Significance
A recent exciting development in cancer therapy involves using a patient’s own immune system to attack cancers. The attack is carried out by CD8+ T cells that destroy the malignant cells; after infiltrating tumors, however, these cells become hyporesponsive (“exhausted”) and exhibit diminished ability to control the tumors. In this study, we describe the transcriptional network that mediates CD8+ T cell exhaustion. We show that an initiating NFAT induces secondary transcription factors of the TOX and NR4A families, which are critical for CD8+ T cell exhaustion downstream of NFAT. We identify positive feedback loops connecting TOX and NR4A and suggest that diminished expression or activity of these transcription factors may be useful to improve effector function in cancer immunotherapy.
Keywords: TOX, TOX2, NR4A, NFAT, CD8+ T cell hyporesponsiveness
Abstract
T cells expressing chimeric antigen receptors (CAR T cells) have shown impressive therapeutic efficacy against leukemias and lymphomas. However, they have not been as effective against solid tumors because they become hyporesponsive (“exhausted” or “dysfunctional”) within the tumor microenvironment, with decreased cytokine production and increased expression of several inhibitory surface receptors. Here we define a transcriptional network that mediates CD8+ T cell exhaustion. We show that the high-mobility group (HMG)-box transcription factors TOX and TOX2, as well as members of the NR4A family of nuclear receptors, are targets of the calcium/calcineurin-regulated transcription factor NFAT, even in the absence of its partner AP-1 (FOS-JUN). Using a previously established CAR T cell model, we show that TOX and TOX2 are highly induced in CD8+ CAR+ PD-1high TIM3high (“exhausted”) tumor-infiltrating lymphocytes (CAR TILs), and CAR TILs deficient in both TOX and TOX2 (Tox DKO) are more effective than wild-type (WT), TOX-deficient, or TOX2-deficient CAR TILs in suppressing tumor growth and prolonging survival of tumor-bearing mice. Like NR4A-deficient CAR TILs, Tox DKO CAR TILs show increased cytokine expression, decreased expression of inhibitory receptors, and increased accessibility of regions enriched for motifs that bind activation-associated nuclear factor κB (NFκB) and basic region-leucine zipper (bZIP) transcription factors. These data indicate that Tox and Nr4a transcription factors are critical for the transcriptional program of CD8+ T cell exhaustion downstream of NFAT. We provide evidence for positive regulation of NR4A by TOX and of TOX by NR4A, and suggest that disruption of TOX and NR4A expression or activity could be promising strategies for cancer immunotherapy.
The goal of cancer immunotherapy is to harness the immune system to destroy tumors in cancer patients. Two approaches have achieved remarkable success: “immune checkpoint blockade” therapies involving treatment of cancer patients with blocking antibodies to inhibitory cell surface receptors, including the cytotoxic T lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1), and the programmed cell death protein 1 ligand (PD-L1) (1, 2); and the use of T cells expressing chimeric antigen receptors (CARs) that recognize tumor antigens (3–5). Whereas anti–CTLA-4 seems to act by depleting intratumoral regulatory T cells (1, 2), antibodies to PD-1 or PD-L1 act by overcoming a hyporesponsive state, termed “exhaustion” or “dysfunction,” that develops downstream of PD-1 in tumor-infiltrating CD8+ T cells (6). Exhausted CD8+ T cells exhibit decreased effector function (decreased cytokine production and cytolytic activity) and up-regulate numerous inhibitory receptors, including PD-1, CTLA-4, T cell immunoglobin and mucin-domain containing-3 (TIM3), lymphocyte activation gene 3 (LAG3), and T cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT). Anti–PD-1/PD-L1 therapy not only “rejuvenates” the exhausted CD8+ T cells themselves but also, allows expansion of a more “stem-like” CD8+ T cell population that expresses the transcription factor T cell factor 1 (TCF1) (7–9). However, only a subset of patients achieves complete remission with checkpoint blockade therapies, a problem that can potentially be countered by using combinations of antibodies to multiple inhibitory receptors (1, 2). Likewise, CAR T cell therapy has been remarkably effective against hematopoietic cancers, such as B cell chronic lymphocytic leukemia, but it has not been very effective against solid tumors, apparently because the CAR T cells become exhausted, much like T cells responsive to standard peptide/major histocompatibility complex ligands (5, 6). The malignant cells can escape surveillance by down-regulating the tumor antigen recognized by the CAR.
Several mouse models of CD8+ T cell hyporesponsiveness (here termed “exhaustion”) have been described (10). In each case, exhausted cells are defined—as in humans—by diminished production of the cytokines interferon-γ (IFN-γ), tumor necrosis factor (TNF), and interleukin-2 and increased expression of inhibitory receptors, including PD-1, TIM3, and LAG3. The model systems include chronic infection with lymphocytic choriomeningitis virus (LCMV) (11–13), antitumor responses to transplanted (14) or spontaneously arising tumors (14–17), and a CAR T cell mouse model of antitumor responses previously developed in our laboratory (18) in which mice were inoculated with tumors expressing human CD19 (hCD19) and adoptively transferred with CD8+ T cells expressing a second-generation CAR against hCD19.
