Significance
The deuterium/protium (2H/1H) ratio of microbial lipids varies substantially and appears to be correlated with the mode of metabolism in the host organism, with lipids from chemoautotrophs and photoautotrophs 2H-depleted and heterotrophs in many cases 2H-enriched. Such patterns suggest that the H-isotope ratios of lipids could be used to infer the metabolism of environmental organisms, with applications ranging from Earth history to global carbon cycling and ecology. Here, we learn to understand this information by using metabolic flux analysis. We show that the full range of lipid 2H/1H ratios from a diverse set of aerobic heterotrophs can be quantitatively explained by fluxes through various NADP+-reducing reactions due to differences in their kinetic isotope effects.
Keywords: hydrogen isotopes, fatty acids, fractionation, metabolic flux analysis, NADPH
Abstract
The hydrogen-isotopic compositions (2H/1H ratios) of lipids in microbial heterotrophs are known to vary enormously, by at least 40% (400‰) relative. This is particularly surprising, given that most C-bound H in their lipids appear to derive from the growth medium water, rather than from organic substrates, implying that the isotopic fractionation between lipids and water is itself highly variable. Changes in the lipid/water fractionation are also strongly correlated with the type of energy metabolism operating in the host. Because lipids are well preserved in the geologic record, there is thus significant potential for using lipid 2H/1H ratios to decipher the metabolism of uncultured microorganisms in both modern and ancient ecosystems. But despite over a decade of research, the precise mechanisms underlying this isotopic variability remain unclear. Differences in the kinetic isotope effects (KIEs) accompanying NADP+ reduction by dehydrogenases and transhydrogenases have been hypothesized as a plausible mechanism. However, this relationship has been difficult to prove because multiple oxidoreductases affect the NADPH pool simultaneously. Here, we cultured five diverse aerobic heterotrophs, plus five Escherichia coli mutants, and used metabolic flux analysis to show that 2H/1H fractionations are highly correlated with fluxes through NADP+-reducing and NADPH-balancing reactions. Mass-balance calculations indicate that the full range of 2H/1H variability in the investigated organisms can be quantitatively explained by varying fluxes, i.e., with constant KIEs for each involved oxidoreductase across all species. This proves that lipid 2H/1H ratios of heterotrophic microbes are quantitatively related to central metabolism and provides a foundation for interpreting 2H/1H ratios of environmental lipids and sedimentary hydrocarbons.
For nearly two decades, scientists have worked to interpret the record of 2H/1H (deuterium/protium) ratios in sedimentary lipids (1, 2). The persistence of these isotopic signals over geologic time makes them potentially useful tracers of biogeochemical processes (3), but this requires an understanding of how such signals are generated in extant organisms. In plants and other photoautotrophs, relatively constant 2H/1H fractionations are observed between lipids and source water, making plant leaf waxes useful as proxies for the past hydrologic cycle (4). In contrast, isotopic analyses of microbial lipids from environmental samples, including hot springs (5, 6), marine particulate organic carbon (7, 8), and marine sediments (9), have revealed unexpectedly large ranges of 2H/1H of up to 400‰ in the conventional H notation. Zhang et al. (10) first showed in laboratory cultures that the magnitude of this fractionation is strongly correlated with the type of metabolism used during growth, ranging from slightly 2H-enriched aerobic heterotrophs to moderately 2H-depleted photoautotrophs and strongly 2H-depleted chemoautotrophs. These correlations highlight the potential for lipid H values to provide meaningful insights into the metabolism of both modern and ancient microorganisms.
More recent work has focused on understanding the biochemical origins of H-isotopic variability in lipids and has revealed additional complexity (6, 8, 12–17). Thus far, aerobic heterotrophs show the largest range of variation and the most 2H-enriched lipids, with lipid/water fractionations () from −150‰ to 200‰ (10, 15). Lipid 2H/1H ratios vary systematically with the growth substrate and are generally more 2H-depleted when bacteria are grown on sugars relative to tricarboxylic acid (TCA) cycle substrates. Anaerobic heterotrophs exhibit similar isotope patterns when nitrate is the terminal electron acceptor, but are more 2H-depleted and do not vary with growth substrate when sulfate is the electron acceptor (10, 15, 18). Photoautotrophs produce fatty acids with intermediate 2H-depletion ranging from −250‰ to −150‰ (14, 16, 17). And chemoautotrophs, at least in some cases, produce very strongly 2H-depleted fatty acids with fractionations between −200‰ and −400‰ (10, 15, 19, 20), but exceptions are known (16). Other factors thought to potentially influence fractionations include different biosynthetic pathways (1, 21, 22), salinity in algae and cyanobacteria, growth phase, and temperature (12–15, 23–25). Still, variations due to these environmental influences are rather small compared with those associated with different types of metabolism.
Isotopic differences in the hydrogen that is transferred from organic substrates to the redox cofactor NADP+ have been suspected as a possible source of variance in lipid H (10). This inference is based mainly on the fact that NADPH is the single largest source of hydrogen (50%) in the highly conserved fatty acid biosynthesis pathway. Although NADPH provides the reducing power for numerous anabolic reactions in all organisms, it is reduced only by a few reactions. In heterotrophs, reduced NADPH is generated by dehydrogenase reactions in central metabolism (Fig. 1), as well as by transhydrogenases that interconvert NADH and NADPH (11). All of these reactions involve concerted hydride transfers and so can plausibly transmit isotopic information (i.e., in comparison with electron-only transfers). When studied in vitro, each of these enzymes also exhibits a different kinetic isotope effect (KIE) (26, 27). Although it is difficult to predict isotopic fractionation in vivo due to committed pathways (28), it is nevertheless likely that varying fluxes through dehydrogenases and transhydrogenases can lead to differences in the isotopic composition of NADPH, and thus of fatty acids (29). The essential question though has been whether this effect is the main one controlling lipid H or merely a bit player among many competing effects. To address this question directly, we compared central metabolic fluxes and isotopic fractionations in five versatile aerobic heterotrophs as well as five Escherichia coli knockout mutants deficient in specific dehydrogenases and transhydrogenases.
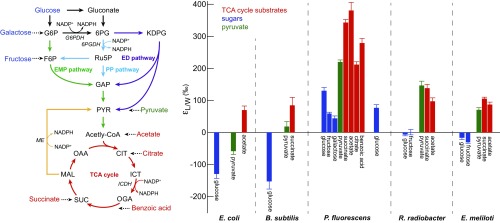
Fig. 1.
(Left) Schematic representation of the central metabolic pathways that are involved in glucose catabolism in aerobic heterotrophs—namely, the EMP pathway (green), the PP pathway (light blue), the ED pathway (violet), and the TCA cycle (red). The major NADPH-generating reactions are highlighted (11). The dehydrogenase enzymes that catalyze these reactions are G6PDH, 6PGDH, ICDH, and ME (yellow). For simplicity, not all reactions and intermediates are shown. (Right) Summary of lipid values from E. coli, B. subtilis, P. fluorescens, R. radiobacter, and E. meliloti grown on different sugars, TCA cycle intermediates, pyruvate, and benzoic acid. values are calculated from the abundance-weighted mean H values of fatty acids and that of culture medium. Error bars represent the corresponding abundance-weighted SDs. Typical replicate precision is ‰, larger than typical analytical uncertainties of ‰.
Results
Bacterial Strains and Fatty Acids.
Five common, aerobic heterotrophic bacteria with widely varying carbon fluxes and different strategies to balance NADPH consumption and production were grown in batch cultures on different substrates (31, 32). The model organism E. coli MG1655, a facultative anaerobic -Proteobacterium, was grown on glucose, pyruvate, and acetate. The versatile Pseudomonas fluorescens WCS365, an obligately aerobic -Proteobacterium, was cultured on glucose, fructose, galactose, pyruvate, succinate, acetate, citrate, and benzoic acid. An additional P. fluorescens strain, 2-79, was grown on glucose. The two Rhizobiaceae, Ensifer meliloti and Rhizobium radiobacter from the phylum -Proteobacterium, were grown on glucose, fructose, pyruvate, succinate, and acetate. The gram-positive model organism Bacillus subtilis, a facultative anaerobe from the phylum Firmicutes, was cultured on glucose, pyruvate, and succinate. All glucose cultures were grown in triplicate. One large culture (300 mL) was used to measure isotopic fractionation, and two smaller (30 mL) cultures were amended with 20% or 100% -glucose for measurements of metabolic flux. Additional details of these bacteria are provided in Materials and Methods.
