Abstract
The Golgi apparatus is a membrane-bound organelle that serves as the center for trafficking and processing of proteins and lipids. To perform these functions, the Golgi forms a multilayer stacked structure held by GRASP55 and GRASP65 trans-oligomers and perhaps their binding partners. Depletion of GRASP proteins disrupts Golgi stack formation and impairs critical functions of the Golgi, such as accurate protein glycosylation and sorting. However, how Golgi destruction affects other cellular activities is so far unknown. Here, we report that depletion of GRASP proteins reduces cell attachment and migration. Interestingly, GRASP depletion reduces the protein level of α5β1 integrin, the major cell adhesion molecule at the surface of HeLa and MDA-MB-231 cells, due to decreased integrin protein synthesis. GRASP depletion also increases cell growth and total protein synthesis. These new findings enrich our understanding on the role of the Golgi in cell physiology and provide a potential target for treating protein-trafficking disorders.
INTRODUCTION
The Golgi apparatus is an essential cellular membrane organelle at the center of the secretory pathway (Klute et al., 2011). Its basic structure is a stack of flat, closely arranged cisternae. Each mammalian cell contains ∼100 Golgi stacks, which often line up and laterally link to form a ribbon localized in the perinuclear region (Rambourg et al., 1987; Klumperman, 2011). The primary function of the Golgi is to process membrane and secretory proteins. Cargo proteins synthesized at the endoplasmic reticulum (ER) are transferred by COPII vesicles to the cis-Golgi (Brandizzi and Barlowe, 2013). While traveling across the Golgi stack, they undergo posttranslational modifications, including glycosylation, phosphorylation, and proteolysis (Goldfischer, 1982). In the trans-Golgi network (TGN), proteins are sorted for delivery to proper destinations, such as endosomes, lysosomes, the plasma membrane, or outside the cell by constitutive or regulated secretion (Marsh and Howell, 2002). Proteins derived from one-third of the human genes travel through the secretory pathway (Pfeffer, 2013), and proper functioning of the Golgi is required for a variety of cellular activities and homeostasis.
The exact mechanism of Golgi stack formation is not fully understood, but the two Golgi reassembly and stacking proteins (GRASPs), GRASP55 and GRASP65, are the only Golgi stacking proteins identified so far (Barr et al., 1997; Shorter et al., 1999; Xiang and Wang, 2010; Zhang and Wang, 2015). GRASP55 and GRASP65 localize at the medial-/trans- and cis-Golgi, respectively, form trans-oligomers through the N-terminal GRASP domain to hold the cisternae together into stacks (Wang et al., 2005), and link Golgi stacks into a ribbon (Puthenveedu et al., 2006). The C-terminal serine-proline–rich domains of GRASP55 and GRASP65 are more divergent, but both are phosphorylated in mitosis to dissociate the protein trans-oligomers and disassemble the Golgi structure. In telophase, Golgi tubules and vesicles are divided equally between daughter cells, where they are reassembled into stacks and ribbons upon dephosphorylation of GRASP proteins (Vielemeyer et al., 2009; Tang et al., 2012). Therefore, GRASPs are essential proteins for maintaining an intact and dynamic Golgi structure (Zhang and Wang, 2015, 2016). In addition to GRASPs, GRASP-interacting proteins such as GM130 and Golgin 45 may also facilitate Golgi stack formation (Lee et al., 2014).
The discovery of the GRASP proteins provides us with a molecular tool to control Golgi stacking and thus to probe the functional significance of Golgi structure formation. We have inhibited Golgi stacking by microinjecting GRASP65 antibodies (Wang et al., 2003, 2008), by depleting both GRASPs in cells via knockdown (Tang et al., 2010; Xiang and Wang, 2010), and by GRASP knockout (KO) (Bekier et al., 2017). These caused accelerated trafficking of several marker proteins, including CD8, Vesicular stomatitis virus G-protein, cathepsin D, and integrins (Wang et al., 2008; Xiang et al., 2013; Bekier et al., 2017). Furthermore, GRASP depletion significantly decreased both global N-linked glycosylation and glycan complexity and changed the glycolipid composition at the cell surface (Xiang et al., 2013; Bekier et al., 2017). A plausible explanation is that stacking reduces the accessibility of coat proteins to Golgi membranes, which decreases the rate of vesicle budding and transport but ensures accurate glycosylation and sorting. The Golgi harbors various glycosyltransferases and glycosidases in different subcompartments, but unlike the ER, which contains a high concentration of folding chaperones that retain improperly modified cargoes (Helenius and Aebi, 2001; Hebert and Molinari, 2007; Pearse and Hebert, 2010), the Golgi lacks a rigorous system to control the fidelity of its biosynthetic processes. An ordered structure and a controlled cargo flow through the Golgi are likely required to carry out precise, sequential modifications as cargo proteins pass between cisternae (Zhang and Wang, 2015; Huang and Wang, 2017). In addition, GRASP depletion also caused missorting of the lysosomal enzyme cathepsin D to the extracellular space (Xiang et al., 2013), suggesting that stacking may ensure that sorting occurs only when cargo molecules reach the TGN. These findings demonstrate that Golgi stack formation is fundamentally important for Golgi function (Zhang and Wang, 2016).
Proper glycosylation and sorting of proteins in the Golgi are required for many cell activities, such as cell attachment, migration, proliferation, and embryonic development (Zheng et al., 1994; Ono and Hakomori, 2004; Janik et al., 2010; Hang et al., 2017). In addition, Golgi structural defects have been linked to human diseases, including Alzheimer’s (Aridor and Balch, 1999; Joshi et al., 2014; Evin, 2015), Huntington’s (Hilditch-Maguire et al., 2000), Parkinson’s (Mizuno et al., 2001), autoimmune diseases (Fritzler et al., 1984; Bizzaro et al., 1999), cancer (Yoshimura et al., 1996; Roberts et al., 1998; Dennis et al., 1999; Diaz-Corrales et al., 2004; Krishnan et al., 2005), viral infections (Ng et al., 2003), and congenital disorders of glycosylation and Wiskott–Aldrich syndrome (Durand and Seta, 2000; Freeze and Ng, 2011). However, the causal relationship between Golgi defects and disease pathogenesis remains largely unexplored. Therefore, it is crucial to understand how Golgi structure disruption affects essential cellular activities and physiology such as cell attachment, migration, and growth.
Cell attachment depends on a set of cell adhesion proteins called integrins; α5β1 integrin is the main form of integrin complex in HeLa and MDA-MB-231 cells (Mierke et al., 2011; Yu et al., 2011; Jia et al., 2016). As highly glycosylated transmembrane proteins, α5 and β1 integrins are processed by the Golgi and transported to the plasma membrane for their proper functions. When transported through the TGN, α5 integrin undergoes posttranslational cleavage for activation, wherein pro-integrin α5 (∼170 kDa) is cleaved by proprotein convertases (PCs) into heavy (∼130 kDa) and light (∼19 kDa) chains that are linked by a disulfide bond (Paule et al., 2012). Because GRASP depletion accelerates α5-integrin trafficking and processing (Xiang et al., 2013), it is reasonable to expect that it may also affect α5- and β1-integrin glycosylation and sorting as well as cell attachment.
In this study, we found that Golgi destruction by GRASP depletion reduces α5β1-integrin protein level, which results in compromised cell activities such as decreased cell adhesion and migration. Using radioactive labeling and pulse–chase assays, we showed that decreased protein level of α5β1 integrin is due to reduced protein synthesis rather than accelerated protein degradation. Surprisingly, Golgi unstacking accelerated overall protein synthesis and cell proliferation. Our study revealed a direct link between Golgi structure, function, and cellular activities.
RESULTS
GRASP depletion reduces cell adhesion
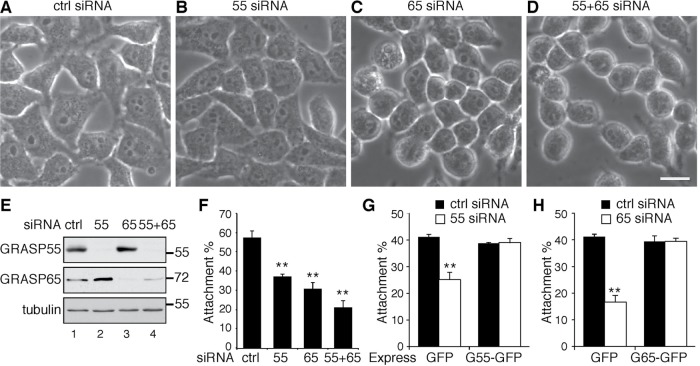
The proper glycosylation and localization of cell surface proteins are important for cell adhesion and migration. We observed that, when examined under a phase-contrast microscope, HeLa cells depleted of GRASP65 or both GRASPs spread less well on the dish, and they also formed clusters and appeared to be rounder compared with cells transfected with control or GRASP55 small interfering RNA (siRNA) (Figure 1, C and D vs. A and B, and E). To further assess the effect of GRASP depletion on cell adhesion, we detached the cells using EDTA and placed the cells on fibronectin-coated plates. After incubation with serum-free medium at 37°C for 30 min, the attached cells were counted. Compared with the control, in which 55 ± 4% of cells were attached to the dish, only 38 ± 1% of GRASP55-depleted cells and 31 ± 3% of GRASP65-depleted cells bound to the dish (Figure 1F). When both GRASP55 and GRASP65 were depleted, the percentage of attached cells was further reduced to 22 ± 4% (Figure 1F). This effect was rescued by the exogenous expression of green fluorescent protein (GFP)-tagged GRASP65 or GRASP55, but not GFP alone (Figure 1, G and H). Similar results were obtained in HeLa cells in which GRASPs were knocked out by CRISPR/Cas9-mediated genome editing (Supplemental Figure 1) (Bekier et al., 2017). These results demonstrate that Golgi unstacking reduces cell adhesion.
