Significance
Cytochrome P450 monooxygenase enzymes metabolize drugs, carcinogens, and endogenous molecules in the Eukarya, the Bacteria, and the Archaea. The notion that viral genomes contain P450 genes was not considered until discovery of the giant viruses. We have uncovered multiple and unique P450 genes in giant viruses from the deep ocean, terrestrial sources, and human patients. P450s were also found in a herpesvirus and a mycobacteriophage, and we report a crystal structure of the phage P450. Our findings herald an era of research regarding the evolution of this important gene family, with implications for understanding the biology and the origin of the giant viruses themselves. The findings also present potential drug targets for giant viruses that may be human pathogens.
Keywords: cytochrome P450, virus, evolution, domains of life, redox partner
Abstract
Genes encoding cytochrome P450 (CYP; P450) enzymes occur widely in the Archaea, Bacteria, and Eukarya, where they play important roles in metabolism of endogenous regulatory molecules and exogenous chemicals. We now report that genes for multiple and unique P450s occur commonly in giant viruses in the Mimiviridae, Pandoraviridae, and other families in the proposed order Megavirales. P450 genes were also identified in a herpesvirus (Ranid herpesvirus 3) and a phage (Mycobacterium phage Adler). The Adler phage P450 was classified as CYP102L1, and the crystal structure of the open form was solved at 2.5 Å. Genes encoding known redox partners for P450s (cytochrome P450 reductase, ferredoxin and ferredoxin reductase, and flavodoxin and flavodoxin reductase) were not found in any viral genome so far described, implying that host redox partners may drive viral P450 activities. Giant virus P450 proteins share no more than 25% identity with the P450 gene products we identified in Acanthamoeba castellanii, an amoeba host for many giant viruses. Thus, the origin of the unique P450 genes in giant viruses remains unknown. If giant virus P450 genes were acquired from a host, we suggest it could have been from an as yet unknown and possibly ancient host. These studies expand the horizon in the evolution and diversity of the enormously important P450 superfamily. Determining the origin and function of P450s in giant viruses may help to discern the origin of the giant viruses themselves.
One of the key moments in biology was the discovery of a new heme protein more than 50 y ago, termed cytochrome P450 (CYP; P450), and, further, that P450 proteins could catalyze monooxygenase activity (1–3). Today, cytochrome P450 enzymes constitute one of the largest enzyme superfamilies known, occurring broadly in the Bacteria, Archaea, and Eukarya (4), the three classical domains of life proposed by Woese and Fox (5). P450 enzymes play essential roles in steroid/sterol biosynthesis and degradation, vitamin biosynthesis, detoxification and activation of many drugs and environmental pollutants, enzymatic activation of carcinogens, and the biosynthesis of vast arrays of secondary metabolites (6). The numbers of P450 genes encoded in genomes of eukaryotic species generally range from a few to several hundred. Anaerobic microbes often have no P450 genes, but some bacteria (e.g., the Actinomycetes) have tens. More than 800,000 P450 sequences are known, and it is estimated that more than 1 million will be known by 2020 (7).
Our understanding of the distribution of P450s in nature began to change in 2004 with another key moment in modern biology, the publication of the genome of Acanthamoeba polyphaga mimivirus (APMV). That paper defined a new class of virus, the nucleocytoplasmic large DNA virus (8). The APMV genome is predicted to contain 1,000+ genes, with fewer than 300 having predicted functions (8, 9). New giant viruses continue to be discovered in and captured with amoebae (10, 11) and in metagenomes (12, 13). Giant virus taxa of concern include the Mimiviridae, Tupanvirus, Marseilleviridae, Pandoraviridae, Mollivirus, Pithovirus, Orpheovirus, Faustovirus, Kaumoebavirus, and Klosneuvirinae (14). The viruses fall in a proposed new order, the Megavirales (15, 16). A core set of genes involved in viral replication and lysis is most often conserved in these viruses (10), yet most genes in the giant viruses (70–90%) do not have any obvious homolog in existing virus databases.
Strikingly, many of the giant viruses possess genes coding for proteins typically involved in cellular processes, including protein translation, DNA repair, and eukaryotic metabolic pathways previously thought not to occur in viruses (17). One such gene in Mimivirus (MIMI_L808) was annotated as encoding a lanosterol demethylase, which is an activity of CYP51, a well-known P450 essential in sterol biosynthesis (18). (Here, the term Mimivirus is used to refer to the original APMV, to the genus Mimivirus, and also to sublineage group A in the Mimiviridae.) Subsequently, that Mimivirus sequence was cloned and expressed, and the recombinant protein was shown spectrophotometrically to be a P450 (19). The unexpected finding of a P450 in Mimivirus and the growing number of known giant viruses prompted us to search for P450 genes throughout the virosphere. We now report that genes for multiple and unique P450s are common in several taxa of giant viruses. We also uncovered P450 genes in viruses other than giant viruses. The discoveries open a new realm in the diversity and distribution of P450s in nature, especially in the enigmatic giant viruses.
Results
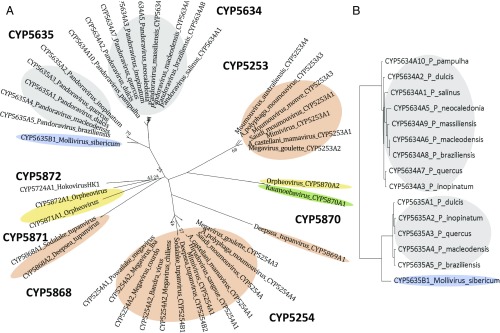
We identified P450 genes in multiple viruses, uncovered by examination of available genomes in the Megavirales, and all other available virus genomes. Fig. 1 shows the phylogeny of viruses in the Megavirales. We assigned genes as encoding P450s based on the presence of consensus motifs found in nearly all P450s, including the EXXR motif in the K helix, the conserved cysteine heme-thiolate P450 motif, and overall domain identification using Pfam models. Our search revealed P450 genes in viruses in multiple branches of the Megavirales, including in the Mimiviridae, in the Pandoraviridae, in Mollivirus, in Kaumoebavirus, in the tupanviruses, and in Orpheovirus (Fig. 1 and Tables 1 and 2). The molecular phylogeny of P450s in the giant viruses is shown in Fig. 2, based on inferred amino acid sequences. Additionally, P450 genes were found encoded in phage and a herpesvirus (Table 2).
Fig. 1.
Megavirales phylogeny constructed from the RNAPol2 gene. This ML phylogenetic tree shows the relationship of key members of the Megavirales viruses. The clades that contain encoded cytochrome P450 genes are highlighted in colors that correspond to the cytochrome P450 phylogeny in Fig. 2. Bootstrap support values are derived from 250 replicates, using the HIVB (HIV between-patient) model of amino acid substitution. Only values below 90% are shown.
Table 1.
