Abstract
COPII-coated vesicles are the primary mediators of ER-to-Golgi trafficking. Sar1, one of the five core COPII components, is a highly conserved small GTPase, which, upon GTP binding, recruits the other COPII proteins to the ER membrane. It has been hypothesized that the changes in the kinetics of SAR1 GTPase may allow for the secretion of large cargoes. Here we developed a cell-free assay to recapitulate COPII-dependent budding of large lipoprotein cargoes from the ER. We identified fatty-acid binding protein 5 (FABP5) as an enhancer of this budding process. We found that FABP5 promotes the budding of particles ∼150 nm in diameter and modulates the kinetics of the SAR1 GTPase cycle. We further found that FABP5 enhances the trafficking of lipoproteins and of other cargoes, including collagen. These data identify a novel regulator of SAR1 GTPase activity and highlight the importance of this activity for trafficking of large cargoes.
INTRODUCTION
Lipoproteins traffic insoluble lipids within an apolipoprotein shell. Large lipoproteins, including chylomicrons, very low-density lipoproteins (VLDLs), and its lipolytic conversion product, LDL, are produced in the intestine and liver, respectively, and transport the majority of cholesterol and triglyceride in the bloodstream. High LDL levels are a major risk factor for atherosclerosis and coronary heart disease (Budoff, 2016), one of the leading causes of mortality in developed countries (Heron, 2012); therefore, increased understanding of the production and processing of lipoproteins bears directly on human health.
The pathophysiology of rare genetic diseases can serve as a powerful entry point for understanding of genes that are most physiologically relevant to a biological pathway. Chylomicron retention disease/Anderson’s disease (CMRD) results in an inability to transport newly synthesized chylomicrons out of intestinal epithelial cells and, in some patients fatty liver and hypocholesterolemia, suggesting that the disease affects lipoprotein transport in the liver as well (Nemeth et al., 1995; Charcosset et al., 2008; Silvain et al., 2008; Peretti et al., 2009; Georges et al., 2011), making it relevant to the study of general lipoprotein production.
CMRD is caused by mutations in SAR1B (Jones et al., 2003; Charcosset et al., 2008; Silvain et al., 2008; Peretti et al., 2009; Georges et al., 2011), part of the COPII coat, the primary mediator of protein traffic from the endoplasmic reticulum (ER) to the Golgi apparatus (Hicke and Schekman, 1989; Salama et al., 1993; Yoshihisa et al., 1993; Schekman and Novick, 2004). Strikingly, despite disrupting this fundamental cellular pathway, CMRD only affects limited tissues. Loss of function of other COPII components is associated with other highly specific phenotypes in both humans and animal models (Boyadjiev et al., 2006; Lang et al., 2006; Bianchi et al., 2009; Schwarz et al., 2009; Merte et al., 2010; Sarmah et al., 2010; Niu et al., 2012).
Traffic-specific phenotypes associated with COPII lesions may help explain another significant question in the protein trafficking field, namely, how very large cargoes are transported (Bonfanti et al., 1998; Stephens and Pepperkok, 2002; Canty and Kadler, 2005). Although cargo-free, self-assembled COPII vesicles are 40–80 nm in diameter, procollagen bundles form stiff 300-nm rods, and other cargoes (e.g., chylomicrons) can be 600 nm in size. Certain COPII components appear to have evolved specialized functions to allow for expansion of vesicle size and transport of large cargoes, whereas other proteins such as TANGO1 or KLHL12 may enhance COPII-dependent large cargo budding by influencing the SAR1 GTPase cycle (Siddiqi et al., 2003, 2010; Boyadjiev et al., 2006; Lang et al., 2006; Sarmah et al., 2010; Melville et al., 2011; Saito et al., 2011, 2014, 2017; Wilson et al., 2011; Siddiqi and Mansbach, 2012; Santos et al., 2016; Tanabe et al., 2016).
The SAR1 GTPase cycle plays a critical role in COPII coat formation, and may play a key role in regulation of vesicle size in large cargo secretion (Venditti et al., 2012; Ma and Goldberg, 2016). The cycle involves two major processes: exchange of GDP for GTP mediated by the guanine exchange factor (GEF) SEC12 (prolactin regulatory element binding [PREB] in mammals; Barlowe and Schekman, 1993; Weissman et al., 2001) and GTPase activity mediated by the GTPase-activating protein (GAP) SEC23. The GAP activity of SEC23 is substantially stimulated by the proline-rich “active fragment” of SEC31, which binds, overlapping the junction of SEC23/SAR1 (Bi et al., 2007; Fromme et al., 2007).
TANGO1 exemplifies one mechanism by which the SAR1 GTPase cycle may be regulated for secretion of large proteins. In collagen secretion, TANGO1 dampens SAR1 GTPase activity by preventing SEC31 from binding to SEC23 (Ma and Goldberg, 2016), introducing a pause that may allow for the loading of large cargoes and genesis of large vesicular carriers (Raote et al., 2017). As the budding event for large cargoes has not been reconstituted with pure components, other factors may come into play.
Previous work on the mechanism of lipoprotein traffic from the ER demonstrated a role for COPII vesicles (Gusarova et al., 2003). In this study, we have refined the cell-free vesicle budding reaction and discovered a novel requirement for the capture of lipoprotein particles. Using the unique level of control afforded by in vitro reconstitution, we have identified FABP5 as a factor that enhances large lipoprotein transport. We further found that FABP5 binds Sec12/PREB and modulates the guanine exchange of SAR1, as well as interacting with the active fragment of SEC31 and altering the GTPase activity of SAR1. We found that the effect of FAPB5 extends to collagen trafficking. We propose that FABP5 is a novel regulator of the SAR1 GTPase cycle and plays a significant role in large protein trafficking.
RESULTS
In vitro reconstitution of VLDL budding
To provide a tool for understanding the cellular requirements for budding of large lipoprotein cargoes, we attempted to reconstitute VLDL budding in a cell-free assay. We modified previously published cell-free reactions designed to detect the formation of transport vesicles, including ones that carry vesicles as large as collagen, that bud from ER membranes (Figure 1A; Kim et al., 2005; Merte et al., 2010; Gorur et al., 2017; Yuan et al., 2017).
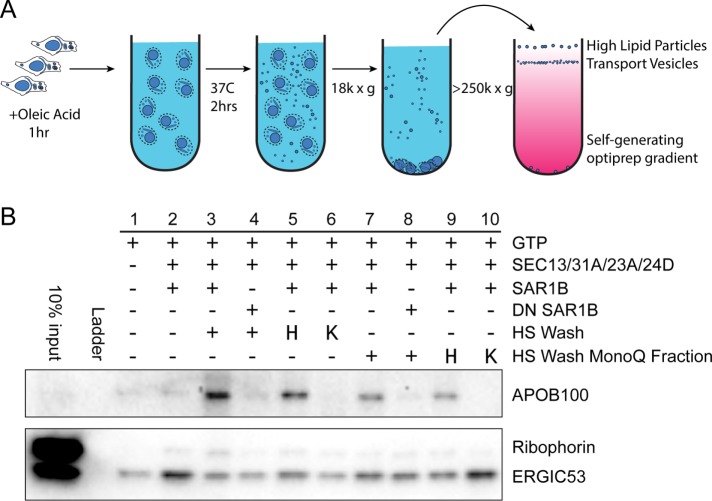
FIGURE 1:
Budding of APOB lipoprotein cargoes in a cell-free reaction. (A) Scheme of cell-free vesicle budding reaction. In brief, McArdle RH7777 cells were incubated with oleic acid. Treated cells were used to prepare donor membranes which were then incubated at 37°C with GTP and purified recombinant human COPII proteins. McArdle membranes were washed with high salt, the high-salt wash (HSW) was dialyzed and added to a vesicle budding reaction where indicated, heated (“H”) or treated with proteinase K, then heated (“K”) as indicated. Vesicles in 18,000 × g supernatant fractions from budding reactions were isolated by density gradient flotation. (B) Fractions from the top of an OptiPrep gradient were analyzed by immunoblot. APOB serves as a marker for large VLDL cargoes and ERGIC53 serves as a marker for small traditional COPII cargoes. Ribophorin serves as a marker for ER contamination.
Donor ER membrane was prepared from McArdle-RH7777, cultured Rattus norvegicus liver hepatoma cells. Membranes were incubated at 37°C with nucleotides and purified recombinant human COPII proteins for 2 h to allow the formation of transport vesicles. Budded vesicles were separated from membrane by centrifugation at 18,000 × g to sediment donor membrane, and then the supernatant fraction was applied to an OptiPrep flotation gradient. After a high-speed centrifugation step at 350,000 × g for 3 h, we collected fractions from the top, and analyzed their contents by immunoblotting (Figure 1B). We used a protease protection assay to confirm that vesicle contents were protease resistant in the absence of detergent.
We blotted for the APOB100 isoform of apolipoprotein B, the primary protein component of VLDL as a marker of VLDL, ER–Golgi intermediate compartment 53 kDa protein (ERGIC53) as a marker of canonical COPII vesicles, and ribophorin as a marker of ER contamination. Buoyant lipid vesicles floated to the top of the gradient, as shown by the COPII-dependent enrichment of ERGIC53; however, with COPII alone, little APOB100 was detected (Figure 1B, lane 2). We hypothesized that the addition of cytosol might allow for budding of large VLDL; however, in our hands we did not see a consistent increase in APOB100 signal upon cytosol addition. As an alternative to cytosol, we added an enriched fraction of peripheral membrane proteins obtained by a high-salt wash (HSW) of McArdle membranes. This HSW was desalted by dialysis and added to the budding reaction. We found that HSW enhanced budding of APOB-positive cargoes, but not other cargoes, and possibly even had a deleterious effect on other cargoes (Figure 1B, lane 3).
