Abstract
Lipid droplets (LDs) in all eukaryotic cells are coated with at least one of the perilipin (Plin) family of proteins. They all regulate key intracellular lipases but do so to significantly different extents. Where more than one Plin is expressed in a cell, they associate with LDs in a hierarchical manner. In vivo, this means that lipid flux control in a particular cell or tissue type is heavily influenced by the specific Plins present on its LDs. Despite their early discovery, exactly how Plins target LDs and why they displace each other in a “hierarchical” manner remains unclear. They all share an amino-terminal 11-mer repeat (11mr) amphipathic region suggested to be involved in LD targeting. Here, we show that, in vivo, this domain functions as a primary highly reversible LD targeting motif in Plin1–3, and, in vitro, we document reversible and competitive binding between a wild-type purified Plin1 11mr peptide and a mutant with reduced binding affinity to both “naked” and phospholipid-coated oil–water interfaces. We also present data suggesting that a second carboxy-terminal 4-helix bundle domain stabilizes LD binding in Plin1 more effectively than in Plin2, whereas it weakens binding in Plin3. These findings suggest that dual amphipathic helical regions mediate LD targeting and underpin the hierarchical binding of Plin1–3 to LDs.
INTRODUCTION
Lipid droplets (LDs) form in almost all cell types and constitute the primary organelles for lipid storage. They consist of a neutral lipid core surrounded by an amphipathic phospholipid monolayer (Fujimoto and Parton, 2011; Tauchi-Sato et al., 2002) that reduces surface tension at the oil–water (cytosol) interface (Thiam et al., 2013b). This unique surface topology is recognized by a host of proteins involved in LD formation and fate (Guo et al., 2008; Pol et al., 2014; Kory et al., 2016; Ben M’barek et al., 2017; Thiam and Beller, 2017; Thul et al., 2017). Among these proteins, those directly associated with the droplet surface typically use amphipathic helices (AHs), lipid anchors, or monotopic hairpins to do so (Kory et al., 2016; Bersuker and Olzmann, 2017).
The most abundant LD coat protein in mammalian cells is the perilipin (Plin) family of five proteins, Plin1-5 (Greenberg et al., 1991). All Plins share a series of 11-mer repeats (11mr) toward the amino terminal that are involved in LD targeting through their ability to form AHs (McManaman et al., 2003; Nakamura and Fujimoto, 2003; Targett-Adams et al., 2003; Orlicky et al., 2008; Bulankina et al., 2009; Rowe et al., 2016; Copic et al., 2018). Plin1–3 and 5 share similar 11mr motifs of ∼100 amino acids, whereas Plin4 is distinguished by more copies of the 11mrs, up to 1200 amino acids in length (Copic et al., 2018). Toward their carboxyl terminus, all Plins are predicted to have a 4-helix bundle (4HB) domain (Chong et al., 2011), based on the homology of this region in Plins 1, 2, 4, and 5 compared with that of Plin3, for which the crystal structure was resolved many years ago (Hickenbottom et al., 2004). In Plin3, the 4HB is zipped together by four short β-sheets (Hickenbottom et al., 2004).
Helix bundles conceal hydrophobic residues of helices from the aqueous environment, but the bundle can open up, thereby deploying amphipathic helical structures that can then associate with lipid membranes (Narayanaswami et al., 2010). This behavior suggests that the 4HB of Plins could also be involved in LD binding, and some experimental support for the involvement of these regions of Plins exists (Garcia et al., 2003; McManaman et al., 2003; Subramanian et al., 2004; Chong et al., 2011; Rowe et al., 2016).
What then do Plins do once associated with LDs? They probably all have a role in reducing surface tension and stabilizing LDs, but they are also key regulators of lipid fluxes into and out of LDs. By decorating the surface of LDs, they are ideally placed to regulate the access of hydrolytic lipases, such as adipose tissue triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) to their lipid substrates in the core or possibly on the surface of LDs, and several elegant studies have elucidated the mechanisms by which Plins regulate ATGL and HSL (Granneman et al., 2007; Wang et al., 2009a, 2011; Kimmel et al., 2010; Kimmel and Sztalryd, 2016; Brasaemle and Wolins, 2012; Zechner et al., 2012; Itabe et al., 2017). Studies have also now suggested that exactly how Plins regulate these lipases differs (Granneman et al., 2009; Wang et al., 2011; Patel et al., 2014). This has important physiological implications, as where multiple Plins are present in cells, they bind in a hierarchical manner (Wolins et al., 2006; Straub et al., 2008; Beller et al., 2010). For example, in white adipocytes, Plin1 is ultimately the only Plin at the LD surface, while other Plins are expressed and associate with the LD surface at earlier stages of droplet formation (Wolins et al., 2006; Skinner et al., 2009; Brasaemle and Wolins, 2012). This is important physiologically, as evidenced by the phenotype of Plin1-null mice, in which basal lipolysis is increased and stimulated lipolysis relatively blunted (Martinez-Botas et al., 2000; Tansey et al., 2001). In humans with loss-of-function Plin1 mutations, Plin2 is up-regulated in white adipose tissue (Kozusko et al., 2015), but it fails to alleviate the severe metabolic consequences of impaired Plin1 function (Gandotra et al., 2011; Kozusko et al., 2015). Plin2 and 3 are widely expressed, and mouse knockout studies suggest that Plin2 is particularly important in hepatocytes, which do not normally express Plin1 (Libby et al., 2016). Plin3 is reported to localize to droplets emerging from the endoplasmic reticulum (ER) (Skinner et al., 2009; Pol et al., 2014), and a recent report suggests that Plin3-null mice manifest increased beiging of white adipocytes (Lee et al., 2018).
Given the physiological importance of the hierarchical binding of Plin1–3 to LDs, we sought to understand the molecular basis of this phenomenon. Consistent with works from several groups (McManaman et al., 2003; Nakamura and Fujimoto, 2003; Targett-Adams et al., 2003; Orlicky et al., 2008; Bulankina et al., 2009; Rowe et al., 2016), we found that the amino-terminal 11-mer AH repeat regions of Plin1–3 are sufficient for LD localization. However, importantly, we found that this binding is highly reversible for all three Plins. In contrast, the binding of the C-terminal 4HB-containing regions varies more substantially and either stabilizes or seems to destabilize net binding of the full-length proteins. Thus, we propose a cooperative binding mechanism that accounts for the observed hierarchical binding properties of this primordial family of LD coat proteins and is critical for optimal in vivo co-ordination of lipid storage and release.
RESULTS
Comparative localization of Plin1–3 in mammalian cell lines
To directly compare the localization of Plin1–3, we studied the localization of amino-terminal fluorescently (green fluorescent protein [GFP] or mCherry) tagged full-length (FL) Plins, as well as complementary fragments of the proteins containing their predicted 11mr and 4HB domains, respectively denoted Plin-N and Plin-C (Figure 1A). The Plin-N and Plin-C boundaries were designed based on the known structure of the 4HB of Plin3 (Hickenbottom et al., 2004) and our own prior characterization of the 11mr regions of Plins 1-3 (Rowe et al., 2016). In the case of Plin-N, each of these constructs also included the putative PAT domain, and in the case of the Plin-C domains, each included the full carboxy terminus of the proteins as well. Binding of these domains was primarily studied in oleate-loaded human hepatoma–derived (Huh7) and HeLa cells in which only Plins2 and 3 were endogenously expressed, Plin1 being mainly expressed in adipocytes (Supplemental Figure S1A).
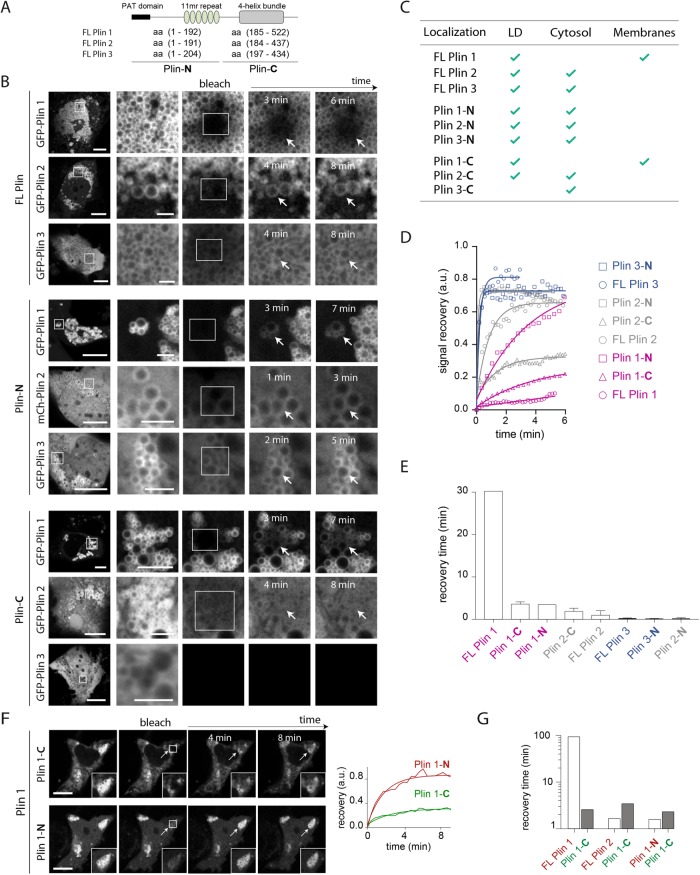
FIGURE 1:
(A) Simplified schematic illustration of the 11mr and 4HB domains of Plin1–3 alongside their respective amino acid numbers. PAT domain refers to the conserved amino-terminal region of the protein (black rectangle). Amino acids from 1 to 192, 191, and 204, respectively, in Plin1–3 are termed Plin(1–3)-N. Amino acids from 193, 192, and 205 to the end of each protein, respectively, in Plin1–3 are termed Plin(1–3)-C. (B) FRAP analysis of FL Plins and their complementary fragments containing the predicted 11mr and 4HB domains in a mammalian Huh7 cell line. The protein constructs were fused to GFP on the amino-terminus, except for Plin2-N, which had an mCherry tag instead. The left-hand panel shows the steady-state localization of each protein before photobleaching. The inset squares indicate the bleached region in the second panel from the left, and then subsequent panels show this region at the indicated times thereafter. Representative image sequence is shown. Each experiment was repeated at least three times. Scale bars: 10 µm. (C) Relative steady-state localization (LD/cytosol/membranes) of tagged Plin1–3 and fragments in Huh7 cells. We considered proteins to be 1) present on LDs when they formed a fluorescent ring around LDs; 2) cytosolic when they were associated with a diffuse fluorescence signal in the cytoplasm; and 3) membrane associated when they were associated with a reticular fluorescence signal in the cytoplasm—subsequent analyses showed that the reticular fluorescence signal colocalized with the ER marker Sec61. A minimum of 20 cells was analyzed for each construct to validate localization of the proteins. (D) Quantitative analysis of the recovery kinetics of Plins and fragments thereof to the LD surface following FRAP. Data are normalized to both pre- and postbleach intensities. Curves are exponential fits of the data. Each FRAP experiment was repeated three times, and representative recovery rates are shown. (E) Histogram of characteristic recovery times of the different peptides for experiments in B and D. The characteristic time “tau,” referred to herein as “recovery time,” is obtained by the following exponential fit: 1 − exp(−t/tau). The value of tau is the average value of all experiments performed in B and D. (F) Representative images of a FRAP image sequence of the GFP-tagged Plin1-C and mCherry-tagged Plin1-N when coexpressed in Huh7 cells. Scale bars: 10 µm. The normalized fluorescence recovery of the bleached LD cluster is shown over time. Curves correspond to exponential fits. (G) Histogram of the characteristic recovery times of the coexpressed GFP-Plin1-C, respectively, with mCherry-Plin1 and 2 and Plin 1-N. Black bars correspond to the recovery time of Plin 1-C in dual FRAP analysis against FL Plin1, FL Plin2, and Plin1-N that correspond to white bars. This experiment was repeated three times, and the value of tau corresponds to the representative images/quantification shown.
