Significance
Cyclin-dependent kinases are the main drivers of the mitotic cell cycle. Here, we show that the activity of the main cell-cycle regulator in the model plant Arabidopsis, CDKA;1, also governs one of the most important processes in meiosis: the formation of meiotic cross-overs. We show that CDKA;1 activity especially affects the major class of meiotic cross-overs, known as class I cross-overs. We find that lowering kinase activity leads to a progressive loss of cross-overs, of which cross-overs near the chromosome ends are the last to disappear. Conversely, an increase of kinase activity increases the rate of cross-over formation.
Keywords: meiosis, cyclin-dependent kinase, cross-over interference, class I cross-overs, meiotic recombination
Abstract
Little is known how patterns of cross-over (CO) numbers and distribution during meiosis are established. Here, we reveal that cyclin-dependent kinase A;1 (CDKA;1), the homolog of human Cdk1 and Cdk2, is a major regulator of meiotic recombination in Arabidopsis. Arabidopsis plants with reduced CDKA;1 activity experienced a decrease of class I COs, especially lowering recombination rates in centromere-proximal regions. Interestingly, this reduction of type I CO did not affect CO assurance, a mechanism by which each chromosome receives at least one CO, resulting in all chromosomes exhibiting similar genetic lengths in weak loss-of-function cdka;1 mutants. Conversely, an increase of CDKA;1 activity resulted in elevated recombination frequencies. Thus, modulation of CDKA;1 kinase activity affects the number and placement of COs along the chromosome axis in a dose-dependent manner.
CDKs in conjunction with their cyclin partners are the major driving force of progression through the cell cycle. CDKA;1, the single homolog of mammalian Cdk1 and Cdk2 present in Arabidopsis, is the main cell-cycle regulator of mitosis in Arabidopsis (1). Moreover, CDKA;1 appears to be a key regulator of meiosis as seen in hypomorphic mutants with very little kinase activity that are completely sterile due to several defects in early meiosis, including unequal distribution of chromosomes and premature termination of meiosis (2, 3). Interestingly, there is accumulating evidence that a reduction of Cdk activity is the underlying cause of the Ph1 locus in wheat that is responsible for the faithful pairing and recombination of homologs versus homeologous chromosomes in tetraploid and hexaploid wheat (4–6). Wheat is also an interesting example of a species with a distinct recombination pattern. In the case of wheat, the majority of cross-overs is strongly localized at chromosome ends leaving the pericentromeric regions largely devoid of recombination events (7). Because such patterns limit breeding approaches, there is a rising interest in the question as to how such patterns may be shaped (8, 9).
Results
A Hypomorphic CDKA;1 Allele Causes Reduction of Cross-Overs.
To overcome the early meiotic defects and the subsequent sterility of plants with little CDKA;1 activity allowing the analysis of CDKA;1 function in meiotic recombination, we scanned through a collection of transgenic lines expressing mutant alleles of CDKA;1 in cdka;1 background that were generated in our laboratory. The aim of this approach was to identify plant with intermediate CDKA;1 levels and hence less severe phenotypes as seen in the previously isolated hypomorphic mutants in which for instance a conserved Threonine in the T loop was exchanged with an Aspartate (CDKA;1T161D, or short D), or the exchange of a conserved Threonine and Tyrosine in the P loop with Aspartate and Glutamate (CDKA;1T14D;Y15E, short DE) (2, 3). Among these mutants, we found one allele, CDKA;1DBD, which grew similar to the wild type, especially compared with the small size of the two hypomorphic mutants D and DE (2, 3). CDKA;1DBD is a fusion of CDKA;1 with a genomic part of CYCLINB1;1 encoding a mutated form of the destruction box (Methods; DBD: destruction box-dead). Importantly, cdka;1DBD plants were partially fertile and a few viable seeds could be obtained per silique, indicating a less affected meiosis in comparison with the fully sterile D and DE plants (2, 3). Notably, cdka;1DBD is recessive and hence does not represent a neomorphic allele. The intermediate kinase activity levels of two independent cdka;1DBD transformant lines, i.e., between wild-type levels and the levels found in the D and DE mutants, were confirmed by in vitro kinase assays (SI Appendix, Fig. S1). Furthermore, the kinase assays also suggested that the two tested cdka;1DBD lines differ in kinase activity, with line #1 showing a higher kinase activity than line #2.
Cytological analysis of male meiocytes (pollen mother cells) of both cdka;1DBD lines revealed that early stages of meiotic prophase I are largely unaffected and homologous chromosomes pair, leading to cross-over (CO) formation (Fig. 1A and SI Appendix, Fig. S2). Chiasma formation between homologous chromosomes was verified in line #1 with the help of fluorescent in situ hybridization using 5S and 45S rDNA probes (SI Appendix, Fig. S3).
