Abstract
Cellular actin networks exhibit diverse filamentous architectures and turnover dynamics, but how these differences are specified remains poorly understood. Here, we used multicolor total internal reflection fluorescence microscopy to ask how decoration of actin filaments by five biologically prominent Tropomyosin (TPM) isoforms influences disassembly induced by Cofilin alone, or by the collaborative effects of Cofilin, Coronin, and AIP1 (CCA). TPM decoration restricted Cofilin binding to pointed ends, while not interfering with Coronin binding to filament sides. Different isoforms of TPM provided variable levels of protection against disassembly, with the strongest protection by Tpm3.1 and the weakest by Tpm1.6. In biomimetic assays in which filaments were simultaneously assembled by formins and disassembled by CCA, these TPM isoform–specific effects persisted, giving rise to filaments with different lengths and treadmilling behavior. Together, our data reveal that TPM isoforms have quantitatively distinct abilities to tune actin filament length and turnover.
INTRODUCTION
Mammalian cells build a wide range of filamentous actin structures with properties uniquely tailored to different biological functions such as cell migration, cytokinesis, and intracellular transport. This functional diversity depends on the ability of cells to assemble actin building blocks into architectures that persist for long periods of time (e.g., stereocilia and sarcomeres) or are dynamically assembled and disassembled (e.g., lamellipodia or filopodia) (Watanabe and Mitchison, 2002; Rzadzinska et al., 2004; Narayanan et al., 2015). How cells are able to assemble actin structures with such vastly different turnover rates, side by side in a shared cytoplasm, has remained an important open question. Resolving this mystery requires gaining a deeper understanding of the rules that underlie molecular self-assembly, as well as how different actin-associated proteins work in concert and affect one another.
A key triad of proteins ubiquitously involved in promoting actin filament disassembly are Cofilin/ADF (actin depolymerizing factor), Coronin, and AIP1 (actin-interacting protein 1), collectively referred to herein as “CCA.” These proteins have been shown to work in concert to rapidly disassemble actin filaments by promoting robust severing, depolymerization, and barbed-end capping (Brieher et al., 2006; Kueh et al., 2008; Chen et al., 2015; Gressin et al., 2015; Jansen et al., 2015; Nadkarni and Brieher, 2014). The CCA three-component mechanism can also induce rapid filament disassembly, even under assembly-promoting conditions, where the concentration of actin monomers exceeds the critical concentration of actin assembly. To date, the effects of CCA have been analyzed only on bare (undecorated) actin filaments, whereas filaments in vivo are decorated by many other proteins.
One of the most prominent families of actin filament–interacting proteins are the Tropomyosins (TPMs), which decorate almost all known cellular actin structures in organisms ranging from yeast to humans (Gunning et al., 2015; Hitchcock-DeGregori and Barua, 2017). TPMs are coiled-coil proteins that form stable parallel dimers, which in turn polymerize head-to-tail into long strands along the sides of actin filaments (Gunning et al., 2015). Some TPMs have been shown to compete with Cofilin for binding to actin filaments in cosedimentation assays and/or to inhibit Cofilin-induced actin disassembly in bulk fluorescence experiments (DesMarais et al., 2002; Robaszkiewicz et al., 2016; Gateva et al., 2017; Ostrowska et al., 2017). Two recent TIRF studies directly showed that TPM and Cofilin decoration is mutually exclusive, which provides key insights into the molecular basis of how TPMs attenuate Cofilin-induced F-actin disassembly (Jansen et al., 2015; Mikati et al., 2015; Christensen et al., 2017; Gateva et al., 2017). However, important mechanistic questions remain about how different TPM isoforms influence the binding and activity of Cofilin on F-actin as well as the more potent cellular actin-disassembly activity of CCA.
Mammals have four separate TPM genes that are alternatively spliced to produce 18 different TPM isoforms in nonmuscle cells (Gunning et al., 2015). These TPM isoforms localize to most known cellular actin structures and drive a wide range of actin-based processes. However, it is not clear what the precise mechanistic role of each TPM isoform is, or how TPM isoforms differ in their abilities to control actin filament turnover dynamics and/or the binding of other actin-associated proteins to filaments.
In this study, we focus on five of the most prominent TPM isoforms (Tpm1.6, Tpm1.7, Tpm2.1, Tpm3.1, and Tpm4.2). These isoforms are expressed in nonmuscle cells and have critial roles in cell motility, cell morphogenesis, cytokinesis, neurogenesis, and intracellular transport (Gunning et al., 2015). We use multicolor total internal reflection fluorescence (TIRF) microscopy to examine how each TPM isoform spatially influences Cofilin binding to actin filaments, Cofilin- and CCA-mediated severing of filaments, and, ultimately, filament length. These effects are tested both on preformed actin filaments in the absence of actin monomers and on treadmilling filaments undergoing simultaneous actin assembly at their barbed ends and disassembly at their pointed ends.
RESULTS
TPM isoforms restrict Cofilin binding to pointed ends of actin filaments and inhibit severing along filament sides
To investigate how human Tpm1.6, Tpm1.7, Tpm2.1, Tpm3.1, and Tpm4.2 isoforms influence actin filament disassembly by Cofilin, we first compared their effects on filament severing by human Cofilin-1. Previous studies have shown that each of the five TPM isoforms binds to actin filaments in a cooperative manner and has an affinity of ∼0.5 μM, with the exception of Tpm2.1, which has a much lower affinity of ∼5 µM (Janco et al., 2016). Further, these binding characteristics for TPM isoforms are similar for muscle and nonmuscle F-actin (Janco et al., 2016), and we used muscle actin for all of our experiments herein.
