Abstract
The intergenic spacer (IGS) of rDNA is frequently built of long blocks of tandem repeats. To estimate the intragenomic variability of such knotty regions, we employed PacBio sequencing of the Cucurbita moschata genome, in which thousands of rDNA copies are distributed across a number of loci. The rRNA coding regions are highly conserved, indicating intensive interlocus homogenization and/or high selection pressure. However, the IGS exhibits high intragenomic structural diversity. Two repeated blocks, R1 (300–1250 bp) and R2 (290–643 bp), account for most of the IGS variation. They exhibit minisatellite-like features built of multiple periodically spaced short GC-rich sequence motifs with the potential to adopt non-canonical DNA conformations, G-quadruplex-folded and left-handed Z-DNA. The mutual arrangement of these motifs can be used to classify IGS variants into five structural families. Subtle polymorphisms exist within each family due to a variable number of repeats, suggesting the coexistence of an enormous number of IGS variants. The substantial length and structural heterogeneity of IGS minisatellites suggests that the tempo of their divergence exceeds the tempo of the homogenization of rDNA arrays. As frequently occurring among plants, we hypothesize that their instability may influence transcription regulation and/or destabilize rDNA units, possibly spreading them across the genome.
Keywords: Cucurbita moschata, ribosomal DNA intergenic spacer, DNA-minisatellite, intragenomic structural heterogeneity, non-canonical DNA conformations
1. Introduction
In eukaryotes, the 35S nuclear ribosomal DNA (rDNA) units are organized in tandem repeats located at one or more chromosomal loci, the so-called nucleolar organizing region (NOR).1 The rDNA unit is composed of the coding regions for the 18S, 5.8S and 26S ribosomal RNA (rRNA); the internal transcribed spacers (ITS1 and ITS2); and the intergenic spacer (IGS). The IGS, located between the 3’ end of the 26S rRNA gene and the 5’ end of the 18S rRNA gene, comprises the 3′ external transcribed spacer (3′-ETS), the non-transcribed spacer, and the 5’-ETS). In general, the IGS is built of multiple blocks of tandem and dispersed repeats that may be arranged in highly complex patterns, resulting in variations between related species, populations and even within an individual.2 The presence of conserved structural features in the IGS, such as repeating elements, sequences with self-complementarity having the potential to generate a conserved secondary structure, the transcription initiation site (TIS) and the transcription termination site, all led to the recognition of the IGS as a functionally important region.1
The presence of highly variable repeats in the IGS of most higher eukaryotes suggests that these fulfil an important function.1 Their function as possible enhancers of RNA polymerase I (Pol I) transcription has been studied predominantly in interspecific hybrids.3 The rDNA loci with a longer IGS, containing more homologous repeats upstream of TIS, transcriptionally dominate over those with a shorter IGS in Triticum-Aegilops,4Triticum-Secale5 and Tragopon mirus2 but not in Solanum,6Brassica7 and Arabidopsis.8 Repeats from Arabidopsis thaliana can, however, substitute for Xenopus repeats to enhance Pol I transcription.9 Additionally, repeats within the 5′-ETS constitute a recognition site for DNA-interacting proteins, which may stimulate the transcription of rDNA.10 In Tragopogon,2Solanum11 and Vigna radiate,1 transcriptionally dominant rDNA variants harbour more repeats in the 5′-ETS. Repeats localized upstream of the TIS may contain multiple termination signals for Pol I.12 In many species, individual rDNA repeats do not evolve independently but in a concerted manner due to the process of sequence homogenization, resulting in the co-existence of numerous nearly identical units in the same genome.13 However, accumulating data suggest that multiple rDNA repeat units with different sequence similarities and lengths, mainly due to variable repeats in IGS, can be simultaneously present in the same genome.2,14 The origin of such diversity has not been explained. Nevertheless, only a subset of rRNA genes with distinct structures is transcriptionally active, a phenomenon known as nucleolar dominance in hybrids.7 The interplay of DNA methylation, histone modification, and chromatin remodelling activities guided by ncRNA derived from variable IGSs is required for the specific establishment of chromatin organization in silent 35S rDNA in animals15 and plants.16
Cucurbita pepo, Cucurbita moschata, and Cucurbita maxima are the most economically important cultivated species within Cucurbita with relatively small genome sizes 0.55, 0.43 and 0.46 pg/1C, respectively (Kew Angiosperm DNA C-values database). Isozyme assays and high (relative to other Cucurbitaceae) chromosome numbers (n = 20–24) have led to the suggestion that the genus is of allopolyploid origin.17 In C. moschata (2n = 40), the 45S rDNA occupies large parts of five chromosome pairs.17 However, sequencing data for IGS are currently not available for this species, and only a single IGS has been completely sequenced in each related species C. pepo and C. maxima, showing that duplicated promoter-like sequences with TIS for Pol I are separated by multiple tandem repeats,18 some of which may function in enhancing transcription, as shown for the related species Cucumis sativus.10 In C. pepo, restriction polymorphism showed that 3,400 rDNA units fall into seven classes that are distinct in length and/or nucleotide sequence.19 Therefore, nothing is currently known about either the structure of IGS in C. moschata or the distribution of 35S rDNA loci in C. pepo and C. maxima chromosomal complements.
To elucidate the regulation of the cellular rRNA level, we initiated a study of plant IGSs, particularly the regions of initiation and termination of transcription. To estimate the intragenomic structural variability of the IGS, we leveraged the advantage of long reads provided by PacBio sequencing methodology, which is relatively insensitive to highly structured, GC-rich and repetitive, DNA, characteristic of IGS. As a model plant, we selected C. moschata, a species with rDNA units distributed among multiple rDNA loci, suggesting high structural variability. We focussed on two extremely GC-rich repetitive regions located upstream of genic and spacer promoter-like sequences and evaluated their heterogeneity in the content and the distribution of sequence motifs with the potential to adopt non-canonical DNA structures, which are believed to participate in transcription regulation. We further evaluated the frequency of single nucleotide polymorphisms (SNPs) and length polymorphisms caused by short insertions, deletions or variable numbers of subrepeats along the entire rDNA unit, with a focus on comparing genic regions with an IGS and ITS.
2. Materials and methods
2.1. Plant material, DNA extraction, and Southern and slot blot hybridizations
Musquee de Provence (Muscat de Provence) cultivar of C. moschata Duch. ex Poir. and Rouge vif d’Etampes cultivar of C. pepo L. were grown in a garden. Fresh young leaves were ground in liquid nitrogen to a fine powder, which was mixed with 2.5 volumes (v/w) of extraction buffer [1.1 M NaCl, 1.4% cetyltrimethylammonium bromide, 71 mM Tris-HCl (pH 8.0), 14 mM ethylenediaminetetraacetic acid (EDTA), and 0.1% 2-mercaptoethanol] and incubated for 60 min at 60°C with occasional gentle mixing. The extract was gently mixed with an equal volume of chloroform:octanol (24:1). After centrifugation [13,000 × g, 20 min, room temperature (RT)], the DNA was precipitated with 2/3 volume of isopropanol, washed with 75% ethanol/10 mM NH4OAc several times for 6 h, dissolved in TE, treated with RNase A (100 µg.ml−1, SIGMA) for 60 min at 37°C and protease K (200 µg. ml−1, SIGMA) for 20 h at 52°C, extracted with phenol-chloroform and precipitated with ethanol. The quantity and quality of DNA were checked by spectrophotometry and gel electrophoresis. For Southern analysis, genomic DNA (2 µg) was digested with restriction endonucleases (10 U; 2 × 4 h), separated by gel electrophoresis and blotted onto nylon membranes (GE Healthcare). The hybridization was carried out in modified Church–Gilbert buffer.20 The 26S rDNA probe was a 220-bp PCR product derived from the 3′ end of the 26S rRNA gene.21 This PCR product was also used as a standard for slot blot analysis. The IGS rDNA probes, covering the putative spacer promoter and R2 region, represent the PCR product amplified from GenBank accession MG744572 (cl7) by primers CF and CR (see PCR amplifications) and digested with NdeI. The hybridization signals were visualized by PhosphorImaging (Typhon; GE Healthcare) and quantitatively analysed by ImageQuant (GE Healthcare).
2.2. Fractionalization of genomic DNA and PacBio sequencing
To enhance the content of rDNA in the sample, the rDNA was separated from appropriately restricted bulk genomic DNA by preparative electrophoresis. One milligram of DNA was digested with EcoRV (3 × 8 h; 3 × 1,000 U), and the completeness of digestion was checked by Southern blot hybridization against 26S rDNA (Supplementary Fig. S1A). The digested DNA (300 µg) was size-fractionated by electrophoresis in a 0.5% preparative agarose gel using Bio-Rad Model 491 Prep-Cell (Serva). When the xylene cyanol dye marker reached the bottom of the gel, individual fractions (1.3 ml/4 min) were collected under electrophoretic conditions of 70 V/5.5 h (fractions 1–83), followed by 100 V (fractions 84–174). Each fraction was lyophilized and dissolved in 100 µl TE + 1 mM DTT, and 2-µl aliquots were analysed by Southern hybridization to the 26S rDNA probe. The fractions with the highest ratio between the hybridization signal and total input DNA (Supplementary Fig. S1B; framed) were collected, desalted with NucAwayTM Spin Columns (Ambion), lyophilized and checked for DNA quantity and quality (Supplementary Fig. S1C).
