Abstract
Mitochondria exist in an equilibrium between fragmented and fused states that shifts heavily toward fission in response to cellular damage. Nuclear-to-cytoplasmic cyclin C relocalization is essential for dynamin-related protein 1 (Drp1)–dependent mitochondrial fission in response to oxidative stress. This study finds that cyclin C directly interacts with the Drp1 GTPase domain, increases its affinity to GTP, and stimulates GTPase activity in vitro. In addition, the cyclin C domain that binds Drp1 is contained within the non–Cdk binding second cyclin box domain common to all cyclin family members. This interaction is important, as this domain is sufficient to induce mitochondrial fission when expressed in mouse embryonic fibroblasts in the absence of additional stress signals. Using gel filtration chromatography and negative stain electron microscopy, we found that cyclin C interaction changes the geometry of Drp1 oligomers in vitro. High–molecular weight low–GTPase activity oligomers in the form of short filaments and rings were diminished, while dimers and elongated filaments were observed. Our results support a model in which cyclin C binding stimulates the reduction of low–GTPase activity Drp1 oligomers into dimers capable of producing high–GTPase activity filaments.
INTRODUCTION
Mitochondrial fission and fusion help facilitate responses to physiological processes such as cell division, autophagy, metabolic changes, oxidative stress response, synaptic transmission, and apoptosis (Bossy-Wetzel et al., 2003; Liesa et al., 2009; Westermann, 2010; Chan, 2012). GTPases of the dynamin superfamily act as the molecular motors that convert energy from GTP hydrolysis to either divide or fuse membranes (van der Bleik et al., 2013; Richter et al., 2015; Lee and Yoon, 2016; Ramachandran, 2018). Dynamin-related protein 1 (Drp1) facilitates mitochondrial fission in response to all stimuli studied to date. Drp1 is a cytosolic protein that exists in dynamic equilibrium between dimers and tetramers (Smirnova et al., 2001). During fission, Drp1 localizes to the mitochondrial outer membrane (MOM) through one of several receptors (Fis1, Mff, MIEF1, MiD49/51), forming elongated filaments encircling the mitochondria, which constrict upon GTP hydrolysis to cleave the organelle (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010; Palmer et al., 2011, 2013; Zhao et al., 2011; Lee and Yoon, 2016). Drp1 dysregulation has been implicated in in several pathological conditions, including neurodegenerative disorders such as Parkinson’s or Huntington’s disease and several cancers (Liesa et al., 2009).
Drp1 is required for fission in response to a variety of prolife or prodeath stimuli. For example, Drp1 mediates mitochondrial fragmentation during mitosis to facilitate organelle partitioning at cell division. Conversely, Drp1-dependent mitochondrial fission in response to oxidative stress represents an early event in the programmed cell death (PCD) pathway (Youle and van der Bliek, 2012; Jezek et al., 2018). Under these conditions, Drp1 helps recruit the proapoptotic protein Bax following treatment with the PCD-inducing kinase inhibitor staurosporine (Karbowski et al., 2002). These results indicate that specific signals directed toward Drp1 play a role in determining the type of scission event that occurs. How the cell differentiates one type of fission from another may in part be determined by the method by which Drp1 is activated. For example, phosphorylation of Ser616 by Cdk1–cyclin B is essential for mitochondrial fission during mitosis but not following oxidative stress (Taguchi et al., 2007; Liesa et al., 2009; Wang et al., 2015). On the other hand, we previously identified an essential role for the nuclear transcription factor cyclin C in stimulating oxidative stress–induced mitochondrial fission, which is conserved from yeast to mammals (Cooper et al., 2014; Wang et al., 2015). Under normal conditions, cyclin C regulates transcription through association with the Mediator complex of RNA polymerase II (Bourbon, 2008). In contrast to other cyclins, its concentration does not change during cell cycle progression (Leopold and O’Farrell, 1991; Lew et al., 1991; Cooper et al., 1997). Rather, oxidative stress induces cyclin C nuclear release to the mitochondria, where it facilitates fission through Drp1 and Mff (Wang et al., 2015). This report describes the molecular role of cyclin C in stimulating Drp1 activity.
RESULTS AND DISCUSSION
Cyclin C binds directly to the Drp1 GTPase domain through its C-terminal cyclin box
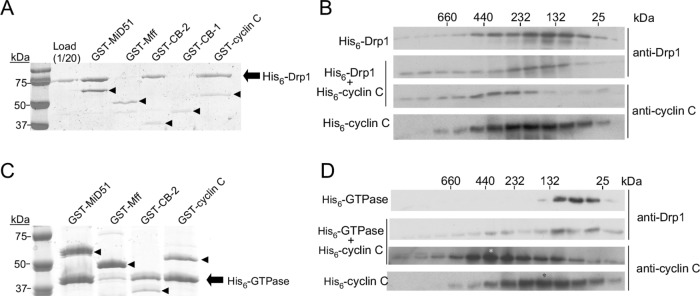
Stress-induced translocation of cyclin C from the nucleus to the cytosol initiates mitochondrial hyperfission, which is mediated by Drp1 and the hFis1 and Mff MOM receptors (Wang et al., 2015). Coimmunoprecipitation studies indicated that cyclin C and Drp1 interact, although it was unclear whether this association was direct. To determine whether cyclin C and Drp1 interact directly, pull-down experiments were performed with recombinant Drp1 and GST-tagged cyclin C (see Materials and Methods for details). Consistent with earlier studies (Palmer et al., 2011; Loson et al., 2014; Richter et al., 2014; Liu and Chan, 2015; Osellame et al., 2016; Yu et al., 2017), we observed that Drp1 bound GST-MiD51(D118N) but not GST-Mff(DTM) (Figure 1A). Repeating this experiment with recombinant GST–cyclin C indicated that these proteins directly interact. In addition, these results suggest that additional stress-induced factors or posttranslational modifications are not required for this association.
FIGURE 1:
Drp1 directly binds cyclin C through the GTPase domain. (A) Binding reactions containing Drp1 incubated with either GST–MiD51(DN118), GST–Mff(DTM), GST–cyclin box 2 (CB2), GST–cyclin box 1 (CB1), or GST–cyclin C as indicated were collected by glutathione beads and then separated by SDS–PAGE followed by Coomassie staining. Drp1 and the GST fusion proteins are indicated by the arrow and arrowheads, respectively. Molecular weight markers (kDa) are indicated on the left. (B) Western blotting of SEC fractions of 1 µM His6–Drp1, 1 µM His6–cyclin C, or both. Blots were probed as indicated on the right. The elution profile of molecular weight standards is indicated at the top. (C) As in A except that the GTPase domain of Drp1 was assayed for GST–cyclin C binding. (D) As in B except that a 1 µM Drp1 GTPase domain was added. The asterisks indicate the fraction of peak protein abundance.
