Abstract
The architecture of the cytoskeleton and its remodeling are tightly regulated by dynamic reorganization of keratin-rich intermediate filaments. Plakin family proteins associate with the network of intermediate filaments (IFs) and affect its reorganization during migration, differentiation, and response to stress. The smallest plakin, periplakin (PPL), interacts specifically with intermediate filament proteins K8, K18, and vimentin via its C-terminal linker domain. Here, we show that periplakin is SUMOylated at a conserved lysine in its linker domain (K1646) preferentially by small ubiquitin-like modifier 1 (SUMO1). Our data indicate that PPL SUMOylation is essential for the proper reorganization of the keratin IF network. Stresses perturbing intermediate-filament and cytoskeletal architecture induce hyper-SUMOylation of periplakin. Okadaic acid induced hyperphosphorylation-dependent collapse of the keratin IF network results in a similar hyper-SUMOylation of PPL. Strikingly, exogenous overexpression of a non-SUMOylatable periplakin mutant (K1646R) induced aberrant bundling and loose network interconnections of the keratin filaments. Time-lapse imaging of cells expressing the K1646R mutant showed the enhanced sensitivity of keratin filament collapse upon okadaic acid treatment. Our data identify an important regulatory role for periplakin SUMOylation in dynamic reorganization and stability of keratin IFs.
INTRODUCTION
The orchestration of numerous architectural proteins is crucial for the coordination of efficient cellular cytoskeleton assembly, its movement, and in the maintenance of tissue integrity. The Plakin family consists of seven large multidomain proteins often called cytolinker proteins. Plakins serve as adaptors inter-connecting cytoskeletal intermediate filaments (IFs) and are integral components of intercellular junctional complexes (Ruhrberg and Watt, 1997). The interplay of plakins helps in the formation of a dense intracellular framework of filaments that is integral to efficient cellular communication and modulation of biological processes such as cell adhesion, migration, differentiation, and signaling. However, mutations or defects in plakin family genes, both inherited or acquired, lead to drastic disruptions of tissue integrity and affect the stability of the cornified envelope of skin epidermis, the normal functioning of muscular and nervous systems but induce no developmental lethality (Sonnenberg and Liem, 2007).
Plakins harbor multiple interacting domains and exhibit a tripartite structure: an N-terminal globular plakin domain, a central coiled-coil rod domain and a carboxyl terminus with a variable number of tandem plakin repeat domains (PRD) repeats (types A, B, and C) responsible for association with IFs (Virata et al., 1992). A short highly homologous linker-subdomain (L) often resides between the B and C type repeats (Leung et al., 2001; Jefferson et al., 2004). The smallest member of the plakin family, periplakin (PPL), was identified as a major constituent of the cornified envelope precursor and is targeted to cell membranes and desmosomes in keratinocytes (DiColandrea et al., 2000). PPL forms a heterodimeric complex with another plakin family member envoplakin, and together they serve as strong antigens for the autoimmune disease paraneoplastic pemphigus (Ruhrberg et al., 1997). Periplakin is a unique member of the plakin family because it lacks a complete PRD and carries just the linker domain at the C-terminus.
Interestingly, the small C-terminal linker domain is sufficient for specific interactions with IF proteins K8, K18, and vimentin (Kazerounian et al., 2002). The linker domain served as a hub for the unique interactions of PPL with lens fiber cell-specific IF proteins CP49 and filensin (Yoon and FitzGerald, 2009). Although PPL-dependent K8 reorganization was demonstrated to be critical for maintenance of epithelial integrity in migrating cells, its downregulation perturbed keratin rearrangement and cable formation resulting in impaired wound closure (Long et al., 2006). Interactions with the membrane-binding protein annexin A9 (Boczonadi and Maatta, 2012), desmosome associated protein kazrin (Groot et al., 2004), and nuclear protein periphilin (Kazerounian and Aho, 2003) suggest that PPL is not just an IF-associated protein but an important player in cellular cytoskeleton homeostasis. Activities of the signaling receptor proteins such as high-affinity immunoglobulin G (IgG) receptor FcγRI (Beekman et al., 2004), G-protein receptor MOP-1, melanin-concentrating hormone receptor-1 (Milligan et al., 2004) and Protein Kinase B have been shown to be modulated by interaction with PPL (van den Heuvel et al., 2002).
Down-regulation of periplakin has been correlated with the development and progression of many types of cancer, specifically, bladder, colon, and esophageal squamous cell carcinoma (Nishimori et al., 2006; Matsumoto et al., 2014; Otsubo et al., 2015; Li et al., 2017). Moreover, PPL was also identified as a metastasis suppressor in TNBC (triple-negative breast cancer) (Choi et al., 2013). All these reports hint toward the emerging new roles for PPL as a crucial regulator of various biological processes through its remarkably versatile interactome.
Protein–protein interactions are often regulated by posttranslational modification and plakins are no exception to this. Phosphorylation of plectin and desmoplakin has been shown to play an important regulatory role in their interaction with IFs (Bouameur et al., 2013; Kroger et al., 2013). SUMOylation, one of the most prominent posttranslational modifications in eukaryotic biology, represents the covalent attachment of one or more small ubiquitin-like modifier (SUMO) proteins to specific lysine residues on a target protein. It has been shown to regulate functional activity, protein-protein interaction and intracellular dynamics of target proteins. Consequently, SUMOylation modulates several biological pathways including nuclear signaling, transport, transcription, genome integrity and DNA replication/repair. Moreover, SUMOylation is known to regulate functions of several cytoskeletal proteins including K8 and vimentin.
While several SUMOylation targets have prominent function in the nucleus, nonnuclear SUMO substrates including cytoskeletal filament proteins like keratins, vimentin, septins, and other cytoplasmic proteins have been identified but only a few have been characterized for the roles of SUMOylation on their function (Snider et al., 2011; Ribet et al., 2017). Importantly, SUMOylation has emerged as a critical player of keratin dynamics, and keratin and vimentin stably interact with PPL and help in the assembly of the robust intermediate filament network. However, how the association of plakins with keratin cytoskeleton and keratin SUMOylation may affect functions of the plakin family of cytolinkers, particularly of periplakin, is not understood and remains to be revealed.
We report here that periplakin is SUMOylated at a conserved lysine 1646 (K1646) residue present in its linker domain. Periplakin exhibits strong paralogue specificity toward SUMO1 for covalent modification. We observed that apoptotic and mechanical stresses affecting keratin cyto-architecture-induced PPL hyper-SUMOylation. The extent of PPL hyper-SUMOylation increased further when keratin filament network collapse was induced by okadaic acid (OA) or orthovanadate (OV) mediated hyperphosphorylation of keratin. Interestingly, lack of periplakin SUMOylation (K1646R mutant) induces perturbations in the robust organization of the keratin intermediate filament and thus contributes to the enhanced sensitivity to OA-induced keratin filament collapse.
RESULTS
The cytolinker protein periplakin is a target for SUMOylation
To our knowledge, the SUMOylation status of any plakin family protein has not been established. PPL is the smallest plakin family cytoskeletal linker protein that connects intermediate filaments to cellular junctions and other membrane locations. In our preliminary interaction analysis, periplakin interacted strongly with SUMOylation machinery (unpublished data). To further analyze the nature of the interaction of periplakin with SUMOylation machinery, we first examined whether PPL is a target of SUMOylation and can be covalently modified. HEK293T cells were transfected with GFP-tagged conjugatable (SUMO1GG/SUMO2GG) or nonconjugatable (SUMO1G/SUMO2G) forms along with full-length hemagglutinin (HA)-tagged PPL. Immunoprecipitation of PPL with anti-HA antibodies identified a slow migrating band consistent with the size of GFP-SUMO-modified full-length PPL representing SUMOylation of PPL (Figure 1A). However, this slow migrating band was observed only with SUMO1GG but not with SUMO2GG transfection (Figure 1B). This suggested that PPL is SUMOylated and exhibits paralogue specificity for SUMO1 over SUMO2. The same slow migrating band was detected when immunoprecipitates described above were blotted with anti-GFP antibodies (Figure 1C). The molecular weight of this band corresponds to the predicted shift induced by the covalent linkage of GFP-SUMO-1 to PPL. The extent of modification detected was very low (<5%) as observed with many SUMO targets (Hay, 2005). To examine the SUMOylation of endogenous PPL we first generated PPL specific polyclonal antibodies in rabbit and confirmed the specificity of the antiserum in different cell lines. Immunofluorescence experiments using our anti-PPL antiserum showed reactivity at the plasma membrane and at cell-cell contacts (Supplemental Figure S1A) as expected for PPL. The antiserum specifically recognized an ∼200-kDa size band in Western blots on various cell lysates (Supplemental Figure S1, B and C). Using the PPL-specific IgGs we immunoprecipitated periplakin from MCF7a cell lysates and probing with anti-SUMO1 and anti-PPL antibodies again identified a slow migrating band corresponding to the size of SUMO-modified PPL (Figure 1D). The reversible nature of the modification is consistent with the observation that the SUMO-modified band was detected only in immunoprecipitation performed in the presence of the SUMO-isopeptidase (SENP) inhibitor N-ethylmaleimide (NEM) (Figure 1E). Our immunoprecipitation data establish that both endogenous and exogenously expressed PPL are SUMOylated.
