Abstract
During the course of a bacterial infection, cells are exposed simultaneously to a range of bacterial and host factors, which converge on the central transcription factor nuclear factor (NF)-κB. How do single cells integrate and process these converging stimuli? Here we tackle the question of how cells process combinatorial signals by making quantitative single-cell measurements of the NF-κB response to combinations of bacterial lipopolysaccharide and the stress cytokine tumor necrosis factor. We found that cells encode the presence of both stimuli via the dynamics of NF-κB nuclear translocation in individual cells, suggesting the integration of NF-κB activity for these stimuli occurs at the molecular and pathway level. However, the gene expression and cytokine secretion response to combinatorial stimuli were more complex, suggesting that other factors in addition to NF-κB contribute to signal integration at downstream layers of the response. Taken together, our results support the theory that during innate immune threat assessment, a pathogen recognized as both foreign and harmful will recruit an enhanced immune response. Our work highlights the remarkable capacity of individual cells to process multiple input signals and suggests that a deeper understanding of signal integration mechanisms will facilitate efforts to control dysregulated immune responses.
INTRODUCTION
During the course of a bacterial infection, host cells are confronted with a complex and dynamic environment brimming with diverse innate immune stimuli. In addition to stimulation with several types of pathogen-associated molecular patterns (PAMPs), such as lipopolysaccharide (LPS), host cells also simultaneously encounter cytokines and chemokines, such as those produced by stromal and infiltrating immune cells. From this complex environmental milieu, cells must make a decision about how to proceed—whether to initiate, limit, perpetuate, or amplify the immune response. Although the cellular response to isolated innate immune stimuli has been extensively characterized, we still lack a comprehensive understanding of how cellular decision-making emerges in the context of complex combinations of stimuli, a situation more akin to what a cell may perceive in its native environment.
Cells have the potential to integrate their response to multiple stimuli at several levels from signaling and transcription to cytokine production. In fact, many studies have shown that combinations of innate immune stimuli induce synergistic cellular responses, at the level of either transcription or cytokine secretion—meaning that a system treated with two stimuli can respond more strongly than would be predicted from the response to either stimulus individually (Napolitani et al., 2005; Natarajan et al., 2006; Bagchi et al., 2007; Qiao et al., 2013; Lin et al., 2017). While these synergistic responses reflect the ability of cells to engage in complex processing of extracellular signals, it is less clear how such responses are orchestrated by a limited set of upstream signaling pathways, several of which are activated by multiple types of stimuli.
Recent advances in live-cell imaging have revealed extensive heterogeneity in the activation of signaling pathways in single cells (Purvis et al., 2012; Purvis and Lahav, 2013; Albeck et al., 2013; Dalal et al., 2014; Ryu et al., 2016). Such studies have found that cells can transmit information about the type and dose of a stimulus in the temporal activation patterns of several signaling proteins, including nuclear factor (NF)-κB, a central regulator of innate immune responses (Tay et al., 2010; Batchelor et al., 2011; Sung et al., 2014). While the bulk of these studies have been carried out under single input conditions, a recent study characterized NF-κB dynamics in response to a combination of Toll-like receptor (TLR) inputs, LPS and PAM3CSK4 (Kellogg et al., 2017). In response to dual stimulation, cells were found to display NF-κB dynamics that were characteristic of either LPS or PAM3CSK4, indicating that cells respond to either individual stimulus, but not both together. This work further suggests that the fraction of cells in each group may be the basis for downstream synergistic responses rather than the integration of both stimuli at the single-cell level. However, it remains unclear whether this form of cellular decision-making in response to multiple stimuli is generalizable, either across NF-κB–activating stimuli or to other signaling pathways.
NF-κB activation occurs in response to PAMPs, such as LPS and PAM3CSK4, but can also be triggered in response to cytokines, most notably tumor necrosis factor (TNF), a critical proinflammatory cytokine produced by host cells during bacterial infection. Although LPS and TNF engage distinct receptors, they both initiate signaling cascades that lead to the degradation of an NF-κB inhibitor, allowing NF-κB to translocate to the nucleus and induce expression of a host of downstream cytokines and chemokines. Furthermore, in single cells both TNF and LPS have been shown to induce distinct patterns of NF-κB signaling dynamics that have been linked to differences in the gene expression of cytokines and chemokines (Lee et al., 2014; Lane et al., 2017). Moreover, LPS has been shown to induce NF-κB to produce TNF (Xaus et al., 2000; Covert et al., 2005). Therefore, in a physiological setting (e.g., during the course of a bacterial infection), it would be very likely for cells to be exposed to LPS and TNF simultaneously. Furthermore, it has been proposed that, for full innate immune activation to occur, cells must register both the presence of a foreign microbe “stranger” and that this presence is causing distress or “danger” signals, such as TNF, to be released by other cells (Matzinger, 1994; Gallucci and Matzinger, 2001). How do cells integrate and process different stimuli when both occur simultaneously, and how does the cellular response to a combined stimulus differ from the response to either stimulus individually (Figure 1A)?
FIGURE 1:
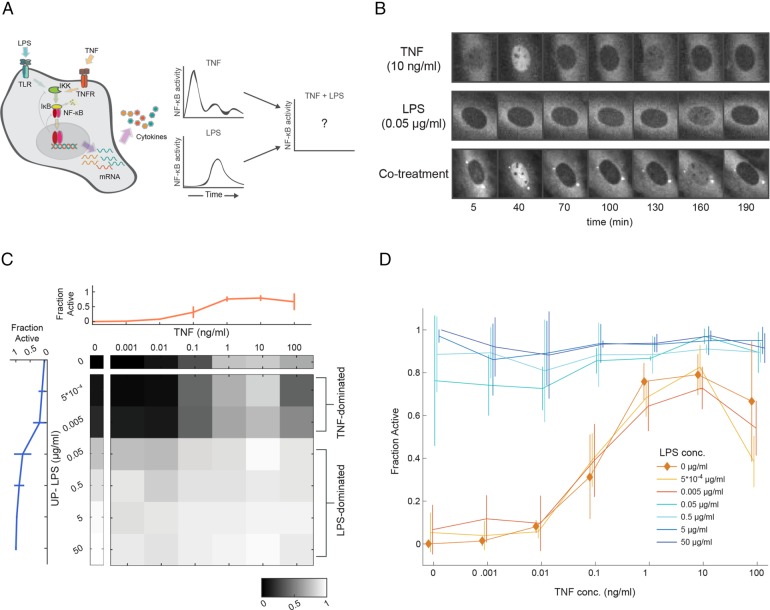
NF-κB population activation in response to single and combined stimuli. (A) Schematic depicting the NF-κB–activating stimuli that were measured in this study, along with what is known about single-cell NF-κB dynamics for each condition. (B) RelA−/− 3T3 fibroblasts expressing a p65-dsRed construct were stimulated with TNF (10 ng/ml), LPS (0.05 μg/ml), and a combination of the two. p65 reporter localization in representative cells for each condition are shown. (C) Heatmap showing the fraction of the cell population for which NF-κB is activated (as described by visible nuclear translocation at any time point during imaging) for all concentrations of TNF, LPS, and combinations. The LPS (blue) and TNF (orange) single stimulus concentrations are also shown with error bars (mean ± SD), N = 5908 cells total. (D) The active fraction is plotted as a function of TNF concentration, at several different concentrations of LPS. The trace representing TNF stimulation without LPS present is highlighted with diamonds. At LPS concentrations below 0.05 μg/ml, the population responds in the graded response of TNF and at or above that concentration, population activation is dominated by the LPS response (lines are mean ± SD).
