Significance
The pattern of synaptic inputs and outputs of neurons is established during development and is maintained throughout adulthood. An unknown set of cellular mechanisms establishes and maintains these synaptic connections. We used Caenorhabditis elegans to discover genes that act in the pathway of axodendritic polarity of a particular well-studied neuron, the cholinergic motorneuron DA9. We discovered that heterochronic genes regulate neuronal polarity during development and in postmitotic neurons in adults. This work reveals that developmental timing genes regulate the polarity of these C.elegans motor neurons. Understanding at a mechanistic level how neuronal polarity pathways are maintained in adulthood suggests potential strategies for therapeutic intervention in neurodegenerative diseases and models of cognitive decline in vivo.
Keywords: heterochronic, miRNA, neuronal polarity
Abstract
Many neurons display characteristic patterns of synaptic connections that are under genetic control. The Caenorhabditis elegans DA cholinergic motor neurons form synaptic connections only on their dorsal axons. We explored the genetic pathways that specify this polarity by screening for gene inactivations and mutations that disrupt this normal polarity of a DA motorneuron. A RAB-3::GFP fusion protein that is normally localized to presynaptic terminals along the dorsal axon of the DA9 motorneuron was used to screen for gene inactivations that disrupt the DA9 motorneuron polarity. This screen identified heterochronic genes as major regulators of DA neuron presynaptic polarity. In many heterochronic mutants, presynapses of this cholinergic motoneuron are mislocalized to the dendrite at the ventral side: inactivation of the blmp-1 transcription factor gene, the lin-29/Zn finger transcription factor, lin-28/RNA binding protein, and the let-7miRNA gene all disrupt the presynaptic polarity of this DA cholinergic neuron. We also show that the dre-1/F box heterochronic gene functions early in development to control maintenance of polarity at later stages, and that a mutation in the let-7 heterochronic miRNA gene causes dendritic misplacement of RAB-3 presynaptic markers that colocalize with muscle postsynaptic terminals ectopically. We propose that heterochronic genes are components in the UNC-6/Netrin pathway of synaptic polarity of these neurons. These findings highlight the role of heterochronic genes in postmitotic neuronal patterning events.
The formation of synaptic connections between neurons and their targets is a key feature of the nervous system. Caenorhabditis elegans neurons with reproducible morphologies, locations, neurotransmitter types, and patterns of connectivity are generated using a variety of spatially reiterated patterns of neuroblast divisions during development (1). As one of many specialized features of each neural type, axons and dendrites of particular neurons are programmed to become functionally and molecularly distinct specialized signaling centers. These regional specializations are often maintained throughout life. C. elegans neuronal polarity is also specified in some regions by the spatial placement of molecular cues, such as the UNC-6/Netrin extracellular positioning protein that is expressed by asymmetrically localized signaling centers and read by the UNC-5/Netrin receptor protein in neurons and other polarized tissues (2). Much of the C. elegans neuronal polarity generated during development is maintained throughout adulthood.
The C. elegans cholinergic motor neuron DA9 has been used to study synaptic polarity (3). Netrin signaling plays a key role in the establishment and maintenance of DA9 neuron polarity (4). Upon binding to its receptor, UNC-5 expressed in the DA9 neuron netrin, triggers a signaling cascade that leads to reorganization of DA9 membrane and cytoskeletal components (5). Genes that act in the same DA9 neuronal polarity pathway as the UNC-5/Netrin receptor and UNC-6/Netrin include cytoskeletal regulators of actin and microtubules (6, 7). Discovering additional genetic components that specify DA9 presynaptic polarity may reveal genes that are important for the maintenance of neuronal polarity in other neural classes as well. Understanding how polarity pathways are integrated into developmental pathways may reveal new avenues for therapeutics that maintain signaling networks. Defining genes that alter the maintenance of neuronal polarity might be important in neurodegenerative diseases and aging.
Heterochronic genes control the timing of developmental events to regulate a number of C. elegans neural developmental events. For example, the heterochronic gene lin-14 regulates the timing of synaptic remodeling during larval stage 1 (L1) in the GABAergic Dorsal D (DD) motorneurons (8). LIN-14 also regulates the timing of axon guidance in the AVM, PVM, and PVT neurons (9, 10), and axon degeneration in the PLM neurons (11). Another C. elegans heterochronic gene, the conserved let-7 miRNA gene, mediates the age-dependent decline in axon regeneration (12).
RAB-3 is a Ras GTPase localized to presynaptic vesicles of many C. elegans neurons (13). In the DA class of motoneurons, RAB-3::GFP is specifically localized to the dorsal axon of those neurons, where it mediates the cholinergic presynaptic signaling via neuromuscular junctions to neurite-like outgrowths of dorsal muscles (14). Here we report results from an RNAi screen to identify genes that disrupt the normally dorsal polarity of presynaptic RAB-3::GFP localization in the C. elegans DA9 neuron. We interrogated a cherry-picked library of 135 RNAi-based gene inactivations, including genes that cause premature aging (15). Among the most potent gene inactivations that disrupt RAB-3::GFP polarity in the DA9 neuron was blmp-1. blmp-1 encodes a transcription factor, which regulates the timing of a dorsal migration of the distal tip cells of the hermaphrodite gonad by inhibiting precocious unc-5 and lin-29 expression (16). In addition to blmp-1, we demonstrate that other heterochronic genes regulate the normally dorsal localization of DA9 neuron presynaptic signaling specializations. We propose that the heterochronic genes that regulate the gonad migration are also significant components in the same UNC-6/Netrin pathway of synaptic polarity of DA9 neurons during development.
Results
An RNAi Screen Reveals Heterochronic Gene Control of Synapse Specification.
DA9 is a cholinergic motor neuron that is comprised of easily identified compartments. Its cell body resides ventrally and extends an anteriorly oriented dendrite and a posteriorly oriented axon that via a commissure extends into the dorsal nerve cord (14). In the dorsal nerve cord, DA9 forms ∼25 en passant presynaptic specializations to neuromuscular junctions in neuron-like processes with postsynaptic specializations that extend from C. elegans dorsal muscle cells (SI Appendix, Fig. S1B). The ventral and dorsal compartments of DA9 are molecularly distinct, and their pattern is well characterized (4). The axon and dendrite of DA9 neuron are highly specialized: the axon is long and contains presynaptic vesicle proteins and active zone markers, whereas the dendrite is short and expresses dendritic postsynaptic signaling proteins (4, 17). Thus, this simple neuron serves as an excellent model to uncover genes and pathways that establish and maintain neuronal polarity.
To identify gene inactivations that disrupt the normal specialization of synapse localization, we visualized presynaptic specializations in the C. elegans cholinergic motor neuron DA9 (3) and scored for RNAi gene inactivations that disrupt this polarity. To directly visualize the DA9 synaptic specializations, a RAB-3::GFP fusion protein that is normally localized to presynaptic puncta in C. elegans neurons was expressed in the DA9 neuron under the control of a DA9-specific promoter itr-1 (18). To identify genetic pathways that specify synaptic placement in the DA9 motorneuron, we conducted an RNAi screen for gene inactivations that disrupt the normal localization of RAB-3 to the dorsal region of DA9 that mediates cholinergic synaptic signaling to dorsal muscles. We used a cherry-picked RNAi library composed of 135 RNAi clones that shorten the lifespan of long-lived daf-2/insulin receptor mutant C. elegans (15). These gene inactivations that cause premature aging were candidates to accelerate the loss of neural function, including synaptic polarization, which occurs during aging. These progeric gene inactivations had also been characterized for defects in movement and many of the identified genes were annotated to mediate neural control of locomotion (15).