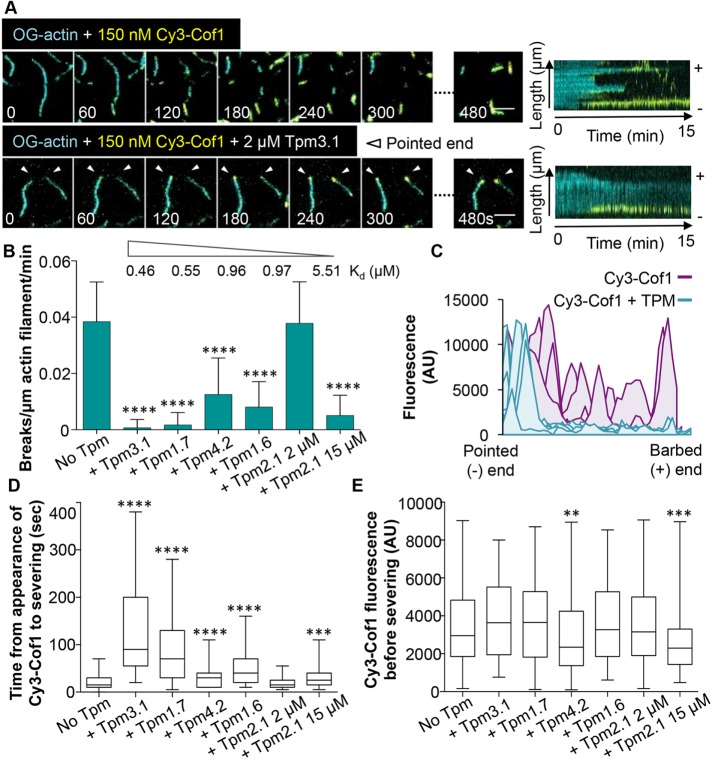
In TIRF microscopy assays, we first assembled sparsely tethered Oregon Green (OG)-labeled actin filaments in the presence or absence of different TPM isoforms (2 µM), and then washed out free actin monomers and flowed in Cy3-labeled human Cofilin-1 (Cy3-Cof1). Our previous studies have shown that Cy3-Cof1 has the same severing activities as unlabeled Cofilin-1 (Jansen et al., 2015; Chin et al., 2016). In the absence of TPM, Cy3-Cof1 cooperatively decorated filaments along their lengths and induced severing events at the bounderies between bare and Cofilin-decorated regions, as previously described (Suarez et al., 2011) (Figure 1A and Supplemental Figure S1A). However, predecoration of filaments with any TPM isoform except for Tpm2.1 confined Cy3-Cof1 binding to the pointed ends (Figure 1A, white arrowheads, and Supplemental Figure S1A). Identification of the pointed ends was unequivocal, as we tracked the assembly of filaments during their growth phase before flowing out actin monomers and flowing in Cy3-Cof1 (Supplemental Figure S1A). Over time, larger Cy3-Cof1 patches began to appear along the sides of actin filaments in the presence of Tpm4.2 or Tpm1.6 (Supplemental Movie 1), but not Tpm3.1 or Tpm1.7. In the presence of Tpm2.1, Cy3-Cof1 immediately began to appear along the entire length of filaments. These observations, consistent with the lower binding affinity of Tpm2.1 for F-actin (Kd = 5.51 μM) (Janco et al., 2016), suggest that 2 µM Tpm2.1 may not decorate filaments as effectively as other TPM isoforms (Figure 1B).
FIGURE 1:
TPMs restrict Cof1 binding to the pointed end and block severing. OG-labeled actin filaments were preassembled in the presence or absence of the indicated human TPM isoform (2 µM). Then, excess actin monomers were flowed out and human Cy3-Cof1 (150 nM) was flowed in. (A) Representative time points and kymographs from TIRF microscopy movies with and without Tpm3.1. The pointed end of each filament is indicated by white arrowheads. For representative time points from TIRF reactions containing other TPM isoforms, see Supplemental Figure S1. Scale bars: 5 μm. (B) From time-lapse imaging (as in A and Supplemental Figure S1), severing rates were measured (n = 120, from three independent experiments). The Kd of each TPM isoform for actin filament binding (as determined by Janco et al., 2016) is indicated above the bar graph. (C) Accumulation of Cy3-Cof1 fluorescence on actin filaments in the absence (purple curves) or presence (teal curves) of Tpm3.1. (D) Distributions of time intervals between first appearance of a Cy3-Cof1 spot on an actin filament and severing event near that Cy3-Cof1 spot. The box-and-whisker plots represent minimum, median, and maximum values for each condition. (E) Distributions of fluorescence intensities of Cy3-Cof1 spots immediately before severing events. The box-and-whisker plots represent minimum, median, and maximum values for each condition. For B, D, and E, statistical difference from control reactions (No TPM) was determined using one-way analysis of variance (ANOVA). ****, p < 0.0001; ***, p < 0.001; **, p < 0.01.
Movie S1.
Effects of TPMs on actin disassembly by Cof1. Oregon Green (OG)-labeled actin filaments (teal) were preassembled in the presence or absence of the indicated TPM isoforms (2 μM). Then, excess actin monomers were flowed out, and the same TPM isoform (2 μM) was flowed in together with Cy3-labeled human Cy3-Cof1 (150 nM). Scale bar, 10 m. Time stamp in min:sec.
The same TPM isoforms that blocked Cy3-Cof1 binding to filament sides also strongly inhibited severing by Cof1 (Figure 1, B and C, and Supplemental Figure S1A). Levels of inhibition ranged from nearly complete abrogation of severing by 2 µM Tpm3.1 to no inhibition of severing by 2 µM Tpm2.1. However, at 15 µM Tpm2.1 (well above its Kd of 5.51 μM), Cy3-Cof1 decoration was restricted to pointed ends, and severing was strongly inhibited (Figure 1B and Supplemental Figure S1A). Thus, for all five TPM isoforms, inihibitory effects on Cy3-Cof1 binding and Cof1-mediated severing scale with binding affinities for F-actin (listed in Figure 1B) (Janco et al., 2016).
Further analysis of our two-color TIRF imaging data showed that each TPM isoform significantly increased the time between Cy3-Cof1 binding to filament sides and subsequent severing events (Figure 1D). In addition, we observed specifically that addition of Tpm4.2 and Tpm2.1 (15 μM) resulted in Cy3-Cof1 patches of lower fluorescence intensity before severing, whereas Tpm3.1, Tpm1.7, and Tpm1.6 did not (Figure 1E). From these observations, we draw two conclusions. First, decoration by all five TPM isoforms increases the time required for Cofilin to induce severing. Second, for filaments decorated by Tpm4.2 and Tpm2.1, reduced levels of Cy3-Cof1 in patches on filament sides will induce fragmentation.