Cells were harvested in midlog phase commonly between optical density at 600 nm (OD600) of 1 and 1.5. Fatty acids were solvent-extracted, derivatized as methyl esters, and quantified by gas chromatography/mass spectrometry (GC/MS). Abundances of identified fatty acids ranged from 0.2 to 28 mg/g cell dry weight (CDW). Relative abundances are listed for each strain and culture condition in SI Appendix, Table S1. The most abundant fatty acids in E. coli and P. fluorescens were hexadecanoic (16:0) and cis-9-hexadecenoic (16:1) acids. Cis-9,10-methylene-hexadecanoic acid (cyc-17) was an additional abundant fatty acid in E. coli. By far, the biggest contribution to the fatty acid profile with 80% came from cis-9- and cis-11-octadecenoic acids (18:1) in E. meliloti and R. radiobacter. Except for the low abundance of hexadecanoic acid (16:0), only iso- and anteiso-branched fatty acids were identified in B. subtilis. Fatty acid abundances varied widely between strains but were generally consistent in the same strain grown on different substrates. Identified fatty acids and their abundances are in good agreement with published fatty acid profiles for these species (10, 33–36).
Variations in Fractionation between Species.
H values of individual fatty acids are listed in SI Appendix, Table S2. They varied considerably between cultures (−232‰ to 284‰) but on average only by 36‰ between different fatty acids from the same culture. No clear trends were apparent between fatty acids within the same culture. Consequently, we can rule out lipid biosynthetic processes such as desaturation, elongation, and different precursors for normal-, anteiso-, and iso-fatty acids as major sources of lipid 2H/1H variation in our experiments. To focus on the larger 2H/1H variations due to metabolism, and to facilitate quantitative comparison between strains that produce different fatty acids, we calculated an abundance-weighted mean H value and SD for each culture (SI Appendix, Table S2), as have previous studies (e.g., ref. 16). Because the microbes studied here make only a few different fatty acids, the use of individual fatty acid H values instead of weighted-mean values makes no significant difference to our conclusions about patterns of fractionation.
values derived from abundance-weighted mean values are summarized in Fig. 1. The range of fractionations is enormous and spans −154‰ to 380‰. This range is similar to previous reports (10, 15, 16), despite the fact that the bacteria investigated here have more limited metabolic diversity. B. subtilis and E. coli yielded the most 2H-depleted lipids, with ranging from −154‰ to 84‰. P. fluorescens exhibited the greatest variability, with the most 2H-enriched lipids (+44‰ to +380‰). Even the two closely related P. fluorescens strains 2-79 and WCS365 exhibited measurable differences (54‰). R. radiobacter and E. meliloti showed intermediate 2H/1H fractionation ranging from −30‰ to 146‰. values for E. coli and P. fluorescens are in good agreement with reported values (10, 15).
Our data show a strong relationship between the organic carbon source and lipid values. Growth on TCA cycle intermediates (acetate, succinate, and citrate) typically produced the most positive fractionations (+70‰ to +380‰), followed by pyruvate (−57‰ to 220‰) and sugars (−154‰ to 132‰, glucose, fructose, and galactose), the same order as has previously been observed (10). An exception to this pattern was R. radiobacter, with the highest values for cells grown on pyruvate, followed by succinate, acetate, glucose, and fructose. Nevertheless, we excluded the substrate isotopic composition as a major source of lipid 2H/1H variability because the H values of the two carbon sources (acetate and glucose) differed by only 16‰ and therefore cannot explain the large range of lipid H values. Water incorporation was tested by manipulating the isotopic composition of culture water. For this purpose, B. subtilis, E. meliloti, R. radiobacter, and P. fluorescens were grown on glucose by using culture medium with values ranging from −87‰ to 911‰. Correlations between lipid H values and growth medium water as well as derived fractionation curves were consistent with the majority (>80) of lipid H coming from growth water rather than organic substrate (SI Appendix, Fig. S2).
Carbon Flux Varies Widely between Species.
Two of the three parallel cultures for each strain were grown with -glucose labels for metabolic flux analysis based on the detection of mass isotopomer patterns in proteinogenic amino acids as described previously (31, 37, 38). With this procedure, carbon fluxes were determined for all major reactions in the Entner–Doudoroff (ED), pentose phosphate (PP), Embden–Meyerhof–Parnas (EMP; glycolysis), and TCA cycle pathways (SI Appendix, Table S3). These metabolic fluxes reflected reaction rates of enzymatic conversion in millimole per hour and per gram of biomass and are summarized in SI Appendix, Table S4. The ED and EMP pathways do not directly generate NADPH, but are major pathways in the catabolism of glucose and thus compete with glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH) for substrates (Fig. 1). Physiological parameters for each culture including growth rate, glucose uptake rates, and acetate secretion rates are also listed in SI Appendix, Table S4.
To compare fluxes quantitatively between species with different growth rates, all fluxes were normalized to the glucose uptake rate (SI Appendix, Table S4) and are presented in Table 1. The relative contribution of each pathway (ED, PP, EMP, and TCA cycle) to glucose catabolism varied widely among the investigated species. E. coli and B. subtilis relied heavily on the EMP pathway for glucose catabolism, with NADPH produced mainly by G6PDH and 6PGDH in the PP pathway. In fact, the PP fluxes through 6PGDH in these two strains were the highest observed among all species. TCA cycle fluxes were significantly lower in these organisms than in all of the other species due to extensive secretion of acetate. In contrast, in both rhizobia, E. meliloti and R. radiobacter, the ED pathway was the nearly exclusive route for glucose metabolism with high fluxes through G6PDH. Both rhizobia species exhibited much higher TCA cycle fluxes than either E. coli or B. subtilis, while PP flux was minor. Major enzymes for NADPH generation in these strains were therefore G6PDH and isocitrate dehydrogenase (ICDH). In P. fluorescens, the ED pathway and TCA cycle were used almost exclusively to metabolize glucose. Low G6PDH fluxes were observed because of periplasmatic conversion of glucose to gluconate and 2-ketogluconate, and subsequent uptake according to published flux distributions (30) was assumed. Another relevant difference in P. fluorescens metabolism is the operation of a cyclic flux to metabolize glucose that recycles glyceraldehyde-3-phosphate back to glucose-6-phosphate (30). The reactions that comprise this cycle belong to the ED, the reverse EMP, and the PP pathways. In general, the determined fluxes are similar to those reported for the same species (31).
Table 1.
Hydrogen isotope data and selected metabolic fluxes for the five wild-type species and five E. coli knockout mutants all grown on glucose
| Relative flux,§ of glucose uptake | ||||||||||
| Culture | ,* ‰ | ,† ‰ | ,‡‰ | G6PDH (ED+PP) | 6PGDH (PP) | ICDH (TCA) | ME | EMP¶ | ED | NADPH,# balance |
| Wild types | ||||||||||
| E. coli | ||||||||||
| B. subtilis | ||||||||||
| P. fluorescens 2-79 | ∥ | |||||||||
| P. fluorescens WCS365 | ∥ | |||||||||
| R. radiobacter | ||||||||||
| E. meliloti | ||||||||||
| E. coli knockout mutants** | ||||||||||
| JW1841 | ||||||||||
| JW3985 | ||||||||||
| PntAB | ||||||||||
| UdhA | ||||||||||
| UdhA–PntAB | ||||||||||
H of culture medium before inoculation.
Abundance-weighted mean H values of all measured fatty acids.
Fractionation between H of culture medium and abundance-weighted mean H value of fatty acids.
Fluxes through the four NADPH-generating reactions (G6PDH, 6PGDH, ICDH, and ME) and EMP and ED pathways as a percentage of the total glucose uptake flux.
Negative fluxes indicate reversed EMP flux.
Negative values indicate NADPH underproduction relative to anabolic demand; positive values indicate NADPH overproduction.
For Pseudomonas species, published relative flux distributions (30) were assumed due to the periplasmatic conversion of glucose to gluconate and 2-keto-gluconate.
Mutants carry the following gene deletions: G6PDH in JW1841, glucose-6-phosphate isomerase in JW3985, membrane-bound transhydrogenase in PntAB, soluble transhydrogenase in UdhA, and both transhydrogenae genes in UdhA–PntAB.