FIGURE 1:
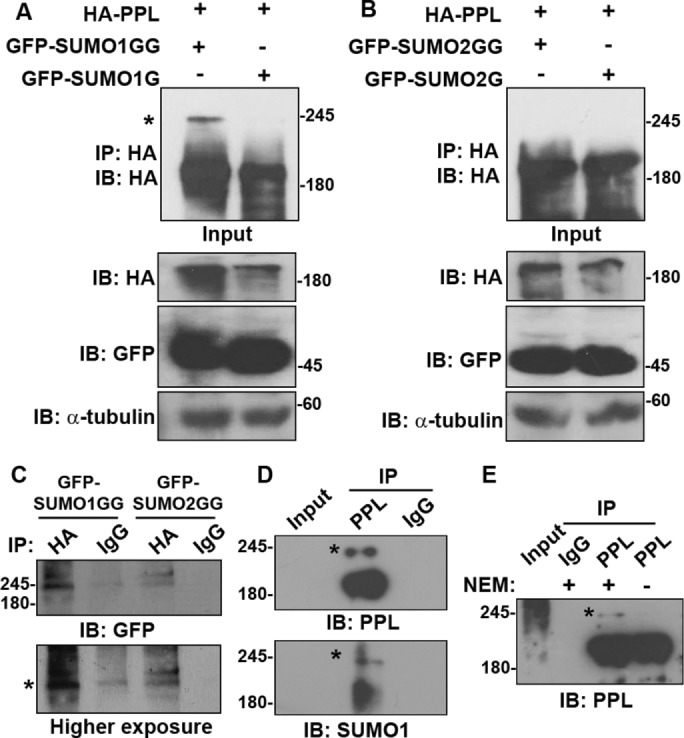
PPL is conjugated preferentially by SUMO1. (A) Lysates prepared from HEK293T cells coexpressing HA-PPL full-length and GFP-SUMO1G or GFP-SUMO1GG were subjected to IP using anti-HA antibody. IP was analyzed for the presence of any modified PPL bands by WB using anti-HA antibody. Input fractions and loading controls are shown. An asterisk indicates SUMO-modified bands. (B) Same as in A but GFP-SUMO2 or SUMO2GG was coexpressed along with PPL. (C) Lysates prepared from HEK293T cells coexpressing HA-PPL full-length and GFP-SUMO1G or GFP-SUMO1GG were subjected to IP using anti-HA and rabbit IgG antibodies. IP was analyzed for the presence of any modified PPL bands by WB using anti-GFP antibody. An asterisk indicates SUMO-modified bands. (D) IP of endogenous PPL from MCF7a lysates with anti-PPL antibody and detection of PPL SUMOylation with anti-PPL (top panel) and anti-SUMO1 (bottom panel) antibodies. IP with rabbit IgG serves as negative control. (E) IP of PPL as performed in D but in the presence or absence of the deSUMOylation inhibitor NEM. IP with rabbit IgG serves negative control. An asterisk indicates SUMO-modified bands. IP images are a representative from at least n = 3 repeat experiments.
Periplakin is modified by SUMO1 in the C-terminal linker domain
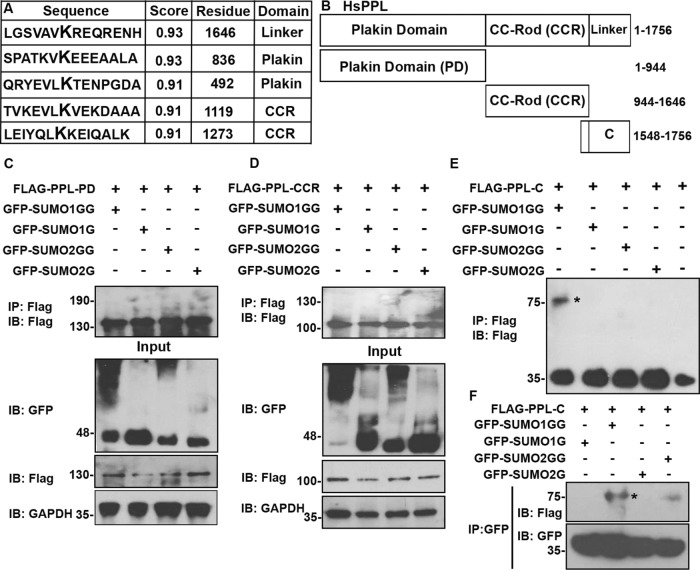
After establishing that PPL is modified by SUMO1 we next attempted to identify the site of SUMO modification on PPL. We used three different prediction algorithms SUMOplot (www.abgent.com), GPS-SUMO (SUMOsp.biocuckoo.org), and JASSA (www. jassa.fr) to predict potential SUMOylation sites in PPL. All three algorithms predicted five high-probability SUMO modification sites in PPL distributed throughout its three domains (Figure 2A). We have noted above that the level of PPL full-length SUMOylation was minimal. So to map SUMOylation sites on PPL, we made domainwise Flag-tagged constructs for expressing all the three domains in cells: the N-terminal plakin domain (PD), the central coiled-coil rod (CCR) domain, and the C-terminal linker (C) subdomain (Figure 2B). One of the two highest probability SUMOylation sites lies at the junction of the rod and C-terminal linker domain. To retain the consensus SUMOylation site, the linker domain construct was extended to have overlapping residues with rod domain. Moreover, various reports that demonstrate specific interactions of keratin8, vimentin, PKB, and G-protein–coupled receptors with the periplakin C-terminal region have highlighted the critical importance of these overlapping residues from the rod domain (Milligan et al., 2004).
FIGURE 2:
PPL is SUMOylated in its C-terminal linker domain. (A) In silico predicted high-probability SUMOylation sites on PPL using available SUMO site prediction software. (B) Schematic representation of PPL domain constructs used in transfection experiments. 3x-FLAG tagged PPL full-length (1–1756), PPL-PD (1–944), PPL-CCR (944–1646), and PPL-C-terminal linker domain (PPL-C, 1548–1756). (C) IP of PPL using anti-Flag antibodies from HEK293T cell lysates coexpressing FLAG-PPL-PD and conjugatable GFP-SUMO1GG or 2GG or nonconjugatable GFP-SUMO1G or 2G. Immunoblotting with anti-Flag antibodies to detect SUMO modification of PPL-PD. Input fractions were probed for detection of expressed proteins. The bottom panel is the loading control with anti-GAPDH immunoblotting. (D) Similarly to C but here the FLAG-tagged PPL-CCR was coexpressed with conjugatable GFP-SUMO1GG or 2GG or nonconjugatable GFP-SUMO1G or 2G. (E) Similarly to C but here PPL-C was coexpressed with conjugatable GFP-SUMO1GG or 2GG or nonconjugatable GFP-SUMO1G or 2G. IP and immunoblotting was performed with anti-Flag antibodies. An asterisk indicates SUMO-modified bands. (F) IP with anti-GFP antibodies on lysates obtained as in E and immunoblotting of IP sample with anti-FLAG (top panel) and anti-GFP (bottom panel) to detect PPL-C modification. An asterisk indicates SUMO-modified bands. IP images are a representative from at least n = 3 repeat experiments.
Transient overexpression of individual domains of PPL in HeLa cells showed variations in their expression levels (Supplemental Figure S2A). As reported earlier, the C-terminal linker domain localization was comparable to full-length protein in the cell, that is, mostly bound to the intermediate filament network. Strikingly, CCR and PD constructs showed very distinct subcellular localization as compared with PPL (fl) (Karashima and Watt, 2002) (Supplemental Figure S2B).
To identify the site(s) of SUMOylation HEK293T cells were cotransfected with GFP-SUMO1G/SUMO1GG or GFP-SUMO2G/SUMO2GG along with Flag-PPL-PD, Flag-PPL-CCR, and Flag-PPL-C constructs independently. Immunoprecipitation (IP) was performed with anti-Flag antibodies on lysates prepared from these transfections. Subsequent immunoblotting of these immunoprecipitates with anti-Flag antibodies did not reveal any slow migrating band with PPL-PD and PPL-CCR domain constructs (Figure 2, C and D). In the case of C-terminal domain construct, a distinct slower migrating band corresponding to the SUMO-modified PPL-C (Figure 2E, highlighted by an asterisk) was observed with GFP-SUMO1GG. In a complementary coimmunoprecipitation experiment with an anti-GFP antibody, yet again a distinct slow migrating band corresponding to GFP-SUMO1GG-modified linker domain was detected with anti-Flag antibodies (Figure 2F). These data suggest that the primary site for SUMO modification is possibly the one that lies at the junction of rod and linker domain of PPL. Thus, we propose that PPL is covalently modified by SUMO1 in its linker domain.
Periplakin is SUMOylated at the conserved lysine (K1646) residue of the linker domain
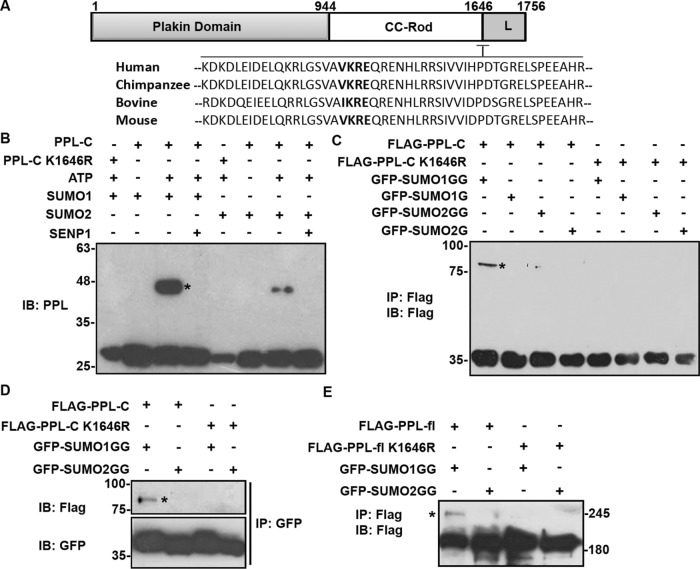
Domainwise mapping of PPL SUMOylation and predictions using algorithms suggested that K1646 present at the start of the linker domain can serve as the primary acceptor residue for SUMOylation. Sequence alignment of the region surrounding the putative SUMOylation site (K1646) in PPL from higher organisms showed that the consensus Ψ-K-X-E (VKRE) motif is highly conserved (Figure 3A, in bold). This conservation may be indicative of its potential relevance in functional regulation. To test whether the PPL C-terminus K1646 residue is a site for covalent SUMO modification, we mutated the lysine 1646 to arginine (K1646R), which renders the residue unconjugatable for SUMO (SUMOylation-deficient). We first addressed the ability of the mutant to be conjugated by SUMO in an in-vitro SUMOylation assay.