To determine whether the APOB100 budding observed upon the addition of HSW was COPII dependent, we utilized a dominant-negative form of SAR1B, and found that it indeed inhibited APOB budding (Figure 1B, lane 4). This suggested that a factor in the HSW was enhancing COPII activity to allow for VLDL secretion. To determine whether the active factor in HSW was protein, we heated the HSW in a boiling water bath before aliquots were added to the budding reaction. Surprisingly, extreme heating did not inhibit the activity of this fraction (Figure 1B, lane 5). To distinguish a small molecule effector from a thermostable protein, we treated the HSW fraction with proteinase K before heating. Proteinase-treated HSW no longer enhanced APOB budding (Figure 1B, lane 6), suggesting that the active factor was indeed protein.
In an attempt to further purify the active protein component from crude HSW, we subjected the fraction to MonoQ anion exchange chromatography. The activity was contained in the flowthrough fraction (Figure 1B, lane 7), and once again the activity was proteinase sensitive, heat resistant, and sustained a budding reaction that was inhibited by dominant-negative SAR1B (Figure 1B, lanes 8–10).
FABP5 enhances lipoprotein secretion
To identify the protein component of HSW responsible for APOB budding activity, we utilized tandem mass spectrometry to identify proteins in the heated HSW that did not bind to the MonoQ column. From this analysis we detected several candidate proteins, one of which was a fatty-acid binding protein, FABP5, chosen in part because FABP5 is a known heat-stable protein that has been implicated in chylomicron secretion (Siddiqi et al., 2003; Siddiqi and Mansbach, 2012). We subcloned the rat FABP5 gene from McArdle cell mRNA into pGEX-2T vectors, and recombinantly expressed and purified the protein from Escherichia coli. We found that the addition of FABP5 to budding reactions enhanced APOB budding (Figure 2A). The effect was dose dependent and most effective in the presence of COPII (Figure 2B). The addition of FABP5 also appeared to enhance budding of ERGIC53 and SEC22B, although not the severalfold change seen with APOB.
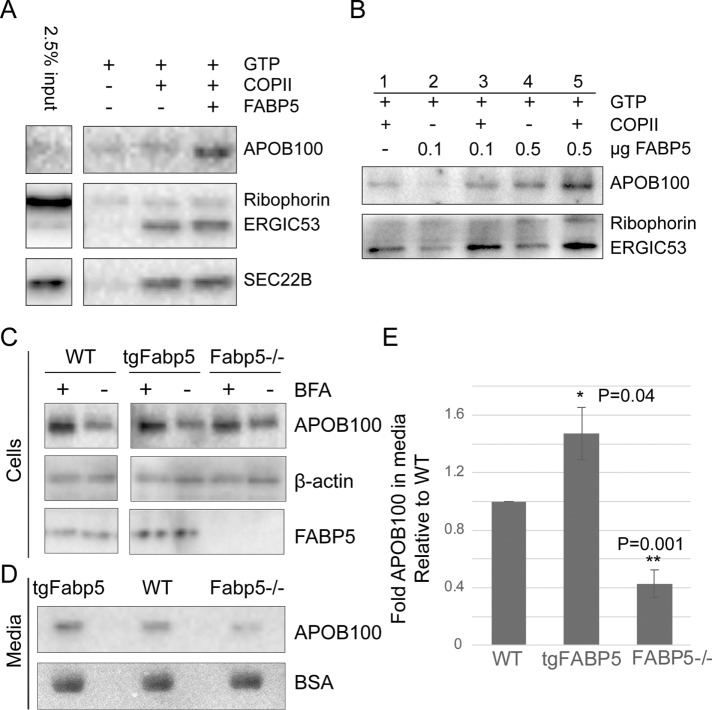
FIGURE 2:
FABP5 enhances lipoprotein secretion. (A) Immunoblot of top fraction of OptiPrep gradient from budding reactions with McArdle donor membrane. (B) Immunoblot of budding reaction for dose response to FABP5. (C) Immunoblot of APOB in wild-type McArdle cells (WT), McArdle transformed by lentivirus to overexpress FABP5 (tgFabp5) or CRISPR/CAS9-mediated knockout cells (Fabp5−/−). β-Actin used as a loading control. Brefeldin A (BFA) added to inhibit secretion and show baseline levels of cellular APOB. (D) APOB secreted into the medium of those cells after 3 h and Ponceau-S stain of BSA in membrane for loading control. (E) Quantification of collagen secreted into media (n = 3).
To test the cellular function of FABP5, we used CRISPR/Cas9 to generate McArdle cell lines lacking FABP5 expression. We also used a tetracycline-inducible lentivirus expression system to overexpress FABP5. We found that whereas all three cells lines had similar levels of APOB when secretion was blocked with brefeldin A (BFA; Figure 2C), the amount of APOB100 detected in the medium was increased or decreased with the increase or decrease of FABP5 levels (Figure 2, D and E). Consistent with this observation, the amount of intracellular APOB100 remaining in cells after 3 h of incubation with oleic acid showed the opposite trend, although the changes were not statistically significant (P = 0.1). These data are consistent with what we found in the cell-free budding reactions, suggesting that FABP5 enhances the secretion of large VLDL cargoes.
FABP5 increases budded vesicle size
We next examined the size of the APOB-containing carriers in relation to traditional COPII cargo carriers. During optimization of the cell-free vesicle budding assay, we observed that with low concentrations of OptiPrep in self-forming gradients (Figure 3A), we were able to separate APOB100-positive fractions from the fractions containing other cargoes, presumably due to the high lipid content and buoyancy of VLDL (Figure 3B).
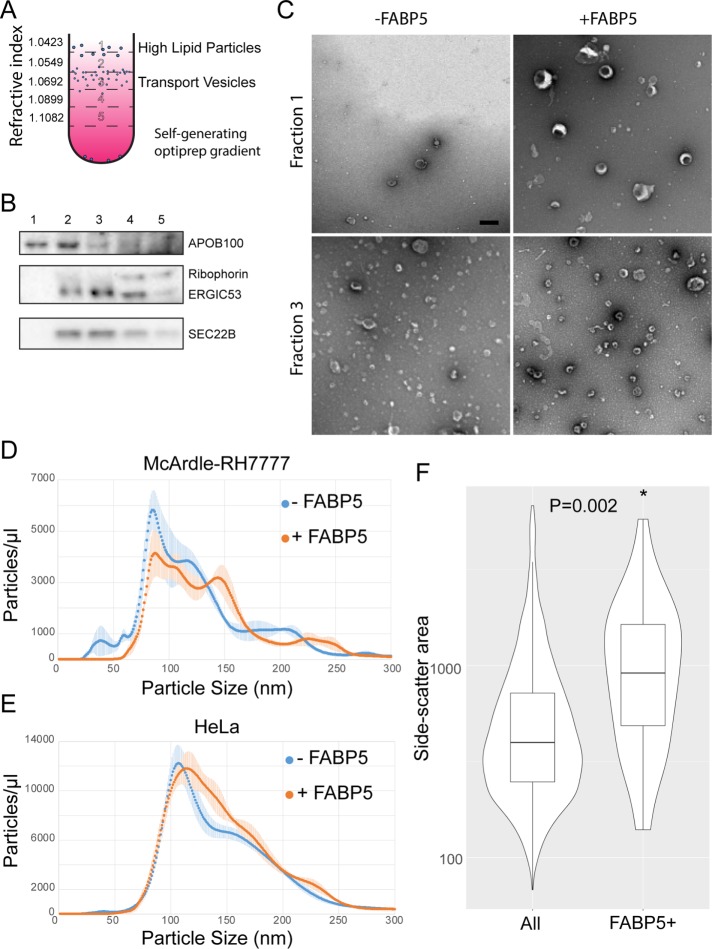
FIGURE 3:
FABP5 leads to a subpopulation of cargoes of increased size. (A) Optiprep flotation scheme to separate high lipid VLDL particles from small budded cargo vesicles. (B) Immunoblot showing which fractions correspond to which vesicular cargo. (C) Negative-stain EM of fractions 1 and 3 from budding reactions using McArdle donor membranes, GTP, COPII, and FABP5. Scale bar: 200 nm. (D) Concentration of different size particles from budding reactions using McArdle and (E) HeLa donor membranes (n = 3) measured by Brownian motion with a NanoSight particle analyzer. (F) Relative size of FABP5-positive particles represented by side scatter of particles in flow cytometry with a LSR Fortessa with output from budding reactions monitored using fluorescently labeled FABP5 and SAR1.
To visualize and measure the particles contained in fractions 1 and 3, we used negative staining and transmission electron microscopy (TEM). Images of the ERGIC53-containing fractions displayed characteristic cup-shaped vesicular particles ∼100 nm in diameter, consistent with COPII budded vesicles (Figure 3C, bottom). These appeared unchanged by the addition of FABP5. In the APOB100-containing fractions, there were vesicular particles ∼150 nm in diameter only in the FABP5-containing reaction (Figure 3C, top). These data suggest that the FABP5-enhanced APOB100-containing cargoes are vesicular and larger than traditional COPII vesicles.
Whereas TEM provides a snapshot on the size and appearance of a fairly small number of vesicles, it does not provide much information on a population level. To test what portion of vesicles were large within the overall population of budded membranes, we utilized a NanoSight NS300 particle analyzer, which employs a measure of Brownian motion to compute nanoparticle size distributions. We found that FABP5-containing budding reactions had an extra population peak of ∼150 nm, consistent with the TEM data, and that this peak accounted for between 5 and 10% of the overall vesicle population (Figure 3D).