FIGURE 1:
GRASP depletion reduces cell attachment. (A–D) Phase-contrast images of HeLa cells transfected with indicated siRNAs. Scale bar: 20 µm. Note that GRASP-depleted cells are generally rounder than control siRNA-treated cells. (E) Western blots of HeLa cells transfected with indicated siRNAs. GRASP55 and GRASP65 were effectively depleted. (F) Cell attachment was reduced by GRASP depletion. A total of 3 × 105 control (ctrl) or GRASP siRNA-treated cells were seeded on fibronectin-coated plates and incubated in serum-free medium for 30 min. After the removal of unbound cells, the number of attached cells was counted. Results are presented as mean ± SEM; statistical analysis was performed by comparison with control siRNA-treated cells using Student’s t test. **, p < 0.01. (G) The reduced attachment of GRASP55-depleted cells was rescued by the expression of GRASP55-GFP but not GFP. (H) The reduced attachment of GRASP65-depleted cells was rescued by expressing GRASP65-GFP.
GRASP depletion reduces cell migration and invasion
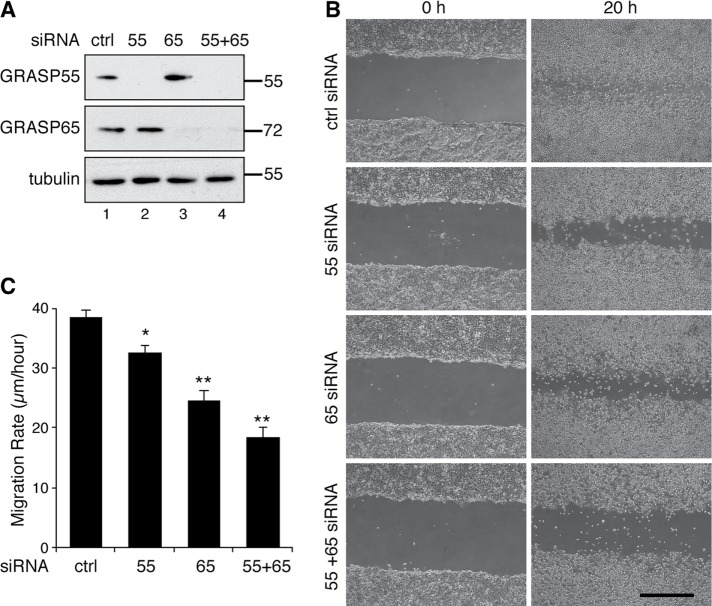
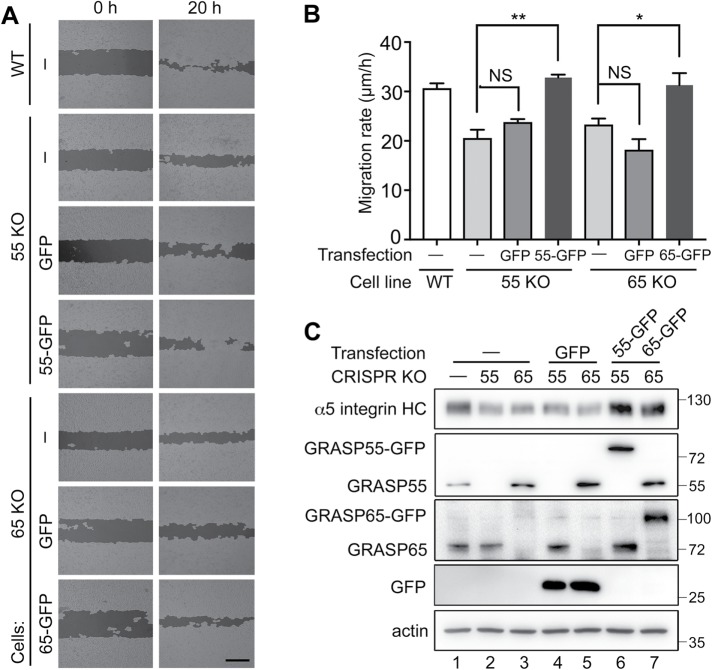
To determine the effect of Golgi unstacking on cell migration, we first used a well-established wound-healing assay using the human breast cancer cell line MDA-MB-231 (Liang et al., 2007). Both GRASP55 and GRASP65 could be readily depleted by siRNA treatment in this cell line (Figure 2A). Seventy-two hours after siRNA transfection, fully confluent MDA-MB-231 cells were starved in serum-free medium for 24 h. A scratch was made in the confluent layer using a 200-µl pipette tip, and the cells were cultured in complete growth medium for another 20 h to allow the cells to migrate into the wounded region. When examined under a microscope, cells that had been transfected with control siRNA almost completely covered the wound area, while GRASP-depleted cells only covered a portion of the wound region (Figure 2B). Control siRNA-treated cells migrated ∼39 ± 1 μm/h, while cells depleted of GRASP55, GRASP65, or both migrated at 33 ± 1, 25 ± 2, and 19 ± 2 μm/h, respectively, significantly slower than the control cells (Figure 2C). The same wound-healing assay was done with GRASP-KO HeLa cells, and consistent results were obtained (Figure 3 and Supplemental Figure 2). Cells depleted of GRASP55, GRASP65, or both migrated at 22 ± 2, 16 ± 2, and 15 ± 2 μm/h, respectively, significantly slower than the 32 ± 3 μm/h of wild-type (WT) cells (Supplemental Figure 2). This defect was rescued by the expression of GFP-tagged GRASP65 or GRASP55, but not GFP alone (Figure 3, A–C). Interestingly, the level of the cell adhesion molecule α5 integrin appeared to be lower in GRASP-KO cells and was rescued by the expression of GRASP55-GFP and GRASP65-GFP in the corresponding knockout cells (Figure 3C, lanes 6 and 7 vs. lanes 4 and 5).
FIGURE 2:
GRASP depletion reduces cell migration. (A) Western blots of MDA-MB-231 cells transfected with the indicated siRNAs. GRASP55 and GRASP65 were effectively depleted. (B) GRASP depletion reduces MDA-MB-231 cell migration. Cells transfected with indicated siRNAs were analyzed in a wound-healing assay. Images were taken at 0 h and 20 h after scratching. Scale bar: 500 µm. (C) Quantitation of the migration rate in B. Statistical analysis was performed by comparison with control siRNA-treated cells using Student’s t test. *, p < 0.05; **, p < 0.01.
FIGURE 3:
GRASP expression rescues the cell migration defects in GRASP-KO cells. (A) GRASP expression rescues the decreased cell migration in GRASP-KO HeLa cells. Cells transfected with indicated constructs were tested in a wound-healing assay, and images were processed by WimScratch: Wound Healing Assay Image Analysis Solution (Release 4.0). Images were taken at 0 and 20 h after scratching. Scale bar: 500 µm. (B) Quantitation of A. The reduced migration of GRASP55-KO and GRASP65-KO cells was rescued by expressing GRASP55-GFP and GRASP65-GFP, respectively, but not by GFP alone. Results are presented as mean ± SEM; statistical analysis was performed by comparison with WT control (ctrl) using Student’s t test. *, p < 0.05; **, p < 0.01. (C) Western blot of HeLa cells transfected with indicated constructs. α5-Integrin heavy chain (α5 integrin HC), GRASP55, GRASP65, GFP, and actin were blotted. The reduced protein levels of α5 integrin in GRASP55-KO and GRASP65-KO cells were rescued by expressing GRASP55-GFP or GRASP65-GFP, respectively, but not by GFP alone.