Correlation between Metabolic Flux and 2H/1H Fractionation.
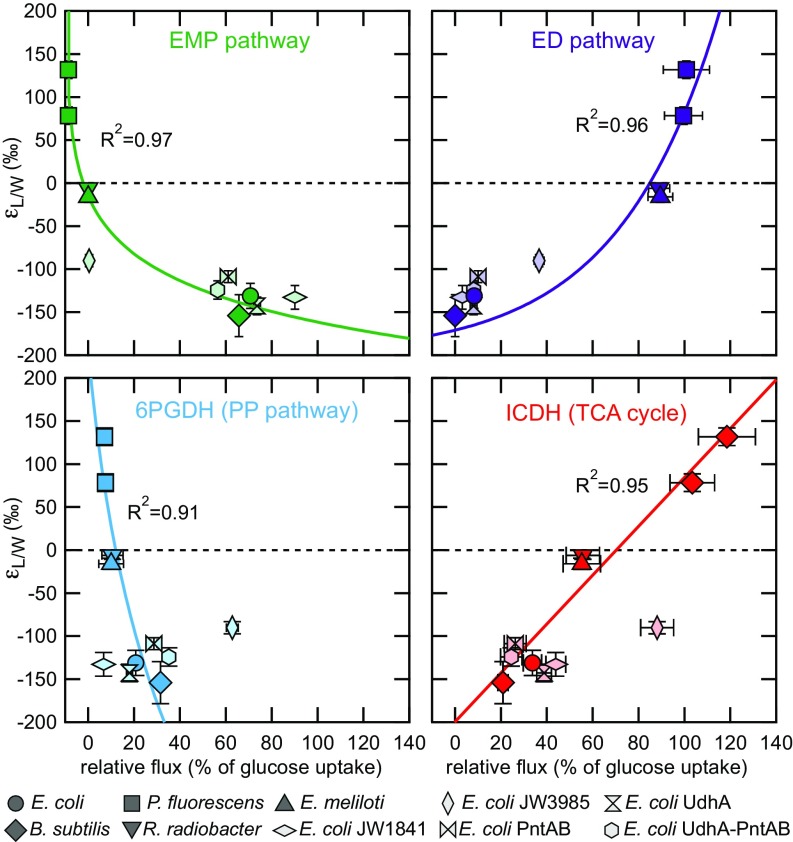
Measured H values are summarized in Table 1. Calculated values varied by >280‰ among the five species and show strong correlations ( > 0.9; Fig. 2) with relative carbon fluxes through the two NADPH dehydrogenases ICDH and 6PGDH, as well as through the ED and EMP pathways that do not directly produce NADPH but compete for substrates. EMP pathway and 6PGDH fluxes were negatively correlated with , while ED pathway and ICDH fluxes were strongly positively correlated with fractionation. For the two other NADPH-generating dehydrogenases, malic enzyme (ME) and G6PDH, no clear correlation to exists (SI Appendix, Fig. S3). The latter result is probably due to the fact that in P. fluorescens the largest fraction of the G6PDH flux is bypassed via extracellular conversion of glucose to gluconate (30, 39). The observed relationships (between isotopic fractionation and metabolic fluxes) are not expected to be linear, because these pathway fluxes are not independent from each other. Indeed, all fluxes in such reaction networks are at least partially cross-correlated. As just one example, the correlation with EMP flux (Fig. 2) was unexpected because this pathway does not generate NADPH. However, a relative increase in EMP flux must be accompanied by a corresponding decrease in PP + ED flux (Fig. 1), both of which do directly contribute to NADPH reduction. Regardless, our results clearly demonstrate that fluxes through metabolic pathways play a key role in modulating lipid hydrogen isotope ratios. Our data indicate that high ED and TCA fluxes relative to PP and EMP are associated with more 2H-enriched lipids, and vice versa, presumably due to differences in the KIEs associated with NADP+ reduction in these pathways. Next, we address the question of whether NADPH-balancing mechanisms such as transhydrogenases also affect values.
Fig. 2.
Correlations between lipid/water fractionation () and the relative carbon flux through 6PGDH and ICDH as well as the EMP and ED pathways. Relationships were derived from E. coli, B. subtilis, P. fluorescens strains WCS368 and 2-79, R. radiobacter, and E. meliloti. In addition, five E. coli knockout mutants lacking specific dehydrogenase and transhydrogenase genes (text) are shown in light colors. Plotted values are calculated between the culture medium and abundance-weighted mean H values of fatty acids. The error bars represent the corresponding abundance-weighted SD. Note that some error bars are smaller than marker sizes.
NADPH Overproduction/Underproduction Correlates with Fractionation.
To maintain growth, the supply of reduced NADPH from catabolic reactions needs to be closely balanced with the demand for NADPH in >100 anabolic reactions (32). Due to fluctuating environmental conditions, this can be difficult to maintain, and bacteria must therefore use effective NADPH-balancing mechanisms. Decoupling of NADPH production from catabolism via transhydrogenase enzymes is one important mechanism. Transhydrogenases interconvert NADPH (used mainly for reductions during biosynthesis) and NADH (used mainly for energy conservation during respiration) via intact hydride transfer, allowing the host to balance catabolic and anabolic fluxes (11). Transhydrogenases are common in metabolically versatile microbes, although certainly not ubiquitous. Depending on the direction of the transhydrogenation reaction, associated KIEs may lead to 2H enrichment or depletion in NADPH and thus fatty acids. Comparison of the sum of all NADPH producing fluxes with the anabolic demands provides a quantitative estimate of NADPH balance (Table 1 and SI Appendix, Table S4). E. coli and B. subtilis were the only two species with a negative NADPH balance, meaning that catabolic production of NADPH does not meet the anabolic demand. In E. coli, the rest of the required NADPH is thought to come from the membrane-bound transhydrogenase PntAB (40), which transfers from NADH to NADP+.
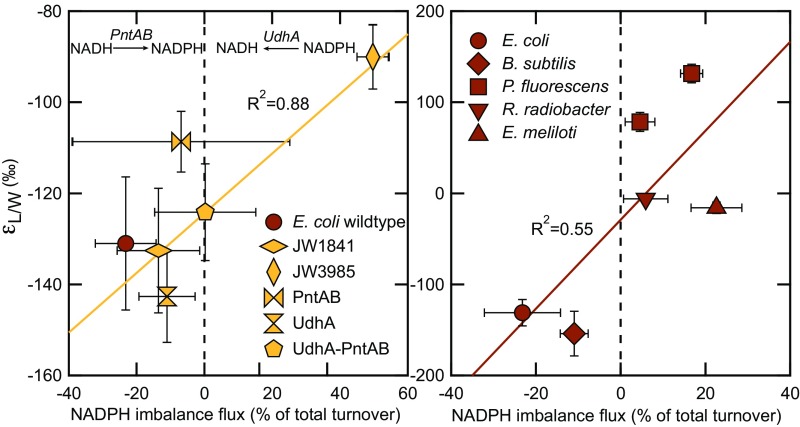
In contrast, the other three species exhibited a positive NAPDH balance, and so potentially use soluble transhydrogenases to consume NADPH. However, the specific mechanism(s) for dealing with overproduction and underproduction of NADPH in these organisms is not currently known, apart from E. coli (32, 40). Comparing all five species, a weak positive correlation is observed between the NADPH flux imbalance and lipid/water isotopic fractionation (Fig. 3, Right).
Fig. 3.
The relationship between lipid/water fractionation () and relative NADPH imbalance flux in five E. coli knockout mutants (plus wild type; Left) and in six wild-type species (Right). Negative values indicate NADPH underproduction; positive values indicate NADPH overproduction. E. coli balances NAPDH levels via membrane-bound (PntAB) and soluble (UdhA) transhydrogenases. For all other species, the mechanism to balance NADPH levels is currently unknown. Not included in the linear regression is the mutant PntAB because the NADPH underproduction in this mutant is not balanced by PntAB.
