Significance
FBXW7 is a typical tumor suppressor by targeting many oncoproteins for ubiquitylation and degradation, whereas LSD1 has oncogenic activity. Whether and how FBXW7 and LSD1 interact, with what biological consequence, are unknown. Here, we report that LSD1 is a pseudosubstrate of FBXW7. Upon binding with FBXW7, LSD1, instead of being degraded, disrupts FBXW7 dimerization and promotes FBXW7 self-ubiquitylation and degradation via proteasome and lysosomal pathways in a manner independent of its demethylase activity. As such, LSD1 abrogates biological functions of FBXW7 in growth suppression, nonhomologous end-joining repair, and radioprotection. Our study reveals a novel oncogenic mechanism of LSD1 by targeting FBXW7 and provides a sound strategy to reactivate FBXW7 by PROTAC-based LSD1 degradation in human cancers harboring wild-type FBXW7 with overexpressed LSD1.
Keywords: degradation, DNA damage repair, SCF E3 ligase, ubiquitylation
Abstract
FBXW7 acts as a typical tumor suppressor, with loss-of-function alterations in human cancers, by promoting ubiquitylation and degradation of many oncoproteins. Lysine-specific demethylase 1 (LSD1) is a well-characterized histone demethylase. Whether LSD1 has demethylase-independent activity remains elusive. Here we report that LSD1 directly binds to FBXW7 to destabilize FBXW7 independent of its demethylase activity. Specifically, LSD1 is a pseudosubstrate of FBXW7 and LSD1–FBXW7 binding does not trigger LSD1 ubiquitylation, but instead promotes FBXW7 self-ubiquitylation by preventing FBXW7 dimerization. The self-ubiquitylated FBXW7 is subjected to degradation by proteasome as well as lysosome in a manner dependent on autophagy protein p62/SQSTM1. Biologically, LSD1 destabilizes FBXW7 to abrogate its functions in growth suppression, nonhomologous end-joining repair, and radioprotection. Collectively, our study revealed a previously unknown activity of LSD1, which likely contributes to its oncogenic function. Targeting LSD1 protein, not only its demethylase activity, might be a unique approach for LSD1-based drug discovery for anticancer application.
LSD1 (also known as KDM1A) was the first histone demethylase identified as a transcriptional repressor to regulate gene expression via catalyzing the demethylation of mono- and dimethylated histone 3 lysine 4 (H3K4Me1, H3K4Me2) (1). Subsequent studies demonstrated that LSD1 also demethylates H3K9 (Me1 and Me2) to trigger gene activation programs (2). In addition to histone 3, recent studies showed that LSD1 also acts as demethylase of nonhistone proteins, such as p53 (3), E2F1 (4), and HIF-1α (5). Furthermore, LSD1 was shown to cooperate with multiple transcriptional regulators, such as CoREST (2), NuRD (6), and SIRT1/HDAC (7). Through these activities, LSD1 serves as a master regulator of gene expression and protein function and is actively involved in many biological processes.
LSD1 is overexpressed in many human cancers which is associated with poor patient survival (8). Increased preclinical data validated LSD1 as an attractive cancer target, resulting in extensive drug discovery efforts (9). So far, most of LSD1 studies focused on its demethylase activity and associated biological functions. Likewise, all drug discovery efforts on LSD1 targeting were directed to find the demethylase inhibitors (9). However, whether LSD1 has other activities/functions independent of its demethylase activity, and what is the underlying mechanism remain largely unknown, although a recent study showed that ZNF217 naturally interacts with LSD1 to coordinately regulate gene expression independently of its demethylase functions in a prostate cancer model (10).
The tumor suppressor FBXW7 functions as a well-characterized substrate recognition subunit of SCF (SKP1–CUL1–F-box protein) E3 ubiquitin ligase (11, 12). SCFFBXW7 promotes ubiquitylation and degradation of a large number of oncogenic substrates such as Cyclin E, c-JUN, c-MYC, NOTCH-1, and MCL-1 to suppress growth and survival of cancer cells (11, 13). Consistently, FBXW7 inactivation by point mutation, genomic deletion, or promoter hypermethylation (14) has been frequently observed in many types of cancers (15). In a number of mouse models, tissue-specific ablation of Fbxw7 accelerated tumorigenesis in the lung (16), intestine (17), and pancreas (18), further supporting its tumor suppressor function. Biochemically, FBXW7 is subjected to posttranslational modifications. While phosphorylation at Ser205 by ERK (19) triggers the proteasomal degradation of FBXW7 mediated by self-ubiquitylation (20), FBXW7 dimerization enhances the binding affinity and confers more accessible substrate lysine residues for ubiquitin conjugation (21, 22). However, whether FBXW7 dimerization is subjected to regulation by other FBXW7 binding proteins remains largely elusive.
Almost all FBXW7 substrates contain a conserved phosphomotif, termed as CPD (Cdc4 phosphodegron), which facilitates FBXW7 binding upon phosphorylation (23) and subsequent ubiquitylation by SCFFBXW7 via the K48 linkage for degradation (24). One exception is our recent finding that FBXW7 promotes XRCC4 polyubiquitylation via the K63 linkage, not for degradation, but for facilitating nonhomologous end-joining (NHEJ) repair (25). A recent report also showed that FBXW7 promotes γ-catenin polyubiquitylation via the K63 linkage (26). In this report, we presented yet another exception. LSD1 is a pseudosubstrate of FBXW7, not being ubiquitylated by FBXW7, but unexpectedly promotes FBXW7 ubiquitylation. Specifically, LSD1, in a manner independent of its demethylase activity, directly binds with FBXW7 in the CPD-dependent manner to disrupt FBXW7 dimerization, which triggers monomeric FBXW7 for self-ubiquitylation, followed by degradation via proteasome and lysosome systems, eventually leading to FBXW7 inactivation. Our study revealed a demethylase-independent activity of LSD1 in triggering FBXW7 degradation and established a LSD1–FBXW7 interaction axis to regulate FBXW7-mediated growth suppression, NHEJ repair, and radiosensitivity. Our study also has translational implication to provide a rationale for FBXW7 reactivation through targeting LSD1 protein for degradation, rather than inhibiting its enzymatic activity, in human cancers with LSD1 overexpression.
Results and Discussion
FBXW7 Binds to LSD1, but Fails to Promote Its Ubiquitylation.