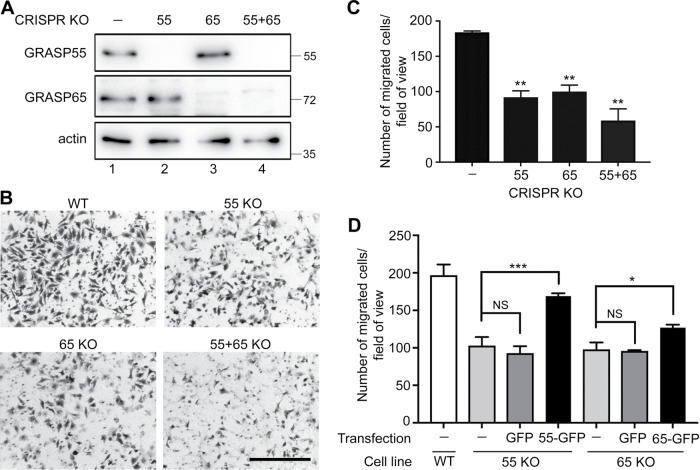
To confirm the result that GRASP depletion reduces cell migration, we performed a Transwell assay using WT and GRASP-KO HeLa cells (Isaji et al., 2006). In this assay, the same number of cells was seeded in the inserts with serum-free medium, and full growth medium with serum was added in the lower chamber as a chemoattractant. After a 20 h incubation, cells on the lower side of the membrane were fixed, stained with crystal violet, and imaged. The results showed that the migration rate was significantly reduced by the knockout of GRASP55 or GRASP65, while double deletion of both GRASPs had a more severe effect (Figure 4, A–C). In the control, 184 ± 3 cells migrated to the lower side of the membrane per field of view, while GRASP55, GRASP65, and double-knockout cells had 94 ± 15, 100 ± 15, and 59 ± 28 cells migrate to the bottom side, respectively (Figure 4C). Exogenous expression of GFP-tagged GRASP65 or GRASP55 rescued the migration defect in GRASP-KO cells (Figure 4D and Supplemental Figure 3). On the basis of the results from the wound-healing and Transwell assays, we conclude that GRASP depletion results in decreased cell migration and invasion.
FIGURE 4:
GRASP depletion reduces cell migration and invasion. (A) Western blots of WT and GRASP-KO HeLa cells for GRASP55, GRASP65, and actin. (B) GRASP depletion reduces cell migration and invasion. Indicated WT and GRASP-KO HeLa cells were analyzed in a Transwell assay. Images were taken after a 20-h migration. Scale bar: 500 µm. (C) Quantitation of the migration rate in B. Results are presented as mean ± SEM; statistical analysis was performed by comparison with WT cells using Student’s t test. (D) GRASP expression rescues the decreased cell migration in GRASP-KO cells. GRASP55-KO or GRASP65-KO cells transfected with indicated constructs were analyzed by a Transwell assay. Example images are shown in Supplemental Figure 3. Note that defects in cell invasion in GRASP55-KO and GRASP65-KO cells were rescued by the expression of GRASP55-GFP and GRASP65-GFP, respectively, but not by GFP alone. Results are presented as mean ± SEM; statistical analysis was performed by comparison with WT cells (ctrl) using Student’s t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Golgi unstacking reduces α5β1-integrin protein level in the cell
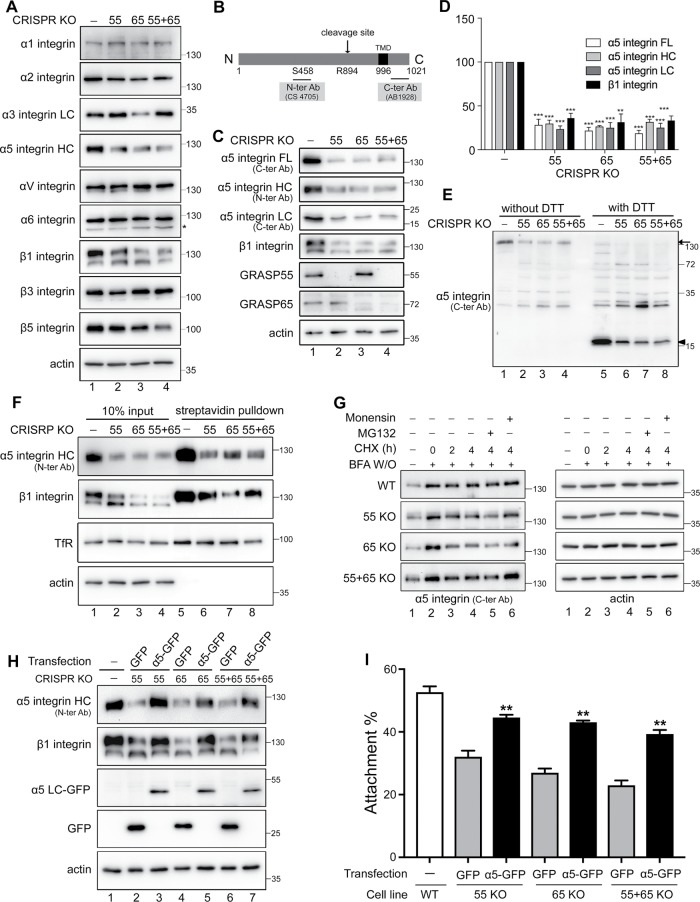
The fact that Golgi unstacking reduces cell adhesion and migration (Figures 1–4) suggests that it may affect cell adhesion molecules such as integrin, as shown in Figure 3C. Therefore, we examined the effects of GRASP depletion on the protein level of several integrins that are known to be expressed in HeLa cells, including α1, α2, α3, α5, αV, α6, β1, β3, and β5 (Bergman et al., 1995; Liu et al., 2009; Scheffer et al., 2013; Wan et al., 2014; Ratcliffe et al., 2016), by Western blot. As shown in Figure 5A, α1, αV, and α6 levels did not change in GRASP-KO cells compared with WT HeLa. α3 Integrin was reduced only in GRASP65 KO cells, while the β3 level was slightly reduced only in GRASP55 KO cells. Consistent with the results shown in Figure 3C, both α5 and β1 integrins were robustly reduced in GRASP single- or double-knockout cells, suggesting that Golgi unstacking reduces cell adhesion by decreasing the α5β1-integrin level.
FIGURE 5:
GRASP knockout reduces α5β1-integrin protein level. (A) GRASP knockout reduces α5β1 level but has only modest effects on other integrins. Western blots of WT and GRASP-KO HeLa cells for indicated integrins and actin. Note that the protein levels of α1, αV, and α6 did not change after GRASP depletion in contrast to the robust reduction of α5 and β1 integrins. (B) Schematic α5-integrin domain structure and epitopes of indicated antibodies. (C) GRASP knockout reduces the α5β1-integrin level. The full-length (FL), heavy-chain (HC), and light-chain (LC) forms of α5 integrin, β1 integrin, GRASP55, GRASP65, and actin were blotted. Note the reduced protein level of α5 integrin on all three blots as well as β1 integrin compared with that in WT cells. (D) Quantitation of C normalized with actin. Statistical analysis was performed by comparison with WT cells. (E) Western blot of α5 integrin on nonreducing (without DTT) and reducing gels (with DTT). Arrow and arrowhead indicate full length and light chain, respectively. (F) GRASP knockout reduces α5β1-integrin level at the cell surface. GRASP-KO cells were surface biotinylated, this was followed by streptavidin pull-down and Western blotting of α5 integrin, β1 integrin, TfR, and actin. Note that only the highly glycosylated forms of integrins were pulled down; this was clearer with β1 integrin (top band). (G) GRASP knockout accelerates α5-integrin maturation. WT and GRASP-KO HeLa cells were pretreated with BFA for 2 h and washed out and then released into CHX for 0, 2, or 4 h or CHX for 4 h with MG132 or monensin. As GRASP-KO cells have a lower level of integrin level, we exposed those gels longer, so all cell lines had a similar signal at the 0 time point to start with, and the reduction of the protein was assessed over time. (H) Western blot of indicated HeLa cells transfected with α5 integrin-GFP or GFP. α5-Integrin heavy chain (α5-integrin HC), β1 integrin, GFP, and actin were blotted. The reduced protein levels of α5 and β1 integrins in GRASP-KO cells were rescued by expressing α5 integrin-GFP, but not by GFP alone. (I) α5-Integrin expression rescues the decreased cell attachment of GRASP-KO cells. Cells transfected with indicated constructs were analyzed in an attachment assay, as described in Figure 1. The quantitation results are shown. **, p < 0.01; ***, p < 0.001.
