Significance
SR9009 has been described as an agonist for circadian nuclear receptors REV-ERBα and -β, and its pleotropic biological effects have been attributed to REV-ERBs in numerous publications, as well as websites selling the compound illicitly to the lay public. Here, a conditional REV-ERBα/β double-deletion mouse model was generated and used to test the specificity of SR9009 and a related compound. SR9009 was found to exert REV-ERB–independent effects on proliferation, metabolism, and gene transcription in 2 different cell types depleted of REV-ERBs. Therefore, effects of these drugs cannot be exclusively attributed to its actions on REV-ERBs, and it is imperative that investigators using the drug be made aware of this.
Keywords: REV-ERB, circadian rhythms, SR9009, ligand, specificity
Abstract
The nuclear receptors REV-ERBα and -β link circadian rhythms and metabolism. Like other nuclear receptors, REV-ERB activity can be regulated by ligands, including naturally occurring heme. A putative ligand, SR9009, has been reported to elicit a range of beneficial effects in healthy as well as diseased animal models and cell systems. However, the direct involvement of REV-ERBs in these effects of SR9009 has not been thoroughly assessed, as experiments were not performed in the complete absence of both proteins. Here, we report the generation of a mouse model for conditional genetic deletion of REV-ERBα and -β. We show that SR9009 can decrease cell viability, rewire cellular metabolism, and alter gene transcription in hepatocytes and embryonic stem cells lacking both REV-ERBα and -β. Thus, the effects of SR9009 cannot be used solely as surrogate for REV-ERB activity.
The cell-autonomous circadian clock regulates a plethora of physiological processes in the human body (1–3). The importance of proper clock maintenance is highlighted by linkages between circadian desynchrony and a variety of illnesses, including sleep and metabolic disorders, cardiovascular diseases, and cancer (1, 2, 4–7). This has fueled recent interest in the circadian clock as a therapeutic target (8–12). The nuclear receptors (NRs) REV-ERBα and -β, encoded by Nr1d1 and Nr1d2, link circadian rhythms and metabolism (1, 13). REV-ERBs lack the canonical NR activation domain, and thus function as a transcriptional repressor (14). Mice lacking REV-ERBα have defective circadian clocks (15, 16) and display a variety of metabolic abnormalities in many tissues (16–20), which have been reviewed elsewhere (21).
Like other NRs, REV-ERBs can be regulated by ligands, including naturally occurring heme (22, 23), which modulate their repressive activity. Attempts to pharmacologically target REV-ERBs by the use of putatively specific synthetic agonists, particularly SR9009 (24), have suggested a wide range of beneficial effects in healthy as well as diseased animal models and cell systems (11, 24–34). The beneficial effects of SR9009 on weight and exercise in mice (24, 27) have led to an interest in the use of the compound by humans, with online advertisements selling SR9009 highlighting REV-ERBs as the molecular target (e.g., https://www.simplyanabolics.com/sarms/sr9009-stenabolic/). This, in turn, has prompted newly developed assays that detect its surreptitious use (31, 33, 35).
Most studies utilizing SR9009 did not include genetic evidence that REV-ERBs are the actual or exclusive molecular target of SR9009. Nevertheless, many of these reports conclude that the effects of SR9009 are mediated via REV-ERBs. Thus, is it critical to ascertain whether the effects of SR9009 are mediated by REV-ERBs. Here we report the generation and validation of a model of genetic deletion of both REV-ERBα and -β in mice. Using this system to derive hepatocytes and embryonic stem cells lacking REV-ERBs, we demonstrate that SR9009 has strong effects on cell viability, metabolism, and gene expression in the genetic absence of the REV-ERBs. Therefore, the effects of SR9009 cannot be used as proxy for REV-ERB activity.
Results
Generation and Characterization of a Mouse Model for Conditional Deletion of REV-ERBα and -β.
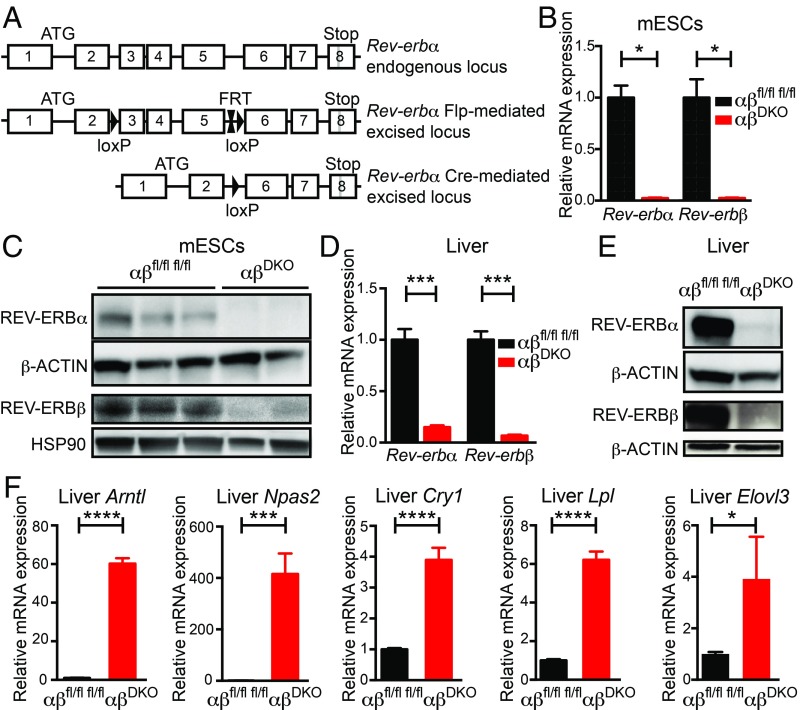
Although a putative conditional Cre-based deletion of REV-ERBα in the mouse has been used in several publications (16, 36–38), this model leads to in-frame deletion of the DNA-binding domain of REV-ERBα and thus results in a mutant protein rather than a complete deletion of REV-ERBα (13). Therefore, we engineered a different mouse model in which the DNA-binding domain-containing exons 3–5 of Rev-erbα are floxed, resulting in an out-of-frame deletion upon Cre-based recombination (Materials and Methods and Fig. 1A). We subsequently crossed this model with an existing Rev-erbβ floxed model (16) to generate a conditional double-knockout (DKO) C57BL/6 model of both REV-ERBα and -β.