All proteins localized to the LD surface to at least some extent, except for Plin3-C, which was observed only in the cytosol (Figure 1B and Supplemental Figure S1B). Whereas FL Plin1 was almost entirely localized around LDs and on the ER membranes (Supplemental Figure S1, D and F), FL Plin2 also displayed some cytosolic localization, and FL Plin3 was predominantly cytosolic (Figure 1B). Regarding the fragments, in addition to targeting LDs, they were all soluble to some extent in the cytosol and in the nucleus, especially Plin3-N (Supplemental Figure S1C); the only exception being Plin1-C, which was either observed on LDs or ER membrane (Supplemental Figure S1, B and E), like FL Plin1. In contrast, Plin2-C colocalized with LDs in a minority of cells but was otherwise predominantly cytosolic (Supplemental Figure S1, B and C). These data are summarized in Figure 1C.
For validation of these data in a different cell line, all of the GFP-tagged constructs were also expressed in a second mammalian cell line (HeLa) in which similar localization patterns were observed (Supplemental Figure S1G).
Differential binding stability of Plin1–3 on LDs
To assess the stability of Plins and fragments thereof on the surface of LDs, we performed fluorescence recovery after photobleaching (FRAP) analyses by bleaching the fluorescence signal in a voxel incorporating a cluster of LDs and then recording its recovery (Figure 1B). From the observed recovery rates, we distinguished three patterns (Figure 1, D and E): 1) proteins that recovered rapidly included FL Plin3 and Plins2-N and 3-N; 2) FL Plin2 and Plin1-N had intermediate recovery times; 3) Plins1-C, 2-C, and particularly FL Plin1 had the slowest recovery rates. The data suggest that FL Plin1 is considerably more stably associated with the LD surface than FL Plin2, which is in turn more stably associated than FL Plin3. The data are consistent with overall affinity reflecting the combined binding stability of the Plin1–3 fragments. For instance, for Plin2 and Plin3, whose N-terminals manifest similar recovery rates (Figure 1, D and E), the C-terminal containing the 4HB ultimately determines the differential LD binding affinity between the FL proteins. In fact, FL Plin3 behaved very similarly to Plin3-N alone, in keeping with the suggestion that Plin3-C does not associate with the droplet surface.
We recorded slower recovery rates for Plin1-C and 2-C than their corresponding N-terminals containing the 11mrs, which suggests that they both stably bind LDs and might have slow “on” and/or “off” rates. However, these 4HB-containing domains manifested different net behaviors: Plin2-C was predominantly cytosolic and only bound to LDs in a minority of cells, whereas Plin1-C mostly bound to LDs and ER membrane (Figure 1C and Supplemental Figure S1, B–F). We hypothesize that the 4HB of Plin3 is stabilized or “zipped together” by the β-sheets in the associated αβ domain and thus remains largely in the closed conformation seen in its crystal structure (Hickenbottom et al., 2004) (Supplemental Figure S2A). The β-sheets are also predicted to be present in Plin2, whereas they are not present in Plin1 (Supplemental Figure S2B). Consequently, we speculate that the 4HB of Plin1 might have a greater propensity to unfold and hence afford Plin1-C a higher membrane/LD on rate than that of Plin2-C, but both could have low off rates.
The high LD and membrane affinity of Plin1-C was striking. To confirm this result, we coexpressed Plin1-C with Plin1-N (Figure 1F), FL Plin1 (Supplemental Figure S2C), or FL Plin2 (Supplemental Figure S2D) and performed dual FRAP analyses. These data confirmed that Plin1-C associated with LDs more stably than all the other peptides/proteins besides FL Plin1 (Figure 1G).
Evidence indicating that Plin1–3 compete for LD surface occupancy in vivo
The FRAP analyses provide trends of the relative LD binding affinities of the various peptides studied (Figure 1, D and E). However, a limitation of this analysis is that the sources of the signal recovery are not necessarily identical for all conditions. Signal recovery could occur by protein exchange between LDs, or directly from the cytosol, or possibly by lateral diffusion from the ER to LDs. Additionally, precise control over the protein expression levels in individual cells is lacking (Goedhart et al., 2011). Nevertheless, we took advantage of the variability of cellular expression levels of the proteins to document their competitive binding in coexpression experiments (Kory et al., 2015). In these experiments, cells were transfected with the different plasmids and expressed the proteins at different levels. We then took cells with variable amounts of each expressed protein and analyzed them.
Initially, we coexpressed FL mCherry-Plin1, as a reference protein, with GFP-tagged versions of FL Plin2 (Figure 2A), FL Plin3, Plin1-N, or the Plin1-C region (Figure 2, B–D). We quantified the loss of the GFP-tagged fragments as a fraction of the total fluorescence present on the LD surface and used the resulting profile to estimate a critical concentration of the FL Plin1 on LDs required to displace half of the competing protein (see Figure 2, A–D, graphs). Plin1 completely displaced all other proteins from LDs at high expression levels, to a greater extent than any other FL Plins or fragments thereof. Switching the GFP and mCherry tags on FL Plin1 and 2 led to very similar results (Supplemental Figure S2E); endogenous Plin2 and 3 levels are not affected by such manipulations (Supplemental Figure S2F) and may behave as GFP-Plin2 and GFP-Plin3. All competing proteins were displaced into the cytosol, except for Plin1-C, which relocated to the ER membrane (Figure 2D). These experiments corroborate the previous data, insofar as they suggest that Plin1 competes with the other peptides with a range of displacement efficiencies that are similarly ordered (Figure 2, E and F) as in the single FRAP analysis studies (Figure 1). These findings were also in agreement with dual FRAP analyses in cells coexpressing FL Plin1 and FL Plin2 (Supplemental Figure S3A) or the Plin1 fragments (Supplemental Figure S3, B–D).
