Significance
This study investigates how evolutionary changes in enzyme activity occur. Multiple species of Drosophila flies have adapted to food with different levels of alcohol. This study uncovers genetic changes responsible for these repeated adaptive events, focusing on the main enzyme responsible for alcohol metabolism, Alcohol dehydrogenase. Better alcohol metabolism could be achieved either through changes to the enzyme itself or through changes in DNA regulatory sequences that affect how many enzyme molecules are produced. In four different cases, it was found that regulatory changes were the most frequent contributors to evolution. These findings have important implications because most studies of enzyme evolution focus exclusively on changes to protein sequence, and thus a significant source of adaptive changes may be overlooked.
Keywords: evolution, enzyme, regulatory, Drosophila, alcohol dehydrogenase
Abstract
The quantitative evolution of protein activity is a common phenomenon, yet we know little about any general mechanistic tendencies that underlie it. For example, an increase (or decrease) in enzyme activity may evolve from changes in protein sequence that alter specific activity, or from changes in gene expression that alter the amount of protein produced. The latter in turn could arise via mutations that affect gene transcription, posttranscriptional processes, or copy number. Here, to determine the types of genetic changes underlying the quantitative evolution of protein activity, we dissected the basis of ecologically relevant differences in Alcohol dehydrogenase (Adh) enzyme activity between and within several Drosophila species. By using recombinant Adh transgenes to map the functional divergence of ADH enzyme activity in vivo, we find that amino acid substitutions explain only a minority (0 to 25%) of between- and within-species differences in enzyme activity. Instead, noncoding substitutions that occur across many parts of the gene (enhancer, promoter, and 5′ and 3′ untranslated regions) account for the majority of activity differences. Surprisingly, one substitution in a transcriptional Initiator element has occurred in parallel in two species, indicating that core promoters can be an important natural source of the tuning of gene activity. Furthermore, we show that both regulatory and coding substitutions contribute to fitness (resistance to ethanol toxicity). Although qualitative changes in protein specificity necessarily derive from coding mutations, these results suggest that regulatory mutations may be the primary source of quantitative changes in protein activity, a possibility overlooked in most analyses of protein evolution.
A central goal of evolutionary biology is to identify the precise genetic and molecular basis of phenotypic evolution. Enormous efforts have been made to elucidate the mechanisms of change in a wide variety of traits. There is now a large body of empirical studies of the evolution of particular characters, and of the genes and proteins that specify them (1–6). Beyond the particulars of individual cases, however, it is crucial to understand whether there are general genetic rules or tendencies to certain kinds of evolutionary changes. One such general principle that has emerged from empirical studies and theoretical considerations is that the evolution of morphological traits in animals largely occurs through mutations within cis-regulatory sequences of developmental regulatory genes and the target loci they control (3, 4, 7–10).
In contrast, the genetic and molecular factors governing the evolution of protein function are not so sharply circumscribed. Much research has been focused on the evolution of qualitatively distinct protein activities, and there is massive empirical evidence that important functional differences between species have resulted from changes in the primary sequences of proteins directly involved in, for example, animal vision (11), respiration (12), digestion (13), host defense (14), and other physiological processes. Quantitative differences in protein activity, on the other hand, are widespread in populations and between species, yet we know little about the precise genetic basis of real-world cases of adaptation among such traits (4, 15).
Obviously, the overall activity of a protein is a product of its specific activity and the amount that is produced. Specific activity is determined by the amino acid sequence. However, protein level may be affected by many different facets of gene expression and structure (16), including (i) the rate of transcription as determined by the strength of enhancers and promoters; (ii) posttranscriptional processes such as RNA splicing; (iii) translational efficiency which may be influenced by 5′ and 3′ untranslated regions (UTRs), mRNA secondary structure, and codon usage; and (iv) the activity of trans-acting factors that mediate these processes.
It follows that mutations within any part of a gene could potentially affect protein activity. Indeed, mutations that affect gene expression levels have been found in virtually every part of metazoan gene structure in standing genetic variation (17, 18). However, it is not yet clear how each has actually contributed to adaptive levels of protein activity in nature and across evolutionary history (8). To sort among the possible contributors to protein activity differences, we need a better grasp of the patterns of causative substitutions that contribute to adaptive evolution.
Here, we sought a model trait whose evolution could be attributed to a particular protein and where functional divergence was plausibly the result of adaptation. The Alcohol dehydrogenase (Adh) gene of Drosophila is a classic evolutionary and molecular genetic model that meets these criteria (19–21). The typical Drosophila fly species feeds on fermenting fruit and various species have independently adapted to different levels of alcohol in their diet (21, 22). Some lineages have switched to lower-alcohol habitats such as fresh fruit or fungi, while others now inhabit high-alcohol habitats such as breweries and wine cellars (22). The Adh gene is critical for alcohol metabolism, and the quantity of ADH enzymatic activity (which is the product of the amount of proteins made and their specific activity) in a fly species is correlated both with the presence of alcohol in the breeding habitat and with flies’ tolerance of alcohol (22). In addition, Adh is a convenient experimental model for the study of adaptation because, unlike many other gene-level traits, its activity can be measured with a direct biochemical assay (NAD+-dependent ethanol oxidation) that works across species.
We use genetic mapping to determine which part(s) of the Adh gene contribute to quantitative activity differences in four pairs of Drosophila lineages. We find that multiple parts of the gene contribute to activity differences, but with only a relatively minor contribution from protein coding changes. We raise the possibility that regulatory mutations could play an underappreciated role in the evolution of quantitative biochemical traits.
Results
ADH Activity and Expression Differ Between and Within Species.
