Abstract
DYT1 dystonia is caused by an in-frame deletion of a glutamic acid codon in the gene encoding the AAA+ ATPase TorsinA (TorA). TorA localizes within the lumen of the nuclear envelope/endoplasmic reticulum and binds to a membrane-spanning cofactor, lamina associated polypeptide 1 (LAP1) or lumenal domain like LAP1 (LULL1), to form an ATPase; the substrate(s) of TorA remains ill-defined. Here we use budding yeast, which lack Torsins, to interrogate TorA function. We show that TorA accumulates at nuclear envelope-embedded spindle pole bodies (SPBs) in a way that requires its oligomerization and the SUN (Sad1 and UNc-84)-domain protein, Mps3. We further show that TorA physically interacts with human SUN1/2 within this system, supporting the physiological relevance of these interactions. Consistent with the idea that TorA acts on a SPB substrate, its binding to SPBs is modulated by the ATPase-stimulating activity of LAP1. TorA and TorA-ΔE reduce the fitness of cells expressing mps3 alleles, whereas TorA alone inhibits growth of cells lacking Pom152, a component of the nuclear pore complex. This genetic specificity is mirrored biochemically as TorA, but not TorA-ΔE, binds Pom152. Thus, TorA–nucleoporin interactions might be abrogated by TorA-ΔE, suggesting new experimental avenues to interrogate the molecular basis behind nuclear envelope herniations seen in mammalian cells lacking TorA function.
INTRODUCTION
DYT1 dystonia is an early-onset, heritable movement disorder caused by an autosomal dominant mutation removing a glutamic acid codon (∆E) in the DYT1/TOR1A gene that encodes the AAA+ ATPase TorsinA (TorA) (Ozelius et al., 1997). How expression of the TorA-∆E allele, and several others (Ozelius et al., 1997; Zirn et al., 2008; Xiao et al., 2009; Rebelo et al., 2015), causes dystonia remains ill-defined and requires additional molecular insight into the function of TorA. It remains unclear whether TorA is a typical AAA+ ATPase, which act as oligomeric ring assemblies that perform mechanical work on protein or protein-complex substrates (Hanson and Whiteheart, 2005), as there is no clear consensus as to the identity of TorA substrates.
Challenges with identifying TorA substrates likely relate to the fact that TorA is an unusual member of the AAA+ family. For example, it is uniquely localized within the contiguous lumen of the nuclear envelope (NE)–endoplasmic reticulum (ER) system (Hewett et al., 2000; Kustedjo et al., 2000). Further, whereas TorA has canonical AAA+ ATPase motifs (Ozelius et al., 1997), its ATP-binding pocket lacks the arginine finger (Brown et al., 2014; Sosa et al., 2014) required for ATP hydrolysis (Ogura et al., 2004). To form a functional enzyme, TorA must bind to one of two membrane-spanning cofactors, lamina associated polypeptide 1 (LAP1) or lumenal domain like LAP1 (LULL1), that contribute this critical arginine finger to TorA’s ATP-binding pocket (Zhao et al., 2013; Brown et al., 2014; Sosa et al., 2014; Demircioglu et al., 2016); TorA-∆E is unable to bind to LAP1 or LULL1, suggesting that dystonia is caused by a loss of ATPase function (Naismith et al., 2009; Zhu et al., 2010; Zhao et al., 2013; Demircioglu et al., 2016). Of note, LAP1 and LULL1 have distinct spatial distributions (Goodchild and Dauer, 2005), with LULL1 having access to the entire ER (Goodchild et al., 2015) and LAP1 being restricted to the inner nuclear membrane (INM) (Senior and Gerace, 1988). This suggests the compelling possibility that these cofactors may influence TorA ATPase activity (or potential substrates) in distinct subdomains of the ER, including the NE.
The idea that TorA has a NE-specific role is bolstered by the appearance of NE herniations or blebs found in the developing neurons of TorA–/– and TorA∆E/∆E mice (Goodchild et al., 2005; Tanabe et al., 2016). In most cases, these herniations, which have been visualized in several other model systems where TorA function is perturbed, bloom from an electron-dense structure at the INM of enigmatic origin (Naismith et al., 2004; Kim et al., 2010; Jokhi et al., 2013; VanGompel et al., 2015; Laudermilch et al., 2016; Tanabe et al., 2016). Recently, electron tomography of HeLa cells lacking all four Torsin isoforms revealed that the electron-dense structures are morphologically similar to nuclear pore complexes (NPCs) and can be labeled with anti-nucleoporin antibodies (Laudermilch et al., 2016). As similar herniations form over defective NPCs in budding yeast in response to perturbations in NPC assembly and/or the triggering of NPC quality control pathways (Wente and Blobel, 1993, 1994; Aitchison et al., 1995; Murphy et al., 1996; Siniossoglou et al., 1996; Zabel et al., 1996; Emtage et al., 1997; Ryan and Wente, 2002; Webster et al., 2014, 2016; Onischenko et al., 2017; Thaller and Lusk, 2018; Zhang et al., 2018), it seems reasonable that TorA might play a role in the biogenesis of NPCs. It remains unclear, however, what components of the NPC could be targeted by TorA, although gp210/Nup210 is an obvious candidate given its massive (Wozniak et al., 1994) and structurally conserved lumenal domain rich in repetitive immunoglobulin (Ig)-like domains (Upla et al., 2017).
It is equally likely, however, that TorA might indirectly influence NPC assembly, for example, by altering the function of proteins that contribute to lipid metabolism (Grillet et al., 2016) or by binding and modulating other NE components like LINC (Linker of Nucleoskeleton and Cytoskeleton) complexes, which have been implicated in NPC biogenesis from yeast to humans (Liu et al., 2007; Lu et al., 2008; Friederichs et al., 2011; Talamas and Hetzer, 2011; Chen et al., 2014). LINC complexes are composed of trimers of INM SUN (Sad1 and UNc-84) and outer nuclear membrane KASH (Klarsicht, Anc-1, and Syne homology) domain containing proteins that interact within and span the perinuclear space, physically coupling the cytoskeleton and nucleoskeleton (Padmakumar et al., 2005; Crisp et al., 2006; Sosa et al., 2012). Indeed, the concept that TorA might remodel LINC complexes remains a compelling narrative, supported by several biochemical, genetic, and cell biological data (Nery et al., 2008; Vander Heyden et al., 2009; Jungwirth et al., 2011; Atai et al., 2012; Saunders et al., 2017; Dominguez Gonzalez et al., 2018).
Here, we take advantage of a heterologous budding yeast genetic system to interrogate the function of TorA. While yeast lack a TorA orthologue, they express several evolutionarily conserved NE components including NPCs, LEM-domain integral INM proteins, and at least one SUN-domain protein, Mps3 (Webster and Lusk, 2016). Our data are consistent with a model in which human TorA can interact with and impact the function of yeast NE proteins with conserved lumenal domains, including Mps3. In fact, we show binding between human SUN proteins and TorA within the yeast system, suggesting that SUN proteins are likely TorA substrates. Moreover, both genetic and physical interaction data support that TorA can interact with the likely functional homologue of Nup210, Pom152. That this interaction is lost with TorA-∆E points to a new perspective on how to interpret the underlying causes of the NE abnormalities associated with early onset dystonia.
