Significance
The sea urchin calcite spicules and vertebrate blood vessels are quite distinct in their function, yet both have a tubular structure and are controlled by the vascular endothelial growth factor (VEGF) pathway. Here we study the downstream program by which VEGF signaling drives sea urchin spiculogenesis and find remarkable similarities to the control of vertebrate vascularization. The similarities are observed both in the upstream gene regulatory network, in the downstream effector genes, and the cellular processes that VEGF signaling controls at the site of the calcite spicule formation. We speculate that sea urchin spiculogenesis and vertebrate vascularization diverged from a common ancestral tubulogenesis program that was uniquely coopted for biomineralization in the echinoderm phylum.
Keywords: gene regulatory networks, biomineralization, tubulogenesis, VEGF signaling, evolution
Abstract
Biomineralization is the process by which living organisms use minerals to form hard structures that protect and support them. Biomineralization is believed to have evolved rapidly and independently in different phyla utilizing preexisting components. The mechanistic understanding of the regulatory networks that drive biomineralization and their evolution is far from clear. Sea urchin skeletogenesis is an excellent model system for studying both gene regulation and mineral uptake and deposition. The sea urchin calcite spicules are formed within a tubular cavity generated by the skeletogenic cells controlled by vascular endothelial growth factor (VEGF) signaling. The VEGF pathway is essential for biomineralization in echinoderms, while in many other phyla, across metazoans, it controls tubulogenesis and vascularization. Despite the critical role of VEGF signaling in sea urchin spiculogenesis, the downstream program it activates was largely unknown. Here we study the cellular and molecular machinery activated by the VEGF pathway during sea urchin spiculogenesis and reveal multiple parallels to the regulation of vertebrate vascularization. Human VEGF rescues sea urchin VEGF knockdown, vesicle deposition into an internal cavity plays a significant role in both systems, and sea urchin VEGF signaling activates hundreds of genes, including biomineralization and interestingly, vascularization genes. Moreover, five upstream transcription factors and three signaling genes that drive spiculogenesis are homologous to vertebrate factors that control vascularization. Overall, our findings suggest that sea urchin spiculogenesis and vertebrate vascularization diverged from a common ancestral tubulogenesis program, broadly adapted for vascularization and specifically coopted for biomineralization in the echinoderm phylum.
Biomineralization is the process in which soft organic tissues use minerals to produce shells, skeletons, and teeth for various functions, such as protection and physical support (1). This process occurs within diverse organisms from the five kingdoms of life: bacteria, protista, fungi, animals, and plants (1). Biomineralization is thought to have evolved independently and rapidly in different phyla, through the use of preexisting components and the evolution of specialized biomineralization proteins that utilize different minerals and shape them in different forms (2, 3). To understand the biological control and evolution of biomineralization, it is essential to study the gene regulatory networks (GRNs) that control this process and unravel their origin. However, the structure and the function of the GRNs that control biomineralization processes have only been studied for a few examples, mostly in vertebrates’ bone and teeth formation (4, 5).
The model of GRN that controls skeletogenic cell specification in the sea urchin embryo is one of the most elaborate of its kind (6). Studies of the sea urchin larval skeleton have significantly contributed to the field of biomineralization by illuminating the pathway of mineral uptake and deposition in live embryos (7–10). However, the skeletogenic GRN was mostly studied at the early stages of skeletogenesis before the spicules are formed, and therefore it is still unclear how the skeletogenic GRN controls spicule formation and biomineralization.
The sea urchin skeleton is made of two rods of calcite generated by the skeletogenic mesodermal (SM) cells (Fig. 1A). The SM cells ingress into the blastocoel, fuse through their filopodia, and form a ring with two lateral cell clusters (see red cells in Fig. 3A). In these clusters, the cells construct a syncytial cytoplasmic cord into which they secrete vesicles of calcium carbonate that form the calcite spicules (Fig. 1 B–F) (7–9, 11−12). The spicule cord has a tubular structure (8, 9, 13) and the forming calcite rods have a circular shape (Fig. 1 D–F) (9). SM cell migration, lateral cluster formation, and sea urchin spiculogenesis were shown to depend on the vascular endothelial growth factor (VEGF) pathway (6, 14–16). The sea urchin VEGF3 gene is expressed in two lateral ectodermal domains positioned in close proximity to the SM lateral cell clusters and its receptor, VEGFR-10-Ig, is expressed exclusively in the SM cells (see also Fig. 3A) (14, 15). For simplicity, throughout the paper, we use the annotation VEGF for VEGF3 and VEGFR for VEGFR-10-Ig (14). Perturbations of the VEGF pathway completely abolish calcite spicule formation in the sea urchin embryo (14, 15), but the downstream cellular and molecular mechanisms activated by sea urchin VEGF signaling, are largely unknown.
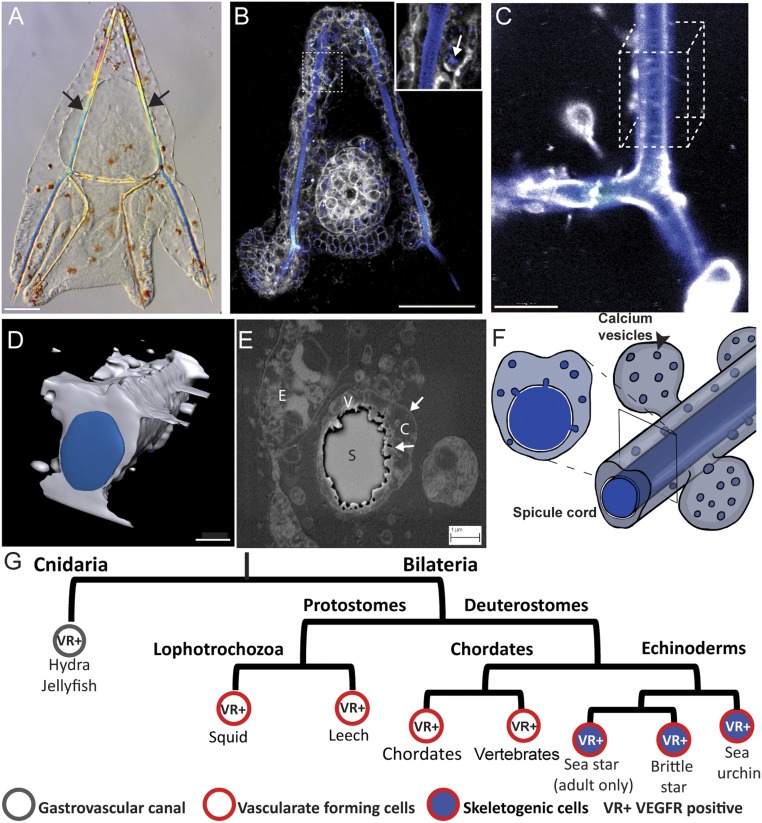
Fig. 1.
Spiculogenesis in the sea urchin embryo and VEGFR expression in tubular organs in metazoan. (A) Sea urchin larva at 3 dpf, showing its two calcite spicules (arrows). (Scale bar, 50 µm.) (B) Live sea urchin embryo at 2 dpf stained with the membrane tracker FM4-64 (gray) and green-calcein that binds to calcium ions (false-colored blue). Enlargement shows the calcium vesicles in the skeletogenic cells (arrow). (Scale bar, 50 µm.) (Enlargement magnification, 400×.) (C) Confocal image of the spicule in live embryo at 3 dpf stained with blue calcein (blue) and FM4-64 (gray). (Scale bar, 10 µm.) (D) Three-dimensional model of the spicule structure based on 50 confocal z-stacks of the cube in C. (Scale bar, 3 µm.) (E) Scanning electron micrograph of a cross-section of the spicule at 2 dpf showing the double membrane cytoplasmic cord that surrounds the spicule and the vesicles inside the cord. C, cytoplasm; E, ectodermal cell; S, spicule; V, vesicles, arrows point to the membranes. (Scale bar, 1 µm.) (F) Schematic model of the spicule and vesicle secretion, showing the calcium vesicles and spicule in blue and cytoplasm in gray. Image courtesy of Yarden Ben-Tabou de-Leon (artist). (G) Partial phylogenetic tree presenting VEGFR expression in cells that generate tubular structures in different phyla throughout the animal kingdom.