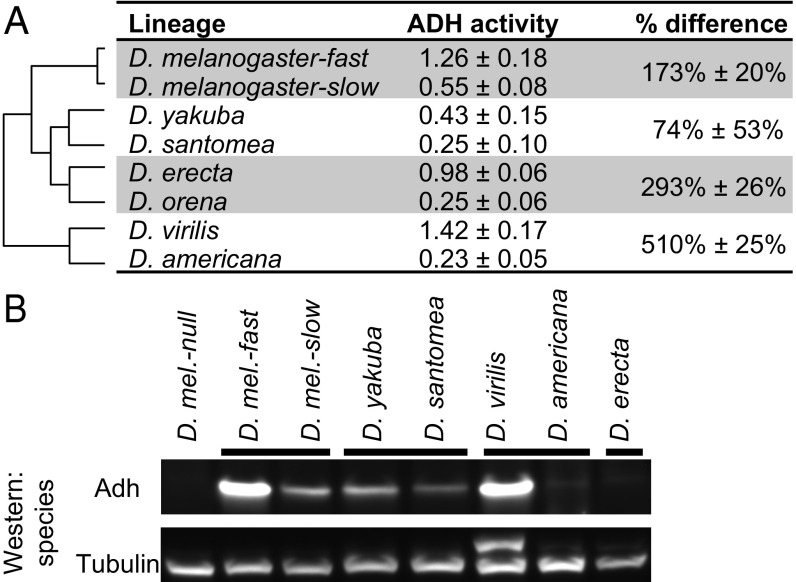
The total level of ADH activity, as measured in crude extracts of adult flies, differs among several pairs of Drosophila species as well as within Drosophila melanogaster (Fig. 1) (detailed methods are presented in SI Appendix). Specifically, flies of D. melanogaster strain Florida-9 (fast allele) showed 173% (2.7-fold) higher ADH activity than flies of strain Canton-S (slow allele). Drosophila yakuba flies showed 74% higher activity than those from its sister species Drosophila santomea, Drosophila virilis flies had 510% higher activity than those from its sister species Drosophila americana, and Drosophila erecta had 293% higher activity than those from its sister species Drosophila orena.
Fig. 1.
ADH activity and protein level differ among pairs of Drosophila alleles and species. (A) Cladogram shows relationships between species. Data from ref. 23. ADH activity is in units of ∆Abs340 per minutes per milligram soluble protein (mean ± SD). Percent difference = ([activity(high)/activity(low) − 1] × 100%), mean ± SD. Different assay conditions were used for the D. erecta/D. orena pair and for the D. virilis/D. americana pair (SI Appendix, Methods), so the means of, for example, D. santomea and D. orena, should not be compared. These data are also presented in different form in Fig. 2 A and B and SI Appendix, Fig. S1. (B) Differences in ADH protein levels between pairs of strains and species are also apparent in Western blot. Differences in band intensity among pairs (but not within pairs) are potentially affected by sequence divergence in the region to which the antibody was raised (SI Appendix, Fig. S2). The additional band in D. virilis in the anti-tubulin blot is likely the result of cross-reactivity with the polyclonal antibody.
These large differences in enzyme activity prompted us to determine their underlying mechanistic causes. In principle, higher activity could be the consequence of greater enzyme specific activity, the production of more enzyme, or both. To determine whether the differences observed might be due to the production of different amounts of Adh protein, we used Western blots with an anti-Adh antibody to examine the relative amounts of Adh protein in whole-fly extracts. In three cases, the species or strain with higher ADH activity produced more Adh protein, indicating that differences in protein expression level are at least partly responsible [Fig. 1B; Adh protein was not detected in the D. erecta/D. orena pair, likely due to amino acid divergence in the epitopes against which the antibody was raised (SI Appendix, Fig. S2)].
ADH Activity Evolution Originates Primarily from the Adh Gene.
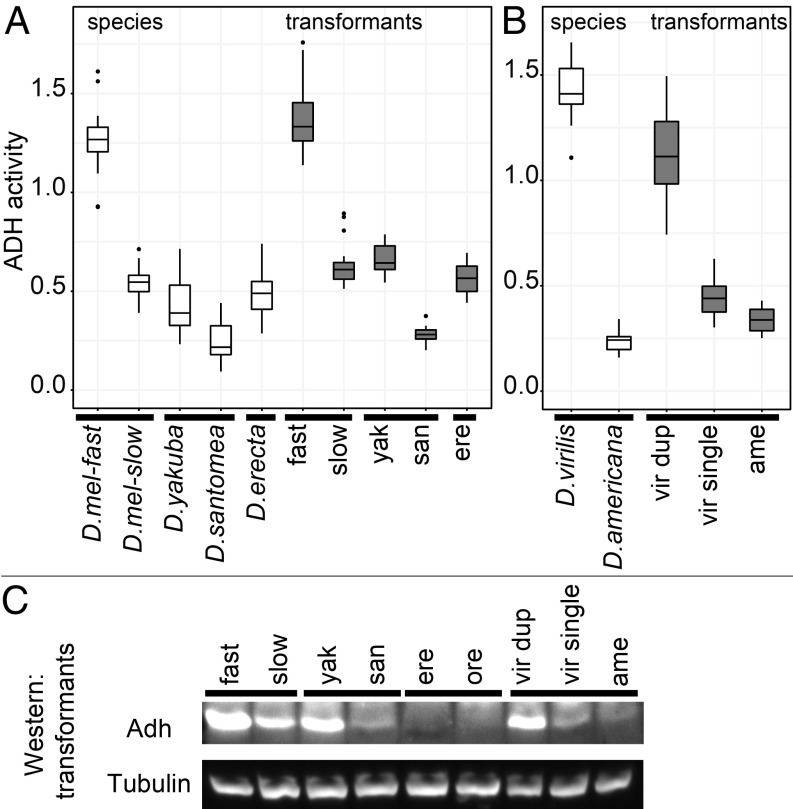
Differences in ADH activity could be due to substitutions at the Adh locus and/or to trans-acting factors outside of the locus. To determine if ADH activity differences originated from substitutions within the Adh gene, we cloned the Adh alleles from each species or strain and then transformed them back into a specific D. melanogaster attP-PhiC31 genomic landing site in a uniform Adh null genetic background (24). Cloned loci were ∼8 kb with identical boundaries in each pair, containing all known sequences required for adult expression (SI Appendix, Methods). In each case, the transgenic Adh alleles largely recapitulated the between- and within-species differences in Adh activity (Fig. 2 A and B). A similar pattern of relative differences in protein level was seen in Western blots (Fig. 2C). We could therefore use Adh transgenes to determine the contribution of protein coding versus noncoding substitutions to evolutionary differences in ADH activity.
Fig. 2.
ADH enzyme activity and protein level differ between and within species. (A) ADH enzyme activity of Drosophila strains and species (white box plots) is largely recapitulated when cloned Adh loci are transformed into D. melanogaster (gray box plots). (B) Transformants of D. virilis and D. americana Adh loci also largely recapitulate the species difference and show a major contribution from tandem duplication. Data from ref. 25. (C) Relative differences in levels of ADH protein between pairs of transformants are also apparent in Western blot. Low signal intensity in ere and ore is plausibly due to sequence divergence in the region to which the antibody was raised (SI Appendix, Fig. S2).
