Abstract
Actomyosin contractility can influence the canonical Wnt signaling pathway in processes like mesoderm differentiation and tissue stiffness during tumorigenesis. We identified that increased nonmuscle myosin II activation and cellular contraction inhibited Wnt target gene transcription in developing Drosophila imaginal disks. Genetic interactions studies were used to show that this effect was due to myosin-induced accumulation of cortical F-actin resulting in clustering and accumulation of E-cadherin to the adherens junctions. This results in E-cadherin titrating any available β-catenin, the Wnt pathway transcriptional coactivator, to the adherens junctions in order to maintain cell–cell adhesion under contraction. We show that decreased levels of cytoplasmic β-catenin result in insufficient nuclear translocation for full Wnt target gene transcription. Previous studies have identified some of these interactions, but we present a thorough analysis using the wing disk epithelium to show the consequences of modulating myosin phosphatase. Our work elucidates a mechanism in which the dynamic promotion of actomyosin contractility refines patterning of Wnt transcription during development and maintenance of epithelial tissue in organisms.
INTRODUCTION
The Wnt signaling pathway (Wingless [Wg] in Drosophila), is highly conserved across metazoans and essential during development and tissue homeostasis for the regulation of proliferation and patterning (Clevers and Nusse, 2012). Wnt signaling achieves proper biological outcomes through extensive crosstalk with other signaling pathways. Recent studies have begun to elucidate how mechanical forces may also play critical roles in regulating signaling pathways during development (Farge, 2011). Here we present a detailed characterization of the effects of activated nonmuscle myosin on Wnt signaling.
The canonical Wnt/Wg pathway centers on the stabilization and localization of the key effector protein, β-catenin (β-cat) (Armadillo [Arm] in Drosophila). β-Cat is continuously produced in most cells for its roles in both the formation and maintenance of adherens junctions (AJs) and as a transcriptional activator for Wnt signaling (Valenta et al., 2012). AJs are major epithelial cell–cell adhesion complexes that maintain tissue integrity in response to external forces like morphogenesis (Harris and Tepass, 2010). AJs mainly form an apical-lateral beltlike structure around cells, holding neighbouring cells together through the homophilic binding of the transmembrane protein E-cadherin (E-cad). β-Cat binds to the cytoplasmic tail of E-cad and to α-catenin, which interacts with the actin cytoskeleton. Thus AJs act as mechanical force integration sites across cells and at a tissue level (Lecuit and Yap, 2015).
In the absence of a Wnt ligand, cytoplasmic β-cat is targeted for degradation by a multiprotein destruction complex that assembles on the scaffolding protein Axin and includes kinases that phosphorylate β-cat, targeting it for ubiquitination and subsequent proteasomal digestion. On Wnt/Wg binding to its coreceptors Frizzled (Fz) and LRP/Arrow, Dishevelled (Dvl/Dsh) is recruited to Fz and triggers the recruitment of the destruction complex to the membrane. This event disrupts the destruction complex, allowing β-cat to accumulate and translocate to the nucleus, where it acts with T-cell factor (TCF)/lymphoid enhancer factor (LEF) transcription factors to initiate target gene expression (Daniels and Weis, 2005). Disruptions of the core components or regulatory proteins have been found in numerous cancers and developmental disorders (Clevers and Nusse, 2012).
Recent studies suggest that canonical Wnt signaling, actomyosin contractility, and mechanical forces are integrated in the regulation of development and homeostasis. Wnt activation can drive mechanical strain-induced cell proliferation (Benham-Pyle et al., 2015, 2016) as well as activation of nonmuscle myosin II (NMII) leading to morphogenesis (Zimmerman et al., 2010). Conversely, force induction and subsequent cytoskeletal rearrangements can regulate Wnt signaling, but these studies have typically focused on extracellular matrix (ECM) stiffness in stem cells or on tumorigenic situations (Schlessinger et al., 2009; Samuel et al., 2011; Fernández-Sánchez et al., 2015; Przybyla et al., 2016).
Several earlier studies have examined the relationship between E-cadherin and Wnt signaling in distinct contexts. For example, Heasman et al. (1994) report that overexpression of cadherins during dorsal ventral patterning in Xenopus can phenocopy inhibition of Wnt, which can occur through depletion of β-catenin. A subsequent study found that binding of cadherins to β-catenin antagonizes their signaling activities (Fagotto et al., 1996). In addition, overexpression of E-cad in Drosophila could mimic a wingless loss of function phenotype (Sanson et al., 1996). Gottardi et al. (2001) found that the tumor suppressor function of E-cadherin is linked to its inhibition of the oncogenic activity of β-catenin in SW480 colon cancer cells. They further show that transcriptionally active β-catenin can be depleted by E-cadherin binding, such that elevated E-cad can directly impact transcription downstream of Wnt and that this effect is observed only with E-cad that can bind to β-catenin. Collectively these previous studies establish in diverse contexts a link between E-cad and Wnt/β-catenin signaling.
Recently our lab identified multiple components of myosin phosphatase in a kinome and phosphatome RNA interference (RNAi) screen to identify novel phosphoregulators of Wnt signaling in developing Drosophila larvae (Swarup et al., 2015). Myosin phosphatase is the major inhibitor of NMII in cells. It consists of two major proteins, either one of two targeting subunits, the myosin phosphatase targeting protein MYPT1/2 (Myosin binding subunit [MBS] in Drosophila) or MYPT3 (Drosophila Mypt-75D) and the catalytic protein phosphatase type 1β (PP1β) subunit (encoded by flapwing [flw] in Drosophila) (Vereshchagina et al., 2004). Myosin phosphatase inactivates NMII by dephosphorylating Thr-18 and Ser-19 (Drosophila Thr-20 and Ser-21), the two critical activation residues of the regulatory light chain (encoded by spaghetti squash (sqh) in Drosophila) of NMII (Karess et al., 1991; Hirata et al., 2009).
NMII is the major actin-binding motor protein that drives actomyosin cytoskeletal contraction. Its activation controls a diverse range of mechanisms included cell shape, adhesion, migration, cell cycle, and cell division (Vicente-Manzanares et al., 2009). NMII regulatory light chain phosphorylation and the resulting contractile force activity can be induced by numerous kinases, some of which also phosphorylate and inhibit myosin phosphatase (Vicente-Manzanares et al., 2009). Several upstream Rho GTPases that activate myosin kinases and thus stimulate NMII can inhibit Wg activity in Drosophila, but a fully defined mechanism is not known (Greer et al., 2013). Here we demonstrate in a systematic manner that actomyosin accumulation due to NMII stimulation can modulate Wnt signaling and tissue patterning by preferentially stabilizing cell–cell adhesion at the AJs to maintain tissue integrity at the expense of transcription and patterning. In doing so, we confirm and extend previous observations from distinct contexts and unite them through a series of connections that we establish through genetic and cell biological examination.
RESULTS
Myosin phosphatase promotes activation of Wg signaling
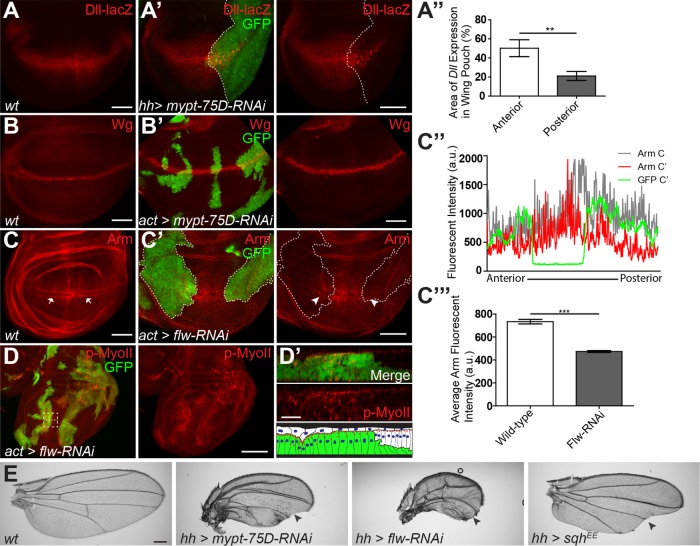
Components of myosin phosphatase were identified in an RNAi screen due to their ability to modulate Wg target gene expression in the wing imaginal disk (Swarup et al., 2015). The Wg target gene Distal-less (Dll) is expressed in a broad domain within the wing pouch (Figure 1A) (Zecca et al., 1996). Expression of mypt-75D-RNAi or flw-RNAi in the posterior domain of the wing imaginal disk using hedgehog (hh)-Gal4 (referred to as hh>mypt-RNAi) caused a significant reduction in the breadth of the Dll transcription domain, compared with the control anterior side of the disk (Figure 1, A′ and A″, and Supplemental Figure S1A). Adult flies had a dramatic reduction in the size of the posterior wing blade as well as notches and loss of wing bristles, hallmarks of reduced Wg signaling (Figure 1E). The use of hh-Gal4 caused dramatic tissue distortions and clefts, so we also utilized actin flip-out clones to generate random misexpression clones in the wing disk. The Wg ligand is expressed in a band two to three cells wide along the dorsoventral (D/V) boundary (Figure 1B), which was unaffected in green fluorescent protein (GFP)-marked actin flip-out clones expressing mypt-75D-RNAi or flw-RNAi (Figure 1B′ and Supplemental Figure S1B), indicating that reduced myosin phosphatase was not disrupting ligand production to inhibit Wg signaling.
FIGURE 1:
Myosin phosphatase regulates NMII and Wg activity during wing development. (A, A′) Dll-lacZ expression in control (hh-Gal4 driving gfp) (A) and hh-Gal4 driving gfp and mypt-75D-RNAi (A′) third-instar wing imaginal disks. (A″) Dll expression area of the wing pouch, in the anterior (GFP negative), and posterior (GFP and MYPT-75D-RNAi positive) domains (n = 7). (B, B′) Wg protein expression in wild type (B) and GFP-marked actin flip-out clones driving mypt-75D-RNAi (B’). (C–C″′) Arm stabilization pattern in wild type (C arrows) and in flip-out clones driving flw-RNAi (C′ arrowheads). Fluorescence intensity plot of Arm and GFP along the D/V boundary of the wing pouch (C″), with average Arm intensity compared in wild type and gfp with flw-RNAi expressing tissue (C″′). (D, D′) p-MyoII stained in flw-RNAi flip-out clones. Cross-section seen in D′ is the magnified dashed line area of D. (E) Adult wings of wild type, and hh-Gal4 driving mypt-75D-RNAi, flw-RNAi, or sqhEE (arrowheads mark loss of bristles and wing margins). Data presented as mean ± SEM; **p = 0.0029, ***p < 0.0001. Scale bars: (A–C) 50 µm, (D) 100 µm, (D′) 20 µm, (E) 300 µm.
We next examined the stability of the key effector, Arm, which is ubiquitously expressed and accumulates at the highest concentrations in the cytoplasm and nucleus in two visible bands of cells flanking the Wg-producing cells (Figure 1C, arrows) (Marygold and Vincent, 2003). Flip-out clones expressing flw-RNAi (Figure 1C′) or hh > mypt-75D-RNAi (Supplemental Figure S1C) both caused reduced stabilized Arm, as seen by loss of bands of stabilized Arm in clones (arrowheads in Figure 1C′). Quantification of fluorescence intensity plots across the D/V boundary of the wing imaginal disk pouch showed a significant reduction in stabilized Arm in flw-RNAi expressing cells (Figure 1, C″ and C″′).
