Summary
Sclerotinia sclerotiorum is a devastating necrotrophic fungal pathogen that infects over 400 species of plants worldwide. Reactive oxygen species (ROS) modulations are critical for the pathogenic development of S. sclerotiorum. The fungus applies enzymatic and non‐enzymatic antioxidants to cope with the oxidative stress during the infection processes. Survival factor 1 was identified and characterized to promote survival under conditions of oxidative stress in Saccharomyes cerevisiae. In this research, a gene named SsSvf1 was predicted to encode a survival factor 1 homologue in S. sclerotiorum. SsSvf1 transcripts showed high expression levels in hyphae under oxidative stress. Silencing of SsSvf1 resulted in increased sensitivity to oxidative stress in culture and increased levels of intracellular ROS. Transcripts of SsSvf1 showed a dramatic increase during the initial stage of infection and the gene‐silenced strains displayed reduced virulence on oilseed rape and Arabidopsis thaliana. Inhibition of plant ROS production partially restores virulence of SsSvf1 gene‐silenced strains. SsSvf1 gene‐silenced strains exhibited normal oxalate production, but were impaired in compound appressorium formation and cell wall integrity. The results suggest that SsSvf1 is involved in coping with ROS during fungal‐host interactions and plays a crucial role in the pathogenicity of S. sclerotiorum.
Keywords: oxidative stress, Sclerotinia sclerotiorum, Survival factor 1, virulence
Introduction
Sclerotinia sclerotiorum (Lib.) de Bary is one of the most devastating fungal pathogens with a worldwide distribution. This pathogen can infect more than 400 plant species and lead to significant losses in many cultivated crops including oilseed, sunflower, soybean and the common bean (Boland and Hall, 1994; Bolton et al., 2006; Purdy, 1979).
S. sclerotiorum has been considered a model necrotrophic fungal pathogen, which kills host tissue via the secretion of oxalic acid (OA) (Cessna et al., 2000; Favaron et al., 2004; Kim et al., 2008; Williams et al., 2011) and cell wall degrading enzymes (Martel et al., 1996; Poussereau et al., 2001; Riou et al., 1991; Yajima et al., 2009; Yu et al., 2016; Zuppini et al., 2005). Recent evidence indicates that this fungus secretes effector proteins to suppress host defence in the initial stage of infection (Guyon et al., 2014; Lyu et al., 2016; Yang et al., 2018; Zhu et al., 2013). These strategies are mainly applied by biotrophic and hemibiotrophic fungal pathogens. In addition, biotrophic growth at the leading edge of fungal colonization was suggested for S. sclerotiorum (Kabbage et al., 2015). These studies indicate that the pathogenesis of S. sclerotiorum is more complex than we thought and more evidence is needed to detail the underlying molecular mechanism.
Rapid generation of reactive oxygen species (ROS) including hydrogen peroxide (H2O2), the superoxide anion (O2 −), and hydroxyl radical (OH•) is an early resistance response in many plant/pathogen interactions (Lamb and Dixon, 1997). Such oxidative bursts have direct and powerful antimicrobial activity including inhibition of the spore germination of a number of fungal pathogens (Mousavi and Robson, 2004; Peng and Kuc, 1992). In response, fungal pathogens apply specific enzymes and non‐enzyme‐mediated antioxidant mechanisms to handle ROS (Aguirre et al., 2006). Several studies have shown that ROS modulation is critical for the hyphae of S. sclerotiorum to successfully colonize host plant tissue (Kim et al., 2011; Williams et al., 2011; Yarden et al., 2014); however, evidence for the molecular mechanisms of ROS detoxification and tolerance in S. sclerotiorum are still sparse.
The Survival factor 1 (SVF1) gene was first identified in Saccharomyces cerevisiae in a screen for mutations that could be functionally complemented by exogenous expression of the human anti‐apoptotic gene Bcl‐xL (Brace et al., 2005; Vander Heiden et al., 2002). However, SVF1 and Bcl‐xL have distinct roles in regulating cell survival (Brace et al., 2005). S. cerevisiae cells lacking Svf1 protein showed hypersensitivity to direct chemical precursors of ROS, suggesting that SVF1 is necessary for survival under oxidative stress (Brace et al., 2005). Deeper research has shown that Svf1‐mediated cell survival under conditions of oxidative stress by affecting the sphingolipid metabolism in S. cerevisiae (Brace et al., 2007). To date, the role of Svf1 in the oxidative stress response and pathogenicity of filamentous fungal pathogens has remained unknown.
Here, a gene in S. sclerotiorum (SS1G_01919) named SsSvf1 (Sclerotinia sclerotiorum Survival factor 1) was predicted to encode a yeast Svf1 homologous protein. The function of SsSvf1 was determined via a reverse genetic approach, and its role in oxidative stress response and pathogenicity was investigated. The research may help clarify the function of Svf1 in fungal plant pathogens and the pathogenicity of S. sclerotiorum in more detail.
Results
SsSvf1 encodes a survival factor‐1 homologue in S. sclerotiorum
The S. sclerotiorum SsSvf1 gene consists of four exons and three introns, and encodes a protein with 381 amino acids. Conserved Domain Database (CDD) analysis of the protein sequence revealed that a Svf‐like domain was predicted at amino acid position T52–I380 (Marchler‐Bauer et al., 2017). Alignment of amino acid sequences of the N‐terminal and C‐terminal of the Svf1 domains in SsSvf1 and yeast Svf1 exhibit great similarity (Brace et al., 2005) (Fig. 1). BLASTP searches using the amino acid sequence of SsSvf1 as a query showed that the homologous sequences are widely present in fungi including some important plant pathogens such as Botrytis cinerea (XP_001548941), Gibberella zeae (XP_011323561) and Colletotrichum higginsianum (XP_018161001).
Figure 1.
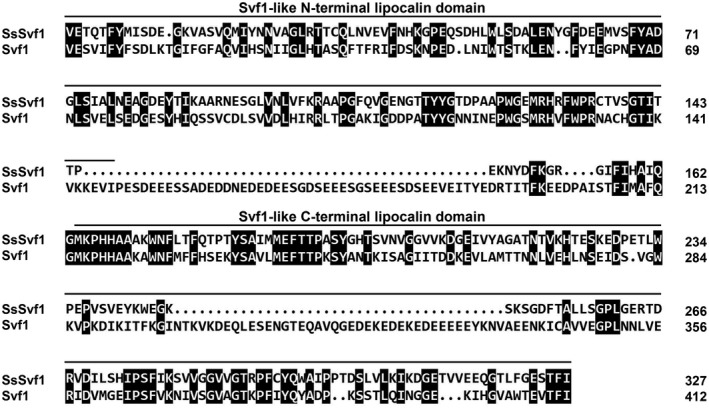
Alignment of amino acid sequences of Svf1 domains in SsSvf1 and yeast Svf1 protein (KZV12585) using ClustalX. Shading indicates sequence similarities of 100% (dark).