The PacBio RS library insert length was 2.1 kb on average (Supplementary Fig. S2A). During sequencing, the library inserts were passed by the polymerase ∼12 times on average (Supplementary Fig. S2B), thus creating a mean read quality of the insert sequences of 99.61% (Supplementary Fig. S2C). Using CLC bio (http://cgs.hku.hk/portal/index.php/software-for-next-generation-sequencing), reads of inserts (ROIs) were mapped to a reference sequence assembled from GenBank accessions for the C. pepo 26S rRNA gene (AF479108.1), IGS (X55960.1), 18S rRNA gene (AF206895.1) and ITS1-5.8S_rDNA_gene–ITS2 region (consensus sequence derived from AM981169.1; AM981168.1; AJ488214.1; KF835490.1; FJ915107.2; FJ915103.2; FJ915102.2 and EF595858.1) (Supplementary Fig. S3). The reference sequence was terminally delimited by a unique EcoRV restriction site located within the 5.8S rRNA gene. The mean insert length of the mapped ROIs was ∼2.1 kb (Supplementary Fig. S2D). More than 100 ROIs, covering predominantly repetitive regions in the IGS, were difficult to map and were assigned manually based on dot-plot figures specific for pumpkin IGS. We therefore extracted the consensus sequence derived from all successfully mapped ROIs, and all available ROIs were again mapped to this consensus. When compared with the initial mapping, additional ROIs were mapped, particularly those homologous to IGS repeat. After the second mapping, we extracted the resulting consensus sequence (Supplementary Fig. S4) and evaluated the frequency and distributions of abundant (>10%) SNPs.
The read quality of each ROI, harbouring the R2 repeat, was estimated from the number of subreads. Only those subreads spanning the entire R2 region were considered for calculations (Supplementary Fig. S2E). To evaluate the capacity of the PacBio to determine the correct length of GC-rich and highly repeated regions, the lengths of the longest R1 and R2 consensus sequences, determined by dot-plot analyses, were compared with the lengths in each corresponding subread (Supplementary Fig. S5A and B).
2.3. Sequence analysis
The copy numbers and lengths of monomeric units as well as the overall lengths of repetitive regions were determined by the Tandem Repeats Finder22 (https://tandem.bu.edu/trf/trf.html). The lengths of repetitive regions were also estimated from (i) the length of the DNA with an uninterrupted GC content higher than 75% determined by the bend.it® server (http://pongor.itk.ppke.hu/dna/bend_it.html#/bendit_intro) (window size 31) and (ii) dot plot diagrams constructed by the YASS: genomic similarity search tool (http://bioinfo.lifl.fr/yass/yass.php). This approach was also used to determine duplicated promoter-like regions delimited by the conserved boundary sequences CCATCACCCATT and TGGGCATATGCTTGG. The size variability at repetitive regions was also evaluated from distances between the conserved boundary unique sequences GGAGGYTAACC and TACCAACA for R1 or TGGGCATATGCTTGG and CCATCACCCATT for R2. The variability in the distances between duplicated TIS sequences ATATAGGGGG was estimated as well.
The potential to form G-quadruplexes (pG4) was evaluated with QGRS Mapper23 (http://bioinformatics.ramapo.edu/QGRS/analyze.php). Because the sequence propensity for forming Z-DNA was on the order of d(GC)n > d(CA)n > d(CGGG)n > d(AT)n,24 we separately evaluated the frequency and distribution of (CG)n repeats longer than 7 nt, more complex (PuPy)n, or CG3 motifs. The frequency and distribution of short dispersed repeated motifs were evaluated manually.
Phylogenetic relationships were constructed from the SNP distribution among the promoter-like sequence variants linked to the R1 and/or R2 regions in the corresponding ROIs. The sequences of genic and spacer promoters were aligned altogether using the MUSCLE programme, and the phylogenetic tree was constructed with the MEGA 7 programme. The evolutionary history was inferred using a maximum likelihood method based on the Tamura-Nei model.25 The confidence of inferred evolutionary relationships was assessed by bootstrap analysis with 500 repetitions.
For comparative statistics, the Mann-Whitney (http://vassarstats.net/utest.html) non-parametric U-test and Student’s parametric t-test (Microsoft Excel) were used.
2.4. Fluorescence in situ hybridization
Chromosomes from squash root tips were prepared according to previous methods,26 except for enzymatic treatment, which lasted 30–45 min. Squashes were treated with RNase A (100 µg/ml) in a humidified chamber (37°C/1 h), rinsed in 2× standard saline citrate (SSC) at RT for 3 × 5 min, treated with pepsin (50 µg/ml) at RT for 5–7 min, rinsed in 2× SSC (3 × 5 min), fixed in 3.7% formaldehyde/phosphate-buffered saline (PBS) for 10 min, rinsed in 2× SSC (3 × 5 min), dehydrated in an ethanol series (50, 70 and 100%) for 2 min each, and air-dried. The 18S probe (GenBank X51576.1) was labelled with SpectrumGreen dUTP using a Nick Translation Kit (Abbott Molecular, IL, USA). The 5S probe (GenBank JX101915.1) was labelled with Amersham FluoroLinkTM Cy3-dUTP (GE Healthcare, Chalfont, St Giles, England) using the Nick Translation Mix from Roche. The hybridization mix (40% formamide, 50% dextran sulphate, 2× SSC and 100–150 ng of each probe in a 30 µl volume) was heated (10 min/75°C), cooled on ice, pipetted onto a slide and covered with a plastic coverslip. The slide was incubated in a humidified chamber (75°C/5 min, 65°C/2 min, 55°C/2 min, 45°C/2 min and 37°C/17 h) and washed in 2× SSC (42°C/2 × 5 min), 0.1× SSC (42°C/2 × 5 min), 2× SSC (42°C/2 × 5 min), 2× SSC (RT/5 min) and 4× SSC/0.1% Tween 20 (RT/7 min) with a final brief wash in PBS. The chromosomes were counterstained in Vectashield (Vector Laboratories, Inc., Burlingame, CA, USA) containing 1.5 µg/ml 4’, 6-diamidino-2’-phenylindole (DAPI). Fluorescence images were captured using an Olympus AX 70 fluorescence microscope equipped with a digital camera. Images were analysed and processed using ISIS software (MetaSystems, Altlussheim, Germany).
2.5. PCR amplifications, cloning and sequencing
For PCR amplification of R2 repetitive regions, primers CF (5′-ACTTGAAAGAATGACGCCGGT-3′) and CR (5′-CAAGAAAACAACAACTCCACATGTAA-3′) (Supplementary Fig. S4) were designed. The PCR mixture was composed of 0.4 µM each primer, 0.5 ng/µl of template genomic DNA, 0.2 mM of each deoxyribonucleotide triphosphate, 1× KAPA Taq Buffer (2.5 mM MgCl2), 5% DMSO and 0.02 U/µl KAPA Taq DNA Polymerase (KAPABIOSYSTEMS). The cycling conditions were as follows: initial denaturation (95°C/180 s), followed by 35 cycles (95°C/30 s, 57°C/30 s, 72°C/60 s) and then 72°C/5 min. The PCR products were checked by agarose-gel electrophoresis, ligated into the pDrive Cloning Vector (QIAGEN) and sequenced by Sanger methodology.
2.6. Data availability
The PacBio FASTQ data generated from this study were submitted to NCBI under the BioProject ID PRJNA400686 (http://www.ncbi.nlm.nih.gov/bioproject/400686). The data derived from Sanger sequencing have been submitted to GenBank under accessions MG744571-MG744575.