RESULTS AND DISCUSSION
TorA accumulation at SPBs requires oligomerization
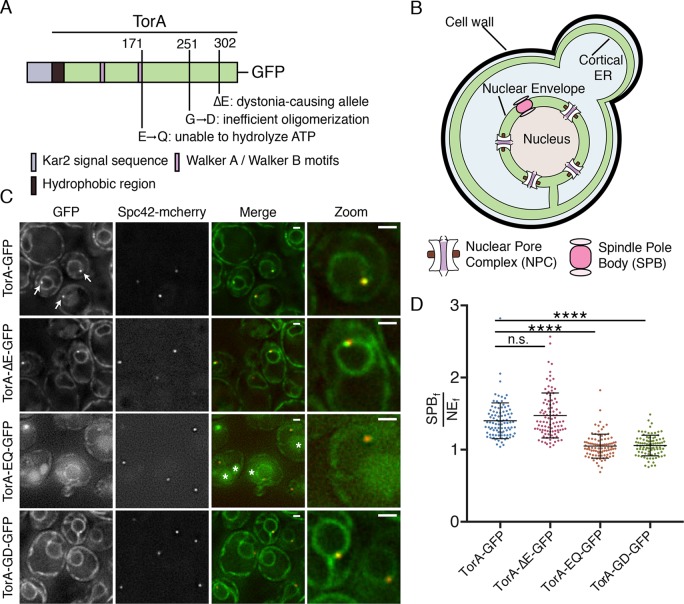
Several prior studies have expressed TorA in budding yeast, but no clear functional insight for TorA has emerged (Valastyan and Lindquist, 2011; Zacchi et al., 2014; Adam et al., 2017; Zacchi et al., 2017). However, given that these studies were largely performed before the discovery of the LAP1 and LULL1 cofactors required for TorA ATPase activity, we felt it was worthwhile to revisit this system using the fully functional enzyme complex. We took advantage of TorA constructs generated by Valastyan and Lindquist (2011) that were engineered with a yeast-specific promoter, a Kar2 signal sequence and a C-terminal green fluorescent protein (GFP) (Figure 1A). To reduce variation in cell-to-cell expression levels, we modified these constructs to allow for chromosomal integration and generated yeast strains expressing TorA-GFP and several TorA alleles including TorA-∆E-GFP, TorA-EQ-GFP, and TorA-GD-GFP (Figure 1A).
FIGURE 1:
TorA-GFP and TorA∆E-GFP, but not TorA-GD-GFP, accumulate at SPBs. (A) Schematic of TorA-GFP constructs—relevant motifs and locations (numbers are amino acid residue positions from the start methionine of the human gene) of amino acid changes are shown. (B) Diagram (not to scale) of a budding yeast cell schematizing the NE/ER system with NPCs and SPBs. (C) Deconvolved fluorescence micrographs of strains expressing the indicated TorA-GFP constructs and Spc42-mCherry. Green and red fluorescence images alongside a merge and magnification of one cell (zoom) are shown. Arrows point to NE foci that colocalize with Spc42-mCherry. TA-EQ-GFP might accumulate in vacuoles (asterisks). Bar is 1 µm. (D) Plot of SPBf/NEf of indicated TorA-GFP constructs in individual cells from three biological replicates (n = 32/replicate) with mean and SD. Kruskal–Wallis one-way ANOVA with post hoc Dunn’s test. ****p < 0.0001.
We first examined the localization of TorA-GFP in logarithmically growing wild-type (wt) cells. As previously published (Valastyan and Lindquist, 2011), TorA-GFP was found in a perinuclear (i.e., NE) and cortical distribution, consistent with the morphology of the budding yeast ER (Figure 1, B and C). We remarked, however, that TorA-GFP accumulated in one or two foci at the NE (Figure 1C, arrows). This localization was particularly striking in cells expressing low levels of TorA-GFP, best observed on growing cells to saturation (Supplemental Figure S1A). In these cases, we observed TorA-GFP in one or two puncta per cell with a nearly undetectable pool in the rest of the NE/ER, raising the possibility that TorA preferentially binds to a NE-specific structure.
The focal accumulation of TorA-GFP at the NE was reminiscent of spindle pole bodies (SPBs), the yeast centrosome equivalents that span both membranes of the NE (Jaspersen and Ghosh, 2012)(Figure 1B). To test this idea, we examined the localization of TorA-GFP in a strain expressing an mCherry-tagged core component of the SPB, Spc42. As shown in Figure 1C and Supplemental Figure S1A, we observed clear coincidence between virtually all Spc42-mCherry and TorA-GFP NE-foci, confirming that TorA-GFP likely associates with SPBs. In these logarithmically growing cells, we also compared the mean fluorescence of TorA-GFP at the SPB (SPBf) with the broader NE (NEf) on an individual cell basis to provide a metric of relative SPB enrichment (SPBf/NEf), which ranged from 1.04 to 2.82 and had an average SPBf/NEf of 1.40 (Figure 1D).
We next tested whether TorA-∆E, TorA-EQ, and TorA-GD would also enrich at SPBs. While TorA-ΔE-GFP was produced at lower levels than TorA-GFP (Supplemental Figure S1B), it nonetheless accumulated at SPBs (mean SPBf/NEf of 1.47), similar to its wt counterpart. In contrast, TorA-EQ-GFP did not enrich at SPBs (mean SPBf/NEf = 1.05), although we struggled to find conditions in which TorA-EQ-GFP was stably expressed—note that even the NE/ER signal was low and there was green fluorescence in the vacuole (see asterisks in Figure 1C and Supplemental Figure S1A) that could indicate its degradation. Interestingly, in the absence of LAP1 or LULL1, TorA-EQ can aggregate in vitro (Sosa et al., 2014), which may explain its potential targeting for degradation in our system. Strikingly, however, a TorA-GD mutant, which cannot oligomerize due to disruption of the “back interface” (Chase et al., 2017), localized but failed to accumulate at SPBs (mean SPBf/NEf = 1.06) despite being expressed at levels similar to TorA-ΔE and TorA-EQ (Supplemental Figure S1B). This was particularly obvious when comparing these strains grown to saturation (Supplemental Figure S1A). Taken together, these results suggest that TorA might interact with a SPB component in a manner that could be amplified by its oligomerization.
LAP1-LD expression releases TorA from SPBs
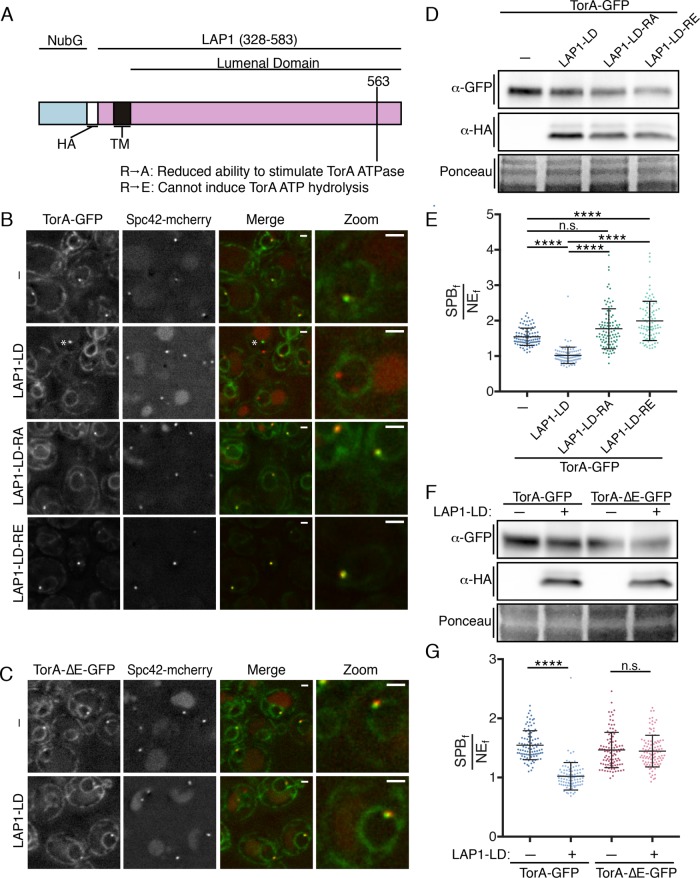
There is a general consensus that TorA can self-assemble into an oligomer in its ATP-bound form (Vander Heyden et al., 2009; Jungwirth et al., 2010; Chase et al., 2017). Interestingly, recent work also indicates that binding of the LAP1 lumenal domain (LAP1-LD) to a TorA hexamer and subsequent ATP hydrolysis causes oligomer disassembly (Chase et al., 2017). We took advantage of these observations to attempt to recapitulate a putative ATPase cycle in vivo using TorA-GFP SPB accumulation as a proxy for a substrate interaction. While efforts to produce full-length LAP1 in yeast were unsuccessful, by replacing the N-terminus of LAP1 with a fragment of ubiquitin (NubG), we could express the NubG-LAP1-LD (indicated as LAP1-LD, below) under the control of a galactose-inducible (GAL1) promoter (Figure 2A), which reached peak levels after ∼5 h of growth in the presence of galactose (Supplemental Figure S1C).
