Abstract
Dendritic spines (DS) are actin-rich postsynaptic terminals of neurons that are critical for higher-order brain functions. Maturation of DS is accompanied by a change in actin architecture from linear to branched filamentous structures. Presumably, the underlying cause of this is a switch in a mode of actin assembly from formin-driven to Arp2/3-mediated via an undefined mechanism. Here we present data suggesting that neuron-specific actin-binding drebrin A may be a part of such a switch. It is well documented that DS are highly enriched in drebrin A, which is critical for their plasticity and function. At the same time, mDia2 is known to mediate the formation of filopodia-type (immature) spines. We found that neuronal drebrin A directly interacts with mDia2 formin. Drebrin inhibits formin-mediated nucleation of actin and abolishes mDia2-induced actin bundling. Using truncated protein constructs we identified the domain requirements for drebrin–mDia2 interaction. We hypothesize that accumulation of drebrin A in DS (that coincides with spine maturation) leads to inhibition of mDia2-driven actin polymerization and, therefore, may contribute to a change in actin architecture from linear to branched filaments.
INTRODUCTION
Continuous assembly, disassembly, and reorganization of actin cytoskeleton is necessary for proper cell shape and function. For example, in neurons, actin remodeling in dendritic spines (DS) and axonal growth cones is essential for synaptic plasticity and connectivity (Bertling and Hotulainen, 2017). Stability and dynamics of actin cytoskeleton are regulated by a large number of accessory proteins, and understanding their interplay in different cellular contexts presents a great challenge (Rottner et al., 2017).
Formins are actin nucleating and elongating factors that play important roles in many cellular processes (Chesarone et al., 2010; Breitsprecher and Goode, 2013). Some of the formins can also bind microtubules and actin filaments’ sides, further contributing to the regulation of cytoskeletal dynamics (Gaillard et al., 2011). On a structural level, the functional form of formins is an antiparallel dimer (Shimada et al., 2004; Xu et al., 2004) that stabilizes transient actin dimers and trimers, aiding F-actin nucleation. Formins are also processive actin-elongating factors that tightly associate with the barbed ends (B-ends) of filaments (Pruyne et al., 2002; Kovar et al., 2006). Most often, formin-assisted actin polymerization is accelerated in the presence of profilin–actin complexes (physiological state of monomeric actin) compared with G-actin alone (Sagot et al., 2002). This effect is due to the presence of poly-proline motifs within FH1 domains of formins that serve as “traps” for profilin–actin complexes, increasing their local concentration near filaments B-ends (Courtemanche and Pollard, 2012). Thus, under physiological conditions, formins assist actin elongation through FH1-dependent G-actin–profilin transfer while processively moving with growing B-ends of actin filaments.
Owing to their potent effect on actin assembly, formins are most often autoinhibited in vivo (Watanabe et al., 1999; Alberts, 2001). Autoinhibition in different formins can be released by GTPases, often in conjunction with phosphorylation or other regulatory mechanisms (Kühn and Geyer, 2014). How fully activated formins are regulated in different cell types remains an open question. Available data suggest that formin-mediated actin nucleation and the duration of processive assembly runs can be fine-tuned in order to yield diverse actin structures. It has been reported that protein regulators such as Bud6, CLIP170, Adenomateous polyposis coli protein, and some tropomyosins can aid formin-mediated actin nucleation (Chesarone et al., 2010; Breitsprecher and Goode, 2013; Alioto et al., 2016). In contrast to that, only a few regulators that attenuate formins activity toward actin have been identified to date. The ubiquitous actin regulators such as profilin and capping protein are known to inhibit formin-mediated nucleation and elongation, respectively (Paul and Pollard, 2008; Shekhar et al., 2016). Spire, which is an actin nucleating protein, works together with Capu formin in vivo, but inhibits its activity toward actin in vitro (Quinlan et al., 2007; Quinlan, 2013; Montaville et al., 2014). Formins are inhibited also by Hof1, Smy1, and Bud14 that function in yeast (Chesarone et al., 2009; Chesarone-Cataldo et al., 2011; Graziano et al., 2014). However, our understanding of how formins’ activities are regulated in mammalian, and especially neuronal cells, is highly incomplete.
Formins emerged as an important class of regulators that are important in neuronal development and function. It was documented that formins are involved in shaping axonal growth cones (DAAM) (Matusek et al., 2008; Gombos et al., 2015), dendritic spine development (mDia2) (Hotulainen et al., 2009), and expansion (FMNL2) (Chazeau et al., 2014), as well as in linking receptors to the actin cytoskeleton (delphilin) (Miyagi et al., 2002). In line with the critical importance of formins in neuronal function, heterozygous deletion of the gene encoding formin 2 leads to intellectual disability in humans (Almuqbil et al., 2013). However, how formins’ activities are incorporated with those of other neuronal actin regulators is not well understood.
In DS, actin-rich postsynaptic terminals of neurons, formins must exert their function in the presence of a high concentration of drebrin, which is one of the key actin regulators in DS (Koganezawa et al., 2017). Drebrin is an actin-stabilizing protein that alters the morphology of actin filaments and attenuates their severing by cofilin (Sharma et al., 2011; Mikati et al., 2013; Grintsevich and Reisler, 2014). Drebrin loss is reportedly associated with impaired higher-order brain functions, and it is a hallmark of Alzheimer’s disease and some other complex neurodegenerative disorders (Shim and Lubec, 2002; Ivanov et al., 2009; Ma et al., 2015). Thus, both formins and drebrin are critical for neuronal function and their missregulation is linked to brain disease. However, how drebrin and formins affect each other’s activities is unknown. To address this question, we investigated the effect of brain-specific drebrin A on actin assembly and remodeling by diaphanous formin 2 (mDia2).
RESULTS
Drebrin A inhibits mDia2-driven actin assembly
To bypass an autoinhibition of mDia2 formin, we employed its truncated construct that is missing the N-terminal part of the sequence (seq. 1–520) and contains only FH1, FH2, and the C-terminal tail (mDia2-FFC, seq. 521–1171) (Li and Higgs, 2005). As expected, mDia2-FFC greatly accelerated the rate of actin polymerization compared with actin alone (Supplemental Figure S1, compare green and black traces). Next, we examined the effects of full-length neuronal drebrin A (DrbA-FL; Figure 1A) on mDia2-driven actin polymerization using a standard bulk pyrene fluorescence assay. We found strong inhibition of actin polymerization by drebrin in the absence (Figure 1, B and C) and in the presence (Figure 1, D and E) of profilin. Drebrin-induced inhibition of mDia2-driven actin polymerization was concentration-dependent. The dependencies of polymerization rates on drebrin concentration yielded IC50 of 40 and 70 nM, (with and without profilin, respectively). Next, we tested the C-terminal truncated form of drebrin (Drb1–300; Figure 1F) that has affinity to F-actin, similar to that of DrbA-FL, and induces the same morphological changes in actin filaments (increased length of helical repeats) (Sharma et al., 2012). This drebrin construct (previously defined as the actin-binding core of drebrin, sequence 1–300) also inhibited mDia2-driven actin polymerization, but at least one order of magnitude weaker than DrbA-FL (Figure 1, G and H). The inhibitory effect of Drb1–300 was also concentration-dependent (Figure 1G) and was observed both in the absence and in the presence of profilin. The dependency of mDia2-driven actin polymerization rates on Drb1–300 concentration yielded IC50 close to 1 µM. It is important to note that the extent of inhibition of formin-driven actin assembly is lower with Drb1–300 construct than with DrbA-FL.