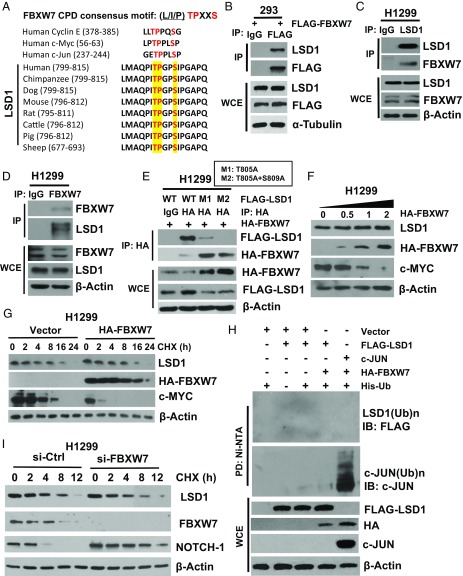
FBXW7 is a typical tumor suppressor that binds to a broad range of oncogenic substrates through their Cdc4 phosphodegron (CPD, L/I/PTPXXS) for subsequent polyubiquitylation via K48 linkage and proteasome degradation (23). We recently showed that FBXW7 also binds to XRCC4 for subsequent polyubiquitylation via K63 linkage, not for degradation, but for facilitating Ku70/80 heterodimer and XRCC4/Lig4/XLF complex binding to achieve effective NHEJ repair (25). However, whether FBXW7 would regulate the process of epigenetic modification via binding to its modifier(s) as ubiquitylation substrate(s) is largely unknown. Here we report the detection of an evolutionarily conserved CPD motif (ITPGPS) within the codons 804–809 of lysine-specific demethylase 1 (LSD1) (Fig. 1A). Indeed, ectopic expressed FLAG-FBXW7 readily pulled down endogenous LSD1 (Fig. 1B). More significantly, two proteins at the endogenous levels bound to each other under unstimulated physiological conditions (Fig. 1 C and D and SI Appendix, Fig. S1 A–B).
Fig. 1.
FBXW7 binds to LSD1, but fails to promote its ubiquitylation. (A) Sequence alignment of the phosphodegron sequences recognized by FBXW7 on LSD1, along with those found in known FBXW7 substrates, as indicated. (B–E) The FBXW7–LSD1 binding: the HEK293 cells were transiently transfected with FLAG-FBXW7, followed by IP with FLAG-Ab or control IgG, and IB with indicated Abs (B). H1299 cells were subjected to IP with LSD1-Ab (C) or FBXW7-Ab (D) or control IgG, followed by IB with indicated Abs. H1299 cells were cotransfected with HA-FBXW7 and FLAG-LSD1-WT, M1 (T805A) or M2 (T805A+S809A), followed by IP with FLAG-Ab or control IgG, and IB with indicated Abs (E). (F and G) FBXW7 overexpression has no effect on LSD1 levels. H1299 cells were transfected with increasing amounts of HA-FBXW7 (F) or fixed amount of HA-FBXW7, followed by CHX treatment for indicated periods of time (G), and IB with indicated Abs. (H) FBXW7 fails to promote LSD1 ubiquitylation. HEK293 cells were transfected with indicated plasmids, lysed under denatured condition at 6M guanidinium solution, followed by Ni-beads pull down, and IB to detect polyubiquitylation of LSD1 or c-JUN. (I) FBXW7 knockdown has no effect on LSD1 levels. H1299 cells were transfected with siRNA targeting LSD1, followed by CHX treatment for indicated periods of time and IB with indicated Abs.
To map the interaction domains between two proteins, we made a series of truncation mutants with FBXW7 into two domains (the N terminus and C-terminal WD40 domain), and LSD1 into three domains (N-terminal, AOL-N, and C-terminal), fused with FLAG or HA-epitope tags. FLAG tag-based pull-down assay revealed that FBXW7 interacts with LSD1 through the WD40 domain (SI Appendix, Fig. S1C), whereas LSD1 interacts with FBXW7 via its C-terminal region (AA415–852), where the evolutionarily conserved CPD motif (804–809) is localized (SI Appendix, Fig. S1D). We further defined whether the binding was indeed dependent on the CPD motif on LSD1 by generating the single (T805A) or double (T805A/S809A) LSD1 mutant, designated as M1 or M2. The mutants either largely or completely lost the ability to bind to FBXW7 (Fig. 1E). Interestingly, the FBXW7 levels are much higher when cotransfected with binding-dead LSD1 mutants than with the binding-capable wild-type LSD1 (Fig. 1E), suggesting that LSD1 may negatively regulate FBXW7 upon binding. Collectively, FBXW7 binds to LSD1 under physiological condition in a manner dependent on the CPD motif.
Given FBXW7 is a well-established E3 ligase, we next determined whether FBXW7 would reduce LSD1 protein levels by promoting its ubiquitylation and degradation. Unexpectedly, ectopic expression of FBXW7 in a dose-increasing manner failed to reduce endogenous LSD1 (Fig. 1F and SI Appendix, Fig. S1E), failed to shorten LSD1 protein half-life (Fig. 1G and SI Appendix, Fig. S1F), and failed to promote LSD1 ubiquitylation (Fig. 1H) with c-MYC, NOTCH-1 and c-JUN included as positive controls of FBXW7 substrates (27–29). Likewise, siRNA-based FBXW7 knockdown or genetic FBXW7 deletion had no effect on LSD1 protein half-life, nor protein levels, while extending the protein half-life or protein levels in known FBXW7 substrates, such as c-MYC and NOTCH-1 (Fig. 1I and SI Appendix, Fig. S1 G and H). Furthermore, in an in vitro demethylase activity assay using purified FBXW7 and LSD1 proteins with an artificially dimethyl-modified H3K4 peptide as the substrate, we found that FBXW7 had no effect on enzymatic activity of LSD1 (SI Appendix, Fig. S1I). Taken together, these results clearly demonstrated that LSD1 is a pseudosubstrate of FBXW7, which binds to FBXW7, but failed to be ubiquitylated and degraded by FBXW7; and FBXW7 does not affect LSD1 demethylase activity.
LSD1 Negatively Regulates FBXW7 Stability through Both Proteasomal and Lysosomal Pathways.