FIGURE 3:
PPL is modified by SUMO1 at conserved lysine (K1646) residue. (A) Multiple sequence alignment of periplakin linker domain from different species using Clustal omega software shows that the SUMO motif (marked bold) is highly conserved in all these species. (B) Anti-PPL blot of an in vitro reconstituted reaction including recombinant E1, E2, and SUMO1 with PPL-C or PPL-C K1656R mutant. (C) IP with anti-Flag antibody from HEK293T lysates expressing WT Flag-PPL-C or K1646R mutant with WT-GFP-SUMO1GG/2GG or nonconjugatable GFP-SUMO1G/2G mutants and subsequent analysis by Western blotting using anti-Flag antibody. (D) GBP (GFP binding protein) pull down of HEK293T lysates expressing WT Flag-PPL-C or K1646R mutant with WT-GFP-SUMO1GG/2GG, or nonconjugatable GFP-SUMO1G/2G mutants and analysis by Western blotting using anti-Flag antibody. (E) HEK293T cells were transfected with Flag-PPL full-length (wild type or K1646R mutant) and WT-GFP-SUMO1GG or 2GG followed by immunoprecipitation and immunoblotting with anti-Flag antibody. An asterisk indicates SUMO-modified bands. IP images are a representative from n = 3 repeated experiments.
In vitro SUMOylation reactions performed with recombinant PPL-C (wild type) and PPL-C K1646R mutant showed that PPL-C is SUMO-modified and this modified band is lost in the presence of recombinant catalytic domain of SENP1 (Figure 3B). Importantly, the SUMO-modified band was absent when reactions were carried out with the PPL-C K1646R mutant. Furthermore, the appearance of SUMO-modified PPL-C band was ATP-dependent confirming covalent conjugation of SUMO to PPL-C. In the in vitro SUMOylation reactions we noticed that SUMO2GG could also modify the PPL-C, but the extent of modification was considerably lower. We further observed in in bacto SUMOylation assays (Supplemental Figure S3A) with quaternary plasmids pSUMO1 and pSUMO2 that PPL-C exhibits paralogue specificity toward SUMO1 over SUMO2 (Weber et al., 2014). The preference of PPL for SUMO1 over SUMO2 for modification even under artificial conditions like in vitro and in bacto SUMOylation strongly suggested a specific interaction of PPL and SUMO1 leading to paralogue specificity. We noticed the presence of a strong SUMO-interacting motif (SIM) [I/V-X-I/V-I/V] adjacent to the SUMO acceptor lysine K1646. Several SUMO substrates are known to possess SIMs, which can play a crucial role in protein–protein interaction as well as in the SUMO conjugation and deconjugation cycles (Takahashi et al., 2005). We asked whether the putative SIM in the vicinity of SUMOylation site had any significance in PPL SUMOylation. Residues of the predicted SIM in PPL-C terminus were all mutated to alanine and subjected to in bacto SUMOylation. We observed no significant difference between SUMO modification of PPL-C and PPL-C SIM mutant (Supplemental Figure S3B). Immunoprecipitation experiments performed with cell lysates expressing SUMO1GG/G or SUMO2GG/G and wild type or PPL-C K1646R mutant identified SUMO modification of wild-type PPL-C with SUMO-1GG in a paralogue-specific manner. These observations further substantiated the fact that K1646 is the site for SUMO modification on PPL (Figure 3, C and D). We further demonstrated that single site SUMO modification of PPL occurs at K1646R with full-length PPL. In vivo SUMO conjugation performed with full-length PPL (wild type) and the single point mutant PPL (PPL-K1646R) identified a single slow migrating band in SUMO1GG-expressing conditions. While no modification was seen with SUMO2GG, the single point mutant (PPL-fl K1646R) did not show any SUMOylation under similar conditions (Figure 3E). Together, our data identify Lys1646 in the linker domain of PPL as major acceptor residue for SUMO conjugation. Furthermore, PPL SUMOylation shows paralogue specificity both in vitro and in vivo. Importantly, the PPL-C linker domain mimics localization and SUMOylation observations made with PPL full length and this formed the basis for using PPL-C for subsequent functional experiments.
Stresses affecting cytoskeletal dynamics and IF architecture enhance periplakin SUMOylation
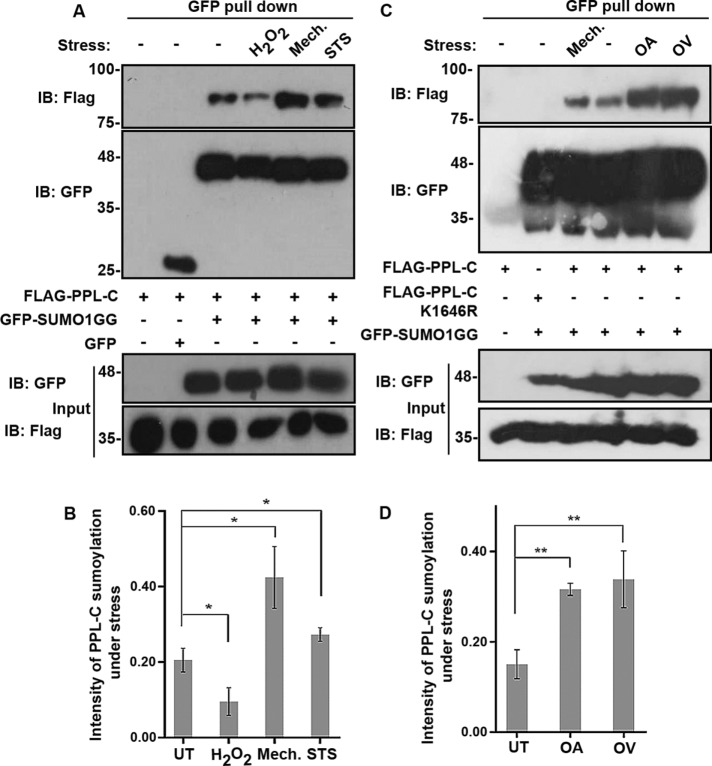
SUMOylation of many substrate proteins has been reported to be induced as a response and adaption to cellular stress (Tempe et al., 2008). A few proteins have also been reported to be conditionally SUMOylated under specific stimuli (Mabb et al., 2006). More importantly, SUMOylation of keratin has recently been implicated in keratin IF network dynamics in response to stress (Snider et al., 2011). In addition to posttranslational modification, interlinking of IFs by plakin family proteins is thought to be a major factor contributing to such cytoskeletal network remodeling (Jefferson et al., 2004). We asked whether PPL SUMOylation is altered in response to various types of cellular stress and how this would affect IF rearrangement. To analyze how SUMOylation status of periplakin is affected under various stresses, we assessed the relative extent of SUMOylation of PPL-C during apoptotic, oxidative and mechanical stresses. GFP binding protein (GBP) can be used for efficient purification of GFP tagged protein in a manner similar to anti-GFP antibodies (Rothbauer et al., 2006; Rothbauer et al., 2008). GBP was subcloned and expressed as GST-tagged proteins, and the complex formed between GST-GBP and GFP-tagged proteins present in lysates was purified using GSH-sepharose beads (Supplemental Figure S4A). Lysates prepared from cells transfected with Flag-PPL-C and GFP-SUMO constructs were used for pull down, and the presence of a SUMO-modified PPL-C bands was confirmed using anti-Flag antibodies. Consistent with our previous observations, periplakin (PPL-C) was SUMOylated only when cotransfected with GFP-SUMO1GG and not in any of the other relevant control conditions. While PPL-C SUMOylation was reduced slightly in case of oxidative stress, stresses significantly affecting the cytoskeletal filament reorganization like apoptotic and mechanical stresses showed increased PPL-C SUMOylation (Figure 4, A and B). Periplakin has been extensively characterized as a keratin intermediate filament-associated protein, and it is known that hyperphosphorylation-dependent (induced by the serine/threonine phosphatase inhibitor OA) reorganization of keratin IFs affects the distribution of periplakin in cells.
FIGURE 4:
Stresses directly affecting keratin filament architecture enhance PPL SUMOylation. (A) GFP pull down of HEK293T lysates expressing WT Flag-PPL-C with GFP-SUMO1GG. Lysates were prepared after treating cells with the indicated stresses or no stress (–), followed by immunoblotting with anti-Flag and anti-GFP antibodies. Input fractions were immunoblotted with the indicated antibodies to detect presence of expressed proteins. (B) Quantification of SUMO-conjugated PPL-C under normal control untreated (UT) conditions and various stresses like oxidative (H2O2), apoptotic (staurosporine, STS), and mechanical stress (Mech.). Data represented are mean ± SD of three independent experiments. Asterisk indicates statistical significance p < 0.05. (C) Same as in A but cells treated with phosphatase inhibitors OA and sodium OV. (D) Bar diagrams representing relative intensities of PPL SUMOylation obtained under OA and OV treatment conditions. Data represent mean ± SD of three independent experiments. Asterisks indicate statistical significance p < 0.01.
Furthermore, periplakin associates with keratin via its C-terminal linker domain and this association is unperturbed by OA-induced cytoskeletal granule formation (Long et al., 2006). We also assessed PPL SUMOylation in the presence of the tyrosine phosphatase inhibitor sodium OV, which is also known to induce considerable reorganization of keratin filaments. It is observed that OA-induced keratin reorganization is slow, taking hours to complete, whereas OV-induced keratin filament network breakdown is rapid (10–20 min) and reversible (Strnad et al., 2002). Although the reorganization kinetics of the keratin network, under the influence of these drugs differs, we nevertheless observed hyper-SUMOylation of PPL-C as a result of each of the drug treatments (Figure 4, C and D). Importantly, the extent of SUMOylation observed, in this case, was significantly higher than that seen in mechanical stress. However, consistent with our previous observation, SUMO2 conjugation to PPL-C under all the tested stress conditions was relatively minor, emphasizing the fact that periplakin shows strong paralogue specificity for SUMO1 over SUMO2 (Supplemental Figure S4, B and C).