FIGURE 1:
Neuronal drebrin A inhibits mDia2-FFC-mediated actin assembly in a concentration-dependent manner. (A) Domain structure of DrbA-FL. Actin Depolymerizing Factor homology domain (ADFhd) is shown in green; helical-charged domain is shown in orange; Actin Binding Domain (ABD) is shown in blue; intrinsically disordered C-terminal region is shown as a solid black line. The amino acid numbering is based on mouse drebrin isoform A (UniProtKB: Q9QXS6; methionine-1 was not present in the protein sequence). (B) DrbA-FL strongly inhibits mDia2-FFC-mediated actin assembly. mDia2-FFC drives rapid assembly of actin (1 µM) in the absence of DrbA-FL (black trace). DrbA-FL inhibits mDia2–FFC-driven actin assembly in a concentration-dependent manner (dark-blue to light-blue traces; 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, and 0.7 µM DrbA-FL, respectively). (C) Quantification of the data shown in B. The concentration of DrbA-FL yielding half-maximum inhibition (IC50) of mDia2-FFC-mediated actin assembly is 70 nM. (D) DrbA-FL inhibits mDia2-FFC-mediated actin (1 µM) assembly in the presence of profilin (5 µM). The concentrations of DrbA-FL used were the same as in B. (E) Quantification of the data shown in D. The concentration of DrbA-FL yielding half-maximum inhibition (IC50) of mDia2-FFC-mediated actin assembly is 40 nM. (F) Domain structure of Drb1–300. The disordered C-terminal region of drebrin has been truncated. (G) The rates of mDia2-FFC-mediated actin assembly decrease as Drb1–300 concentration is increased (black trace: no drebrin added; dark blue to light blue: 0.45, 0.7. 2, 4, 5, 6, 8, and 10 µM Drb1–300, respectively). (H) Quantification of data shown in G. Drb1–300 concentration yielding half-maximum inhibition (IC50) of mDia2-FFC-mediated actin assembly is 810 nM. Conditions: [Actin] = 1 µM (10% pyrene-maleimide labeled); mDia2-FFC = 30 nM (B, C, D, E, F, and H). Additionally, 5 µM profilin was present in D and E. Buffer: KMEH7 (see Materials and Methods).
Next, we set out to determine which aspect(s) of formin-driven actin polymerization is (are) affected by drebrin. The increased lag phase in actin polymerization observed in our pyrene fluorescence assays (Figure 1, B, D, and G) suggested inhibition of mDia2-driven actin nucleation by drebrin A. To determine the effect of drebrin on mDia2-driven actin nucleation, we used direct total internal reflection fluorescence (TIRF) microscopy observation. We incubated actin (15% Alexa488SE-labeled on lysines), profilin, and formin with and without drebrin constructs for 10 min and then imaged the resulting mixtures on a polylysine-coated surface. We observed a reduction in the number of filaments formed in the presence of DrbA-FL compared with the control (Figure 2, compare A and B). At the same concentration, Drb1–300 also inhibited actin nucleation but to a significantly (P = 0.007, unpaired Student’s t test) smaller extent (Figure 2, compare B and C). Thus, drebrin inhibits mDia2 formin-mediated actin nucleation.
FIGURE 2:
Effects of drebrin on actin nucleation and elongations assessed in TIRF microscopy assays. (A–C) Representative images showing drebrin inhibition of mDia2-mediated nucleation of profilin–actin complexes. Actin polymerization (15% Alexa488SE-labeled) was started by the addition of polymerizing salts and the indicated proteins. After 10 min, F-actin was diluted with imaging buffer and immobilized on polyK-treated coverslip. Representative fields are shown. Conditions: [Actin] = 2 µM; [Profilin] = 10 μM; mDia2-FFC = 30 nM; [DrbA-FL] or [Drb1–300] = 0.7 µM. (D) Rates of actin filaments elongation determined from time-lapse TIRF microscopy experiments in the absence and presence of drebrin constructs. Left panel: without formin; Right panel: in the presence of mDia2-FFC formin construct. The rates that are not representative of the mDia2-driven actin assembly are under the dashed line (bottom-right panel). Actin filaments that were elongating at slower rates than those in the “actin-profilin-DrbA-FL” sample are encircled (right panel). Conditions: [Actin] = 0.5 µM; [profilin] = 2.5 µM; [mDia2-FFC] = 1.5–3 nM; [DrbA-FL] or [Drb1–300] = 0.7 µM. (E) Population size of fast and slow F-actin elongation events observed in the samples (graphic representation of data in D, right panel). We used “FFC only” control (D, right panel, green dots) to define the fast population. In this sample, two populations of elongating filaments are evident: main (fast, n = 21 events) and minor (slow, n = 3 events) populations. The mean rate of the fast population in “FFC only” samples was 17.3 ± 2.7 subunits/s (n = 21 events). We defined fast elongation events in all three samples as those with elongation rates = mean ± 2 SD of the fast population in “FFC only” samples (white part of the bar). Populations of slow and intermediate elongating events are represented by a gray part of the bar. Note that we observed an increase in population size of slow elongating filaments with both DrbA-FL and Drb1–300. The numbers of filaments switching their elongation rates within observation time are shown for all samples in the bottom panel. Rare instances of filaments capped by photo-induced actin dimers (depolymerizing under our conditions) were excluded from the analysis.
Drebrin effects on actin elongation in the absence of formin
To further clarify the effects of drebrin on mDia2-driven actin polymerization, we assessed drebrin’s effect on the processive actin elongation by mDia2 in time-lapse single-filament TIRF assays.
First, we set out to determine whether neuronal drebrin A has any effects on elongation of actin filaments in the absence of mDia2 formin. Intriguingly, our single-filament TIRF microscopy assays revealed a previously undocumented effect of DrbA-FL on F-actin dynamics. We found that DrbA-FL inhibits F-actin elongation (Supplemental Figure S2A). We also found, that this inhibitory effect of DrbA-FL on F-actin elongation is preserved in the presence of profilin (Figure 2D, left panel, compare green and red dots). Specifically, the rate of filament elongation with profilin–actin complexes decreased approximately twofold (from 4.3 ± 0.7 subunits/s [n = 45] to 2.3 ± 0.5 subunits/s [n = 19] for 0.5 µM actin). This suggests that neuronal drebrin A is a leaky capper of actin filaments that allows addition of actin monomers to growing ends but slows the rate of this process.
To clarify whether the observed inhibitory effect of DrbA-FL on actin elongation is due to the drebrin-induced morphological changes in actin filaments, we employed Drb1–300, which induces the same morphological changes in F-actin as DrbA-FL (Sharma et al., 2011, 2012). Interestingly, Drb1–300 construct did not show inhibition of actin filament elongation in time-lapse TIRF assays when used at the same concentration as DrbA-FL (Figure 2D, left panel, compare green and blue dots). We also tested a higher concentration of Drb1–300 (1.4 µM). Based on previously determined Kd values and stoichiometries, this concentration yields the same occupancy on actin as 0.7 µM of DrbA-FL (Ishikawa et al., 1994; Grintsevich et al., 2010; Sharma et al., 2011). Again, we did not observe any inhibition of actin elongation with Drb1–300. Furthermore, in both independent experiments described above, actin elongation rates in Drb1–300 containing samples were (on average) slightly higher (∼15%) than the corresponding controls (actin-profilin samples) assayed on the same day with the same set of reagents (Figure 2D, left panel, compare green and blue dots). Thus, DrbA-FL but not Drb1–300 inhibit actin elongation from profilin–actin complexes.
To confirm that drebrin impacts the B-ends of actin filaments, we carried out annealing experiments (Supplemental Figure 2SB). We observed that filament annealing was greatly inhibited in the presence of DrbA-FL and much less with Drb1–300. This result is consistent with DrbA-FL inhibition of B-end elongation. Together, our results reveal that drebrin is a leaky capper of actin filaments and this activity depends on the intrinsically disordered C-terminal region of drebrin that is missing in the Drb1–300 construct.
Drebrin effect on actin elongation in the presence of mDia2 formin
Next, we hypothesized that the ability of drebrin to interfere with B-ends of actin filaments may have an effect on formin-mediated processive elongation of F-actin. We tested this hypothesis in time-lapse single-filament TIRF assays (see Supplemental Movies S1–S3). In the presence of profilin–actin and mDia2-FFC, we expect two main populations of actin filaments—elongating fast (formin-bound, predominant population) and slowly (formin-free). In the presence of DrbA-FL we observed greater heterogeneity in elongation rates of actin filaments nucleated by mDia2-FFC from profilin–actin complexes (Figure 2D, right panel, and Supplemental Figure S3). Actin filaments that were switching their elongation rates within the observation time (Figure 2E, bottom panel) were also included in the analysis, and therefore, infrequently, some of the filaments yielded more than one elongation rate (event; see examples in Supplemental Figure S3). When two phases of elongation were observed within the same filament (Supplemental Figure S3), their rates were not averaged but treated as two separate elongation events.