P450 genes in the Mimiviridae
| Taxon/species | Virus source | Gene family and subfamily |
| Mimiviridae A | ||
| Mimivirus | United Kingdom | CYP5253A: CYP5254A |
| Mamavirus | France | CYP5253A: CYP5254A |
| Lentillevirus | France | CYP5253A: CYP5254A |
| Oyster virus | Brazil | CYP5253A: CYP5254A |
| Kroon virus | Brazil | CYP5253A: CYP5254A |
| Hirudovirus | Tunisia | CYP5253A: CYP5254A |
| Niemeyer virus | Brazil | CYP5253A: CYP5254A |
| Mimivirus Bombay | India | CYP5253A: CYP5254A |
| Mimivirus Shirakomae | Japan | CYP5253A: CYP5254A |
| Mimivirus Kasaii | Japan | CYP5234A: CYP5254A |
| Samba virus | Brazil | CYP5253A: CYP5254A |
| Amazonia virus | Brazil | CYP5253A: CYP5254A |
| Mimiviridae B | ||
| Moumouvirus | France | CYP5253A: CYP5254B |
| Moumouvirus monve | France | CYP5253A: CYP5254B |
| Saudi moumouvirus | Saudi Arabia | CYP5253A: CYP5254B |
| Moumouvirus goulette | France | CYP5253A: CYP5254B |
| Moumouvirus australiensis | Australia | CYP5253A: CYP5254B |
| Mimiviridae C | ||
| Megavirus chiliensis | Chile | CYP5254A |
| Mimivirus (LB111) | Tunisia | CYP5254A |
| Megavirus courdo 7 | France | CYP5254A |
| Powai Lake Megavirus | India | CYP5254A |
Table 2.
P450 genes in the Pandoraviridae and other viruses
| Taxon/species | Virus source | Gene family and subfamily |
| Pandoraviridae | ||
| Clade A | ||
| P. salinus | Chile | CYP5634A |
| P. dulcis | Australia | CYP5634A: CYP5635A |
| P. inopinatum | Germany | CYP5634A*: CYP5635A |
| P. quercus | France | CYP5634A: CYP5635A |
| P. celtis | France | CYP5634A: CYP5635A |
| P. pampulha | Brazil | CYP5634A |
| Clade B | ||
| P. neocaledonia | New Caledonia | CYP5634A |
| P. macleodensis | Australia | CYP5634A: CYP5635A |
| P. brazilinsis | Brazil | CYP5634A: CYP5635A |
| P. massiliensis | Brazil | CYP5634A |
| Mollivirus sibericum | Siberia | CYP5635B |
| Orpheovirus IHUMI-LCC2 | France | CYP5870A: CYP5871A: CYP5872A |
| Kaumoebavirus | Saudi Arabia | CYP5870A |
| Hokovirus HKV1 | Hong Kong | CYP5724A |
| Tupanvirus (soda lake) | Brazil | CYP5254B: CYP5868A |
| Tupanvirus (deep ocean) | Brazil | CYP5254B: CYP5868A: CYP5869A |
| Mycobacteria phage | ||
| Adler phage | M. abscessus | CYP102L1 |
| Herpesvirus | ||
| Ranid herpesvirus 3 | R. temoraria | CYP5723A1 |
Bold indicates probable known host-derived genes.
Two separate gene predictions joined.
Fig. 2.
Molecular phylogeny of virus cytochrome P450 genes. (A) Unrooted ML phylogeny of predicted amino acid sequences for viral P450s shows the close relationships between Pandoraviridae CYP5634 and CYP5635, and among Mimiviridae CYP5253s and CYP5254s. Note the CYP5254s in tupanviruses, and the CYP5870s from the Kaumoebavirus and Orpheovirus. (B) Detailed phylogeny of selected Pandoraviridae CYP5634s and CYP5635s and Mollivirus CYP5635. Color blocks are the same as in Fig. 1.
Mimiviridae: AT-Rich P450 Genes.
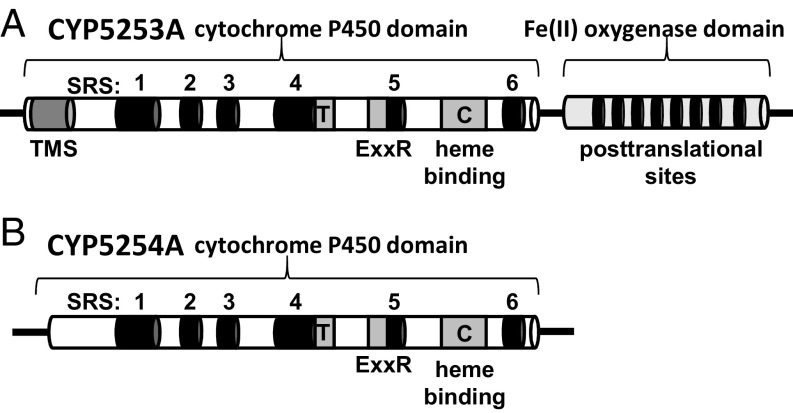
The Mimiviridae family currently is divided into three sublineages: group A, the mimiviruses and Mamaviruses; group B, the moumouviruses; and group C, the megaviruses (20). Searching the Mimiviridae genomes uncovered sequences similar to those first found in the Mimivirus (APMV) genome (19) (i.e., MIMI_L808, now named CYP5253A1, and MIMI_L532, named CYP5254A1). The CYP5253 gene codes for a unique chimeric protein with a predicted N-terminal membrane anchor in a P450 domain of about 500 amino acids, which is fused at the C terminus to a distinct 200-amino acid domain (Fig. 3). The P450 domain shares, at most, 28% identity with known P450s, while the C-terminal domain exhibits low identity with members of the 2-oxoglutarate/Fe(II)–dependent dioxygenase superfamily (2OG-FeII). The CYP5254A1 gene encodes a smaller P450 domain without an encoded recognizable membrane anchor. All of the Mimiviridae P450 genes are AT-rich (69–75%), like the Mimiviridae genomes overall (SI Appendix, Table S1).
Fig. 3.
Domain regions in Mimivirus CYP5353A1 (A) and CYP5254A1 (B). C, absolute conserved cysteine residue present in the heme binding domain; ExxR, conserved glutamate-residue-residue arginine motif found in the K helix; SRS, substrate recognition site; T, conserved threonine in the P450 I helix; TMS, transmembrane segment. PTMs include one N-glycosylation site, one protein kinase C phosphorylation site, four caesin kinase II phosphorylation sites, and three myristoylation sites.
Chimeric CYP5253A genes were discovered in all Mimiviridae group A and group B viruses but not in group C viruses, while CYP5254 family genes occur in all group A, B, and C viruses. Known viruses in lineage C that are only distantly related to Mimivirus, including Cafeteria roenbergensis virus (CroV), Phaeocystis globosa virus, and Pyramimonas orientalis virus, do not have P450 genes.
There is a surprisingly high degree of identity (98–100%) between inferred CYP5253A1 protein homologs in the group A mimiviruses isolated from different parts of the world. Likewise, there is >95% identity among the group B (Moumouvirus) CYP5253A1 genes. Nucleotide sequences also are highly identical within subgroups. There is a lower degree of identity between the apparent orthologs in different subgroups of the Mimiviridae. Thus, the inferred sequences of full-length CYP5253A proteins (P450 domain plus the putative 2OG/FeII domain) share from 55 to 88% identity between groups A and B.
The orthologs of CYP5254s also are highly similar within subgroups, but show less identity between groups. The CYP5254 identity falls below 55% between mimiviruses and the moumouviruses, placing the Moumouvirus CYP5254 in a new P450 gene subfamily, CYP5254B, while the genes in the other groups are in subfamily CYP5254A. While both Mamavirus and Lentillevirus appear to have a CYP5253A gene, there is some uncertainty for the gene prediction in these viruses.
