Abstract
To ensure faithful genome propagation, mitotic cells alternate one round of chromosome duplication with one round of chromosome separation. Chromosome separation failure thus causes genome reduplication, which alters mitotic chromosome structure. Such structural alterations are well documented to impair mitotic fidelity following aberrant genome reduplication, including in diseased states. In contrast, we recently showed that naturally occurring genome reduplication does not alter mitotic chromosome structure in Drosophila papillar cells. Our discovery raised the question of how a cell undergoing genome reduplication might regulate chromosome structure to prevent mitotic errors. Here, we show that papillar cells ensure mitotic fidelity through interphase cohesin regulation. We demonstrate a requirement for cohesins during programmed rounds of papillar genome reduplication known as endocycles. This interphase cohesin regulation relies on cohesin release but not cohesin cleavage and depends on the conserved cohesin regulator Pds5. Our data suggest that a distinct form of interphase cohesin regulation ensures mitotic fidelity after genome reduplication.
INTRODUCTION
When cycling cells skip chromosome separation and then reenter S-phase, the genome is reduplicated. Such cycles are referred to as endocycles. Endocycles generate polyploid cells, which are common throughout nature (Fox and Duronio, 2013; Orr-Weaver, 2015). Following developmental endocycles or in pathological conditions, some polyploid cells return to mitosis (Levan and Hauschka, 1953; Fox et al., 2010; Davoli and de Lange, 2012). Division of such genome-reduplicated cells can generate genome instability through a variety of mechanisms, such as multipolar division or the formation of diplochromosomes, a mitotic chromosome structure that is a form of polyteny in which all products of replication are held together in one chromosome. Such diplochromosomes lead to mitotic errors when cells divide (Vidwans et al., 2002; Hassel et al., 2014; Schoenfelder et al., 2014; Chen et al., 2016; Stormo and Fox, 2016, 2017). Diplochromosomes have also been observed in tumor models in mice (Davoli et al., 2010) and following chemotherapeutic drug treatments in human cell culture (Blakeslee and Avery, 1937; Sumner, 1998).
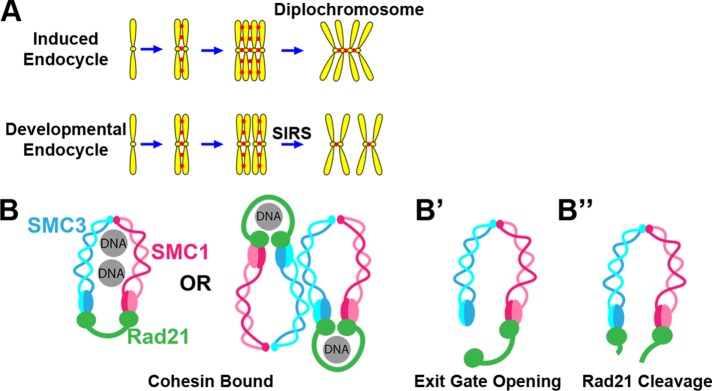
Previously, we developed parallel models of naturally occurring and experimentally induced endocycled Drosophila cell types (Stormo and Fox, 2016). One cell type, the rectal papillar precursors of the hindgut, undergo developmentally programmed endocycles before returning to mitosis (hereafter “papillar cells”). The second cell type, wing imaginal disc cells, can be induced to endocycle by transient heat-shock driven expression of the endocycle regulator Cdh1/fizzy-related (hereafter- “HS>fzr cells”). Both papillar and HS>fzr cells return to mitosis after endocycling, but chromosome configuration at anaphase onset is very different. In papillar cells, chromatids undergo preanaphase chromosome Separation Into Recent Sister pairs (SIRS) (Figure 1A) (Stormo and Fox, 2016). In contrast, chromatids in HS>fzr cells are arranged in diplochromosomes as anaphase begins (Figure 1A). Likely because of these structural differences, papillar cell mitosis is relatively error free, whereas HS>fzr cell mitosis is highly error prone. These results raised the question of what molecular mechanism accounts for the difference in chromosome structure between cells capable or incapable of SIRS.
FIGURE 1:
Potential cohesin regulation and impact on mitotic chromosome structure in two distinct Drosophila cell types that undergo endocycles. (A) Depiction of the outcome of two endocycles. In an induced endocycle, all sister chromatids are attached generating a diplochromosome. During a developmental endocycle, only recent sisters are attached at mitosis. (B) Two potential simplified depictions of the cohesin complex entrapping a pair of sister chromatids. (B′) DNA release by exit gate opening. (B″) DNA release by Rad21 cleavage.
One candidate regulator of reduplicated chromosome structure is the cohesin complex. Cohesins are responsible for holding sister chromatids together beginning at S-phase (when chromosomes are first duplicated) until anaphase (when chromosomes are separated). The cohesin complex consists of three main components: SMC1, SMC3, and Rad21. Several models have been developed for how cohesins bind DNA (Figure 1B) (Ivanov and Nasmyth, 2005; Haering et al., 2008; Nasmyth and Haering, 2009; Eng et al., 2015; Skibbens, 2016; Stigler et al., 2016). The cohesin complex interacts with DNA in multiple discrete steps. Cohesins can be loaded onto chromatin throughout the cell cycle via an “entry gate” (Murayama and Uhlmann, 2015), which may be most critical in G1 (Lengronne et al., 2006). G1 loaded cohesins are stabilized by replication fork passage and are maintained through G2 (Yeh et al., 2008; Rhodes et al., 2017). In many species, including Drosophila, the majority of cohesins are removed from chromosome arms early in mitosis by the prophase pathway, which opens an “exit gate” (Figure 1, B vs. B′) (Sumara et al., 2002; Shintomi and Hirano, 2009; Eichinger et al., 2013). Finally, any remaining cohesins, mostly at the centromere, are removed at the metaphase-to-anaphase transition by Separase-mediated Rad21 cleavage (Figure 1B″) (Uhlmann et al., 1999).