Similarly to other cyclins, cyclin C consists of two five-helix bundles (Hoeppner et al., 2005; Schneider et al., 2011). The N-terminal 170-amino acid (aa) helix bundle composes the canonical cyclin box fold (referred to here as cyclin box 1 or CB1) that predominantly mediates Cdk association (Hoeppner et al., 2005). Thus, CB1 facilitates the transcriptional function of cyclin C. The C-terminal 113-aa domain contains the second five-helix bundle (cyclin box 2 or CB2), whose function has not been delineated. To test the requirement for either cyclin box domain in Drp1 association, we repeated the pull-down experiments with Drp1 and either GST-tagged CB1 or CB2. These studies revealed that Drp1 bound GST–CB2 but not GST–CB1 (Figure 1A). Thus, Drp1 directly binds cyclin C through the C-terminal cyclin box 2. To our knowledge, this finding is the first function ascribed to the cyclin box 2 domain for any cyclin.
To determine whether cyclin C preferentially binds to a particular oligomeric state of Drp1, we performed pull-down experiments with two Drp1 mutants. The G363D mutation predisposes Drp1 to form only lower-order species (Tanaka et al., 2006; Chang et al., 2010; Clinton et al., 2016), while the mutant Drp1R376E is unable to form any multimer (Strack and Cribbs, 2012; Clinton et al., 2016). Similarly to the wild type, both mutant Drp1 derivatives interacted with GST–cyclin C or GST–CB2 in pull-down experiments (Supplemental Figures S1A and S1B). These results indicate that cyclin C binds Drp1 in multiple oligomeric states. The ability of Drp1 to interact with cyclin C was further tested using size exclusion chromatography (SEC). The elution profile revealed that Drp1 was monodispersed with the molecular weight predominantly of dimeric and tetrameric units (Figure 1B). This result is consistent with several structural studies with Drp1 and other dynamins demonstrating the presence of a GTPase dimerization domain (Chappie et al., 2010; Kishida and Sugio, 2013; Francy et al., 2017). The addition of cyclin C (no tag) resulted in a shift in the Drp1 elution profile to the higher–molecular weight fractions. In addition, cyclin C was observed in the higher–molecular weight fractions. Taken together with the pull-down experiments, these results indicate that cyclin C interacts directly with Drp1.
To identify the Drp1 domain that binds cyclin C, we performed pull-down assays with GST–cyclin C and truncation derivatives corresponding to the GTPase domain (aa 1–337), GTPase with the middle domain (aa 1–501), and the ∆GED domain (excluding the C-terminal GED domain, aa 1–634; Chang et al., 2010). GST–MiD51 positively precipitated the GTPase domain (Figure 1C), consistent with the cryo-EM structure results (Kalia et al., 2018). Both GST–CB2 and GST–cyclin C bound the GTPase domain alone, suggesting that cyclin C interacts with Drp1 through this region. Consistent with this conclusion, GST–CB2 failed to associate with a Drp1 derivative lacking the GTPase domain (Stalk + variable region, Supplemental Figure S1C). Finally, the presence of His6–cyclin C shifted the GTPase domain elution profile to higher molecular weights, with a peak at ∼350 kDa (Figure 1D). His6–cyclin C itself eluted with a broad profile that also shifted to a higher molecular weight, with a new peak corresponding to the multimeric GTPase population. These results indicate that cyclin C associates directly with the Drp1 GTPase domain through the cyclin box 2 domain.
Cyclin box-2 is sufficient to induce mitochondrial fragmentation in the absence of other stress signals
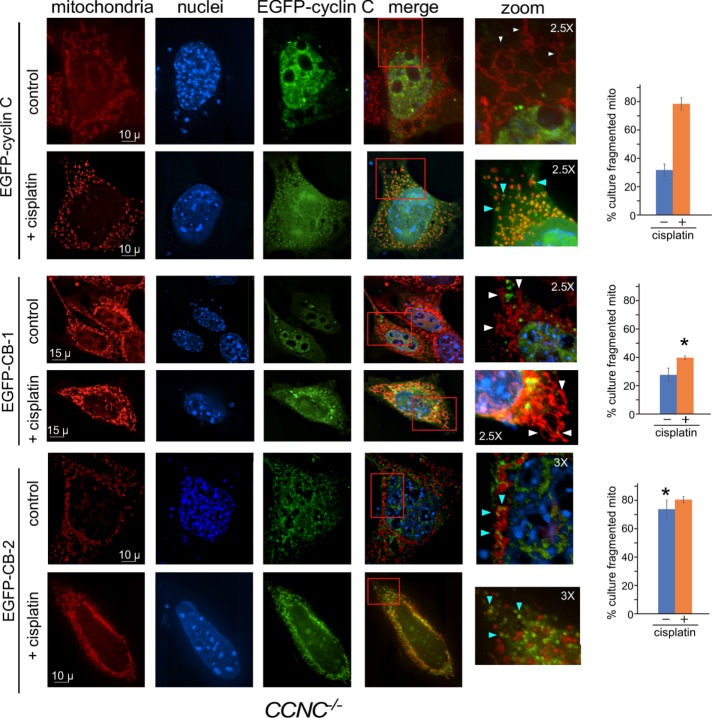
Our findings indicate that GST–CB2 binds Drp1 in vitro. To determine whether CB2 is necessary and/or sufficient for mitochondrial fragmentation in vivo, we transiently transfected CCNC null (CCNC–/–) mouse embryonic fibroblast (MEF) cells with plasmids expressing EGFP–cyclin C, EGFP–CB1, or EGFP–CB2. Previous studies demonstrated that EGFP–cyclin C complemented the PCD deficiency phenotype in CCNC–/– cells (Wang et al., 2015). EGFP subcellular localization and mitochondrial morphology were monitored by fluorescence microscopy in unstressed and cisplatin-treated cultures. Consistent with earlier observations (Wang et al., 2015), EGFP–cyclin C is predominantly nuclear, with cells displaying filamentous mitochondria (Figure 2, top panels). As expected, cisplatin treatment induced EGFP–cyclin C nuclear release, where it colocalized with fragmented mitochondria. Similarly, EGFP-CB1 accumulated predominantly in the nucleus in the absence of stress (Figure 2, middle panels). Although cisplatin treatment induced its nuclear release, mitochondrial morphology remained unchanged, indicating that the CB1 domain alone is unable to induce fission. Conversely, EGFP–CB2 was cytoplasmic prior to cisplatin treatment, with cells displaying a significant increase in mitochondrial fragmentation (Figure 2, bottom panels; see Supplemental Figure S2 for additional images). This result is consistent with our previous findings that addition of recombinant cyclin C to permeabilized MEF cells is sufficient to induce mitochondrial fission in the absence of stress (Wang et al., 2015). Treating EGFP–CB2–expressing cells with cisplatin did not significantly enhance mitochondrial fragmentation. These results indicate that CB-2 is both necessary and sufficient to mediate mitochondrial fragmentation. Interestingly, the subcellular localization pattern of EGFP–CB2 was different in cells treated with CP versus the controls. In stressed cells, EGFP–CB2 covered the fragmented mitochondria (blue arrowheads), while unstressed cells exhibited smaller foci on the periphery of the organelle. In addition, although the percentages of the population exhibiting fission based on our criteria were similar, the overall extent to which the mitochondria fragmented appeared greater in stressed cells. These findings suggest that complete mitochondrial fragmentation requires a stress signal in EGFP–CB2–expressing cells. To verify that CB2 was inducing fission through Drp1, we attempted coimmunoprecipitation experiments between EGFP–CB2 and endogenous Drp1 in both CCNC–/– MEFs and HeLa cells (to increase transfection efficiency). We were unable to detect this interaction in unstressed cells. Previously, this interaction was detected when both full-length cyclin C and GFP–Drp1 were overexpressed (Wang et al., 2015). Our inability to detect this interaction with CB2 only may reflect changes in complex stability in the absence of CB1. Alternatively, our finding that cyclin C is released from Drp1 multimers (see below) may suggest that CB2 is more readily expelled during filament formation, thus reducing its interaction window.