We previously used our CAR T cell model to show that CAR T cells lacking all three members of the NR4A nuclear receptor family of transcription factors (NR4A1, NR4A2, and NR4A3) were far more effective at suppressing the growth of hCD19+ tumors compared with wild-type (WT) CAR T cells (18). In this study, we show that, similarly, CAR-expressing tumor-infiltrating T cells [tumor-infiltrating lymphocytes (TILs)] deficient in two high-mobility group (HMG)-box transcription factors, TOX and TOX2, are more effective at promoting hCD19+ tumor regression compared with WT CAR TILs or CAR TILs singly deficient in either TOX or TOX2 alone. We have also defined the role of TOX and NR4A transcription factors in the transcriptional network that mediates CD8+ T cell exhaustion. Briefly, we show that TOX and TOX2 as well as NR4A family members are highly induced in CD8+ CAR+ PD-1high TIM3high (“exhausted”) TILs by the calcium/calcineurin-regulated transcription factor NFAT, even in the absence of its partner AP-1 (FOS-JUN). Tox DKO CAR TILs resemble NR4A-deficient CAR TILs in showing increased cytokine expression and decreased expression of inhibitory receptors; they also display increased accessibility of chromatin regions that are enriched for motifs that bind nuclear factor κB (NFκB) and basic region leucine zipper transcription factors, which are classically associated with T cell activation and effector function. Together, these data indicate that TOX and NR4A transcription factors are critical for the transcriptional program of CD8+ T cell exhaustion downstream of NFAT. Finally, we show that NR4A and TOX transcription factors positively regulate each other’s expression. Interfering with TOX and NR4A expression or activity could be a promising strategy for cancer immunotherapy.
Results and Discussion
Striking Up-Regulation of TOX and NR4A Family Transcription Factors in Multiple Models of Exhaustion.
In all published comparisons of RNA-sequencing (RNA-seq) data from exhausted vs. control CD8+ T cells, we observed consistent up-regulation of messenger RNAs (mRNAs) encoding the HMG-box transcription factors TOX, TOX2, and one or more nuclear receptors belonging to the NR4A family (NR4A1, NR4A2, and NR4A3) (SI Appendix, Fig. S1). Tox, Tox2, and Nr4a2 mRNAs were up-regulated in exhausted compared with effector cells in the LCMV model (13), and Tox, Tox2, and all Nr4a mRNAs were up-regulated in exhausted compared with naïve CD8+ T cells (12) (SI Appendix, Fig. S1A). Similarly, Tox, Nr4a2, and Nr4a3 mRNAs were up-regulated in MHC class I-restricted, ovalbumin-specific compared with LCMV GP33-41–specific H-2Db–restricted TCR TILs infiltrating a B16-OVA melanoma (14), and Tox, Tox2, and all Nr4a mRNAs were up-regulated in CD8+ T cells infiltrating a malignant liver lesion compared with CD8+ T cells responding to acute Listeria infection (SI Appendix, Fig. S1B). In our CAR T cell model (18), Tox, Tox2, and Nr4a2 mRNAs were up-regulated in PD-1highTIM3high CAR TILs compared with endogenous PD-1lowTIM3low TILs (SI Appendix, Fig. S1C).
We also examined cells expressing CA-RIT-NFAT1, a constitutively active version of NFAT1 that cannot interact with AP-1 (19, 20); CA-RIT-NFAT1 induces a transcriptional program very similar to exhaustion both in vitro and in vivo (20, 21), whereas another mutated version of CA-RIT-NFAT1 that cannot bind DNA (DBD mut) does not (21). Notably, Tox, Tox2, Nr4a2, and Nr4a3 mRNAs were up-regulated in CD8+ T cells retrovirally expressing CA-RIT-NFAT1 compared with cells expressing the DBD mut (21) (SI Appendix, Fig. S1D). These data indicate that TOX and NR4A transcription factors can both be induced in exhausted CD8+ T cells by NFAT in the absence of AP-1. Finally, human CD8+ T cells that had infiltrated patient melanomas and were expressing high levels of TOX and TOX2 mRNAs also expressed high levels of PDCD1, HAVCR2, and LAG3 mRNAs (SI Appendix, Fig. S1E); the PD-1highTIM3high subset of these human TILs cells also show increased expression of NR4A mRNAs (18).
Strong Positive Correlation of TOX and TOX2 Expression with PD-1 Expression on CAR TILs.
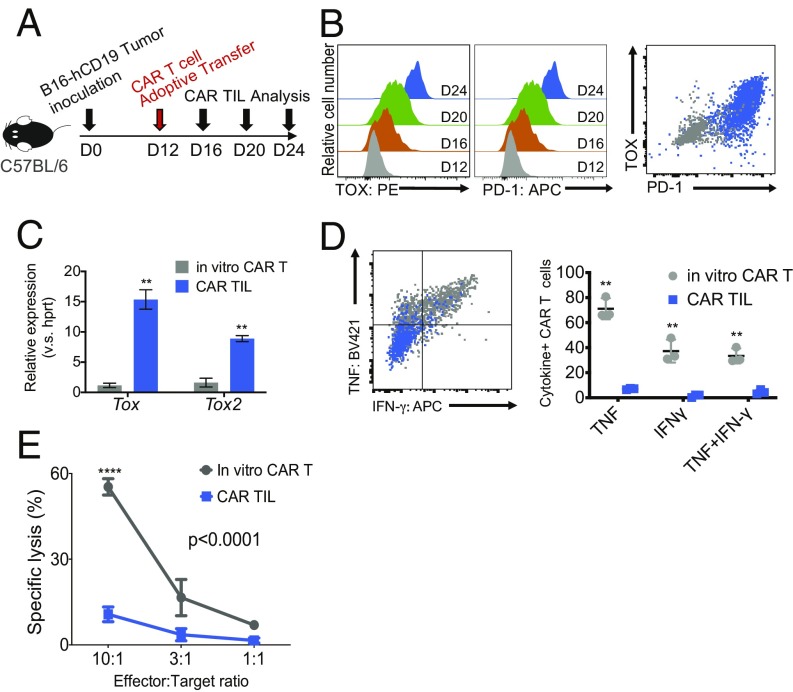
We previously showed that NR4A family members were key transcriptional effectors of the CD8+ T cell exhaustion program downstream of NFAT (18). Given the increase in Tox and Tox2 mRNA expression in exhausted CD8+ T cells in all of the above models (SI Appendix, Fig. S1), we examined the roles of TOX and TOX2 in CD8+ T cell exhaustion. CAR T cells were transferred into C57BL/6 mice bearing B16-OVA-hCD19 tumors (hereafter termed B16-hCD19 tumors) 12 d after tumor inoculation, and CAR TILs in the tumor were analyzed 4, 8, and 12 d later (days 16, 20, and 24) (Fig. 1A). The CAR TILs showed a striking increase in both TOX and PD-1 expression over time (Fig. 1B). Compared with retrovirally transduced (pretransfer) CAR T cells (“in vitro CAR T cells”), CAR TILs showed high expression of mRNAs encoding both Tox and Tox2 at day 24 (Fig. 1C) (there are no good antibodies for staining of TOX2). Compared with in vitro CAR T cells, CAR TILs at day 24 also showed strikingly diminished production of the effector cytokines IFN-γ and TNF after in vitro stimulation with EL4-hCD19 cells (Fig. 1D) as well as decreased target cell lysis of MC38-hCD19 cells (Fig. 1E). These data emphasize that TOX protein and Tox and Tox2 mRNA expression are highly induced in CD8+ CAR TILs and that their induction correlates strongly with PD-1 expression and the acquisition of a “dysfunctional” (i.e., hyporesponsive) phenotype of diminished cytokine production and cytolytic activity.