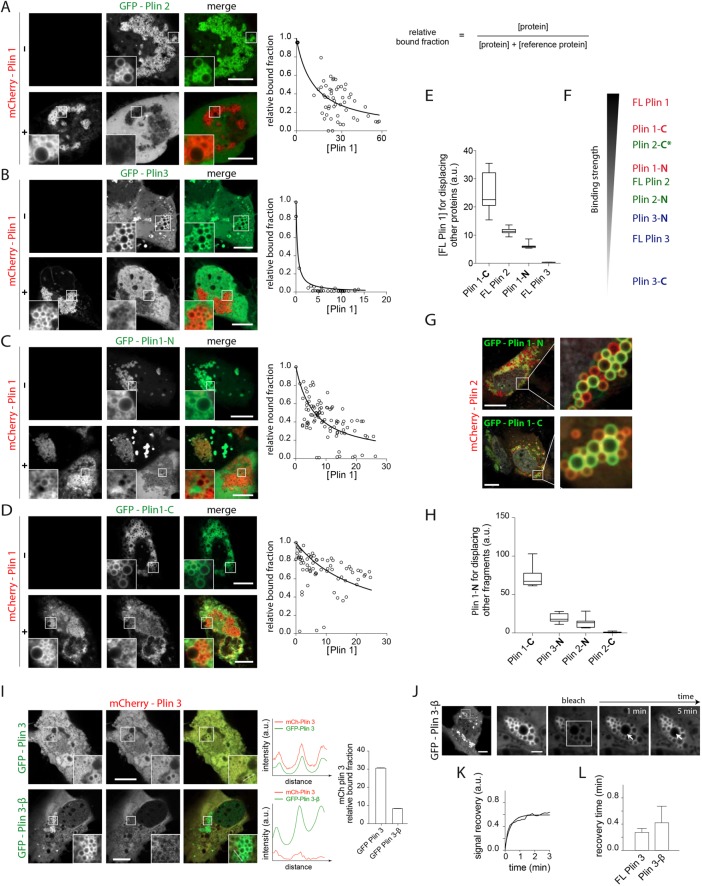
FIGURE 2:
mCherry-FL Plin1 coexpression and LD colocalization with GFP-tagged versions of (A) FL Plin2, (B) FL Plin3, and (C, D) Plin1 fragments in Huh7 cells. The inset squares indicate the colocalization region of corresponding proteins to the LD surface. Scale bars: 10 µm. This experiment was repeated at least three times with more than 15 cells analyzed for each condition. The relative bound fraction level is reported in the right panel and indicates the relative amount of a protein bound to LDs when mCherry-FL Plin1 (the reference protein) is expressed at a given level; it is calculated as follows: [protein]/([protein] + [reference protein]), and represented against [reference protein]. Each experimental dot corresponds to an average of the signal on 10–20 LDs. (E) The critical concentration of FL Plin1 required for displacing half of the competing proteins is reported. Plin1-C required much more FL Plin1 to be displaced, as compared with FL Plin2 and 3 or Plin1-N. Results are presented as box-and-whisker plots of the critical concentration assessed from at least three different sets of data. The central box represents the interquartile ranges (25th to 75th percentile), the middle line represents the median, and the horizontal lines represent the minimum and the maximum value of observation range. Values are expressed as median ± IR. (F) Classification of the relative binding strength to the LD surface of Plins and fragments thereof based on their recovery rates from FRAP and competition experiments. Plin 2-C, labeled with the asterisk, constitutes a particular case, as it has a low LD on rate, suggested from coexpression experiments, and a high off rate, observed from the FRAP experiment. (G) mCherry-FL Plin2 coexpression and colocalization with GFP-tagged versions of the Plin1 fragments. Plin2 is not fully displaced by the fragments and displays frequent differential LD enrichment with the fragments. The experiments were repeated twice and three times, respectively. Scale bars: 10 µm. (H) Critical concentrations for the displacement of proteins competing against mCherry-Plin1-N, shown in Supplemental Figure S3H. Plin2-N and Plin3-N were displaced by similar concentrations, whereas Plin1-C required considerably more Plin1-N to displace it. Plin2-C was barely detected on LDs in this coexpression experiment. Results are presented as box-and-whisker plots of the critical concentration obtained from at least three sets of data. (I) GFP-Plin3-β (without β-sheets) coexpression and LD colocalization with mCherry-FL Plin3 in Huh7 cells. FL Plin3 is readily displaced from LDs when coexpressed with GFP-Plin3-β. The inset squares indicate the colocalization region of corresponding proteins to the LD surface. In control experiments, GFP-Plin3 did not displace mCherry-Plin3 as observed with GFP-Plin3-β. Right panel shows the intensity profile of each line section drawn in the inset. Signal on LDs is displayed as peaks; mCherry-Plin3 is almost absent on LDs only when coexpressed with GFP-Plin3-β. Far right panel compares the relative bound fraction of mCherry-Plin3 when coexpressed at similar level with GFP-Plin3 or GFP-Plin3-β. Scale bars: 10 µm. (J) FRAP analysis of GFP-Plin3-β in a Huh7 cell. A representative image sequence is shown with typical recovery kinetic (K). FRAP experiment was repeated three times. Scale bars: 10 µm. (L) Histogram of characteristic recovery times of FL Plin3 and Plin3-β in individual FRAP experiments. The characteristic recovery times correspond to means ± SD.
We also observed intermediate situations in which Plin1 and Plin2 were enriched on different LDs, suggesting that Plin2 displacement by Plin1 might occur gradually on different LDs (Supplemental Figure S3E). We next assessed the binding affinity between FL Plin2 and FL Plin3 (Supplemental Figure S3F). As predicted, Plin2 fully displaced Plin3. FL Plin3 was also displaced by Plin1-N to level similar to that of FL Plin2; it was even more efficiently displaced by Plin1-C (Supplemental Figure S3, F and G), confirming the highest LD binding affinity of this 4HB-containing domain after FL Plin1 (Figure 2F). We then probed whether the individual domains of Plin1 were sufficient to fully displace FL Plin2, but the data suggest that these domains cannot do so. Instead, we found that FL Plin2 was either on the same LDs as the fragments or there were distinct LDs enriched with either FL Plin2 or the Plin1 fragments (Figure 2G). Such separation was not seen between Plin1–3 fragments (Supplemental Figure S3H), suggesting that the N-11mr– and the C-4HB–containing domain jointly regulate Plin binding to LD subsets.
The FRAP analysis of the individually expressed Plin1-C and 2-C fragments (summarized in Figure 1D) suggests similar recovery rates, whereas their steady-state localization is very different (Figure 1B and Supplemental Figure S1, C and E). To explore this in more detail, we used mCherry-Plin1-N as the reference protein and coexpressed it with GFP-tagged versions of the other fragments that localized to LDs (Supplemental Figure S3H). Higher expression of Plin1-N displaced the other fragments from the LD surface to some extent. Plin1-C tended to remain at the interface and had the highest critical concentration (Supplemental Figure S3H), whereas Plin2-C was barely detectable on the LD surface when coexpressed with Plin1-N (Supplemental Figure S3H), or even with Plin3-N or FL Plin3 (Supplemental Figure S3I); for this Plin2-4HB–containing domain, crowding the LD surface with proteins seems to be sufficient to fully prevent binding to the droplet surface, consistent with our previous suggestion that the 4HB-containing domain of Plin2 has a much lower LD on rate than that of Plin1. Regarding the 11mr-containing domains of Plin2 and 3, they were more readily displaced than Plin1-C, and at very similar levels (Figure 2H).
In summary, the kinetic (FRAP data in Figure 1) and steady-state (Figure 2) data suggest that the 11mr-containing domains of Plin1 and 2 have similar binding affinities, both apparently higher than that of Plin3. The 4HB-containing domain of Plin3 does not bind LDs on its own in either of the cell types we studied. The 4HB-containing domain of Plin2 only associates with LDs when LD surface protein coverage allows it to do so, whereas that of Plin1 is almost always associated with membranes.
A truncation mutant of Plin3-C manifests enhanced LD localization
The 4HB structure of Plin3 is zipped together by short β-sheets (Supplemental Figure S2A; Hickenbottom et al., 2004). To test the hypothesis that this prevents unfolding and subsequent membrane association of the helix bundle, we generated a truncated version of Plin3 (Plin3 1-413, referred to herein as Plin3-β) lacking the β-sheets, which follow the sequences for the helices constituting the 4HB and zip the helix bundle together. When expressed in oleate-loaded Huh7 cells, GFP-Plin3-β consistently localized around LDs to a greater extent than GFP-Plin3, whose signal was often very faint around the LDs (Supplemental Figure S2H). When coexpressed with mCherry-Plin3, GFP-Plin3-β almost entirely displaced mCherry-Plin3 from LDs (Figure 2I and Supplemental Figure S2I), strongly suggesting that the Plin3-β mutation enhances LD surface association. This observation is not a result of the GFP/mCherry tags themselves, as GFP-Plin3 largely just colocalized with coexpressed mCherry-Plin3 (Figure 2I). This result is consistent with the suggestion that the β-sheets in the 4HB of Plin3 influence unfolding and hence the propensity to associate with hydrophobic membranes. However, in individual FRAP experiments, Plin3-β recovery was rapid and similar to that of FL Plin3 (Figure 2, J–L). We deduce that the Plin3-β mutation only marginally increases LD binding affinity and that this difference is below the resolution of the FRAP analysis in this setting.
In vitro analysis of the binding mode of Plins on an oil–water interface
Theoretically, a protein with both membrane binding and soluble domains has the same membrane lateral diffusivity as the binding domain alone, because the viscosity of the membrane is higher than that of the aqueous phase (Supplemental Text) (Figure 3A), but the soluble domain of the protein might enhance membrane “fall off” the protein into the aqueous phase (Figure 3A). If, however, the soluble domain also interacts with the membrane, the diffusivity of the protein will be reduced, and this additional membrane binding domain is also predicted to reduce the protein membrane fall-off (Figure 3A). We hypothesize that the 11mr-containing domains of Plin1–3 and Plin1–3-N constitute the primary LD localization motif and that this LD association is consistently stabilized by the 4HB-containing domain in Plin1, under optimal conditions in Plin2, but not in Plin3, at least under the conditions we have tested.
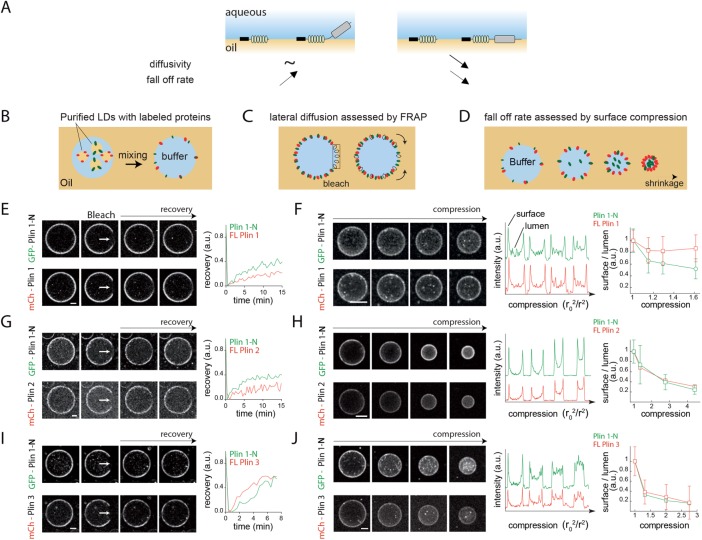
FIGURE 3:
(A) Schematic illustration of Plin binding to LDs. The FL protein contains a membrane binding domain and a second soluble domain that potentially interacts with the membrane, which would slow down diffusion and the fall-off rate. (B) Schematic illustration of the in vitro system. Purified LDs in buffer are mixed with TO to generate buffer in oil emulsion droplets. LD protein content is relocated in this manner to the resulting oil–water interface. (C) Schematic illustration of the lateral mobility of the proteins studied by photobleaching. (D) Schematic illustration of membrane fall-off in shrinking buffer in oil droplets containing two differently labeled proteins at the droplet interface. (E) Lateral recovery of mCherry-FL Plin1 and GFP-Plin1-N at the oil–water interface of an artificial droplet as sketched in C. Representative image sequences are shown. Scale bar: 30 µm. Mean fluorescence recovery of the bleached drop surface area over time is shown (right). The experiment was reproduced three times, and a representative situation is shown. (F) Representative images of mCherry-FL Plin1 and GFP-Plin1-N fall-off from the oil–water interface during shrinkage of the artificial drop (as sketched in D). Scale bar: 30 µm. Fluorescence intensity profiles (middle) in the equatorial focal plane of the artificial droplet are plotted against the droplet compression factor, (r2 (time 0)/r2 (respective time point); r = drop radius). In the far right panel, the mean ± SD surface/lumen signal during compression is reported. The experiment was reproduced three times, and a representative situation is shown. (G) Lateral recovery rates of mCherry-FL Plin2 or mCherry-FL Plin3 (I) compared with the GFP-Plin1-N at the oil–water interface are reported. Representative images are shown. Scale bar: 30 µm. Mean fluorescence recovery on the droplet bleached surface area over time is shown (right). The experiments were reproduced three times for G and twice for I. (H) mCherry-FL Plin2 or mCherry-FL Plin3 (J) and GFP-Plin1-N fall-off during shrinkage are shown. Scale bars: 30 µm. Fluorescence intensity profiles are plotted against the drop compression factor (middle). In the far right panel, the mean surface/lumen signal ± SD during compression is reported. The experiments were reproduced more than three times for H and at least twice for J.
