Significance
Deletion or loss-of-function mutations of the maternally inherited allele of UBE3A, which encodes an E3 ubiquitin ligase, lead to Angelman syndrome (AS), a developmental neurological disorder with severe intellectual disability. The consequences of UBE3A dysfunction are not well understood. Here, we demonstrate that UBE3A ubiquitinates PTPA, an activator of protein phosphatase 2A. Maternal loss of Ube3a in an AS mouse model leads to significant increases in PTPA level and PP2A activity. Genetic reduction of PTPA or pharmacologic inhibition of PP2A in an AS mouse model alleviated the deficits in dendritic spine morphology and synaptic transmission and improved behavioral phenotypes. These data suggest a critical role of UBE3A-PTPA-PP2A signaling in the pathogenesis of UBE3A-related disorders.
Keywords: UBE3A, Angelman syndrome, ubiquitin, protein phosphatase 2A, spine morphology
Abstract
Deficiency in the E3 ubiquitin ligase UBE3A leads to the neurodevelopmental disorder Angelman syndrome (AS), while additional dosage of UBE3A is linked to autism spectrum disorder. The mechanisms underlying the downstream effects of UBE3A gain or loss of function in these neurodevelopmental disorders are still not well understood, and effective treatments are lacking. Here, using stable-isotope labeling of amino acids in mammals and ubiquitination assays, we identify PTPA, an activator of protein phosphatase 2A (PP2A), as a bona fide ubiquitin ligase substrate of UBE3A. Maternal loss of Ube3a (Ube3am−/p+) increased PTPA level, promoted PP2A holoenzyme assembly, and elevated PP2A activity, while maternal 15q11–13 duplication containing Ube3a down-regulated PTPA level and lowered PP2A activity. Reducing PTPA level in vivo restored the defects in dendritic spine maturation in Ube3am−/p+ mice. Moreover, pharmacological inhibition of PP2A activity with the small molecule LB-100 alleviated both reduction in excitatory synaptic transmission and motor impairment in Ube3am−/p+ mice. Together, our results implicate a critical role of UBE3A-PTPA-PP2A signaling in the pathogenesis of UBE3A-related disorders and suggest that PP2A-based drugs could be potential therapeutic candidates for treatment of UBE3A-related disorders.
The imprinted gene UBE3A encodes the E3 ubiquitin ligase UBE3A, which conjugates polyubiquitin chains to specific lysine residues in its substrates, regulating the expression and function of these proteins (1, 2). Deletion or loss-of-function mutations of the maternally inherited allele of UBE3A result in Angelman syndrome (AS), a neurodevelopmental disorder characterized by severe developmental delay, intellectual disability, motor dysfunction, and seizures (3, 4). On the other hand, maternal duplication or triplication of the chromosome 15q11–13 region, where UBE3A resides, is associated with autism spectrum disorder (ASD) (5). These studies demonstrate that appropriate dosage of UBE3A is critical for normal brain development and function.
The AS mouse model with loss of the maternal allele of Ube3a (Ube3am−/p+) recapitulates many of the phenotypes of AS, including motor dysfunction, seizure susceptibility, and cognitive impairment (6). Ube3am−/p+ mice have significantly reduced spine density and aberrant spine morphology, deficits that are highly correlated with abnormalities in synaptic transmission and/or synaptic plasticity (7–10). Ube3am−/p+ mice also display impaired experience-dependent dendritic spine maintenance and synaptic maturation in cortical circuits, consistent with sensory processing abnormalities in individuals with AS (8, 10, 11). These findings demonstrate that UBE3A plays a critical role in normal dendritic spine development, as well as neural circuit wiring and plasticity. However, the mechanisms that link changes in UBE3A level to neurodevelopmental disorders are not well understood.
Accounting for 1% of total cellular protein, protein phosphatase 2A (PP2A) is highly conserved and responsible for most of cellular serine/threonine phosphatase activity (12). Its holoenzyme is a heterotrimer, consisting of a core dimer of a catalytic C subunit (PP2Ac) and a scaffolding A subunit (PR65), and one regulatory B subunit. The regulatory B subunit belongs to one of four families containing PR55/B (B55), PR61/B′ (B56), PR48/PR72/PR130/B″, or PR93/PR110/B‴, and determines the substrate specificity and enzymatic activity of PP2A (13). In the nervous system, PP2A is crucial for neuronal growth and differentiation, cytoskeleton assembly, dendritic spine morphology, and synaptic plasticity (14, 15). However, it remains largely unknown how regulatory factors function together to modulate PP2A activity in vivo.
Here, we show that PTPA (phosphotyrosyl phosphatase activator), an activator of PP2A, is a ubiquitin ligase substrate of UBE3A. In Ube3am−/p+ mice, elevating PTPA protein level increased the methylation of the catalytic subunit of PP2A and promoted PP2A holoenzyme assembly. Genetically reducing PTPA expression using Ptpa+/− mice or pharmacological inhibition of PP2A activity using LB-100 significantly rescued the deficits in spine morphology and excitatory synaptic transmission in Ube3am−/p+ mice. Chronic treatment of Ube3am−/p+ mice with PP2A inhibitor LB-100 also significantly rescued behavioral deficits. In summary, our study identifies the PTPA/PP2A complex as a UBE3A substrate and suggests that it serves as a target for therapeutic intervention in UBE3A-related disorders.
Results
UBE3A Negatively Regulates PP2A Activity in the Brain.