FIGURE 2:
LAP1–LD-mediated release of TorA-GFP from SPBs likely requires ATP hydrolysis. (A) Diagram of the LAP1-LD constructs with N-terminal NubG fragment, HA epitope, transmembrane (TM) segment, and position of indicated mutations. (B) Deconvolved fluorescence micrographs of strains expressing TorA-GFP and Spc42-mCherry with or without (-) the production of LAP1-LD constructs. Green and red fluorescence alongside a merge and magnification of a cell (zoom) are shown. Asterisk points to a rare NE focus that is not a SPB only seen on LAP1-LD expression. (C) As in B with TorA-ΔE-GFP. Bar is 1 µm. (D) Western blot of TorA-GFP (α-GFP) and LAP1-LD constructs (α-HA) with Ponceau stain to assess loads. (E) Plot of SPBf/NEf of experiment in B of individual cells from three biological replicates (n = 32/replicate) with mean (middle line) and SD. Kruskal–Wallis one-way ANOVA with post hoc Dunn’s test. ****p < 0.0001. (F) Western blot of TorA/∆E-GFP (α-GFP) and LAP1-LD (α-HA) levels with reference to Ponceau stain of total protein loads. (G) As in E with indicated expression constructs.
Remarkably, at the highest levels of LAP1-LD expression, TorA-GFP was no longer visibly concentrated at SPBs (Figure 2, B and E; mean SPBf/NEf = 1.02). Importantly, this reduction in SPBf/NEf values was due to lower SPBf and not higher NEf, as TorA-GFP levels remained unaltered on production of the LAP1-LD (Figure 2D). Further, and consistent with the idea that the ability of LAP1-LD to reduce TorA association with the SPB is direct, LAP1-LD expression at similar levels (Figure 2F) had no effect on the SPB accumulation of TorA-∆E-GFP, which is unable to stably interact with the LAP1-LD (Naismith et al., 2009; Zhu et al., 2010; Zhao et al., 2013; Brown et al., 2014) (Figure 2, C and G).
LAP1 acts as a cofactor for TorA by contributing a critical arginine residue (at position 563) to the TorA–ATP-binding pocket (Brown et al., 2014; Sosa et al., 2014); changing this “arginine finger” to either alanine or glutamic acid reduces or abolishes ATP hydrolysis, respectively (Brown et al., 2014) (Figure 2A). We therefore tested whether abrogating LAP1’s ability to stimulate TorA ATP hydrolysis impacted its accumulation at SPBs by expressing LAP1-LD-RA and -RE mutants at similar levels as LAP1-LD (Figure 2D). In striking contrast to LAP1-LD, expression of LAP1-LD-RA, but in particular LAP1-LD-RE, leads to increased TorA-GFP accumulation at the SPB (SPBf/NEf values reached 3.90, mean of 2.00; Figure 2, B and E). These data suggest that the LAP1-LD mutants lead to the “trapping” of a potential TorA substrate at the SPB. In addition, as the SPBf/NEf values of TorA-GFP in the LAP1-LD mutants exceed those of TorA-GFP when expressed alone, these data raise the intriguing possibility that there is a cofactor endogenous to yeast capable of stimulating ATP hydrolysis. Such a model predicts that inhibition of this putative cofactor would mirror the hyperaccumulation of TorA at SPBs observed on expressing the LAP1-RA/E alleles. Regardless, in the aggregate, these data support that TorA-GFP is capable of undergoing an ATP-hydrolysis cycle by binding and releasing a substrate that is a likely component of the SPB.
TorA and TorA-∆E specifically interact with NE proteins
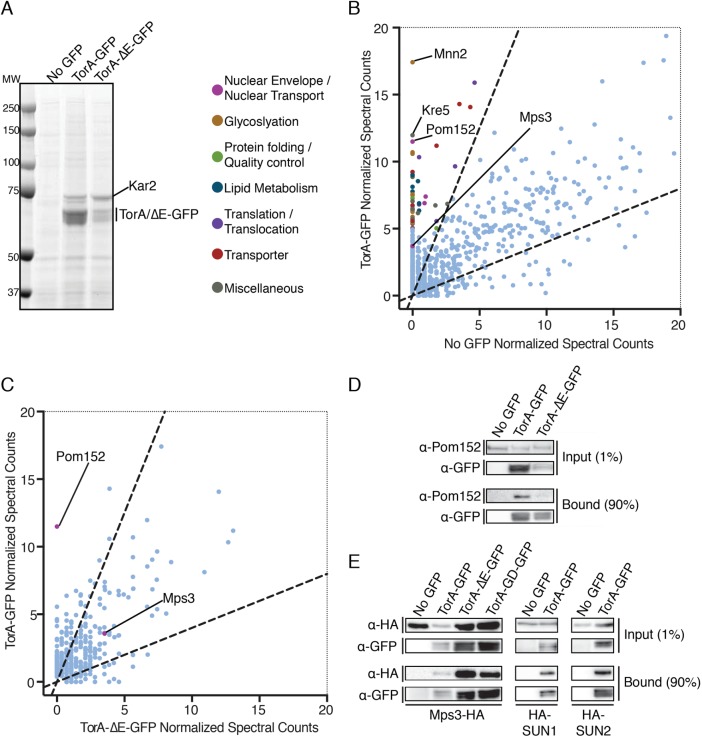
The ability of the “back interface” mutation to disrupt accumulation of TorA-GFP at the SPB and the release of TorA-GFP from the SPB on expression of LAP1-LD suggest that TorA may bind to a substrate at the SPB. To facilitate the identification of potential TorA-binding partners, we affinity-purified TorA-GFP and TorA-∆E-GFP from cryolysates derived from wt cells using anti-GFP nanobody-coupled magnetic beads. As shown in Figure 3A, we pulled out both TorA-GFP and TorA-∆E-GFP alongside at least one major binding partner of ∼70 kDa, which we identified by mass spectrometry (MS) to be Kar2 (the orthologue of the ER chaperone, binding immunoglobulin protein [BiP]). This result is consistent with previous work and suggests that Kar2 plays a critical role in ensuring ER translocation and/or stability of these human proteins (Zacchi et al., 2014). It also remains formally possible that TorA forms a functional complex with Kar2 and contributes to protein quality control pathways in the ER.
FIGURE 3:
TorA and TorA-ΔE interact with a shared set of proteins, but TorA specifically binds to Pom152. (A) Coomassie-stained SDS–PAGE gel of bound proteins eluted from α-GFP nanobody beads incubated with cell extracts expressing TorA-GFP, TorA-ΔE-GFP, or “no GFP.” Position of MW standards are on the left. (B) Magnification of plot (full plot in Supplemental Figure S2A) comparing average normalized MS spectra from three replicates identified in TorA-GFP vs. no GFP control. Dashed lines demarcate 2.5-fold enrichment. Proteins with an average of five or more spectra that were 2.5-fold enriched in TorA-GFP sample or absent in no GFP sample are colored based on the classifications indicated. (C) As in B (full plot in Supplemental Figure S2C) but comparing average normalized spectra (from two replicates) between TorA-GFP and TorA-ΔE-GFP affinity purifications. (D) Western blots of input and bound fractions from affinity purifications of TorA-GFP, TorA-∆E-GFP with no GFP control. (E) Western blots of input and bound fractions from affinity purifications of TorA-GFP, TorA-∆ but comparing average normalized spectra (from two replicates) between TorA-GFP and TorA-ΔE-GFP affinity purifications.