To test this hypothesis, the GFP- or mCherry-tagged Plin1-N or FL Plin1–3 was individually expressed in cells, and LDs bound by these proteins were then purified. This isolated LD fraction was then encapsulated in buffer in triolein (TO) artificial emulsion droplets and mixed (Figure 3B); this manipulation resulted in the transfer of the proteins to the buffer in oil inverted droplet surface, as described previously (Kory et al., 2015) (Figure 3B). FRAP analysis was then used to probe the lateral mobility of GFP- and mCherry-tagged proteins (Figure 3C), and droplet binding stability was evaluated by comparing the tendency of the proteins to transfer off the buffer–oil interface into the buffer lumen in shrinking droplets (due to evaporation of water) (Figure 3D).
FL mCherry-Plin1 recovered more slowly than GFP-Plin1-N, confirming that Plin1-C contributes to the binding (Figure 3E). This result was supported by comparing the surface-bound fraction in shrinking buffer in oil droplets with both peptides at their surfaces (surface/lumen signal in Figure 3F): FL Plin1 remained at the interface, while the surface concentration of Plin1-N decreased. These two sets of experiments were repeated with the same peptides but with switched tags to show that the GFP and mCherry tags did not influence the data (Supplemental Figure S4, A–D). FL Plin2 also diffused more slowly than Plin1-N, suggesting that its 4HB is also involved in binding to the artificial oil–water interface (Figure 3G). In this experiment, we think that the 4HB in Plin2 does unfold and localize at this naked interface, which is optimal for the binding of Plin2-C. However, during surface shrinkage (Figure 3H), Plin2 fell off the interface at a level similar to that of Plin1-N (Figure 3H). These data suggest that surface shrinkage increased protein crowding, which favored refolding of the 4HB in Plin2-C and solubilization of FL Plin2 within the aqueous droplet lumen.
FL Plin3 behaved similarly to Plin1-N, supporting our previous observations that its 4HB is not involved in LD binding (Figure 3, I and J). Finally, we also used Plin1-C as the reference protein and compared it with FL Plin1 and FL Plin2 (Supplemental Figure S4, C–F). The data were similar to what we had seen with Plin1-N, for example, FL Plin1 and 2 diffuses more slowly on the initially naked oil–water interface than the single 4HB domain, but droplet shrinkage causes similar fall-off rates for this domain and FL Plin2.
Collectively, these data affirm the suggestion that the 11mrs in Plin-N sense the droplet surface and that the 4HBs in Plin-C regulate the relative binding affinity and stability of Plin1–3 on LDs, thus accounting, at least in part, for the observed net hierarchical binding of Plin1–3.
The 11mr of Plin1 associates reversibly with an artificial oil–water interface
The involvement of the 11mr regions in LD localization of all known Plins is increasingly well established by several independent groups, including our own (McManaman et al., 2003; Nakamura and Fujimoto, 2003; Targett-Adams et al., 2003; Orlicky et al., 2008; Bulankina et al., 2009; Rowe et al., 2016; Copic et al., 2018). We also showed that these regions could mediate targeting of human Plin1–3 to LDs in Saccharomyces cerevisiae, which strongly suggests that this process did not require interaction with additional proteins. Copic et al. (2018) have very recently reported similar findings using 11mr regions of Plin4. They also showed that purified fragments of the Plin4 11mr regions could associate with LDs in vitro. We previously purified a fragment of Plin1 from amino acids 93–192, which includes its predicted 11mr region (Figure 1A), and showed that this domain is unstructured in solution but folds into α-helices in the presence of detergent micelles (Rowe et al., 2016). Its behavior was compared with that of a mutant in which one neutral leucine residue was substituted with a negatively charged aspartate residue (L143D) on the hydrophobic face of the predicted AH.
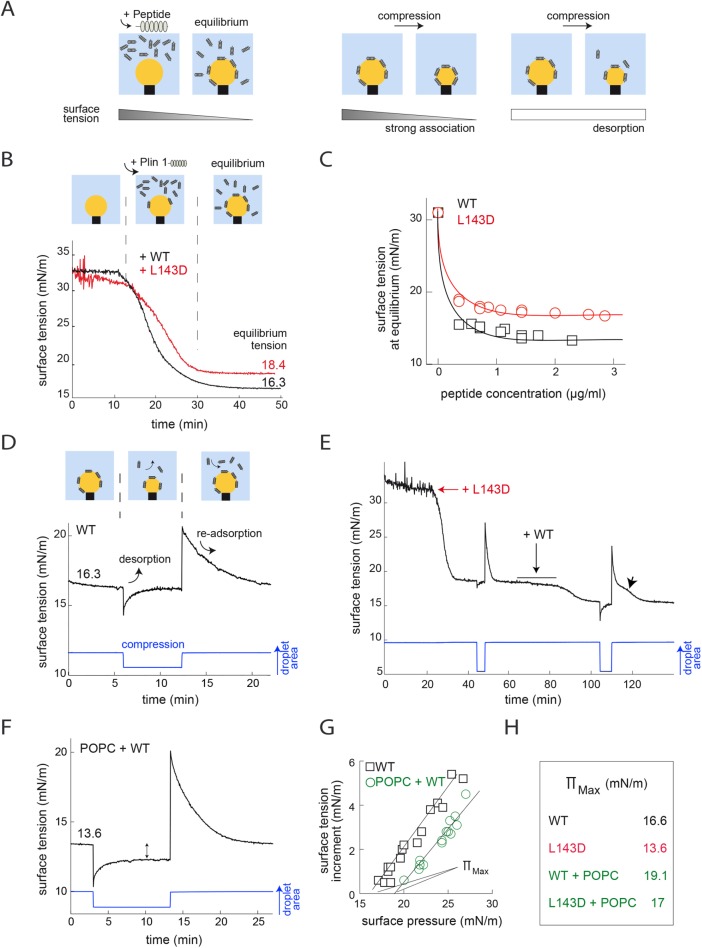
Lipoprotein particles are physically similar to lipid droplets in terms of having a central neutral lipid core surrounded by a phospholipid monolayer. In this case, apolipoproteins associate with the lipoprotein surface (and its core in some instances), where they regulate lipases in a manner highly analogous to that of the Plins. How they interact with the droplet interface has been extensively characterized using oil droplet tensiometry (Small et al., 2009; Mitsche and Small, 2011; Meyers et al., 2012; Rathnayake et al., 2014). Given these similarities, we sought to document the behavior of the wild-type (WT) and L143D mutant 11mr region of Plin1 in this experimental paradigm (Figure 4A). Initially, a naked TO in water droplet was generated, with an interfacial tension of ∼32 mN/m—this value translates an energy cost per unit surface, thus a tension, of oil in contact with water. Equal amounts of the WT or L143D peptides were then added to the solution, and surface tension fell from ∼32 mN/m to an equilibrium tension of ∼16.3 mN/m for WT and 18.4 mN/m for L143D, meaning that the peptides adsorbed to the interface to reach an equilibrium density (Figure 4B)—by absorbing to the interface, the peptides masked the interface, which decreased the tension at the oil–water interface. Varying the peptide concentrations revealed that equilibrium tension was consistently lower for WT than for the L143D mutant peptide (Figure 4C), meaning that, for a given concentration, the WT peptide was always consistently better at masking the oil–water interface and decreasing the surface tension.
FIGURE 4:
(A) Schematic illustration of oil droplet tensiometry showing an oil droplet, whose volume can be adjusted, at the end of a J-tube in an aqueous buffer. When added to the buffer, purified peptide reduces surface tension. Shrinking the droplet reduces its surface area and increases the concentration of surface peptide, further altering surface tension and/or forcing the peptide off the surface. (B) Plin1 11mr-containing domain (aa 93–192) WT (black line) and mutant (L143D, red line) peptides decrease the interfacial tension of a TO–water (TO–W) interface, but less for the mutant. (C) Equilibrium surface tension vs. the concentration of peptide in the bulk phase. WT produces a lower surface tension than the L143D mutant at all concentrations. At the lowest concentrations (less than 0.5 µg/ml), the values may be too low, because equilibrium probably had not been reached. (D) Having reached a stable equilibrium (∼16.3 mN/M) following injection of the Plin1 WT peptide, the droplet area (size) was rapidly reduced to produce a surface compression and reexpanded after a few minutes. Changes in surface tension are displayed during this procedure. After the rapid compression, surface tension comes back to the initial equilibrium tension. Data from further compressions are included in Supplemental Figure S5A. (E) Competition for the TO–W interface between Plin1 93–192 WT and mutant L143D. Initial (red arrow) addition of 10 µg of mutant L143D to the TO–W interface promptly reduced surface tension to ∼18.6 mN/m (γeq). The area was then reduced by ∼30%, causing the tension to fall rapidly to ∼17.6 mN/m. It then quickly returned to equilibrium. The area was then reexpanded, and tension spiked to ∼27.1 mN/m before falling back to γeq. An equivalent amount of Plin1 WT peptide was then also injected within a few minutes (+WT arrow), and surface tension slowly fell further to a new γeq of ∼15.6 mN/m, indicating that WT displaced the mutant peptide. The surface tension profile following a repeat compression and reexpansion was somewhat different from that recorded during a similar compression in the presence of mutant peptide alone, insofar as there was a “shoulder” (arrowhead) in the recovery period—we interpret this as reflecting initial rapid reassociation of both mutant and WT peptides with the interface, with the mutant then entirely displaced by the WT peptide over time. (F) This image is similar to D, but here phospholipid (POPC) has been added to the buffer before addition of Plin1 WT peptide and then a compression/reexpansion perturbation. Note that the equilibrium surface tension is lower when POPC is added, that is, ∼13.6 mN/m. After the rapid compression, the new equilibrium surface tension is lower than the initial equilibrium tension, in contrast to D. Data from further compressions for both the WT and L143D mutant peptide are included in Supplemental Figure S5B. (G, H) The maximum pressure the peptide can withstand without being ejected from the interface is referred to as Πmax. Data from a number of rapid-compression experiments plotting the maximum Π (Πo) obtained for a given compression are plotted against the change in (Δγ) after compression. The extrapolations to Δγ = 0 give Πmax for each peptide on the two interfaces. (G) The Πmax for the WT peptide is shown for the TO–W and POPC–TO–W interfaces. Πmax is higher on the POPC–TO–W interface, suggesting that the presence of POPC helps to retain the peptide at the interface. Exclusion pressure is calculated by extrapolating the regression lines to a surface tension increment of zero. These data are reported in H.