In addition to the five wild-type strains (above), we cultured five E. coli knockout mutants with perturbed NADPH metabolism. Lipid H values and metabolic fluxes for these strains are given in Table 1, Fig. 2 (light colors), and SI Appendix, Table S4. In strain JW3985, glucose-6-phosphate isomerase in the EMP pathway was deleted, which led to an extreme NADPH overproduction of and 2H-enriched lipids relative to the E. coli wild type. All other E. coli mutants exhibited balanced or negative NADPH levels and similar fatty acid H values relative to the E. coli wild type. In JW1841, G6PDH was deleted, and in the mutants PntAB and UdhA, the membrane-bound (PntAB) and soluble (UdhA) transhydrogenases were knocked out, respectively. In the UdhA–PntAB double-knockout mutant, both transhydrogenases were deleted. Except for mutant PntAB, these mutants balanced NADPH overproduction by the soluble transhydrogenase UdhA and NADPH underproduction by the membrane-bound transhydrogenase PntAB. Fig. 3, Left shows the positive correlation between values of the E. coli knockout mutants and wild-type and the NADPH transhydrogenase fluxes. This correlation indicates 2H-depletion by PntAB and 2H-enrichment by UDhA, respectively. A more detailed description of each individual E. coli knockout mutant and the observed correlation is provided in SI Appendix.
Quantitative Model for Lipid 2H/1H Fractionation.
The strong influence of relative flux distributions on lipid 2H values is presumably due to varying KIEs of the involved oxidoreductases. However, it remains uncertain whether such KIEs are the same in different organisms. To investigate this quantitatively, we constructed a simple mass-balance model to relate lipid H values to sources of hydrogen and to metabolic fluxes. Conceptually, we describe the fatty acid isotope ratio (RFA≡[2H]/[1H]) as a mixture of isotopically distinct pools of hydrogen that come from water (Rwater), acetyl-CoA (Racetate), and NADPH (). The isotopic mass balance is a linear combination of the isotope ratios of each pool multiplied by their relative contribution:
| [1] |
Contributions of each source are derived from the known stoichiometry of fatty acid biosynthesis, where of C-bound H in a fatty acid is derived from NADPH, from water, and the rest from acetyl-CoA (1, 29). We used these values for our model and neglected any possible additional hydrogen exchange between metabolites and water. NADPH can be further subdivided into isotopically distinct sources from G6PDH (), 6PGDH (), ICDH (), and ME (). Contributions from the two transhydrogenases have to be considered as separate cases. In the case of NADPH underproduction, the membrane-bound transhydrogenase transfers from NADH to NADP+ with flux 0 and represents therefore an additional source of hydrogen (). The isotopic mass balance is then a linear combination of the isotope ratios of each source multiplied by their fractional flux contribution (J’s represent absolute fluxes).
| [2] |
In the case of NADPH overproduction, the soluble transhydrogenase is active with flux 0, while the membrane-bound transhydrogenase is not ( = 0). In contrast to NADPH underproduction, there are now two sinks, one that transfers to fatty acids and an additional one that transfers from NADPH to . This represents a branchpoint in the reaction network (SI Appendix, Fig. S1). In addition to the four dehydrogenases, the fractionation between the two sinks for NADPH (described as term) and the branching ratio of NADPH between anabolism and conversion to NADH now affect as well. Following the approach of Hayes (28), Eq. 2 then becomes:
| [3] |
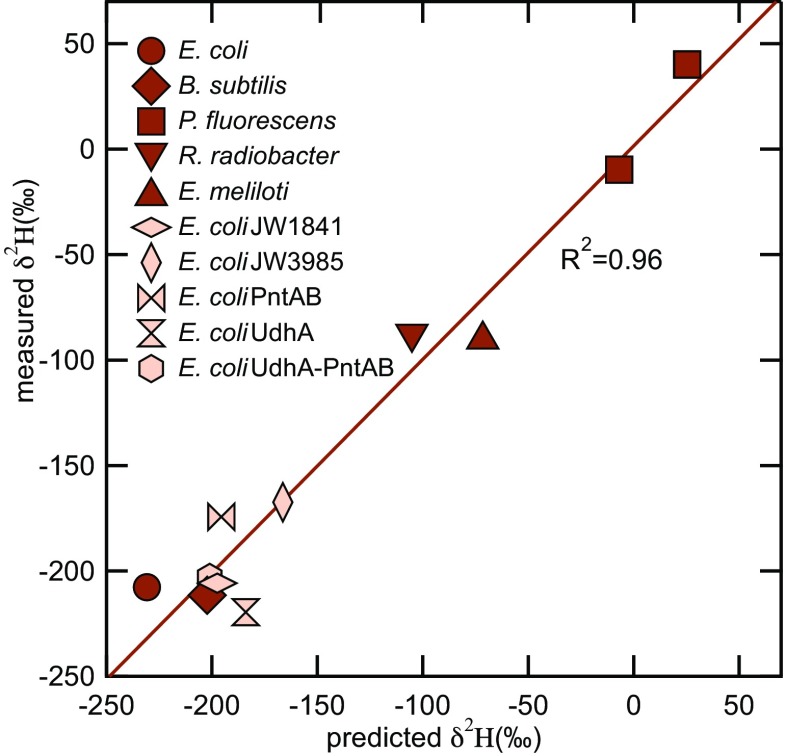
A more detailed derivation of Eqs. 1–3 is provided in SI Appendix. Absolute fluxes ( terms) for dehydrogenases and transhydrogenases were taken from the experimentally determined metabolic fluxes. The isotope ratios of NADPH from the four dehydrogenase and the membrane-bound transhydrogenase reactions were treated as constant free parameters to be fit. They reflect the isotope compositions of the substrate-bound that is transferred to NADPH plus any isotope effects associated with this transfer. In the case of the membrane-bound transhydrogenase, the isotopic composition of NADH that supplies probably varies for all of the same reasons that varies; however, we have no way to constrain this at present and so treat as a constant free parameter as well. is another fitted parameter. and account for the transhydrogenases PntAB and UdhA from E. coli, respectively. For the other four species, separate and values were fitted, reflecting the fact that other mechanisms than transhydrogenases are involved in balancing NADPH levels (32). The average H value of growth water in our experiments was 80‰, and the exchange between organic H and water was suggested to 2H-deplete lipids relative to the water (3). Therefore, we assumed a H value of −180‰ for the H derived from water (Rwater). Because we have no way to distinguish isotopically between H from water and acetyl-CoA, we made the same assumption for the H derived from acetyl-CoA (Racetate). The best-fit solution to the eight free parameters was found by minimizing the rms difference between experimental data and predicted lipid H values across all glucose-grown cultures. In Table 2, the best-fit values are listed as isotope ratios, H values, and equivalent fractionations ( values) to facilitate comparison with literature-derived in vitro KIEs. The best-fit values yielded an rms error of just 17.3‰ and a linear correlation between measured and modeled lipid H values of = 0.96 (Fig. 4 and Table 2). Given that we assume constant isotope ratios for each H source in the model, the excellent fit indicates that changes in metabolic fluxes alone can plausibly explain virtually all of the isotopic variability in fatty acids. The model does not prove that isotopic fractionations associated with dehydrogenase and transhydrogenases are constant between organisms, but it is consistent with that conclusion. Given the existence of eight fitted parameters in the model, there are of course large uncertainties in their estimated values. Nevertheless, the conclusion that H from 6PGDH and G6PDH was strongly 2H-depleted relative to that from ICDH appears robust (SI Appendix, Fig. S4), also that both transhydrogenases exhibited normal KIEs (i.e., the light isotope reacts more quickly).
Table 2.
Comparison of best-fit parameters with literature deuterium KIEs associated with NADPH dehydrogenases and transhydrogenases
| Best fit values† | Literature values | ||||||||
| Enzyme | Pathway | Parameter‡ | Ratio | H,§ ‰ | ,¶ ‰ | ,∥‰ | In vitro KIE | Ref. | |
| G6PDH | PP and ED | 7.05 10−5 | — | 2.97 | (45) | ||||
| 6PGDH | PP | 1.08 10−4 | — | 1.61 | (46) | ||||
| ICDH | TCA cycle | 2.99 10−4 | — | +1,085 | 0 | 1 | (26, 42, 47) | ||
| ME | Anaplerotic node | 0.00 | — | — | — | 1.47 | (45) | ||
| PntAB | — | 3.46 10−5 | — | to | 1.8–4.5 | (27) | |||
| unknown | — | 0.00 | — | — | — | — | — | — | |
| UdhA | — | — | 0.84 | — | — | — | — | ||
| sTH or other | — | — | 0.57 | — | — | — | — | ||
Best-fit parameters were derived by minimizing the rms difference between experimental data and predicted lipid H values from Eqs. 1–3. Uncertainties associated with these values are discussed in SI Appendix, Fig. S4.
and account for the membrane-bound (PntAB) and soluble (UdhA) transhydrogenase from E. coli, respectively. account for the soluble transhydrogenases (sTH) or alternative mechanisms to balance NADPH levels in all other species. accounts for a currently unknown mechanism to balance NADPH levels in B. subtilis.