We observed that hyperSUMOylation of PPL-C was induced by OA treatment of cells—a condition under which keratin is reported to be hyperphosphorylated (Yatsunami et al., 1993; Blankson et al., 1995). It is, therefore, possible that PPL SUMOylation could represent an adaptation to alterations in keratin filament reorganization. During mitosis, rapid changes have been observed in keratin phosphorylation status and which is directly correlated to its ability to reorganize relatively fast (Windoffer and Leube, 2004). This prompted us to examine the relative levels of SUMOylated PPL-C in different phases of cell division. PPL-C and GFP-SUMO1 coexpressing cells were synchronized and arrested in different phases of the cell cycle (Supplemental Figure S4D); however, no significant change was observed in PPL-C SUMOylation when cells were arrested in different phases of the cell cycle (Supplemental Figure S4E). These data suggested that normal physiological changes in the phosphorylation status of keratin do not directly influence PPL SUMOylation. Rather, PPL hyper-SUMOylation is likely a result of stress-dependent regulated alterations of keratin filaments seen in several human diseases. Our observations suggest that SUMOylation of PPL is specifically induced as a response to stress-dependent keratin reorganization.
Periplakin SUMOylation is required for higher-order organization and dynamics of intermediate filament architecture
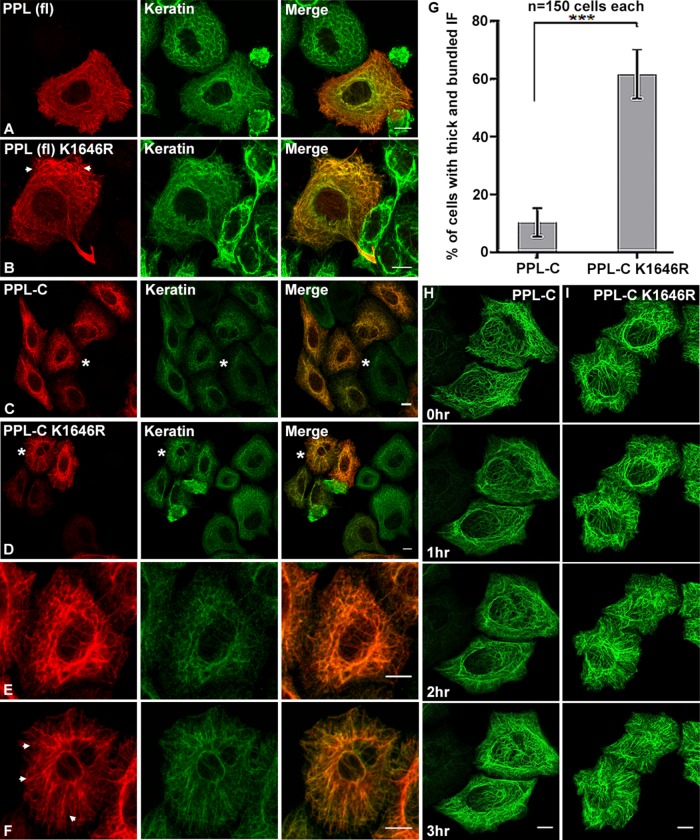
It is reported that SUMOylation can affect localization of target proteins thereby regulating their functions. To examine whether SUMOylation could regulate PPL localization, we compared the subcellular distribution of the PPL full-length wild type and K1646R mutant in cells. The FLAG-tagged full-length PPL wild type, and mutant proteins showed no observable differences in their subcellular localization and overlapping completely with the keratin and vimentin IF network (Figure 5A and Supplemental Figure S5A). A comprehensive look at the keratin network in the PPL (fl) K1646R mutant-expressing cells we did observe excessive peripheral bundling (arrowheads in Figure 5B). The quantification of the same has been shown in Supplemental Figure S5F. The subtle phenotype observed with keratin is probably due to the other domains of PPL engaging in their regular interactions involved in keratin reorganization (Boczonadi et al., 2007). To explore further the role of PPL SUMOylation on keratin intermediate filament, we resorted to the C-terminal linker domain of the PPL, which is known to mediate the interaction with K8 and vimentin (Kazerounian et al., 2002). We have demonstrated that the localization of the isolated PPL-C domain is identical to that of PPL full length (Supplemental Figure S2B). When the wild-type or K1646R mutant PPL-C were expressed in cells, no difference in their colocalization with keratin (Figure 5, C and D) and vimentin rich IF network (Supplemental Figure S5A) was observed. Similar colocalization patterns of wild-type PPL-C and the K1646R mutant with IFs suggested that periplakin SUMOylation is dispensable for its association with the keratin filaments. Subsequently, wild-type PPL-C and the K1646R mutant were utilized for different cellular studies relevant to PPL localization and association with the IF network. Although the colocalization of periplakin with keratin was unperturbed, cells expressing the non-SUMOylatable version of PPL-C (K1646R) displayed striking differences in the keratin filament bundles and network pattern (cells marked by asterisks and enlarged, Figure 5, C–F). The aberrant keratin filament assemblies, hereafter called “aberrant bundles,” appeared thick and ramified and were distinctly different from interconnected, crossed assemblies formed with wild-type (WT) PPL-C. While the keratin IF network was more robust and interconnected in wild-type, it was loose and more bundled in PPL-C K1646R mutant-expressing cells. This phenotype was noticed independently of whether the anti-pan keratin (recognizing keratins 4, 5, 6, 8, 10, 13, and 18) or specifically anti-keratin8 antibodies were used to visualize the keratin network (Supplemental Figure S5B). Keratin8 has been shown to specifically interact with PPL C-terminal tail (Kazerounian et al., 2002). Upon quantification, it was established that almost 60% of cells expressing mutant PPL-C had significantly more aberrant IF filament assemblies as compared with ∼10% of cells expressing wild-type PPL-C (Figure 5G). This observation suggests that SUMOylation does not alter PPL’s ability to form a complex with keratin or with other IFs but seems critical for the lateral organization of keratins into a robust filamentous network in the cell. Our data suggest that periplakin SUMOylation is a key event to maintain the intermediate filament integrity and robustness. Cellular stresses probably serve as sensors that hyperSUMOylate periplakin to exert its effects on keratin network reorganization.
FIGURE 5:
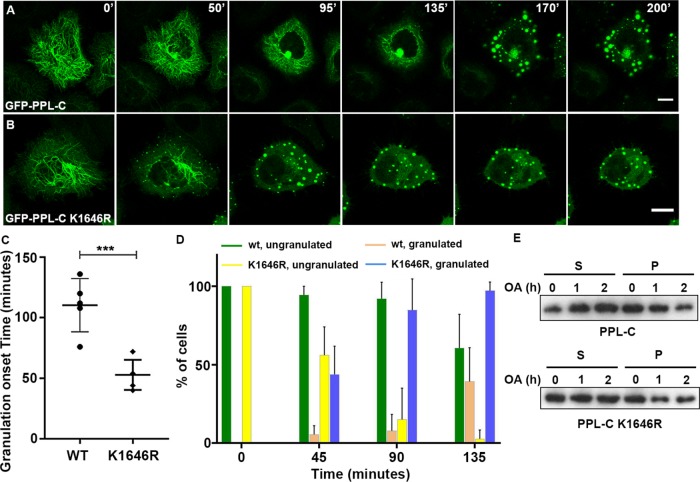
SUMOylation-deficient PPL colocalizes with keratin but affects keratin dynamics and forms aberrant IF bundles. (A, B) Fluorescence light microscopy images of HeLa cells transfected with FLAG-tagged WT or K1646R full-length PPL. Cells were stained for PPL (anti-Flag antibodies, red) and endogenous pan cyto-keratin (keratin antibodies, green). Arrowheads in B highlight peripheral keratin bundles in PPL (fl) K1646R-expressing cells. (C, D) HeLa cells were transfected with FLAG-tagged PPL-C or PPL-C K1646R. Cells were stained for PPL (anti-Flag antibodies, red) and endogenous pan cyto-keratin (keratin antibodies, green). (E, F) Zoomed view of cells marked by asterisks in C and D highlighting the striking differences in filament organization of PPL-C or PPL-C K1646R. Arrowheads in F highlight aberrant bundles in PPL-C K1646R-expressing cells. (G) Percentage of HeLa cells displaying aberrant keratin bundles after transfection with WT or K1646R mutant PPL. Asterisks indicate statistical significance p < 0.01 (n = 150 cells for each PPL-C or PPL-C K1646R from N = 5 independent experiments). (H, I) Time-lapse images at indicated time points for a representative HeLa cell expressing GFP-tagged PPL-C or PPL-C K1646R. Scale bars = 10 μm.
The association of PPL-C and keratin is very strong and is unperturbed by treatments affecting the integrity of the IFs. OA-induced keratin filament collapse and keratin granule formation showed complete colocalization of keratin and PPL-C inside keratin granules in both wild-type and K1646R mutant cases (Supplemental Figure S5C). Our data thus further establish the strong and unperturbed nature of PPL-keratin interaction and this association is independent of phosphorylation status and long filamentous architecture of the keratin assemblies. Additionally, it also reinforces the notion that the PPL-keratin interaction is independent of PPL SUMOylation status. While PPL-C colocalized completely with the keratin IF network, the wild-type and K1646R mutant PPL-C did not colocalize with the compositionally distinct actin network (Supplemental Figure S5D).