FIGURE 2:
Fluorescence microscopy of CCNC–/– MEF cells transfected with full-length EGFP–cyclin C (top panels), EGFP-CB1 (middle panels), or EGFP-CB2 (bottom panels) before and after cisplatin treatment, as indicated. The cyclin box-2 domain is necessary and sufficient to induce mitochondrial fission. Boxes in the merge panels indicate the magnified regions on the right. White and blue arrowheads in the zoom images indicate fused and fragmented mitochondria, respectively. Quantitation of cells exhibiting fragmented mitochondria (three independent cultures) is provided on the right. Asterisks indicate statistical differences (p < 0.01) from EGFP–cyclin C values. Scale bars are shown. Magnification in zoomed images is indicated.
Cyclin C increases Drp1 oligomerization
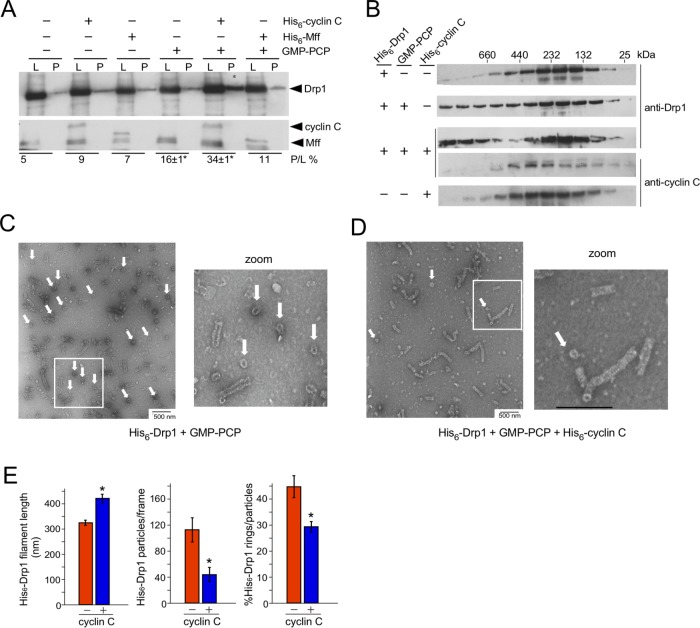
Our results indicate that cyclin C stimulates mitochondrial fission through Drp1 association. To mechanistically explore this observation further, we employed sedimentation assays to measure the impact cyclin C has on Drp1 oligomerization. In these experiments, we used a lower Drp1 concentration (1 µM) than normally employed in these assays to allow even subtle changes in oligomerization to be detected. Under these conditions, the percentage of His6–Drp1 pelleting was not increased above background (5%) following incubation with either His6–Mff (9%) or His6–cyclin C (7%; Figure 3A). The addition of GMP–PCP, a nonhydrolyzable analogue of GTP, stimulated oligomerization in these assays, as determined by increased His6–Drp1 in the pellet fraction (16 ± 1%). Combining His6–cyclin C and GMP–PCP increased the concentration of pelleted Drp1 another twofold (34 ± 1%). Interestingly, cyclin C itself was absent in the pellet fraction. These results indicate that although cyclin C enhances oligomerization in the presence of GMP–PCP, it does not remain associated with the Drp1 filament under these conditions.
FIGURE 3:
Cyclin C stimulates Drp1 aggregate dissolution and filament formation. (A) Oligomerization was monitored using Western blot analysis of fractions following sedimentation. His6–cyclin C, His6–Mff, and/or GMP–PCP were incubated with His6–Drp1 as indicated and then subjected to high-speed centrifugation. His6–Drp1 and His6–cyclin C in the resulting pellets (P) and the load (L) were visualized by Western blot and then quantified (below each lane) by calculating the ratio of His6–Drp1 in the pellet (P) toy the amount loaded (L). GMP–PCP and GMP–PCP + His6–cyclin C experiments were repeated three times (mean ± SEM; *p < 0.02 from no addition control). (B) Western blot analysis of SEC fractions obtained following incubation of His6–Drp1, His6–cyclin C, or both with or without GMP–PCP. Primary antibodies used are indicated on the right. (C, D) TEM images of His6–Drp1 incubated with GMP–PCP/Mg2+ with or without His6–cyclin C as indicated. White arrows indicate Drp1 rings. (E) Quantitation of Drp1 filament formation from TEM images counting 1485 and 493 particles without and with His6–cyclin C, respectively, across 11 frames of identical magnification. Total particles include filaments and rings. Bars are mean ± SEM. Asterisks indicate p < 0.02.
Next, SEC was employed to monitor Drp1 and cyclin C complex formation in the presence of GMP–PCP. Western blot analysis of fractions obtained from this study found that, consistent with earlier findings (Macdonald et al., 2014; Hatch et al., 2016), the majority of isolated His6–Drp1 was found in the ∼130–250 kDa molecular weight range, suggesting primarily dimer/tetramer formation (Figure 3B). In the presence of GMP–PCP, His6–Drp1 was distributed evenly into higher–molecular weight fractions, indicating enhanced oligomer formation. The addition of His6–cyclin C and GMP–PCP caused two changes to the His6–Drp1 elution profile. First, the even distribution of His6–Drp1 in the higher–molecular weight fractions observed with His6–Drp1 alone was absent. Instead, His6–Drp1 concentrations increased in the highest–molecular weight fraction (>660 kDa) but decreased in the 440–660 kDa window. The increased His6–Drp1 concentrations in the higher–molecular weight fractions following His6–cyclin C and GMP–PCP addition is consistent with the increased sedimentation observed in Figure 3A. Second, the proportion of His6–Drp1 in the dimeric fractions increased following His6–cyclin C addition compared with the His6–Drp1 profile with GMP–PCP. Also in agreement with our sedimentation studies, His6–cyclin C was not distributed to the higher–molecular weight His6–Drp1 fractions, suggesting that it is not part of these higher-order structures.