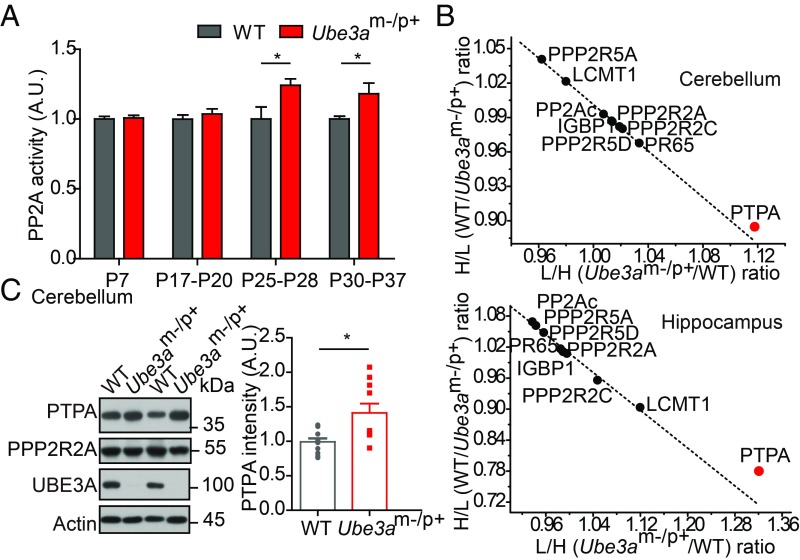
To investigate whether UBE3A regulates the activity of PP2A, we systematically assayed PP2A-specific phosphatase activity in four developmental stages, namely postnatal (P) 7, P17–P20, P25–P28, and P30–P37, by immunoprecipitating it from brain lysates from littermate wild-type (WT) or Ube3am−/p+ mice (SI Appendix, Fig. S1A). We found that PP2A activity was significantly increased in Ube3am−/p+ brain lysates, compared with WT brain lysates, from the third postnatal week onwards (Fig. 1A). We further evaluated the effect of UBE3A overexpression on PP2A activity in HEK293T cells. Consistently, overexpression of UBE3A dramatically reduced PP2A activity (SI Appendix, Fig. S1 B and C). Conversely, overexpression of the ligase-dead mutant (UBE3A-C838A) of UBE3A significantly increased PP2A activity. As an additional control, expression of GFP did not affect PP2A activity, indicating that the regulation between UBE3A and PP2A activity was specific. Together, these findings demonstrate that UBE3A negatively regulates PP2A activity at early developmental stages.
Fig. 1.
UBE3A negatively regulates PP2A activity, as well as the protein level of its activator PTPA. (A) PP2A activity from brain tissues in WT, Ube3am−/p+ at P7 (n = 3); P17–P20 (n = 3); P25–P28 (n = 4); and P30–P37 (n = 7) (n represents the number of mice). (B) Quantitative mass spectrometry analysis of the protein levels of PP2A-related proteins in the cerebellum and hippocampus based on SILAM. Black dots, proteins with no changes in levels; red dots, PTPA. (C) PTPA protein level was increased in the cerebellum of Ube3am−/p+ mice at P30. n = 8 mice per condition. In all quantifications, error bars indicated mean ± SEM; *P < 0.05, Student’s unpaired t test.
Using Western blotting, we found that both UBE3A and PP2A were expressed in neurons and glial cells (SI Appendix, Fig. S1D), consistent with the published literature (2, 16). Using duplex in situ hybridization, we found that they were coexpressed in the same cells in hippocampus, cerebellum, and cerebral cortex, and at high levels in neurons of the hippocampus and cerebellum (SI Appendix, Fig. S1E). Since UBE3A has been reported to function as an E3 ligase or a transcriptional coregulator (17), we next tested mRNA and protein levels of PP2A subunits in cerebellar and hippocampal lysates of the Ube3am−/p+ mice. Although PP2A activity was negatively regulated by UBE3A, the mRNA and protein levels of PP2A subunits were not significantly affected in Ube3am−/p+ mice (SI Appendix, Fig. S2 A–C). These results suggest that PP2A is unlikely to be a direct substrate of UBE3A.
UBE3A Regulates the Protein Level of PP2A Activator PTPA.
We thus used stable isotope labeling of amino acids in mammals (SILAM) combined with quantitative mass spectrometry (18) to systematically screen for proteins whose levels are altered in Ube3am−/p+ mice at P14–P17 (SI Appendix, Fig. S2D). Mass spectrometry analysis also did not show significant changes in any PP2A subunits in cerebellar and hippocampal lysates (Fig. 1B, and SI Appendix, Table S1, and Dataset S1). Interestingly, we found that the level of an activator of PP2A, known as PTPA (phosphotyrosyl phosphatase activator, encoded by Ppp2r4), was elevated, compared with that in WT mice (Fig. 1B). The levels of other known PP2A regulators, including IGBP1 and LCMT1, were unchanged. Consistent with PTPA being a potential substrate of UBE3A, its protein level, rather than its mRNA level, increased significantly at P14 and P30 in the cerebellum and hippocampus of Ube3am−/p+ mice, compared with littermate WT mice (Fig. 1C and SI Appendix, Fig. S3 A–D). Consistently, in the human 15q11–13 maternal duplication mice (matDp/+) with elevated UBE3A protein level (19), we found a significant reduction in the level of PTPA (SI Appendix, Fig. S3E). These results indicate that PTPA is a potential ubiquitin ligase substrate of UBE3A.