H values were calculated as H = (/ − 1) 1,000.
Enrichment factors were estimated as = ( − 1) 1,000 and = (H + 1)/( + 1) − 1.
Enrichment factors are calculated from in vitro KIEs according to = (1/KIE − 1) 1,000.
Fig. 4.
Comparison of predicted H values vs. experimental data.
Discussion
Our results demonstrate that the large variance in H-isotopic fractionations by a diverse group of aerobic heterotrophs can be entirely and quantitatively explained by varying metabolic fluxes. This confirms that the reduction of NADP+ by different dehydrogenases and transhydrogenases is a key biochemical control on lipid H due to varying KIEs and suggests that the isotopic fractionations exhibited by these reactions are perhaps nearly constant across different organisms. In the following, we discuss this finding with respect to each of the relevant enzymes and then consider what other factors might contribute to variability. We finish by showing that our model is able to successfully predict H values for some organisms and conditions beyond those directly investigated here.
Dehydrogenases.
In the studied species, four dehydrogenase enzymes comprise the major sources of NADPH: G6PDH, 6PGDH, ME, and ICDH (11). Each of the four dehydrogenases transfers an intact hydride moiety () to NADPH with a characteristic isotope composition that depends on both that of the substrate H that is transferred to NADPH and the KIE associated with the transfer. Comparison of our results with published in vitro KIEs for these dehydrogenases suggests that both factors are important (26). Enrichment factors for and are in excellent agreement with values derived from KIEs described in the literature for the reactions catalyzed by G6PDH and 6PGDH, respectively (Table 2). The strong negative correlation between PP pathway flux and lipid/water fractionation indicates a decrease in the H value of NADPH with increasing fluxes through G6PDH and 6PGDH, and so is also consistent with this conclusion (Fig. 2). In contrast, the apparent lack of a normal KIE for ICDH in vitro is unusual (26) (Table 2). The rate-limiting step in this reaction is not considered to be C–H bond cleavage, but rather breaking of the C–O bond or product release (41, 42), which explains the lack of 2H-depletion associated with ICDH. Nevertheless, a strong positive correlation between lipid H and ICDH flux and a positive value for of 918‰ require a mechanism for enrichment of 2H in NADPH from this source (Fig. 2 and Table 2). As reported, three reaction steps upstream of isocitrate which potentially 2H-enrich the hydrogen that is transferred to NADPH by ICDH can explain this observation (10). The first step is the condensation of acetyl-CoA and oxaloacetate by citrate synthase. Proton abstraction from the methyl group is associated with a significant normal KIE (43). Following the fate of this methylene group in the TCA cycle, a second H is removed by succinate dehydrogenase, which is associated with a very large normal KIE of 5.4 and constitutes a second 2H-enrichment step (44). After fumarase adds one H from O to this C–H position, which may dilute the 2H enrichment somewhat, a further H is removed after one turn of the TCA cycle by aconitase (41). The more than 150‰ lower of P. fluorescens during growth on citrate compared with acetate is further evidence of significant 2H-enrichment by these TCA cycle reactions (Fig. 1). During growth on acetate, high citrate synthase activity is expected that channels acetyl-CoA into the TCA cycle and consequently through all of the prior 2H-enrichment steps described above. In contrast, during growth on citrate, little to no citrate synthase activity is expected. Thus, only the removal of H from the methylene group of citrate by aconitase causes 2H-enrichment. The transfer from malate to NADPH by ME may also be affected by the 2H-enrichment of the TCA cycle. On the other hand, ME is associated with a small normal in vitro KIE that should result in a negative correlation between and ME flux and a 2H-depleting value. In fact, no clear correlation is observed, and is 0, indicating that ME fluxes are not required to explain lipid 2H/1H variations in this dataset (Table 2 and SI Appendix, Fig. S3). The reason for this result could be related to the large uncertainties associated with the rather small ME fluxes (e.g., ME flux for E. coli PntAB is 10 31‰) that complicates the detection of the underlying isotope trend (Table 1). Moreover, the 2H-enrichment in the TCA cycle combined with a possible 2H-depletion by the transfer to NADPH by ME may result in only small isotope fractionation, which makes detection even more challenging. Another possible explanation for our result is the unknown cofactor specificity of ME (32). It is possible that in some of the investigated species ME utilizes instead of NADP+.
Transhydrogenases
Transhydrogenase enzymes serve to interconvert NADPH and NADH via intact hydride transfer. Such enzymes exist in all metabolically flexible organisms and serve the important role of allowing the host to properly balance catabolic and anabolic fluxes (11). The in vitro deuterium KIEs associated with transhydrogenases are large and can be up to 4.5 (27). E. coli contains two different transhydrogenase enzymes, a membrane-bound version (PntAB) that is energy-dependent and a soluble version (UdhA) (40). Normal KIEs will lead to 2H-enrichment of residual reactants and 2H-depletion of products. We thus expect overproduction of NADPH to correlate with 2H-enriched lipids and underproduction of NADPH to correlate with 2H-depleted lipids, a pattern that is observed in our data for E. coli (Fig. 3). In addition, the predicted flux weighted H for PntAB is in excellent agreement with literature in vitro KIEs (’s in Table 2). Compared with PntAB, the estimated value that accounts for the soluble transhydrogenase UdhA is rather small but appears to be consistent with a normal KIE and thus our hypothesis (Table 2). A similar result was obtained for the electron-bifurcating transhydrogenase NfnAB in the sulfate-reducing bacteria Desulfovibrio alaskensis (17). Leavitt et al. showed that this transhydrogenase is important for 2H/1H fractionation. While E. coli wild-type and knockout mutants rely on the two transhydrogenases (40), the other investigated species potentially have other strategies to balance NADPH levels (32). In P. fluorescens and E. meliloti, no significant transhydrogenase activity during growth on glucose was measured. In these organisms, mechanisms to avoid NADPH imbalance were suggested to be the dual cofactor specificity of G6PDH and 6PGDH, as well as the existence of isoenzymes with distinct cofactor specificities (32). Moreover, B. subtilis was the only investigated culture apart from E. coli strains that exhibited a negative NADPH balance, but lacked a membrane-bound transhydrogenase to supply the missing NADPH (32) (Table 1). The fitted isotope ratio that accounts for NADPH underproduction in this culture () is zero and points therefore also toward a yet-unknown mechanism to balance NADPH levels (Table 2). Regardless, all other investigated species are genetically capable of expressing both a membrane-bound and a soluble transhydrogenase variant (32). The observed positive correlation between and NADPH imbalance flux (Fig. 3) as well as the predicted flux weighted value from the model (Table 2) support the interpretation that soluble transhydrogenase activity contributes to lipid H values.
Other Potential Sources of 2H/1H Variability.
The redox-active H on NADPH is moderately acidic and can potentially exchange with water. Indeed, labeling experiments with in different transformed human cell lines revealed that ∼40% to ∼70% of the redox active hydrogen of NADPH comes from water (48). It was shown that NADPH-dependent flavin enzymes such as glutathione reductase that hold the hydride in a N–H bond instead of a C–H bond catalyze this exchange. Thus, it is possible that hydrogen exchange of NADPH with water also happened in our investigated species and mutes the divergent 2H/1H ratios that are generated by different metabolic pathways. The rate at which this happens should be a function of the turnover (residence) time of NADPH, with a longer cellular residence time for NADPH expected to yield less extreme fractionations because of more significant hydrogen exchange. For the six wild-type strains, we calculated NADPH turnover times from production rates based on metabolic fluxes and intracellular NADPH concentrations, assuming the cell is in steady state (SI Appendix, Fig. S5). The data showed no significant correlation with fractionation, suggesting that such exchange effects on were minor, possibly because these organisms all grow quickly. Another source of potential variation in is the cofactor specificity of enoyl–ACP reductase that transfers from NADPH to the fatty acid. Most organisms rely solely on NADPH for this reaction, but in some organisms such as E. coli and B. subtilis, both NADH and NADPH can be used (49, 50). We do not currently know whether the H values of NADH and NADPH are correlated or not, though it seems plausible that they could be. Further 2H/1H variation can be introduced by other -dependent dehydrogenases such as NADP+-dependent glyceraldehyde-3-phosphate dehydrogenase, glucose dehydrogenase, or methylenetetrahydrofolate dehydrogenase (MTHFD), for which we did not account (11, 22). It is unlikely that these dehydrogenases are relevant in the investigated species (32), but can contribute significantly to NADPH levels in certain organisms. For example, in transformed human cell lines, 2H-labeling experiments showed that up to of NADPH is produced by MTHFD in folate metabolism (22).