We confirmed that the reduction of Arm was not due to cell death, by staining for the apoptotic marker cleaved caspase-3 (C.Casp-3), and found that apoptosis was not elevated following knockdown of flw or mypt-75D (Supplemental Figure S1, B and C). Knockdown of the other targeting subunit, MBS, gave results similar to flw-RNAi or mypt-RNAi but could induce cell death and was therefore not used in further experiments (Supplemental Figure S2). Taken together these results demonstrate a previously uncharacterized role for myosin phosphatase in the promotion of Wg signaling in Drosophila.
Increased NMII activity inhibits Wg signal activation
The key role of myosin phosphatase is to dephosphorylate and inactivate NMII. We confirmed that in this context knockdown of flw or mypt-75D lead to hyperactive phospho-NMII, as has been shown previously (Vereshchagina et al., 2004; Sun et al., 2011; Urbano et al., 2018). The wing imaginal disk consists of tightly packed columnar epithelial cells where Wg signaling occurs and a thin layer of squamous peripodial epithelium above its apical surface (Figure 1D′, flat white cells in cartoon) (Widmann and Dahmann, 2009). RNAi clones of either flw or mypt-75D had increased phosphorylated Sqh (p-MyoII) (Figure 1D and Supplemental Figure S1D). flw-RNAi cells had elevated levels of p-MyoII, were constricted and formed clefts (Figure 1D′ and Supplemental Figure S1E), and had reduced apical surface area, indicated by E-cad::GFP (Supplemental Figure S1E), another sign of increased NMII activity, and a proxy to force generation (Xie and Martin, 2015).
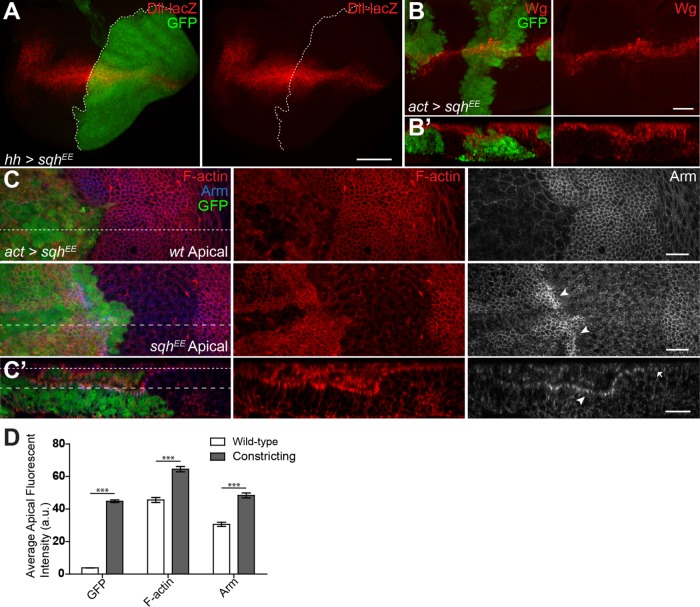
We next asked whether directly activating NMII could phenocopy the loss of Wg signaling seen with myosin phosphatase knockdown. We used an activated phosphomimetic NMII regulatory light chain (SqhEE) (Winter et al., 2001). This transgene has been used to study activated NMII functions in numerous studies (Vereshchagina et al., 2004; Kirchner et al., 2007; Zimmerman et al., 2010). A recent study shows that while SqhEE is not a fully functional phosphomimetic, it does possess more basal activity than nonphosphorylated Sqhwt (Vasquez et al., 2016). Thus, while the reagent is not an ideal way to study fully activated NMII, its behavior in our assays is consistent with increased actomyosin contractility. Using hh-Ga4 to express SqhEE induced notched wings in adults (Figure 1E) and led to reduced Wg target gene expression (Figure 2A). Like myosin phosphatase knockdown, SqhEE could induce tissue constriction (Figure 2, B′ and C′) but did not appear to affect overall Wg protein levels (Figure 2, B and B′). Cells with increased NMII activity also had elevated levels of F-actin along the apical surface (Figure 2, C and D). Activated NMII binds actin and stabilizes filaments as it pulls them together, reducing their turnover rate and causing an overall increase in F-actin in the cell (Murthy et al., 2005). SqhEE expression could also dramatically enrich Arm at the apical surface of the constricting cells (Figure 2C, arrowheads). The increased levels of AJ Arm at the apical surface of constricting cells was also clearly present in cross-sectional analysis of the wing disk, when compared with adjacent wild-type cells (Figure 2, C′ and D), and resembles enriched apical Arm seen previously in flw-RNAi expressing cells (Yang et al., 2012). This suggested that an apical enrichment or sequestration of Arm in cells with increased NMII activity may underlie the reduction in Wg targets.
FIGURE 2:
NMII regulates Wg activity during wing development. (A) hh>sqhEE, gfp stained for Dll-lacZ expression. (B, B′) Total Wg in sqhEE flip-out clones, and cross-section showing cell constriction. (C, C′) GFP flip-out clones driving sqhEE stained for F-actin and Arm. (C′) Cross-section shows apical F-actin and Arm (C′, arrowhead vs. arrow) (dashed lines of C corresponding to section location and depth of C′). (D) Average fluorescence intensity of the apical surface of the wild-type and constricting columnar epithelial cells shown in C′. Data presented as mean ± SEM; ***p < 0.0001. Scale bars: (A) 50 µm, (B–C′) 20 µm.
NMII activation alters Arm localization independently of the destruction complex
As our results indicate that myosin phosphatase likely affects Wg signaling through inactivation of NMII, in subsequent experiments knockdown of myosin phosphatase was used as a proxy for specifically stimulating NMII, as has been done successfully by others (Kirchner et al., 2007; Urbano et al., 2018). Since increased NMII activity led to disrupted Arm localization and a loss of Wg target gene expression, we next asked whether NMII could affect destruction complex proteins. To do this, we used Drosophila salivary gland (SG) cells, as their large cells are ideal for studying protein localization in vivo, and we previously used SGs to characterize the subcellular localization of Wg pathway components (Hall and Verheyen, 2015).
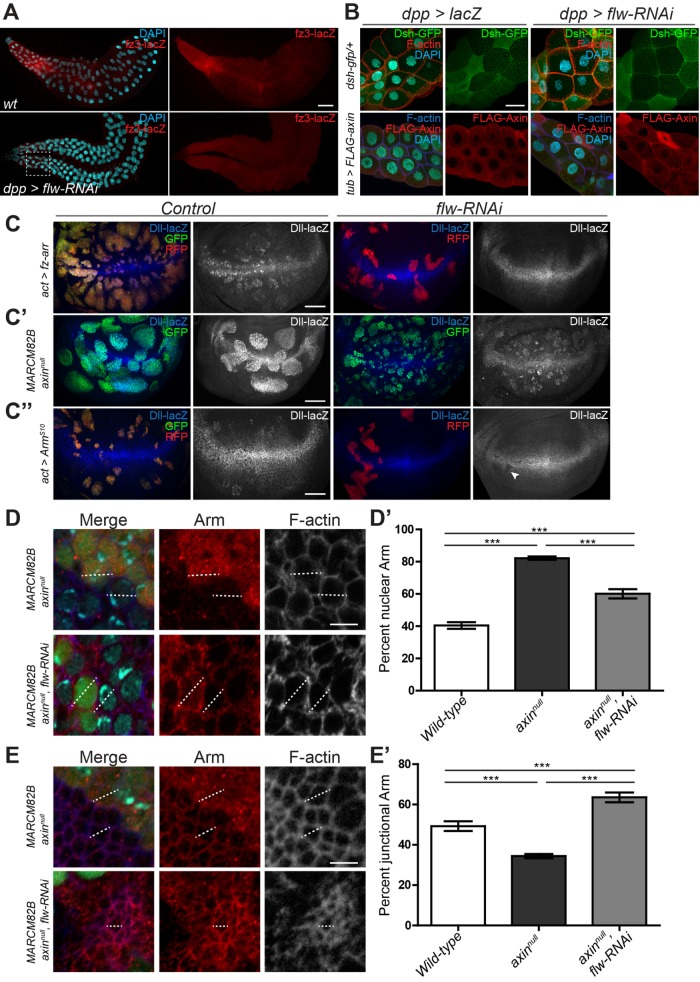
Within the SG, the Wg reporter fz3-lacZ is expressed in a gradient which is highest at the proximal end of the gland (Figure 3A). dpp-Gal4 is expressed uniformly throughout the SG in the Wg receiving cells (Hall and Verheyen, 2015). Knockdown of flw using dpp-Gal4 resulted in a marked reduction in fz3-lacZ expression (Figure 3A), indicating that activated NMII can also inhibit Wg signaling in the SG. After Wg binds to its receptors in the proximal cells of the salivary gland, Dsh, Axin, and other the components of the destruction complex are recruited to the membrane, presumably by Fz (Figure 3B), causing inactivation of the complex (Bilic et al., 2007). While there is some variability in staining in the SG, this was observed in both control and experimental conditions. flw-RNAi appeared to have no effect on the cell surface distribution of either Dsh-GFP or FLAG-Axin (Figure 3B), compared with other proteins previously identified to affect recruitment to the membrane (Hall and Verheyen, 2015), indicating that Wg’s regulation of the destruction complex still occurs in cells with elevated NMII activity. These results suggest that increased NMII activity affects Wg signaling downstream of the receptors.
FIGURE 3:
NMII activity inhibits Wg activation by reducing nuclear Arm independently of the destruction complex. (A) Salivary glands from control or dpp>flw-RNAi stained for fz3 expression to demonstrate the graded response to Wg in expression of the fz3-lacZ reporter. The white box highlights the general area within glands that we examined in B. (B) Localization of Dsh-GFP and FLAG-Axin in the proximal cells of the salivary gland, represented by the general dashed line area of A. (C–C″) Effects of flw-RNAi on ectopic Dll-lacZ in wing imaginal disks: (C) RFP marked flip-out clones expressing Fz-Arr and GFP or flw-RNAi (C′) GFP-positive axinnull MARCM clones and flw-RNAi in axinnull MARCM clones. (C″) ArmS10 flip out clones with GFP or flw-RNAi (arrowhead identifies distinct loss of Dll expression). (D–E′) Effects of flw-RNAi on Arm distribution in GFP-marked axinnull cells, at nuclear (D) and apical sections (E). (D, E) DAPI was used to identify nuclei, and F-actin to mark the edges and junctions of the cell. (D′) Percentage of nuclear Arm in cells was measured as an intensity plot (dotted line D) in wild type (n = 16), axinnull (n = 20), and axinnull, flw-RNAi cells (n = 15). (E′) Percentage of junctional Arm in cells was measured as an intensity plot (dotted line E) in wild-type (n = 13), axinnull (n = 13), and axinnull, flw-RNAi cells (n = 13). Data presented as mean ± SEM; ***p < 0.0001. Scale bars: (A) 100 µm, (B–C″) 50 µm, (D) 5 µm.