TPM isoform–specific effects on Cor1B, Cof1, and AIP1 (CCA)-induced actin filament disassembly
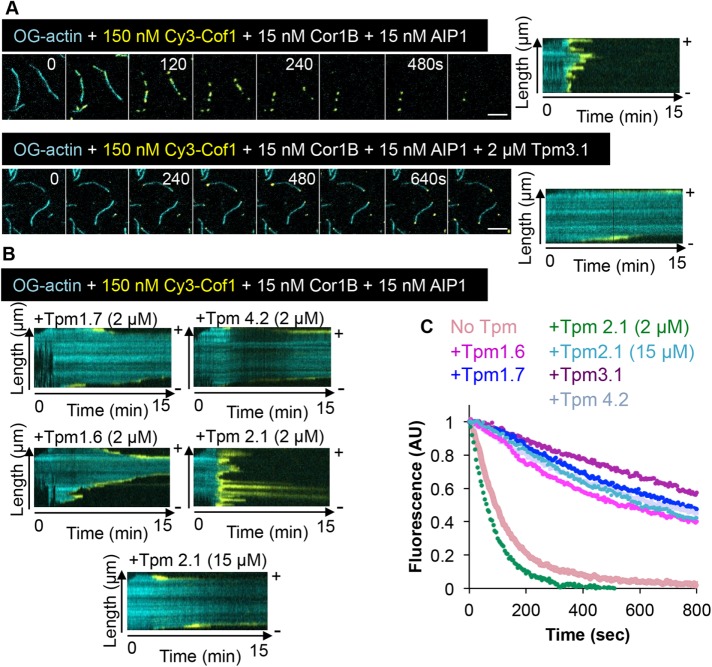
Previous studies have shown that Cor1B and AIP1 strongly enhance Cof1-mediated severing and disassembly of actin filaments (Brieher et al., 2006; Jansen et al., 2015) and that this trio (which we refer to as CCA) works in concert through an ordered mechanism (Jansen et al., 2015). Cor1B binds to filaments first, and this leads to accelerated recruitment of Cof1 to filament sides. Then, Cof1 recruits AIP1, and this immediately results in a severing event. Further, the CCA trio induces rapid severing and disassembly, even under assembly conditions, because it blocks growth at the new barbed ends generated by severing. Given that these three proteins are ubiquitous in animal cells, we tested how CCA disassembly activity is affected by TPM decoration of filaments. For these experiments, we used a molar ratio of 10:1:1 (Cof1, Cor1B, and AIP1, respectively), which is similar to the reported ratio in cells (Brieher et al., 2006; Jansen et al., 2015; Chin et al., 2016).
CCA alone (no TPMs) led to the rapid decoration of filaments by Cy3-Cof1 and the rapid disassembly of actin (Figure 2, A and C, and Supplemental Movie 2). However, in the presence of TPMs, CCA-induced disassembly was attenuated (Figure 2, A–C, and Supplemental Movie 2), with the level of protection scaling approximately with TPM affinities for F-actin. Interestingly, in the combined presence of TPMs and CCA, Cy3-Cof1 binding was observed at both the barbed and pointed ends of filaments. These Cof1-bound regions underwent rapid disassembly, leading to filaments gradually disappearing over time (Figure 2B). This behavior is different from what we observed for Cy3-Cof1 alone (without Cor1B and AIP1), where Cy3-Cof1 decoration was restricted to the pointed ends by TPM, and the filaments shortened much less rapidly. Thus, the presence of Cor1B and AIP1 enable Cof1-mediated disassembly at filament ends in the presence of TPMs.
FIGURE 2:
TPM isoform-specific protection of actin filaments from the collaborative disassembly effects of Cor1B, Cof1, and AIP1. (A) OG-actin filaments were first assembled in the presence or absence of 2 µM Tpm3.1, then the indicated concentrations of Cor1B, Cy3-Cof, and AIP1 were flowed in. Shown are representative time points and kymographs from TIRF imaging. Scale bars: 5 μm. (B) Kymographs from movies, as in A, but in the presence of different TPM isoforms (concentrations indicated). (C) Disassembly rates of actin filaments (OG labeled) in TIRF reactions measured by monitoring the decrease in OG-actin fluorescence over time (n = 60 filaments, pooled from three independent experiments). Filaments were assembled in the presence of different TPM isoforms (2 µM unless otherwise indicated), and disassembly was induced at time zero by washing out actin monomers and flowing in 150 nM Cof1, 15 nM Cor1B, and 15 nM AIP1.
Movie S2.
Effects of TPMs on actin disassembly by Cof1, Cor1B and AIP1. Oregon Green (OG)-labeled actin filaments (teal) were preassembled in the presence or absence of the indicated TPM isoforms (2 μM). Then, excess actin monomers were flowed out, and the same TPM isoform (2 μM) was flowed in together with 150 nM Cy3-Cof1 (yellow), 15 nM Cor1B, and 15 nM AIP1. Scale bar, 10 m. Time stamp in min:sec.
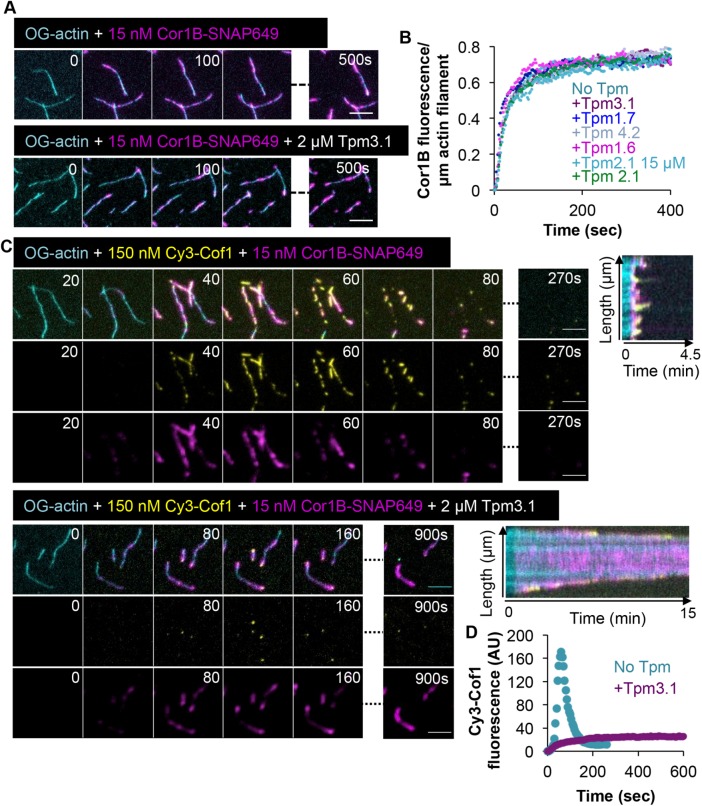
Because Cor1B binding to F-actin leads to the rapid recruitment of Cof1 on filament sides in the absence of TPMs (Jansen et al., 2015; Mikati et al., 2015), our results raise the possibility that TPM blocks Cor1B binding to F-actin. To test this model, we repeated the experiments using labeled Cor1B (Cor1B-SNAP649). These experiments showed that Cor1B-SNAP649 readily bound to filament sides in the presence of any of the five different TPM isoforms (Figure 3, A and B). To further characterize these effects, we performed three-color imaging using labeled actin, Cor1B-SNAP649, and Cy3-Cof1 (with unlabeled AIP1 and TPM). We focused on Tpm3.1, as it is the isoform with the strongest protective effects against CCA. Cor1B-SNAP649 readily bound to filament sides, both in the presence and absence of Tpm3.1, whereas Cy3-Cof1 was observed to bind filament sides only in the absence of Tpm3.1 (Figure 3, C and D). Thus, TPM decoration does not interfere with Cor1B binding to filament sides, yet it blocks Cof1B-induced recruitment of Cof1 to the same sites. Therefore, TPMs may provide a mechanism in cells to counteract the rapid actin-disassembly effects of CCA, while still allowing Coronin to bind filaments and perform its other functions, for example, in bundling F-actin and regulating Arp2/3 complex activities (Goode et al., 1999; Humphries et al., 2002; Cai et al., 2007).