Fig. 1.
TOX transcription factors are highly expressed in exhausted CAR TILs in solid tumors. (A) Experimental scheme for analyzing tumor-infiltrating CAR T cells (CAR TILs); 5 × 105 B16-hCD19 tumor cells were inoculated subcutaneously into C57BL/6 mice. Twelve days later, 1.5 × 106 CAR T cells were adoptively transferred into the tumor-bearing mice by intravenous injection. CAR TILs were isolated every 4 d after CAR T cell transfer. (B–E) Gray color (dots, histograms, and shading) indicates T cells retrovirally transduced with the CAR and analyzed in vitro, and the orange, green, and blue colors indicate CAR TILs analyzed the indicated number of days after CAR T cell transfer. (B, Left and Center) Expression of TOX and PD-1 was analyzed by flow cytometry (PE: phycoerythrin, APC: allophycocyanin). Shown are histograms for TOX and PD-1 expression of CAR TILs on days 12, 16, 20, and 24 after tumor inoculation; “in vitro CAR T” refers to CAR T cells before adoptive transfer. (B, Right) Combined flow cytometry plot showing PD-1 and TOX expression on in vitro-transduced CAR T cells (gray) as well as CAR TILs isolated on day 24 (blue). (C) mRNA expression levels of Tox and Tox2 (relative to Hprt) in bulk CAR TILs on day 24. (D, Left) Representative flow cytometry plot showing TNF and IFN-γ expression after EL4-hCD19 cells restimulation in in vitro-transduced CAR T cells (gray) and CAR TILs isolated on day 24 (blue). (D, Right) Quantification of cytokine production (three mice per group). The data from C and D were analyzed by Student’s t test. **P ≤ 0.01. (E) In vitro-transduced CAR T cells and CAR TILs were cocultured with tumor cells expressing MC38-hCD19, and target cell lysis was measured 5 h later. The data are representative of two biologically independent experiments. The data from the two groups in E were analyzed by two-way ANOVA with a Bonferroni multiple comparisons test. ****P ≤ 0.0001.
CAR T Cells Doubly Deficient in TOX and TOX2 Show Enhanced Antitumor Effects.
To assess the potentially redundant roles of TOX and TOX2 in CAR TIL function in vivo, we transduced CD8+ T cells from WT or Tox2−/− mice with the CAR retrovirus as well as with four different short hairpin RNAs (shRNAs) against Tox (SI Appendix, Fig. S2 A and B). This strategy was preferred to the use of Tox−/− Tox2−/− mice, because Tox−/− mice have a profound defect in CD4+ T cell development (22). We pooled the four shRNAs before transduction, because this strategy gave the most effective knockdown of Tox mRNA (SI Appendix, Fig. S2C). Tox mRNA levels were substantially diminished in shTOX-transduced cells (SI Appendix, Fig. S2D).
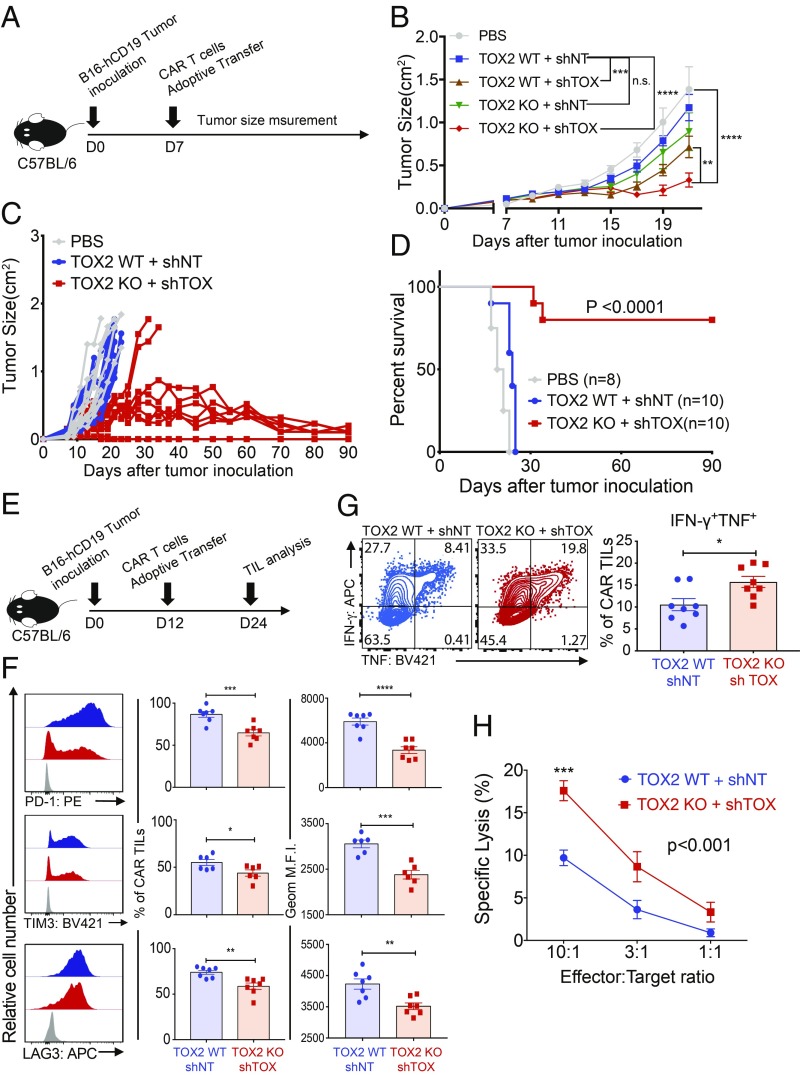
We compared tumor growth in C57BL/6 (immunocompetent) mice bearing B16-hCD19 tumors after injection with phosphate-buffered saline (PBS) or adoptive transfer with CAR T cells (Fig. 2A). The CAR T cells were from WT or Tox2−/− mice and transduced with nontargeting shRNA (shNT) or shRNA to deplete TOX (shTOX) (SI Appendix, Fig. S2A), yielding the four groups WT + shNT, WT + shTOX, TOX2 knockout (KO), and TOX2 KO + shTOX (Fig. 2B). The mice receiving Tox DKO CAR T cells showed more significant tumor regression than any of the other three groups (Fig. 2B), indicating that double deficiency of both TOX and TOX2 in CAR TILs is more effective at slowing tumor growth than individual deficiency of either TOX or TOX2 alone. For all subsequent experiments, we compared Tox DKO CAR T cells with WT CAR T cells without analyzing CAR T cell individually deficient in TOX or TOX2. When compared directly with mice receiving control (WT + shNT) CART cells in longer-term experiments, a striking proportion of mice receiving Tox DKO CAR TILs showed complete regression of B16-hCD19 tumors by ∼90 d (Fig. 2 C and D).