Next, to fully identify the TorA interactome, we subjected the entire TorA-GFP- and TorA-ΔE-GFP- bound fractions to shotgun LC-MS/MS. As expected from these highly sensitive approaches, we identified hundreds of proteins, most of which were nonspecific and present in the “no GFP” control (complete peptide lists can be found in Supplemental Table S1, “all normalized spectra” tab). We therefore plotted mean-normalized spectral counts identified from three independent affinity purifications and considered proteins identified from peptides that were at least 2.5-fold enriched in the TorA-GFP-bound fractions relative to the no GFP control (Supplemental Figure S2A, dashed lines) to be likely interactors. To facilitate visualizing proteins less abundant than Kar2, we have magnified the region of the plot with the majority of specific proteins (Supplemental Figure S2A, box) and have color-coded those with an average of more than five peptides based on our own functional classification scheme (Figure 3B). Strikingly, whereas the top two hits identified were Mnn2 and Kre5 (two highly abundant enzymes involved in glycosylation), the third protein with the most peptides (with none found in the no GFP control) was Pom152 (Figure 3B).
Pom152 is the functional homologue of Nup210 and is the only component of the NPC with a large (>10 kDa) lumenal domain. We could also detect at least one additional nucleoporin, Nup192, and two nuclear transport factors, but these had fewer peptides with some in the no GFP control (Supplemental Table 1, TorA vs. no GFP tab). Thus, despite the specific interaction with Pom152, it was unlikely that TorA-GFP was binding to NPCs. Consistent with this, using a drug-inducible NPC-clustering strain (Colombi et al., 2013), we did not observe enrichment of TorA-GFP within the NPC cluster (Supplemental Figure S1, D and E). It is most likely then that Pom152 binds to TorA within a pool that is outside of NPCs, perhaps NPC assembly intermediates, or even at SPBs (Sezen et al., 2009; Katta et al., 2015; Rüthnick et al., 2017).
To identify putative factors that might specifically interact with TorA-GFP but not with TorA-ΔE-GFP, we directly compared normalized spectral counts from two independent TorA-GFP and TorA-ΔE-GFP affinity purifications (Figure 3C and Supplemental Figure S2C). Here, Pom152 immediately stood out. In fact, we did not identify any peptides for Pom152 in either replicate of TorA-ΔE-GFP (Supplemental Figure S2B, Supplemental Table 1, TorA-ΔE vs. no GFP tab), a result that we further confirmed by Western blotting using anti-Pom152 antibodies of additional affinity purifications (Figure 3D). Whereas there were several other proteins that also show specificity to TorA-GFP (and a few to TorA-ΔE-GFP as well; see Supplemental Table S1), fewer peptides for each were identified, and the potential functional relevance of these proteins with respect to TorA or TorA-ΔE is not obvious and difficult to ascribe.
Remarkably, the only other lumenal domain-containing NE protein identified in all replicates of both TorA-GFP and TorA-ΔE-GFP pullouts was the SUN-domain-containing protein, Mps3 (Figure 3, B and C). Critically, Mps3 is the only component of the SPB identified in any replicate of TorA-GFP or TorA-ΔE-GFP purifications, strongly supporting the specificity of this interaction. We further confirmed TorA and TorA-ΔE interactions with Mps3 by Western blotting by detecting a hemagglutinin (HA) epitope-tagged version of Mps3 in bound fractions (Figure 3E). Interestingly, we also found that TorA-GD-GFP can specifically bind to Mps3-HA, suggesting that, while oligomerization might be required to amplify interactions with the SPB (Figure 1, C and D), TorA might interface with Mps3 as a monomer. Finally, to examine whether interactions with Mps3 reflected physiological interactions with human SUN-domain proteins, we expressed HA-tagged versions of human SUN1 and SUN2 in budding yeast. As shown in Figure 3E, both HA-SUN1 and HA-SUN2 could be specifically copurified with TorA-GFP. Thus, taken together, our data support a model in which TorA-GFP can functionally interact with SUN domain-containing proteins and strongly suggest that they might be TorA substrates.
TorA/TorA-ΔE specifically impact the fitness of mps3 and pom152 strains
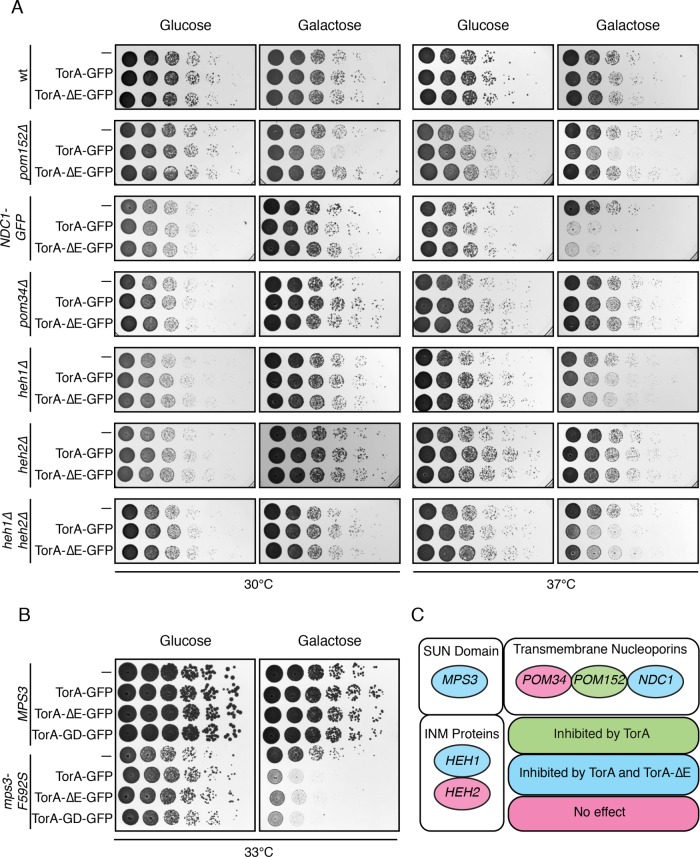
The localization of TorA in an oligomerization and ATPase activity-dependent manner to the SPB, combined with the identification of Pom152 and Mps3 as likely TorA binding partners, raise the possibility that TorA could influence the function of these (or other) NE proteins. We therefore tested whether TorA expression impacted the fitness of yeast strains with alleles of POM152 and MPS3. For these experiments, we placed TorA and TorA-ΔE under the control of the conditional GAL1 promoter as we observed progressive loss of TorA-GFP expression on serial culturing in some strain backgrounds. Consistent with our hypothesis and biochemistry, strains null for POM152 were specifically sensitive to the expression of TorA but not TorA-ΔE (Figure 4A, galactose panels). This result suggests that TorA acts as a dominant negative in the absence of Pom152, perhaps by binding and inhibiting an essential factor that lacks the capacity to interact with TorA-ΔE. Alternatively, particularly in light of our biochemical analysis, we favor a model where Pom152 could directly interact with TorA in a way that modulates its activity.
FIGURE 4:
Conditional expression of TorA-GFP or TorA∆E-GFP differentially impacts the fitness of mps3 and pom152 strains. (A, B) The indicated strains were 10-fold serially diluted onto glucose- or galactose-containing plates, which repress or induce the expression of TorA/TorA-ΔE, respectively. Colony size was assessed after growth at the indicated temperatures for 1 or 2 d. (C) Summary of genetic backgrounds inhibited by TorA/TorA∆E expression.