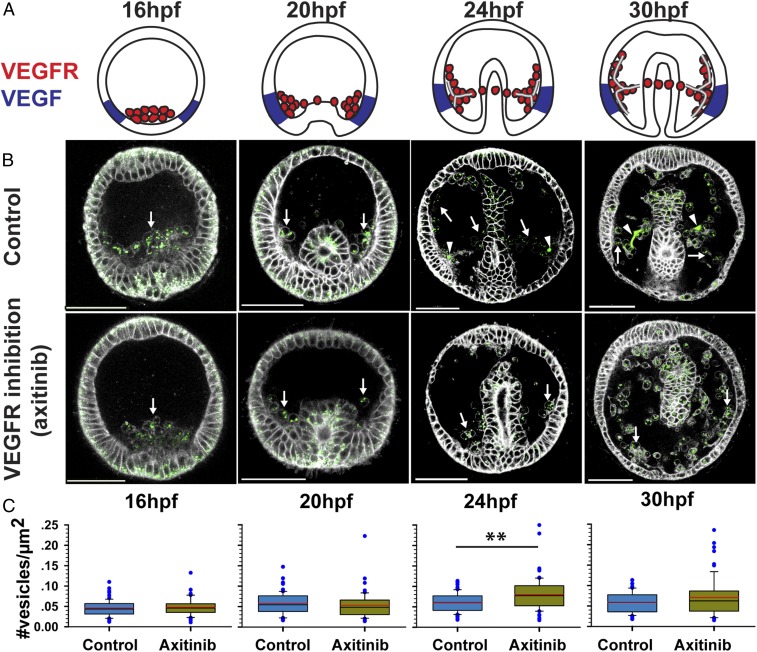
Fig. 3.
Possible role of VEGF signaling in calcium vesicle secretion. (A) Schematic diagrams showing Pl-VEGFR expression in the skeletogenic cells (red) and Pl-VEGF ectodermal expression (blue) at different developmental times (similar times in A–C). Image courtesy of Yarden Ben-Tabou de-Leon (artist). (B) confocal images of calcein staining (green) and FM4-64 membrane marker (white) show the presence of calcium vesicles in the SM cells (white arrows) in normal and VEGFR inhibited embryos (axitinib). Arrowheads indicate the spicules in control embryos. (Scale bars, 50 µm.) (C) Vesicle number per square micrometer in the SM cells in control and VEGFR inhibition. Each box plot shows the median (black line), average (red line) of the first and the third quartiles (edges of boxes) and outliers (n = 3, exact number of cells in each condition is provided in SI Appendix, Table S1). **P = 0.001.
The activation of VEGF signaling early in embryogenesis is probably a key to understanding the evolution of the larval skeleton in echinoderms. While all echinoderm classes generate calcite endoskeletons in their adult form (17), some echinoderm classes lack the larval skeleton (sea stars) or have a significantly reduced skeletal structure [sea cucumbers (17)]. Nevertheless, the embryonic mesodermal GRN is highly similar in all echinoderm classes, regardless of the presence or absence of a larval skeleton (18). Indeed, VEGFR expression is one of the only differences in the mesoderm regulatory state between echinoderm embryos that produce larval skeletons [brittle stars and sea urchins (14, 19)] and the sea star embryo, which does not (14, 15, 18–21). Furthermore, VEGFR expression is observed in the adult skeletogenesis centers and VEGF is expressed in the adult supporting cells in all studied echinoderm classes [sea urchin (20) brittle stars and sea stars (19)]. Together, these observations suggest that VEGF signaling is a prominent part of the echinoderm biomineralization program and its embryonic activation might be associated with the gain of echinoderm larval skeletons (Fig. 1G).
While in echinoderms the VEGF pathway controls spiculogenesis, in many other phyla it controls the formation of other tubular structures, and particularly, VEGF signaling regulates blood vessel formation in several bilaterian phyla (Fig. 1G). In vertebrates, VEGF guides the migration of hemangioblasts, the progenitors of endothelial and hematopoietic cells, and drives the formation of blood vessels during embryogenesis (vascularization), in adult ischemic tissues, and in cancer (angiogenesis) (22, 23). In ascidians, VEGF signaling promotes the regeneration of blood vessels, which suggests that its role in vascularization is conserved inside chordates (24). Within the protostomes, VEGFR is essential for blood vessel formation in two lophotrocozoan species, the squid and the leech (25, 26). In cnidarians VEGFR is expressed in two tubular organs: the gastrovascular canal and the tentacles (27, 28). The participation of VEGF signaling in generating tubular organs in different phyla raises the possibility that biomineralization in echinoderms is evolutionarily related to these other tubulogenesis programs. Here we studied the mechanisms that VEGF signaling activates during sea urchin spiculogenesis, including calcium vesicle accumulation and secretion, transcriptional targets and their function, and VEGF regulation of the upstream skeletogenic transcription factors. Our findings reveal intriguing similarities to the control systems that regulate vertebrate vascularization that could support a common origin of these two distinct tubulogenesis programs.
Results
VEGF–VEGFR Recognition Is Conserved Between Human and Sea Urchin.
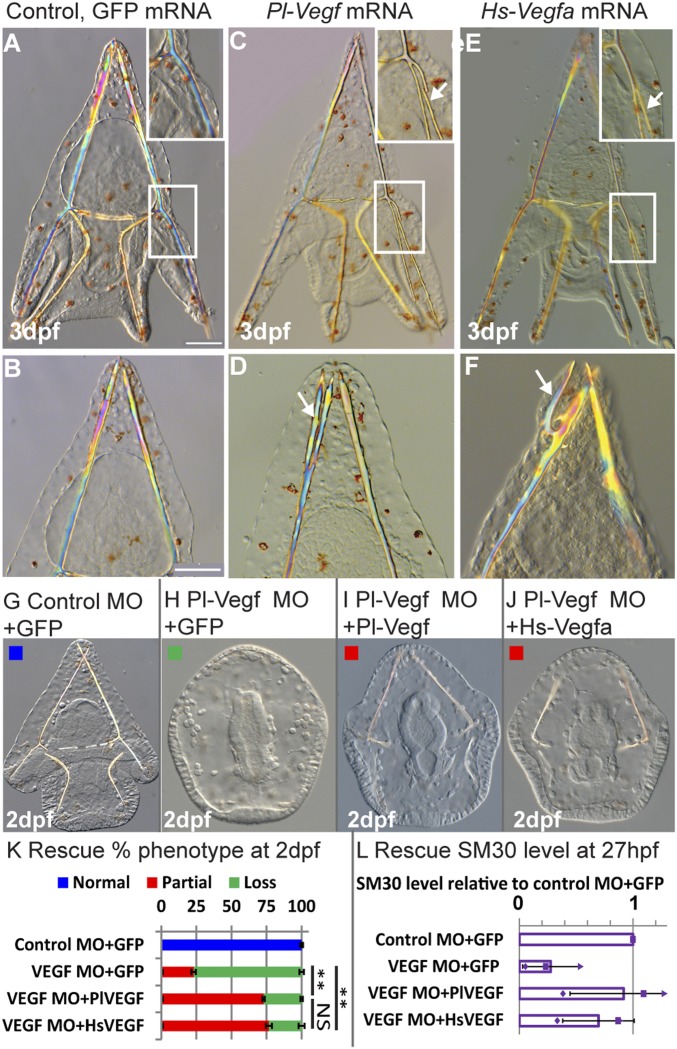
Six hundred million years of divergent evolution between vertebrates and echinoderms have generated significant differences in VEGF (SI Appendix, Fig. S1A) and VEGFR sequences; distinctly, the sea urchin VEGFR has 10 Ig domains while vertebrate VEGF receptors have 7 (14). Our models of the structure of sea urchin VEGF and VEGF–VEGFR complexes show similarities to the structure of the human proteins (SI Appendix, Fig. S1B). However, the question remains: Are these proteins functionally similar? To test this we experimentally studied the recognition between human VEGF and sea urchin VEGFR by overexpressing human VEGF in sea urchin embryos. We injected the mRNA of one of the most abundant forms of human VEGF, Hs-VEGFa(165) (29) into the eggs of the Mediterranean sea urchin, Paracentrotus lividus (Fig. 2 A–F). Hs-VEGFa(165) overexpression results in the formation of ectopic spicule branching 3 days postfertilization (dpf) (Fig. 2 E and F), similar to the phenotype of sea urchin Pl-VEGF overexpression (Fig. 2 C and D). To verify that the observed phenotype is due to human VEGF specific-activation of the sea urchin VEGF pathway, we conducted a rescue experiment using a sea urchin VEGF splicing morpholino antisense oligonucleotide (MO), tested before in this species (14). Embryos injected with control MO and GFP mRNA show normal skeletal rods at 2 dpf, while embryos injected with Pl-VEGF MO and GFP mRNA show severe skeletal loss and a reduction in the level of the VEGF-target, SM30 (Fig. 2 G, H, K, and L). VEGF control of SM30 was studied in refs. 14 and 15. Coinjection of Pl-VEGF MO with either human or sea urchin mRNA partially rescues the knockdown skeletogenic phenotype and SM30 expression level in a similar way (Fig. 2 I–L). Thus, human VEGF is able to rescue sea urchin VEGF knockdown with the same efficiency as sea urchin VEGF, indicating that VEGF–VEGFR recognition is conserved despite the large evolutionary distance between these two organisms.