Amino Acid Replacements Account for only a Minor Fraction of Activity Evolution.
To directly determine the relative contribution of protein coding sequences to overall activity differences, we made a set of constructs that substituted the amino acid sequence from one species or strain into the allele from the other species or strain, leaving all noncoding substitutions unchanged. In these experiments, it was critical to be able to reliably detect small increments of differences between transgenic flies. To do so, we scaled up the sensitive ADH activity assay, measuring multiple batches of flies from multiple transgenic lines. We estimate that we could detect activity differences of around 4 to 8% after correcting for multiple testing (SI Appendix, Methods).
The number of amino acid replacements between strains or species was small. Just one amino acid difference separates the slow and fast D. melanogaster alleles (a lysine-to-threonine substitution at position 192; K192T), while three and four amino acid differences distinguish the santomea/yakuba and orena/erecta alleles, respectively (SI Appendix, Fig. S2). To measure any potential difference between the D. virilis and D. americana coding regions, we first had to consider the tandem duplication of the entire Adh gene and flanking region that occurs in D. virilis. The tandem copies are identical except for three substitutions in the 3′ noncoding region that have been shown to not affect activity (25). This allowed us to delete one duplicate from the construct, resulting in a single copy that had orthologous synteny with D. americana. We could then substitute the one amino acid change (virilis: L51, americana: I51) into this single-copy virilis Adh locus and determine if it contributed significantly to the species difference.
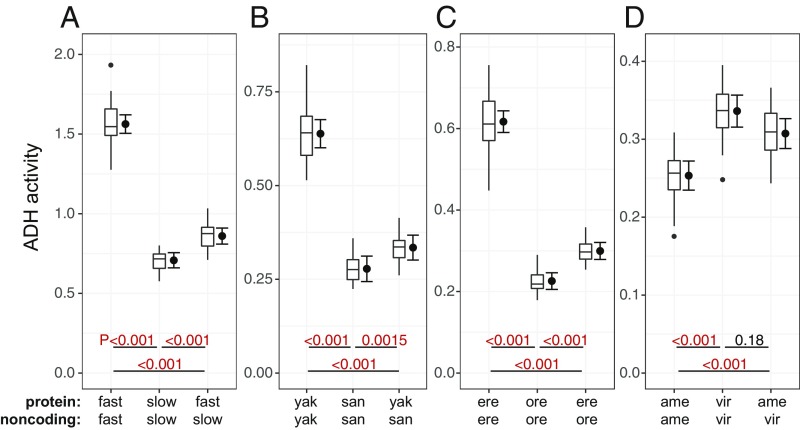
We found that the swapping of amino acid residues had the effect of changing ADH activity by at most 22% (the D. melanogaster K192T substitution) (Fig. 3 and Table 1, percent difference). In the case of D. virilis–D. americana, the single amino acid substitution had no significant effect [P = 0.09 (Table 1); after correction for multiple pairwise comparisons, P = 0.26 (Fig. 3D)]. Thus, amino acid replacements within the ADH protein contributed 0 to 25% of the overall difference in ADH activity between the loci we compared (Table 1, percent of total). It follows that 75 to 100% of ADH activity differences are the result of noncoding substitutions.
Fig. 3.
ADH activity differences are mostly cis-regulatory. (A–D) Activity is shown for transformant lines carrying either unmodified Adh alleles or swap alleles, which have the amino acid sequence of one allele and the noncoding sequence of the other. Noncoding sequence predicts ADH activity substantially better than protein sequence does. Activity data shown in Figs. 3 and 4 were collected simultaneously and are presented in full in SI Appendix, Fig. S3. P values shown here were adjusted for all pairwise comparisons with DF 18–22. Box plots show the distribution of data, while error bars show 95% confidence intervals of the mean.
Table 1.
Contribution of gene regions to four cases of ADH activity divergence
| Construct | Parameter | Whole locus, % | Amino acid swap | 5′-Flanking | 5′ UTR | Coding | 3′ UTR | 3′-Flanking |
| mel fast/slow | % difference | 121 | 22% | 15% | 49% | 18% | 11% | −1% |
| % of total | 25% | 17% | 50% | 21% | 13% | −2% | ||
| P | ** | ** | ** | ** | ** | 0.95 | ||
| yak/san | % difference | 130 | 20% | 52% | 12% | 8% | 23% | 2% |
| % of total | 22% | 50% | 14% | 9% | 24% | 2% | ||
| P | 0.0016 | ** | ** | 0.031 | ** | 0.90 | ||
| ere/ore | % difference | 173 | 17% | 76% | 14% | 57% | −18% | 6% |
| % of total | 16% | 56% | 13% | 45% | −19% | 6% | ||
| P | ** | ** | ** | ** | ** | 0.085 | ||
| vir/ame (single) | % difference | 33 | 10% | 7% | 24% | −4% | 10% | −5% |
| % of total | 32% | 24% | 75% | −15% | 34% | −18% | ||
| P | 0.090 | 0.28 | ** | 0.63 | 0.027 | 0.43 |
Percent difference ([fold_change(region) − 1] × 100%) denotes the net difference in ADH activity observed by substituting the “high” allele for the “low” allele at that region. Percent of total ([log_fold_change(region)/log_fold_change(total)]) denotes the geometric contribution of each region to the total. P values are from sequential multiple comparisons (mvt method) with degrees of freedom (Satterthwaite method) between 18 and 22. Significant values (P < 0.05) are shown in bold. **P < 0.001. These data are presented in different forms in Figs. 3 and 4 and SI Appendix, Fig. S3.
Multiple Parts of the Adh Gene Contribute to Activity Differences.