We next tested whether conditional expression of TorA impacted the growth of a mps3Δ strain complemented by either MPS3 or a temperature-sensitive allele (mps3-F592S) that disrupts the SUN domain and the function of Mps3 (Jaspersen et al., 2006). Interestingly, while expression of TorA or TorA-ΔE did not alter growth of MPS3-containing cells, both specifically impacted the fitness of strains expressing the mps3-F592S allele (Figure 4B), suggesting that TorA can perturb a compromised SPB and/or impaired SPB insertion mechanism. TorA-GD had a similar effect, further bolstering the idea that TorA need not oligomerize to impact SPB function. As Mps3 likely contributes to both SPB insertion and NPC biogenesis into the NE (Jaspersen et al., 2002, 2006; Jaspersen and Ghosh, 2012; Friederichs et al., 2011; Chen et al., 2014), we tested whether shared components of the SPB and NPC (e.g., Ndc1) (Winey et al., 1993; Chial et al., 1998) are also impacted by TorA expression. For this experiment, we took advantage of our prior observation that C-terminal tagging of Ndc1 with GFP partially impairs its function (Yewdell et al., 2011) and, consistent with this, TorA and TorA-ΔE inhibited the growth of Ndc1–GFP-expressing cells, particularly at 37°C (Figure 4A). We also observed a slight impact on the fitness of heh1Δ cells but not those lacking HEH2 or POM34 (Figure 4, A and C). Thus, while TorA also genetically interacts with MPS3 and HEH1, in these cases, the interactions are not specific to TorA and are also seen with the TorA-ΔE allele, again consistent with our biochemical analysis (Figure 4C).
mps3 and pom152 alleles differentially affect SPB accumulation of TorA
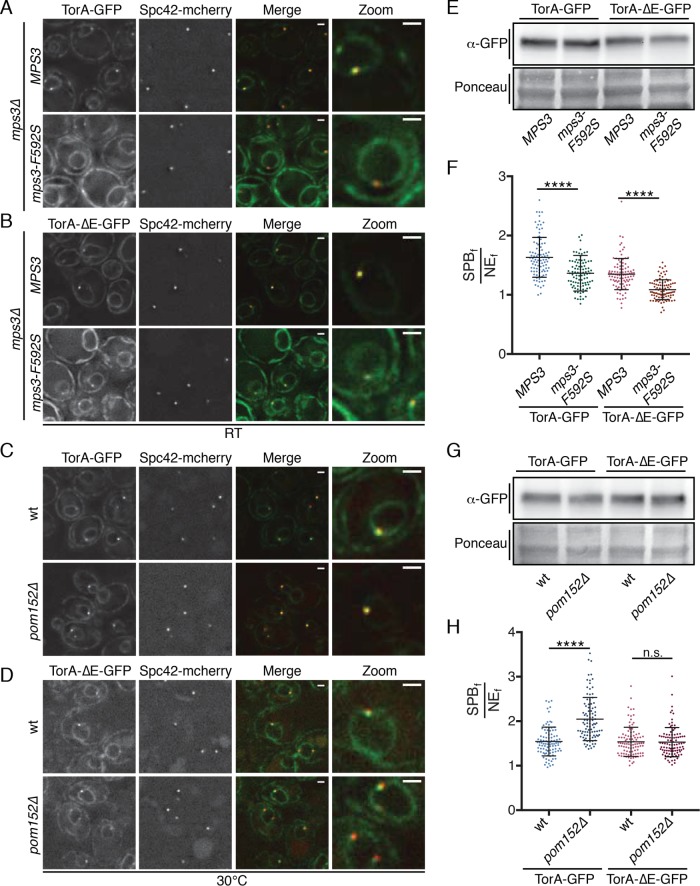
Finally, we tested whether disruption of MPS3 and POM152 function altered the distribution of TorA-GFP at the SPB. Interestingly, we observed lower SPBf/NEf ratios of both TorA-GFP and TorA-ΔE-GFP at the SPB in mps3-F592S cells compared with the wt MPS3 counterpart, even at permissive growth temperatures (room temperature [RT]) (Figure 5, A, B, and F). These data open up the possibility that TorA (and TorA-∆E) is recruited to the SPB through a direct interaction with the Mps3 SUN domain. In contrast to the loss of SPB recruitment observed in mps3-F592S cells, we observed higher SPBf/NEf values for TorA-GFP (up to 3.53, mean SPBf/NEf of 2.05) in pom152Δ cells (Figure 5, C and H), which resembles the hyperaccumulation of TorA-GFP at SPBs seen on expression of the LAP1-LDs deficient in stimulating TorA-GFP ATP hydrolysis (Figure 2B). Perhaps most interestingly, this effect was specific, and we observed no change in SPBf/NEf levels of TorA-∆E-GFP in pom152Δ cells (Figure 5, D and H). These data thus closely mirror the genetic and biochemical analysis and support the concept that Pom152 could impact TorA function analogously to its established activator, LAP1.
FIGURE 5:
TorA and TorA-ΔE do not enrich at SPBs in mps3 strains, while TorA specifically hyperaccumulates in cells lacking POM152. (A–D) Deconvolved fluorescence micrographs of cells expressing TorA-GFP or TorA-∆E-GFP with Spc42-mCherry in the indicated genetic backgrounds. Green and red fluorescence alongside a merge and magnification of one cell (zoom) are shown. Bar is 1 µm. (E) Western blot of whole cell extracts from the indicated strains detecting TorA/Tor∆E-GFP levels (α-GFP) with Ponceau stain to show relative protein loads. (F) Plot of SPBf/NEf of TorA- and TorA-∆E-GFP in individual cells from three biological replicates (n = 32/replicate) with mean and SD. Kruskal–Wallis one-way ANOVA with post hoc Dunn’s test. ****p < 0.0001. (G, H) As in E and F.
Outlook
In conclusion, we have employed the budding yeast model to interrogate the function of TorA. By uncovering an interaction between TorA and SPBs that can be altered by the introduction of the LAP1-LD, we have developed a visual in vivo assay to investigate the molecular determinants and perhaps the function (in the future) of a TorA-mediated ATP hydrolysis cycle. Perhaps most critically, our data point to the discovery of a likely substrate at the NE with the most obvious candidate being the SUN domain-containing Mps3. This interpretation is strengthened by our use of the yeast system in that there are no other conserved membrane proteins that anchor the SPB (as the mammalian centrosome does not span the NE), and budding yeast lack an obvious KASH domain partner (Mps2 is considered to be “KASH-like”[Friederichs et al., 2011], but we did not identify any peptides to Mps2 in our affinity purifications). Indeed, the Mps3 SUN domain shares ∼30% sequence identity with its mammalian counterparts (Jaspersen et al., 2006) and importantly, we show that human SUN1 and SUN2 can copurify with TorA in the yeast system. There are also several additional studies that would support the conclusion that SUN proteins are TorA substrates (Nery et al., 2008; Vander Heyden et al., 2009; Jungwirth et al., 2011; Atai et al., 2012; Saunders et al., 2017; Dominguez Gonzalez et al., 2018). In the future, we look forward to the direct reconstitution of a SUN protein–TorA complex, although it is possible the interaction between TorA and SUN-domain-containing proteins is not with the SUN domain per se, as we also detect structural homology (with ∼99% confidence per Phyre2 [Kelley et al., 2015]) between the recently identified auto-inhibitory domain in either SUN1 (Xu et al., 2018) or SUN2 (Nie et al., 2016) with the analogous region preceding the Mps3 SUN domain (Supplemental Figure S2, D–F).
Clearly, an additional priority going forward is to determine whether Pom152, and, most critically, its functional homologue Nup210, directly interacts with TorA. The most exciting feature of this putative interaction, be it direct or indirect, is that it is specific for TorA and is disrupted by the ΔE mutation. These data are compelling as they lend support to the concept that defects in NPC biogenesis could arise in the absence of a putative TorA-Nup210 interaction and, in turn, contribute to the formation of the NE herniations that are the cellular hallmark of the dystonia disease phenotype. Perhaps most interestingly, our data hint at the existence of another cofactor that could be capable of stimulating TorA hydrolysis. While there is nothing obvious about the structure of the Pom152 lumenal domain, which consists of repetitive Ig motifs (Wozniak et al., 1994; Upla et al., 2017), that resembles the RecA fold of the LAP1 or LULL1 lumenal domains (Brown et al., 2014; Sosa et al., 2014), our data nonetheless raise the possibility that Pom152 could contribute to TorA-mediated ATP hydrolysis. Thus, we hope that this study facilitates direct testing of these hypotheses in mammalian systems and provides additional avenues to uncover the molecular basis of early onset DYT1 dystonia.