SsSvf1 is required for response to oxidative stress
The expression of SsSvf1 under oxidative stress conditions was analysed to explore the role of the SsSvf1 gene in response to oxidative stress of S. sclerotiorum. As shown in Fig. 2A, the expression level of SsSvf1 was much higher in hyphae treated with H2O2 (5 mM and 10 mM). To determine the function of SsSvf1, a gene‐silencing vector was constructed based on pSilent‐1 (Nakayashiki et al., 2005) as described in methods. The vector was linearized and transformed into the wild‐type strain of S. sclerotiorum via PEG (polyethylene glycol) methods (Rollins, 2003). Several transformants were obtained, and silencing of SsSvf1 in the transformants was evaluated by real‐time Reverse Transcription‐Polymerase Chain Reaction (RT‐PCR) (Fig. S1). The expression levels of SsSvf1 in SiSvf1‐230 and SiSvf1‐213 were 15% and 2% of that in the wild‐type strain, respectively. Thus, these two strains were chosen for deeper research.
Figure 2.
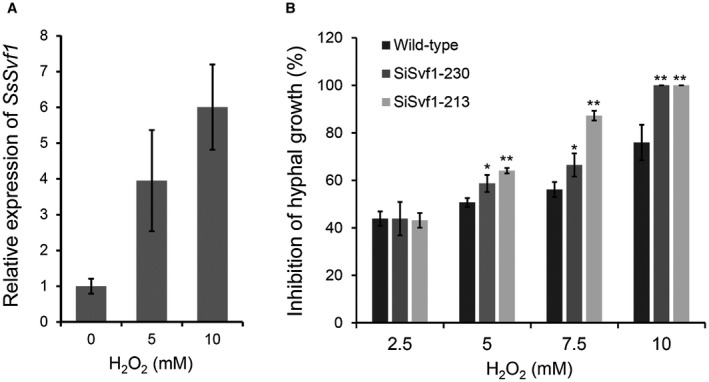
Functional analysis of SsSvf1 in response to H2O2. (A) Relative expression level of SsSvf1 in hyphae treated with H2O2 (5 mM and 10 mM). Total SsSvf1 cDNA abundance in the samples was normalized using tub1 gene as a control. The relative expression of SsSvf1 in the untreated strain was set as one. Bars indicate standard deviation. (B) Percent growth inhibition of wild‐type strain and SsSvf1 gene‐silenced strains on potato dextrose agar (PDA) medium with H2O2. The strains were inoculated on PDA plates amended with H2O2 at concentrations of 0 mM to 10 mM. Percentage inhibition of hyphal growth was calculated at 36 hpi. Bars indicate standard deviation. Asterisks denote significant differences (one‐way analysis of variance [ANOVA]): *P < 0.05; **P < 0.01.
The hyphal growth of the wild‐type and SsSvf1 gene‐silenced strains on potato dextrose agar (PDA) containing 0 mM to 10 mM H2O2 were compared. The results showed that these two gene‐silenced strains displayed wild‐type levels of susceptibility to 2.5 mM H2O2, while being more sensitive at higher H2O2 concentrations (Fig. 2B). SiSvf1‐230 and SiSvf1‐213 were also more sensitive to menadione, a chemical inducer of oxidative stress, than the wild‐type strain (Fig. 3). The results indicated that SsSvf1 was required for managing oxidative stress in S. sclerotiorum.
Figure 3.
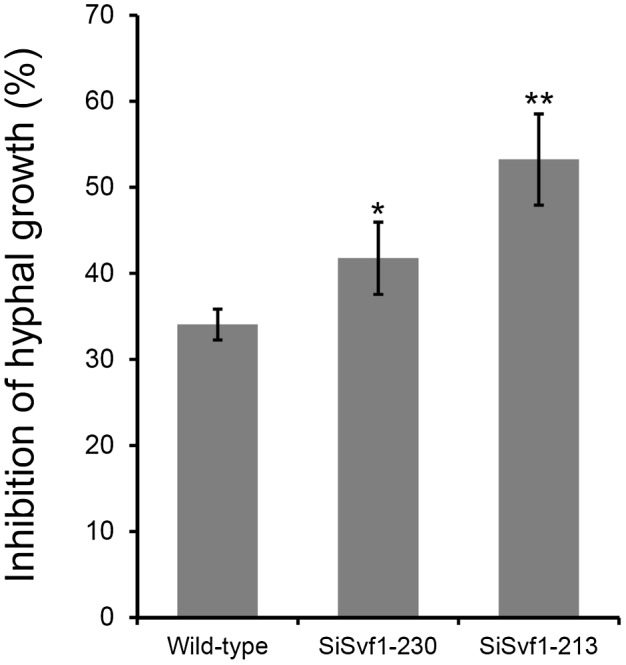
The inhibition of menadione to the hyphal growth of SsSvf1 gene‐silenced strains and wild‐type strain. Bars indicate standard deviation. Asterisks denote significant differences (one‐way analysis of variance [ANOVA]): *P < 0.05; **P < 0.01.
SsSvf1 gene‐silenced strains show overproduction of ROS
Svf1 inhibits ROS generation in S. cerevisiae (Brace et al., 2005). To understand whether silencing of SsSvf1 would lead to altered ROS generation, we detected ROS production during hyphal growth in SsSvf1 gene‐silenced strains. The strains were stained with nitroblue tetrazolium (NBT), which specifically detects superoxide. More dark‐blue formazan precipitates were seen in the hyphae of SiSvf1‐230 and SiSvf1‐213, which indicated that SsSvf1 gene‐silenced strains produce more superoxide than the wild‐type strain (Fig. 4A). The ROS production in the hyphae tips was then quantified via mean pixel intensity. The results showed that SiSvf1‐230 and SiSvf1‐213 exhibited greatly increased superoxide production (Fig. 4B).
Figure 4.
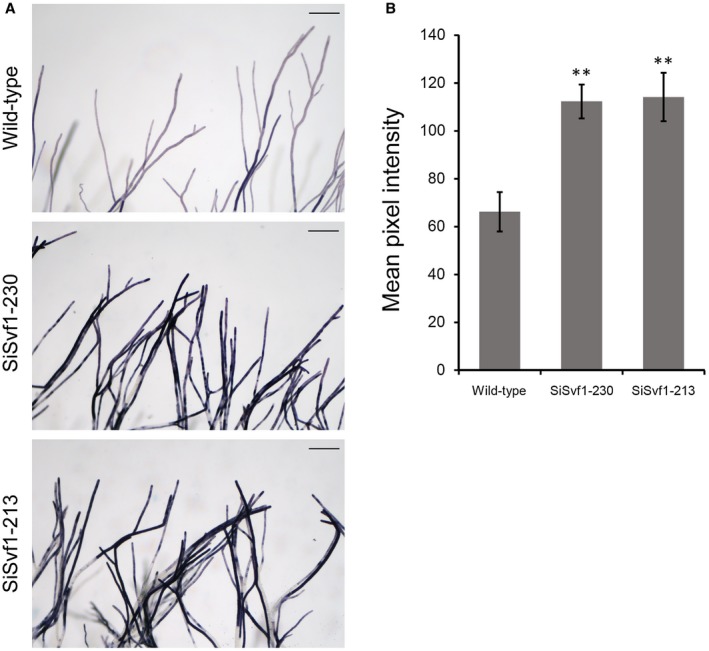
Reactive oxygen species (ROS) accumulation in SsSvf1 gene‐silenced strains. (A) Detection of ROS production in the hyphae of wild‐type and SsSvf1 gene‐silenced strains. The strains were inoculated on potato dextrose agar (PDA) plates and stained with 0.5 mg/mL nitroblue tetrazolium (NBT) at 2 dpi. Photographs were taken with light microscopy. Blue staining indicates the accumulation of superoxide. Bars = 200 μM. (B) Mean pixel intensity in the hyphal tips stained with NBT to detect superoxide. Bars indicate standard deviation. Asterisks denote significant differences (one‐way analysis of variance [ANOVA]): **P < 0.01.