To determine where NMII acts within the pathway, we induced ectopic Wg signaling at different points within the signaling cascade and asked whether NMII could suppress ectopic target gene activation. An activated Fz-Arrow fusion protein induced ectopic Dll expression, which could be dramatically suppressed by flw-RNAi (Figure 3C), supporting the model that increased NMII inhibits Wg activity below the level of the receptors. To determine whether NMII affects the destruction complex itself we generated GFP-positive axinnull MARCM clones, as Axin is the scaffolding protein on which the destruction complex assembles (Zeng et al., 1997). Clones lacking Axin had high levels of ectopic Dll and were large due to increased proliferation (Figure 3C′). Expression of flw-RNAi in axinnull MARCM clones could not suppress ectopic Dll, but clones were generally smaller and did not show the smoothed edges seen in the axinnull clones (Figure 3C’), suggesting that NMII can affect aspects of the axinnull phenotype. We next tested expression of a degradation-resistant ArmS10 (Pai et al., 1997). flw-RNAi suppressed ectopic and even some endogenous Dll expression in clones with ArmS10 (Figure 3C″, arrowhead).
Considering cells with increased NMII activity had increased Arm at the apical surface (Figure 2, C and D) suggests that NMII may suppress ectopic ArmS10 activity by inhibiting its ability to enter the nucleus or be retained there. To confirm this, we looked at the relative distribution of Arm in axinnull tissue to eliminate any effect that NMII may have on destruction complex effectiveness and Arm turnover rates. Using F-actin to mark the edges of individual cells and diamidino-2-phenylindole (DAPI) to stain nuclei, intensity plots were drawn across individual cells to look at the distribution of Arm (Figure 3D, dotted line). axinnull cells had roughly double the amount of Arm in the nucleus as wild type (Figure 3D′). Introduction of flw-RNAi resulted in a significant decrease in nuclear Arm in an axinnull background but which was still higher than in wild-type cells (Figure 3D′). These results are consistent with the level of Wg activity and maintained ectopic Dll seen in axinnull cells with flw-RNAi (Figure 3C′). We next looked at these clonal cells in more apical sections to measure AJ associated Arm (Figure 3E). Surprisingly these showed an almost inverse relationship of Arm distribution across the cell surface compared with nuclear sections. axinnull cells had significantly less AJ-associated Arm than both wild-type and axinnull cells with flw-RNAi (Figure 3E′). axinnull, flw-RNAi–expressing cells, although appearing to still have a higher overall Arm intensity across the entire cell than wild type, still maintained the highest ratio of junction-associated Arm, compared with both wild-type and axinnull cells (Figure 3E′), which is similar to what was seen with SqhEE tissue (Figure 2, C and D). Furthermore, similar results were also seen in Myc-tagged ArmS10 clones, as the coexpression of flw-RNAi shifted ArmS10 to the periphery of the cell (Supplemental Figure S3). These findings suggest that increased NMII activity results in reduced entry or retention of Arm in the nucleus, possibly by subcellular redistribution.
NMII activation increases retention of adherens junction proteins
The regulation of Arm localization by NMII could be mediated by any one of the processes that NMII normally influences, including other downstream signaling pathways. We systematically examined key cellular functions of NMII to determine how NMII can modulate Wg signaling.
Loss of myosin phosphatase and increased NMII activity can stimulate JNK (Drosophila Basket [Bsk]) activity in the developing wing disk (Kirchner et al., 2007), and JNK has been shown to promote Wnt signaling (Wu et al., 2008). Using dpp-Gal4 expressed along the anterior/posterior (A/P) boundary of the wing disk (Supplemental Figure S4, A and E) to express flw-RNAi reduced Dll expression, but in this context did not cause elevated JNK activity, seen by expression of JNK target gene puc (Martín-Blanco et al., 1998) (Supplemental Figure S4, A, A′, C, E, E′, and G). Expression of a dominant negative BskDN, inhibiting JNK, did not affect Dll or puc in the wing pouch (Supplemental Figure S4, B and F). Importantly when coexpressed with flw-RNAi, Dll was still reduced and puc was unaltered (Supplemental Figure S4, D and H), indicating that NMII’s ability to suppress Wg signaling is not mediated through JNK. We next investigated NMII’s role in controlling integrin clustering for the formation of focal adhesion and ECM attachment (Vicente-Manzanares et al., 2009). Loss of function clones for the sole Drosophila βPS integrin subunit (Brown, 1993) had no effect on expression of the Wg target Sens (Supplemental Figure S4, I and I′).
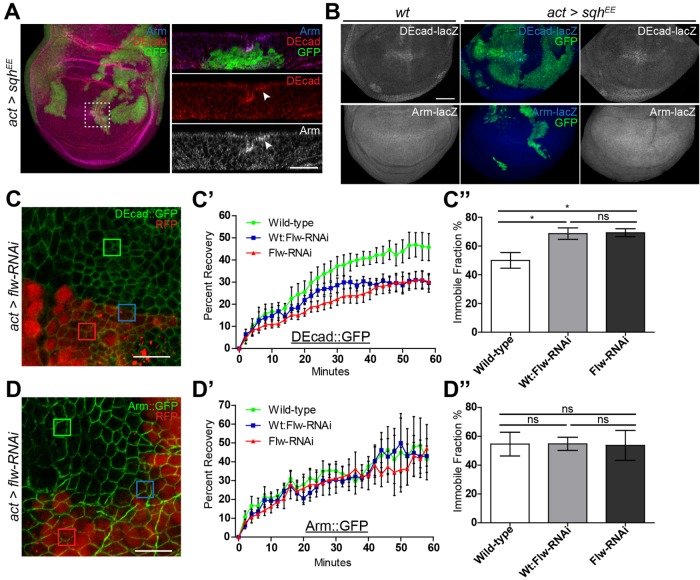
Engl et al. (2014) studied the dynamics of NMII in suspension cell doublets. They showed that as a result of NMII activation (as cells begin to constrict) E-cad increases at the AJ along with other proteins, including β-cat/Arm, which as a result maintains and reinforces cell–cell adhesion. Our finding of increased apical Arm in cells with elevated NMII activity suggested that Wg signaling may be modulated by NMII’s ability to control E-cad clustering and retention during cell constriction. In clones expressing activated NMII (SqhEE), Drosophila E-cad (DE-cad) and Arm are enriched along the apical surface at the AJ of constricting cells (Figure 4A, arrowhead). Activated myosin did not affect transcription of these genes, as seen by expression of lacZ reporters in SqhEE flip out clones (Figure 4B), suggesting that Arm and DE-cad proteins were accumulating at the AJ in cells with increased NMII activity. Such an effect was previously shown, although no link to Wg signaling was tested (Hong et al., 2013; Wu et al., 2014). We performed fluorescence recovery after photobleaching (FRAP) analysis to measure the turnover of DE-cad and Arm at the AJ. We induced random small flip-out clones in the wing disk 72 h after egg laying, which allowed us to maintain all cell genotypes within a relatively close focal plane while imaging, prior to the onset of NMII-induced constriction. FRAP of ubiquitously expressed DE-cad::GFP or Arm::GFP was measured along cell interfaces of wild-type cells (Figure 4, C and D, green box), cells expressing elevated NMII via flw-RNAi (Figure 4, C and D, red box) and at the interface of wild-type/flw-RNAi cells (Figure 4, C and D, blue box). FRAP revealed DE-cad recovery rates are significantly reduced at any flw-RNAi cell interfaces (Figure 4C′) and contained a significantly higher immobile fraction (Figure 4C″), while Arm was unaltered (Figure 4, D′ and D″). The inability to detect a change in Arm may be do the random positioning of the clones within the wing disk, as total Arm, and its distribution within a cell vary dramatically dependent on a cell’s position within the disk. The altered DE-cad recovery and immobile fraction rate, unaltered transcription coupled with protein accumulation, are likely due to the increased force generated by NMII which can stabilize cortical F-actin, which in turn stabilizes DE-cad at the AJ (Goldenberg et al., 2013; Hong et al., 2013; Engl et al., 2014; Wu et al., 2014). This accumulated DE-cad at the AJ can subsequently bind and accumulate Arm, as has been shown in other contexts previously (Fagotto et al., 1996; Gottardi et al., 2001).
FIGURE 4:
NMII activation increases retention of adherens junction proteins. (A, B) GFP-marked actin flip-out clones driving sqhEE stained for (A) DE-cad and Arm (arrowheads identify apical increases) and (B) expression of DE-cad or arm. (C–D″) FRAP analysis of DE-cad::GFP and Arm::GFP in wing imaginal disks with RFP-marked flw-RNAi expressing flip-out clones. (C, D) DE-cad::GFP and Arm::GFP wing imaginal disk with squares indicating bleached regions of the wing disk. Green squares represents wild-type cell interfaces, blue for wt:flw-RNAi cell interface, and red for flw-RNAi cell interfaces. (C′) AJ DE-cad::GFP (n = 6) recovery curves of FRAP analyses show cells adjacent to or expressing flw-RNAi have significantly slower recovery rates than wild type (p = 0.0029), (C″) and greater immobile protein fractions, *p < 0.05. (D′) flw-RNAi had no effect on AJ Arm::GFP (n = 6) recovery curves from FRAP analyses (p = 0.4794), (D″) or immobile protein fractions. Data presented as mean and mean curve ± SEM. Scale bars: (A) 20 µm, (B) 50 µm, (C, D) 10 µm.
NMII mediates DE-cad accumulation and sequesters Arm to the AJs, inhibiting Wg signaling
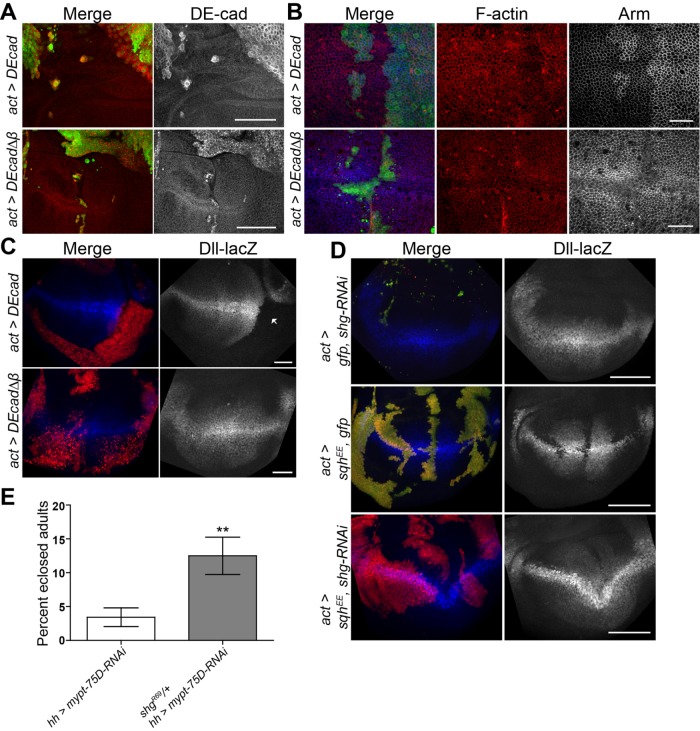
To further study the role of DE-cad in regulation of Arm, we expressed full-length DE-cad and a truncated version of DE-cad lacking the Arm binding domain (DEcad∆β). Flip-out clones in which these constructs were expressed were very unhealthy and difficult to characterize. Nonetheless we were able to make several observations. Ectopic wild-type DE-cad was uniformly enriched along the cell periphery at the AJs, while DEcad∆β expression was also seen in puncta (Figure 5A), since DE-cad that cannot bind Arm is endocytosed and accumulates in vesicles (Langevin et al., 2005). Expressing either transgene had no effect on levels or distribution of F-actin (Figure 5B). However, ectopic DE-cad dramatically increased Arm at the AJ (Figure 5B) and could strongly suppress Dll expression (Figure 5C, arrow). DE-cad’s ability to suppress Wg signaling has been previously reported (Sanson et al., 1996) and is likely due to the higher binding affinity of Arm to DE-cad over TCF binding for transcriptional activation (Torres et al., 2007). Thus any cells exhibiting increased levels of DE-cad will titrate freely available Arm to the AJ, stabilizing DE-cad and increasing cellular adhesion. This is supported by the fact that expression of DEcad∆β resulted in decreased levels of AJ Arm and did not suppress Dll expression (Figure 5, B and C).