Integrins are heterodimers consisting of α and β subunits, both of which are type I transmembrane proteins with a small cytosolic tail and a large extracellular domain. The best-characterized integrin complex in HeLa cells is the α5β1 integrin (Yu et al., 2011; Jia et al., 2016; Hang et al., 2017), which mediates cell adhesion by binding to fibronectin, and this requires proper N-glycosylation of α5β1 integrin (Guo et al., 2002; Isaji et al., 2006). Given the reduced binding of GRASP-depleted cells to fibronectin-coated plates (Figure 1 and Supplemental Figure 1), we further examined the effects of GRASP depletion on the protein level of α5 integrin using two antibodies that recognize the N- or C-terminus of the protein, or the heavy (∼130 kDa) or light (∼19 kDa) chain of the mature protein, respectively (Figure 5B). Depletion of GRASP proteins by siRNA, or knockout of GRASPs by CRISPR/cas9, both significantly reduced α5β1-integrin protein levels (Figure 5, C and D, and Supplemental Figure 4, A and B), whereas the protein level of other cell surface proteins, such as transferrin receptor (TfR) and insulin-like growth factor-1 receptor (IGF-R) did not change (Figure 5F, and Supplemental Figure 4, A and B). In contrast to GRASP depletion, disruption of the Golgi structure by nocodazole (Noc) or brefeldin A (BFA) treatment had no effect on the protein level of α5β1 integrin (Supplemental Figure 4C). In addition, disruption of the Golgi ribbon by Golgin 84 depletion had no effect on integrin level (Supplemental Figure 6 in Xiang et al., 2013). These results indicate a unique role of Golgi stacking rather than ribbon linking in regulating integrin-mediated cell adhesion and migration.
Through the use of both N- and C-terminal antibodies, we confirmed that α5-integrin heavy-chain, light-chain, and full-length proteins were equally decreased in GRASP-knockout HeLa cells (Figure 5, B–D). In addition, we analyzed the samples under both reducing and nonreducing conditions. Under the nonreducing condition, both heavy and light chains are held together by a disulfide bond to form mature α5 integrin; while under the reducing condition, the heavy and light chains of α5 integrin are unlinked when the disulfide bond is disrupted by dithiothreitol (DTT). As shown in Figure 5E, both mature α5 integrin and its individual components were equally reduced by GRASP knockout.
As a cell surface receptor, mature α5β1 integrin is mainly localized at the plasma membrane and is dynamically internalized and recycled. Given that only those molecules at the surface are functional in cell adhesion, we performed a cell surface biotinylation and streptavidin pull-down assay. The results showed that the cell surface level of α5β1 integrin was robustly reduced in GRASP-knockout cells compared with the level in control cells (Figure 5F). Another plasma membrane protein, TfR, was unaffected by GRASP knockout and was also pulled down, while the cytosolic protein actin was not detected in the pull-down. As an internal control, only the glycosylated upper band of β1 integrin was isolated (Figure 5F). These results demonstrate that GRASP depletion reduces α5β1-integrin level at the cell surface, which subsequently decreases cell adhesion.
α5 Integrin is synthesized by the ER and transported through the Golgi to the plasma membrane. In the TGN, it is cleaved by PCs to become mature. To determine whether GRASP depletion reduces α5-integrin level by inducing ER-associated protein degradation (ERAD) or by affecting its maturation, we blocked newly synthesized α5 integrin in the ER by BFA treatment for 2 h and released it for indicated times in the presence of cycloheximide (CHX) and MG132 or monensin. MG132 inhibits ERAD, while monensin blocks the trafficking of α5 integrin from the Golgi stack to the TGN and thus inhibits α5-integrin cleavage. In this experiment, we determined the level of the full-length uncleaved form of α5 integrin on reducing gels. As GRASP KO cells have lower integrin levels, we exposed those gels longer, and thus all cell lines had a similar signal at the 0 time point to start with, and the reduction of the protein was assessed over time. As shown in Figure 5G, the decrease of full-length α5 integrin in GRASP-knockout cells was faster than in WT cells. The reduction of full-length α5 integrin could be the end result of one of two processes. One is ERAD; this possibility was ruled out, because the α5-integrin level was not rescued by MG132 (Figure 5G, lane 5 vs. lane 4). The other possibility is that α5 integrin is cleaved by PCs in the TGN to mature; this explanation was supported by the inhibition of the reduction by monensin (Figure 5G, lane 6 vs. lane 4). These results demonstrate that GRASP KO does not cause ERAD of α5 integrin; rather, it accelerates integrin trafficking and maturation, which is consistent with our previous observation in GRASP-depleted cells (Xiang et al., 2013).
Expression of α5β1 integrin rescues the attachment and migration defects of GRASP-KO cells
So far, we have confirmed that GRASP depletion reduces α5β1-integrin level (Figure 5) and decreases cell attachment and migration (Figures 1–4). Reexpression of GRASP in the related GRASP-KO cells not only restored the α5β1-integrin level (Figure 3C) but also the defects in cell attachment (Figure 1, G and H, and Supplemental Figure 1) and migration (Figures 3 and 4 and Supplemental Figures 1 and 3). To determine whether the reduction of α5β1 integrin is the main reason for the decreased cell attachment and migration in GRASP-depleted cells, we exogenously expressed α5 integrin-GFP, which is known to restore the protein level of both α5 and β1 integrins (Isaji et al., 2006, 2009). As expected, expression of α5-GFP, but not GFP alone, increased the protein levels of both α5 and β1 integrins in GRASP single- and double-knockout cells to levels comparable to those of WT cells (Figure 5H). Significantly, restoration of α5β1-integrin level in GRASP-KO cells largely rescued the reduced cell adhesion (Figure 5I) and migration (Supplemental Figure 5) phenotype. These results demonstrate that GRASP depletion decreases cell adhesion and migration via the reduction of α5β1 integrin.
GRASP depletion reduces α5β1-integrin protein synthesis
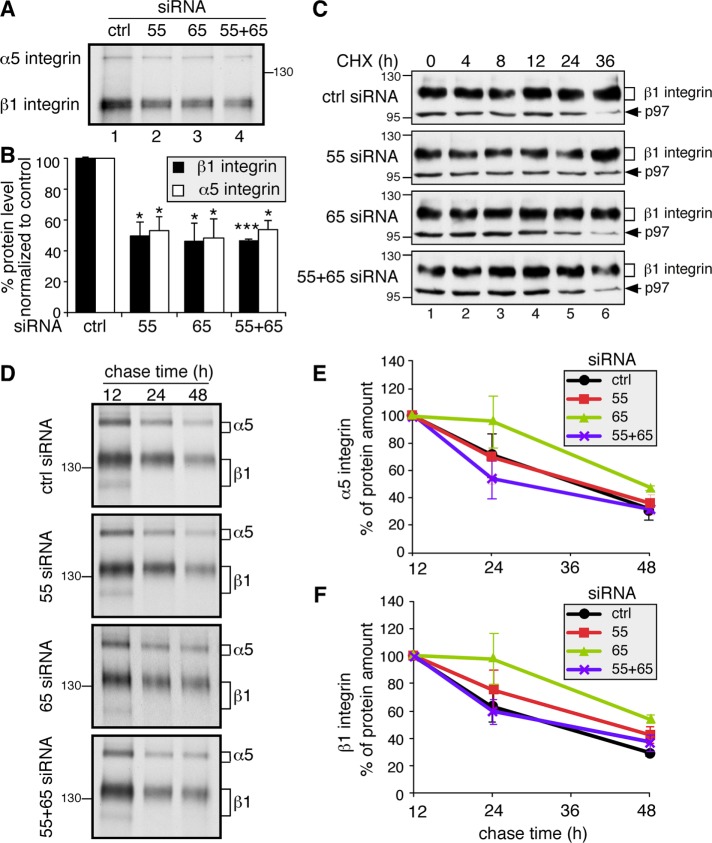
To determine whether the reduction of α5β1-integrin level is due to decreased protein synthesis or increased degradation, we assessed integrin synthesis by measuring the incorporation of 35S-labeled methionine and cysteine. Cells transfected with control or GRASP siRNAs were incubated in a medium containing 35S-labeled methionine and cysteine for 1 h, and α5 and β1 integrins were immunoprecipitated and analyzed by SDS–PAGE and autoradiography. The newly synthesized α5 and β1 integrins existed as single bands of 135 and 110 kDa, respectively (Figure 6A), corresponding to their immature ER forms. Notably, the amounts of α5 and β1 integrins were reduced by ∼50% following GRASP depletion (Figure 6B), consistent with the observation that the α5- and β1-integrin protein levels were reduced in GRASP-KO cells in a steady state (Figure 5, A–E).