Tupanviruses.
Two new viruses related to the Mimiviridae, named the tupanviruses, were discovered in a soda lake in Brazil and in the deep ocean (3,000-m depth) (12). These viruses differ in morphology from the mimiviruses (12). Tupanviruses also differ from other Mimiviridae in their P450 gene complement. The deep sea Tupanvirus has three P450 genes, while the soda lake virus has two (Table 2). Neither virus has a gene encoding an ortholog of the chimeric CYP5253A1. However, both viruses have a CYP5254 gene, classified with the Moumouvirus CYP5254 subfamily (i.e., CYP5254B). Both also have a gene that comprises another new family and subfamily, CYP5868A. The deep sea Tupanvirus has a third P450 gene in a new family, CYP5869A1.
Pandoraviridae: GC-Rich P450 Genes.
Giant viruses isolated in 2013 off the coast of Chile and from a freshwater pond in Australia constituted a new family, the Pandoraviridae, with no genomic or morphological resemblance to other viruses. The family was defined by Pandoravirus dulcis, Pandoravirus salinus (21), and Pandoravirus inopinatum [from an Acanthamoeba strain LaHel culture from the eye of a patient with keratitis in Germany (22)]. More pandoraviruses have been described recently (23, 24), including Pandoravirus celtis in 2019 (25). The pandoraviruses group in two clades, A and B (23) and the genomes range up to 2.5 Mb, predicted to encode from 1,414 (Pandoravirus massiliensis) to nearly 2,700 (Pandoravirus braziliensis) proteins (24). All of the genomes are GC-rich (58–64%) (SI Appendix, Table S1).
Most Pandoravirus genomes contain sequences predicted to encode membrane-bound P450s, in two new P450 families, CYP5634 and CYP5635. All members of both Pandoravirus clades have a CYP5634 sequence (Table 2), although P. inopinatum has two short sequences that, based on gene order (discussed below), appear to be CYP5634A gene fragments rather than a complete sequence. Some members of both clades also have a CYP5635A sequence (Table 2).
Mollivirus P450.
Two new giant viruses were obtained from the Late Pleistocene Siberian permafrost Mollivirus sibericum (26) and Pithovirus sibericum (27). The Mollivirus genome contains a P450 gene, whereas the Pithovirus does not. Mollivirus has a GC-rich genome (60%) and is related phylogenetically and morphologically to the pandoraviruses. The inferred Mollivirus P450 protein has a putative membrane anchor, and shares closer identity with Pandoravirus CYP5635As (48–51%) than with the CYP5634As. The Mollivirus P450 gene thus constitutes a new CYP5635 subfamily, CYP5635B (Table 2).
Kaumoebavirus and Orpheovirus P450s.
A P450 gene also appears in the genome of Kaumoebavirus, a giant virus (465 genes) phylogenetically related to faustoviruses and the Asfarviridae (28). Kaumoebavirus was isolated from sewage in Saudi Arabia, and cultured from the amoeba Vermamoeba vermiformis rather than an Acanthamoeba species. The predicted Kaumoebavirus gene, CYP5870A1, encodes a 398-residue P450 lacking a membrane anchor.
Another virus isolated by coculture with V. vermiformis is Orpheovirus IHUMI-LCC2 (11). Orpheovirus is related phylogenetically to pithoviruses (11), but while the latter does not have P450 genes, Orpheovirus has three. One Orpheovirus P450 is closely related to CYP5870A1 in Kaumoebavirus (Fig. 2), and is thus classified as CYP5870A2. The other two Orpheovirus P450s are not closely related to any other virus P450 genes (Fig. 2) and are classified to new P450 gene families, with genes CYP5871A1 and CYP5872A1 (Table 2).
In addition to the isolated viruses, assembly bins in environmental metagenomes have been identified as belonging to giant viruses, including Klosneuvirus, Indirivirus, Hokovirus, and Catovirus (14). A novel AT-rich (70%) P450 gene was identified in Hokovirus HK1, isolated in Hong Kong. This P450 is unrelated to other virus P450s, and thus defines a separate new family, CYP5724A1 (Table 2). Other orpheoviruses are also implied in metagenomes (13), but, as yet, these viral genomes are incompletely characterized.
Non-Giant Virus P450s.
In examining the 8,100+ completely sequenced viral genomes and metagenomic data, we identified P450 genes in two viruses not in the Megavirales. A P450 gene designated CYP5723A1 (GenBank accession no. ARR28948) was identified in the genome of a double-stranded DNA herpesvirus, Ranid herpesvirus 3, a pathogen that causes proliferative dermatitis in the frog Rana temoraria (29). CYP5723A1 is predicted to code for a soluble protein that shares 24–27% sequence identity with members of the cholesterol 7α-monooxygenases (CYP7s), hinting at a possible function in lipid or steroid metabolism.
The genome of the Mycobacterium phage Adler, which infects Mycobacteroides (previously Mycobacterium) abscessus subsp. bolletii F1660, has a gene predicted to encode a P450 (GenBank accession no. AHB79207; genome sequence locus tag: CH35_gp012). The predicted 471-amino acid protein defines a new subfamily and gene, CYP102L1, within the broader CYP102 family. A P450 subsequently found in the mycobacterial host is 99% identical to the phage P450; therefore, it is also classified as CYP102L1. Additional members of the new CYP102L subfamily (CYP102L1–8) have now been found in other Mycobacterioides species (SI Appendix, Fig. S1). The phage CYP102L1 shows lower sequence identity with genes in other CYP102 subfamilies (CYP102A–Z) (SI Appendix, Table S2). It shares only 34% identity yet retains remarkable structural similarity with the P450 domain of CYP102A1 (P450BM3), an extensively characterized P450 (discussed below) (SI Appendix, Fig. S2).
Analysis of P450 Gene Synteny.
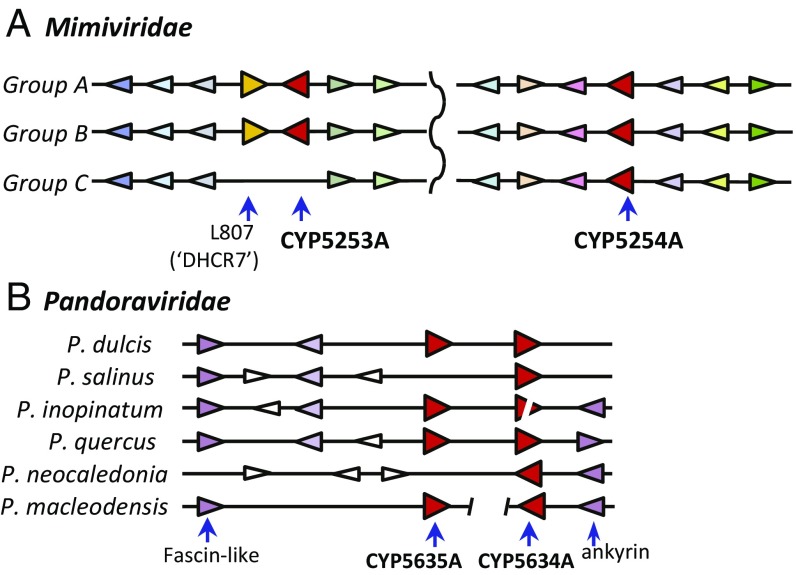
We assessed the evolutionary relatedness among P450 genes in the Megavirales using analyses of shared gene order (synteny). In the Mimiviridae, the CYP5253 and CYP5254 genes are in different locations in the sequenced genomes. The same 10 genes flank CYP5254 in viruses in groups A, B, and C. Likewise, the genes flanking the CYP5253A genes are conserved in the mimiviruses (group A) and the moumouviruses (group B) (Fig. 4A). Group C megaviruses are missing CYP5253A and also the adjacent gene R807; however, the synteny around the CYP5253A and R807 loci in groups A and B is preserved in group C genomes. The absence of genes encoding both R807 and CYP5253A is an intriguing difference from the other Mimiviridae, suggesting that these genes were lost together from a Megavirus ancestor. The CYP5254B genes in both tupanviruses share gene order like that around CYP5254B1 in Moumouvirus (SI Appendix, Fig. S3), suggesting a closer evolutionary history with the group B mimiviruses than with group A or C viruses.