FIGURE 3:
TPM decoration permits Cor1B binding but inhibits Cof1 recruitment. (A) Representative time points from TIRF microscopy imaging of Cor1B-SNAP649 binding to preassembled OG-actin filaments that were either undecorated (top) or decorated with 2 µM Tpm3.1 (bottom). (B) Accumulation of Cor1B-SNAP649 fluorescence on individial OG-actin filaments (n = 45, pooled from three independent experiments) decorated by 2 µM of the indicated TPM isoform. (C) Representative time points and kymographs from TIRF microscopy imaging, in which OG-labeled actin filaments were assembled in the presence or absence of Tpm3.1 (2 µM), and then the indicated concentrations of Cy3-Cof1 and Cor1B-SNAP-649 were flowed in. (D) Accumulation over time of Cy3-Cof1 fluorescence on actin filaments in the presence or absence of Tpm3.1 (2 µM). The average fluorescence signal is shown (n = 45 filaments, pooled from three independent experiments). Scale bars in all panels: 5 μm.
TPM effects on actin filament length and turnover dynamics in a biomimetic treadmilling system
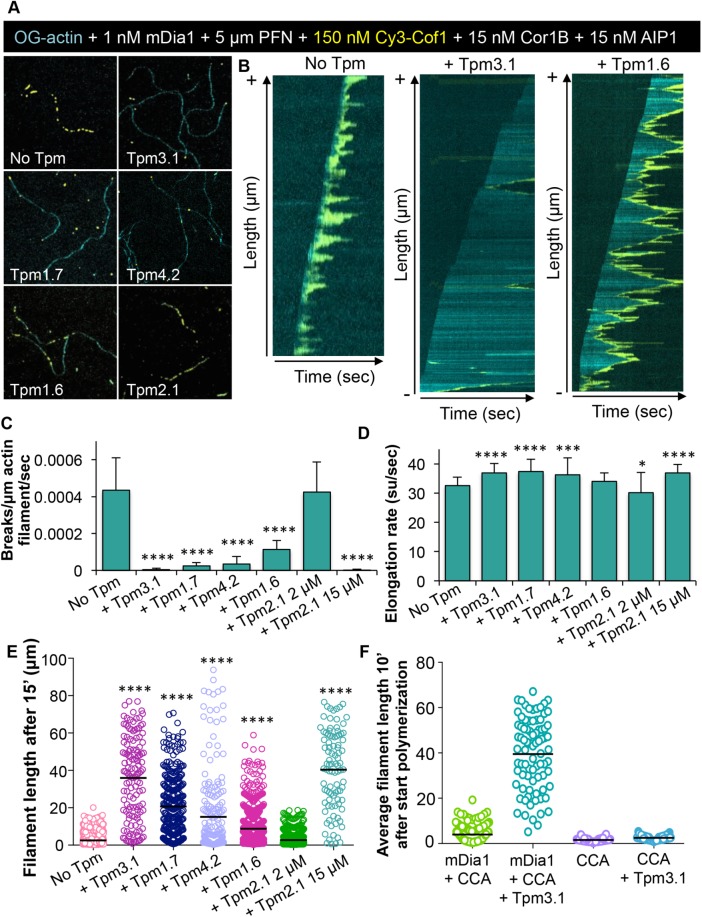
We next tested the effects of TPM decoration on reconstituted filaments that were being rapidly polymerized at their barbed ends by the formin mDia1 and simultaneously disassembled at their pointed ends by CCA. In the absence of TPMs, formin-elongated filaments showed rapid barbed-end growth balanced by rapid pointed-end disassembly, making filaments relatively short (Figure 4, A–D, and Supplemental Movie 3). The addition of TPMs to this system had little effect on formin-mediated elongation rates at barbed ends (Figure 4D), but strongly inhibited filament severing (Figure 4, A–C). As a result, TPM addition led to much longer filaments (Figure 4E and Supplemental Figure S2A). Moreover, the effects on filament length correlated with the strength of the TPM isoform in blocking CCA-induced severing and disassembly. Interestingly, we never observed binding of Cy3-Cof1 to the barbed end of growing filaments, which differs from our observations on preassembled (nongrowing) filaments. This is most likely because the barbed ends of growing filaments consist of ATP/ADP-Pi–bound actin, and Cofilin binds preferentially to ADP-actin (Blanchoin and Pollard, 1998). These results suggest that the barbed-end effects of Cofilin and CCA may be most relevant in vivo on nongrowing filaments and/or filaments that do not have processive elongators on their barbed ends.
FIGURE 4:
TPM isoforms differentially tune the length of actin filaments undergoing simultaneous assembly and turnover. (A) Example of actin filaments being elongated by the formin mDia1 (1 nM) and profilin (PFN; 5 µM), while simultaneously being disassembled by Cy3-Cof1, Cor1B, and AIP1 (150, 15, and 15 nM, respectively) in the presence or absence of different TPM isoforms (2 µM unless otherwise indicated). (B) Kymographs of filaments from TIRF movies as shown in A. (C) Effects of different TPM isoforms on filament severing rate by the three-component system (Cy3-Cof1, Cor1B, and AIP1). Severing rates were obtained from kymographs as in B (n = 80 filaments per condition). (D) Filament barbed-end elongation rates, quantified from reactions as in A containing mDia1, profilin, Cy3-Cof1, Cor1B, AIP1, and different TPM isoforms (n = 80 filaments per condition; su = subunit). (E) Average length of filaments measured 15 min after reactions were initiated. (F) Average length of filaments measured 10 min after reactions were initiated. CCA indicates reactions contain Cor1B, Cof1, and AIP1. Statistical significance from the control condition (No TPM) was determined using one-way ANOVA. ****, p < 0.0001; ***, p < 0.001; *, p < 0.05.
Movie S3.
Effects of TPMs on treadmilling filaments. Oregon Green (OG)-labeled actin filaments (teal) were assembled in the presence of formin mDia1 (1 nM), G-actin (1 μM; 10% OG-labeled), and human profilin (5 μM), while simultaneously being disassembled by 150 nM Cy3-Cof1 (yellow), 15 nM Cor1B, and 15 nM AIP1 in the presence or absence of the indicated TPM isoforms (2 μM). Scale bar, 10 m. Time stamp in min:sec.
