Significance
Considering that Alzheimer’s disease (AD) is a chronic disease progressing over a long period of time, even a slight increase of BACE1 expression may have a profound effect on Aβ accumulation. We describe a previously unknown mechanism that negatively regulates BACE1 and BACE1-AS expression and demonstrate its pivotal role in the progression of Aβ and Tau pathologies and cognitive impairment in two mouse models of AD. Given the recent failures of the clinical trials using enzymatic inhibitors of BACE1, it is critical to explore alternative approaches such as down-regulating BACE1 and BACE1-AS transcription. Our finding that NRF2 negatively regulates BACE1 and BACE1-AS therefore suggests a potential for disease modification by NRF2-activating phytochemicals or synthetic small molecules in AD.
Keywords: NRF2, BACE1, Alzheimer’s disease, 3xTg-AD mice, 5xFAD mice
Abstract
BACE1 is the rate-limiting enzyme for amyloid-β peptides (Aβ) generation, a key event in the pathogenesis of Alzheimer’s disease (AD). By an unknown mechanism, levels of BACE1 and a BACE1 mRNA-stabilizing antisense RNA (BACE1-AS) are elevated in the brains of AD patients, implicating that dysregulation of BACE1 expression plays an important role in AD pathogenesis. We found that nuclear factor erythroid-derived 2-related factor 2 (NRF2/NFE2L2) represses the expression of BACE1 and BACE1-AS through binding to antioxidant response elements (AREs) in their promoters of mouse and human. NRF2-mediated inhibition of BACE1 and BACE1-AS expression is independent of redox regulation. NRF2 activation decreases production of BACE1 and BACE1-AS transcripts and Aβ production and ameliorates cognitive deficits in animal models of AD. Depletion of NRF2 increases BACE1 and BACE1-AS expression and Aβ production and worsens cognitive deficits. Our findings suggest that activation of NRF2 can prevent a key early pathogenic process in AD.
Alzheimer’s disease (AD) is the most common type of dementia and is characterized by accumulation of amyloid-β (Aβ) plaques and neurofibrillary tangles, synaptic and neuronal loss, and cognitive decline. BACE1 is the only β-secretase responsible for the production of Aβ and therefore plays a key role in the pathogenesis of AD (1–3). A long noncoding RNA transcribed from the opposite strand of BACE1 (BACE1-AS) stabilizes BACE1 mRNA by forming a heteromeric RNA duplex (4). BACE1 mRNA and protein levels as well as BACE1-AS transcript are abnormally elevated in postmortem brain tissue from patients with AD (4–8). A small increase in BACE1 induces a dramatic increase in Aβ production (9), and inhibitors of BACE1 enzyme activity are being pursued as a therapeutic strategy for AD (10). Genetic reduction of BACE1 or BACE1-AS levels reduces Aβ plaque pathology in mouse models of AD (4, 11–13), suggesting that identification of transcriptional repressors of BACE1 gene expression could provide an avenue for intervention in AD.
Nuclear factor erythroid-derived 2-related factor 2 (NRF2/NFE2L2) is a transcription factor that binds to the antioxidant response elements (AREs) and regulates a variety of cytoprotective and detoxification genes (14). In the inactive state, kelch-like ECH-associated protein1 (KEAP1) binds to NRF2 and retains it in the cytoplasm where it is degraded by proteasomes (15, 16). NRF2 activators, such as sulforaphane and tert-butylhydroquinone (tBHQ), modify cysteine residues of KEAP1, leading to conformational change and disrupting the KEAP1-NRF2 interaction, and accumulated NRF2 then translocates to the nucleus and transactivates target genes by binding to their AREs (17, 18). NRF2 levels are reduced, and NRF2 is localized predominately in the cytoplasm of hippocampal neurons of AD patients (19). In addition, altered expression of NRF2 target genes is associated with Aβ pathology in AD animal models (20–22). Here we show that NRF2 is a negative regulator of BACE1 expression that can ameliorate Aβ pathology and cognitive deficits in mouse models of AD.
Results
Association of NRF2 and BACE1 with AD.
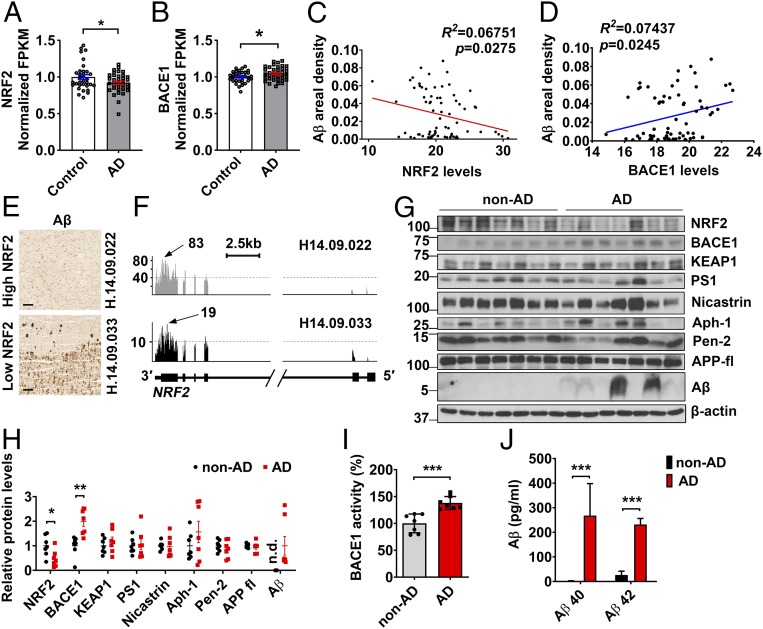
To investigate the association of NRF2 with Alzheimer’s disease, we first analyzed RNA-sequencing (RNA-seq) data from the Allen Brain Institute’s Aging, Dementia, and TBI Study. Since several samples, however, showed the abnormal pathological features, we selected AD samples (n = 38) revealing AD neuropathological features, i.e., Aβ42 secretion, Aβ42/Aβ40 ratio, and presence of amyloid plaques, and control samples (n = 34) not showing AD-related markers. Using these selected samples, we found that the number of NRF2 reads is significantly reduced in AD patients compared with controls, whereas BACE1 is elevated in AD (Fig. 1 A and B). The number of reads for KEAP1, γ-secretase complex [Presenilin 1(PSEN1), Nicastrin, APH-1α, and PEN-2], and APP were not different between control and AD brains (SI Appendix, Fig. S1). Linear regression analysis revealed a significant negative correlation between NRF2 levels and Aβ plaque accumulation (Fig. 1C). In contrast, a positive correlation was found between BACE1 levels and Aβ plaque accumulation (Fig. 1D). While the samples showing high NRF2 read numbers reveal lower Aβ plaque load, low read numbers of NRF2 transcript are related to higher Aβ plaque amounts (Fig. 1 E and F).