For assessment of the reversibility of peptide association, the droplet surface area was rapidly reduced to modulate the surface protein density (Figure 4A) and then reexpanded. This perturbation immediately reduced surface tension before it almost returned to the initial equilibrium value (Figure 4D). Reexpansion of the droplet surface area resulted in a rapid increase in surface tension followed by a gradual return to the equilibrium tension. This pattern was largely repeated with further larger compressions and was also similarly seen with the L143D mutant peptide (Supplemental Figure S5A). We interpret these data as indicating that the initial surface compression results in a fall in surface tension as the protein density at the interface rapidly rises (Figure 4A, middle panel), but this is then relieved by expulsion of some of the peptide from the interface to restore equilibrium (Figure 4A, right panel). Reexpansion increases surface tension, but the peptides then reassociate with the interface, restoring surface tension back to equilibrium. For comparison, when a similar experiment is done using apolipoprotein B, compression leads to a net decrease of tension and no protein desorption occurs, whereas with peptides derived from apolipoprotein C, similar behavior is observed (Wang et al., 2009b; Meyers et al., 2012, 2015).
Collectively, these data demonstrate reversible binding of the Plin1 11mr to the oil–water interface, as seen in vivo (Figure 1, B and D); highlight the impact of changes in the peptide sequence on the nascent amphipathic helices; and finally, confirm that this domain cannot withstand high surface protein densities, which induce its desorption.
The 11mr of Plin1 displaces the L143D mutant
To assess the potential for 11mr peptides to “compete” for surface occupancy, we initially added the L143D mutant peptide and, as before, surface tension fell to ∼18.4 mN/m (Figure 4E). Compression of the interface barely changed the equilibrium tension (Figure 4E), as already observed in Figure 4D. WT peptide was then added, and the equilibrium tension fell further to ∼15.6 mN/m, as if the WT peptide was alone on the surface (Figure 4B), suggesting that it displaced the L143D mutant from the interface. Indeed, compressing and reexpanding the droplet resulted in a different surface tension–restoring profile (Figure 4E, arrowhead). We interpret this last observation as suggesting that, initially, both the WT and L143D mutant peptides reassociate with the expanded surface, so surface tension falls rapidly toward ∼18.4 mN/m, but over time, the WT entirely displaces the L143D peptide, restoring surface tension to ∼15.6 mN/m.
Proteins displace the 11mr from the droplet surface more effectively than phospholipids
To more closely reproduce the in vivo situation, we added phospholipids (palmitoyloleoyl phosphatidylcholine [POPC]) to the TO–water interface (TO–W), reducing surface tension from ∼32 to ∼26 mN/m (Supplemental Figure S5B). Addition of the WT Plin1 peptide (Supplemental Figure S5B) prompted a further reduction in surface tension to a new equilibrium of ∼13.4 mN/m, which was lower than that observed with the peptide alone (on TO–W interface) (Figure 4B). Here, too, the L143D Plin1 mutant peptide (Supplemental Figure S5B) reduced surface tension less effectively to ∼16.5 mN/m. Interestingly, when the droplet surface was compressed and reexpanded, the surface tension profiles were slightly different from what was observed in the absence of POPC: following compression and the initial fall in surface tension, surface tension rose but remained at a new equilibrium below the baseline equilibrium (compare equilibrium tension after compression in Figure 4, D and F, and Supplemental Figure S5, A and B). We interpret these data as indicating that the peptide is only partially removed from the interface upon surface compression and can then rapidly “snap” back onto the interface when it is reexpanded. This phenomenon was more striking with the WT than with the mutant peptide (Supplemental Figure S5B) and was also apparent in competition experiments undertaken after addition of phospholipids to the TO–W interface (Supplemental Figure S5C).
We next repeated the compression experiments with a range of concentrations of the WT and mutant peptides, which allowed us to estimate the maximum lateral surface pressure exerted by compression that the peptides can withstand without being ejected (Figure 4G); we called this pressure Πmax. We found that Πmax of the WT and mutant (Figure 4G and Supplemental Figure S5D) was consistently higher in the presence of POPC. This result suggests that phospholipids aid retention of the peptides on the interface, at least in this context. As anticipated, the exclusion pressure for L143D was lower than that for WT peptide before and after the addition of POPC (Figure 4H and Supplemental Figure S5D).
DISCUSSION
Lipid storage is carefully regulated to both alleviate potential lipotoxicity and to provide a rapidly available energy source when needed in either a cell-autonomous manner or, in metazoans, by other more highly oxidative tissues. In vertebrates, white adipocytes constitute the primary lipid storage cell type for the whole organism, whereas other more oxidative tissues like the liver and skeletal muscle have progressively lower tendencies to accumulate lipids. In each of these different tissue/cell types, cellular lipid fluxes are finely coordinated in keeping with their respective physiological functions. Lipid flux is regulated at multiple levels both outside (i.e., blood flow and intravascular lipolysis) and within individual cell types (i.e., intracellular lipolysis). Plins play key roles in regulating intracellular lipid storage and lipolysis and are significant contributors in determining the fate of lipids in cells. A key aspect of how they contribute to these tissue-specific differences in lipid flux is through differential tissue distribution, that is, they are expressed at different levels in different tissues. Then, where more than one member of the Plin family is expressed in a particular cell type, they bind to LDs in a differential manner (Wolins et al., 2006; Straub et al., 2008; Beller et al., 2010; Hsieh et al., 2012). Importantly, binding to the LD surface is also a major determinant of overall expression levels of Plin1 and 2, as both are subject to proteosomal degradation when not bound to LDs, whereas Plin3 is stable in the cytoplasm (Xu et al., 2005, 2006; Masuda et al., 2006). This posttranscriptional regulation of Plin expression may contribute to the specificity of LD association, which is another important feature of Plin biology.
Our data address two main questions: How do Plin1–3 recognize and localize on LDs? What determines their apparently hierarchical and competitive binding to LDs? In keeping with our own prior work in yeast suggesting that the 11mr regions of human Plin1–3 was sufficient to mediate LD localization in this ancient cell type (Rowe et al., 2016) and in which the 11mrs were unlikely to interact with other host proteins given the evolutionary distance between humans and S. cerevisiae, the data suggest that the 11mr acts as a general and primary LD-sensing motif. These data are also consistent with several other reports (McManaman et al., 2003; Nakamura and Fujimoto, 2003; Targett-Adams et al., 2003; Orlicky et al., 2008; Bulankina et al., 2009; Rowe et al., 2016) and with data suggesting that other 11mr domain–containing proteins such as α-synuclein can also localize to LDs (Cole et al., 2002; Brasaemle, 2007; Thiam et al., 2013a). We envisage the 11mr regions behaving as nascent helices that only fold into an AH in the presence of “surface voids” on the LDs. Importantly, at least under the conditions we have tested, the binding affinities of the 11mr regions seem similar among Plin1–3 (Figures 1, D and E, and 2H). However, in vitro experiments with the point mutant L143D, which associates less effectively with an artificial droplet interface than the WT peptide, indicate that differences in the amino acid sequences of the nascent AHs can result in peptides “forcing” other peptides with lower membrane affinity off the interface. There may well be subtle differences in the binding affinity of this region among Plin1–3 related to the differences in the compositions of their AHs. For example, it has been recently proposed that tryptophan or phenylalanine are important residues for LD binding selectivity (Prevost et al., 2018). Two of these residues are present in the Plin1 11mr, but there are none in either the Plin2 or Plin3 11mr regions. However the Plin2 and 3 11-mers localize effectively to LDs (Figure 1), meaning that although these bulky hydrophobic residues might aid LD binding selectivity, they are not essential; this conclusion is supported by the LD binding of the Plin4 11mrs, which also lack these amino acids (Copic et al., 2018).
The association of the 11mr regions with LDs is significantly enhanced by the presence of an additional membrane binding domain, which encompasses the 4HB region, as illustrated by both in vivo and in vitro experiments. These findings are consistent with prior reports suggesting that this region of Plin1 was involved in LD targeting (Garcia et al., 2003) and help to reconcile these data with data highlighting the importance of the 11mr regions. However, whereas the 4HB region of Plin1 is consistently associated with LDs, and when not on LDs, it associates with the ER (Figure 1C and Supplemental Figure S1, E and F), that of Plin2 is less consistently associated with LDs and is readily displaced by protein crowding induced by coexpression of other proteins or peptides that localize to LDs (Supplemental Figure S3, H and I). In our studies, the Plin3 4HB-containing region (Plin3-C) did not localize to LDs in Huh7 or HeLa cells, in keeping with our prior work in S. cerevisiae (Rowe et al., 2016). The 4HB of Plin4 was also recently shown not to localize to LDs (Copic et al., 2018). The behavior of these 4HBs is generally consistent with their known or predicted structures (Hickenbottom et al., 2004). Indeed, except for Plin1, the 4HB is zipped together by four-stranded β-sheets in a smaller αβ domain (Hickenbottom et al., 2004) (Supplemental Figure S2, A and B). Plin1 has one extra exon at its C-terminus (404–522) in vertebrate evolution, and the homology to other Plins consequently stops sharp at position 403 (Supplemental Figure S2B) (Patel et al., 2014). The β-sheet is hence most likely absent in the Plin1 structure, and the 4HB is therefore probably not stabilized as in other Plins (Supplemental Figure S2A), thereby potentially explaining its instability in solution and greater propensity to open up and associate with phospholipid membranes. Our data suggest that the Plin2 4HB is likely to open up in cells, though exactly how this is regulated is not yet clear to us. We also cannot formally exclude the possibility that the 4HB of Plin3 can open up in specific circumstances.