Current research efforts aim to understand the diversity of cohesin regulation across different cell types and developmental stages (Nasmyth and Haering, 2009; Skibbens, 2016). How cohesin regulation is adapted for genome reduplicated cells is poorly understood. Diplochromosomes (e.g., HS>fzr cells) form in cells that have undergone two rounds of replication and cohesion establishment with presumably no intervening removal of the cohesins (Vidwans et al., 2002; Stormo and Fox, 2016). It is unknown how papillar cells can avoid mitotic chromosome separation defects, as they also have diplochromosome-like polytene chromosomes prior to undergoing SIRS.
Here we investigate the role of cohesins on the structure of chromosomes in cells undergoing endocycles, using our two model cell types. Unlike other endocycled cells, such as the Drosophila salivary gland, these two cell types return to mitosis, which allows direct visualization of chromosome structure and the effects of cohesin regulation on mitosis. We find that in SIRS-capable papillar cells, cohesin exit gate opening during endocycles prevents formation of diplochromosomes. This interphase cohesin exit gate opening depends on the conserved cohesin regulator Pds5. These findings reveal new interphase cohesin regulation during endocycles and shed light on the structural regulation of chromosomes in genome reduplicated cells.
RESULTS
Cohesin cleavage is sufficient to separate reduplicated chromatids
Previous studies in genome-reduplicated cells have found cohesins to be dispensable for chromosome structure (Pauli et al., 2008). However, these studies focused on nonmitotic cells. Our previous work (Stormo and Fox, 2016) showed a major difference in mitotic fidelity between genome-reduplicated cells that are capable of SIRS and those that are not. We showed that tetraploid cells that are SIRS-deficient retain conjoined diplochromosomes at metaphase. However, we did not explore whether differential regulation of cohesins is responsible for the decreased mitotic fidelity in such cells with persistent diplochromosomes. We therefore examined the role of the cohesin complex in chromosome structure of mitotic polyploid cells, using our two previously established models.
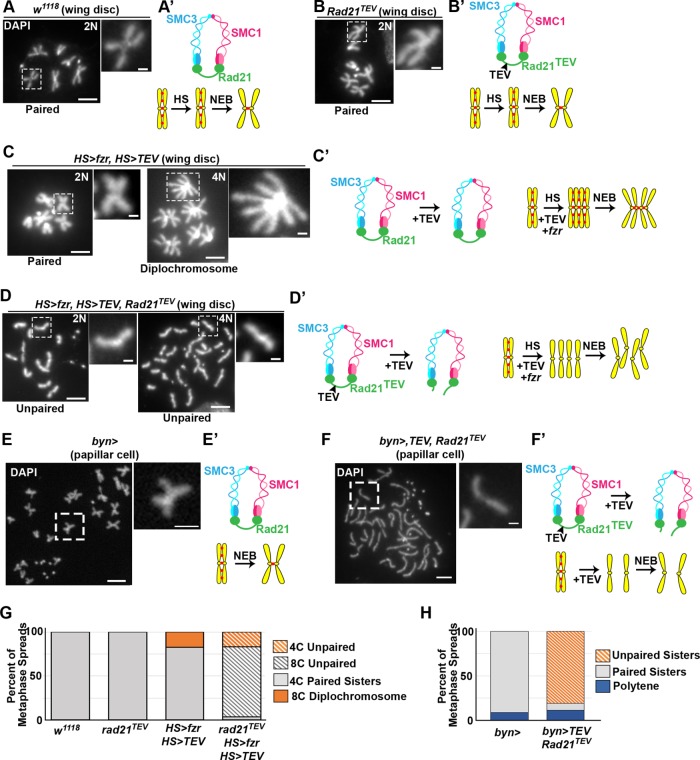
We first tested whether cleavage of the Rad21 cohesin subunit is sufficient to dissociate the conjoined diplochromosome configuration found in HS>fzr wing cells. To do this, we took advantage of an established system that enables heat-shock-inducible Rad21 cleavage (Rad21TEV, Materials and Methods). We first confirmed that heat shock (Figure 2, A and G) and Rad21TEV alone has no effect on chromosome structure (Figure 2, B and G). TEV-protease expression also has no effect on diploid or polyploid mitotic chromosomes when Rad21 is wild type (Figure 2, C and G). We next combined induced endocycles and cohesin cleavage by driving expression of both HS>fzr and HS>TEV transgenes using a single heat shock in a rad21TEV animals. In these animals endocycling still occurs, resulting in tetraploid cells, but these chromosomes lack cohesion between sisters and instead unpaired chromatids are visible (Figure 2, D and G). These data strongly suggest that diplochromosomes are held together by cohesin in the same manner as wild-type mitotic chromosomes. Further, we find that cohesin cleavage is sufficient to dissociate the conjoined chromatids found in diplochromosomes.
FIGURE 2:
Chromatids are held together by the canonical cohesin complex in endocycled cells. Representative wing imaginal disc cell (A–D) and papillar cell (E, F) chromosome spreads. Corresponding diagrams depict cohesin state. 4’6-Diamidino-2-phenylindole (DAPI) marks DNA. Ploidy is indicated (N value, top right). (A) Heat-shocked wild-type (w1118). (B) Cells in which rad21TEV is the only source of Rad21. (C) HS>fzr plus HS>TEV protease. (D) HS>fzr plus HS>TEV protease in a rad21TEV background. (E) Wild-type (byn>gal4 alone). (F) byn>gal4 driving upstream activating sequence (UAS)>TEV in a rad21TEV background. Scale bar = 5 µm (main images), 1 µm (insets). (G) Quantification of metaphase spreads in A–D. From left to right, N = 77, 11, 34, and 59 cells per genotype. (H) Quantification of metaphase spreads in E and F. From left to right, N = 22, 52 cells pergenotype.
We next performed a similar experiment in papillar cells, which lack conjoined metaphase chromosomes. It was possible that these cells are able to undergo SIRS because they lack standard cohesins. We tested whether papillar chromatid pairs are held together by cohesins. As in the wing disc, following Rad21TEV cleavage all chromosome cohesion is lost, and we observe individual chromatids (Figure 2, E vs. F and H). These data show that in both cells with induced endocycles, which result in diplochromosomes, and in papillar cell endocycles, which result in paired sisters, Rad21 cleavage is sufficient to separate the products of replication at metaphase.