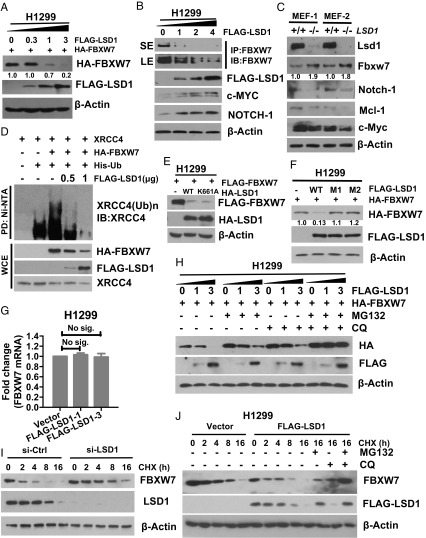
Given that LSD1 is not a bona fide substrate of FBXW7, and the level of FBXW7 was much lower when cotransfected with wild-type LSD1 (which bound to FBXW7) than that cotransfected with two LSD1 mutants (which failed to bind to FBXW7) (Fig. 1E), we hypothesized that LSD1 might negatively regulate FBXW7. Indeed, ectopic LSD1 expression in a dose-dependent manner remarkably reduced or eliminated exogenous FBXW7 protein (Fig. 2A and SI Appendix, Fig. S2A), as well as endogenous FBXW7 with consequent accumulation of FBXW7 substrates c-MYC and NOTCH-1 (27, 28) (Fig. 2B). In paired HCT116 cells (WT vs. FBXW7-null), ectopic LSD1 expression decreased or increased the levels of endogenous FBXW7 and its substrates c-MYC and NOTCH-1, respectively, only in wild-type cells (SI Appendix, Fig. S2B). Reciprocally, in two pairs of MEF cells derived from littermate embryos of LSD1-fl/fl mice (30), Ad-Cre–mediated Lsd1 deletion caused Fbxw7 accumulation and reduction of its substrates, Notch-1, Mcl-1, and c-Myc (Fig. 2C). Consequently, the E3 ubiquitin activity of FBXW7 was impaired in a dose-dependent manner upon LSD1 cotransfection, as reflected by reduced polyubiquitylation of FBXW7 substrate XRCC4 (25) (Fig. 2D) and a well-known substrate c-JUN (SI Appendix, Fig. S2C). Furthermore, LSD1 binding to FBXW7 in the presence of MG132 and chloroquine (CQ) that prevented FBXW7 degradation (see below) significantly abrogated FBXW7 binding with its substrates, such as c-MYC, cyclin E, and NOTCH-1 (SI Appendix, Fig. S2D), whereas LSD1 failed to bind to cyclin E (SI Appendix, Fig. S2E), indicating that the pseudosubstrate could compete with real substrates for FBXW7 binding. These findings collectively indicate that LSD1 negatively regulates FBXW7 levels and consequently inhibits its E3 activity, as well as competes with FBXW7 substrates for FBXW7 binding.
Fig. 2.
LSD1 destabilizes FBXW7 via both proteasome and lysosome pathways. (A and B) LSD1 overexpression decreases the protein levels of FBXW7. H1299 cells were transfected with increasing amounts of FLAG-LSD1 with (A) or without (B) HA-FBXW7 cotransfection, followed by IB (A) or IP/IB (B) with indicated Abs to detect levels of exogenous (A) or endogenous (B) FBXW7. SE, short exposure; LE, long exposure. (C) Genetic Lsd1 depletion increases the protein levels of Fbxw7. Two pairs of MEF cells with indicated genotypes were subjected to IB with indicated Abs. (D) LSD1 inhibits FBXW7 E3 ligase activity. The HEK293 cells were transfected with indicated plasmids, harvested 15 min post-IR and lysed under denatured condition at 6 M guanidinium solution, followed by Ni-bead pull down and IB to detect polyubiquitylation of XRCC4. (E) LSD1 reduction of FBXW7 is independent of its demethylase activity. H1299 cells were cotransfected with FLAG-FBXW7 and HA-LSD1-WT or a demethylase dead mutant K661A, followed by IB with indicated Abs. (F) LSD1 reduction of FBXW7 is dependent on its binding to FBXW7. H1299 cells were cotransfected with FLAG-FBXW7 and HA-LSD1-WT or either one of the binding site mutants M1/M2, followed by IB with indicated Abs. (G) LSD1 has no effect on FBXW7 mRNA. H1299 cells were transfected with increasing amounts of FLAG-LSD1 (1 μg and 3 μg) for 48 h and then harvest cells for total RNA isolation and qRT-PCR analysis. Shown is mean ± SEM from three independent experiments. (H–J) LSD1 reduces FBXW7 levels and shortens FBXW7 half-life, blocked by MG132 and CQ. Cells were transfected with indicated plasmids (H and J) or siLSD1 (I) for 48 h, followed by CHX treatment for indicated periods of time (I and J) in the absence or presence of MG132 or CQ alone or in combination for 6 h before harvesting for IB. Note that selected blots were quantified by densitometry scan using ImageJ software. The values in the blots were calculated after normalization of β-actin with the control sample setting at 1.
Since LSD1 is a well-known protein demethylase, we next determined whether LSD1 enzymatic activity is required for its inhibitory effect on FBXW7, using both genetic and small molecular inhibitor approaches. We first generated a K-to-A mutant at codon 661, which was previously reported as an enzyme-dead mutant (31). Interestingly, just like the wild-type form, LSD1-K661A mutant was able to reduce FBXW7 levels (Fig. 2E), and LSD1-induced FBXW7 reduction cannot be blocked by LSD1 inhibitors, compound 6b (32), or GSK2879552 (33) (SI Appendix, Fig. S2F). In contrast, two LSD1 binding mutants LSD1-M1 or LSD1-M2 had no effect on FBXW7 levels (Fig. 2F). These results clearly demonstrated that LSD1 acts as a negative regulator of FBXW7 in the manner that is independent of its demethylase activity, but dependent on its FBXW7 binding. Furthermore, the qRT-PCR analysis excluded the possibility that LSD1-mediated FBXW7 reduction occurred at the transcription levels (Fig. 2G and SI Appendix, Fig. S2G), strongly suggesting a posttranslational event.
We then determined whether proteasome inhibitor, MG132, or lysosomal inhibitor CQ, would rescue FBXW7 reduction by LSD1. While either compound showed a partial rescue, the combination of the two caused a full rescue (Fig. 2H). Likewise, the LSD1-K661A–induced FBXW7 reduction was partially rescued by either compound and completely by the combination (SI Appendix, Fig. S2H). Finally, we determined whether LSD1 affected the FBXW7 protein half-life. While siRNA-based LSD1 knockdown significantly extended the protein half-life of endogenous FBXW7 (Fig. 2I), ectopic LSD1 expression shortened it of both ectopically expressed and endogenous FBXW7, which was again abrogated by the combination of MG132 and CQ (Fig. 2J and SI Appendix, Fig. S2 I and J). Taken together, these results clearly indicated that LSD1 destabilizes FBXW7 by shortening its protein half-life via both proteasomal and lysosomal degradation pathways.
LSD1 Promotes FBXW7 Self-Ubiquitylation by Disrupting Its Dimerization.