Fig. 1.
Generation and validation of REV-ERBα/β DKO model. (A) Scheme depicting wild-type, floxed, and deleted Rev-erbα loci (details in Materials and Methods). (B) Relative Rev-erbα/β mRNA expression in double-floxed control vs. DKO mESCs (n = 3 lines per genotype). *P < 0.01, by 2-sided Student’s t test. Data are presented as mean ± SEM. (C) Immunoblot for REV-ERBα and REV-ERBβ from double-floxed control (n = 3) vs. DKO (n = 2) mESCs. (D) Relative Rev-erbα/β mRNA expression in double-floxed control (n = 5) vs. DKO (n = 3) livers. ***P < 0.0001, by 2-sided Student’s t test. Data are presented as mean ± SEM. (E) Immunoblot for REV-ERBα and REV-ERBβ from double-floxed control vs. DKO livers. (F) Relative mRNA expression of REV-ERB target genes in double-floxed control vs. DKO (n = 3) livers (n = 5). *P = 0.05, ***P < 0.001, and ****P < 0.0001 by 2-sided Student’s t test. Data are presented as mean ± SEM.
The DKO model was validated in several different ways. First, mouse embryonic stem cells (mESCs), in which clock genes such as Rev-erbs are expressed but have no known circadian role (2, 39, 40), were obtained from double-floxed embryos and transfected with vectors for Cre:GFP (DKO) or GFP (floxed controls). Expression of Rev-erbα and -β mRNA (Fig. 1B) and proteins (Fig. 1C) were abolished in the derived monoclonal DKO mESC lines. Of note, unlike the previous conditional Rev-erbα allele, whose recombination led to a mutant protein (13), we did not observe altered forms of REV-ERBα protein (SI Appendix, Fig. S1A). As a second model, 10-wk-old double-floxed mice received tail vein injections of AAV8-TBG-Cre-GFP (or AAV8-TBG-GFP as control), generating a hepatocyte-specific (41) DKO model. Two weeks following the injection, expression of REV-ERBα and -β mRNA (Fig. 1D) and protein (Fig. 1E) were near detection limits in the livers of double-floxed mice, again without the appearance of additional forms of the protein (SI Appendix, Fig. S1B). The loss of REV-ERBα/β function in DKO livers was supported by the robust hepatic derepression of Arntl, Npas2, Cry1, Lpl, and Elovl3 (Fig. 1F), all of which are well-established REV-ERB–repressed genes in the liver (13). These 2 model systems were used to interrogate the role of REV-ERBs in the effects of SR9009.
SR9009 Affects Cell Viability and Proliferation Independent of REV-ERBs.
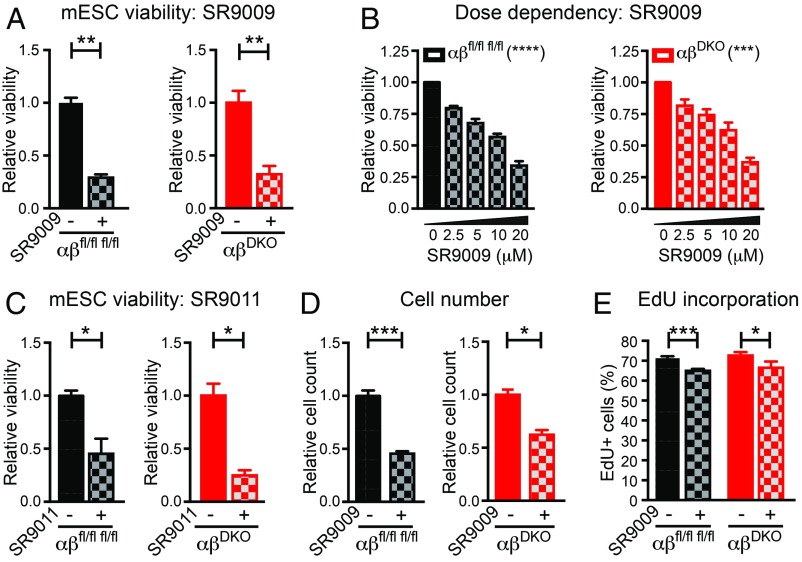
mESCs are similar to cancer cells in terms of their high proliferative capacity. Consistent with a previous report studying cancer cell lines (11), we found that treatment with SR9009 for 2 d at a concentration (10 μM) used in many published studies (24, 28, 32, 34, 42–44) reduced the viability of immortalized mouse and human cell lines, including some cancer cells (SI Appendix, Fig. S2A) but not fibroblasts (SI Appendix, Fig. S2B). In addition, SR9009 decreased the viability of wild-type mESCs in a dose-dependent manner (SI Appendix, Fig. S2C). SR9009 also reduced cell viability of double-floxed control mESCs and, remarkably, had similar effects on REV-ERB DKO mESCs (Fig. 2A). This effect was dose-dependent (Fig. 2B) and apparent after 1 d of treatment (SI Appendix, Fig. S3). SR9011, a related compound that is also a putative REV-ERB agonist (24), decreased the viability of control and DKO mESCs to similar extents as well (Fig. 2C). In addition to the cell viability test, which measures ATP levels, cell number was also directly measured and found to be similarly reduced in control and DKO mESCs treated with SR9009 (Fig. 2D). Moreover, the percentage of cells in S-phase was reduced in mESCs treated with SR9009, again to a similar extent in control and DKO mESCs (Fig. 2E).