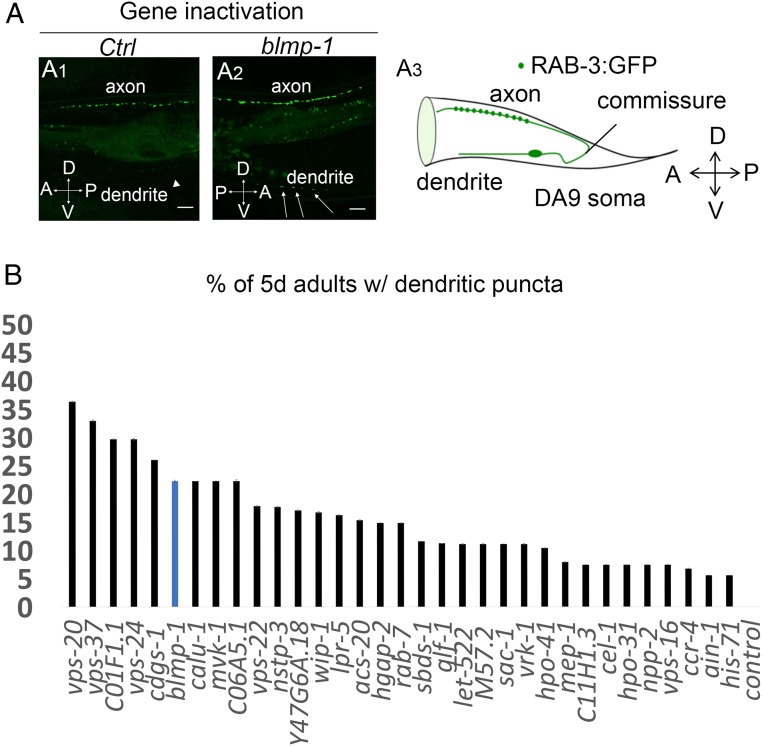
To enhance neuronal sensitivity to RNAi in our screen, we crossed the RAB-3::GFP presynaptic reporter into a strain carrying the RNAi hypersensitive mutations nre-1(hd20)lin-15b(hd126). We scored animals for ectopic placement of synaptic vesicles in the ventral dendrite of DA9 and for changes in the density and fluorescence of synaptic puncta in the dorsal axon (SI Appendix, Fig. S1A). We screened 5-d-old adult animals to be able to score for defective maintenance of synapse placement during aging, as well as defective initial placement of synapses during development. One of the most effective gene inactivations that disrupted RAB-3 localization was blmp-1. Inactivation of BLMP-1 potently affected RAB-3 localization (Fig. 1A). Like the blmp-1 gene inactivation by RNAi, a blmp-1 mutation also caused ectopic placement of presynaptic puncta in the dendrite of DA9 neuron (Fig. 2 C and D) [33% ectopic puncta in blmp-1(s71) mutants vs., 2% ectopic puncta in wild type]. Perhaps as a consequence of the inappropriate ventral placement of puncta, the intensity of synaptic puncta on the dorsal side, where they are normally specifically localized, was lower in the blmp-1 mutant compared with wild type (SI Appendix, Fig. S3). In addition to blmp-1, of the 135 progeria genes tested, we identified 19 other gene inactivations that cause ectopic dendritic localization of RAB-3 synaptic components in more than 20% of animals tested (Fig. 1B and SI Appendix, Table S1); many of these genes mediate endosomal vesicular trafficking (SI Appendix, Fig. S2 and Table S1). We focused on blmp-1 because it is a heterochronic gene that regulates developmental timing events. C. elegans blmp-1 (the ortholog of mammalian B lymphocyte-induced maturation protein 1) encodes a zinc finger and SET domain-containing protein (16, 19). In humans, BLIMP-1 acts as a repressor of β-IFN gene expression (20). C. elegans BLMP-1 regulates the dorsal migration of distal tip cells of the gonad during development (16, 19). Blmp-1 is a transcriptional regulator that affects the late events in developmental timing circuits, including those of migratory cells.
Fig. 1.
Identification of gene inactivations that modify GFP::RAB-3 localization in the DA9 motor neuron. (A1–A2) blmp-1 RNAi causes misplacement of presynaptic marker RAB-3 in the dendrite of the DA9 neuron. Representative linescans of Pitr1:rab-3:gfp;nre-1(hd20)lin-15b(hd126) transgenic animals on empty vector or control (ct) RNAi and blmp-1 RNAi. Arrows indicate ectopic dendritic GFP::RAB-3 presynaptic puncta. Arrowhead indicates the dendrite in animals treated with control RNAi. A, anterior; P, posterior; D, dorsal; V, ventral. Images were taken at the 5-d-old adult stage. (Scale bars, 10 μm.) (A3) On the left side, the DA9 cell body extends a dendrite anteriorly along the ventral side and an axon extends via a commissure to the dorsal side. En passant presynaptic terminals (green dots) are formed within a discrete axonal region. A, anterior; D, dorsal; P, posterior; V, ventral. (B) Gene inactivations that cause ectopic dendritic localization of the RAB-3 presynaptic marker in the DA9 neuron. The plotted x axis corresponds to the median values of three independent experiments. Data are represented relative to the empty vector control as the mean ± SEM of three biological replicates.
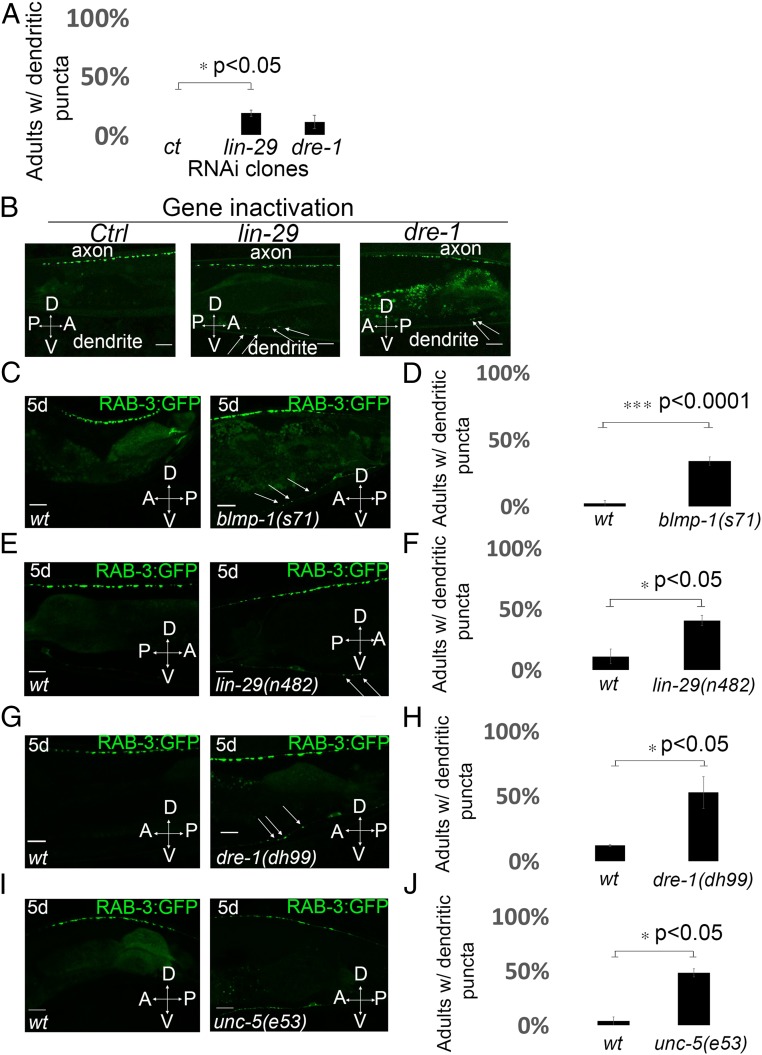
Fig. 2.
Heterochronic mutations disrupt RAB-3::GFP localization in the DA9 neuron. (A) Quantification of ectopic dendritic localization phenotype of the RAB-3 puncta in transgenic animals treated with RNAi of heterochronic genes. n = 32–39 worms in each condition. *P < 0.05, two-tailed Student’s t test. Error bars represent SEM. (B) Representative linescans of Pitr1:rab-3:gfp;nre-1(hd20)lin-15b(hd126) transgenic animals after knockdown of heterochronic genes by RNAi at 20 °C. Animals were scored for ectopic localization of GFP::RAB-3 puncta in the dendrite. Arrows mark the ectopic RAB-3 presynaptic puncta in the dendrite of the DA9 neuron. A, anterior; D, dorsal; P, posterior; V, ventral. (Scale bars, 10 μm.) (C and D) Loss of blmp-1 causes ectopic dendritic localization of RAB-3 presynaptic marker in 5-d-old adult animals. (C) Confocal images of synaptic vesicle associated protein GFP::RAB-3 pattern in wild-type and blmp-1(s71) mutants. Arrows indicate ectopic dendritic RAB-3::GFP presynaptic puncta. (Scale bars, 10 μm.) (D) Quantification of ectopic dendritic localization phenotype of the RAB-3 puncta in wild-type and blmp-1(s71) mutants. n = 45–47 worms per genotype. ***P < 0.0001, two-tailed Student’s t test. Error bars are SEM. (E and F) lin-29(n482) mutants show dendritic GFP::RAB-3 puncta localization in 5-d-old adults. (E) Confocal images of the pattern of synaptic vesicle associated protein GFP::RAB-3 in wild type and lin-29(n482) mutants. Arrows indicate ectopic dendritic RAB-3::GFP presynaptic puncta. (Scale bars, 10 μm.) (F) Quantification of ectopic dendritic localization phenotype of the RAB-3 puncta in wild type and lin-29(n482) mutants. n = 43–47 worms per genotype. *P < 0.05, two-tailed Student’s t test. Error bars are SEM. (G and H) dre-1(dh99) lof mutants show ectopic dendritic GFP::RAB-3 puncta in 5-d-old adult animals. (G) Linescans of representative wild-type or dre-1(dh99) mutant 5-d-old adults expressing GFP::RAB-3 presynaptic puncta. Arrows indicate ectopic dendritic GFP::RAB-3 presynaptic puncta. (Scale bars, 10 μm.) (H) Quantification of ectopic dendritic localization phenotype of the RAB-3 puncta in wild type and dre-1(dh99) mutants. n = 34–36 worms per genotype. *P < 0.05, two-tailed Student’s t test. Error bars are SEM. (I and J) Loss of unc-5 function causes ectopic dendritic GFP::RAB-3 puncta localization in 5-d-old adult animals. (I) Representative linescans of wild-type and unc-5(e53) mutant animals expressing GFP::RAB-3. (Scale bars, 10 μm.) (J) Quantification of the phenotype of GFP::RAB-3 localization in DA9 in unc-5(e53) mutant animals. n = 27 animals per genotype. *P < 0.05, two-tailed Student’s t test. Error bars are SEM.