Applicability of Model to Other Substrates and Organisms.
Growth substrate is one of the dominant controlling variables for lipid H values in aerobic heterotrophs, as shown in Fig. 1 and previous reports (10, 16, 25). Even though we did not carry out flux measurements for cultures growing on carbon sources other than glucose, we can test our model by predicting H values from published flux data for growth on those substrates. During growth on fructose, both Rhizobia and P. fluorescens produced lipids with on average 25‰ lower H values than when glucose was the growth substrate (Fig. 1). In comparison with glucose metabolism where ED is the main pathway, flux data shows that Pseudomonas putida channels fructose only 54 through ED, whereas the rest is processed in the EMP and PP pathways (30). Modeling lipid H values for both carbon sources (fructose and glucose) predicts a 22‰ lower H value for fructose, which is in excellent agreement with our culture data. Fluxes of gluconate catabolism in E. coli were found to be essentially opposite to glucose catabolism, with high ED pathway fluxes and almost exclusive production of NADPH by 6PGDH, ICDH, and PntAB due to NADPH underproduction (51, 52). Zhang et al. (10) measured lipid/water fractionation of −123‰ in E. coli during growth on gluconate, a 2H/1H fractionation that our model accurately predicts with an NADPH underproduction of . The high 2H enrichments in cultures grown on direct precursors or intermediates of the TCA cycle are particularly interesting. In E. coli, flux measurements have been carried out during growth on acetate (51), under conditions similar to our cultures that yielded fatty acids with values of 60‰ (Fig. 1). Combining these data, our model can account for the 2H-enriched lipids only with an overproduction of NADPH in the range of . Indeed, overproduction of NADPH by E. coli is suggested during growth on acetate (40, 52), which is consistent with our prediction. Nevertheless, overproduction of NADPH by 75 seems to be slightly excessive and probably indicates that other 2H-enrichment processes might be important. A difference between glucose and acetate metabolism is the use of the glyoxylate cycle, a variation of the TCA cycle that allows E. coli to grow on simple carbon sources like acetate (51, 53). In this cycle, isocitrate is converted into glyoxylate and succinate instead of -ketoglutarate. Glyoxylate is subsequently combined with acetyl-CoA to produce malate, catalyzed by malate synthase. This bypasses the decarboxylation steps of the TCA cycle, allowing acetate to be used to build carbon skeletons. The operation of this cycle may result in higher 2H-enrichment in the lipids than when only the TCA cycle is active. Similar to citrate synthase, malate synthase abstracts a proton from acetyl-CoA and is associated with a large intramolecular normal KIE of 3.8 (26). 2H-enrichment in the residual -group can be expected with further 2H-enrichment by aconitase (41). Moreover, isotope dilution by H incorporation in fumarate does not happen in this cycle.
Culture-based studies of anaerobic nitrate-reducing bacteria suggest that they exhibit a similar correlation of H-isotopic fractionation on substrate, as do aerobes, but with slightly more 2H-depleted lipids and less variation (10, 16). Sulfate reducers seem not to vary their fractionation between growth substrates at all (16). We propose that the reduced variance in in anaerobes could reflect a lower metabolic versatility of these organisms related to the reduced energy yield of anaerobic respiration. Flamholz et al. (54) showed that prokaryotes use different glycolytic pathways depending on the energy yield of the overall respiratory pathway. Genomic analysis revealed that anaerobes rely heavily upon the higher ATP yield of the EMP pathway, whereas the ED pathway is common among facultative anaerobes and even more widespread among aerobes (54). When we rerouted fluxes of the ED pathway through the EMP (50) and PP (50) pathways in silico in the investigated wild-type species, predicted lipid H values in P. fluorescens strains were 90‰ lower, and variations in in the wild-type species diminished from 272‰ to 200‰, mirroring the trends observed between aerobes and nitrate reducers (16). However, further metabolic flux measurements will be necessary to fully understand 2H/1H variation in those organisms. Likewise, by applying our results to the study of heterotrophs in natural environments, the additional processes of fatty acid assimilation and degradation will need to be considered. H-isotopic fractionation has not yet been studied in either process, although we expect such fractionations (if any) to be smaller than those reported here. Still, the possibility that assimilated fatty acids may reflect the isotopic composition of the food source rather than the host metabolism needs to be carefully considered.
Concluding Remarks.
Our results demonstrate that metabolic fluxes through specific dehydrogenase and transhydrogenase enzymes play a key role in controlling H values of lipids from diverse aerobic heterotrophs. It remains to be seen whether this is also true for other organisms growing under different conditions. Regardless, lipid H values hold the potential to provide useful insights into metabolism. To date, metabolic information is almost exclusively available for fast-growing, culturable microorganisms. However, metabolic processes catalyzed by environmental bacteria within their natural habitats are challenging to investigate and thus largely unknown. Lipid 2H/1H ratios can provide information on how NADPH is generated in these uncultured microorganisms and could therefore tell us which metabolic pathways (e.g., PP pathway, EMP pathway, or TCA cycle) are active in a certain environment. With this information, we might be able to reconstruct the metabolic lifestyle of these microorganisms. In addition, H values could have numerous applications in other fields, including ecology, industry, and biomedicine. Examples range from the use of lipid 2H/1H ratios to assess in situ metabolic interactions of symbionts, to study the metabolic response to environmental stressors, or perhaps even to investigate human diseases such as cancer that are associated with large changes in NADPH metabolism (55). Measurement of 2H/1H ratios of other biomolecules could provide additional constraints on metabolism. H values of amino acids are promising, particularly those that require NAD(P)H for biosynthesis (proline, isoleucine, methionine, etc.). Fogel et al. (56) showed recently that measurement of H values of individual amino acids from an E. coli culture is possible and proposed the use of these signals as a new biosignature for microbial pathways. Measurement of amino acid H values in environmental samples would also enable the identification of the host organism via the isolation and sequencing of specific proteins. We are clearly far from realizing all that potential, but our findings provide a mechanistic understanding of the information content hidden in H values and lay the basis and direction for future research.
Materials and Methods
Strains, Media, and Growth Conditions.
E coli MG1655, B. subtilis PY79, P. fluorescens WCS365 and 2–79 (Newman laboratory), R. radiobacter C58 (German Collection of Microorganisms and Cell Cultures, DSMZ 5172) E. meliloti Young 2003 (DSMZ 1981), and the E. coli knockout mutants JW1841 (Keio strain collection), JW3985 (Keio strain collection), PntAB (Sauer laboratory), UdhA (Sauer laboratory), and UdhA-PntAB (Sauer laboratory) were grown in batch cultures on a rotary shaker at 200 rpm. B. subtilis and E. coli were grown at C and all other cultures at C. M9 minimal medium was prepared and used as described in Fuhrer et al. (31) with the exception that the vitamin solution was only added to B. subtilis and R. radiobacter cultures. A concentration of 50 g/mL kanamycin was added to strains JW3985 and JW1841. Sterile glucose, fructose, galactose, acetate, pyruvate, succinate, citrate, or benzoic acid was supplemented to each culture at a final concentration of 4 g/L. For isotope analysis, the carbon source was added to 300 mL of medium in 1,000-mL flasks. For -labeling experiments, glucose was either added entirely in the form of 100% 1--glucose to 30-mL medium in 125-mL flasks or as a mixture of 20% (wt/wt) U--glucose and 80 (wt/wt) natural glucose. Cell growth was monitored by measuring OD600. Glucose and acetate concentrations were determined with a commercially available enzyme kit (Megazyme). Intracellular NAD(P)H and NAD(P)+ concentrations were measured according to Kern and Newman (57). CDW was inferred from three different OD600 points (usually 0.5, 1, and 1.5). At each point, three 25-mL samples were taken, spun down, washed with 0.9 NaCl, transferred in predried and weighed aluminum vessels, dried in an oven at C for 24 h, and weighted. All cultures were harvested in midexponential phase at OD600 between 1 and 1.5, and samples dedicated for flux analysis were washed twice with 0.9% NaCl at 4°C. All samples were stored at −80°C until isotope and flux analysis was carried out.