Following the observation of periplakin SUMOylation-dependent altered keratin network organization in fixed preparations, using time-lapse fluorescence microscopy we examined whether PPL SUMOylation played any role in the cycle of keratin assembly-disassembly or influenced the dynamics of keratin movement. GFP-tagged full-length PPL constructs when overexpressed showed mislocalization of proteins and formed large aggregates. They also failed to display the fibrillar cytoskeleton staining seen earlier with FLAG- and HA-tagged PPL full-length constructs even when very low plasmid concentration was used for transfection. On the other hand, PPL-C associates with keratin generating cellular staining patterns that are replicas of keratin filament like patterns showed by full-length PPL (Figure 5, A–D) and has been characterized as the minimal domain of PPL required for IF binding. We thus employed time-lapse fluorescence microscopy of GFP-tagged PPL-C-expressing HeLa cells and noticed that GFP-PPL-C normally localized to IF with no signs of the random aggregate formation. The GFP-PPL-C signal appeared to be in continuous motion akin to that of the keratin network (Kolsch et al., 2010). We suggest that PPL-C remains associated with the keratin network during cycles of assembly and disassembly of keratin (Figure 5H and Supplemental Movie 1). We observed peripheral seeding and inward motion of filaments, as reported for keratin (Windoffer et al., 2011), in both wild-type and K1646R mutant PPL-C-expressing cells. However as observed earlier with fixed sample preparations, a distinct bundling of filaments forming thickened cytoskeletal cables was observed with the K1646R mutant (Figure 5F). While the overall pattern of motion of PPL-C signal was largely unchanged irrespective of expression of wild-type or the K1646R mutant, distinctly slower keratin motility was observed in cells transfected with the K1646R mutant (Figure 5I and Supplemental Movie 2). In addition, with an expression of the K1646R mutant, increased IF bundling was noticed and time-dependent instability of the perinuclear cage was evident. Expression levels of GFP-PPL-C and GFP-PPL-C K1646R were confirmed by Western blotting (Supplemental Figure S5E). Studies of keratin dynamics have revealed faster filament movement at the cellular periphery, which is the site for nucleation of keratin filament precursors (Moch et al., 2013). A dynamic equilibrium appears to exist between nucleation and keratin filament formation. Keratin dynamics at nucleating sites in PPL-C K1646R mutant-expressing cells were comparatively much slower than the filament dynamics observed in wild-type PPL-C-expressing cells. We predict that excessive aberrant bundles seen in cells expressing the SUMOylation-deficient PPL-C could affect the dynamic equilibrium of keratin filaments, which further affect their nucleation at the cellular periphery and overall kinetics of filament movement.
Movie S1.
Movie of GFP-PPL-C transfected HeLa cells, recorded for 3.5hr at 100X.
Movie S2.
Movie of GFP-PPL-C K1646R transfected HeLa cells, recorded for 3.5hr 100X.
PPL SUMOylation regulates keratin disassembly dynamics under stress
We have established through our data that PPL is hyper-SUMOylated under conditions leading to IF reorganization. Importantly, in the absence of PPL SUMOylation, we observed altered cyto-architecture that showed reduced network rigidity and the inability of keratin to assemble into robust interconnected filaments. Previous reports have suggested that plakin cytolinker proteins can act as cyto-protectors by playing a key role in keratin filament reorganization particularly in response to stress. Studies on epiplakin and plectin reveal important roles for plakins in the regular kinetics of keratin disassembly under conditions of hyperphosphorylation induced by okadaic acid (Osmanagic-Myers et al., 2006; Spazierer et al., 2008). We noticed that PPL was hyper-SUMOylated under conditions of stress without altering its association with keratin but apparently affecting keratin dynamics. This prompted us to ask what regulatory effects PPL SUMOylation could have on the keratin filament network particularly under filament destabilizing stresses. Since the association of keratin with PPL-C wild-type (or the K1646R mutant) is unaffected under various stresses, we monitored the GFP-PPL-C signal as a readout of keratin filament dynamics. Imaging of fixed samples prepared from OA-treated GFP-PPL-C wild-type or non-SUMOylatable variant- (K1646R) expressing cells showed GFP signals completely merging with keratin filaments as well as the OA-induced granules (Supplemental Figure S6, A and B). Importantly, in contrast to GFP-PPL-C K1646R transfected cells, the untransfected cells in the same field displayed a comparatively unaltered keratin filament network (Supplemental Figure S6, C and D, labeled with asterisks), implying inefficient SUMOylation as the possible cause of defects seen with GFP-PPL-C mutant-expressing cells.
GFP-PPL-C wild-type or K1646R mutant-expressing cells were examined by time-lapse microscopy in the presence of OA and followed for 3 h (Strnad et al., 2001). A representative cell (Figure 6A) showed that at the start of the experiment, cells expressing PPL-C wild-type, displayed a well-spread filamentous network that appeared to be retracted toward the nucleus. At later time points, a progressive breakdown of filaments was observed, with granules starting to first form at the cellular periphery by ∼130 min post–OA treatment. The onset of the granule formation is followed by the coalition of these peripheral granules into a dense perinuclear ring. The perinuclear ring eventually fragmented into numerous granules of various sizes by 3 h of OA treatment (Supplemental Movie 3). In contrast, the PPL-C 1646R mutant-expressing cells showed thicker bundled filaments, displayed a rapid breakdown of filaments and showed first signs of visible granulation by ∼50 min post OA addition (Figure 6B). While significant granulation was observed by 90 min of OA treatment in K1646R mutant conditions, granulation in wild-type had just begun. An early collapse of the keratin network and an unstable perinuclear cage appear to be hallmarks of K1646R-expressing cells (Supplemental Movie 4). We observed similar phenotype in additional cell lines HEK293T (Supplemental Movies 7 and 8) and MCF7a (Supplemental Movies 9 and 10). When a lower concentration of OA was used, the kinetics of keratin network breakdown and granule formation was observed at a later time point with PPL-C wild-type expression (∼140 min, Supplemental Movie 5) yet the PPL-C K1646R mutant displayed a significantly earlier onset of granulation (∼80 min). Our observations with a lower concentration of OA indicated that the PPL-C K1646R mutant imparts susceptibility to the process of IF filament assembly (Supplemental Figure S6E and Supplemental Movie 6). Quantitation of granulation onset time and the percentage of cells with a granulation phenotype with time highlight the importance of PPL-C SUMOylation in keratin disassembly kinetics (Figure 6, C and D). The breakdown of keratin IFs increases the detergent-soluble pools of keratin (Snider and Omary, 2016). Given that PPL-C (wt or K1646R mutant) stably associates with keratin, we monitored detergent-soluble and insoluble periplakin levels during OA treatment and observed a gradual increase in the levels of PPL-C in the soluble fraction mirroring the granulation kinetics. The non-SUMOylatable PPL-C, in contrast, has increased abundance in the soluble fraction at t = 0 and t = 1 h and saturates around the t = 2 h time point (Figure 6E). Quantification of soluble and pellet fractions for wild-type and mutant PPL-C also reflected the same (Supplemental Figure S6F).
FIGURE 6:
SUMOylation deficient PPL is sensitive to phosphatase-inhibitor-induced IF collapse. (A) Time-lapse images at indicated time point for a representative HeLa cell expressing GFP-tagged PPL-C treated with okadaic acid. Scale bars = 10 μm. (B) Time-lapse images at indicated time point for a representative HeLa cell expressing GFP-tagged PPL-C K1646R mutant treated with okadaic acid. Scale bars = 10 μm. (C) Quantification of okadaic acid treated cells (n = 48 cells from N = 5 independent experiments) highlighting onset of granule formation. Asterisks indicate statistical significance p < 0.01. (D) Quantification of okadaic acid treated cells (n = 48 cells from N = 5 independent experiments) at different time points indicating percentage of cells having soluble granules. (E) GFP-tagged PPL-C or PPL-C K1646R mutant transfected cells were treated with OA or left untreated for the indicated time points. Total detergent-soluble (S) and insoluble (P) cell fractions obtained from cells were immunoblotted with GFP antibodies.
Movie S3.
Movie of GFP-PPL-C transfected HeLa cells, recorded for 3.5hr in the presence of 0.5μM OA.
Movie S4.
Movie of GFP-PPL-C K1646R transfected HeLa cells, recorded for 3.5hr in the presence of 0.5μM OA.
Movie S5.
Movie of GFP-PPL-C transfected HeLa cells, recorded for 3.5hr at 100X in the presence of 0.25μM OA.
Movie S6.
Movie of GFP-PPL-C K1646R transfected HeLa cells, recorded for 3.5hr at 100X in the presence of 0.25μM OA.
Movie S7.
Movie of GFP-PPL-C transfected HEK293T cells, recorded for 2hr in the presence of 0.5μM OA.
Movie S8.
Movie of GFP-PPL-C K1646R transfected HEK293T cells, recorded for 2hr in the presence of 0.5μM OA.
Movie S9.
Movie of GFP-PPL-C transfected MCF7a cells, recorded for 2hr in the presence of 0.5μM OA.
Movie S10.
Movie of GFP-PPL-C K1646R transfected MCF7a cells, recorded for 2hr in the presence of 0.5μM OA.
DISCUSSION
SUMOylation exerts control in diverse cellular processes, including DNA replication and repair, transcriptional regulation, nuclear transport, and cell-cycle progression. Although independent proteomic screens have identified cytoskeletal proteins as putative SUMO-1 or SUMO-2/3 targets, only a few proteins have been characterized as authentic SUMO substrates (Rosas-Acosta et al., 2005; Kaminsky et al., 2009). Functional implications of SUMOylation on major cytoskeleton architecture regulators such as components of IF, actin, tubulin, and septins are known (Alonso et al., 2015; Ribet et al., 2017) but there are only predictions for other cytoskeletal proteins. Plakin family proteins are integral to the stability of keratin intermediate filaments. While the SUMOylation of IF proteins like keratin and vimentin shed more light on the effects of SUMOylation on their functions, it is not known whether the plakin family members, the stable interactors of IF, show such modifications. A recent report identified plectin a plakin family protein in a proteomic screen with SUMO2 (Wen et al., 2014). However, covalent SUMO modification of any plakin protein has not been reported—possibly due to their high molecular weight, relatively low levels of SUMOylation as well as its reversible nature. To this end, our data constitute the first report entailing the characterization of SUMO-modification of a plakin family protein, periplakin (PPL). We observed that the steady-state cellular levels of PPL SUMOylation are extremely low. This may indicate that PPL is transiently SUMOylated through a regulated process, for example during the assembly-disassembly cycles of keratin filaments or that only a small fraction of PPL molecules are modified by SUMO in an assembled filament. The low level of PPL SUMOylation perhaps is a consequence of SUMOylation of small fraction of PPL through the tight regulation exerted by keratin filament dynamics.
PPL is selectively modified by SUMO1 and domainwise mapping to identify the PPL SUMOylation site narrowed it to the C-terminal linker domain. A highly conserved lysine (K1646) that marks the junction of the coil-coiled rod and linker domains is the site for SUMO1 modification. A predicted canonical SUMO-interacting motif (SIM) like sequence in the linker domain of PPL is dispensable for PPL SUMOylation.