Fig. 2.
SR9009 affects mESC viability and proliferation independent of REV-ERBs. (A) Relative viability of double-floxed control vs. DKO mESCs (n = 3 per condition), treated with 10 μM SR9009 for 2 d. **P ≤ 0.005, by 2-sided Student’s t test. (B) SR9009 dose-response showing relative viability of double-floxed control vs. DKO mESCs (n = 4 per condition), ***P < 0.001, ****P < 0.0001, by one-way ANOVA. (C) Relative viability of double-floxed control vs. DKO mESCs (n = 3 per condition), treated with 10 μM SR9011 for 2 d. *P < 0.05, by 2-sided Student’s t test. (D) Relative cell number of double-floxed control vs. DKO mESCs (n = 3 per condition), treated with 10 μM SR9009 for 2 d. *P = 0.015, ***P = 0.0006, by 2-sided Student’s t test. (E) EdU incorporation of double-floxed control (n = 5 per condition) vs. DKO (n = 4 per condition) mESCs treated with 10 μM SR9009 for 2 d and incubated with 10 μM EdU for 1 h before flow cytometry. *P = 0.05, ***P = 0.0003, by 2-sided Student’s t test. Data are presented as mean ± SEM.
To rule out the possibility that these results were influenced by the long-term depletion of both REV-ERBs, we acutely transfected double-floxed mESC lines with Cre:GFP vs. GFP (control) overexpressing plasmids and performed cell sorting for (Cre:)GFP+ (SI Appendix, Fig. S4 A and B). Treatment of these cells with SR9009 led to a comparable reduction in viability between control (GFP+) and DKO (Cre:GFP+) mESCs (SI Appendix, Fig. S4C), indicating that the REV-ERB–independent cytotoxicity of SR9009 was not a result of compensatory effects. Thus, the effect of SR9009 on mESC proliferation was REV-ERB–independent.
SR9009 Affects Mitochondrial Respiration Independent of REV-ERBs.
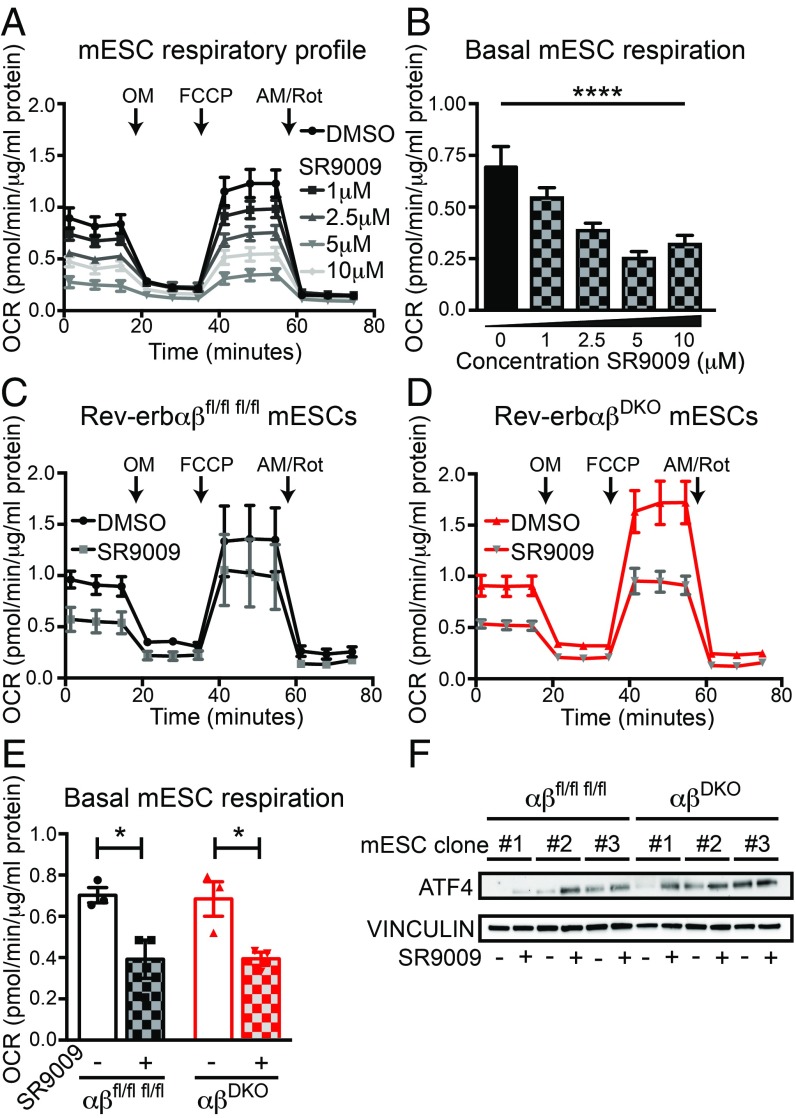
Because SR9009 has been reported to regulate mitochondrial metabolism (27), we hypothesized that the antiproliferative effects of SR9009 might be related to hampered mitochondrial respiration. Indeed, SR9009 had powerful, metabolic effects on mESC mitochondria, decreasing both their stimulated (Fig. 3A) and basal (Fig. 3B) respiration in a dose-dependent manner. Importantly, SR9009 impaired stimulated (Fig. 3 C and D) as well as basal (Fig. 3E) respiration in mitochondria from control and DKO mESCs to a similar extent, indicating that this metabolic effect was REV-ERB–independent. We also noted an SR9009-dependent increase in activating transcription factor 4 (ATF4), a marker of mitochondrial stress (45), which occurred to a similar degree in both control and DKO mESCs (Fig. 3F). Thus, SR9009 causes mitochondrial stress and alters mitochondrial function in a REV-ERB–independent manner.
Fig. 3.
