Abstract
The dynein-2 complex drives retrograde ciliary protein trafficking by associating with the intraflagellar transport (IFT) machinery, containing IFT-A and IFT-B complexes. We recently showed that the dynein-2 complex, which comprises 11 subunits, can be divided into three subcomplexes: DYNC2H1–DYNC2LI1, WDR34–DYNLL1/DYNLL2–DYNLRB1/DYNLRB2, and WDR60–TCTEX1D2–DYNLT1/DYNLT3. In this study, we demonstrated that the WDR34 intermediate chain interacts with the two light chains, DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2, via its distinct sites. Phenotypic analyses of WDR34-knockout cells exogenously expressing various WDR34 constructs showed that the interactions of the WDR34 intermediate chain with the light chains are crucial for ciliary retrograde protein trafficking. Furthermore, we found that expression of the WDR34 N-terminal construct encompassing the light chain–binding sites but lacking the WD40 repeat domain inhibits ciliary biogenesis and retrograde trafficking in a dominant-negative manner, probably by sequestering WDR60 or the light chains. Taken together with phenotypic differences of several WDR34-knockout cell lines, these results indicate that incorporation of DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2 into the dynein-2 complex via interactions with the WDR34 intermediate chain is crucial for dynein-2 function in retrograde ciliary protein trafficking.
INTRODUCTION
Microtubule motor proteins regulate a wide variety of cellular processes, including the positioning and dynamics of membrane-bound organelles, trafficking of proteins and carrier vesicles containing cargo molecules, and organization and remodeling of microtubules during cell division. In general, the kinesin motors drive plus end–directed movement of proteins and organelles along microtubules, whereas dyneins drive minus end–directed movement (Hirokawa et al., 2010).
Cilia are microtubule-based appendages extending from the surface of various eukaryotic cells, and contain a number of unique proteins, including ion channels and G protein–coupled receptors (GPCRs). Cilia play crucial roles in sensing extracellular stimuli, and in integrating a variety of signal transduction pathways, such as those involving Hedgehog (Hh; Briscoe and Thérond, 2013; Mukhopadhyay and Rohatgi, 2014). Although the ciliary membrane is continuous with the plasma membrane, the composition of proteins and lipids in the cilioplasm and on the ciliary membrane are substantially different from that of the cytoplasm and the plasma membrane, as the transition zone located at the base of cilia restricts the movement of proteins by serving as a diffusion/permeability barrier (Garcia-Gonzalo and Reiter, 2017; Gonçalves and Pelletier, 2017). Therefore, the dysregulation of ciliary protein trafficking as well as impaired biogenesis and functions of cilia result in diverse hereditary disorders accompanying a broad spectrum of clinical manifestations, which are collectively called the ciliopathies (Brown and Witman, 2014; Braun and Hildebrandt, 2017). These include Bardet-Biedl syndrome (BBS), Joubert syndrome, Meckel syndrome, and short-rib thoracic dysplasia (SRTD).
The bidirectional trafficking of ciliary proteins along the microtubule-based axoneme is mediated by the intraflagellar transport (IFT) machinery, which contain the two large multisubunit complexes IFT-A and IFT-B, and the BBSome complex (Taschner and Lorentzen, 2016; Nachury, 2018; Nakayama and Katoh, 2018). Anterograde protein trafficking from the base to the tip of cilia is mediated by the IFT-B complex driven by kinesin-2 motor proteins (Funabashi et al., 2018), whereas retrograde trafficking is mediated by the IFT-A complex driven by the dynein-2 complex (Ishikawa and Marshall, 2011; He et al., 2016). The BBSome connects the IFT machinery to cargo GPCRs (Nachury, 2018; Nakayama and Katoh, 2018; Wingfield et al., 2018); recent studies demonstrated that the BBSome regulates export of ciliary membrane proteins across the ciliary gate (Liu and Lechtreck, 2018; Nozaki et al., 2018; Ye et al., 2018).
We, as well as others, recently demonstrated the architecture of these IFT complexes; the IFT-A complex is composed of six subunits, with which an adaptor protein, TULP3, is associated (Hirano et al., 2017); the IFT-B complex is composed of 16 subunits, which can be divided into the core (IFT-B1) subcomplex composed of 10 subunits and the peripheral (IFT-B2) subcomplex composed of six subunits, and these two subcomplexes are connected by composite interactions involving four subunits (Boldt et al., 2016; Katoh et al., 2016; Taschner et al., 2016). The BBSome complex is composed of eight subunits (Loktev et al., 2008; Katoh et al., 2015).
Dynein-2 itself is also a multisubunit complex that is composed of 11 subunits (Asante et al., 2014). Among them, a heavy chain (DYNC2H1), two intermediate chains (WDR34 and WDR60), a light intermediate chain (DYNC2LI1), and a light chain (TCTEX1D2) are unique to the dynein-2 complex (Asante et al., 2014). Mutations in any of the dynein-2–specific subunits are known to cause SRTD (Schmidts, 2014; McInerney-Leo et al., 2015; Reiter and Leroux, 2017). The other six subunits are light chains (DYNLL1, DYNLL2, DYNLRB1, DYNLRB2, DYNLT1, and DYNLT3) that are shared by the dynein-1 and dynein-2 complexes (Asante et al., 2014; Hou and Witman, 2015; Roberts, 2018). DYNLL1 and DYNLL2, DYNLRB1 and DYNLRB2, and DYNLT1 and DYNLT3, play interchangeable roles (Hamada et al., 2018).
We recently delineated the overall architecture of the dynein-2 complex (Hamada et al., 2018), using the visible immunoprecipitation (VIP) assay, which we developed as a simple and flexible strategy enabling visual detection of not only binary, but also one-to-many and many-to-many protein interactions without the need for SDS–PAGE and immunoblotting (Katoh et al., 2015, 2018). In our predicted architectural model, the dynein-2 holocomplex can be divided into three subcomplexes: the H1 subcomplex composed of DYNC2H1 and DYNC2LI1; the WDR34 subcomplex composed of WDR34, DYNLL1/DYNLL2, and DYNLRB1/DYNLRB2; and the WDR60 subcomplex composed of WDR60 and the TCTEX1D2–DYNLT1/DYNLT3 dimer (see Figure 1A). The WDR34 and WDR60 subcomplexes interact with each other, and in turn interact with the H1 subcomplex (see Figure 1A; Hamada et al., 2018). We also established knockout (KO) cell lines of WDR60 and TCTEX1D2, and showed that the interaction between WDR60 and the TCTEX1D2–DYNLT1/DYNLT3 dimer is crucial for retrograde trafficking of ciliary proteins (Hamada et al., 2018).