Fig. 4.
Microsynteny around giant virus cytochrome P450 genes. (A) Mimiviridae: groups A, B, and C display high degrees of shared gene order extending 10 genes around both CYP5253A (Left) and CYP5254A (Right). Genes that are highly similar to each other are colored the same across the various genomes. Completely shared synteny is evident across all three clades for CYP5254A. Clade C viruses are missing both CYP5253A (L808 ortholog) and gene R807 [a putative 7-dehydrocholesterol reductase (DHCR7)]. (B) Pandoraviridae: Note shared synteny around P450s in pandoraviruses in clades A and B. The figure also shows that CYP5634 and CYP5635 are adjacent, and illustrates that CYP5634A1 may be a pseudogene in P. inopinatum. Genes in white are not similar, while purple-colored genes are orthologous. Other identifiable genes are indicated (fascin-like domain and ankyrin).
Shared synteny around the P450 genes is also found in the pandoraviruses, with examples shown in Fig. 4B. Where there are two P450 genes in a Pandoravirus genome, they are closely linked; for example, in P. dulcis, CYP5634A2 and CYP5635A1 are separated by just 162 base pairs (bp). The CYP5634A and CYP5635A loci in the various Pandoravirus genomes all are near to a fascin-like domain and/or an ankyrin gene, demonstrating shared synteny that indicates a common evolutionary history of these P450 genes. Location analysis of the two CYP5634-related sequences in P. inopinatum shows they are adjacent gene fragments separated by 160 bp and a stop codon. The two stretches together would code for a 356-amino acid protein, 84% identical to the CYP5634A in P. dulcis. Although Mollivirus is related to the pandoraviruses, the gene order around the Mollivirus CYP5634B appears not to be shared with the Pandoravirus CYP5634A genes.
Conserved synteny around Kaumoebavirus CYP5870A1 and Orpheovirus CYP5870A2 shows an evolutionary relatedness of these genes (SI Appendix, Fig. S4). In both viruses, the CYP5870A genes are adjacent to a sequence (BNJ_00167) predicted to code for a sterol desaturase. The encoded protein shares 35–41% identity with eukaryotic sterol C5 desaturase enzymes, although it is missing ∼60 N-terminal and 90 C-terminal amino acids found in eukaryotic enzymes. Sterol C5 desaturase catalyzes the dehydrogenation of a sterol precursor C5(6) bond in cholesterol-, phytosterol-, and ergosterol-producing organisms. The linkage of CYP5850A1 and BNJ_00167 suggests a possible functionality in sterol metabolism, not unlike the linkage of Mimivirus gene L808 (CYP5253A1) and R807 (discussed below).
Concerning Host Origin of Giant Virus P450s: P450 Genes Encoded in A. castellenii.
The majority of giant viruses have been isolated from or captured with an amoeba, most often either A. polyphaga or A. castellanii. To determine whether P450s in giant viruses are related to P450s in one of these contemporary hosts, we searched the completely sequenced A. castellanii genome (30) and identified 27 P450 genes. Two A. castellanii genes are predicted to encode CYP51 and CYP710D2, P450s involved in ergosterol synthesis, the major membrane sterol found in amoeba. The remaining 25 A. castellanii P450s were classified to new gene families or subfamilies, with no known function for these at present. A. castellanii P450 sequences shared no more than 25% identity with any virus P450.
Spectrophotometric Analysis of Recombinant P450 Proteins.
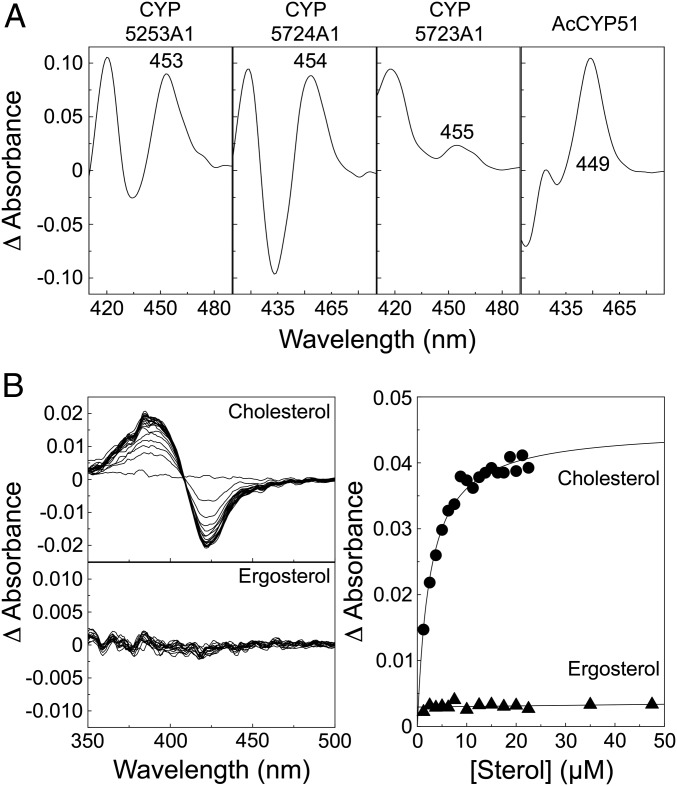
We repeated expression and isolation of recombinant Mimivirus CYP5253A1 protein several times. As before (19), a native P450 spectrum [in the carbon monoxide (CO)-bound, reduced state] was obtained with the expressed chimera (Fig. 5A). Recombinant proteins of Hokovirus CYP5724A1 and Ranid herpesvirus 3 CYP5723A1 also exhibited P450 spectral properties (Fig. 5A). We also expressed Mimivirus CYP5253A1 genetically engineered without the putative 2OG/FeII fusion domain. The recombinant P450 domain alone did not yield a native P450 spectrum but, instead, exhibited a peak at 420 nm, which represents cytochrome P420, a conformation that would be inactive in an enzyme assay. This suggests that the 2OG/FeII domain contributes to maintaining the correct P450 protein conformation and/or state of the thiolate ligand to the heme (31).
Fig. 5.