To investigate whether His6-cyclin C was inducing actual His6–Drp1 filament formation or via nonspecific aggregates, a mutant form of Drp1 (Drp1-4A) was used that is defective in filament formation (Frohlich et al., 2013; Liu and Chan, 2015). The addition of His6–cyclin C did not change the His6–Drp1-4A SEC elution profile (Supplemental Figure S3A), even though cyclin C associates with the mutant. This result suggests that cyclin C stimulates normal Drp1 filament formation and not nonspecific aggregation. Finally, Mff addition did not alter Drp1 oligomerization ability (Supplemental Figure S3B). Taken together, these results indicate that cyclin C enhances Drp1 oligomerization but is not retained in these larger filaments. To further explore this latter possibility, GST–cyclin C was used as bait in pull-down experiments with Drp1 or Drp1-4A. Without GTP or GMP–PCP, GST–cyclin C associated similarly with either Drp1 or Drp1-4A (Supplemental Figure S3C; compare lanes 1 and 4). However, GMP–PCP addition caused a reduction in GST–cyclin C association with Drp1 from that with oligomerization-defective Drp1-4A mutants (compare lanes 2 and 5). These results are consistent with the model in which cyclin C association with Drp1 is reduced in higher-order structures. Surprisingly, GST–cyclin C was equally poor at binding Drp1 or Drp1-4A in the presence of GTP (lanes 3 and 6). These results may represent differences in GTPase domain structure in the presence of these two guanine nucleotides (Wenger et al., 2013).
Cyclin C enhances Drp1 filament formation
The SEC results just described suggest that cyclin C stimulates Drp1 oligomerization at the expense of intermediate–molecular weight species. To further examine this result, we directly measured the impact of cyclin C on Drp1 oligomerization using transmission electron microscopy. His6–Drp1 was incubated with GMP–PCP with or without His6–cyclin C. Typical for His6–Drp1 + GMP–PCP in solution (Macdonald et al., 2014, 2016), helical filaments and rings (arrows, Figure 3C) were observed. However, in the presence of His6–cyclin C and GMP–PCP, the incidence of ring formation greatly diminished, while linear His6–Drp1 filaments were 25% longer than with His6–Drp1 alone (Figure 3D; quantified in Figure 3E). In addition, the percentage of filaments observed in the upper ranges of sizes increased with the addition of His6–cyclin C (Supplemental Figure S3D). Consistent with these observations, the total number of electron-dense particles decreased in the presence of His6–cyclin C, as did the percentage of rings to total particles (Figure 3D). The cyclin C–dependent increase in His6–Drp1 filament length and reduction in smaller rings are consistent with the SEC results, in which His6–Drp1 eluted in the higher– and lower–molecular weight fractions but was reduced in the intermediate fractions in the presence of His6–cyclin C. There was also a small but statistically significant increase in filament diameter in the presence of His6–cyclin C (Supplemental Figure S3E). We are unsure whether this small change in filament diameter represents a change in Drp1 activity. Taken together, these results indicate that cyclin C supports higher-order His6–Drp1 filament formation and reduces the presence of ring aggregates.
Cyclin C increases Drp1 GTPase activity by enhancing GTP affinity
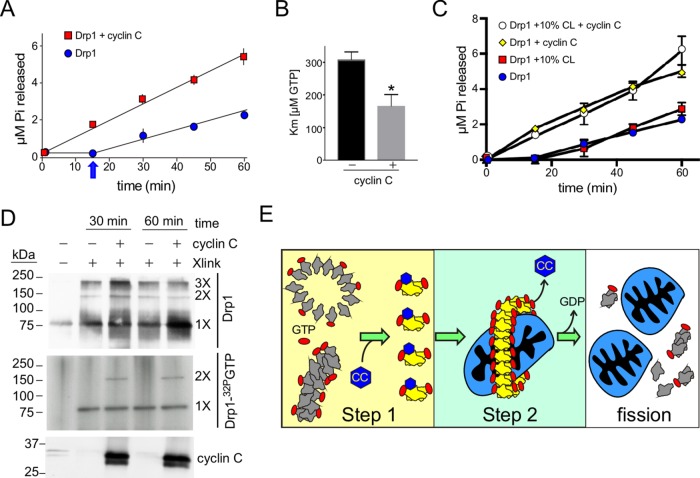
Drp1 is a mechanochemical enzyme that utilizes the energy from GTP hydrolysis to constrict and cleave mitochondrial membranes. In solution, Drp1 GTPase activity is low but is stimulated by specific lipids (Bustillo-Zabalbeitia et al., 2014; Macdonald et al., 2014, 2016; Clinton et al., 2016; Francy et al., 2017) or proteins (Ji et al., 2015; Hatch et al., 2016). Because cyclin C stimulates Drp1 oligomerization in solution, we tested whether cyclin C influences GTPase activity itself. First, we measured the effect of cyclin C on the kinetic parameters (Km and Kcat) of Drp1 GTPase activity. The catalytic efficiency of Drp1 hydrolysis (1.6 ± 0.08 min–1, Figure 4A) is increased ∼50% by addition of cyclin C (Kcat 2.4 ± 0.02). We found that the Km of Drp1 for GTP was 309 ± 23 µM (Figure 4B), which is consistent with earlier studies (Bustillo-Zabalbeitia et al., 2014; Macdonald et al., 2016). In the presence of cyclin C, there was a nearly twofold increase in the binding affinity of Drp1 to GTP (Km 166 ± 35 µM GTP). In addition, the lag observed with Drp1-only reactions (arrow, Figure 4A) was absent on cyclin C addition, suggesting that the appearance of assembly-ready dimers was enhanced. The elevated GTPase activity is also consistent with the enhanced filament formation observed in the presence of cyclin C (Figure 3).
FIGURE 4:
Cyclin C stimulation of Drp1 GTPase activity. (A) Kinetics of GTP hydrolysis by Drp1 with the indicated additions. The arrow indicates the lag phase normally observed with Drp1 GTPase activity studies. (B) Km of GTP binding by Drp1 with and without cyclin C. Mean ± SEM indicated (n = 3). Asterisk indicates p < 0.02. (C) GTP hydrolysis was followed for the samples as indicated. Liposomes with or without 10% cardiolipin (CL) were included as indicated. (D) UV cross-linking of reactions containing Drp1 and cyclin C as indicated with either GTP (top panel) or 32P-GTP (middle panel). GTP samples were separated by SDS–PAGE and subjected to Western blot analysis probing for Drp1 (top panel) or cyclin C (bottom panel). 32P-GTP samples were separated by PAGE with the gel dried and subjected to autoradiography (middle panel). 1×, 2×, and 3× represent predicted multimeric states of Drp1 based on molecular weight (kDa, indicated on the left). (E) Proposed two-step model for cyclin C–dependent stimulation of Drp1 activity. Step 1. In the absence of cyclin C, Drp1 forms low–GTPase activity oligomers or rings. Cyclin C binding induces a conformational change resulting in enhanced GTP binding and dissolution of the low–GTPase activity aggregates into dimers able to promote formation of high–GTPase activity filaments. Step 2. High–GTPase oligomers exclude cyclin C as filaments form with high efficiency. Subsequent GTP hydrolysis induces mitochondrial scission and dissolution of the filaments, providing the building blocks for the next round of division.