C. elegans neuronal polarity in particular postmitotic neurons is regulated by the asymmetrically expressed spatial cue, UNC-6/netrin, which is detected by the UNC-5/Netrin receptor. The polarity of the DA9 neuron is disrupted by mutations in unc-5/Netrin receptor or the unc-6/Netrin gene (21). These genes mediate initial placement of synaptic components early in development, as well as the maintenance of that polarity throughout adulthood (21). The netrin receptor protein UNC-5 is expressed in the DA9 neuron early in development and accumulates in dendrites to exclude presynaptic components from the dendrite (4). UNC-5 is also expressed throughout adulthood and it is required to maintain neuronal polarity (4). Temporally regulated expression of the netrin receptor UNC-5 in the distal tip cell, a somatic cell that guides the migration of the developing germline, is also required for the dorsal migration of the distal tip cells in the hermaphrodite gonad during development (22, 23). Given that heterochronic genes control the timing of distal tip cell migration in the hermaphrodite gonad by the timely expression of the netrin guidance receptor unc-5, we hypothesized that heterochronic genes might affect the polarity of DA9 neurons in adults.
Given the strong effect of BLMP-1 on synaptic polarity and the precedent for heterochronic gene control of netrin signaling, we examined other heterochronic mutations, including lin-42, daf-12, lin-29, and dre-1, which affect developmental timing in the gonadal distal tip cells (16). Inactivation by RNAi of heterochronic genes (such as lin-29 and dre-1) caused the assembly of ectopic ventral presynapses, as reported by ectopic placement of RAB-3::GFP puncta in the ventral dendrite of the DA9 neuron (Fig. 2B, quantified in Fig. 2A). In the lin-29 heterochronic mutant, RAB-3::GFP was similarly mislocalized to the dendrite in 40% of animals tested, far more than wild type (Fig. 2E, quantified in Fig. 2F). lin-29 encodes a zinc-finger transcription factor (24) that is first expressed in the early L4 larval stage, where it mediates seam cell differentiation; LIN-29 is also expressed in particular neurons but its role there is unknown (25). LIN-29 expression is also maintained in the adult (26). The RAB-3::GFP localization defect in the DA9 neuron in adults hints at a late-stage neural role for LIN-29 function.
C. elegans dre-1 encodes a conserved F box protein that functions in an SCF E3-ubiquitin ligase complex to regulate the transition to adult hypodermal programs (27). BLMP-1 is a substrate of the DRE-1 complex (19). We tested whether a dre-1 mutation affected the localization of the RAB-3 presynaptic marker. We generated a CRISPR mutant allele of dre-1(dh99) in the RAB-3::GFP reporter strain. At the adult stage, the dre-1 mutant mislocalized RAB-3::GFP in the dendrite of DA9 (Fig. 2 G and H) [52% ectopic dendritic presynaptic puncta in dre-1(dh99) mutants vs. 11% ectopic puncta in wild type].
Mutations in some heterochronic genes cause reiterations of earlier developmental events, a retarded phenotype, whereas other heterochronic mutants cause a skipping of developmental events, a precocious phenotype (28, 29). Although dre-1(dh99) and blmp-1(s71) cause retarded and precocious distal tip cell migrations, respectively, they have the same DA9 polarity defect: RAB-3::GFP puncta were mislocalized in the DA9 dendrite in both blmp-1(s71) and dre-1(dh99) mutant animals (SI Appendix, Fig. S4B). The double-mutant dre-1(dh99);blmp-1(s71) showed a frequency of ectopic dendritic RAB-3::GFP puncta similar to either single mutant, suggesting that these genes act in the same pathway for DA9 polarity (SI Appendix, Fig. S4).
One possible explanation for the effect of heterochronic mutations on the polarity of the DA9 neuron is that heterochronic genes act in the same pathway as the netrin receptor, unc-5 in the DA9 neuron, similar to its regulation in the gonad during the migration of distal tip cells. In unc-5–null mutants, presynaptic specialized puncta were displaced to the ventral side of animals in 48% of animals tested (Fig. 2I, quantified in Fig. 2J and described ref. 4). To test whether BLMP-1 acts in the same pathway as the UNC-5/Netrin receptor to affect synaptic polarity of DA9 neuron, we analyzed unc-5(e53);blmp-1(s71) double-mutant 5-d-old adult animals. The unc-5(e53);blmp-1(s71) double mutant showed a much stronger DA9 ventral synaptic puncta defect than either single mutant: 11% of unc-5(e53) mutants show axon guidance defect and abnormal clustering of RAB-3::GFP presynapses, the blmp-1(s71) mutant shows no axon guidance defect, whereas 60% of unc-5(e53);blmp-1(s71) double mutant shows an axon guidance defect and abnormal placement of RAB-3::GFP presynaptic specializations (SI Appendix, Fig. S5). This aberrant axon positioning and placement of DA9 presynapses in unc-5(e53);blmp-1(s71) double mutants was also present at the L4 developmental stage (SI Appendix, Fig. S6). This result indicates that there is a strong genetic interaction between the two blmp-1 and unc-5–null mutations during axon guidance of the DA9 neuron, suggesting that these genes act in parallel pathways.
Developmental Timing Genes Control RAB-3 Localization at Early Developmental Stages.
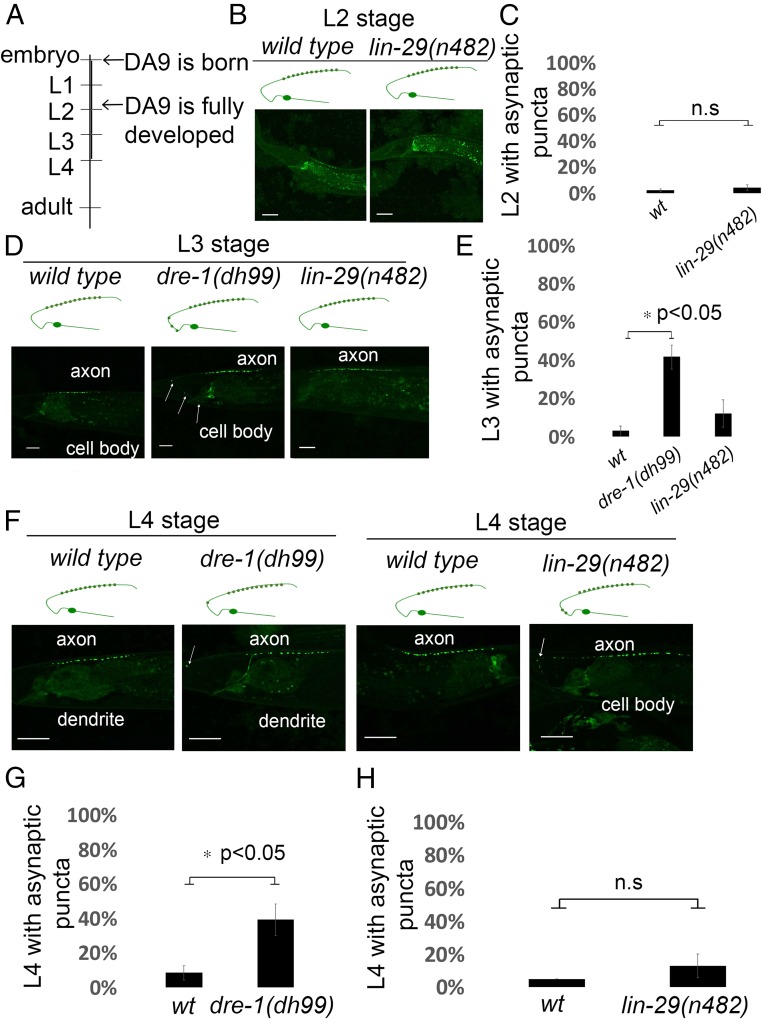
We also observed the placement of DA9 synaptic specializations in wild-type and heterochronic mutants at earlier larval stages to discern whether the heterochronic genes specify the initial placement of synapses or the maintenance of those specializations. The DA9 neuron is generated during embryonic development and by the end of the L2 larval stage is fully developed (Fig. 3A). We first observed the dorsal RAB-3::GFP placement at the L2 stage in wild type or lin-29(n482). We observed no difference in the dorsal localization of GFP::RAB-3 puncta at either the L2 or L3 stages in the lin-29 mutant compared with wild type (Fig. 3 B–E). This is not surprising because the earliest expression of LIN-29 is during the L4 larval stage (26). At the L4 larval stage, lin-29(n482) mutants showed a small but not significant shift of GFP::RAB-3 puncta into the dorsal asynaptic area compared with wild-type animals (Fig. 3F, quantified in Fig. 3H).