FIGURE 5:
NMII activation inhibits Wg signaling through DE-cad. (A, B) GFP-marked actin flip-out clones expressing DE-cad or DE-cad∆β, stained for (A) DE-cad, or (B) F-actin and Arm. (C) RFP-marked actin flip-out clones expressing DE-cad or DE-cad∆β, stained for Dll expression. (D) Dll-lacZ expression in RFP-marked clones of the indicated genotypes. (E) Eclosion percentage of hh>mypt-75D-RNAi and hh>mypt-75D-RNAi heterozygous for shg Drosophila. Data presented as mean ± SEM; **p = 0.0022; n ≥145. Scale bars: (A, C, D) 50 µm, (B) 20 µm.
We next examined the effects of reduction of overall DE-cad on Wg signaling. RNAi against shotgun (shg), which encodes DE-cad, appeared to induce cell death (based on the punctate appearance of clones) and failed to induce any viable clones within the wing disk pouch (Figure 5D). Clones of activated NMII (SqhEE) caused a strong suppression of Dll expression, which was rescued by coexpression of shg-RNAi (Figure 5D). Furthermore, widespread cell death was suppressed when shg-RNAi was coexpressed with SqhEE, as seen by viability of flip-out clones. Further evidence for a genetic interaction was seen when we observed that hh>mypt-75D-RNAi adult flies had a much lower than expected viability and eclosion rate, which was rescued by heterozygosity for the shgR69 null allele (Godt and Tepass, 1998) (Figure 5E).
NMII’s effect on the Wg pathway is mediated through F-actin stability
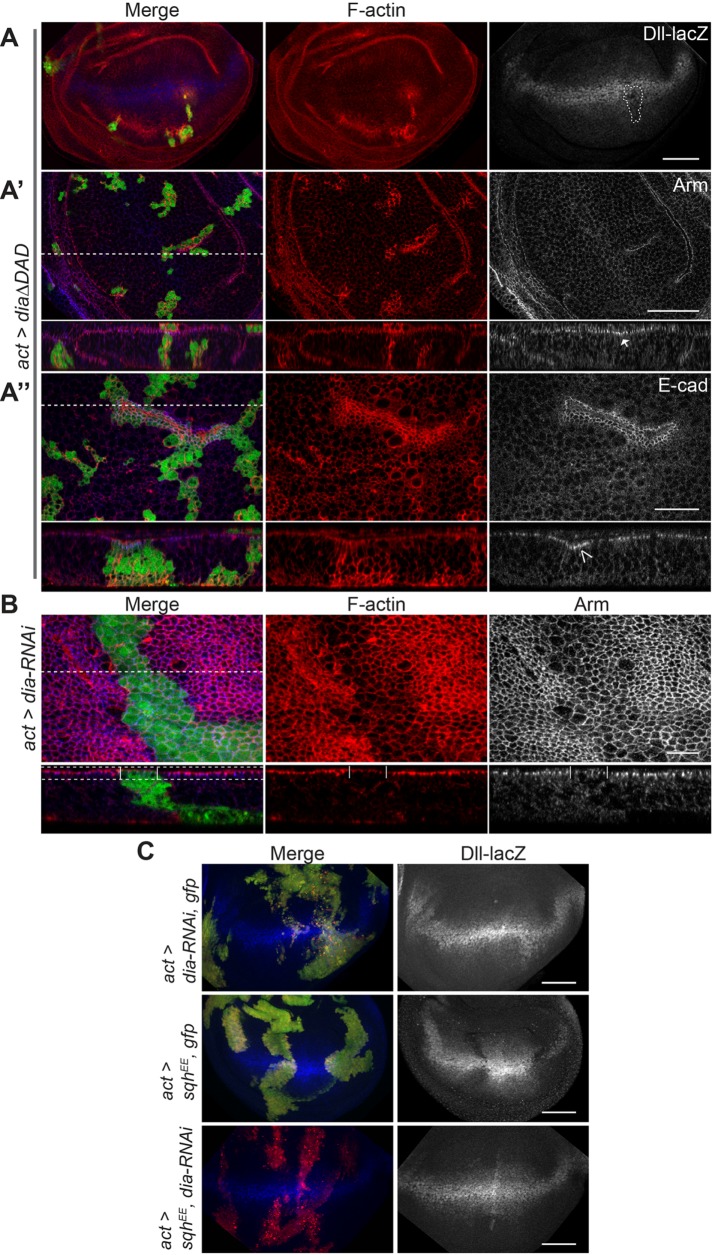
Engl et al. (2014) demonstrated that increased NMII activity results in decreased F-actin turnover, which can guide E-cad clustering and retention (Hong et al., 2013). To confirm whether F-actin levels in a developing tissue could also affect Wg activation we used the formin protein Diaphanous (Dia), which promotes the polymerization of filamentous actin (Afshar et al., 2000). Mitotic clones expressing a constitutively active Dia protein lacking its autoinhibitory domain (Dia∆DAD) had dramatic increases in levels of F-actin (Figure 6, A and A″) and partially phenocopied the effects of increased NMII activation. Clones had mildly decreased levels of Dll (Figure 6A, arrowhead), a correspondingly mild increase in apical Arm, and enriched DE-cad along the apical surface of the cells (Figure 6, A′, arrow, and A″, open arrowhead). Furthermore, cells in larger clones appeared to constrict, likely due to filament cross-linking and bundling (Sun et al., 2010) (Figure 6A″, open arrowhead).
FIGURE 6:
NMII inhibits Wg signaling by increased F-actin. (A–A″) GFP-marked actin flip-out clones expressing dia∆DAD stained to detect (A) F-actin and Dll-lacZ, (A′) F-actin and Arm, and (A″) F-actin and DE-cad. dia∆DAD induces increased apical AJ Arm (A′, arrow) and DE-cad (A″, open arrowhead), and cell contractions (A″, open arrowhead). (B) GFP-marked actin flip-out clones expressing dia-RNAi stained for F-actin and Arm. Apical sections visualized from dashed box area shown in cross-section, with clone area marked by vertical lines. (C) GFP-marked actin flip-out clones of indicated genotypes stained to detect Dll-lacZ. Dashed lines correspond to location visualized in cross-section images. Scale bars: (A, A′, C) 50 µm, (A″) 20 µm, (B) 10 µm.
To confirm whether the effect of NMII on Wg is directly mediated through F-actin stability and constriction, we tested whether reduced F-actin could alleviate NMII’s suppression of Wg signaling, using dia-RNAi. Cell with low levels of Dia had lower levels of F-actin, as well as increased apical cell surfaces, marked by Arm, mimicking what is seen with reduction of NMII activity (Rauskolb et al., 2014) (Figure 6B). Reduction of F-actin via dia-RNAi had no major effect on endogenous Wg signaling, as seen by wild-type Dll expression levels, although the overall disk morphology is distorted (Figure 6C). In an activated myosin background (SqhEE), which strongly inhibits Dll, the coexpression of dia-RNAi could restore wild-type Dll expression (Figure 6C). These results confirm that NMII can influence Wg pathway activity through its function to bind and stabilize actin leading to accumulation and constriction of filamentous actin.
NMII regulates Wnt in mammalian cells by sequestering β-cat to E-cad containing AJs
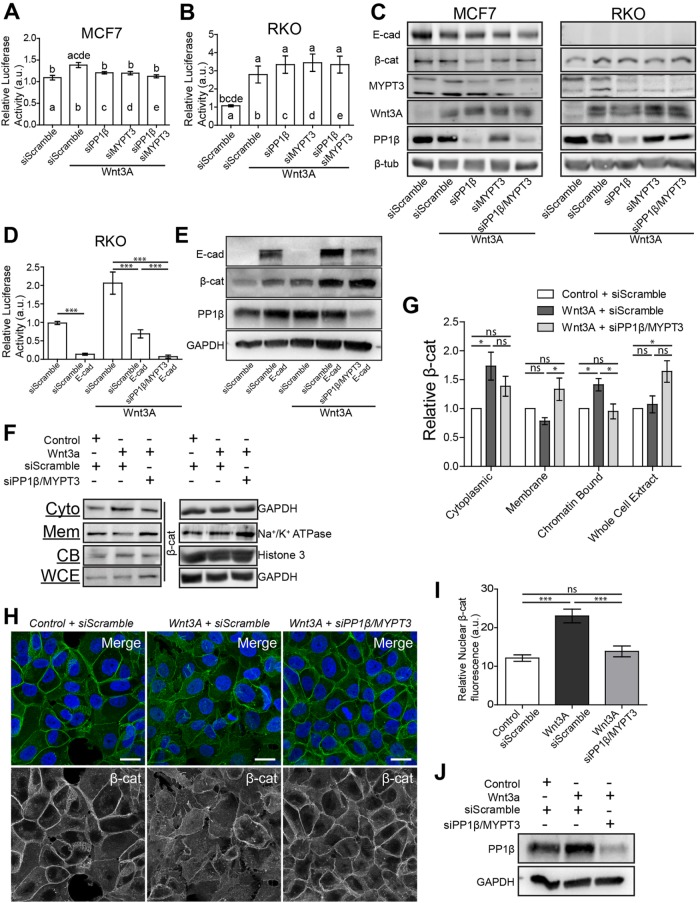
We next examined the effects of increased NMII activation on Wnt signaling in human cell lines. Wnt pathway activity was induced in MCF7 and RKO cells by transfection of Wnt3A, and the response was measured using a TCF-responsive TOPFLASH transcriptional reporter (Korinek et al., 1997). MCF7 are epithelial cells that have well defined adherens junctions (de Beco et al., 2009), while RKO cells are mutant for E-cad and completely lack E-cad–based adherens junctions (Figure 7C). The only β-cat present in RKO cells is solely for the regulation of Wnt target gene activation (Gagliardi et al., 2008). Transfection of Wnt3A resulted in a significant increase in reporter activity in both cell lines, although RKO cells had a more robust response (Figure 7, A and B). To increase NMII activation within these cell lines, we transfected in small interfering RNA (siRNA) against individual myosin phosphatase components. siPP1β could reduce total PP1β by ∼70% (Figure 7C), while siMYPT3 reduced MYPT3 (the orthologue of Mypt-75D) by ∼60% (Figure 7C). Knockdown of these components reduced the amount of the other myosin phosphatase protein, suggesting complex formation is essential for stability (Figure 7C). In MCF7 cells, knockdown of PP1β or MYPT3 could reduce Wnt activation significantly, and cotransfection of siPP1β and siMYPT3 reduced transcriptional activity back to baseline levels similar to that in cells with no Wnt3A (Figure 7A). In RKO cells, there was no significant change following knockdown of either or both subunits; in fact, there was a slight increase in Wnt transcriptional activation (Figure 7B). The reintroduction of E-cad into RKO cells by transfection resulted in a strong suppression of endogenous Wnt activity and increased the amount of β-cat found in cells, which is roughly equal to levels seen when exposed to Wnt3A alone (Figure 7, D and E). Treatment of RKO cells with Wnt3A induced TOPFLASH expression. Transfection of E-cad brought transcriptional activity roughly back to baseline levels, while further increasing the amount of total β-cat (Figure 7, D and E). Importantly, following knockdown of PP1β and MYPT3, Wnt-treated RKO cells transfected with E-cad showed further suppression of Wnt transcriptional activation (Figure 7D). This indicates the importance of E-cad for NMII-based regulation of Wnt activity (Figure 7, B and D). These results indicate that increased NMII activation from reduction of myosin phosphatase components can inhibit Wnt activity in mammalian cells as well but only in cells that contain E-cad adherens junctions.