Opening the SMC3-Rad21 exit gate is required for SIRS and subsequent mitotic fidelity
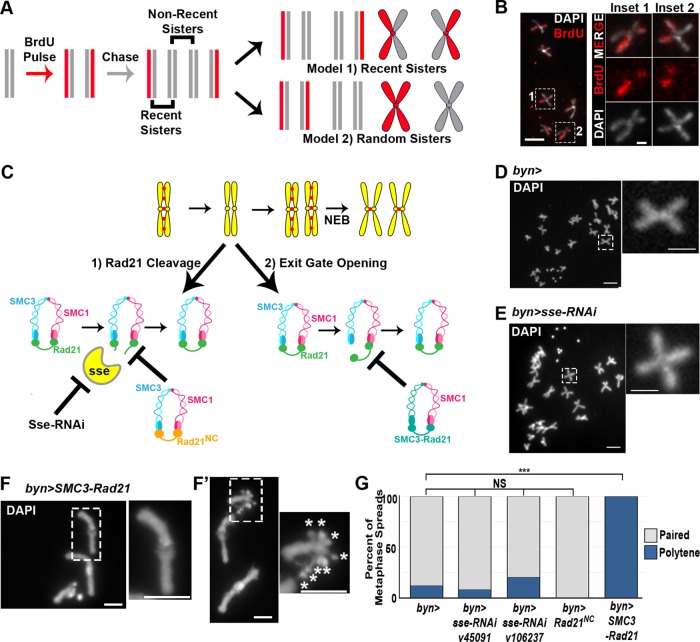
Cohesion regulation during mitotic cell cycles ensures that chromosomes are attached specifically to their sisters and not to other chromosomes, regardless of proximity within the nucleus or homology. Moreover, following SIRS, papillar chromosomes lose polytene structure and are attached to only a subset of their sister chromatids prior to anaphase (Figure 1A). We previously suggested that papillar chromatids were attached via cohesion with only the most recent sister chromatid (Stormo and Fox, 2016), which we had evidence for based on the symmetric appearance of random or radiation-induced chromosome breaks at the same location on adjacent chromatids (Bretscher and Fox, 2016). To further examine whether papillar chromatids were attached in recent rather than random sister pairs (Figure 3A, model 1 vs. model 2), we pulse-labeled chromatids with 5-bromo-2′-deoxyuridine (BrdU) (Figure 3A, Materials and Methods). Our data are consistent with papillar chromatids pairing with their recent sisters (Figure 3B).
FIGURE 3:
Cohesin exit gate opening is required for SIRS. (A) Model of BrdU labeling scheme in papillar cells. Vertical lines, DNA strands. X-shapes, chromatids. Gray, unlabeled DNA, red, BrdU labeled DNA. If papillar chromosomes are composed of recent sisters, then each chromosome would have one labeled chromatid and one unlabeled chromatid (model 1). If sister chromatids are arranged randomly at metaphase, then chromosomes within the same cell would be composed of labeled and unlabeled chromosomes (model 2). Also see Materials and Methods. (B) Representative group of labeled papillar chromosomes. DAPI (DNA, white), BrdU, red. Insets, close ups of 2 chromosomes. Inset locations indicated by hatched rectangles in low magnification image for all panels in this figure. (C) Model depicting when we hypothesize cohesins are removed in papillar cells and two potential mechanisms: exit gate opening and Rad21 cleavage. Experimental methods to block these two mechanisms are shown. (D–F) Representative metaphase chromosome spreads of papillar cells. (D) byn>gal4 control. (E) byn>gal4 plus separase-RNAi driven throughout development. (F) byn>gal4 plus SMC3-Rad21 driven throughout development. Asterisk denotes X-chromosome centromeres. (G) Quantification of percentage of metaphase spreads in each class of the indicated genotypes. From left to right, N = 17, 64, 10, 8, 10 cells per genotype.
We next sought to uncover the mechanism that enables papillar chromatids to establish cohesion with only their most recent sister. We hypothesized that, during papillar endocycles, cohesin is removed between each sister chromatid after each round of replication. Such interphase cohesin regulation could occur through one of two pathways. First, cohesins could be removed from papillar chromosomes during each endocycle by Rad21 destruction, similarly to its destruction at anaphase (Figure 3C, “Rad21 Cleavage”). Second, cohesins could be removed during each endocycle by exit gate opening (Figure 3C, “Exit gate opening”), similarly to how nonpericentric cohesin is removed in prophase.