Previous studies found that increasing amounts of the mitochondrial enriched lipid cardiolipin (CL) stimulated Drp1 GTPase activity (Bustillo-Zabalbeitia et al., 2014; Clinton et al., 2016). GTPase reactions were repeated with 10 and 25% CL with respect to total lipid composition. Liposomes containing 10% CL provided little or no increase in Drp1 GTPase activity (Figure 4C), while 25% increases hydrolysis severalfold (Supplemental Figure S4A). Addition of 10% CL also had no effect on cyclin C-dependent Drp1 GTPase stimulation. If the amount of CL was increased to 25%, cyclin C addition did not further enhance GTPase activity (Supplemental Figure S4A). Although several possibilities exist, these results formally suggest that cyclin C and CL function in the same pathway to stimulate Drp1 GTPase activity.
Our results suggested the possibility that cyclin C association alters the conformation of Drp1 to enhance GTP binding. To examine this possibility, a cross-linking approach was utilized to determine whether a GTP bound-Drp1 conformation could be identified that was dependent on cyclin C. Radioactive GTP (32P–GTP) was incubated with Drp1 with or without cyclin C. To increase the chance of identifying an altered conformational state, the reactions were conducted for an extended period (30 or 60 min) before UV irradiation was applied to cross-link GTP to Drp1 (Naylor et al., 2006). The cross-linked reactions were separated by SDS–PAGE and the presence of Drp1 and 32P–GTP–Drp1 was determined by Western blot and autoradiography, respectively. Western blot analysis revealed that Drp1 species with mobilities consistent with monomer, dimer, and trimer formation in all reactions (Figure 4D). The trimer species is not predicted from earlier Drp1 studies and most likely reflects the use of the UV nonspecific cross-linking method that can capture both physiological and nonphysiological complex formation. As expected, autoradiography revealed a 32P–GTP signal with the Drp1 monomer in all reactions. However, a predicted Drp1 dimer species exhibiting 32P–GTP binding was observed only when cyclin C was added. These results are consistent with a model in which cyclin C association stimulates a conformational change in Drp1 that enhances its GTP-binding ability. This model is consistent with our finding that cyclin C interacts with the GTPase domain and enhances GTP binding. Therefore, we tested whether cyclin C altered GTP binding and GTP hydrolysis of the GTPase domain itself. The experiment described in Figure 4A was repeated with the GTPase domain with and without cyclin C addition. These studies revealed that GTPase activity was stimulated (Supplemental Figure S4B) with a corresponding enhancement in GTP binding ability (Supplemental Figure S4C). Finally, cyclin C did not alter GTPase activity in Drp1 mutants that were unable to form oligomers (dimeric-only mutant Drp1-4A, Supplemental Figure S4D) or (monomeric restricted Drp1K631E, Supplemental Figure S4, D and E). Taken together, these findings support our model in which cyclin C interacts with the GTPase domain to stimulate Drp1’s ability to produce high–GTPase activity filaments.
In vitro studies revealed that Drp1 forms filaments and rings in the presence of GMP-PCP (Koirala et al., 2013; Macdonald et al., 2014; Basu et al., 2017; Kalia et al., 2018). Conditions inducing long Drp1 helical filaments while reducing ring formation, such as low ionic strength or the presence of cardiolipin-enriched liposomes, also increased its GTP binding affinity (Koirala et al., 2013; Macdonald et al., 2016). Moreover, liposome studies reported that precocious self-assembly of Drp1 in solution interferes with Mff association (Clinton et al., 2016). Rather, they found that Drp1 dimers were more effectively recruited by lipid-bound Mff. This observation is consistent with an earlier study, based on mutations that produced exclusive dimers (C505A) or higher-order structures (C300A) in solution, that found that dimers possessed an enhanced ability to produce remodeling-competent higher-order polymers on membranes (Macdonald et al., 2014). Similarly to these studies, we found that cyclin C diminished ring formation while increasing both dimer concentration and overall filament length.
We propose a model in which cyclin C stimulates fission through a two-step process. First, cyclin C association induces a conformational change that increases GTP binding and promotes disassociation of these inactive complexes into a dimer species capable of forming active filaments (Step 1, Figure 4E). This model is consistent with the change in Drp1 SEC elution profiles, in which cyclin C addition increased the abundance of larger and smaller multimers at the expense of intermediate-sized oligomers. In addition, the rapid production of dimers is also consistent with the ability of cyclin C to eliminate the lag observed in GTPase assays. Therefore, our finding that cyclin C induces breakdown of low GTPase-activity oligomers in favor of more reactive dimer species provides a compelling mechanism for how stress-induced hyperfission is orchestrated. This model is also consistent with our yeast studies, which found that the presence of cyclin C in the cytoplasm increased the production of functional mitochondrial Dnm1 foci at the expense of nonproductive mitochondria-associated oligomers (Cooper et al., 2014).
Finally, both sedimentation and SEC results indicated that higher-order filaments were devoid of cyclin C (Step 2). This result suggests that the conformational change associated with Drp1 filament formation displaces cyclin C. Different GTPase-domain dimerization strategies have been proposed to account for Drp1 conformational differences when bound to cardiolipin as opposed to other lipids (Francy et al., 2017). This reorientation has been suggested to enhance GTP binding through intermolecular GTPase domain association. One possibility is that the presence of cyclin C facilitates this conformational switch, allowing Drp1 to more readily bind GTP. It is still not clear whether GTP binding is a prerequisite for disassociating low-GTPase oligomers or whether GTP association occurs in the dimer state. Further studies are required to address this question.
All cyclins contain a repeat of the five-helix bundle termed the cyclin box fold (Hoeppner et al., 2005). Our finding that the second cyclin box domain (CB2), and not the first cyclin box domain (CB1) which binds Cdk8, is responsible for the mitochondrial function of cyclin C was surprising. Cyclin C does not have a canonical nuclear localization signal and appears to be maintained in the nucleus by association with Cdk8 or Med13 through CB1 (Cooper et al., 2014; Khakhina et al., 2014). Conversely, CB2 is not targeted to the nucleus but can interact with Drp1 and induce mitochondrial fission in the absence of other stress signals. These results suggest a model in which the nuclear and mitochondrial functions of cyclin C are separable. We were unable to find other studies that identified a functional role specifically for this second domain in any cyclin, although Cdk-independent roles for cyclins have been reported (Neuman et al., 1997; McMahon et al., 1999). Given that a second, more divergent cyclin box exists in all family members, more Cdk-independent roles for this protein family may be forthcoming.