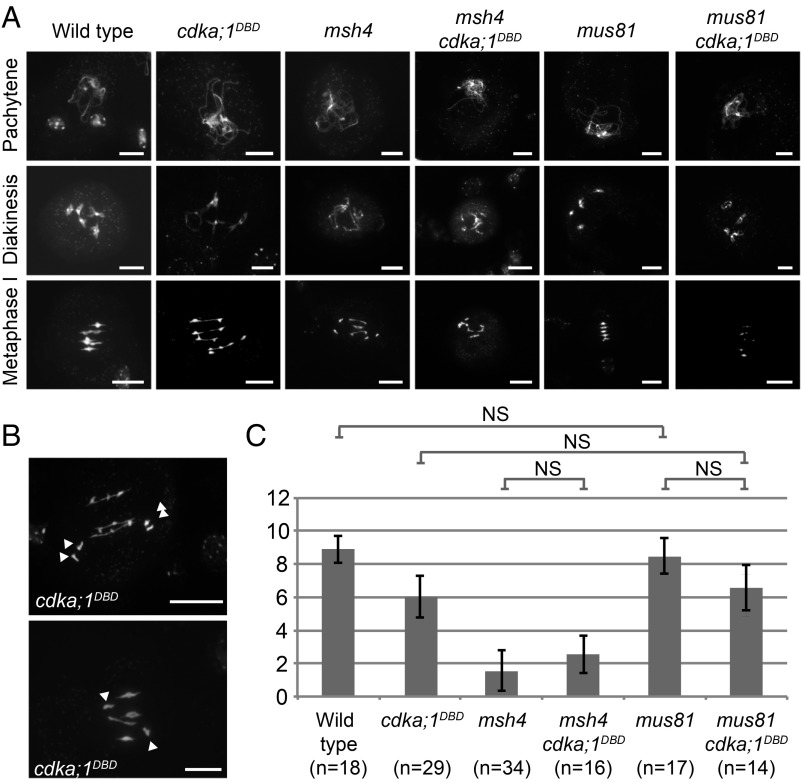
Fig. 1.
Meiotic prophase and CO formation in cdka;1DBD and double mutants. All images show DAPI-labeled DNA. (A) Prophase progression in the wild type, cdka;1DBD, msh4, mus81, and double mutants of both msh4 and mus81 with cdka;1DBD. All combinations show pachytene stages with apparently complete pairing (Top Row). Differences occur at diakinesis and metaphase I in which chiasma numbers differ, as witnessed by the number of ring and rod bivalents. (B) Two more examples of metaphase I in cdka;1DBD. (Top) Precocious segregation of two homolog pairs (white arrowheads). (Bottom) The presence of a univalent pair (two arrowheads highlight univalent pair). (Scale bars, 10 µm.) (C) Estimated chiasma numbers in metaphase I cells of the above-shown wild type, single, and double mutants. Single and double mutants all refer to cdka;1DBD inset line #1. Error bars represent SDs. NS: nonsignificant difference (α = 0.05) in a Bonferroni-corrected two-sample Kolmogorov–Smirnov test.
Differences with wild-type meiosis became noticeable at diplotene, with the occasional occurrence of univalents in 7 out of 28 cells in cdka;1DBD #1. Univalents were also observed in metaphase I with a frequency of 12 out of 40 cells. In addition, in metaphase I cells, an average of 2.5 rod- and 1.5 ring bivalents in cdka;1DBD #1 were observed (31 cells) while the wild type typically had 1.1 rod- and 3.9 ring bivalents per cell. Rod- and ring bivalents are indicative of the presence of at least one chiasma and two chiasmata, respectively. Together with the presence of univalents, the reduction in ring-bivalent frequency in cdka;1DBD #1 indicated a reduction of chiasma number. In addition, we observed several chromosomes at equidistant positions from the metaphase plane in 8 out of 35 cells (Fig. 1B). This could be an indication for an asynchronous segregation of homologous chromosomes. During later stages, meiosis progressed largely regularly except for the occasional formation of micronuclei and spores with unbalanced chromosome numbers that are suspected to result from random univalent segregation at anaphase I (SI Appendix, Fig. S2).