Fig. 2.
CAR TILs with combined deficiency of TOX and TOX2 (Tox DKO CAR TILs) promote tumor regression and prolong survival of tumor-bearing mice. (A) Experimental scheme for monitoring tumor growth and survival; 5 × 105 B16-hCD19 tumor cells were inoculated, and 3 × 106 CAR T cells were adoptively transferred 7 d later. Tumor sizes were measured by calipers every 2 d. (B) CAR T cells deficient in both TOX and TOX2 were the most efficient at controlling tumor growth. (C) Time course of tumor growth in individual mice adoptively transferred with WT or Tox DKO CAR T cells. (D) Kaplan–Meier curves showing survival of each group of mice over time. The data from D were analyzed using a log rank Mantel–Cox test. (E) Experimental scheme for phenotypic analysis; 5 × 105 tumor cells were inoculated into C57BL/6 mice, the mice were adoptively transferred with the indicated CAR T cells (1.5 × 106) 12 d later, and CAR TILs were isolated on day 24. (F) Expression of the inhibitory receptors PD-1, TIM3, and LAG3 was analyzed by flow cytometry. Gray histograms show staining with isotype control antibody. (G) CAR TILs were restimulated with phorbol 12-myristate 13-acetate (PMA)/ionomycin, and expression of TNF and IFN-γ was analyzed by intracellular staining and flow cytometry. The data from F and G were analyzed by Student’s t test. (H) To measure the cytolytic activity of CAR TILs in vitro, the indicated CAR TILs were pooled from 10 Rag1−/− mice bearing B16-hCD19 tumors 12 d after adoptive transfer. Cytolytic activity was assessed by coculture with MC38-hCD19 tumor cells as targets. In all bar graphs, each dot represents CAR TILs from a single recipient mouse. The data from B and H were analyzed by two-way ANOVA with a Bonferroni multiple comparisons test. The data are obtained from two independent biological experiments. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; n.s. = not significant.
To analyze the phenotypic and genome-wide changes in Tox DKO and WT CAR T cells and correlate them with antitumor responses, we transferred CAR T cells 12 d after tumor inoculation and isolated CAR TILs 12 d later (Fig. 2E and SI Appendix, Fig. S4A); this modified protocol allowed us to isolate a sufficient number of CAR TILs for genome-wide assessment of gene transcription and chromatin accessibility. At day 24 after tumor inoculation, there was a mild increase in the percentage and number of Tox DKO compared with WT CAR TILs, which did not reach statistical significance (SI Appendix, Fig. S4B), but no significant increase in Ki67 expression (SI Appendix, Fig. S4C). In contrast, expression of the inhibitory receptors PD-1, TIM3, and LAG3 was significantly decreased in Tox DKO CAR TILs compared with WT CAR TILs (Fig. 2F) based on geometric mean fluorescence intensity (MFI) calculated over the entire population. Moreover, in tests of effector function, we found that the percentage of cells coexpressing IFN-γ and TNF was significantly higher in Tox DKO CAR TILs compared with WT TILs (Fig. 2G). Finally, expression of the transcription factors TCF1 (SI Appendix, Fig. S4D) and EOMES (SI Appendix, Fig. S4E) was decreased while T-bet expression was slightly but not significantly increased in Tox DKO CAR TILs compared with WT CAR TILs; TCF1+ T cells are thought to represent the CD8+ T cell subset that recovers effector function after PD-1/PD-L1 blockade (7–9). Thus, the decrease of TCF1-expressing cells observed in Tox DKO compared with WT CAR TILs (SI Appendix, Fig. S4D) indicates that the reversal of the exhausted phenotype in Tox DKO CAR TILs may not involve their reversion to a stem-like state characterized by increased TCF1.