In vivo, most cell types with small droplets do not express Plin1, so Plin2 or Plin3 can occupy the LD surface. In cells without or with very few LDs, Plin3 will remain in the cytoplasm ready to associate with new LDs emerging from the ER (Pol et al., 2014), whereas Plin2 expression will be very low and will depend on the presence of LDs. Plin1 expression is regulated by PPARγ and it is, for example, expressed only at a relatively late stage of adipocyte differentiation (Arimura et al., 2004), when cells possess fairly large LDs to which it can bind. In adipocytes with smaller LDs, we suspect that Plin2 displaces Plin3 from the surface of LDs by virtue of the greater tendency of its 4HB to unfold and associate with the LD surface. Later on, when Plin1 is expressed, it duly displaces Plin2 from the LD surface. This is particularly important physiologically, as evidenced by the increase in basal lipolysis observed in Plin1-null mice, in which Plin2 replaces Plin1 on LDs (Tansey et al., 2001). In humans, loss-of-function Plin1 mutations also alter lipolytic regulation and result in severe metabolic disease despite compensatory up-regulation of Plin2 (Tansey et al., 2003; Kozusko et al., 2015).
In summary, our data suggest that the nascent amphipathic helices encoded by the 11mr regions, present in all Plins, constitute the initial LD-sensing domain. The subsequent stability of Plin1–3 on LDs is then heavily influenced by the differential propensities of their 4HB domains to unfold and bind to the LD surface. Ultimately, we suggest that the combined affinities of these two domains underpin the hierarchical LD binding that we and others have observed and that is essential for optimal fatty-acid traffic into and out of cellular LDs.
MATERIALS AND METHODS
Cloning of Plin1–3 and peptide fragments
Human cDNAs encoding FL Plin1 (aa 1–522), Plin2 (aa 1–437), and Plin3 (aa 1–434) were PCR amplified using Phusion DNA polymerase (Thermo Fisher) from the templates pcDNA-Myc Plin1–3 (Patel et al., 2014) and subcloned in frame into pEGFPC1 (Clontech) or pMCherry. The cDNAs encoding the fragments for Plin1 (aa 1–191 and aa 185–522), Plin2 (aa 1–191 and aa 184–437), and Plin3 (aa 1–204 and aa 197–434) were PCR amplified from the pcDNA Plin1–3-Myc (Rowe et al., 2016) and also subcloned into pEGFCPC1 and pMCherry. The restriction endonucleases employed for the Plin1–3 subcloning strategies are as follows HindIII/Sal1 for Plin1, EcoR1/Sal1 for Plin2, and EcoR1/BamH1 for Plin3. Plin3-β (aa 1–413) was amplified by PCR from the template pEGFP Plin3 and subcloned into pEGFPC1. All Plin constructs were cloned such that the GFP or mCherry fluorescent tag was N-terminal to the gene and thus the peptide or protein.
Expression and purification of the Plin1 11mr region
Recombinant plasmids pET22b-CPD-SalI expressing Plin1 amino acids (93–192) were transformed into the Escherichia coli expression strain NiCo21 (New England Biolabs, Hitchin, UK). The L143D Plin1 mutant peptide was generated as described previously (Rowe et al., 2016). Overnight cultures were diluted into 1 l of lysogeny broth supplemented with 0.2% glucose and grown with shaking at 37°C to A600 = 1.0. Isopropyl-d-1-thiogalactopyranoside was added (1 mM final concentration), and cultures were further grown for 80 min at 37°C. Cell pellets from a 500-ml culture were resuspended in 14 ml of B-PER (Thermo Scientific) with 10% glycerol, 4 mg of lysozyme (Sigma), 500 U of Pierce Universal Nuclease (Thermo Scientific), and complete EDTA-free protease inhibitors (Roche Applied Science). Cell lysates were rotated at room temperature for 15 min to ensure full cell lysis, centrifuged at 28,000 × g for 30 min, and filtered (Minisart 0.22 μm; Sartorius, Göttingen, Germany). Affinity purification was performed using 0.5–1.0 ml of nickel nitrilotriacetic acid-agarose beads (Qiagen) while rotating for 1 h at 4°C, before bound fusion proteins were subjected to high-stringency washes (40 mM imidazole, 0.5 M NaCl) to reduce nonspecific protein binding, and transferred to the CPD reaction buffer (20 mM Tris, 60 mM NaCl, 250 mM sucrose, 3 mM imidazole, pH 7.4). Cleaved products were eluted into the supernatant using 50–100 μM inositol hexakisphosphate by gentle rotation for 2 h at 4°C. Size-exclusion chromatography was performed using an AKTA pure chromatography system and SuperdexTM 75 10/300 GL column (GE Healthcare). Protein concentrations were determined by amino acid analysis. For confirmation of ≥95% purity, equivalent amounts of isolated proteins were resolved on an 18% Tris-glycine gel (Life Technologies) and stained with quick Coomassie stain (Generon, Maidenhead, UK). The purified proteins were analyzed by mass spectrometry (LC-MS) revealing a 90 amino acid peptide sequence and apparent mass of ∼9400 Da. We were able to distinguish the WT from the mutant L143D peptide based on retention time and mass profile. We also identified a cryptic cleavage site close to the C-terminus that removed amino acid residues 181–192 from the intact peptide. Further dynamic light-scattering studies (0.5 mg/ml protein samples in 10 mM potassium phosphate buffer, pH 7.4) using a Zetasizer NanoS instrument (Malvern Ltd, UK) showed negligible levels of aggregation and confirmed earlier findings that these proteins were essentially monomeric by the initial gel-filtration elution profile (unpublished data).
Cell culture
Huh7 cells were maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS; Life Technologies), Both 4.5 g/l d-glucose, 0.1 g/l sodium pyruvate (Life Technologies), and 1% penicillin–streptomycin (Life Technologies). The cells were cultured at 37°C under a 5% CO2 atmosphere. Confluent monolayers of cells were resuspended after trypsinization and plated into 35 mm cell-culture MatTek dishes (with glass coverslips at the bottom) (MatTek, Ashland, MA).
HeLa (human cervical carcinoma obtained from the American Type Culture Collection) cells were maintained in DMEM supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, penicillin–streptomycin, 1% sodium pyruvate, and 1% nonessential amino acids.
Transfection and cotransfection
When indicated, Huh7 cells (60–70% confluence) were exposed for 1 h to 500 µM oleic acid coupled to bovine serum albumin (BSA) (1% vol/vol) to induce LD formation, and then cells were transfected with 3 µg of plasmid DNA/ml using Polyethylenimine HCl MAX (Polysciences) following the manufacturer’s instructions. For coexpression competition experiments, mCherry- or GFP-tagged plasmid constructs in equal concentrations (1.5–2 µg for each one) were transfected into cells. Cells were imaged at 24 h after transfection.
HeLa cells were seeded onto coverslips in 12-well tissue culture plates with a density of 65,000 cells per well and transfected using Lipofectamine LTX (Thermo Fisher Scientific). Oleic acid (400 µM) conjugated to BSA, along with either Bodipy 558/568 C12 or 493/502 (1:2500 dilution), was supplemented at 4 h after transfection for 20 h to promote lipid droplet formation and staining. Cells were fixed with 4% formaldehyde for 15 min; this was followed by three washes in phosphate-buffered saline (PBS). Cells were mounted on microscope slides with ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific), and the fluorescently tagged protein localization was determined using a Leica TCS SP8 confocal microscope with a 63× immersion oil objective (1.3 NA). GFP fluorescence was excited at 488 nm, and emission was detected between 490 and 550 nm, while M-cherry–tagged protein fluorescence was excited at 588, and emission was detected between 600 and 650 nm. LDs were detected with either Bodipy 493/502 (excitation: 502 nm; emission: 515–560 nm) or Bodipy 558/568 C12 (excitation: 558 nm; emission: 590–700 nm).
FRAP experiments
For FRAP experiments, we bleached the signal on a collection of drops and monitored the increase of signal during recovery. The background signal, for example, from the cytosol, was removed from the recorded signal, which was at the end normalized by intrinsic bleaching of nonbleached areas. We next used GraphPad Prism to fit the FRAP recovery curves with a nonlinear regression and the exponential one-phase association model. The characteristic recovery time that corresponds to the time it takes for fluorescence intensity to reach half the maximum of the plateau level is obtained by the software.
Image quantification and statistics
Images were analyzed using ImageJ software. For quantification of the percent LD-targeted signal for a given protein, the image was background corrected and the total fluorescence signal on LDs was determined as a ratio to the total fluorescence signal in the whole cell. In coexpression experiments, the fluorescence signal on LDs was calculated by subtracting the fluorescence signal elsewhere in each cell. Protein concentrations on LDs were derived from the mean fluorescence measured on LDs in each channel; each experimental point shown in Figures 2 and 3 corresponds to the average of the signal on 10–20 LDs. Values from 15–20 cells were combined, and the SD was calculated for statistical analysis. The critical concentration was determined by fitting the data with the following function: y = 1/(1 + x/c), where c represents the critical concentration at which half of the competing protein is displaced.