Was the elevation of PTPA responsible for the dysregulation of PP2A activity in Ube3am−/p+ mice? Using whole brain lysates, we did not observe a change of PP2A activity between WT and Ube3am−/p+ mice at P14 (Fig. 1A). However, when we used lysates from specific brain regions to measure PP2A activity, we found that PP2A activity was increased in the cerebellum and hippocampus of the Ube3am−/p+ mice at P14, compared with littermate WT mice (SI Appendix, Fig. S3F). Neither PTPA level nor PP2A activity was changed at P7 in Ube3am−/p+ mice (SI Appendix, Fig. S3 G and H). The temporal similarity in changes of PP2A activity and protein level of its activator suggests that PTPA elevation may be responsible for the increased PP2A activity in Ube3a-deficient mice during the second postnatal week.
UBE3A Ubiquitinates PTPA.
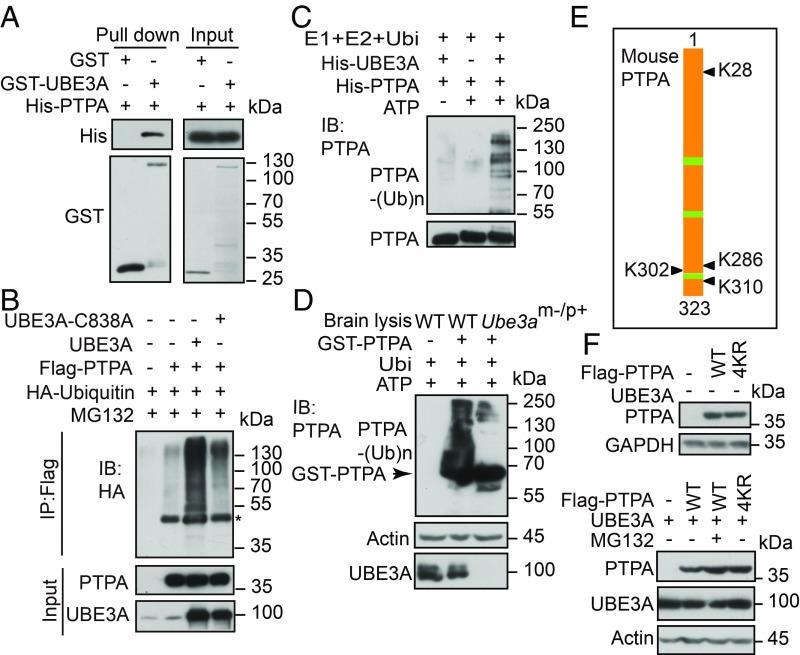
To further test the hypothesis that PTPA is a potential ubiquitin ligase substrate of UBE3A, we first examined the physical interaction between UBE3A and PTPA by GST pull-down assay using purified proteins. Glutathione Sepharose resins charged with GST-UBE3A or GST were incubated with His-PTPA and the bound proteins were eluted and subjected to Western blotting analysis. We found that PTPA directly interacted with GST-UBE3A, but not with GST (Fig. 2A). Ubiquitination assay with cell lysates using HA-tagged ubiquitin showed that PTPA was efficiently ubiquitinated in the presence of UBE3A, compared with cells expressing the ligase-dead mutant (UBE3A-C838A) (Fig. 2B). Using K48-linked polyubiquitin antibody, we found that PTPA was ubiquitinated by UBE3A in K48-linked polyubiquitin chains (SI Appendix, Fig. S4A). In the in vitro ubiquitination assay, purified PTPA was ubiquitinated by UBE3A in the presence of E1 and E2 enzymes, an effect that was blocked by omitting ATP or UBE3A (Fig. 2C). Further ubiquitination experiments using purified GST-PTPA and brain lysates showed that GST-PTPA can be efficiently ubiquitinated by brain lysates derived from WT, but not Ube3am−/p+ mice (Fig. 2D), directly supporting the hypothesis that UBE3A is the specific E3 ligase mediating PTPA ubiquitination. Using mass spectrometry analysis, we identified four lysine residues (K28, K286, K302, and K310) in PTPA that were significantly ubiquitinated in cells overexpressing UBE3A, but not in control cells (Fig. 2E and SI Appendix, Fig. S4B). Consistently, a mutant form of PTPA (PTPA-4KR), in which all four lysine residues (K) were replaced with arginine (R), rendered PTPA much less susceptible to ubiquitination (SI Appendix, Fig. S5A). Additionally, in the presence of UBE3A, both PTPA-4KR and wild-type PTPA in the presence of proteasome inhibitor MG132, were more stable than wild-type PTPA alone (Fig. 2F). To determine if all four K residues contribute to the ubiquitination of PTPA, we constructed single K mutants of PTPA (K28R, K286R, K302R, and K310R) and performed the ubiquitination assay in transfected cells. We found that the ubiquitination of each PTPA mutant was less than that of the WT form, and that each lysine contributed similarly to the ubiquitination of PTPA (SI Appendix, Fig. S5B). Together, these results demonstrate that UBE3A regulates PTPA at the protein level through the ubiquitin-proteasome system (UPS).
Fig. 2.