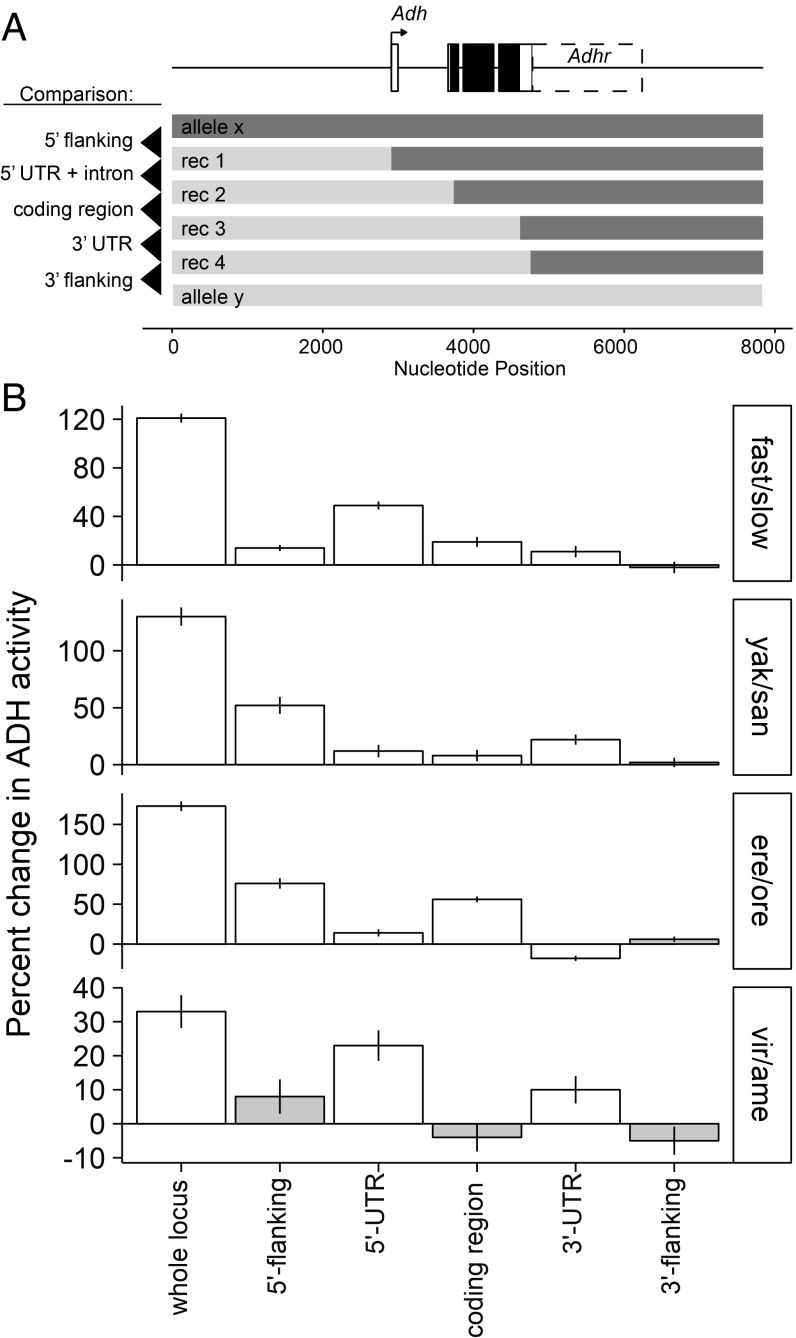
Since the amino acid sequence contributed only a small portion of the observed differences in activity, we next sought to determine what part or parts of the Adh gene were contributing to activity divergence. We divided the gene into five parts: 5′ flanking, 5′ UTR, coding sequence (including introns), 3′ UTR, and 3′ flanking (based on gene coordinates from Flybase Release 5; SI Appendix, Methods). Recombinant constructs were then engineered in vitro where the left half of one allele was fused to the right half of another at the precise junctions between regions (Fig. 4A).
Fig. 4.
ADH activity evolved through changes in many parts of the gene. (A) Model of Adh locus and recombinant genotypes used for mapping ADH activity differences to five regions of the gene. The primary adult-expressed Adh isoform is shown; other rare 3′ isoforms also include the Adhr coding sequence. For D. virilis and D. americana, the locus structure is slightly different (SI Appendix, Fig. S3, which also shows additional mapping within the 5′ flanking region). (B) Multiple regions contribute to activity evolution in each of four lineages, and the distribution of regional contributions is different in each species pair. Data are presented as proportional changes to emphasize the distribution of region effects. The underlying ADH activity measurements and statistical analysis are presented in SI Appendix, Fig. S3. Column height shows percent difference in ADH activity from substituting a particular region, mean ± SE, with white fill indicating regions with statistically significant effects on ADH activity.
In general, we observed that multiple regions contributed to enzyme activity differences within or between species (Fig. 4, Table 1, and SI Appendix, Fig. S3). For the D. melanogaster allelic polymorphism, four of the five regions were found to contribute to the overall enzyme activity difference (Fig. 4B, Top). The largest effect came from the 5′ UTR, representing a 49% difference in activity and 50% of the total difference between wild-type Adh-fast and Adh-slow alleles (Fig. 4B and Table 1). The coding region contributed a significant 18% difference in activity, similar to the 22% difference observed from the amino acid swap construct where the K192T substitution is engineered into the Adh-slow allele (Table 1). The 3′-flanking region contributed no significant difference in activity (P = 0.95).
The same four regions contributed to ADH activity divergence among species. All regions except the 3′-flanking region contributed significantly to activity differences between D. yakuba and D. santomea as well as between D. erecta and D. orena (Fig. 4 and Table 1). In D. virilis and D. americana, apart from the tandem duplication, the two UTRs were the only regions that showed significant contributions to the activity difference. However, additional recombination mapping conducted within the 5′-flanking region of D. virilis and D. americana uncovered two segments with significant but opposite effects on activity (SI Appendix, Fig. S3F; compare ame, rec1, rec1b, and rec1c). Thus, regions that did not contribute a net difference may still have evolved activity-altering substitutions. Together, these results show that multiple, noncoding parts of the Adh gene have repeatedly made the majority contribution to enzyme activity evolution.
Repeated contribution from the same gene regions could be the result of parallel evolution, where the same nucleotide changes occur in each species. If this were the case, it would mean that quantitative evolution is constrained to a few mutational paths. Instead, the magnitude of change originating from the four regions was different in most instances (Fig. 4 and Table 1). First, the 5′ UTR is the predominant contributor in D. melanogaster, while the 5′-flanking region is the primary contributor in both the D. erecta–D. orena and the D. yakuba–D. santomea comparisons. Second, the 3′ UTR in the D. erecta–D. orena comparison contributes in the opposite direction than in the other species. Third, the 57% contribution from the coding region in D. erecta–D. orena is only partially explained by a 17% contribution from swapping amino acid substitutions, revealing a regulatory contribution from the coding sequence and/or its introns (Table 1). Finally, gene duplication is the main contributor to the D. virilis–D. americana activity difference. Duplication constitutes a larger single effect (more than twofold) than any region or mutation in any of the other species. These results are not consistent with a highly constrained set of causative sites but rather with quantitative differences in ADH activity originating from unique sets of multiple substitutions in each lineage.