A global increase in SUMOylation has been reported in response to various stresses. Recently keratin hyperSUMOylation under stress has been reported to alter filament dynamics and naturally occurring mutations affecting sumoylation have been identified as a marker for human liver cirrhosis. PPL SUMOylation levels demonstrate stress-dependent changes with apoptotic and mechanical stresses inducing hyper-SUMOylation of PPL in comparison to basal conditions. More importantly, stresses that induced collapse of the keratin network (Okadaic acid and Orthovanadate) resulted in highest levels of PPL hyper-SUMOylation. We consistently observed decreased PPL SUMOylation upon oxidative stress in concurrence with reports suggesting that ROS (reactive oxygen species), at low concentrations, resulting in the rapid disappearance of most SUMO conjugates (Manza et al., 2004; Bossis and Melchior, 2006). Together, this change in PPL SUMOylation levels may suggest an interesting mechanism of SUMOylation/hyper-SUMOylation of many cytoskeletal proteins to coordinate stress-induced structural changes occurring in the cytoarchitecture. Our results suggest that PPL and keratin functions are regulated similarly by a unifying mechanism of SUMOylation/hyper-SUMOylation. In addition, PPL SUMOylation was seen to be unaltered during cell cycle progression, reinforcing the notion that stress-mediated keratin reorganization and not keratin phosphorylation acts as a cue to induce PPL modification.
As summarized in Figure 7, we suggest that preventing SUMOylation of periplakin does not alter the canonical PPL-keratin association. Analysis of keratins in cells expressing non-SUMOylatable periplakin indicates that periplakin SUMOylation prevents keratin filaments from forming aberrant bundle structures that appeared stressed, and is required for the organization of keratin filaments. In place of reticular, robust and interconnected keratin network normally found in cells, keratin filaments were bundled in thick groups, appeared loose and lacked network rigidity. Our observation with PPL SUMOylation is in congruence with a recent observation noted with SUMOylation of the nonpolar cytoskeletal proteins septins (Ribet et al., 2017). SUMOylation of septins plays a pivotal role in septin filament bundling and cell division but not in septin–septin interactions. We hypothesize SUMOylated PPL to provide structural flexibility and to bring about possible regulation of the IF network and to work efficiently as a cytolinker.
FIGURE 7:
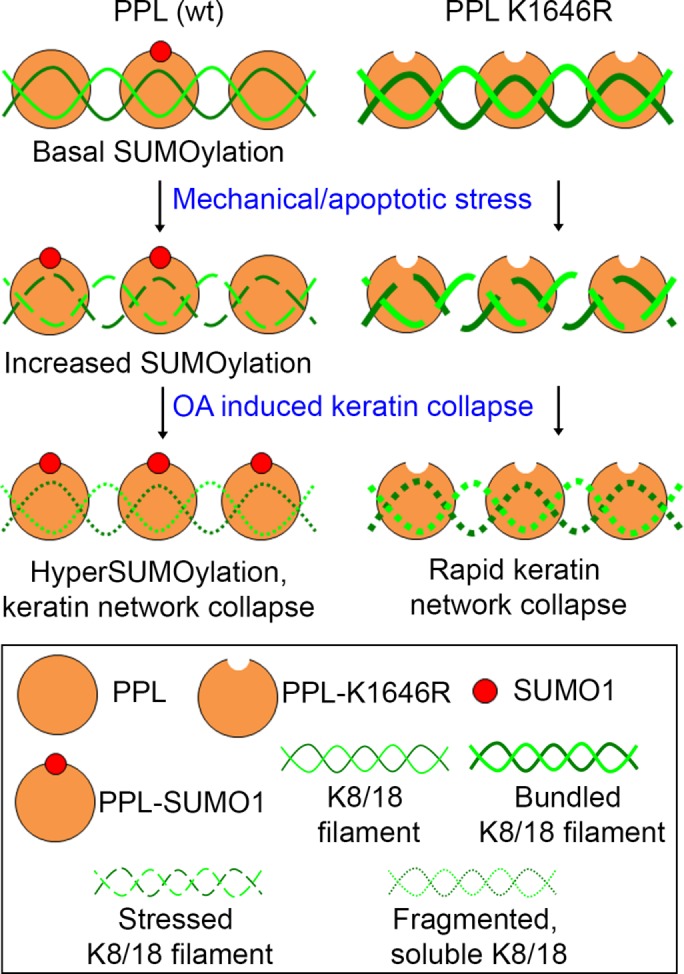
SUMOylation of periplakin is required to maintain homeostasis during the conditions of cytoskeletal stress. IF-associated protein PPL is basally modified by SUMO1 at a conserved residue at its C-terminal linker domain. Mechanical and apoptotic stresses affecting cytoskeletal dynamics increase PPL SUMOylation and induce bundling of keratin in the absence of SUMOylation. Okadaic acid-mediated phosphorylation-induced filament reorganization/collapse further enhances periplakin SUMOylation. In the absence of PPL SUMOylation, OA induces rapid collapse and solubilization of stressed keratin bundles.
Importantly, observations with time-lapse imaging help us suggest that lack of PPL SUMOylation imparts less flexibility and stability, leading to a keratin filament network more susceptible to stress. Keratin filament dynamics at the cellular periphery in PPL-C K1646R mutant-expressing cells was a direct indication of aberrant keratin bundles, a potential outcome of the limited substrate availability at the nucleation step of the keratin cycle (Windoffer et al., 2011).
PPL-C K1646R mutant-expressing cells when treated with okadaic acid observe precocious soluble granule formation. Approximately 80% of cells expressing the PPL-C mutant produced soluble granules by 90 min of OA treatment in contrast to ∼20% of cells expressing PPL-C wild type. OA treatment results in the hyperphosphorylation of keratin and increases its solubility leading to granule formation. It can be speculated in pathological conditions that induce IF dynamics stress; PPL will be hyper-SUMOylated to provide better resistance and maintain the dynamics of IFs. Thus, we propose that SUMOylation of periplakin is critical for the correct assembly of keratin higher-order structures by providing flexibility to the network to rapidly assemble and disassemble as per the need of the cell. Concordantly, SUMOylation has been identified as a potential regulator for the morphological and biochemical changes occurring during keratinocyte differentiation (Deyrieux et al., 2007).
In addition to interaction with IFs, the periplakin C-terminus has been shown to interact with several other proteins. These binding partners include the nuclear protein periphilin and a few membrane-bound receptors like GPCRs and Ig superfamily receptors that appear to be regulated by binding to periplakin. While it is not known whether binding of PPL with IFs and interactions with other proteins are mutually exclusive, it is conceivable that different periplakin fractions in the cell may engage in unique functions and interactions. PPL is known interactor of signaling receptors and downstream molecules; it will be of greater interest to know whether PPL sumoylation as a stress sensor can impinge on any signaling pathway. Being an important interactor and regulator of IF dynamics, it is not surprising that PPL levels are associated with the development of many types of carcinoma. However, the molecular mechanism underlying the regulation of PPL levels with cancer progression and malignancy is largely unknown. Loss of the required balance between global SUMOylation and deSUMOylation events has been reported in various types of cancer (Seeler and Dejean, 2017).
A possible far-reaching role for the SUMOylation status of PPL can be envisaged in carcinomas. In summary, we suggest that periplakin SUMOylation acts as an important organizer of keratin intermediate filament architecture. An enhancement in PPL SUMOylation level provides a handle to reorganize the IF network to respond to cytoskeletal stresses. While SUMOylation in general influences different components of the cellular cytoskeleton, our study elaborates on how the changes in SUMOylation of periplakin maintain homeostasis in conditions of cytoskeletal stress.
MATERIALS AND METHODS
Plasmid constructs and antibodies
Plasmids pEGFP-C1-SUMO1G, pEGFP-C1-SUMO1GG, pEGFP-C1-SUMO2G, and pEGFP-C1-SUMO2GG were a kind gift from Jomon Joseph, NCCS-Pune. PPL domains were amplified with appropriate primers from pCI-Neo-PPL full-length clone as a template (kind gift from Fiona Watt, King’s College London). PPL (fl), PPL K1646R (fl), PPL-PD, PPL-CCR domain, PPL linker domain (PPL-C), and PPL K1646R (PPL-C mutant) were cloned in pCMV3tag1a vector and confirmed by sequencing. All PPL full-length and domain constructs were subcloned in pEGFP-C1 and pGEX6P1 vectors. PPL K1646R and PPL-SIM mutants were made using Q5 Site-Directed Mutagenesis Kit (NEB) according to the manufacturer’s protocol. pSUMO1/2 was a gift from Primo Schaer (Department of Biomedicine, University of Basel; Addgene plasmid # 52258, 52259). Detailed cloning information is available upon request.
Polyclonal antibodies were raised against the C-terminal region of periplakin, SUMO1, GST and Hexa-Histidine tag in rabbits. Antibodies were affinity purified over antigen columns and used in Western blotting detecting periplakin (1:4000), human-SUMO1 (1:3000), GST (1:500) and Hexa-Histidine tag (His)6 (1:100). The following antibodies were also used for Western blotting: anti-GFP (1:6000; Santa Cruz Biotechnology, sc-9996), anti-HA (1:2000; Sigma H6908), anti-Flag (1:2000; Sigma F7425), anti-Flag (1:4000; Sigma F1804), anti-GAPDH (1:4000; Abgenex10011), anti–α-tubulin (1:3000; DHSB), goat anti-rabbit IgG-HRP (1:10,000; GeNei 114038001A), goat anti-mouse IgG-HRP (1:10,000; GeNei 114068001A) and protein A HRP linked antibody (1:8000; Sigma GENA9120). The following antibodies were also used for immunofluorescence: anti-Cytokeratin (1:300; C11 Biolegend), anti-keratin8 (1:500; Sigma 5301) and Alexa Fluor–conjugated secondary antibodies (1:1000; Invitrogen).