We wanted to assess whether this larger size vesicle peak was due to the presence of FABP5 alone or required the large APOB100 cargo. We therefore performed NanoSight analysis on a budding reaction where HeLa cells, which do not secrete lipoproteins (or collagen), were used as a source of donor membrane. In this reaction, we did not see a distinct peak at 150 nm, but we did see a shoulder of increased size in the curve (Figure 3E), suggesting that FABP5 may increase the range of COPII sizes, but that cargo may also be an important vesicle size determinant.
Given its role as a fatty-acid carrier protein, it seemed reasonable to consider that FABP5 may interact directly with membranes to influence the COPII vesicle budding reaction. To test this, we fluorescently labeled FABP5 and SAR1B using amine-reactive dyes NHS-Alexa 680 and NHS-rhodamine, respectively. We performed a budding reaction, and then used a LSRFortessa cell analyzer to select SAR1-positive membrane particles. Of those particles, we determined which were FABP5 positive. The number of FAPB5-positive SAR1-negative particles was negligible. We found that although only a small percentage of particles were FABP5 positive (<2%), those particles were larger on average than FABP5-negative particles (Figure 3F). This suggests that FABP5 directly interacts with or helps to form large COPII vesicles. The low percentage of FABP5-positive particles among the population of large particles, as observed by NanoSight, may reflect a transient interaction of this peripheral protein with membranes.
FABP5 alters the kinetics of SAR1 GTPase activity
We next set out to determine the mechanism by which FABP5 interacts with COPII to increase vesicle size. Because the influence of SEC31 on GTP hydrolysis by SAR1 has been implicated in the COPII packaging of large cargoes (Venditti et al., 2012; Ma and Goldberg, 2016; Raote et al., 2017), we hypothesized that FABP5 may interact with the GTPase stimulatory active fragment of SEC31A (Fromme et al., 2007). We performed immunoprecipitation (IP) reactions with a StrepII-tagged fragment of SEC31A and found that regardless of the presence of SAR1, FABP5 coprecipitated with a large or small fragment of SEC31 (Figure 4A, lanes 1–4 and lanes 7 and 8), suggesting a direct interaction between the two proteins.
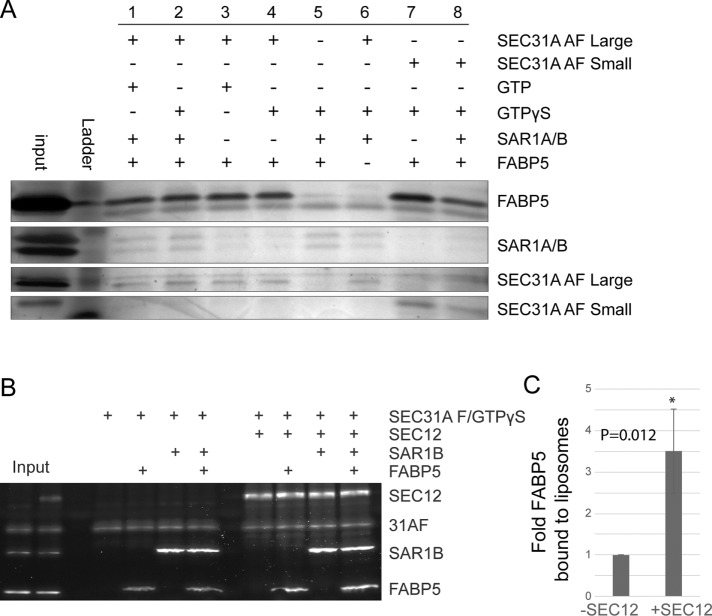
FIGURE 4:
FABP5 interacts with the active fragment of SEC31A and SEC12. (A) Silver stain of SDS–PAGE gel from strep II–tag pull-down reaction of strep-tagged SEC31 active fragments with COPII components and FABP5. (B) SYPRO Ruby stain of FABP5 and COPII components recruited to floated synthetic liposomes (n = 3). (C) Quantification of FABP5 recruited to liposome in the absence or presence of SEC12.
We considered the possibility that FABP5 interacting with SEC31 may influence the recruitment of other COPII components. The recruitment of COPII components to membranes was assessed using a liposome flotation assay with or without FABP5. Purified recombinant human COPII proteins were incubated with synthetic liposomes and the nonhydrolyzable GTP analogue GTPγS at 37°C for 20 min and applied to the bottom of a sucrose density gradient. After a brief high-speed centrifugation step, floated liposomes were isolated from the top of the sucrose gradient and the COPII proteins present in the liposome fraction were analyzed by SDS–PAGE and SYPRO Ruby staining. We did not observe a consistent difference in recruitment of COPII components in the presence of FABP5 (unpublished data); however, we did find that in the presence a soluble large fragment of SEC12, the GEF for SAR1, FABP5 was ∼3.5-fold more efficiently recruited to liposomes (Figure 4, B and C, and Supplemental Figure 1A).
Considering that FABP5 is recruited to membrane by the GEF of SAR1 and interacts with an activator of SAR1’s GAP activity, we considered that FABP5 might have an effect of the GTPase cycle of SAR1. To test this we used a tryptophan fluorescence-based assay performed with purified proteins (Antonny et al., 2001; Futai et al., 2004; Fromme et al., 2007). In this assay, the nucleotide-bound state of Sar1 is monitored by relative fluorescence measurements. The intrinsic tryptophan fluorescence of SAR1-GTP is significantly higher than that of SAR1-GDP.
We first wanted to test whether FABP5 altered the kinetics of SAR1 GTP loading. To this end, we measured the fluorescence of reactions containing purified soluble SAR1 (missing the amphipathic helix), GTP, and combinations of FABP5 or SEC12. We found, as expected, that SEC12 significantly increased the efficiency of GTP loading into SAR1 (Figure 5A). We further found that FABP5 also increased GTP loading efficiency, although not to the same extent as SEC12. When both FABP5 and SEC12 were present, the GTP loading efficiency increased further than either one alone.
FIGURE 5:
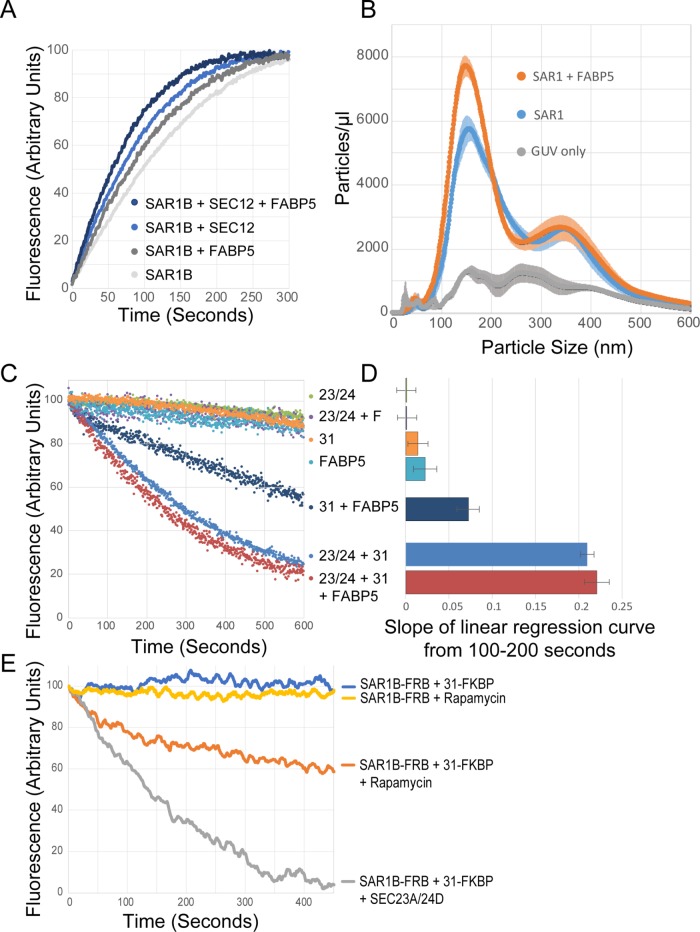
FABP5 alters the GTPase cycle of SAR1. (A) Tryptophan fluorescence assay measuring loading of GTP into SAR1 in the presence of FABP5 and PREP. (B) Concentration of particles at specific sizes generated from GUVs incubated with SAR1 and FABP5. (C) Tryptophan fluorescence assay measuring GTPase activity of SAR1 in the presence of indicated COPII components. (D) Quantification of the slope of linear regression curves fitted by the least-squares method for GTPase activity from 100 to 200 s (n = 3). (E) Tryptophan fluorescence assay measuring GTPase activity of SAR1 in the presence of indicated tagged COPII components in rapamycin-induced FRB/FKBP12 binding reaction.
Because FABP5 increases the rate of GTP loading of SAR1, we decided to test whether FABP5 could increase SAR1 activity with membrane. We developed a simple assay with giant unilamellar vesicles (GUVs) and the NanoSight particle analyzer. We found that incubation of GUVs with SAR1 and GTP alone was sufficient to produce particles detectable by NanoSight (Figure 5B). We utilized this to test whether FABP5 alone could alter SAR1’s ability to generate vesicles. We found that the addition of FABP5 to this reaction significantly increased the number of detected particles, although it did not appear to alter the size of those particles, as observed when using cell membranes instead of GUVs. These data suggest that FABP5 directly enhances SAR1 activity; however, that enhancement does not, in and of itself, alter the size of budded vesicles.