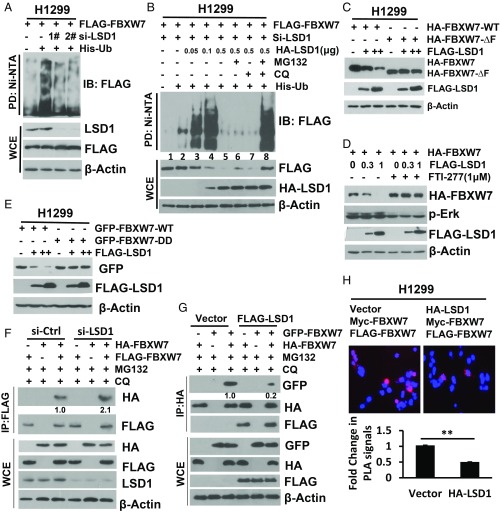
Having defined that LSD1 bound to and destabilized FBXW7, we next explored the underlying mechanism. Previous studies have shown that FBXW7 is subject to self-ubiquitylation while in the monomeric form, followed by degradation (19, 20). We, therefore, determined potential effect of LSD1 on FBXW7 self-ubiquitylation. While LSD1 knockdown significantly reduced FBXW7 self-ubiquitylation (Fig. 3A), LSD1 ectopic expression (in cells with endogenous LSD1 depleted) remarkably affected the levels of polyubiquitylated FBXW7 in a manner dependent on LSD1 dose. Specifically, while at the low doses (with 0.05–0.1 μg of plasmid DNA transfected) LSD1 promoted FBXW7 self-ubiquitylation (Fig. 3B, lanes 2–4), it triggered its degradation at the higher dose (with 0.5 μg of plasmid DNA transfected), leading to elimination of polyubiquitylated FBXW7 (Fig. 3B, lanes 5–7). Although the detailed underlying mechanism is unknown at the present time, the fact that degradation of polyubiquitylated FBXW7 induced by the high dose of LSD1 can be fully rescued only by the combinational treatment of MG132 and CQ, not by either treatment alone (Fig. 3B, lane 8), suggests that the process is mediated via both proteasome and lysosome pathways.
Fig. 3.
LSD1 promotes FBXW7 self-ubiquitylation by disrupting its dimerization. (A and B) LSD1 manipulation regulates FBXW7 self-ubiquitylation. Cells were transfected with two independent sets (1 and 2) of siRNAs targeting LSD1 for 24 h, then transfected with indicated plasmids for another 48 h (A), or one siLSD1 siRNA, then transfected with a fixed amount of FBXW7 along with an increasing amount of LSD1 in the absence or presence of MG132 or CQ (B), followed by Ni-bead pull down and IB for FLAG-FBXW7. (C–E) LSD1 reduction of FBXW7 blocked by ΔF mutation, DD mutation, and ERK inhibition. Cells were cotransfected with LSD1 and FBXW7-WT or ΔF (C), DD (E), or pretreated with ERK inhibitor FTI-277 (D), followed by IB. (F and G) LSD1 disrupts FBXW7 dimerization. H1299 cells were transfected with si-LSD1 (F) or FLAG-LSD1 (G), along with FBXW7 with different tags as indicated, followed by IP with FLAG-Ab (F) or HA-Ab (G) and IB with the indicated Abs. (H) In situ PLA to measure FBXW7 dimerization. The HEK293 cells were cotransfected with indicated plasmids, seeded on coverslips, and treated with CQ and MG132. Cells were stained with primary antibody, followed by staining using the In Situ Fluorescence kit. Cell images were acquired (Top), and positive-stained cells were quantified from at least 30 cells. Shown are the mean PLA intensity per cell (mean ± SD) (Bottom). Statistical analysis was performed using GraphPad Prism 8.0 software. **P < 0.01. Note that selected blots were quantified by densitometry scan using ImageJ software. The values in the blots were calculated after normalization of β-actin with the control sample setting at 1.
We also employed an in vitro ubiquitylation assay to further confirm FBXW7 polyubiquitylation promoted by LSD1. Indeed, purified LSD1-WT as well as the enzymatic-dead LSD1-K661A mutant promoted FBXW7 polyubiquitylation, but had no effect on FBXW7-ΔF (SI Appendix, Fig. S3A), a F-box deleted mutant lack of ligase activity. Consistently, when cotransfected, LSD1 reduced the levels of FBXW7, but not its FBXW7-ΔF mutant (Fig. 3C).
It has been shown that ERK kinase phosphorylates FBXW7 at Thr205 to trigger FBXW7 degradation by promoting its self-ubiquitylation (19). To determine a potential effect of this phosphorylation, we generated a phosphorylation-dead T205A mutant and found that the mutant bound to LSD1 as equally as well as the wild-type form (SI Appendix, Fig. S3B), indicating that the FBXW7–LSD1 binding is independent of the phosphorylation on this site. Interestingly, we detected a higher level of HA-FBXW7-T205A, compared with HA-FBXW7-WT (SI Appendix, Fig. S3B, lane 3 vs. lane 2) upon LSD1 cotransfection, suggesting a background level of FBXW7 degradation by LSD1, if ERK is not inhibited to block self-ubiquitylation. Indeed, when cells were treated with ERK inhibitor FTI-227 (34), LSD1-induced FBXW7 degradation was completely abrogated (Fig. 3D). Taken together, our results clearly demonstrated that LSD1 promotes FBXW7 self-ubiquitylation in a manner independent of its demethylase activity for subsequent degradation via proteasomal and lysosomal pathways.
We next investigated how LSD1 triggers FBXW7 self-ubiquitylation. Previous studies showed that the monomeric form of FBXW7 is prone to self-ubiquitylation, whereas the dimeric form is stable and critical for its E3 ubiquitin ligase activity toward its substrates (20, 35). To test whether LSD1 promoted FBXW7 self-ubiquitylation by interfering with its dimerization, we generated a dimerization-deficient mutant, with mutations on the dimerization domain, including A252D, L253D, L256D, and I257D, designated as FBXW7-DD, as previously reported (35), and found that compared with wild-type FBXW7, the FBXW7-DD mutant had a shorter protein half-life (SI Appendix, Fig. S3C), weaker ligase activity toward its substrate XRCC4, but stronger for self-ubiquitylation. (SI Appendix, Fig. S3D). Interestingly, unlike wild-type FBXW7, LSD1 had no effect on FBXW7-DD (Fig. 3E). We next determined the effect of LSD1 on FBXW7 dimerization, using different tagged-FBXW7 (HA-FBXW7 vs. FLAG-FBXW7 or HA-FBXW7 vs. GFP-FBXW7), and found that LSD1 knockdown moderately increased (Fig. 3F, and SI Appendix, Fig. S3E), whereas LSD1 ectopic expression significantly reduced FBXW7 dimerization, as demonstrated by two independent dimerization assays: (i) coimmunoprecipitation (Fig. 3G) and (ii) proximity ligase assay (PLA) (36) (Fig. 3H). Since β-TrCP was reported as another F-box protein capable of forming homodimer (37), we investigated potential effect of LSD1 and found that unlike FBXW7, LSD1 had no effect on β-TrCP dimerization (SI Appendix, Fig. S3F), indicating its specificity to FBXW7. Finally, we used an in vitro dimerization assay with affinity-purified FLAG-tagged LSD1 (wild type vs. a binding-dead mutant, M2) and FBXW7 (HA tagged or GFP fused), and found that LSD1-WT, but not LSD1-M2, disrupted FBXW7 dimerization in a dose-dependent manner, and no dimerization was found in FBXW7-DD as a positive control (SI Appendix, Fig. S3G), suggesting that the direct binding of LSD1 to FBXW7 is required for LSD1-mediated disruption of FBXW7 dimerization. Taken together, we concluded that LSD1 specifically disrupted FBXW7 dimerization to trigger its self-ubiquitylation in a monomeric form for subsequent degradation.