Fig. 2.
VEGF–VEGFR recognition is conserved between human and sea urchin. (A–F) Ectopic spicule branching on the postoral rods and body rods were observed at 3 dpf in Pl-VEGF mRNA-injected embryos (arrows in C and D; biological replicates: n = 3, 52/114 embryos, 46%) and in Hs-VEGFa(165) mRNA-injected embryos (arrows in E and F; n = 4, 39/85, 46%) but not in control GFP-injected embryos (A and B; n = 4, 0/114, 0%). (Scale bars, 50 µm.) (Enlargement magnification, 200×.) (G–L) Rescue experiment at 2 dpf. (G–J) Embryos injected with 800 µM Control MO and 650 ng/µL GFP mRNA (G, normal skeleton), Pl-VEGF MO and GFP mRNA (H, skeletal loss), Pl-VEGF MO and Pl-VEGF mRNA (J, partial skeletal gain), Pl-VEGF MO and Hs-VEGF mRNA (J, partial skeletal gain). (Magnification, 200×.) (K) Quantification of rescue phenotypes, color code is indicated in the representative images, G–J (n = 3, **P < 0.001; NS, nonsignificant, Fisher test; number of embryos scored is provided in SI Appendix, Table S1). (L) SM30 mRNA levels in different treatments compared with control MO+GFP mRNA, showing average ratio (bars) and individual measurements, (QPCR). Error bars indicate SD.
Possible Role of VEGF Signaling in Calcium Vesicle Secretion.
In both tubulogenesis and biomineralization, vesicle formation and secretion play an important role. During vascular tubulogenesis, vesicles are formed in endothelial cells through pinocytosis, an apical surface is established between two adjacent cells, and vesicles are secreted into the intercellular domain to form the lumen [cord hollowing (30, 31)]. When VEGF signaling is inhibited, the apical surface still forms but the lumen is not generated (32). Relatedly, during sea urchin biomineralization, calcium is accumulated through endocytosis (11, 33) and concentrated as amorphous calcium-carbonate in intracellular vesicles that are then secreted into the spicule cord where crystallization occurs (Fig. 1F) (7, 9, 13, 33); however, the role of VEGF signaling in these processes has not been previously investigated.
To study the role of VEGF in calcium vesicle accumulation and secretion, we compared these processes between normal embryos and embryos treated with the VEGFR inhibitor, axitinib, at different developmental stages (Fig. 3). Axitinib binds specifically to the kinase domain of human VEGF receptor (34) that is highly conserved between humans and sea urchins (SI Appendix, Fig. S2 A and B). Axitinib treatment results in analogous phenotypes to those observed in VEGF and VEGFR knockdown in P. lividus, similar to its effect in other sea urchin species (SI Appendix, Fig. S2C) (15). We used calcein staining to mark calcium carrying vesicles (7) and FM4-64 to mark cell membranes in live sea urchin embryos (Fig. 3B). Calcein binds to calcium in all of the calcium phases and therefore marks calcium ions, calcium-carbonate, amorphous and crystallite calcium-carbonate (7, 11, 12, 33).
Calcium-carrying vesicles are visible in all cells of the embryo, in agreement with previous studies (7, 11). VEGF inhibition does not prevent calcium vesicle accumulation in the SM cells throughout skeletogenesis (arrows in Fig. 3B), yet the calcite spicules form only in normal embryos (arrowheads in Fig. 3B). The number of calcium vesicles per area within the SM cells does not change with VEGFR inhibition before spicule formation (Fig. 3C) [16 h postfertilization (hpf) and 20 hfp, P > 0.05, unpaired two-tailed t test]. However, just after spicule initiation, at 24 hpf, there is a significant increase in vesicle number in the SM cells in VEGFR-inhibited embryos compared with control embryos, implying that vesicle secretion might be repressed by VEGFR inhibition (Fig. 3C) (P = 0.001, unpaired two-tailed t test). A similar trend is observed at 30 hpf but is not statistically significant (P > 0.05, unpaired two tailed t test). Overall, these observations imply that calcium-carrying vesicles are present in the skeletogenic cells in VEGFR inhibition, yet the spicules do not form, possibly because vesicle secretion is inhibited. Still, the cellular and molecular processes involved in vesicle secretion downstream of VEGF signaling require further investigation.
VEGF Controls the Expression of Biomineralization and Vascularization Genes.
The formation of a tubular structure and vesicle secretion in various systems require the activation of an extensive molecular tool kit that regulates cytoskeletal remodeling and other cellular mechanisms (31, 35). To identify the molecular mechanisms that sea urchin VEGF activates during spiculogenesis, we explored the change in gene expression in response to VEGFR inhibition using RNA sequencing (RNA-seq) before, during, and after the spicules are formed (16, 20, 24, and 30 hpf) (see SI Appendix, Fig. S3 A and B for experimental design). Interestingly, a major transcriptional response to VEGFR inhibition is detected only after the first stage of SM cell migration and lateral cluster formation, at the onset of spicule formation (24 hpf) (SI Appendix, Fig. S3C). Relatedly, VEGF guidance of endothelial cell migration during angiogenesis is mediated through direct regulation of cytoskeleton remodeling proteins, such as the small GTPase RhoA and RhoA Kinase (ROCK) (36–40). Possibly, similar posttranscriptional mechanisms are activated by sea urchin VEGF signaling to guide the initial SM cell migration.
In an additional RNA-seq experiment, we detected a recovery of the expression of the majority of VEGF-responsive transcripts at 24 and 30 hpf, 4 and 6 h after axitinib was washed, respectively (SI Appendix, Fig. S3 B and D). The observed recovery in gene expression is in agreement with the full recovery of the skeleton at the pluteus stage after axitinib wash (15). We combined the results of the time course and wash experiments at 24 and 30 hpf to achieve high confidence in the prediction of VEGF targets [SI Appendix, Fig. S3E and Dataset S1 (differentially expressed genes only) and Dataset S3 (quantitative RNA-seq data for all transcripts)]. VEGFR inhibition significantly affects the expression of hundreds of genes at 24 and 30 hpf, enriched with gene ontology (GO) terms related to growth factor signaling, biomineralization, cell fate specification, and interestingly, vasculogenesis and circulatory system development (SI Appendix, Fig. S4 and Dataset S3). We studied the spatiotemporal expression and verified the response to VEGFR inhibition of a few key VEGF target genes that participate in these biological processes.
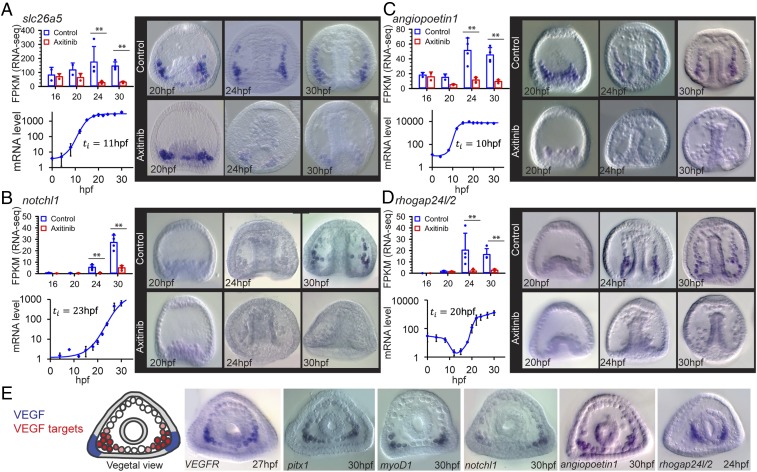
The formation of the sea urchin calcite spicules requires uptake and homeostasis of carbonate ions (41), as well as the production of spicule matrix proteins that control calcium-carbonate nucleation (42). Accordingly, within VEGF targets we observed enrichment of genes with GO terms related to carbonate homeostasis and calcium binding, such as members of solute carrier HCO3− transporter families (e.g., Pl-slc26a5) (Fig. 4A), the enzyme carbonic anhydrase like 7 (Pl-caral7) (SI Appendix, Fig. S5A), and different spicule matrix proteins (e.g., Pl-SM30E) (SI Appendix, Fig. S5B), in agreement with refs. 14 and 15. These biomineralization genes are initially expressed broadly in the SM cells independently of VEGF signaling (20 hpf) (Fig. 4A and SI Appendix, Figs. S5 A and B, and S6 A–C). The expression of these genes then localizes to the SM cell clusters and becomes critically dependent on the VEGF pathway from 24 hpf onward (Fig. 4A and SI Appendix, Figs. S5 A and B, and S6 A–C). This implies that the biomineralization proteins encoded by these genes could still be present in the skeletogenic cells in VEGFR inhibition at 24 hpf, which precludes them from explaining the complete skeletal loss in this condition. Possibly, these biomineralization proteins are necessary but not sufficient for spicule formation, or they require additional posttranscriptional activation that depends on VEGF signaling.