We next wanted to test whether FABP5 altered the other half of the SAR1 GTPase cycle—the GTPase activity itself. Again, we used the GTPase fluorescence assay. We found that FABP5 alone was insufficient to stimulate GTP hydrolysis, and we did not see a change in the rate of GTP hydrolysis when FABP5 was added to the complete COPII reaction (Figure 5, C and D). However, the combination of FABP5 and the active fragment of Sec31 stimulated hydrolysis, albeit at a lower level than the SEC23A/SEC24D heterodimer combined with the SEC31A active fragment (Figure 5D), suggesting that FABP5 can indeed have a role in SAR1 GTPase activity, but that that role is overshadowed by the more prominent role of SEC23A in in vitro conditions. We hypothesized that the role of FABP5 in SAR1 GTPase activity may be due to bringing the active domain of SEC31 into proximity with SAR1. To address the possibility that this may be sufficient to induce GTP hydrolysis, we utilized a FRB/FKBP/rapamycin approach to induce dimerization between SAR1 and SEC31, and found partial GTPase activation quite similar to that seen with FABP5 (Figure 5E; Chen et al., 1995; Inobe and Nukina, 2016).
The effects of FABP5 on COPII activity observed above do not point to a role unique in lipoprotein trafficking. Therefore, we considered that, rather than having a role specific to lipoprotein trafficking as we initially assumed, FABP5 may have a more generalized role in protein trafficking. To test this, we used HeLa cells as a source of donor membrane for a budding reaction. HeLa membranes work well for budding, even in the absence of cytosol, and are not known to be capable of secreting large cargo molecules, thus making them a good model for generalized secretion. We performed a budding reaction using HeLa donor membrane and found that FABP5 enhanced COPII-dependent budding of ERGIC53 (Figure 6A). FABP5 also altered the budding of SEC22B, although not in the same manner as ERGIC53, and less consistently. These data suggest that FABP5 does indeed act on cargoes other than lipoproteins, and acts in a cargo-specific manner.
FIGURE 6:
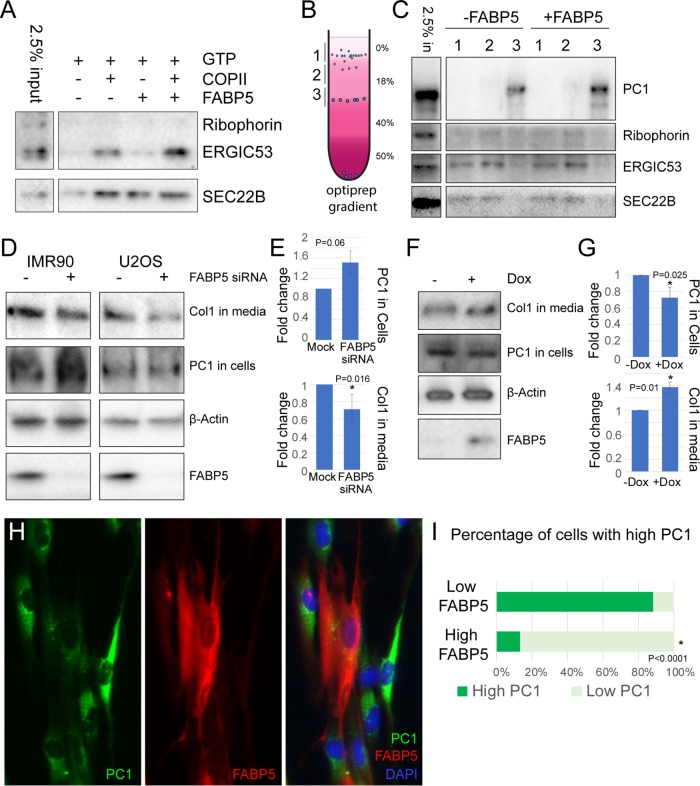
FABP5 affects collagen budding and secretion. (A) Immunoblot of response of traditional COPII cargoes ERGIC53 and SEC22B to FABP5 added to budding reactions with HeLa donor membranes. Ribophorin used as a marker of ER contamination. (B) Schematic of OptiPrep gradient flotation of budding reaction using IMR90 cell donor membrane. (C) Immunoblot of cargoes from budding reaction, with ribophorin as a control for ER contamination. (D) Immunoblot of type I collagen in media and procollagen in cells after 30 min of ascorbic acid treatment in IMR90 and U2OS cells after siRNA knockdown of FABP5. Actin was used as loading control. (E) Quantification by Student’s two-tailed t test; p values indicated (n = 3). (F) Immunoblot of type I collagen in media and procollagen in cells after 30 min of ascorbic acid treatment in IMR90 after overnight doxycycline treatment to overexpress FABP5. Actin was used as loading control. (G) Quantification by Student’s two-tailed t test; p values indicated (n = 4). (H) Immunofluorescently labeled IMR90 cells. Nuclei are stained with DAPI. (I) Quantification of collagen and FABP5 levels observed in 100 cells.
FABP5 stimulates collagen secretion
Given an effect of FABP5 on other cargo, we examined collagen packaging, which we previously showed to be stimulated by ubiquitylation of the SEC31 subunit (Jin et al., 2012; Gorur et al., 2017). We performed a budding reaction using membranes from IMR90 human lung fibroblasts, which secrete high levels of collagen. The budding reaction was performed in the presence of collagenase to minimize a background of non–membrane-encased collagen likely arising from membranes that rupture during the preparation procedures. We found FABP5 stimulated the production of a population of procollagen-containing vesicles that sedimented at a higher equilibrium density position of an OptiPrep gradient (Figure 6, C and D).
Although the data from the cell-free budding reaction suggest that FABP5 could alter collagen trafficking, we sought a test of this protein in cells by small interfering RNA (siRNA) to knockdown of FABP5 expression in IMR90, as well as U2OS, a human osteosarcoma. We used immunoblotting to analyze the levels of processed collagen secreted into the media, as well as of intracellular procollagen, which should accumulate if secretion is diminished. We found that, upon knockdown of FABP5, processed collagen in the medium decreased, whereas intracellular procollagen increased for both cell lines (Figure 6, D and E). Conversely, when we overexpressed FABP5 through a lentivirus-mediated tetracycline-inducible system in IMR90 cells, the processed collagen in the medium increased, while the intracellular procollagen decreased (Figure 6, F and G).
We used immunofluorescence microscopy to visualize the subcellular localization of procollagen and FABP5 in the inducible IMR90 cells. Although the cytosolic nature of FABP5 made visualization difficult, we were able to observe a low level of collagen remaining in cells that expressed a high level of FABP5 (Figure 6, H and I). This observation is consistent with what we observed in the immunoblots of procollagen in cells (Figure 6, D–G), that collagen is more efficiently secreted from cells in the presence of FABP5.
DISCUSSION
As the primary mediator of ER-to-Golgi trafficking, COPII must contend with myriad shapes, sizes, and quantities of cargo in order to maintain cellular homeostasis. We have identified FABP5 as a novel regulator of that COPII function, especially pertaining to the large cargoes that pose a special challenge to COPII. We have found that FABP5 appears to modulate the GTPase cycle of SAR1 and allows for formation of large COPII vesicles.
A model of FABP5’s role in COPII-mediated trafficking of large cargoes
Our data present three potential areas where COPII, and its ability to respond to diverse cellular needs for secretion of cargoes of different quantities and sizes, may be regulated. One is through the regulation of GTPase activity, as also seen with other proteins like Sedlin and TANGO1 (Venditti et al., 2012; Ma and Goldberg, 2016), where the presence of FABP5 stimulates a low level of SAR1 GTPase activity, even in the absence of SEC23, the SAR1 GAP (Figure 5C).
A second potential area of regulation is through modulation of GDP/GTP exchange, which FABP5 enhances (Figure 5A), and this leads to an increase in SAR1’s ability to bud particles from a GUV (Figure 5B). It has been found previously that SAR1 binds membranes of high curvature (Hanna et al., 2016), therefore, a slight enhancement in the initiation of membrane curvature as may occur if FABP5 is present, may lead to a forward feedback loop and a further increase in SAR1 recruitment.
A third mechanism is through the size of the cargoes themselves. The lack of a size change in particles budded from GUVs, despite a seeming increase in SAR1 activity (Figure 5B), suggests the FABP5-mediated increase in SAR1 activity alone is insufficient to increase vesicle size. A caveat to this observation is that the GUV-derived vesicles appear to be naturally larger than ER-budded vesicles, so if FABP5 is having a more direct effect, it may be masked by the larger size of the GUV vesicles. Furthermore, the vesicles budded from HeLa membranes show an increase in larger particles, but not a distinct peak at 150 nm as is seen in McArdle membranes, further suggesting that the cargoes themselves may play a role in regulating COPII size.
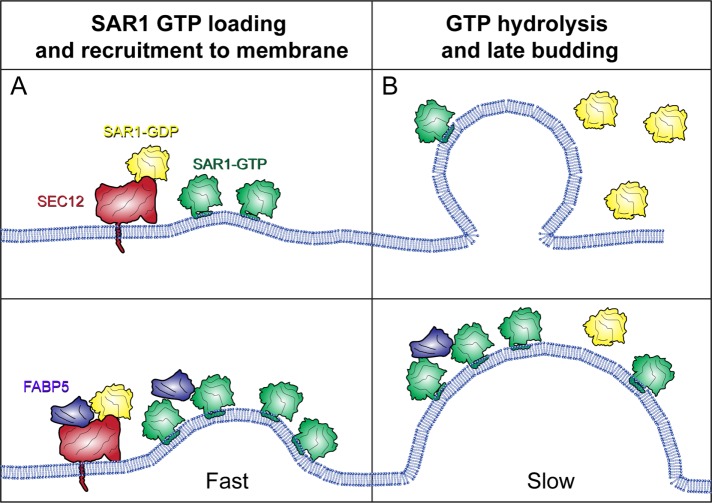
To explain these data, we propose that SEC12 recruits both SAR1 and FABP5 to the membrane. The presence of FABP5 increases the GEF activity of SEC12, leading to more activated SAR1, and therefore more SAR1 at the membrane. Because SAR1 also responds to membrane curvature (Hanna et al., 2016), this may lead to a further increase in the amount of SAR1 bound, and a faster initiation of COPII budding (Figure 7A). For later steps of COPII biogenesis, FABP5 may have the opposite effect, stimulating the GTPase activity of SAR1, but at a slower rate than that achieved by SEC23, therefore acting in a mechanism similar to TANGO1, slowing the kinetics of SAR1 to allow sufficient time for loading of large cargoes into vesicles (Figure 7B).