MATERIALS AND METHODS
Plasmid generation
All plasmids used in this study are listed in Supplemental Table S2 under the “Plasmids” tab. To generate pMC13/pMC25, pMC14/pMC26, and pMC22/pMC28, the coding sequences of TorA-GFP, TorA-∆E-GFP, and TorA-EQ-GFP (Valastyan and Lindquist, 2011) were subcloned into p406ADH1/p406GAL1 using XhoI and XbaI. The promoter and coding region of pMC13, pMC14, and pMC22 were then subcloned into pRS403 (American Type Culture Collection) using EagI and SacI to generate pMC15, pMC16, and pMC24. The Gibson Assembly Mastermix (NEB) was used to insert the coding sequence for amino acids 328–583 of human LAP1 (LAP1-LD), which was PCR-amplified by Q5 DNA polymerase, into pDSL-NX (Dualsystems Biotech; 2µ, TRP1), which was used to generate NubG (N-terminus of Ubiquitin) fusions behind the control of the CYC1 promoter, to generate pDT07. Subsequently, pMC12 was made by subcloning the NubG-HA-LAP1-LD coding sequence from pDT07 into p406GAL1 using SpeI and SmaI. pMC31/pMC32, pMC33, and pMC35 encode the TorA-GD, LAP1-LD-R563A, and LAP1-LD-R563E point mutants, respectively, which were generated using Quikchange Site-Directed Mutagenesis with Pfu Turbo (Agilent Technologies). To produce pMC43 and pMC45, the coding sequences for human SUN1 (amino acids 287–811) and full-length SUN2 (May and Carroll, 2018) (TCP1104, TCP442) were subcloned into pMC38 (p406GAL1 containing NubG-HA coding region) using AscI and PacI.
Yeast strain generation, growth, and genetic analysis
All yeast strains are derived from a wt W303 genetic background and are listed in Supplemental Table S2, “Yeast Strains” tab. Gene knockouts and fluorescent protein/epitope-tagging of endogenous genes were performed using a PCR-based integration approach (Longtine et al., 1998; Van Driessche et al., 2005). To integrate TorA-GFP or LAP1-LD constructs into the genome at the URA3 or HIS3 loci, plasmids were digested with BstBI or BmtI, respectively, prior to transformation using standard methods (Sikorski and Hieter, 1989; Burke et al., 2000). As indicated in Supplemental Table S2, some strains were generated by mating and subsequent sporulation and tetrad dissection using standard methods (Burke et al., 2000). Yeast strains were grown at 30°C in YPA (1% yeast extract, 2% peptone, 0.025% adenine) with 2% dextrose (YPAD), 2% raffinose (YPAR), or 2% galactose (YPAG).
To test genetic interactions, relative growth of yeast strains was assessed by plating 10-fold serial dilutions of overnight cultures onto YPD or YPG plates. Plates were incubated at 30°C, 33°C, or 37°C and imaged at the times indicated in the figure legends.
Preparation of whole-cell protein extracts and Western blotting
To generate whole-cell protein extracts for Western blotting, ∼2 OD600 of cells were collected by centrifugation, washed once with 1 mM EDTA, and lysed for 10 min on ice with 250 µl of 2 M NaOH. Proteins were subsequently precipitated for 30 min after the addition of 250 µl 50% trichloroacetic acid. Samples were centrifuged at 18,000 × g for 10 min at 4°C to pellet precipitated proteins, washed once with 1 ml –20°C acetone, resuspended in 40 µl 5% SDS followed by 40 µl of 2× SDS Laemmli sample buffer containing 100 mM dithiothreitol (DTT), and heat-denatured for 5 min at 95°C.
Protein samples were separated on a 4–20% gradient gel (Bio-Rad) and transferred using the Mini Trans-Blot Cell (Bio-Rad) at 100 V for 60 min onto 0.2 µm nitrocellulose (Bio-Rad). Protein samples collected during affinity purification experiments (below) were separated on a 4–12% gel (NuPAGE) and transferred using Mini Blot Module (Invitrogen) at 25 V for 70 min onto 0.2 µm nitrocellulose (Bio-Rad). Nitrocellulose was subsequently blocked with 5% skim milk in Tris-buffered saline with 0.1% Tween-20 (TBST) for 1 h at RT, incubated with primary antibodies for 1 h at RT, extensively washed in TBST, incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. After washing, proteins were detected by ECL. Antibodies used in this study are listed in Supplemental Table 2, “Antibodies” tab.
Affinity purification of TorA and TorA-∆E-GFP
To identify the interactome of TorA-GFP and TorA-∆E-GFP, 1-l cultures of W303 (no GFP control), MC99 (TorA-GFP), or MC108 (TorA-∆E-GFP) were grown to an OD600 of ∼2 and pelleted at 4000 × g for 15 min at 4°C. Cell pellets were transferred to a 50-ml conical tube, washed once with dH2O, weighed, and suspended in 100 µl freezing buffer (20 mM HEPES, pH 7.5, 1.2% polyvinylpyrrolidone, protease inhibitor cocktail [1:200]) per gram of cell pellet to make a slurry. A 20-gauge needle was used to puncture a hole into the bottom of a 50-ml conical tube, and a plunger was used to force the slurry, drop-wise, through the hole into a second 50 ml conical tube filled with liquid nitrogen. Frozen cell pellets were cryomilled with a ball mill (Retsch) to generate a frozen powder of cell lysate (Alber et al., 2007). For each affinity purification, 800 µl of extraction buffer (100 mM HEPES, pH 7.5, 300 mM KCl, 1 mM EDTA, 100 mM MgOAc, 1% Triton X-100, 10 mM 2-mercaptoethanol) was added to 200 mg of lysate powder. The sample was vortexed to resuspend prior to centrifugation at 14,000 rpm at 4°C for 10 min to clear the lysate. The supernatant was added to 10 µl GFP-Trap magnetic-agarose beads (ChromoTek) previously equilibrated in 1 ml lysis buffer. After a 1-h incubation at 4°C, the unbound fraction was collected, and the beads were washed 5× in 1 ml extraction buffer. To elute bound proteins, beads were resuspended in 20 µl lithium dodecyl sulfate sample buffer (NuPAGE), denatured at 70°C for 10 min, and centrifuged at 18,000 × g for 1 min prior to physical separation of beads from the elution using a magnet. The elution was transferred to a fresh microcentrifuge tube, reduced by the addition of 1 µl 1 M DTT (final concentration of 50 mM DTT), and heated again for 10 min at 70°C. Bound proteins were identified by MS or Western blotting.
Mass spectrometry
Eluted proteins were loaded onto a 4–12% gel (NuPAGE) until the dye front entered the gel. Proteins were visualized using Imperial Protein Stain (Thermo Fisher Scientific). After washing with dH2O, a clean razor blade was used to excise the protein band (∼10 µg). The gel piece was transferred to a clean 1.5 ml tube, and an in-gel tryptic digest was performed. Peptides were separated on a Waters nanoACQUITY ultra-high-pressure liquid chromatograph prior to detection on either a Waters/Micromass AB QSTAR Elite (two replicates for wt cells [no GFP] and those expressing TorA-GFP; one replicate for cells expressing TorA-∆E-GFP) or a Thermo Fisher Scientific LTQ-Orbitrap XL Fusion (third replicate for wt [no GFP] and TorA-GFP samples and second replicate for TorA-∆E-GFP) mass spectrometer.
Mascot was used to analyze all MS/MS data (Matrix Science, London, UK; version 2.6.0). Mascot searched the SwissProt_2017_01 database (selected for Saccharomyces cerevisiae, unknown version, 7904 entries) assuming strict trypsin enzyme digestion, a fragment ion mass tolerance of 0.020 Da, and allowed oxidation of methionine and propionamide of cysteine.