To assess the cytolytic activity of the CAR TILs, we used Rag1−/− mice to avoid interference from endogenous CD8+ T cells. We transferred Tox DKO and WT CAR T cells into Rag1−/− mice 12 d after inoculation with B16-hCD19 tumor cells, pooled CAR TILs from 10 to 12 recipient mice, and cocultured them with MC38-hCD19 tumor cells as targets. Consistent with the hypothesis that TOX restrains CD8+ effector function, the cytolytic activity of Tox DKO CAR TILs was significantly greater than that of WT CAR TILs (Fig. 2H).
The Transcriptional Interplay Among Transcription Factors of the NFAT, TOX, and NR4A Families.
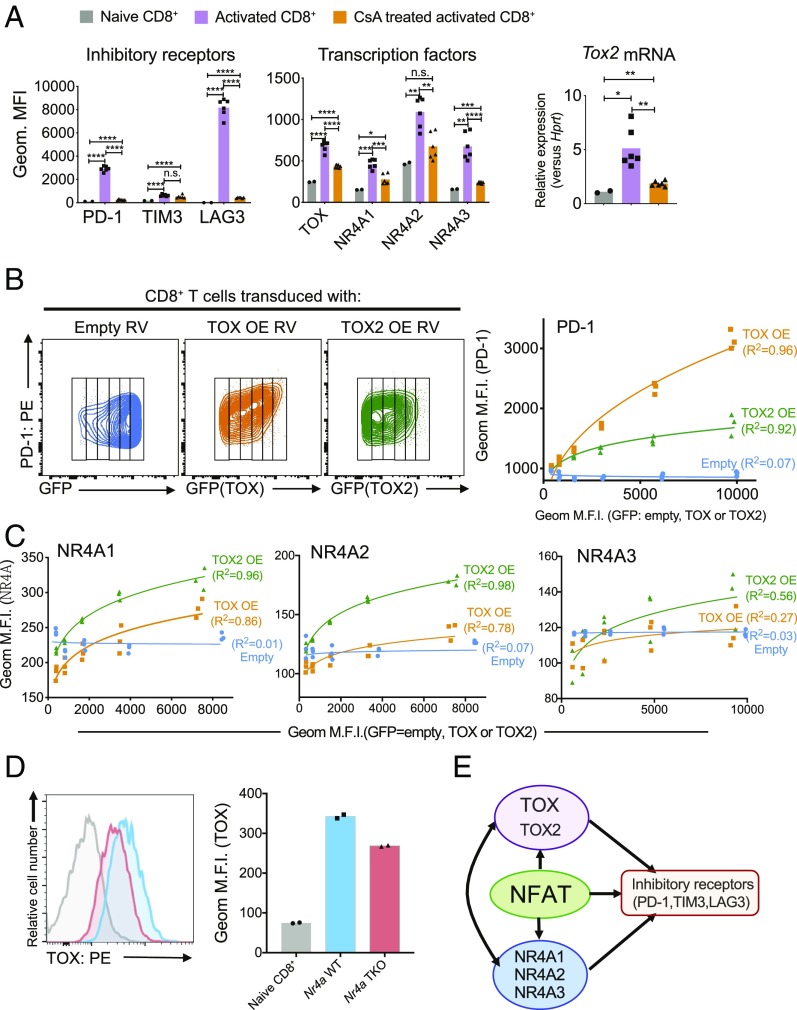
We next explored the potential functional interplay among NFAT, NR4A, and TOX transcription factors (Fig. 3). The inhibitory receptors PD-1 and LAG3 and the transcription factors TOX, NR4A1, NR4A2, and NR4A3 were all induced at the protein level in CD8+ T cells acutely activated for either 16 or 60 h with anti-CD3 and anti-CD28 as was mRNA encoding TOX2 (Fig. 3A and SI Appendix, Fig. S5A, compare gray and purple bars). We used the calcineurin inhibitor cyclosporin A (CsA) to assess the involvement of NFAT transcription factors, which are induced by calcium influx and calcineurin activation (23, 24). With the exception of TIM3, in which expression was not significantly different in cells activated in the presence or absence of CsA, induction of all surface receptors and TOX and NR4A transcription factors was either completely or partially inhibited by CsA (Fig. 3A and SI Appendix, Fig. S5A, compare purple and orange bars).
Fig. 3.
NFAT, TOX, and NR4A transcription factors cooperate to control the expression of inhibitory receptors on CD8+ T cells and positive regulatory loops connecting TOX and NR4A. (A) Splenic CD8+ T cells from C57BL/6 mice were incubated with or without CsA for 30 min and then stimulated with anti-CD3 and anti-CD28 for 60 h. The expression of (Left) inhibitory receptors PD-1, TIM3, and LAG3 and (Center) transcription factors TOX, NR4A1, NR4A2, and NR4A3 was analyzed by flow cytometry. (Right) The expression of Tox2 mRNA was measured by qPCR and normalized to the level of Hprt mRNA expression. Naïve CD8+ cell from splenic CD8+ cells are used as a control. The data were obtained from two biological experiments. (B and C) The indicated retroviruses (RVs), either empty or encoding TOX or TOX2 [TOX overexpression (TOX OE) and TOX2 overexpression (TOX2 OE)], were used to transduce splenic CD8+ T cells from C57BL/6 mice, after which PD-1 (B) as well as NR4A1, NR4A2, and NR4A3 (C) expression levels were analyzed by flow cytometry as a function of TOX or TOX2 expression estimated as GFP expression from an IRES-GFP cassette (B, Left). Geometric MFIs of PD-1 (B, Right) and NR4A1, NR4A2, and NR4A3 (C) were plotted in the graphs for each increment of GFP (i.e., TOX or TOX2) expression. Blue, empty RV; orange, TOX OE RV; green, TOX2 OE RV. Transduction with empty retrovirus does not change the levels of PD-1 or NR4A expression, whereas transduction with TOX or TOX2 retrovirus leads to a significant increase. The data are representative of two biologically independent experiments. (D) TOX expression was analyzed by flow cytometry in Nr4a WT (blue) or Nr4a TKO (red) CD8+ T cells. Naïve CD8+ T cells were used as a control. (E) Schematic illustrating the proposed roles of NFAT, NR4A, and TOX/TOX2 transcription factors in CD8+ T cells. Each dot is representative of CD8+ T cells from individual mice. The data are representative of two biologically independent experiments. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; n.s. = not significant. PE = phycoerythrin.