SsSvf1 gene‐silenced strains show reduced virulence and a lower efficiency of appressoria formation
The expression level of SsSvf1 during wild‐type infection was evaluated, and the results indicated that the expression of SsSvf1 showed a strong increase during the initial stage of infection (Fig. 5A). To carry out functional analysis of the role of SsSvf1 in the pathogenicity of S. sclerotiorum, SsSvf1 gene‐silenced strains were inoculated on detached oilseed rape leaves and on Arabidopsis thaliana plants. As shown in Fig. 5B, both gene‐silenced strains caused small lesions on the oilseed rape leaves. The smaller lesions were also observed on A. thaliana inoculated with the SsSvf1 gene‐silenced strains (Fig. 5B). The results indicate that SsSvf1 is important for the pathogenicity of S. sclerotiorum.
Figure 5.
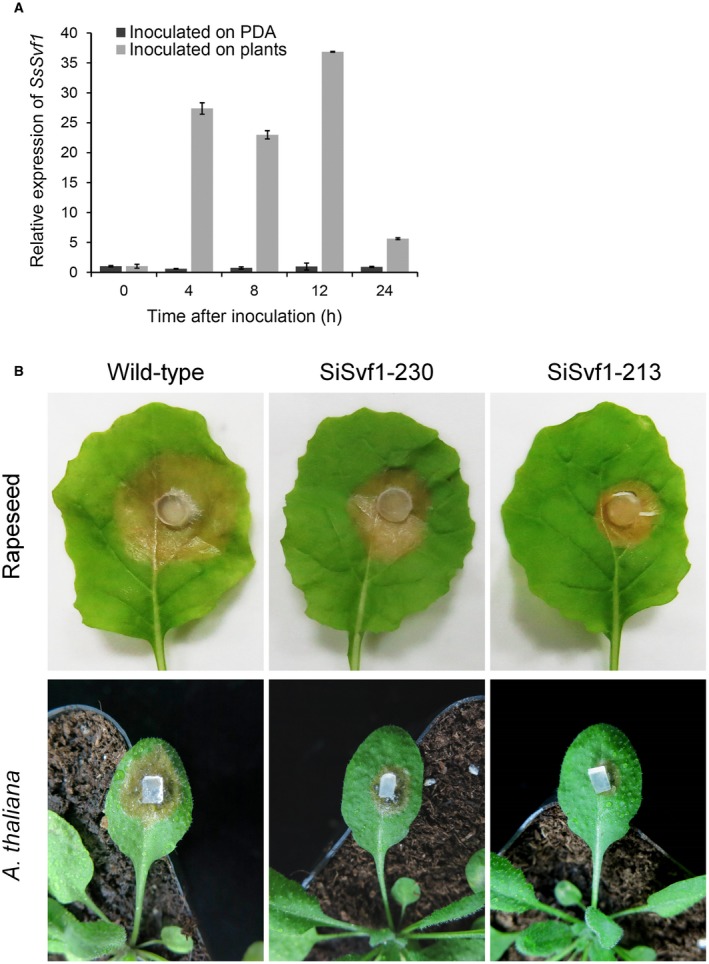
Functional characterization of SsSvf1 in pathogenicity of S. sclerotiorum. (A) Relative expression of SsSvf1 in wild‐type hyphae after contact with Arabidopsis thaliana and growing on potato dextrose agar (PDA) plates. The Tub1 gene in each sample was used as an internal control. The relative expression of SsSvf1 in hyphae stage or in hyphae inoculated on plants at 0 h was set as one. Bars indicate standard deviation. (B) Pathogenicity of SsSvf1 gene‐silenced strains on detached leaves of rapeseed and on A. thaliana plants. Each strain was investigated with five rapeseed leaves or A. thaliana plants each time. One representative replicate from three experiments is shown.
The OA and compound appressoria were key factors in the pathogenesis of S. sclerotiorum infection; thus, the OA accumulation and compound appressorium formation were compared between the wild‐type and SsSvf1 gene‐silenced strains. Our results showed that SsSvf1 gene‐silenced strains secreted similar levels of OA as the wild‐type strain (Fig. S2). To assay appressoria formation, both the wild‐type and SsSvf1 gene‐silenced strains were inoculated on parafilm‐overlaid growth medium and on rapeseed leaves. The results showed that the wild‐type strain formed complex and frequent appressoria, but SiSvf1‐213 rarely produced appressoria on parafilm or rapeseed leaves (Fig. 6A). This indicated that SsSvf1 is associated with compound appressoria formation in S. sclerotiorum. To confirm this, the rapeseed leaves were wounded with a dissecting needle and then inoculated with the wild‐type strain and SiSvf1‐213. The results showed that SiSvf1‐213 caused larger lesions by 48 h post‐inoculation on wounded rapeseed leaves than on intact leaves (Fig. 6B). SiSvf1‐213 was also inoculated on wounded leaves of A. thaliana and similar phenotypes were observed (Fig. 6C).
Figure 6.
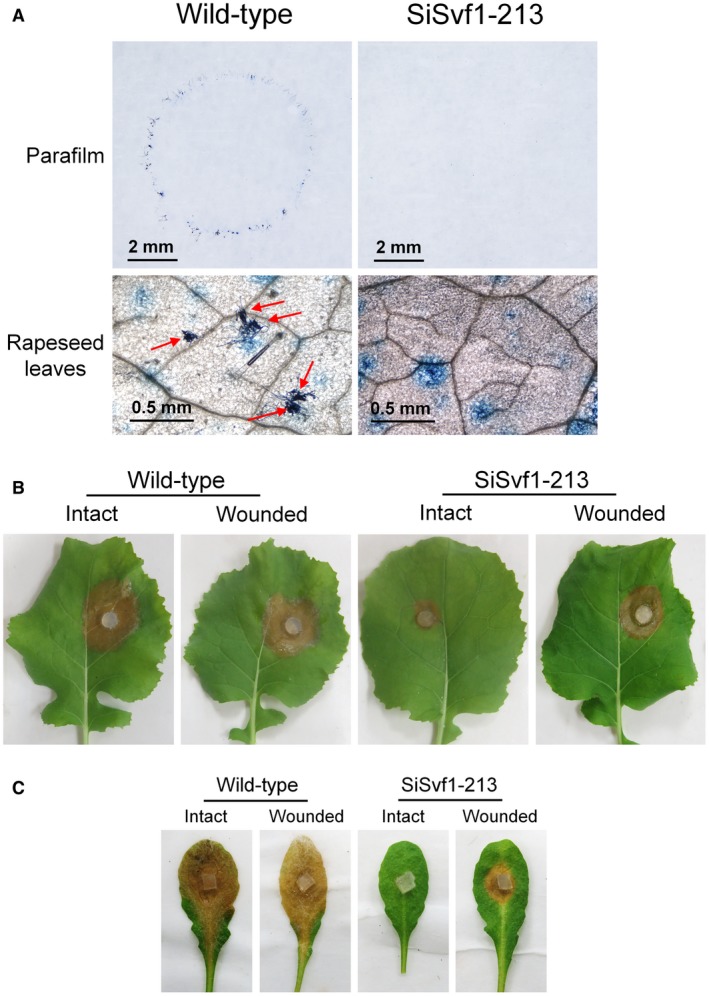
Appressoria formation of SsSvf1 gene‐silenced strains. (A) Compound appressoria formation of wild‐type strain and SiSvf1‐213 on parafilm and on rapeseed leaves inoculated with mycelial plugs. The photographs were taken at 8 hpi. Arrowheads indicate appressoria formation. (B) Pathogenicity of wild‐type strain and SiSvf1‐213 on wounded leaves of rapeseed. (C) Pathogenicity assay on wounded leaves of A. thaliana. Photographs were taken at 48 hpi. One representative replicate from the three experiments is shown.