Finally, in our reconstituted actin turnover system, we observed that the combination of Tpm3.1 and mDia1 yielded extremely long filaments (39.49 ± 15.8 μm) (Figure 4F and Supplemental Figure S2B). This was in contrast to reactions containing only Tpm3.1 without mDia1 (2.5 ± 0.9 μm) or mDia1 without Tpm3.1 (3.96 ± 3.6 μm) or reactions lacking both Tpm3.1 and mDia1 (1.6 ± 0.7 μm). These results suggest that mDia1 and Tpm3.1 can synergize under assembly-promoting conditions in protecting filaments from the disassembly effects of CCA.
DISCUSSION
Actin filaments can assemble into many different architectures, characterized by unique dimensions, turnover dynamics, and lifetimes ranging from seconds, to minutes, or even to hours and days. Our results here suggest that some of this diversity in the length and dynamics of actin structures can be achieved by the influence of different TPM isoforms on actin-disassembly mechanisms. Most cellular actin structures are decorated by one or more TPM isoforms, although our knowledge of the specific isoforms that decorate each in vivo actin structure is still incomplete (Gunning et al., 2015). Previous studies have demonstrated convincingly that TPMs can antagonize actin disassembly induced by Cofilin or by Cofilin and AIP1 (Bryce et al., 2003; Yu and Ono, 2006; Mikati et al., 2015; Robaszkiewicz et al., 2016; Christensen et al., 2017; Gateva et al., 2017; Ostrowska et al., 2017). Further, these studies have shown that TPM and Cofilin compete for binding on actin filaments. However, a number of important questions about the underlying mechanism and the TPM isoform-specific effects have remained. By using multiwavelength TIRF imaging and directly visualizing the effects of five prominent mammalian nonmuscle TPM isoforms on both Cofilin- and CCA-mediated actin disassembly and on both preformed and actively growing filaments, we have gained new insights into these mechanisms and TPM differences.
In our multiwavelength TIRF analysis, we used labeled actin and labeled Cofilin. In contrast, a previous TIRF study on TPM isoforms by Gateva and coworkers used labeled TPMs and Cofilin with unlabeled actin filaments (Gateva et al., 2017), and therefore did not directly visualize and quantify effects on severing. Our experimental conditions allowed us to quantify severing events and simultaneously monitor where Cofilin binds on filaments in real time in the presence of different TPMs. We found that filament decoration by any one of the five mammalian TPM isoforms spatially restricts human Cy3-Cof1 binding to the pointed ends of filaments and strongly impedes severing. These effects are similar to those reported for Schizosaccharomyces pombe TPM and ADF/Cofilin (Christensen et al., 2017), suggesting there is a conserved functional relationship between TPMs and Cofilin in species as diverse as fission yeast and mammals. We also observed that the presence of TPM isoforms increased the delay between Cy3-Cof1 binding to filaments and resulting severing events. The relative strength of these TPM protective effects against Cof1 severing scaled with the reported binding affinities of the TPM isoforms for F-actin (Janco et al., 2016). Tpm3.1 showed the strongest protective effects against Cof1-mediated severing, in agreement with previous in vivo studies showing that overexpression of Tpm3.1 leads to cellular F-actin structures being more resistant to Latrunculin A-induced disassembly (Percival et al., 2000; Creed et al., 2008). Because different TPMs decorate different cellular actin networks and function in highly variable physiological processes, it has been suggested that they may have qualitatively distinct effects on actin assembly and turnover dynamics. However, with respect to actin disassembly (Cof1 and CCA mediated), our data indicate that the differences between TPM isoforms are primarily quantitative rather than qualitative.
It is also important to note that some of our results differ from the effects of TPM isoforms reported in the Gateva et al. study (2017). Using TIRF assays, we found that the TPM isoforms with the strongest protective effects against Cof1 were (in order from strongest to weakest): Tpm3.1, Tpm1.7, Tpm1.6, Tpm4.2, and Tpm2.1. In contrast, Gateva et al. used bulk assays to quantitatively compare Cof1-induced F-actin disassembly, and observed that Tpm3.1 was the least protective against Cof1 and that Tpm1.6 was the strongest. It is possible that these discrepancies arise from differences in the assays and/or differences in the preparation of TPM isoforms. Gateva and coworkers used bulk assays to assess the ability of TPMs to protect filaments from Cof1-induced disassembly. In contrast, we used TIRF assays to directly visualize the effects of TPM isoforms in protecting filaments from Cof1 binding and severing. Another important difference is that, in our assays, TPMs were present during the initial actin-filament assembly phase, permitting TPMs to bind F-actin before conversion of ADP+Pi actin to ADP-actin. This may help facilitate filament decoration by some TPM isoforms. In addition, while both studies used TPM isoforms expressed in Escherichia coli, Gateva and coworkers modified their TPMs to include short N-terminal “acetylation mimic” motifs, whereas we did not. Acetylation can play an important role in regulating TPM interactions with F-actin (Monteiro et al., 1994); however, in most cell types, it remains unclear what fraction of each TPM isoform is acetylated versus nonacetylated in vivo. Therefore, it is important to continue studying the activities of acetylated and nonacetylated TPMs. Indeed, a direct comparison of our results and those of Gateva et al. (2017) raises the intriguing possibilty that acetylation more tightly controls the activities of Tpm2.1 and Tpm3.1 than the activities of some of the other TPM isoforms.
Our study also examines for the first time the effects of TPM isoforms on actin disassembly induced by Cof1, Cor1B, and AIP1 (CCA), a ubiquitous group of actin binding proteins that work together to induce rapid F-actin disassembly, even under assembly-promoting conditions (Brieher et al., 2006; Jansen et al., 2015). Our data show that all five mammalian TPM isoforms protected filament sides from CCA-induced severing and restricted decoration of Cy3-Cof1 to the pointed ends of actin filaments. On the other hand, TPM decoration allowed Cor1B binding to filament sides, while hampering Cor1B's ability to recruit Cof1 to filaments and acclerate severing. Thus, all five TPM isoforms obstruct binding of Cof1, but not Cor1B, to F-actin. This agrees with recent electron microscopy studies suggesting that Cor1B and Cof1 have neighboring yet distinct binding sites on F-actin and that TPM and Cof1 binding sites on F-actin overlap (Ge et al., 2014; von der Ecken et al., 2015, 2016).
In contrast to Cof1 alone, CCA was effective in promoting pointed-end disassembly of TPM-decorated filaments. These observations have important implications for the in vivo regulation of actin turnover. They suggest that combining TPM decoration and CCA may provide a powerful mechanism for preserving the filamentous architecture of an actin network (e.g., polarized cables, cytokinetic rings, stress fibers, and stereocilia), while permitting turnover at the pointed ends of the network.