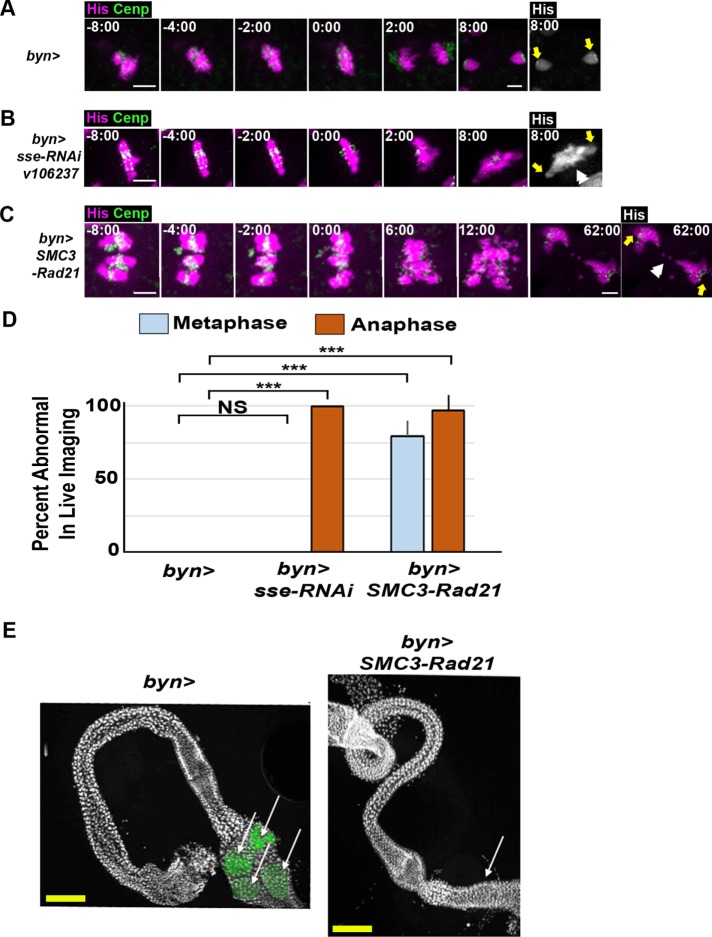
We first tested whether Rad21 destruction takes place during papillar endocycles. Normally, Rad21 is cleaved by Separase at the onset of anaphase. Our previous work found no evidence of mitosis during papillar endocycles (Fox et al., 2010), but we reasoned that it was possible for Separase to be regulated in a noncanonical manner in papillar cells, so that it was active during endocycles. To test this hypothesis, we first knocked down separase using two separate RNA interference (RNAi) lines (Materials and Methods). We then examined the structure of chromosomes in those cells during the first mitosis after papillar endocycles and SIRS. We find that in the absence of Separase, papillar chromosomes undergo SIRS normally, as chromosomes are arranged in pairs at metaphase that are indistinguishable from wild type (Figure 3, D vs. E and G). To ensure that knockdown of separase was successful, we performed live imaging on separase RNAi papillar cells undergoing mitosis. separase RNAi papillar chromosomes form a normal metaphase plate with neatly aligned pairs of chromatids similarly to wild-type cells (Figure 4, A vs. B, –2:00). However, mitosis is aberrant in separase RNAi cells because, as expected, chromatid separation fails, resulting in a DNA bridge (Figure 4, A vs. B, 2:00, 8:00, and D). As an alternative approach, we expressed a previously established noncleavable Rad21 that lacks the Separase cleavage site (UAS>rad21NC; Materials and Methods; Urban et al., 2014). Here, too, chromosome structure at the first mitosis is unaffected (Figure 3G). Together, these data strongly suggest that papillar cells do not require Rad21 cleavage to remove cohesion during endocycles.
FIGURE 4:
Cohesin exit gate opening is required for mitotic fidelity and organ development in papillar cells. (A–C) Time lapse of papillar cell mitosis from the indicated genotypes. His-2av-GFP, magenta; CenpC-tomato, green; time, minutes from anaphase onset. Final panel depicts histone channel only; yellow arrows show spindle pole position. White double arrowhead indicates DNA bridge. Scale bar = 5 µm (main images), 1 µm (insets). (D) Quantification of the percentage of cells with aberrant metaphase and anaphases of the indicated genotypes (+SEM, from left to right, N = 12, 8, 27 metaphase and N = 12, 11, 39 cells per genotype). (E) Representative images of the adult hindgut of animals of the indicated genotypes (from N = 12 control and 8 SMC3-Rad21 animals). DAPI (DNA, white); papillae pseudocolored (green). Arrows indicate the location (or absence) of papillar structures. Yellow scale bar = 12.5 µm.
As an alternative to cohesin cleavage, we reasoned that cohesin turnover via exit gate opening could account for the interphase cohesin regulation that we hypothesize is responsible for paired sister chromatids at papillar metaphase. To test this hypothesis, we used a previously established transgenic construct in which Rad21 and SMC3 are fused via a linker region (Eichinger et al., 2013) (Figure 3C). This fusion can load onto chromosomes normally but cannot be removed from chromatin because the fusion closes the exit gate. This construct is still cleaved normally at anaphase.
To test whether exit gate opening during endocycles was required for normal papillar chromosome structure, we expressed UAS>SMC3-Rad21 and looked at chromosomes during the first metaphase after SIRS. Chromosome structure is often substantially altered in these cells. Specifically, chromosomes persist as polytene chromosomes, suggesting SIRS does not take place (Figure 3, F, F′, and G). These chromosome phenotypes do not appear to disrupt papillar endocycles, as in metaphase spreads where the X centromeres are separate (as we described before for papillar cells; Stormo and Fox, 2016), we can count 8 X centromeres, indicating these cells are octoploid (Figure 3F′, asterisks in inset). The finding that cohesins can persist on papillar chromosomes without disrupting multiple endocycle S-phases is consistent with the observation that cohesin can remain associated with chromosomes during DNA replication (Rhodes et al., 2017). The chromosomal phenotype in papillar cells of SMC3-Rad21 animals suggests cohesin exit gate opening, most likely during endocycles, is important for SIRS.
We were surprised that, at the first metaphase post-SIRS, SMC3-Rad21 papillar cells displayed the haploid number of observed distinct chromosomes, as opposed to the diploid number. This implies that blocking cohesin exit gate opening can also promote ectopic homologue–homologue pairing. We had instead expected that homologues would remain separate at mitosis despite persistent sister-chromatid cohesion, because homologous chromosomes are not normally cohesed but are instead associated by somatic pairing mechanisms. In Drosophila, somatic homologue pairing is antagonized by condensins (Smith et al., 2013). At mitosis, condensins overcome the attractive forces of somatic pairing to drive homologues apart, but sisters remain attached by cohesins. We suspect SMC3-Rad21 expression antagonizes this condensin activity. Interestingly, cohesin and condensin II can antagonize each other's functions in alignment of sister chromatids in cultured Drosophila cells (Senaratne et al., 2016).
We next analyzed the consequence of the disrupted mitotic chromosome structure phenotype of SMC3-Rad21 on papillar cell mitosis by performing live imaging. SMC3-Rad21 papillar cells fail to form a proper metaphase plate with pairs of sister chromatids bioriented. Instead, polytene chromosomes are still evident until anaphase (Figure 4C, –2:00). Subsequently, anaphase of SMC3-Rad21 papillar cells is highly error prone (Figure 4C, 6:00, D). Given these mitotic defects, we examined the consequence of such error-prone divisions on tissue development by examining adult hindgut structure. Unlike in control animals, which display four rectal papillar structures in adults, SMC3-Rad21 animals completely lack obvious rectal papillar structures (Figure 4E). This suggests that SMC3-Rad21 cells do not survive the extremely aberrant mitotic divisions and thus fail to produce cells which are normally required for their construction (Fox et al., 2010; Schoenfelder et al., 2014). In contrast, the rest of the hindgut, which is formed by multiple rounds of diploid mitoses (Fox and Spradling, 2009; Sawyer et al., 2017) and also expresses SMC3-Rad21 under the same byn>Gal4 driver, appears unaffected. We hypothesize that the lack of tissue-level phenotype in diploid cells of SMC3-Rad21 animals is due to Separase-mediated cohesin cleavage of chromatids at anaphase in these cells. These results suggest that, in papillar cells, the distinct processes of genome reduplication followed by SIRS creates an additional chromosome structural challenge for mitotic fidelity. We propose that as these cells undergo endocycles, cohesin exit gate opening is important for later dissolution of homologue–homologue pairing, as well as for maintaining that chromatids are only cohesed with their most recent sister. Failure to properly regulate cohesins prior to mitosis leads to severe mitotic infidelity and organ malformation.