Inhibition of plant ROS production partially restores virulence of SsSvf1 gene‐silenced strain
SsSvf1 gene‐silenced strains showed increased sensitivity to ROS, which is an early plant response to pathogen infection. Thus, the virulence of SsSvf1 gene‐silenced strain was next tested on oilseed rape leaves with reduced generation of ROS. The rapeseed leaves were sprayed with diphenyleneiodonium (DPI) to inhibit activity of the plant NADPH oxidases, and the leaves were then inoculated with the wild‐type strain and SsSvf1‐213, respectively. As shown in Fig. 7, larger lesions were produced by SiSvf1‐213 in leaves treated with DPI than in leaves treated with water.
Figure 7.
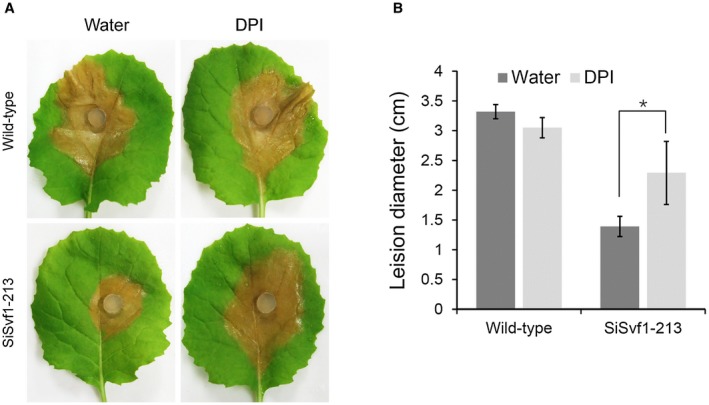
Pathogenicity of SsSvf1 gene‐silenced strains restored by diphenyleneiodonium (DPI) treatment. (A) Pathogenicity assay. The rapeseed leaves were inoculated with mycelium plugs of wild‐type strain and SiSvf1‐213 after treatment with or without DPI (5 μM) over three independent experiments. Photographs were taken at 48 hpi and the figure shows representative photographs. (B) The size of the expending lesions. Bars indicate standard deviation. Asterisks denote significant differences (one‐way analysis of variance [ANOVA]): *P < 0.05.
SsSvf1 gene‐silenced strains are impaired in cell wall integrity of hyphae
The cell wall integrity was compared between the wild‐type and SsSvf1 gene‐silenced strains. The results showed that SiSvf1‐230 and SiSvf1‐213 were more sensitive to SDS (sodium dodecyl sulphate) than the wild‐type strain, suggesting a weakened cell wall for SsSvf1 gene‐silenced strains (Fig. 8). To evaluate cell wall alteration, the effects of specific cell wall perturbation agents to the SsSvf1 gene‐silenced strains were determined. The results showed that SiSvf1‐230 and SiSvf1‐213 displayed wild‐type levels of susceptibility to calcofluor white (CFW) while being more sensitive to Congo red (CR) (Fig. 8). This suggests that SsSvf1 gene‐silenced strains were impaired in some aspect of hyphal cell wall integrity. Since the cell wall integrity pathway and hyperosmotic stress response share common functional aspects and are positively coordinated (Alonso‐Monge et al., 2001; Rodríguez‐Peña et al, 2010), the hyphal growth of SsSvf1 gene‐silenced strains under hyperosmotic stress were determined. The inhibition of hyphal growth was significantly greater for SiSvf1‐230 and SiSvf1‐213 than the wild‐type strain when growing on medium with sodium chloride or sorbitol (Fig. 8).
Figure 8.
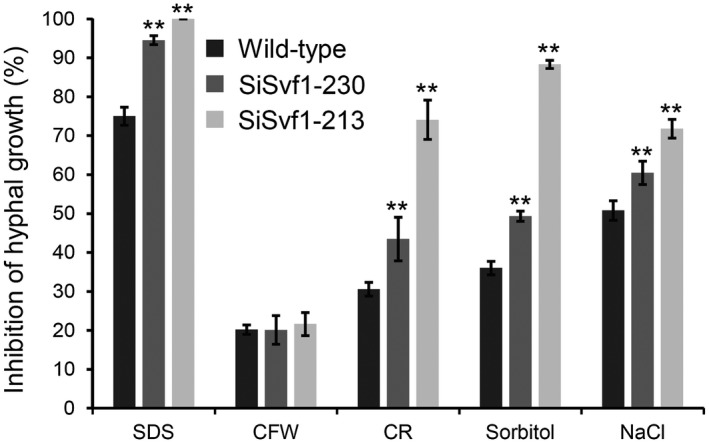
Sensitivity of SsSvf1 gene‐silenced strains to cell wall perturbation agents and hyperosmotic stress. The strains were inoculated on potato dextrose agar (PDA) plates amending 0.02% sodium dodecyl sulphate (SDS), 200 μM calcofluor white (CFW), 0.4 g/L Congo red (CR), 1 M sorbitol, and 0.4 M NaCl. Percentage inhibition of hyphal growth was calculated at 36 hpi. Bars indicate standard deviation. Asterisks denote significant differences (one‐way analysis of variance [ANOVA]): **P < 0.01.
Discussion
Fungi use specific enzymatic and non‐enzymatic processes to regulate antioxidant response (Aguirre et al., 2006). To date, only a few proteins have been reported related to the anti‐oxidative stress responses in S. sclerotiorum. SsSOD1 encodes a Cu/Zn superoxide dismutase (SOD) in S. sclerotiorum, and the gene‐deletion mutant exhibited increased sensitivity to oxidative stress (Xu and Chen, 2013). Our previous report also showed that a putative BAX inhibitor‐1 in S. sclerotiorum was involved in withstanding H2O2 (Yu et al., 2015). In this study, SsSvf1 is predicted to encode a S. cerevisiae Svf1 homologue in S. sclerotiorum. SsSvf1 shows increased expression under oxidative stress and the gene‐silenced strains are more sensitive to H2O2 and menadione. To date, Svf1 was only identified and characterized in S. cerevisiae; it was required for survival in response to oxidative stress (Brace et al., 2005). Our data demonstrates the function of SVF1 homologues in filamentous fungi for the first time and suggests an important role of Svf1 in response to oxidative stress in S. sclerotiorum.