3. Results
3.1. Considerably high numbers of rDNA units distributed across multiple chromosomal loci suggest the coexistence of rDNA structural variants in the pumpkin genome
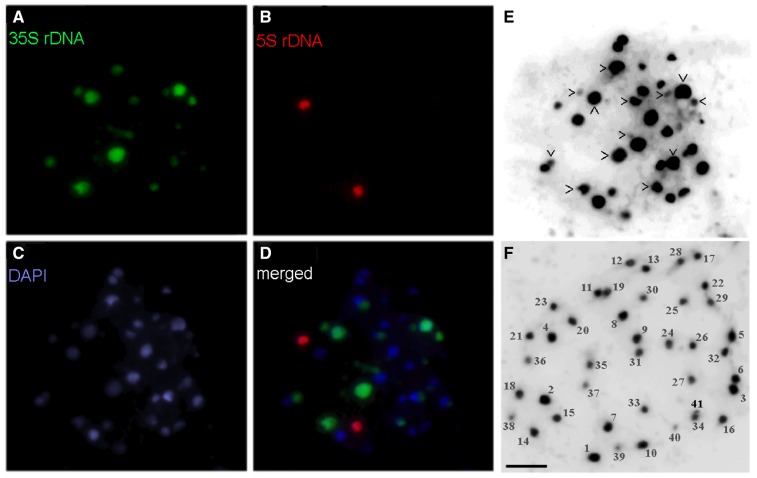
Digestion of C. moschata genomic DNA with EcoRV resulted in almost completely restricted rDNA into a population of ∼10 kb long units that migrated during electrophoresis as a prominent band detected either by staining with ethidium bromide or by hybridization with rDNA probe (Supplementary Fig. S1A), suggesting considerably high rDNA copy numbers in the genome. For a detailed evaluation of rDNA intragenomic heterogeneity, the DNA-species forming this band were subjected to PacBio sequencing. Approximately one quarter (4,200) of the ROIs were successfully mapped to a 35S rDNA reference sequence, indicating ∼3-fold enhancement of the rDNA content in the sample in comparison to the native pumpkin genomic DNA, in which a fraction of ∼7.8% (6,500 copies) was estimated to be composed of 35S rDNA (Supplementary Table S1) distributed across five chromosomal loci of highly variable sizes as follows from fluorescence in situ hybridization (FISH) analyses performed previously by Waminal et al.17 (2011). To evaluate the variability in the chromosomal distribution of 35S rDNA within Cucurbita, we performed a similar analysis with C. pepo. As in C. moschata, hybridization signals at two loci (four signals) were significantly stronger than the remaining signals (Fig. 1A) frequently present on extremely small chromosomes (Fig. 1E). Although the number of strong 35S rDNA signals was invariant, the total number of signals slightly varied (12–13) among ten here analysed metaphases, suggesting potential aneuploidy in C. pepo, which was supported by some metaphase preparations with counted 41 chromosomes (Fig. 1F). In addition, higher total number of 35S rDNA sites in C. pepo, than in C. moschata suggests some instability of 35S rDNA across Cucurbita. Similarly, two loci and one locus of the 5S rDNA detected in C. moschata17 and C. pepo (Fig. 1B), respectively, suggests that instability in rDNA is rather common in Cucurbita. Nevertheless, the distribution of 35S rDNA across multiple loci in both species might indicate the coexistence of 35S rDNA structural variants within Cucurbita.
Figure 1.
FISH at metaphase chromosomes of C. pepo. Four very strong and nine significantly weaker signals for 35S rDNA (green; panels A and D) complemented with two strong signals for 5S rDNA (red; panels B and D) were detected. Chromosomes were counterstained with DAPI (C). To count all very small chromosomes hardly detected with colour, chromosomes were highlighted in inverted black and white format (panels E and F). Two metaphase preparations are shown, and panel E corresponds to panels (A–D). The chromosomes that harbour 35S rDNA are marked by arrowheads. Panel (F) shows metaphase preparation with all chromosomes well separated, counted and numbered from 1 through 41, approximately in the order of their relative sizes. Note that odd total number of chromosomes suggests an aneuploidy in this metaphase, which is in agreement with the odd number of rDNA signals shown in panels (A, D and E), although showing different metaphase. Scale bars (A–F) = 10 µm.
It is necessary to stress that such a mode of rDNA sample purification, based on a specific restriction pattern, may eliminate 35S rDNA pseudogenes or unusual IGS variants that are sometimes found dispersed in genomes outside regular NORs.27 In theory, such a method is suitable for the enrichment of samples with each type of repeat, which might be restricted from the genome using an appropriate restriction endonuclease.
Although relatively low numbers of 35S rDNA PacBio ROIs were analysed compared with the high copy numbers of 35S rDNA in the pumpkin genome, we believe that even those ROIs associated with the region of lowest coverage (repeats in IGS) might depict the variability in prominent structural motifs with sufficient accuracy.
3.2. Common sequence blocks in the IGS are shared in considerably variable proportions among individual rDNA copies
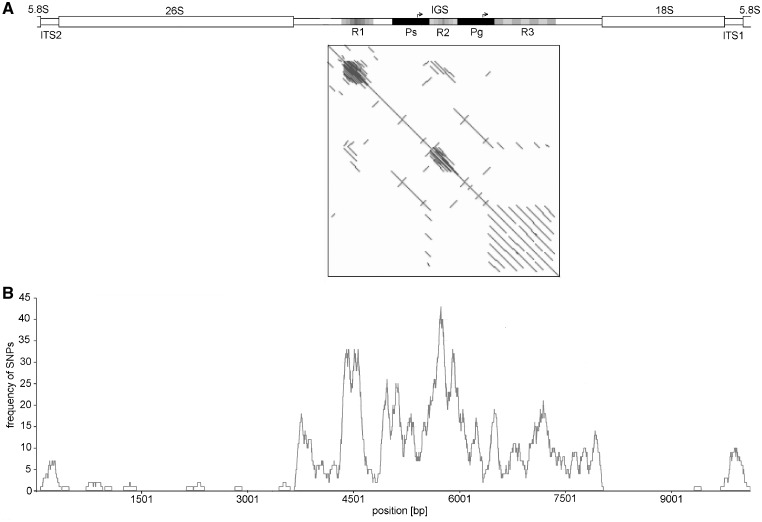
As the IGS represents the region with the highest sequence heterogeneity within the pumpkin rDNA unit (Fig. 2B), we concentrated our further efforts to estimate the intra-individual structural variability of this evolutionarily highly variable region. Each IGS copy is assembled from the three repeat regions R1–R3, which are separated by two copies of a unique duplicated sequence (Pg and Ps) (Fig. 2A). These duplicates are mutually highly related (Fig. 2A; dot plot) and internally moderately variable nucleotide sequences (Supplementary Fig. S6) with sizes ranging from 442 to 459 bp. Both contain a highly conserved sequence motif (TATATAGGGG) related to the known plant TIS for Pol l, suggesting that they may function as putative genic (Pg) and spacer (Ps) Pol I promoters. Interestingly, both duplicates harbour another short, highly conserved motif, CCCCTATAT, which is located ∼40 bp downstream of each (genic and spacer) TIS and represents nearly perfect reverse-complemented reflection of the TIS.
Figure 2.
Repetitive organization of the IGS in the pumpkin 35S rDNA unit. (A) Three regions (R1–R3), each built of tandemly arranged repetitive units, are separated by two mutually highly homologous sequences (Ps and Pg), as demonstrated by the dot-plot diagram constructed from the consensus sequence derived from mapped ROIs. Motifs related to TISs are denoted by arrows. (B) The frequency of abundant (>10%) SNPs plotted along the rDNA unit (100-bp window) shows that the R1 and R2 regions represent the most variable extremes in the rDNA unit. In contrast, all three genes are highly conserved and separated by the moderately variable ITS1 and ITS2. The pattern of all abundant SNPs along the source consensus sequence is shown in Supplementary Fig. S4. It is necessary to stress that the highest variability at repeats R1 and R2 may be artificially overestimated owing to possible incorrect mapping of the corresponding ROIs with variable copy numbers of repeats.
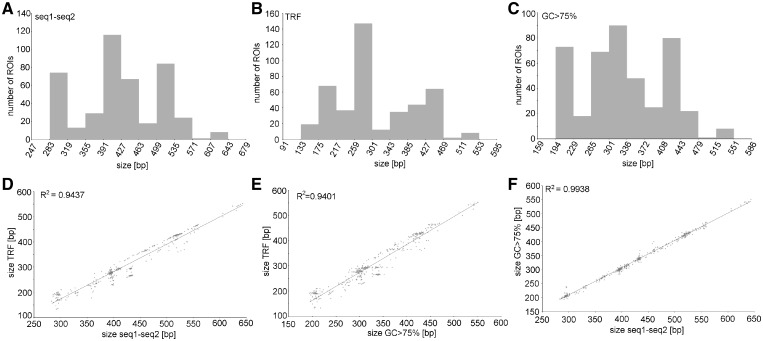
Although duplicated promoter-like sequences are rather invariable in length, repeated regions substantially contribute to the overall length variability of the IGS. Analyses of 134 and 436 ROIs by several independent methods consistently demonstrated extraordinarily high size variability at R1 (Supplementary Fig. S7A and B) and R2 (Fig. 3) tandem repeats, respectively, which ranges with fine graduations of more than hundreds of bp and a multimodal size distribution. Such a broad size variability at R1 and R2 even resulted in reciprocal lengths of both repeats in some IGSs, as demonstrated by ROIs bearing either more abundant R1 or more abundant R2 (Supplementary Fig. S8).
Figure 3.
The multimodal distribution of size variants in the R2 repeated region. Three independent methods were used to evaluate the size variability: (A) Distance between two boundary unique sequences (see Section 2.3). (B) Overall size of all tandem repeats detected in the R2 region by Tandem Repeats Finder. (C) Size of the uninterrupted region with GC content higher than 75%. Significant correlations between the results obtained using these three approaches, are demonstrated in panels (D–F).
In contrast to repeats R1 and R2, located upstream of the genic TIS, the unique repeat R3, located downstream (Fig. 2A), seemed to be substantially more homogeneous in size (Supplementary Fig. S7C), ranging from 2.9 to 3.4 tandemly arranged repeats with a consensus sequence varying from 248 to 261 bp. The exception was a single R3 region composed of 4.3 repeats.