UBE3A ubiquitinates PTPA. (A) GST pull-down assay to validate the direct interaction between UBE3A and PTPA proteins in vitro. (B) PTPA was ubiquitinated by UBE3A but not its ligase-dead mutant Ube3a-C838A in HEK293T cells. Immunoblots were incubated with anti-HA antibodies. *, nonspecific bands. (C) In vitro ubiquitination of PTPA mediated by UBE3A analyzed by immunoblotting. (D) In vitro ubiquitination of PTPA using WT and Ube3am−/p+ mice brain lysates. (E) The lysine residues in PTPA ubiquitinated by UBE3A analyzed by LC-MS/MS. Rectangles in green indicate ATP binding sites. (F) PTPA mutation (4KR) blocked UBE3A-mediated PTPA degradation. Three independent experiments were performed.
Regulation of Spine Morphology by Interaction between PTPA and UBE3A.
If PTPA were a substrate of UBE3A, would it mimic some of the effects of UBE3A loss of function? To address this question, we examined the effect of overexpressing PTPA on dendritic spines in cultured pyramidal neurons and found no significant effect on spine density (SI Appendix, Fig. S6 A and B). We further categorized dendritic spines into three groups (mushroom, stubby, and thin), according to their morphology, with mushroom and stubby spines representing mature spines and thin spines being more dynamic and immature (20). We found that the proportion of immature, filopodia-like thin spines significantly increased in PTPA-overexpressing neurons, compared with control neurons, with a concomitant reduction in mature spines (SI Appendix, Fig. S6C). Quantitative measurement of spine morphology showed a significant increase in spine length and reduction in spine width in neurons overexpressing PTPA, compared with control neurons (SI Appendix, Fig. S6D). These results are similar to previous observations in the visual cortex of Ube3am−/p+ mice (10).
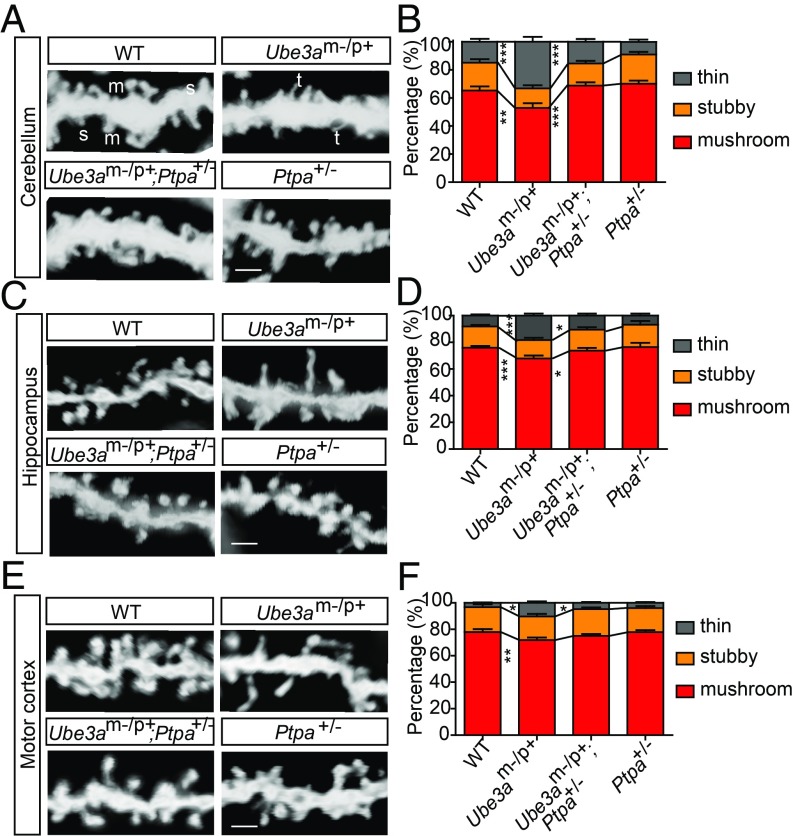
To investigate whether a reduction in PTPA level could reverse the spine defects in Ube3am−/p+ mice, we generated Ptpa mutant mice using CRISPR/Cas9 technology (see Materials and Methods for details). Homozygous mutants were embryonic lethal, while heterozygotes displayed a significant reduction in PTPA (SI Appendix, Fig. S6E). Using Golgi staining, we compared spine density and morphology in cerebellar Purkinje cells and the apical dendrites of hippocampal CA1 pyramidal neurons between WT and three different mutant mouse genotypes (Ube3am−/p+, Ptpa+/−, and Ube3am−/p+;Ptpa+/−) at P30. Similar to the results in cultured pyramidal neurons, we observed a higher frequency of immature, thin spines and a reduction of mature spines in neurons from these two brain regions of Ube3am−/p+ mice, compared with WT mice (Fig. 3 A–D). Importantly, these changes in spine morphology were completely reversed in double heterozygous mice (Ube3am−/p+;Ptpa+/−), deficient in both UBE3A and PTPA. Consistent with the spine subtype results, neurons from Ube3am−/p+ mice displayed longer and thinner spines, effects that were also rescued in Ube3am−/p+;Ptpa+/− double heterozygous mice (SI Appendix, Fig. S7 A and B). In terms of spine density, both Ube3am−/p+ and Ube3am−/p+;Ptpa+/− mice showed reduced spine density, compared with WT mice (SI Appendix, Fig. S7 C and D). In the basal dendrites of layer 2/3 pyramidal neurons of motor cortex and of primary somatosensory cortex, alterations in spine subtypes of Ube3am−/p+ mice were also rescued in Ube3am−/p+;Ptpa+/− mice (Fig. 3 E and F and SI Appendix, Fig. S7 E and F). Together, these results suggest that PTPA plays an important role in mediating the effects of UBE3A on spine morphology.