Movie S1.
Examples of F-actin elongation events assisted by mDia2-FFC in the presence of profilin-actin complexes. [Actin]=0.5μM, [profilin]=2.5μM, [mDia2-FFC]=1.5nM; Bar=10μm. Movie speed: 3 frames/s.
Movie S2.
Representative movie of F-actin elongation assisted by mDia2-FFC in the presence of profilin-actin complexes and DrbA-FL. [Actin]=0.5μM, [profilin]=2.5μM, [DrbA-FL]=0.7μM, [mDia2-FFC]=3nM; Bar=10μm. Movie speed: 3 frames/s.
Movie S3.
Representative movie of F-actin elongation assisted by mDia2-FFC in the presence of profilin-actin complexes and Drb1-300. [Actin]=0.5μM, [profilin]=2.5μM, [1-300]=0.7μM, [mDia2-FFC]=3nM; Bar=10μm. Movie speed: 3 frames/s.
As expected, in the “mDia2-FFC only” sample (Figure 2D, right panel, green dots) we detected two populations of F-actin-elongating events—fast (main, formin-bound population, n = 21 events) and slow (minor, formin-free population, n = 3). Therefore, we used these values to define the fast and slow populations of actin-elongating events in drebrin-containing samples (Figure 2E and its legend). We defined fast elongation events in all three samples as those with elongation rates within mean ± 2 SD (Figure 2D, right panel, green dots) of the fast population in “mDia2-FFC only” (Figure 2E, white segments of the bars). Relative population sizes of the fast and slow actin elongation events are shown in Figure 2E. Comparison of mDia2-driven processive elongation of F-actin in the absence and presence of drebrin constructs revealed that even at saturating concentrations of DrbA-FL, a fraction of filaments exhibited rates similar to those observed in drebrin-free samples (17.4 ± 2.6 subunits/s [n = 16 events] vs. 17.3 ± 2.7 subunits/s [n = 21 events], respectively). The samples containing drebrin constructs exhibited several differences compared with the drebrin-free samples. First, the fraction of slow F-actin elongation events was greater than in the drebrin-free samples (Figure 2E, gray parts of the bars). This effect was most pronounced in the presence of DrbA-FL (50% of events exhibited slow or intermediate actin elongation rates). Second, with DrbA-FL, we observed two populations of slow elongating F-actin. Specifically, some of these filaments grew at rates similar to those observed in the absence of formin, probably due to mDia2 displacement from the B-ends (compare Figure 2D, red dots in the right and left panels). At the same time, we also observed a fraction of filaments that were elongating at slower rates (encircled in Figure 2D, right panel) than those in the “actin-profilin–DrbA-FL” sample (Figure 2D, left panel). Therefore, we speculate that DrbA-FL may form an inhibitory complex with formin at the B-end, attenuating mDia2-driven F-actin elongation. Thus, drebrin A inhibits formin-driven nucleation of actin and decreases the fraction of filaments growing at the rates characteristic of mDia2-driven processive elongation compared with a drebrin-free control.
Domain requirements for inhibition of mDia2-driven actin assembly by drebrin
We set out to determine the domain requirements for the observed inhibition of mDia2 by drebrin. To this end, we employed C-terminal truncations of drebrin and mDia2 formin constructs. Structurally, the dimeric FH2 domain of formins is necessary and sufficient for actin nucleation (Sagot et al., 2002). However, it was recently shown that C-terminal regions of formins (tails) aid actin nucleation and improve the processivity of formins (Chhabra and Higgs, 2006; Vizcarra et al., 2014). Notably, formins’ tails are considered a “hot spot” for interactions with other proteins (such as Spire, tubulin, tumor suppressor adenomatous polyposis coli protein, and Bud6) (Pechlivanis et al., 2009; Gaillard et al., 2011; Vizcarra et al., 2011; Breitsprecher et al., 2012; Tu et al., 2012). Therefore, we employed mDia2, with and without its C-terminal tail (mDia2-FFC and mDia2-FF constructs), to test for a possible modulation of its function by drebrin.
We used pyrene fluorescence assays to assess the extent of mDia2-FFC and mDia2-FF formin inhibition by DrbA-FL and Drb1–300 constructs (Figure 3). We used concentrations of drebrin constructs sixfold to ninefold higher than their IC50 values (Figure 1) to saturate the effect. We observed that DrbA-FL (0.25 µM) inhibits actin polymerization driven by mDia2-FFC to a greater extent than Drb1–300 construct at 7 µM (Figure 3A and Supplemental Figure S4). This is consistent with the presence of a second mDia2-binding site in the C-terminal part of drebrin sequence (amino acids 301–706). Truncation of C-terminal “tail” region of mDia2-FFC (mDia2-FF construct) completely abolished inhibition by Drb1–300 in profilin-free samples (Figure 3B). It should be noted that in the presence of profilin, some inhibition of mDia2-FF-mediated actin assembly was observed with saturating concentrations of Drb1–300 (Supplemental Figure S4). We speculate that high concentrations of Drb1–300 (7 µM) may affect the processivity of formin, which would explain some inhibition of mDia2-FF in the presence of profilin. This is in line with our single-filament TIRF microscopy assays revealing a greater fraction of slow actin elongation events in the presence of Drb1–300 construct compared with a no-drebrin control (Figure 2E). At the same time, inhibition of mDia2-FF-mediated actin assembly by DrbA-FL was greatly weakened, but still observed at all concentrations tested (0.25–1 µM) (Figure 3B, Supplemental Figure S4B, and unpublished data). The results of our bulk polymerization assays suggest that drebrin may have two binding sites on formin—one within the formin’s tail region and another one in the FH1-FH2 sequence.
FIGURE 3:
Domain requirements for drebrin-induced inhibition of mDia2-driven actin assembly. Graphic representations of proteins present in the reactions (except for drebrin constructs) are shown in each panel. The FH1, FH2, and C-terminal tail domains of mDia2 are shown in blue, gray, and orange, respectively. Green spheres represent actin. Each panel demonstrates the effects of DrbA-FL and Drb1–300 on the rates of actin assembly by mDia2-FFC (A) or its C-terminal tail missing version, mDia2-FF (B) (shown as an orange line in the schematics). In each panel (A, B) white, gray, and black bars represent actin assembly rates when no drebrin, DrbA-FL, or Drb1–300, respectively, are added. (A) Actin assembly mediated by mDia2-FFC is inhibited strongly in the presence of DrbA-FL. This inhibition is weaker in the presence of Drb1–300. (B) Actin assembly mediated by mDia2-FF is partially inhibited in the presence of DrbA-FL but not in the presence of Drb1–300. Note that the error bar in the no-drebrin condition is too small to be visible. Representative sets of data are shown. Error bars mean ± SD (n = 3 replicates). Conditions: [Actin] = 1 µM (10% pyrene-maleimide); [mDia2-FFC] = 30 nM; [DrbA-FL] = 0.25 µM; [Drb1–300] = 7 µM. Buffer: KMEH7.