FIGURE 6:
GRASP depletion reduces α5- and β1-integrin synthesis but has no effect on their turnover. (A) GRASP depletion reduces protein synthesis of α5 and β1 integrin. An equal number of HeLa cells transfected with indicated siRNAs were labeled with Trans 35S-Label [35S] for 1 h. Immunoprecipitated α5β1 integrin was analyzed by gel electrophoresis and autoradiography. The protein synthesis rates of both α5 and β1 integrin decreased in GRASP-depleted cells. (B) Quantitation of A. Statistical analysis was performed by comparison with control (ctrl) siRNA-treated cells. *, p < 0.05; ***, p < 0.001. (C) Western blot of GRASP-depleted HeLa cells treated with CHX for the indicated times. At 72 h posttransfection with the indicated siRNA, cells were treated with 100 µM CHX for 0, 4, 8, 12, 24, and 36 h; lysed; and analyzed by Western blot for β1 integrin and p97 on the same gel. As GRASP knockdown cells have a lower level of integrins, we exposed those gels longer, so all cell lines had a similar signal at the 0 time point to start with, and the reduction of the protein was assessed over time. (D) GRASP depletion does not increase α5- and β1-integrin degradation. HeLa cells transfected with indicated siRNAs were labeled with Trans 35S-Label [35S] for 1 h and chased for 12, 24, and 48 h. Immunoprecipitated α5β1 integrins were analyzed by SDS–PAGE and autoradiography. (E) Quantification of α5 integrin in D. Note that there is no significant difference in the degradation rate of α5 integrin between control siRNA-treated and GRASP-depleted cells. (F) Quantification of β1 integrin in D. There is no significant difference in the degradation rate of β1 integrin between control siRNA-treated and GRASP-depleted cells.
To test the possibility that GRASP depletion may also affect the stability of α5β1 integrin, we blocked protein synthesis by CHX treatment and assessed the α5β1-integrin level over time. These proteins were stable, with no significant reduction within 36 h of CHX treatment, and were not affected by GRASP knockdown (Figure 6C) or knockout (Supplemental Figure 6). These results are consistent with previous reports that integrins are stable proteins in the cell (Lobert et al., 2010; Bottcher et al., 2012). To further confirm these results by an alternative approach, we performed the same metabolic labeling and pulse–chase assay as described earlier. HeLa cells transfected with control or GRASP siRNAs were incubated in a medium containing 35S-labeled methionine and cysteine for 1 h; chased for 12, 24, or 48 h; and lysed. Then α5 and β1 integrin were immunoprecipitated and analyzed by nonreducing SDS–PAGE and autoradiography. We found that, within 48 h, the degradation rates of both α5 and β1 integrin were not significantly affected by GRASP depletion (Figure 6, D–F). Therefore, we conclude that GRASP depletion reduces the synthesis of α5β1 integrin but does not affect their stability.
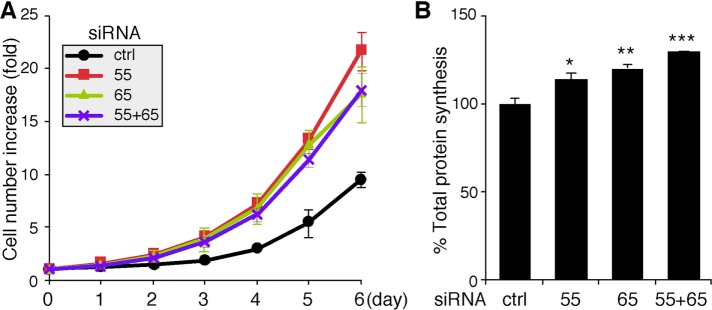
GRASP depletion enhances total protein synthesis and cell growth
To determine how GRASP depletion–mediated Golgi destruction affects cell growth, we examined the growth rate of GRASP-depleted cells using an established crystal violet assay (Tang et al., 2010). Cells were transfected with indicated control or GRASP siRNAs and seeded into 24-well plates at 24 h after transfection. After incubation for 24 h, the number of cells was assessed for 6 successive days. As shown in Figure 7A, the increase in the cell number was significantly higher in cells treated with GRASP siRNAs compared with those treated with control siRNA.
FIGURE 7:
Depletion of GRASP55 and/or GRASP65 enhances cell growth and total protein synthesis. (A) Growth rate of HeLa cells transfected with the indicated siRNAs, as measured using crystal violet staining. The measurement began at 48 h after transfection. (B) Total protein synthesis is enhanced in GRASP-depleted cells. HeLa cells transfected with indicated siRNAs were labeled with Trans 35S-Label [35S] for 1 h. Equal amounts of total proteins were precipitated with TCA, and [35S]methionine/cysteine incorporation was assessed by scintillation counting. Statistical analysis was performed by normalization and comparison with control (ctrl) siRNA-treated cells using Student’s t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We then determined the total amount of protein synthesis occurring in these cells. Cells treated with control or GRASP siRNAs were labeled using 35S-labeled methionine and cysteine for 1 h and lysed in detergent. Equal amounts of total protein were then precipitated by trichloroacetic acid (TCA), and the total amount of radioactivity incorporated was measured using a scintillation counter. As shown in Figure 7B, compared with control siRNA-treated cells, the total protein synthesis increased when GRASPs were depleted. This result is consistent with the observation that Golgi unstacking increases cell growth, suggesting a link between protein synthesis, trafficking, posttranslational modification, and cell growth.
DISCUSSION
In this study, we found that disruption of the Golgi structure by GRASP knockdown or knockout impacted multiple cell activities, including decreasing cell attachment, migration, and invasion, which could be explained by the decreased level of α5β1 integrin. We also found that GRASP depletion increased cell growth, which may be attributed to the increased overall protein synthesis and accelerated protein trafficking. Based on the literature, this is the first systematic study that links Golgi structure formation and function with cellular activities, including cell attachment, migration, and growth. It is reasonable to speculate that Golgi fragmentation found in diseases may cause global defects in protein trafficking, glycosylation, and secretion that impact essential cell activities. Thus, studying the cellular activities under Golgi destruction and discovering the factors that drive these changes will help us understand disease pathogenesis. In the long term, molecular tools may be developed to restore normal Golgi structure and function under disease conditions and thus to delay disease development.
Using GRASP-knockdown and GRASP-KO cells, we demonstrated that disrupting the Golgi stacks reduced cell adhesion and migration. As integrins are key proteins in these cellular activities, we determined the protein level of nine integrins that are known to be expressed in HeLa cells. Among these integrin subunits, α5 and β1, the major and best-characterized integrins in HeLa cells, exhibited the most dramatic reduction in GRASP-KO cells, while other integrins did not change or only modestly decreased. This demonstrates that different integrins are regulated differently in their expression, as previously described (Judware and Culp, 1997; Palomino et al., 2005). We then used two different antibodies and different gel systems to further characterize α5β1 integrin in GRASP-KO cells. We found that both mature integrins and the individual components in the complex were all reduced by GRASP depletion. More importantly, restoration of the α5β1-integrin level in GRASP-KO cells rescued the reduced cell adhesion and migration phenotype. These results indicate that the reduced cell adhesion and migration after GRASP depletion is mostly attributable to the reduced α5β1-integrin expression.
Our results are consistent with previous reports that a higher expression level of α5 integrin or increased trafficking to the plasma membrane increases cell attachment and migration (Wan et al., 2014; Breuksch et al., 2017). Given that the Golgi plays a critical role in protein glycosylation and sorting (Zhang and Wang, 2016; Huang and Wang, 2017) and that proper glycosylation of α5β1 integrin is important for its function and activation (Zheng et al., 1994; Guo et al., 2002; Gu and Taniguchi, 2004; Hang et al., 2017), we expected that Golgi structural defects might affect integrin glycosylation, trafficking, and degradation. There are two possible reasons for the reduced α5β1-integrin level in GRASP-depleted cells: one is increased degradation, the other is decreased synthesis. Given that protein glycosylation has been thought to be a key mechanism to help protein folding and maintain protein stability (Live et al., 1996; Shental-Bechor and Levy, 2008; Sola and Griebenow, 2009; Lee et al., 2015), we originally expected that GRASP depletion might affect integrin stability. However, the results demonstrated that GRASP depletion did not trigger integrin degradation by ERAD or its turnover; instead, it reduced α5β1-integrin synthesis, although the underlying mechanism remains unknown. In addition, GRASP depletion also accelerated α5-integrin trafficking and maturation, which is consistent with our previous findings (Xiang et al., 2013).
In contrast to the reduced integrin synthesis, total protein synthesis was increased by GRASP depletion, while some other proteins at the cell surface, such as IGFR and TfR, were unaffected. The observation of increased protein synthesis in this study could also be attributed to the increased demand of the cells for more proteins with increased trafficking and cell growth. It has been demonstrated that, at the onset of mitosis, phosphorylation of GRASPs and golgins by mitotic kinases results in Golgi disassembly, which is required for cell cycle progression (Jesch et al., 2001; Sutterlin et al., 2002; Duran et al., 2008; Xiang and Wang, 2010; Guizzunti and Seemann, 2016), and GRASP depletion accelerates cell cycle progression (Tang et al., 2010). It is conceivable that cells with unstacked Golgi grow faster, as protein synthesis and cargo transport are enhanced. Some signaling pathways may be selectively activated or inhibited to control the synthesis of different proteins. Exploiting the underlying mechanism will be a future direction of this study.
In summary, we found that disrupting the Golgi structure by GRASP knockdown or knockout results in decreased cell adhesion and migration due to decreased synthesis of α5β1 integrin. These cells grow faster, possibly because of accelerated protein trafficking and increased overall protein synthesis.