Fig. 1.
Association of NRF2 and BACE1 with AD. (A–F) RNA-seq and neuropathological protein quantification data obtained from the Allen Brain Institute’s Aging, Dementia, and TBI Study. (A and B) NRF2 and BACE1 normalized fragments per kilobase million (FPKM) in the parietal cortex of control (n = 34) and AD patients (n = 38). (C and D) Linear regression analysis between areal percentage covered by Aβ and NRF2 (C) and BACE1 (D) levels by RNA-seq in parietal cortex of both control and AD patients (n = 72). (E) Aβ immunohistochemistry images on AD patients’ parietal cortex selected for the RNA-seq study. (Scale bar, 200 μm.) (F) Coverage reads of normalized RNA-seq throughout the NRF2 locus in the parietal cortex of AD patients characterized in E. Arrows indicate the highest read coverage point. (G) Western blot analysis of NRF2, BACE1, KEAP1, PS1 (Presenilin 1), Nicastrin, Aph-1, Pen-2, full-length APP(APP-fl), and Aβ levels in the brains of AD patients (n = 7) and nondemented controls (non-AD, n = 7). Research subject demographics and amyloid plaque data are shown in SI Appendix, Table S1. (H) Results of quantitative analysis of G. (I) BACE1 enzymatic activities in non-AD and AD patients’ brain extracts. (J) Levels of Aβ40 and Aβ42 were quantified in non-AD (n = 5) and AD (n = 5) patients’ brain by ELISA. Values are the means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001; two-tailed Student’s t test (A, B, and H–J).
Then, we determined the levels of NRF2 and BACE1 in postmortem brain tissue from AD patients and age-matched neurologically normal subjects (SI Appendix, Methods). In AD subjects, NRF2 protein levels were reduced by 50% compared with nondemented controls, whereas BACE1 levels were significantly elevated, as were levels of Aβ (Fig. 1 G and H). These BACE1 increases corresponded with increases in BACE1 activity and Aβ production in AD brains (Fig. 1 I and J). In accord with RNA-seq data, the protein levels of KEAP1, γ-secretase complex, and APP were not different between control and AD (Fig. 1 G and H). These data revealed that NRF2 expression is strongly reduced in AD patients.
NRF2 Negatively Regulates the Transcription of BACE1 and BACE1-AS.
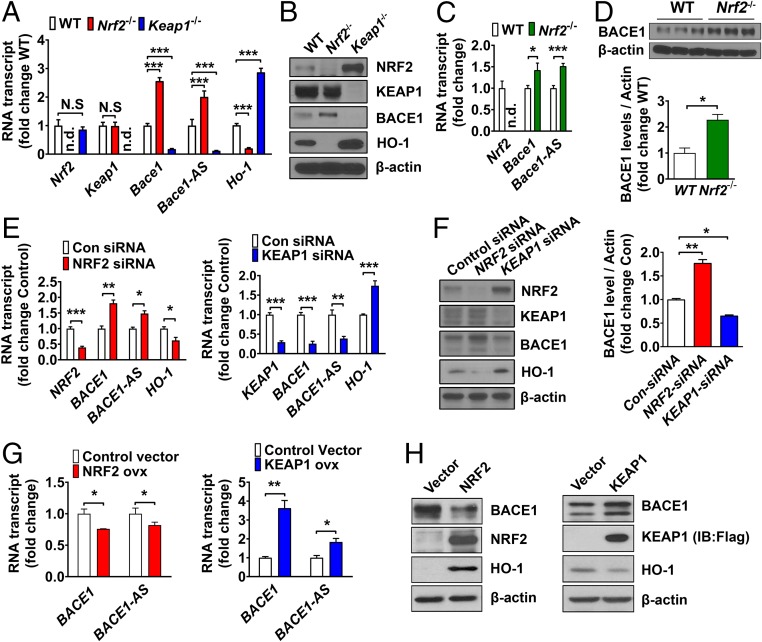
We next analyzed the expression of Bace1 in mouse embryonic fibroblasts (MEFs) from Keap1−/−, Nrf2−/−, and wild-type (WT) mice. We found that the levels of Bace1 and Bace1-AS transcript were increased in Nrf2−/− MEFs compared with WT MEFs, whereas Bace1 and Bace1-AS transcript levels were decreased in Keap1−/− MEFs that exhibit NRF2 accumulation as a result of defective KEAP1-mediated NRF2 degradation (Fig. 2 A and B). As expected, expression of the known NRF2 target gene Heme oxygenase 1(Ho-1) was substantially reduced in Nrf2−/− MEFs and increased in Keap1−/− MEFs (Fig. 2 A and B). Bace1 and Bace1-AS expression was significantly increased in the brains of 12-month-old Nrf2−/− mice compared with WT mice, suggesting that NRF2 negatively regulates Bace1 and Bace1-AS gene expression (Fig. 2 C and D). Knockdown of NRF2 in human neuronal SH-SY5Y cells using RNA interference technology increased the expression of BACE1 and BACE1-AS and reduced HO-1 expression (Fig. 2 E and F). Conversely, reduction of KEAP1 increased NRF2 levels, suppressed BACE1 and BACE1-AS expression, and increased HO-1 expression (Fig. 2 E and F). On the other hand, ectopic expression of NRF2 led to decreased BACE1 and BACE1-AS levels, whereas BACE1 and BACE1-AS levels were increased when KEAP1 was overexpressed (Fig. 2 G and H). Consistent with these findings, we also observed that two pharmacological NRF2 inducers, sulforaphane and tBHQ, significantly down-regulated BACE1 and BACE1-AS and up-regulated the NRF2 target gene HO-1 (SI Appendix, Fig. S2 A–D). Together, these results suggest that NRF2 negatively regulates BACE1 and BACE1-AS gene transcription.
Fig. 2.