To address the decision-making capacity of a cell in response to both a host and pathogen signal, specifically in the case where both signals converge on the central regulator NF-κB (Pahl, 1999), we stimulated cells over a broad concentration range (encompassing six orders of magnitude) for TNF and LPS, both separately and together. We then measured NF-κB nuclear translocation in individual cells over time, coupled this with single-molecule RNA-FISH (smFISH) measurements of mRNA expression after signaling had been induced, and, finally, used bulk cytokine measurements to characterize the downstream consequences of these single-cell behaviors on the population.
RESULTS
Cellular NF-κB activation in response to combined stimuli is largely determined by the concentration of LPS
Our reporter for single-cell NF-κB activation was the nuclear translocation of a p65-dsRed fusion protein, expressed in a mouse 3T3 fibroblast cell line as described previously (Nelson et al., 2004; Lee et al., 2009). We demonstrated previously that the NF-κB response to TNF or LPS was predominantly related to the number of active cells in the population rather than a change in the activity of each individual cell (Tay et al., 2010; Gutschow et al., 2013). Our first goal was therefore to determine the percentage of cells that were activated by either TNF or LPS separately, as well as by both together. Both stimuli activate NF-κB with different kinetics (Figure 1B), therefore our live-cell imaging approach was essential to allow us follow individual cells over time and capture activation status independent of the timing of the response—a cell was classified as active if the reporter was visually observed in the nucleus at any time over a 6-h time course. We chose a concentration range for each stimulus that spanned the full range of NF-κB activation, varying over six orders of magnitude (Figure 1C). Consistent with our previous measurements, cell activation in the presence of TNF alone began at 0.01 ng/ml and gradually rose to a peak activation fraction of ∼80% starting at 1 ng/ml. In contrast, we observed that LPS stimulation is better described as “all or none,” as concentrations of 0.05 μg/ml or higher lead to activation of ∼80% of the cells, while lower concentrations are essentially inert. When concentrations of 0.05 μg/ml LPS are used, the translocation of NF-κB takes on average 2 h; this lag has previously been demonstrated to be TLR4 dependent (Gutschow et al., 2013). When both LPS and TNF were introduced together, NF-κB activation fractions reflected those seen with LPS stimulation alone. At LPS concentrations of 0.05 μg/ml or higher, almost all of the cells were activated regardless of TNF concentration. At lower LPS concentrations, the percentage of active cells largely depended on the TNF concentration, with minimal contribution to activity by LPS (Figure 1, C and D). In terms of NF-κB activation, above a certain threshold concentration the PAMP stimulus is saturating and pushes the bulk of the population toward NF-κB activation, independent of the TNF stimulus concentration.
NF-κB activation dynamics in response to stimulus combinations
Our results regarding NF-κB activation status in response to combined stimuli suggest that cells are LPS-like in their NF-κB response under most concentrations of ligand. However, the single-cell dynamics of p65-dsRed translocation have been shown to differ substantially under LPS and TNF stimulation conditions (Tay et al., 2010; Sung et al., 2014; Kellogg et al., 2017). Therefore, we next considered whether the presence of combined stimuli might be better represented in the dynamics of NF-κB rather than simply in its activation fraction. We compared NF-κB dynamics across individual and combined stimulus conditions to determine whether in the presence of both stimuli the average NF-κB dynamics of the population are more LPS or TNF-like.
Differences in phase and frequency of NF-κB oscillations in single cells make it challenging to average this time-series data; therefore, to compare NF-κB time-series data across conditions, we used dynamic time warping (DTW) and DTW barycenter averaging (DBA) (Figure 2A; see Materials and Methods). DTW is analogous to the alignment of biological sequences, but instead of nucleotides or amino acids, DTW aligns real-valued time-series data (Aach and Church, 2001). DBA determines a mean trace for sequences aligned using DTW (Petitjean et al., 2011). This approach allowed us to computationally align the single-cell NF-κB traces for a given condition and determine the mean of the population (Figure 2A). Both time-warped and non–time-warped single-cell NF-κB dynamics along with the mean trace for the population are shown for all concentrations of both TNF and LPS individually, as well as in combination (Figure 2B and Supplemental Figure S1).