Evidence for the molecular mechanism underlying Svf1 function in oxidative stress response is still sparse. S. cerevisiae SVF1 gene expression in mammalian cells increases apoptotic resistance (Brace et al., 2005). Deep research showed that yeast Svf1 regulates cell survival by affecting sphingolipid metabolism (Brace et al., 2007). Sphingolipids are important regulators of apoptosis in plant and animal cells and some sphingolipid metabolites may promote cell survival following induction of death by various treatments (Alden et al., 2011; Maceyka et al., 2002). In this research, the relationship between SsSvf1 and sphingosine metabolism in S. sclerotiorum is unclear. The sequences of SsSvf1 were used in a search for potential structural homologues via the HHpred server (Söding et al., 2005; Zimmermann et al., 2018). The top‐ranked hit is a putative lipocalin in Nitrosomonas europaea with an E‐value of 4.94e‐24 and a probability score of 99.91. A search with WoLF PSORT server showed that SsSvf1 is most likely located in the cytoplasm, suggesting a role for SsSvf1 in lipid metabolism in the cytoplasm. It is interesting to note that lipocalin has been reported to be a protective factor against H2O2 toxicity (Roudkenar et al., 2008).
Svf1 inhibited ROS generation in S. cerevisiae, and the cells lacking SVF1 have increased levels of ROS (Brace et al., 2005). Our results showed that silencing of SsSvf1 led to overproduction of ROS in the hyphae suggesting that SsSvf1 also regulates ROS generation of S. sclerotiorum. To date, only a few genes have been demonstrated to play important roles in regulating the internal redox environment in S. sclerotiorum. NADPH oxidases (Nox) are a primary source for endogenous generators of ROS (Nauseef, 2008; Takemoto et al., 2007). Silencing of the S. sclerotiorum Nox‐encoded gene Ssnox1 expression can impair superoxide production (Kim et al., 2011). The γ‐glutamyl transpeptidase (Ggt) catalyzes the first step in glutathione (GSH) metabolism and recycling; it is an important factor in maintaining cellular redox homeostasis (Lee and Bostock, 2007). Deletion of the Ggt‐encoded gene Ss‐Ggt1 in S. sclerotiorum resulted in H2O2 being hyper‐accumulated in sclerotia (Li et al., 2012). However, more evidence is required to elucidate the mechanism whereby SsSvf1 is involved in regulating ROS generation in S. sclerotiorum, because SsSvf1 exhibits no similarity with any known enzymes.
S. sclerotiorum secretes OA to elicit a host programmed cell death (PCD) response, which provides nutrients that are beneficial for this necrotrophic fungal pathogen (Kim et al., 2008). The PCD response requires ROS generation, the production of which may be stimulated by necrotrophs (Marino et al., 2012). Enhanced ROS generation stimulates the necrosis induced by S. sclerotiorum (Govrin and Levine, 2000). Thus, ROS modulation is critical for successful infection by this fungus. In this research, the virulence of SsSvf1 gene‐silenced strains were impaired on different hosts, while showing partial recovery on hosts in which the ROS generation was inhibited. Although DPI was also used to inhibit NADPH oxidases in B. cinerea and S. sclerotiorum, direct evidence that it can inhibit ROS production in these two fungi cannot be found (Kim et al., 2011; Segmüller et al., 2008). Our results suggest that SsSvf1 plays an important role in anti‐oxidation during S. sclerotiorum infection. Recent evidence indicates that S. sclerotiorum has a short biotrophic phase during the early stages of infection, and it uses OA to suppress the oxidative burst during initial stages (Kabbage et al., 2015; Williams et al., 2011). The inhibition of ROS generation by DPI can benefit the fungus during compatible interactions, although more evidence is needed.
As multi‐cellular infectious structures, compound appressoria play important roles in S. sclerotiorum pathogenesis (Huang et al., 2008; Liang et al., 2015a, b; Xiao et al., 2014). Compound appressoria might help the fungus adhere to and penetrate the host cuticle and also secrete enzymes and toxins (Huang et al., 2008; Jamaux et al., 1995). In S. sclerotiorum, appressorium development might depend on the cAMP‐PKA signalling pathway and be associated with OA accumulation (Jurick and Rollins 2007; Liang et al., 2015a, b). In this research, SsSvf1 gene‐silenced strains showed impaired activity in appressorial formation, and their virulence was partially restored on wounded leaves of rapeseed, indicated that SsSvf1 was important for the appressoria formation. Svf1 function was only reported in S. cerevisiae, a non‐pathogenic and non‐appressoria forming fungus that did not form appressorium. Thus, the mechanism of SsSvf1 involved in appressorial formation is still unknown. A closer investigation showed that SsSvf1 gene‐silenced strains were more sensitive to some cell wall damaging agents suggesting a possible defect in the cell wall components of the hyphae for these strains. The combined evidence suggests that the reduced efficiency in appressorial formation is due to the impaired integrity of the hyphae cell wall. Defects in the cell wall components influencing appressorial formation have been reported for many important pathogenic fungi, including Magnaporthe grisea (Jeon et al., 2008; Skamnioti et al., 2007; Xu, 2000) and Colletotrichum graminicola (Albarouki and Deising 2013).
The molecular mechanism of SsSvf1 in maintaining the cell wall integrity of hyphae in S. sclerotiorum remains unclear. S. cerevisiae Svf1 affected the localized generation of a pool of phytosphingosine, which might activate the Ypk1p pathway that play a role in the cell wall integrity pathway (Brace et al., 2007; Liu et al., 2005; Schmelzle et al., 2002). Our results showed that the SsSvf1 gene‐silenced strains were highly sensitive to CR but exhibited a similar level of sensitivity to CFW versus the wild‐type strain. The CFW inhibits chitin polymer assembly in the cell wall, but CR affects glucan polysaccharide assembly (Daher et al., 2011; Puttikamonkul et al., 2010; Ram and Klis, 2006). The results suggest that the chitin content was likely unaffected in SsSvf1 gene‐silenced strains. Glucan is an important component of the cell wall and is critical for maintaining cell integrity (Ram et al., 1998). However, additional studies on the alteration of glucan in SsSvf1 gene‐silenced strains are needed.
In summary, our results demonstrate that SsSvf1 encodes a S. cerevisiae survival factor 1 homologue in S. sclerotiorum. The gene was characterized and found to play a crucial role in response to oxidative stress, cell wall integrity and pathogenicity of S. sclerotiorum. This finding helps to explain the pathogenicity of S. sclerotiorum and molecular mechanism involved in the antioxidant response in plant fungal pathogens.
Experimental Procedures
Fungal strains and cultured conditions
S. sclerotiorum strain 1980 was used as the wild‐type strain and was routinely cultured on PDA medium. Transformant strains were cultured on PDA with 100 μg/mL hygromycin B (Calbiochem, San Diego, CA).
RNA isolation and cDNA synthesis
Total RNA was extracted from frozen mycelium of the wild‐type strain or transformants, or inoculated A. thaliana leaf sample with Rrizol reagent (Huashun Biogengineering Co, Shanghai, China) according to the manufacturer's instructions. The total RNA was then treated with DNase 1 (RNase free) (Takara, Dalian, China) to remove DNA contaminants. Approximately 1 μg of treated RNA was used to synthesize the cDNA with the ReventAidTM First Strand cDNA Synthesis Kit (MBI Fermentas, Flamborough, ON, Canada) following the manufacturer's instructions.