NRF2 regulates the transcription of BACE1 and BACE1-AS. (A and B) Transcript (A) and protein (B) levels of Nrf2, Keap1, Bace1, Bace1-AS, and Ho-1 in WT, Nrf2−/−, and Keap1−/− MEF cells. (C and D) Transcript levels (C) of Nrf2, Bace1, and Bace1-AS, and BACE1 protein levels (D) in cerebral cortex lysates of 12-month-old WT mice and Nrf2−/− mice. (E and F) Transcript (E) and protein (F) levels of NRF2, BACE1, BACE1-AS, and HO-1 were determined in SH-SY5Y cells transfected with control, NRF2, or KEAP1 siRNA. (G) Transcription levels of BACE1 and BACE1-AS are reduced in SH-SY5Y cells overexpressing NRF2, whereas BACE1 and BACE1-AS transcripts are increased when overexpressed with KEAP1. (H) Protein levels of BACE1, NRF2, HO-1, and β-actin were determined from the same samples as in G. NRF2 overexpression reduced BACE1 expression (Left), and NRF2 inhibition by KEAP1 overexpression increased BACE1 levels (Right). For all graphs, values are the means ± SEM; *P < 0.05, **P < 0.01, and ***P < 0.001; two-tailed Student’s t test (A and C–G). N.S, nonsignificant; n.d., not detectable. For all panels, n = 3 separate cultures.
NRF2 Binds to the AREs of BACE1 and BACE1-AS Promoters and Suppresses Their Expression.
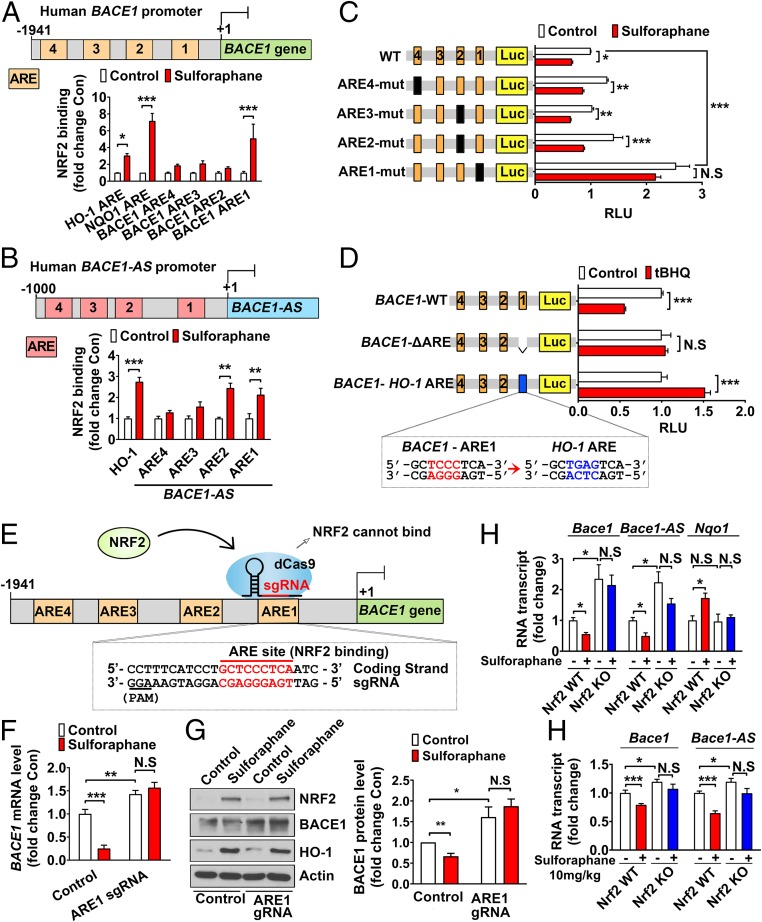
We next investigated the possibility that BACE1 and BACE1-AS are direct target genes of the transcription factor NRF2. Sequence analysis using USCS Genome Browser revealed four, four, three, and one putative AREs in the proximal promoter regions of human BACE1, human BACE1-AS, mouse Bace1, and mouse Bace1-AS, respectively (SI Appendix, Figs. S3–S6). We performed chromatin immunoprecipitation (ChIP) assays and found that NRF2 binding to ARE1 in human BACE1 promoter was increased in sulforaphane-treated SH-SY5Y cells compared with controls, with no significant changes seen at the other ARE sites in human BACE1 promoter (Fig. 3A). In human BACE1-AS promoter, NRF2 binding to ARE1 and ARE2 was significantly enhanced by sulforaphane treatment (Fig. 3B). Sulforaphane increased binding of NRF2 to the ARE sites of the HO-1 and NAD(P)H:quinone oxidoreductase 1 (NQO1) promoters (Fig. 3 A and B). By in vivo ChIP analysis, we also detected prominently enhanced binding of NRF2 to ARE3 of the mouse Bace1 promoter and ARE1 of mouse Bace1-AS promoter in the brain tissues of sulforaphane-treated WT mice compared with vehicle-treated mice (SI Appendix, Fig. S7 A and B).
Fig. 3.
NRF2 directly binds to the ARE sites in the BACE1 promoter, and NRF2 activation reduces BACE1 expression. (A) Endogenous NRF2 binding affinity to ARE1 in human BACE1 promoter and NFR2 binding to the HO-1 and NQO1 promoters are increased in SH-SY5Y cells treated with sulforaphane (1 μM). (B) Endogenous NRF2 binding to ARE1 and ARE2 in the human BACE1-AS promoter and to the HO-1 promoter is increased in SH-SY5Y cells treated with sulforaphane. (C) Firefly luciferase reporter plasmids carrying the WT or ARE mutant human BACE1 promoters were cotransfected with the Renilla luciferase reporter plasmid (pRLTK ΔARE) into HEK293T cells, and the cells were treated with or without sulforaphane. pRLTK ΔARE was used for normalizing transfection efficiency (RLU, relative luciferase units). (D) Relative luciferase activity of BACE1-WT and modified vectors was measured in the HEK293T cells after treatment with tBHQ (10 μM) for 24 h. (E) The ARE1 location in human BACE1 promoter and CRISPRi strategy with a guide RNA (sgRNA) to target dCas9 to the ARE1 sequence. (F and G) CRISPRi of ARE1 in human BACE1 promoter blocks NRF2-induced decrease in BACE1 mRNA (F) and protein (G). SH-SY5Y cells were transfected with ARE1 sgRNA only or pdCas9 vector and ARE1 sgRNA. The ARE1 sgRNA was designed with a 20-bp complementary region including human BACE1 promoter ARE1. To select a pure population of dCas9-expressing cells, SH-SY5Y cells were treated with puromycin and sulforaphane. (H) Transcription levels of Bace1, Bace1-AS, and Nqo1 were measured in WT and Nrf2−/− MEFs treated with vehicle (−) or 4 μM sulforaphane (+) for 24 h. (I) Bace1 and Bace1-AS transcript levels in hippocampus of WT mice and Nrf2−/− mice treated with vehicle (0.67% dimethyl sulfoxide) or 10 mg/kg sulforaphane every other day for 4 wk (+). There were six WT mice and eight Nrf2−/− mice in each treatment group. Values are the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001; two-tailed Student’s t test (A–D, F, and G) or two-way ANOVA (H and I) (N.S, nonsignificant) (n = 3, separate cultures).