FIGURE 2:
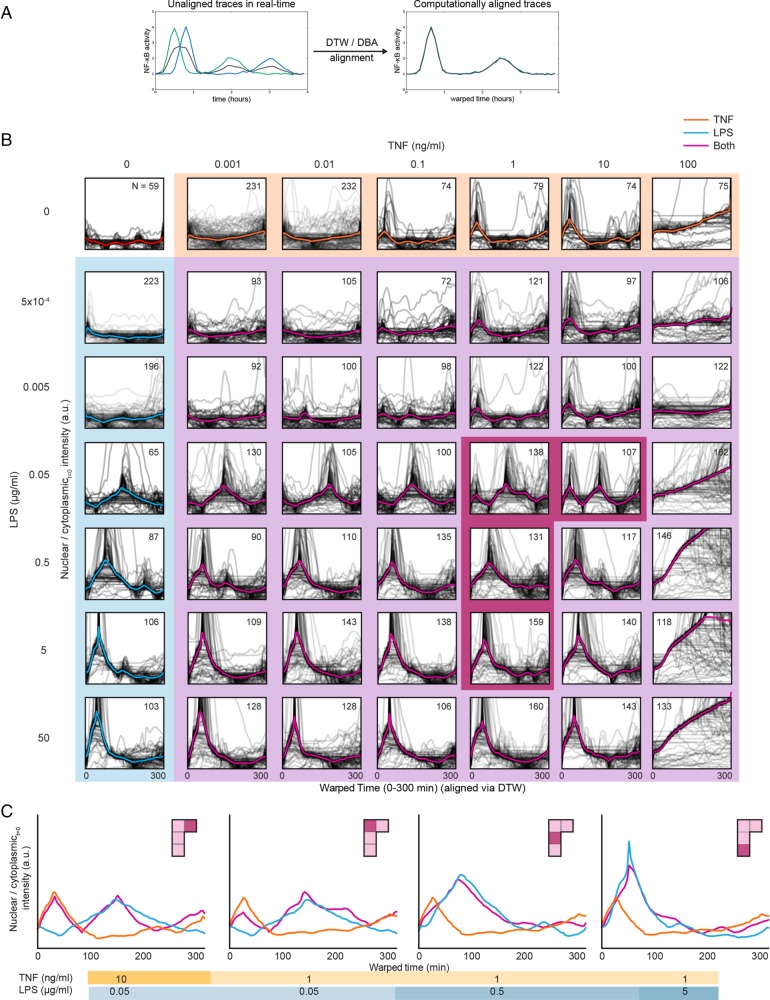
NF-κB activation dynamics are distinct under stimulus combinations. (A) Schematic of how DTW and DBA can be used to align single-cell traces of NF-κB activity. Two single-cell traces are shown in blue and green and the mean in black. (B) NF-κB nuclear translocation dynamics were aligned using DTW to increase the visibility of semi-time-independent features (e.g., oscillations) that are lost using traditional averaging techniques. Single-cell traces are shown in gray, and the mean traces of the aligned single-cell traces are thicker and in color. Plots highlighted in orange are treated with TNF alone, blue highlights indicate treatment by LPS alone and purple highlights indicate combinations. Plots highlighted in dark magenta are shown in further detail in C. (C) Mean traces of ligand combinations that exhibited a particularly notable interaction effect between the LPS and TNF stimuli. These are shown, left to right, in order from high to low ratios of TNF to LPS.
We found that the dynamics of p65-dsRed nuclear translocation varied substantially across concentrations and with respect to TNF–LPS combinations (Figure 2B). In several cases, the combined dynamics strongly resembled those of the TNF-only or LPS-only condition. This was expected when either TNF or LPS was present at a low concentrations; however, when TNF and LPS are both at high concentrations, the reporter translocation dynamics resembled the LPS-only condition. We note that the highest concentration of TNF (100 ng/ml) led to prolonged and higher levels of nuclear translocation followed by a loss of cell viability; as a result, we do not consider these conditions further. Taken together with our finding that LPS concentration largely determines the fraction of cells in which NF-κB is activated (Figure 1, C and D), this result seems to confirm our earlier observation that the effect of LPS often supercedes the role of TNF in activating NF-κB.
As mentioned in the Introduction, some studies have found that the output of certain combinations of innate immune stimuli result in either additive or synergistic responses (Napolitani et al., 2005; Natarajan et al., 2006; Bagchi et al., 2007; Qiao et al., 2013; Lin et al., 2017), while in another case individual cells in a population were shown to respond to one or other stimuli but not both (Kellogg et al., 2017). Under the stimulation conditions used in this study the mean NF-κB trace generally reflected that of either LPS or TNF stimulation alone. However, we identified four concentration pairs, highlighted with magenta in Figure 2B, where the mean NF-κB trace resembled a composite of the dynamics observed after stimulus with TNF and LPS alone—resulting in either two strong peaks or a single extended peak of NF-κB activity (Figure 2C) (determined by comparison to a linear superposition model; see Materials and Methods and Supplemental Figure S2). Two combinations involved the 0.05 μg/ml LPS concentration, with TNF concentrations of 1 and 10 ng/ml (left two plots of Figure 2C). In both of these cases, the population-level response to LPS alone was a peak of p65-dsRed nuclear translocation at ∼150 min, while the TNF response exhibited a first strong peak at around 30 min (with subsequent nonsynchronous peaks of decreasing amplitude in individual cells). The combined response exhibited one strong peak at each of these times. The remaining two combinations occurred at a TNF concentration of 1 ng/ml with LPS concentrations of 0.5 and 5 μg/ml (right two plots of Figure 2C). Under single stimulus conditions as the concentration of LPS was increased to 0.5 and 5 μg/ml, the time to first peak of NF-κB translocation decreased to 90 and then 60 min, as previously shown (Gutschow et al., 2013). For the two combined cases in which these LPS concentrations were added with 1 ng/ml TNF, a single long peak was observed in the population, which appeared to combine the peaks obtained when either stimulus was used separately. Taken together, these results suggest that stimulation conditions exist where the NF-κB signaling response to both LPS and TNF is an integration of features of both single stimulus responses rather than responding to only one or the other stimulus.
The interaction effect for NF-κB activation dynamics occurs at the single-cell level
With respect to single-cell behaviors, our and others’ work has shown that a population-level response can be composed of distinct phenotypes produced by two or more cellular subpopulations (Lahav et al., 2004; Nelson et al., 2004; Santos et al., 2007; Tay et al., 2010; Purvis et al., 2012; Albeck et al., 2013). We therefore wondered whether the combined stimulus responses we observed in Figure 2C could be explained either by phenotypic heterogeneity (e.g., one group of cells only responds to TNF, while the other only responds to LPS) or else by the interaction between TNF- and LPS-downstream signaling in individual cells (all cells respond to both TNF and LPS). In the former case, we would see distinct populations of cells in the combined stimulus experiments that resembled the response to one or the other single stimuli. In the latter, single cells would exhibit translocation responses that mirrored the population response.