Real‐time RT‐PCR
To determine the expression levels of SsSvf1, real‐time RT‐PCR using SYBR Green I technology on a CFX96TM Realtime System (BioRad, Hercules, CA, USA) was performed. The housekeeping gene Tub1 encoding β‐tubulin was used as an internal control and amplified with Rt‐tubfp/Rt‐tubrp primers. The primer pair Rt‐Svf1fp/Rt‐Svf1rp was designed according to the cDNA sequence of SsSvf1. Real‐time RT‐PCR was conducted in a 20 μL reaction mixture containing 10 μL of SYBR Green Realtime PCR Master Mix (Toyobo, Osaka, Japan), 4 pmoL concentration of each primer, 1 μL of cDNA, and nuclease‐free water. The following amplification programme was applied: 95 °C for 2 min (1 cycle), followed by 95 °C for 20 s, 57 °C for 15 s and 72 °C for 20 s (40 cycles). Each sample was analysed in three biological replications, and the average cycle threshold was calculated to evaluate the relative expression. The primers used here are shown in Table 1.
Table 1.
Sequence of primers used in the research.
| Primer | Primers sequence (5′‐3′) |
|---|---|
| Rt‐tubfp | GTGAGGCTGAGGGCTGTGA |
| Rt‐tubrp | CCTTTGGCGATGGGACG |
| Rt‐Svf1fp | TGTCGTAGGAACTGAGGAGCC |
| Rt‐Svf1rp | GTACTCTCTTGAGCCTTCCATTG |
| SiSvf1Xho I | CCGCTCGAGTCTGATGCGCTCGAAAACTATG |
| SiSvf1Hind III | CGCAAGCTTATGGCGTGAATAAAAATACCGG |
| SiSvf1Kpn I | CGGGGTACCTCTGATGCGCTCGAAAACTATG |
| SiSvf1Bgl II | GGAAGATCTATGGCGTGAATAAAAATACCGG |
SsSvf1 gene‐silenced vector construction and transformant
The SsSvf1 gene‐silencing vector was constructed based on plasmid pSilent‐1 (Nakayashiki et al., 2005). To amplify the sense and antisense fragments of SsSvf1, the primers SiSvf1Xho I/SiSvf1Hind III (sense fragment) and SiSvf1Kpn I/SiSvf1Bgl II (antisense fragment) were used. The amplified sense and antisense fragments were inserted into the corresponding multiple cloning sites of pSilent‐1 to generate the RNA silencing vector pSiSvf1. The vector was then linearized with Spe I and used to transform the wild‐type strain of S. sclerotiorum according to the method of Rollins (2003).
ROS detection assay
The ROS detection assay was performed according to Kim et al. (2011). The strains were inoculated on PDA plates for 2 days and the plates were then immersed in 0.5 mg/mL NBT (10 mM potassium phosphate buffer, pH 7.5) aqueous solution for 2 h with gentle shaking. The hyphae were observed with light microscopy. ROS accumulation in hyphae tips was quantified via mean pixel intensity with ImageJ software according to Egan et al. (2007).
Pathogenicity assay
A. thaliana Columbia‐0 and Brassica napus Zhongyou 821 were used for the pathogenicity assay according to Yu et al. (2017). To analyse the pathogenicity of the strains on host plants in which ROS production was inhibited, leaves of B. napus were sprayed with 5 μM DPI dissolved in water and then were inoculated with mycelia‐colonized agar (6 mM in diameter) obtained from the growing colony margins of the strain. The lesions were measured at 48 hpi. In each experiment, each strain was inoculated on five plants or leaves and the experiment was repeated three times.
OA concentration assays
The OA concentration assay was performed according to Yu et al. (2017) with minor modifications. The wild‐type strain and SiSvf1‐213 were cultured on PDA medium for 2 days. Subsequently, four mycelium plugs (6 mM in diameter) of each strain were cultured in 50 mL of potato dextrose broth (PDB) for 3 days with shaking at 150 rpm. OA accumulation was determined by high‐performance liquid chromatography according to Zhang et al. (2010). The concentration was expressed as milligrams of OA per gram of dry mycelia. Each strain was repeated three times.
Compound appressorium assay
To assay the compound appressorium formation of the wild‐type and SsSvf1 gene‐silenced strains, 2‐day‐old culture plugs (6 mM in diameter) were inoculated onto parafilm and rapeseed leaves. The plugs were removed at 8 hpi for those on parafilm. To observe the appressorium, the parafilm surface were stained with 5% trypan. The leaves were cleared with ethanol/acetic acid (3:1 v/v) solution for 12 h and then stained with 5% trypan blue for 12 h.
Cell wall integrity assay
The sensitivity of the hyphal growth to different cell wall inhibitors were tested to evaluate the cell wall integrity of the wild‐type and SsSvf1 gene‐silenced strains. Two‐day‐old culture plugs (6 mM in diameter) of each strain were inoculated onto PDA amended with 0.02% SDS, 400 μg/mL CR, or 200 μM CFW. The colony diameters were measured after incubation for 36 h to determine the inhibition of hyphal growth; unmodified plates served as the control.
Supporting information
Fig. S1 Expression level of SsSvf1 in different isolates containing pSiSvf1. Tub1 gene in each strain was the internal control. The relative expression of SsSvf1 in the wild‐type strain was set as one. Bars indicate standard deviation.
Fig. S2 Oxalic acid (OA) accumulation in wild‐type strain and SiSvf1‐213. Each strain was cultured in potato dextrose broth (PDB) for 3 days, and the resulting liquid culture was analysed for OA accumulation.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (No. 31671973), Chongqing Research Program of Basic Research and Frontier Technology (No. cstc2017jcyjAX0096), Fundamental Research Funds for the Central Universities, China (No. XDJK2019B034 and XDJK2017A006), Open Funds of the State Key Laboratory of Agricultural Microbiology (AMLKF201707). We thank anonymous reviewers for their kind suggestions.