FIGURE 1:
Region of WDR34 involved in DYNLL2 binding. (A) Schematic representation of the architecture of the dynein-2 complex predicted from our previous study (Hamada et al., 2018). (B–D) Determination of the region of WDR34 involved in its interaction with DYNLL2. Lysates prepared from HEK293T cells cotransfected with expression vectors for an EGFP-fused WDR34 construct, as schematically shown in B, and mChe-fused DYNLL2 were immunoprecipitated with GST-tagged anti-GFP Nb prebound to glutathione–Sepharose beads, and subjected to the VIP assay (C) or immunoblotting analysis (D). IP, immunoprecipitation; IB, immunoblotting.
Previous studies in Chlamydomonas reinhardtii and in Trypanosoma brucei showed that the other intermediate chain WDR34 also participates in retrograde IFT (Rompolas et al., 2007; Blisnick et al., 2014). On the other hand, conditional KO of Caenorhabditis elegans DYCI-1, which is a homologue of not only DYNC1I1/DYNC1I2 but also WDR34/WDR60 and has the characteristics of an IFT-dynein subunit, was shown to result in reduced ciliary length (Li et al., 2015), and embryonic fibroblasts from Wdr34-KO mice were shown to have very short cilia (Wu et al., 2017). In this study, we characterized the details of the interaction between WDR34 and DYNLL1/DYNLL2, and that between WDR34 and DYNLRB1/DYNLRB2. Our analyses using WDR34-KO cell lines, and those exogenously expressing WDR34 constructs lacking light chain–binding sites, showed the importance of these interactions in retrograde ciliary protein trafficking. We further showed that WDR34 truncation mutants that retain light chain–binding ability inhibit ciliogenesis in a dominant-negative manner.
RESULTS
Interactions of WDR34 with DYNLL2 and DYNLRB1
We recently delineated the overall architecture of the dynein-2 complex (Hamada et al., 2018). In the holocomplex, the light chains DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2 interact with the intermediate chain WDR34, resulting in the WDR34 subcomplex. However, in this previous study, we did not address the interaction modes of the light chains with WDR34.
Using the VIP assay, we first determined the region of the WDR34 protein involved in its interaction with DYNLL1/DYNLL2. When the WDR34 protein was divided into the C-terminal WD40 repeat domain and the N-terminal region (see Figure 1B), the latter region fused to EGFP coimmunoprecipitated mCherry (mChe)–fused DYNLL2 (Figure 1C, compare column 3 with column 4). When the WDR34 protein was truncated from the N-terminus, the WDR34(72–536) construct retained the ability to interact with DYNLL2 (column 5), whereas the WDR34(106–536) construct lost this ability (column 6). The VIP assay using mChe-WDR34(Δ73–105) (column 7) confirmed that the region of WDR34 encompassing residues 73–105 participates in its interaction with DYNLL2. By narrowing down the region of WDR34, we found that the region encompassing residues 80–93 is important for the interaction of WDR34 with DYNLL2 (column 9). The VIP results were confirmed by conventional immunoblotting analysis (Figure 1D). Thus, we conclude that WDR34 interacts with DYNLL2 via a region encompassing residues 80–93, as summarized in Figure 1B.
We then addressed which region of the WDR34 protein participates in its interaction with DYNLRB1/DYNLRB2. The VIP assay (Figure 2B) and immunoblotting analysis (Figure 2C) using various deletion constructs of WDR34 (Figure 2A) showed that a region encompassing residues 106–131 of WDR34 is required for its interaction with DYNLRB1, as summarized in Figure 2A.
FIGURE 2:
Region of WDR34 involved in DYNLRB1 binding. (A–C) Determination of the region of WDR34 involved in its interaction with DYNLRB1. Lysates prepared from HEK293T cells cotransfected with expression vectors for an EGFP-fused WDR34 construct, as schematically shown in A, and mChe-fused DYNLRB1 were immunoprecipitated with GST-tagged anti-GFP Nb prebound to glutathione–Sepharose beads, and subjected to the VIP assay (B) or immunoblotting analysis (C). (D) Schematic representation of the organization of the WDR34 domains and sequence alignment of the N-terminal region of vertebrate WDR34 and Chlamydomonas FAP133. Residues conserved in all species are shown in black boxes and those conserved in at least four species are in gray boxes, and sequences involved in DYNLL and DYNLRB binding are boxed in yellow and orange, respectively.
Figure 2D shows a sequence alignment of the WDR34 proteins of ciliated organisms. The regions involved in interactions with DYNLL2 and DYNLRB1 are relatively conserved across vertebrate species, and are in close vicinity to but are distinct from each other. Within these regions, there is the sequence AQVQT. This sequence is reminiscent of the conserved motif (K/R)XTQT in the dynein-1 intermediate chain DYNC1I1, which is required for the binding of DYNC1I1 to DYNLL1 (Lo et al., 2001). A corresponding sequence in Chlamydomonas FAP133/WDR34 was also proposed to participate in its binding to LC8/DYNLL (Rompolas et al., 2007).
Defects in retrograde ciliary protein trafficking in WDR34-KO cells
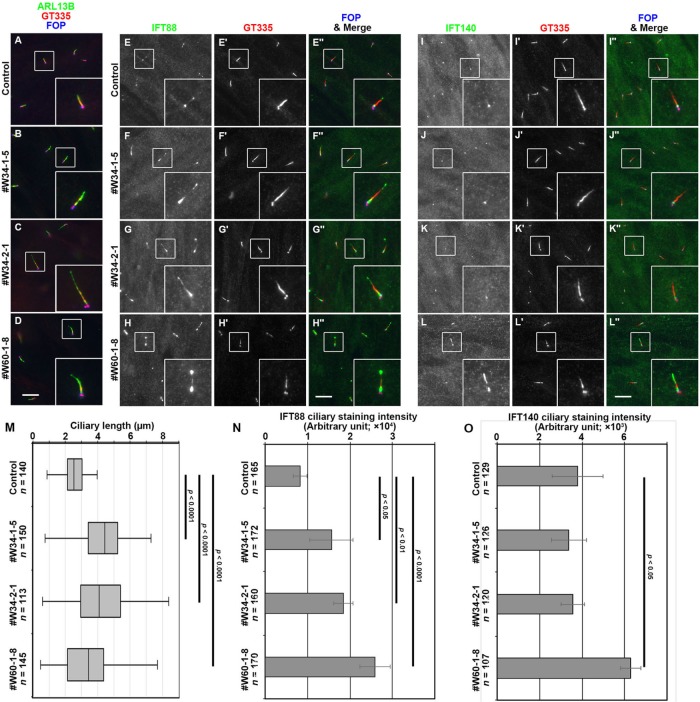
We established WDR34-KO cell lines using the CRISPR/Cas9 system with our original modifications (Katoh et al., 2017; see Supplemental Figure S1 for a detailed genetic characterization of the two selected WDR34-KO cell lines [#W34-1-5 and #W34-2-1], which were established for distinct target sequences). We then compared the phenotypes of these WDR34-KO cell lines with those of the WDR60-KO cell line (#W60-1-8), which we previously established (Hamada et al., 2018). When immunostained for ARL13B and FOP (also known as FGFR1OP) to visualize the ciliary membrane and the basal body, respectively, we noticed that WDR34-KO cells have significantly longer cilia than those of control retinal pigmented epithelial 1 (RPE1) cells (Figure 3, A–C; also see Figure 3M). WDR60-KO cells also demonstrated a tendency to grow slightly longer cilia (Figure 3D).