SR9009 reduces mitochondrial respiration in mESCs independent of REV-ERBs. (A) OCR in mitochondrial stress test in wild-type mESCs treated with different doses of SR9009 for 2 d. (B) Basal mitochondrial respiration in wild-type mESCs (n = 8 per dose), ****P < 0.0001, by one-way ANOVA. (C) OCR of double-floxed control and (D) DKO mESCs. Measurements in A–D were performed with a Seahorse XF96 Flux Analyzer, under basal conditions followed by the sequential addition of OM (2.5 μM), FCCP (0.5 μM), and AM/Rot (0.5 μM), as indicated (n = 3 per condition). Cells were either pretreated with DMSO or with 10 μM SR9009 for 2 d before the assay. Data are presented as mean ± SEM. (E) Basal mitochondrial respiration in double-floxed control and DKO mESCs (n = 3 per condition). *P < 0.05, by a one-sided Student’s t test. (F) Immunoblot for ATF4 from DKO vs. double-floxed control mESCs treated with 10 μM SR9009 for 24 h (n = 3 per condition).
SR9009 Regulates Gene Expression in Hepatocytes Independently of REV-ERBs.
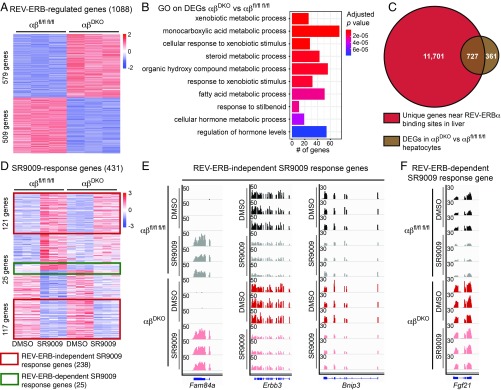
Having shown that canonical REV-ERB target genes were markedly derepressed in REV-ERB DKO liver, we derived hepatocytes from these livers for ex vivo studies. We first confirmed deletion of Rev-erbs (SI Appendix, Fig. S5A) and derepression of its known target genes (SI Appendix, Fig. S5B). Comparison of control vs. DKO transcriptomes revealed 1,088 genes that were differentially expressed (fold-change > 1.5, P < 0.01) (Fig. 4A), with clock output (SI Appendix, Fig. S5C) and metabolic target (SI Appendix, Fig. S5D) genes being largely derepressed. Gene ontology on the differentially expressed genes (DEGs) was in line with the known role of REV-ERB in the liver (Fig. 4B). In addition most DEGs (66.8%) overlapped with the REV-ERBα cistrome in the liver (13) (Fig. 4C). From these results we conclude that the hepatocyte REV-ERB DKO hepatocyte is a valid model to probe the effects of putative REV-ERB agonists for specificity.
Fig. 4.
SR9009 affects gene expression in a REV-ERB–independent manner. (A) Heatmap of z-score–normalized read counts showing 1,088 DEGs between double-floxed control and DKO hepatocytes (579 up vs. 509 down). (B) Gene ontology analysis on genes depicted in A. (C) Overlap between REV-ERBα cistrome in the liver (13) and the DEGs depicted in A. (D) Heatmap of z-score–normalized read counts showing DEGs anchored on double-floxed control hepatocytes following treatment with SR9009 (10 μM for 8 h) vs. DMSO and compared with treatment of DKO hepatocytes. A total of 431 genes were selected based on fold-change > 1.5 and P < 0.01. (E) Normalized RNA-seq read counts (RPM) for REV-ERB–independent SR9009-regulated genes (Fam84a, Erbb3, and Bnip3) and (F) one REV-ERB–dependent (Fgf21) SR9009 response gene. Visualization shown using Integrative Genomics Viewer.