Lipid Extraction, Derivatization, and Quantification.
Frozen cell pellets were lyophilized overnight. A total of 40 mg of the dried biomass was transesterified with hexane/methanol/acetyl chloride at 100 °C for 10 min, followed by extraction of the resultant fatty acid methyl esters (FAMEs) in hexane and concentration under a stream of (58). FAMEs were identified via GC/MS on a Thermo Scientific Trace DSQ by injecting the sample in splitless mode. Chromatographic separation was achieved on a 30-m × 0.25-mm capillary column (ZB-5 ms, 0.25 m film thickness; Zebron). Peaks were identified by comparing mass spectra with standards and library data. Quantification was achieved simultaneously by a flame ionization detector. Peak areas of unknown analytes were compared with that of an internal standard (hexadecanoic acid 2-methylpropyl ester) that was added to the sample before injection.
Isotope Analysis.
The H values of FAMEs were measured by a gas chromatograph coupled to an isotope-ratio mass spectrometer (Thermo-Scientific Delta+XP) using a pyrolysis interface. Chromatographic separation was carried out with a 30-m × 0.25-mm capillary column (ZB-5 ms, 1 m film thickness; Zebron) with identical settings as for GC/MS analysis. Peaks could therefore be identified by retention order and relative height. For each sample, triplicate measurements were performed to determine H values which were reported as arithmetic means in per mil relative to the Vienna Standard Mean Ocean Water (VSMOW) standard and were corrected for the added methyl H in the derivatization. For the determination of H of culture medium, 1-mL samples were taken from each culture before inoculation. Samples were analyzed on a Los Gatos Research DLT-100 liquid water isotope analyzer and calibrated against up to four working standards with H values ranging from −73‰ to 458‰. These standards were in turn calibrated against the VSMOW, Greenland Ice Sheet Precipitation, and Standard Light Antarctic Precipitation international standards (59). Average precision was 0.6‰. Net fractionations between lipids and water were calculated as =(+1)/(+1) − 1.
Metabolic Flux Analysis.
The pellets were hydrolyzed in 1.5 mL of 6 M HCl for 24 h at C in sealed 2-mL Eppendorf tubes and subsequently dried overnight at C under a constant air stream. The hydrolyzate was dissolved in 50 L of 99.8 pure dimethyl formamide, and 30 L of precipitate-free supernatant were transferred into a new Eppendorf cup. Derivatization was achieved by adding 30 L of N-methyl-N-(tert-butyldimethylsilyl)-trifluoroacetamide and following incubation at 85 °C with shaking at 550 rpm for 60 min (60). The -labeling patterns of proteinogenic amino acids were determined on a 6890N Network GC system with a 5975 inert XL mass selective detector (Agilent Technologies Inc.); mass distribution vectors of the proteinogenic amino acids were corrected for the natural abundance of all stable isotopes as described (61, 62); and the relative metabolic flux ratios were calculated by using the Fiat Flux software (63). The determined linear system of mass balances, relative flux ratios, quantitative physiology data, and biomass requirements was then solved with the fmincon function from MatLab using the Netto module from FiatFlux (63) to obtain the net metabolic fluxes as described (38). All fluxes are provided in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank Jared Leadbetter for use of laboratory facilities and for providing helpful discussion and assistance with microbial cultures; Fenfang Wu and Stephanie Connon for laboratory assistance; and Megan Bergkessel for assistance with the plate reader and for providing P. fluorescens strains. This work was supported by National Science Foundation Award EAR-1529120 (to A.L.S.); and Swiss National Science Foundation Early Postdoc Mobility Fellowship P2EZP2 159080 (to R.S.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818372116/-/DCSupplemental.
References
- 1.Sessions A. L., Burgoyne T. W., Schimmelmann A., Hayes J. M., Fractionation of hydrogen isotopes in lipid biosynthesis. Org. Geochem. 30, 1193–1200 (1999). [Google Scholar]
- 2.Sessions A. L., Factors controlling the deuterium contents of sedimentary hydrocarbons. Org. Geochem. 96, 43–64 (2016). [Google Scholar]
- 3.Sessions A. L., Sylva S. P., Summons R. E., Hayes J. M., Isotopic exchange of carbon-bound hydrogen over geologic timescales. Geochim. Cosmochim. Acta 68, 1545–1559 (2004). [Google Scholar]
- 4.Sachse D., et al. , Molecular paleohydrology: Interpreting the hydrogen-isotopic composition of lipid biomarkers from photosynthesizing organisms. Annu. Rev. Earth Planet Sci. 40, 221–249 (2012). [Google Scholar]
- 5.Naraoka H., Uehara T., Hanada S., Kakegawa T., .C–D distribution of lipid biomarkers in a bacterial mat from a hot spring in Miyagi Prefecture, NE Japan. Org. Geochem. 41, 398–403 (2010). [Google Scholar]
- 6.Osburn M. R., Sessions A. L., Pepe-Ranney C., Spear J. R., Hydrogen-isotopic variability in fatty acids from Yellowstone National Park hot spring microbial communities. Geochim. Cosmochim. Acta 75, 4830–4845 (2011). [Google Scholar]
- 7.Jones A. A., Sessions A. L., Campbell B. J., Li C., D/H ratios of fatty acids from marine particulate organic matter in the California Borderland Basins. Org. Geochem. 39, 485–500 (2008). [Google Scholar]
- 8.Heinzelmann S. M., et al. , Seasonal changes in the D/H ratio of fatty acids of pelagic microorganisms in the coastal North Sea. Biogeosciences 13, 5527–5539 (2016). [Google Scholar]
- 9.Li L., Sessions A. L., Kinnaman F. S., Hydrogen-isotopic variability in lipids from Santa Barbara Basin sediments. Geochim. Cosmochim. Acta 73, 4803–4823 (2009). [Google Scholar]
- 10.Zhang X., Gillespie A. L., Sessions A. L., Large D/H variations in bacterial lipids reflect central metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 106, 12580–12586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spaans S. K., Weusthuis R. A., van der Oost J., Kengen S. W. M., NADPH-generating systems in bacteria and archaea. Front. Microbiol. 6, 742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirghangi S. S., Pagani M., Hydrogen isotope fractionation during lipid biosynthesis by Haloarcula marismortui. Geochim. Cosmochim. Acta 119, 381–390 (2013). [Google Scholar]
- 13.Dirghangi S. S., Pagani M., Hydrogen isotope fractionation during lipid biosynthesis by Tetrahymena thermophila. Org. Geochem. 64, 105–111 (2013). [Google Scholar]
- 14.Dawson K. S., Osburn M. R., Sessions A. L., Orphan V. J., Metabolic associations with archaea drive shifts in hydrogen isotope fractionation in sulfate-reducing bacterial lipids in cocultures and methane seeps. Geobiology 13, 462–477 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Heinzelmann S. M., et al. , Impact of metabolism and growth phase on the hydrogen isotopic composition of microbial fatty acids. Front. Microbiol. 6, 408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osburn M. R., Dawson K. S., Fogel M. L., Sessions A. L., Fractionation of hydrogen isotopes by sulfate- and nitrate-reducing bacteria. Front. Microbiol. 7, 1166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leavitt W. D., Flynn T. M., Suess M. K., Bradley A. S., Transhydrogenase and growth substrate influence lipid hydrogen isotope ratios in Desulfovibrio alaskensis G20. Front. Microbiol. 7, 918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Sachs J. P., Marchetti A., Hydrogen isotope fractionation in freshwater and marine algae: II. Temperature and nitrogen limited growth rate effects. Org. Geochem. 40, 428–439 (2009). [Google Scholar]
- 19.Campbell B. J., Li C., Sessions A. L., Valentine D. L., Hydrogen isotopic fractionation in lipid biosynthesis by -consuming Desulfobacterium autotrophicum. Geochim. Cosmochim. Acta 73, 2744–2757 (2009). [Google Scholar]
- 20.Valentine D. L., Sessions A. L., Tyler S. C., Chidthaisong A., Hydrogen isotope fractionation during / acetogenesis: Hydrogen utilization efficiency and the origin of lipid-bound hydrogen. Geobiology 2, 179–188 (2004). [Google Scholar]