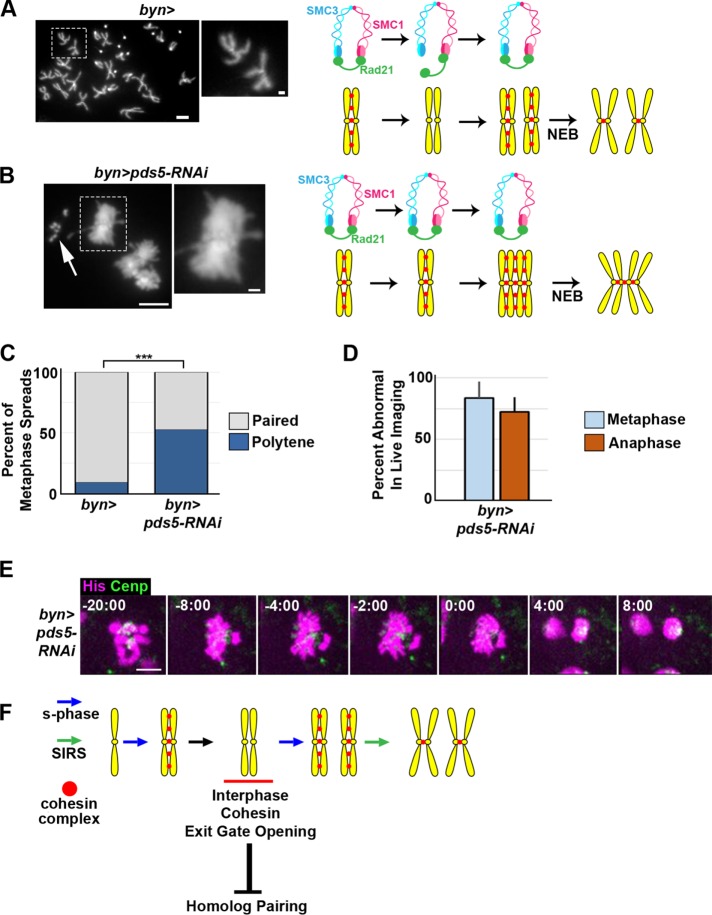
Pds5 is required for interphase chromosome regulation
Our data suggest that cohesin removal in cells undergoing endocycles followed by SIRS is crucial for subsequent mitotic fidelity. We next sought to determine the regulation of cohesin exit gate opening. From a candidate analysis of known cohesin regulators, our most striking result was with Pds5. In multiple organisms including Drosophila (Dorsett et al., 2005), Pds5 is required for sister chromatid cohesion. At the first metaphase post-SIRS, pds5 animals exhibit ectopic homologue pairing and chromatids remain in a polytene configuration at metaphase (Figure 5, A vs. B and C), as in SMC3-Rad21 animals. However, pds5 papillar chromosomes contrasted with those of SMC3-Rad21 flies in one important respect. In cells expressing SMC3-Rad21, chromosome arms are often closely aligned in a classic polytene configuration (Figure 3, F and F′). In contrast, following knockdown of pds5, chromosome arms separate, but all centromeres remain together, a configuration more similarl to cells with diplochromosomes (Figure 5, B and C). As with SMC3-Rad21 flies, live imaging shows a failure to separate chromatids in pds5 animals, as evidenced by clusters of DNA that move in tandem and a failure to form a metaphase plate (Figure 5, D and E). Also as with SMC3-Rad21 flies, papillar ploidy appears unaffected by pds5 RNAi, as evidenced by our ability to count ∼8 separate fourth chromosomes in otherwise polytene pds5 RNAi cells (Figure 5B, arrow). These results strongly suggest that pds5-mediated cohesin exit gate opening at centromeres is key in interphase cohesin regulation during the premitotic endocycles of papillar cells. Deficiencies in this mechanism contribute to mitotic errors in cells with genome reduplication.
FIGURE 5:
Pds5 is required for cohesin regulation during papillar endocycles. (A, B) Representative metaphase chromosome spreads of wild-type and pds5-RNAi papillar cells. Corresponding diagrams depict cohesin state. DAPI marks DNA. (C) Quantification of percentage of metaphase spreads in each class. N = 44 control and 19 pds5 RNAi cells. (D) Quantification of mean error rate in live imaging of pds5-RNAi animals (+SEM, from N = 9 cells). (E) Time lapse of papillar cell mitosis in a bynGal4 plus UAS pds5-RNAi animal. His-2av-GFP, magenta; CenpC-tomato, green; time, minutes from anaphase onset. Scale bar = 5 µm (main images), 1 µm (insets). (F) Proposed model of cohesin regulation and function in papillar cells during endocycles, SIRS, and the first subsequent mitosis.
DISCUSSION
Cohesins typically hold together all products of S-phase through interphase until cleavage of the complex at anaphase. Here our results imply that cohesin removal mechanisms can be repurposed in cells undergoing endocycles to prevent subsequent mitotic infidelity. In Drosophila papillar cells, our results suggest that chromosomes lose cohesion and then reestablish it with the most recent sister chromatid during endocycles (Figure 5F). We thus propose that this cohesin regulation occurs during interphase but involves a repurposing of the cohesin exit gate opening mechanism normally used during prophase. Our work also reveals a role for cohesin exit gate opening in antagonizing pairing between homologous chromosomes in cells that endocycle (Figure 5F).