FIGURE 3:
Accumulation of IFT88 within cilia in WDR34-KO cells. Control RPE1 cells (A, E, I), the WDR34-KO cell lines #W34-1-5 (B, F, J) and #W34-2-1 (C, G, K), and the WDR60-KO cell line #W60-1-8 (D, H, L) were serum-starved for 24 h to induce ciliogenesis, and triple immunostained with either an anti-ARL13B (A–D), anti-IFT88 (E–H), or anti-IFT140 (I–L) antibody, a GT335 antibody (A–D and E′–L′), which can react with polyglutamylated tubulins, and an anti-FOP antibody (A–D and E′′–L′′). Scale bars, 10 µm. (M) Ciliary lengths of individual control cells and of the WDR34-KO cell lines #W34-1-5 and #W34-2-1, and the WDR60-KO cell line #W60-1-8 were measured and are shown as box-and-whisker plots. The box represents the 25th–75th percentiles (interquartile region [IQR]) with the median indicated, and the whiskers show the minimum and maximum within 1.5× IQR from the 25th and 75th percentiles, respectively. The total numbers of ciliated cells analyzed (n) are shown. p, one-way analysis of variance (ANOVA) followed by Tukey post hoc analysis. (N, O) The staining intensity for IFT88 (N) or IFT140 (O) within cilia in control, WDR34-KO, and WDR60-KO cells were measured and are expressed as bar graphs. Values are means ± SD of four (N) or three (O) independent experiments. In each experiment, 30–52 (N) and 33–52 (O) ciliated cells were observed, and the total numbers of ciliated cells observed (n) are shown. p, one-way ANOVA followed by Tukey post hoc analysis.
When immunostained for IFT88, an IFT-B subunit, the staining was found mainly at the ciliary base and weakly at the distal tip (Figure 3E). WDR60-KO cells showed considerable accumulation of IFT88 within cilia, in particular at the distal tip (Figure 3, H and N), as described previously (Hamada et al., 2018). The WDR34-KO cell lines also demonstrated significant accumulation of IFT88 within cilia (Figure 3, F, G, and N), although to a lesser extent than WDR60-KO cells.
We also stained control RPE1, WDR34-KO, and WDR60-KO cells with an antibody against IFT140 (an IFT-A subunit). In control RPE1 and WDR34-KO cells, IFT140 immunostaining was predominantly found at the base of cilia (Figure 3, I–K and O), whereas WDR60-KO cells demonstrated slight but significant accumulation of IFT140 within cilia (Figure 3, L and O), as described previously (Hamada et al., 2018).
These observations suggest that, in the absence of WDR34, retrograde trafficking of the IFT machinery is moderately impaired. To determine whether ciliary protein trafficking is indeed affected in WDR34-KO cells, we then analyzed the localization of Smoothened (SMO) and GPR161 under basal conditions and those when Hh signaling is stimulated; both SMO and GPR161 are GPCRs, which positively and negatively, respectively, regulate Hh signaling. Under basal conditions in control cells, SMO is absent from cilia (Figure 4A; also see Figure 4Q), whereas GPR161 is localized within cilia (Figure 4I; also see Figure 4R). When control cells are stimulated with Smoothened Agonist (SAG), which is a small molecule activator of Hh signaling, a significant amount of SMO enters cilia (Figure 4E; also see Figure 4Q), whereas GPR161 exits cilia (Figure 4J; also see Figure 4R). On the other hand, in the WDR34-KO cell lines #W34-1-5 and #W34-2-1, a substantial amount of SMO was found within cilia under basal conditions (Figure 4, B and C; also see Figure 4Q); ciliary SMO localization was further increased under SAG-stimulated conditions (Figure 4, F and G; also see Figure 4Q). In contrast, a substantial amount of GPR161 was retained within cilia even in the presence of SAG (Figure 4, compare N and O with J and K; also see Figure 4R). These observations together indicate that retrograde trafficking of ciliary GPCRs is significantly impaired in WDR34-KO cells, although not so robustly as in WDR60-KO cells (see Figure 4, Q and R).
FIGURE 4:
Accumulation of SMO and GPR161 within cilia in WDR34-KO cells. (A–L) Control RPE1 cells (A, E, I, M), the #W34-1-5 cell line (B, F, J, N), the #W34-2-1 cell line (C, G, K, O), and the #W60-1-8 cell line (D, H, L, P) were serum-starved for 24 h and further incubated for 24 h without (−SAG; A–D, I–L) or with (+SAG; E–H, M–P) 200 nM SAG. The cells were triple immunostained with antibody against SMO (A–H) or GPR161 (I–P), a GT335 antibody (A′–P′), and an anti-FOP antibody (A′′–P′′). Scale bars, 10 µm. (Q, R) Staining intensities of SMO (Q) and GPR161 (R) in control, WDR34-KO, and WDR60-KO cells were measured, and relative intensities of the cells are expressed as bar graphs. Values are means ± SD of three independent experiments. In each set of experiments, 31–57 (Q) and 30–53 (R) ciliated cells were analyzed and the total numbers of ciliated cells analyzed (n) are shown. p, one-way ANOVA followed by Tukey post hoc analysis for comparison among the cell lines, and the Student’s t test for comparison between the cells with and without SAG treatment.
Recent studies showed that the BBSome participates in export of ciliary membrane proteins, including GPCRs, across the ciliary gate (Liu and Lechtreck, 2018; Nozaki et al., 2018; Ye et al., 2018). We therefore examined whether the BBSome localization was affected by the absence of the dynein-2 intermediate chain. In control RPE1 cells, the BBS9 immunostaining was evenly distributed within cilia of ∼40% of ciliated cells (Supplemental Figure S2, A and E) as previously reported (Seo et al., 2011; Nozaki et al., 2018). In the WDR34-KO and WDR60-KO cell lines, however, the BBS9 immunostaining was not uniform, often concentrated at the distal tip of elongated cilia (Supplemental Figure S2, B–D). Furthermore, compared with control RPE1 cells, the proportion of BBS9-positive cilia was significantly increased in the WDR34-KO and WDR60-KO cell lines (Supplemental Figure S2E). These observations indicate that retrograde trafficking of the BBSome together with the IFT machinery is also impaired in the absence of WDR34 or WDR60. Given that the BBSome can interact with peptides from intracellular regions of ciliary GPCRs in vitro (Klink et al., 2017), it is possible that the accumulation of GPCRs within cilia in WDR34-KO and WDR60-KO is due, at least in part, to impaired retrograde trafficking of the BBSome.