Characterization of recombinant cytochromes P450. (A) Dithionite-reduced, CO difference spectra of the purified viral cytochromes P450 and the control A. castellanii CYP51 (AcCYP51) were obtained. The concentrations of P450 used were as follows: CYP5253A1 (2.5 μM), CYP5724A1 (2.5 μM), CYP5723A1 (1.25 μM), and AcCYP51 (1.25 μM), respectively. (B) P450-sterol binding assays: Stock solutions of 2.5 mM cholesterol (●) and ergosterol (▲) were titrated against 4 μM CYP5253A1 in 50 mM Tris⋅HCl (pH 8) and 20% (vol/vol) glycerol, with the difference spectrum determined after each addition of sterol, which were used to construct ligand saturation curves.
P450 Substrate Binding and Catalytic Activity.
In the original report of the Mimivirus genome, the CYP5253A1 was annotated as a lanosterol 14α-demethylase (i.e., CYP51), although the P450 domain of that chimera has only 23% identity to any known CYP51. The preliminary analysis of the recombinant CYP5253A1 protein failed to detect any lanosterol 14α-demethylase activity using rat cytochrome P450 reductase (POR) (19). In a more complete and exhaustive analysis to assess possible CYP5253A1 sterol demethylase activity, we performed side-by-side experiments with the purified recombinant A. castellanii CYP51 (Fig. 5A), using all three major CYP51 substrates favored by animals, fungi, and plants/amoebae: lanosterol, eburicol, and obtusifoliol, respectively. The A. castellanii CYP51 was shown to demethylate all three sterol substrates with mean velocities of 0.69 ± 0.03 min−1 (lanosterol), 2.47 ± 0.19 min−1 (eburicol), and 3.32 ± 0.02 min−1 (obtusifoliol). However, Mimivirus CYP5253A1 showed no activity toward these CYP51 substrates under any of the conditions used (SI Appendix, Fig. S5A).
The gene adjacent to CYP5253A1 in Mimivirus (and in Moumouvirus) is gene R807, predicted to code for a 7-dehydrocholesterol reductase (7DHC). 7DHC catalyzes the final step of cholesterol synthesis, removal of the C(7–8) double bond converting 7-dehydrocholesterol to cholesterol (32). The linkage of Mimivirus R807 (7DHC) and L808 (CYP5253A1) suggests that these two virally encoded proteins might function with sterols, particularly cholesterol. Thus, we investigated whether CYP5253A1 could interact with cholesterol. Recombinant CYP5253A1 gave a type I binding spectrum with cholesterol (Fig. 5B) with an apparent binding constant (Ks) value of 2.86 ± 0.45 μM. In contrast, and as before (19), we failed to find any evidence that CYP5253A1 binds the amoeba membrane sterol ergosterol.
The binding spectrum with cholesterol suggests it could be a substrate for CYP5253A1. However, with in vitro reconstituted systems using purified recombinant CYP5253A1 and purified mammalian POR to drive P450 activity, we did not detect metabolites of cholesterol or 7-dehydrocholesterol by gas chromatography - mass spectrometry (SI Appendix, Fig. S5B). Our earlier work has shown that P450s can be selective for their potential redox partner proteins (33). Accordingly, we undertook a bioinformatic search of all giant viral genomes that contain P450 genes but found no virally encoded candidate redox partner proteins that might drive P450 activity. Therefore, we used three fundamental heterologous redox partner systems utilized universally in research to drive P450 enzyme activity to probe viral P450 cholesterol metabolism. These were eukaryotic POR, from both fungal (Aspergillus fumigatus) and mammalian (Homo sapiens) sources; bacterial (Escherichia coli) flavodoxin and flavodoxin reductase; and plant (Spinacia oleracea) ferredoxin and ferredoxin reductase. Cholesterol was not metabolized by recombinant viral CYP5253A1 with any of these commonly used P450 redox partners under any of the conditions employed.
We also assessed possible activity of expressed and purified Hokovirus CYP5724A1. This P450 shows up to 40% identity with P450s known to be involved in sterol and fatty acid metabolism, leading us to hypothesize that CYP5724A1 may also be either a sterol or fatty acid metabolizing P450. Hence, we assessed whether the sterols lanosterol, eburicol, obtusifoliol, cholesterol, and 7-dehydrocholesterol or the fatty acids lauric acid, palmitic acid, and oleic acid could be substrates for this P450. No metabolites were observed for any of these potential substrates with any assay conditions and redox partners (e.g., SI Appendix, Fig. S5B).
Structure and Function of Mycobacteriophage CYP02L1.
Heterologous expression of Mycobacterium phage Adler CYP102L1 in E. coli resulted in the production of recombinant heme-containing protein at 1 μmol/L E. coli culture. Purified CYP102L1 gave a reduced CO-Soret peak at 448 nm (Fig. 6A). The protein was crystallized in an open state, and the structure was determined by single-wavelength anomalous diffraction and refined with a maximum resolution of 2.5 Å. The overall structure of the CYP102L1 exhibits the typical P450 fold consisting of α-helical and β-sheet domains as seen in all other known archaeal, bacterial, and eukaryotic P450 structures (Fig. 6B). The heme cofactor is located between the α-helical domain and the β-sheet domain to create a substrate binding pocket. The CYP102L1 has a conserved cysteine (Cys419) that serves as the fifth axial thiolate ligand to the heme iron, as is typical in most P450s. The structure solved is strikingly similar to that for the P450 domain of the Bacillus megaterium CYP102A1 (Fig. 6C and SI Appendix, Fig. S2).
Fig. 6.
Structural features of CYP102L1. (A) Dithionite-reduced CO difference spectra of the purified Adler phage CYP102L1 (P450 concentration = 1.25 μM) showing a Soret spectral maximum at 448 nm. (B) Open crystal structure of CYP102L1 (Protein Data Bank ID code 6N6Q) exhibits the typical P450-fold consisting of α-helical and β-sheet domains as seen in all other known P450 structures. Heme is shown as a red stick model. (C and D) Comparison of the crystal structures of Adler phage CYP102L1 (green) and B. megaterium CYP102A1 (P450 BM3; blue). Although sequence identity remains low (34%), a number of residues that have been implicated in CYP102A1 function are conserved between both proteins, including A82 (92), F87 (97), F261 (275), A264 (278), T268 (282), A328 (245), A330 (347), F393 (412), and I401 (420), with CYP102L1 residues in parentheses. The CYP102A1 residues are involved in substrate binding affinity and turnover (A82 and I401), substrate selectivity (F87, T268, A328, and A330), and electron transfer (F261, A264, T268, and F393). Additional residues and references documenting residue involvement are cited in SI Appendix.
Type I spectral shift changes were observed following the addition of the fatty acids lauric acid (C12), myristic acid (C14), or palmitic acid (C16) to recombinant CYP102L1, suggesting these fatty acids are substrates (SI Appendix, Fig. S6). Metabolism experiments using the exogenous CYP102A1 reductase domain, eukaryotic POR, and bacterial redox systems as electron donor proteins for CYP102L1 have yielded inconsistent results with palmitic acid as a potential substrate. It is possible that there is an as yet unknown redox system that could be necessary for phage CYP102L1 function. Conversely, phage CYP102L1 may be the first CYP102 described to date that does not function in fatty acid metabolism.
In Vivo Expression of Giant Virus P450 Genes.
An important question concerning giant virus P450 genes is whether they are transcribed and translated in the host. Data gleaned from mass spectrometry-based proteomic studies of Mimivirus virions identified MIMI_L532 (CYP5254A1) among 114 Mimivirus proteins detected in the virus particle (34), confirmation that a giant virus P450 was transcribed and translated in a host. Subsequent transcriptomic studies showed that more than 90% of Mimivirus genes are transcribed upon infection, including the Mimivirus gene L808 (the chimeric CYP5253A1), which is an “early transcribed” gene, peaking at about 3 h postinfection (35).