In vitro experiments
For purification of LDs from cells expressing fluorescently tagged LD proteins, cells from five 150 cm dishes were harvested, washed once in ice-cold PBS, and lysed using a 30 G needle in 1 ml Tris-EDTA (20-20 mM, respectively) buffer containing complete protease and phosphatase inhibitor tablets at pH 7.5. For LD isolation, 1 ml of cell lysates was mixed with 1 ml of 60% sucrose in Tris-EDTA buffer supplemented with protease inhibitors, overlaid with 20, 10, and 0% buffered sucrose on top of one another in 5 ml Ultra-Clear centrifuge tubes (Beckman). Gradients were centrifuged for 16 h at 100,000 × g and 4°C, using an SW60 rotor in a Beckman L8-70 centrifuge, and 300 µl was collected from the top as the LD fraction.
In vitro experiments were performed in HKM buffer: 50 mM HEPES, 120 mM potassium acetate, and 1 mM MgCl2 (in Milli-Q water) at pH 7.4. To create buffer in oil drops, 4 µl of a buffer-diluted LD fraction was mixed with 40 µl of TO by vortexing, as previously done (Kory et al., 2015). About 100 drops were formed and imaged in the field of observation. For shrinking experiments, aqueous drops bounded by the proteins were imaged for 10–30 min on glass plates during water evaporation. For the diffusion experiments, we bleached part of the droplet surface, and the fraction of recoverable fluorescence was calculated with respect to initial fluorescence.
Lipids
Triolein (TO) was purchased from Nu-Chek Prep (Elysian, MN). It was >99% pure and its interfacial tension at 25.0°C was 32 ± 1 mN/m. POPC from Avanti Polar Lipids (Alabaster, AL) was stored at −20°C in chloroform (25.0 mg/ml). Small unilamellar vesicles (SUVs) of POPC, ∼30 nm in diameter, were made at 2.5 mg/ml in a standard 5 mM sodium phosphate buffer (pH 7.4) by sonication, as previously described (Mitsche et al., 2010; Meyers et al., 2013).
Interfacial tension (γ) and surface pressure (Π) measurements
We modified an oil-drop tensiometer (Mitsche et al., 2010; Mitsche and Small, 2013; Meyers et al., 2013) designed by Teclis Instruments (Tassin, France) to measure the interfacial tension (γ) of lipid–water interfaces. γ is the energy required to create one new square centimeter of surface (i.e., γ = ergs/cm2 or mN/m). All experiments were conducted at 25.0 ± 0.2°C in a thermostated system and repeated at least twice.
For creation of TO–W interfaces (Mitsche and Small, 2011), TO drops (16.0 µl) were formed at the tip of a J-needle submerged in 7.0 ml of bulk buffer. Their surface area was ∼30 mm2 (diameter = 3.1 mm). The buffer was 5 mM sodium phosphate at pH 7.4. Before interfacial studies, aliquots of the WT and mutant Plin1 peptides were thawed and solubilized with hexafluoro-2-propanol at a concentration of 2.5% (vol/vol). The TO–W interface stabilized at γTO = 32.0 ± 1 mN/m. Adsorption of amphipathic molecules (i.e., phospholipid, Plin peptides) to this interface shields much of the TO from the aqueous phase and decreases γ to a nearly constant value defined as equilibrium tension (γeq). For creation of POPC–TO–W interfaces, TO drops of 16 µl were formed in bulk buffer containing 1.0 mg of POPC SUV. After POPC adsorbed to the TO drop, the buffer was exchanged with 250 ml of POPC-free buffer, originally described for egg yolk phosphatidyl choline (Mitsche et al., 2010), to wash out >99.9% of the original buffer and all POPC SUVs suspended in the bulk phase. After the washout, γ was usually 25–27 mN/m, corresponding to ∼1.1 µmol POPC/m2 for POPC–TO–W interfaces. Varied amounts of Plin1 peptide were added to the bulk phase to obtain different protein concentrations ranging from 0.2 to 4 µg/ml. As peptide adsorbed to TO–W and POPC–TO–W interfaces, γ was monitored continuously as it fell to an equilibrium value (γeq). Surface pressure (Π) was defined as the difference in γ between a pure TO–W interface (γTO = 32.0 mN/M) and the interface with bound POPC and/or peptide (Π) = γTO – γ). The initial pressures (Πi) of POPC–TO–W interfaces represent the difference between γTO and γ after POPC adsorption (Πi = γ TO − γPOPC) and was between 5 and 7 mN/m. Πi of the TO–W interface was 0 mN/m. After peptide adsorption, the equilibrium pressure of all interfaces was calculated as Πeq = γTO − γeq. The change in pressure (ΔΠ) at these interfaces induced by adsorption was ΔΠ = Πeq − Πi.
Pressure/area-mediated desorption and readsorption processes
Following adsorption of peptide to either a TO–W or a POPC–TO–W interface, the drop underwent a series of compressions and reexpansions with the goal of determining whether peptide completely or only partially desorbed from the respective interface. Once γ approached a γeq, the TO drop (16 µl) was compressed by rapidly decreasing the volume by different ratios: 6.25% (1 µl), 12.5% (2 µl), 25% (4 µl), 37.5% (6 µl), 50% (8 µl), or when possible, 62.5% (10 µl). This sudden decrease in volume induced a decrease in drop surface area, resulting in a sudden compression and abrupt decrease in γ. The oil drop was held at this reduced volume for 5–10 min, with γ recorded continuously. If peptide readily desorbed, γ increased to a γeq observed as a desorption curve. After 5–10 min, the interface was expanded by increasing the volume of the drop back to its initial volume (16 µl). As the surface area increased upon expansion, γ abruptly increased. If peptide adsorbed from the bulk phase and adhered to the newly formed extra surface, γ decreased to the initial γeq, observed as a readsorption curve. This process of stress compression and reexpansion was repeated after the bulk buffer was exchanged with 150 ml of 5 mM phosphate buffer (pH 7.4) devoid of peptide.
Values of Πmax
The desorption and readsorption protocol provided information about not only the nature of ejection of the peptide from either interface but also the Π at which such ejection occurs. Πmax is the maximal pressure (Π) that a peptide can withstand before all or part of the molecule is ejected from the surface. Once a γeq had been reached following adsorption of peptide to a TO–W or POPC–TO–W interface, a series of experiments were conducted in which the drop area was decreased abruptly, thereby decreasing γ and increasing Π to a given value, Πo. The change in tension (∆γ) over the following 5–10 min as peptide desorbed from the surface was plotted against Πo. Regression of a linear fit to the plot reveals Πmax as the point at which ∆γ = 0, such that no peptide desorbs from the surface upon compression.
Exclusion pressure measurements
Exclusion pressure (ΠEX) for each variant is the surface pressure above which that peptide cannot bind and insert into POPC–TO–W interfaces (Mitsche et al., 2010). In other words, ΠEX is the pressure of a POPC–TO–W interface at which addition of peptide to the bulk phase leads to no adsorption-induced change in surface pressure (ΔΠ = 0 mN/m).
Quantification and statistical analysis
The data for the critical concentrations for displacing competing proteins experiments are presented using Tukey box-and-whisker plots, where the central box represents the interquartile ranges (IR; 25th to 75th percentile), the middle line represents the median, and the horizontal lines represent the minimum and the maximum values of the observation range. Values are expressed as median ± IR.
The statistical evaluation of FL Plin3 displacement from LDs by the Plin1 11mr or FL Plin2 (in Supplemental Figure S3G) was done with a Mann-Whitney nonparametric test using Prism software, p values <0.05 were considered significant. All values shown in the text and figures are mean ± SD, where N = 4, from independent experiments.
Supplementary Material
Acknowledgments
We thank W. Urbach and F. Pincet for helpful discussions. D.B.S. is supported by the Wellcome Trust (WT 107064), the MRC Metabolic Disease Unit, the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, and NIHR Rare Disease Translational Research Collaboration. A.R.T. is supported by the ANR-MOBIL, ANRS (The Agence Nationale de Recherches sur le Sida et les hépatites virales), Programme Emergence de la Ville de Paris, PSL, and ATIP-Avenir. D.A. is supported by ANRS.
Abbreviations used:
- 11mr
11-mer repeat
- 4HB
4-helix bundle
- AH
amphipathic helix
- ATGL
adipose tissue triglyceride lipase
- BSA
bovine serum albumin
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- FL
full length
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- HSL
hormone-sensitive lipase
- LD
lipid droplet
- Plin
perilipin
- POPC
palmitoyloleoyl phosphatidylcholine
- TO–W
triolein–water interface
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0534) on January 16, 2019.