Mitochondrial division occurs in response to a variety of signals including cell damage, mitophagy, and cell division (Horbay and Bilyy, 2016). Although the role of Drp1-induced fission in mitophagy is still debated (Deng et al., 2008; Poole et al., 2008; Mendl et al., 2011; Yamashita et al., 2016), mitochondrial fission during cell division and following cell damage has been clearly established. While not a commitment point, mitochondrial division following cellular stress is often an early event in the programmed cell death pathway (Jezek et al., 2018). Conversely, fission during mitosis allows efficient distribution of the organelle into daughter cells, enhancing homeostasis. To achieve mitochondrial fragmentation, Drp1 is activated by different pathways. For example, cyclin B-Cdk1 phosphorylation stimulates Drp1 activity during the G2 phase (Taguchi et al., 2007; Liesa et al., 2009), while oxidative stress triggers cyclin C release from the nucleus to induce mitochondrial fragmentation (Cooper et al., 2014; Wang et al., 2015). However, cyclin C is not required for fission during mitosis and the activating phosphorylation by Cdk1 does not occur following stress (Wang et al., 2015). These results indicate that although the outcome of mitochondrial fission is the same, the signaling pathways are very different. These observations suggest that the cell uses these different regulatory modes to provide “value added” information to the fission process. Consistent with this notion, cyclin B–Cdk1 activity also stimulates actin cable assembly (Miao et al., 2016) in G2 and induces degradation of the mitofusion Mfn1 (Park and Cho, 2012). When combined with Drp1 stimulation, these events help ensure proper mitochondrial inheritance. On the other hand, we found that cyclin C also helps recruit the proapoptotic protein Bax to the mitochondrion in a fission complex–dependent manner (J. Jezek and R. Strich, unpublished data). Therefore, cyclin B–Cdk1 phosphorylation or the translocation of cyclin C confers different information to the cell, enabling the coordination of cellular events with these growth or death stimuli.
MATERIALS AND METHODS
Antibodies and reagents
Western blot analyses were conducted with anti-Drp1 (BD Biosciences #611738, 1:500), cyclin C (Thermo Fisher Scientific PA5-16227, 1:1000), GTPase domain (Abcam Ab56788, 1:500), Mff (Abcam Ab81127, 1:500), and His6 (Abcam Ab18184, 1:1000). Secondary antibodies harboring alkaline phosphatase (Sigma A7434) were used at a 1:5000 dilution. Western blot signals were visualized by chemiluminescence (Tropix) and quantified using a Kodak digital imaging station.
Plasmids
The full-length Drp1-short (a.k.a. Drp1-3) isoform was expressed from pET21b (Novagen) as described in Liu and Chan (2015) and was a gift from D. Chan, California Institute of Technology. His6-tagged Mff (DTM) was expressed in pET28a (Novagen) as described in Clinton et al. (2016) was a gift from J. Mears, Case Western Reserve University. The DN118 GST-MiD51 derivative expression plasmid (Richter et al., 2014) was a gift from M. Ryan, Monash University. The DTM GST-Mff expression plasmid (Macdonald et al., 2016) was a gift of R. Ramachandran, Case Western Reserve University. His6-tagged human cyclin C was generated by subcloning the human CCNC cDNA into the BamHI and XmaI restriction sites of pEQ30 (Qiagen). GST-tagged human cyclin C was generated by subcloning the human CCNC cDNA into the BamHI–XmaI sites in pGEX4T-1 (GE Healthcare). For eukaryotic expression, the human CCNC cDNA was subcloned into the PstI and XmaI sites in pEGFP-C1 (Clontech). All Drp1 mutants (4A, G363D, and R376E), the N-terminal 337–amino acid GTPase domain, and cyclin C (cyclin box 1, aa 1–170, and cyclin box 2, aa 171–284) were generated using the Q5 site-directed mutagenesis kit (New England Biolabs). The DRP1 stalk+variable domain derivative was expressed in pET15b and was a gift of H. Sesaki, Johns Hopkins University.
Expression and purification of recombinant proteins
Rosetta2 BL21 E. coli cells expressing either GST or His6 recombinant proteins were grown to an OD600 of 0.6–1.0 in terrific broth (Tartoff and Hobbs, 1987) and expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 18 h at 16°C. The cell pellets were lysed by sonication in HCBT150 (150 mM KCl, 50 mM HEPES, 5 mM tris(2-carboxyethyl)phosphine-HCl [TCEP.HCl]) and 10% glycerol supplemented with a protease inhibitor cocktail (Sigma). The soluble lysates were clarified by ultracentrifugation (1 h at 100,000 × g) and incubated with 1 ml of glutathione agarose (Sigma) or Cobalt (Clontech) resin for 1 h at 4°C with rocking. The resin was washed with 50 column volumes (CV) of HCB150. The bound protein was eluted with HCB150 supplemented with either 10 mM glutathione or 250 mM imidazole for GST- and His6-tagged proteins, respectively. For purification of all His6-tagged proteins, lysis and wash buffers were supplemented with 20 mM imidazole. For purification of GST– or His6–cyclin C, the HCB150 buffer was supplemented with 0.1% Triton X-100 for cell lysis. After elution, the protein-containing fractions were exchanged into HSB150 (without glutathione or imidazole) using Zeba 7K desalting columns (Thermo Fisher). For the pull-down experiments, the His6 tag was removed from Drp1 or the GTPase domain using Prescission protease (GE) followed by removal of the Prescission protease using glutathione agarose resin and His6 tag using Cobalt resin. The His6 tag was removed from the stalk+variable domain of Drp1 using thrombin, followed by removal of thrombin using Benzimidine sepharose resin.
GST pull-down assays
Purified Drp1 (tag removed) and derivatives were incubated with the bait (GST-tagged cyclin C, Mff, or MiD51) proteins in 500 µl HCB150 at ambient temperature for 1 h. Equal amounts of glutathione agarose slurry were added to each reaction and incubated further for 15 min. The unbound fraction was removed by centrifugation and discarded. The beads were washed twice with 500 µl of HCB150. The bound fraction was eluted by boiling the beads in 50 µl SDS-loading dye and resolved by SDS–PAGE. The gels were either stained with Coomassie or subjected to Western blot analysis. Protein determinations were accomplished using the microBradford assay.
Cosedimentation assays
His6–Drp1 (1 µM) was incubated with or without 2 mM of the GTP analogue, GMP–PCP, and 2 mM Mg2+ in the absence or presence of His6–cyclin C (1 µM) or His6–Mff (5 µM) in 500 µl final volume for 1 h at room temperature. The binding reaction was centrifuged at 100,000 × g for 30 min at 24°C. After the supernatant was collected, the pellet was resuspended in 500 µl of HCB150. Equal volumes of pellet and supernatant were resolved on SDS–PAGE, blotted, and probed with anti-His6 antibodies. The amount of the protein in the pellet as a fraction of the total load was determined by densitometric analysis of the blots from a triplicate data set using imageJ analysis software.