Meiotic progression in cdka;1DBD #2 is largely similar to cdka;1DBD #1, but differs in the number of observed chiasmata at metaphase I. Contrary to #1, all observed metaphase I cells (n = 20) showed the presence of univalent pairs and most observed bivalents (28 out of 32; 88%) are rods. With an average of 1.7 chiasmata per meiosis, cdka;1DBD #2 shows a lower chiasma frequency than cdka;1DBD #1 in which we observed 6.1 chiasmata per meiosis (n = 29). Taken together, both cdka;1DBD lines show a similar meiotic phenotype of reduced CO formation, with an apparent difference in chiasma frequency. Judged from chiasma frequency, cdka;1DBD #1 shows the best rescue of the cdka;1 mutant phenotype consistent with our kinase assays. Because of its better rescue, we used cdka;1DBD #1 for all consecutive experiments and refer to it as “cdka;1DBD.”
Class I COs Are Sensitive to Low CDKA;1 Activity.
The majority (85%) of COs in Arabidopsis are class I COs, which exhibit interference and are more distantly spaced to each other than expected by chance (10). The formation of class I COs involves, among others, the complex composed by the proteins MUT S HOMOLOG4 (MSH4) and MSH5. In addition, the complex formed by the DNA mismatch repair proteins MUT L HOMOLOG1 (MLH1) and MLH3 allows class I CO resolution. A smaller subset of COs (an estimated 10–15% in Arabidopsis) belongs to the class II that shows no interference and act in a pathway dependent on the endonuclease MMS AND UV SENSITIVE81 (MUS81) (11).
The very low number of COs in cdka;1DBD #2 suggested that likely type I COs are affected by CDKA;1 activity. To address further which class of COs is reduced in cdka;1DBD, we constructed double mutants of cdka;1DBD with msh4 as well as with mus81. By counting ring- and rod-bivalent frequencies in chromosome spreads (Fig. 1 A and B and SI Appendix, Fig. S2), we estimated chiasmata frequencies at metaphase I and tested for significant differences between the wild type and (double-) mutants using a Bonferroni-corrected Kolmogorov–Smirnov test (α = 0.05) (Fig. 1C). In both the wild type as well as a cdka;1DBD mutant background we note a significant reduction in the number of chiasmata when msh4 is mutated (i.e., when no class I COs can be formed). This suggests that the majority of the remaining chiasmata in cdka;1DBD are class I COs. Since we detected no difference in chiasmata number between the msh4 single- and the msh4 cdka;1DBD double mutants (both show about two chiasmata), the class II CO pathway seems functional in a cdka;1DBD background. Conversely, we did not find a significant reduction of chiasmata in cdka;1DBD mus81 double mutants in comparison with cdka;1DBD single mutants, indicating that the number of type II COs is at least not largely increased in cdka;1DBD.
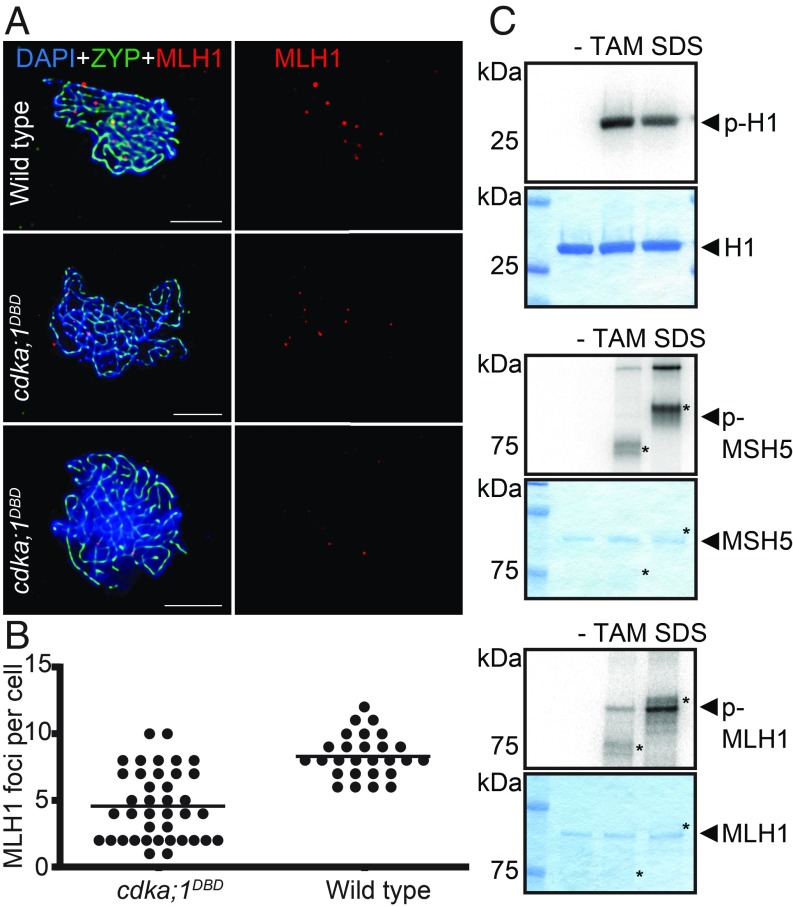
To directly assess a possible reduction of class I COs, we counted foci of the class I CO factor MLH1 in immunocytochemistry experiments. This showed a significant decrease in the number of MLH1 foci from on average 8.3 in the wild type to 4.5 in cdka;1DBD (t test, two-sided, P = 3.13 × 10−10) (Fig. 2 A and B). The observation that the number of MLH1 foci is lower than the estimated number of chiasmata as shown in Fig. 1C (8.3 vs. 8.9 in wild type and 6.0 vs. 4.5 in cdka;1DBD may be explained by the presence of class II COs that are not labeled by MLH1).