Cell culture and transfections
HEK293T, HeLa, and MCF7a cell lines were cultured in DMEM (Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (Invitrogen) and 1% antibiotics (Sigma) in a humidified incubator at 37°C under 5% CO2 conditions. The cells were grown in 100-mm cell culture plates (1 × 107 cells) or in six-well plates (3 × 105 cells per well) for immunoprecipitation and Western blot analysis. After 12 h of plating, cells were transfected using polyethylenimine (PEI) 25-kDa linear (23966-2g; Polysciences Corporation Ltd.) or Effectene Transfection reagent (Qiagen) following manufacturer’s instructions. For cell synchronization, cells were arrested in S phase by using a double thymidine block (early S-phase block) protocol. Cells were washed with 1x phosphate-buffered saline (PBS), and 2.5 mM thymidine was added for 18 h (first block). After the first block thymidine was removed by washing with 1xPBS and fresh DMEM was added for 9 h to release cells from the block, 2.5 mM thymidine was added again for 17 h (second block). Cells were arrested in mitosis by synchronizing first in thymidine for 24 h (S-phase block) followed by 3 h release and then blocking for 12 h in 100 ng/ml nocodazole (mitotic block). Synchronization of cells into different cell cycle phases was assessed by fluorescence-activated cell sorting (FACS).
Immunoprecipitation
Cells were collected using a cell scraper and washed with ice-cold PBS twice and resuspended in chilled RIPA buffer (50 mM Tris, pH 7.5, 10 mM EDTA, 1 mM EGTA [ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid], 150 mM NaCl, 1% Triton, and 0.1% sodium deoxycholate) followed by 10 min incubation on ice. Cells were lysed by sonication at 4°C with 30% amplitude (3 s ON and 3 s OFF for 2 min) using VibraCell Sonics. For IP assays and Western blotting (WB) analysis, the supernatant was collected by centrifuging samples at 14,000 rpm for 30 min at 4°C. Protein concentration was determined using Bradford reagent (Bio-Rad). For IP, indicated antibodies were bound to 10 μl of Protein-A Sepharose beads (Agarose Beads Technologies) overnight at 4°C with constant mixing. The samples were centrifuged at 400 × g for 1 min in a refrigerated centrifuge followed by five washes with chilled PBS. Bound proteins were eluted using 0.2 M glycine, pH 2.5, and boiled in 6X SDS Laemmli sample buffer.
GFP-trap pull downs
GBP clone was a kind gift from Heinrich Leonhardt (Ludwig-Maximilians-University of Munich). GBP cDNA was subcloned into the pGEX-6P1 vector to express GST-tagged GBP. GFP-SUMO-1GG/G and -2GG/G were coexpressed with FLAG-PPL-C (six-well or 60-mm dish format) in HEK293T cells. Cleared lysates prepared from GFP-SUMO variants expressing cells were mixed with GST-GBP and the complex formed was pulled down on glutathione-Sepharose beads. Bound material was eluted and processed in Laemmli buffer for Western blotting.
Western blotting
Eluted proteins were separated on SDS–PAGE and then transferred to polyvinylidene difluoride (PVDF) membrane. The membranes were incubated in blocking buffer 5% skimmed milk in PBS with 0.1% Tween-20 (PBS-T) for 1 h at room temperature. After incubation with indicated primary antibodies, membranes were given three washes of 10 min each with PBS-T, followed by incubation with HRP conjugated secondary antibody for 1 h at room temperature. Detection was performed using enhanced chemiluminescence substrate (ECL Plus; ThermoFisher Scientific).
In vitro and in bacto SUMOylation
In vitro SUMOylation reactions contained E1 enzyme (0.25 µg GST-SAE2/SAE1), E2 enzyme (1.0 μg (His)6-Ubc9), SUMO-1 or SUMO-2 protein (4 μg), and GST tagged C-terminus of periplakin (PPL-C) wild type and or K1646R mutant in reaction buffer (50 mM Tris, pH 7.5, 5 mM MgCl2, 5 mM ATP, 5 mM dithiothreitol). SUMOylation reactions were incubated at 37°C overnight and terminated with 6X Laemmlli buffer. SUMOylation was analyzed by resolving reaction products on SDS–PAGE. The deconjugation reactions were performed at 25°C for 20 min in the presence of GST-SENP1 (0.2 µg).
Different PPL-C constructs were cloned in pGEX6P1 vector and cotransformed with pSUMO1 or pSUMO2 (Addgene). Double transformants were selected on Luria broth (LB) agar plates having 50 mg/l of ampicillin and 25 mg/l of streptomycin. For in bacto SUMOylation, mid–log phase cultures were induced overnight with 200 µM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 25°C. Cells were harvested by centrifugation, and soluble protein fractions were extracted by sonication in buffer “A” (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Triton, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride). Crude lysates were then cleared by centrifugation 20,000 × g at 4°C for 30 min. Proteins were affinity purified on Glutathione beads (ABT) followed by four washes with buffer “A” containing 400 mM NaCl. Proteins bound to beads were extracted by boiled in Laemmli buffer and subsequently analyzed on SDS–PAGE.
In vivo SUMOylation assay
HEK293T cells were cotransfected with either pEGFP-SUMO1G/2G or pEGFP-SUMO1GG/2G with PPL constructs. Forty-eight hours posttransfection, total cell lysates were prepared in IP lysis/wash buffer supplemented with 40 mM NEM (Sigma 04259). Total cell lysates were incubated with 4 µg of anti-PPL/anti-GFP antibody in IP lysis buffer and were incubated with constant rotation at 4°C overnight. Following this, 20 µl of Protein A/G plus Agarose bead slurry was mixed and rotated further for 4 h at 4°C. The immunoprecipitated samples were collected by centrifugation and washed several times with PBS and eluted and boiled in Laemmli buffer for further analysis.
Induction of cellular stresses
For inducing different cellular stress, cells were first grown under conditions described earlier and transfected as per requirement. Okadaic Acid Ammonium salt (0.5 µM; Calbiochem 459616) was used for 3 h for inducing intermediate filament collapse stress (unless stated differently), staurosporine (1 µM; S5921 Sigma) for 3 h for apoptotic stress, and H2O2 (1 mM, Merck) for 45 min for oxidative stress. Cells were grown to 100% confluence and mechanical stress was induced by inflicting 10 scratches/strokes per 60 mm dish creating a cell free gap and in a confluent cell monolayer by scratch assay (scratching using 200 μl sterile tip). The cells were then gently washed twice with 2 ml PBS to remove the detached cells. Cells were then replenished with fresh medium. Cells were incubated for 1 h before samples were prepared for pull down.
Immunofluorescence microscopy
HeLa and MCF7a cells grown on glass coverslips were transfected with indicated constructs. Cells were fixed using 4% formaldehyde (Sigma) for 15 min followed by 10 min incubation with rehydration buffer (10 mM Tris, 150 mM NaCl, 0.1% Triton). Cells were then blocked with 2% normal goat serum (NGS) for 1 h followed by incubation with primary antibodies diluted in PBS containing 2% NGS overnight at 4°C. Cells were then washed thrice with PBS and incubated for 45 min at room temperature with Alexa Fluor–conjugated secondary antibodies (Invitrogen). Cells were again washed thrice with PBS and Hoechst-33342 dye (Sigma) used to stain DNA was added in the final wash (1:7000). Coverslips were mounted on glass slides using Vectashield mounting medium (Vector Laboratories).
Time-lapse live-cell imaging
HeLa cells were seeded on glass bottom 30-mm dishes (Ibidi, Germany) and transfected with GFP-tagged PPL-C wild type or K1646R mutant clones. Live cell recordings were performed using Olympus Confocal Laser Scanning Microscope–FV3000 under controlled conditions with constant temperature (37°C), humidity, and CO2 supply. Maximum intensity projection images at different time points were obtained from videos (recorded continuously for 3.5 h with 2 min intervals) using CellSens software.
Statistical analysis
Image analysis, processing, and band quantitation were done using ImageJ software or GelQuant.NET. The experiments were independently repeated at least three times, and the values are expressed as mean ± SD. The p values were calculated using Student’s t test; p ≤ 0.05 was considered statistically significant. Graphs were plotted using GraphPad Prism.
Supplementary Material
Acknowledgments
We thank Jomon Joseph (NCCS-Pune) for generously providing reagents, helpful discussions, and critical comments on the manuscript. We also thank Shanker Jha, Vivek Pandey, Usha Singh, and Saurabh Mehta for their constant support, encouragement, and critical comments on the manuscript. This work was supported by Science and Engineering Research Board (SERB), India, grant # EMR/2016/001819, intramural funding from the Department of Biological Sciences, Indian Institute of Science Education, and Research Bhopal, and University Grant Commission (UGC) fellowship to M.G.
Abbreviations used:
- GBP
GFP binding protein
- GFP
green fluorescent protein
- HA
hemagglutinin
- IF
immunofluorescence
- IFs
intermediate filaments
- IP
immunoprecipitation
- NEM
N-ethylmaleimide
- OA
okadaic acid
- OV
orthovanadate
- PEI
polyethylenimine
- PPL
periplakin
- PRD
plakin repeat domains
- SENP
sentrin-specific protease
- SIM
SUMO-interacting motif
- STS
staurosporine
- SUMO1
small ubiquitin-like modifier 1
- SUMO2
small ubiquitin-like modifier 2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-04-0244) on December 5, 2018.