Ubiquitylated FBXW7 Is Degraded by Both Proteasome and p62-Mediated Lysosome Pathways.
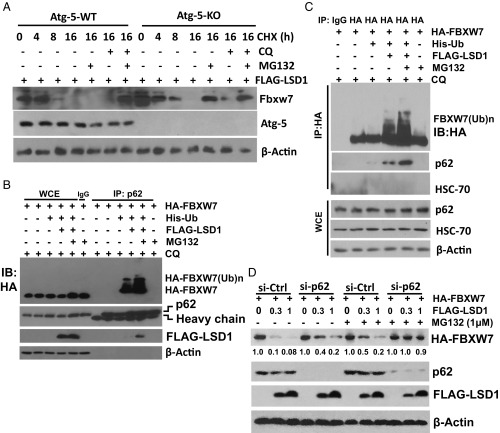
Since lysosome was involved in the degradation of polyubiquitylated FBXW7, and also in the process of autophagy, we next determined whether FBXW7 degradation occurs upon autophagy induction. Indeed, autophagy induced by serum starvation or rapamycin treatment reduced FBXW7 levels in the presence of proteasome inhibitor MG132 (SI Appendix, Fig. S4 A and B). To further determine the involvement of autophagy, we next used paired Atg5+/+ and Atg5−/− MEF cells (38) with treatment of cycloheximide (CHX) to block new protein synthesis. In Atg5+/+ cells, only the combination of MG132 and CQ, but neither drug alone, blocked LSD1-induced FBXW7 degradation, whereas in Atg5−/− cells, which are defective in autophagy induction, MG132 alone was sufficient to block FBXW7 degradation (Fig. 4A). Thus, it appears that autophagy plays a role in LSD1-induced FBXW7 degradation, only when FBXW7 is in polyubiquitylated form, induced by LSD1, and maintained by MG132 treatment.
Fig. 4.
Ubiquitylated FBXW7 is degraded by both proteasome and p62-mediated lysosome pathways. (A) Autophagy involvement in FBXW7 degradation. Paired MEF cells with Atg5-WT or KO were transfected with FLAG-LSD1, then switched 48 later to fresh medium containing CHX with or without MG132 and CQ treatment for indicated periods, and harvested for IB. (B and C) p62 binds to ubiquitylated FBXW7. Cells were transfected with indicated plasmids for 48 h and treated with or without MG132 and CQ in the last 6 h, lysates were subjected to IP with p62-Ab (B) or HA-Ab (C), and followed by IB. (D) p62 knockdown partially reverses LSD1 reduction of FBXW7. Cells were transfected with si-p62 or si-control for 48 h, then transfected with indicated plasmids for another 48 h, in the absence or presence of MG132 in the last 6 h before harvesting for IB. The blot was quantified by densitometry scan using ImageJ software. The values in the blots were calculated after normalization of β-actin with the control sample setting at 1.
To further confirm the role of autophagy, and to define associated mechanism of action, we focused on p62/SQSTM1, a key molecular receptor mediating autophagic clearance of polyubiquitylated proteins (39). No binding was detected between endogenous p62 and ectopically expressed FBXW7 under LSD1 cotransfected condition (SI Appendix, Fig. S4C). Remarkably, the binding between endogenous p62 and ubiquitylated FBXW7, but not nonubiquitylated FBXW7, was readily detected, if ubiquitin was included in cotransfection, and transfected cells were treated with CQ (to block autophagy), or in combination with MG132 (to block proteasome degradation as well, which achieved an even greater binding), regardless of using p62 pull down (Fig. 4B) or FBXW7 pull down (Fig. 4C). In FBXW7 pull-down complex, we detected p62, but not HSC-70 (Fig. 4C), a molecule reported to mediate selective protein degradation in chaperone mediated-autophagy pathway (40), thus excluding that possibility. Finally, we found that p62 knockdown partially or completely blocked LSD1-mediated FBXW7 degradation, alone or in combination with MG132, respectively (Fig. 4D), indicating a causal role of p62 in degradation of ubiquitylated FBXW7 via selective autophagy. Taken together, we concluded that ubiquitylated FBXW7 was degraded via both proteasome and p62-mediated lysosomal pathways, which was blocked by the combination of the inhibitors of both proteasome (e.g., MG132) and lysosome (e.g., CQ).
LSD1 Inhibits the Cellular Functions of FBXW7.
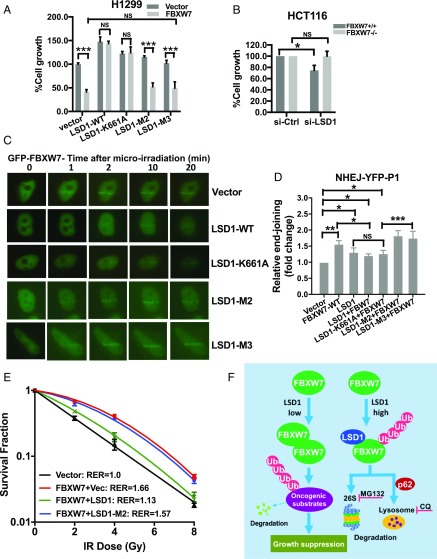
LSD1 is overexpressed in multiple human cancers with an oncogenic role and was validated as an attractive cancer target (41). Given that the vast majority of LSD1 functional studies were focused on its enzymatic activity, extensive drug discovery efforts have been made to identify its inhibitors of demethylase activity (9, 32, 42). Indeed, some of these inhibitors have been advanced to phase I/IIa clinical trials (41). Having established a demethylase-independent function of LSD1 in FBXW7 degradation, we next determined its biological consequence with a main focus on the regulation of FBXW7 functions in growth suppression (12), DNA damage repair, and radiation protection (25). We first showed that ectopic FBXW7 expression or LSD1 knockdown inhibited growth, whereas FBXW7 knockdown or ectopic LSD1 expression promoted it in lung cancer cells (SI Appendix, Fig. S5 A–D), indicating their tumor-suppressive or oncogenic activities, respectively. Ectopic expression of wild-type LSD1 (LSD1-WT) also stimulated clonogenic survival of lung cancer cells, and this stimulating activity was reduced in two LSD1 mutants with almost complete loss in a triple mutant (LSD1-M3), abrogating both enzymatic activity and FBXW7 binding ability (SI Appendix, Fig. S5 E and F), suggesting that both demethylase activity and FBXW7 binding ability of LSD1 are required for its fully oncogenic activity. Similarly, in a cotransfection experiment, ectopic expression of LSD1-WT significantly stimulated cell growth in HA-FBXW7–expressing cells, and this effect was reduced in either the enzymatic-dead LSD1-K661 or the FBXW7 binding-deficient LSD1-M2 mutant, and completely abrogated in LSD1-M3 mutant (SI Appendix, Fig. S5 G–I). More interestingly, under serum-deprivation conditions (2% serum), LSD1 and its enzymatic-dead mutant completely override growth suppressive function of FBXW7, which was totally abrogated in FBXW7 binding-deficient mutant or the double mutant (Fig. 5A), suggesting that under unfavorable growth conditions, the ability to degrade FBXW7 is more important for LSD1 to stimulate growth. We further compared the effect of LSD1 knockdown on paired HCT116 cells with or without FBXW7 deleted (43) and found that LSD1 knockdown only mediated growth suppression in FBXW7+/+ cells (Fig. 5B), suggesting a FBXW7-dependent mechanism. Taken together, these results suggested that oncogenic activity of LSD1 is mediated, at least in part, by promoting FBXW7 degradation.