We generated a series of point mutants for the human BACE1 promoter-driven reporters and compared their promoter activities. Disruption of ARE1 of the human BACE1 promoter by site-directed mutagenesis (mutations of conserved nucleotides in the consensus sequence: TGANNNNGC) led to an ∼2.5-fold enhancement of promoter activity compared with the WT promoter (Fig. 3C). Moreover, when the ARE1 site was mutated, sulforaphane-mediated NRF2 activation barely suppressed human BACE1 promoter transcription, whereas WT and other ARE mutants showed significant responses to the treatment of sulforaphane (Fig. 3C). To confirm that the AREs of mouse Bace1 and Bace1-AS promoters were functional, we constructed WT and point mutant mouse Bace1 and Bace1-AS promoter-driven reporters and tested their activities in mouse neuronal HT22 cells. We found that the reporter activities of WT mouse Bace1and Bace1-AS promoters were significantly decreased by NRF2 induction, while ARE3-mutated mouse Bace1 and ARE1-mutated Bace1-AS promoters did not respond to NRF2 induction (SI Appendix, Fig. S7 C and D).
To investigate how NRF2 can function as both a transcriptional activator and a repressor, we changed the ARE1 sequence of the BACE1 promoter (GCTCCCTCA) into the ARE sequence of the HO-1 promoter (GCTGAGTCA) and compared the promoter activities. The WT BACE1 promoter showed significantly reduced activity in response to the NRF2 inducer tBHQ (Fig. 3D). Activity of the BACE1 promoter lacking ARE1 (BACE1-ΔARE1) was not affected by tBHQ, whereas activity of the BACE1 promoter in which ARE1 was replaced with the ARE of HO-1 (BACE1-HO-1 ARE) was significantly increased by tBHQ treatment (Fig. 3D). These results indicate that differences in nonconserved sequences within the ARE sequence (TGANNNNGC) determine whether NRF2 acts as a transcriptional activator or repressor.
We next asked whether the NRF2 binding element of the BACE1 promoter in the human genome responds to endogenous NRF2. CRISPR interference (CRISPRi) technology (23, 24) was used to direct a catalytically inactive Cas9 (dCas9) to the ARE1 of BACE1 promoter with guide RNA (gRNA) to interfere with NRF2 binding to the site (Fig. 3E). We found that CRISPRi abolished NRF2-mediated repression of BACE1 expression (Fig. 3 F and G). Sulforaphane, an NRF2 inducer, reduced Bace1 and Bace1-AS transcripts and increased Nqo1 transcript levels in WT MEFs, while these effects of sulforaphane were lost in Nrf2−/− MEFs (Fig. 3H). Sulforaphane-mediated reduction of Bace1 and Bace1-AS transcript levels did not occur in brain tissues of Nrf2−/− mice (Fig. 3I). Together, these results demonstrate that NRF2 inducers suppress the transcription of Bace1 and Bace1-AS genes in an NRF2-dependent manner.
We also determined whether NRF2 affects the expression of γ-secretase components or the activity of γ-secretase. When we transfected SH-SY5Y cells with NRF2 or KEAP1 cDNA expression plasmids, the protein levels of five different components of γ-secretase complex were not changed (SI Appendix, Fig. S8 A and B). Further, we examined γ-secretase activity using luciferase-based γ-secretase activity assay. Overexpression of NRF2 or KEAP1 did not alter γ-secretase activity (SI Appendix, Fig. S8 A and B). Also, sulforaphane, an NRF2 activator, did not affect γ-secretase activity, whereas DAPT, a γ-secretase direct inhibitor, significantly reduced the γ-secretase activity (SI Appendix, Fig. S8B). These data suggest that NRF2 activation may not affect the γ-secretase pathway to alter amyloidosis.
NRF2-Mediated Reduction of Bace1 and Bace1-AS Expression Is Independent of ROS Regulation.
Since a number of NRF2-regulated genes control oxidative stress, and Bace1 expression can be regulated by reactive oxygen species (ROS), we determined whether the NRF2-mediated suppression of Bace1 and Bace1-AS expression depends on the ROS level. The ROS level was ∼10-fold up-regulated in Nrf2−/− MEFs compared with WT, whereas ROS levels in Keap1−/− MEFs were similar to WT MEFs (SI Appendix, Fig. S9A). However, Keap1−/− cells exhibited increased expression of Nrf2 and Ho-1 and reduced expression of Bace1 and Bace1-AS compared with WT cells (Fig. 2 A and B). Treatment with the antioxidant N-acetyl cysteine (NAC) significantly reduced ROS levels (SI Appendix, Fig. S9B), without affecting levels of Bace1 and Bace1-AS transcripts in cells lacking NRF2 (SI Appendix, Fig. S9 C and D). Down-regulation of NRF2 increased ROS levels in SH-SY5Y cells (SI Appendix, Fig. S9E), whereas NAC treatment did not affect the NRF2 knockdown-induced transcription of BACE1 and BACE1-AS (SI Appendix, Fig. S9 F and G). Interestingly, NAC treatment increased transcript levels of BACE1 and BACE1-AS, although ROS levels were significantly reduced by NAC treatment. Collectively, the data (SI Appendix, Fig. S9) suggest that NRF2-mediated reduction of BACE1 and BACE1-AS expression is independent of ROS regulation.
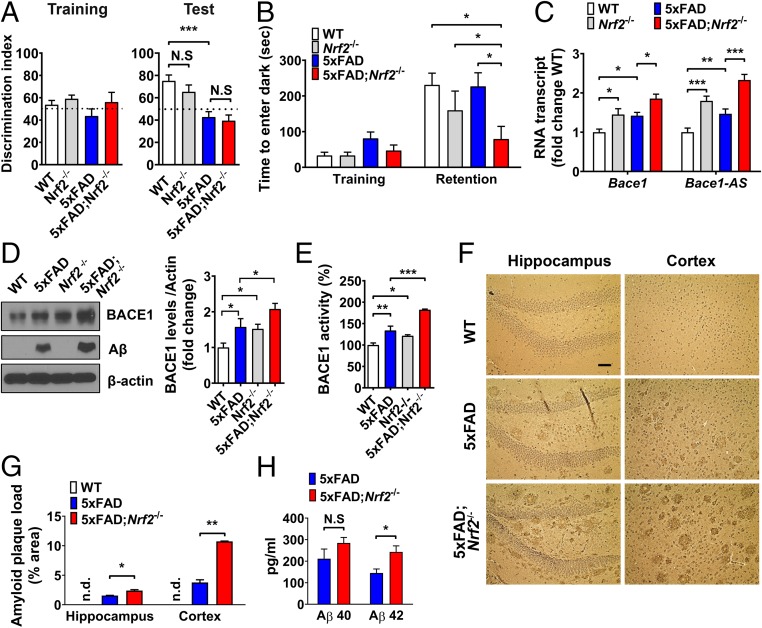
Deficiency of Nrf2 Accelerates Bace1 Expression, Aβ Pathology, and Cognitive Decline in 5xFAD Mice.