Does such interphase cohesin regulation that we propose to occur in papillar cells also occur in other polyploid cell types? Drosophila ovarian nurse cells partially separate chromatids but do not proceed to a full mitosis (Hammond and Laird, 1985; Dej and Spradling, 1999) and thus may also undergo some degree of interphase cohesin regulation. In nonmitotic polyploid cells such as salivary gland cells, the cohesin complex is present and dynamic (Gause et al., 2010; Cunningham et al., 2012). However, because these cells are nonmitotic, chromatids are never separated enough to observe constraint by cohesins. Our work here suggests that interphase cohesin regulation is definitely not a property of all polyploid cells, as cells with induced endocycles contain diplochromosomes, where four chromatids remain cohesed (Figure 2). The difference between naturally occurring and ectopically induced endocycles may account for mitotic defects associated with the latter. Additionally, we also uncover a surprising role for cohesin exit gate opening in antagonizing homologue pairing. While lack of this anti-pairing mechanism may contribute to the phenotypes we see in papillar cells with compromised exit gate opening, we note that cells with ectopic diplochromosomes do not pair (Figure 2) and exhibit similar mitotic defects to papillar cells with defective exit gate opening (Stormo and Fox, 2016). Future work can examine the connection between cohesin exit gate opening and known homologue pairing regulators such as condensins. Further, we note that while we heavily favor an interphase model of cohesin regulation, it is possible that only the cohesins between recent sister chromatids, and not any other cohesins, somehow resist the prophase pathway specifically during SIRS. Such a mechanism would still likely involve some differential marking (such as acetylation) of the cohesins between recent sisters, which would likely have to occur in the last endocycle (i.e., would still be an interphase mechanism).
With respect to molecular mechanisms of cohesin regulation, our results also revealed differences between pds5 knockdown and SMC3-Rad21 expression in papillar cells. We propose that SMC3-Rad21 represents a situation where cohesin complexes cannot be removed by the prophase pathway at onset of mitosis, and therefore chromosome arms remain attached. In contrast, pds5 knockdown blocks cohesin release during endocycles but not during prophase. Our results reveal a differential sensitivity for cohesin regulation at chromosome arms and centromeres in this RNAi condition. This could reflect that Pds5 in papillar cells is more essential for centromeric cohesion than for arm cohesion. Along these lines, kinase activity of polo kinase as well as Aurora B phosphorylation of SA1 and 2 participate with Pds5 and Wapl in the prophase pathway to remove arm cohesion (Sumara et al., 2002; Gimenez-Abian et al., 2004; Hauf et al., 2005; Kueng et al., 2006; Shintomi and Hirano, 2009), and it is possible that in pds5 RNAi animals these other arm cohesin regulators are still able to separate papillar polytene chromosome arms.
Future work can address regulation of pericentric cohesins during papillar cell endocycles. In mitotic cells, pericentromeric cohesin is not removed during the prophase pathway, because that region is protected by Shugoshin (Moore et al., 1998; Lee et al., 2004; Watanabe, 2005). Shugoshin directly antagonizes Wapl, a partner of Pds5 (Hara et al., 2014). If Pds5 is required to remove cohesins during endocycles, then how does it bypass Shugoshin? One possibility is that Shugoshin is not present in these cells during endocycles. If so, then this would allow the prophase pathway to clear cohesins from the entire chromosome, including centromeres, during each endocycle.
In disease, continued study of chromosome structure after genome reduplication is important because diplochromosomes are induced by common cancer therapeutics such as topoisomerase inhibitors (Hande, 1998; Sumner, 1998). Our data suggest that if cells prone to diplochromosomes regulated cohesins differently, so that only paired recent sisters were present at metaphase, then the rate of mitotic errors and aneuploidy in these cells would dramatically drop. Given our identification here of interphase cohesin regulation during papillar endocycles, papillar cells represent a valuable system for further study of cohesin regulation. Additionally, the importance of mitotic genome reduplicated cells in disease suggests that understanding chromosome structure in these cells may give insight into new therapies.
MATERIALS AND METHODS
Drosophila stocks
Stocks were obtained from the Bloomington Drosophila Stock Center (stock number in parentheses): w1118 (3605); His-2av-GFP (24163); vtdex14 (26165); vtdex8, rad21.271TEV-myc (27613); UAS>NLS-V5-TEV-NLS (27605); HS>NLS-V5-TEV-NLS (27612); pds5-RNAi (35632, previously validated by Kusch (2015) to cause meiotic recombination phenotypes); Gal80TS (7018); the Vienna Drosophila Stock Center (Dietzl et al., 2007): sse-RNAi (v45091, previously validated to phenocopy sse mutants in larval neuroblasts; Cipressa et al., 2016); sse-RNAi (v106237); or were kind gifts: tomato-Cenp-C; HS>fzr (Sigrist and Lehner, 1997); byn>gal4 (Singer et al., 1996); UAS>SMC3-vtd-GFP (Eichinger et al., 2013); UAS>rad21NC (Urban et al., 2014).
Drosophila culture and genetics
All flies were raised on standard media (Archon Scientific, Durham, NC). All experiments involving a UAS transgene (including RNAi) were performed at 29°C to maximize Gal4-mediated transgene expression. Heat shocks to induce fzr or TEV expression were performed on third instar larvae. Animals for these experiments were heat shocked in a vial at 37°C (water bath) for 20 min. In experiments involving inducible transgenic Rad21 cleavage, endogenous Rad21 was removed using two null rad21 mutant alleles in trans (vtdex14 and vtdex8). These mutant alleles were rescued by a ubiquitously expressed rad21 transgene containing a tobacco etch virus (TEV) cleavage site (rad21TEV, Pauli et al., 2008). This construct was then cleaved either in all cells (using a heat shock promoter, HS>TEV) or specifically in our cell type of choice using a UAS promoter (UAS>TEV). Animals were examined 10 h after HS>fzr expression, as this is the time where we previously established that mitosis resumes after heat shock in these animals.