Fig. 3.
PTPA is required for UBE3A-mediated spine maturation. (A) Representative images of dendritic spine of Golgi stained Purkinje cells from WT, Ube3am−/p+, Ube3am−/p+;Ptpa+/−, and Ptpa+/− mice. m, mushroom; s, stubby; t, thin. (Scale bar, 2 μm.) (B) Quantitation of dendritic spine subtypes. n = 60–80 dendrites per condition, n = 3 mice per genotype. (C) Representative images of apical dendrites of hippocampal CA1 pyramidal neurons in WT, Ube3am−/p+, Ube3am−/p+;Ptpa+/−, and Ptpa+/− mice. (Scale bar, 2 μm.) (D) Quantitation of dendritic spine subtypes. n = 20–40 dendrites per condition, n = 3 mice per genotype. (E) Representative images of basal dendrites of layer 2/3 pyramidal neurons of the motor cortex in WT, Ube3am−/p+, Ube3am−/p+;Ptpa+/−, and Ptpa+/− mice. (Scale bar, 2 μm.) (F) Quantitation of dendritic spine subtypes of motor cortex. n = 30–40 dendrites per condition, n = 3 mice per genotype. In all quantifications, error bars indicate mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA with Bonferroni posttest.
UBE3A Inhibits PTPA-Mediated PP2A Assembly and Activity.
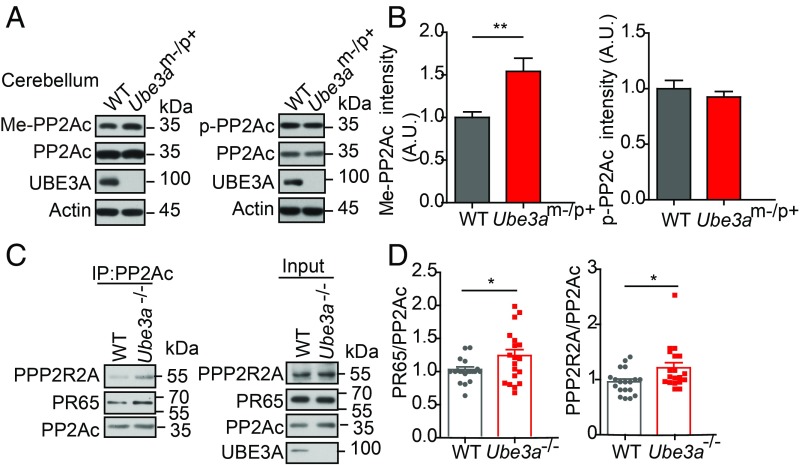
Having shown an interaction between PTPA and UBE3A at both the biochemical and functional levels, we next asked whether these interactions extend to the PP2A complex, as PTPA is an activator of PP2A. PTPA interacts with the catalytic subunit PP2Ac, and is known to regulate its phosphorylation and/or methylation to promote PP2A holoenzyme assembly in nonneuronal cells (21, 22). To test the link between PTPA up-regulation and enhanced PP2A activity in Ube3am−/p+ mice, we first examined PP2A activity in Ptpa mutant mice (Ptpa+/−), and showed that PP2A activity was reduced in Ptpa+/− mice (SI Appendix, Fig. S8A). Co-IP in HEK293T cells expressing Flag-PTPA also showed that PTPA interacts with PP2Ac (SI Appendix, Fig. S8B). Using an antibody that recognized methylated PP2Ac (Leu309) (23), we found that the level of methylated PP2Ac (Me-PP2Ac) was significantly increased in both the cerebellum and hippocampus of the Ube3am−/p+ mice (Fig. 4 A and B and SI Appendix, Fig. S8 C and D). Conversely, matDp/+ mice displayed a reduction in carboxylmethylation of PP2Ac in the hippocampus (SI Appendix, Fig. S8 E and F). In contrast to PP2Ac methylation, phosphorylated PP2Ac (p-PP2Ac) was not significantly different between WT and Ube3am−/p+ mice. These results suggest that methylation of PP2Ac, not its phosphorylation, is specifically regulated by UBE3A in the brain.
Fig. 4.
UBE3A regulates the PP2A activity by regulating its holoenzyme assembly. (A) Representative images showing that PP2Ac methylation level was increased in the cerebellum of Ube3am−/p+ mice. (B) Quantification of Me-PP2Ac and p-PP2Ac level, normalized to that of PP2Ac. A.U., arbitrary units. n = 11 mice per condition. (C and D) Co-IP analysis of the amount of the PP2A holoenzyme in WT and Ube3a−/− mice. Quantification shown in D as the ratio of PR65/PPP2R2A intensity to that of PP2Ac intensity. In all quantifications, error bars indicate mean ± SEM; *P < 0.05, **P < 0.01, Student’s unpaired t test.
Since PP2Ac methylation was known to enhance binding of the PR55/B subunit to the PP2A core dimer (24), we further examined whether PP2A holoenzyme assembly is affected by UBE3A deficiency. Our co-IP analysis showed that higher amounts of PR65 and PPP2R2A subunits were coimmunoprecipitated with the PP2Ac subunit in lysates from Ube3a−/− mice, compared with WT littermates (Fig. 4 C and D). Furthermore, PPP2R2A immunoprecipitated more PP2A core dimer in Ube3a−/− mice (SI Appendix, Fig. S8G). The immunoprecipitates enriched with the PP2Ac antibody were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Consistently, we found more PR65 and PR55/B subunits in the immunoprecipitates of the Ube3am−/p+ mice, compared with WT littermates (Dataset S2). Taken together, these findings indicate that UBE3A negatively regulates PP2A activity through UPS degradation of PTPA, which enhances PP2Ac methylation to promote PP2A assembly.