The D. melanogaster Alleles Differ by Six Causative Substitutions.
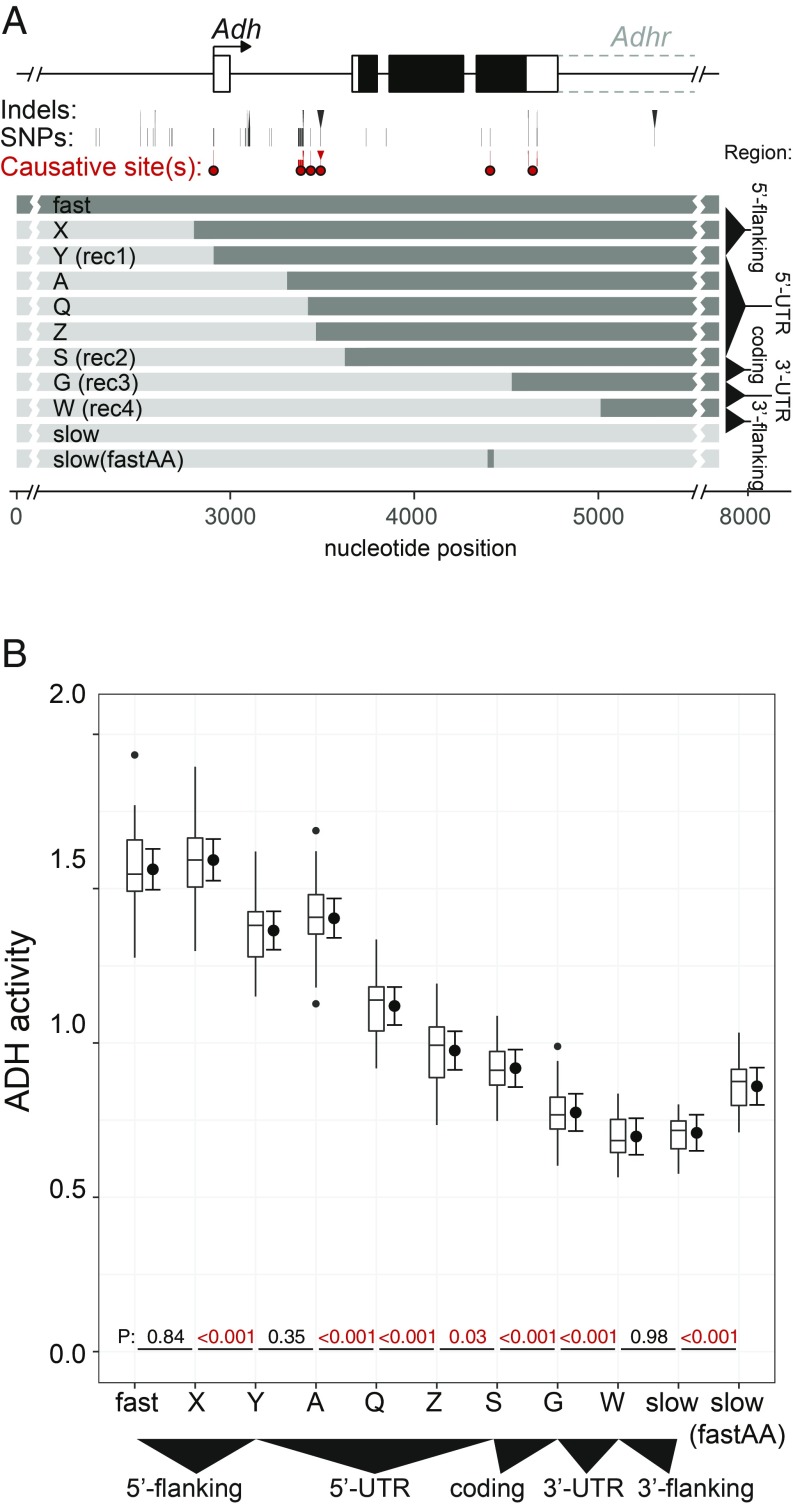
To better understand the distribution and effect sizes of mutations contributing to this trait, we next determined the causative nucleotide substitutions underlying the D. melanogaster Adh-fast and Adh-slow activity difference. Previous studies had identified two causative substitutions in the 5′-UTR intron and in the coding region (26, 27). The 3′ UTR was also implicated as a causative region but the causative site(s) were not mapped (28). Our five-region map additionally implicated the 5′-noncoding region. We therefore attempted to verify the two previously ascertained sites and determine the specific nucleotide changes behind the other unknown sites.
Fine-scale mapping of the D. melanogaster alleles confirmed the two known sites and uncovered four additional causative sites (Fig. 5 and SI Appendix, Fig. S4). These six sites must be nonequivalent in molecular function, as one causative site is in the 5′-noncoding region, three are in the 5′ UTR, one is an amino acid change in the coding region, and (at least) one is in the 3′ UTR. A detailed description of mapping results is presented in SI Appendix. Three observations are worth note. First, all six higher-activity variants appear to be derived (SI Appendix, Fig. S6), suggesting a history of directional selection. Second, three causative substitutions in the 5′ UTR occur within 100 bp, suggesting a previously undescribed regulatory element. Finally, the causative substitution in the 5′-noncoding region occurs in the core promoter. The causative C/T substitution is in a binding site for Initiator, a transcription factor that positions RNA polymerase II (29). Remarkably, a parallel C/T substitution at this site distinguishes the D. yakuba sequence from the D. santomea sequence (SI Appendix, Fig. S6).
Fig. 5.
At least six causative substitutions play a role in the D. melanogaster fast and slow Adh alleles. (A) Diagram of D. melanogaster allele and location of six causative substitutions. Single nucleotide polymorphism (SNP) and indel substitutions are marked below the gene model. Substitutions or groups of substitutions with significant effects are marked below by dots. Mapping these specific substitutions was achieved using the recombinant constructs shown, with additional refinement of the coding region and 3′ UTR in mapping experiments presented in SI Appendix, Fig. S4. The map is also presented as a pairwise sequence alignment in SI Appendix, Fig. S5. (B) ADH activity of melanogaster fast/slow recombinants reveals six causative sites. P values are from sequential multiple comparisons with DF 33–35.
Evolution of Adh Activity Affects Resistance to Ethanol.