Interaction of WDR34 with light chains is required for normal retrograde trafficking
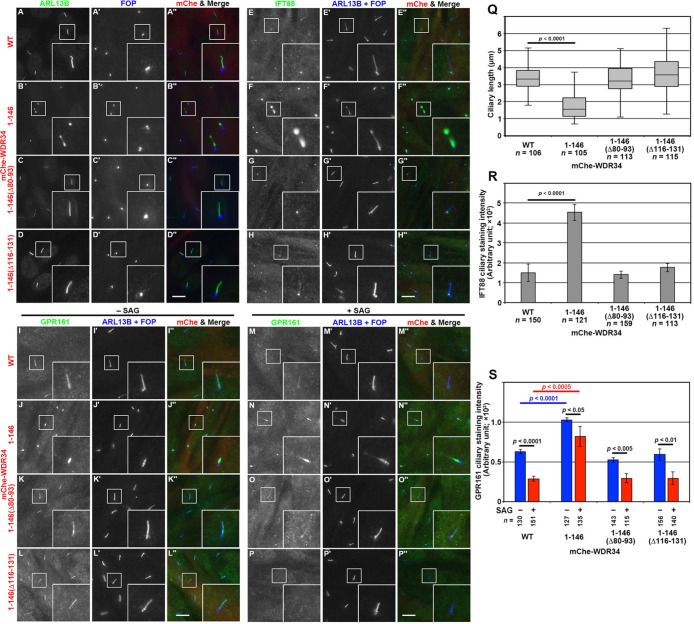
We then analyzed the effects of exogenous expression of WDR34 in WDR34-KO cells, to rule out the possibility that the defect observed in WDR34-KO cells was due to off-target effects of the CRISPR/Cas9 system. When mChe-WDR34(WT) was stably expressed in the WDR34-KO cell line #W34-1-5, ciliary lengths were significantly reduced (Figure 5B) compared with WDR34-KO cells expressing mChe as a negative control (Figure 5A; also see Figure 5G). When mChe-tagged WDR34(Δ80–93) and WDR34(Δ116–131), which lack the binding sites for DYNLL2 and DYNLRB1 (see Figures 1 and 2), respectively, were stably expressed in WDR34-KO cells, elongated cilia were not restored to the length of control cells (Figure 5, E and F; also see Figure 5G). The stable expression of mChe-WDR34(147–536), which lacks the N-terminal region for light chain binding, also had no apparent effects with respect to ciliary length (Figure 5D; also see Figure 5G). On the other hand, we unexpectedly found that stable expression of mChe-WDR34(1–146), which is the N-terminal region encompassing the light chain–binding sites (see Figures 1 and 2), in WDR34-KO cells resulted in considerable shortening of cilia, with a large proportion of cells having vestigial cilia (Figure 5C; also see Figure 5G). This concern is addressed later in the article.
FIGURE 5:
Expression of WDR34(1–146) in WDR34-KO cells inhibits ciliogenesis. The #W34-1-5 cell line stably expressing mChe (A), mChe-fused WDR34(WT) (B), WDR34(1–146) (C), WDR34(147–536) (D), WDR34(Δ80–93) (E), or WDR34(Δ116–131) (F) was cultured for 24 h under serum-starved conditions and double immunostained for ARL13B (A–F) and FOP (A′–F′). Scale bar, 10 µm. (G) Ciliary lengths of individual control cells and of the #W34-1-5 cell line expressing the WDR34 construct indicated were measured and are shown as box-and-whisker plots as described in the legend to Figure 3M.
We then analyzed the effects of the stable expression of WDR34(WT) or its mutant lacking the binding site for DYNLL2 or DYNLRB1, on the localization of IFT88 and GPR161 in WDR34-KO cells. As shown in Figure 6, A and B (also see Figure 6M), the accumulation of IFT88 within cilia of WDR34-KO cells was significantly inhibited by the exogenous expression of mChe-WDR34(WT). In contrast, the expression of mChe-tagged WDR34(Δ80–93) or WDR34(Δ116–131) did not restore the normal localization of IFT88; rather, IFT88 accumulation tended to be promoted by the exogenous expression of WDR34(Δ80–93) or WDR34(Δ116–131) (Figure 6, C and D; also see Figure 6M). GPR161 accumulation within cilia in WDR34-KO cells under basal and SAG-stimulated conditions was restored by the stable expression of mChe-WDR34(WT) (Figure 6, E and F, and I and J), whereas it was enhanced by the expression of mChe-tagged WDR34(Δ80–93) or WDR34(Δ116–131) (Figure 6, G and H, and K and L; also see Figure 6N). These observations together indicate that interactions of WDR34 with both DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2 are essential for dynein-2–mediated retrograde trafficking of ciliary proteins.
FIGURE 6:
Interactions of WDR34 with both DYNLL2 and DYNLRB1 are required for dynein-2–mediated retrograde trafficking. The #W34-1-5 cell line stably expressing mChe (A, E, I), mChe-fused WDR34(WT) (B, F, J), WDR34(Δ80–93) (C, G, K), or WDR34(Δ116–131) (D, H, L) was cultured for 24 h under serum-starved conditions and further incubated for 24 h in the absence (−SAG; E–H) or presence (+SAG; I–L) of 200 nM SAG. The cells were triple immunostained for either IFT88 (A–D) or GPR161 (E–L) and ARL13B+FOP (A′–L′). Scale bars, 10 µm. (M, N) The staining intensity for IFT88 (M) and GPR161 (N) within cilia in the #W34-1-5 cell line expressing the mChe-fused WDR34 construct indicated were measured and are expressed as bar graphs. Values are shown as means ± SD of three independent experiments. In each set of experiments, 31–50 (M) and 31–60 (N) ciliated cells were analyzed and the total numbers of ciliated cells analyzed (n) are shown. p, one-way ANOVA followed by Tukey post hoc analysis for comparison among the cell lines expressing different WDR34 constructs, and Student’s t test for comparison between the cells with and without SAG treatment.