Increased phosphorylation of Ca2+/calmodulin-dependent protein kinase CaMKIIα at T286, a substrate for PP2A in the cytoplasm (25), was reported in hippocampus of Ube3am−/p+ mice (26). Although we observed an increase in phosphorylated CaMKIIα in Ube3am−/p+ mice, the phosphorylation level of CaMKIIα did not change in the hippocampus of Ptpa+/− mice (SI Appendix, Fig. S8 H and I). These results suggest that CaMKIIα might not be downstream of the UBE3A-PTPA-PP2A axis.
Pharmacological Inhibition of PP2A Activity Rescues Cellular and Behavioral Phenotypes in Ube3am−/p+ Mice.
Previous findings on Ube3a-deficient mice have shown extensive defects in spine morphology and physiology, as well as behavioral impairment. Therefore, we examined motor behaviors of WT and mutant mice (Ube3am−/p+, Ptpa+/−, and Ube3am−/p+;Ptpa+/− mice) at P30. Although Ube3am−/p+ mice performed worse than WT mice, we did not observe an improvement in Ube3am−/p+;Ptpa+/− mice compared with Ube3am−/p+ mice (SI Appendix, Fig. S9A). Since the regulation of PP2A by UBE3A occurred postnatally, we surmised that down-regulation of PTPA during the embryonic period may exert a pleiotropic effect on normal development.
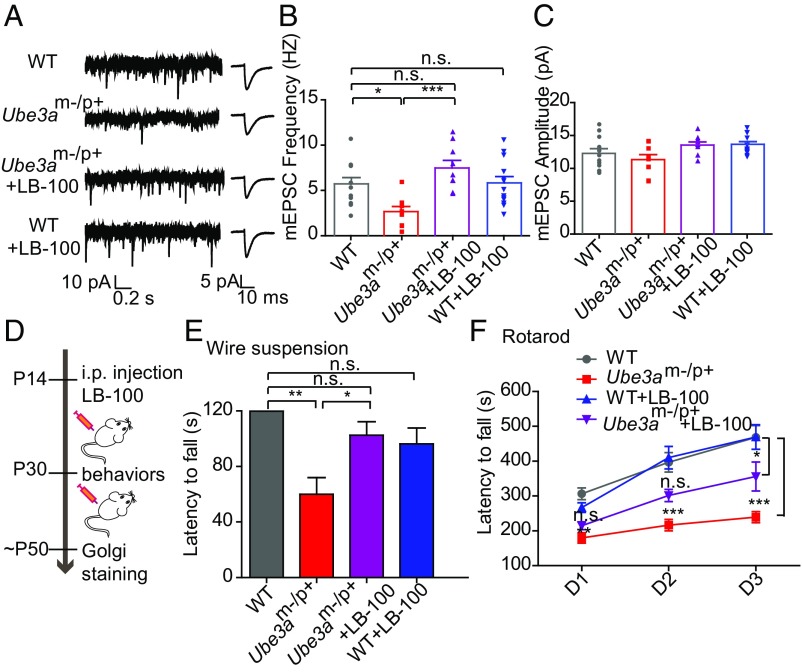
Since PP2A activity was significantly elevated in Ube3a mutant mice from P14 onwards, we asked whether pharmacological inhibition of PP2A activity at later developmental stages in Ube3am−/p+ mice could ameliorate cellular and behavioral defects. We thus examined whether PP2A activity could be reversed in Ube3am−/p+ mice using LB-100, a small molecular inhibitor of PP2A. Treatment of acute brain slices with LB-100 (300 nM) showed that the elevated PP2A activity in Ube3am−/p+ mice could be rescued by LB-100 (SI Appendix, Fig. S9B). Then, using the same treatment, we examined excitatory synaptic transmission in the primary motor cortex (M1), by whole-cell recording of miniature excitatory postsynaptic currents (mEPSC) in layer 2/3 pyramidal neurons. Consistent with previous findings (8), we found that average mEPSC frequencies in Ube3am−/p+ mice at P30 were significantly lower than those of WT littermates, whereas average mEPSC amplitudes were not affected (Fig. 5 A–C). Incubation of brain slices with PP2A inhibitor LB-100 elevated the mEPSC frequencies in Ube3am−/p+ mice to a level similar to that of the WT mice (Fig. 5 A–C). These results confirmed that the synaptic deficits in Ube3am−/p+ mice were due to elevated PP2A activity.
Fig. 5.
LB-100, an inhibitor of PP2A activity, restores changes in excitatory synapses and motor deficits in Ube3am−/p+ mice. (A) Representative mEPSC traces (Left) and average waveforms (Right) from acute brain slices of motor cortex of P30 mice, conditions as indicated. (B and C) Quantification of mEPSC frequencies and amplitudes. n represents the number of cells: WT, n = 12; Ube3am−/p+, n = 9; Ube3am−/p++LB-100, n = 9; WT+LB-100, n = 13; one-way ANOVA with Tukey’s posttest. (D) Schematic of experimental design. i.p., intraperitoneal. (E) Wire suspension test. n = 7 mice per condition; one-way ANOVA with Tukey’s posttest. (F) Rotarod test. n = 8 mice per condition; two-way ANOVA with Bonferroni posttest. Error bars indicate mean ± SEM; ***P < 0.001, **P < 0.01, *P < 0.05, n.s., not significant.