- 21.Chikaraishi Y., Suzuki Y., Naraoka H., Hydrogen isotopic fractionations during desaturation and elongation associated with polyunsaturated fatty acid biosynthesis in marine macroalgae. Phytochemistry 65, 2293–2300 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Fang J., et al. , Hydrogen isotope fractionation in lipid biosynthesis by the piezophilic bacterium Moritella japonica DSK1. Chem. Geol. 367, 34–38 (2014). [Google Scholar]
- 23.Romero-Viana L., Kienel U., Wilkes H., Sachse D., Growth-dependent hydrogen isotopic fractionation of algal lipid biomarkers in hypersaline Isabel Lake (Mexico). Geochim. Cosmochim. Acta 106, 490–500 (2013). [Google Scholar]
- 24.Sachse D., Sachs JP., Inverse relationship between D/H fractionation in cyanobacterial lipids and salinity in Christmas Island saline ponds. Geochim. Cosmochim. Acta 72, 793–806 (2008). [Google Scholar]
- 25.Heinzelmann S. M., et al. , Comparison of the effect of salinity on the D/H ratio of fatty acids of heterotrophic and photoautotrophic microorganisms. FEMS Microbiol. Lett. 362, 1–6 (2015). [DOI] [PubMed] [Google Scholar]
- 26.OLeary M. H., Multiple isotope effects on enzyme-catalyzed reactions. Annu. Rev. Biochem. 58, 377–401 (1989). [DOI] [PubMed] [Google Scholar]
- 27.Jackson J. B., Peake S. J., White S. A., Structure and mechanism of proton-translocating transhydrogenase. FEBS Lett. 464, 1–8 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Hayes J. M., Fractionation of the isotopes of carbon and hydrogen in biosynthetic processes. Rev. Mineral Geochem. 43, 225–277 (2001). [Google Scholar]
- 29.White S. W., Zheng J., Zhang Y. M., Rock S. J., The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Nikel P. I., Chavarría M., Fuhrer T., Sauer U., de Lorenzo V., Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 290, 25920–25932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuhrer T., Fischer E., Sauer U., Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187, 1581–1590 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuhrer T., Sauer U., Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J. Bacteriol. 191, 2112–2121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikemoto S., et al. , Cellular fatty-acid composition in Pseudomonas species. J. Gen. Appl. Microbiol. 24, 199–213 (1978). [Google Scholar]
- 34.Kaneda T., Fatty acids in the genus Bacillus. I. Iso- and anteiso-fatty acids as characteristic constituents of lipids in 10 species. J. Bacteriol. 93, 894–903 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouzar H., Jones J. B., Hodge N. C., Differential characterization of Agrobacterium species using carbon-source utilization patterns and fatty acid profiles. Phytopathology 83, 733–739 (1993). [Google Scholar]
- 36.Basconcillo L. S., McCarry B. E., Comparison of three GC/MS methodologies for the analysis of fatty acids in Sinorhizobium meliloti: Development of a micro-scale, one-vial method. J. Chromatogr. B 871, 22–31 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Fischer E., Sauer U., Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270, 880–891 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Fischer E., Zamboni N., Sauer U., High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived constraints. Anal. Biochem. 325, 308–316 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Eisenberg R. C., Butters S. J., Quay S. C., Friedman S. B., Glucose uptake and phosphorylation in Pseudomonas fluorescens. J. Bacteriol. 120, 147–153 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauer U., The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279, 6613–6619 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Thomson J. F., Nance S. L., Bush K. J., Szczepanik P. A., Isotope and solvent effects of deuterium on aconitase. Arch Biochem. Biophys. 117, 65–74 (1966). [DOI] [PubMed] [Google Scholar]
- 42.O’Leary M. H., Limburg J. A., Isotope effect studies of role of metal-ions in isocitrate dehydrogenase. Biochemistry 16, 1129–1135 (1977). [DOI] [PubMed] [Google Scholar]
- 43.Lenz H., et al. , Stereochemistry of si-citrate synthase and ATP-citrate-lyase reactions. Eur. J. Biochem. 24, 207–215 (1971). [DOI] [PubMed] [Google Scholar]
- 44.Retey J., et al. , Stereochemical studies of exchange and abstraction of succinate hydrogen on succinate dehydrogenase. Eur. J. Biochem. 14, 232–242 (1970). [DOI] [PubMed] [Google Scholar]
- 45.Hermes J. D., Roesker C. A., O’Leary M. H., Cleland W. W., Use of multiple isotope effects to determine enzyme mechanisms and intrinsic isotope effects—malic enzyme and glucose-6-phosphate-dehydrogenase. Biochemistry 21, 5106–5114 (1982). [DOI] [PubMed] [Google Scholar]
- 46.Rendina A. R., Hermes J. D., Cleland W. W., Use of multiple isotope effects to study the mechanism of 6-phosphogluconate dehydrogenase. Biochemistry 23, 6257–6262 (1984). [DOI] [PubMed] [Google Scholar]
- 47.Ramachandran N., Durbano M., Colman R. F., Kinetic isotope effects in the NAD- and NADP-specific isocitrate dehydrogenases of pig heart. FEBS Lett. 49, 129–133 (1974). [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z., Chen L., Liu L., Su X., Rabinowitz J. D., Chemical basis for deuterium labeling of fat and NADPH. J. Am. Chem. Soc. 139, 14368–14371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergler H., Fuchsbichler S., Högenauer G., Turnowsky F., The enoyl-[acyl-carrier-protein] reductase (FabI) of Escherichia coli, which catalyzes a key regulatory step in fatty acid biosynthesis, accepts NADH and NADPH as cofactors and is inhibited by palmitoyl-CoA. Eur. J. Biochem. 242, 689–694 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Heath R. J., Su N., Murphy C. K., Rock C. O., The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J. Biol. Chem. 275, 40128–40133 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Gerosa L., et al. , Pseudo-transition analysis identifies the key regulators of dynamic metabolic adaptations from steady-state data. Cell Syst. 1, 270–282 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Haverkorn van Rijsewijk B. R. B., Kochanowski K., Heinemann M., Sauer U., Distinct transcriptional regulation of the two Escherichia coli transhydrogenases PntAB and UdhA. Microbiology 162, 1672–1679 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Zhao J., Shimizu K., Metabolic flux analysis of Escherichia coli K12 grown on -labeled acetate and glucose using GC-MS and powerful flux calculation method. J. Biotechnol. 101, 101–117 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Flamholz A., Noor E., Bar-Even A., Liebermeister W., Milo R., Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc. Natl. Acad. Sci. U.S.A. 110, 10039–10044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fogel M. L., Griffin P. L., Newsome S. D., Hydrogen isotopes in individual amino acids reflect differentiated pools of hydrogen from food and water in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 113, 4648–4653 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kern S. E., Newman D. K., “Measurement of phenazines in bacterial cultures” in Pseudomonas Methods and Protocols, A. Filloux, J.-L. Ramos, Eds. (Springer, New York, 2014), pp. 303–310. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez-Ruiz J., Belarbi E. H., Sanchez J., Alonso D. L., Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol. Tech. 12, 689–691 (1998). [Google Scholar]
- 59.Coplen T. B., Normalization of oxygen and hydrogen isotope data. Chem. Geol. 72, 293–297 (1988). [Google Scholar]
- 60.Dauner M., Sauer U., GC-MS analysis of amino acids rapidly provides rich information for isotopomer balancing. Biotechnol. Progr. 16, 642–649 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Nanchen A., Fuhrer T., Sauer U., Determination of Metabolic Flux Ratios from 13C-Experiments and Gas Chromatography-Mass Spectrometry Data: Protocol and Principles (Methods in Molecular Biology Series, Humana, Totowa, NJ, 2007), vol. 358, pp. 177–197. [DOI] [PubMed] [Google Scholar]
- 62.Zamboni N., Fendt S. M., Rühl M., Sauer U., .-based metabolic flux analysis. Nat. Protoc. 4, 878–892 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Zamboni N., Fischer E., Sauer U., FiatFlux–a software for metabolic flux analysis from -glucose experiments. BMC Bioinf. 6, 209 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.