MATERIALS AND METHODS
Reagents, plasmids, and antibodies
All reagents were purchased from Sigma-Aldrich, Roche, Calbiochem, and Fisher unless otherwise stated. Most of the cDNA constructs used in this paper were described previously: pEGFP-N1-GRASP55 (WT), pEGFP-N1-GRASP65 (WT), and pEGFP-N1 (Xiang and Wang, 2010; Zhang et al., 2018). pEGFP-N1-α5-integrin (WT) cDNA construct was a kind gift from Jianguo Gu (Tohoku Medical and Pharmaceutical University). Antibodies used in this study include monoclonal antibodies against β-actin (Proteintech Group, cat. no. 66009-1-Ig), GFP (Proteintech, cat. no. 66002-1-lg), α1 integrin (Santa Cruz, cat. no. SC-271034), α2 integrin (Santa Cruz, cat. no. SC-74466), α3 integrin (Santa Cruz, cat. no. SC-374242), α5 integrin (DSHB, cat. no. BIIG2), α6 integrin (Santa Cruz, cat. no. SC-19622), αV integrin (Santa Cruz, cat. no. SC-376156), β1 integrin (DSHB, cat. no. P5D2), β3 integrin (Santa Cruz, cat. no. SC-46655), β5 integrin (Santa Cruz, cat. no. SC-398214), TfR (Invitrogen, cat. no. 13-6800), and α-tubulin (DSHB, Cat# AA4.3); polyclonal antibodies against human GRASP55 (Proteintech Group, cat. no. 10598-1-AP), IGF-Rβ (Santa Cruz, cat. no. SC-713), α5-integrin C-terminus (Millipore, cat. no. AB1928), α5-integrin N-terminus (Cell Signaling, cat. no. 4705), β1-integrin C-terminus (Abcam, cat. no. EP1041Y), β1-integrin C-terminus (Cell Signaling, cat. no. 4706), and human GRASP65 (gift from Joachim Seemann, University of Texas Southwestern Medical Center).
Cell culture, transfection, and treatment
For cell cultures, HeLa and MDA-MB-231 cells were grown in DMEM (Invitrogen) containing 10% super calf serum (Gemini, cat. no. 100-510) and glutamine at 37°C in a 5% CO2 incubator. For knockdown of GRASP55 and/or GRASP65, HeLa cells were plated at 40% confluency in six-well plates and 3 µl of a 50 µM siRNA stock was added to 250 µl of Opti-MEM. In a separate tube, 5 µl of transfectamine RNAiMAX (Invitrogen) was mixed with 250 µl of Opti-MEM and incubated for 5 min at room temperature. The two mixtures were combined and incubated at room temperature for 20 min and then added to the cells in 2 ml DMEM containing 10% super calf serum. siRNAs for human GRASP55 (AACTGTCGAGAAGTGATTATT) and GRASP65 (CCTGAAGGCACTACTGAAAGCCAAT) (Xiang and Wang, 2010; Xiang et al., 2013) were purchased from Ambion and Invitrogen, respectively. Control nonspecific siRNAs were purchased from Ambion. GRASP-KO cell lines were described previously (Bekier et al., 2017).
For expression of exogenous GRASP proteins, HeLa cells of ∼60% confluency were transfected with the indicated GRASP constructs (Tang et al., 2010; Xiang and Wang, 2010). For a 10-cm plate, 10 µg of pEGFP-N1-GRASP65 (WT) construct or 15 µg of pEGFP-N1-GRASP55 (WT) construct was mixed with 30 µl of polyethylenimine (PEI) and 1 ml of serum-free medium for 15 min at room temperature and then added to the cells in 9 ml of DMEM containing 10% super calf serum. For the control treatment, 10 µg of pEGFP-N1 construct was mixed with 30 µl of PEI and 1 ml of serum-free medium for 15 min and then added to the cells in 9 ml of DMEM containing 10% super calf serum.
For restoration of the α5β1-integrin level in GRASP-KO cells, cells at ∼60% confluency in a 6-cm plate were transfected with 8 µg of α5-GFP cDNA construct by Lipofectamine 2000 (Invitrogen, cat. no. 11668019) following the manufacture’s instructions, and pEGFP-N1 was used as a control. In three independent experiments, the transfection rate of α5-GFP in GRASP55, GRSAP65, and double-knockout cells was 66 ± 1%, 67 ± 3%, and 71 ± 1%, respectively. This was tested at 72 h posttransfection, a time point used for the cell adhesion and migration assays.
For drug treatment, 5 µg/ml BFA, 500 ng/ml Noc, 100 µg/ml CHX, 50 µM MG132, or 2 µM monensin was directly applied to the medium alone or in combination for indicated times. Cells were lysed and analyzed by Western blotting.
Cell attachment assay
For cell attachment, a total of 3 × 105 control or GRASP siRNA-treated cells were seeded on fibronectin-coated plates and incubated in serum-free medium for 30 min. Twelve-well plates were coated with fibronectin from human plasma (Sigma, F0895) at a concentration of 1 µg/ml in Tris-buffered saline (pH 7.5) at 4°C for 16 h and blocked with 2% bovine serum albumin (BSA) for 30 min. HeLa cells transfected with the indicated siRNAs, or GRASP-KO HeLa cells with or without GRASP or integrin rescue, were detached using 20 mM EDTA in phosphate-buffered saline (PBS), pelleted, and resuspended in serum-free DMEM. The cells were seeded on the plate (3 × 105 cell per well, three wells for each condition) and cultured in serum-free DMEM at 37°C for 30 min. Cells were then gently washed four times with PBS, and the attached cells were treated with 20 mM EDTA, collected by centrifugation, resuspended in 200 µl of growth medium, and counted using a hemocytometer. Attachment was presented as the percentage of attached cells out of the total number of cells.
Wound-healing assay
The wound-healing assay used here was performed using the protocol described by Jun-Lin Guan and colleagues (Liang et al., 2007). MDA-MB-231 or HeLa cells were transfected with the indicated plasmids; and at 48 h after transfection, the cells were replated in 12-well plates at 80% confluence in complete growth medium. Twenty-four hours later, after the cells became confluent and formed a monolayer, the medium was replaced with serum-free DMEM. Twenty-four hours afterward, the cell monolayer was scraped in a straight line to create a “scratch” with a 200-μl pipette tip. The cells were washed once with DMEM, and images were taken using an inverted bright-field microscope with a 4× lens and an IMAGING ERTIGA 1300R camera. The cells were then allowed to grow in complete growth medium for 20 h, and images of the same area were taken. Images were analyzed with ImageJ software (National Institutes of Health) or WimScratch: Wound Healing Assay Image Analysis Solution (Release 4.0; available from www.wimasis.com/en/products/9/WimScratch) (Jameson et al., 2013).
Transwell assay
HeLa cells were detached with 20 mM EDTA and resuspended in serum-free DMEM. Cells were seeded in Transwell inserts (BD BioCoat control 8.0-µm-pore-size inserts, cat. no. 0877121) at a concentration of 5 × 104 in 250 μl of serum-free DMEM. In the lower chamber, 750 μl of full growth medium was added as a chemoattractant. After incubation at 37°C for 20 h, cells on the upper side were gently removed by scraping with a cotton swab. The membranes in the inserts were fixed with 4% paraformaldehyde (PFA) for 30 min and stained with 0.5% crystal violet for 1 h, and cells that had migrated to the lower side were visualized under a Leica MZ FLIII stereomicroscope. Images were analyzed with ImageJ software (National Institutes of Health).
Cell growth rate
HeLa cells transfected with the indicated siRNAs were trypsinized 24 h after transfection. Cells (1 × 104) were plated in 24-well plates and incubated in complete growth medium. One day later, the cell number was measured every 24 h for 6 d using crystal violet staining, as described previously (Feinstein and Linstedt, 2007). Briefly, cells were washed with PBS, fixed in 4% PFA for 30 min, stained with 0.5% crystal violet (in 30% methanol) for 15 min, and extensively washed with H2O. The crystal violet was extracted using 1 ml of 10% acetic acid for 5 min, and the optical density was measured at 590 nm. The cell number at different times was normalized to the cell number from the first measurement (day 0).
Radioactive labeling and immunoprecipitation
This method was previously described (Xiang et al., 2013). Briefly, HeLa cells grown in 60-mm dishes were incubated in methionine/cysteine-free DMEM (Invitrogen) for 1 h and labeled with 1 ml of medium containing 250 µCi/ml Trans 35S-Label [35S] (MP Biomedicals, cat. no. 15100614) for 1 h. After being washed with PBS, cells were collected either immediately on ice or after incubation in 4 ml of complete growth medium containing 2 mM l-cysteine and l-methionine for the indicated time periods. Cells were lysed in 0.8 ml of lysis buffer (PBS containing 1 mM CaCl2, 0.5 mM MgCl2, 1% Triton X-100, protease inhibitor cocktail, and 1 µM pepstatin A) and centrifuged at 14,000 rpm for 15 min. The protein concentration of the supernatant was measured using a Bio-Rad protein assay (Bio-Rad Laboratories, cat. no. 5000006). The cell lysates (2 mg) were immunoprecipitated with 50 ng of the α5-integrin antibody BIIG2 and 60 ng of the β1-integrin antibody P5D2 overnight at 4°C. The antibodies were precipitated with 30 µl of protein G beads (Roche Diagnostics GmbH, Germany) at 4°C for 2 h. The beads were washed five times with lysis buffer, and the immunoisolated materials were eluted by boiling for 5 min in nonreducing SDS sample buffer. Integrins were resolved using 6.5% nonreducing SDS–PAGE followed by autoradiography.