Fig. 2.
MLH1 localization and phosphorylation. (A) Fluorescent labeling of chromatin (DAPI, blue), the synaptonemal complex protein ZYP1 (anti-ZYP1 antibody, green), and class I CO protein MUTL HOMOLOG1 (anti-MLH1 antibody, red) in wild type (Top) and two cells of cdka;1DBD (Middle and Bottom) to determine the number of MLH1 foci per cell. (Scale bars, 5 μm.) (B) The number of MLH1 foci detected in cell spreads of wild type (Right) and cdka;1DBD (Left). Each dot represents one cell and vertical bars indicate the average. MLH1 foci occur in lower number in cdka;1DBD (average = 4.5) than in wild type (8.3) (t test, two-sided, P = 3.13 × 10−10) . (C) MLH1, but not MSH5 (both proteins marked by arrowheads), can be phosphorylated in vitro by CDKA;1 in combination with the meiotic cyclins TAM and in particular with SDS (both proteins marked by asterisks in their respective lanes). CDKA;1 has no activity without a cyclin partner (lane labeled with “−”). As positive control, histone H1 was used as a generic substrate and subjected to both CDK–cyclin combinations revealing approximately equal activity of these kinase complexes. Each panel shows the kinase activity by autoradiography in the top and Coomassie staining of the loaded proteins in the bottom.
While we cannot rule out an indirect effect, the reduction of class I COs may involve a direct action of CDKA;1 on the class I CO machinery. To explore this hypothesis further, we subjected factors involved in class I CO formation and resolution to in vitro kinase assays with CDKA;1. While MSH4 has no predicted CDKA;1 consensus phosphorylation site and was not assessed, MSH5 has two predicted phosphorylation sites and is a suggested phosphotarget of human Cdk1-CyclinA2 (12). While CDKA;1 in complex with either one of the meiotic-specific cyclins SOLO DANCERS (SDS) or TARDY ASYNCHRONOUS MEIOSIS (TAM) showed high activity against the generic substrate histone H1, MSH5 was not phosphorylated in our in vitro assay using both kinase complexes (Fig. 2). In contrast, we found that MLH1 is, at least in vitro, a target of especially CDKA;1-SDS and to lesser degree of CDKA;1-TAM complexes offering a possibility how CDKA;1 could directly regulate class I COs (Fig. 2C). Interestingly, it was previously found that MLH1 activity is reduced in wheat plants that contain the Ph1 locus (and presumably have reduced Cdk levels) albeit MHL1 loading onto chromatin in wheat appeared to be unaffected by Ph1 (13).
Recombination Patterns Change in cdka;1DBD.
Given the loss of class I COs in cdka;1DBD, we studied the recombination landscape of cdka;1DBD in a backcross design to determine the effect of CDKA;1 on CO positioning. To this end, we isolated an allele of cdka;1 in the Landsberg erecta (Ler) background, designated cdka;1–3, by screening an EMS-mutagenized Arabidopsis population for plants with defects in the number of nuclei in pollen since this is the predominant mutant phenotype of heterozygous cdka;1 mutants (14, 15). Next, cdka;1DBD#1 (in a Col-0 genetic background) was crossed with Ler heterozygous for cdka;1–3. The obtained Col-0*Ler F1 hybrid carrying both the Ler and the Col-0 cdka;1 alleles was used as a pollinator in a backcross with female tester lines (Col-0 and Ler) to generate 392 male-derived cdka;1DBD offspring. A wild-type Col-0*Ler F1 was used as pollinator to give rise to a male BC1 (“wild type”). Both populations were genotyped with 39 KASP SNP markers spanning the entire genome (SI Appendix, Fig. S4 and Dataset S1) (16).