We next evaluated the effects of SR9009 in hepatocytes. The a priori expectations for genes regulated by a bona fide REV-ERB agonist would include the following: (i) target genes would not be regulated in the absence of REV-ERBs, and (ii) because agonists potentiate repression, the direction of regulation by agonist should be opposite of that of deletion of the REV-ERBs. Of 431 genes that were differentially expressed upon SR9009 treatment in floxed control hepatocytes (fold-change > 1.5, P < 0.01), the majority (∼55%) were regulated similarly by SR9009 in DKO liver cells (e.g., Fam84a, Erbb3, and Bnip3) (Fig. 4 D and E). Of the 193 genes regulated by SR9009 in control hepatocytes but not significantly changed in the DKO cells treated with SR9009, a minority of 25 were regulated in the opposite way by DKO as would be predicted for a REV-ERB agonist (e.g., Fgf21) (Fig. 4 D and F). These results strongly suggest that the vast majority of gene-expression changes induced by SR9009 in liver cells are due to actions that are independent of REV-ERBs.
Discussion
We generated and validated a true conditional mouse model for complete deletion of REV-ERBα. This model was used, in conjunction with a REV-ERBβ conditional deletion mouse, to generate a REV-ERBα/β DKO mouse whose phenotypes reflect the physiological roles of endogenous REV-ERBs. Studies of REV-ERB DKO ESCs and hepatocytes revealed that the putative REV-ERB agonist SR9009 has numerous effects on proliferation, metabolism, and gene expression, and thus its effects cannot be attributed solely to its regulation of cellular REV-ERB activity.
SR9009 treatment slowed down mESC proliferation with marked decrease in cell numbers after 2 d of treatment. We did not observe overt cell death, as has been reported by others studying cancer cell proliferation (11). That study treated cells for longer times (3–7 d), used SR9009 at a concentration of 20 μM, and mainly quantified cytotoxicity via a metabolic assay [NAD(P)H-based: WST-1]. In the present study we used 10 μM, which is the most commonly used concentration among published studies (24, 28, 32, 34, 42–44), and quantified cytotoxicity via a metabolic assay as well as cell counting and EdU incorporation. Thus, our results demonstrate antiproliferative effects of SR9009 at shorter times of exposure and lower doses, which are seen even when REV-ERBs are drastically depleted from proliferating cells.
We found that SR9009 induces a severe reduction in mitochondrial respiration of mESCs. This reduced mitochondrial functioning in mESCs is in line with a previous report showing swollen mitochondria upon SR9009 treatment (11). We note that others reported that SR9009 increases mitochondrial function in myoblastic C2C12 cells (27). This seeming discrepancy might be explained by differences in bio-energetics between the myoblasts and mESCs. Consistent with the conclusions from mESCs, the REV-ERB–independent effects of SR9009 on hepatocyte gene expression strongly suggest that the transcriptional response to SR9009 in liver does not solely depend upon REV-ERBs.
The mechanisms by which SR9009 and related compounds exert pleiotropic effects on cell proliferation, metabolism, and gene expression in the absence of REV-ERBs remain to be determined. Of note, both SR9009 and SR9011 contain known toxicophores, such as nitrothiophene moieties, as active groups (46, 47). In any case, the fact that many of the effects of SR9009 are REV-ERB–independent has several important implications. First and foremost, the effects of SR9009 and related compounds cannot be used as a surrogate for REV-ERB activity, as has frequently been the case since they were first described (11, 24, 26, 27). Second, because REV-ERBs are not the only target, biological parameters affected by these compounds are not necessarily linked to the circadian clock. Finally, it is critical that this message be conveyed to the scientific community, as well as the lay public, who are using these compounds not only in laboratory experiments but as nutritional supplements based on spurious connections to REV-ERBs.
Materials and Methods
Gene Targeting, Genotyping, and Mice.
Standard gene targeting procedures were performed in C57BL/6 mESCs to generate mice carrying floxed alleles (Exon 3-4-5) at Rev-erbα loci (Genoway). Rev-erbαfl/fl; Rev-erbβfl/fl animals were generated by breeding the Rev-erbαfl/fl to Rev-erbβfl/fl animals also on C57BL/6 background (Institut Clinique de la Souris, Illkirch, France). Genotyping was performed following DNA extraction from mouse tissue with standard PCR assay. Rev-erbα genotyping PCR primers 5′-ATAGAGAAGTCTTCCCAGATCTCCTGCACA-3′ and 5′- ACAGTCTACGGCAAGGCAACACCAA-3′ detect wild-type (411 bp) and floxed (511 bp) gene alleles. Rev-erbβ genotyping PCR primers 5′- GGTTAGGTTTGTGAGTGTCCACAGC-3′ and 5′- GGAAGTGCTCCAACAAGGTAGTGCA-3′ detect wild-type (237 bp) and floxed (376 bp) gene alleles. All animal care and use procedures followed the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania in accordance with the NIH guidelines.
mESC Derivation, Cell Culture, and Treatments.