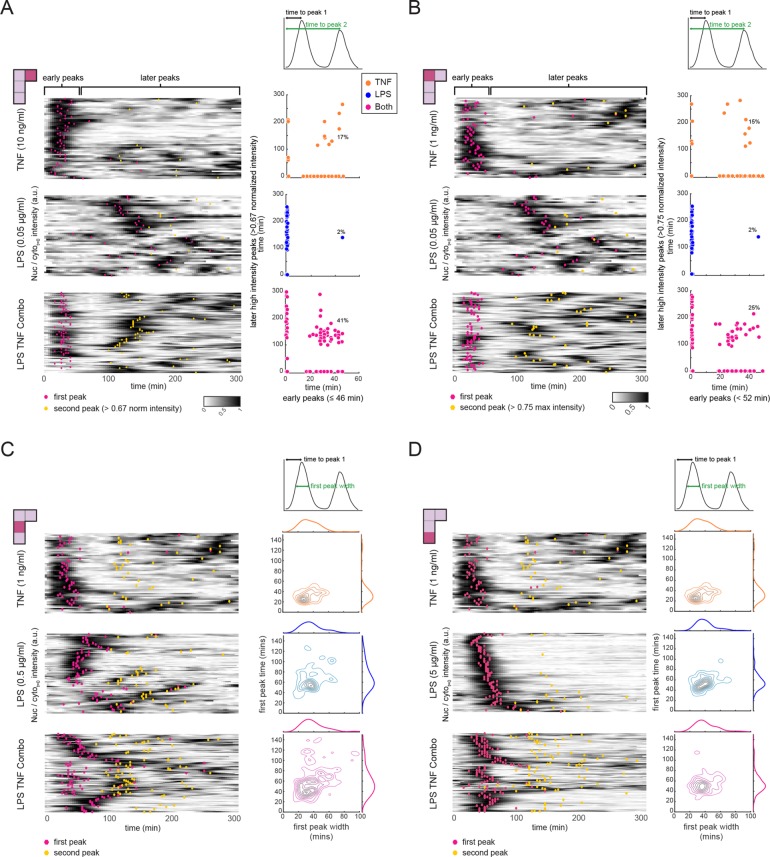
To distinguish between these two possibilities, we examined the NF-κB dynamics of individual cells for the (0.05 μg/ml LPS, 10 ng/ml TNF) condition in combination, as well as for each stimulus alone (Figure 3A). For cells stimulated with TNF, the majority of the first nuclear NF-κB translocations occur within 46 min (median time 37 min, 90% confidence interval 19–52 min), followed by lower intensity nuclear translocations after 46 min. The first LPS peaks occur at a broader distribution of times but later than those found with TNF (median time 124 min, 90% confidence interval of 46–253 min). However, in response to the combined stimulus, a fraction of cells displayed features of both TNF and LPS responses, suggesting that a single cell could respond to both ligands (Figure 3A, left panel). To quantify this observation, we used the fact that LPS and TNF activate NF-κB with different kinetics to determine how the NF-κB response of single cells shifted between single and dual inputs. By comparing the early NF-κB response to that in the later stages we found, as expected, that the NF-κB response during TNF stimulation was higher in the early period of imaging, while for LPS later activation predominated over early responses (Figure 3A, right panel). However, in the combined stimulus while some cells showed responses similar to either TNF or LPS, there was an expansion in the number of cells with NF-κB activation in both early and late periods—from 17% with TNF, 2% with LPS to 41% with the combined stimulus. By comparing features of NF-κB dynamics across stimuli, we found that several features were sufficient to distinguish the combined stimulus group from either of the single stimuli (Supplemental Figure S3A). We then further dissected our combined stimulus experiment, classifying cells in this group as TNF-like, LPS-like, or as dual responders (as shown in Figure 3A, lower right panel). Surprisingly, we found that the feature comparisons for these subgroups within one experimental condition (Supplemental Figure S3B) were very similar to the comparisons of whole-group comparisons shown in Supplemental Figure 3A. This supports the hypothesis that cells undergoing a combined stimulus are responding to TNF, LPS, or both. We observed a similar dual-response phenotype for cells stimulated with 1 ng/ml TNF and 0.05 μg/ml LPS, albeit at different frequencies (Figure 3B). Thus, cells exhibiting a distinct, combined response to dual stimulation were consistently observed across these two combinations. However, when 1 ng/ml TNF was combined with higher doses of LPS (0.5 or 5 μg/ml), the response time for both ligands were less well separated and thus in the combined response the two peaks often merged into a single long peak (Figure 2C, right two panels). This hinders our ability to classify cells as purely dual or single responders. That said, we did find that the joint distribution of first peak time and first peak width for dual-stimulated cells was distinct from and reflected a composite of the TNF and LPS single-stimulated joint distributions (Figure 3, C and D), supporting the idea that the differences between dynamic NF-κB activation profiles arising from combining stimuli are reflected at the single-cell level.
FIGURE 3:
The interaction effect for NF-κB activation dynamics occurs at the single-cell level. NF-κB activation dynamics at the single-cell level for the following concentration combinations (panel A: 0.05 μg/ml LPS, 10 ng/ml TNF; panel B: 0.05 μg/ml LPS, 1 ng/ml TNF; panel C: 0.5 μg/ml LPS, 1 ng/ml TNF; panel D: 5 μg/ml LPS, 1 ng/ml TNF). Cells were treated with TNF alone, LPS alone, or a combination of both ligands. Heatmaps for all three conditions, where each row represents a single-cell trace of NF-κB nuclear translocation over time. Peaks corresponding to NF-κB nuclear localization are dark (see color scale), with local maxima indicated by pink (first peak) or yellow (second peak) dots. For A and B scatter plots relate the time of any early peak (peaks occurring in 46 min or less) to the timing of a strong second peak occurring after 46 min. Under the stimulation conditions used in C and D, the first and second peak converge to form a longer peak in the combined stimulus case. To capture this phenomenon, we plot contour plots of the first peak width vs. first peak time beside each heatmap along with kernel density estimates of each of the features in the marginal axes. For each panel, the single-cell features that are compared can be found in the schematic above the scatter plots.
Combined stimulation induces changes in gene expression and cytokine output
With differences in NF-κB dynamics identified on combined TNF and LPS stimulation our next question was whether these distinct signaling patterns impacted gene expression, given NF-κB’s centrality as a core transcription factor in the innate immune system. Even under dual stimulus conditions, cells in the population display a mix of NF-κB signaling behaviors (Figure 3A); thus, population-level measurements are insufficient to answer this question. Instead, we decided to profile gene expression using smFISH (Battich et al., 2013); this approach allowed us to directly connect NF-κB dynamics with mRNA levels in the same cell (Figure 4A). We treated cells with one of the combined stimulus combinations where the combinatorial NF-κB response was evident (5 μg/ml LPS, 1 ng/ml TNF—note that the specific LPS response has been shown to vary from batch to batch; see Materials and Methods), as well as the corresponding concentrations of each stimulus alone, and selected a 4-h time point to assay gene expression as this was sufficient time for both LPS and TNF dynamics to be observed (Figure 3D). We considered a number of known NF-κB target genes (Cxcl1, Csf2, Ccl5, Il-6, Cxcl10, Csf3, Cxcl5) previously shown to be induced by either or both stimuli (Pahl, 1999; Tian et al., 2005; Sharif et al., 2007) and compared their gene expression across stimuli (Figure 4B and Supplemental Figure S4A).