FIGURE 7:
Model for FABP5 function in COPII-mediated large cargo trafficking. (A) During early stages of budding, FABP5 enhances SEC12’s GEF function, leading to enhanced GTP-loaded SAR1 on the membrane, and therefore high levels of membrane deformation. (B) During later stages of budding, FABP5 and SEC31 may lead to slower kinetics of SAR1 hydrolysis, allowing for cargo loading in a manner similar to TANGO1.
The proposed mechanism would likely have an effect on COPII budding beyond large cargoes. Indeed, we do see an increase in the budding of other cargoes, namely, ERGIC53 and SEC22B, although the effect on these smaller cargoes does not appear to be as pronounced.
The many roles of FABP5
Fatty-acid binding proteins, such as FABP5 (also known as E-FABP or Mal1), are intracellular carriers for fatty acids and lipids and have been shown to play a surprisingly diverse role in multiple cellular processes including important roles in metabolism (Hotamisligil and Bernlohr, 2015), neurogenesis (Matsumata et al., 2016), inflammation (Berger et al., 2012; Gally et al., 2013; Bogdan et al., 2018), and cancer (Levi et al., 2013). FABP5 acts through both binding and transport of fatty acids and other lipids such as retinoic acid (Schug et al., 2007; Bando et al., 2014, 2017), and as an indirect regulator of gene expression through interactions with the PPAR class of nuclear receptors (Berger et al., 2012; Gally et al., 2013; Thumser et al., 2014).
The diverse roles of lipids in cellular processes, may, in part, account for the diversity of roles of proteins that carry them. That fatty-acid binding proteins may play a role in membrane trafficking should not be surprising. FABP5 has recently been found to bind the cytoplasmic domain of calnexin as well (Jung et al., 2017), further suggesting a relationship with the ER.
FABP5 is not the first fatty-acid binding protein to be implicated in lipoprotein trafficking. FABP1 has been proposed to play a role in chylomicron budding, and even that it has a binding partner in SAR1B (Siddiqi and Mansbach, 2012), although the proposed mechanism of action for FABP1 appears unrelated to what we have observed with FABP5.
It is not surprising that FABP5 would have a role in lipoprotein trafficking, as the delivery of fatty acids from the cytoplasm is an integral part of the biogenesis of lipoproteins assembled in the lumen of the ER. Why FABP5 would also have a role in secretion of other large cargoes is less apparent, except insofar as the function of FABP5 relates to the generation of larger vesicles capable of packing unusual cargo complexes.
Many of the roles of FABP5 have functional overlap with its most closely related paralogue, FABP4. In fact, FABP5 knockout (KO) mice do not have reduced levels of APOB-containing lipoprotein in serum (Babaev et al., 2011), as would be expected given the partial effect we observe on lipoprotein and collagen secretion in FABP5 knockout cells. However, FABP4/5 double knockout mice (in an ApoE−/− background) do secrete lower levels of APOB lipoproteins (Boord et al., 2004). This suggests that there may be significant overlap in the function of the two proteins in their role in lipoprotein trafficking as well. The many roles of FABP5 make it difficult to parse what role of the protein is responsible for which function. Our direct biochemical approach has allowed us to focus on one aspect of its function in regulating traffic of lipoprotein particles from the ER.
MATERIALS AND METHODS
Antibodies
Commercially available antibodies used for IP, immunoblotting, and immunofluorescence were as follows: Goat anti–apolipoprotein B (EMD, Hayward, CA; #178467 1:500 for immunoblot); rabbit anti-FABP5 human (Proteintech, Rosemont, IL; #12348-1-AP 1:2000 for immunoblot); rat anti-FABP5 monoclonal antibody (Thermo Fisher Scientific, Waltham, MA; ma5-24029 1:300 for immunofluorescence); rabbit anti–PC1 LF-41 was a gift from L. Fisher (National Institute of Dental and Craniofacial Research, Bethesda, MD [Fisher et al., 1989, 1995; Bernstein et al., 1996]; 1:5000 for immunoblot); rabbit anti-collagen type I (Millipore, Hayward, CA; #AB745 1:300 for immunofluorescence); Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (IgG) (Invitrogen, Carlsbad, CA; #A-21206 1:250 for immunofluorescence); Alexa Fluor 546 goat anti-rat IgG (Invitrogen #A-11081 1:250 for immunofluorescence).
Cell culture, transfection, and drug treatments
Rat liver hepatoma MrArdle-RH7777 and human osteosarcoma U2OS were obtained from the American Type Culture Collection. Human lung fibroblasts IMR-90 were obtained from Coriell Cell Repositories at the National Institute on Aging, Coriell Institute for Medical Research, Camden, NJ. IMR-90 and U2OS were maintained in DMEM plus 10% fetal bovine serum (FBS; GE Healthcare, Chicago, IL). MrArdle-RH7777 were maintained in DMEM plus 20% FBS (GE Healthcare). Cells were kept in a 37°C incubator with 5% CO2. Transfection of DNA constructs into IMR-90 U2OS cells was performed using Lipofectamine 2000 as detailed in the manual provided by Invitrogen. Oleic acid treatment was used at 0.75 mM oleic acid complexed to fatty-acid–free bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, MO). Ascorbate treatment was used at 0.25 mM ascorbic acid (Sigma) and 1 mM ascorbic-2-phosphate (Sigma-Aldrich). This concentration was in addition to the amount of ascorbic acid present in FBS (0.08 mM), as supplied by the manufacturer. Doxycycline (Sigma-Aldrich) was used at 1 µg/ml. BFA (Sigma-Aldrich) was used at 10 µg/ml.
Lentivirus production and adipocyte transduction
Rat FABP5 was subcloned into pLenti-puro (Addgene Cambridge, MA; plasmid #39481). The plasmid containing FABP5 was transfected with Lipofectamine 2000 (Invitrogen), following the manufacturer’s recommendations, into HEK293T cells at 50% confluence the day of transfection along with lentiviral packaging plasmids pVSVg (3.5 µg) and psPAX2 (6.5 µg; Addgene). Transfection was performed using one 10-cm dish. After a 24-h transfection, the medium was changed, and after an additional 24 h, the medium was removed and filtered through a 0.45-µm low-protein binding membrane (VWR International, Radnor, PA). McArdle-RH7777 or IMR-90 was then transduced with the virus with 8 µg/ml polybrene (Sigma-Aldrich). After 24 h, medium was replaced with fresh medium, and after an additional 24 h, 2 µg/ml puromycin (Sigma-Aldrich) was added to select transduced cells.
siRNA transfection
We obtained three distinct siRNAs targeting FABP5 Hs_FABP5_8 FlexiTube siRNA (Qiagen, Hilden, Germany; SI04210941), Hs_FABP5_9 FlexiTube siRNA (Qiagen SI04210948), and Hs_FABP5_5 FlexiTube siRNA (Qiagen SI03145835). IMR90 and U2OS cells were transfected with these siRNAs at a final concentration of 20 nM using the Lipofectamine RNAiMAX reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Owing to the high stability of FABP5 protein, after 48 h the process was repeated, and after a further 48 h cells were harvested for immunoblot.
HSW preparation and purification for tandem mass spectrometry
Plates (3 × 15 cm) of McArdle-RH7777 cells were cultured to 95% confluence, incubated with 10 µg/ml BFA and 0.75 mM oleic acid for 1 h. Cells were treated with 20 µg/ml digitonin (5 min on ice) in B88 (20 mM HEPES, pH 7.2, 250 mM sorbitol, 150 mM potassium acetate, and 5 mM magnesium acetate) and washed with 10 ml 1 M KOAc. The HSW was collected as a supernatant fraction after centrifugation at 300 × g for 5 min. It was then desalted through dialysis in HKM overnight at 4°C at a final protein concentration of 0.5 mg/ml.
For further purification of the active HSW fraction, HSW was, where indicated, heated in a boiling water bath for 10 min. Precipitates were centrifuged at 20,000 × g for 10 min and the supernatant was dialyzed in MonoQ Buffer A (20 mM Tris-HCl, pH 7.5, 1 mM MgOAc,10 mM dithiothreitol) overnight at 4°C and then loaded onto an anion exchange column (Mono Q 10/100 GL) on an AKTA FPLC system (GE Healthcare). Flowthrough (unbound) fraction was collected and stored at −80°C at a final protein concentration of 50 ng/µl.
For proteinase K treatment, HSW was incubated with 10 μg/ml proteinase K (Sigma-Aldrich) and stored on ice for 30 min. The reactions were stopped by heating for 10 min in a boiling water bath.
For tandem mass spectrometry analysis, 2 μg of heated monoQ unbound fraction of HSW were used. In-solution trypsin digestion, desalting, and liquid chromatography with tandem mass spectrometry were performed by the QB3/Chemistry Mass Spectrometry Facility at the University of California, Berkeley (UC Berkeley) (Strader et al., 2006; Cociorva et al., 2007; Hervey et al., 2007; Rebecchi et al., 2011).