FIGURE 4:
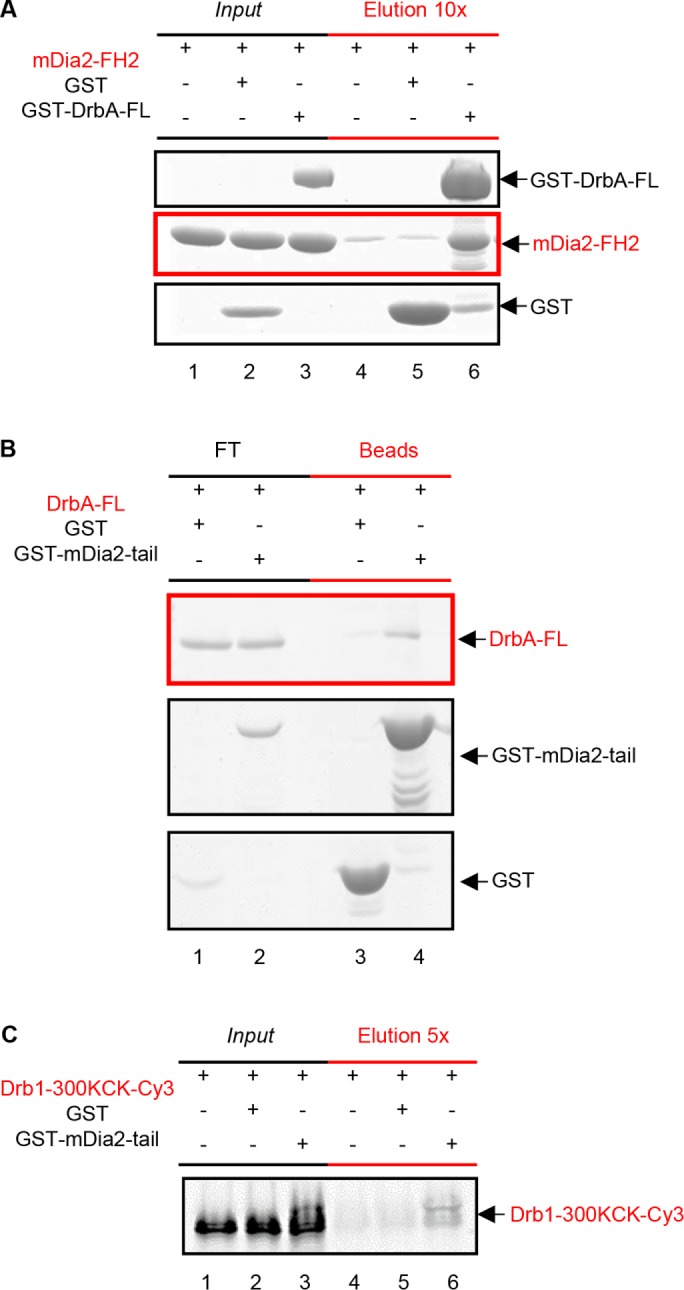
Drebrin A is a novel binding partner of mDia2. (A) GST-DrbA-FL directly binds mDia2-FH2. GST or GST-DrbA-FL (1 µM) was incubated with mDia2-FH2 (2 µM) at 4°C overnight (lanes 1–3, Input). These samples were applied to glutathione-Sepharose beads, incubated for 3 h at 4°C, and then eluted with a volume of glutathione, 1/10th of the input volume (lanes 4–6, Elution 10x). Samples were analyzed by SDS–PAGE and Coomassie Blue staining. Note that mDia2-FH2 does not significantly bind beads or GST alone (lanes 4 and 5, red-lined panel), but coelutes with GST-DrbA-FL (lane 6, red-lined panel). Buffer: KMEH7, 0.5 mM Thesit, 0.05% IGEPAL CA 630. (B) GST-mDia2-tail binds substoichiometric amounts of DrbA-FL. GST or GST-mDia2-tail (2 µM) was incubated with DrbA-FL (1 µM) and glutathione-Sepharose beads for 2 h at 4°C. Flow-through (FT, lanes 1 and 2) and beads (lanes 3 and 4) were analyzed by SDS–PAGE and Coomassie Blue staining. Note that DrbA-FL binds GST-mDia2-tail (lane 4, red-lined panel) to a much greater extent than GST alone (lane 3, red-lined panel). Buffer: KMEH7, 0.5 mM Thesit. (C) GST-mDia2-tail binds sub-stoichiometric amounts of Drb1–300KCK-Cy3. GST or GST-mDia2-tails (2 µM) were incubated with Cy3-labeled Drb1–300 (1 µM) at 4°C overnight (lanes 1–3, Input). These samples were applied to glutathione-Sepharose beads, incubated for 3 h at 4°C, and then eluted with a volume of glutathione, 1/5th of the input volume (lanes 4–6, Elution 5×). Samples were analyzed by SDS–PAGE and then visualized using Cy3 fluorescence. Note that Drb1–300 KCK-Cy3 binds very weakly to beads or GST alone (lanes 4 and 5), but coelutes with GST-mDia2-tail (lane 6). Buffer: KMEH7, 0.5 mM Thesit, 0.05% IGEPAL CA 630.
Drebrin is a novel binding partner of mDia2
We used pull-down assays to probe for direct interaction between mDia2 and drebrin. The FH2 domain and C-terminal tail region of formins are largely responsible for accelerated actin nucleation and formin’s processivity (Sagot et al., 2002; Pring et al., 2003; Vizcarra et al., 2014). Drebrin binding to one or both of these regions would be consistent with its inhibition of formin-mediated actin assembly.
We first tested GST-drebrin interaction with FH2 domain of formin (tail region is truncated). Our data revealed complex formation between FH2 domain of mDia2 and DrbA-FL (Figure 4A). Specifically, incubation of 1 µM of GST-DrbA-FL with 2 µM of mDia2-FH2 resulted in their complex formation with ∼1:1 binding stoichiometry (Figure 4A, lane 6). Each molar equivalent of GST-DrbA-FL bound 0.92 ± 0.21 equivalents of mDia2-FH2 (n = 4 pull-down experiments; mean ± SD). The quantitative pull-down experiment yielded a Kd of ∼0.7 µM for mDia2-FH2 binding to drebrin A (Supplemental Figure S5). In contrast to that, binding of Drb1–300 to mDia2-FH2 was weak at best (Supplemental Figure S6).
As shown in Figure 3B and Supplemental Figure S4B, the truncation of C-terminal tail of mDia2 weakened drebrin’s inhibition of mDia2-FF-driven actin assembly. Thus, we expected drebrin to bind strongly to C-terminal tails of mDia2 formin. However, our pull-down experiments with DrbA-FL and Drb1–300 indicated that drebrin interaction with formin tail alone was weak (Figure 4, B and C). Sub-stoichiometric amounts of DrbA-FL were pulled down with GST-mDia2-tails, which is consistent with a weak interaction between drebrin and the mDia2 tail region (Figure 4B). We also tested the binding of Drb1–300 to GST-mDia2-tail in pull-down experiments. To aid in protein detection, we employed a Drb1–300-KCK construct labeled with Cy3 fluorescent dye. Incubation of 2 µM of GST-mDia2-tail with 1 µM of Drb1–300-KCK-Cy3 revealed weak interaction between these two constructs that was detected via Cy3 fluorescence (Figure 4C). Thus, mDia2 and neuronal DrbA-FL interact directly through two independent binding sites. Based on our data, the strong drebrin binding site is located in the FH2 domain of formin and it likely interacts with the disordered C-terminal region of drebrin. The second (weak) drebrin binding site is located in the tail region of mDia2. Taken together, our data suggest that drebrin interaction with two sites on formin is required for strong inhibition of actin assembly (see Discussion).