Size polymorphisms detected by PacBio sequencing in the R1 and R2 regions were verified in the genome-wide range by PCR and Southern blot analyses. PCR amplification of R2 regions provided multiple products with an overall shape characteristic of tandem repeats (Supplementary Fig. S9). They all were, however, shorter than expected and the origins of the PCR products were, therefore, verified by Sanger sequencing. All cloned PCR products were assembled of the spacer promoter and short repeats of different sizes related to the R2 region.
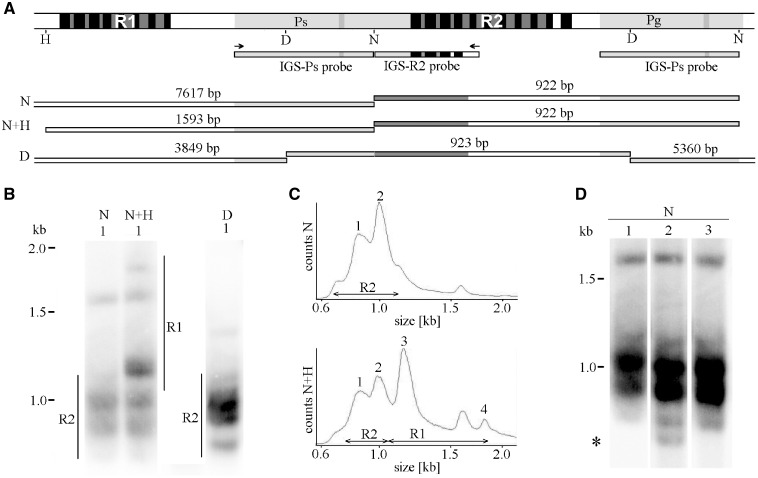
To evaluate the size variability at repeated regions by Southern blot analyses, genomic DNA was digested by restriction enzymes with targets in the vicinity of those repeats, and restriction fragments of interest were visualized by hybridization to the IGS probe(s) (Fig. 4A). Restriction with NdeI or DraI was used for R2 repeats, whereas double digestion with NdeI and HpaI showed overall size heterogeneity in both R1 and R2 regions. Each digest provided multiple rather diffuse hybridization signals (Fig. 4B), indicating the number of size variants present in both R1 and R2 regions and continuously spanning a considerably wide range of size distributions. Such pattern correlated well with the multimodal size distribution of PacBio ROIs (compare Fig. 4C with Fig. 3A and Supplementary Fig. S7A). Evident structural variability across plants within single cultivar (Fig. 4D), suggests fast repeats spreading or contraction in pumpkin IGS. As expected from the restriction map (Fig. 4A), both NdeI and DraI digests also provided long (>5 kb) hybridization signals (Supplementary Fig. S10). The NdeI fragment (7617 bp) harbours the entire R1, whereas the DraI fragment (5,360 bp) contains the entire R3 repeat. More diffuse pattern provided by NdeI confirmed higher size variability in the R1 region than in the R3 region.
Figure 4.
Size polymorphisms at R2(R1) region(s) may be supported on a genome-wide range by Southern blot analyses. (A) Target(s) for IGS hybridization probe(s) are shown on the background of a restriction map for NdeI (N), HpaI (H) and DraI (D) constructed from the consensus sequence for the corresponding IGS regions (Supplementary Fig. S4). The restricted DNA fragments that are expected to hybridize with the probe(s) are drawn below. (B) Southern hybridization of IGS probe(s) to genomic DNA restricted by the quoted RE reveals multiple rather diffuse bands. For better resolution, only short (<2 kb) fragments are shown (long fragments are shown in Supplementary Fig. S10). (C) Densitometric evaluations of hybridization profiles. Note that NdeI digests show size variability in R2 (Bands 1–2), whereas double digestion with NdeI and HpaI shows size variability in both R1 (Bands 3–4) and R2 (1–2) regions. (D) Longer exposition of NdeI digests highlighted structural variability (asterisk) across three plants selected within a single pumpkin cultivar. Plant marked by number 1 was sequenced and analysed in panel (B).
3.3. Structurally divergent DNA minisatellites coexist in the R2 region
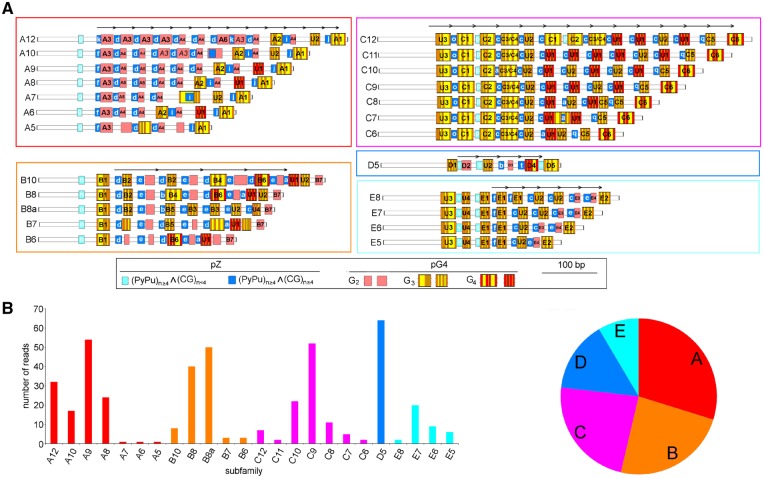
Multimodal distribution of size variants in both the R1 and R2 regions, which were consistently detected by PacBio sequencing and Southern analyses, suggested the coexistence of two or more prominent lengths and/or structural variants in both regions. We further focussed on detailed structural analyses of the R2 repeat because it is adjacent to both promoter-like regions (Fig. 2A), and the multimodal distribution of size variants appeared to be more pronounced compared with the R1 and R3 regions. Each R2-repeat variant was significantly enriched in GC content (Supplementary Fig. S8), and the sense DNA strand was enriched with C at the expense of G (C:G ∼ 7:3). We therefore inspected individual ROIs for the frequency and distribution of sequence motifs associated with GC-rich and inter-strand C/G bias as follows: (i) pG4- and pZ-motifs with the potential to form non-canonical G-quadruplexes and left-handed Z-DNA structures, respectively; (ii) tandemly arrayed partially degenerated short DNA motifs forming microsatellite-like DNA stretches; and (iii) the dispersed highly abundant permutated motifs C3G and GC3. Based on the highly variable number and spatial arrangement of these motifs, all R2 repeats might be classified into five structural families, A–E (Fig. 5A), arranged according to their abundance (Fig. 5B). They all are formed of relatively short (34–79 bp) tandemly arrayed units, each consisting of equal numbers of pZ- and pG4-motif(s) (Supplementary Table S2). Therefore, each repeat unit has a polarised structure with pZ characteristics prevailing towards the spacer promoter, while pG4 structural characteristics are more prominent towards the genic promoter. Intrinsic structures of both pZ- (Supplementary Table S3) and pG4- (Supplementary Table S4) motifs were, however, specific for a particular family. An enormous structural variability in the R2 region was further emphasized by multiple subfamilies with a distinct number of tandemly arranged repetitive units and overall length, which might be defined within the majority of families (Fig. 5). Selected structural features that further discriminated individual families are summarized in Table 1.
Figure 5.
Five prominent repeated families coexist in R2 regions in pumpkin IGSs. (A) Each family (A–E) is characterized by a specific arrangement of two kinds of short sequence motifs, pZ and pG4. Individual abundant pZ- and pG4-variants are distinguished by small and numbered capital letters, respectively, within corresponding rectangles and specified in Supplementary Tables S3 and S4, respectively. C3/C4 means that two pG4-motifs (C3 and C4) are overlapping. Three pG4 prominent variants that are distinct in length of G tracts (G2–G4) are considered. A particular family may consist of several size variants (subfamilies) with a distinct number of basic tandem repeats (arrows). Each subfamily is represented by the most abundant distribution of pG4-motifs. (B) The relative abundance of individual subfamilies (left) and families (right) in the genome is estimated from the number of corresponding ROIs.
Table 1.