Our work has also exposed a working relationship between TPMs and formins in assembling long filaments in the presence of CCA. This observation suggests that specific TPMs and formins may function together in vivo to promote formation of specific actin network architectures consisting of long filaments. Indeed, yeast TPMs are recruited to actin cables and cytokinetic rings, which are nucleated by formins, but excluded from cortical actin patches nucleated by Arp2/3 complex (DesMarais et al., 2002). Further, specific TPMs can pair with specific formins in yeast to drive actin cable assembly (Johnson et al., 2014; Alioto et al., 2016). Similarly, in mammalian cells, TPMs heavily decorate a number of actin structures polymerized by formins, including stress fibers, cytokinetic rings, and neuronal microspikes (Gunning et al., 2015). Finally, although more effort is needed to determine how TPMs and formins work together to control actin organization and dynamics, the expression of 18 different nonmuscle TPM isoforms combined with 15 different formins provides a rich and diverse platform for tuning the architectures and dynamics of cellular actin networks.
MATERIALS AND METHODS
Plasmids
pET bacterial expression plasmids for human Tpm 1.6 (Tm 2), human Tpm 1.7 (Tm 3), human Tpm2.1 (Tm 1), human Tpm3.1 (Tm5NM1), and human Tpm4.2 (Tm4) were graciously sent by Peter Gunning (University of New South Wales, Australia). A plasmid for expressing mouse Coronin-1B (Cor1B) with a C-terminal 8His-Strep tag in mammalian cells (vector pTT5SH8Q2) was kindly provided by Jim Bear (University of North Carolina). A SNAP tag was cloned at the C-terminus to generate Cor1B-SNAP. The plasmid-carrying mouse AIP1 was kindly provided by Naoki Watanabe (Tohoku University, Japan), and the insert was cloned into the same mammalian expression vector as Cor1B. The plasmid for expressing human Cof1 in E. coli was generously provided by David Kovar (University of Chicago). For fluorescence labeling of human Cof1, we used a previously described Cof1 construct that has only one exposed Cys residue (Jansen et al., 2015). All constructs were verified by DNA sequencing.
Protein purification and labeling
Rabbit skeletal muscle actin (RMA) was purified as previously described (Graziano et al., 2013). In brief, RMA was purified by generating an acetone powder from ground muscle tissue, which was stored in aliquots at −80°C. Aliquots of acetone powder were then pulverized using a coffee grinder, resuspended in G-buffer, and cleared by low-speed centrifugation. The actin was polymerized overnight and then pelleted. The pellet was disrupted by dounce homogenizing, dialyzed against G-buffer for 2–3 d, and then gel filtered on a 16/60 S200 column (GE Healthcare). Column fractions were stored at 4°C. Actin was labeled on Cys-374 with OG maleimide as described in Kuhn and Pollard (2007). Briefly, monomeric actin reconstituted from an actin pellet was dialyzed against two changes of G-buffer without dithiothreitol (DTT) for 1 h each. After clarification at 500 × g for 5 min, actin was polymerized by mixing an equal volume of cold 2× label buffer (2× = 50 mM imidazole, pH 7.5, 0.2 M KCl, 4 mM MgCl2, 6 mM NaN3, and 0.6 mM ATP). After 5 min, polymerized actin was diluted to 1 mg/ml with cold 1× label buffer, and then a 10-fold molar excess of OG was added dropwise to the actin while stirring, and the solution was stirred gently overnight. Labeled actin was first clarified at 500 × g for 5 min to remove large aggregates and then centrifuged at 105,000 × g for 2 h to pellet actin filaments. The pellet was resuspended in G-buffer by dounce homogenizing, dialyzed for 2 d against two changes of G-buffer, and gel filtered on a 16/60 S200 column (GE Healthcare). Peak fractions were combined and stored at −20°C. Labeling efficiency of OG-actin was measured by absorbance at 290 and 491 nm, and extinction coefficient E491 = 77,800 M−1cm−1. The absorption at 290 nm was corrected for background fluorescence from the OG dye (correction factor 0.016991).
The formin mDia1 (6xHis-FH1-FH2-C) was inducibly expressed in yeast and purified by sequential Ni2+-NTA and gel-filtration chromatography steps. Cor1B and AIP1 were expressed and purified from transfected HEK293T cells (ATCC). Cells were grown on plates at 37°C under a humidified atmosphere containing 5% CO2 in DMEM, supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum, glucose (4.5 g/l), penicillin (100 U/ml), and streptomycin (100 µg/ml). Cells at 30–40% confluence were transiently transfected using 25 kDa linear polyethylenimine (Polysciences, Warrington, PA). At 72 h posttransfection, cells were harvested in phosphate-buffered saline (PBS), pelleted by centrifugation at 1000 × g for 5 min, and lysed by repeated freeze-thawing in 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 1% (vol/vol) Triton X-100, and a standard cocktail of protease inhibitors (Roche, Germany). After a 30 min incubation on ice, cell lysates were cleared by centrifugation at 20,000 × g at 4°C using an eppendorf tabletop centrifuge and incubated with Ni2+-NTA beads (Qiagen, Valencia, CA) for 90 min at 4°C in the presence of 20 mM imidazole. After being washed with Buffer A (20 mM Tris, pH 7.5, 300 mM NaCl, 50 mM imidazole, and 1 mM DTT), proteins were eluted in Buffer A supplemented with 250 mM imidazole, concentrated, and purified further on a Superose 6 gel-filtration column (GE Healthcare Biosciences, Pittsburgh, PA) equilibrated in Buffer B (20 mM Tris, pH 8.0, 50 mM KCl, and 1 mM DTT). For fluorescence labeling of SNAP-Cor1B, the fusion protein was bound to Ni2+-NTA beads, washed extensively in PBS with 1 mM DTT, and incubated with a fivefold excess of benzylguanine or benzylchloropyrimidine SNAP-Surface 649 (New England Biolabs, Ipswich, MA) for 2 h at room temperature. Next, beads were washed extensively in PBS with 1 mM DTT and eluted in PBS with 250 mM imidazole. Proteins were exchanged into Buffer B on PD-10 columns (GE Healthcare Biosciences) to remove free dye. Labeling efficiencies were determined spectrophotometrically using the absorbance at 650 nm and an extinction coefficient of 250,000 M−1 cm−1 for SNAP-Surface 649, combined with absorbance at 280 nm and an estimated extinction coefficient of 82,850 M−1 cm−1 for Cor1B-SNAP. The absorption at 280 nm was corrected for background fluorescence from the dye (correction factor 0.024). Human Cof1 was expressed in BL21 (DE3) E. coli by growing cells at 37°C in TB medium to log phase, then inducing expression with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 18°C for 16 h. Cells were harvested by centrifugation and stored at −80°C and then lysed by sonication in 20 mM Tris (pH 8.0), 50 mM NaCl, 1 mM DTT, and protease inhibitors. Lysates were cleared by centrifugation at 30,000 × g for 20 min in a Fiberlite F13-14 × 50CY rotor (Thermo Scientific, Rockport, IL), and applied to a 5 ml HiTrap HP Q column (GE Healthcare Biosciences). The flow-through containing Cof1 was collected and dialyzed into 20 mM HEPES (pH 6.8), 25 mM NaCl, and 1 mM DTT. Next, the protein was applied to a 5 ml HiTrap SP FF column (GE Healthcare Biosciences) and eluted with a linear gradient of NaCl (25–500 mM). Fractions containing Cof1 were concentrated and dialyzed into Buffer B, aliquoted, snap-frozen in liquid N2, and stored at −80°C until use. Dye-labeled Cof1 was purified similarly, except that the protein was eluted from the SP FF column with PBS and then incubated with a 10-fold excess of Cy3-maleimide (GE Healthcare Biosciences) for 2 h at room temperature in the presence of 0.3 mM Tris(2-carboxyethyl)phosphine (TCEP). Excess dye was removed by passing the protein over a PD-10 column equilibrated in Buffer B. Final labeling efficiency was 30%. Labeling efficiency was determined as described earlier, using absorbance at 550 nm and an extinction coefficient of 150,000 M−1 cm−1 for Cy3, combined with absorbance at 280 nm and an estimated extinction coefficient of 14,440 M−1 cm−1 for Cof1.