For determination of total protein synthesis, 40 µl of the cell lysate was precipitated with 10 µl of 100% (wt/vol) TCA at 4°C for 10 min. The protein pellets were washed with acetone, dissolved in 0.2 M NaOH, neutralized with 0.2 M HCl, and mixed with Scintiverse BD cocktail (Fisher Scientific, cat. no. B14001). The incorporation of radioactivity was analyzed using scintillation counting and normalized based on the total protein amount.
Cell surface biotinylation
All procedures were performed on ice or at 4°C. Cells grown on 15-cm plates were incubated on ice for 20 min, washed three times with PBS, treated with 6 ml of 0.5 mg/ml NHS-SS-biotin (Fisher, cat. no. P121331) in PBS for 20 min, and quenched by 100 mM glycine/PBS for 10 min. After three washes with PBS, cells were lysed in RIPA buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM EDTA, 0.5% sodium orthovanadate, 0.1% sodium fluoride, and 1× protease inhibitor cocktail [Bimake, cat. no. B14001]). Cell lysates were adjusted to 5.86 mg/ml in lysis buffer, and 3-mg samples were incubated with 50 μl of streptavidin-agarose beads (GE Healthcare, cat. no.17-511-01) at 4°C overnight. After being washed, beads were boiled in SDS sample buffer with 100 mM DTT, and bound proteins were analyzed by SDS–PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and blotted with α5-integrin (Cell Signaling, cat. no. 4705), β1-integrin (Cell Signaling, cat. no. 4706), TfR (Invitrogen, cat. no. 13-6800), and β-actin (Proteintech, cat. no. 66009-1-Ig) antibodies.
Quantification and statistics
In all figures, the quantification results are expressed as the mean ± SEM from three independent experiments, unless otherwise stated. The statistical significance of the results was assessed using a Student’s t test. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
Supplementary Material
Acknowledgments
We thank Cunming Duan (University of Michigan) for IGFR antibodies, Joachim Seemann for GRASP65 antibodies, Sai Srinivas Panapakkam Giridharan (University of Michigan) for TfR antibodies, Jianguo Gu for the α5 integrin-GFP construct, and members of the Wang lab for stimulating discussions and technical support. This work was supported by the National Institutes of Health (grant GM112786), MCubed, and the Fast Forward Protein Folding Disease Initiative of the University of Michigan to Y.W. and a University of Michigan Rackham Predoctoral fellowship to E.A.
Abbreviations used:
- BFA
brefeldin A
- BSA
bovine serum albumin
- CHX
cycloheximide
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- GFP
green fluorescent protein
- GRASPs
Golgi reassembly and stacking proteins
- IGF-R
insulin-like growth factor-1 receptor
- KO
knockout
- Noc
nocodazole
- PBS
phosphate-buffered saline
- PC
proprotein convertase
- PEI
polyethylenimine
- PFA
paraformaldehyde
- siRNA
small interfering RNA
- TCA
trichloroacetic acid
- TfR
transferrin receptor
- TGN
trans-Golgi network
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-07-0462) on January 16, 2019.
REFERENCES
- Aridor M, Balch WE. (1999). Integration of endoplasmic reticulum signaling in health and disease. Nat Med , 745–751. [DOI] [PubMed] [Google Scholar]
- Barr FA, Puype M, Vandekerckhove J, Warren G. (1997). GRASP65, a protein involved in the stacking of Golgi cisternae. Cell , 253–262. [DOI] [PubMed] [Google Scholar]
- Bekier ME, 2nd, Wang L, Li J, Huang H, Tang D, Zhang X, Wang Y. (2017). Knockout of the Golgi stacking proteins GRASP55 and GRASP65 impairs Golgi structure and function. Mol Biol Cell , 2833–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman M, Joukov V, Virtanen I, Alitalo K. (1995). Overexpressed Csk tyrosine kinase is localized in focal adhesions, causes reorganization of alpha v beta 5 integrin, and interferes with HeLa cell spreading. Mol Cell Biol , 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzaro N, Pasini P, Ghirardello A, Finco B. (1999). High anti-Golgi autoantibody levels: an early sign of autoimmune disease? Clin Rheumatol , 346–348. [DOI] [PubMed] [Google Scholar]
- Bottcher RT, Stremmel C, Meves A, Meyer H, Widmaier M, Tseng HY, Fassler R. (2012). Sorting nexin 17 prevents lysosomal degradation of beta1 integrins by binding to the beta1-integrin tail. Nat Cell Biol , 584–592. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Barlowe C. (2013). Organization of the ER-Golgi interface for membrane traffic control. Nat Rev , 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuksch I, Prosinger F, Baehr F, Engelhardt FP, Bauer HK, Thuroff JW, Heimes AS, Hasenburg A, Prawitt D, Brenner W. (2017). Integrin alpha5 triggers the metastatic potential in renal cell carcinoma. Oncotarget , 107530–107542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JW, Granovsky M, Warren CE. (1999). Glycoprotein glycosylation and cancer progression. Biochim Biophys Acta , 21–34. [DOI] [PubMed] [Google Scholar]
- Diaz-Corrales FJ, Asanuma M, Miyazaki I, Ogawa N. (2004). Rotenone induces disassembly of the Golgi apparatus in the rat dopaminergic neuroblastoma B65 cell line. Neurosci Lett , 59–63. [DOI] [PubMed] [Google Scholar]
- Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, Yates J, Zimmerman T, Malhotra V. (2008). The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol Biol Cell , 2579–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand G, Seta N. (2000). Protein glycosylation and diseases: blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin Chem , 795–805. [PubMed] [Google Scholar]
- Evin G. (2015). How accelerated Golgi trafficking may drive Alzheimer’s disease (comment on DOI 10.1002/bies.201400116). Bioessays , 232–233. [DOI] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. (2007). Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol Biol Cell , 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze HH, Ng BG. (2011). Golgi glycosylation and human inherited diseases. Cold Spring Harb Perspect Biol , a005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzler MJ, Etherington J, Sokoluk C, Kinsella TD, Valencia DW. (1984). Antibodies from patients with autoimmune disease react with a cytoplasmic antigen in the Golgi apparatus. J Immunol , 2904–2908. [PubMed] [Google Scholar]
- Goldfischer S. (1982). The internal reticular apparatus of Camillo Golgi: a complex, heterogeneous organelle, enriched in acid, neutral, and alkaline phosphatases, and involved in glycosylation, secretion, membrane flow, lysosome formation, and intracellular digestion. J Histochem Cytochem , 717–733. [DOI] [PubMed] [Google Scholar]
- Gu J, Taniguchi N. (2004). Regulation of integrin functions by N-glycans. Glycoconj J , 9–15. [DOI] [PubMed] [Google Scholar]
- Guizzunti G, Seemann J. (2016). Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression. Proc Natl Acad Sci USA , E6590–E6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HB, Lee I, Kamar M, Akiyama SK, Pierce M. (2002). Aberrant N-glycosylation of beta1 integrin causes reduced alpha5beta1 integrin clustering and stimulates cell migration. Cancer Res , 6837–6845. [PubMed] [Google Scholar]
- Hang Q, Isaji T, Hou S, Wang Y, Fukuda T, Gu J. (2017). A key regulator of cell adhesion: identification and characterization of important N-glycosylation sites on integrin alpha5 for cell migration. Mol Cell Biol , e00558-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. (2007). In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev , 1377–1408. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. (2001). Intracellular functions of N-linked glycans. Science , 2364–2369. [DOI] [PubMed] [Google Scholar]
- Hilditch-Maguire P, Trettel F, Passani LA, Auerbach A, Persichetti F, MacDonald ME. (2000). Huntingtin: an iron-regulated protein essential for normal nuclear and perinuclear organelles. Hum Mol Genet , 2789–2797. [DOI] [PubMed] [Google Scholar]
- Huang S, Wang Y. (2017). Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000Res , 2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaji T, Sato Y, Fukuda T, Gu J. (2009). N-glycosylation of the I-like domain of beta1 integrin is essential for beta1 integrin expression and biological function: identification of the minimal N-glycosylation requirement for alpha5beta1. J Biol Chem , 12207–12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaji T, Sato Y, Zhao Y, Miyoshi E, Wada Y, Taniguchi N, Gu J. (2006). N-glycosylation of the beta-propeller domain of the integrin alpha5 subunit is essential for alpha5beta1 heterodimerization, expression on the cell surface, and its biological function. J Biol Chem , 33258–33267. [DOI] [PubMed] [Google Scholar]
- Jameson KL, Mazur PK, Zehnder AM, Zhang J, Zarnegar B, Sage J, Khavari PA. (2013). IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat Med , 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik ME, Litynska A, Vereecken P. (2010). Cell migration—the role of integrin glycosylation. Biochim Biophys Acta , 545–555. [DOI] [PubMed] [Google Scholar]