Since the PP2A inhibitor LB-100 is a small molecule that can cross the blood–brain barrier (27) and inhibits the activity of PP2A (SI Appendix, Fig. S9C), we intraperitoneally injected mice with LB-100 (1 mg/kg) every 2 days starting at P14 and examined their behavior at P30 (Fig. 5D). Using gait analysis, wire suspension, and rotarod, we found that muscle strength, motor coordination, and learning in Ube3am−/p+ mice were deficient compared with WT mice, and that these deficiencies were largely rescued by LB-100 treatment (Fig. 5 E and F and SI Appendix, Fig. S10A). After behavioral testing, we used Golgi staining to examine the synaptic morphology of layer 2/3 pyramidal neurons in M1 and cerebellar Purkinje cells at around P50. Consistent with the behavioral results, we found that the increase in the proportion of thin spines found in Ube3am−/p+ mice was significantly rescued in mutant mice treated with LB-100 (SI Appendix, Fig. S10 B and C). These results suggest that PP2A inhibitors could be used to ameliorate cellular and behavioral deficits induced by UBE3A deficiency.
Discussion
Given that alterations in UBE3A levels leads to AS and ASD, identifying neuronal substrates of UBE3A E3 ligase is critical for understanding disease progress and for developing methods for clinical interventions. In the present study, we demonstrate that UBE3A directly binds to and degrades the PP2A activator PTPA, a process that is critical for dendritic spines morphogenesis and excitatory synaptic function in the developing brain. First, using quantitative mass spectrometry and biochemical methods, we demonstrate that PTPA protein level is bidirectionally altered in the AS mouse model and in the human 15q11–13 duplication mouse model. Second, we observed corresponding changes downstream of the UBE3A-PTPA pathway, including methylation of PP2Ac, assembly of PP2A holoenzyme, and PP2A activity in the AS mouse model. Third, down-regulation of PTPA or pharmacological inhibition of PP2A rescued spine morphology defects and synaptic transmission in AS model mice, suggesting that UBE3A targets PTPA-PP2A to regulate synaptic function. Collectively, these results suggest that PTPA functions as a substrate of UBE3A, and that dysregulation of the UBE3A-PTPA-PP2A pathway contributes to UBE3A-related neurodevelopmental disorders (SI Appendix, Fig. S11).
The diversity of PP2A substrates and function depends on multiple PP2A holoenzymes and a plethora of endogenous regulators, including PTPA, IGBP1 (α4), LCMT1, PME-1, and TIPRL1 (13). Our data suggest that UBE3A regulates PP2A activity through ubiquitinating its activator PTPA, specially regulating the methylation of PP2Ac and its selective recruitment of PR55/B regulatory subunits. In AS mutant mice, the phosphorylation levels of CaMKIIα at Thr286 and Thr305 in hippocampus are increased, and its PP1/PP2A phosphatase activity is reduced (26). In neurons, CaMKIIα is dephosphorylated by PP1 in the postsynaptic density (PSD) and dephosphorylated in cytosol or synaptosomes by PP2A. The latter pathway is likely to be regulated by the α4 regulator of PP2A (25), because α4 physically interacts with CaMKIIα and a neuronal specific α4 deficiency in hippocampus leads to an increase of CaMKIIα activity in the cytoplasm. Thus, we reasoned that UBE3A-PTPA-PP2A targets other downstream pathways to regulate brain development. Consistently, we found that IGBP1 (α4) level is not affected in AS mice and the reduction of PTPA in hippocampus did not affect the phosphorylation of CaMKIIα. Together, our findings thus provide insights into a specific regulation of PP2A activity in the nervous system.
Abnormal spine development and maturation have been a consistent anatomical observation in neurodevelopmental disorders, including AS and ASD. However, the mechanism through which UBE3A regulates spine development and maturation is not well understood. In this study, we demonstrate that excessive PTPA protein in an AS mouse model contributes to defects in dendritic spine maturation. Importantly, in vivo reduction of PTPA or pharmacological inhibition of PP2A activity prevents these defects in the AS mouse model. Consistently, inhibition of PP2A activity also restores the level in excitatory synaptic transmission in AS model mice. Phenotypically, administration of LB-100 rescues defects in motor function and learning in Ube3am−/p+ mice. In contrast, genetic reduction of PTPA using Ptpa+/− did not rescue the motor defects in Ube3a-deficient mice. One possible explanation is that PTPA has UBE3A-independent functions during embryonic development. Consistently, Ptpa−/− null mice are embryonic lethal, while Ube3am−/p+ mice are viable. UBE3A regulates the PTPA-PP2A pathway from the second postnatal week, a period important for experience-dependent formation and maintenance of dendritic spines (9). Thus, our findings offer an important mechanistic link between impaired PP2A activity and synaptic and behavioral deficits in UBE3A-related neurodevelopmental disorders.
Last but not least, to our knowledge, our results provide evidence that the PTPA/PP2A complex may be a target for treatment of UBE3A-related disorders. LB-100 has recently been tested in a phase I clinical trial for cancer therapy (28) and was found to be safe. Since our experiments demonstrate that LB-100 administration starting from P14 significantly rescued spine maturation, synaptic transmission, and motor function, it provides an exciting direction with much potential for the treatment of AS. Taken together, our findings offer an understanding of the mechanisms underlying the pathology of UBE3A-related neurodevelopmental disorders and suggest a target for therapeutic intervention toward AS and autism.