REFERENCES
- Alonso A, Greenlee M, Matts J, Kline J, Davis KJ, Miller RK. (2015). Emerging roles of sumoylation in the regulation of actin, microtubules, intermediate filaments, and septins. Cytoskeleton (Hoboken) , 305–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman JM, Bakema JE, van de Winkel JG, Leusen JH. (2004). Direct interaction between FcgammaRI (CD64) and periplakin controls receptor endocytosis and ligand binding capacity. Proc Natl Acad Sci USA , 10392–10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankson H, Holen I, Seglen PO. (1995). Disruption of the cytokeratin cytoskeleton and inhibition of hepatocytic autophagy by okadaic acid. Exp Cell Res , 522–530. [DOI] [PubMed] [Google Scholar]
- Boczonadi V, Maatta A. (2012). Annexin A9 is a periplakin interacting partner in membrane-targeted cytoskeletal linker protein complexes. FEBS Lett , 3090–3096. [DOI] [PubMed] [Google Scholar]
- Boczonadi V, McInroy L, Maatta A. (2007). Cytolinker cross-talk: periplakin N-terminus interacts with plectin to regulate keratin organisation and epithelial migration. Exp Cell Res , 3579–3591. [DOI] [PubMed] [Google Scholar]
- Bossis G, Melchior F. (2006). Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell , 349–357. [DOI] [PubMed] [Google Scholar]
- Bouameur JE, Schneider Y, Begre N, Hobbs RP, Lingasamy P, Fontao L, Green KJ, Favre B, Borradori L. (2013). Phosphorylation of serine 4,642 in the C-terminus of plectin by MNK2 and PKA modulates its interaction with intermediate filaments. J Cell Sci , 4195–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YK, Woo SM, Cho SG, Moon HE, Yun YJ, Kim JW, Noh DY, Jang BH, Shin YC, Kim JH, et al. (2013). Brain-metastatic triple-negative breast cancer cells regain growth ability by altering gene expression patterns. Cancer Genomics Proteomics , 265–275. [PubMed] [Google Scholar]
- Deyrieux AF, Rosas-Acosta G, Ozbun MA, Wilson VG. (2007). Sumoylation dynamics during keratinocyte differentiation. J Cell Sci , 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiColandrea T, Karashima T, Maatta A, Watt FM. (2000). Subcellular distribution of envoplakin and periplakin: insights into their role as precursors of the epidermal cornified envelope. J Cell Biol , 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot KR, Sevilla LM, Nishi K, DiColandrea T, Watt FM. (2004). Kazrin, a novel periplakin-interacting protein associated with desmosomes and the keratinocyte plasma membrane. J Cell Biol , 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. (2005). SUMO: a history of modification. Mol Cell , 1–12. [DOI] [PubMed] [Google Scholar]
- Jefferson JJ, Leung CL, Liem RK. (2004). Plakins: goliaths that link cell junctions and the cytoskeleton. Nat Rev Mol Cell Biol , 542–553. [DOI] [PubMed] [Google Scholar]
- Kaminsky R, Denison C, Bening-Abu-Shach U, Chisholm AD, Gygi SP, Broday L. (2009). SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans. Dev Cell , 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karashima T, Watt FM. (2002). Interaction of periplakin and envoplakin with intermediate filaments. J Cell Sci , 5027–5037. [DOI] [PubMed] [Google Scholar]
- Kazerounian S, Aho S. (2003). Characterization of periphilin, a widespread, highly insoluble nuclear protein and potential constituent of the keratinocyte cornified envelope. J Biol Chem , 36707–36717. [DOI] [PubMed] [Google Scholar]
- Kazerounian S, Uitto J, Aho S. (2002). Unique role for the periplakin tail in intermediate filament association: specific binding to keratin 8 and vimentin. Exp Dermatol , 428–438. [DOI] [PubMed] [Google Scholar]
- Kolsch A, Windoffer R, Wurflinger T, Aach T, Leube RE. (2010). The keratin-filament cycle of assembly and disassembly. J Cell Sci , 2266–2272. [DOI] [PubMed] [Google Scholar]
- Kroger C, Loschke F, Schwarz N, Windoffer R, Leube RE, Magin TM. (2013). Keratins control intercellular adhesion involving PKC-alpha-mediated desmoplakin phosphorylation. J Cell Biol , 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CL, Liem RK, Parry DA, Green KJ. (2001). The plakin family. J Cell Sci , 3409–3410. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang G, Wang Y, Elgehama A, Sun Y, Li L, Gu Y, Guo W, Xu Q. (2017). Loss of periplakin expression is associated with the tumorigenesis of colorectal carcinoma. Biomed Pharmacother , 366–374. [DOI] [PubMed] [Google Scholar]
- Long HA, Boczonadi V, McInroy L, Goldberg M, Maatta A. (2006). Periplakin-dependent re-organisation of keratin cytoskeleton and loss of collective migration in keratin-8-downregulated epithelial sheets. J Cell Sci , 5147–5159. [DOI] [PubMed] [Google Scholar]
- Mabb AM, Wuerzberger-Davis SM, Miyamoto S. (2006). PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol , 986–993. [DOI] [PubMed] [Google Scholar]
- Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC. (2004). Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol , 1706–1715. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Ikeda M, Sato Y, Kuruma H, Kamata Y, Nishimori T, Tomonaga T, Nomura F, Egawa S, Iwamura M. (2014). Loss of periplakin expression is associated with pathological stage and cancer-specific survival in patients with urothelial carcinoma of the urinary bladder. Biomed Res , 201–206. [DOI] [PubMed] [Google Scholar]
- Milligan G, Murdoch H, Kellett E, White JH, Feng GJ. (2004). Interactions between G-protein-coupled receptors and periplakin: a selective means to regulate G-protein activation. Biochem Soc Trans , 878–880. [DOI] [PubMed] [Google Scholar]
- Moch M, Herberich G, Aach T, Leube RE, Windoffer R. (2013). Measuring the regulation of keratin filament network dynamics. Proc Natl Acad Sci USA , 10664–10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimori T, Tomonaga T, Matsushita K, Oh-Ishi M, Kodera Y, Maeda T, Nomura F, Matsubara H, Shimada H, Ochiai T. (2006). Proteomic analysis of primary esophageal squamous cell carcinoma reveals downregulation of a cell adhesion protein, periplakin. Proteomics , 1011–1018. [DOI] [PubMed] [Google Scholar]
- Osmanagic-Myers S, Gregor M, Walko G, Burgstaller G, Reipert S, Wiche G. (2006). Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J Cell Biol , 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo T, Hagiwara T, Tamura-Nakano M, Sezaki T, Miyake O, Hinohara C, Shimizu T, Yamada K, Dohi T, Kawamura YI. (2015). Aberrant DNA hypermethylation reduces the expression of the desmosome-related molecule periplakin in esophageal squamous cell carcinoma. Cancer Med , 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Boscaini S, Cauvin C, Siguier M, Mostowy S, Echard A, Cossart P. (2017). SUMOylation of human septins is critical for septin filament bundling and cytokinesis. J Cell Biol , 4041–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. (2005). A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics , 56–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics , 282–289. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, et al. (2006). Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods , 887–889. [DOI] [PubMed] [Google Scholar]
- Ruhrberg C, Hajibagheri MA, Parry DA, Watt FM. (1997). Periplakin, a novel component of cornified envelopes and desmosomes that belongs to the plakin family and forms complexes with envoplakin. J Cell Biol , 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhrberg C, Watt FM. (1997). The plakin family: versatile organizers of cytoskeletal architecture. Curr Opin Genet Dev , 392–397. [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. (2017). SUMO and the robustness of cancer. Nat Rev Cancer , 184–197. [DOI] [PubMed] [Google Scholar]
- Snider NT, Omary MB. (2016). Assays for Posttranslational Modifications of Intermediate Filament Proteins. Methods Enzymol , 113–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Weerasinghe SV, Iniguez-Lluhi JA, Herrmann H, Omary MB. (2011). Keratin hypersumoylation alters filament dynamics and is a marker for human liver disease and keratin mutation. J Biol Chem , 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Liem RK. (2007). Plakins in development and disease. Exp Cell Res , 2189–2203. [DOI] [PubMed] [Google Scholar]
- Spazierer D, Raberger J, Gross K, Fuchs P, Wiche G. (2008). Stress-induced recruitment of epiplakin to keratin networks increases their resistance to hyperphosphorylation-induced disruption. J Cell Sci , 825–833. [DOI] [PubMed] [Google Scholar]
- Strnad P, Windoffer R, Leube RE. (2001). In vivo detection of cytokeratin filament network breakdown in cells treated with the phosphatase inhibitor okadaic acid. Cell Tissue Res , 277–293. [DOI] [PubMed] [Google Scholar]
- Strnad P, Windoffer R, Leube RE. (2002). Induction of rapid and reversible cytokeratin filament network remodeling by inhibition of tyrosine phosphatases. J Cell Sci , 4133–4148. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hatakeyama S, Saitoh H, Nakayama KI. (2005). Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J Biol Chem , 5611–5621. [DOI] [PubMed] [Google Scholar]
- Tempe D, Piechaczyk M, Bossis G. (2008). SUMO under stress. Biochem Soc Trans , 874–878. [DOI] [PubMed] [Google Scholar]
- van den Heuvel AP, de Vries-Smits AM, van Weeren PC, Dijkers PF, de Bruyn KM, Riedl JA, Burgering BM. (2002). Binding of protein kinase B to the plakin family member periplakin. J Cell Sci , 3957–3966. [DOI] [PubMed] [Google Scholar]
- Virata ML, Wagner RM, Parry DA, Green KJ. (1992). Molecular structure of the human desmoplakin I and II amino terminus. Proc Natl Acad Sci USA , 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AR, Schuermann D, Schar P. (2014). Versatile recombinant SUMOylation system for the production of SUMO-modified protein. PLoS One , e102157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Xu Z, Xia L, Liu X, Tu Y, Lei H, Wang W, Wang T, Song L, Ma C, et al. (2014). Important role of SUMOylation of Spliceosome factors in prostate cancer cells. J Proteome Res , 3571–3582. [DOI] [PubMed] [Google Scholar]
- Windoffer R, Beil M, Magin TM, Leube RE. (2011). Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J Cell Biol , 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Leube RE. (2004). Imaging of keratin dynamics during the cell cycle and in response to phosphatase inhibition. Methods Cell Biol , 321–352. [DOI] [PubMed] [Google Scholar]
- Yatsunami J, Komori A, Ohta T, Suganuma M, Yuspa SH, Fujiki H. (1993). Hyperphosphorylation of cytokeratins by okadaic acid class tumor promoters in primary human keratinocytes. Cancer Res , 992–996. [PubMed] [Google Scholar]
- Yoon KH, FitzGerald PG. (2009). Periplakin interactions with lens intermediate and beaded filaments. Invest Ophthalmol Vis Sci , 1283–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.