FIGURE 4:
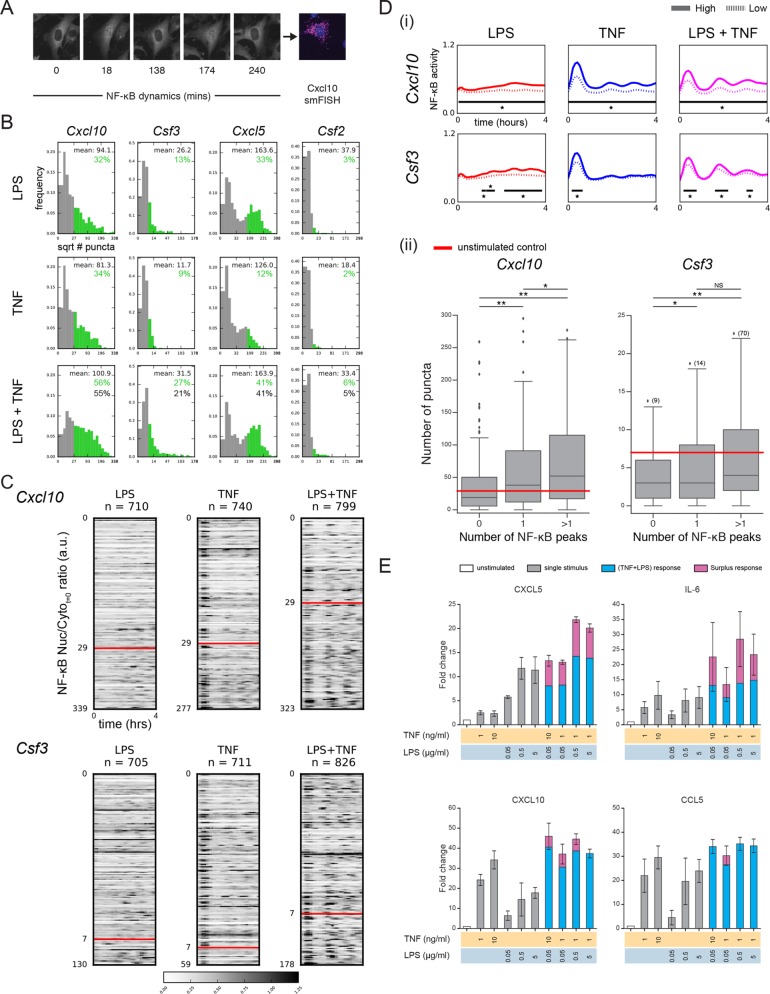
Combined TNF and LPS stimulation translates into differences in gene expression and cytokine secretion. (A) Microscopy images illustrating the combination of live-cell imaging of NF-κB dynamics and smFISH. Images shown are for cells stimulated with both TNF (1 ng/ml) and LPS (5 µg/ml) and smFISH used probes against Cxcl10. (B) Dual stimulation is associated with changes in gene expression for several NF-κB target genes. For each target gene, cells were stimulated with TNF (1 ng/ml), LPS (5 µg/ml), or both. For each condition histograms of the square root of mRNA puncta per cell, as determined by smFISH, are shown. A threshold of expression was set based on the 95th percentile of the unstimulated control and bars are colored green where the signal is above this threshold. Mean of the subpopulation of cells with high transcript expression (green) is shown along with the observed (green) and expected (black) percentage of cells in the population above the threshold. The expected response was calculated using FLPS+FTNF-FLPS*FTNF, where FLPS is the fractional response to LPS and FTNF is the fractional response to TNF. (C) Increased mRNA expression is linked to increased NF-κB dynamics. For Cxcl10 and Csf3, cells in each stimulus condition were rank ordered according to the number of mRNA puncta and heatmaps of NF-κB dynamics for all cells are shown. Peaks corresponding to NF-κB nuclear localization are black (see color scale) and were scaled between 0 and 1.25. For each gene, the threshold used in B is indicated by the red line and was used to separate cells into those with low and high gene expression based on a control measurement. (D) (i) The average NF-κB dynamics of cells with low and high gene expression is distinct. For each condition cells were separated according to the mRNA puncta threshold defined in B and the mean NF-κB trace for each subgroup is shown. High expressing cells are indicated by the unbroken line and low expressing cells by the dotted line. Black bars and * indicate the timepoints at which differences between the two subgroups were significant using the Kolmogorov–Smirnov test with a confidence level of p = 0.01. Note that for Csf3 in the LPS condition two of the bars are close to each other but are in fact separated by a single time point. (ii) For each cell in the dual stimulus condition the number of NF-κB peaks during the time course was determined. Cells were binned into three groups based on the number of peaks and the number of puncta for cells in each bin is shown. Data shown are for Cxcl10 and Csf3. Significance was calculated using a two-sided independent t test (* p < 0.05, ** p < 0.001). Owing to the broad range of Csf3 expression, outliers are not shown but instead indicated by the number in parenthesis next to the diamond. (E) Cytokine secretion by cells treated with a range of stimulus combinations. Fold change of cytokine secretion normalized to the untreated control (white) for an immunoassay performed 6 h after stimulation. For the combined stimulus conditions any signal above an additive effect of the individual ligands is indicated in pink. Increased secretion of cytokines is observed upon dual stimulation compared with either stimulus alone, most notably for CXCL5 and IL6. Secretion is reported in MFI. N = 3 independent experiments.
Four of the genes were either not significantly induced at the time point selected (Csf2, Il-6), or else the dually stimulated cells were not observed to be activated in ratios indicative of a combinatorial response (Cxcl1, Ccl5) (Figure 4B and Supplemental Figure S4A). The other three genes (Cxcl10, Csf3, Cxcl5) exhibited a population of activated cells that seemed likely to contain some dual responders. We therefore considered these three genes in more detail. In particular, we aligned the NF-κB activation time courses for all cells based on the number of mRNA puncta observed in each cell (Figure 4C and Supplemental Figure S4B). We found that the fraction of cells that had mRNA counts above the untreated control (red lines) was higher in the dual-stimulus population. The heatmaps also revealed a general trend toward increased NF-κB responses, as indicated by more frequent occurrence of regions of higher (darker) intensity, in the cells with above-control mRNA counts in the dual-stimulated condition (Figure 4C and Supplemental Figure S4B), which we quantified and found to be significant (Figure 4D and Supplemental Figure S4C). The reverse was also true: as cells increased their NF-κB activity—determined by the number of peaks—the number of cells with transcript levels of Cxcl10 and Csf3 above the unstimulated control increased (Figure 4D). Thus, cells with higher levels of Cxcl10 and Csf3 (and to a lesser extent, Cxcl5) mRNA expression are also more likely to have increased NF-κB activity over time.