Human TPM isoforms were expressed in BL21 (DE3) E. coli by growing cells at 37°C in TB medium to log phase and then inducing expression with 1 mM IPTG at 18°C for 16 h. Cells were harvested by centrifugation, and lysed by sonication in 20 mM Tris (pH 7.5), 0.5 M NaCl, and 5 mM MgCl2. For separation of the heat-stabile TPMs from contaminating proteins, the lysate was incubated at 80°C for 10 min in a waterbath, cooled for 10 min at −20°C, and cleared by centrifugation at 30,000 × g for 20 min. Next, the supernatant was isoelectrically precipitated by dropwise addition of 0.3 M HCl to pH ∼4.7 and centrifuged at 5000 rpm for 15 min at 4°C, after which the pellet was resuspended in 100 mM Tris (pH 7.5), 0.5 M NaCl, 5 mM MgCl2, and 1 mM DTT. This precipitation step was repeated one more time, and the resuspended pellet was dialyzed to 20 mM HEPES (pH 6.8), 50 mM NaCl, and 0.5 mM DTT. Next, dialyzed protein was applied to a 5 ml HiTrap Q HP column (GE Healthcare Biosciences) and eluted with a linear gradient of NaCl (50–600 mM). The fractions containing TPM protein were concentrated and further purified on a Superose 6 gel-filtration column (GE Healthcare Biosciences) equilibrated in 20 mM Tris (pH 7.5), 50 mM KCl, 2 mM MgCl2, and 1 mM DTT). Aliquots were flash-frozen and stored at −80°C. For three of the TPM isoforms (Tpm1.6, Tpm3.1, and Tpm4.2), we verified that the concentration we used in our TIRF assays (2 µM) saturated F-actin binding in cosedimentation assays, as previously reported (Janco et al., 2016).
TIRF microscopy
For all TIRF experiments, coverslips were first cleaned by sonication in detergent for 60 min, followed by successive sonications in 1 M KOH and 1 M HCl for 20 min each and then sonication in ethanol for at least 60 min. Coverslips were then washed extensively with H2O; dried in an N2-stream; layered with 200 µl of 80% ethanol (pH 2.0), 2 mg/ml poly-ethylene glycol (PEG)-silane, and 2 µg/ml biotin-PEG-silane (Laysan Bio, Arab, AL); and incubated for 16 h at 70°C. Flow cells were assembled by rinsing PEG-coated coverslips extensively with H2O, then attaching them to a flow chamber (Ibidi, Martinsried, Germany) using double-sided tape (2.5 cm × 2 mm × 120 µm) and epoxy resin. Lightly tethered actin filaments were used for all TIRF experiments in this study. To accomplish this, flow cells were incubated for 3 min with HBSA (HEK buffer with 1% bovine serum albumin), which was followed by 30 s incubation with 0.1 mg/ml streptavidin in HEK buffer. Flow cells were washed with HBSA and equilibrated with TIRF buffer (10 mM imidazole, pH 7.4, 50 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid [EGTA], 0.2 mM ATP, 10 mM DTT, 15 mM glucose, 20 µg/ml catalase, 100 µg/ml glucose oxidase, and 0.5% methylcellulose [4000 cP]). Reactions were initiated by rapidly diluting actin monomers (1 µM final, 10% OG-labeled, 0.25% biotinylated) into TIRF buffer, after which the mixture was transferred into a flow chamber. After filaments had polymerized to lengths of ∼10–15 μm, the reaction mixture was replaced with TIRF buffer containing the indicated proteins. Multiwavelength time-lapse TIRF imaging was performed using a Nikon-Ti200 inverted microscope equipped with a 150 mW Ar-Laser (Mellot Griot, Carlsbad, CA), a TIRF-objective with a NA of 1.49 (Nikon Instruments, New York, NY), and an EMCCD camera (Andor Ixon, Belfast, Northern Ireland). During measurements, optimal focus was maintained by the perfect focus system (Nikon Instruments).
TIRF data analysis
TIRF data were analyzed using ImageJ software (National Institutes of Health [NIH], Bethesda, MD). Before each analysis, the background was subtracted using the standard background subtraction tool (rolling ball radius 50 pixels). Severing rates for Cofilin in the absence of Cor1B and AIP1 were calculated by measuring the initial lengths of filaments before flow-in, and then by counting severing events observed during the next 600 s after flow-in of the indicated protein combinations. For analysis of Cy3–Cof1 patches, severing events were scored, and then a 1 μm × 1 μm box was drawn around the Cy3–Cof1 patch one frame before severing and saved as a region of interest (ROI). The fluorescence intensity of the Cy3 fluorescence in the boxes was determined using the Measure Integrated Density function. For kinetic analysis of Cor1B-SNAP649 or Cy3–Cof1 binding to filaments, OG-actin filaments were traced based on the signal in the 488-nm channel, saved as an ROI, and then used to determine the fluorescence profiles in the 561 or 640 nm channel using the Plot Z-axis profile tool. Severing rates of the three-component (CCA) system were obtained by monitoring the decrease in OG fluorescence of individual filaments during 600 s after flow-in of the indicated protein combinations. For reconstituted actin filaments simultaneously assembling and undergoing turnover, individual kymographs were generated for each filament. The slope of the kymographs was used to determine barbed-end elongation rates, whereas the number of troughs over the entire filament length over time was used to determine the severing rates. Statistical analyses were performed with Prism 5.0.