Materials and Methods
All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Institute of Neuroscience, Chinese Academy of Sciences and were in accordance with the Society for Neuroscience guidelines. Ube3a-deficient mice were generated by Jiang et al. (6). The UBE3A duplication mouse carries an interstitial duplication of 6 Mb on mouse chromosome 7 that corresponds to human chromosome 15q11–13, as previously described (19).
For details, see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Mu-Ming Poo for critical comments on the manuscript. We thank Dr. Qian Hu, Qian-Ru Ma, Dan Xiang, Xu-Xin Chen from the Optical Imaging Core Facility at the Institute of Neuroscience (ION) for assistance with confocal imaging, and Ms. Ling Han of the ION Animal Facility for assistance with animal care. We thank colleagues at ION and members of the Z.-Q.X. laboratory for helpful discussion and suggestions. This work was supported by grants from the Chinese Academy of Sciences (XDB32060000, QYZDJ-SSW-SMC010), the Ministry of Science and Technology (2016YFA0501002), and the Science and Technology Commission of Shanghai Municipality (16JC1420202).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820131116/-/DCSupplemental.
References
- 1.Rotin D., Kumar S., Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Sell G. L., Margolis S. S., From UBE3A to Angelman syndrome: A substrate perspective. Front. Neurosci. 9, 322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kishino T., Lalande M., Wagstaff J., UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 15, 70–73 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Bird L. M., Angelman syndrome: Review of clinical and molecular aspects. Appl. Clin. Genet. 7, 93–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glessner J. T., et al. , Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459, 569–573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y. H., et al. , Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 21, 799–811 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Dindot S. V., Antalffy B. A., Bhattacharjee M. B., Beaudet A. L., The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum. Mol. Genet. 17, 111–118 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Yashiro K., et al. , Ube3a is required for experience-dependent maturation of the neocortex. Nat. Neurosci. 12, 777–783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato M., Stryker M. P., Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc. Natl. Acad. Sci. U.S.A. 107, 5611–5616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H., Kunz P. A., Mooney R., Philpot B. D., Smith S. L., Maternal loss of Ube3a impairs experience-driven dendritic spine maintenance in the developing visual cortex. J. Neurosci. 36, 4888–4894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walz N. C., Baranek G. T., Sensory processing patterns in persons with Angelman syndrome. Am. J. Occup. Ther. 60, 472–479 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Depaoli-Roach A. A., et al. , Serine/threonine protein phosphatases in the control of cell function. Adv. Enzyme Regul. 34, 199–224 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Sents W., Ivanova E., Lambrecht C., Haesen D., Janssens V., The biogenesis of active protein phosphatase 2A holoenzymes: A tightly regulated process creating phosphatase specificity. FEBS J. 280, 644–661 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Stockwell J., Chen Z., Niazi M., Nosib S., Cayabyab F. S., Protein phosphatase role in adenosine A1 receptor-induced AMPA receptor trafficking and rat hippocampal neuronal damage in hypoxia/reperfusion injury. Neuropharmacology 102, 254–265 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Yu U. Y., Yoo B. C., Ahn J. H., Regulatory B subunits of protein phosphatase 2A are involved in site-specific regulation of tau protein phosphorylation. Korean J. Physiol. Pharmacol. 18, 155–161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sontag E., et al. , Altered expression levels of the protein phosphatase 2A ABalphaC enzyme are associated with Alzheimer disease pathology. J. Neuropathol. Exp. Neurol. 63, 287–301 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Nawaz Z., et al. , The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol. Cell. Biol. 19, 1182–1189 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krüger M., et al. , SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Nakatani J., et al. , Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell 137, 1235–1246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtmaat A. J., et al. , Transient and persistent dendritic spines in the neocortex in vivo. Neuron 45, 279–291 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Longin S., et al. , An inactive protein phosphatase 2A population is associated with methylesterase and can be re-activated by the phosphotyrosyl phosphatase activator. Biochem. J. 380, 111–119 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y., et al. , PTPA activates protein phosphatase-2A through reducing its phosphorylation at tyrosine-307 with upregulation of protein tyrosine phosphatase 1B. Biochim. Biophys. Acta 1833, 1235–1243 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Chasseigneaux S., et al. , Cytoplasmic SET induces tau hyperphosphorylation through a decrease of methylated phosphatase 2A. BMC Neurosci. 15, 82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryant J. C., Westphal R. S., Wadzinski B. E., Methylated C-terminal leucine residue of PP2A catalytic subunit is important for binding of regulatory Balpha subunit. Biochem. J. 339, 241–246 (1999). [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita T., et al. , Regulation of CaMKII by alpha4/PP2Ac contributes to learning and memory. Brain Res. 1082, 1–10 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Weeber E. J., et al. , Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J. Neurosci. 23, 2634–2644 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lecca S., et al. , Rescue of GABAB and GIRK function in the lateral habenula by protein phosphatase 2A inhibition ameliorates depression-like phenotypes in mice. Nat. Med. 22, 254–261 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Chung V., et al. , Safety, tolerability, and preliminary activity of LB-100, an inhibitor of protein phosphatase 2A, in patients with relapsed solid tumors: An open-label, dose escalation, first-in-human, phase I trial. Clin. Cancer Res. 23, 3277–3284 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.