FIGURE 7:
NMII activation recruits β-cat to cell membranes inhibiting Wnt signaling. (A, B) Wnt-responsive luciferase TOPFLASH assay measuring TCF/LEF reporter activity in (A) MCF7 and (B) RKO cells following Wnt3A transfection and siPP1β and siMYPT3 transfection. Letters above each column represent significant difference from corresponding columns, (p < 0.01). (C) Western blot analysis of total cellular levels of E-cad, β-cat, MYPT3, Wnt3A and PP1β. β-Tub was used as a loading control. (D) TOPFLASH assay of RKO cells transfected with combinations of E-cad, Wnt3A and siPP1β with siMYPT3. (E) Western blot analysis of total cellular levels of E-cad, β-cat, and PP1β of RKO cells from conditions in D. GAPDH was used as a loading control. (F) Western blot analysis of β-cat in cytoplasmic (Cyto), membranous (Mem), chromatin-bound (CB), and whole cell extract (WCE) fractions in MCF7 cells. GAPDH, Na+/K+ ATPase, and Histone 3 were used as loading controls for corresponding fractions. Complete fraction, see Supplemental Figure S5. (G) Quantification of relative levels of β-cat from each fraction taken from F, normalized to Control + siScramble conditions (n = 4 biological replicates). (H) MCF7 cells stained for β-cat and DAPI. (I) Average fluorescence intensity of nuclear β-cat of MCF7 cells in Control + siScramble (n = 13), Wnt3A + siScramble (n = 13), and Wnt3A + siPP1β/siMYPT3 (n = 13) transfected cells. Values normalized to the maximum fluorescence intensity equal to 100 for each cell quantified. (J) Western blot analysis of total PP1β for cells analyzed in H and I. GAPDH was used as a loading control. All data are presented as mean ± SEM. ns = not significant, *p < 0.05, ***p < 0.0001. Scale bars: 20 µm.
Although reduction of PP1β or MYPT3 was able to inhibit Wnt signaling in MCF7 cells there was no dramatic change in overall β-cat levels (Figure 7C), suggesting that there may be a localization defect as seen in Drosophila. To address this, we performed cell fractionation and examined subcellular protein distribution. Fractionation efficiency was established using antibodies against established markers of each compartment (Supplemental Figure S5). Transfection of Wnt3A induced a significant increase of cytoplasmic and chromatin-bound (transcriptionally active) β-cat, while membrane associated (AJ) β-cat slightly decreased (Figure 7, F–H). The decrease in membranous β-cat is likely due to the fact that Wnt can induce mild EMT effects in MCF7 and other epithelial cancer lines (Green et al., 2013). Similar effects were seen with E-cad (Supplemental Figure S5). Whole cell extracts showed only a minor increase in total β-cat (Figure 7, F and G). A striking inverse in distribution was seen after increasing NMII activation via siPP1β and siMYPT3 transfection. Cytoplasmic levels of β-cat decreased, and there was a significant reduction in chromatin-bound levels back to baseline, matching results seen in TOPFLASH and immunofluorescence assays (Figure 7, A and F–I). Cells with activated NMII had a significant increase in membrane associated β-cat levels, as well as increased total β-cat (Figure 7, F and G). Considering the increase in total β-cat, yet lack of transcriptional activation, and redistribution of the protein within the cell, we propose that in mammalian cells with elevated NMII activity, freely available b-cat is enriched at AJs to enforce cell–cell adhesion at the cost of transcriptional activation of Wnt targets.
NMII activation modulates Wnt signaling during development and homeostasis to maintain cell–cell adhesion
We next tested this model in Drosophila. We generated mitotic recombinant clones that can form and maintain AJs without the need for Arm using functionally validated fusion proteins in which DE-cad is fused to α-cat (DEcad::αCat), as well as a truncated fusion of the proteins lacking their Arm binding domains (DEcad∆β::αCat∆VH1) (Desai et al., 2013). Actin flip-out clones expressing either transgene led to increased levels of DE-cad and did not affect levels of F-actin (Supplemental Figure S6, A–D). DEcad::αCat could still bind Arm, forming puncta within the cells and resulting in a suppression of Dll expression (Supplemental Figure S6, A and C). DEcad∆β::αCat∆VH1, which cannot bind Arm, did not affect Arm distribution or Dll expression (Supplemental Figure S6, B and D), so we utilized this transgene for further experiments.
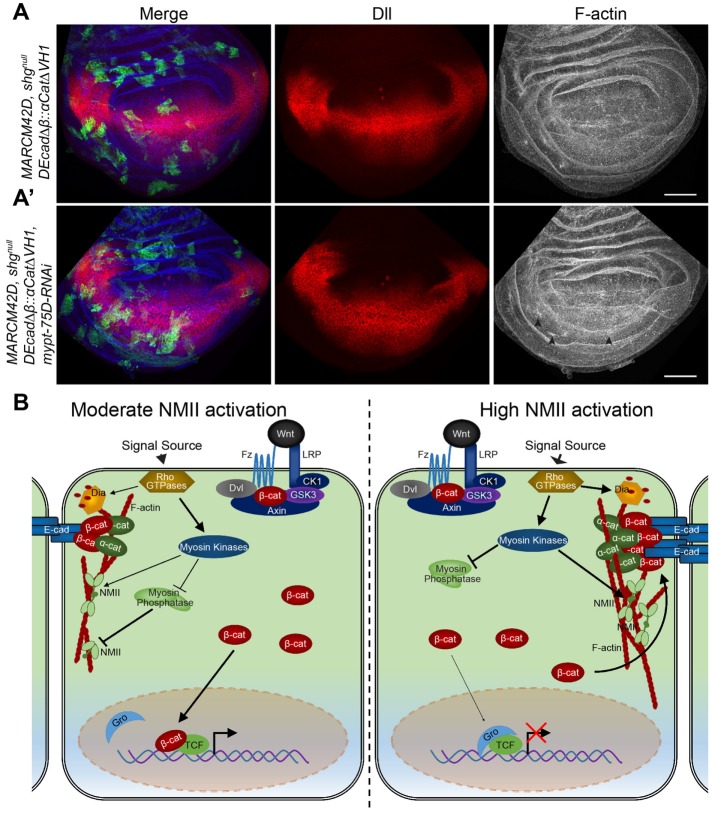
Mitotic clones of the shgR69 null allele are nonviable and were quickly extruded from wing disk (unpublished data). When DEcad∆β::αCat∆VH1 was expressed in these cells using the MARCM technique, clones could divide and grow (Figure 8A), confirming that DEcad∆β::αCat∆VH1 could form AJs without binding Arm. Clonal tissue had normal Dll protein levels and F-actin (Figure 7A). Importantly, the expression of mypt-75D-RNAi in shg clones had no effect on Dll but did induce minor accumulations in F-actin (Figure 8A′, arrowheads). These results were mimicked when DEcad∆β::αCat∆VH1 was coexpressed with SqhEE, namely clonal tissue still constricted and had elevated F-actin, but the reduced Dll expression was significantly rescued (Supplemental Figure S6E and Figure 5D). These results indicate that NMII activation can only suppress Wg signaling in this epithelium when DE-cad binds to Arm.
FIGURE 8:
NMII activation inhibits Wnt signaling in a dynamic manner across developing tissue by Arm titration to AJs. (A, A′) GFP-marked shgnull MARCM clones expressing (A) DE-cad∆β::α-cat∆VH1 (n = 18) and (A′) DE-cad∆β::α-cat∆VH1 with mypt-75D-RNAi (n = 8), stained for Dll and F-actin (arrowheads show F-actin accumulation). (B) Model for regulation of Wnt signaling by activation of NMII resulting in AJ accumulation and stabilization in response to contractile forces; see text for details. Scale bars: (A, A′) 50 µm.
DISCUSSION
The link between mechanical forces and canonical Wnt signaling has been extensively studied in contexts including mesoderm differentiation during cardiomyogenesis (Happe and Engler, 2016) and the effects of tissue stiffness of the ECM on stem-cell behavior and mechanical strain-induced proliferation during carcinogenesis (Benham-Pyle et al., 2015, 2016; Przybyla et al., 2016). These studies have looked at the interplay between cell–cell adhesion and E-cadherin and β-catenin, as well as with the Hippo pathway transcription activator Yap. They all utilize stretch/ECM stiffness mechanisms to increases tension driving proliferation and transcriptional initiation, demonstrating that increased cell–matrix adhesion and concurrent cell–cell tension drive these responses across a monolayer (Benham-Pyle et al., 2015, 2016; Przybyla et al., 2016). Less research has focused on how mechanical forces and maintenance of cell–cell adhesion may directly influence Wnt activation in normal developing epithelia. Our work shows that in a cell autonomous manner, increased NMII activation in epithelial cells induces contraction and accumulation of cortical F-actin, and as a result E-cad accumulates and titrates freely available Arm/β-cat to the AJs in order to maintain cell–cell adhesion. The resulting decreased levels of cytoplasmic Arm/β-cat causes insufficient nuclear translocation and reduced Wnt target gene transcription (Figure 8B).
We show that NMII activation can inhibit the nuclear accumulation of Arm causing a suppression of overall transcriptional initiation, even in genotypes lacking Arm degradation machinery or expressing Arm that is resistant to degradation. These results identify a distinct mechanism in regulating Wnt signaling, independent of the adhesion-based enhanced destruction complex activity turnover of Arm/β-cat (Maher et al., 2009). Our results are complementary to those of Greer et al. (2013), who identified that RhoGEF and GTPase activity (upstream activators of NMII) could suppress Arm localization or activation in developing Drosophila. We further excluded the possibility that NMII suppresses Wnt through several known interaction mechanisms, like JNK and ECM/Integrin signaling (Wu et al., 2008; Przybyla et al., 2016).
Cells with elevated NMII activity and adjacent cells that were under increased contraction exhibited elevated Arm and DE-cad at AJ, while transcription rates of the genes encoding these proteins appeared normal. DE-cad (shg) was previously identified as a Wnt target gene in wing disks (Widmann and Dahmann, 2009). Its maintained transcription in flw-RNAi cells may be due to compensation by another transcriptional mechanism. Furthermore, the recovery rate of DE-cad to the AJ was reduced and contained a higher immobile fraction of DE-cad, indicating accumulation and retention. Although we did not detect any significant changes in Arm recovery rates or immobile fraction at the AJs, this may be due to positional effect of our measurements across the wing disk. Total Arm and its distribution within a cell vary dramatically across the wing disk. To maintain healthy tissue to generate accurate measurements, we induced flw-RNAi clones at random positions in the tissue, which may explain the high variance. As FRAP experiments encompassed the entire AJ and membrane, the recovery rates are likely an indirect measure of vesicle trafficking of Arm and DE-cad (Goldenberg et al., 2013). This is bolstered by the fact that decreased actomyosin levels have been shown to increase AJ endocytosis in developing Drosophila (Goldenberg et al., 2013). The accumulation of E-cad is likely an active mechanosensitive mechanism to bolster cell–cell adhesion, potentially through plasma membrane clustering and vesicle-based redistribution (Hong et al., 2013; Engl et al., 2014; Lecuit and Yap, 2015).