Fig. 5.
LSD1 inhibits the cellular functions of FBXW7. (A and B) H1299 (A) or paired HCT116-FBXW7+/+ or −/− (B) cells were transfected with indicated plasmids or siLSD1 for 48 h, then switched to fresh medium containing 2% FBS for 72 h (A), or switched to fresh medium containing 10% FBS for 72 h, followed by ATPlite proliferation assay. (C) LSD1 impairs the recruitment of GFP-FBXW7 to DNA damage sites. HeLa cells were transfected with GFP-FBXW7 and FLAG-LSD1-WT or its mutants as indicated, followed by laser-beam microradiation. Localization of GFP-FBXW7 to DNA damage sites were visualized and photographed. (Scale bar: 10 μm.) (D) LSD1 impairs FBXW7-mediated NHEJ. Cells were transfected with indicated plasmids for 48 h, then transfected with linearized pEYFP plasmid (by NheI digestion), and analyzed by qPCR 12 h later using one set of primers (designated as P1), flanking the ligated pEYFP region. Shown is mean ± SEM from three independent experiments with normalization to an uncut flanking DNA sequence. *P < 0.05, **P < 0.01; ***P < 0.001. (E) LSD1 impairs FBXW7-mediated radioprotection. HCT116-FBXW7−/− cells were transfected with indicated plasmids, then irradiated with indicated doses, followed by clonogenic survival assay. Data are presented as mean ± SEM of three repeats. (F) Proposed working model for LSD1 regulation of FBXW7. In cells with low levels of LSD1, FBXW7 forms active dimer to promote ubiquitylation and degradation of oncoprotein substrates to suppress the growth. In cells with high levels of LSD1, LSD1 binding of FBXW7 prevents its dimerization, leading to FBXW7 self-ubiquitylation, followed by degradation by proteasome and p62-dependent lysosome pathways. NS, no significance.
Recently we reported that FBXW7 maintains genome integrity via promoting XRCC4 polyubiquitylation to facilitate NHEJ repair (25). And as shown in Fig. 2D, LSD1 significantly inhibited FBXW7 E3 ligase activity toward XRCC4. Therefore, we next determined whether LSD1 interferes with NHEJ repair via degrading FBXW7. In a laser microirradiation assay to detect the recruitment of the GFP-FBXW7 fusion protein to DNA damage sites when cotransfect with LSD1 or its mutants, we found that LSD1-WT and its enzymatic-dead mutant (LSD1-K661A) significantly blocked and delayed FBXW7 recruitment, whereas the FBXW7 binding-deficient mutant or double mutant (LSD1-M2/M3) had no effect (Fig. 5C), indicating a LSD1–FBXW7 binding-dependent event. Furthermore, using a linearized plasmid-based NHEJ assay (25), we found that ectopic FBXW7 expression had repair-promoting activity, as did LSD1, consistent with a previous report (44). Interestingly, FBXW7 combination with LSD1-WT or LSD1-K661A had reduced activity, which was completely rescued by LSD1-M2 or LSD1-M3 (Fig. 5D and SI Appendix, Fig. S5J, note that two independent sets of primers, designated as P1 and P2 were used to detect NHEJ activity), again suggesting a FBXW7 binding-dependent event.
Finally, we determined whether LSD1 affects the radioprotection effect of FBXW7 (25). Indeed, ectopic FBXW7 expression conferred significant radioprotection with a resistance enhancement ratio (RER) of 1.66. This effect was significantly abrogated by coexpression of LSD1, but not FBXW7 binding-deficient mutant, LSD1-M2 (Fig. 5E), further suggesting a LSD1/FBXW7 interaction-dependent event. Taken together, we conclude that LSD1 destabilization of FBXW7 has biological consequence by abrogating FBXW7 cellular functions.
FBXW7 is a well-studied F-box protein that binds its oncogenic substrates via a consensus binding motif for targeted polyubiquitylation via the K48 linkage and subsequent proteasome degradation (12). One exception is FBXW7-mediated polyubiquitylation of XRCC4, which is through the K63 linkage, not for degradation, but for facilitating the NHEJ repair (25). In this study, we made several findings: First, FBXW7 has a pseudosubstrate, LSD1. While FBXW7 selectively binds to LSD1 via its consensus binding motif, it failed to promote LSD1 polyubiquitylation, failed to reduce LSD1 levels, and failed to shorten its protein half-life. Second, instead of being degraded by FBXW7, LSD1 actually promotes FBXW7 self-ubiquitylation by disrupting FBXW7 dimerization, although how it does so through induced conformational change or via another mechanism will await structural demonstration when cocrystallization of two proteins becomes available. Third, LSD1 promotes FBXW7 polyubiquitylation in a manner independent of its demethylase activity, but dependent on its FBXW7 binding. Fourth, ubiquitylated FBXW7 is degraded by an expected proteasome pathway, as well as by a previously unknown lysosome pathway in a manner dependent on p62, as evidenced by the observation that LSD1-induced FBXW7 degradation can only be completely abrogated by the combination of inhibitors for both pathways. Why cells need to develop two independent pathways to degrade polyubiquitylated FBXW7 is an interesting open question. It is possible that both are safeguarding each other to ensure timely elimination of FBXW7 when cell proliferation is essential, given that FBXW7 is such a powerful tumor suppressor. Finally, LSD1 abrogates FBXW7 functions in growth suppression, NHEJ repair, and radiation protection, establishing a demethylase-independent oncogenic mechanism of LSD1 via FBXW7 targeting.