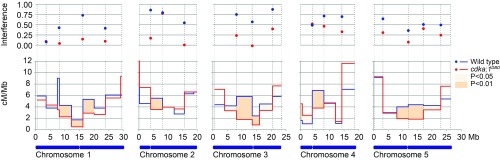
We first compared the recombination rate for all genetic intervals between the wild type and cdka;1DBD (Fig. 3) to assess qualitative differences between both populations. Of the 34 intervals, 12 intervals that are adjacent to centromeres show a significant reduction in recombination frequencies in cdka;1DBD in comparison with wild-type meiosis (χ2 test; P < 0.05).
Fig. 3.
Comparison of recombination rates and CO interference between wild type and cdka;1DBD for all five chromosomes. A blue horizontal bar depicts each chromosome and an indentation indicates the centromere position. For each chromosome, recombination rates are shown for the wild type (blue) and cdka;1DBD (red) in the lower graph as recombination rate (y axis, cM per Mb) for 34 intervals along the physical chromosome (precise marker positions are given in SI Appendix, Fig. S4). Intervals that significantly differ in recombination rates are indicated in orange (χ2 test; see legend). Note that recombination in cdka;1DBD is lower in all regions in which recombination rates significantly differ, except for the top of chromosome 4. Significant differences are only detected in regions adjacent to centromeres. The upper graph shows interference strength (Interference = 1 − c.o.c.) for 22 marker intervals (indicated by dashed vertical lines). A dot indicates interference strength between the intervals to the left and right of it. Note that some of the 34 smaller intervals were merged for interference calculations (see Methods and SI Appendix, Fig. S4).
Using the joinmap algorithm, we then estimate the total genetic map length of wild-type meiosis to be 532 cM (using the Kosambi mapping function), consistent with what was previously obtained for male meiosis in Col-0*Ler hybrids (16). The cdka;1DBD genetic map was 26% shorter, giving rise to a length of 394 cM (SI Appendix, Fig. S5). Our cytological observations indicated the presence of an average of 4.5 MLH1 foci (i.e., class I COs) per meiosis. If an average of about 2 COs formed through the class II CO pathway is added to these, a genetic map length of (50 × 6.5 =) 325 cM is expected. The slightly larger length of the experimental map could be explained by assuming that cells with low CO numbers (and higher likeliness of generating aneuploid gametes) are selected against, and do not give rise to offspring.
We hypothesized that if class I COs form a smaller proportion of the total of COs in cdka;1DBD, the remainder of COs in cdka;1DBD should show a reduction in interference. In wild-type meiosis, the CO distribution on four out of five chromosomes significantly differs from the Poisson distribution, whereas in cdka;1DBD plants only one chromosome significantly deviates from Poisson (χ2 test, P < 0.01). This suggests CO placement is indeed closer to random in cdka;1DBD (and thus less affected by interference) than in the wild type (SI Appendix, Fig. S6 and Table S1). Furthermore, estimating CO interference strength using the coefficient of coincidence (c.o.c.) showed lower interference values for 14 out of 17 intervals in comparison with the wild type (Fig. 3 and SI Appendix, Fig. S4).
Physical Chromosome Length Does Not Determine Map Length in cdka;1DBD.
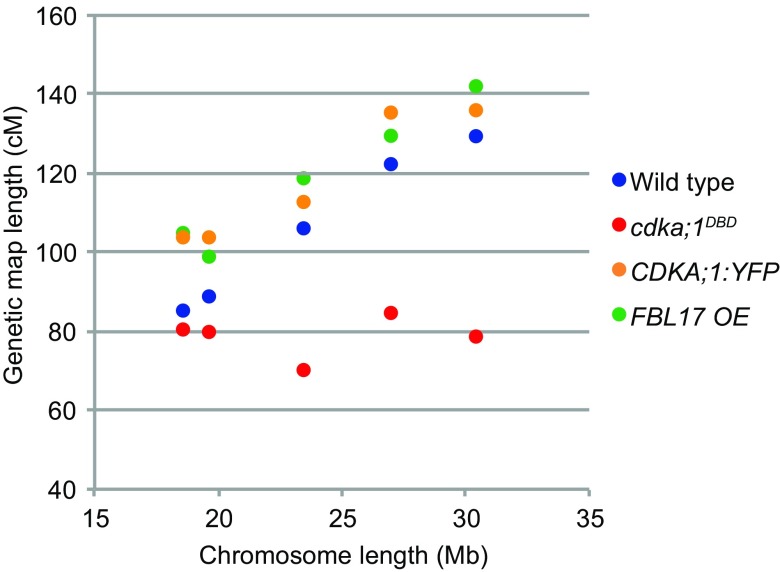
In the wild type, the genetic map length of the five chromosomes ranges from 85 to 130 cM and strongly correlates with the physical chromosome length, from 18.6 to 30.4 Mb (R2 = 0.98). Remarkably, we found that this relationship becomes uncoupled in cdka;1DBD (R2 = 0.01) and all chromosomes have approximately the same genetic length, i.e., between 79 and 84 cM. The short chromosomes 2 and 4 retain their genetic map length, whereas the large chromosomes 1, 3, and 5 had a strongly reduced genetic length (Fig. 4). This recombination decrease is strongly positively correlated with chromosome length (R2 = 0.84) (SI Appendix, Table S1). The longest chromosome reduces 39.2% in length whereas the shortest only 5.1%. To further verify that in cdka;1DBD there is no proportional decrease in CO frequencies for all chromosomes, we also compared the number of detected COs per chromosome pair between wild type and cdka;1DBD. With a proportional decrease, one would expect more nonrecombinant chromosomes to occur among the offspring. Shorter chromosomes in cdka;1DBD are not enriched for nonrecombinant chromatids, and all five chromosomes have near-identical fractions of nonrecombinant chromosomes in the offspring: 40, 41, 45, 41, and 39% for chromosomes 1 through 5, respectively (χ2 test, df = 4, P = 0.833) (SI Appendix, Table S2).