To elucidate whether Nrf2 deficiency mediates increased Bace1 expression, Aβ pathology, and associated cognitive deficits in AD, we ablated Nrf2 in 5xFAD mice (5xFAD;Nrf2−/−). The novel object recognition test showed no significant differences between 5xFAD and 5xFAD;Nrf2−/− mouse groups, but 5xFAD;Nrf2−/− mice showed substantial cognitive impairment in the passive avoidance test compared with 5xFAD mice (Fig. 4 A and B). In brain tissue, Bace1 and Bace1-AS expression levels were higher in Nrf2−/− mice than in WT mice, and 5xFAD;Nrf2−/− mice showed higher levels of Bace1 and Bace1-AS transcripts and BACE1 protein compared with Nrf2−/− and 5xFAD mice (Fig. 4 C and D). There was a significant increase in Aβ levels in 5xFAD;Nrf2−/− mice compared with 5xFAD mice, with Aβ levels below the limit of detection in WT and Nrf2−/− mice (Fig. 4D). As previously reported (5), a positive correlation was found between BACE1 expression levels and BACE1 activity in these mice groups (Fig. 4E). Aβ plaque loads in the hippocampus and cortex of 5xFAD;Nrf2−/− were significantly greater than in age-matched 5xFAD mice (Fig. 4 F and G and SI Appendix, Fig. S10). Brain deposition of Aβ42 was also significantly elevated in 5xFAD;Nrf2−/− mice compared with 5xFAD, whereas Aβ40 levels were not different in both mice groups (Fig. 4H). In the 5xFAD and 5xFAD;Nrf2−/− mice, the amounts of Aβ accumulation reflects Bace1 levels of each genotype. These findings suggest that Nrf2 deficiency accelerates cognitive decline through the induction of Bace1 and Bace1-AS resulting in accelerated Aβ pathology in AD mice.
Fig. 4.
Nrf2 deficiency increases BACE1 expression and exacerbates Aβ-associated cognitive deficits in 5xFAD mice. Nine-months-old WT, 5xFAD, Nrf2−/−, and 5xFAD;Nrf2−/− mice were examined (n = 8∼12 per group). (A) In the novel object recognition test, mice in each group spent the same time exploring the two objects during the training session. The 5xFAD and 5xFAD;Nrf2−/− mice failed to spend more time with the novel object than the familiar object in the test session. (B) In the passive avoidance test, only 5xFAD;Nrf2−/− mice exhibited impaired learning and memory function. (C) Levels of Bace1 and Bace1-AS transcripts in cerebral cortical tissue samples of the indicated genotypes of mice. (D) Levels of BACE1 and Aβ proteins in cerebral cortical tissue samples of the indicated genotypes of mice. (E) BACE1 enzymatic activity measured in the same samples as C and D. (F) Light microscopic images of Aβ immunoreactivity with hematoxylin counterstaining in cortex and hippocampus of WT, 5xFAD, and 5xFAD;Nrf2−/− mice. (Scale bar, 50 μm.) (G) Aβ plaque loads in cortex and hippocampus of WT, 5xFAD, and 5xFAD;Nrf2−/− mice. (H) The levels of Aβ1–40 and Aβ1–42 of the cerebral cortex samples measured by ELISAs. Values are the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001; one-way ANOVA with Tukey’s (A, B, D, and E), two-way ANOVA (C and G), or two-tailed Student’s t test (H) (N.S, nonsignificant; n.d., not detectable).
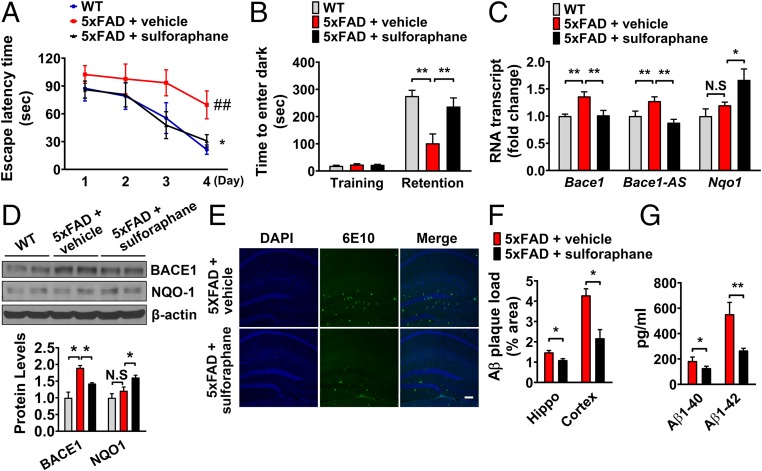
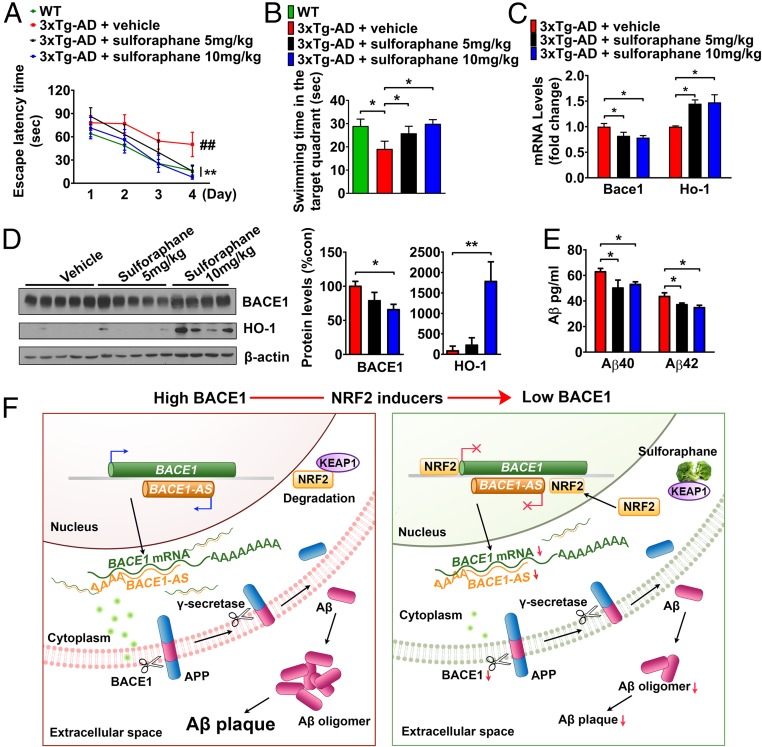
NRF2 Inducer Sulforaphane Ameliorates Cognitive Deficits and Aβ Accumulation by Reducing Bace1 Expression in 5xFAD and 3xTg-AD Mice.