To assess the role of TOX factors, we retrovirally transduced naïve CD8+ T cells with empty viruses or viruses encoding TOX or TOX2 and bearing an IRES-GFP cassette, and we evaluated the expression of inhibitory receptors and NR4A family proteins 5 d later (Fig. 3 B and C and SI Appendix, Fig. S5 B and C). To ensure quantitative analysis of the flow cytometry data, we divided the flow cytometry plot into “slices” representing different levels of TOX or TOX2 expression (Fig. 3 B, Left) and calculated the geometric MFI of expression of inhibitory receptors and NR4A family proteins in each slice (Fig. 3 B and C and SI Appendix, Fig. S5C). The data showed unambiguously that TOX and TOX2 were both individually capable of inducing expression of PD-1 (Fig. 3B and SI Appendix, Fig. S5B), TIM3 (SI Appendix, Fig. S5C), LAG3 (SI Appendix, Fig. S5C), and all three NR4A proteins (Fig. 3C). TOX was more effective than TOX2 at inducing PD-1 expression (Fig. 3B); the two TOX proteins almost equivalently induced TIM3 and LAG3 expression (SI Appendix, Fig. S5C), and TOX2 was more effective than TOX at inducing NR4A1, NR4A2, and NR4A3 expression, although the effect of TOX2 on NR4A3 expression was very mild (Fig. 3C). Finally, by measuring TOX protein expression in activated CD8+ T cells from WT and Nr4a TKO mice (18), we showed that NR4A proteins partly contribute to the induction of TOX expression (Fig. 3D); similarly, by expressing TOX and TOX2 in CD8+ T cells from WT and Nr4a TKO mice (18), we showed that NR4A and TOX proteins both contribute to PD-1 expression, with TOX2 again having a lesser effect (SI Appendix, Fig. S5D).
Together, these data point to a model in which NFAT induces TOX, TOX2, and NR4A family proteins; all three transcription factor families contribute to the expression of inhibitory receptors, and there is clear evidence of positive feedback regulation between NR4A and TOX (Fig. 3E).
Transcriptional Profiling and Chromatin Accessibility Landscape of Tox DKO CAR TILs.
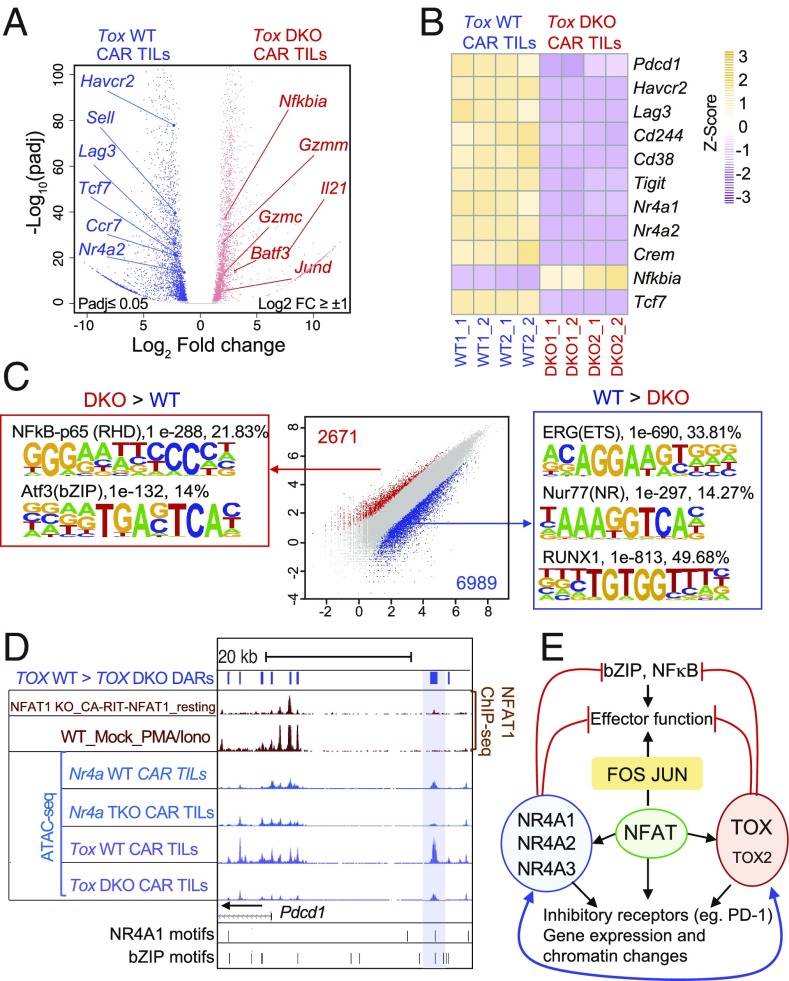
To confirm these findings at a genome-wide level in vivo, we assessed the transcriptional and chromatin accessibility of WT and Tox DKO CAR TILs by RNA-seq and Assay for Transposase Accessible Chromatin with high-throughput sequencing (ATAC-seq), respectively (Fig. 4; principal component analysis and hierarchical clustering of the RNA-seq samples are shown in SI Appendix, Fig. S6A, and genome browser views of differentially accessible ATAC-seq peaks in the Tox and Tox2 loci are shown in SI Appendix, Fig. S6 B and C). Consistent with their increased ability to promote tumor regression (Fig. 2), Tox DKO CAR TILs showed evidence of increased effector function compared with WT CAR TILs as judged by decreased expression of multiple inhibitory receptors (Pdcd1, Havcr2, Lag3) and increased expression of granzymes, Il21, and multiple bZIP transcription factors (Fig. 4 A and B and SI Appendix, Table S1). Notably, Tox DKO CAR TILs also showed decreased expression of Nr4a1 and Nr4a2 compared with WT CAR TILs (Fig. 4 A and B), confirming the cross-regulation of NR4A by TOX observed in cultured cells (Fig. 3).