Transcriptomic analysis showed that P450 genes are among those transcribed in a host (23) infected also by P. salinus, Pandoravirus quercus, Pandoravirus neocaledonia, or Pandoravirus macleodensis. The transcribed genes included those for the CYP5634s and CYP5635s (23). While stringent strand-specific RNA-sequencing experiments and particle proteome analyses predicted that P450 genes are transcribed and translated into protein, it is not clear that they are incorporated into the virion (23). Likewise, the M. sibericum P450 is expressed in vivo upon infection of amoebae (26, 27).
Retention of Mimivirus P450 Genes.
A study in which Mimivirus was subcultured 150 times in axenic amoebae resulted in 155 of the 1,018 coding sequences being lost from the genome (36). It was hypothesized that the genes lost were “useless genes” (i.e., nonessential genes), while the retained genes are essential to the viral life cycle (36). Examining the gene lists showed that both CYP5253A1 and CYP5254A1 are retained. Both the amino acid sequence and the nucleotide sequence showed 100% identity for CYP5253A1 before and after 150 rounds of replication. However, in CYP5254A1, there was a single amino acid change after subculturing, at residue 64 (Arg to Lys), due to a single point mutation (AGA to AAA).
P450 enzymes, including the viral P450s described herein, require incorporation of heme for protein architecture and function. To date, no genes that encode for heme-biosynthetic enzyme machinery have been found in any giant virus genome. Therefore, viruses encoding P450s must utilize heme synthesized by the host. The retention of Mimivirus P450s following 150 passages of subculture, and a dependence on the host for heme, indicates these P450s must be important in Mimivirus biology, although the exact role is unclear.
Discussion
Through searching all available viral genomes, we discovered a surprising number, diversity, and distribution of genes encoding unique P450 proteins. Most were found in the giant viruses, but we also uncovered P450 genes in a phage and a herpesvirus. In other organisms, P450 enzymes have broad metabolic roles in altering endogenous and exogenous chemicals in synthetic and degradation pathways not thought to be relevant in viruses. Our findings fundamentally challenge this viewpoint and raise critical questions concerning the origin and the function(s) of P450s in viruses, and of P450s in general.
Viral P450 Structure and Function.
CYP102L1 in mycobacteriophage Adler.
The P450 gene found in this mycobacteriophage defined an additional subfamily and gene, CYP102L1. To date, only one CYP102 protein has been examined structurally: CYP102A1 (P450 BM3). The crystal structure of phage CYP102L1 adds a second CYP102 structure to this gene family. The CYP102L1 structure closely resembles that of CYP102A1, suggesting that other CYP102s might have retained similar structural architecture. The high degree of protein sequence identity suggests the phage CYP102L1 was directly derived from the host (SI Appendix, Fig. S7). This apparent shuttling of a P450 gene on a small double-stranded DNA virus resembles the occurrence of many P450 genes found encoded in natural plasmids; for example, CYP101A1 (P450cam) on a plasmid affords the bacterium Pseudomonas putida the ability to utilize camphor as a sole carbon source.
CYP5253A1 in Mimivirus.
Unlike the phage CYP102L1, the P450 genes in the giant viruses do not bear any close sequence identity to P450s in contemporary hosts, or to any other known P450s. The chimeric CYP5253A1, found in Mimiviruses, is the most unusual, and there is no precedent for a P450 structure fusing a putative 2OG/FeII-dependent dioxygenase domain and a P450 domain. We do not know how the P450 domain and the putative 2OG domain may be functionally linked. The fact that Mimivirus CYP5253A showed a native P450 spectrum only when it was expressed as a recombinant fusion protein, and not as the P450 domain alone, suggests there could be a functional connection between the P450 and the 2OG/FeII-dependent dioxygenase domains. In other P450 fusion proteins (nitric oxide synthetases, P450BM3), both moieties participate in a common functional outcome.
The type I binding spectrum demonstrates that cholesterol binds to Mimivirus recombinant CYP5253A1 as a substrate would bind, suggesting that cholesterol may be a substrate of this P450, although transformation of cholesterol was not detected. While an enzymatic function was not observed for the recombinant giant virus P450s examined, a biological role in giant viruses is clearly implied by experimental observations that in both Mimiviridae and Pandoravirdiae, the virus P450s are among genes transcribed and translated in the host upon infection. The P450s in Mimivirus are also retained through multiple rounds of viral gene descent, suggesting some essential role.
Origin of the Virus P450s.
The sequence similarity of the P450 in mycobacteriophage Adler and a P450 in the Mycobacteroides host indicates that the phage almost certainly acquired this gene from the host. However, the situation is quite different in the giant viruses. Our uncovering and annotation of P450s in A. castellanii allow us to say that the P450s in giant viruses of amoebae are remarkably unlike any of the P450s in a common Acanthamoeba host. This suggests that, if acquired, most giant virus P450s would be derived from some unknown host, possibly early in giant virus evolution. It is also possible that such an early host may have been an evolutionary dead-end, while the virus continued. Genetic mutations during virus evolution could have erased similarity to P450 genes in a presumed host. The high degree of identity between apparently orthologous P450 genes in different viruses from different parts of the globe is not consistent with rapid evolution, or with an ancient acquisition. However, metagenomic analysis of ballast water from four ships (cargo carriers) in Korea revealed the presence of multiple viruses, including pandoraviruses and megaviruses (37), suggesting this is a contemporary avenue contributing to a global distribution of highly similar giant viruses.
Deciphering the origin of P450s in the Megavirales is complicated by the presence of genes for multiple P450s differing within and between taxa, and the loss of P450 genes from some. The finding that the P450 genes in the pandoraviruses are closely linked and share gene order suggests they arose by duplication of an ancestral P450 gene before the divergence of the two Pandoraviridae clades. CYP5634 and CYP5635 appear to define the P450 complement of pandoraviruses. However, while CYP5634A occurs in all pandoraviruses known to date, CYP5635A apparently was lost from some members of both clades.
As with the pandoraviruses, the occurrence of apparently orthologous P450 genes in mimiviruses from dramatically different environments around the globe (i.e., soils, other terrestrial sources, the deep ocean) indicates that these genes define the P450 complement in the Mimiviridae. Unlike P450s in the Pandoravirus, the CYP5253 and CYP5254 genes in the Mimiviridae are dramatically different from one another and reside in different locations in the genomes of species where both occur. The differences between CYP5253 and CYP5254 gene products in the Mimiviridae suggest that these genes could have separate origins. The Tupanvirus CYP5254B is more like the CYP5254B in Moumouvirus than the CYP5254A in other viruses in the Mimiviridae. The lack of CYP5253 genes in the megaviruses suggests that the different lineages in the Mimiviridae all may have diverged from a common ancestor, but that megaviruses lost a CYP5253 precursor.