We next examined whether suppression of Bace1 expression by sulforaphane resulted in a delay of cognitive decline by reducing Aβ production in vivo. Two different animal models of AD, 5xFAD and 3xTg-AD mice, were treated every other day with sulforaphane for 8 wk. In the Morris water maze and passive avoidance tests, administration of sulforaphane improved the impaired learning and memory of 5xFAD and 3xTg-AD mice (Figs. 5 A and B and 6 A and B). Bace1 and Bace1-AS transcript levels were up-regulated in the brain tissues of 5xFAD mice compared with WT mice and were reduced in 5xFAD mice that had been treated with sulforaphane (Fig. 5 C and D). As in 5xFAD mice, Bace1 mRNA and protein levels were significantly decreased in brain tissues of sulforaphane-treated 3xTg-AD mice (Fig. 6 C and D). Brain deposition of Aβ was also significantly reduced in sulforaphane-treated 5xFAD and 3xTg-AD mice (Figs. 5 E–G and 6E). Tau is an important mediator of Aβ neurotoxicity, and Aβ clearance leads to reduction of Tau pathology in AD mice (25–28). To determine whether NRF2 activation influences tau phosphorylation, levels of phosphorylated Tau (pTau) were analyzed in sulforaphane- and vehicle-treated 3xTg-AD mice. Levels of pTau (PHF-1, Tau phospho Ser-198, and Tau phospho Thr-217) were significantly reduced in brain tissues of sulforaphane-treated 3xTg-AD mice compared with vehicle-treated mice (SI Appendix, Fig. S11A). Unexpectedly, total Tau level was increased in 10 mg/kg sulforaphane-treated 3xTg-AD mice. Immunohistochemical analysis confirmed that pTau (pS198) and Aβ (6E10) levels were decreased in sulforaphane-treated 3xTg-AD mice in a dose-dependent manner (SI Appendix, Fig. S11B).
Fig. 5.
The NRF2 inducer sulforaphane ameliorates cognitive deficits and Aβ accumulation by reducing Bace1 expression in 5xFAD mice. Three-month-old 5xFAD mice and age-matched nontransgenic littermates (WT) were treated with sulforaphane (10 mg/kg every other day) or vehicle for 2 month (n = 6∼8 per group). (A) Acquisition of memory of the location of the hidden platform (escape latency) in the water maze on four consecutive days of training. Values are the mean ± SEM. ##P < 0.01, WT versus 5xFAD; *P < 0.05 5xFAD versus 5xFAD+sulforaphane (one-way ANOVA with Tukey’s post hoc test). (B) Latency times in training and memory retention trials in a passive avoidance task. (C) Transcript expression of Bace1, Bace1-AS, and Nqo1 in cerebral cortex tissue from WT mice, and control- and sulforaphane-treated 5xFAD mice. (D) Immunoblot analysis of BACE1, NQO1, and β-actin proteins in cerebral cortex samples from WT mice, and control- and sulforaphane-treated 5xFAD mice. (E) Staining of amyloid plaques in the hippocampus of control- and sulforaphane-treated 5xFAD mice. (Scale bar, 200 μm.) (F) The numbers of Aβ plaques/mm2 in hippocampus and cortex were quantified. (G) Aβ1–40 and Aβ1–42 levels in cerebral cortex samples were measured using specific ELISAs. Values are the means ± SEM. *P < 0.05 and **P < 0.01; two-way ANOVA with Bonferroni posttests (B–D) or two-tailed Student’s t test (F and G) (N.S, nonsignificant).
Fig. 6.
Administration of sulforaphane reduces Bace1 expression in brain and prevents memory impairments in the 3xTg AD mice. Seven-month-old 3xTg-AD mice were treated with vehicle (PBS) or sulforaphane (5 or 10 mg/kg, every other day) for 2 months. Mice were trained and tested on the spatial memory version of the Morris water maze (MWM). (A) Sulforaphane rescued spatial memory acquisition deficits during the 4 d of training. All mice were trained to criterion in the MWM task (indicated by solid lines at 120-s escape latency). Vehicle-treated 3xTg-AD mice require more training to reach criterion in the MWM compared with WT mice and sulforaphane-treated 3xTg-AD mice. Values are means ± SEM (n = 8). ##P < 0.01, WT versus 3xTg-AD + vehicle; **P < 0.01, 3xTg-AD + vehicle versus 3xTg-AD + sulforaphane (5 or 10 mg/kg). (B) Mice were given a memory retention probe trial with the escape platform removed at 24 h after the last training trial. The 3xTg-AD mice treated with sulforaphane exhibited a dose-dependent increased time in the target quadrant. No significant differences in the time spent in the target quadrant were seen with sulforaphane (10 mg/kg) treatment compared with WT mice. (C) Quantitative real-time PCR analyses of Bace1 and Ho-1 expression in cerebral cortex of vehicle- and sulforaphane-treated 3xTg-AD mice. (D) BACE1 and HO-1 protein levels in the cerebral cortex of vehicle- and sulforaphane-treated 3xTg-AD mice. (E) Levels of Aβ40 and Aβ42 were quantified in cortical tissue samples from sulforaphane- and vehicle-treated 3xTg-AD mice by ELISA. (F) Diagram showing the mechanism by which the NRF2/ARE pathway negatively regulates BACE1 and BACE1-AS gene transcription. NRF2 activators disrupt the KEAP1-NRF2 interaction, and activated-NRF2 translocates to the nucleus. NRF2 directly binds to AREs in the BACE1 and BACE1-AS promoters and represses their transcription. Values are the mean ± SEM. *P < 0.05 and **P < 0.01; one-way ANOVA with Tukey’s (B–E).