Fig. 4.
Gene expression and chromatin accessibility profiles of Tox DKO CAR TILs compared with WT CAR TILs. (A) Volcano plots of genes differentially expressed in WT compared with Tox DKO CAR TILs. Selected differentially expressed genes with an adjusted P value ≤ 0.05 and log2 fold change >1 or −1 are indicated. (B) Heat maps (transcripts per kilobase million normalization with z score) showing expression of representative genes in WT vs. Tox DKO CAR TILs in individual replicates. (C) Scatterplot of pairwise comparisons of ATAC-seq density (Tn5 insertions per kilobase) in WT vs. Tox DKO CAR TILs. Differentially accessible regions and associated de novo motif analysis are shown. (D) Genome browser view of the Pdcd1 locus incorporating ChIP-seq (NFAT1, KO_CA-RIT-NFAT1_resting and WT_Mock_PMA_Iono; GSE64409) and ATAC-seq (Nr4a WT, Nr4a TKO CAR TILs; GSE123739 and Tox WT, Tox DKO CAR TILs) samples. The blue bar shows the “exhaustion-specific” enhancer located ∼23 kb 5′ of the Pdcd1 transcription start site. DARs = differentially accessible regions; PMA/Iono = phorbol 12-myristate 13-acetate/ionomycin. (E) Diagram illustrating the proposed involvement of NFAT, NR4A, and TOX proteins in the transcriptional program of CD8+ T cell exhaustion. The RNA-seq and ATAC-seq samples were obtained from two independent biological experiments.
Analysis of ATAC-seq data revealed enrichment for consensus nuclear receptor binding motifs, Runt [Runt-related transcription factor (RUNX)-binding] motifs, and E26 transformation-specific (ETS) motifs in regions more accessible in WT compared with Tox DKO CAR TILs (Fig. 4 C, Center, blue dots and Right; more information about the de novo motif analysis is in SI Appendix, Table S2). Thus, accessible regions lost in Tox DKO CAR TILs include regions capable of binding nuclear receptors of the NR4A and RUNX families in WT CAR TILs. The depletion of NR4A binding motifs in Tox DKO compared with WT CAR TILs is consistent with the positive regulation of NR4A protein expression by TOX and TOX2 (Fig. 3C). The depletion of RUNX-binding motifs (Fig. 4 C, Center, blue dots and Right) is potentially consistent with a previous report showing increased levels of RUNX1 and RUNX3 in CD8 single-positive thymocytes of TOX-transgenic mice (22). However, neither Tox DKO nor Nr4a TKO CAR TILs showed a significant decrease in Runx1 and Runx3 mRNA compared with WT CAR TILs. Furthermore, in a comparable mouse model of antitumor responses, RUNX3 deficiency impaired TIL accumulation (25). In our hands, however, Tox DKO CAR TILs were recovered at similar levels compared with WT CAR TILs at day 12 after transfer (SI Appendix, Fig. S4B). The functional consequence of the loss of ETS-binding motifs is not clear but is under investigation in our laboratory.
Notably, regions that were more accessible in TOX/TOX2-deficient CAR TILs compared with the WT were enriched for binding motifs for NFκB and bZIP transcription factors that are classically activated in stimulated effector T cells (Fig. 4 C, Left and Center, red dots). A similar enrichment for NFκB and bZIP consensus motifs was also observed in regions with increased accessibility in Nr4a TKO compared with WT CAR TILs (18). Although the actual identities of the bZIP and NFκB/Rel transcription factors that occupy these differentially accessible sites are not known, both Tox DKO and Nr4a TKO CAR TILs show increased expression of Batf and Jund mRNAs relative to WT CAR TILs, suggesting that further investigation of the roles of the bZIP transcription factors BATF and JUND in CD8+ T cell exhaustion would be worthwhile. The picture with regard to NFκB is more complex, because Nfkbia mRNA encoding the NFκB inhibitor IκB is increased in Tox DKO (but not Nr4a TKO) compared with WT CAR TILs, a result apparently inconsistent with the increased accessibility of consensus NFκB binding motifs in these cells. Additional analysis is needed to resolve this point.
The potential interplay among NFAT, NR4A, and TOX transcription factors is illustrated in a genome browser view of the −23-kb Pdcd1 enhancer (12, 18, 26); the enhancer shows decreased accessibility in Nr4a TKO and Tox DKO CAR TILs compared with WT CAR TILs, and chromatin immunoprecipitation sequencing (ChIP-seq) analyses show that it is capable of binding CA-RIT-NFAT1 (albeit very weakly) in retrovirally transduced NFAT1-deficient cells (21) (Fig. 4D). This enhancer does not, however, bind endogenous NFAT1 in activated, nonexhausted cells as judged by NFAT1 ChIP-seq. We confirmed the binding of ectopically expressed, 3xflag-tagged TOX and TOX2 proteins to this “exhausted related” Pdcd1 enhancer by ChIP and qPCR (SI Appendix, Fig. S7). We propose that this region represents an “enhanceosome” (27), which in exhausted CD8+ T cells, is capable of cooperatively binding members of all three families of transcription factors: NFAT, TOX, and NR4A. ChIP is currently not sensitive enough to prove this point formally in the small numbers of experimentally available TILs.