Fig. 4.
Genetic map length as a function of chromosome length for wild type, cdka;1DBD, CDKA;1:YFP, and FBL17 OE. Physical chromosome length (in Mb) is shown on the x axis, ordering chromosomes from left to right from the smallest chromosome 4 (18.8 Mb) through chromosomes 2, 3, and 5 to the largest chromosome 1 (right, 30 Mb). Corresponding genetic map lengths are on the y axis. Wild type shows a strong correlation between physical and genetic map length, which in cdka;1DBD is absent. In CDKA;1:YFP and FBL17 OE the correlation is strong, and map lengths are longer than wild type.
Taken together, our data suggest that all chromosomes receive identical CO numbers. Thus, we conclude that CO assurance is still in place in cdka;1DBD mutants and seems to operate rather independent from Cdk activity and the proper number of class I COs. Conversely, CDKA;1 action appears to link CO number with the physical chromosome length.
CDKA;1 Controls Class I COs in a Dose-Dependent Manner.
Finally, we wondered what the effect of increased CDKA;1 activity on the CO landscape would be. We therefore generated two Col-0*Ler hybrids containing transgenes increasing CDKA;1 activity. The first construct delivers, in addition to the wild-type CDKA;1 levels, a YFP-tagged CDKA;1 fusion protein, hence doubling the amount of CDKA;1 in planta as previously shown (17). The second construct FBL17 OE (over)expresses the F-box protein F BOX-LIKE FBL17 from a CDKA;1 promoter in addition to the endogenously expressed FBL17 (18). FBL17 mediates the proteolytic destruction of CDKA;1 inhibitors and hence causes higher CDKA;1 activity levels (18–21). These Col-0*Ler F1 hybrids were backcrossed as pollen donors to a male sterile (ms1) Ler line [CDKA;1:YFP population; n = 220; FBL17 OE population (n = 205) (Dataset S1)]. The CDKA;1:YFP and FBL17 OE populations had total genetic map lengths of 592 and 596 cM, showing recombination increases by 11 and 12%, respectively (Fig. 4). These lengths are significantly larger than the wild-type genetic map (one-tailed t test, unequal variance; P = 0.000374 and P = 0.000437, respectively). All chromosomes experience increases in CO frequencies ranging between 11 and 23%.
Here we have shown that CDKA;1 regulates CO formation in a dose-dependent manner and is especially crucial for the formation of class I COs. Lowering CDKA;1 activity reduces the total number of formed COs, and leads to an altered distribution of COs leading to reduced recombination in the centromere-proximal regions. In contrast, the number of distally placed COs is not altered in cdka;1DBD in the intervals studied. Interestingly, a CO reduction only predominantly occurs on the long chromosomes 1, 3, and 5, leaving the average number of CO on chromosomes 2 and 4 unaffected.
Given the preservation of MLH1 as well as Cdk1-type proteins, it is tempting to speculate that the control of class I CO number and positioning is conserved across eukaryotes. Moreover, it has been found that Cdk2 is often colocalized with MLH1 in mice (22). Thus, modulation of Cdk activity offers a possibility of altering CO number and/or positioning. Variations in the levels of CDKA;1-type proteins in species with agronomical importance may allow the in- or decrease of genetic map lengths using certain allelic combinations, with hardly any problems in meiosis and importantly, no obvious sporophytic growth defects as shown here.