Size-exclusion chromatography
Full-length His6–Drp1 and derivatives (1 µM) were incubated with or without 1 µM His6-tagged human cyclin C or His6–Mff for 1 h at room temperature in a final volume of 500 µl in HCB150. GTP or the GMP–PCP analogue was added (2 mM final concentration) along with 2 mM Mg2+. After incubation, the samples were loaded onto a Superose 6 column (GL 10 × 300) at a rate of 0.5 ml/min using the AKTA purifier (GE Healthcare). One-milliliter fractions were manually collected, resolved by SDS–PAGE, and then blotted and probed with appropriate antibodies.
Cellular mitochondrial dynamics assays
A CCNC–/– null MEF culture was transfected with 5 µg plasmid DNA expressing EGFP fused to either full-length cyclin C, CB1, or CB2 in pEGFP-C1. After 24 h incubation, the cultures were seeded in triplicate 6-well culture dishes on lysine-coated cover slips (in complete DMEM supplemented with 10% fetal bovine serum and antibiotics) for 24 h. One triplicate set was treated with cisplatin (20 µM) for 24 h. After the medium was removed and the cells were washed with PBS, they were fixed with 4% paraformaldehyde (PFA) for 10 min at 37°C. After the cells were washed with PBS, the coverslips were mounted on slides using DAPI-Vectashield mounting medium. EGFP signals were detected with fluorescence microscopy (Nikon 90-i). Mitochondrial morphology was scored as described previously (Wang et al., 2015). Briefly, mitochondrial fragmentation was quantitated by calculating the percentage of the population with >10 mitochondria with a length of ≥10 μM. SDs of the means were calculated using Student’s t test.
GTPase assays
The GTPase assays were performed essentially as described (Ji et al., 2015; Clinton et al., 2016). All reactions were done in triplicate. Drp1 (1 µM) was incubated without or with 1 µM His6–cyclin C for 1 h at room temperature. To measure GTPase activity of Drp1 in the presence of liposomes, His6–Drp1 and His6–cyclin C were incubated at a final concentration of 2 µM each at room temperature for 1 h. An equal volume of liposomes corresponding to 200 µM total lipid was added to the protein mixture and incubated for 15 min. The liposome–Mff complex mixture was added to the Drp1–cyclin C–His mixture in equal volume and incubated further for 15 min. Liposomes were prepared as described in Clinton et al. (2016) with solutions of 1,2-dioleoyl-sn-glycero-3-phosphocholine and cardiolipin mixed at stoichiometric ratios of 90:10 or 75:25 as indicated in the text. The amount of free phosphate was determined by increased absorbance of malachite green at OD650 as described (Baykov et al., 1988). To determine kcat and Km values, three GTP concentrations (0.5, 1.0, and 2.0 mM) were measured at 0, 15, 30, 45, and 60 min. The lag time was ignored when observed, and only values composing the linear response were used in the calculation. Calculations were performed using GraphPad Prism version 7.0 for Mac OS X, GraphPad Software, La Jolla, CA (www.graphpad.com).
GTP binding assays
GTP binding assays were performed as described in Naylor et al. (2006) with the following modifications. His6–Drp1 (1 µM) was incubated with His6–cyclin C (1 µM) as indicated for 1 h at room temperature. GTP or radiolabeled GTP [α-32P]-GTP was added (0.5 µM) and the samples were incubated at room temperature for 30 or 60 min. The samples were cross-linked by UV irradiation (5 min) and then resolved by SDS–PAGE. The radioactive and nonradioactive gels were visualized by autoradiography and Western blot analysis, respectively.
Negative-stain transmission electron microscopy
His6-Drp1 (1 µM) was incubated with GMP–PCP/Mg2+ (2 mM) with or without 1 µM His6–cyclin C for 1 h at room temperature in a final volume of 100 µl. Five microliters of the mixture was dotted onto carbon-coated grids in triplicate for 1 min and then stained with 2% uranyl acetate for 1 min. The grids were washed twice by dipping in ddH2O for 30 s each and examined with a JEOL 1010 electron microscope at 100 KeV fitted with a Hamamatsu digital camera and AMT Advantage image capture software. The number of electron-dense particles (filaments and rings), filament length, and diameter were measured in ImageJ across 11 frames of identical magnification totaling 1485 and 493 particles without and with cyclin C addition, respectively.
Supplementary Material
Acknowledgments
We thank D. Chan (California Institute of Technology, Pasadena, CA), J. Mears (Case Western Reserve University, Cleveland, OH), M. Ryan (Monash University, Melbourne, Australia), R. Ramachandran (Case Western Reserve University, Cleveland, OH) and H. Sesaki (Johns Hopkins University, Baltimore, MD) for plasmids. We thank T. Baumgart and Chun Liu (University of Pennsylvania, Philadelphia, PA) for help in preparing liposomes. We thank the Imaging core at the Fox Chase Cancer Center for electron microscopy. We thank J. Jezek and P. Y. K. Sundarasivarao for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health awarded to K.F.C. (GM113196) and R.S. (GM113052) and by the New Jersey Cancer Commission (DHFS16PPC067) to V.G. This work was also supported by a grant from the New Jersey Health Foundation to R.S.
Abbreviations used:
- aa
amino acid
- CB1
cyclin box 1
- CB2
cyclin box 2
- GMP-PCP
guanylyl β,γ-methylene-diphosphonate
- GTP
guanosine triphosphate
- MEF
mouse embryonic fibroblast
- MOM
mitochondrial outer membrane
- PCD
programmed cell death
- SEC
size exclusion chromatography
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-07-0463) on December 5, 2018.