Full-length drebrin A inhibits F-actin bundling by mDia2-FH2
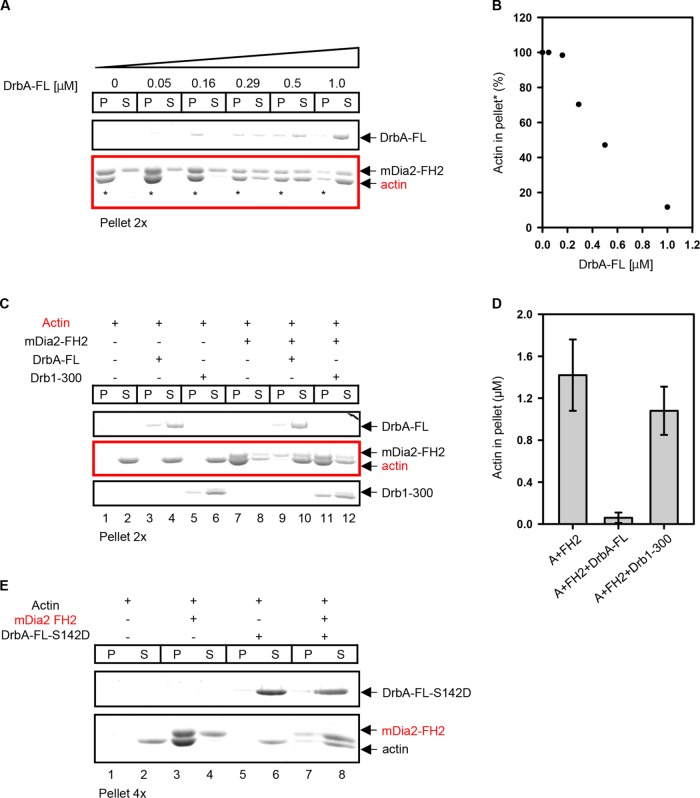
To better understand the functional consequences of DrbA-FL binding to the FH2 domain of mDia2, we employed cosedimentation assays. It was documented earlier that mDia2 formin has actin side-binding activity. This activity leads to a formation of F-actin bundles and is mediated by the mDia-FH2 domain (Harris et al., 2006). In light of a direct interaction of drebrin with the FH2 domain of formin, we hypothesized that DrbA-FL (but not Drb1–300) may affect actin filaments bundling by mDia2. Our low-speed centrifugation assays confirmed this hypothesis. Specifically, DrbA-FL abolished F-actin bundling by mDia2 in a concentration-dependent manner (Figure 5, A and B). The effect of Drb1–300 construct on actin bundling by mDia2-FH2 was dramatically weakened compared with that of the DrbA-FL (Figure 5, C and D).
FIGURE 5:
Drebrin A inhibits bundling of filamentous actin by mDia2. (A) DrbA-FL inhibits mDia2–FH2-induced F-actin bundling in a concentration-dependent manner. Increasing amounts of DrbA-FL (0, 0.05, 0.15, 0.29, 0.50, and 1 µM) were added to mDia2-FH2 (0.8 µM) and F-actin (2 µM). Low-speed pellets were concentrated twofold to aid in protein quantification. Note that as DrbA-FL concentration increases, the amount of actin in the low-speed pellet decreases (lanes denoted with asterisks, red box), whereas the amount of actin in the supernatant increases. (B) Amounts of F-actin bundled by mDia2-FH2 in the presence of DrbA-FL. Quantification of the data shown in the red box panel of A (in lanes marked with asterisk). (C) C-terminal truncation of drebrin sequence dramatically reduces its inhibitory effect on mDia2–FH2-induced F-actin bundling. Either DrbA-FL (0.85–1 µM) or Drb1–300 (1.9 µM) was added to mDia2-FH2 (0.8 µM) and phalloidin-stabilized F-actin (2 µM). Pellets were concentrated twofold to aid in quantification. Note that no bundling (actin in pellet) is detected with actin alone and both drebrin constructs (lanes 1–6, red panel). In the presence of mDia2-FH2 actin was found in the pellet (lane 7, red panel). DrbA-FL strongly inhibits mDia2-FH2-mediated bundling such that almost no actin was detected in the pellet (lane 9, red panel). However, Drb1–300 did not affect mDia2-FH2-mediated bundling to any considerable extent (lane 11, red panel). (D) Quantification of the amounts of actin in low-speed pellets in the presence of mDia2-FH2 (FH2) and drebrin constructs. Representative gel is shown in C. Error bars: mean ± SD (n = 3 experiments). (E) Phosphomimetic mutant of drebrin A (DrbA-FL-S142D) inhibits mDia2-FH2 F-actin bundling as well as DrbA-FL. DrbA-FL-S142D (1 µM) was added to mDia2-FH2 (0.8 µM) and F-actin (2 µM). Low-speed pellets were concentrated fourfold to aid in their quantification. Note that F-actin alone is not bundled (actin can be detected in the supernatant, lane 2). mDia2-FH2 bundles F-actin (actin can be detected in the pellet, lane 3, red panel). When DrbA-S142D is introduced, mDia2-FH2-mediated bundling of F-actin is inhibited (actin is mostly in the low-speed supernatant; compare lanes 7 and 8, red panel).
We wished to gain more insight into debrin-induced inhibition of mDia2-FH2-mediated bundling. Under our experimental conditions, F-actin bundling was not detected with any of the drebrin constructs tested by low-speed cosedimentation (Figure 5C, lanes 3–6). However, our TIRF assays show that Drb1–300 can lightly bundle F-actin. Such activity of a similar drebrin construct was reported previously (Worth et al., 2013). It was predicted that a deletion of C-terminal sequence of drebrin (as well as phosphorylation at S142) exposes an actin binding site (which is otherwise masked by the C-terminus of drebrin) and allows for bundling. We were concerned that different effects of DrbA-FL versus Drb1–300 constructs (on actin bundling by mDia2-FH2) may arise from the differences in their own actin bundling properties. Therefore, we employed a phosphomimetic mutant of drebrin (DrbA-FL-S142D) that was shown to induce F-actin bundling similar to the Drb1–300 construct (Worth et al., 2013). We set out to determine whether DrbA-FL-S142D would inhibit mDia2-FH2–induced actin bundling in the same manner as unphosphorylated drebrin. We found that similar to DrbA-FL, its S142D phosphomimetic mutant abolishes F-actin bundling by mDia2-FH2 (Figure 5E). This is in a good agreement with the evidence for direct interaction of drebrin with the FH2 domain of mDia2 formin.
Notably, high-speed centrifugation showed that mDia2-FH2 and DrbA-FL can cobind F-actin leading to a formation of a tertiary complex (Figure 6A). We also found that DrbA-FL causes partial displacement of mDia2-FH2 from actin filaments but Drb1–300 does not (Figure 6B). Therefore, we asked whether DrbA-FL inhibits mDia2-FH2–induced bundling by partially displacing it from actin filaments. We found that in the presence of DrbA-FL, the amount of mDia2-FH2 that was still bound to F-actin (∼0.5 µM) would be sufficient to induce extensive bundling (Supplemental Figure S7). Specifically, in the presence of DrbA-FL, binding of 0.5 µM of mDia2-FH2 to actin (Figure 6B) resulted in 2.8 ± 2.4% of bundled F-actin (in low-speed pellet, Figure 5D, middle bar). In contrast to that, 0.5 µM mDia2-FH2 added to bare F-actin yielded >70% bundling (see Supplemental Figure S7B, red arrow). Thus, partial displacement of mDia2-FH2 by DrbA-FL cannot account for the observed dramatic inhibition of actin bundling.
FIGURE 6:
mDia2-FH2 and drebrin constructs codecorate F-actin. (A) High-speed sedimentation of DrbA-FL and Drb1–300 with F-actin and mDia2-FH2. DrbA-FL (0.85 µM) or Drb1–300 (1.9 µM) was added to mDia2-FH2 (0.8 µM) and F-actin (2 µM). Note that both mDia2-FH2 and drebrin constructs are detected in high-speed pellets, with the formin construct band just above the actin band (lanes 9 and 11). (B) DrbA-FL, unlike Drb1–300, partially displaces mDia2-FH2 (FH2) from F-actin. Quantification of data shown in A. Error bars: mean ± SD (n = 3 experiments). (C) Proposed working model of mDia2–drebrin A interaction. FH1, FH2, and C-terminal tail of mDia2 formin are shown in dark blue (line), gray, and orange (line), respectively. The N-terminal sequence of drebrin is shown as a tricolored box, and the intrinsically disordered C-terminal part of drebrin is shown as a black line. Strong drebrin binding to mDia2 requires two interaction sites. High-affinity binding between the FH2 domain of formin and the C-terminal part of drebrin guides its interaction with the second interacting site within formin tails. Drebrin interaction with isolated mDia2-tails is weak but it is improved in mDia2-FFC construct due to the increase of local drebrin concentration near the formin’s tails.
High-speed centrifugation with Drb1–300 revealed its codecoration of F-actin with mDia2-FH2, similar to that observed with DrbA-FL. Because Drb1–300 did not show strong binding to mDia2-FH2 in pull-down assays, these results suggest that the N-terminal sequence of drebrin (Drb1–300) and mDia2-FH2 may occupy nonoverlapping binding sites on F-actin. Structural analysis of such complexes, which is beyond the scope of this study, is needed to test this prediction. Thus, our data are consistent with the possibility that inhibition of mDia2-induced actin bundling by drebrin A is mediated through its interaction with the FH2 domain of mDia2 formin.