In summary, our study supports the following working model: In normal cells with a low level of LSD1, FBXW7 forms a dimer to act as an effective E3 ligase to promote ubiquitylation for proteasomal degradation of oncoprotein substrates, leading to suppression of cell outgrowth. In cancer cells with overexpressed LSD1, LSD1 binds to FBXW7 in a demethylase-independent manner to block FBXW7 dimerization and trigger its self-ubiquitylation, followed by degradation via proteasome and p62-dependent lysosome pathways, leading to accelerated growth (Fig. 5F).
This study identifies FBXW7 as having a pseudosubstrate, which actually promotes its degradation via the p62-dependent lysosomal pathway, in addition to the known proteasome pathway. Our study also has a translational implication. The efforts to discover LSD1 demethylase inhibitors, some of them currently in phase I/IIa clinical trials (41), should be extended to small molecules that specifically bind to LSD1, not necessary to inhibit its demethylase activity, for PROTAC-based degradation (45). This type of LSD1-specific PROTAC drugs that trigger LSD1 degradation should have broad utility for the treatment of human cancers harboring a wild-type FBXW7 with LSD1 overexpression, as well as for immunotherapy in combination with PD-L1 blockade (46).
Materials and Methods
Generation of MEF Cells.
MEF cells were derived from the Fbxw7flox/flox mouse strain, a gift from I. Aifantis, NYU School of Medicine, New York, NY (47) or Lsd1flox/flox mouse strain, a gift from S. Orkin, Harvard Medical School, Boston, MA (30). The Atg5+/+ and Atg5−/− MEF cells were a gift from N. Mizushima, Riken BioResource Center Cell Bank (cellbank@brc.riken.jp). The MEF cells were maintained as described in ref. 48. Briefly, MEFs were generated from day E13.5 embryos and cultured in DMEM with 15% FBS, 2 mM l-glutamine, and 0.1 mM MEM nonessential amino acids at 37 °C in a 5% CO2 humidified chamber. MEF cells were infected with adenoviruses expressing Cre recombinase (Ad-Cre) to delete the Fbxw7 or Lsd1 allele along with Ad-GFP as a control. All animal studies were approved and conducted in accordance with the guidelines established by the Committee on Use and Care of Animals at the University of Michigan (UM approval RPO00006919).
Site-Directed Mutagenesis.
Various LSD1 or FBXW7 mutants were generated using the QuikChange XL Site-Directed Mutagenesis Kit (Agilent). Mutants were designated as follows: LSD1-M1, T805A; LSD1-M2, T805A+S809A; LSD1-M3, K661A+T805A+S809A; and FBXW7-DD, A252D+L253D+L256D+I257D (35).
The In Vivo and In Vitro Ubiquitylation Assay.
HEK293 or H1299 cells were transfected with various plasmids or siRNAs. Approximately 48 h later, the transfected cells were treated with MG132 (10 μM) for 4 h before harvesting. The in vivo ubiquitylation assays were performed after Ni-NTA bead purification of ubiquitylated proteins, as described in ref. 49. For the in vitro ubiquitylation assays, HA-tagged FBXW7-WT (or FBXW7-ΔF) E3 was precipitated by HA beads from HEK293 cells and eluted with HA peptide, FLAG-tagged LSD1-WT (or LSD1-K661A) was pulled down by FLAG beads and eluted with FLAG peptide (Sigma), followed by incubation of FBXW7 with or without LSD1 in the presence of E1 and E2 in a ubiquitin reaction buffer [1.5 mM MgCl2, 5 mM KCl, 1 mM DTT, 20 mM Hepes (pH 7.4)] for 60 min under constant vortexing at 30 °C. Polyubiquitinated FBXW7 was resolved by SDS/PAGE and detected by immunoblotting (IB) with anti-HA Ab.
The In Vitro Demethylase Activity Assay.
LSD1 demethylase activity on free histones was carried out by the in vitro assay, as described in ref. 1. Briefly, peptides corresponding to the N-terminal 21 amino acids of histone H3 with dimethylated K4 residue (H3K4-Me2, AA1-21) were incubated with purified FLAG-LSD1 with or without purified HA-FBXW7 in the histone demethylase activity (HDM) assay buffer [50 mM Tris (pH 8.5), 50 mM KCl, 5 mM MgCl2, 0.5% BSA, and 5% glycerol] for 1 h at 37 °C. The demethylase activity of LSD1 was evaluated by dot blotting using H3K4-Me2 specific antibody.
FBXW7 Dimerization Assay.
For in vivo dimerization assay, FBXW7 and LSD1 plasmids with different tags were transfected individually or in combinations. The cell lysates were immunoprecipitated (IP) with anti-FLAG or anti-HA antibodies, followed by IB with various antibodies.
For in vitro dimerization assay, FLAG-LSD1 (WT/M2) and HA-FBXW7 constructs were transfected into HEK293 cells. Cells were treated with nocodazole (50 ng/mL) 48 h posttransfection for 8 h, and anti-FLAG/HA immunoprecipitation was performed to affinity-purify LSD1/FBXW7 proteins. The proteins were eluted out using 3XFLAG/HA peptide (from Sigma), and eluted HA-FBXW7 proteins were incubated with increasing doses (0, 0.3, 1, or 3 mM) of eluted FLAG-LSD1 (WT/M2) proteins as described previously (20, 35), followed by GFP-FBXW7 immunoprecipitation. The resolving proteins were resolved by SDS/PAGE.
The In Situ PLA.
The assay was performed as described previously (36, 50). Briefly, the HEK293 cells were cotransfected with plasmids expressing FBXW7-FLAG and FBXW7-MYC combined with HA-LSD1, along with the vector for 24 h. Cells were seeded on coverslips and treated with CQ and MG132. Cells were then washed with ice-cold PBS, fixed, permeabilized, and incubated with mouse anti-MYC (Sigma-Aldrich) and rabbit anti-FLAG (Sigma-Aldrich) primary antibodies and then stained using Duolink In Situ Fluorescence kit (Sigma-Aldrich), according to the manufacturer’s recommendation. The images were taken using a fluorescence microscope with the appropriate filter. The Duolink PLA signal is recognized as discrete fluorescent spots in various locations of the studied cells. PLA images were collected using Olympus Cellsense Dimension acquisition software from several fields of view per condition. Fluorescent intensity was quantified from at least 30 cells in each group using Duolink ImageTool (Sigma-Aldrich).
Microirradiation.
Cells were seeded onto glass-bottom dishes (MatTek) and transfected with pEGFP-FBXW7α with or without cotransfection of FLAG-LSD1 (WT and mutants). GFP-positive cells were then irradiated with a 365-nm UVA laser focused through a 60 ×/1.2-W objective using a Zeiss Axiovert equipped with LSM 520 Meta. UVA (4.36 J/m2) was introduced to an area of ∼12 × 0.1 mm. Images were captured with indicated time intervals.