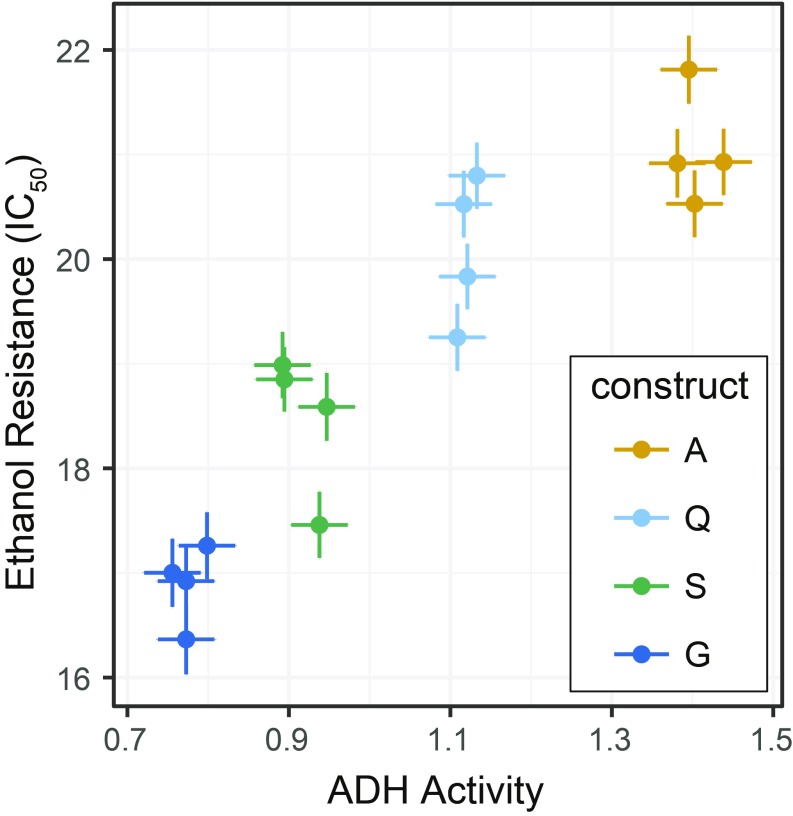
Our observation that six causative mutations affect activity in the same direction is consistent with possible directional selection on each site. This raises the question of whether small increments of ADH activity (i.e., 1.1- to 1.2-fold) are subject to selection. Although a proposed role for ADH in protecting against ethanol toxicity has been debated for decades, the influence of naturally occurring Adh sequence divergence on flies’ resistance to ethanol has not been clearly established (20, 30–32). The transgenic lines developed for this study allowed us to directly test the hypothesis that ADH activity level affects resistance to ethanol. We exposed adult flies from four different melanogaster recombinant genotypes (A, Q, S, and G in Fig. 5) that differ in steps of ∼20% in ADH activity to varying concentrations of ethanol. The proportion of flies that were incapacitated or dead after 24 h increased logistically with ethanol dose (SI Appendix, Fig. S7). This allowed us to quantify ethanol resistance as the incapacitating concentration (IC50) at which 50% of flies were unable to right themselves after 24 h of ethanol exposure.
Ethanol resistance showed a significant positive correlation with ADH activity (Pearson product-moment correlation, r = 0.91, n = 16 lines, P < 0.005) (Fig. 6). In pairwise comparisons, both coding and noncoding substitutions were found to contribute to resistance. Genotypes S and G differ by the K192T coding substitution, with the higher-activity S genotype showing significantly higher resistance (sequential comparison of IC50 from binomial glmm, n = 4 lines per construct, P = 0.017). Genotypes A, Q, and S differ by noncoding substitutions in the first intron (Fig. 5). Genotype Q showed significantly higher resistance than S (P = 0.017), while the resistance of genotypes A and Q was not significantly different from one another (P = 0.109), although the mean difference was in the expected direction (Fig. 6). These results show that both coding and noncoding substitutions that affect ADH activity directly contribute to the ecologically relevant trait of ethanol resistance.
Fig. 6.
Substitutions responsible for Adh activity evolution also affect resistance to ethanol toxicity, and ethanol resistance positively correlates with ADH activity for both coding and noncoding substitutions. Survival was found to decrease logistically with higher ethanol doses (SI Appendix, Fig. S7). Here, resistance is presented as IC50, the concentration at which 50% of flies were incapacitated. ADH activity of each line was measured separately on flies not exposed to ethanol (data from Fig. 5). Mean and SE of the four replicate lines per construct are shown.
Discussion
We dissected the genetic bases of differences in ADH protein activity among several Drosophila species. Our results demonstrate that substitutions in both coding and noncoding sequences, as well as gene duplication, contribute to activity divergence, and that fairly small increments of ADH activity (1.1- to 1.2-fold) measurably affect organismal fitness (ethanol resistance). However, amino acid substitutions account for only a minority (0 to 25%) of between- and within-species differences in enzyme activity, with the majority of activity differences resulting from noncoding substitutions within various regulatory sequences (enhancer, promoter, and 5′ and 3′ UTRs). These findings raise general issues concerning the relative contribution of amino acid versus regulatory substitutions in the evolution of protein activity under natural selection. They also raise questions about the expected effect sizes of coding substitutions, regulatory substitutions, and gene duplication.
A Limited Contribution of Amino Acid Changes to Enzyme Activity Evolution.
In principle, quantitative changes in enzyme activity could occur through changes in specific activity, in the amount of protein produced, or both. We found evidence for both mechanisms. Amino acid swap experiments identified coding substitutions that contributed to activity differences in several cases, and gene recombination experiments revealed the contribution of multiple noncoding segments to activity differences. Mutations that increase the specific activity of a protein might be expected to be preferred over regulatory mutations that alter protein production, as they come “for free” (i.e., without the metabolic cost of synthesizing additional protein). However, we found that amino acid substitutions explained only incremental changes in protein activity (≤22%) and accounted for only a minor fraction (0 to 25%) of overall activity differences. Our results raise the question of why coding changes are not a more significant source of quantitative activity differences between or within species.
There is a large body of empirical and theoretical work concerning the evolution of enzyme specificity and activity. It has been widely noted that most enzymes do not exhibit the maximum theoretical catalytic efficiency (33, 34). Rather, structural and physicochemical constraints appear to limit most enzymes to more “moderate” efficiencies. Such constraints could explain the limited ability of amino acid substitutions to contribute to ADH activity differences. For example, mutations that increase specific activity (the rate of reaction) may cause other deleterious effects such as altered solubility, stability, or substrate specificity and would be selected against in nature.