Fig. 3.
Testing heterochronic mutants for DA9 neuron GFP::RAB-3 polarity defects at early developmental stages. (A) A diagram depicting the timeline of the DA9 neuron development. Phenotypes of heterochronic mutants were scored at larval stages L2, L3, and L4. (B and C) Localization of GFP::RAB-3 in wild-type and lin-29(n482) animals at the L2 larval stage. In both wild-type and lin-29(n482) mutant animals, RAB-3 is localized at the dorsal side in the axonic region of DA9. (B) Schematic diagrams showing phenotypes are displayed above each image. (Scale bars, 10 μm.) (C) Quantification of the localization of GFP::RAB-3 of wild-type and lin-29(n482) animals at L2 larval stage. Images were generated by a confocal microscope. n = 37–47 worms per genotype. n.s is not significant. Error bars are SEM. (D and E) Animals expressing GFP::RAB-3 specifically in the DA9 (wyIs85) in wild type, dre-1(dh99), and lin-29(n482) mutants at L3 larval stage. In dre-1(dh99) mutants there is localization of GFP::RAB-3 in the normally asynaptic domain of DA9 as shown in D. Arrows indicate ectopic GFP::RAB-3 in DA9. Linescans were generated by a confocal microscope. Schematic diagrams showing phenotypes are displayed above each image. (Scale bars, 10 μm.) (E) Quantification of the GFP::RAB-3 localization at the asynaptic domain at L3 larval stage. n = 26–38 worms per genotype. *P < 0.05, two-tailed Student’s t test. Error bars are SEM. (F–H) Localization of RAB-3 puncta in wild type, dre-1(dh99), and lin-29(n482) mutants at L4 larval stage. (F) Representative confocal images of animals expressing the synaptic vesicle marker GFP::RAB-3 in wild type, dre-1(dh99), and lin-29(n482) mutants. Arrows indicate the ectopic GFP::RAB-3 puncta at the asynaptic domain of DA9. (Scale bars, 10 μm.) Schematic diagrams showing phenotypes are displayed above each image. (G) Quantification of GFP::RAB-3 localization along the DA9 in wild type and dre-1(dh99) at L4 larval stage. n = 54–60 worms per genotype. *P < 0.05, two-tailed Student’s t test. Error bars are SEM. (H) Quantification of GFP::RAB-3 localization along the DA9 in wild type and lin-29(n482) at L4 larval stage. n = 60 worms per genotype. n.s is not significant, two-tailed Student’s t test. Error bars are SEM.
Given that dre-1 mutant animals had the most penetrant RAB-3 dendritic localization defect at the adult stage, we determined at which larval stage the defect arises. We observed that the dre-1 mutant showed defects in RAB-3 dendritic localization at the L3 stage (Fig. 3D, quantified in Fig. 3E). Presynaptic puncta were mislocalized in the normally asynaptic ventral domain of the DA9 neuron in 40% of dre-1(dh99) L3 stage mutant animals compared with 3% in L3 stage wild type (Fig. 3 D and E). After the L3 stage, RAB-3 synaptic puncta continued to mislocalize at the asynaptic region of DA9 in the dre-1 mutant (Fig. 3F). RAB-3 was mislocalized in 39% of dre-1(dh99) mutant animals at L4 larval stage compared with 8% in L4 stage wild-type animals (Fig. 3F, quantified in Fig. 3G). Thus, the synaptic mislocalization of dre-1 mutants arises before the L3 larval stage.
lin-29 activity is required to trigger hypodermal seam cell terminal differentiation at the L4 stage (26); lin-29 mutants display a retarded phenotype or delayed transition to adulthood (28, 30). Mutations in other heterochronic genes, such as lin-4, lin-14, lin-28, and lin-42 alter the timing of seam cell terminal differentiation but act at earlier larval stages (28, 31). Mutations in lin-14, lin-28, and lin-42 cause precocious adult stage differentiation, resulting in “larvae” with adult cuticle (28). Sequential activities of heterochronic genes at earlier larval stages culminates in the adult-stage expression of LIN-29 transcription factor. We tested whether additional heterochronic genes also specify the pattern of presynapses in DA9 neuron in animals that are 5 d old.
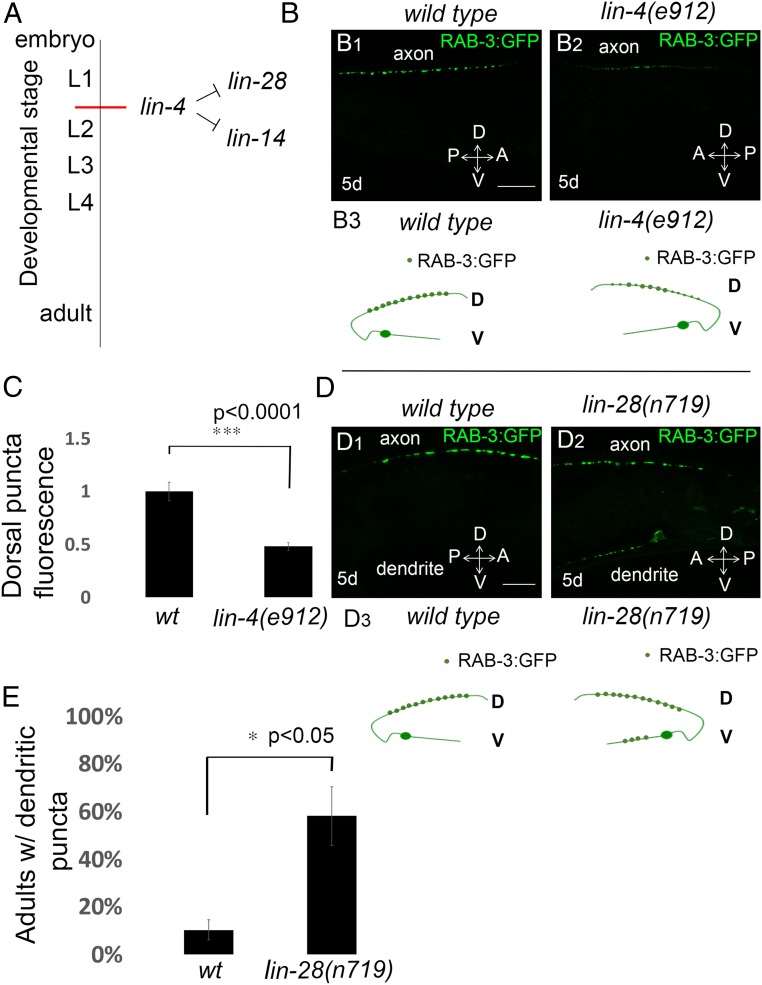
The lin-4 miRNA directs hypodermal seam cell differentiation at the end of the first larval stage through negative regulation of its mRNA targets lin-28/lin-14 (Fig. 4A) (28, 30, 32). We assessed whether a lin-4–null mutant, lin-4(e912) causes ectopic dendritic localization of RAB-3 presynaptic marker or affected dorsal RAB-3 puncta. The lin-4(e912) mutation caused a 50% reduction in dorsal GFP::RAB-3 compared with wild type (Fig. 4B, quantified in Fig. 4C). We also tested whether lin-28 heterochronic mutation affects the localization of RAB-3 presynapses in the DA9 neuron. A lin-28 mutation caused inappropriate positioning of presynaptic puncta in the dendrite of DA9 neurons in 58% of animals tested compared with wild type (Fig. 4D, quantified in Fig. 4E). The above genetic results suggested that heterochronic genes differentially regulate RAB-3 presynaptic localization.
Fig. 4.
Heterochronic mutants differentially regulate synapse localization in the DA9 neuron at the adult stage. (A) A diagram showing the timing of expression and the lin-28 and lin-14 mRNA targets of lin-4 miRNA during larval development. (B) Dorsal presynapses have decreased fluorescence in the dorsal side in lin-4(e912) mutants compared with wild type. Note the reduced GFP::RAB-3 intensity in the dorsal processes in lin-4(e912) mutants (B2), compared with wild-type animals (B1). D, dorsal processes; V, ventral processes; Schematic diagrams are illustrated in B3. (Scale bars, 10 μm.) (C) Average intensity quantification of fluorescence intensity for dorsal GFP::RAB-3 at 5 d old adult stage; n = 27 worms in each genotype. ***P < 0.0001, Student’s t test. Error bars are SEM. (D) Wild-type animals expressing GFP::RAB-3 in DA9 motor neurons (wyIs85) at 5-d-old adult stage (D1). lin-28(n719) mutants show ectopic dendritic localization of GFP::RAB-3 puncta at 5-d-old adult stage (D2). Schematic diagrams showing phenotypes are displayed below each image. Green dots, GFP::RAB-3; D, dorsal processes; V, ventral processes. Schematic diagrams are illustrated in D3. (Scale bars, 10 μm.) (E) Quantification of ectopic dendritic RAB-3 puncta localization phenotype. n = 38–39 worms in each genotype. *P < 0.05, Error bars are SEM.