Fig. 4.
VEGF-pathway regulates the expression of biomineralization, regulatory and vascularization genes. (A–D) In each panel we present: gene-expression level for control and VEGFR inhibited embryos showing average expression level and individual measurements [measured by RNA-seq in fragments per kilobase of transcript per million mapped reads (FPKM); **P < 0.05 after FDR correction; 16 hpf and 20 hpf, n = 2; 24 hpf and 30 hpf, n = 4)]; temporal expression profile and initiation time (quantitative PCR, n = 3, error bars correspond to SD); spatial gene expression in control and in VEGFR inhibited embryos (whole-mount in situ hybridization, n = 3, lateral view). (E, Left) Illustration of Pl-VEGF expression (blue) and VEGF target gene expression (red). (Right) Examples of VEGF target gene expression in a vegetal view. Image courtesy of Yarden Ben-Tabou de-Leon (artist). (Magnification, 200×.)
We identified several VEGF targets that have vertebrate homologs essential for vascularization or angiogenesis: for example, notchl1 (22); angiopoietin1 (43) (Sp-fred); the cytoskeleton remodeling genes, thsd7a (44) (Sp-thsd7b), rhogap24l/2 (45, 46); as well as the previously reported VEGFR itself (14, 15). The Notch pathway plays a prominent role in angiogenetic sprouting (22); angiopoietins are predominantly expressed at vascular supporting cells and control blood vessel number and diameter (43); human Thrombospondin type I, Thsh7A (Sp-Thsh7B) inhibits endothelial cell migration and tube formation (44). The sea urchin homologs of these genes are expressed in the skeletogenic cells and depend on VEGF activity from 24 hpf onward (Fig. 4 B–D and SI Appendix, Figs. S5 C and D, and S6 D–H). Overall, VEGF signaling becomes essential to the localized expression of both biomineralization and vascularization genes in the lateral SM clusters, which is the site of spicule formation, at the onset of spiculogenesis.
Pl-Rhogap24l/2 Is Required for Normal Spiculogenesis.
We wanted to study the role of the VEGF-target, Pl-rhogap24l/2, a cytoskeleton remodeling gene (Fig. 4D), becausee cytoskeleton remodeling is critical for tubulogenesis (31), vascularization (36–40), and vesicle secretion (35) in other systems. Pl-rhogap24l/2 is homologous to the family of vertebrate genes encoding the Rho-GAPs (GTPase-activating proteins), arhgap24, arhgap22, and arhgap25 (45–48) (see Pl-rhogap24l/2 phylogenetic tree in SI Appendix, Fig. S7). Isoforms of arhgap24 and arhgap22 are among the highest expressed Rho-GAPs in endothelial cells (45, 46) and arhgap25 is expressed primarily in hematopoietic cells (49). Mammalian Arhgap22, -24, and -25 are activated by RhoA, and subsequently inactivate RAC1 (47). Hence, they contribute to the counteracting interactions between RhoA and RAC1, essential for correct cytoskeleton rearrangement (46). Particularly, knockdown of Hs-arhgap24 in human umbilical vein endothelial cells (HUVEC) culture inhibits blood vessel formation (45).
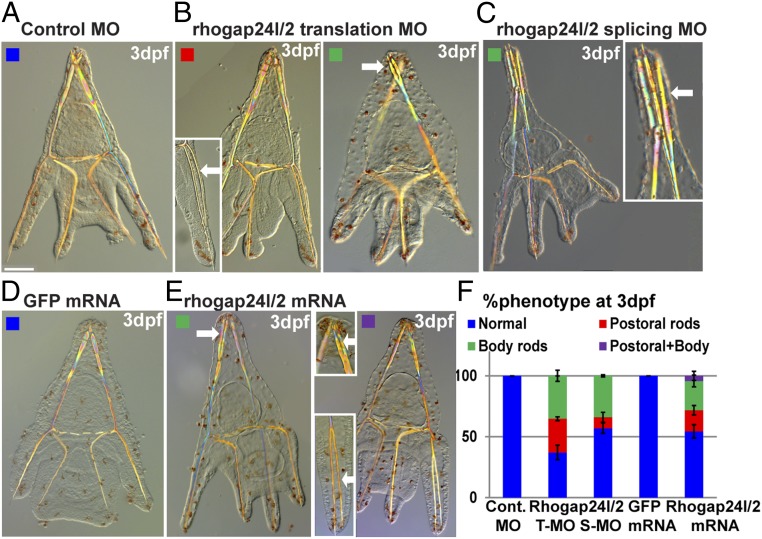
To explore the role of Pl-rhogap24l/2 in sea urchin skeletogenesis, we studied the phenotype of Pl-rhogap24l/2 knockdown by the injection of translation and splicing MOs (Fig. 5 A–C and SI Appendix, Fig. S8) and overexpression by the injection of mRNA of Pl-rhogap24l/2 (Fig. 5 D and E). These opposing perturbations result in an ectopic spicule branching of both the body and postoral rods at 3 dpf (Fig. 5F). Possibly, the sea urchin Pl-Rhogap24l/2 contributes to the oscillations between RhoA and RAC1 activities (46, 48), and therefore either up- or down-regulating it interferes with the balance between these small GTPases and impairs spicule branching. Thus, the VEGF target, Pl-rhogap24l/2, a sea urchin homolog of endothelial Rho-GAP genes, is activated by VEGF signaling at the SM cells and its function is necessary for normal skeletogenesis.
Fig. 5.
VEGF target, Pl-Rhogap24l/2, is essential for normal skeletogenesis. (A–E) Pl-rhogap24l/2 perturbations. (A) Embryo injected with control MO shows normal skeleton at 3dpf. Pl-rhogap24l/2 knockdown using either translation MO (B) or splicing MO (C) results in ectopic branching at the postoral and body rods at 3 dpf. (D) GFP overexpression results in normal skeleton while (E) overexpression of Pl-rhogap24l/2 results in ectopic branching at the postoral and body rods, sometimes within the same embryo. (Scale bar, 50 μm.) (Enlargement magnification, 200×.) (F) Quantification of Pl-rhogap24l/2 perturbations, color code is indicated in the representative images (A–E) (n = 3–5, exact number of embryos scored is provided in SI Appendix, Table S1).
Spiculogenesis GRN.
Our results show that VEGF signaling activates its targets specifically at the lateral SM clusters located most proximally to the ectodermal VEGF-secreting cells (Fig. 4E). Therefore, VEGF signaling could be controlling the initiation of spiculogenesis at the SM cell clusters via its regulation of differential gene expression in this subset of skeletogenic cells. However, VEGF is a signaling molecule and the transcriptional activation of its targets has to be mediated through a skeletogenic transcription factor. Therefore, we wanted to investigate which transcription factors are expressed in the skeletogenic cell clusters at the time of spiculogenesis and whether their expression is VEGF-dependent.
The skeletogenic GRN was studied in great detail mostly at the early stages of sea urchin development (6). Some of the early skeletogenic transcription factors turn off within the skeletogenic cells hours before the spicules form (50–53) or were shown to be unaffected by VEGF perturbation (Tbr) (14). We therefore focused on the remaining six skeletogenic transcription factors: Ets1/2, Erg, Hex, Tel, FoxO, and Alx1 (6); each of them was shown to have a role in sea urchin skeletogenesis (6, 54). Intriguingly, vertebrate homologs of five of these transcription factors are expressed in endothelial cells and regulate different aspects of endothelial cell function [Ets1, Ets2, Erg (Erg and Fli), Hex, Tel, and FOXO1 and FOX3a] (55–61).