DISCUSSION
In this study we documented that drebrin is a previously unknown inhibitor of actin assembly driven by mDia2. Additionally, we demonstrated using single-filament TIRF microscopy assays that neuronal drebrin A inhibits actin elongation in the absence of formins. We gained insights into the mechanism of drebrin’s inhibition of mDia2 function by demonstrating a direct interaction between these two proteins in pull-down assays. Furthermore, we clarified the domain requirements for mDia2 inhibition by drebrin using their truncated constructs. On the basis of our data, we propose a model of drebrin–formin interaction in which FH2 domain of formin interacts with the intrinsically disordered C-terminal part of drebrin molecule. At the same time, our data suggest the presence of a second (weaker) drebrin-binding site in the C-terminal “tails” of mDia2 (see model in Figure 6C).
We documented that drebrin attenuates several aspects of actin dynamics in the absence and presence of mDia2 formin. First, we showed that drebrin inhibits actin nucleation. The results of our pull-down experiments, TIRF microscopy and pyrene fluorescence assays suggest that drebrin’s association with both the C-terminal tail and FH2 domain of mDia2 contributes to this effect. This conclusion is based on the observations (Figure 2, A–C) that both DrbA-FL and Drb1–300 inhibit actin nucleation but to different extents. Specifically, full-length drebrin exhibits stronger inhibition of mDia2, consistent with the presence of an FH2 domain-binding site in its C-terminal sequence (aa 301–706) in addition to that in the aa 1–300 region (Figure 4). We also found that truncation of the tail region of mDia2 (mDia2-FF construct) dramatically reduces but does not abolish the inhibition of formin-driven actin assembly by DrbA-FL, which is consistent with the presence of a drebrin-binding site outside the mDia2-tail region (Figure 3B and Supplemental Figure S4B). We hypothesize that strong drebrin binding to formin requires two interaction sites (Figure 6C). Our data are consistent with the scenario in which high-affinity binding between FH2 domain of formin and the C-terminal part of drebrin guides its interaction with the second interacting site within formin tails. Therefore, drebrin interaction with isolated mDia2-tails is weak but it is improved in mDia2-FFC construct due to the increase of local drebrin concentration near the formin’s tails. Experiments with mDia2-FF formin construct (Figure 3) suggest that drebrin interaction with formin tails is largely responsible for the observed inhibition of mDia2-driven actin assembly.
We also documented that neuronal drebrin A inhibits actin elongation in the absence of formin. It is unlikely that this effect would be mediated via characteristic drebrin-induced morphological changes in actin filaments. We deduced this from our experiments with Drb1–300 construct that induces the same changes in F-actin morphology as DrbA-FL (Sharma et al., 2012) but does not inhibit actin elongation (Figure 2D, left panel). Thus, it is possible that intrinsically disordered C-terminal part of drebrin A can transiently interact with the B-end of actin filament and interfere with monomers addition. In vivo, such activity of debrin A may interfere with the binding of B-end interacting proteins to actin filaments.
In the presence of mDia2 and drebrin constructs we observed a greater heterogeneity of single-filament elongation rates than in the no-drebrin control (Figure 2, D and E). We hypothesize that such increased heterogeneity may arise from several factors (or their combination). Because we observed a larger fraction of slow F-actin–elongating events with both full-length drebrin A and Drb1–300, it is possible that drebrin-induced changes in actin morphology may affect the duration of formin’s processive runs. Additionally, decoration of actin filaments by drebrin may bring its inhibitory sequence(s) into close proximity to barbed-end bound formin. Specifically, we observed a fraction of filaments elongating at slower rates (encircled in Figure 2D, right panel) than those in the “actin-profilin-DrbA-FL” sample, which suggests a formation of an inhibitory complex between drebrin (bound to F-actin) and formin at the B-end. The above factors (or their combination) may attenuate the processivity of mDia2. It should be noted that it is unknown whether the drebrin sequence(s) that inhibit(s) mDia2 is/are also responsible for its leaky F-actin capping activity.
We also documented strong inhibition of actin bundling by mDia2-FH2 in the presence of drebrin A, but the physiological role of this inhibition is yet to be determined. Thus, the role of mDia2-induced actin bundling in neuronal function and development is yet to be defined.
Several lines of evidence suggest that a cross-talk between drebrin and formins can be physiologically important for dendritic spine formation. It was previously documented that mDia2-induced actin nucleation/polymerization is critical for the development and morphology of DS (Hotulainen et al., 2009). During DS development, filopodium-type protrusions (lacking spine heads) progress to the spine types that can support synaptic transmission and are usually composed of a neck and bulbous head. Hotulainen et al. (2009) proposed a model of actin remodeling during dendritic spine development. Their model suggests that spine formation starts with the initiation and elongation of dendritic filopodium, where actin polymerization is driven by mDia2 formin localized in the filopodium tip. Based on the loss of spine heads on overexpression of constitutively active mDia2, it was proposed that during spine head formation, the mechanism of actin assembly changes from mDia2-driven (unbranched) to Arp2/3-mediated (branched), which is consistent with a critical role of Arp2/3 in spine head formation (Hotulainen et al., 2009). However, what triggers such a change in actin architecture during DS development is unclear. Our results point toward the possible role of drebrin A in this process. We speculate that accumulation of drebrin A in DS, occurring during spine maturation, can lead to the inhibition of mDia2-driven actin polymerization. This may aid other actin regulators in promoting actin branching. It is also important to note that, even at saturating concentrations of drebrin, we observed a fraction of formin-bound filaments that were elongating with rates similar to drebrin-free controls. This might be physiologically significant for the reported formin-driven expansion of mature spines, where drebrin could limit formin-driven actin polymerization but does not block it completely—allowing for the formation of spike-like protrusions in spine heads (Chazeau et al., 2014).
In conclusion, this study provides new insights into formin regulation of actin assembly in neuronal cells and its interplay with drebrin.
MATERIALS AND METHODS
Protein expression, purification, and labeling
Rabbit skeletal actin (Spudich and Watt, 1971), profilin (Harris et al., 2004), Drb1–300, and GST-Drb1–300 construct (Grintsevich et al., 2010) were prepared as described previously. Actin was labeled with pyrene maleimide and Alexa488-SE using published protocols (Grintsevich et al., 2017). Drb1–300-KCK was labeled with Cy3 as previously described (Sharma et al., 2012; Grintsevich et al., 2017).
The mDia2-FFC (amino acids 521–1171, mouse) containing plasmid was a generous gift from H. Higgs (Dartmouth College). The mDia2-FF construct was created via mutagenesis, with primers designed to introduce a stop codon after the FH2 domain (residue 1034). The mDia2-FH2 construct (amino acids 612–1034, mouse) was a kind gift from M. Quinlan (UCLA). The mDia2-FFC, FF, and tail (seq. 1008–1171) constructs were expressed as N-terminal GST fusion constructs in Escherichia coli Rosetta2 cells (Li and Higgs, 2003). Purification of the formin constructs was carried out essentially as described (Li and Higgs, 2003), with the following modifications. Formin constructs were purified on a glutathione-Sepharose 4B (GE Healthcare) column, and GST-tag was cleaved with thrombin overnight at 4°C. Cleaved formin constructs were further purified using gel-filtration on HiLoad 16/60 Superdex 75 (Amersham Biosciences).