Selected features characterizing individual R2 families
| Feature | Specification | Family | ||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | ||
| Overall sizea,b,c | mean [bp] | 341; 294; 433 | 303.5; 284.5; 396.5 | 424; 427; 519 | 207; 189; 295 | 264; 215; 358 |
| min. [bp] | 199; 186; 286 | 216; 205; 318 | 309; 310; 401 | 194; 133; 283 | 200; 170; 291 | |
| max. [bp] | 446; 435; 539 | 415; 403; 511 | 551; 553; 643 | 239; 201; 327 | 310; 275; 401 | |
| max. – min. [bp] | 247; 249; 253 | 199; 198; 193 | 242; 243; 242 | 45; 68; 44 | 110; 105; 110 | |
| Tandem unita,d | consensus mean size [bp] | 34 | 79 [39]e | 42 | 38 | 37 |
| copy numbers range | 4.8–12.6 | 2.0–4.3 [7.1]e | 6.4–13.5 | 3.6 | 2.4–3.8 | |
| pZ-motifsa,f | copy numbers range | 5–12 | 6–10 | 6–12 | 5 | 5–8 |
| no of variants (family-specific) | 7(4) | 6(1) | 5(3) | 5(4) | 5(2) | |
| the most abundant variant(s) | (GC)6 | (GC)6; (CG)6C | (CG)5C | (GC)5 | (CG)5C; (CG)6C | |
| pG4-motifsa,g,c | copy numbers rangeh | 2–3 | 4–7 | 9–14 | 4 | 5–8 |
| no of variants (family-specific) | 8(6) | 10(7) | 9(6) | 6(5) | 8(4) | |
| the most abundant varianti | G2 | G2; G3 | G4 | G2 | G3 | |
| Microsatellite | consensus | nd | c4g; c4gc3(gc)3j | cgc3 | nd | c4g; ccacgc |
| abundance [% of ROIs] | nd | 59; 30k | 92 | nd | 97; 76 | |
| copy number mean | nd | 7.6; 5.6 | 43.2 | nd | 26.2; 4.3 | |
| copy number min. | nd | 6.6; 5.6 | 13.0 | nd | 10.2; 4.3 | |
| copy number max. | nd | 19.6; 8.6 | 62.2 | nd | 33.6; 5.3 | |
| Ratio GC3/C3Gc | mean | 1.5 | 1.1 [0.9]e | 1.0 | 1.2 | 1.3 |
| RTAnT-sizec,l | mean; min.; max. [bp] | 39; 21; 49 | 12; 11; 14 | 8; 0; 8 | 10; 9; 10 | 9; 8; 9 |
For individual subfamilies, see also Supplementary Table S5.
Three values for each descriptive statistical parameter are derived from three independent methods used for repeat size estimations and representing: (i) region with GC content >75%, (ii) Tandem Repeats Finder and (iii) distance between conserved unique bordering sequences, respectively.
For comparative statistics, see also Supplementary Table S6.
For the consensus, see also Supplementary Table S2.
Values for the B8a subfamily are in square brackets, if significantly different from other B-subfamilies.
Frequency of abundant pZ-motifs are comprehensively specified in Supplementary Table S3.
Frequency of abundant pG4-variants are comprehensively specified in Supplementary Table S4.
Only motifs with G3 and G4 tracts are considered.
Only variants with G-tracts of different lengths (G2–G4) are distinguished.
Longer motif is specific for the B8a-subfamily.
Computed from the number of ROIs in B8 and B8a subfamilies, respectively.
Length of the spatially linked AT-region (RTAnT) located between the putative spacer promoter and R1 minisatellite (Supplementary Fig. S4).
Families A and C represent highly heterogeneous populations of size variants built of variable numbers of monomeric units, each composed of single pZ- and single pG4-motifs. Both motifs, however, significantly differ between families in structure. In Family A, the majority of pG4-motifs are formed of short G2 tracts, resulting in the shortest monomeric unit. In contrast, G3 and G4 tracts are characteristic for Family C, resulting in a substantially longer unit with higher potential to form G4-conformations. Nevertheless, in both families, pG4 variants with greater potential to form G4 conformations are accumulated towards the genic promoter. In Family A, the number of such pG4 variants appears to be independent of the considerably variable total number of pZ- and pG4-motifs in individual size subfamilies. Such an arrangement may result in a polarized structure of the R2 region as a whole. It is necessary to stress that the pG4 variant U1 with the highest potential to form G-quadruplexes is significantly more abundant in Family C in comparison to other families (Supplementary Table S4).
Family B is exceptional in carrying the longest basic monomeric repeated units, each built of alternating arrangements of two pZ- and two pG4-variants. Such an arrangement results in partial homology within each unit. Although both abundant subfamilies B8 and B8a share the same number of pZ-motifs, they differ substantially in the occurrence of specific pG4-motifs (Fig. 5 and Supplementary Table S4) and relative content of GC3 and C3G motifs (Table 1).
Family D is the shortest and the most homogeneous in size, built of a unique copy number of basic monomeric units, each composed of single pZ- and single pG4-motifs, both with specific structural features (Supplementary Tables S3 and S4).
In size, the moderately variable Family E is built of monomers that are closely related to those found in Family C; however, they are, on average, shorter due to the absence of pG4 motifs with G4 tracts that are frequent in Family C. In addition, two highly conserved pZ-variants (Supplementary Table S3) as well as four pG4-variants (Supplementary Table S4) are specific for this family.
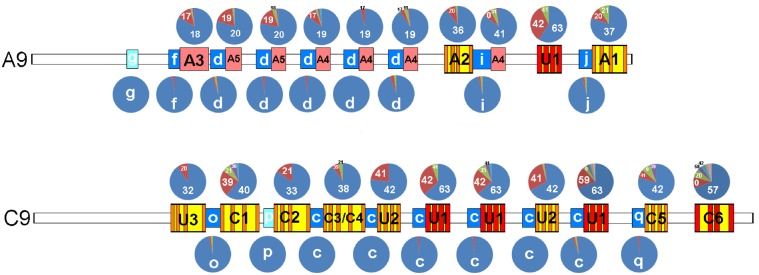
Considerably high structural variability occurred within almost each pG4 position along each subfamily (Fig. 6). In contrast, variability in adjacent pZ motifs appeared rather negligible, suggesting a diverse mode of evolution. Substantial variability within each family was further demonstrated using comprehensive descriptive statistics performed for the number of quantitative features within each corresponding subfamily (Supplementary Table S5). Finally, significant differences among all five R2 families were supported by comparative statistics in all evaluated structural characteristics with only a few exceptions (Supplementary Table S6).
Figure 6.
Structural variability of pG4- and pZ-motifs in distinct positions along the R2 minisatellites. The relative abundance of particular pG4- and pZ-variants are represented by charts located above and under the corresponding positions, respectively, in selected A9- and C9-sub-families (for more annotation, see Fig. 5). The pG4-variants are distinguished by numerals representing the in silico computed potential (score) to form a G4-conformation. Each pG4-variant may, however, be composed of several sequence motifs with the same score. Only variants that occurred more frequently than in one ROI are distinguished.
3.4. Correlation between the structural variability in R2 repeats and adjacent regions of pumpkin IGSs
Suggesting the concerted evolution of IGSs as a unit, we questioned whether the structural variability at the R2 region was reflected in the distal R1 region with comparable multimodal length variability (Supplementary Fig. S7A and B). Both regions are GC rich and share some motifs, particularly C4GC, in similar abundances, suggesting a common origin. Nevertheless, a rather low overall sequence similarity between regions was detected by dot plot analyses (Fig. 2A and Supplementary Fig. S8A) and further supported by the occurrence of predominantly different short GC-rich motifs in both regions. (i) The pZ-motifs prevail in R2 as (CG)n variants (Fig. 5), whereas more complex (PyPu)n variants are common in R1, and therefore, except for the (CG)4C motif, there is no shared pZ-motif variant in either region (compare Supplementary Table S3 with Supplementary Table S7). (ii) No pG4-motif variant with G4 stretches is shared by the two regions (Supplementary Table S8). (iii) The ratio between G3C and C3G motifs is mostly reversed between R1 and R2 repeats.
The mutual distribution of the pZ- and pG4-motifs along the entire R1 region and its overall length and size variability suggest that five basic structural variants, S, T, U, V and X (Supplementary Fig. S11), coexist in the R1 region. Families S, U and X are short with low size variability, whereas families T and V are highly variable in size but substantially different in the mutual distribution of pZ- and pG4-motifs. Family T is composed of a large number of pZ-motifs that are regularly distributed along the entire length and complemented with a few pG4-motifs adjacent to the spacer promoter. In contrast, Family V is full of pG4-motifs that are regularly arranged along the entire region and complemented with three pZ-motifs adjacent to the spacer promoter. Among the short R1 families, S is distinct, having a lower number of pZ-motifs in comparison to the pG4-motifs, whereas both the U and X families contain high accumulations of pZ-motifs, significantly exceeding the number of pG4-motifs. The U family has a lower number of pZ-motifs than the X family. Common, as well as specific pZ-motifs (Supplementary Table S7) are present in all R1 families.
To evaluate whether distinct R1 families are specifically linked to corresponding R2 families, we analysed a limited number of ROIs composed of both R1 and R2 regions (or the larger part) linked to a spacer promoter. We showed that for each R1 family, all corresponding ROIs fall into a unique R2 family and vice versa (Supplementary Table S9). It is necessary to stress that the R2 satellite families A and C, which are rather long and variable in size, are linked to the short and size-invariant R1 families S and U, respectively. In contrast, the short and less variable in size R2 Families D and B are linked to the long and highly variable in size R1 families V and T, respectively (Supplementary Fig. 12).