mESCs were derived from Rev-erbαfl/fl; Rev-erbβfl/fl mice as previously described (48). Blastocysts were collected at embryonic day 3.5 and cultured on a feeder layer of Mitomycin C (MedChem Express)-treated mouse embryonic fibroblasts (MEFs) in KnockOut Serum Replacement (KOSR) mESC medium: DMEM-high glucose (Gibco) supplemented with 15% KOSR (Gibco), 1% nonessential amino acids (Gibco), 1% penicillin/streptomycin (Gibco), 0.1 μM β-mercapto-ethanol (Sigma), 103 IU ESGRO leukemia inhibitory factor (LIF; EMD Millipore), 1 μM PD0325901 (Axon Medchem), and 3 μM CHIR99021 (Axon Medchem) until the blastocyst hatches and forms. Then mESCs were passaged and cultured as a normal mESC line in standard mESC medium: DMEM with 15% FBS (HyClone), 1% nonessential amino acids (Gibco), 1% penicillin/streptomycin (P/S; Gibco), 0.1 μM β-mercapto-ethanol (Sigma), and 103 IU ESGRO LIF (EMD Millipore). For all downstream assays, ESCs were transferred to gelatin-coated tissue culture plates and treated with SR9009 (EMD Millipore) or SR9011 (Excessbio) for 2 d, dissolved in DMSO. The identity and purity of SR9009 was confirmed by mass spectrometry (R. Miller, J. Durtra, T. Chappie; Pfizer). DMSO was always used as a control.
HeLa, U2OS, HEK293FT cells, and MEFs were all cultured in DMEM (Gibco) supplemented with 10% FBS (Tissue Culture Biologicals) and 1% P/S. AML12 cells were cultured in DMEM/F12 medium, supplemented with 1% ITS-G (Gibco), 40 ng/mL dexamethasone, and 10% FBS (Tissue Culture Biologicals). Viability was assessed using a CellTiter-Glo luminescent cell viability assay (Promega).
Gene Transduction in Mouse Liver and Hepatocyte Isolation.
Adeno-associated viruses encoding Cre-GFP or GFP under the liver-specific TBG promoter (AAV8-TBG-Cre-GFP, and AAV8-GFP used as a negative control) were prepared by the Vector Core of the Pennsylvania Diabetes Research Center, as described previously (49). The 5e11 virus particles were injected into each mouse by tail-vein injection. Two weeks later mice were harvested for analysis.
Hepatocytes were isolated using a 2-step collagenase/DNase digestion protocol (50) and plated on collagen-coated cell culture plates in DMEM (Gibco) supplemented with 10% FBS (Tissue Culture Biologicals) and 1% P/S.
Immunoblotting.
Samples were lysed in RIPA buffer supplemented with complete protease inhibitor mixture (Roche) and with phosSTOP (Roche). Lysates were resolved by gel electrophoresis (Bio-Rad), transferred to PVDF membrane (Immubulon-P, Millipore), and probed with the following antibodies: anti–REV-ERBα (1:1,000, Abcam #ab174309), anti–REV-ERBβ (1:1,000, SantaCruz #sc-398252), anti-HSP90 (1:1,000, CST #4874S), anti-ATF4 (1:1,000, CST #11815), anti–β-ACTIN-HRP (1:5,000, CST #5125S), anti–VINCULIN-HRP (1:5,000, CST #E18799), anti-rabbit IgG, HRP-linked (1:10,000, CST #7074S), anti-mouse IgG, HRP-linked (1:10,000, CST #7076S).
Quantitative RT–PCR.
Total RNA was extracted from cells and tissues using RNAeasy (Qiagen) according to the manufacturer’s instructions, and treated with DNase (Qiagen) before reverse transcription. cDNA was generated using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR reactions were performed using PowerSYBR Green PCR Master Mix (Applied Biosystems) with specific primers on a QuantStudio 6 Flex instrument (Applied Biosystems). mRNA expression was normalized to the housekeeping gene 36B4 for liver samples and Ppib for all mESCs and hepatocytes. Primer sequences for qRT–PCR: 36B4-fw, 5′-TCATCCAGCAGGTGTTTGACA-3′; 36B4-rev, 5′-GGCACCGAGGCAACAGTT-3′; Ppib-fw, 5′-GCAAGTTCCATCGTGTCATCAAG-3′; Ppib-rev, 5′-CCATAGATGCTCTTTCCTCCTG-3′; Rev-erbα-fw, 5′-GTCTCTCCGTTGGCATGTCT-3′; Rev-erbα-rev, 5′-CCAAGTTCATGGCGCTCT-3′; Rev-erbβ-fw, 5′-TTCTACTGTGTAAAGTCTGTGGG-3′; Rev-erbβ-rev, 5′-CTGGATGTTTTGCTGAATGCTC-3′; Arntl-fw, 5′-TAGGATGTGACCGAGGGAAG-3′; Arntl-rev, 5′-TCAAACAAGCTCTGGCCAAT-3′; Npas2-fw, 5′-ATGTTCGAGTGGAAAGGAGAC-3′; Npas2-rev, 5′-CAAGTGCATTAAAGGGCTGTG-3′; Cry1-fw, 5′-AGCGCAGGTGTCGGTTATGAGC-3′; Cry1-rev, 5′-ATAGACGCAGCGGATGGTGTCG-3′; Lpl-fw, 5′-TTTTCTGGGACTGAGGATGG-3′; Lpl-rev, 5′-GCCAGCTGAAGTAGGAGTCG-3′; Elovl3-fw, 5′- ATGCAACCCTATGACTTCGAG-3′; Elovl3-rev, 5′-ACGATGAGCAACAGATAGACG-3′.