REFERENCES
- Arimura N, Horiba T, Imagawa M, Shimizu M, Sato R. (2004). The peroxisome proliferator-activated receptor gamma regulates expression of the perilipin gene in adipocytes. J Biol Chem , 10070–10076. [DOI] [PubMed] [Google Scholar]
- Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jackle H, Kuhnlein RP. (2010). PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab , 521–532. [DOI] [PubMed] [Google Scholar]
- Ben M’barek K, Ajjaji D, Chorlay A, Vanni S, Foret L, Thiam AR. (2017). ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev Cell , 591–604 e597. [DOI] [PubMed] [Google Scholar]
- Bersuker K, Olzmann JA. (2017). Establishing the lipid droplet proteome: mechanisms of lipid droplet protein targeting and degradation. Biochim Biophys Acta , 1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle DL. (2007). Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res , 2547–2559. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Wolins NE. (2012). Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem , 2273–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, Burger KN, Honing S. (2009). TIP47 functions in the biogenesis of lipid droplets. J Cell Biol , 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong BM, Russell TD, Schaack J, Orlicky DJ, Reigan P, Ladinsky M, McManaman JL. (2011). The adipophilin C terminus is a self-folding membrane-binding domain that is important for milk lipid secretion. J Biol Chem , 23254–23265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Murphy DD, Grider T, Rueter S, Brasaemle D, Nussbaum RL. (2002). Lipid droplet binding and oligomerization properties of the Parkinson’s disease protein alpha-synuclein. J Biol Chem , 6344–6352. [DOI] [PubMed] [Google Scholar]
- Copic A, Antoine-Bally S, Gimenez-Andres M, Garay CLT, Antonny B, Manni MM, Pagnotta S, Guihot J, Jackson CL. (2018). A giant amphipathic helix from a perilipin that is adapted for coating lipid droplets. Nat Commun , 1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, Parton RG. (2011). Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol , a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra S, Le Dour C, Bottomley W, Cervera P, Giral P, Reznik Y, Charpentier G, Auclair M, Delepine M, Barroso I, et al. (2011). Perilipin deficiency and autosomal dominant partial lipodystrophy. N Engl J Med , 740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A, Sekowski A, Subramanian V, Brasaemle DL. (2003). The central domain is required to target and anchor perilipin A to lipid droplets. J Biol Chem , 625–635. [DOI] [PubMed] [Google Scholar]
- Goedhart J, van Weeren L, Adjobo-Hermans MJ, Elzenaar I, Hink MA, Gadella TW., Jr (2011). Quantitative co-expression of proteins at the single cell level—application to a multimeric FRET sensor. PLoS One , e27321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Granneman RL, Greenberg AS, Obin MS, Zhu Z. (2007). Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem , 5726–5735. [DOI] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Krishnamoorthy R, Rathod M. (2009). Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J Biol Chem , 34538–34544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C. (1991). Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem , 11341–11346. [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. (2008). Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature , 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickenbottom SJ, Kimmel AR, Londos C, Hurley JH. (2004). Structure of a lipid droplet protein; the PAT family member TIP47. Structure , 1199–1207. [DOI] [PubMed] [Google Scholar]
- Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. (2012). Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J Cell Sci , 4067–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itabe H, Yamaguchi T, Nimura S, Sasabe N. (2017). Perilipins: a diversity of intracellular lipid droplet proteins. Lipids Health Dis , 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. (2010). Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res , 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, Sztalryd C. (2016). The perilipins: major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu Rev Nutr , 471–509. [DOI] [PubMed] [Google Scholar]
- Kory N, Farese RV, Jr, Walther TC. (2016). Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol , 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, Thiam AR, Farese RV, Jr, Walther TC. (2015). Protein crowding is a determinant of lipid droplet protein composition. Dev Cell , 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozusko K, Tsang V, Bottomley W, Cho YH, Gandotra S, Mimmack ML, Lim K, Isaac I, Patel S, Saudek V, et al. (2015). Clinical and molecular characterization of a novel PLIN1 frameshift mutation identified in patients with familial partial lipodystrophy. Diabetes , 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Sohn JH, Han JS, Park YJ, Jeon YG, Ji Y, Dalen KT, Sztalryd C, Kimmel AR, Kim JB. (2018). Perilipin 3 deficiency stimulates thermogenic beige adipocytes through PPARalpha activation. Diabetes , 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby AE, Bales E, Orlicky DJ, McManaman JL. (2016). Perilipin-2 deletion impairs hepatic lipid accumulation by interfering with sterol regulatory element-binding protein (SREBP) activation and altering the hepatic lipidome. J Biol Chem , 24231–24246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Botas J, Anderson JB, Tessier D, Lapillonne A, Chang BH, Quast MJ, Gorenstein D, Chen KH, Chan L. (2000). Absence of perilipin results in leanness and reverses obesity in Leprdb/db mice. Nat Genet , 474–479. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Itabe H, Odaki M, Hama K, Fujimoto Y, Mori M, Sasabe N, Aoki J, Arai H, Takano T. (2006). ADRP/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J Lipid Res , 87–98. [DOI] [PubMed] [Google Scholar]
- McManaman JL, Zabaronick W, Schaack J, Orlicky DJ. (2003). Lipid droplet targeting domains of adipophilin. J Lipid Res , 668–673. [DOI] [PubMed] [Google Scholar]
- Meyers NL, Larsson M, Olivecrona G, Small DM. (2015). A pressure-dependent model for the regulation of lipoprotein lipase by apolipoprotein C-II. J Biol Chem , 18029–18044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers NL, Wang L, Gursky O, Small DM. (2013). Changes in helical content or net charge of apolipoprotein C-I alter its affinity for lipid/water interfaces. J Lipid Res , 1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers NL, Wang L, Small DM. (2012). Apolipoprotein C-I binds more strongly to phospholipid/triolein/water than triolein/water interfaces: a possible model for inhibiting cholesterol ester transfer protein activity and triacylglycerol-rich lipoprotein uptake. Biochemistry , 1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsche MA, Small DM. (2011). C-terminus of apolipoprotein A-I removes phospholipids from a triolein/phospholipids/water interface, but the N-terminus does not: a possible mechanism for nascent HDL assembly. Biophys J , 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsche MA, Small DM. (2013). Surface pressure-dependent conformation change of apolipoprotein-derived amphipathic alpha-helices. J Lipid Res , 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsche MA, Wang L, Small DM. (2010). Adsorption of egg phosphatidylcholine to an air/water and triolein/water bubble interface: use of the 2-dimensional phase rule to estimate the surface composition of a phospholipid/triolein/water surface as a function of surface pressure. J Phys Chem B , 3276–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Fujimoto T. (2003). Adipose differentiation-related protein has two independent domains for targeting to lipid droplets. Biochem Biophys Res Commun , 333–338. [DOI] [PubMed] [Google Scholar]
- Narayanaswami V, Kiss RS, Weers PM. (2010). The helix bundle: a reversible lipid binding motif. Comp Biochem Physiol A Mol Integr Physiol , 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlicky DJ, Degala G, Greenwood C, Bales ES, Russell TD, McManaman JL. (2008). Multiple functions encoded by the N-terminal PAT domain of adipophilin. J Cell Sci , 2921–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Yang W, Kozusko K, Saudek V, Savage DB. (2014). Perilipins 2 and 3 lack a carboxy-terminal domain present in perilipin 1 involved in sequestering ABHD5 and suppressing basal lipolysis. Proc Natl Acad Sci USA , 9163–9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Gross SP, Parton RG. (2014). Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol , 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost C, Sharp ME, Kory N, Lin Q, Voth GA, Farese RV, Jr, Walther TC. (2018). Mechanism and determinants of amphipathic helix-containing protein targeting to lipid droplets. Dev Cell , 73–86 e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnayake SS, Mirheydari M, Schulte A, Gillahan JE, Gentit T, Phillips AN, Okonkwo RK, Burger KN, Mann EK, Vaknin D, et al. (2014). Insertion of apoLp-III into a lipid monolayer is more favorable for saturated, more ordered, acyl-chains. Biochim Biophys Acta , 482–492. [DOI] [PubMed] [Google Scholar]
- Rowe ER, Mimmack ML, Barbosa AD, Haider A, Isaac I, Ouberai MM, Thiam AR, Patel S, Saudek V, Siniossoglou S, Savage DB. (2016). Conserved amphipathic helices mediate lipid droplet targeting of perilipins 1–3. J Biol Chem , 6664–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JR, Shew TM, Schwartz DM, Tzekov A, Lepus CM, Abumrad NA, Wolins NE. (2009). Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem , 30941–30948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DM, Wang L, Mitsche MA. (2009). The adsorption of biological peptides and proteins at the oil/water interface. A potentially important but largely unexplored field. J Lipid Res (suppl), S329–S334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub BK, Stoeffel P, Heid H, Zimbelmann R, Schirmacher P. (2008). Differential pattern of lipid droplet-associated proteins and de novo perilipin expression in hepatocyte steatogenesis. Hepatology , 1936–1946. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Garcia A, Sekowski A, Brasaemle DL. (2004). Hydrophobic sequences target and anchor perilipin A to lipid droplets. J Lipid Res , 1983–1991. [DOI] [PubMed] [Google Scholar]
- Tansey JT, Huml AM, Vogt R, Davis KE, Jones JM, Fraser KA, Brasaemle DL, Kimmel AR, Londos C. (2003). Functional studies on native and mutated forms of perilipins. A role in protein kinase A-mediated lipolysis of triacylglycerols. J Biol Chem , 8401–8406. [DOI] [PubMed] [Google Scholar]
- Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, Reitman ML, Deng CX, Li C, Kimmel AR, et al. (2001). Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci USA , 6494–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targett-Adams P, Chambers D, Gledhill S, Hope RG, Coy JF, Girod A, McLauchlan J. (2003). Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J Biol Chem , 15998–16007. [DOI] [PubMed] [Google Scholar]
- Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. (2002). The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem , 44507–44512. [DOI] [PubMed] [Google Scholar]
- Thiam AR, Antonny B, Wang J, Delacotte J, Wilfling F, Walther TC, Beck R, Rothman JE, Pincet F. (2013a). COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc Natl Acad Sci USA , 13244–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Beller M. (2017). The why, when and how of lipid droplet diversity. J Cell Sci , 315–324. [DOI] [PubMed] [Google Scholar]
- Thiam AR, Farese RV, Jr, Walther TC. (2013b). The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol , 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul PJ, Tschapalda K, Kolkhof P, Thiam AR, Oberer M, Beller M. (2017). Targeting of the Drosophila protein CG2254/Ldsdh1 to a subset of lipid droplets. J Cell Sci , 3141–3157. [DOI] [PubMed] [Google Scholar]
- Wang H, Bell M, Sreenivasan U, Hu H, Liu J, Dalen K, Londos C, Yamaguchi T, Rizzo MA, Coleman R, et al. (2011). Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem , 15707–15715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu L, Dalen K, Dorward H, Marcinkiewicz A, Russell D, Gong D, Londos C, Yamaguchi T, Holm C, et al. (2009a). Activation of hormone-sensitive lipase requires two steps, protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J Biol Chem , 32116–32125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Martin DD, Genter E, Wang J, McLeod RS, Small DM. (2009b). Surface study of apoB1694–1880, a sequence that can anchor apoB to lipoproteins and make it nonexchangeable. J Lipid Res , 1340–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins NE, Brasaemle DL, Bickel PE. (2006). A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett , 5484–5491. [DOI] [PubMed] [Google Scholar]
- Xu G, Sztalryd C, Londos C. (2006). Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim Biophys Acta , 83–90. [DOI] [PubMed] [Google Scholar]
- Xu G, Sztalryd C, Lu X, Tansey JT, Gan J, Dorward H, Kimmel AR, Londos C. (2005). Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J Biol Chem , 42841–42847. [DOI] [PubMed] [Google Scholar]
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. (2012). FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling. Cell Metab , 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.