In addition, there are genetic constraints operating on protein evolution that affect the probability of activity-enhancing substitutions to arise. For example, only a limited set of amino acids may affect specific activity, and therefore the target size for such mutations is small. Moreover, the capacity of substitutions to affect specific activity without altering functional specificity appears to depend on how they interact with other residues, and thus the available mutational paths of protein evolution are further constrained by epistasis (35).
These constraints, and the observation that the vast majority of enzymes have only moderate theoretical efficiency, has prompted the proposal that the catalytic activities of most enzymes in nature have already been optimized in the course of evolution (34). For example, RuBisCO (d-ribulose 1,5-bisphosphate carboxylase/oxygenase), perhaps the most important enzyme on the planet, exhibits notable inefficiencies but appears to be optimized for the local environments in which it operates (36). If moderate efficiency and local optimality are general features of enzyme evolution, then extant enzymes in nature may be able to attain relatively little additional overall activity through new coding mutations and natural selection.
A Major Role for Noncoding Regulatory Substitutions in Activity Evolution.
In contrast to the small contribution of amino acid replacements to activity evolution between and within species, we found that noncoding regions across the Adh locus contributed the majority of activity differences, with each lineage displaying a different distribution of effects. Because these regions included the promoter and 5′ upstream regions, the 5′ UTR, and the 3′ UTR, we infer that all these regions’ ultimate effects are on protein expression levels, that is, are regulatory in nature. The consistent observation of regulatory sequence involvement in activity divergence raises the question of why noncoding substitutions predominate in ADH activity evolution and what that may signify about activity evolution in general.
In comparison with the structural and genetic constraints on protein sequences described above, weaker constraints operate on regulatory sequences. One important distinction concerns the potential pleiotropic effects of coding versus noncoding mutations. Whereas a mutation that alters protein specific activity may also change protein solubility, stability, or substrate specificity, most mutations that affect gene transcription, RNA splicing, and translation only impact the rate/level of protein expression and not protein structure. In addition to fewer constraints, the mutational target size for regulatory mutations is likely to be much larger, as seen here, encompassing sites in multiple regulatory regions.
Based on these arguments, we suggest that regulatory substitutions are likely to be the primary source of quantitative change in protein activity in nature.
This inference further implies that quantitative enzyme activity traits might follow a pattern of evolution similar to that of quantitative gene expression traits. Studies of gene expression variation within populations have suggested two patterns that are consistent with this claim. First, although variation in upstream (trans) regulators can change a gene’s expression, changes in the gene itself (in cis) are much more common contributors to expression variation, and because the measured phenotypes are RNA levels, the causes are necessarily noncoding/regulatory (17, 18). Second, genetic variation for gene expression is often associated with substitutions in or near promoters (17). Promoters may thus be a common source of quantitative evolutionary events. For Adh, we identified one surprising case of parallel evolution of a substitution in the core promoter Initiator site. The Initiator site is the most common core promoter element in bilaterian animals and is largely sequence-invariant in Drosophila (37), and yet biologically meaningful activity divergence can result from mutations to the Initiator site.
The Effect Size of Mutations in Quantitative Evolution.
Another possible explanation for the observed excess of regulatory changes in Adh evolution might be that such substitutions convey different effect sizes than coding substitutions. However, the effect sizes of coding and noncoding sequence variants that we were able to determine directly were similar. Mutations in both coding and regulatory sequences that have larger effect sizes should be possible in principle. It seems likely, however, that mutations with modest effects on gene expression (i.e., modification of existing regulatory sequences) may simply be more probable and thus more common.
We did observe one large-effect sequence variant: the tandem duplication of the entire Adh gene in D. virilis. This duplicate showed ∼2.7-fold higher activity than a single-copy transgene (25), an order of magnitude larger than the point substitutions. Although the mechanism producing excess transcription from tandem duplicates is not known (25), such large effects of whole-gene tandem duplication (greater than twofold increases in gene expression) have been seen in transgenic experiments (25), mutation accumulation lines (38), and a rare disease (39). Thus, the effect size of duplicates appears to be much greater than the typical point substitution contributing to Adh evolution. However, because duplication mutations are much rarer than point mutations (40, 41), most gene activity evolution is likely to result from sequential point substitutions, with occasional but large increases from gene duplication.
Small Changes in ADH Activity Affect Fitness.
Our study establishes strong evidence that modest changes in enzyme activity can have measurable effects on organismal phenotype. Changes in gene expression are often discussed in terms of fold changes, yet we were able to measure clear phenotypic consequences from <20% differences in gene activity. A direct relationship between ethanol resistance and ADH activity has also been seen for Adh alleles engineered to carry rare codons (31). These observations are consistent with the hypothesis that changes in ADH enzyme activity provided a selective advantage accompanying habitat shifts to high-alcohol food sources (22).
Conclusion: The Evolution of More Versus Different.
The idea that an enzyme’s activity is the product of its concentration and its structure is as old as the description of enzyme kinetic laws themselves (42). Qualitative differences in enzyme activity, that is, shifts in substrate specificity, almost certainly require changes to the protein structure. Similarly, null mutations that abolish enzyme activity appear to generally require coding mutations (4). Quantitative differences, in contrast, derive from both protein structure and protein level. Our results suggest that this regulatory dimension is the primary mode of quantitative evolution. This pattern makes sense in light of our growing understanding of gene structure and evolution, and in particular the sprawling regulatory architecture of higher eukaryotic genes. Thus, the many demonstrated cases of amino acid changes with functional effects may point to an even larger number of quantitative regulatory substitutions just below the surface.
Methods
We investigated the genetic basis of Adh enzyme activity divergence in Drosophila using transgenes. Adh alleles consisting of 7 to 8 kb of genomic sequence were PCR-amplified, cloned, and sequenced (GenBank accession nos. MH614199–MH614205 and KU559568.1). Adh transgenes were inserted into Adh-null D. melanogaster flies using the PhiC31-attP system as described in SI Appendix, Methods. This approach facilitates identification of small differences in enzyme activity because genetic variants are inserted into the same chromosomal site in identical genetic background. ADH enzyme activity was measured from homogenates of whole flies using a high-throughput protocol described in SI Appendix, Methods. The experimental design had a nested structure: ADH activity was measured from a large number of fly samples from a small number (i.e., two to four) of replicate transgenic lines and was therefore analyzed using a mixed-effects model. Ethanol resistance was measured as survival after 24-h exposure to X% ethanol of 4-d-old males from recombinant transgenic lines. Resistance experiments also had a nested structure and were analyzed using a generalized linear mixed model as described in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Sara Stemberger for help constructing recombinants; Jorge Vieira for sharing draft D. americana genome data; Ron Bassar, Brianna Heggeseth, and Nicholas Keuler for advice on statistics; Fiona Ukken and Pei-Wen Chen for technical advice; and members of the S.B.C. laboratory and many others who provided guidance and suggestions. D.W.L. was supported by a Howard Hughes Medical Institute – Life Sciences Research Foundation postdoctoral fellowship and startup funds from Williams College. J.R.A. was supported by a Williams College Summer Science Research Fellowship. S.B.C. was supported by a Howard Hughes Medical Institute Investigatorship and funds from the Wisconsin Alumni Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MH614199–MH614205 and KU559568.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904071116/-/DCSupplemental.