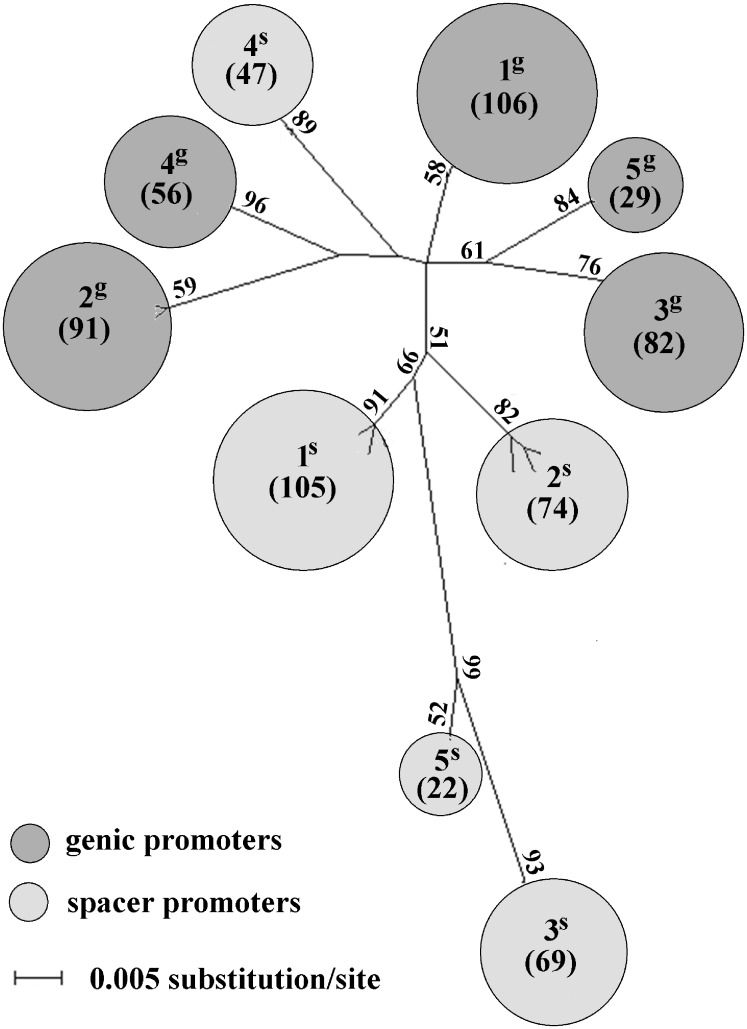
As both R1 and R2 repetitive regions show signs of common evolution, we extended our effort to evaluate structural variability at adjacent non-repetitive promoter regions. The phylogenetic analysis well separated the genic and spacer promoter duplicates. In addition, both genic and spacer promoters were split into five clusters 1g–5g and 1s–5s, respectively, supported by bootstrap values ranging from 52 to 96% (Fig. 7). All promoter sequences forming particular cluster were linked to a unique family from the R1 and R2 regions (Supplementary Table S10) and vice versa, suggesting a common evolution of repeated and unique sequences in pumpkin IGS.
Figure 7.
ML phylogenetic tree constructed from the SNP distribution along putative promoters. Genic and spacer promoter variants, determined in Supplementary Fig. S6, are well separated. In addition, both genic and spacer promoter sequences are split into the same number (five) of clusters of 1g–5g and 1s–5s, respectively. Owing to high number of sequences, individual clusters are represented by circles with areas proportional to the number (specified in brackets) of corresponding promoters, specified in more details in Supplementary Table S10. Only bootstrap values higher than 50% are shown.
Finally, the size ranges of RTAnT-motifs, located between the spacer promoter and R1 repeat (Supplementary Fig. S4) are highly specific for each corresponding spatially linked R1/R2 family (Table 1).
Classification of the pumpkin IGSs into multiple families appears to be highly legitimate because structural variants of R1 (Supplementary Fig. S11) and R2 (Fig. 5) repeats, defined based on the arrangement of distinct structural blocks, correlate well with (i) adjacent promoter variants (Fig. 7; Supplementary Table S10) that are independently defined from the distribution of SNPs and (ii) the size of A/T-rich region RTAnT (Table 1).
4. Discussion
4.1. The minisatellite-like organization suggests genetic instability and accelerated structural evolution of R2 (R1) regions, which might affect transcription from adjacent Pol I promoters
The repeat unit size, copy number, sequence purity, and GC content of R2/R1 repeats in pumpkin IGSs are typical for so-called DNA minisatellites. The origin of the extended copy number variability at R2/R1 regions may, therefore, be explained by mechanisms that are common for satellites where the number of repeated units expands or contracts at high frequencies by mechanisms such as intra- and inter-locus recombination and/or replication slippage.28 In pumpkin IGSs, minisatellites that are highly variable in length (Families A and C) coexist with those with sizes that are rather stable (Family D), suggesting distinct mechanisms of evolution.
The occurrence of tandem repeats in the vicinity of Pol I promoters appears to be a common feature of IGSs across flowering plants (Supplementary Table S11). Their structures (size of monomeric unit, copy number, nucleotide sequence and GC content) vary; however, with wide ranges even between closely related species. Nevertheless, significantly higher GC content, compared with adjacent promoter-like sequences, appeared to be the most striking feature of IGS satellites and the highest GC content within plants is characteristic for Cucurbita. Microsatellites (the most striking example represents Capsicum), minisatellites (Olea) and satellites (Hordeum) have evolved upstream of TIS, whereas only minisatellites (Eruca) and satellites (Cucurbita) were found downstream of TIS. The distance between TIS and adjacent satellites varies widely, and the corresponding nucleotide sequence is extremely variable even in the short ‘core’ Pol I region adjacent to TIS.
The instability of satellites with mutation frequencies from 10−2 to 10−5 per generation is within the range of frequencies of genetic point mutations (10−8 to 10−9) and epigenetic switches (10−1 to 10−2).29 Therefore, these repetitive elements share both genetic and epigenetic features. As tandem repeats can reversibly expand or contract without a complete loss of information, they resemble epigenetic transitions, which do not lead to severe changes in function or completely novel features. Generally, tandem repeats are frequently located in gene promoters where their variations can act as mediators of rapid and fine-tuned phenotypic changes, perhaps through alterations in chromatin structure.29,30 The activation of Pol I transcription appears to be mediated by transcription factors that interact with both the promoter and the upstream located mini(satellites) (enhancers), although their structural features vary with a wide range between maize,31Xenopus32 and mice33 (Supplementary Table S11). In pumpkin, R2 minisatellites are built of rather atypical repeat units, each composed of two kinds of asymmetrically distributed GC-rich and short microsatellite-like sequences with the potential, although considerably variable, to form Z- or G4-non-canonical DNA structures. There are direct and indirect pieces of evidence that both G4- and Z-DNA conformations are formed in vivo: (i) functionally important genomic regions, including promoters, rDNA and sites with low nucleosome occupancy are enriched in these motifs.34,35 (ii) Specific proteins with affinity to pZ- or pG4- or both-conformations participate in their regulation either by promotion or destabilization.36 Namely, two abundant nucleolus-specific proteins, nucleolin and nucleophosmin, show specific affinity to G4-conformations in rDNA.37 Related proteins might recognize the satellites in pumpkin IGSs that are rich in both motifs, although their distributions and proportions vary widely. During transcription, these proteins may interact with G4 structures formed by the non-template G-rich strand to prevent renaturation of the duplex and to render the template strand available for multiple rounds of transcription. The stability of the melted region may correlate with variable numbers or densities of pG4-motifs. (iii) Hoogsteen base pairs may transiently form in canonical double-stranded DNA,38 and G4 conformations can be formed in supercoiled duplex DNA and DNA in chromatin in vivo.39 As local disruption of the regular double helix DNA structure is associated with B-Z transition,40 G4-formation may be promoted by closely linked pZ-motifs. Diversely methylated pZ-motifs may undergo a B-Z transition with varying efficiency,41 and thereby increase the epigenetically determined structural heterogeneity of distinct IGS variants.
The occurrence of two significantly larger FISH signals in both C. moschata and C. pepo may suggest that these loci are transcriptionally active and form dominant NORs. However, FISH analyses did not show any secondary constriction at these loci in either C. moschata17 or C. pepo (Fig. 1). We previously showed that there is no unequivocal correlation between the size of rDNA loci and their transcription activity, and even loci with highly reduced rDNA copy numbers may be transcriptionally dominant, forming active NORs.42
Specific structural features of the short repeat unit together with its variable copy numbers may result in decreased sequence complexity along considerably long and variable DNA stretches associated with altered GC content, bendability, intrinsic curvature and, consequently, chromatin structure.43 Altered chromatin structure established at such repeats may consequently partially spread into adjacent regulatory elements and gradually sterically attract/repulse certain factors that participate in transcription or replication.