Expression of the WDR34 N-terminal region encompassing light chain–binding sites inhibits ciliary biogenesis and retrograde trafficking in a dominant-negative manner
As described above, we observed that stable expression of mChe-WDR34(1–146) in WDR34-KO cells resulted in a considerable decrease in ciliary length (compare Figure 5, A and C; also see Figure 5G). We then expressed mChe-WDR34(1–146) in control RPE1 cells, by anticipating that its expression would inhibit ciliogenesis in a dominant-negative manner. As expected, the majority of RPE1 cells stably expressing mChe-WDR34(1–146) had short or vestigial cilia (compare Figure 7, A and B; also see Figure 7Q). In striking contrast, stable expression of mChe-tagged WDR34(1–146; Δ80–93) or WDR34(1–146; Δ116–131) did not significantly affect ciliary length (Figure 7, C and D; also see Figure 7Q). It is thus likely that the exogenous expression of mChe-WDR34(1–146) has dominant-negative effects on ciliogenesis, at least in part, via its interaction with the light chains.
FIGURE 7:
WDR34(1–146) dominant negatively inhibits ciliogenesis and retrograde trafficking. RPE1 cells stably expressing mChe-fused WDR34(WT) (A, E, I, M), WDR34(1–146) (B, F, J, N), WDR34(1–146, Δ80–93) (C, G, K, O), or WDR34(1–146, Δ116–131) (D, H, L, P) were cultured for 24 h under serum-starved conditions and immunostained for ARL13B (A–D) and FOP (A′–D′), or IFT88 (E–H) and ARL13B+FOP (E′–H′). In I–P, after serum starvation, the cells were further cultured for 24 h in the absence (I–L) or presence (M–P) of 200 nM SAG and immunostained for GPR161 (I–P) and ARL13B+FOP (I′–L′). Scale bars, 10 µm. (Q) Ciliary lengths of individual cells expressing the WDR34 construct indicated were measured and are shown as box-and-whisker plots as described in the legend for Figure 3M. (R) Staining intensities for IFT88 within cilia were measured and are expressed as bar graphs. Values are means ± SD of three independent experiments. In each experiment, 30–52 ciliated cells were observed, and the total numbers of ciliated cells observed (n) are shown. p, one-way ANOVA followed by Tukey post hoc analysis. (S) Staining intensities for GPR161 within cilia were measured and are expressed as bar graphs. Values are means ± SD of three independent experiments. In each set of experiments, 31–57 ciliated cells were analyzed and the total numbers of ciliated cells analyzed (n) are shown. p, one-way ANOVA followed by Tukey post hoc analysis for comparison among the cell lines expressing different WDR34 constructs, and the Student’s t test for comparison between the cells with and without SAG treatment.
We then analyzed whether exogenous expression of the WDR34(1–146) construct inhibits retrograde ciliary protein trafficking in a dominant-negative manner. Whereas staining of IFT88 was found mainly at the ciliary base and faintly at the distal tip in RPE1 cells stably expressing WDR34(WT) (Figure 7E), stable expression of WDR34(1–146) led to a substantial accumulation of IFT88 around the tip of short cilia and a considerable increase in total IFT88 staining intensity within cilia (Figure 7F; also see Figure 7R). By contrast, the stable expression of WDR34(1–146; Δ80–93) or WDR34(1–146; Δ116–131) did not significantly affect the localization of IFT88 (Figure 7, G and H; also see Figure 7R). In addition, exogenous expression of the WDR34(1–146) construct in RPE1 cells increased ciliary GPR161 levels under basal and SAG-stimulated conditions compared with the expression of WDR34(WT) (Figure 7, I, J, M, and N; also see Figure 7S). By contrast, the expression of WDR34(1–146; Δ80–93) or WDR34(1–146; Δ116–131) had no apparent effect on GPR161 localization (Figure 7, K, L, O, and P; also see Figure 7S). Thus, the dominant-negative effects of the WDR34(1–146) construct on ciliary biogenesis and retrograde trafficking are dependent on its interaction with the light chains.
The above results showing that WDR34(1–146) expression suppresses ciliogenesis are compatible with a previous study of Wdr34-KO mice showing that their embryonic fibroblasts have very short cilia (Wu et al., 2017); these mice were established by deleting exons 3–8 of the Wdr34 gene and are expected to express a truncated WDR34 protein, p.Asn126Lys*. In addition, Vuolo et al. (2018) reported that WDR34-KO cell lines established from RPE1 cells were severely compromised with respect to ciliogenesis; the KO cell lines (designated clones #1 and #2) were expected to express truncated proteins, p.Arg114Serfs*51 and p.Ser147Leufs*17, respectively. Thus, the truncated WDR34 proteins in the previously reported KO cells retain at least one of the light chain–binding sites.
To reproduce the observations in the previous studies, we finally established two other WDR34-KO cell lines (#W34-3-3 and W34-4-14), which are expected to express truncated WDR34 proteins, p.Val119Alafs*4 and p.Thr151Thrfs*41, respectively, from one of the two disrupted alleles (see Supplemental Figure S3). As expected, large proportions of the newly established KO cells had vestigial cilia (Figure 8, D and E; also see Figure 8F). Taken together with the data shown in Figures 5 and 7, we conclude that expression of truncated WDR34 proteins results in severely impaired ciliogenesis.
FIGURe 8:
Difference in the ciliary length among different WDR34-KO cell lines. Control RPE1 cells (A) and the WDR34-KO cell lines, #W34-1-5 (B), #W34-2-1 (C), #W34-3-3 (D), and #W34-4-14 (E), were serum-starved for 24 h to induce ciliogenesis and triple stained with an anti-ARL13B antibody, a GT335 antibody, and an anti-FOP antibody. Scale bar, 10 µm. (F) Ciliary lengths of individual cells were measured and are shown as box-and-whisker plots as described in the legend for Figure 3M.
DISCUSSION
In this study, we first determined the regions of the WDR34 protein responsible for its interactions with DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2. Similar to binding of the TCTEX1D2–DYNLT1/DYNLT3 dimer to the WDR60 N-terminal region (Hamada et al., 2018), DYNLL2 and DYNLRB1 were found to interact with the WDR34 N-terminal region upstream of the WD40 repeat domain, but at distinct sites (Figures 1 and 2). The DYNLL2-binding site in WDR34 contains a conserved sequence reminiscent of the DYNLL1 binding site in the dynein-1 intermediate chain DYNC1I1 (Lo et al., 2001). It is thus likely that dynein-1 and dynein-2 adopt the same binding mode of DYNLL1/DYNLL2 to the intermediate chains.