To validate MS/MSbased peptide and protein identifications, Scaffold (version Scaffold_4.8.7; Proteome Software, Portland, OR), a program that incorporates the ProteinProphet algorithm (Nesvizhskii et al., 2003), was used with the following parameters: peptide identifications needed at least 95.0% probability by the Scaffold Local FDR algorithm; protein identifications required at least 22.0% probability to achieve an FDR less than 2.0% and at least two identified peptides; proteins that could not be distinguished by MS/MS analysis alone due to similar peptides were grouped to satisfy the principles of parsimony.
Scaffold (version Scaffold_4.8.7, Proteome Software, Portland, OR) was used to generate normalized spectral counts for each replicate. To identify proteins at least 2.5-fold enriched in TorA-GFP or TorA-∆E-GFP relative to the no GFP control, normalized peptide counts for individual proteins were averaged; TorA/∆E-GFP average for each protein was divided by the corresponding average no GFP control. Proteins present in TorA/∆E-GFP but absent in the no GFP control were also considered to be 2.5-fold enriched.
Structure prediction
To identify domains in Mps3 that are structurally similar to other SUN domain proteins, the region between the transmembrane domain and the established SUN domain (Jaspersen et al., 2006) (amino acids 171–458) was threaded into Phyre 2.0 (Kelley et al., 2015). PyMOL was used to superimpose the predicted structure onto mouse Sun1 crystal structure (Xu et al., 2018).
Microscopy
A Deltavision widefield deconvolution microscope (Applied Precision/GE Healthcare) with a 100×, 1.40 numerical aperture objective (Olympus), solid-state illumination, and a CoolSnap HQ2 CCD camera (Photometrics) was used to acquire all images. Unless otherwise stated in the text, all micrographs presented are of cells grown to mid–log phase at 30°C. To prepare cells for imaging, they were gently pelleted by centrifugation, suspended in complete synthetic medium, and pipetted onto a glass slide. Z-stacks with a 0.25 µm step were acquired with the exception of cells expressing TorA-EQ-GFP, where only a single plane was imaged to prevent signal loss due to photobleaching because of its low levels. To cluster NPCs, MCCPL561 was treated with 10 µg/ml rapamycin for 30 min prior to imaging as described in Colombi et al. (2013).
Image processing, analysis, and statistics
All images presented were deconvolved via softWoRx (Applied Precision/GE Healthcare). Subsequent processing was performed using Fiji/ImageJ (Schindelin et al., 2012) and Photoshop (Adobe). Image analysis (detailed below) was performed on raw (i.e., nondeconvolved) images using Fiji/ImageJ.
To determine the relative accumulation of TorA-GFP at the SPB, the GFP fluorescence at the SPB (SPBf) was divided by the NE GFP signal (NEf). To calculate SBPf, a 6 × 6 pixel square was used to measure the mean GFP fluorescence at the GFP accumulation that colocalized or overlappedlapped with the SPB, as visualized with Spc42-mCherry. To obtain a NEf value, a portion of the NE was outlined and the mean GFP fluorescence was measured. After subtracting mean background fluorescence, SPBf was divided by NEf for each cell. To simplify the analysis, only cells with a single SPB were analyzed. Thirty-two cells per genotype per biological replicate were analyzed, and each experiment represents three independent biological replicates, totaling 96 cells per genotype per experiment.
The SPBf/NEf distribution in TorA–GFP-expressing cells failed the D’Agostino and Pearson normality test; therefore, the nonparametric Kruskal–Wallis one-way analysis of variance (ANOVA) was used to compare multiple genotypes within an experiment simultaneously, and a post hoc Dunn’s test was used to identify significant differences.
Supplementary Material
Acknowledgments
We are grateful for invaluable discussions and advice from Christian Schlieker. Thanks also to Megan King and members of the Lusk and King groups for discussion and assistance provided by the Keck Proteomics Facility at Yale University. This work was generously supported by the Dystonia Medical Research Foundation and the National Institutes of Health (NIH): RO1 GM105672 to C.P.L. M.C. is also funded by NIH-5F31HL134272 and NIH-5T32GM007223 and K.W.B. by a National Science Foundation Graduate Research Fellowship DGE-1122492.
Abbreviations used:
- BiP
binding immunoglobulin protein
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HA
hemagglutinin
- KASH
Klarsicht, Anc-1, and Syne homology
- LAP1
lamina associated polypeptide 1
- LINC
linker of nucleoskeleton and cytoskeleton
- LULL1
lumenal domain like LAP1
- NE
nuclear envelope
- NPC
nuclear pore complex
- RT
room temperature
- SPB
spindle pole body
- SUN
Sad1 and UNc-84
- TorA
TorsinA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-09-0585) on January 9, 2019.
REFERENCES
- Adam I, Jossé L, Tuite MF. (2017). Human TorsinA can function in the yeast cytosol as a molecular chaperone. Biochem J , 3439–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. (1995). Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol , 1659–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. (2007). Determining the architectures of macromolecular assemblies. Nature , 683–694. [DOI] [PubMed] [Google Scholar]
- Atai NA, Ryan SD, Kothary R, Breakefield XO, Nery FC. (2012). Untethering the nuclear envelope and cytoskeleton: biologically distinct dystonias arising from a common cellular dysfunction. Int J Cell Biol , 634214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RS, Zhao C, Chase AR, Wang J, Schlieker C. (2014). The mechanism of Torsin ATPase activation. Proc Natl Acad Sci USA , E4822–E4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. (2000). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Chase AR, Laudermilch E, Wang J, Shigematsu H, Yokoyama T, Schlieker C. (2017). Dynamic functional assembly of the Torsin AAA+ ATPase and its modulation by LAP1. Mol Biol Cell , 2765–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Smoyer CJ, Slaughter BD, Unruh JR, Jaspersen SL. (2014). The SUN protein Mps3 controls Ndc1 distribution and function on the nuclear membrane. J Cell Biol , 523–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial HJ, Rout MP, Giddings TH, Winey M. (1998). Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J Cell Biol , 1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombi P, Webster BM, Fröhlich F, Lusk CP. (2013). The transmission of nuclear pore complexes to daughter cells requires a cytoplasmic pool of Nsp1. J Cell Biol , 215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol , 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demircioglu FE, Sosa BA, Ingram J, Ploegh HL, Schwartz TU. (2016). Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia. Elife , e17983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez Gonzalez B, Billion K, Rous S, Pavie B, Lange C, Goodchild R. (2018). Excess LINC complexes impair brain morphogenesis in a mouse model of recessive TOR1A disease. Hum Mol Genet , 2154–2170. [DOI] [PubMed] [Google Scholar]
- Emtage JL, Bucci M, Watkins JL, Wente SR. (1997). Defining the essential functional regions of the nucleoporin Nup145p. J Cell Sci (Pt 7), 911–925. [DOI] [PubMed] [Google Scholar]
- Friederichs JM, Ghosh S, Smoyer CJ, McCroskey S, Miller BD, Weaver KJ, Delventhal KM, Unruh J, Slaughter BD, Jaspersen SL. (2011). The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet , e1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Buchwalter AL, Naismith TV, Holbrook K, Billion K, Dauer WT, Liang CC, Dear ML, Hanson PI. (2015). Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J Cell Sci , 2854–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. (2005). The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol , 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT. (2005). Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron , 923–932. [DOI] [PubMed] [Google Scholar]
- Grillet M, Dominguez Gonzalez B, Sicart A, Pöttler M, Cascalho A, Billion K, Hernandez Diaz S, Swerts J, Naismith TV, Gounko NV, et al. (2016). Torsins are essential regulators of cellular lipid metabolism. Dev Cell , 235–247. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. (2005). AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol , 519–529. [DOI] [PubMed] [Google Scholar]
- Hewett J, Gonzalez-Agosti C, Slater D, Ziefer P, Li S, Bergeron D, Jacoby DJ, Ozelius LJ, Ramesh V, Breakefield XO. (2000). Mutant torsinA, responsible for early-onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum Mol Genet , 1403–1413. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Ghosh S. (2012). Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes. Nucleus , 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Giddings TH, Winey M. (2002). Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol , 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Martin AE, Glazko G, Giddings TH, Jr, Morgan G, Mushegian A, Winey M. (2006). The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J Cell Biol , 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ, Budnik V. (2013). Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep , 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth M, Dear ML, Brown P, Holbrook K, Goodchild R. (2010). Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum Mol Genet , 888–900. [DOI] [PubMed] [Google Scholar]