References
- Aguirre, J. , Hansberg, W. and Navarro, R. (2006) Fungal responses to reactive oxygen species. Med. Mycol. 44(Supplement_1), S101–S107. [DOI] [PubMed] [Google Scholar]
- Albarouki, E. and Deising, H.B. (2013) Infection structure‐specific reductive iron assimilation is required for cell wall integrity and full virulence of the maize pathogen Colletotrichum graminicola . Mol. Plant‐Microbe Interact. 26, 695–708. [DOI] [PubMed] [Google Scholar]
- Alden, K.P. , Dhondt‐Cordelier, S. , McDonald, K.L. , Reape, T.J. , Ng, C.K.Y. , McCabe, P.F. and Leaver, C.J. (2011) Sphingolipid long chain base phosphates can regulate apoptotic‐like programmed cell death in plants. Biochem. Bioph. Res. Co. 410, 574–580. [DOI] [PubMed] [Google Scholar]
- Alonso‐Monge, R. , Real, E. , Wojda, I. , Bebelman, J.P. , Mager, W.H. and Siderius, M. (2001) Hyperosmotic stress response and regulation of cell wall integrity in Saccharomyces cerevisiae share common functional aspects. Mol. Microbiol. 41, 717–730. [DOI] [PubMed] [Google Scholar]
- Boland, G.J. and Hall, R. (1994) Index of plant hosts of Sclerotinia sclerotiorum . Can. J. Plant Pathol. 16, 93–108. [Google Scholar]
- Bolton, M.D. , Thomma, B.P.H.J. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Brace, J.L. , Lester, R.L. , Dickson, R.C. and Rudin, C.M. (2007) SVF1 regulates cell survival by affecting sphingolipid metabolism in Saccharomyces cerevisiae . Genetics, 175, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace, J.L. , VanderWeele, D.J. and Rudin, C.M. (2005) Svf1 inhibits reactive oxygen species generation and promotes survival under conditions of oxidative stress in Saccharomyces cerevisiae . Yeast, 22, 641–652. [DOI] [PubMed] [Google Scholar]
- Cessna, S.G. , Sears, V.E. , Dickman, M.B. and Low, P.S. (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell, 12, 2192–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher, J.Y. , Koussa, J. , Younes, S. and Khalaf, R.A. (2011) The Candida albicans Dse1 protein is essential and plays a role in cell wall rigidity, biofilm formation, and virulence. Interdiscip. Perspect. Infect. Dis. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, M.J. , Wang, Z.Y. , Jones, M.A. , Smirnoff, N. and Talbot, N.J. (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA, 104, 11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaron, F. , Sella, L. and D'Ovidio, R. (2004) Relationships among endo‐polygalacturonase, oxalate, pH, and plant polygalacturonase‐inhibiting protein (PGIP) in the interaction between Sclerotinia sclerotiorum and soybean. Mol. Plant‐Microbe Interact. 17, 1402–1409. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Guyon, K. , Balagué, C. , Roby, D. and Raffaole, S. (2014) Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum . BMC Genom. 15, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. , Buchenauer, H. , Han, Q. , Zhang, X. and Kang, Z. (2008) Ultrastructural and cytochemical studies on the infection process of Sclerotinia sclerotiorum in oilseed rape. J. Plant Dis. Protect. 115, 9–16. [Google Scholar]
- Jamaux, I. , Gelie, B. and Lamarque, C. (1995) Early stages of infection of rapeseed petals and leaves by Sclerotinia sclerotiorum revealed by scanning electron microscopy. Plant Pathol. 44, 22–30. [Google Scholar]
- Jeon, J. , Goh, J. , Yoo, S. , Chi, M.H. , Choi, J. , Rho, H.S. , Park, J. , Han, S.S. , Kim, B.R. , Park, S.Y. , Kim, S. and Lee, Y.H. (2008) A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae . Mol. Plant‐Microbe Interact. 21, 525–534. [DOI] [PubMed] [Google Scholar]
- Jurick, W.M. II and Rollins, J.A. (2007) Deletion of the adenylate cyclase (sac1) gene affects multiple developmental pathways and pathogenicity in Sclerotinia sclerotiorum . Fungal Genet. Biol. 44, 521–530. [DOI] [PubMed] [Google Scholar]
- Kabbage, M. , Yarden, O. and Dickman, M.B. (2015) Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Chen, C. , Kabbage, M. and Dickman, M.B. (2011) Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases. Appl. Environ. Microb. 77, 7721–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.S. , Min, J.Y. and Dickman, M.B. (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant‐Microbe Interact. 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Lamb, C. and Dixon, R.A. (1997) The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Lee, M.H. and Bostock, R.M. (2007) Fruit exocarp phenols in relation to quiescence and development of Monilinia fructicola infections in Prunus: A role for cellular redox? Phytopathology, 97, 269–277. [DOI] [PubMed] [Google Scholar]
- Li, M. , Liang, X. and Rollins, J.A. (2012) Sclerotinia sclerotiorum γ‐glutamyl transpeptidase (Ss‐Ggt1) is required for regulating glutathione accumulation and development of sclerotia and compound appressoria. Mol. Plant‐Microbe Interact. 25, 412–420. [DOI] [PubMed] [Google Scholar]
- Liang, X. , Liberti, D. , Li, M. , Kim, Y.T. , Hutchens, A. , Wilson, R. and Rollins, J.A. (2015a) Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Moomaw, E.W. and Rollins, J.A. (2015b) Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 16, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K. , Zhang, X. , Lester, R.L. and Dickson, R.C. (2005) The sphingoid long chain base phytosphingosine activates AGC‐type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 280, 22679–22687. [DOI] [PubMed] [Google Scholar]
- Lyu, X. , Shen, C. , Fu, Y. , Xie, J. , Jiang, D. , Li, G. and Cheng, J. (2016) A small secreted virulence‐related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 12, e1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka, M. , Payne, S.G. , Milstien, S. and Spiegel, S. (2002) Sphingosine kinase, sphingosine‐1‐phosphate, and apoptosis. Biochim. Biophys. Acta. 1585, 193–201. [DOI] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Bo, Y. , Han, L. , He, J. , Lanczycki, C.J. , Lu, S. , Chitsaz, F. , Derbyshire, M.K. , Geer, R.C. , Gonzales, N.R. , Gwadz, M. , Hurwitz, D.I. , Lu, F. , Marchler, G.H. , Song, J.S. , Thanki, N. , Wang, Z. , Yamashita, R.A. , Zhang, D. , Zheng, C. , Geer, L.Y. and Gwadz, M. (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, D. , Dunand, C. , Puppo, A. and Pauly, N. (2012) A burst of plant NADPH oxidases. Trends Plant Sci. 17, 9–15. [DOI] [PubMed] [Google Scholar]
- Martel, M.B. , Létoublon, R. and Fěvre, M. (1996) Purification of endo polygalacturonases from Sclerotinia sclerotiorum: multiplicity of the complex enzyme system. Curr. Microbiol. 33, 243–248. [DOI] [PubMed] [Google Scholar]
- Mousavi, S.A. and Robson, G.D. (2004) Oxidative and amphotericin B‐mediated cell death in the opportunistic pathogen Aspergillus fumigatus is associated with an apoptotic‐like phenotype. Microbiology, 150, 1937–1945. [DOI] [PubMed] [Google Scholar]
- Nakayashiki, H. , Hanada, S. , Quoc, N.B. , Kadotani, N. , Tosa, Y. and Mayama, S. (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42, 275–283. [DOI] [PubMed] [Google Scholar]
- Nauseef, W.M. (2008) Biological roles for the NOX family NADPH oxidases. J. Biol. Chem. 283, 16961–16965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, M. and Kuc, J. (1992) Peroxidase‐generated hydrogen peroxide as a source of antifungal activity in vitro and on tobacco leaf disks. Phytopathology, 82, 696–699. [Google Scholar]