REFERENCES
- Basu K, Lajoie D, Aumentado-Armstrong T, Chen J, Koning RI, Bossy B, Bostina M, Sik A, Bossy-Wetzel E, Rouiller I. (2017). Molecular mechanism of DRP1 assembly studied in vitro by cryo-electron microscopy. PLoS One , e0179397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baykov AA, Evtushenko OA, Avaeva SM. (1988). A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem , 266–270. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Barsoum MJ, Godzik A, Schwarzenbacher R, Lipton SA. (2003). Mitochondrial fission in apoptosis, neurodegeneration and aging. Curr Opin Cell Biol , 706–716. [DOI] [PubMed] [Google Scholar]
- Bourbon HM. (2008). Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res , 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. (2014). Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PLoS One , e102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. (2012). Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet , 265–287. [DOI] [PubMed] [Google Scholar]
- Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, Blackstone C. (2010). A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem , 32494–32503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. (2010). G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature , 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton RW, Francy CA, Ramachandran R, Qi X, Mears JA. (2016). Dynamin-related Protein 1 oligomerization in solution impairs functional interactions with membrane-anchored mitochondrial fission factor. J Biol Chem , 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Khakhina S, Kim SK, Strich R. (2014). Stress-induced nuclear-to-cytoplasmic translocation of cyclin C promotes mitochondrial fission in yeast. Dev Cell , 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Smith JB, Strich R. (1997). Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p). EMBO J , 4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. (2008). The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA , 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francy CA, Clinton RW, Frohlich C, Murphy C, Mears JA. (2017). Cryo-EM studies of Drp1 reveal cardiolipin interactions that activate the helical oligomer. Sci Rep , 10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. (2013). Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J , 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. (2008). The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell , 2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch AL, Ji WK, Merrill RA, Strack S, Higgs HN. (2016). Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol Biol Cell , 3109–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner S, Baumli S, Cramer P. (2005). Structure of the mediator subunit cyclin C and its implications for CDK8 function. J Mol Biol , 833–842. [DOI] [PubMed] [Google Scholar]
- Horbay R, Bilyy R. (2016). Mitochondrial dynamics during cell cycling. Apoptosis , 1327–1335. [DOI] [PubMed] [Google Scholar]
- Jezek J, Cooper KF, Strich R. (2018). Reactive oxygen species and mitochondrial dynamics: the yin and yang of mitochondrial dysfunction and cancer progression. Antioxidants (Basel) , E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji WK, Hatch AL, Merrill RA, Strack S, Higgs HN. (2015). Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife , e11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia R, Wang RY, Yusuf A, Thomas PV, Agard DA, Shaw JM, Frost A. (2018). Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature , 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. (2002). Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol , 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhina S, Cooper KF, Strich R. (2014). Med13p prevents mitochondrial fission and programmed cell death in yeast through nuclear retention of cyclin C. Mol Biol Cell , 2807–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida H, Sugio S. (2013). Crystal structure of GTPase domain fused with minimal stalks from human dynamin-1-like protein (Dlpl) in complex with several nucleotide analogues. Curr Top Pept Protein Res , 67–77. [Google Scholar]
- Koirala S, Guo Q, Kalia R, Bui HT, Eckert DM, Frost A, Shaw JM. (2013). Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc Natl Acad Sci USA , E1342–E1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Yoon Y. (2016). Mitochondrial fission and fusion. Biochem Soc Trans , 1725–1735. [DOI] [PubMed] [Google Scholar]
- Leopold P, O’Farrell PH. (1991). An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell , 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Dulic V, Reed SI. (1991). Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell , 1197–1206. [DOI] [PubMed] [Google Scholar]
- Liesa M, Palacin M, Zorzano A. (2009). Mitochondrial dynamics in mammalian health and disease. Physiol Rev , 799–845. [DOI] [PubMed] [Google Scholar]
- Liu R, Chan DC. (2015). The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol Biol Cell , 4466–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loson OC, Liu R, Rome ME, Meng S, Kaiser JT, Shan SO, Chan DC. (2014). The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure , 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PJ, Francy CA, Stepanyants N, Lehman L, Baglio A, Mears JA, Qi X, Ramachandran R. (2016). Distinct splice variants of Dynamin-related protein 1 differentially utilize mitochondrial fission factor as an effector of cooperative GTPase activity. J Biol Chem , 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PJ, Stepanyants N, Mehrotra N, Mears JA, Qi X, Sesaki H, Ramachandran R. (2014). A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell , 1905–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon C, Suthiphongchai T, DiRenzo J, Ewen ME. (1999). P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc Natl Acad Sci USA , 5382–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendl N, Occhipinti A, Muller M, Wild P, Dikic I, Reichert AS. (2011). Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci , 1339–1350. [DOI] [PubMed] [Google Scholar]
- Miao Y, Han X, Zheng L, Xie Y, Mu Y, Yates JR, 3rd, Drubin DG. (2016). Fimbrin phosphorylation by metaphase Cdk1 regulates actin cable dynamics in budding yeast. Nat Commun , 11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. (2006). Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J Biol Chem , 2177–2183. [DOI] [PubMed] [Google Scholar]
- Neuman E, Ladha M, Lin N, Upton T, Miller S, DiRenzo J, Pestell R, Hinds P, Dowdy S, Brown M, Ewen M. (1997). Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol , 5338–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame LD, Singh AP, Stroud DA, Palmer CS, Stojanovski D, Ramachandran R, Ryan MT. (2016). Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J Cell Sci , 2170–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. (2010). Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol , 1141–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Elgass KD, Parton RG, Osellame LD, Stojanovski D, Ryan MT. (2013). Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J Biol Chem , 27584–27593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. (2011). MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep , 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YY, Cho H. (2012). Mitofusin 1 is degraded at G2/M phase through ubiquitylation by MARCH5. Cell Div , 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. (2008). The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA , 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R. (2018). Mitochondrial dynamics: the dynamin superfamily and execution by collusion. Semin Cell Dev Biol , 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter V, Palmer CS, Osellame LD, Singh AP, Elgass K, Stroud DA, Sesaki H, Kvansakul M, Ryan MT. (2014). Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. J Cell Biol , 477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter V, Singh AP, Kvansakul M, Ryan MT, Osellame LD. (2015). Splitting up the powerhouse: structural insights into the mechanism of mitochondrial fission. Cell Mol Life Sci , 3695–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider EV, Bottcher J, Blaesse M, Neumann L, Huber R, Maskos K. (2011). The structure of CDK8/CycC implicates specificity in the CDK/cyclin family and reveals interaction with a deep pocket binder. J Mol Biol , 251–266. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell , 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack S, Cribbs JT. (2012). Allosteric modulation of Drp1 mechanoenzyme assembly and mitochondrial fission by the variable domain. J Biol Chem , 10990–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. (2007). Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem , 11521–11529. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Kobayashi S, Fujiki Y. (2006). Peroxisome division is impaired in a CHO cell mutant with an inactivating point-mutation in dynamin-like protein 1 gene. Exp Cell Res , 1671–1684. [DOI] [PubMed] [Google Scholar]
- Tartoff KD, Hobbs CA. (1987). Improved media for growing plasmid and cosmid clones. Bethesda Res Lab Focus , 12. [Google Scholar]
- van der Bleik AM, Shen Q, Kawajiri S. (2013). Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol , a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Yan R, Cooper KF, Strich R. (2015). Cyclin C mediates stress-induced mitochondrial fission and apoptosis. Mol Biol Cell , 1030–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger J, Klinglmayr E, Frohlich C, Eibl C, Gimeno A, Hessenberger M, Puehringer S, Daumke O, Goettig P. (2013). Functional mapping of human dynamin-1-like GTPase domain based on x-ray structure analyses. PLoS One , e71835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. (2010). Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol , 872–884. [DOI] [PubMed] [Google Scholar]
- Yamashita SI, Jin X, Furukawa K, Hamasaki M, Nezu A, Otera H, Saigusa T, Yoshimori T, Sakai Y, Mihara K, Kanki T. (2016). Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J Cell Biol , 649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. (2012). Mitochondrial fission, fusion, and stress. Science , 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Liu T, Jin SB, Ning C, Lendahl U, Nister M, Zhao J. (2017). MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff. Sci Rep , 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. (2011). Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J , 2762–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.