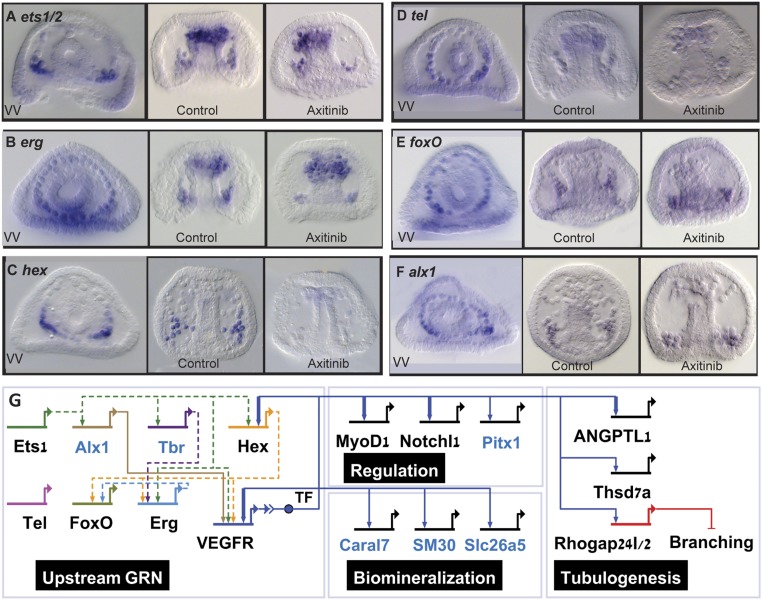
The sea urchin genes, ets1/2, erg, hex, tel, foxo, and alx1 are expressed in the skeletogenic cells at the time of spicule formation and the expression of hex depends on VEGF signaling (23–25 hpf; Fig. 6 A–F and SI Appendix, Fig. S6 K–P). Unlike the localized expression of VEGF targets at the lateral skeletogenic clusters, the expression of some of these transcription factors extends to the oral (erg, tel, and foxo) and aboral (erg, foxo, and alx1) parts of the skeletogenic ring of cells (see vegetal views in Fig. 6 A–F). Of these, the most likely mediators of VEGF signaling are the ETS factors—Ets1/2, Erg, and Tel—of which, ets1/2 is specifically localized at the SM lateral clusters at 24 hpf (Fig. 6A). The expression of the genes ets1/2, erg, hex, and tel is also detected in non-SM cells at the tip of the archenteron that differentiate into hematopoietic and myogenic cells (Fig. 6 A–D). Thus, the combination of transcription factors expressed at the SM cell clusters at the time of spicule formation includes the VEGF-independent expression of the transcription factors Ets1/2, Erg, Tel, FoxO, Alx1 (Fig. 6), and Tbr1 (14), as well as VEGF targets, Hex, Pitx1, and MyoD1 (Fig. 6C and SI Appendix, Figs. S5 E and F and S6 I, J, and M). We summarize our results in a partial model of the spiculogenesis GRN portraying the similarities and differences to vertebrates’ vascularization GRN (Fig. 6G).
Fig. 6.
The regulatory state and VEGF downstream GRN in the skeletogenic cell clusters during the initiation of spiculogenesis. (A–F) Spatial expression and response to VEGFR inhibition at the initiation of spiculogenesis (23–25 hpf) of the genes encoding the transcription factors, Ets1/2 (A), Erg (B), Hex (C), Tel (D), FoxO (E), and Alx1 (F), n ≥ 3. VV, vegetal views in control embryos. (Magnification, 200×.) (G) Model of the regulatory network at the skeletogenic clusters at the time of spiculogenesis (24 hpf). The regulatory interactions between the upstream transcription factors were studied only at earlier stages (6) and are therefore indicated by dashed lines. TF refers to the transcription factor that mediates VEGFR signaling. Blue gene names indicate skeletogenic specific genes and black names indicate genes common to both spiculogenesis and vascularization (56–60, 88–90). Arrows indicate activation and bars represent repression. Thick links indicate regulatory interactions observed in vertebrates’ hemangioblast differentiation (90) or in human endothelial cells (88, 89) (Dataset S4).
Discussion
To gain a mechanistic understanding of the biological control of biomineralization and its rapid, parallel evolution, we need to study and compare the GRNs that control it. The larval skeleton of the sea urchin is an important model that contributed significantly both to the understanding of GRN structure and function (6) and to the field of biomineralization (7–9). The sea urchin embryo builds its biominerals inside a tubular cavity, using VEGF signaling, a key driver of vascular systems and tubular organs in other animals (Fig. 1G). Here we unraveled some of the cellular and molecular mechanisms activated by VEGF signaling to drive sea urchin biomineralization. Our findings reveal multiple parallels to the mechanisms that drive vertebrates’ vascularization, raising the possibility that the sea urchin biomineralization evolved through a cooption of an ancestral vascular-tubulogenesis program.
Our studies portray the transcriptional gene network that sea urchin VEGF activates and the similarities between the spiculogenesis GRN and the GRN that controls vascularization (Fig. 6G). VEGF signaling regulates the expression of hundreds of genes at the onset of spicule formation, including regulatory, biomineralization, and vascularization related genes (SI Appendix, Figs. S3 C–E and S4). The 10 transcriptional targets we focused on here require VEGF signaling for their localized expression at the SM cell clusters, the subset of skeletogenic cells that generate the spicules (Fig. 4 and SI Appendix, Fig. S5). To identify the possible mediators of the transcriptional response to VEGF signaling, we studied the skeletogenic regulatory state: the combination of regulatory genes coexpressed within this cell lineage (62, 63). The regulatory state is considered to be the hallmark of GRN and cell-type homology that lingers throughout evolution (62, 64). Remarkably, a common set of five transcription factors (Ets1/2, Erg, Hex, Tel, and FoxO) and three signaling pathways (VEGFR, Notch, and Angiopoetin) essential for vascularization (22, 43, 56–60) are expressed at the sea urchin SM clusters at the time of spicule formation (Fig. 6G, black gene names). Indeed, these transcription factors and signaling pathways are also involved in other developmental processes across metazoans, but their coexpression in a cell lineage is unique to vertebrates’ endothelial cells and vascularization (56–60). Furthermore, within VEGF targets that we studied here, three genes that participate in vertebrate vascularization are linked to the process of tubulogenesis (Angiopoetin, Thsd4a, and Rhogap24l/2), which is the common morphogenetic event between these different cell types. Overall, the similarity between the spiculogenesis GRN and the GRN that drives endothelial cell specification and vascularization suggests a common evolutionary origin of these two networks.
Ours and previous findings point toward vesicle secretion and cytoskeleton remodeling as drivers of internal cavity formation during sea urchin spiculogenesis and vertebrates’ vascularization, possibly downstream of VEGF signaling. Lumen formation in vascularization depends on vesicle secretion (30, 31) and is regulated by VEGF-activated cytoskeleton remodeling proteins (31, 38–40). Here we show that VEGFR inhibition leads to an increase in calcium vesicle number in the skeletogenic cells at the time of spicule formation in normal embryos, which implies that vesicle secretion could require VEGF signaling (Fig. 3). We also demonstrate that the VEGF transcriptional target, rhogap24l/2, a cytoskeleton remodeling gene, is necessary for normal skeletogenesis (Fig. 5). As stated above, mammalian homologs of this gene are the most enriched Rho-GAPs in endothelial cells and, particularly, Arhgap24 is essential for blood vessel formation in endothelial cell culture (45, 46). Furthermore, the cytoskeleton remodeling proteins, ROCK1 and CDC42, known mediators of VEGF signaling that are essential for vascular tubulogenesis (38–40, 65–67), are critical for spicule formation in the sea urchin embryo (68, 69). Thus, common cytoskeleton remodeling proteins are essential for both spicule formation in the sea urchin embryo and for vascularization in vertebrates. However, to fully understand the molecular control of calcium vesicle secretion and spicule cavity formation, it is critical to further study VEGF control of cytoskeleton remodeling proteins and their exact functions.