Discussion
Although previous studies have shown the protective function of NRF2 against Aβ neurotoxicity, the role of NRF2 has been focused on the induction of antioxidant genes and autophagy-related genes (21, 29–31). Our findings reveal a previously unknown molecular mechanism that negatively regulates the expression of BACE1 and BACE1-AS by directly binding to the ARE sites of the mouse and human BACE1 and BACE1-AS promoters (Fig. 6F). Nrf2-deficient AD (5xFAD;Nrf2−/−) mice exhibited significantly elevated Bace1 and Bace1-AS expression, increased Aβ plaque pathology, and more severe cognitive impairment compared with 5xFAD and Nrf2−/− mice. Pharmacological activation of NRF2 suppressed BACE1 and BACE1-AS expression and Aβ production and ameliorated cognitive deficits and AD-related pathologies in 5xFAD and 3xTg-AD mice. While drugs that inhibit BACE1 enzyme activity are being developed (32–34), our findings suggest that down-regulation of BACE1 and BACE1-AS transcription is another viable approach. Bace1-deficient mice do not generate Aβ (2, 35), and the discovery of an APP mutation that reduces β-cleavage and protects against AD supports inhibition of BACE1 activity as a promising therapeutic strategy for AD (10, 36, 37).
The expression of BACE1 is regulated by complex mechanisms at both the transcription and translational levels, all of which appear to have a role in elevating BACE1 levels and activity in AD (38). Moreover, BACE1-AS, a natural antisense transcript to the BACE1 gene, increases BACE1 mRNA stability by forming a RNA duplex with the sense transcript; this duplex masks the binding site for miR-485–5p, thereby preventing the miRNA-mediated mRNA decay and translational repression of BACE1 (4, 39).
Aging is the strongest risk factor for AD. Evidence in several species shows that transcriptional activity of NRF2 declines with aging (40, 41), and it was recently reported that an NRF2 activator enhances lifespan in mice (42). Moreover, repressed NRF2 signaling contributes to the premature aging phenotype of Hutchinson–Gilford progeria syndrome (HGPS), while reactivation of NRF2 decreases ROS and restores cellular HGPS defects (43). NRF2 is predominantly cytoplasmic in hippocampal neurons, and NRF2-mediated transcription is suppressed in neurons of AD patients (19). The expression of both NRF2 and target genes of the NRF2-ARE pathway is reduced in old AD animals (29). NRF2 deficiency leads to enhanced autophagic dysfunction (21), phosphorylated-Tau (22, 44), and vulnerability to oxidative stress (45). Conversely, up-regulation of NRF2-ARE pathway protects neurons against oxidative proteotoxic stress (29, 46–48).
In most cases, NRF2 acts as a transcriptional activator that binds to the ARE site in the target gene promoter and increases the expression of the target gene. However, there are several reports suggesting that NRF2 negatively regulates the expression of certain genes (49, 50). One unresolved issue is how NRF2 is able to function as both activator and a repressor. Variation of four nucleotides between TGA and GC of the ARE (TGANNNNGC) might be associated with NRF2 acting as a transcriptional repressor. We tested this possibility by generating reporter vectors in which we converted the ARE1 sequence of human BACE1 promoter (GCTCCCTCA) into the ARE sequence of the HO-1 promoter (GCTGAGTCA), a gene induced by NRF2. NRF2 activation significantly increased the activity of BACE1 promoter substituted with HO-1 ARE but inhibited the original BACE1 promoter activity. Further studies are warranted to detail how these sequence differences contribute to the transcriptional repressor function of NRF2. NRF2 might also induce epigenetic changes in the BACE1 and BACE1-AS promoters. The Encyclopedia of DNA Elements (ENCODE) integrative analysis showed that there is an EZH2 binding site near to the ARE1 of BACE1 promoter (51). EZH2 is the epigenetic modifier forming H3K27me3 which is commonly associated with silencing of genes. Whether NRF2, located near ARE1 of BACE1 promoter, might recruit or interact with EZH2 to silence BACE1 expression requires further study.
Our findings suggest that, in addition to up-regulation of antioxidant enzymes, activators of NRF2 can modify a specific pathogenic molecular pathway involved in AD pathogenesis. The NRF2 inducer sulforaphane ameliorates AD-related cognitive deficits by reducing Bace1 and Bace1-AS expression and subsequently Aβ generation in both 5xFAD and 3xTg-AD mice. Consumption of vegetables is associated with reduced risk for AD (52), and sulforaphane is present in relatively high amounts in vegetables such as broccoli and leafy greens (53). Our findings suggest a potential for disease modification by NRF2-activating phytochemicals or synthetic small molecules in AD.
Materials and Methods
Animals and Behavioral Tests.
The sources, breeding protocols, housing conditions, experimental treatment procedures, and behavioral testing methods are detailed in SI Appendix, Methods. All animal experiments and procedures were approved by the Institutional Animal Care and Use Committee of Sungkyunkwan University.
Immunohistochemistry, Immunoblot, qPCR, and Chromatin Immunoprecipitation.
Methods for mouse brain tissue preparation, immunostaining, immunoblot analysis, quantitative analysis of mRNA expression, and chromatin immunoprecipitation are detailed in SI Appendix, Methods.
Cell Cultures and Experimental Treatments.
SH-SY5Y cells and HEK293T cells were purchased from ATCC, and Keap1−/− MEFs were graciously gifted by Masayuki Yamamoto, Tohoku University, Sendai, Japan. HT22, mouse hippocampal neuronal cells, were kindly gifted by David Schubert, Salk Institute for Biological Studies, La Jolla, CA. Methods for culture maintenance and experimental manipulations are detailed in SI Appendix, Methods.
Biochemical Assays.
Methods for luciferase, BACE1, and γ-secretase activity assays and for ELISA and ROS analyses are detailed in SI Appendix, Methods.
Statistical Analysis.
All statistical analyses were performed with Prism7 (GraphPad Software, San Diego, CA), using two-tailed Student’s t test, one-way ANOVA with Tukey’s, or two-way ANOVA. Data are expressed as mean ± SEM. Groups were considered significantly different when P < 0.05 (*P < 0.05, **P < 0.01, and ***P < 0.001).
SI Appendix contains additional data, including SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This research was supported by grants (2019R1A2C3011422, 2012R1A5A2A28671860, 2017M3C7A1048268, 2018M3C7A1021851) funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF); the Ministry of Education, Science and Technology, Republic of Korea; and, in part, by the Intramural Research Program of the National Institute on Aging, National Institutes of Health (NIH).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. V.M.Y.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819541116/-/DCSupplemental.