A recent minimetagenomic approach detected 15 new assembled giant virus genomes in terrestrial soils, along with one additional genome obtained from the bulk metagenome (38). Five of these new genomes contain P450 genes. Satyrvirus, which is in a clade with Tupanvirus, has a CYP5254B like that found in Tupanvirus. Edafosvirus also has a CYP5254B similar to the Tupanvirus, and has one P450 that is plant-like and one that is fungus-like. Harvfovirus and Hyperionvirus both have a CYP5253 gene for a P450-2OG/FeII chimera that defines an additional subfamily, CYP5253B. Each virus also has another P450 gene, related to plant P450s; in Hyperionvirus, that P450 appears to be acquired from a moss. Terrestrivirus has a P450 gene weakly similar to a eukaryote P450 gene (CYP4). This suggests that in some environments, there are giant viruses that have acquired some P450 genes from eukaryotes.
Questions regarding the origin of giant virus P450 genes intersect with questions regarding the origin of the giant viruses themselves. Giant viruses all have genomes containing hundreds to thousands of genes, often including translation-associated genes that are common in cellular life. They have been proposed as a separate fourth domain of life (39), and possibly even a basal group that contributed to the origin of eukaryotes (40). Either hypothesis is consistent with the idea that the giant viruses are very ancient, and may have been derived from early cellular ancestors, losing many genes in the course of evolution. If the giant viruses began as large entities with many genes, then the P450 genes could be remnants of that original condition. If we accept the idea that the Megavirales originated near the time when cellularity first occurred, then one possibility is that the P450s originated in some entity ancestral to the Mimiviridae, Pandoraviridae, and others.
Conversely, others suggest that the giant viruses are derived from viral antecedents that developed large gene complements through acquisition (41–43), perhaps through waves of gene gain and loss (44). Phylogenetic analyses of genes for transfer RNA (tRNA) synthetases and core genes (14, 16) lend support to the idea that the Megavirales arose from a common ancestor that contained many fewer genes (16). If the hypothesis that giant viruses evolved by acquisition of genes is correct, then one could conclude that the P450 genes in the Mimiviridae, Pandoraviridae, and Mollivirus lineages were acquired from some host(s) in their evolution.
It is impossible to know at present whether a single ancestral P450 gene may have been acquired from an early host, and then duplicated and diverged during virus evolution, or whether there was acquisition and subsequent divergence of multiple different P450 genes. Given the differences between the Mimiviridae and Pandoraviridae P450s, and the phylogenetic separation between the Megavirales taxa bearing P450 genes, it is perhaps more plausible that P450 genes were acquired multiple times. This would be similar to a proposed independent acquisition of some tRNA synthetases in the giant viruses (14).
As yet, there is no consensus regarding the origin and evolution of the giant viruses (45, 46), which continue to be discovered (47, 48). It is noteworthy that the giant viruses with genes for multiple translation-associated proteins tend to be the ones that also contain cytochrome P450 genes. Further understanding of the viral P450s, their function, and their evolution may help to distinguish between giant virus evolution by genome growth, or genome reduction.
Materials and Methods
Viral P450 Sequence Datasets.
The P450 domain is readily identifiable bioinformatically. Candidate P450 sequences were retrieved from the National Center for Biotechnology Information sequence databases using BLAST. Inferred protein sequences were aligned to the Pfam P450 hidden Markov model (PF00067) using ClustalOmega (49), and scored with T-Coffee (50). Alignments were visualized in BioEdit (51). Virus P450 genes were named based on the established guidelines of the International Cytochrome P450 Nomenclature Committee (52). Virus names are those in the original reports or GenBank. Pairwise amino acid identities reported in the text are based on the length of the shortest protein in the alignment. Identity values used in the tables are based on multiple sequence alignments and are usually not more than 1% different from values stated in the text.
Phylogenetic Analyses of Viral P450s.
Phylogenetic trees of P450 domains were estimated by maximum likelihood (ML) using RAxML (v8.2.6) with the PROTCATLG model of amino acid substitution (53). Replicate ML trees were constructed, and the best-scoring tree was subjected to rapid bootstrapping in RAxML to assess bifurcation support. Virus phylogenies were constructed from the conserved RNA Pol2 (RNAP2) gene using the PROTCATHIVB model of amino acid substitution.
P450 Expression and Sterol Binding.
Sequences predicted to encode viral CYPs were synthesized by Eurofins MWG Operon or by DNA2.0 (ATUM) and cloned into the expression vector pCWori+ and expressed in E. coli (8). These included the full-length Mimivirus CYP5253A1 comprising both the heme and C-terminal chimeric domains, the Mimivirus CYP5253A1 heme domain alone, Hokovirus CYP5724A1, and Ranid herpesvirus 3 CYP5723A1 (SI Appendix, Fig. S8). Specific P450 heme contents were ∼17 nmol/mg of protein for those P450s expressed and purified, which equates to almost 100% heme incorporation.
Mycobacteriophage Adler CYP102L1 was cloned into expression vector pET28a and expressed in E. coli. The A. castellanii CYP51 was expressed and purified by methods used previously (54). A six-histidine tag was engineered at the C terminus of each P450, and proteins were purified according to the method of Arase et al. (55) using Ni2+-nitrilotriacetic acid agarose (54). The purification of recombinant CYP102L1 is described in detail in SI Appendix. Protein purity was assessed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SI Appendix, Fig. S9). Reduced CO difference spectra were measured by established methods (56). For P450 ligand binding experiments, the spectral difference was recorded after the incremental addition of substrate, and the Ks of the P450/substrate complex was determined (57). Reconstitution assays for P450 catalytic activity are described in detail in SI Appendix. Chemicals were the highest grade from Sigma Chemical Company unless otherwise stated. Eburicol and obtusifoliol were a gift from David Nes, Texas Tech University, Lubbock, TX.
Isolation, Crystallization, and Structure Determination of Adler Phage CYP102L1.
The Adler phage CYP102L1 gene was synthesized by GenScript and used to express protein for crystallization. Protein was expressed and purified by experimental procedures described in SI Appendix. Crystals of CYP102L1 were grown in hanging drops. Diffraction-quality crystals were obtained using an additive screen that yielded the final condition of 0.1 M sodium cacodylate (pH 6.5), 1.26 M ammonium sulfate, and 4.2% (wt/vol) dextran sulfate. Data were collected at the Stanford Synchrotron Radiation Lightsource Beamline 9-2. The structure was determined by single-wavelength anomalous diffraction, and a hybrid substructure search was performed using Fe-searched scattering type, with S as an additional scattering type. The details and the final refinement statistics are given in SI Appendix, Table S3. The coordinates and associated structure factors have been deposited with the Protein Data Bank (ID code 6N6Q).
Supplementary Material
Acknowledgments
We thank Dr. David Nes (Texas Tech University) for providing sterols and Dr. Matthieu Legendre and Dr. Chantal Abergel (CNRS, Marseille) for access to the P. celtis sequences. Drs. Irina Arkhipova, Mark Hahn, Judith Luborsky, and Ann Bucklin commented on the manuscript. The research was supported by a USA-UK Fulbright Scholarship and a Royal Society grant (to D.C.L.), the Boston University Superfund Research Program [NIH Grant 5P42ES007381 (to J.J.S. and J.V.G.) and NIH Grant 5U41HG003345 (to J.V.G.)], the European Regional Development Fund and Welsh Government Project BEACON (S.L.K.), the Woods Hole Center for Oceans and Human Health [NIH Grant P01ES021923 and National Science Foundation Grant OCE-1314642 (to J.J.S.)], and NIH Grant R01GM53753 (to T.L.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and associated structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 6N6Q).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901080116/-/DCSupplemental.