Finally, we decided to determine whether changes in NF-κB dynamics and gene expression are translated into differential cytokine production. Paracrine signaling is known to play a critical role in population behavior during innate immune signaling (Shalek et al., 2014; Xue et al., 2015); therefore, to do this we selected the Luminex platform, a population-based assay (in lieu of single-cell immunofluorescence, as the latter prevents paracrine signaling due to the requirement for Brefeldin A, which blocks protein secretion from the cell). Using this multiplexed experimental platform, we interrogated cytokine production for a panel of cytokines, including the genes profiled by smFISH, across a range of single and dual stimulus conditions (Figure 4E and Supplemental Figure S5). For those cytokines produced at measurable levels, we found that each of the combined stimulations led to increased expression of CXCL5 and IL-6. For CXCL10 there was increased expression for all but one combination, while for CCL5 only one combination produced an increased response. Overall, although our secretion measurements are made at the population level we find that under certain conditions a trend of increased population-level cytokine secretion is in qualitative agreement with the increased NF-κB dynamics under combinatorial stimulation; differences in the magnitude of these effects could be due to any of a number of regulatory steps found in between NF-κB activation and cytokine secretion, such as posttranscriptional and posttranslational processing and cell–cell communication.
DISCUSSION
NF-κB can be activated by a diverse range of stimuli, from PAMPs to cytokines and stress stimuli. While significant advances have been made in defining the single-cell NF-κB response to individual stimuli (Nelson et al., 2004; Tay et al., 2010; Sung et al., 2014; Kellogg et al., 2017), much less is known about how the cellular response to a given stimulus changes when additional activators are also present in the environment. Our results demonstrate that together TNF and LPS elicit a combinatorial response reflected in sustained NF-κB dynamics. These results are in contrast to a recent study that found that in response to two different PAMPs, LPS and PAM3CSK4, single cells fail to show a combinatorial response and instead only respond to one or the other ligand (Kellogg et al., 2017). Taken together, these two studies suggest that for two non-self inputs, such as PAMPs, only certain cells in the population respond to either input perhaps enabling a division of labor in the population; while for a combination of self and non-self inputs, such as TNF and LPS, both inputs are processed. The differences between our two studies could stem from the fact that both LPS and PAM3CSK4 signal through a common set of intermediates to activate NF-κB, while in contrast TNF and LPS converge on NF-κB using different upstream components. As a consequence, NF-κB activation by multiple PAMPs may be sensitive to limitations in the availability of these shared components as well as to common negative feedback mechanisms in ways that NF-κB activation by TNF and LPS are not. These contrasting results also underscore the need to evaluate the processing capacity of NF-κB on a stimulus by stimulus (as well as by combination of stimuli) level.
Moreover, since previous studies of this kind have focused on population measurements, it has been unclear whether synergistic cytokine responses emerge as a consequence of a uniform change in expression in all cells in the population. Our results establish that individual cells process the presence of multiple stimuli in distinct ways—and even at different levels of cellular phenotype, from signaling and transcription to cytokine production. For instance, at the level of NF-κB signaling, our results suggest that individual cells have the capacity to respond to both LPS and TNF simultaneously. However, the decoding of such signaling occurs in a heterogeneous and complex manner, in terms of both gene expression and cytokine production. For instance, increased NF-κB dynamics is linked with increased Cxcl10 expression, while for Csf3 this connection is weaker with only a fraction of cells with multiple NF-κB peaks displaying changes in gene expression, suggesting that genes are differentially sensitive to any increase in NF-κB activity. Furthermore, several features of individual cells, such as the activation state of other transcription factors in addition to NF-κB, their absence/presence at a gene promoter, the differential priming or binding of enhancer regions and epigenetic effects, along with differences in posttranslational processes, may be responsible for the differences we observe in how NF-κB signaling determines gene expression and cytokine production (Giorgetti et al., 2010; Sen and Smale, 2010; Franco et al., 2015; West et al., 2015). A comprehensive understanding of the relationship between signaling, transcription, and cytokine production will require all three of these cellular processes to be measured in the same single cell. We anticipate that this type of approach will open a new window into how cells determine the most appropriate responses to complex environments.
MATERIALS AND METHODS
Cell culture
Cell culture was performed as previously described (Gutschow et al., 2013). The cells were polyclonal p65−/− mouse 3T3 fibroblasts, courtesy of the Baltimore Lab (Beg et al., 1995), infected with lentivirus to express p65-dsRed and H2B-GFP. Cells were cultured in DMEM (Invitrogen 11965-092) supplemented with 2 mM l-glutamine (Gibco25030), 100 U/ml penicillin, 100 μg/ml streptomycin (Life Technologies 15140), and 10% fetal bovine serum (FBS) (Omega Scientific, FB-11, lot 105247 [cell culture], lot 100203 [Luminex]). Cells were treated with recombinant mouse TNF (11271156001; Roche, Indianapolis, IN) and/or Invivogen LPS-EB Ultrapure (Invivogen tlrl-pelps (microscopy experiments) and later, a further purified version, tlrl3-pelps (Luminex), from Escherichia coli O111:B4). We note that we have found this preparation of Ultrapure LPS to show variable NF-κB responses between lots. As a result, the smFISH analysis was carried out on cells stimulated with TNF (1 ng/ml) and LPS (5 µg/ml) as with the batch of LPS used for the smFISH experiments the NF-κB responses most closely resembled those shown in Figure 3B, which were obtained using a different lot of LPS and a combination of TNF (1 ng/ml) and LPS (0.05 µg/ml). For microscopy, solutions were prepared in imaging media (DMEM prepared without riboflavin, folic acid, or phenol red, with 1% FBS) and kept on ice, and then warmed to 37°C just before stimulation.
Microscopy
Cells were imaged on wells of a glass-bottom, 96-well plate (164588; Nunc [Thermo Scientific], Waltham, MA) that had been precoated with 10 μg/ml human fibronectin (FC010; Millipore, Billerica, MA). The day before imaging, ∼7000 cells/well were seeded onto the plate in DMEM with 10% FBS. One hour before stimulation, the wells were switched to imaging media (DMEM prepared without riboflavin, folic acid, or phenol red, with 1% FBS). Microscopy was performed on a Nikon Eclipse Ti fluorescence microscope, using a 20× air/0.75 numerical aperture objective. The camera was a Photometrics CascadeII: 1024 electron-multiplying charge-coupled device. Image acquisition was controlled by Micro-Manager (Edelstein et al., 2014). Images were acquired using fluorescein isothiocyanate (FITC) and tetramethylrhodamine isothiocyanate filter sets (Semrock, Lake Forest, IL) every 5–6 min. Temperature (37°C), CO2 (5%), and humidity were held constant. Breathe-Easy sealing membrane (Sigma Z380059) was used to minimize evaporation from the wells.