DrbA-FL was expressed and purified as described (Sharma et al., 2011), with the following modifications. Before elution from the glutathione-Sepharose column, preparations were subjected to a wash (50 ml total volume) with buffer A (20 mM HEPES, pH 7.4, 20 mM MgCl2, 5 mM ATP, 150 mM KCl, 0.1% IGEPAL CA 630) in addition to high-salt (1 M NaCl) and glycerol (10%) washes. Specifically, GST-drebrin–bound glutathione-Sepharose beads were incubated with 10 ml of buffer A for 25 min and then washed with the rest of buffer A. The wash with buffer A was found to be important for consistency of pyrene-actin polymerization assays in the presence of formin. After protein elution from the glutathione-Sepharose column and overnight dialysis at 4°C against drebrin prep buffer (21.4 mM Na2HPO4, 3.8 mM KH2PO4, pH 7.5, 300 mM NaCl, and 5 mM β-mercaptoethanol [β-ME]), GST-tag was cleaved with PreScissionProtease for 2.5 h at 4°C. After the cleavage, DrbA-FL was loaded onto HisTrap column (1 ml) and treated with benzonaze for 15 min at 4°C. Before elution, DrbA-FL was washed with buffer A without IGEPAL CA 630. Drebrin was eluted with the gradient (0–100%) of imidazole (10 mM Tris, pH 8, 300 mM NaCl, 5% glycerol, 3 mM β-ME, and 0–250 mM imidazole). The final gel-filtration step was carried out on a HiLoad 16/60 Superdex 75 column (Amersham Biosciences). Drebrin was concentrated, dialyzed against the DDB buffer (5 mM Tris, 100 mM KCl, and 1 mM dithiothreitol [DTT]), and stored at –80°C. GST-DrbA-FL was purified using the same scheme except for the omission of the HisPur Cobalt gravity column, GST-tag cleavage step, and gel-filtration. GST-DrbA-FL preparations contained a small fraction of C-terminally truncated GST-DrbA that could not be efficiently separated due to GST dimerization.
Actin polymerization assays
Mg-ATP G-actin (10% pyrene-maleimide labeled) was prepared by incubating Ca-ATP-G-actin with ME exchange buffer (0.05 mM MgCl2 and 0.2 mM ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid [EGTA]) for 3 min. Actin polymerization was initiated immediately after Ca/Mg exchange by the simultaneous addition of polymerization buffer KMEH7 (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM HEPES, pH 7, 1 mM DTT, and 0.2 mM ATP) and actin-binding proteins (profilin, drebrin, and formin constructs). The average delay between the beginning of actin polymerizations and the first measurement was ∼35 s. Changes in pyrene fluorescence signal were monitored over time (1–4 h) using a microplate-reading fluorimeter (TECAN). SigmaPlot software was used to fit actin polymerization kinetics to exponential expressions yielding polymerization rates and their standard deviations.
TIRF microscopy
To assess actin nucleation efficiency of mDia2 in the presence of drebrin constructs, we carried out the reactions, diluted them, and immobilized actin filaments on the polylysine-coated surface. Specifically, coverslips were treated with 1 mg/ml polylysine for 3 min, rinsed with mQ water, and air-dried. Mg-ATP-G-actin (2 µM 15% Alexa488-SE labeled) was polymerized in KMEH7 in the presence of profilin (10 µM) and mDia2-FFC (30 nM) with and without drebrin constructs (0.7 µM). After 10 min at room temperature, samples were diluted 67-fold in KMEH7, supplemented with 100 mM DTT and 1 µM phalloidin, and applied onto polyK-coated coverslips. Random fields were imaged using a DMI6000 TIRF microscope (Leica). Actin filaments were counted manually.
Time-lapse TIRF microscopy was performed as described (Gurel et al., 2014; Grintsevich et al., 2017). In brief, untethered actin filaments were imaged on a Pluronic coated surface. Flow chambers (V ∼12 μl) were treated with two chamber volumes (CV) of 1% Pluronic F127 solution (Sigma; P2443) for 3 min and then equilibrated with 2 CV of 1× TIRF-imaging buffer (10 mM HEPES, 1 mM MgCl2, and 50 mM KCl, 0.2 mM EGTA [pH 7] supplemented with 50 mM DTT, 0.2 mM ATP, 20 mM glucose, and 0.5% methyl cellulose). G-actin–profilin mixtures (0.5 µM 15% Alexa488-SE labeled actin, 2.5 µM human profilin-1) were incubated for 3 min at room temperature (RT) with ME exchange buffer. Then, Mg-ATP-G-actin in complex with profilin was mixed with polymerizing salts supplemented with mDia2FFC (1–3 nM), with and without drebrin constructs (0.7 µM). The resulting mixture (4 CV) was introduced into the flow chamber. Actin mixtures were supplemented with 0.05 mg/ml caseine, 0.25 mg/ml glucose oxidase, and 50 μM catalase to minimize protein damage due to free radical formation and photobleaching during the imaging. Filaments were imaged using a DMI6000 TIRF microscope (Leica). Images were acquired every 10 s. All time-lapse TIRF data were analyzed using ImageJ (Fiji) software (National Institutes of Health, Bethesda, MD).
Pull-down assays
Pull-down experiments with GST-DrbA-FL or GST-mDia2-tail constructs were performed as follows. GST-tagged constructs or GST were incubated overnight at 4°C with untagged proteins of interest in IT-KMEH7 buffer (10 mM HEPES, pH 7, 1 mM EGTA, 1 mM MgCl2, 50 mM KCl, 1 mM DTT, 0.05% IGEPAL, 0.5 mM Thesit, and 0.9–2.2 mM Tris, pH 8). The resulting samples (Inputs, Figure 4, A and C) were applied onto glutathione-Sepharose beads equilibrated with IT-KMEH7 and nutated for 3 h at 4°C. Beads were pelleted using a bench-top centrifuge (max 2000 × g; ISC BioExpress minicentrifuge C-1301P) and then washed one time with five bead volumes of IT-KMEH7 (125 µl). Proteins were eluted with 10 mM glutathione elution buffer (1/10th or 1/5th of input volumes for Figure 4, A and C, respectively).
Pull-down experiments with GST-mDia2-tails and untagged DrbA-FL (Figure 4B) were carried out essentially as described above with minor modifications. GST-tagged mDia2-tail or GST were incubated with DrbA-FL for 10 min at 25°C in T-KMEH7 buffer (10 mM HEPES, pH 7, 1 mM EGTA, 1 mM MgCl2, 50 mM KCl, 1 mM DTT, and 0.5 mM Thesit). Samples were applied onto glutathione-Sepharose beads (equilibrated with T-KMEH7) and nutated at 4°C for 2 h. Beads were precipitated by centrifugation (14,000 rpm [20,817 × g], 2 min, 4°C). Beads were then washed three times with eight column volumes of T-KMEH7 (24 column volumes total). After washes, beads were pelleted by centrifugation and resuspended in one column volume of T-KMEH7. Supernatants and bead suspensions were supplemented with SDS–PAGE sample-loading buffer and boiled for 3 min.
All inputs and eluates were analyzed by SDS–PAGE. Drb1–300-KCK-Cy3 was detected via Cy3 fluorescence. Gels were stained with Coomassie Blue. Quantification of the gels was done using ImageJ software.
High- and low-speed pelleting assays
For high-speed pelleting assay, Mg-ATP F-actin (5 µM) was prepared by incubating Ca-ATP-G-actin in ME buffer for 3 min, followed by overnight incubation in a polymerization buffer (10 mM HEPES, pH 7, 1 mM EGTA, 1 mM MgCl2, 50 mM KCl, 1.4–1.8 mM Tris, pH 8, 1 mM DTT, and 0.2 mM ATP) at 4°C. Actin stock was then diluted to 2 µM with KMEH7 (10 mM HEPES, pH 7, 1 mM EGTA, 1 mM MgCl2, and 50 mM KCl) in the presence or absence of mDia2-FH2 and drebrin constructs (at concentrations that would correspond to ≥80% occupancy on F-actin: [DrbA-FL] = 0.85 µM and [Drb1–300] = 1.9 µM). Reaction mixtures were incubated at room temperature for 10 min and then subjected to high-speed centrifugation (TLA100 rotor, 80,000 rpm, 4°C, 20 min).
For low-speed pelleting experiments with mDia2-FH2, we followed the established protocol (Harris et al., 2006). To pellet F-actin bundles, protein mixtures were incubated at room temperature for 1 h and then subjected to low-speed centrifugation (16,000 × g, 4°C, 5 min) (Harris et al., 2006). For some of these assays, F-actin was stabilized with phalloidin (1:1 M ratio), as indicated in the figure legends.
High- and low-speed supernatants and pellets were analyzed by SDS–PAGE. Gels were stained with Coomassie Blue. ImageJ software was used to determine the intensities of the gel bands.