References
- 1.Klingenberg M., Pigments of rat liver microsomes. Arch. Biochem. Biophys. 75, 376–386 (1958). [DOI] [PubMed] [Google Scholar]
- 2.Omura T., Sato R., A new cytochrome in liver microsomes. J. Biol. Chem. 237, 1375–1376 (1962). [PubMed] [Google Scholar]
- 3.Estabrook R. W., Cooper D. Y., Rosenthal O., The light reversible carbon monoxide inhibition of the steroid C21-hydroxylase system of the adrenal cortex. Biochem. Z. 338, 741–755 (1963). [PubMed] [Google Scholar]
- 4.Nelson D. R., Goldstone J. V., Stegeman J. J., The cytochrome P450 genesis locus: The origin and evolution of animal cytochrome P450s. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese C. R., Fox G. E., Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. U.S.A. 74, 5088–5090 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guengerich F. P., Munro A. W., Unusual cytochrome P450 enzymes and reactions. J. Biol. Chem. 288, 17065–17073 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson D. R., Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 1866, 141–154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raoult D., et al. , The 1.2-megabase genome sequence of Mimivirus. Science 306, 1344–1350 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Legendre M., Santini S., Rico A., Abergel C., Claverie J. M., Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol. J. 8, 99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colson P., La Scola B., Levasseur A., Caetano-Anollés G., Raoult D., Mimivirus: Leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Microbiol. 15, 243–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreani J., et al. , Orpheovirus IHUMI-LCC2: A new virus among the giant viruses. Front. Microbiol. 8, 2643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrahão J., et al. , Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 9, 749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreani J., Verneau J., Raoult D., Levasseur A., La Scola B., Deciphering viral presences: Two novel partial giant viruses detected in marine metagenome and in a mine drainage metagenome. Virol. J. 15, 66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz F., et al. , Giant viruses with an expanded complement of translation system components. Science 356, 82–85 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Abergel C., Legendre M., Claverie J. M., The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev. 39, 779–796 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Aherfi S., Colson P., La Scola B., Raoult D., Giant viruses of amoebas: An update. Front. Microbiol. 7, 349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claverie J. M., Abergel C., Mimivirus and its virophage. Annu. Rev. Genet. 43, 49–66 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Aoyama Y., Yoshida Y., Sato R., Yeast cytochrome P-450 catalyzing lanosterol 14 alpha-demethylation. II. Lanosterol metabolism by purified P-450(14)DM and by intact microsomes. J. Biol. Chem. 259, 1661–1666 (1984). [PubMed] [Google Scholar]
- 19.Lamb D. C., et al. , The first virally encoded cytochrome P450. J. Virol. 83, 8266–8269 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desnues C., et al. , Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl. Acad. Sci. U.S.A. 109, 18078–18083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philippe N., et al. , Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341, 281–286 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Antwerpen M. H., et al. , Whole-genome sequencing of a pandoravirus isolated from keratitis-inducing Acanthamoeba. Genome Announc. 3, e00136-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legendre M., et al. , Diversity and evolution of the emerging Pandoraviridae family. Nat. Commun. 9, 2285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aherfi S., et al. , A large open pangenome and a small core genome for giant pandoraviruses. Front. Microbiol. 9, 1486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legendre M., et al. , Pandoravirus celtis illustrates the microevolution processes at work in the giant Pandoraviridae genomes. Front. Microbiol. 10, 430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legendre M., et al. , In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. U.S.A. 112, E5327–E5335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legendre M., et al. , Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. U.S.A. 111, 4274–4279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajrai L. H., et al. , Kaumoebavirus, a new virus that clusters with Faustoviruses and Asfarviridae. Viruses 8, E278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Origgi F. C., et al. , Ranid herpesvirus 3 and proliferative dermatitis in free-ranging wild common frogs (Rana temporaria). Vet. Pathol. 54, 686–694 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Clarke M., et al. , Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 14, R11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y., et al. , Investigations of heme ligation and ligand switching in cytochromes P450 and P420. Biochemistry 52, 5941–5951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilton D. C., Munday K. A., Skinner S. J., Akhtar M., The biological conversion of 7-dehydrocholesterol into cholesterol and comments on the reduction of double bonds. Biochem. J. 106, 803–810 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei L., Waterman M. R., Fulco A. J., Kelly S. L., Lamb D. C., Availability of specific reductases controls the temporal activity of the cytochrome P450 complement of Streptomyces coelicolor A3(2). Proc. Natl. Acad. Sci. U.S.A. 101, 494–499 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renesto P., et al. , Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J. Virol. 80, 11678–11685 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legendre M., et al. , mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 20, 664–674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer M., et al. , Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. U.S.A. 108, 10296–10301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang J., Park S. Y., Lee S., Lee T. K., High diversity and potential translocation of DNA viruses in ballast water. Mar. Pollut. Bull. 137, 449–455 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Schulz F., et al. , Hidden diversity of soil giant viruses. Nat. Commun. 9, 4881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raoult D., TRUC or the need for a new microbial classification. Intervirology 56, 349–353 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Forterre P., Gaïa M., Giant viruses and the origin of modern eukaryotes. Curr. Opin. Microbiol. 31, 44–49 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Iyer L. M., Balaji S., Koonin E. V., Aravind L., Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117, 156–184 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Yutin N., Wolf Y. I., Koonin E. V., Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 466–467, 38–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abrahão J. S., Araújo R., Colson P., La Scola B., The analysis of translation-related gene set boosts debates around origin and evolution of mimiviruses. PLoS Genet. 13, e1006532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filée J., Multiple occurrences of giant virus core genes acquired by eukaryotic genomes: The visible part of the iceberg? Virology 466–467, 53–59 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Koonin E. V., Yutin N., Multiple evolutionary origins of giant viruses. F1000 Res. 7, 1840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colson P., et al. , Ancestrality and mosaicism of giant viruses supporting the definition of the Fourth TRUC of microbes. Front. Microbiol. 9, 2668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson W. H., et al. , Genomic exploration of individual giant ocean viruses. ISME J. 11, 1736–1745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cottrell M. T., Kirchman D. L., Virus genes in Arctic marine bacteria identified by metagenomic analysis. Aquat. Microb. Ecol. 66, 107–116 (2012). [Google Scholar]
- 49.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang J. M., Di Tommaso P., Notredame C., TCS: A new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol. Biol. Evol. 31, 1625–1637 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Hall T. A., BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- 52.Nelson D. R., et al. , P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6, 1–42 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamb D. C., et al. , Azole antifungal agents to treat the human pathogens Acanthamoeba castellanii and Acanthamoeba polyphaga through inhibition of sterol 14α-demethylase (CYP51). Antimicrob. Agents Chemother. 59, 4707–4713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arase M., Waterman M. R., Kagawa N., Purification and characterization of bovine steroid 21-hydroxylase (P450c21) efficiently expressed in Escherichia coli. Biochem. Biophys. Res. Commun. 344, 400–405 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Omura T., Sato R., The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 239, 2379–2385 (1964). [PubMed] [Google Scholar]
- 57.Lamb D. C., et al. , The mutation T315A in Candida albicans sterol 14alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J. Biol. Chem. 272, 5682–5688 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.