Early studies of arm in Drosophila revealed the dual role of β-catenin in both Wg signaling and at the AJ through interactions with E-cad and α-catenin (Harris and Peifer, 2005). These studies were complemented by work done in Xenopus and cultured mammalian cells that collectively show that elevated E-cad can suppress Wnt/Wg signaling (Heasman et al., 1994; Fagotto et al., 1996; Sanson et al., 1996; Gottardi et al., 2001). This effect is likely due to the higher binding affinity of Arm/β-cat to E-cad over TCF for transcriptional activation (Cox et al., 1996; Torres et al., 2007). In this study, we show that the activation status of NMII can directly impact the stabilization of the β-cat/E-cad complex and thus affect Wnt signaling readouts. We validated this model by expressing wild-type DE-cad or mutant DE-cad, lacking the Arm binding sequences. Our results demonstrated that ectopic DE-cad caused high levels of Arm to be enriched along the AJ and strongly suppressed Wg target gene expression, while mutant DE-cad led to reduced AJ Arm and did not affect Wg targets. Importantly neither of these transgenes had any effect on F-actin, showing that E-cad acts downstream of F-actin in this NMII activation pathway to suppress Wnt signaling. In addition, the reduction of DE-cad in wing disk tissue was able to rescue Wg activity defects and viability of flies expressing activated NMII.
In our in vivo work, we were able to confirm the model by Engl et al. (2014) and Hong et al. (2013) that NMII activation recruits and stabilizes DE-cad, Arm, and other AJ core proteins in developing tissue by stabilization and accumulation of F-actin and in our context resulting in a suppression of Wg activation. The expression of a constitutively active formin protein phenocopied the ability of activated NMII to inhibit Wg target gene expression, and the reduction of F-actin was able rescue the effect of increased NMII activity on Wg target gene expression.
Building on our Drosophila work, we were able to confirm that NMII has similar effects on Wnt in human cells that contain E-cad–based AJs. Stimulation of NMII was able to suppress Wnt transcriptional activation in MCF7 cells containing AJ but had no effect in RKO cells. Interestingly, although RKO cells lack E-cad, they exhibit elevated levels of N-cadherin (Yan et al., 2015), indicating that NMII stimulation likely does not affect Wnt activity through N-cad–based cell–cell adhesion. Furthermore, reintroduction of E-cad into RKO cells not only supressed Wnt activity but also further sensitized these cells to NMII activation, indicating a specific role for contractile forces and E-cad–based adhesion. Importantly, increased NMII activity following knockdown of myosin phosphatase induced a significant redistribution of β-cat from the nuclear/chromatin-bound fraction to the membrane and increased overall β-cat protein levels within the cells. Although these cells had significantly higher levels of β-cat, their inability to initiate Wnt target gene transcription indicates that β-cat is being sequestered at the AJ, as was seen in immunofluorescence analysis.
We validated this model by generating clonal tissue in which we replaced endogenous E-cad with a fusion protein that does not require Arm/β-cat for the formation of complete AJ. In these cells, when NMII activity was stimulated there was no suppression of Wnt target gene transcription, confirming NMII inhibits Wnt by titrating Arm/β-cat to the adherens junctions.
This may be a physiological regulatory mechanism in developing tissue for proliferation, patterning, and morphogenesis and later in the homeostasis of epithelia. As tissues proliferate, change shape, and respond to physical cues, the cells respond to all these factors and induce variable levels of NMII activation. To maintain overall tissue integrity as cells change their shape, cell–cell adhesion must be increased. This results in the sequestration of Arm/β-cat to increase AJ adhesion and inhibit canonical Wnt’s ability to promote patterning. In essence, the preservation of tissue integrity overrides Wnt-inducible gene expression in epithelia. Recently there have been comparable instances of this in other developmental signaling pathways, through distinct mechanisms. Increased NMII activation and cytoskeleton tension have been widely identified to inhibit Hippo signaling, while stimulating JNK activation in epithelia (Kirchner et al., 2007; Khoo et al., 2013; Rauskolb et al., 2014). Rauskolb et al. (2014) studied the tension in wing imaginal disks at different developmental time points and found that in older third-instar wing disks (which are the same times that we examined) there is reduced cytoskeletal tension, compared with younger disks. Modulation of myosin through knockdown of Rho-associated kinase had similar effects to those we observed with dia-RNAi, namely decreased F-actin intensity and increased the apical surface area of the cell. They also show that elevated tension leads to recruitment of Warts to junctions via Ajuba, and this leads to elevated Yorkie (Yki) activity. They did not examine effects on junctional Arm or Wg signaling. In another study, Wittkorn et al. (2015) demonstrate regulation of Wg transcription by Yki in eye imaginal disks. This might suggest that our observed effects could be due to enhanced Wg expression as a result of increased Yki activity following myosin activation. However, in our work, we see no effect on Wg protein levels, suggesting that such regulation is not occurring in the wing imaginal disk. Therefore, while links between junctional tension and Yki, and Hippo and Wg signaling have been made in Drosophila, no clear mechanism links these two studies. Thus, it is likely that modulating myosin in disks, as we have done in our study, may also affect Hippo signaling, but we feel there is no contradiction to our conclusions with respect to the effect of Hippo on Wg signaling as previously described. With this study, we demonstrate that canonical Wnt signaling is another key developmental pathway that is regulated by NMII.
It will be interesting to determine whether these dynamic contractile forces exerted on developing tissue are also critical for the maintenance of stem-cell niches. For instance, in intestinal crypts, Wnt activity is refined to the very basal cells of the crypt for the maintenance of stem cells, where cells are apically constricted to form the concave base of the crypt (Buske et al., 2012). Monitoring Wnt activity in crypt cells as well as proliferation and differentiation rates when exposed to increased or decreased levels of NMII activity, or even removing the tissue to grow on a flat surface, may provide insights into the relevance of why and how tissue structures arise due to the forces that are exerted on cells across a tissue in order to regulate homeostasis. The modeling of stem-cell niche formation in crypts has suggested that the loss of curvature regulation can result in tissue consisting of Paneth cells of the crypt and undifferentiated cells, which is seen in some cases of intestinal adenoma and carcinoma (Buske et al., 2012). Our results here have provided new and supportive evidence that the interactions between biochemical signaling, Wnt in this case, and mechanical forces, guiding cell shape and adhesion, are critical for the normal development and maintenance of healthy epithelial tissue in an organism.
MATERIALS AND METHODS
Drosophila husbandry, crosses, and clone generation
Fly strains and crosses were raised on standard medium at 25°C unless stated otherwise. w1118 was used as wild type. In assays examining the interactions between two or more UAS transgenes, control crosses were performed with UAS-lacZ or UAS-GFP to rule out effects due to titration of Gal4. Heat-shock inducible actin flip-out clones were generated by crossing either RFP-marked flip-out or GFP-marked flip-out strains to corresponding lines, and then larvae were heat-shocked at 37°C for 12.5 or 15 min, respectively, 48–72 h after egg laying (AEL) (depending on the assay), and incubated at 29°C until dissection. Mosaic analysis with a repressible cell marker (MARCM) clones were generated by crossing MARCM lines to corresponding lines and larvae were heat-shocked at 37°C for 1.5 h, 48 h (MARCM82B), or 72 h (MARCM42D) AEL and incubated at 29°C until dissection.
The following fly strains were used: 1) Dll-lacZ (BL10981), 2) dpp-Gal4 (BL 1553), 3) y1 sc* v1; P{TRiP.HMS00521}attP2 (mbs-RNAi) (BL 32516), 4) UAS-bskDN (BL 6409), 5) y1 w67c23; P{lacW}shgk03401/CyO (shg-lacZ) (BL 10377), 6) w*; P{FRT(whs)}G13 shg1/CyO; P{Ubi-p63E-shg.GFP} (ubi-shg-GFP) (BL 58471), 7) UAS-armS10 (BL 4782), 8) arm-GFP (BL 8555), 9) UAS-diaΔDAD (BL 56752), 10) y1 v1; P{TRiP.HM05027}attP2 (UAS-dia-RNAi) (BL 28541), 11) mys1,FRT19A/FM7c (βPSnull) (BL 23862) (obtained from the Bloomington Drosophila Stock Center), 12) UAS-flw-RNAi) (VDRC 104677, 29622), 13), UAS-mypt-75D-RNAi (VDRC 109909), 14) UAS-mbs-RNAi (VDRC 105762), 15) UAS-shg-RNAi (VDRC 27082), (obtained from the Vienna Drosophila Resource Center), 16) ;;pucE69-lacZ (Ring and Martinez Arias, 1993), 17) fz3-lacZ/FM7a (Sato et al., 1999), 18), yw, arm-lacZ, FRT19A;; eyFLP/TM6B (arm-lacZ) (Vincent et al., 1994), 19) hh-GAL4 (Port et al., 2011), 20) UAS-sqhE20E21 (UAS-sqhEE) (Winter et al., 2001), 21) UAS-DEcad::αcat∆VH1 (Desai et al., 2013), 22) shgR69, FRT42D/CyO (shgnull) (Godt and Tepass, 1998), 23) hsFLP;; Act>CD2>Gal4, UAS-GFP/SM6∼TM6 (GFP-marked flip-out) (Bruce Edgar, Zentrum für Molekulare Biologie der Universität Heidelberg, Germany), 24) y,w,hsflp,UAS-GFP,tub-Gal4;;FRT82B tubGal80 (MARCM82B) (Bruce Edgar, Zentrum für Molekulare Biologie der Universität Heidelberg, Germany), 25);;UAS-GFP,hsflp122,FRT42,tub-GAL80,tub-GAL4/TM6B (MARCM42D) (Jessica Treisman, NYU School of Medicine), 26) UAS-DEcad∆β::αcat∆VH1 (Ulrich Tepass, University of Toronto, Department of Cell & Systems Biology), 27) Dsh-GFP/CyO (Jeffrey Axelrod, Dept. of Pathology, Stanford University School of Medicine), 28) yw; tub>FLAG-axin/CyO (Marcel Wehrli, Oregon Health & Science University), 29) UAS-fz-myc-arr, (Marcel Wehrli, Oregon Health & Science University), 30) axinS044230,FRT82B/TM6B (axinnull) (Marcel Wehrli, Oregon Health & Science University), 31) en-Gal4,UAS-gfp (Konrad Basler, Institute of Molecular Life Science, University of Zurich, Switzerland), 32) y,w,hsflp122; sp/CyO; Act>CD2>GAL4,UAS-RFP/TM6B (RFP-marked flip-out), 33) shgR69,FRT42D,mypt-75D-RNAi; UAS-DEcad∆β::αcat∆VH1/SM6a∼TM6B.