For papillar cell experiments, we used the hindgut specific byn>gal4 driver to express transgenes. For all transgenes except for UAS TEV, we expressed these transgenes throughout development. To avoid prolonged UAS-TEV expression, we used previously established methods that rely on Gal80ts to repress Gal4 expression (Fox and Spradling, 2009; Fox et al., 2010) to confine expression of UAS-TEV to the period of endocycles (second larval instar) and not mitosis (which occurs much later: hours 24–48 post–puparium formation at 22°C).
Chromosome cytology
Chromosome preparations were performed as previously described (Gatti et al., 1994; Fox et al., 2010). We used enriched for metaphase cells by first incubating tissue in colcemid (Sigma, St. Louis, MO) at 50 μg/ml for 20 min in phosphate-buffered saline (PBS). Imaging was performed on a Zeiss Axio Imager 2 with a 63× oil immersion lens.
Live imaging
Live-imaging preparations were prepared as previously described (Prasad et al., 2007; Fox et al., 2010). Imaging was performed on a spinning disk confocal (Yokogawa CSU10 scanhead) on an Olympus IX-70 inverted microscope using a 60×/1.3 NA UPlanSApo Silicon oil, 488 and 568 nm Kr-Ar laser lines for excitation, and an Andor Ixon3 897 512 electron-multiplying charge-coupled device camera. The system was controlled by MetaMorph 7.7. Images were analyzed in ImageJ (Schneider et al., 2012).
BrdU feeding and staining
To determine whether papillar chromatids are associated with most recent sisters, we fed 1 mg/ml BrdU dissolved in PBS + food coloring for 1 h during the second instar stage, when papillar cells endocycle twice to reach 8C ploidy. The goal of this experiment was to occasionally label only the second-to-last S-phase in these cells so that one-fourth of all DNA strands at mitosis were BrdU labeled at mitosis. If one-fourth of DNA strands contain BrdU, we would expect one half of chromatids to be labeled and each chromosome to contain one labeled chromatid if recent sisters are paired (Figure 3A, Recent Sisters). In contrast, if sister chromatids are randomly paired, then we would see chromosomes in which neither chromatid was labeled, as well as chromosomes in which both chromatids were labeled, in the same cell (Figure 3A, Random Sisters). To ensure pulse labeling of chromosomes, larvae were washed in PBS after feeding, and animals with no food coloring in their gut were discarded.
To image BrdU in metaphase spreads, chromosome cytology and BrdU antibody staining (Rat anti-BrdU, Serotec 1:100, clone 3J9) was performed based on Sullivan and Karpen (2001) with slight modifications. In brief, hour 24–48 post–puparium formation (at 22°C) animals were dissected. Dissected tissue was incubated in 0.5% sodium citrate for 15 min then fixed on a coverslip in 11:11:2 methanol:acetic acid:H2O. Fix was removed and replaced with 10 µl of 45% acetic acid. The coverslip was then squashed on a positively charged slide (VWR, Radnor, PA) and then frozen in liquid nitrogen until the coverslip could be removed using a razor blade. Slides were then transferred to 95% ethanol at –20°C. All subsequent steps were performed directly on the slide, and tissue denaturation and BrdU antibody staining were performed as described previously (Fox and Spradling, 2009).
Statistics
All statistics were computed in Prism 7 (GraphPad, La Jolla, CA). Metaphase spreads were blinded and then scored. Metaphase spreads were compared with wild type using a chi-squared test on total counts. For live imaging, cells were averaged within animals, and then mean and standard error were calculated by averaging between animals. Means were compared using one-way analysis of variance (ANOVA). NS, not significant for p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001.
Acknowledgments
The following stock centers provided reagents used in this study: The Bloomington Stock Center (NIH P4OD018537) and the Vienna Drosophila Resource Center ( www.vdrc.at). We thank Stefan Heidmann and Raquel Oliveira for additional stocks. We thank Danny Lew and Beth Sullivan along with the Fox lab for reading the manuscripts and providing helpful comments. This project was supported by both National Institute of General Medical Sciences grant GM118447 and a Pew Scholar Award (Pew Charitable Trusts) to D.F.
Abbreviations used:
- BrdU
5-bromo-2′-deoxyuridine
- SMC
structural maintenance of chromosome
- TEV
tobacco etch virus
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-10-0582) on November 21, 2018.
REFERENCES
- Blakeslee AF, Avery AG. (1937). Methods of inducing doubling of chromosomes in plants by treatment with colchicine*. J Hered , 393–411. [Google Scholar]
- Bretscher HS, Fox DT. (2016). Proliferation of double-strand break-resistant polyploid cells requires Drosophila FANCD2. Dev Cell , 444–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Stout JR, Dharmaiah S, Yde S, Calvi BR, Walczak CE. (2016). Transient endoreplication down-regulates the kinesin-14 HSET and contributes to genomic instability. Mol Biol Cell , 2911–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipressa F, Morciano P, Bosso G, Mannini L, Galati A, Raffa GD, Cacchione S, Musio A, Cenci G. (2016). A role for Separase in telomere protection. Nat Commun , 10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MD, Gause M, Cheng Y, Noyes A, Dorsett D, Kennison JA, Kassis JA. (2012). Wapl antagonizes cohesin binding and promotes Polycomb-group silencing in Drosophila. Development , 4172–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, de Lange T. (2012). Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell , 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. (2010). Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell , 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej KJ, Spradling AC. (1999). The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development , 293–303. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature , 151–156. [DOI] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. (2005). Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development , 4743–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger CS, Kurze A, Oliveira RA, Nasmyth K. (2013). Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chromosomes during interphase and mitosis. EMBO J , 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng T, Guacci V, Koshland D. (2015). Interallelic complementation provides functional evidence for cohesin–cohesin interactions on DNA. Mol Biol Cell , 4224–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. (2013). Endoreplication and polyploidy: insights into development and disease. Development , 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Gall JG, Spradling AC. (2010). Error-prone polyploid mitosis during normal Drosophila development. Genes Dev , 2294–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Spradling AC. (2009). The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell , 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Bonaccorsi S, Pimpinelli S. (1994). Looking at Drosophila mitotic chromosomes. In: Methods in Cell Biology, ed. Lawrence SBG, Eric AF , Amsterdam: Elsevier/Academic Press, 371–391. [DOI] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D. (2010). Dosage-sensititve regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol , 4940–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Abían JF, Sumara I, Hirota T, Hauf S, Gerlich D, la Torre de C, Ellenberg J, Peters J-M. (2004). Regulation of sister chromatid cohesion between chromosome arms. Curr Biol , 1187–1193. [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas A-M, Arumugam P, Metson J, Nasmyth K. (2008). The cohesin ring concatenates sister DNA molecules. Nature , 297–301. [DOI] [PubMed] [Google Scholar]