Many of the skeletogenic regulatory genes of the sea urchin embryo are expressed in the adult skeletogenic cells and in the embryonic mesoderm of all studied echinoderms (18–20, 70, 71), which makes the evolution of this program within echinoderms quite intriguing. As stated above, all echinoderm classes generate calcite endoskeletons in their adult form but a full larval skeleton is generated only in the sea urchin and brittle star embryos (Fig. 1G) (17, 18). Like the larval spicules, the adult skeletal elements grow in a syncytial cavity generated by a set of skeletogenic cells (72). Many of the skeletogenic regulatory genes, including VEGFR, Alx1, Ets1, Hex, and Erg (but not Tbr, Tel, and FoxO) are expressed in the adult skeletogenic cells of both the sea urchin and the sea star (20). Therefore, the sea urchin larval skeleton was proposed to be an evolutionary gain through the precocious activation of the adult echinoderm skeletogenic program that is believed to be the echinoderm ancestral skeletogenic GRN (20). In the light of our findings, it is tempting to propose that the adult skeletogenic program evolved from the ancient VEGF-driven tubulogenesis program. Second, this program was activated independently in the sea urchins and in the brittle stars embryos for making their larval skeletons (18). The high morphological similarity between the brittle star and the sea urchin larval skeletons could suggest another evolutionary scenario. Possibly, the adult skeletogenic program was precociously activated in the common ancestor of the four echinoderm classes: sea star, brittle stars, sea urchin, and sea cucumbers. Then the larval skeleton was lost in the sea star, at least partially through the loss of VEGFR embryonic expression, and reduced in the sea cucumber. Either way, most likely, the plesiomorphic skeletogenic GRN of the adult echinoderm evolved from the ancient VEGF-driven tubulogenesis program.
The distinct differences in the function of blood vessels and calcite spicules raise the question about the role of the ancient tubulogenesis program: Was it driving transport and circulation or solid biomineral formation? VEGF signaling participates in various developmental processes in different organisms and some are related to biomineralization: for example, blood cell (hemocyte) development in protostomes (73–75) and osteoblast differentiation in vertebrates (76, 77). However, in all of the studied systems where VEGF signaling is regulating tubulogenesis outside the echinoderm phylum, the tubular structure is used for circulation and transport (Fig. 1G) (24–28). Indeed, blood vessels are essential for bone development in vertebrates (78, 79) and they carry calcium vesicles to the site of bone formation (80, 81), but this is within their role in transporting critical elements to the site where they are needed. Moreover, the vertebrates’ biomineralization program is quite distinct from the biomineralization program of echinoderm, including major differences in upstream GRNs, various phylum specific biomineralization proteins, and the use of calcium-phosphate and not calcium-carbonate to form the biomineral (4, 5). Additionally, in between the vertebrates and echinoderm in the deuterostome branch there are other classes that do not produce biominerals but have VEGF-dependent blood vessels, like the ascidians (24). These differences imply that the biomineralization programs evolved independently in vertebrates and echinoderms, after they diverged from their common ancestor (2, 3). Thus, we cannot completely rule out the alternative, but most likely the ancestral VEGF-driven tubulogenesis program was used to distribute nutrients and blood cells and was coopted for biomineralization uniquely in echinoderms.
We speculate that the ancestral tubulogenesis program generated simple blood vessels, like the capillary and unlike the complex multilayered structure of vertebrates’ arteries and veins (82). Indeed, the diameter of the spicule cord is about 4 µm (Fig. 1 C–E), which is of the order of the average capillary size (82). However, while the SM cells generate the spicule cord but keep their round mesenchymal shape (Fig. 1G), the vertebrates’ endothelial cells form an epithelial layer that constitutes the blood vessel (30, 31). Thus, there are differences not only in the filling of the tubes but also in the shape of the skeletogenic and endothelial cells and their function that must have evolved through major changes in the ancestral GRN.
Relatedly, there are apparent differences between the spiculogenesis and the endothelial GRNs that our model underrepresents (Fig. 6G). Multiple gene duplications have occurred in vertebrates leading to several paralogs of every sea urchin gene: for example, Ets1/2 duplication into Ets1 and Ets2, Erg duplication into Erg and Fli, and the multiple vertebrates’ paralogs of FoxO, VEGF, and VEGFR (55–61). However, the conserved VEGF–VEGFR recognition between human and sea urchin implies that these genes originated from the same protein families and are functionally related. These gene duplications might have supported the evolution of the complex vascularization system of vertebrates with its specialized arteries and veins. Distinctive to the sea urchin skeletogenic cells are the activation of the transcription factors Tbr, Alx1, and Pitx1 and the expression of the echinoderm-specific spiculo-matrix proteins and other biomineralization related genes (42) (Fig. 6G, blue genes). These and probably other differences between the sea urchin skeletogenic GRN and vertebrates’ vascularization GRN had apparently contributed to the divergent outcome of these two morphogenetic programs.
In the GRN diagram we divided VEGF targets into three different functional modules: regulation, biomineralization, and tubulogenesis (Fig. 6G). While in the skeletogenic cells these modules are probably tightly linked, it could be that they represent the actual building blocks from which the sea urchin skeletogenic GRN evolved. Computational simulations of the evolution of biological networks have shown that changing goals speed up evolution and leads to a more modular network structure, where different blocks are responsible for different tasks (83, 84). This could be an example of the way that phylum specific biomineralization programs rapidly evolved through the insertion of novel biomineralization modules into ancestral developmental GRNs.
Materials and Methods
Embryos were cultured in artificial sea water (Red Sea Fish Farm LTD) at 18 °C. The exact number of biological replicates and of embryos scored for each condition in the experiments described in this work is available in SI Appendix, Table S1.
Imaging.
Embryos were fixed and prepared for scanning electron microscopy, as described in SI Appendix. Light microscopy of live embryos was done using either a Zeiss Axioimager M2 or Nikon A1R confocal microscope. A 3D model of the spicule was generated using 52 confocal stacks by the software Imaris 7.6.5. Calcein staining was done using green calcein (C0875, Sigma) or blue calcein (M1255, Sigma). We used FM4-64 (T13320, Life Technologies) to stain membranes in live embryos. Further information is provided in SI Appendix.
VEGF and VEGFR protein models were constructed based on known structures of human proteins from the Protein Data Bank, as described in SI Appendix. Protein sequence alignments were done using ClustalOmega.
mRNA Injection.
cDNA of 30 hpf P. lividus embryos were used as a template for the cloning of Pl-VEGF and Pl-rhogap24l/2 cDNAs (primer list is provided in Dataset S5). Hs-VEGFa(165) is a gift from Gera Neufeld and Ofra Kessler, Cancer Research and Vascular Biology Center, The Bruce Rappaport Faculty of Medicine, Technion, Israel Institute of Technology, Haifa, Israel. mRNAs were generated and microinjected into sea urchin eggs. For the VEGF rescue experiment, mRNA was microinjected into sea urchin eggs along with random or VEGF splicing MO (14). Statistical analyses were done using IBM SPSS statistics v21. Injection solutions and further details are provided in SI Appendix.
Calcium Vesicle Quantification.
VEGFR inhibition was done using by axitinib (AG013736, Selleckchem), in a final concentration of 150 nM. Experiments were conducted in three biological replicates for each time point. Cell area was measured in Fiji and vesicles per cell area were counted manually by three different people. Statistical analyses were done using IBM SPSS statistics 21, as described in SI Appendix.
RNA-Seq Experiments.
Total RNA isolation from control and VEGFR inhibited embryos was carried out using the RNeasy Mini Kit (50) from Qiagen (#74104). Illumina libraries were constructed using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (E7420). Sequencing was carried out at the Center for Genomic Technologies, The Hebrew University of Jerusalem, on an Illumina NextSeq machine (Illumina) with a 100 paired-end (PE) run. Further information is provided in SI Appendix. Cleaned PE reads were assembled using Trinity (v2.0.2) PE de novo assembly (85). Trinity-genes were annotated using mouse Ensembl data and the sea urchin Strongylocentrotus purpuratus RNA-Seq assembly (http://www.echinobase.org/Echinobase). Trinity-genes were quantitated by EdgeR using R3.0.2. We defined significant effect of time and treatment contrasts, at P value < 0.05 threshold, after false discovery rate (FDR) correction. Functional enrichment analysis was conducted using GOseq using S. purpuratus annotation (http://www.echinobase.org/Echinobase). Further details of the analyses are provided in SI Appendix.
Accession Numbers.
Raw data read sequences are available at the European Nucleotide Archive (ENA) of the European Bioinformatics Institute (EBI) under accession no. PRJEB10269 (86). The assembled transcriptome sequences are also available at the EBI (Study PRJEB10269, accession range HACU01000001–HACU01667838) (86).
Rhogap24l/2 Phylogenetic Analysis.