As expected from the role of dynein-2 in intraciliary protein trafficking, WDR34-KO RPE1 cell lines (#W34-1-5 and W34-2-1) showed defects in retrograde trafficking that were similar to, but somewhat less severe than, those of WDR60-KO cells (Figures 3 and 4). When WDR34(WT) was exogenously expressed in WDR34-KO cells, defects in retrograde trafficking were reversed, excluding the possibility that the phenotype observed in our WDR34-KO cells resulted from off-target effects of the CRISPR/Cas9 system. In striking contrast, the exogenous expression of WDR34 mutants that were deficient with respect to their binding to DYNLL1/DYNLL2 or DYNLRB1/DYNLRB2 did not improve, but rather had a tendency to exacerbate, the retrograde trafficking defects of WDR34-KO cells (Figure 6), demonstrating that normal functions of dynein-2 in retrograde ciliary protein trafficking require the incorporation of both DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2 into the dynein-2 complex via their interactions with WDR34.
During the course of the rescue experiments of WDR34-KO cells, we unexpectedly found that the WDR34(1–146) construct, which encompasses the light chain–binding sites but lacks the WD40 repeat domain, impairs ciliary biogenesis and retrograde trafficking in a dominant-negative manner (Figures 5, C and G, and 7). The dominant-negative effects on both ciliogenesis and trafficking are dependent on the binding of WDR34(1–146) to the light chains DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2 (Figure 7). These observations are not incompatible with the phenotypes of our WDR34-KO cell lines (Figure 3, A–C and M); namely, the #W34-1-5 and #W34-2-1 cell lines have a p.Val24Alafs*74 and p.Gly44Alafs*60 mutation, respectively, at least in one allele (Supplemental Figure S1, B and C), and thus both KO cell lines are expected to express very short WDR34 proteins lacking the binding sites for both DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2.
On the other hand, two newly established WDR34-KO cell lines (#W34-3-3 and #W34-4-14), which are expected to express truncated WDR34 proteins encompassing the light chain–binding sites but lacking the WD40 repeat domain, demonstrated a severe ciliogenesis defect (Figure 8). With respect to ciliogenesis, these two cell lines phenocopied embryonic fibroblasts from Wdr34-KO mice reported by (Wu et al., 2017) and WDR34-KO RPE1 cell lines reported by (Vuolo et al., 2018); these KO cells are also expected to express truncated WDR34 proteins containing at least one of the binding sites for the light chains.
We do not know the exact reason why the truncated WDR34 proteins had dominant-negative effects on ciliary biogenesis as well as on retrograde trafficking, but a few explanations are possible, as follows. 1) The WDR34(1–146) construct might in some way affect the function of the dynein-2 complex by locking WDR60, as it retains the ability to interact with the WDR60 subcomplex. 2) The dominant effects might also be implicated in sequestration of the light chains, as the WDR34(1–146) construct lacking the light chain–binding site did not show dominant effects. In this context, it is noteworthy that, as reported previously, a mutation in LC8 (the sole DYNLL orthologue) of Chlamydomonas (Pazour et al., 1998) and conditional KO of DLC-1 (the sole DYNLL orthologue) in C. elegans (Li et al., 2015) resulted in short cilia/flagella. Thus, the lack of or decreased level of the light chains might impair ciliary biogenesis and retrograde trafficking. In the context of these two possibilities, a recent in vitro study by Toropova et al. is also noteworthy (Toropova et al., 2017). On the basis of their cryoelectron microscopic analysis and motor activity analysis of purified DYNC2H1 and its mutant lacking the ability to form a dimer, they proposed an intriguing hypothesis: when dynein-2 is trafficked anterogradely as a cargo of the IFT trains, it adopts an autoinhibited conformation to prevent a tug-of-war between kinesin-2 and dynein-2, whereas, at the ciliary tip, some localized signal activates dynein-2 to adopt a functional conformation to be able to power the retrograde IFT. This hypothesis was recently supported by a cryoelectron tomographic study of anterograde IFT trains in Chlamydomonas flagella (Jordan et al., 2018). It is therefore tempting to speculate that the sequestration of WDR60 or the light chains might cancel the autoinhibition, even while dynein-2 is being anterogradely trafficked, thereby disrupting the anterograde IFT trains driven by the kinesin-2 motor. 3) In addition to their roles as subunits of the dynein-2 complex, DYNLL1/DYNLL2 and DYNLRB1/DYNLRB2 might also be implicated in ciliogenesis by serving as dynein-1 subunits, as they are shared by the dynein-1 and dynein-2 complexes (Asante et al., 2014; Hou and Witman, 2015; Roberts, 2018). Ciliary biogenesis is presumed to initiate upon docking of the mother centriole with ciliary vesicles, which are most likely delivered by dynein-1–driven minus end–directed movement of vesicles from the Golgi apparatus or recycling endosomes toward the mother centriole (Schmidt et al., 2012). Hence, it is possible that the lack of light chains as dynein-1 subunits might impede the initiation process of ciliogenesis. However, we think this possibility low, because our analyses showed that assembly of mitotic spindles (Supplemental Figure S4, A–D) and subcellular distribution of organelles, including the Golgi apparatus and lysosomes (Supplemental Figure S4, E–H), were unaffected by exogenous WDR34(1–146) expression, although these processes were reported to be impaired in cells from Dync1h1-KO mice (Harada et al., 1998).
Compatible with the dominant-negative, severe phenotype of cells expressing truncation mutants of WDR34, almost all of the reported mutations of the WDR34 gene in SRTD individuals are missense substitutions; the majority of them are located in the WD40 repeat domain, and a minor proportion of them are found in the N-terminal region (Huber et al., 2013; Schmidts et al., 2013; You et al., 2017; Zhang et al., 2018). The only exception reported to date is a p.Gln158* mutation, with a p.Lys36Arg mutation in the other allele, in an SRTD individual (Schmidts et al., 2013). Although there were no data with respect to cilia in that case report, it is expected that cells of this individual have very short cilia.
MATERIALS AND METHODS
Plasmids, antibodies, and reagents
WDR34 constructs in the fluorescent protein vectors used in this study are listed in Supplemental Table S1. Other constructs for the dynein-2 subunits were described previously (Hamada et al., 2018). Antibodies used in this study are listed in Supplemental Table S2. Glutathione S-transferase (GST)–tagged anti-GFP Nanobody (Nb) prebound to glutathione–Sepharose 4B beads were prepared as described previously (Katoh et al., 2015). SAG was purchased from Enzo Life Sciences.
VIP assay and immunoblotting analysis
VIP assays were carried out as described previously (Katoh et al., 2015, 2016; for detailed protocol, see Katoh et al., 2018) with a minor modification; namely, lysates were prepared from cells transfected with expression vectors for fluorescent fusion proteins using HMDEKN cell lysis buffer (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM dithiothreitol, 0.5 mM EDTA, 25 mM KCl, 0.05% NP-40; Nishijima et al., 2017). After observation under a fluorescence microscope (BZ-8000, KEYENCE), the immunoprecipitated beads bearing fluorescent fusion proteins were subjected to immunoblotting analysis as described previously (Katoh et al., 2015, 2016).
