar]
- 59. Murphy DA, Ely HA, Shoemaker R, et al. Detecting gene rearrangements in patient populations through a 2-step diagnostic test comprised of rapid IHC enrichment followed by sensitive next-generation sequencing. Appl Immunohistochem Mol Morphol 2017;25:513–23. 10.1097/PAI.0000000000000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hung YP, Fletcher CDM, Hornick JL. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology 2018;73:634–44. 10.1111/his.13666 [DOI] [PubMed] [Google Scholar]
- 61. Rudzinski ER, Lockwood CM, Stohr BA, et al. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am J Surg Pathol 2018;42:927–35. 10.1097/PAS.0000000000001062 [DOI] [PubMed] [Google Scholar]
- 62. Gatalica Z, Xiu J, Swensen J, et al. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol 2019;32:147–53. 10.1038/s41379-018-0118-3 [DOI] [PubMed] [Google Scholar]
- 63. Davis JL, Lockwood CM, Stohr B, et al. Expanding the spectrum of pediatric NTRK-rearranged mesenchymal tumors. Am J Surg Pathol 2019;43:435–45. 10.1097/PAS.0000000000001203 [DOI] [PubMed] [Google Scholar]