- Poussereau, N. , Gente, S. , Rascle, C. , Billon‐Grand, G. and Fèvre, M. (2001) aspS encoding an unusual aspartyl protease from Sclerotinia sclerotiorum is expressed during phytopathogenesis. FEMS Microbiol. Lett. 194, 27–32. [DOI] [PubMed] [Google Scholar]
- Purdy, L.H. (1979) Sclerotinia sclerotiorum: history, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology, 69, 875–880. [Google Scholar]
- Puttikamonkul, S. , Willger, S.D. , Grahl, N. , Perfect, J.R. , Movahed, N. , Bothner, B. , Park, S. , Paderu, P. , Perlin, D.S. and Cramer, R.A. Jr (2010) Trehalose 6‐phosphate phosphatase is required for cell wall integrity and fungal virulence but not trehalose biosynthesis in the human fungal pathogen Aspergillus fumigatus . Mol. Microbiol. 77, 891–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram, A.F. and Klis, F.M. (2006) Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 1, 2253–2256. [DOI] [PubMed] [Google Scholar]
- Ram, A.F. , Kapteyn, J.C. , Montijn, R.C. , Caro, L.H.P. , Douwes, J.E. , Baginsky, W. , Mazur, P. , van den Ende, H. and Klis, F.M. (1998) Loss of the plasma membrane‐bound protein Gas1p in Saccharomyces cerevisiae results in the release of β 1, 3‐Glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 180, 1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou, C. , Freyssinet, G. and Fevre, M. (1991) Production of cell wall‐degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum . Appl. Environ. Microb. 57, 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Peña, J.M. , García, R. , Nombela, C. and Arroyo, J. (2010) The high‐osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast, 27, 495–502. [DOI] [PubMed] [Google Scholar]
- Rollins, J.A. (2003) The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant‐Microbe Interact. 16, 785–795. [DOI] [PubMed] [Google Scholar]
- Roudkenar, M.H. , Halabian, R. , Ghasemipour, Z. , Roushandeh, A.M. , Rouhbakhsh, M. , Nekogoftar, M. , Kuwahara, Y. , Fukumoto, M. and Shokrgozar, M.A. (2008) Neutrophil gelatinase‐associated lipocalin acts as a protective factor against H2O2 toxicity. Arch. Med. Res. 39, 560–566. [DOI] [PubMed] [Google Scholar]
- Schmelzle, T. , Helliwell, S.B. and Hall, M.N. (2002) Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell Biol. 22, 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segmüller, N. , Kokkelink, L. , Giesbert, S. , Odinius, D. , van Kan, J. and Tudzynski, P. (2008) NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea . Mol. Plant‐Microbe Interact. 21, 808–819. [DOI] [PubMed] [Google Scholar]
- Skamnioti, P. , Henderson, C. , Zhang, Z. , Robinson, Z. and Gurr, S.J. (2007) A novel role for catalase B in the maintenance of fungal cell‐wall integrity during host invasion in the rice blast fungus Magnaporthe grisea . Mol. Plant‐Microbe Interact. 20, 568–580. [DOI] [PubMed] [Google Scholar]
- Söding, J. , Biegert, A. and Lupas, A.N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, D. , Tanaka, A. and Scott, B. (2007) NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet. Biol. 44, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Vander Heiden, M.G. , Choy, J.S. , VanderWeele, D.J. , Brace, J.L. , Harris, M.H. , Bauer, D.E. , Prange, B. , Kron, S.J. , Thompson, C.B. and Rudin, C.M. (2002) Bcl‐x L complements Saccharomyces cerevisiae genes that facilitate the switch from glycolytic to oxidative metabolism. J. Biol. Chem. 277, 44870–44876. [DOI] [PubMed] [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.J. , Britt, R. and Dickman, M.B. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X. , Xie, J. , Cheng, J. , Li, G. , Yi, X. , Jiang, D. and Fu, Y. (2014) Novel secretory protein Ss‐Caf1 of the plant‐pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant‐Microbe Interact. 27, 40–55. [DOI] [PubMed] [Google Scholar]
- Xu, J.R. (2000) Map kinases in fungal pathogens. Fungal Genet. Biol. 31, 137–152. [DOI] [PubMed] [Google Scholar]
- Xu, L. and Chen, W. (2013) Random T‐DNA mutagenesis identifies a Cu/Zn superoxide dismutase gene as a virulence factor of Sclerotinia sclerotiorum . Mol. Plant‐Microbe Interact. 26, 431–441. [DOI] [PubMed] [Google Scholar]
- Yajima, W. , Liang, Y. and Kav, N.N. (2009) Gene disruption of an arabinofuranosidase/β‐xylosidase precursor decreases Sclerotinia sclerotiorum virulence on canola tissue. Mol. Plant‐Microbe Interact. 22, 783–789. [DOI] [PubMed] [Google Scholar]
- Yang, G. , Tang, L. , Gong, Y. , Xie, J. , Fu, Y. , Jiang, D. , Li, G. , Collinge, D.B. , Chen, W. and Cheng, J. (2018) A cerato‐platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum . New Phytol. 217, 739–755. [DOI] [PubMed] [Google Scholar]
- Yarden, O. , Veluchamy, S. , Dickman, M.B. and Kabbage, M. (2014) Sclerotinia sclerotiorum catalase SCAT1 affects oxidative stress tolerance, regulates ergosterol levels and controls pathogenic development. Physiol. Mol. Plant Pathol. 85, 34–41. [Google Scholar]
- Yu, Y. , Xiao, J. , Du, J. , Yang, Y. , Bi, C. and Qing, L. (2016) Disruption of the gene encoding endo‐β‐1, 4‐xylanase affects the growth and virulence of Sclerotinia sclerotiorum . Front. Microbiol. 7, 1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Xiao, J. , Yang, Y. , Bi, C. , Qing, L. and Tan, W. (2015) Ss‐Bi1 encodes a putative BAX inhibitor‐1 protein that is required for full virulence of Sclerotinia sclerotiorum . Physiol. Mol. Plant Pathol. 90, 115–122. [Google Scholar]
- Yu, Y. , Xiao, J. , Zhu, W. , Yang, Y. , Mei, J. , Bi, C. , Qian, W. , Qing, L. and Tan, W. (2017) Ss-Rhs1, a secretory Rhs repeat-containing protein, is required for the virulence of Sclerotinia sclerotiorum. Mol. Plant Pathol. 18, 1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Wu, M. , Li, G. , Jiang, D. and Huang, H. (2010) Effect of mitovirus infection on formation of infection cushions and virulence of Botrytis cinerea. Physiol. Mol. Plant Pathol. 75, 71–80. [Google Scholar]
- Zhu, W. , Wei, W. , Fu, Y. , Chen, J. , Xie, J. , Li, G. , Yi, X. , Kang, Z. , Dickman, M.B. and Jiang, D. (2013) A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS One, 8, e53901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, L. , Stephens, A. , Nam, S.Z. , Rau, D. , Kübler, J. , Lozajic, M. , Gabler, F. , Söding, J. , Lupas, A.N. and Alva, V. (2018) A completely Reimplemented MPI bioinformatics toolkit with a new HHpred server at its Core. J. Mol. Biol. 430, 2237–2243. [DOI] [PubMed] [Google Scholar]
- Zuppini, A. , Navazio, L. , Sella, L. , Castiglioni, C. , Favaron, F. and Mariani, P. (2005) An endopolygalacturonase from Sclerotinia sclerotiorum induces calcium‐mediated signaling and programmed cell death in soybean cells. Mol. Plant‐Microbe. Interact. 18, 849–855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Expression level of SsSvf1 in different isolates containing pSiSvf1. Tub1 gene in each strain was the internal control. The relative expression of SsSvf1 in the wild‐type strain was set as one. Bars indicate standard deviation.
Fig. S2 Oxalic acid (OA) accumulation in wild‐type strain and SiSvf1‐213. Each strain was cultured in potato dextrose broth (PDB) for 3 days, and the resulting liquid culture was analysed for OA accumulation.