Establishment of KO cell lines using the CRISPR/Cas9 system
The strategy for disrupting the WDR34 gene in hTERT-RPE1 cells (American Type Culture Collection; CRL-4000) by the CRISPR/Cas9 system using homology-independent repair (the version 2 method) was described in detail previously (Katoh et al., 2017; also see Nozaki et al., 2018; Takahara et al., 2018; Takei et al., 2018). Single-guide RNA (sgRNA) sequences targeting the human WDR34 gene (see Supplemental Table S3) were designed using CRISPOR (http://crispor.tefor.net/). Double-stranded oligonucleotides for the target sequences were inserted into peSpCAS9(1.1)-2 × sgRNA (Addgene 80768), an all-in-one sgRNA expression vector. hTERT-RPE1 cells grown on a 12-well plate were transfected with the sgRNA vector and the pDonor-tBFP-NLS-Neo(universal) donor knock-in vector (Addgene 80767) using X-tremeGENE9 transfection reagent (Roche Applied Science). After selection of transfected cells in the presence of 600 µg/ml G418, colonies of the cells with blue nuclear fluorescence signals were isolated. Single cell sorting using SH800S (Sony) was performed at the Medical Research Support Center, Graduate School of Medicine, Kyoto University. To confirm disruption of the WDR34 gene, genomic DNA was extracted from the isolated cells, and processed for PCR using KOD FX Neo DNA polymerase (Toyobo). Three PCR primer sets (Supplemental Table S3) were used to distinguish the following three states of donor vector integration: forward integration, reverse integration, and no integration with a small indel (Supplemental Figure S1A). The PCR products were subjected to direct sequencing to confirm disruption of both WDR34 alleles; a small deletion resulting in a frameshift in one allele and a forward integration of the donor knock-in vector in the other allele (Supplemental Figure S1, B and C).
Preparation of cells stably expressing the mChe-fused WDR34 construct
Lentiviral vectors for the WDR34 constructs were prepared as described previously (Takahashi et al., 2012). Briefly, HEK293T cells were transfected with pRRLsinPPT-mChe-WDR34 or its deletion construct along with the packaging plasmids (pRSV-REV, pMD2.g, and pMDL/pRRE), which are kind gifts from Peter McPherson, McGill University (Thomas et al., 2009). The culture medium was replaced 8 h after transfection. Culture media containing lentiviral particles were then collected at 24, 36, and 48 h after transfection, passed through a 0.45-µm filter, and centrifuged at 32,000 × g at 4°C for 4 h. Precipitated viral particles were resuspended in Opti-MEM (Invitrogen) and stored at –80°C until use. Cells expressing the mChe-fused WDR34 construct were prepared by the addition of the lentiviral suspension to the culture medium, and processed for immunofluorescence analysis.
Immunofluorescence analysis
hTERT-RPE1 cells were cultured in DMEM/F-12 (Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS) and 0.348% sodium bicarbonate. To induce ciliogenesis, cells were grown to 100% confluence on coverslips, and starved for 24 h in Opti-MEM containing 0.2% bovine serum albumin.
For immunofluorescence analysis, cells were fixed and permeabilized with 3% paraformaldehyde at 37°C for 5 min and subsequently in 100% methanol for 5 min at −20°C, and washed three times with phosphate-buffered saline. The fixed/permeabilized cells were blocked with 10% FBS and stained with antibodies diluted in 5% FBS. The stained cells were observed using an Axiovert 200M microscope (Carl Zeiss). Statistical analyses were performed using JMP Pro 13 software (SAS Institute).
Supplementary Material
Acknowledgments
We thank Peter McPherson for providing plasmids for the production of recombinant lentiviruses, Nobuhiro Nakamura for providing anti–golgin-97 antibody, and Helena Akiko Popiel for critical reading of the manuscript. This work was supported in part by grants from the Japan Society for the Promotion of Science (Grants no. 15H04370 and 15K14456 to K.N. and no. 15K07929 and 18H02403 to Y.K.), from the Astellas Foundation for Research on Metabolic Disorders to K.N., and from the Uehara Memorial Foundation to Y.K.
Abbreviations used:
- BBS
Bardet-Biedl syndrome
- FBS
fetal bovine serum
- GPCR
G protein–coupled receptor
- GST
glutathione S-transferase
- Hh
Hedgehog
- hTERT-RPE1
human telomerase reverse transcriptase-immortalized retinal pigment epithelial 1
- IFT
intraflagellar transport
- KO
knockout
- mChe
mCherry
- Nb
nanobody
- SAG
Smoothened Agonist
- sgRNA
single guide RNA
- SMO
Smoothened
- SRTD
short-rib thoracic dysplasia
- VIP
visible immunoprecipitation
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0678) on January 16, 2019.