- Jungwirth MT, Kumar D, Jeong DY, Goodchild RE. (2011). The nuclear envelope localization of DYT1 dystonia torsinA-ΔE requires the SUN1 LINC complex component. BMC Cell Biol , 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta SS, Chen J, Gardner JM, Friederichs JM, Smith SE, Gogol M, Unruh JR, Slaughter BD, Jaspersen SL. (2015). Sec66-dependent regulation of yeast spindle-pole body duplication through Pom152. Genetics , 1479–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc , 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CE, Perez A, Perkins G, Ellisman MH, Dauer WT. (2010). A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc Natl Acad Sci USA , 9861–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustedjo K, Bracey MH, Cravatt BF. (2000). Torsin A and its torsin dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem , 27933–27939. [DOI] [PubMed] [Google Scholar]
- Laudermilch E, Tsai PL, Graham M, Turner E, Zhao C, Schlieker C. (2016). Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study. Mol Biol Cell , 3964–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, Burke B, Roux KJ. (2007). Functional association of Sun1 with nuclear pore complexes. J Cell Biol , 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- Lu W, Gotzmann J, Sironi L, Jaeger VM, Schneider M, Lüke Y, Uhlén M, Szigyarto CA, Brachner A, Ellenberg J, et al. (2008). Sun1 forms immobile macromolecular assemblies at the nuclear envelope. Biochim Biophys Acta , 2415–2426. [DOI] [PubMed] [Google Scholar]
- May CK, Carroll CW. (2018). Differential incorporation of SUN-domain proteins into LINC complexes is coupled to gene expression. Valastyan , e0197621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. (1996). GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell , 1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith TV, Dalal S, Hanson PI. (2009). Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J Biol Chem , 27866–27874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith TV, Heuser JE, Breakefield XO, Hanson PI. (2004). TorsinA in the nuclear envelope. Proc Natl Acad Sci USA , 7612–7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Zeng J, Niland BP, Hewett J, Farley J, Irimia D, Li Y, Wiche G, Sonnenberg A, Breakefield XO. (2008). TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci , 3476–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem , 4646–4658. [DOI] [PubMed] [Google Scholar]
- Nie S, Ke H, Gao F, Ren J, Wang M, Huo L, Gong W, Feng W. (2016). Coiled-coil domains of SUN proteins as intrinsic dynamic regulators. Structure , 80–91. [DOI] [PubMed] [Google Scholar]
- Ogura T, Whiteheart SW, Wilkinson AJ. (2004). Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol , 106–112. [DOI] [PubMed] [Google Scholar]
- Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K. (2017). Natively unfolded FG repeats stabilize the structure of the nuclear pore complex. Cell , 904–917.e919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de Leon D, Brin MF, Raymond D, Corey DP, et al. (1997). The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet , 40–48. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. (2005). The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci , 3419–3430. [DOI] [PubMed] [Google Scholar]
- Rebelo S, da Cruz E, Silva EF, da Cruz E, Silva OA. (2015). Genetic mutations strengthen functional association of LAP1 with DYT1 dystonia and muscular dystrophy. Mutat Res Rev Mutat Res , 42–47. [DOI] [PubMed] [Google Scholar]
- Ryan KJ, Wente SR. (2002). Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet , 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüthnick D, Neuner A, Dietrich F, Kirrmaier D, Engel U, Knop M, Schiebel E. (2017). Characterization of spindle pole body duplication reveals a regulatory role for nuclear pore complexes. J Cell Biol , 2425–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CA, Harris NJ, Willey PT, Woolums BM, Wang Y, McQuown AJ, Schoenhofen A, Worman HJ, Dauer WT, Gundersen GG, Luxton GW. (2017). TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J Cell Biol , 657–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A, Gerace L. (1988). Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J Cell Biol , 2029–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezen B, Seedorf M, Schiebel E. (2009). The SESA network links duplication of the yeast centrosome with the protein translation machinery. Genes Dev , 1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics , 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. (1996). A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell , 265–275. [DOI] [PubMed] [Google Scholar]
- Sosa BA, Demircioglu FE, Chen JZ, Ingram J, Ploegh HL, Schwartz TU. (2014). How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife , e03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. (2012). LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell , 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamas JA, Hetzer MW. (2011). POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol , 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe LM, Liang CC, Dauer WT. (2016). Neuronal nuclear membrane budding occurs during a developmental window modulated by Torsin paralogs. Cell Rep , 3322–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller DJ, Lusk CP. (2018). Fantastic nuclear envelope herniations and where to find them. Biochem Soc Trans , 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, et al. (2017). Molecular architecture of the major membrane ring component of the nuclear pore complex. Structure , 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan JS, Lindquist S. (2011). TorsinA and the torsinA-interacting protein printor have no impact on endoplasmic reticulum stress or protein trafficking in yeast. PLoS One , e22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J. (2005). Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast , 1061–1068. [DOI] [PubMed] [Google Scholar]
- Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. (2009). LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell , 2661–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGompel MJ, Nguyen KC, Hall DH, Dauer WT, Rose LS. (2015). A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol Biol Cell , 1752–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jäger J, Lusk CP. (2014). Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell , 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Lusk CP. (2016). Border safety: quality control at the nuclear envelope. Trends Cell Biol , 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Thaller DJ, Jäger J, Ochmann SE, Borah S, Lusk CP. (2016). Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J , 2447–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. (1993). A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol , 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. (1994). NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol , 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Hoyt MA, Chan C, Goetsch L, Botstein D, Byers B. (1993). NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J Cell Biol , 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Blobel G, Rout MP. (1994). POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J Cell Biol , 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Bastian RW, Perlmutter JS, Racette BA, Tabbal SD, Karimi M, Paniello RC, Blitzer A, Batish SD, Wszolek ZK, et al. (2009). High-throughput mutational analysis of TOR1A in primary dystonia. BMC Med Genet , 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li W, Ke H, Feng W. (2018). Structural conservation of the autoinhibitory domain in SUN proteins. Biochem Biophys Res Commun , 1337–1343. [DOI] [PubMed] [Google Scholar]
- Yewdell WT, Colombi P, Makhnevych T, Lusk CP. (2011). Lumenal interactions in nuclear pore complex assembly and stability. Mol Biol Cell , 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. (1996). Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol , 1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi LF, Dittmar JC, Mihalevic MJ, Shewan AM, Schulz BL, Brodsky JL, Bernstein KA. (2017). Early-onset torsion dystonia: a novel high-throughput yeast genetic screen for factors modifying protein levels of torsinAΔE. Dis Model Mech , 1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi LF, Wu HC, Bell SL, Millen L, Paton AW, Paton JC, Thomas PJ, Zolkiewski M, Brodsky JL. (2014). The BiP molecular chaperone plays multiple roles during the biogenesis of torsinA, an AAA+ ATPase associated with the neurological disease early-onset torsion dystonia. J Biol Chem , 12727–12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Neuner A, Rüthnick D, Sachsenheimer T, Lüchtenborg C, Brügger B, Schiebel E. (2018). Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J Cell Biol , 877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Brown RS, Chase AR, Eisele MR, Schlieker C. (2013). Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci USA , E1545–E1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Millen L, Mendoza JL, Thomas PJ. (2010). A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J Biol Chem , 37271–37280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirn B, Grundmann K, Huppke P, Puthenparampil J, Wolburg H, Riess O, Müller U. (2008). Novel TOR1A mutation p.Arg288Gln in early-onset dystonia (DYT1). J Neurol Neurosurg Psychiatry , 1327–1330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.