Immunofluorescence, wing mounting, and imaging
Third-instar larvae were dissected in phosphate-buffered saline (PBS). Wing imaginal disks and salivary glands were fixed in 4% paraformaldehyde at room temperature for 20 min followed by three washes in PBS for 5 min. Tissue was blocked (2% bovine serum albumin diluted in PBS 0.1% Triton X-100 [PBST]) for 45 min at room temperature, followed by incubation with primary antibodies overnight at 4°C. Tissue was then washed three times for 5 min with PBST and incubated with secondary antibodies at room temperature for 1.5 h. Phalloidin-rhodamine, or -647 (ThermoFisher Scientific), or phalloidin-fluorescein (Sigma-Aldrich) and DAPI (ThermoFisher Scientific) were added at this point, if required. A final series of three PBST washes were performed, followed by mounting in 70% glycerol in PBS. The following primary antibodies and dilutions were used: mouse anti-β-galactosidase (1:2000; Promega), mouse anti-Wg (1:100; DSHB), mouse anti-Arm (1:50; DSHB), rabbit anti-cleaved Caspase 3 (1:100; Cell Signaling), guinea pig anti-Sens (1:500, a gift from Hugo Bellen, Baylor College of Medicine and Howard Hughes Medical Institute), mouse anti-Dll (1:300; a gift from Ian Duncan, Washington University), rabbit anti-Phospho-Myosin Light Chain 2 (Ser19) (p-MyoII) (1:25; Cell Signaling), mouse anti-GFP (1:500; Cell Signaling), rabbit anti-FLAG (1:200; Sigma), rat anti-DEcad (extracellular domain) (1:50; DSHB). Secondary antibodies (Jackson ImmunoResearch) were used at a 1:200 dilution.
A minimum of 20 disks or glands were mounted per slide for a given genotype. Adult wings were dissected in 95% ethanol and mounted in Aquatex (EMD Chemicals). A minimum of 12 wings were mounted per genotype for analysis. Microscopy images were taken with an A1R laser scanning confocal microscope (Nikon) and adult wings were imaged with a Zeiss Axioplan 2 microscope.
Live imaging and FRAP
Third-instar wing imaginal disks were dissected and mounted in SFX-Insect serum-free insect cell culture medium (Hyclone) supplemented with methyl cellulose (Sigma-Aldrich) at a concentration of 4% wt/vol to increase viscosity to prevent disk drifting while imaging. FRAP assays were carried out on an A1R laser scanning confocal microscope (Nikon), with 60 × objective, 5 × artificial zoom. GFP within the region of interest (ROI) was photobleached with a 405- and 488-nm UV laser at 100% power for 15 s. GFP recovery specifically along the AJs was then recorded by time-lapse imaging over 60 min at 2-min intervals. Focal planes were maintained by manual focus during time lapse. Any samples that exhibited phototoxicity or additional photobleaching in control regions during the time lapse were excluded. GFP recovery rates of individual ROI data were normalized with pre-FRAP equal to 100% and post-FRAP equal to 0%. AJ immobile fractions of proteins were calculated as pre-FRAP fluorescence intensities, minus the end value of recovered fluorescence intensity of individual ROI.
Image processing, measurements, and statistical analysis
Following image acquisition, images were processed using NIS Elements (Nikon) and Adobe Photoshop CS6. Immunofluorescence images are presented as maximum intensity projections (MIP) of Z-steps spanning the entire tissue, unless indicated otherwise.
Distribution of Arm was determined by single line fluorescence intensity plot across individual cells with NIS Elements (Nikon). Cell edges were determined by peak F-actin fluorescence and increased DAPI across the plot line marked the nucleus. The percentage of nuclear Arm was determined as the value of Arm within the nuclear area over the total Arm across the intensity plot of the cell. The percentage of junctional Arm was determined along the apical surface of cells by measuring the sum of Arm intensity along the peak F-actin fluorescence range (cell edges) over the total Arm across the intensity plot of the cell. MCF7 nuclear β-cat values were determined by analysis of the MIP of the nuclear range of the cell, with each cells maximum β-cat fluorescence normalized to 100 across a single line fluorescence intensity plot across individual cells. Values were then averaged within a DAPI-positive range of the intensity plot. Both Arm and β-cat fluorescence analysis removes any variability due to variation in fluorescence intensity that may occur across a tissue and different genotype, normalizing values to within each cell analyzed. Cell surface area was quantified using ImageJ (National Institutes of Health [NIH]) software. All data quantifications were performed in Microsoft Excel or GraphPad Prism, and figures were made using Adobe Illustrator CS6.
Statistical analyses were performed using GraphPad Prism. Significant differences between two genotypes were determined by two-tailed Student’s t tests. One-way analysis of variance was performed for multiple comparisons, with Tukey’s multiple comparison as a posttest. All quantified data are presented as mean ± SEM, and p < 0.05 was considered statistically significant. Significance depicted as *p < 0.05, **p < 0.01, ***p < 0.0001, ns = not significant.
Plasmid constructs and oligonucleotides
The following plasmids were used in this study: pCMV-Myc (control vector) (Clontech), TOPFLASH (Korinek et al., 1997), FOPFLASH (Korinek et al., 1997), pRL-CMV (Renilla Luciferase) (Promega), pcDNA-Wnt3A a gift from Cara Gottardi (Feinberg School of Medicine, Northwestern University). The following siRNA oligonucleotides were used in this study: PPP1R16A (MYPT3): ID s39809 (ThermoFisher Scientific), PPP1CB (PP1β): ID s10935 (ThermoFisher Scientific), siRNA Negative control #1 (Scramble) (ThermoFisher Scientific).
Cell culture
Cells were cultured in six-well plates at 37°C in 5% CO2. RKO (CRL-2577; American Type Culture Collection [ATCC]) cells were grown in DMEM (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (hi-FBS; Invitrogen). MCF7 (HTB-22; ATCC) cells were grown in DMEM:F-12 (Life Technologies) supplemented with 10% hi-FBS (Invitrogen). Reverse transfections of siRNA complexes was performed in cells seeded at 50% confluence with Lipofectamine RNAiMAX (ThermoFisher Scientific) in Opti-MEM (Life Technologies), according to the manufacturer’s instructions. Twenty-four hours after seeding, transfections of plasmid DNA was performed with Lipofectamine 3000 and P3000 reagent (ThermoFisher Scientific), according to manufacturer’s instructions. When required, the final amount of DNA used for transfection was kept constant by the addition of control vector DNA. All cells were harvested 48 h after transient DNA plasmid transfection for subsequent assays.
Lysate collection and immunoblotting
Lysates of whole cell extract of MCF7 and RKO cells transfected with respective plasmids and siRNA were generated by collecting and treating the cells with cell lysis buffer (Cell Signaling Technology) supplemented with protease inhibitors (Roche). Lysates were then sonicated for several seconds on ice, followed by a 16,300 × g centrifugation for 10 min at 4°C. The supernatant was removed and protein concentrations were determined by bicinchoninic acid (BCA) assay (ThermoFisher Scientific). MCF7 cellular fraction lysates were generated by the Subcellular Protein Fractionation assay (ThermoFisher Scientific) according to manufacturer’s instructions. Protein concentrations were equalized within individual fractions, as determined by BCA assay (ThermoFisher Scientific). Lysates were boiled for 10 min with Laemmli buffer and then separated on 8–12% SDS–PAGE gels. Proteins were transferred onto nitrocellulose membranes and probed against the following primary antibodies: rabbit anti-β-catenin (1:1000; Cell Signaling), rabbit anti-E-cadherin (1:1000; Cell Signaling), mouse anti-PPP1R16A (MYPT-3) (1:500; abcam), mouse anti-Protein Phosphatase 1 beta (PP1β) (1:1000; abcam), rabbit anti-Wnt3A (1:1000; Cell Signaling), mouse anti-β-tubulin (1:1000; ABM), rabbit anti-GAPDH (1:3000; Cell Signaling), mouse anti-Na+/K+ ATPase (1:50; DSHB), rabbit anti-Histone H3 (1:1000; Cell Signaling). Membranes were visualized using the enhanced chemiluminescence (ECL) Western blotting substrate (Pierce) with a LAS4000 luminescence imager (Fujifilm). The protein levels were determined using ImageJ (NIH) software to perform densitometry. Transfections and Western blotting was performed in triplicate.
Transcriptional reporter assay
Luciferase assays were performed in MCF7 and RKO cells with the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer’s instructions. TOPFLASH or negative control FOPFLASH reporter gene plasmids, with the control reporter plasmid encoding Renilla luciferase (to normalize transfection efficiencies and for monitoring cell viability), were transfected with each expression vector as indicated to determine overall Wnt pathway activity through TCF/LEF reporter activity. The values shown represent the mean ± SEM from four biological replicate transfections, performed each time in triplicate. TOPFLASH values were normalized to the FOPFLASH reporter activity equal to 1 for each individual transfections series.
Supplementary Material
Acknowledgments
We thank the following individuals and stock centers for reagents, comments, and fly strains: Ulrich Tepass, Cara Gottardi, Ken Irvine, the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, and the Developmental Studies Hybridoma Bank. We particularly thank Stacey Ogden for allowing Eric Hall to complete experiments after joining her lab for his postdoctoral fellowship. We also thank Thalia Wiens for help with experiments for the manuscript revision. This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR) (grant number FRN 133522).
Abbreviations used:
- AJ
adherens junction
- Arm
Armadillo, Drosophila b-catenin
- b-cat
b-catenin
- bsk
basket
- CB
chromatin-bound
- C.casp3
cleaved caspase 3
- cyto
cytoplasmic
- DAPI
diamidino-2-phenylindole
- DE-cad
Drosophila E-cadherin
- DEcadDb
Drosophila E-cadherin with deletion of b-cat binding domain
- dia
diaphanous, Drosophila formin
- Dll
Distal-less
- dpp
decapentaplegic
- D/V
dorso-ventral
- Dvl/Dsh
Dishevelled
- E-cad
E-cadherin
- ECM
extracellular matrix
- EMT
epithelial-to-mesenchymal transition
- F-actin
filamentous actin
- flw
flapwing, encodes catalytic protein phosphatase type 1b subunit
- FRAP
fluorescence recovery after photobleaching
- Fz
Frizzled
- fz3
Frizzled 3
- GFP
green fluorescent protein
- hh
hedgehog
- JNK
Jun N terminal kinase
- LEF
lymphoid enhancer factor
- MARCM
Mosaic analysis with a repressible cell marker
- MBS
myosin binding subunit, Drosophila MYPT1/2
- mem
membranous
- MYPT3
human myosin phosphatase targeting protein 3, orthology of mypt-75D
- mypt-75D
myosin phosphatase targeting protein, Drosophila MYPT3
- NMII
nonmuscle myosin II
- p-MyoII
phosphorylated Sqh
- PP1b
protein phosphatase type 1b
- puc
puckered
- RNAi
RNA interference
- Sens
Senseless
- SG
salivary gland
- shg
shotgun, encodes Drosophila E-cadherin
- siRNA
small interfering RNA
- sqh
spaghetti squash, regulatory light chain of myosin II
- TCF
T-cell factor
- WCE
whole cell extracts
- Wg
Wingless
- Yki
Yorkie
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-06-0345) on December 12, 2018.