REFERENCES
- Asante D, Stevenson NL, Stephens DJ. (2014). Subunit composition of the human cytoplasmic dynein-2 complex. J Cell Sci , 4774–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blisnick T, Buisson J, Absalon S, Marie A, Cayet N, Bastin P. (2014). The intraflagellar transport dynein complex of trypanosomes is made of a heterodimer of dynein heavy chains and of light and intermediate chains of distinct functions. Mol Biol Cell , 2620–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt K, van Reeuwijk J, Lu Q, Koutroumpas K, Nguyen TM, Texier Y, van Beersum SEC, Horn N, Willer JR, Mans D, et al (2016). An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat Commun , 11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Hildebrandt F. (2017). Ciliopathies. Cold Spring Harb Perspect Biol , a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Thérond PP. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol , 416–429. [DOI] [PubMed] [Google Scholar]
- Brown JM, Witman GB. (2014). Cilia and diseases. BioScience , 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Okazaki M, Sugawa M, Nakayama K. (2018). Interaction of heterotrimeric kinesin-II with IFT-B-connecting tetramer is crucial for ciliogenesis. J Cell Biol , 2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Reiter JF. (2017). Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb Perspect Biol , a028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J, Pelletier L. (2017). The ciliary transition zone: finding the pieces and assembling the gate. Mol Cells , 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y, Tsurumi Y, Nozaki S, Katoh Y, Nakayama K. (2018). Interaction of WDR60 intermediate chain with TCTEX1D2 light chain of the dynein-2 complex is crucial for ciliary protein trafficking. Mol Biol Cell , 1628–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. (1998). Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol , 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Agbu S, Anderson KV. (2016). Microtubule-motors drive Hedgehog signaling in primary cilia. Trends Cell Biol , 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Katoh Y, Nakayama K. (2017). Intraflagellar transport-A complex mediates ciliary entry and retrograde trafficking of ciliary G protein-coupled receptors. Mol Biol Cell , 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. (2010). Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron , 610–638. [DOI] [PubMed] [Google Scholar]
- Hou Y, Witman GB. (2015). Dynein and intraflagellar transport. Exp Cell Res , 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber C, Wu S, Kim AS, Sigaudy S, Sarukhanov A, Serre V, Baujat G, Le Quan Sang K-M, Rimoin DL, Cohn DH, et al (2013). WDR34 mutations that cause short-rib polydactyly syndrome type III/severe asphyxiating thoracic dysplasia reveal a role for the NF-κB pathway in cilia. Am J Hum Genet , 926–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. (2011). Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol , 222–234. [DOI] [PubMed] [Google Scholar]
- Jordan MA, Diener DR, Stepanek L, Pigino G. (2018). The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional antrogarde movement in cilia. Nat Cell Biol , 1250–1255. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Michisaka S, Nozaki S, Funabashi T, Hirano T, Takei R, Nakayama K. (2017). Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated homology-independent knock-in system. Mol Biol Cell , 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Nakamura K, Nakayama K. (2018). Visible immunoprecipitation (VIP) assay: a simple and versatile method for visual detection of protein-protein interactions. Bio-protocol , e2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Nozaki S, Hartanto D, Miyano R, Nakayama K. (2015). Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J Cell Sci , 2351–2362. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K. (2016). Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J Biol Chem , 10962–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink BU, Zent E, Juneja P, Kuhlee A, Rausner S, Wittinghofer A. (2017). A rocombinant BBSome core complex and how it interacts with ciliary cargo. eLife , e27434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Peishan YP, Ou G. (2015). Somatic CRISPR–Cas9-induced mutations reveal roles of embryonically essential dynein chains in Caenorhabditis elegans cilia. J Cell Biol , 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Lechtreck KF. (2018). The Bardet-Biedl syndrome protein complex is an adaptor expanding the cargo of range of intraflagellar transport trains for ciliary export. Proc Natl Acad Sci USA , E934–E943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KW-H, Naisbitt S, Fan J-S, Sheng M, Zhang M. (2001). The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J Biol Chem , 14059–14066. [DOI] [PubMed] [Google Scholar]
- Loktev AV, Zhang Q, Beck JS, Searby CC, Scheetz TE, Bazan JF, Slusarski DC, Sheffield VC, Jackson PK, Nachury MV. (2008). A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell , 854–865. [DOI] [PubMed] [Google Scholar]
- McInerney-Leo AM, Harris JE, Marshall MS, Gardiner B, Kinning E, Leong HY, McKenzie F, Ong WP, Vodopiutz J, Wicking C, et al (2015). Whole exome sequencing is an efficient, sensitive and specific method for determining the genetic cause of short-rib thoracic dystrophies. Clin Genet , 550–557. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Rohatgi R. (2014). G-protein-coupled receptors, Hedgehog signaling and primary cilia. Sem Cell Dev Biol , 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV. (2018). The molecular machines that traffic signaling receptors into and out of cilia. Curr Opin Cell Biol , 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y. (2018). Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J Biochem , 155–164. [DOI] [PubMed] [Google Scholar]
- Nishijima Y, Hagiya Y, Kubo T, Takei R, Katoh Y, Nakayama K. (2017). RABL2 interacts with the intraflagellar transport B complex and CEP19 and participates in ciliary assembly. Mol Biol Cell , 1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Kobayashi T, Nakayama K. (2018). BBS1 is involved in retrograde trafficking of ciliary GPCRs in the context of the BBSome complex. PLoS One , e0195005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. (1998). A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J Cell Biol , 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR. (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol , 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ. (2018). Emerging mechanisms of dynein transport in the cytoplasm versus the cilium. Biochem Soc Trans , 967–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Pedersen LB, Patel-King RS, King SM. (2007). Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J Cell Sci , 3653–3665. [DOI] [PubMed] [Google Scholar]
- Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G. (2012). Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol , 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts M. (2014). Clinical genetics and pathobiology of ciliary chondrodysplasias. J Pediatr Genet , 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidts M, Vodopiutz J, Christou-Savina S, Cortés CR, McInerney-Leo AM, Emes RD, Arts HH, Tüysüz B, D’Silva J, Leo PJ, et al (2013). Mutations in the gene encoding IFT dynein complex component WDR34 cause Jeune asphyxiating thoracic dystrophy. Am J Hum Genet , 932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Zhang Q, Bugge K, Breslow DK, Searby CC, Nachury MV, Sheffield VC. (2011). A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet , e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara M, Katoh Y, Nakamura K, Hirano T, Sugawa M, Tsurumi Y, Nakayama K. (2018). Ciliopathy-associated mutations of IFT122 impair ciliary protein trafficking but not ciliogenesis. Hum Mol Genet , 516–528. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K. (2012). Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci , 4049–4057. [DOI] [PubMed] [Google Scholar]
- Takei R, Katoh Y, Nakayama K. (2018). Robust interaction of IFT70 with IFT52–IFT88 in the IFT-B complex is required for ciliogenesis. Biol Open , bio033241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E. (2016). The intraflagellar transport machinery. Cold Spring Harb Perspect Biol , a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Weber K, Mourão A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E. (2016). Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J , 773–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonnière L, McKinney RA, McPherson PS. (2009). Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem , 12410–12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova K, Mladenov K, Roberts AJ. (2017). Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat Struct Mol Biol , 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuolo L, Stevenson NL, Heesom KJ, Stephens DJ. (2018). Dynein-2 intermediate chains play crucial but distinct roles in primary cilia formation and function. eLife , e39655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield JL, Lechtreck KF, Lorentzen E. (2018). Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem , EBC20180030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Li J, Peterson A, Tao K, Wang B. (2017). Loss of dynein-2 intermediate chain Wdr34 results in defects in retrograde ciliary protein trafficking and Hedgehog signaling in the mouse. Hum Mol Genet , 2386–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Nager AR, Nachury MV. (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J Cell Biol , 1847–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S-H, Lee Y-S, Lee C-P, Lin C-P, Lin C-Y, Tsai C-L, Chang Y-L, Cheng P-J, Wang T-H, Chang S-D. (2017). Identification of a c.544C>T mutation in WDR34 as a deleterious recessive allele of short rib-polydactyly syndrome. Taiwan J Obstet Gynecol , 857–862. [DOI] [PubMed] [Google Scholar]
- Zhang W, Taylor SP, Ennis HA, Forlenza KN, Duran I, Li B, Ortiz Sanchez JA, Nevarez L, Nickerson DA, Bamshad M, et al (2018). Expanding the genetic architecture and phenotypic spectrum in the skeletal ciliopathy. Hum Mut , 152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.