Supplementary Material
Acknowledgments
We thank Margot E. Quinlan for comments on the manuscript. This work was supported by US Public Health Service grant no. GM077190 to E.R.
Abbreviations used:
- DS
dendritic spines
- B-ends
barbed ends of actin filaments
- F-actin
filamentous actin
- G-actin
monomeric actin
- IC 50
concentration that yields half-maximum inhibition
- TIRF
total internal reflection fluorescence
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0639) on January 9, 2019.
REFERENCES
- Alberts AS. (2001). Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem , 2824–2830. [DOI] [PubMed] [Google Scholar]
- Alioto SL, Garabedian MV, Bellavance DR, Goode BL. (2016). Tropomyosin and profilin cooperate to promote formin-mediated actin nucleation and drive yeast actin cable assembly. Curr Biol , 3230–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almuqbil M, Hamdan FF, Mathonnet G, Rosenblatt B, Srour M. (2013). De novo deletion of FMN2 in a girl with mild non-syndromic intellectual disability. Eur J Med Genet , 686–688. [DOI] [PubMed] [Google Scholar]
- Bertling E, Hotulainen P. (2017). New waves in dendritic spine actin cytoskeleton: from branches and bundles to rings, from actin binding proteins to post-translational modifications. Mol Cell Neurosci , 77–84. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. (2013). Formins at a glance. J Cell Sci (Pt 1), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. (2012). Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science , 1164–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazeau A, Mehidi A, Nair D, Gautier JJ, Leduc C, Chamma I, Kage F, Kechkar A, Thoumine O, Rottner K, et al (2014). Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J , 2745–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M, Gould CJ, Moseley JB, Goode BL. (2009). Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell , 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. (2010). Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol , 62–74. [DOI] [PubMed] [Google Scholar]
- Chesarone-Cataldo M, Guérin C, Yu JH, Wedlich-Soldner R, Blanchoin L, Goode BL. (2011). The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev Cell , 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. (2006). INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem , 26754–26767. [DOI] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD. (2012). Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J Biol Chem , 7812–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J, Ramabhadran V, Neumanne E, Gurel P, Blanchoin L, Vantard M, Higgs HN. (2011). Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell , 4575–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombos R, Migh E, Antal O, Mukherjee A, Jenny A, Mihály J. (2015). The formin DAAM functions as molecular effector of the planar cell polarity pathway during axonal development in drosophila. J Neurosci , 10154–10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, Yu HY, Alioto SL, Eskin JA, Ydenberg CA, Waterman DP, Garabedian M, Goode BL. (2014). The F-BAR protein Hof1 tunes formin activity to sculpt actin cables during polarized growth. Mol Biol Cell , 1730–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE, Galkin VE, Orlova A, Ytterberg AJ, Mikati MM, Kudryashov DS, Loo JA, Egelman EH, Reisler E. (2010). Mapping of drebrin binding site on F-actin. J Mol Biol , 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE, Ge P, Sawaya MR, Yesilyurt HG, Terman JR, Zhou ZH, Reisler E. (2017). Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat Commun , 2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE, Reisler E. (2014). Drebrin inhibits cofilin-induced severing of F-actin. Cytoskeleton , 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel PS, Ge P, Grintsevich EE, Shu R, Blanchoin L, Zhou ZH, Reisler E, Higgs HN. (2014). INF2-mediated severing through actin filament encirclement and disruption. Curr Biol , 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. (2004). The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem , 20076–20087. [DOI] [PubMed] [Google Scholar]
- Harris ES, Rouiller I, Hanein D, Higgs HN. (2006). Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem , 14383–14392. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, Lappalainen P. (2009). Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol , 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Hayashi K, Shirao T, Xue Y, Takagi T, Sasaki Y, Kohama K. (1994). Drebrin, a development-associated brain protein from rat embryo, causes the dissociation of tropomyosin from actin filaments. J Biol Chem , 29928–29933. [PubMed] [Google Scholar]
- Ivanov A, Esclapez M, Ferhat L. (2009). Role of drebrin A in dendritic spine plasticity and synaptic function: implications in neurological disorders. Commun Integr Biol , 268–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koganezawa N, Hanamura K, Sekino Y, Shirao T. (2017). The role of drebrin in dendritic spines. Mol Cell Neurosci , 85–92. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell , 423–435. [DOI] [PubMed] [Google Scholar]
- Kühn S, Geyer M. (2014). Formins as effector proteins of Rho GTPases. Small GTPases , e29513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. (2003). The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol , 1335–1340. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. (2005). Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem , 6986–6992. [DOI] [PubMed] [Google Scholar]
- Ma L, Li Y, Wang R. (2015). Drebrin and cognitive impairment. Clinica Chimica Acta (Pt B), 121–124. [DOI] [PubMed] [Google Scholar]
- Matusek T, Gombos R, Szécsényi A, Sánchez-Soriano N, Czibula A, Pataki C, Gedai A, Prokop A, Raskó I, Mihály J. (2008). Formin proteins of the DAAM subfamily play a role during axon growth. J Neurosci , 13310–13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikati MA, Grintsevich EE, Reisler E. (2013). Drebrin-induced stabilization of actin filaments. J Biol Chem , 19926–19938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi Y, Yamashita T, Fukaya M, Sonoda T, Okuno T, Yamada K, Watanabe M, Nagashima Y, Aoki I, Okuda K, et al (2002). Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit. J Neurosci , 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaville P, Jégou A, Pernier J, Compper C, Guichard B, Mogessie B, Schuh M, Romet-Lemonne G, Carlier MF. (2014). Spire and Formin 2 synergize and antagonize in regulating actin assembly in meiosis by a ping-pong mechanism. PLoS Biol , e1001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. (2008). The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol , 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechlivanis M, Samol A, Kerkhoff E. (2009). Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J Biol Chem , 25324–25333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. (2003). Mechanism of formin-induced nucleation of actin filaments. Biochemistry , 486–496. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science , 612–615. [DOI] [PubMed] [Google Scholar]
- Quinlan ME. (2013). Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development , 4417–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. (2007). Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol , 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Faix J, Bogdan S, Linder S, Kerkhoff E. (2017). Actin assembly mechanisms at a glance. J Cell Sci , 3427–3435. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. (2002). An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol , 626–631. [DOI] [PubMed] [Google Scholar]
- Sharma S, Grintsevich EE, Hsueh C, Reisler E, Gimzewski JK. (2012). Molecular cooperativity of drebrin1-300 binding and structural remodeling of F-actin. Biophys J , 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Grintsevich EE, Phillips ML, Reisler E, Gimzewski JK. (2011). Atomic force microscopy reveals drebrin induced remodeling of f-actin with subnanometer resolution. Nano Lett , 825–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar S, Pernier J, Carlier MF. (2016). Regulators of actin filament barbed ends at a glance. J Cell Sci , 1085–1091. [DOI] [PubMed] [Google Scholar]
- Shim KS, Lubec G. (2002). Drebrin, a dendritic spine protein, is manifold decreased in brains of patients with Alzheimer’s disease and Down syndrome. Neurosci Lett , 209–212. [DOI] [PubMed] [Google Scholar]
- Shimada A, Nyitrai M, Vetter IR, Kühlmann D, Bugyi B, Narumiya S, Geeves MA, Wittinghofer A. (2004). The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol Cell , 511–522. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem , 4866–4871. [PubMed] [Google Scholar]
- Tu D, Graziano BR, Park E, Zheng W, Li Y, Goode BL, Eck MJ. (2012). Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6. Proc Natl Acad Sci USA , E3424–E3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Bor B, Quinlan ME. (2014). The role of formin tails in actin nucleation, processive elongation, and filament bundling. J Biol Chem , 30602–30613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, Quinlan ME, Eck MJ. (2011). Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci USA , 11884–11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. (1999). Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol , 136–143. [DOI] [PubMed] [Google Scholar]
- Worth DC, Daly CN, Geraldo S, Oozeer F, Gordon-Weeks PR. (2013). Drebrin contains a cryptic F-actin-bundling activity regulated by Cdk5 phosphorylation. J Cell Biol , 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell , 711–723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.