References
- 1.King M. C., Wilson A. C., Evolution at two levels in humans and chimpanzees. Science 188, 107–116 (1975). [DOI] [PubMed] [Google Scholar]
- 2.Carroll S. B., Grenier J. K., Weatherbee S. D., From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design (Blackwell Publishing, Malden, MA, 2001). [Google Scholar]
- 3.Stern D. L., Evolution, Development & the Predictable Genome (Roberts and Co. Publishers, Greenwood Village, CO, 2011). [Google Scholar]
- 4.Martin A., Orgogozo V., The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 67, 1235–1250 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Remington D. L., Alleles versus mutations: Understanding the evolution of genetic architecture requires a molecular perspective on allelic origins. Evolution 69, 3025–3038 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Siddiq M. A., Hochberg G. K. A., Thornton J. W., Evolution of protein specificity: Insights from ancestral protein reconstruction. Curr. Opin. Struct. Biol. 47, 113–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll S. B., Evolution at two levels: On genes and form. PLoS Biol. 3, e245 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wray G. A., The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Carroll S. B., Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134, 25–36 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Loehlin D. W., Werren J. H., Evolution of shape by multiple regulatory changes to a growth gene. Science 335, 943–947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoyama S., Tada T., Zhang H., Britt L., Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 105, 13480–13485 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Projecto-Garcia J., et al. , Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc. Natl. Acad. Sci. U.S.A. 110, 20669–20674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Zhang Y. P., Rosenberg H. F., Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30, 411–415 (2002). [DOI] [PubMed] [Google Scholar]
- 14.ffrench-Constant R. H., Daborn P. J., Le Goff G., The genetics and genomics of insecticide resistance. Trends Genet. 20, 163–170 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Rockman M. V., The QTN program and the alleles that matter for evolution: All that’s gold does not glitter. Evolution 66, 1–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso C. R., Wilkins A. S., The molecular elements that underlie developmental evolution. Nat. Rev. Genet. 6, 709–715 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Gaffney D. J., et al. , Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 13, R7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King E. G., Sanderson B. J., McNeil C. L., Long A. D., Macdonald S. J., Genetic dissection of the Drosophila melanogaster female head transcriptome reveals widespread allelic heterogeneity. PLoS Genet. 10, e1004322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson S. M., McDonald J. F., Biochemical and molecular analysis of naturally occurring Adh variants in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 80, 4798–4802 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinstra P. W. H., Evolutionary genetics of the Drosophila alcohol dehydrogenase gene-enzyme system. Genetica 92, 1–22 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Ashburner M., Speculations on the subject of alcohol dehydrogenase and its properties in Drosophila and other flies. BioEssays 20, 949–954 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Merçot H., Defaye D., Capy P., Pla E., David J. R., Alcohol tolerance, ADH activity, and ecological niche of Drosophila species. Evolution 48, 746–757 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Linde K., Houle D., Spicer G. S., Steppan S. J., A supermatrix-based molecular phylogeny of the family Drosophilidae. Genet. Res. 92, 25–38 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loehlin D. W., Carroll S. B., Expression of tandem gene duplicates is often greater than twofold. Proc. Natl. Acad. Sci. U.S.A. 113, 5988–5992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhary M., Laurie C. C., Use of in vitro mutagenesis to analyze the molecular basis of the difference in Adh expression associated with the allozyme polymorphism in Drosophila melanogaster. Genetics 129, 481–488 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurie C. C., Stam L. F., The effect of an intronic polymorphism on alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 138, 379–385 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stam L. F., Laurie C. C., Molecular dissection of a major gene effect on a quantitative trait: The level of alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 144, 1559–1564 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen S. K., Tjian R., TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell 82, 565–575 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Middleton R. J., Kacser H., Enzyme variation, metabolic flux and fitness: Alcohol dehydrogenase in Drosophila melanogaster. Genetics 105, 633–650 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlini D. B., Experimental reduction of codon bias in the Drosophila alcohol dehydrogenase gene results in decreased ethanol tolerance of adult flies. J. Evol. Biol. 17, 779–785 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Siddiq M. A., Loehlin D. W., Montooth K. L., Thornton J. W., Experimental test and refutation of a classic case of molecular adaptation in Drosophila melanogaster. Nat. Ecol. Evol. 1, 25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Even A., et al. , The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Newton M. S., Arcus V. L., Patrick W. M., Rapid bursts and slow declines: On the possible evolutionary trajectories of enzymes. J. R. Soc. Interface 12, 20150036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starr T. N., Thornton J. W., Epistasis in protein evolution. Protein Sci. 25, 1204–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savir Y., Noor E., Milo R., Tlusty T., Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc. Natl. Acad. Sci. U.S.A. 107, 3475–3480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vo Ngoc L., Wang Y. L., Kassavetis G. A., Kadonaga J. T., The punctilious RNA polymerase II core promoter. Genes Dev. 31, 1289–1301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konrad A., et al. , Mutational and transcriptional landscape of spontaneous gene duplications and deletions in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 115, 7386–7391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayward C. P. M., et al. , The duplication mutation of Quebec platelet disorder dysregulates PLAU, but not C10orf55, selectively increasing production of normal PLAU transcripts by megakaryocytes but not granulocytes. PLoS One 12, e0173991 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katju V., Bergthorsson U., Copy-number changes in evolution: Rates, fitness effects and adaptive significance. Front. Genet. 4, 273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondrashov F. A., Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279, 5048–5057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michaelis L., Menten M. L., Die Kinetik der Invertinwirkung [in German]. Biochem. Z. 49, 333–369 (1913) [translated by R. S. Goody, K. A. Johnson, Biochemistry 50, 8264–8269 (2011)]. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.