RNA-Sequencing.
RNA-sequencing reads were aligned to the mouse genome (mm9) using Hisat2 (51) with default parameters. Only unique mapped reads were considered for further analysis. Normalized expression value, fragments per kilobase of exon per million reads mapped (FPKM), was calculated for each gene using StringTie (52). Genes with FPKM larger than 1 in at least one sample were considered. For differential expression analysis, raw read counts were measured within Ensembl genes (NCBIM37.67) using featureCounts (53), and then the DESeq2 (54) pipeline was used with P < 0.01 and fold-change > 1.5. Gene ontology analysis was performed using the clusterProfiler R package (55) with expressed genes as background. HOMER (56) was used to obtain bigWig files, which were visualized with Integrative Genomics Viewer.
Cellular Respirometry.
mESCs were cultured in mESC medium on Seahorse 96-well plates (Agilent Technologies) precoated with 0.1% gelatin. Culture medium was switched to basal medium (unbuffered DMEM, supplemented with 2 mM glutamine) 30 min before the start of the assay and used for the total duration of the measurements. For the mitochondrial stress test oligomycin (OM; 2.5 μM), FCCP (0.5 μM), and rotenone and antimycin A (AM/Rot; 0.5 μM) were serially injected, using a Seahorse XF96 extracellular flux analyzer (Agilent Technologies). Oxygen consumption rate (OCR) values of 8 replicates per condition were normalized to the protein concentration per well, as a proxy for cell number, via BCA analysis (Thermo Scientific). In general, the average of 3 baseline OCR levels was calculated, as well as the average of 3 measurements after each compound injection. Basal respiration was calculated as the last rate measurement before OM injection – nonmitochondrial respiration rate.
Flow Cytometry.
mESCs were transfected with Cre:GFP and GFP overexpressing plasmids via the use of Lipofectamine 3000 (Thermo Fisher Scientific) and were sorted with a BD FACS Jazz (BD Bioscience). pCAG-Cre:GFP and pCAG-GFP were a gift from Connie Cepko, Department of Genetics, Harvard Medical School, Boston, MA (Addgene plasmid #13776; http://www.addgene.org/13776/; RRID:Addgene_13776 and #11150; http://www.addgene.org/11150/; RRID:Addgene_11150). An iClick EdU AndyFluor 647 Flow Cytometry Assay Kit (ABP Biosciences) was used to assess EdU incorporation. mESCs were treated with DMSO vs. 10 μM SR9009 for 2 d and incubated with 10 μM EdU for 1 h before fixation of the cells. EdU detection was performed following the manufacturer’s guidelines. Cells were additionally stained with 20 μg/mL PI in 0.1% Triton-X100 supplemented with 0.2 mg/mL RNase for 40 min at 37°. Cells were analyzed with a BD FACS Canto (BD Bioscience). Data were analyzed with FlowJo software (v8.7).
Statistical Analysis.
Statistical analyses were performed using Prism (GraphPad Software). All data are presented as mean ± SEM. One/two-sided t tests were used when comparing drug treatment versus DMSO for single concentrations. When comparing the effect of multiple concentrations, a one-way ANOVA was used.
Supplementary Material
Acknowledgments
We thank R. Miller, J. Dutra, and T. Chappie (Pfizer) for evaluating the purity of SR9009; and D. Guan for helping with tail vain injections. This work was supported by NIH Grant R01 DK45586 (to M.A.L.); the JPB Foundation (M.A.L.); a Netherlands Heart Institute postdoctoral fellowship (to P.D.); American Diabetes Association training Grant 1-18-PDF-126 (to M.A.); and NIH Grant F30 DK104513 (to M.J.E.).
Footnotes
Conflict of interest statement: M.A.L. is an Advisory Board Member for Pfizer, Inc. and Eli Lilly and Co., and a consultant to Novartis; he has received research funding from Pfizer, Inc. for studies other than the present work and holds stock in KDAc, Inc.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE123312).
See QnAs on page 12121.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904226116/-/DCSupplemental.