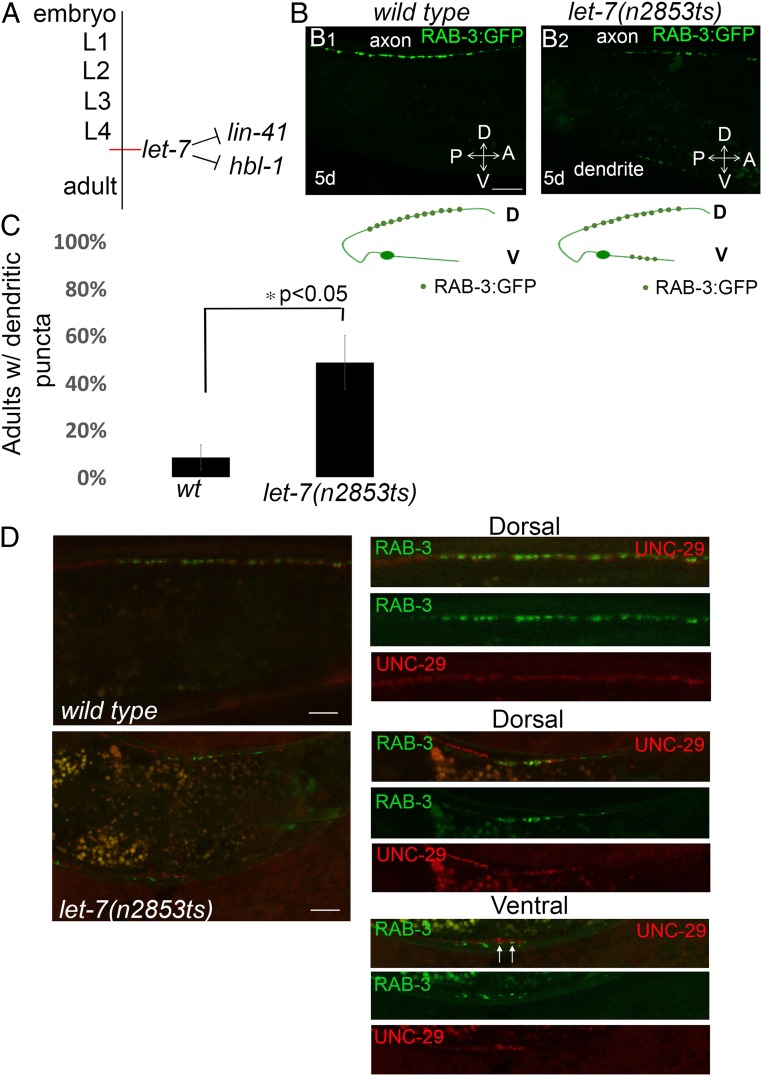
We also tested whether other heterochronic mutants that control the later larval-to-adult transition also control neuronal polarity of the DA9 neuron at the adult stage. The microRNA let-7 directs hypodermal seam cell divisions from developmental stage L4 to adulthood through negative regulation of its mRNA targets hbl-1 and lin-41 (Fig. 5A) (33–36). Similar to the lin-29 mutant, let-7 mutant animals show retarded phenotypes: the let-7 mutants display incomplete hypodermal seam cell divisions and poor or no adult alae, whereas in lin-29 mutants these events fail entirely (33, 35, 37). We tested the RAB-3::GFP localization at the adult stage in the temperature-sensitive mutant let-7(n2853ts). We observed that in let-7(n2853ts) mutants synaptic vesicle proteins accumulated in the ventral processes of DA9 that is normally devoid of synaptic terminals [48% of let-7(n2853ts) adults at the nonpermissive temperature compared with 8% in wild type (Fig. 5B, quantified in Fig. 5C)]. This result indicates that the let-7 miRNA is required for the correct positioning of RAB-3 presynaptic marker in adults.
Fig. 5.
Ectopic ventral RAB-3 puncta in let-7(n2853ts) mutants colocalize with postsynaptic muscle acetylcholine receptors (UNC-29::RFP). (A) A diagram showing the timing of expression of let-7 miRNA and its targets lin-41 and hbl-1 during larval development. (B) Wild-type animals expressing GFP::RAB-3 in DA9 motor neurons (wyIs85) at 5-d adult stage (B1). let-7(n2853ts) mutants show ectopic dendritic localization of GFP::RAB-3 puncta in 5-d-old adult animals (B2). Schematic diagrams showing phenotypes are displayed below each image. Green dots, GFP::RAB-3; D, dorsal processes; V, ventral processes. (Scale bars, 10 μm.) (C) Quantification of ectopic dendritic RAB-3 puncta localization phenotype. n = 36 worms in each genotype. *P < 0.05, Student’s t test. Error bars are SEM. (D) Colocalization of DA9 presynaptic terminals (GFP::RAB-3) with postsynaptic muscle acetylcholine receptors (UNC-29::RFP). Wild-type or let-7(n2853ts) mutant animals coexpressing GFP::RAB-3 and UNC-29::RFP were imaged at 5-d-old adult stage. (Scale bars, 10 μm; scale bars apply to Right panels as well.) Arrows show the colocalization of presynaptic and postsynaptic terminals ectopically.
To test if the ectopic accumulation of presynaptic vesicles reveal functional synapses, we examined whether the ectopic synaptic vesicle puncta colocalize with muscle-specific acetylcholine receptor UNC-29, which constitutes the postsynaptic side of a motor neuron to muscle cholinergic synapse. To visualize the acetylcholine receptors, we used a transgenic strain expressing a knockin UNC-29-tagRFP L-AChR subunit (38). Using confocal linescans, we observed that the ectopic presynaptic puncta that accumulate in the ventral side of DA9 neuron colocalized with postsynaptic UNC-29 acetylcholine receptor in let-7(n2853ts) mutant animals (Fig. 5D). These results suggest that these ectopic accumulations of synaptic components in DA9 dendritic regions are functional synapses.
One explanation for the effect of heterochronic mutations on the polarity of the DA9 neuron is that LIN-29 may be expressed in DA9 neurons and the other heterochronic genes may act upstream of LIN-29. LIN-29 is expressed in multiple ventral cord neurons (25), where it may regulate the expression of the netrin receptor, similar to its regulation in the gonad during the migration of distal tip cells. LIN-29 expression in probable neurons, including the tail region near DA9, is located has been noted (26) and we verified this (SI Appendix, Fig. S7).
A cdk-5 Mutation Alters Localization of the RAB-3 Presynaptic Marker in the DA9 Neuron.
We screened another heterochronic mutant in our collection, in the transcription factor gene pqm-1, for defects in RAB-3 localization in the DA9 motorneuron. We analyzed this heterochronic mutant because pqm-1 regulates adult-specific vitellogenesis in a LIN-29–dependent manner (39). The strain RB711 carries a 1,470-bp deletion of the pqm-1(ok485) locus that was generated by UV mutagenesis. When we crossed the strain with the GFP::RAB-3 presynaptic reporter, we found that the pqm-1(ok485) lesion did not affect GFP::RAB-3 localization but surprisingly identified a locus unlinked to pqm-1 that disrupted the normal pattern of synapse formation in DA9. By SNP polymorphism mapping between N2 and Hawaiian strains (40), we mapped this mutation to the third chromosome. Whole-genome sequencing identified a 155-bp deletion that deletes the last codon of exon 4 of the gene cdk-5 [Chr III (13,779,739-13,782,893)] with an insertion of two bases (GA) (SI Appendix, Fig. S8) that causes a frameshift. This allele was named cdk-5(mg696). cdk-5 encodes a cyclin-dependent kinase-5 (CDK-5) that is known to regulate synaptogenesis in DA9 and DD motorneurons (3, 41).
Does the cdk-5 gene function in the heterochronic genetic pathway to specify DA9 polarity? We generated the double-mutant cdk-5(mg696);blmp-1(s71). cdk-5(mg696) mutant animals showed RAB-3::GFP localization in both dorsal and ventral sides in 100% of animals tested (SI Appendix, Fig. S9). We did not observe any difference in synaptic polarity of DA9 neuron in cdk-5(mg696);blmp-1(s71) mutants compared with cdk-5(mg696) mutants (SI Appendix, Fig. S9). This result indicates that BLMP-1 acts in the same pathway as CDK-5 to modulate DA9 synaptic polarity. We also tested for a genetic interaction between cdk-5 and unc-5 by observing a cdk-5(mg696);unc-5(e53) double mutant. We observed that in ∼50% of cdk-5(mg696);unc-5(e53) animals tested there was a “shift” in RAB-3 localization only to the ventral side, compared with cdk-5(mg696) mutants (SI Appendix, Fig. S10). This result indicates that unc-5 lof mutants affect the ability of CDK-5 to regulate DA9 synaptic polarity, and that like the other heterochronic mutants, cdk-5 acts in a pathway parallel to unc-5.