In summary, we have demonstrated that CD8+ T cell exhaustion involves a striking functional interplay with pronounced positive feedback loops among transcription factors of the NFAT, TOX, and NR4A families (Fig. 4E). Interference with the expression or function of TOX and NR4A or with the ability of NFAT to induce these transcription factors in the absence of AP-1 may prove to be a valuable therapeutic strategy for cancer immunotherapy in the future.
Materials and Methods
Mice.
C57BL/6J, B6.SJL-PtprcaPepcb/BoyJ, and Rag1−/− mice were purchased from The Jackson Laboratories. C57BL/6N mice were purchased from Charles River Laboratories. The Tox2−/− gene-disrupted mouse strain was generated in the A.B. laboratory by deleting exons 3–7, which contain the HMG-box domain. The Tox2−/− mice were generated in the A.B. laboratory (National Cancer Institute, NIH). Nr4a gene-disrupted strains (18, 28) were obtained from Takashi Sekiya, National Center for Global Health and Medicine in Japan, Chiba, Japan, and Akihiko Yoshimura, Keio University School of Medicine, Tokyo, Japan, with permission from P. Chambon, University of Strasbourg, Strasbourg, France. Mice were age matched and used for experiments when they were between 8 and 12 wk old. Both female and male mice were used for experiments. All mice were bred and maintained in the animal facility at La Jolla Institute for Immunology (LJI). All experiments were performed in compliance with the LJI Institutional Animal Care and Use Committee guidelines.
Data Availability.
RNA-seq and ATAC-seq data are available in the Gene Expression Omnibus database under the SuperSeries reference number (GSE130540).
Additional materials and methods are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank C. Kim, M. Haynes, S. Sehic, D. Hinz, and C. Dillingham of the LJI Flow Cytometry Core Facility for cell sorting; J. Day, K. Tanguay, and C. Kim of the LJI Next Generation Sequencing Facility for next generation sequencing; S. Schoenberger for B16-OVA cells; A. W. Goldrath for MC-38 cells; and the Department of Laboratory Animal Care and the animal facility for excellent support. This work was funded by NIH Grants AI108651 (to L.-F.L.), AI140095 (to L.-F.L.), the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH (A.D., Y.H.W., and A.B.), AI109842 (to P.G.H. and A.R.), and AI040127 (to P.G.H. and A.R.); and NIH S10 Equipment Grants OD016262 (to LJI) and RR027366 (to LJI). H.S. was supported by American Association for Cancer Research-Genentech Immuno-oncology Research Fellowship 18-40-18-SEO and the Donald J. Gogel Cancer Research Irvington Fellowship. J.C. was supported by NIH T32 Predoctoral Training Grant GM007752 and a PhRMA Foundation Paul Calabresi Medical Student Research Fellowship. E.G.-A., D.S.-C., and I.F.L.-M. were supported by fellowships from the University of California Institute for Mexico and the United States and El Consejo Nacional de Ciencia y Tecnología (UCMEXUS/CONACYT).
Footnotes
Conflict of interest statement: The La Jolla Institute of Immunology has a pending patent, PCT/US2018/062354, covering the use and production of engineered immune cells to disrupt NFAT-AP1 pathway transcription factors, including TOX and NR4A family members, with H.S., J.C., and A.R. listed as inventors.
Data deposition: The ATAC-seq and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE130540).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905675116/-/DCSupplemental.
References
- 1.Sharma P., Allison J. P., Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 161, 205–214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian S. L., Drake C. G., Pardoll D. M., Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maude S. L., et al. , Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davila M. L., et al. , Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 6, 224ra25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.June C. H, O’Connor R. S., Kawalekar O. U., Ghassemi S., Milone M. C., CAR T cell immunotherapy for human cancer. Science 37, 457–495 (2018). [DOI] [PubMed] [Google Scholar]
- 6.McLane L. M., Abdel-Hakeem M. S., Wherry E. J., CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Im S. J., et al. , Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Utzschneider D. T., et al. , T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Wu T., et al. , The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pereira R. M., Hogan P. G., Rao A., Martinez G. J., Transcriptional and epigenetic regulation of T cell hyporesponsiveness. J. Leukoc. Biol. 102, 601–615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sen D. R., et al. , The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauken K. E., et al. , Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott-Browne J. P., et al. , Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45, 1327–1340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mognol G. P., et al. , Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl. Acad. Sci. U.S.A. 114, E2776–E2785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schietinger A., Greenberg P. D., Tolerance and exhaustion: Defining mechanisms of T cell dysfunction. Trends Immunol. 35, 51–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schietinger A., et al. , Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity 45, 389–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philip M., et al. , Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J., et al. , NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macián F., García-Rodríguez C., Rao A., Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 19, 4783–4795 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macián F., et al. , Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109, 719–731 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Martinez G. J., et al. , The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity 42, 265–278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aliahmad P., Kaye J., Development of all CD4 T lineages requires nuclear factor TOX. J. Exp. Med. 205, 245–256 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan P. G., Chen L., Nardone J., Rao A., Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17, 2205–2232 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Hogan P. G., Lewis R. S., Rao A., Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu. Rev. Immunol. 28, 491–533 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milner J. J., et al. , Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sen D. R., et al. , The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carey M., The enhanceosome and transcriptional synergy. Cell 92, 5–8 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Sekiya T., et al. , Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 14, 230–237 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ATAC-seq data are available in the Gene Expression Omnibus database under the SuperSeries reference number (GSE130540).
Additional materials and methods are provided in SI Appendix, Materials and Methods.