Image analysis
Flat-fielding (correcting for uneven illumination of the field of view) and time-lapse registration (correcting for small imprecision of the stage movement) of images were performed with custom MATLAB code (MathWorks) as previously described (Hughey et al., 2015). Segmentation, cell tracking, peak finding, and curation of the data were also performed using custom MATLAB software or CellTK (https://github.com/braysia/CellTK) (Kudo et al., 2018). Cells were manually defined as active, with the criteria that nuclei were uniformly brighter than the initial cytoplasmic intensity, with visible nucleoli. We measured mean fluorescence intensity over the initial cytoplasmic intensity over time. For visualization of all traces together, dynamics data were aligned and averaged using DTW DBA (Petitjean et al., 2011). DTW (Aach and Church, 2001) is a sequence alignment algorithm for quantitative time series. The algorithm is equivalent to the Smith–Waterman algorithm (Smith and Waterman, 1981; Pahl, 1999) and Needleman–Wunsch algorithm (Needleman and Wunsch, 1970) used in aligning nucleotide and peptide sequences; however, rather than using a substitution matrix, DTW penalizes for the squared difference in value between two points. The same approach can be used for aligning multiple time series, but is not practical due to increasing computational costs. DBA avoids this issue by aligning each real time series to one reference time series. The aligned time series are averaged and the resulting averaged time series is used as the new reference time series. The procedure is repeated until the averaged time series stops appreciably changing. Code is available at https://github.com/CovertLab/mboc_synergy_analysis.
Linear superposition
Several studies have suggested that the output of certain combinations of stimuli can be represented as a weighted sum of the outputs of each individual stimulus—a concept called linear superposition (Geva-Zatorsky et al., 2010; Bollenbach and Kishony, 2011; Wood et al., 2012; Rothschild et al., 2014; Chevereau and Bollenbach, 2015). To determine whether this concept may apply to the four conditions in which NF-κB dynamics resembled a composite of those found using the individual stimuli, we implemented a one-parameter linear superposition model (α * LPS trace + [1-α]* TNF trace), where 0 ≤ α ≤ 1.0. This was applied to the mean traces rather than DTW-aligned traces to preserve dynamic information. Parameters were chosen based on cosine distance between experimental and linear superposition traces.
Model predictions were then compared with our experimentally measured output (Supplemental Figure 2A). Model parameters indicated the relative strength of the TNF and LPS signals (Supplemental Figure 2B); the parameter values were closest to one another (indicating nearly equal strength) in the four concentration pairs found to have composite dynamics, highlighted with magenta in Figure 2A and Supplemental Figure S2B.
smFISH: imaging and analysis
We stimulated cells with the indicated ligands and imaged p65 dynamics as detailed above. At the end of the imaging period (240 min) cells were fixed in 4% paraformaldehyde (PFA). The Quantigene ViewRNA ISH kit (QVC00001; Thermo Fisher) was used in conjunction with the following probes against mouse transcripts: Cxcl5 (VB6-3198753-VC), Cxcl10 (VB6-10663-VC), Csf3 (VB6-3197408-VC), IL-6 (VB6-13850-VC), Csf2 (VB6-3197528-VC), Ccl5 (VB6-12823), and Cxcl1 (VB6-10681). Single-cell gene expression activity was quantified using an automated image analysis procedure. For each field, a z-stack of 21 images in the far-red channel were obtained. Two additional images were taken for nuclear markers (FITC, expressed, and 4′,6-diamidino-2-phenylindole [DAPI], stain). All images were normalized using a blank reference image to correct for shading. The FISH z-stacks were merged into one image representing the maximum intensity at any given position. Noise in the nuclear marker images was reduced by curvature anisotropic smoothing, and the nuclei were segmented using adaptive thresholding. FISH puncta were also segmented using adaptive thresholding. The identified puncta were associated with the closest nuclei in the DAPI (stain) channel, then with the nuclei in the FITC (expressed) channel. Finally, the nuclei in the FITC channel were aligned and associated with the nuclei visible in the last frame of the dynamics images (also in the FITC channel).
Secreted cytokine quantification
Supernatant was collected for Luminex assay at 6 h (run on a 38-plex assay). Biological triplicates were collected, and each sample was run in technical duplicates. The Luminex–eBioscience/Affymetrix Magnetic Bead Kit assay was performed in the Human Immune Monitoring Center at Stanford University. Mouse 38-plex kits were purchased from eBiosciences/Affymetrix and used according to the manufacturer’s recommendations with modifications as described below. Briefly: Beads were added to a 96-well plate and washed in a Biotek ELx405 washer. Samples were added to the plate containing the mixed antibody-linked beads and incubated at room temperature for 1 h followed by overnight incubation at 4°C with shaking. Cold and room temperature incubation steps were performed on an orbital shaker at 500–600 rpm. Following the overnight incubation, plates were washed in a Biotek ELx405 washer and then biotinylated detection antibody added for 75 min at room temperature with shaking. The plate was washed as above and streptavidin-PE was added. After incubation for 30 min at room temperature, a wash was performed as above and reading buffer was added to the wells. Each sample was measured in duplicate. Plates were read using a Luminex 200 instrument with a lower bound of 50 beads per sample per cytokine. Custom assay Control beads by Radix Biosolutions are added to all wells. All stimulated samples were compared with unstimulated control (baseline) samples using median fluorescence intensity (MFI).
Supplementary Material
Acknowledgments
We thank K. C. Huang, David Schneider, Denise Monack, Justin Sonnenburg, and members of the Covert lab for helpful discussions and comments on the manuscript. Some of the computing for this project was performed on the Sherlock cluster. We thank Stanford University and the Stanford Research Computing Center for providing computational resources and support that contributed to these research results. We also gratefully acknowledge an Allen Discovery Center Award, a National Institutes of Health (NIH) R21 (5R21AI104305-02), the Stanford Center for Systems Biology (NIH P50GM107615), an NIH Pioneer Award (5DP1LM01150-05), and an Allen Distinguished Investigator Award to M.W.C., Siebel, Weiland Family, and Rensselaer Engineering Fellowships to M.V.G., a National Institute of Standards and Technology (NIST) Training Grant and Agilent Bioengineering Fellowship to J.C.M., and Bio-X and Achievement Rewards for College Scientists (ARCS) Fellowships to J.J.H.
Abbreviations used:
- DBA
DTW barycenter averaging
- DTW
dynamic time warping
- LPS
lipopolysaccharide
- NF-κB
nuclear factor κB
- PAMPs
pathogen-associated molecular patterns
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
- smFISH
single-molecule RNA-FISH
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-07-0423) on November 21, 2018.