- Jesch SA, Lewis TS, Ahn NG, Linstedt AD. (2001). Mitotic phosphorylation of Golgi reassembly stacking protein 55 by mitogen-activated protein kinase ERK2. Mol Biol Cell , 1811–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Howlader MA, Cairo CW. (2016). Integrin-mediated cell migration is blocked by inhibitors of human neuraminidase. Biochim Biophys Acta , 1170–1179. [DOI] [PubMed] [Google Scholar]
- Joshi G, Chi Y, Huang Z, Wang Y. (2014). Abeta-induced Golgi fragmentation in Alzheimer’s disease enhances Abeta production. Proc Natl Acad Sci USA , E1230–E1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judware R, Culp LA. (1997). Concomitant down-regulation of expression of integrin subunits by N-myc in human neuroblastoma cells: differential regulation of alpha2, alpha3 and beta1. Oncogene , 1341–1350. [DOI] [PubMed] [Google Scholar]
- Klumperman J. (2011). Architecture of the mammalian Golgi. Cold Spring Harb Perspect Biol , 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klute MJ, Melancon P, Dacks JB. (2011). Evolution and diversity of the Golgi. Cold Spring Harb Perspect Biol , a007849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Bane SM, Kawle PD, Naresh KN, Kalraiya RD. (2005). Altered melanoma cell surface glycosylation mediates organ specific adhesion and metastasis via lectin receptors on the lung vascular endothelium. Clin Exp Metastasis , 11–24. [DOI] [PubMed] [Google Scholar]
- Lee HS, Qi Y, Im W. (2015). Effects of N-glycosylation on protein conformation and dynamics: Protein Data Bank analysis and molecular dynamics simulation study. Sci Rep , 8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Tiwari N, Dunlop MH, Graham M, Liu X, Rothman JE. (2014). Membrane adhesion dictates Golgi stacking and cisternal morphology. Proc Natl Acad Sci USA , 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Park AY, Guan JL. (2007). In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc , 329–333. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhao F, Gu W, Yang H, Meng Q, Zhang Y, Yang H, Duan Q. (2009). The roles of platelet GPIIb/IIIa and alphavbeta3 integrins during HeLa cells adhesion, migration, and invasion to monolayer endothelium under static and dynamic shear flow. J Biomed Biotechnol , 829243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Live DH, Kumar RA, Beebe X, Danishefsky SJ. (1996). Conformational influences of glycosylation of a peptide: a possible model for the effect of glycosylation on the rate of protein folding. Proc Natl Acad Sci USA , 12759–12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobert VH, Brech A, Pedersen NM, Wesche J, Oppelt A, Malerod L, Stenmark H. (2010). Ubiquitination of alpha 5 beta 1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev Cell , 148–159. [DOI] [PubMed] [Google Scholar]
- Marsh BJ, Howell KE. (2002). The mammalian Golgi–complex debates. Nat Rev , 789–795. [DOI] [PubMed] [Google Scholar]
- Mierke CT, Frey B, Fellner M, Herrmann M, Fabry B. (2011). Integrin alpha5beta1 facilitates cancer cell invasion through enhanced contractile forces. J Cell Sci , 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, Hattori N, Kitada T, Matsumine H, Mori H, Shimura H, Kubo S, Kobayashi H, Asakawa S, Minoshima S, Shimizu N. (2001). Familial Parkinson’s disease. Alpha-synuclein and parkin. Adv Neurol , 13–21. [PubMed] [Google Scholar]
- Ng ML, Tan SH, See EE, Ooi EE, Ling AE. (2003). Proliferative growth of SARS coronavirus in Vero E6 cells. J Gen Virol , 3291–3303. [DOI] [PubMed] [Google Scholar]
- Ono M, Hakomori S. (2004). Glycosylation defining cancer cell motility and invasiveness. Glycoconj J , 71–78. [DOI] [PubMed] [Google Scholar]
- Palomino WA, Fuentes A, Gonzalez RR, Gabler F, Boric MA, Vega M, Devoto L. (2005). Differential expression of endometrial integrins and progesterone receptor during the window of implantation in normo-ovulatory women treated with clomiphene citrate. Fertil Steril , 587–593. [DOI] [PubMed] [Google Scholar]
- Paule S, Aljofan M, Simon C, Rombauts LJ, Nie G. (2012). Cleavage of endometrial alpha-integrins into their functional forms is mediated by proprotein convertase 5/6. Hum Reprod , 2766–2774. [DOI] [PubMed] [Google Scholar]
- Pearse BR, Hebert DN. (2010). Lectin chaperones help direct the maturation of glycoproteins in the endoplasmic reticulum. Biochim Biophys Acta , 684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. (2013). A prize for membrane magic. Cell , 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. (2006). GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol , 238–248. [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Hermo L, Segretain D. (1987). Tridimensional structure of the Golgi apparatus of nonciliated epithelial cells of the ductuli efferentes in rat: an electron microscope stereoscopic study. Biol Cell , 103–115. [DOI] [PubMed] [Google Scholar]
- Ratcliffe CD, Sahgal P, Parachoniak CA, Ivaska J, Park M. (2016). Regulation of cell migration and beta1 integrin trafficking by the endosomal adaptor GGA3. Traffic , 670–688. [DOI] [PubMed] [Google Scholar]
- Roberts JD, Klein JL, Palmantier R, Dhume ST, George MD, Olden K. (1998). The role of protein glycosylation inhibitors in the prevention of metastasis and therapy of cancer. Cancer Detect Prev , 455–462. [DOI] [PubMed] [Google Scholar]
- Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, Florin L. (2013). Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. J Virol , 3435–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shental-Bechor D, Levy Y. (2008). Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci USA , 8256–8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. (1999). GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J , 4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola RJ, Griebenow K. (2009). Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci , 1223–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Hsu P, Mallabiabarrena A, Malhotra V. (2002). Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell , 359–369. [DOI] [PubMed] [Google Scholar]
- Tang D, Yuan H, Vielemeyer O, Perez F, Wang Y. (2012). Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol Open , 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Wang Y. (2010). The Role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic , 827–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielemeyer O, Yuan H, Moutel S, Saint-Fort R, Tang D, Nizak C, Goud B, Wang Y, Perez F. (2009). Direct selection of monoclonal phosphospecific antibodies without prior phosphoamino acid mapping. J Biol Chem , 20791–20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhu F, Zasadil LM, Yu J, Wang L, Johnson A, Berthier E, Beebe DJ, Audhya A, Weaver BA. (2014). A Golgi-localized pool of the mitotic checkpoint component Mad1 controls integrin secretion and cell migration. Curr Biol , 2687–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G. (2005). Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem , 4921–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. (2003). A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J , 3279–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wei JH, Bisel B, Tang D, Seemann J. (2008). Golgi cisternal unstacking stimulates COPI vesicle budding and protein transport. PLoS One , e1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. (2010). GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol , 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang X, Nix DB, Katoh T, Aoki K, Tiemeyer M, Wang Y. (2013). Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat Commun , 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M, Ihara Y, Matsuzawa Y, Taniguchi N. (1996). Aberrant glycosylation of E-cadherin enhances cell-cell binding to suppress metastasis. J Biol Chem , 13811–13815. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang F, Liu H, Adams G, Aikhionbare F, Liu D, Cao X, Fan L, Hu G, Chen Y, et al. (2011). ACAP4 protein cooperates with Grb2 protein to orchestrate epidermal growth factor-stimulated integrin beta1 recycling in cell migration. J Biol Chem , 43735–43747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang L, Lak B, Li J, Jokitalo E, Wang Y. (2018). GRASP55 senses glucose deprivation through O-GlcNAcylation to promote autophagosome-lysosome fusion. Dev Cell , 245–261 e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y. (2015). GRASPs in Golgi structure and function. Front Cell Dev Biol , 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y. (2016). Glycosylation quality control by the Golgi structure. J Mol Biol , 3183–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Fang H, Hakomori S. (1994). Functional role of N-glycosylation in alpha 5 beta 1 integrin receptor. De-N-glycosylation induces dissociation or altered association of alpha 5 and beta 1 subunits and concomitant loss of fibronectin binding activity. J Biol Chem , 12325–12331. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.