Vesicle budding reaction
Vesicle budding reactions were performed as previously described (Kim et al., 2005; Gorur et al., 2017; Yuan et al., 2017) with the following modifications: McArdle donor ER membrane was prepared by permeabilizing cells (95% confluent in 3 × 10-cm dishes) that had been incubated with 10 µg/ml BFA and 0.75 mM oleic acid for 1 h. Cells were treated with 20 µg/ml digitonin (5 min on ice) in B88 and washed with 1 M KOAc in B88 then B88 and resuspended in B88-0 (20 mM HEPES, pH 7.2, 250 mM sorbitol, and 150 mM potassium acetate) to a final concentration of OD600 2–6. Each 100-µl reaction contained an ATP regeneration system (1 mM ATP, 40 mM creatine phosphate, and 0.2 mg/ml creatine phosphokinase), 3 mM GTP, purified human COPII proteins (0.2 µg SAR1B for McArdle-RH7777, 1 µg for other cell types, 1 µg SEC23A/24D, and 1 µg SEC13/31A), 1 µg FABP5 (or 10 µg of HSW protein) as indicated, and a final concentration of 0.5 OD600/ml donor ER membrane in B88-0. IMR90 membranes were mixed with 0.1 U/µl collagenase (Sigma-Aldrich). Donor membranes were sedimented by centrifugation (7000 × g for IMR90, 13,000 × g for other cell types) at 4°C. For McArlde-RH7777 donor membrane, the supernatant was then mixed with 60% OptiPrep (Sigma-Aldrich) gently until homogeneous, for a final concentration of 25% OptiPrep. This was overlaid with 18% OptiPrep and B88. The OptiPrep gradient was centrifuged at 250,000 × g for 90 min at 4°C (Beckman TLS-55 with 11 × 34-mm tubes). For Figure 3, A–C, the supernatant was mixed with OptiPrep to a final concentration of 20%. Samples were centrifuged at 300,000 × g in a TLA100 rotor for 6 h. Fractions were taken from the top for analysis. For HeLa donor membranes, the supernatant was centrifuged at 100,000 × g for 30 min at 4°C. For IMR90 membranes, supernatant was mixed with OptiPrep to a final concentration of 50% OptiPrep. This was overlaid with 200 μl 40% OptiPrep, 200 μl 18% OptiPrep, and 50 μl B88. The OptiPrep gradient was centrifuged at 250,000 × g for 90 min at 4°C (Beckman TLS-55 with 11 × 34-mm tubes). Aliquots (100 μl) were taken from the top for immunoblot.
Generation of CRISPR/Cas9 KO cell lines
McArdle-RH7777 cells were transfected with a pX330 vector-derived plasmid (Ran et al., 2013) containing the targeting sequence from FABP5 (CACCGGTGGAAAGCCACGGGTTTG), and a PGK promotor-driven Venus construct (reconstructed by Liangqi Xie from the Robert Tjian laboratory at UC Berkeley). After a 24-h transfection, fluorescence-activated cell sorting was performed to inoculate single transfected cells in each well of 96-well plates. After 2 wk, single colonies were expanded and validated by immunoblot and DNA sequencing of the targeted area. Validated positive colonies were employed for the experiments.
Immunoblotting
Standard immunoblotting procedures were followed. In brief, samples were resolved on 4–20% polyacrylamide gels (15-well, Invitrogen; 26-well, Bio-Rad Laboratories), and transferred to polyvinylidene difluoride (PVDF; EMD Millipore) at constant 0.5 A for 4 h, or 0.15 A for 16 h for blots containing collagen. The PVDF membrane was incubated with antibodies (primary overnight at 4°C h and secondary for 1 h at RT), and bound antibodies were visualized by the enhanced chemiluminescence method (Thermo Fisher Scientific) on a ChemiDoc Imaging System (Bio-Rad Laboratories, Hercules, CA) with ImageLab software v4.0 (Bio-Rad Laboratories).
Protein purification
Human SAR1, rat FABP5, the cytosolic domain of SEC12, and SEC31 active fragment proteins were overexpressed in E. coli and purified as cleaved GST fusions, as described for hamster Sar1 purifications (Kim et al., 2005). In brief, a bacterial lysate was first centrifuged at 43,000 × g for 15 min, and then the supernatant fraction was further centrifuged at 185,000 × g for 1 h. The supernatant was incubated with prewashed glutathione-agarose (1 ml slurry/l bacteria; Thermo Fisher Scientific) for 1 h at 4°C. Agarose was washed with wash buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween, 5 mM MgCl2, and 100 µM GDP), and protein was eluted by cleaving with 20 U/ml thrombin (Roche) in TCB (50 mM Tris, pH 8, 250 mM KoAc, 5 mM CaCl2, 5 mM MgCl2, and 100 µM GDP). Purified fractions were pooled and stored at −80°C at final protein concentrations of 2 µg/µl (for SAR1) and 1 µg/µl (for FABP5, SEC12, and SEC31 fragments).
Human SEC13/31A and SEC23A/24D were purified from lysates of baculovirus-infected insect cells, as described previously (Kim et al., 2005). In brief, insect cell lysates were centrifuged at 185,000 × g for 1 h, and 30% ammonium sulfate was added to the supernatant fraction at 4°C. The precipitant was collected by centrifugation at 30,000 × g for 30 min and solubilized in no-salt buffer (20 mM HEPES, pH 8, 10% glycerol, 250 mM sorbitol, 0.1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid [EGTA], 5 mM β-mercaptoethanol, and 10 mM imidazole). The solubilized 30% ammonium sulfate precipitant was cleared at 30,000 × g for 20 min, and the supernatant was incubated with prewashed Ni-NTA resin (1.25 ml slurry/l insect cells; Thermo Fisher Scientific) for 1 h at 4°C. Ni-NTA was washed with 20 mM HEPES, pH 8, 10% glycerol, 250 mm sorbitol, 500 mM KoAc, 0.1 mM EGTA, 5 mM β-mercaptoethanol, and 50 mM imidazole and eluted with 250 mM imidazole. Ni-NTA–eluted SEC13/31A protein was further purified using an anion exchange column (MonoQ) on an AKTA FPLC system (GE Healthcare). Purified fractions were pooled and stored at −80°C at final protein concentrations of 500 ng/µl (for SEC23/24) and 250 ng/µl (for full-length SEC31).
Flow cytometry
Vesicle budding reactions were scaled up 2× for flow cytometry analyses. Budding reactions were performed as described above, with FABP5 and SAR1 labeled with Alexa Fluor 680 NHS ester (Invitrogen A37567) and NHS-rhodamine (Invitrogen 46406) according to manufacturer’s protocol. Aliquots (40 μl) were taken from the top of an OptiPrep gradient. Particles (100,000) were collected for each sample. FSC-A and FSC-H were used to gate for single particles (singlets), which were used for further analysis. Gating of each fluorescent channel was determined by comparing a control sample without any fluorescence labeling and a control that was labeled in a single channel. Data were collected on a BD LSR Fortessa (Becton Dickinson, Franklin Lakes, NJ) and analyzed by FlowJo software and Flowing software. Instruments and software were provided by the LKS flow core facility at UC-Berkeley.
Nanoparticle tracking analysis
Sizes of vesicles budded in vitro were estimated using a NanoSight NS300 instrument equipped with a 405-nm laser (Malvern Instruments, Malvern, United Kingdom). Particles were analyzed in the scatter mode without a filter. Silica 100-nm microspheres (Polysciences, Warrington, PA) were analyzed to check instrument performance and determine the viscosity coefficient of B88. Aliquots (20 μl) of vesicles were collected from the top of the flotation gradient as described in the vesicle budding reaction section and diluted 50× with 980 µl filtered B88 (0.02 µm; Whatman). The samples were automatically introduced into the sample chamber at a constant flow rate of 50 (arbitrary manufacturer unit, ∼10 µl/min) during five repeats of 60-s captures at camera level 11 in scatter mode with Nanosight NTA 3.1 software (Malvern Instruments). The particle size was estimated with detection threshold 5 using the Nanosight NTA 3.1 software, after which “experiment summary” and “particle data” were exported. Particle numbers in each size category were calculated from the particle data, in which “true” particles with track length >3 were pooled, binned, and counted with Excel (Microsoft).
Electron microscopy of budded vesicles
For morphological analysis of the budded vesicles, the top 40 µl of the OptiPrep gradient from a 2× scaled budding reaction (as described above) was fixed with 2% paraformaldehyde (PFA) and 0.2% glutaraldehyde in B88 buffer for 15 min at 4°C. Samples were spread onto a nickel grid coated with Formvar (Plano, Wetzlar, Germany). Excess liquid was blotted off with filter paper, the grid was stained with 1% aqueous uranyl acetate, and excess staining solution was blotted off. Dried specimens were examined on a ZEISS EM 900 transmission electron microscope.
Coimmunoprecipitation assay
Purified protein (1 µg SAR1A, 1 µg SAR1B, 1 µg FABP5, 1 µg active fraction SEC31A, and 3 mM GTP or GTPγS) was added to 100 µl HKM buffer (20 mM HEPES, pH 7.2, 150 mM KOAc, 1 mM Mg[OAc]2) and reactions were incubated at 37°C for 30 min. An aliquot (20 µl) of equilibrated Strep-Tactin Sepharose slurry (IBA Lifesciences, Goettingen, Germany; 2-1201-010) was added to each reaction, and the protein-bead mixture was incubated for 1 h at 4°C with gentle mixing. Beads were washed five times in HKM by centrifugation at 3000 × g for 5 min at 4°C. Protein was eluted by incubation with 2.5 mM desthiobiotin (Sigma D1411) in HKM, and then analyzed by SDS–PAGE followed by silver staining with a Pierce Silver Stain Kit (Thermo Fisher 24612) according to the manufacturer’s protocol.