Behavioral Changes in 5-d-Old Heterochronic Mutants.
Worms can move forward toward attractive stimuli and backward away from aversive stimuli using distinct neural circuits. The motorneuron DA9 is required for backward locomotion. To test whether heterochronic mutants display defects in backward locomotion that may be associated with the DA9 polarity defects, we delivered tapping stimuli to the head of 5-d-old animals and recorded their backward locomotion. Without tapping, wild-type animals displayed a sinusoidal movement caused by waves of muscle contraction along the body axis (SI Appendix, Fig. S11A). blmp-1(s71) animals are smaller than wild type and displayed a sinusoidal wave with similar amplitude to wild type (SI Appendix, Fig. S11B). lin-29(n482) and dre-1(dh99) mutant animals also exhibited altered sinusoidal forward movement similar to wild type (SI Appendix, Fig. S11 C and D). After tapping the head, wild-type animals moved backward along the same trajectory as the preceding forward movement, but they also turned before they changed their direction of movement within 10 s after the initial tapping stimulus (SI Appendix, Fig. S11 F–F″). blmp-1(s71) mutants showed ∼70% reduced backward speed compared with wild type (SI Appendix, Fig. S11E). blmp-1 mutant animals also exhibited changes in backward locomotion: blmp-1(s71) animals moved backward along the same trajectory as the preceding movement but they did not turn and their path was shorter than wild type. This change in backward locomotion could be explained by the altered contraction of dorsal body wall muscles in response to a decrease in DA9 motorneuron synapses on the dorsal side. lin-29(n482) mutants after the tapping stimulus also responded with large reversals and displayed a slightly larger amplitude of sinusoidal movement compared with wild-type animals (SI Appendix, Fig. S11 H–H″). Backward locomotion was also altered in dre-1(dh99) mutants. Whereas dre-1 mutants turned after the tapping stimulus similar to wild-type animals, they also “coiled” before they changed their direction of movement (SI Appendix, Fig. S11 I–I″). We also measured the reversal distance and duration (SI Appendix, Fig. S12). The reversal speed of blmp-1(s71) mutant animals was reduced compared with wild type, lin-29(n482), and dre-1(dh99) mutants; they displayed a similar duration of their reversal behavior, but they covered a shorter reversal distance (SI Appendix, Fig. S12).
Discussion
We have identified heterochronic genes as regulators of synaptic polarity of the DA9 motorneuron. Mutations or gene inactivation by RNAi of heterochronic genes altered the localization of RAB-3 presynaptic marker in the DA9 neuron. RNAi of the blmp-1 heterochronic gene or other heterochronic genes, including lin-29 and dre-1, caused dendritic localization of the normally dorsal presynaptic marker RAB-3 (Figs. 1A and 2 A and B). Loss-of-function mutations in blmp-1 or lin-29 also caused late-onset DA9 synaptic polarity defects (Fig. 2 C–F), whereas dre-1 loss-of-function mutants had ectopic RAB-3 localization as early as the L3 developmental stage (Fig. 3 D–F). We also found that the let-7 miRNA and lin-28/RNA binding heterochronic genes control maintenance of DA9 synaptic polarity (Figs. 4 and 5). Heterochronic genes are known to function in neurons (8–12). We propose that heterochronic genes impact synapses in the DA9 neuron and alter axodendritic polarity. The appearance of ectopic synaptic vesicles in the dendrite in DA9 neuron in heterochronic mutants could indicate an increased synaptogenesis at the ventral side. Further analysis that will test whether the ectopic dendritic synaptic vesicles are able to release neurotransmitters at the ventral side and activate the body wall muscles will further establish their role in synaptic transmission. The additional genetic components of DA9 neuronal polarity were also uncovered from this screen (SI Appendix, Table S1) and may reveal how the UNC-5 receptor signal is transduced to control neuronal axodendritic polarity.
Heterochronic genes act as an intrinsic developmental timer that regulates the pattern of cell lineage and other developmental events during larval development (29). In the hermaphrodite, L1 larvae hatch with 202 somatic and 20 pharyngeal neurons. Only three (of eight) motorneuron classes are present in the first larval stage: the cholinergic DA and DB and the GABAergic class DD. Later on, 80 postembryonic neurons are born to make a total of 302 somatic neurons (42). Interestingly, the timing of neurogenesis for most of the postembryonic neurons is during L1 to L2 and L2 to L3 molts (42). The PDE neurons of the posterior deirid are an example of neurogenesis during the molt. The postembryonic V5 ectoblasts in C. elegans produce a mixture of neurons and hypodermal cells (43). During the L2 stage, the lineage of V5 becomes distinct from that of the other V cells and gives rise to V5.pa cell, a neuroblast that generates PDE in late L2 (44). Intriguingly, some of the heterochronic genes described above regulate developmental events during these molts. For example, the lin-4 heterochronic gene regulates transition from L1 to L2 larval stage (28, 30, 32). Furthermore, mutations in lin-28 gene cause skipping of L2 fates leading to precocious expression of adult cell fates (45). While there is some evidence that the heterochronic gene lin-4 regulates the timing of neuroblast divisions in C. elegans (1), a linkage between the heterochronic pathway and neurogenesis has not been established.
In invertebrates, such as Drosophila, a distinct pattern of transcription factors is expressed in the neuroblasts as they divide to give rise to neurons at each developmental stage (46, 47). In C. elegans, neurogenesis is regulated by a distinct set of transcription factors whose activity is mediated by a battery of downstream genes (1). One of the key elements of neurogenesis is the UNC-6/Netrin and UNC-5 receptor dorsal pathfinding system. During neurogenesis, neurons that turn dorsally activate transcription of the netrin receptor UNC-5 (48). Is the UNC-5/netrin signaling pathway linked to the activity of transcription factors that control neurogenesis in C. elegans? Transcriptional control of the netrin receptor UNC-5 might be regulated by modulating the activity of particular transcription factors (49). It is plausible that maintenance of neuronal polarity of DA9 neuron is controlled by a repertoire of genes that are implicated in the initial neurogenesis, such as transcription factors. There might be a coupling of signals regulated by transcription factors that act early during neurogenesis with heterochronic genes to control polarity of DA9 neuron via unc-5 signaling.
It is not known how the synaptic polarity phenotype of the heterochronic mutants is coupled to UNC-6/Netrin signaling. There may be mRNA targets of heterochronic lin-4 or let-7 miRNAs expressed in DA9 neuron. Interestingly, let-7 controls the temporal cell fate in the mushroom body lineage in Drosophila by targeting the transcription factor chinmo (50). And let-7 is enriched in human and mouse brain samples (51). It is plausible that this conserved miRNA may control the fate of neural differentiation programs in vertebrates.
During gonadal migrations, the hermaphrodite distal tip cells migrate along the larval body wall before they form a mature gonad. Distal tip cells are born during the L1 molt and remain stationary until mid L2 stage (52). Then they move in opposite directions along the ventral body muscles, stopping in mid L3 stage before they move dorsally (52). Distal tip cells stop migrating at the end of L4 (52). Increased expression of UNC-5 in the distal tip cells is required for the ventral-to-dorsal turn of the distal tip cells (53). Heterochronic genes allow UNC-5 to be expressed at the proper time to drive distal tip cell migrations (54). Similarly, during the development of the nervous system, the UNC-5 receptor is expressed and localized to particular segments of developing neurites and it is transcriptionally controlled as neurons turn dorsally (48). In the DA9 neuron UNC-5 is involved in both axon guidance and synaptogenesis and it is required for initial placement and maintenance of presynaptic complexes in C. elegans (4, 55). Our result that blmp-1 lof mutants affect axon guidance of DA9 neuron in adult animals in a UNC-5–dependent manner indicates that the heterochronic signaling pathway interacts with the UNC-6/Netrin and UNC-5 receptor dorsal pathfinding system to guide initial placement of the axon of DA9 at the dorsal side during development. It is plausible that this interaction is required for the presynaptic vesicles to be properly maintained at the correct position later in adulthood.
One form of neural plasticity is neural rewiring, which happens during the molt. One example of neural rewiring is the change in connectivity of the GABArgic DD motor neurons during L1 to L2 transition in development (14). The cyclin-dependent kinase CDK-5 stimulates UNC-104/Kinesin-3–mediated synaptic vesicle transport during synapse remodeling in DD neurons (41). We found that CDK-5 regulates the synaptic polarity of the DA9 neuron and this process is dependent on UNC-5. CDK-5 dysregulation has been linked to Alzheimer’s disease (56), and CDK-5 deregulation leads to neurodegeneration in dopaminergic neurons in Drosophila (57). Because synapse elimination is a key pathological hallmark in several neurodegenerative diseases (58), strategies to delineate the cellular pathways that regulate neuronal polarity might be key to understand the molecular basis of these devastating conditions.