Supplementary Material
Acknowledgments
We are grateful to members of the Goode lab for helpful discussions and to Julian Eskin, David Kast, and Shashank Shekhar for comments on the article. This work was supported by a grant from the National Institutes of Health (R01 GM063691) to B.L.G.
Abbreviations used:
- ADF
actin depolymerizing factor
- AIP1
actin-interacting protein 1
- ANOVA
analysis of variance
- CCA
Cof1, Cor1B, and AIP1
- Cy3-Cof1
Cy3-labeled human Cofilin-1
- DTT
dithiothreitol
- IPTG
isopropyl β- d-1-thiogalactopyranoside
- OG
Oregon Green
- PBS
phosphate-buffered saline
- RMA
rabbit skeletal muscle actin
- ROI
region of interest
- TIRF
total internal reflection fluorescence
- TPM
Tropomyosin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-12-0815) on January 16, 2019.
REFERENCES
- Alioto SL, Garabedian MV, Bellavance DR, Goode BL. (2016). Tropomyosin and profilin cooperate to promote formin-mediated actin nucleation and drive yeast actin cable assembly. Curr Biol , 3230–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. (1998). Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J Biol Chem , 25106–25111. [DOI] [PubMed] [Google Scholar]
- Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. (2006). Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol , 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, et al. (2003). Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell , 1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. (2007). Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell , 915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Courtemanche N, Pollard TD. (2015). Aip1 promotes actin filament severing by cofilin and regulates constriction of the cytokinetic contractile ring. J Biol Chem , 2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin SM, Jansen S, Goode BL. (2016). TIRF microscopy analysis of human Cof1, Cof2, and ADF effects on actin filament severing and turnover. J Mol Biol , 1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JR, Hocky GM, Homa KE, Morganthaler AN, Hitchcock-DeGregori SE, Voth GA, Kovar DR. (2017). Competition between Tropomyosin, Fimbrin, and ADF/Cofilin drives their sorting to distinct actin filament networks. eLife , e23152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creed SJ, Bryce N, Naumanen P, Weinberger R, Lappalainen P, Stehn J, Gunning P. (2008). Tropomyosin isoforms define distinct microfilament populations with different drug susceptibility. Eur J Cell Biol , 709–720. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. (2002). Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J Cell Sci , 4649–4660. [DOI] [PubMed] [Google Scholar]
- Gateva G, Kremneva E, Reindl T, Kotila T, Kogan K, Gressin L, Gunning PW, Manstein DJ, Michelot A, Lappalainen P. (2017). Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr Biol , 705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Durer ZA, Kudryashov D, Zhou ZH, Reisler E. (2014). Cryo-EM reveals different coronin binding modes for ADP- and ADP-BeFx actin filaments. Nat Struct Mol Biol , 1075–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Wong JJ, Butty AC, Peter M, McCormack AL, Yates JR, Drubin DG, Barnes G. (1999). Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J Cell Biol , 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano BR, Jonasson EM, Pullen JG, Gould CJ, Goode BL. (2013). Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. J Cell Biol , 595–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressin L, Guillotin A, Guerin C, Blanchoin L, Michelot A. (2015). Architecture dependence of actin filament network disassembly. Curr Biol , 1437–1447. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. (2015). Tropomyosin—master regulator of actin filament function in the cytoskeleton. J Cell Sci , 2965–2974. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Barua B. (2017). Tropomyosin structure, function, and interactions: a dynamic regulator. Subcell Biochem , 253–284. [DOI] [PubMed] [Google Scholar]
- Humphries CL, Balcer HI, D'Agostino JL, Winsor B, Drubin DG, Barnes G, Andrews BJ, Goode BL. (2002). Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol , 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janco M, Bonello TT, Byun A, Coster AC, Lebhar H, Dedova I, Gunning PW, Bocking T. (2016). The impact of tropomyosins on actin filament assembly is isoform specific. Bioarchitecture , 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S, Collins A, Chin SM, Ydenberg CA, Gelles J, Goode BL. (2015). Single-molecule imaging of a three-component ordered actin disassembly mechanism. Nat Commun , 7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, East DA, Mulvihill DP. (2014). Formins determine the functional properties of actin filaments in yeast. Curr Biol , 1525–1530. [DOI] [PubMed] [Google Scholar]
- Kueh HY, Charras GT, Mitchison TJ, Brieher WM. (2008). Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol , 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. (2007). Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J Biol Chem , 28014–28024. [DOI] [PubMed] [Google Scholar]
- Mikati MA, Breitsprecher D, Jansen S, Reisler E, Goode BL. (2015). Coronin enhances actin filament severing by recruiting cofilin to filament sides and altering F-actin conformation. J Mol Biol , 3137–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro PB, Lataro RC, Ferro JA, de Castro Reinach F. (1994). Functional alpha-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J Biol Chem , 10461–10466. [PubMed] [Google Scholar]
- Nadkarni AV, Brieher WM. (2014). Aip1 destabilizes cofilin-saturated actin filaments by severing and accelerating monomer dissociation from ends. Curr Biol , 2749–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan P, Chatterton P, Ikeda A, Ikeda S, Corey DP, Ervasti JM, Perrin BJ. (2015). Length regulation of mechanosensitive stereocilia depends on very slow actin dynamics and filament-severing proteins. Nat Commun , 6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowska Z, Robaszkiewicz K, Moraczewska J. (2017). Regulation of actin filament turnover by cofilin-1 and cytoplasmic tropomyosin isoforms. Biochim Biophys Acta , 88–98. [DOI] [PubMed] [Google Scholar]
- Percival JM, Thomas G, Cock TA, Gardiner EM, Jeffrey PL, Lin JJ, Weinberger RP, Gunning P. (2000). Sorting of tropomyosin isoforms in synchronised NIH 3T3 fibroblasts: evidence for distinct microfilament populations. Cell Motil Cytoskeleton , 189–208. [DOI] [PubMed] [Google Scholar]
- Robaszkiewicz K, Ostrowska Z, Marchlewicz K, Moraczewska J. (2016). Tropomyosin isoforms differentially modulate the regulation of actin filament polymerization and depolymerization by cofilins. FEBS J , 723–737. [DOI] [PubMed] [Google Scholar]
- Rzadzinska AK, Schneider ME, Davies C, Riordan GP, Kachar B. (2004). An actin molecular treadmill and myosins maintain stereocilia functional architecture and self-renewal. J Cell Biol , 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, Guerin C, Martiel JL, De la Cruz EM, Blanchoin L. (2011). Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol , 862–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. (2016). Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature , 724–728. [DOI] [PubMed] [Google Scholar]
- von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. (2015). Structure of the F-actin-tropomyosin complex. Nature , 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Mitchison TJ. (2002). Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science , 1083–1086. [DOI] [PubMed] [Google Scholar]
- Yu R, Ono S. (2006). Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil Cytoskeleton , 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.