GTPase activity assay
The tryptophan fluorescence GTPase activity assay was performed at 37°C as described (Antonny et al., 2001; Futai et al., 2004; Fromme et al., 2007), using a stirred-cell cuvette. In HKM buffer, we added soluble SAR1B to a final concentration of 1.33 μM and where indicated SEC31 active fragment (2 μM; Fromme et al., 2007), the cytosolic domain of SEC12 (2 μM), or FABP5 (2 μM). Five minutes later, GTP was added to 30 μM. For Figure 5C, SEC23A-SEC24D complex was added to 250 nM after GTP exchange was complete (∼10 min) and FABP5 was added simultaneously where indicated.
Liposome binding assay
The liposome binding assay was performed as described for yeast COPII proteins (Matsuoka et al., 1998; Kim et al., 2005) using 10% cholesterol major-minor mix liposomes with the addition of DGS-NTA(Ni) (Avanti 790404) for binding of his-tagged SEC12. Following a 20 min incubation at 37°C, the protein–liposome mixture was separated by flotation through a sucrose density step gradient (in HKM buffer) achieved by centrifugation at 391,000 × g for 25 min at 22°C.
GUV budding assay
GUVs were prepared by electroformation as previously described using major-minor mix (Matsuoka et al., 1998) synthetic liposomes containing 10% cholesterol (Angelova and Dimitrov, 1986; Bacia et al., 2011). Briefly, lipids were combined in chloroform:methanol (2:1, volume ratio) at 10 mg/ml total concentration and dried as a thin layer on indium-tin-oxide–coated glass slides. The slides were assembled to form a chamber by using 3-mm-thick silicon spacers and holding the slides together with office clips. The chamber was filled with sucrose solution (540 mOsmol/kg) and electroformation was performed over 2 h using a 1.4 V, 10 Hz sinusoidal voltage. GUVs (of 5 μl) were added to 45 μl B88 containing 5 μg SAR1 and 2 μg FABP5 as indicated. Reactions were incubated for 30 min at 37°C and diluted 1:100 for particle analysis on NanoSight NS300 as described above. All lipids were obtained from Avanti Polar Lipids, Alabaster, AL.
Immunofluorescence
Cells growing on glass coverslips were fixed in 4% PFA for 20 min at RT, washed five times with phosphate-buffered saline (PBS), and incubated with permeabilization buffer (PBS containing 0.1% Triton X-100 and 0.2 M glycine) at RT for 15 min. Cells were incubated with blocking buffer (0.5% BSA in PBS) for 30 min at room temperature followed by incubation for 1 h each at RT with primary antibody and then secondary antibody. Antibody incubations were followed by five washes with PBS. Coverslips were mounted in ProLong-Gold antifade mountant with DAPI (Thermo Fisher Scientific) overnight, before imaging. Images were acquired using Zen 2010 software on an LSM 710 confocal microscope system (ZEISS, Oberkochen, Germany). The objectives used were Plan-Apochromat 100×, 1.4 NA.
Supplementary Material
Acknowledgments
We thank the staff at the University of California, Berkeley, shared facilities, including Alison Killilea (Cell Culture Facility) and Kartoosh Heydari (Flow Cytometry Facility). We thank Jeremy Thorner for providing the fluorometer and Shawn Shirazi for 3D printing that was essential for the setup. R.S. is supported as an Investigator of the Howard Hughes Medical Institute and the University of California, Berkeley, Miller Institute of Science. D.M. was supported in part by National Institutes of Health grant #11287155.
Abbreviations used:
- BSA
bovine serum albumin
- CMRD
chylomicron retention disease/Anderson’s disease
- COPII
coat protein complex II
- ER
endoplasmic reticulum
- ERES
ER exit site
- ERGIC
ER–Golgi intermediate compartment
- GAP
GTPase-activating protein
- GEF
guanine exchange factor
- GUV
giant unilamellar vesicles
- HSW
high-salt wash
- LDL
low-density lipoprotein
- TEM
transmission electron microscopy
- VLDL
very low-density lipoprotein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-09-0548) on November 28, 2018.
REFERENCES
- Angelova MI, Dimitrov DS. (1986). Liposome electroformation. Faraday Discuss Chem Soc , 303–311. [Google Scholar]
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. (2001). Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol , 531–537. [DOI] [PubMed] [Google Scholar]
- Babaev VR, Runner RP, Fan D, Ding L, Zhang Y, Tao H, Erbay E, Görgün CZ, Fazio S, Hotamisligil GS, et al. (2011). Macrophage Mal1 deficiency suppresses atherosclerosis in low-density lipoprotein receptor–null mice by activating peroxisome proliferator-activated receptor-γ–regulated genes. Arterioscler Thromb Vasc Biol , 1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacia K, Futai E, Prinz S, Meister A, Daum S, Glatte D, Briggs JAG, Schekman R. (2011). Multibudded tubules formed by COPII on artificial liposomes. Sci Rep , 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando Y, Yamamoto M, Sakiyama K, Inoue K, Takizawa S, Owada Y, Iseki S, Kondo H, Amano O. (2014). Expression of epidermal fatty acid binding protein (E-FABP) in septoclasts in the growth plate cartilage of mice. J Mol Histol , 507–518. [DOI] [PubMed] [Google Scholar]
- Bando Y, Yamamoto M, Sakiyama K, Sakashita H, Taira F, Miyake G, Iseki S, Owada Y, Amano O. (2017). Retinoic acid regulates cell-shape and -death of E-FABP (FABP5)-immunoreactive septoclasts in the growth plate cartilage of mice. Histochem Cell Biol , 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. (1993). SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature , 347–349. [DOI] [PubMed] [Google Scholar]
- Berger WT, Ralph BP, Kaczocha M, Sun J, Balius TE, Rizzo RC, Haj-Dahmane S, Ojima I, Deutsch DG. (2012). Targeting fatty acid binding protein (FABP) anandamide transporters—A novel strategy for development of anti-inflammatory and anti-nociceptive drugs. PLoS One , e50968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein EF, Chen YQ, Kopp JB, Fisher L, Brown DB, Hahn PJ, Robey FA, Lakkakorpi J, Uitto J. (1996). Long-term sun exposure alters the collagen of the papillary dermis. Comparison of sun-protected and photoaged skin by northern analysis, immunohistochemical staining, and confocal laser scanning microscopy. J Am Acad Dermatol , 209–218. [DOI] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J. (2007). Insights into COPII coat nucleation from the structure of Sec23-Sar1 complexed with the active fragment of Sec31. Dev Cell , 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi P, Fermo E, Vercellati C, Boschetti C, Barcellini W, Iurlo A, Marcello AP, Righetti PG, Zanella A. (2009). Congenital dyserythropoietic anemia type II (CDAII) is caused by mutations in the SEC23B gene. Hum Mutat , 1292–1298. [DOI] [PubMed] [Google Scholar]
- Bogdan D, Falcone J, Kanjiya MP, Park SH, Carbonetti G, Studholme K, Gomez M, Lu Y, Elmes MW, Smietalo N, et al. (2018). Fatty acid binding protein 5 controls microsomal prostaglandin E synthase 1 (mPGES-1) induction during inflammation. J Biol Chem , 5295–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfanti L, Mironov AA, Martínez-Menárguez JA, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze HJ, Luini A. (1998). Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell , 993–1003. [DOI] [PubMed] [Google Scholar]
- Boord JB, Maeda K, Makowski L, Babaev VR, Fazio S, Linton MF, Hotamisligil GS. (2004). Combined adipocyte-macrophage fatty acid–binding protein deficiency improves metabolism, atherosclerosis, and survival in apolipoprotein E–deficient mice. Circulation , 1492–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyadjiev SA, Fromme JC, Ben J, Chong SS, Nauta C, Hur DJ, Zhang G, Hamamoto S, Schekman R, Ravazzola M, et al. (2006). Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet , 1192–1197. [DOI] [PubMed] [Google Scholar]
- Budoff M. (2016). Triglycerides and triglyceride-rich lipoproteins in the causal pathway of cardiovascular disease. Am J Cardiol , 138–145. [DOI] [PubMed] [Google Scholar]
- Canty EG, Kadler KE. (2005). Procollagen trafficking, processing and fibrillogenesis. J Cell Sci , 1341–1353. [DOI] [PubMed] [Google Scholar]
- Charcosset M, Sassolas A, Peretti N, Roy CC, Deslandres C, Sinnett D, Levy E, Lachaux A. (2008). Anderson or chylomicron retention disease: molecular impact of five mutations in the SAR1B gene on the structure and the functionality of Sar1b protein. Mol Genet Metab , 74–84. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL. (1995). Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA , 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociorva D, L Tabb D, Yates JR. (2007). Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinf, Chapter 13:Unit 13.4. [DOI] [PubMed] [Google Scholar]
- Fisher LW, Lindner W, Young MF, Termine JD. (1989). Synthetic peptide antisera: their production and use in the cloning of matrix proteins. Connect Tissue Res , 43–50. [DOI] [PubMed] [Google Scholar]
- Fisher LW, Stubbs JT, Young MF. (1995). Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop Scand Suppl , 61–65. [PubMed] [Google Scholar]
- Fromme JC, Ravazzola M, Hamamoto S, Al-Balwi M, Eyaid W, Boyadjiev SA, Cosson P, Schekman R, Orci L. (2007). The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell , 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai E, Hamamoto S, Orci L, Schekman R. (2004). GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J , 4286–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally F, Kosmider B, Weaver MR, Pate KM, Hartshorn KL, Oberley-Deegan RE. (2013). FABP5 deficiency enhances susceptibility to H1N1 influenza A virus-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol , L64–L72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges A, Bonneau J, Bonnefont-Rousselot D, Champigneulle J, Rabès JP, Abifadel M, Aparicio T, Guenedet JC, Bruckert E, Boileau C, et al. (2011). Molecular analysis and intestinal expression of SAR1 genes and proteins in Anderson’s disease (chylomicron retention disease). Orphanet J Rare Dis , 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorur A, Yuan L, Kenny SJ, Baba S, Xu K, Schekman R. (2017). COPII-coated membranes function as transport carriers of intracellular procollagen I. J Cell Biol , 1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarova V, Brodsky JL, Fisher EA. (2003). Apolipoprotein B100 exit from the endoplasmic reticulum (ER) is COPII-dependent, and its lipidation to very low density lipoprotein occurs post-ER. J Biol Chem , 48051–48058. [DOI] [PubMed] [Google Scholar]