Our results support a model that heterochronic genes control DA9 synaptic polarity via unc-5–dependent signaling (SI Appendix, Fig. S13). UNC-6/Netrin expression forms a dorsal-ventral gradient, which is required for DA9 to form the dorsally directed axon and the ventrally directed dendrite (4, 59). Loss of the extracellular cue UNC-6/Netrin or its receptor UNC-5 show ectopic dendritic localization of the RAB-3 presynaptic marker (4). Our work shows that heterochronic genes, which control the migration of gonadal cells during development in an unc-5–dependent manner, also play a role in the maintenance of synaptic polarity of DA9 neuron at the adult stage. Do heterochronic genes act upstream of netrin signaling to control the neuronal polarity or they act intrinsically within the DA9 neuron downstream of unc-6 extracellular cue? Do heterochronic genes affect synaptic polarity in other C. elegans neurons as well? Further investigations into the role of heterochronic genes in mammalian models of polarity, such as cortical pyramidal neurons (60–62), will further establish their impact on axodendritic polarity. Pathways that maintain synaptic polarity may present novel targets for treatment of cognitive decline in the neurodegenerative brain.
Materials and Methods
C. elegans Strains.
N2 Bristol was used as the wild-type reference strain. Worms were raised in OP50 Escherichia coli-seeded NGM plates at 20 °C, except for lin-28(n719), let-7(n2853ts), and let-7(mg279), which were maintained at 15 °C. A complete list of the strains used in this study is described in (SI Appendix, Table S2).
RNAi Strains.
RNAi clones for individual experiments were obtained from the RNAi sublibrary composed of genes that are related to progeria (15) or the full genome Ahringer and Vidal RNAi libraries (63–65), and confirmed by Sanger sequencing. The L4440 empty vector was used in all experiment as the RNAi control. All strains were grown overnight in Luria–Bertani medium with 50 μg/mL ampicillin, concentrated 10× by centrifugation, seeded onto 6-cm agar plates containing 5 mM isopropyl-β-d-thiogalactoside (IPTG), and incubated at room temperature overnight to induce dsRNA expression. L4 larvae expressing Pitr-1::GFP::RAB-3 transgene in the nre-1(hd20)lin-15b(hd126) background (66) were added on the seeded RNAi plates containing Fluorodeoxyuridine (FUDR). The animals were incubated for 5 d at 20 °C before they were imaged.
Epifluorescence Microscopy.
Animals were mounted on 2% agarose pads and immobilized using Polybead Polysterene 0.10-μm Microspheres (Polysciences). Images generated by an epifluorescence microscope were collected using a Zeiss Axio Image Z1 microscope, equipped with a Zeiss AxioCamHRc digital camera, and using Axiovision software. All transgenic worms were imaged at 40× or 63×. Images were processed using ImageJ software. For all fluorescence images, any images shown within the same figure panel were collected together using the same exposure time and then processed identically in ImageJ.
Confocal Microscopy.
Confocal images were acquired using an Olympus FV-1000 confocal microscope with an Olympus PlanApo 60× oil 1.45 NA objective at 2× zoom, a 488-nm Argon laser (GFP), and a 559-nm diode laser (mCherry). Worms were immobilized using polysterene 0.10-μm microspheres (Polysciences) and oriented dorsal up. Maximum-intensity projections of z-series stacks were made using Metamorph 7.1 software (Molecular Devices).
Image Analysis and Quantification.
Confocal scans using fixed acquisition parameter were processed using custom software in IGOR Pro (WaveMetrics) to generate line-scan data. Images of fluorescence slides (Chroma Technology Group) were captured during each imaging session to provide a fluorescence standard for comparing absolute fluorescence levels between animals. Background signal (charge-coupled device dark current and slide autofluorescence) was subtracted before analysis. Automated image analysis provided five image parameters as described in Dittman and Kaplan (67). The “peak” value is the ratio of peak fluorescence to the fluorescent slide standard. “Axon” is the ratio of baseline fluorescence (the minimum fluorescence level over a 5-μm interval within the dorsal cord image) to the fluorescent slide standard. “Synaptic enrichment” (% ∆F/F) is defined as (Fpeak − Faxon)/Faxon. “Width” is the width of each punctum at half the maximum peak fluorescence. “Density” is the number of fluorescence peaks found per 10 µm of cord analyzed. These values were quantified for dorsal GFP::RAB-3 puncta analysis in animals treated with RNAi clones from the sublibrary, including genes in (SI Appendix, Table S1) and for analysis of dorsal GFP::RAB-3 in heterochronic mutants. The dorsal fluorescence “peak” values reported in Fig. 4C and SI Appendix, Fig. S3 are means ± SEM. Quantification of ectopic dendritic puncta was performed manually.
Staging and Analysis.
To precisely synchronize the worms at a developmental stage, gravid adult worms were collected, synchronized by NaOCl bleaching and overnight hatching in M9. Animals at the L1 stage were placed at 20 °C to OP50-containing plates to develop for 9 h and collected as L2 larvae for imaging. To image animals at the L3 larval stage, we placed L1 larvae at OP50-containing plates at 20 °C and collected after 19 h. To image animals at the L4 larval stage, we placed L1 larvae at OP50-containing plates at 20 °C and imaged after 31 h. Then the phenotype of DA9 presynaptic localization was tested using a confocal microscope.
Genome Modification by CRISPR/Cas9.
Guide RNA was selected by searching the desired genomic interval for “NNNNNNNNNNNNNNNNNNRRNGG,” using Ape DNA sequence-editing software (http://jorgensen.biology.utah.edu/wayned/ape/). To generate a guide RNA construct, we used Q5 site-directed mutagenesis, as described previously (68). Repair template oligos were designed as described previously (69, 70). Injections were performed using the editing of dpy-10 (to generate cn64 rollers) as phenotypic co-CRISPR marker (70, 71). Injection mix contained 60 ng/uL each of the co-CRISPR and gene-of-interest targeting Guide RNA/Cas9 construct, and 50 ng/uL each of the co-CRISPR and gene-of-interest repair oligos. Guide RNA and homologous repair template sequences are listed in SI Appendix, Table S3.
Identification of the cdk-5 Deletion.
We performed snip-SNP mapping using SNPs from the Hawaiian strain (40) to identify a region at the III chromosome containing the mutant allele. Deep sequencing was performed using DNA from mutant F2s generated by outcrossing the original mutant strain to the Hawaiian strain. Genomic DNA was prepared using the Gentra Puregene Tissue kit (Qiagen, #158689) according to the manufacturer’s instructions. Genomic DNA libraries were prepared using the NEBNext genomic DNA library construction kit (New England Biololabs, #E6040), and sequenced on an Illumina Hiseq instrument. Deep-sequencing reads were analyzed using Cloudmap (72). Deep sequencing revealed a 155-bp deletion in the cdk-5 gene starting from the last codon of exon 4 [Chr III (13,779,739–13,782,893), Wormbase vWS268] with an insertion of two bases (GA).
Indirect Immunofluorescence.
Worms were prepared using a modification of a freeze crack protocol (73). Briefly, populations of adult worms were washed 3–4× with distilled water. Adult wild-type and mutant nematodes were then added to poly-L-ysine–coated slides and permeabilized by freeze-cracking. Worms were then fixed by methanol/acetone fixation method. Fixed and permeabilized animals were placed in blocking solution (10% serum in antibody buffer) at 4 °C overnight. Samples of fixed worms were incubated with anti–LIN-29 primary polyclonal antibody (26) at a final dilution (1:50) in antibody buffer (10× PBS, 0.5% Triton X-100, 1 mM EDTA, 0.1% BSA, 0.05% sodium azide, pH to 7.2) plus 1% BSA at 4 °C overnight. Slides were washed 3× with antibody buffer and 1× with PBS before adding secondary antibody (Alexa Fluor 647 goat anti-Rabbit IgG, Invitrogen A21245), (1:200) in antibody buffer plus 1% BSA at 4 °C overnight. Slides were washed 3× with antibody buffer and added to PBS solution until mounted with DAPI (Thermo Fisher Scientific). Images were acquired with Zeiss Axio Image Z1 microscope with a 40× and 63× oil-immersion objectives using Axiovision software.
Locomotion Assay and Behavioral Analysis.
Backward locomotion was quantified by placing 5-d-old adults on a freshly spotted NGM plate with OP50 bacteria. After 10 min of free movement at 20 °C, the locomotion behavior of 5-d-old adult worms was recorded on a Zeiss Discovery Stereomicroscope. Individual worms were lightly tapped with a platinum wire. Recordings were 10-s long. Centroid velocity of each animal was analyzed at each frame using object-tracking software in Axiovision (74). The speed of each animal was calculated by averaging the velocity value at each frame. The tapping protocol was repeated for three trials and an average of three trials was taken. Total magnification is 8×. A Mann–Whitney test with nonparametric Gaussian distribution was used for statistical analysis using Prism software. Images of worm tracks were taken on plates with a lawn of OP50 bacteria spotted 24 h before imaging with a Zeiss Discovery Stereomicroscope.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 GM44619. We thank the Caenorhabditis Genetics Center for strains; members of the G.R. laboratory for helpful discussions; Kang Shen (Stanford University) for kindly providing the wyIs85 transgene; A. V. Samuelson for the RNAi library used in the screen; the J. Kaplan laboratory for the use of the Olympus FV-1000 confocal microscope; and Ann Rougvie for kindly providing the α-LIN-29 antibody used in immunofluorescence experiments.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820928116/-/DCSupplemental.