REFERENCES
- Afshar K, Stuart B, Wasserman SA. (2000). Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development , 1887–1897. [DOI] [PubMed] [Google Scholar]
- Benham-Pyle BW, Pruitt BL, Nelson WJ. (2015). Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science , 1024–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benham-Pyle BW, Sim JY, Hart KC, Pruitt BL, Nelson WJ. (2016). Increasing β-catenin/Wnt3A activity levels drive mechanical strain-induced cell cycle progression through mitosis. Elife , e19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilic J, Huang Y-L, Davidson G, Zimmermann T, Cruciat C-M, Bienz M, Niehrs C. (2007). Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science , 1619–1622. [DOI] [PubMed] [Google Scholar]
- Brown NH. (1993). Integrins hold Drosophila together. BioEssays , 383–390. [DOI] [PubMed] [Google Scholar]
- Buske P, Przybilla J, Loeffler M, Sachs N, Sato T, Clevers H, Galle J. (2012). On the biomechanics of stem cell niche formation in the gut—modelling growing organoids. FEBS J , 3475–3487. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. (2012). Wnt/β-catenin signaling and disease. Cell , 1192–1205. [DOI] [PubMed] [Google Scholar]
- Cox RT, Kirkpatrick C, Peifer M. (1996). Armadillo is required for adherens junction assembly, cell polarity, and morphogenesis during Drosophila embryogenesis. J Cell Biol , 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. (2005). Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol , 364–371. [DOI] [PubMed] [Google Scholar]
- de Beco S, Gueudry C, Amblard F, Coscoy S. (2009). Endocytosis is required for E-cadherin redistribution at mature adherens junctions. Proc Natl Acad Sci USA , 7010–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, Tepass U. (2013). Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat Cell Biol , 261–273. [DOI] [PubMed] [Google Scholar]
- Engl W, Arasi B, Yap LL, Thiery JP, Viasnoff V. (2014). Actin dynamics modulate mechanosensitive immobilization of E-cadherin at adherens junctions. Nat Cell Biol , 587–594. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Funayama N, Gluck U, Gumbiner BM. (1996). Binding to cadherins antagonizes the signaling activity of beta-catenin during axis formation in Xenopus. J Cell Biol , 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farge E. (2011). Mechanotransduction in development. Curr Top Dev Biol , 243–265. [DOI] [PubMed] [Google Scholar]
- Fernández-Sánchez ME, Barbier S, Whitehead J, Béalle G, Michel A, Latorre-Ossa H, Rey C, Fouassier L, Claperon A, Brullé L, et al. (2015). Mechanical induction of the tumorigenic β-catenin pathway by tumour growth pressure. Nature , 92–95. [DOI] [PubMed] [Google Scholar]
- Gagliardi M, Piddini E, Vincent J-P. (2008). Endocytosis: a positive or a negative influence on Wnt signalling? Traffic , 1–9. [DOI] [PubMed] [Google Scholar]
- Godt D, Tepass U. (1998). Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature , 387–391. [DOI] [PubMed] [Google Scholar]
- Goldenberg G, Harris TJC, Wu C, Duncan K, Korn E. (2013). Adherens junction distribution mechanisms during cell-cell contact elongation in Drosophila. PLoS One , e79613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi CJ, Wong E, Gumbiner BM. (2001). E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol , 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, La J, Yum KW, Desai P, Rodewald L-W, Zhang X, Leblanc M, Nusse R, Lewis MT, Wahl GM. (2013). Paracrine Wnt signaling both promotes and inhibits human breast tumor growth. Proc Natl Acad Sci , 6991–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer ER, Chao AT, Bejsovec A. (2013). Pebble/ECT2 RhoGEF negatively regulates the Wingless/Wnt signaling pathway. Development , 4937–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall ET, Verheyen EM. (2015). Ras-activated Dsor1 promotes Wnt signaling in Drosophila development. J Cell Sci , 4499–4511. [DOI] [PubMed] [Google Scholar]
- Happe CL, Engler AJ. (2016). Mechanical forces reshape differentiation cues that guide cardiomyogenesis. Circ Res , 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJC, Peifer M. (2005). Decisions, decisions: beta-catenin chooses between adhesion and transcription. Trends Cell Biol , 234–237. [DOI] [PubMed] [Google Scholar]
- Harris TJC, Tepass U. (2010). Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol , 502–514. [DOI] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro CY, Wylie C. (1994). Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell , 791–803. [DOI] [PubMed] [Google Scholar]
- Hirata N, Takahashi M, Yazawa M. (2009). Diphosphorylation of regulatory light chain of myosin IIA is responsible for proper cell spreading. Biochem Biophys Res Commun , 682–687. [DOI] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. (2013). Binding to F-actin guides cadherin cluster assembly, stability, and movement. J Cell Biol , 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess RE, Chang XJ, Edwards KA, Kulkarni S, Aguilera I, Kiehart DP. (1991). The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell , 1177–1189. [DOI] [PubMed] [Google Scholar]
- Khoo P, Allan K, Willoughby L, Brumby AM, Richardson HE. (2013). In Drosophila, RhoGEF2 cooperates with activated Ras in tumorigenesis through a pathway involving Rho1-Rok-Myosin-II and JNK signalling. Dis Model Mech , 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J, Gross S, Bennett D, Alphey L. (2007). The nonmuscle myosin phosphatase PP1beta (flapwing) negatively regulates Jun N-terminal kinase in wing imaginal discs of Drosophila. Genetics , 1741–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. (1997). Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science , 1784–1787. [DOI] [PubMed] [Google Scholar]
- Langevin J, Morgan MJ, Sibarita J-B, Aresta S, Murthy M, Schwarz T, Camonis J, Bellaïche Y, Tepass U, Crair MC, et al. (2005). Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev Cell , 365–376. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Yap AS. (2015). E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol , 533–539. [DOI] [PubMed] [Google Scholar]
- Maher MT, Flozak AS, Stocker AM, Chenn A, Gottardi CJ. (2009). Activity of the beta-catenin phosphodestruction complex at cell-cell contacts is enhanced by cadherin-based adhesion. J Cell Biol , 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A. (1998). puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev , 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Vincent J-P. (2003). Armadillo levels are reduced during mitosis in Drosophila. Mech Dev , 157–165. [DOI] [PubMed] [Google Scholar]
- Murthy K, Wadsworth P, Marquis H, Hostos EL, de Nelson WJ, Roques S, Martel V, Breton-Douillon M, Perret B, Chap H. (2005). Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr Biol , 724–731. [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M. (1997). Negative regulation of Armadillo, a Wingless effector in Drosophila. Development , 2255–2266. [DOI] [PubMed] [Google Scholar]
- Port F, Hausmann G, Basler K. (2011). A genome-wide RNA interference screen uncovers two p24 proteins as regulators of Wingless secretion. EMBO Rep , 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla L, Lakins JN, Weaver VM. (2016). Tissue mechanics orchestrate Wnt-dependent human embryonic stem cell differentiation. Cell Stem Cell , 462–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, Irvine KD. (2014). Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell , 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring JM, Martinez Arias A. (1993). Puckered, a gene involved in position-specific cell differentiation in the dorsal epidermis of the Drosophila larva. Dev Suppl , 251–259. [PubMed] [Google Scholar]
- Samuel MS, Lopez JI, McGhee EJ, Croft DR, Strachan D, Timpson P, Munro J, Schröder E, Zhou J, Brunton VG, et al. (2011). Actomyosin-mediated cellular tension drives increased tissue stiffness and β-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell , 776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson B, White P, Vincent J-P. (1996). Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature , 627–630. [DOI] [PubMed] [Google Scholar]
- Sato A, Kojima T, Ui-Tei K, Miyata Y, Saigo K. (1999). Dfrizzled-3, a new Drosophila Wnt receptor, acting as an attenuator of Wingless signaling in wingless hypomorphic mutants. Development , 4421–4430. [DOI] [PubMed] [Google Scholar]
- Schlessinger K, Hall A, Tolwinski N. (2009). Wnt signaling pathways meet Rho GTPases. Genes Dev , 265–277. [DOI] [PubMed] [Google Scholar]
- Sun SX, Walcott S, Wolgemuth CW. (2010). Cytoskeletal cross-linking and bundling in motor-independent contraction. Curr Biol , R649–R654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yan Y, Denef N, Schupbach T. (2011). Regulation of somatic myosin activity by protein phosphatase 1 controls Drosophila oocyte polarization. Development , 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S, Pradhan-Sundd T, Verheyen EM. (2015). Genome-wide identification of phospho-regulators of Wnt signaling in Drosophila. Development , 1502–1515. [DOI] [PubMed] [Google Scholar]
- Torres VA, Tapia JC, Rodriguez DA, Lladser A, Arredondo C, Leyton L, Quest AFG. (2007). E-cadherin is required for caveolin-1-mediated down-regulation of the inhibitor of apoptosis protein survivin via reduced beta-catenin-Tcf/Lef-dependent transcription. Mol Cell Biol , 7703–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbano JM, Naylor HW, Scarpa E, Muresan L, Sanson B. (2018). Suppression of epithelial folding at actomyosin-enriched compartment boundaries downstream of Wingless signalling in Drosophila . Development , dev155325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Hausmann G, Basler K. (2012). The many faces and functions of β-catenin. EMBO J , 2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez CG, Heissler SM, Billington N, Sellers JR, Martin AC. (2016). Drosophila non-muscle myosin II motor activity determines the rate of tissue folding. Elife , e20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereshchagina N, Bennett D, Szöor B, Kirchner J, Gross S, Vissi E, White-Cooper H, Alphey L. (2004). The essential role of PP1beta in Drosophila is to regulate nonmuscle myosin. Mol Biol Cell , 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J-P, Girdham CH, O’Farrell PH. (1994). A cell-autonomous, ubiquitous marker for the analysis of Drosophila genetic mosaics. Dev Biol , 328–331. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol , 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann TJ, Dahmann C. (2009). Wingless signaling and the control of cell shape in Drosophila wing imaginal discs. Dev Biol , 161–173. [DOI] [PubMed] [Google Scholar]
- Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, Luo L. (2001). Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell , 81–91. [DOI] [PubMed] [Google Scholar]
- Wittkorn E, Sarkar A, Garcia K, Kango-Singh M, Singh A. (2015). The Hippo pathway effector Yki downregulates Wg signaling to promote retinal differentiation in the Drosophila eye. Development , 2002–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SK, Gomez GA, Michael M, Verma S, Cox HL, Lefevre JG, Parton RG, Hamilton NA, Neufeld Z, Yap AS. (2014). Cortical F-actin stabilization generates apical-lateral patterns of junctional contractility that integrate cells into epithelia. Nat Cell Biol , 167–178. [DOI] [PubMed] [Google Scholar]
- Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. (2008). Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell , 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Martin AC. (2015). Intracellular signalling and intercellular coupling coordinate heterogeneous contractile events to facilitate tissue folding. Nat Commun , 7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Yan L, Liu S, Shan Z, Tian Y, Jin Z. (2015). N-cadherin, a novel prognostic biomarker, drives malignant progression of colorectal cancer. Mol Med Rep , 2999–3006. [DOI] [PubMed] [Google Scholar]
- Yang Y, Primrose DA, Leung AC, Fitzsimmons RB, McDermand MC, Missellbrook A, Haskins J, Smylie AS, Hughes SC. (2012). The PP1 phosphatase Flapwing regulates the activity of Merlin and Moesin in Drosophila. Dev Biol , 412–426. [DOI] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. (1996). Direct and long-range action of a wingless morphogen gradient. Cell , 833–844. [DOI] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F. (1997). The mouse fused locus encodes axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell , 181–192. [DOI] [PubMed] [Google Scholar]
- Zimmerman SG, Thorpe LM, Medrano VR, Mallozzi CA, McCartney BM. (2010). Apical constriction and invagination downstream of the canonical Wnt signaling pathway require Rho1 and Myosin II. Dev Biol , 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.