REFERENCES
- Aach J, Church G. (2001). Aligning gene expression time series with time warping algorithms. Bioinformatics , 495–508. [DOI] [PubMed] [Google Scholar]
- Albeck JG, Mills GB, Brugge JS. (2013). Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol Cell , 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A, Herrup EA, Warren H, Trigilio J, Shin H-SS, Valentine C, Hellman J. (2007). MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J Immunol , 1164–1171. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Loewer A, Mock C, Lahav G. (2011). Stimulus-dependent dynamics of p53 in single cells. Mol Syst Biol , 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battich N, Stoeger T, Pelkmans L. (2013). Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Methods , 1127–1133. [DOI] [PubMed] [Google Scholar]
- Beg A, Sha W, Bronson R, Ghosh S, Baltimore D. (1995). Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature , 167–170. [DOI] [PubMed] [Google Scholar]
- Bollenbach T, Kishony R. (2011). Resolution of gene regulatory conflicts caused by combinations of antibiotics. Mol Cell , 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevereau G, Bollenbach T. (2015). Systematic discovery of drug interaction mechanisms. Mol Syst Biol , 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert MW, Leung TH, Gaston JE, Baltimore D. (2005). Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science , 1854–1857. [DOI] [PubMed] [Google Scholar]
- Dalal CK, Cai L, Lin Y, Rahbar K, Elowitz MB. (2014). Pulsatile dynamics in the yeast proteome. Curr Biol , 2189–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. (2014). Advanced methods of microscope control using μManager software. J Biol Methods , e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Nagari A, Kraus W. (2015). TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell , 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci S, Matzinger P. (2001). Danger signals: SOS to the immune system. Curr Opin Immunol , 114–119. [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Dekel E, Cohen AA, Danon T, Cohen L, Alon U. (2010). Protein dynamics in drug combinations: a linear superposition of individual-drug responses. Cell , 643–651. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G. (2010). Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell , 418–428. [DOI] [PubMed] [Google Scholar]
- Gutschow MV, Hughey JJ, Ruggero NA, Bajar BT, Valle SD, Covert MW. (2013). Single-cell and population NF-κB dynamic responses depend on lipopolysaccharide preparation. PLoS One , e53222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey JJ, Gutschow MV, Bajar BT, Covert MW. (2015). Single-cell variation leads to population invariance in NF-κB signaling dynamics. Mol Biol Cell , 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg RA, Tian C, Etzrodt M, Tay S. (2017). Cellular decision making by non-integrative processing of TLR inputs. Cell Rep , 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Jeknic´ S, Macklin DN, Akhter S, Hughey JJ, Regot S, Covert MW. (2018). Live-cell measurements of kinase activity in single cells using translocation reporters. Nat Protoc , 155–169. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. (2004). Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet , 147–150. [DOI] [PubMed] [Google Scholar]
- Lane K, Van Valen D, DeFelice MM, Macklin DN, Kudo T, Jaimovich A, Carr A, Meyer T, Pe’er D, Boutet SC, Covert MW. (2017). Measuring signaling and RNA-Seq in the same cell links gene expression to dynamic patterns of NF-κB activation. Cell Syst , 458–469. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RE, Walker SR, Savery K, Frank DA, Gaudet S. (2014). Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol Cell , 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Denny EM, Sanghvi JC, Gaston JE, Maynard ND, Hughey JJ, Covert MW. (2009). A noisy paracrine signal determines the cellular NF-κB response to lipopolysaccharide. Sci Signal , ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Dutta B, Fraser ID. (2017). Systematic investigation of multi-TLR sensing identifies regulators of sustained gene activation in macrophages. Cell Syst , 25–37. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P. (1994). Tolerance, danger, and the extended family. Annu Rev Immunol , 991–1045. [DOI] [PubMed] [Google Scholar]
- Napolitani G, Rinaldi A, Bertoni F, Sallusto F, Lanzavecchia A. (2005). Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat Immunol , 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan M, Lin K-MM, Hsueh RC, Sternweis PC, Ranganathan R. (2006). A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol , 571–580. [DOI] [PubMed] [Google Scholar]
- Needleman S, Wunsch C. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol , 443–453. [DOI] [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, et al (2004). Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science , 704–708. [DOI] [PubMed] [Google Scholar]
- Pahl H. (1999). Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene , 6853–6866. [DOI] [PubMed] [Google Scholar]
- Petitjean F, Ketterlin A, Gançarski P. (2011). A global averaging method for dynamic time warping, with applications to clustering. Pattern Recognit , 678–693. [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. (2012). p53 dynamics control cell fate. Science , 1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Lahav G. (2013). Encoding and decoding cellular information through signaling dynamics. Cell , 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Giannopoulou EG, Chan CH, Park S-HH, Gong S, Chen J, Hu X, Elemento O, Ivashkiv LB. (2013). Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity , 454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild D, Dekel E, Hausser J, Bren A, Aidelberg G, Szekely P, Alon U. (2014). Linear superposition and prediction of bacterial promoter activity dynamics in complex conditions. PLoS Comput Biol , e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Chung M, Dobrzyński M, Fey D, Blum Y, Lee S, Peter M, Kholodenko BN, Jeon N, Pertz O. (2016). Frequency modulation of ERK activation dynamics rewires cell fate. Mol Syst Biol , 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Verveer PJ, Bastiaens PI. (2007). Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol , 324–330. [DOI] [PubMed] [Google Scholar]
- Sen R, Smale ST. (2010). Selectivity of the NF-κB response. Cold Spring Harb Perspect Biol , a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al (2014). Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature , 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. (2007). Transcriptional profiling of the LPS induced NF-kappaB response in macrophages. BMC Immunol , 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. (1981). Identification of common molecular subsequences. J Mol Biol , 195–197. [DOI] [PubMed] [Google Scholar]
- Sung M-HH, Li N, Lao Q, Gottschalk RA, Hager GL, Fraser ID. (2014). Switching of the relative dominance between feedback mechanisms in lipopolysaccharide-induced NF-κB signaling. Sci Signal , ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. (2010). Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature , 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. (2005). Identification of direct genomic targets downstream of the nuclear factor-kappaB transcription factor mediating tumor necrosis factor signaling. J Biol Chem , 17435–17448. [DOI] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al (2015). Mitochondrial DNA stress primes the antiviral innate immune response. Nature , 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K, Nishida S, Sontag ED, Cluzel P. (2012). Mechanism-independent method for predicting response to multidrug combinations in bacteria. Proc Natl Acad Sci USA , 12254–12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xaus J, Comalada M, Valledor A, Lloberas J, López-Soriano F, Argilés J, Bogdan C, Celada A. (2000). LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-alpha. Blood , 3823–3831. [PubMed] [Google Scholar]
- Xue Q, Lu Y, Eisele MR, Sulistijo ES, Khan N, Fan R, Miller-Jensen K. (2015). Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci Signal , ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.