4.2. Origin and spatial arrangement of multiple 35S rDNA variants in the pumpkin genome
Generally, tandem repeats in individual IGS variants may differ: (i) in the copy number of the common repeat, as observed between species within Capsicum, Solanum and Quercus (Supplementary Table S11). (ii) In the spatial arrangement of common repeats, as found between Tragopogon dubius and Tragopogon porrifolius,2 and (iii) in the occurrence of specific repeats, as has been described for Brassica species.7 Owing to a few IGSs analysed in each species, interspecies heterogeneity detected among highly related species may; however, be the consequence of broad size variability within each species, as documented recently for Arabidopsis14 and here for pumpkin. We describe the coexistence of a number of IGS variants in a single genome, which differ not merely in overall length defined by the number of tandem repeats but also in their intrinsic structure determined by the specific arrangement of distinct short sequence motifs. Such enormous rDNA variability supports the hypothesis of a hybrid/polyploid origin of C. moschata.17 Alternatively, multiple rDNA loci may originate from rDNA translocations in a single genome, and the structural identity of rDNA loci may be maintained as a consequence of inefficient interlocus homogenization. On the whole-genome scale, the mutual arrangement of individual rDNA variants in C. moschata is currently unknown. Considering the equal number (5) of rDNA structural variants (Fig. 5) and number of chromosomal rDNA loci,17 we can suppose that individual variants are distributed separately among multiple chromosomal loci. However, possible homeologous recombinations may lead to homogenization of rDNA at some or all loci during C. moschata diploidization. As a result, individual rDNA families might be partially or completely spatially intermingled. Despite invariable chromosome number in allohexaploid wheat (2n = 6x = 42), the numbers of rDNA chromosomal sites range from 2 to 16 (Plant rDNA database),44 suggesting variable modes of rDNA evolution during diploidization of wheat cultivars. In the recently formed allotetraploid T. mirus, both parental IGS variants have remained preserved.2 Among more ancient Nicotiana allotetraploids, the extent of interlocus rDNA homogenization decreased in the direction N. arentsii > N. tabacum > N. rustica45 and both parental variants remained in N. rustica, whereas complete homogenization of rDNA towards one parental variant occurred in N. arentsii. Using specific FISH probes derived from the parental IGS, both homogenization and preservation of parental rDNA variants were demonstrated in different cultivars of Brassica napus.46 Because of the extraordinarily high GC content of all R2 and R1 variants in pumpkin, they can hardly be distinguished by a similar approach. A significantly higher number of 35S rDNA than 5S rDNA suggests segmental duplications or 35S rDNA translocations during the pumpkin genome evolution. Alternatively, 5S rDNA has partially been eliminated, as may be inferred from only one 5S rDNA locus currently present in C. pepo.
4.3. The global repetitive architecture of C. moschata IGSs is conserved among thousands of rDNA copies but differs in closely related species
Despite the considerably high structural variability within both R1 and R2 minisatellites, the overall repetitive organization of pumpkin IGSs is rather conserved, invariably formed of three prominent repeats intermingled with two highly homologous copies of considerably long non-repetitive sequences, suggesting selection constraints to maintain such complexly structured regions. Although rarely occurring within flowering plants (Supplementary Table S11), similar duplications of promoter-like sequences separated by tandem repeats were previously described in distantly related species Tragopogon (Asteraceae),2Arabidopsis (Brassicaceae),14Quercus (Fagaceae),47Daucus (Apiaceae)48 and Xanthisma (Asteraceae), suggesting recurrent duplication events during dicot evolution. Comprehensive analysis of the structural heterogeneity of the 35S rDNA IGS has, however, been performed only for A. thaliana using PacBio sequencing of PCR amplicons.14 As in pumpkin, high size heterogeneity is due to the variable number of short tandemly arrayed monomeric units, which, however, are significantly more homogeneous in nucleotide sequences compared with repeated units forming five families in pumpkin. In contrast to pumpkin, they are distributed among variable numbers (1–3) of tandem blocks separated by variable numbers of promoter-like duplicates (0–2). Intraspecific variability in the number of promoter-like sequences was also detected in Quercus robur. The number of duplicated promoter-like sequences significantly differs between the related species T. porrifolius (>3 copies) and T. dubius (2 copies). The length of duplicated regions significantly varies between species, ranging from less than 100 bp (probably representing only a core promoter) to several hundred bp.
We presume that together with the non-repetitive sequence surrounding TIS, adjacent tandem repeats were also duplicated, as a whole or in part, during the evolution of the IGS. Alternatively, a unique promoter-like sequence may have been duplicated and translocated inside the ancestral unique tandem repeat. Perhaps due to the action of selective constraints, duplicated unique sequences evolved in a concerted manner to maintain a high degree of homology, both within and between them, in each species. In contrast, duplicated repeated regions could subsequently evolve rather independently either due to the absence of selective constraints or, more likely, their repetitive characteristics, which may have significantly accelerated their diversification. In pumpkin, both modern repetitive regions R1 and R2 share only a few signs: the extremely high GC content and the occurrence of short sequence motifs, including pZ and pG4, which may indicate their common origin. Duplicated satellites in each Q. robur, T. porrifolius and A. thaliana; however, share highly related nucleotide sequences, suggesting a more recent duplication or another mechanism of evolution, such as the impact of selection constraints on each satellite.
The R3 repeat is unrelated to R1 and R2 repeats, made up of significantly longer units with the expected GC content, which seems to be common among dicots where repeats located upstream of TIS are, on average, composed of higher copy numbers of shorter monomers with higher GC content than repeats localized downstream (Supplementary Table S11). In contrast, monocots harbour satellites composed of substantially long repeated units located both upstream and downstream of genic TIS. When compared with R1 and R2 minisatellites, the R3 satellite in C. moschata is significantly more homogeneous in size, composed almost exclusively of three monomeric units, reminiscent of results previously reported for A. thaliana,14 and suggesting rather common features among plants. Despite high size homogeneity within C. moschata, the R3 repeat was significantly amplified in C. maxima, currently forming a tandem of nine units.18 Such interspecific differences might be explained on the basis of the so-called ‘library hypothesis’.49 Accordingly, both size variants of R3 probably evolved in a common ancestor, although perhaps at low copy numbers, followed by subsequent spreading of a shorter variant in C. moschata and C. pepo,18 while the longer variant was amplified in C. maxima. In contrast, the R1 repeat is almost completely deleted in C. maxima, whereas it is preserved in C. pepo. Nevertheless, both highly conserved sequences surrounding the R1 region in C. pepo and C. moschata are completely retained in C. maxima, surrounding the short non-repetitive region with signs of R1 repeats (CG-rich, pZ- and pG4-motifs) (Supplementary Table S2). The pumpkin R1 repeat may, therefore, originated by amplification of the short unique sequence present in the common ancestor rather than by the above mentioned duplication event. Similarly, T. dubius harbours relatively long repeats located downstream of TIS complemented with only one repeat located upstream of TIS, whereas the T. porrifolius IGS is composed of short downstream repeats complemented with multiple satellites located upstream.2
5. Conclusions
We showed that in the genome of C. moschata, five rDNA structural variants are distributed across five chromosomal loci. Structural variability between rDNA variants is determined by the variable repetitive arrangement of a few kinds of short GC-rich sequence blocks with the potential to form non-canonical DNA structures and together to form various minisatellites in the IGS. Because pZ- and pG4-conformations, as well as tandemly arranged repeats in IGSs, are known to participate in transcriptional regulation, we hypothesize that such higher order structural variability of IGS repeats may participate in a graduated affinity of TIFs for the adjacent Pol I promoters, potentially promoting the fine orchestration of rDNA transcription regulation in response to cell protein synthesis demand. We suppose that individual rDNA variants evolved in two or more parental genomes followed by hybridization or, alternatively, within a single genome at separated chromosomal rDNA loci as a consequence of inefficient inter-locus homogenization.
Supplementary Material
Acknowledgements
We thank Radka Vozarova for FISH analyses and helpful comments on the article.
Accession numbers
BioProject ID PRJNA400686 (http://www.ncbi.nlm.nih.gov/bioproject/400686); GenBank MG744571-MG744575.
Funding
The research was funded by the Czech Science Foundation (P501/17/11642S).
Conflict of interest
None declared.