Materials and Methods
Kinase Assays.
The kinase assays were carried out as described in ref. 23.
Generation of Constructs.
The cdka;1DBD plants express a fusion of CDKA;1 to a inactive (dead) destruction box (DB) of CYCLIN B1;1 (CYCB1;1) from the CDKA;1 promoter in a null cdka;1 −/− mutant background (cdka;1 PROCDKA;1:CYCB1;1DBD CDKA;1). The construct was generated using Pfu polymerase (fermentas) to fuse wild-type Arabidopsis CDKA;1 (At3g48750) cDNA and CYCLINB1;1 (At4g37490) genomic DNA by PCR. The fusion was flanked by Gateway attB1 and -2 sites and recombined in pDONR201 (Invitrogen). Primer combinations used to generate the CDKA;1 fragment were AACACAAGTTTGTACAAAAAAGCAGGCTTCAACAATGGATCAGTACGAGAAAG (primer 1) and AGAAGTCATCATAGGCATGCCTCCAAGATCCT (primer 2)and for the CYCLINB1;1 fragment we used GGAGGCATGCCTATGATGACTTCTCGTTCGATTGTTC (primer 3) and GGGGACCACTTTGTACAAGAAAGCTGGGTTCAAACAATCTTTTCTTTCTGTTTCTTCT (primer 4). The two fragments were fused in a final PCR using primers 1 and 4. The destruction box motif 30-RQVLGDIGN-38 was changed by two substitutions (C/Arg > G/Gly and C/Leu > G/Val) to 30-GxxVxxIxN-38 according to ref. 24 by site-directed mutagenesis on the previously described entry clone with PfuTurbo (Stratagene) using primers GTAGCGAAAGGAAGAAACGGTCAAGTTGTTGGTGATATCGGTAATGTTG and GAACAACATTACCGATATCACCAACAACTTGACCGTTTCTTCCTTTCGC to generate the cdka;1DBD variant. After sequencing, the obtained Gateway entry clones were recombined with the binary Gateway destination vector pAM-PAT-GW-ProCDKA;1 (14). Resulting expression vectors conferring phosphinothricin resistance were retransformed into Agrobacterium tumefaciens GV3101-pMP90RK and transformed into heterozygous cdka;1+/− (14) by floral dip. The PRO35S:FBL17 and the PROCDKA;1:CDKA;1:YFP plants were previously described (14, 18).
Plant Growth.
Seeds were disinfected by chlorine dioxide produced from HCl and sodium hypochlorite solution, sown on 1/2 MS Agar plates with sucrose and vernalized for 3–7 d as described by Wijnker et al. (16). Agar plates were then placed in a Percival growth chamber for germination, after which seeds were transplanted to soil under long day conditions with 16-h light (18 °C) and 8-h dark (16 °C), 60% humidity.
Cytogenetic Analysis.
Meiotic cell spreads and slides for fluorescent in situ hybridization were made from whole flower buds, using standard protocols (25). We used plasmid PTa71 of Triticum aestivum (26) as a probe for the coding sequences of the 18S–25S rDNA and plasmid PCt 4.2 containing the Arabidopsis full coding sequence of 5S rDNA (GenBank: M65137.1). These were directly labeled with 7-Diethylaminocoumarin-3-carboxylic acid succinimidyl ester (DEAC; Perkin-Elmer, http://www.perkinelmer.com) and Cyanine dye 3.5 (CY3.5; Amersham, https://www.gelifesciences.com/), respectively, and hybridized using a standard protocol for direct labeling as described in ref. 27. A Zeiss Axioplan microscope equipped with an epifluorescence filterset was used for analysis. Images were captured with a Photometrics Sensys 1,305 × 1,024-pixel CCD camera, and processed with ImageJ (https://imagej.nih.gov/ij/) and Adobe Photoshop.
MLH1 immunolocalization was performed as previously described (28). The following antibodies were used: anti-ZYP1 (rat, 1/1,000 dilution) and anti-MLH1 (rabbit, 1/500). Secondary antibodies were donkey anti-rat IgG FITC conjugated (Agrisera) diluted to 1:50 and goat anti-rabbit IgG Alexa Fluor 555 conjugated (Molecular Probes) diluted to 1:500. Image capture, analysis, and processing were conducted on pachytene cells. The genetic backgrounds of the cells were blinded until MLH1 signals were determined. We only considered ZYP1-associated foci. Image deconvolution was implemented using the iterative algorithm “Richardson–Lucy” (DeconvolutionLab2, ImageJ plugin). This tool improves the discrimination of the signals.
Genetic Analysis.