NHEJ Linearized Plasmid Assay.
For the linearized plasmid-based end-joining assay, pEYFP-N1 (Clontech Laboratories) was linearized by digestion between the promoter and coding sequence of EYFP with NheI. The linear products were gel purified and transfected into serum-starved (overnight) cells. After 12 h, the cells were harvested and lysed to isolate transfected plasmids. The efficiency of end-joining repair was assessed by qPCR of the ligated EYFP region using two sets of primers (termed as P1/P2) for detecting NHEJ activity independently, followed by normalization to an uncut flanking DNA sequence. Their sequences are as follows: P1 set, P1-F 5′-GCTGGTTTAGTGAACCGTCAG-3′ and P1-R 5′-GCTGAACTTGTGGCCGTTTA-3′; P2 set, P2-F 5′-CAACGGGACTTTCC AAAATG-3′ and P2-R 5′-CTCCTCGCCCTTGCTCAC-3′. The primer set for uncut control is NCont-F 5′-TACATCAATGGGCGTGGATA-3′ and Ncont-R 5′-AAGTC CCGTTGATTTTGGTG-3′.
Quantification and Statistical Analysis.
Data with two groups were analyzed by two-tailed Student’s t tests, and data with multiple groups were analyzed by one-way ANOVA, followed by Bonferroni post hoc test using GraphPad Prism statistical programs (GraphPad Prism). Results were expressed as mean ± SEM from three independent assays. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. I. Aifantis for Fbxw7fl/fl mice, Dr. S. Orkin for Lsd1fl/fl mice, and Dr. N. Mizushima for Atg5+/+ and Atg5−/− MEF cells. This work was supported in part by the National Key R&D Program of China (2016YFA0501800 to Y.S.), National Cancer Institution Grant CA156744 (to Y.S.), and National Nature Science Foundation of China Grants 81572718 and 81630076 (to Y.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902012116/-/DCSupplemental.
References
- 1.Shi Y., et al. , Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Wang J., et al. , Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446, 882–887 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Huang J., et al. , p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007). [DOI] [PubMed] [Google Scholar]
- 4.He Y., et al. , LSD1 promotes S-phase entry and tumorigenesis via chromatin co-occupation with E2F1 and selective H3K9 demethylation. Oncogene 37, 534–543 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Lee J. Y., et al. , LSD1 demethylates HIF1α to inhibit hydroxylation and ubiquitin-mediated degradation in tumor angiogenesis. Oncogene 36, 5512–5521 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., et al. , LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Mulligan P., et al. , A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell 42, 689–699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kooistra S. M., Helin K., Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13, 297–311 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Fu X., Zhang P., Yu B., Advances toward LSD1 inhibitors for cancer therapy. Future Med. Chem. 9, 1227–1242 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Sehrawat A., et al. , LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc. Natl. Acad. Sci. U.S.A. 115, E4179–E4188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R. J., Welcker M., Clurman B. E., Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell 26, 455–464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welcker M., Clurman B. E., FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Wang Z., Liu P., Inuzuka H., Wei W., Roles of F-box proteins in cancer. Nat. Rev. Cancer 14, 233–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhoondi S., et al. , Inactivation of FBXW7/hCDC4-β expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res. 12, R105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama K. I., Nakayama K., Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Grim J. E., et al. , Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol. Cell Biol. 32, 2160–2167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sancho R., et al. , F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology 139, 929–941 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q., et al. , Fbxw7 deletion accelerates KrasG12D-Driven pancreatic tumorigenesis via Yap accumulation. Neoplasia 18, 666–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji S., et al. , ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell Res. 25, 561–573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min S. H., et al. , Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol. Cell 46, 771–783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X., et al. , Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 129, 1165–1176 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Welcker M., et al. , Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 27, 2531–2536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nash P., et al. , Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Finley D., Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q., et al. , FBXW7 facilitates nonhomologous end-joining via K63-linked polyubiquitylation of XRCC4. Mol. Cell 61, 419–433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., et al. , FBW7 suppresses cell proliferation and G2/M cell cycle transition via promoting γ-catenin K63-linked ubiquitylation. Biochem. Biophys. Res. Commun. 497, 473–479 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Fryer C. J., White J. B., Jones K. A., Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16, 509–520 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Welcker M., et al. , The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G. Jr, The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Kerenyi M. A., et al. , Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife 2, e00633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M. G., Wynder C., Cooch N., Shiekhattar R., An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Ma L. Y., et al. , Design, synthesis, and structure-activity relationship of novel LSD1 inhibitors based on pyrimidine-thiourea hybrids as potent, orally active antitumor agents. J. Med. Chem. 58, 1705–1716 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Mohammad H. P., et al. , A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28, 57–69 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Kawasaki K., et al. , Ras signaling directs endothelial specification of VEGFR2+ vascular progenitor cells. J. Cell Biol. 181, 131–141 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P., Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Zhou X., et al. , Blockage of neddylation modification stimulates tumor sphere formation in vitro and stem cell differentiation and wound healing in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, E2935–E2944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki H., et al. , Homodimer of two F-box proteins betaTrCP1 or betaTrCP2 binds to IkappaBalpha for signal-dependent ubiquitination. J. Biol. Chem. 275, 2877–2884 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Kuma A., et al. , The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto G., Wada K., Okuno M., Kurosawa M., Nukina N., Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Kaushik S., Cuervo A. M., The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosseini A., Minucci S., A comprehensive review of lysine-specific demethylase 1 and its roles in cancer. Epigenomics 9, 1123–1142 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Stewart C. A., Byers L. A., Altering the course of small cell lung cancer: Targeting cancer stem cells via LSD1 inhibition. Cancer Cell 28, 4–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajagopalan H., et al. , Inactivation of hCDC4 can cause chromosomal instability. Nature 428, 77–81 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Peng B., et al. , Modulation of LSD1 phosphorylation by CK2/WIP1 regulates RNF168-dependent 53BP1 recruitment in response to DNA damage. Nucleic Acids Res. 43, 5936–5947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu S., Cui D., Chen X., Xiong X., Zhao Y., PROTACs: An emerging targeting technique for protein degradation in drug discovery. Bioessays 40, e1700247 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Sheng W., et al. , LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson B. J., et al. , Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 205, 1395–1408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan M., Davis S. W., Saunders T. L., Zhu Y., Sun Y., RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc. Natl. Acad. Sci. U.S.A. 106, 6203–6208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu Q., Tan M., Sun Y., SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 67, 3616–3625 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Gajadhar A., Guha A., A proximity ligation assay using transiently transfected, epitope-tagged proteins: Application for in situ detection of dimerized receptor tyrosine kinases. Biotechniques 48, 145–152 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.