References
- 1.Vassar R., et al. , β-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Cai H., et al. , BACE1 is the major β-secretase for generation of Abeta peptides by neurons. Nat. Neurosci. 4, 233–234 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D. J., Preventing Alzheimer’s disease. Science 337, 1488–1492 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Faghihi M. A., et al. , Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14, 723–730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L.-B., et al. , Elevated β-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat. Med. 9, 3–4 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Coulson D. T., et al. , BACE1 mRNA expression in Alzheimer’s disease postmortem brain tissue. J. Alzheimers Dis. 22, 1111–1122 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Kang M.-J., et al. , HuD regulates coding and noncoding RNA to induce APP→Aβ processing. Cell Rep. 7, 1401–1409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webster J. A., et al. ; NACC-Neuropathology Group , Genetic control of human brain transcript expression in Alzheimer disease. Am. J. Hum. Genet. 84, 445–458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y., Zhou W., Tong Y., He G., Song W., Control of APP processing and Abeta generation level by BACE1 enzymatic activity and transcription. FASEB J. 20, 285–292 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Yan R., Vassar R., Targeting the β secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol. 13, 319–329 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConlogue L., et al. , Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP transgenic mice. J. Biol. Chem. 282, 26326–26334 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Modarresi F., et al. , Knockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesis. Int. J. Alzheimers Dis. 2011, 929042 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadleir K. R., Eimer W. A., Cole S. L., Vassar R., Aβ reduction in BACE1 heterozygous null 5XFAD mice is associated with transgenic APP level. Mol. Neurodegener. 10, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T., Motohashi H., Yamamoto M., Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol. Sci. 34, 340–346 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Itoh K., et al. , Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan H. K., Olayanju A., Goldring C. E., Park B. K., The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 85, 705–717 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Dinkova-Kostova A. T., et al. , Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U.S.A. 99, 11908–11913 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi M., Yamamoto M., Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 46, 113–140 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Ramsey C. P., et al. , Expression of Nrf2 in neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 66, 75–85 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanninen K., et al. , Intrahippocampal injection of a lentiviral vector expressing Nrf2 improves spatial learning in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 106, 16505–16510 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi G., Gan K. A., Johnson D. A., Johnson J. A., Increased Alzheimer’s disease-like pathology in the APP/PS1ΔE9 mouse model lacking Nrf2 through modulation of autophagy. Neurobiol. Aging 36, 664–679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojo A. I., et al. , NRF2 deficiency replicates transcriptomic changes in Alzheimer’s patients and worsens APP and TAU pathology. Redox Biol. 13, 444–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson M. H., et al. , CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi L. S., et al. , Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Götz J., Chen F., van Dorpe J., Nitsch R. M., Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science 293, 1491–1495 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Oddo S., Billings L., Kesslak J. P., Cribbs D. H., LaFerla F. M., Abeta immunotherapy leads to clearance of early, but not late, hyperphosphorylated tau aggregates via the proteasome. Neuron 43, 321–332 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Roberson E. D., et al. , Reducing endogenous tau ameliorates amyloid β-induced deficits in an Alzheimer’s disease mouse model. Science 316, 750–754 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Jin M., et al. , Soluble amyloid β-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc. Natl. Acad. Sci. U.S.A. 108, 5819–5824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanninen K., et al. , Nuclear factor erythroid 2-related factor 2 protects against beta amyloid. Mol. Cell. Neurosci. 39, 302–313 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Pajares M., et al. , Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy 12, 1902–1916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jan A., et al. , eEF2K inhibition blocks Aβ42 neurotoxicity by promoting an NRF2 antioxidant response. Acta Neuropathol. 133, 101–119 (2017). [DOI] [PubMed] [Google Scholar]
- 32.May P. C., et al. , The potent BACE1 inhibitor LY2886721 elicits robust central Abeta pharmacodynamic responses in mice, dogs, and humans. J. Neurosci. 35, 1199–1210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy M. E., et al. , The BACE1 inhibitor verubecestat (MK-8931) reduces CNS β-amyloid in animal models and in Alzheimer’s disease patients. Sci. Transl. Med. 8, 363ra150 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Cebers G., et al. , AZD3293: Pharmacokinetic and pharmacodynamic effects in healthy subjects and patients with Alzheimer’s disease. J. Alzheimers Dis. 55, 1039–1053 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Luo Y., et al. , Mice deficient in BACE1, the Alzheimer’s β-secretase, have normal phenotype and abolished β-amyloid generation. Nat. Neurosci. 4, 231–232 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Jonsson T., et al. , A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488, 96–99 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Kimura R., Devi L., Ohno M., Partial reduction of BACE1 improves synaptic plasticity, recent and remote memories in Alzheimer’s disease transgenic mice. J. Neurochem. 113, 248–261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossner S., Sastre M., Bourne K., Lichtenthaler S. F., Transcriptional and translational regulation of BACE1 expression–implications for Alzheimer’s disease. Prog. Neurobiol. 79, 95–111 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Faghihi M. A., et al. , Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11, R56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suh J. H., et al. , Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc. Natl. Acad. Sci. U.S.A. 101, 3381–3386 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman M. M., Sykiotis G. P., Nishimura M., Bodmer R., Bohmann D., Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging Cell 12, 554–562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strong R., et al. , Longer lifespan in male mice treated with a weakly estrogenic agonist, an antioxidant, an α-glucosidase inhibitor or a Nrf2-inducer. Aging Cell 15, 872–884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubben N., et al. , Repression of the antioxidant NRF2 pathway in premature aging. Cell 165, 1361–1374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jo C., et al. , Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat. Commun. 5, 3496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kensler T. W., Wakabayashi N., Biswal S., Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 47, 89–116 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Calkins M. J., et al. , The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxid. Redox Signal. 11, 497–508 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vargas M. R., Johnson D. A., Sirkis D. W., Messing A., Johnson J. A., Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J. Neurosci. 28, 13574–13581 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., et al. , A polymorphic antioxidant response element links NRF2/sMAF binding to enhanced MAPT expression and reduced risk of Parkinsonian disorders. Cell Rep. 15, 830–842 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thangasamy A., Rogge J., Krishnegowda N. K., Freeman J. W., Ammanamanchi S., Novel function of transcription factor Nrf2 as an inhibitor of RON tyrosine kinase receptor-mediated cancer cell invasion. J. Biol. Chem. 286, 32115–32122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobayashi E. H., et al. , Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 7, 11624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loef M., Walach H., Fruit, vegetables and prevention of cognitive decline or dementia: A systematic review of cohort studies. J. Nutr. Health Aging 16, 626–630 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Fahey J. W., Zhang Y., Talalay P., Broccoli sprouts: An exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc. Natl. Acad. Sci. U.S.A. 94, 10367–10372 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.