References
- 1.Hobert O., Neurogenesis in the nematode Caenorhabditis elegans. WormBook, 1–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ou C. Y., Shen K., Neuronal polarity in C. elegans. Dev. Neurobiol. 71, 554–566 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ou C. Y., et al. , Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell 141, 846–858 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon V. Y., Klassen M. P., Shen K., UNC-6/netrin and its receptor UNC-5 locally exclude presynaptic components from dendrites. Nature 455, 669–673 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arimura N., Kaibuchi K., Neuronal polarity: From extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 8, 194–205 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Adler C. E., Fetter R. D., Bargmann C. I., UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat. Neurosci. 9, 511–518 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C., et al. , MIG-10/lamellipodin and AGE-1/PI3K promote axon guidance and outgrowth in response to slit and netrin. Curr. Biol. 16, 854–862 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Hallam S. J., Jin Y., lin-14 regulates the timing of synaptic remodelling in Caenorhabditis elegans. Nature 395, 78–82 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Zou Y., Chiu H., Domenger D., Chuang C. F., Chang C., The lin-4 microRNA targets the LIN-14 transcription factor to inhibit netrin-mediated axon attraction. Sci. Signal. 5, ra43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aurelio O., Boulin T., Hobert O., Identification of spatial and temporal cues that regulate postembryonic expression of axon maintenance factors in the C. elegans ventral nerve cord. Development 130, 599–610 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Ritchie F. K., et al. , The heterochronic gene lin-14 controls axonal degeneration in C. elegans neurons. Cell Rep. 20, 2955–2965 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Zou Y., et al. , Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science 340, 372–376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nonet M. L., et al. , Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci. 17, 8061–8073 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White J. G., Southgate E., Thomson J. N., Brenner S., The structure of the ventral nerve cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275, 327–348 (1976). [DOI] [PubMed] [Google Scholar]
- 15.Samuelson A. V., Carr C. E., Ruvkun G., Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 21, 2976–2994 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang T. F., et al. , BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 10, e1004428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klassen M. P., Shen K., Wnt signaling positions neuromuscular connectivity by inhibiting synapse formation in C. elegans. Cell 130, 704–716 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Mahoney T. R., et al. , Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol. Biol. Cell 17, 2617–2625 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn M., et al. , DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Dev. Cell 28, 697–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller A. D., Maniatis T., Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 5, 868–879 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Yogev S., Shen K., Establishing neuronal polarity with environmental and intrinsic mechanisms. Neuron 96, 638–650 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Hedgecock E. M., Culotti J. G., Hall D. H., The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron 4, 61–85 (1990). [DOI] [PubMed] [Google Scholar]
- 23.Levy-Strumpf N., Culotti J. G., Netrins and Wnts function redundantly to regulate antero-posterior and dorso-ventral guidance in C. elegans. PLoS Genet. 10, e1004381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rougvie A. E., Ambros V., The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development 121, 2491–2500 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Euling S., Bettinger J. C., Rougvie A. E., The LIN-29 transcription factor is required for proper morphogenesis of the Caenorhabditis elegans male tail. Dev. Biol. 206, 142–156 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Bettinger J. C., Lee K., Rougvie A. E., Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development 122, 2517–2527 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Fielenbach N., et al. , DRE-1: An evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev. Cell 12, 443–455 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Ambros V., Horvitz H. R., Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416 (1984). [DOI] [PubMed] [Google Scholar]
- 29.Moss E. G., Heterochronic genes and the nature of developmental time. Curr. Biol. 17, R425–R434 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Ambros V., A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57, 49–57 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Liu Z., Kirch S., Ambros V., The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development 121, 2471–2478 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Ambros V., Horvitz H. R., The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1, 398–414 (1987). [DOI] [PubMed] [Google Scholar]
- 33.Slack F. J., et al. , The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Grosshans H., Johnson T., Reinert K. L., Gerstein M., Slack F. J., The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell 8, 321–330 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Abrahante J. E., et al. , The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4, 625–637 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Lin S. Y., et al. , The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4, 639–650 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Abbott A. L., et al. , The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9, 403–414 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richard M., Boulin T., Robert V. J., Richmond J. E., Bessereau J. L., Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex. Proc. Natl. Acad. Sci. U.S.A. 110, E1055–E1063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dowen R. H., Breen P. C., Tullius T., Conery A. L., Ruvkun G., A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes Dev. 30, 1515–1528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis M. W., et al. , Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6, 118 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park M., et al. , CYY-1/cyclin Y and CDK-5 differentially regulate synapse elimination and formation for rewiring neural circuits. Neuron 70, 742–757 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White J. G., Southgate E., Thomson J. N., Brenner S., The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 43.Herman M. A., Hermaphrodite cell-fate specification. WormBook, 1–16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hobert O., Specification of the nervous system. WormBook, 1–19 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moss E. G., Lee R. C., Ambros V., The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Bossing T., Udolph G., Doe C. Q., Technau G. M., The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev. Biol. 179, 41–64 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Schmid A., Chiba A., Doe C. Q., Clonal analysis of Drosophila embryonic neuroblasts: Neural cell types, axon projections and muscle targets. Development 126, 4653–4689 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Hamelin M., Zhou Y., Su M. W., Scott I. M., Culotti J. G., Expression of the UNC-5 guidance receptor in the touch neurons of C. elegans steers their axons dorsally. Nature 364, 327–330 (1993). [DOI] [PubMed] [Google Scholar]
- 49.Ruvkun G., “Patterning the nervous system” in C. elegans II, Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R., Eds. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1997), pp. 543–581. [PubMed] [Google Scholar]
- 50.Wu Y. C., Chen C. H., Mercer A., Sokol N. S., Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev. Cell 23, 202–209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sempere L. F., Dubrovsky E. B., Dubrovskaya V. A., Berger E. M., Ambros V., The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev. Biol. 244, 170–179 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Hedgecock E. M., Culotti J. G., Hall D. H., Stern B. D., Genetics of cell and axon migrations in Caenorhabditis elegans. Development 100, 365–382 (1987). [DOI] [PubMed] [Google Scholar]
- 53.Su M., et al. , Regulation of the UNC-5 netrin receptor initiates the first reorientation of migrating distal tip cells in Caenorhabditis elegans. Development 127, 585–594 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Cecchetelli A. D., Cram E. J., Regulating distal tip cell migration in space and time. Mech. Dev. 148, 11–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Killeen M. T., The dual role of the ligand UNC-6/Netrin in both axon guidance and synaptogenesis in C. elegans. Cell Adhes. Migr. 3, 268–271 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cruz J. C., Tsai L. H., Cdk5 deregulation in the pathogenesis of Alzheimer’s disease. Trends Mol. Med. 10, 452–458 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Shukla A. K., Spurrier J., Kuzina I., Giniger E., Hyperactive innate immunity causes degeneration of dopamine neurons upon altering activity of Cdk5. Cell Rep. 26, 131–144.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koffie R. M., Hyman B. T., Spires-Jones T. L., Alzheimer’s disease: Synapses gone cold. Mol. Neurodegener. 6, 63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wadsworth W. G., Hedgecock E. M., Hierarchical guidance cues in the developing nervous system of C. elegans. BioEssays 18, 355–362 (1996). [DOI] [PubMed] [Google Scholar]
- 60.Shoukimas G. M., Hinds J. W., The development of the cerebral cortex in the embryonic mouse: An electron microscopic serial section analysis. J. Comp. Neurol. 179, 795–830 (1978). [DOI] [PubMed] [Google Scholar]
- 61.Noctor S. C., Martínez-Cerdeño V., Ivic L., Kriegstein A. R., Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Barnes A. P., Polleux F., Establishment of axon-dendrite polarity in developing neurons. Annu. Rev. Neurosci. 32, 347–381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamath R. S., Ahringer J., Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Lee S. S., et al. , A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33, 40–48 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Rual J. F., Hill D. E., Vidal M., ORFeome projects: Gateway between genomics and omics. Curr. Opin. Chem. Biol. 8, 20–25 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Schmitz C., Kinge P., Hutter H., Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126). Proc. Natl. Acad. Sci. U.S.A. 104, 834–839 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dittman J. S., Kaplan J. M., Factors regulating the abundance and localization of synaptobrevin in the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 103, 11399–11404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paix A., et al. , Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198, 1347–1356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ward J. D., Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199, 363–377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arribere J. A., et al. , Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., CloudMap: A cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249–1269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller D. M., Shakes D. C., Immunofluorescence microscopy. Methods Cell Biol. 48, 365–394 (1995). [PubMed] [Google Scholar]
- 74.Choi S., Chatzigeorgiou M., Taylor K. P., Schafer W. R., Kaplan J. M., Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron 78, 869–880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.