- Hanna MG, Mela I, Wang L, Henderson RM, Chapman ER, Edwardson JM, Audhya A. (2016). Sar1 GTPase activity is regulated by membrane curvature. J Biol Chem , 1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron M. (2012). Deaths: leading causes for 2008. Natl Vital Stat Rep , 1–94. [PubMed] [Google Scholar]
- Hervey WJ, Strader MB, Hurst GB. (2007). Comparison of digestion protocols for microgram quantities of enriched protein samples. J Proteome Res , 3054–3061. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schekman R. (1989). Yeast Sec23p acts in the cytoplasm to promote protein transport from the endoplasmic reticulum to the Golgi complex in vivo and in vitro. EMBO J , 1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, Bernlohr DA. (2015). Metabolic functions of FABPs—mechanisms and therapeutic implications. Nat Rev Endocrinol , 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inobe T, Nukina N. (2016). Rapamycin-induced oligomer formation system of FRB-FKBP fusion proteins. J Biosci Bioeng , 40–46. [DOI] [PubMed] [Google Scholar]
- Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgärtel C, Schekman R, Rape M. (2012). Ubiquitin-dependent regulation of COPII coat size and function. Nature , 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Jones EL, Bonney SA, Patel HN, Mensenkamp AR, Eichenbaum-Voline S, Rudling M, Myrdal U, Annesi G, Naik S, et al. (2003). Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat Genet , 29–31. [DOI] [PubMed] [Google Scholar]
- Jung J, Wang J, Groenendyk J, Lee D, Michalak M, Agellon LB. (2017). Fatty acid binding protein (Fabp) 5 interacts with the calnexin cytoplasmic domain at the endoplasmic reticulum. Biochem Biophys Res Commun , 202–206. [DOI] [PubMed] [Google Scholar]
- Kim J, Hamamoto S, Ravazzola M, Orci L, Schekman R. (2005). Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J Biol Chem , 7758–7768. [DOI] [PubMed] [Google Scholar]
- Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. (2006). Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet , 1198–1203. [DOI] [PubMed] [Google Scholar]
- Levi L, Lobo G, Doud MK, von Lintig J, Seachrist D, Tochtrop GP, Noy N. (2013). Genetic ablation of the fatty acid-binding protein FABP5 suppresses HER2-induced mammary tumorigenesis. Cancer Res , 4770–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Goldberg J. (2016). TANGO1/cTAGE5 receptor as a polyvalent template for assembly of large COPII coats. Proc Natl Acad Sci USA , 10061–10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumata M, Inada H, Osumi N. (2016). Fatty acid binding proteins and the nervous system: their impact on mental conditions. Neurosci Res , 47–55. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. (1998). COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell , 263–275. [DOI] [PubMed] [Google Scholar]
- Melville DB, Montero-Balaguer M, Levic DS, Bradley K, Smith JR, Hatzopoulos AK, Knapik EW. (2011). The feelgood mutation in zebrafish dysregulates COPII-dependent secretion of select extracellular matrix proteins in skeletal morphogenesis. Dis Model Mech , 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merte J, Jensen D, Wright K, Sarsfield S, Wang Y, Schekman R, Ginty DD. (2010). Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat Cell Biol , 41–46 (suppl, pp. 1–8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Myrdal U, Veress B, Rudling M, Berglund L, Angelin B. (1995). Studies on lipoprotein metabolism in a family with jejunal chylomicron retention. Eur J Clin Invest , 271–280. [DOI] [PubMed] [Google Scholar]
- Niu X, Gao C, Jan Lo L, Luo Y, Meng C, Hong J, Hong W, Peng J. (2012). Sec13 safeguards the integrity of the endoplasmic reticulum and organogenesis of the digestive system in zebrafish. DevBiol , 197–207. [DOI] [PubMed] [Google Scholar]
- Peretti N, Roy CC, Sassolas A, Deslandres C, Drouin E, Rasquin A, Seidman E, Brochu P, Vohl M-C, Labarge S, et al. (2009). Chylomicron retention disease: a long term study of two cohorts. Mol Genet Metab , 136–142. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc , 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raote I, Ortega Bellido M, Pirozzi M, Zhang C, Melville D, Parashuraman S, Zimmermann T, Malhotra V. (2017). TANGO1 assembles into rings around COPII coats at ER exit sites. J Cell Biol , 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecchi KR, Go EP, Xu L, Woodin CL, Mure M, Desaire H. (2011). A general protease digestion procedure for optimal protein sequence coverage and post-translational modifications analysis of recombinant glycoproteins: application to the characterization of human lysyl oxidase-like 2 glycosylation. Anal Chem , 8484–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Maeda M, Katada T. (2017). Regulation of the Sar1 GTPase cycle is necessary for large cargo secretion from the endoplasmic reticulum. Front Cell Dev Biol , 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yamashiro K, Ichikawa Y, Erlmann P, Kontani K, Malhotra V, Katada T. (2011). cTAGE5 mediates collagen secretion through interaction with TANGO1 at endoplasmic reticulum exit sites. Mol Biol Cell , 2301–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yamashiro K, Shimazu N, Tanabe T, Kontani K, Katada T. (2014). Concentration of Sec12 at ER exit sites via interaction with cTAGE5 is required for collagen export. J Cell Biol , 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, Yeung T, Schekman RW. (1993). The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J , 4073–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AJM, Nogueira C, Ortega-Bellido M, Malhotra V. (2016). TANGO1 and Mia2/cTAGE5 (TALI) cooperate to export bulky pre-chylomicrons/VLDLs from the endoplasmic reticulum. J Cell Biol , 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah S, Barrallo-Gimeno A, Melville DB, Topczewski J, Solnica-Krezel L, Knapik EW. (2010). Sec24D-dependent transport of extracellular matrix proteins is required for zebrafish skeletal morphogenesis. PLoS One , e10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Novick P. (2004). 23 genes, 23 years later. Cell , S13–S15. [DOI] [PubMed] [Google Scholar]
- Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. (2007). Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell , 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K, Iolascon A, Verissimo F, Trede NS, Horsley W, Chen W, Paw BH, Hopfner K-P, Holzmann K, Russo R, et al. (2009). Mutations affecting the secretory COPII coat component SEC23B cause congenital dyserythropoietic anemia type II. Nat Genet , 936–940. [DOI] [PubMed] [Google Scholar]
- Siddiqi S, Mansbach CM. (2012). Phosphorylation of Sar1b protein releases liver fatty acid-binding protein from multiprotein complex in intestinal cytosol enabling it to bind to endoplasmic reticulum (ER) and bud the pre-chylomicron transport vesicle. J Biol Chem , 10178–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi SA, Gorelick FS, Mahan JT, Mansbach CM., 2nd (2003). COPII proteins are required for Golgi fusion but not for endoplasmic reticulum budding of the pre-chylomicron transport vesicle. J Cell Sci , 415–427. [DOI] [PubMed] [Google Scholar]
- Siddiqi S, Siddiqi SA, Mansbach CM. (2010). Sec24C is required for docking the prechylomicron transport vesicle with the Golgi. J Lipid Res , 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvain M, Bligny D, Aparicio T, Laforêt P, Grodet A, Peretti N, Ménard D, Djouadi F, Jardel C, Bégué JM, et al. (2008). Anderson’s disease (chylomicron retention disease): a new mutation in the SARA2 gene associated with muscular and cardiac abnormalities. Clin Genet , 546–552. [DOI] [PubMed] [Google Scholar]
- Stephens DJ, Pepperkok R. (2002). Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J Cell Sci , 1149–1160. [DOI] [PubMed] [Google Scholar]
- Strader MB, Tabb DL, Hervey WJ, Pan C, Hurst GB. (2006). Efficient and specific trypsin digestion of microgram to nanogram quantities of proteins in organic-aqueous solvent systems. Anal Chem , 125–134. [DOI] [PubMed] [Google Scholar]
- Tanabe T, Maeda M, Saito K, Katada T. (2016). Dual function of cTAGE5 in collagen export from the endoplasmic reticulum. Mol Biol Cell , 2008–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumser AE, Moore JB, Plant NJ. (2014). Fatty acid binding proteins: tissue-specific functions in health and disease. Curr Opin Clin Nutr Metab Care , 124–129. [DOI] [PubMed] [Google Scholar]
- Venditti R, Scanu T, Santoro M, Di Tullio G, Spaar A, Gaibisso R, Beznoussenko GV, Mironov AA, Mironov A, Zelante L, et al. (2012). Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science , 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman JT, Plutner H, Balch WE. (2001). The mammalian guanine nucleotide exchange factor mSec12 is essential for activation of the Sar1 GTPase directing endoplasmic reticulum export. Traffic , 465–475. [DOI] [PubMed] [Google Scholar]
- Wilson DG, Phamluong K, Li L, Sun M, Cao TC, Liu PS, Modrusan Z, Sandoval WN, Rangell L, Carano RAD, et al. (2011). Global defects in collagen secretion in a Mia3/TANGO1 knockout mouse. J Cell Biol , 935–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Barlowe C, Schekman R. (1993). Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science , 1466–1468. [DOI] [PubMed] [Google Scholar]
- Yuan L, Baba S, Bajaj K, Schekman R. (2017). Cell-free generation of COPII-coated procollagen I carriers. Bio-Protoc , e2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.