- Hammond MP, Laird CD. (1985). Chromosome structure and DNA replication in nurse and follicle cells of Drosophila melanogaster. Chromosoma , 267–278. [DOI] [PubMed] [Google Scholar]
- Hande KR. (1998). Etoposide: four decades of development of a topoisomerase II inhibitor. Eur J Cancer , 1514–1521. [DOI] [PubMed] [Google Scholar]
- Hara K, Zheng G, Qu Q, Liu H, Ouyang Z, Chen Z, Tomchick DR, Yu H. (2014). Structure of cohesin subcomplex pinpoints direct shugoshin–Wapl antagonism in centromeric cohesion. Nat Struct Mol Biol , 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassel C, Zhang B, Dixon M, Calvi BR. (2014). Induction of endocycles represses apoptosis independently of differentiation and predisposes cells to genome instability. Development , 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Roitinger C, Koch B, Dittrich CM, Mechtler K, Peters J-M. (2005). Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol , e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K. (2005). A topological interaction between cohesin rings and a circular minichromosome. Cell , 849–860. [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters J-M. (2006). Wapl controls the dynamic association of cohesin with chromatin. Cell , 955–967. [DOI] [PubMed] [Google Scholar]
- Kusch T. (2015). Brca2–Pds5 complexes mobilize persistent meiotic recombination sites to the nuclear envelope. J Cell Sci , 717–727. [DOI] [PubMed] [Google Scholar]
- Lee JY, Dej KJ, Lopez JM, Orr-Weaver TL. (2004). Control of centromere localization of the MEI-S332 cohesion protection protein. Curr Biol , 1277–1283. [DOI] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. (2006). Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell , 787–799. [DOI] [PubMed] [Google Scholar]
- Levan A, Hauschka TS. (1953). Endomitotic reduplication mechanisms in ascites tumors of the mouse. J Natl Cancer Inst , 1–43. [PubMed] [Google Scholar]
- Moore DP, Page AW, Tang TT, Kerrebrock AW, Orr-Weaver TL. (1998). The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J Cell Biol , 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Uhlmann F. (2015). DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell , 1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. (2009). Cohesin: its roles and mechanisms. Annu Rev Genet , 525–558. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL. (2015). When bigger is better: the role of polyploidy in organogenesis. Trends Genet , 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. (2008). Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell , 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Jang ACC, Starz-Gaiano M, Melani M, Montell DJ. (2007). A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat Protoc , 2467–2473. [DOI] [PubMed] [Google Scholar]
- Rhodes JDP, Haarhuis JHI, Grimm JB, Rowland BD, Lavis LD, Nasmyth KA. (2017). Cohesin can remain associated with chromosomes during DNA replication. Cell Rep , 2749–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JK, Cohen E, Fox DT. (2017). Interorgan regulation of Drosophila intestinal stem cell proliferation by a hybrid organ boundary zone. Development , 4091–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder KP, Montague RA, Paramore SV, Lennox AL, Mahowald AP, Fox DT. (2014). Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development , 3551–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senaratne TN, Joyce EF, Nguyen SC, Wu CT. (2016). Investigating the interplay between sister chromatid cohesion and homolog pairing in Drosophila nuclei. PLoS Genet , e1006169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. (2009). Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl–Pds5 and Sgo1. Genes Dev , 2224–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. (1997). Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell , 671–681. [DOI] [PubMed] [Google Scholar]
- Singer JB, Harbecke R, Kusch T, Reuter R, Lengyel JA. (1996). Drosophila brachyenteron regulates gene activity and morphogenesis in the gut. Development , 3707–3718. [DOI] [PubMed] [Google Scholar]
- Skibbens RV. (2016). Of rings and rods: regulating cohesin entrapment of DNA to generate intra- and intermolecular tethers. PLoS Genet , e1006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HF, Roberts MA, Nguyen HQ, Peterson M, Hartl TA, Wang X-J, Klebba JE, Rogers GC, Bosco G. (2013). Maintenance of interphase chromosome compaction and homolog pairing in Drosophila is regulated by the condensin Cap-H2 and its partner Mrg15. Genetics , 127–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigler J, Çamdere GÖ, Koshland DE, Greene EC. (2016). Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep , 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo BM, Fox DT. (2016). Distinct responses to reduplicated chromosomes require distinct Mad2 responses. Elife , e15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo BM, Fox DT. (2017). Polyteny: still a giant player in chromosome research. Chromosome Res , 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B, Karpen G. (2001). Centromere identity in Drosophila is not determined in vivo by replication timing. J Cell Biol , 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters J-M. (2002). The dissociation of cohesin from chromosomes in prophase is regulated by polo-like kinase. Mol Cell , 515–525. [DOI] [PubMed] [Google Scholar]
- Sumner AT. (1998). Induction of diplochromosomes in mammalian cells by inhibitors of topoisomerase II. Chromosoma , 486–490. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. (1999). Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature , 37–42. [DOI] [PubMed] [Google Scholar]
- Urban E, Nagarkar-Jaiswal S, Lehner CF, Heidmann SK. (2014). The cohesin subunit Rad21 is required for synaptonemal complex maintenance, but not sister chromatid cohesion, during Drosophila female meiosis. PLoS Genet , e1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidwans SJ, DiGregorio PJ, Shermoen AW, Foat B, Iwasa J, Yakubovich N, O'Farrell PH. (2002). Sister chromatids fail to separate during an induced endoreplication cycle in Drosophila embryos. Curr Biol , 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. (2005). Shugoshin: guardian spirit at the centromere. Curr Opin Cell Biol , 590–595. [DOI] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom K. (2008). Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol , 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]