References
- 1. Hemleben V., Zentgraf U.. 1994, Structural organization and regulation of transcription by RNA polymerase I of plant nuclear ribosomal RNA genes, Results Probl. Cell Differ., 20, 3–24. [DOI] [PubMed] [Google Scholar]
- 2. Matyasek R., Dobesova E., Huska D.. 2016, Interpopulation hybridization generates meiotically stable rDNA epigenetic variants in allotetraploid Tragopogon mirus, Plant J., 85, 362–77. [DOI] [PubMed] [Google Scholar]
- 3. Flavell R.B., Odell M., Thompson W.F., et al. 1986, The differential expression of ribosomal-RNA genes, Philos. Trans. Roy. Soc. B., 314, 385-397. doi:10.1098/rstb.1986.0060. [Google Scholar]
- 4. Martini G., O’Dell M., Flavell R.B.. 1982, Partial inactivation of wheat nucleolus organizers by the nucleolus organizer chromosomes from Aegilops-Umbellulata, Chromosoma, 84, 687–700. [Google Scholar]
- 5. Silva M., Pereira H.S., Bento M.. 2008, Interplay of ribosomal DNA loci in nucleolar dominance: dominant NORs are up-regulated by chromatin dynamics in the wheat-rye system. Plos One, 3, e3824. doi:10.1371/journal.pone.0003824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Komarova N.Y., Grabe T., Huigen D.J., et al. 2004, Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids, Plant Mol. Biol., 56, 439–63. [DOI] [PubMed] [Google Scholar]
- 7. Chen Z.J., Pikaard C.S.. 1997, Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. USA., 94, 3442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wanzenbock E.M., Schofer C., Schweizer D., Bachmair A.. 1997, Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana, Plant J., 11, 1007–16. [DOI] [PubMed] [Google Scholar]
- 9. Doelling J.H., Gaudino R.J., Pikaard C.S.. 1993, Functional-analysis of Arabidopsis thaliana ribosomal-RNA gene and spacer promoters in-vivo and by transient expression. Proc. Natl. Acad. Sci. USA., 90, 7528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zentgraf U., Hemleben V.. 1992, Complex-formation of nuclear proteins with the RNA polymerase-I promoter and repeated elements in the external transcribed spacer of Cucumis sativus ribosomal DNA, Nucleic Acids Res., 20, 3685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Komarova N.Y., Grimm G.W., Hemleben V., Volkov R.A.. 2008, Molecular evolution of 35S rDNA and taxonomic status of Lycopersicon within Solanum sect. Petota, Plant Syst. Evol., 276, 59–71. [Google Scholar]
- 12. Schiebel K., von Waldburg G., Gerstner J., Hemleben V.. 1989, Termination of transcription of ribosomal-RNA genes of mung bean occurs within a 175-bp repetitive element of the spacer region, Mol. Gen. Genet., 218, 302–7. [DOI] [PubMed] [Google Scholar]
- 13. Eickbush T.H., Eickbush D.G.. 2007, Finely orchestrated movements: evolution of the ribosomal RNA genes, Genetics, 175, 477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Havlová K., Dvořáčková M., Peiro R., et al. 2016, Variation of 45S rDNA intergenic spacers in Arabidopsis thaliana, Plant Mol. Biol., 92, 457–71. [DOI] [PubMed] [Google Scholar]
- 15. Grummt I., Langst G.. 2013, Epigenetic control of RNA polymerase I transcription in mammalian cells, Biochim. Biophys. Acta, 1829, 393–404. [DOI] [PubMed] [Google Scholar]
- 16. Durut N., Abou-Ellail M., Pontvianne F., et al. 2014, A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis, Plant Cell, 26, 1330–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Waminal N.E., Kim N.S., Kim H.H.. 2011, Dual-color FISH karyotype analyses using rDNAs in three Cucurbitaceae species, Genes Genom., 33, 521–8. [Google Scholar]
- 18. King K., Torres R.A., Zentgraf U., Hemleben V.. 1993, Molecular evolution of the intergenic spacer in the nuclear ribosomal-RNA genes of Cucurbitaceae, J. Mol. Evol., 36, 144–52. [DOI] [PubMed] [Google Scholar]
- 19. Siegel A., Kolacz K.. 1983, Heterogeneity of pumpkin ribosomal DNA, Plant Physiol., 72, 166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matyasek R., Fulnecek J., Leitch A.R., Kovarik A.. 2011, Analysis of two abundant, highly related satellites in the allotetraploid Nicotiana arentsii using double-strand conformation polymorphism analysis and sequencing, New Phytol., 192, 747–59. [DOI] [PubMed] [Google Scholar]
- 21. Lim K.Y., Kovarik A., Matyasek R., Bezdek M., Lichtenstein C.P., Leitch A.R.. 2000, Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units, Chromosoma, 109, 161–72. [DOI] [PubMed] [Google Scholar]
- 22. Benson G. 1999, Tandem repeats finder: a program to analyze DNA sequences, Nucleic Acids Res., 27, 573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kikin O., D’Antonio L., Bagga P.S.. 2006, QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences, Nucleic Acids Res., 34, W676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shin S.I., Ham S., Park J.. 2016, Z-DNA-forming sites identified by ChIP-Seq are associated with actively transcribed regions in the human genome, DNA Res., 23, 477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tamura K., Nei M.. 1993, Estimation of the number of nucleotide substitutions in the control region of mitochondrial-DNA in humans and chimpanzees, Mol. Biol. Evol., 10, 512–26. [DOI] [PubMed] [Google Scholar]
- 26. Jenkins G., Hasterok R.. 2007, BAC ‘landing’ on chromosomes of Brachypodium distachyon for comparative genome alignment, Nat. Protoc., 2, 88–98. [DOI] [PubMed] [Google Scholar]
- 27. Lim K.Y., Skalicka K., Koukalova B., et al. 2004, Dynamic changes in the distribution of a satellite homologous to intergenic 26-18S rDNA spacer in the evolution of Nicotiana, Genetics, 166, 1935–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richard G.F., Paques F.. 2000, Mini- and microsatellite expansions: the recombination connection, EMBO Rep., 1, 122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gemayel R., Vinces M.D., Legendre M., Verstrepen K.J.. 2010, Variable tandem repeats accelerate evolution of coding and regulatory sequences, Annu. Rev. Genet., 44, 445–77. [DOI] [PubMed] [Google Scholar]
- 30. Espley R.V., Brendolise C., Chagne D., et al. 2009, Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples, Plant Cell, 21, 168–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmitz M.L., Maier U.G., Brown J.W.S., Feix G.. 1989, Specific binding of nuclear proteins to the promoter region of a maize nuclear ribosomal-RNA gene unit, J. Biol. Chem., 264, 1467–72. [PubMed] [Google Scholar]
- 32. Caudy A.A., Pikaard C.S.. 2002, Xenopus ribosomal RNA gene intergenic spacer elements conferring transcriptional enhancement and nucleolar dominance-like competition in oocytes, J. Biol. Chem., 277, 31577–84. [DOI] [PubMed] [Google Scholar]
- 33. Kuhn A., Deppert U., Grummt I.. 1990, A 140-base pair repetitive sequence element in the mouse ribosomal-RNA gene spacer enhances transcription by RNA polymerase-I in a cell-free system. Proc. Natl. Acad. Sci. USA., 87, 7527–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yadav V., Hemansi, Kim N., Tuteja N., Yadav P.. 2017, G quadruplex in plants: a ubiquitous regulatory element and its biological relevance, Front Plant Sci., 8,1163, 10.3389/fpls [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang G.L., Vasquez K.M.. 2007, Z-DNA, an active element in the genome, Front. Biosci., 12, 4424–38. [DOI] [PubMed] [Google Scholar]
- 36. Mishra S.K., Tawani A., Mishra A., Kumar A.. 2016, G4IPDB: a database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep., 6, 38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chiarella S., De Cola A., Scaglione G.L., et al. 2013, Nucleophosmin mutations alter its nucleolar localization by impairing G-quadruplex binding at ribosomal DNA, Nucleic Acids Res., 41, 3228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nikolova E.N., Kim E., Wise A.A., O’Brien P.J., Andricioaei I., Al-Hashimi H.M.. 2011, Transient hoogsteen base pairs in canonical duplex DNA, Nature, 470, 498.U484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun D.K., Guo K.X., Shin Y.J.. 2011, Evidence of the formation of G-quadruplex structures in the promoter region of the human vascular endothelial growth factor gene, Nucleic Acids Res., 39, 1256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moradi M., Babin V., Roland C., Sagui C.. 2013, Reaction path ensemble of the B-Z-DNA transition: a comprehensive atomistic study, Nucleic Acids Res., 41, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Temiz N.A., Donohue D.E., Bacolla A., Luke B.T., Collins J.R.. 2012, The role of methylation in the intrinsic dynamics of B- and Z-DNA, PLoS One, 7, e35558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dobešová E., Malinská H., Matyášek R., et al. 2015, Silenced rRNA genes are activated and substitute for partially eliminated active homeologs in the recently formed allotetraploid, Tragopogon mirus (Asteraceae), Heredity, 114, 356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gabrielian A., Bolshoy A.. 1999, Sequence complexity and DNA curvature, Comput. Chem., 23, 263–74. [DOI] [PubMed] [Google Scholar]
- 44. Garcia S., Garnatje T., Kovařík A.. 2012, Plant rDNA database: ribosomal DNA loci information goes online, Chromosoma, 121, 389–94. [DOI] [PubMed] [Google Scholar]
- 45. Dadejova M., Lim K.Y., Souckova S.K.. 2007, Transcription activity of rRNA genes correlates with a tendency towards intergenomic homogenization in Nicotiana allotetraploids, New Phytol., 174, 658–68. [DOI] [PubMed] [Google Scholar]
- 46. Sochorova J., Coriton O., Kuderova A., Lunerova J., Chevre A.M., Kovarik A.. 2017, Gene conversion events and variable degree of homogenization of rDNA loci in cultivars of Brassica napus, Ann. Bot., 119, 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bauer N., Horvat T., Birus I., Vicic V., Zoldos V.. 2009, Nucleotide sequence, structural organization and length heterogeneity of ribosomal DNA intergenic spacer in Quercus petraea (Matt.) Liebl. and Q-robur L, Mol. Genet. Genomics, 281, 207–21. [DOI] [PubMed] [Google Scholar]
- 48. Suzuki A., Tanifuji S., Komeda Y., Kato A.. 1996, Structural and functional characterization of the intergenic spacer region of the rDNA in Daucus carota, Plant Cell Physiol., 37, 233–8. [DOI] [PubMed] [Google Scholar]
- 49. Mestrovic N., Plohl M., Mravinac B., Ugarkovic D.. 1998, Evolution of satellite DNAs from the genus Palorus - experimental evidence for the “library” hypothesis, Mol. Biol. Evol., 15, 1062–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.