The 100 top blast hits for Pl-Rhogap24l/2 were genes from the family of rhogap22, -24, and -25 of different species. We generated a phylogenetic tree of this family using different chordates, hemichordates, and echinoderm genes, as described in SI Appendix, Fig. S7A. This analysis indicates that the Echinoderm predicted RhoGap24/22/25 proteins form a monophyletic clade that separated from other deuterostomes before paralogue formation in vertebrates.
Quantitative PCR.
Quantitative PCR was performed following the procedures outlined in ref. 87, with some modifications described in details in SI Appendix. A complete list of primer sequences is provided in Dataset S5.
Whole-Mount in Situ Hybridization Procedure
RNA DIG probes were generated using ROCHE DIG labeling kit (catalog no. 1277073910) and SP6 polymerase 10810274001 Sigma. Whole-mount in situ hybridization was performed as described in ref. 53, with minor changes described in SI Appendix.
MO Injection.
Translation or splicing of Pl-rhogap24l/2 was blocked by the microinjection of 800 µM Pl-rhogap24l/2 MO into sea urchin eggs (Gene Tools). Translation MO: 5-ATCCTCAAGTATCCGTAGTGTGTGA-3; splicing MO at the 3′ of the second exon: 5-TGTCCTAGAACCGTTATACTCACGT-3 (SI Appendix, Fig. S8). Injections details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Veronica Hinman for helpful discussions; Muki Shpigel and David Ben-Ezra for their help with sea urchin handling; Palle von Huth for help with confocal imaging; Tali Mass and Yulia Polack for help with electron microscopy; Gera Neufeld and Ofra Kessler for the gift of Hs-VEGFa(165) plasmid; Nir Sapir for his advice regarding the statistical analysis; Chuck Ettensohn, Veronica Hinman, Stefan Materna, and Mark Winter for critical review of the manuscript; Majed Layous, Hadeya Zaher, and Nasreen Nakad for technical help with quantitative PCR and whole-mount in situ hybridization; and Yarden Ben-Tabou de-Leon for the illustrations in Figs. 1F, 3A, and 4E. This work was supported by the Israel Science Foundation Grant 41/14 (to Smadar Ben-Tabou de-Leon) and an Eshkol postdoctoral Fellowship of the Israeli Ministry of Science (to M.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw data read sequences were deposited at the European Nucleotide Archive of the European Bioinformatics Institute (accession no. PRJEB10269). The assembled transcriptome sequences are also deposited in the European Bioinformatics Institute (Study PRJEB10269, accession range HACU01000001–HACU01667838).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902126116/-/DCSupplemental.
References
- 1.Lowenstam H. A., Weiner S., On Biomineralization (Oxford University Press, New York, 1989). [Google Scholar]
- 2.Murdock D. J., Donoghue P. C., Evolutionary origins of animal skeletal biomineralization. Cells Tissues Organs 194, 98–102 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Knoll A. H., Biomineralization and evolutionary history. Rev. Mineral. Geochem. 54, 329–356 (2003). [Google Scholar]
- 4.Tucker A., Sharpe P., The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet. 5, 499–508 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Fisher S., Franz-Odendaal T., Evolution of the bone gene regulatory network. Curr. Opin. Genet. Dev. 22, 390–397 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Oliveri P., Tu Q., Davidson E. H., Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. U.S.A. 105, 5955–5962 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidavsky N., et al. , Initial stages of calcium uptake and mineral deposition in sea urchin embryos. Proc. Natl. Acad. Sci. U.S.A. 111, 39–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbins J. R., Tilney L. G., Porter K. R., Microtubules in the formation and development of the primary mesenchyme in Arbacia punctulata. I. The distribution of microtubules. J. Cell Biol. 41, 201–226 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beniash E., Addadi L., Weiner S., Cellular control over spicule formation in sea urchin embryos: A structural approach. J. Struct. Biol. 125, 50–62 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Gong Y. U., et al. , Phase transitions in biogenic amorphous calcium carbonate. Proc. Natl. Acad. Sci. U.S.A. 109, 6088–6093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidavsky N., et al. , Calcium transport into the cells of the sea urchin larva in relation to spicule formation. Proc. Natl. Acad. Sci. U.S.A. 113, 12637–12642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidavsky N., Masic A., Schertel A., Weiner S., Addadi L., Mineral-bearing vesicle transport in sea urchin embryos. J. Struct. Biol. 192, 358–365 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Ingersoll E. P., Wilt F. H., Matrix metalloproteinase inhibitors disrupt spicule formation by primary mesenchyme cells in the sea urchin embryo. Dev. Biol. 196, 95–106 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Duloquin L., Lhomond G., Gache C., Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development 134, 2293–2302 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Adomako-Ankomah A., Ettensohn C. A., Growth factor-mediated mesodermal cell guidance and skeletogenesis during sea urchin gastrulation. Development 140, 4214–4225 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Knapp R. T., Wu C. H., Mobilia K. C., Joester D., Recombinant sea urchin vascular endothelial growth factor directs single-crystal growth and branching in vitro. J. Am. Chem. Soc. 134, 17908–17911 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Raff R. A., Byrne M., The active evolutionary lives of echinoderm larvae. Heredity 97, 244–252 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Cary G. A., Hinman V. F., Echinoderm development and evolution in the post-genomic era. Dev. Biol. 427, 203–211 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Morino Y., et al. , Heterochronic activation of VEGF signaling and the evolution of the skeleton in echinoderm pluteus larvae. Evol. Dev. 14, 428–436 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Gao F., Davidson E. H., Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 6091–6096 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erkenbrack E. M., Petsios E., A conserved role for VEGF signaling in specification of homologous mesenchymal cell types positioned at spatially distinct developmental addresses in early development of sea urchins. J. Exp. Zoolog. B Mol. Dev. Evol. 328, 423–432 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Potente M., Gerhardt H., Carmeliet P., Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Ciau-Uitz A., Pinheiro P., Kirmizitas A., Zuo J., Patient R., VEGFA-dependent and -independent pathways synergise to drive Scl expression and initiate programming of the blood stem cell lineage in Xenopus. Development 140, 2632–2642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiozzo S., Voskoboynik A., Brown F. D., De Tomaso A. W., A conserved role of the VEGF pathway in angiogenesis of an ectodermally-derived vasculature. Dev. Biol. 315, 243–255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida M. A., Shigeno S., Tsuneki K., Furuya H., Squid vascular endothelial growth factor receptor: A shared molecular signature in the convergent evolution of closed circulatory systems. Evol. Dev. 12, 25–33 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Tettamanti G., Grimaldi A., Valvassori R., Rinaldi L., de Eguileor M., Vascular endothelial growth factor is involved in neoangiogenesis in Hirudo medicinalis (Annelida, Hirudinea). Cytokine 22, 168–179 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Krishnapati L. S., Ghaskadbi S., Identification and characterization of VEGF and FGF from Hydra. Int. J. Dev. Biol. 57, 897–906 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Seipel K., et al. , Homologs of vascular endothelial growth factor and receptor, VEGF and VEGFR, in the jellyfish Podocoryne carnea. Dev. Dyn. 231, 303–312 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Robinson C. J., Stringer S. E., The splice variants of vascular endothelial growth factor (VEGF) and their receptors. J. Cell Sci. 114, 853–865 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Xu K., Cleaver O., Tubulogenesis during blood vessel formation. Semin. Cell Dev. Biol. 22, 993–1004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iruela-Arispe M. L., Beitel G. J., Tubulogenesis. Development 140, 2851–2855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strilić B., et al. , The molecular basis of vascular lumen formation in the developing mouse aorta. Dev. Cell 17, 505–515 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Killian C. E., Wilt F. H., Endocytosis in primary mesenchyme cells during sea urchin larval skeletogenesis. Exp. Cell Res. 359, 205–214 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Hu-Lowe D. D., et al. , Nonclinical antiangiogenesis and antitumor activities of axitinib (AG-013736), an oral, potent, and selective inhibitor of vascular endothelial growth factor receptor tyrosine kinases 1, 2, 3. Clin. Cancer Res. 14, 7272–7283 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Segal D., Zaritsky A., Schejter E. D., Shilo B. Z., Feedback inhibition of actin on Rho mediates content release from large secretory vesicles. J. Cell Biol. 217, 1815–1826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Meel R., et al. , The VEGF/Rho GTPase signalling pathway: A promising target for anti-angiogenic/anti-invasion therapy. Drug Discov. Today 16, 219–228 (2011). [DOI] [PubMed] [Google Scholar]