References
- 1.Bass J., Lazar M. A., Circadian time signatures of fitness and disease. Science 354, 994–999 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Dierickx P., Van Laake L. W., Geijsen N., Circadian clocks: From stem cells to tissue homeostasis and regeneration. EMBO Rep. 19, 18–28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass J., Takahashi J. S., Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu E. A., Weaver D. R., Disrupting the circadian clock: Gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany N.Y.) 3, 479–493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hastings M. H., Goedert M., Circadian clocks and neurodegenerative diseases: Time to aggregate? Curr. Opin. Neurobiol. 23, 880–887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelleher F. C., Rao A., Maguire A., Circadian molecular clocks and cancer. Cancer Lett. 342, 9–18 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Dierickx P., et al. , “Circadian rhythms in stem cell biology and function” in Stem Cells and Cardiac Regeneration, Stem Cell Biology and Regenerative Medicine, Madonna R., Ed. (Springer International Publishing, Cham, Switzerland, 2015), pp. 57–78. [Google Scholar]
- 8.Antoch M. P., Kondratov R. V., Pharmacological modulators of the circadian clock as potential therapeutic drugs: Focus on genotoxic/anticancer therapy. Handb. Exp. Pharmacol. 217, 289–309 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elshazley M., et al. , The circadian clock gene BMAL1 is a novel therapeutic target for malignant pleural mesothelioma. Int. J. Cancer 131, 2820–2831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cha H. K., Chung S., Lim H. Y., Jung J.-W., Son G. H., Small molecule modulators of the circadian molecular clock with implications for neuropsychiatric diseases. Front. Mol. Neurosci. 11, 496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sulli G., et al. , Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 553, 351–355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallach T., Kramer A., Chemical chronobiology: Toward drugs manipulating time. FEBS Lett. 589, 1530–1538 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y., et al. , GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348, 1488–1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke L., Downes M., Carozzi A., Giguère V., Muscat G. E., Transcriptional repression by the orphan steroid receptor RVR/Rev-erb beta is dependent on the signature motif and helix 5 in the E region: Functional evidence for a biological role of RVR in myogenesis. Nucleic Acids Res. 24, 3481–3489 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preitner N., et al. , The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Cho H., et al. , Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugge A., et al. , Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerhart-Hines Z., et al. , The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature 503, 410–413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duez H., et al. , Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology 135, 689–698 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Le Martelot G., et al. , REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 7, e1000181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett L. J., Lazar M. A., Nuclear receptor Rev-erbα: Up, down, and all around. Trends Endocrinol. Metab. 25, 586–592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin L., et al. , Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Raghuram S., et al. , Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat. Struct. Mol. Biol. 14, 1207–1213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solt L. A., et al. , Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffin P., et al. , Circadian clock protein Rev-erbα regulates neuroinflammation. Proc. Natl. Acad. Sci. U.S.A. 116, 5102–5107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L., et al. , REV-ERBα ameliorates heart failure through transcription repression. JCI Insight 2, 95177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woldt E., et al. , Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 19, 1039–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayeuf-Louchart A., et al. , Rev-erb-α regulates atrophy-related genes to control skeletal muscle mass. Sci. Rep. 7, 14383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amador A., et al. , Pharmacological and genetic modulation of REV-ERB activity and expression affects orexigenic gene expression. PLoS One 11, e0151014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stujanna E. N., et al. , Rev-erb agonist improves adverse cardiac remodeling and survival in myocardial infarction through an anti-inflammatory mechanism. PLoS One 12, e0189330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazzarino M., Rizzato N., Stacchini C., de la Torre X., Botrè F., A further insight into the metabolic profile of the nuclear receptor Rev-erb agonist, SR9009. Drug Test. Anal. 10, 1670–1681 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Amir M., et al. , REV-ERBα regulates TH17 cell development and autoimmunity. Cell Rep. 25, 3733–3749.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies S. R., et al. , Production of certified reference materials for the sports doping control of the REV-ERB agonist SR9009. Drug Test. Anal. 11, 257–266 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Hwang J., Jiang A., Fikrig E., Rev-erb agonist inhibits chikungunya and O’nyong’nyong virus replication. Open Forum Infect. Dis. 5, ofy315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geldof L., Deventer K., Roels K., Tudela E., Van Eeno P., In vitro metabolic studies of REV-ERB agonists SR9009 and SR9011. Int. J. Mol. Sci. 17, 1676 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eichenfield D. Z., et al. , Tissue damage drives co-localization of NF-κB, Smad3, and Nrf2 to direct Rev-erb sensitive wound repair in mouse macrophages. elife 5, e13024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam M. T. Y., et al. , Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498, 511–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaix A., Lin T., Le H. D., Chang M. W., Panda S., Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 29, 303–319.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yagita K., et al. , Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc. Natl. Acad. Sci. U.S.A. 107, 3846–3851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kowalska E., Moriggi E., Bauer C., Dibner C., Brown S. A., The circadian clock starts ticking at a developmentally early stage. J. Biol. Rhythms 25, 442–449 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., et al. , HNF6 and Rev-erbα integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev. 30, 1636–1644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S., et al. , REV-ERBα integrates colon clock with experimental colitis through regulation of NF-κB/NLRP3 axis. Nat. Commun. 9, 4246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pourcet B., et al. , Nuclear receptor subfamily 1 group D member 1 regulates circadian activity of NLRP3 inflammasome to reduce the severity of fulminant hepatitis in mice. Gastroenterology 154, 1449–1464.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S., et al. , Angiotensin II suppresses Rev-erbα expression in THP-1 macrophages via the ang II type 1 receptor/liver X receptor α pathway. Cell. Physiol. Biochem. 46, 303–313 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Quirós P. M., et al. , Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morley J. O., Matthews T. P., Studies on the biological activity of some nitrothiophenes. Org. Biomol. Chem. 4, 359–366 (2006). [DOI] [PubMed] [Google Scholar]
- 47.de Araújo Neto L. N., et al. , Synthesis, cytotoxicity and antifungal activity of 5-nitro-thiophene-thiosemicarbazones derivatives. Chem. Biol. Interact. 272, 172–181 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Czechanski A., et al. , Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 9, 559–574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang B., et al. , Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller R. A., et al. , Adiponectin suppresses gluconeogenic gene expression in mouse hepatocytes independent of LKB1-AMPK signaling. J. Clin. Invest. 121, 2518–2528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pertea M., et al. , StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.