BC1 offspring were genotyped using a previously described markerset (16) based on codominant KASPar SNP probes (https://www.lgcgroup.com/products/kasp-genotyping-chemistry). Genetic maps were constructed using JoinMap (29) using regression mapping and a fixed order of makers based on known map positions (https://www.kyazma.nl/index.php/JoinMap/).
Interference Strength Calculations.
CO interference was calculated as interference = 1 − c.o.c. The c.o.c. was calculated as the observed number of double-CO events (in two adjacent genetic intervals) divided by the expected number of double CO. Intervals were chosen such that all intervals smaller than 15 cM (as calculated using JoinMap) were merged with their smallest neighboring interval, the only exception being the short arm of chromosome 4, in which a 9.3 interval was maintained, to have all chromosome arms represented. The 22 intervals are indicated in SI Appendix, Fig. S4.
Statistics.
Differences in chiasma frequencies between wild type and five (double) mutant lines [i.e., 5(5 + 1)/2 = 15 comparisons] were calculated using a Bonferroni-corrected two-sample Kolmogorov–Smirnov test. Significance level (α = 0.05) was adjusted to 0.05/15 = 0.0033 and rounded to 0.001. Critical D values were calculated using: critical value D = 1.95√(n1 + n2)/(n1·n2) in which n1 and n2 are the sample sizes of the two samples.
Supplementary Material
Acknowledgments
MLH1 and ZYP1 primary antibodies were kindly provided by Chris Franklin (University of Birmingham, United Kingdom). We acknowledge the help of Hans de Jong (Wageningen University, The Netherlands) with the interpretation of meiotic spread images. This work was funded in part through a European Molecular Biology Organization Long-term Fellowship (ALTF 679-2013) to E.W., a grant by the Spanish National Project (AGL2015-67349-P) to M.P., a Marie-Curie COntrol of Meiotic RECombination network FP7 ITN-606956 grant to P.P.-N., M.P., and A.S., as well as core funding of the University of Hamburg (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820753116/-/DCSupplemental.
References
- 1.Nowack M. K., et al. , Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22, 1030–1040 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Dissmeyer N., et al. , T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19, 972–985 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dissmeyer N., et al. , Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21, 3641–3654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Kaff N., et al. , Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: With deletion mutants and expression profiling. Ann. Bot. 101, 863–872 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths S., et al. , Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439, 749–752 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Greer E., et al. , The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat. Plant Cell 24, 152–162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandhu D., Gill K. S., Gene-containing regions of wheat and the other grass genomes. Plant Physiol. 128, 803–811 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidhu D., Gill K. S., Distribution of genes and recombination in wheat and other eukaryotes. Plant Cell Tissue Organ Cult. 79, 257–270 (2005). [Google Scholar]
- 9.Crismani W., Mercier R., What limits meiotic crossovers? Cell Cycle 11, 3527–3528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercier R., et al. , Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3,whereas the other one is not. Curr. Biol. 15, 692–701 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Higgins J. D., Buckling E. F., Franklin F. C., Jones G. H., Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Yokoo R., et al. , COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín A. C., Shaw P., Phillips D., Reader S., Moore G., Licensing MLH1 sites for crossover during meiosis. Nat. Commun. 5, 4580 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowack M. K., et al. , A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38, 63–67 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Iwakawa H., Shinmyo A., Sekine M., Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45, 819–831 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Wijnker E., et al. , Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant. Nat. Genet. 44, 467–470 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Nowack M. K., et al. , Bypassing genomic imprinting allows seed development. Nature 447, 312–315 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Gusti A., et al. , The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One 4, e4780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X., et al. , A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet. 8, e1002847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H. J., et al. , Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature 455, 1134–1137 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Noir S., et al. , The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-box protein FBL17. Plant Cell 27, 1461–1476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker S. M., et al. , Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13, 336–342 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Harashima H., Schnittger A., Robust reconstitution of active cell-cycle control complexes from co-expressed proteins in bacteria. Plant Methods 8, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weingartner M., et al. , Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell 16, 643–657 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross K. J., et al. , Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res. 5, 551–559 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Gerlach W. L., Bedbrook J. R., Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 7, 1869–1885 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X., et al. , Cross-species bacterial artificial chromosome-fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements. Genetics 180, 1319–1328 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armstrong S. J., Sanchez-Moran E., Franklin F. C., Cytological analysis of Arabidopsis thaliana meiotic chromosomes. Methods Mol. Biol. 558, 131–145 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Stam P., Construction of integrated genetic linkage maps by means of a new computer package: Join Map. Plant J. 3, 739–744 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.