- 37.De Smet F., Segura I., De Bock K., Hohensinner P. J., Carmeliet P., Mechanisms of vessel branching: Filopodia on endothelial tip cells lead the way. Arterioscler. Thromb. Vasc. Biol. 29, 639–649 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Bryan B. A., et al. , RhoA/ROCK signaling is essential for multiple aspects of VEGF-mediated angiogenesis. FASEB J. 24, 3186–3195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroll J., et al. , Inhibition of Rho-dependent kinases ROCK I/II activates VEGF-driven retinal neovascularization and sprouting angiogenesis. Am. J. Physiol. Heart Circ. Physiol. 296, H893–H899 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Sun H., Breslin J. W., Zhu J., Yuan S. Y., Wu M. H., Rho and ROCK signaling in VEGF-induced microvascular endothelial hyperpermeability. Microcirculation 13, 237–247 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Stumpp M., et al. , Acidified seawater impacts sea urchin larvae pH regulatory systems relevant for calcification. Proc. Natl. Acad. Sci. U.S.A. 109, 18192–18197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urry L. A., Hamilton P. C., Killian C. E., Wilt F. H., Expression of spicule matrix proteins in the sea urchin embryo during normal and experimentally altered spiculogenesis. Dev. Biol. 225, 201–213 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Fagiani E., Christofori G., Angiopoietins in angiogenesis. Cancer Lett. 328, 18–26 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Wang C. H., et al. , Thrombospondin type I domain containing 7A (THSD7A) mediates endothelial cell migration and tube formation. J. Cell. Physiol. 222, 685–694 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Su Z. J., et al. , A vascular cell-restricted RhoGAP, p73RhoGAP, is a key regulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 12212–12217 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Buul J. D., Geerts D., Huveneers S., Rho GAPs and GEFs: Controling switches in endothelial cell adhesion. Cell Adhes. Migr. 8, 108–124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura F., FilGAP and its close relatives: A mediator of Rho-Rac antagonism that regulates cell morphology and migration. Biochem. J. 453, 17–25 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Ohta Y., Hartwig J. H., Stossel T. P., FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat. Cell Biol. 8, 803–814 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Csépányi-Kömi R., Sirokmány G., Geiszt M., Ligeti E., ARHGAP25, a novel Rac GTPase-activating protein, regulates phagocytosis in human neutrophilic granulocytes. Blood 119, 573–582 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Rho H. K., McClay D. R., The control of foxN2/3 expression in sea urchin embryos and its function in the skeletogenic gene regulatory network. Development 138, 937–945 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amore G., et al. , Spdeadringer, a sea urchin embryo gene required separately in skeletogenic and oral ectoderm gene regulatory networks. Dev. Biol. 261, 55–81 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Howard-Ashby M., et al. , Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev. Biol. 300, 74–89 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Minokawa T., Rast J. P., Arenas-Mena C., Franco C. B., Davidson E. H., Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr. Patterns 4, 449–456 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Saunders L. R., McClay D. R., Sub-circuits of a gene regulatory network control a developmental epithelial-mesenchymal transition. Development 141, 1503–1513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu F., Patient R., Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ. Res. 103, 1147–1154 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Wei G., et al. , Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood 114, 1123–1130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu F., Walmsley M., Rodaway A., Patient R., Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr. Biol. 18, 1234–1240 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Minami T., et al. , Interaction between hex and GATA transcription factors in vascular endothelial cells inhibits flk-1/KDR-mediated vascular endothelial growth factor signaling. J. Biol. Chem. 279, 20626–20635 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Roukens M. G., et al. , Control of endothelial sprouting by a Tel-CtBP complex. Nat. Cell Biol. 12, 933–942 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Potente M., et al. , Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Invest. 115, 2382–2392 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sperone A., et al. , The transcription factor Erg inhibits vascular inflammation by repressing NF-kappaB activation and proinflammatory gene expression in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 31, 142–150 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Arendt D., et al. , The origin and evolution of cell types. Nat. Rev. Genet. 17, 744–757 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Ben-Tabou de-Leon S., Davidson E. H., Gene regulation: Gene control network in development. Annu. Rev. Biophys. Biomol. Struct. 36, 191–212 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Peter I. S., Davidson E. H., Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sacharidou A., et al. , Endothelial lumen signaling complexes control 3D matrix-specific tubulogenesis through interdependent Cdc42- and MT1-MMP-mediated events. Blood 115, 5259–5269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Montalvo J., et al. , ROCK1 & 2 perform overlapping and unique roles in angiogenesis and angiosarcoma tumor progression. Curr. Mol. Med. 13, 205–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Nieuw Amerongen G. P., van Hinsbergh V. W., Role of ROCK I/II in vascular branching. Am. J. Physiol. Heart Circ. Physiol. 296, H903–H905 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Sepúlveda-Ramírez S. P., Toledo-Jacobo L., Henson J. H., Shuster C. B., Cdc42 controls primary mesenchyme cell morphogenesis in the sea urchin embryo. Dev. Biol. 437, 140–151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Croce J., Duloquin L., Lhomond G., McClay D. R., Gache C., Frizzled5/8 is required in secondary mesenchyme cells to initiate archenteron invagination during sea urchin development. Development 133, 547–557 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Dylus D. V., et al. , Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. Evodevo 7, 2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Erkenbrack E. M., et al. , Ancestral state reconstruction by comparative analysis of a GRN kernel operating in echinoderms. Dev. Genes Evol. 226, 37–45 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Markel K., Roser U., Stauber M., On the ultrastructure and the supposed function of the mineralizing matrix coat of sea urchins (Echinodermata, Echinoida). Zoomorphology 109, 79–87 (1989). [Google Scholar]
- 73.Brückner K., et al. , The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell 7, 73–84 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Cho N. K., et al. , Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell 108, 865–876 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Ivanina A. V., et al. , The role of the vascular endothelial growth factor (VEGF) signaling in biomineralization of the oyster Crassostrea gigas. Front. Maribe Sci. 5, 309 (2018). [Google Scholar]
- 76.Hu K., Olsen B. R., Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair. J. Clin. Invest. 126, 509–526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duan X., et al. , Vegfa regulates perichondrial vascularity and osteoblast differentiation in bone development. Development 142, 1984–1991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watson E. C., Adams R. H., Biology of bone: The vasculature of the skeletal system. Cold Spring Harb. Perspect. Med. 8, a031559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ben Shoham A., et al. , Deposition of collagen type I onto skeletal endothelium reveals a new role for blood vessels in regulating bone morphology. Development 143, 3933–3943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kerschnitzki M., et al. , Transport of membrane-bound mineral particles in blood vessels during chicken embryonic bone development. Bone 83, 65–72 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Kerschnitzki M., et al. , Bone mineralization pathways during the rapid growth of embryonic chicken long bones. J. Struct. Biol. 195, 82–92 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Wiedeman M. P., Dimensions of blood vessels from distributing artery to collecting vein. Circ. Res. 12, 375–378 (1963). [DOI] [PubMed] [Google Scholar]
- 83.Kashtan N., Noor E., Alon U., Varying environments can speed up evolution. Proc. Natl. Acad. Sci. U.S.A. 104, 13711–13716 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kashtan N., Alon U., Spontaneous evolution of modularity and network motifs. Proc. Natl. Acad. Sci. U.S.A. 102, 13773–13778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roopin M., Ben-Tabou de-Leon S., Genome-wide identification of genes activated by VEGF signaling in the sea urchin embryo. European Nucleotide Archive. https://www.ebi.ac.uk/ena/data/view/PRJEB10269. Deposited 8 September 2015.
- 87.Gildor T., Ben-Tabou de-Leon S., Comparative study of regulatory circuits in two sea urchin species reveals tight control of timing and high conservation of expression dynamics. PLoS Genet. 11, e1005435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shin J. W., Huggenberger R., Detmar M., Transcriptional profiling of VEGF-A and VEGF-C target genes in lymphatic endothelium reveals endothelial-specific molecule-1 as a novel mediator of lymphangiogenesis. Blood 112, 2318–2326 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rivera C. G., Mellberg S., Claesson-Welsh L., Bader J. S., Popel A. S., Analysis of VEGF–A regulated gene expression in endothelial cells to identify genes linked to angiogenesis. PLoS One 6, e24887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirmizitas A., Meiklejohn S., Ciau-Uitz A., Stephenson R., Patient R., Dissecting BMP signaling input into the gene regulatory networks driving specification of the blood stem cell lineage. Proc. Natl. Acad. Sci. U.S.A. 114, 5814–5821 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.