Abstract
Ena/VASP tetramers are processive actin elongation factors that localize to diverse F-actin networks composed of filaments bundled by different cross-linking proteins, such as filopodia (fascin), lamellipodia (fimbrin), and stress fibers (α-actinin). Previously, we found that Ena takes approximately threefold longer processive runs on trailing barbed ends of fascin-bundled F-actin. Here, we used single-molecule TIRFM (total internal reflection fluorescence microscopy) and developed a kinetic model to further dissect Ena/VASP’s processive mechanism on bundled filaments. We discovered that Ena’s enhanced processivity on trailing barbed ends is specific to fascin bundles, with no enhancement on fimbrin or α-actinin bundles. Notably, Ena/VASP’s processive run length increases with the number of both fascin-bundled filaments and Ena “arms,” revealing avidity facilitates enhanced processivity. Consistently, Ena tetramers form more filopodia than mutant dimer and trimers in Drosophila culture cells. Moreover, enhanced processivity on trailing barbed ends of fascin-bundled filaments is an evolutionarily conserved property of Ena/VASP homologues, including human VASP and Caenorhabditis elegans UNC-34. These results demonstrate that Ena tetramers are tailored for enhanced processivity on fascin bundles and that avidity of multiple arms associating with multiple filaments is critical for this process. Furthermore, we discovered a novel regulatory process whereby bundle size and bundling protein specificity control activities of a processive assembly factor.
INTRODUCTION
Many important cellular functions depend on formation of actin cytoskeleton networks at the correct time and location with specific architectures and dynamics (Pollard and Cooper, 2009; Campellone and Welch, 2010). For example, filopodia are filamentous actin (F-actin)-rich finger-like protrusions that elongate from the lamellipodium, a dense, branched F-actin network kept short by capping protein (Pollard and Borisy, 2003) at the cell periphery. Filopodia are important for cell motility and environment sensing. Filopodial actin filaments are assembled by actin elongation factors such as formins and Enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) (Mattila and Lappalainen, 2008). The 10–30 filaments in filopodia are bundled primarily by fascin, a globular cross-linking protein containing β-trefoil domains (Vignjevic et al., 2006a; Jansen et al., 2011). Fascin bundles are composed of parallel filaments with narrow spacing, between 8 and 10 nm (Cant et al., 1994; Edwards and Bryan, 1995; Jansen et al., 2011; Yang et al., 2013). During filopodia initiation, Ena/VASP localizes to the edge of the lamellipodium where it can compete with capping protein for barbed ends (Bear et al., 2002; Bear and Gertler, 2009; Svitkina et al., 2003; Barzik et al., 2005; Applewhite et al., 2007; Winkelman et al., 2014) and facilitate generation of long, straight actin filaments. Ena/VASP subsequently localizes to the tips of emerging and mature filopodia, ultimately resulting in fascin-bundled filaments of the same length (Faix and Rottner, 2006; Gupton and Gertler, 2007), presumably ensuring uniform thickness of filopodia required for protrusive force (Svitkina et al., 2003; Winkelman et al., 2014).
Ena/VASP is a multidomain homotetramer with homologues in all metazoan cells (Sebé-Pedrós et al., 2013). A few Ena/VASP homologues have been biochemically characterized, including human VASP (Bachmann et al., 1999; Chereau and Dominguez, 2006; Breitsprecher et al., 2008; Pasic et al., 2008; Hansen and Mullins, 2010), Drosophila Enabled (Winkelman et al., 2014), and Dictyostelium VASP (Breitsprecher et al., 2008). Ena/VASP proteins contain two conserved Ena/VASP homology domains, EVH1 and EVH2 (Figure 1A). The N-terminus EVH1 domain is important for cellular localization and binds to proteins with FPPPP (FP4) repeats, such as lamellipodin, zyxin, and formin (Ball et al., 2001; Bilancia et al., 2014). The C-terminus EVH2 domain consists of three smaller subdomains: G-actin binding domain (GAB) (Bachmann et al., 1999; Ferron et al., 2007), F-actin binding domain (FAB) (Dominguez and Holmes, 2011), and a C-terminal coiled-coil tetramerization domain (Bachmann et al., 1999; Kuhnel et al., 2004). Between the EVH1 and EVH2 domains there is a poly-proline–rich region that binds profilin as well as SH3 domains (Ferron et al., 2007; Hansen and Mullins, 2010).
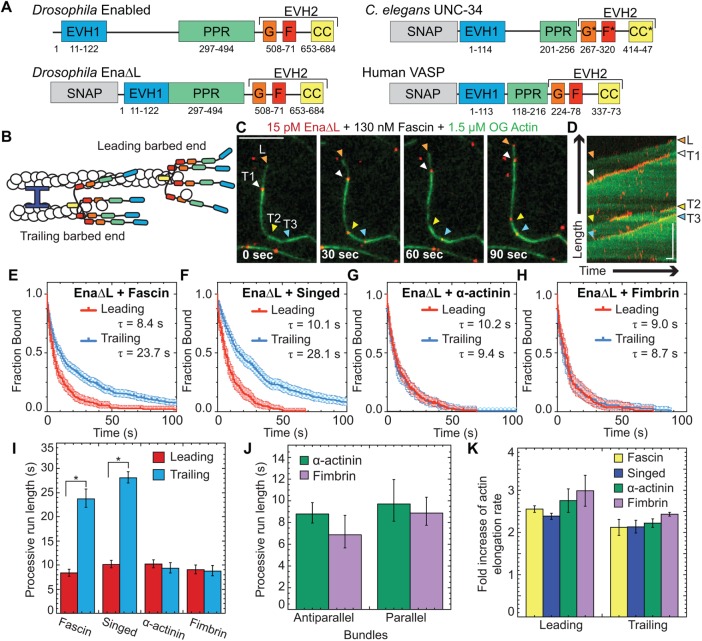
FIGURE 1:
EnaΔL has enhanced processivity on F-actin bundles formed specifically by fascin. (A) Ena/VASP domain organization and constructs used for Ena, UNC-34, and VASP: Self-labeling tag (SNAP), Ena/VASP homology domain 1 (EVH1), polyproline region (PPR), Ena/VASP homology domain 2 (EVH2) includes G-actin binding domain (G), F-actin binding domain (F), coiled coil region (CC). *Putative domain. Two-color TIRFM visualization of 1.5 µM Mg-ATP-actin (15% Oregon green-actin) with 15 pM SNAP(549)-EnaΔL and unlabeled 130 nM human fascin, 250 nM fly fascin Singed, 125 nM a-actinin, or 100 nM fimbrin. (B) Cartoon. Ena/VASPs bound to leading and trailing barbed ends in a fascin bundle. (C, D) Representative experiment of OG-actin with SNAP(549)-EnaΔL and fascin. Arrows indicate leading (orange), first trailing (white), second trailing (yellow), and third trailing (blue) barbed ends. (C) Merged time-lapse micrographs. Scale bar, 5 µm. (D) Merged kymograph of filament length (scale bar, 5 µm) over time (time bar, 10 s). (E–H) Kaplan–Meier curves representing average processive run lengths (τ) for EnaΔL with (E) fascin, (F) Singed, (G) α-actinin, or (H) fimbrin on leading (red) and trailing (blue) barbed ends. Error bars, 95% CI. n ≥ 127. (I) Average processive run lengths for leading (red) and trailing (blue) barbed ends shown in E–H for 2-filament bundles with fascin, Singed, α-actinin, or fimbrin. P values (* <0.0001). Error bars, 95% CI. (J) Average processive run lengths for antiparallel and parallel 2-filament α-actinin (green) or fimbrin (purple) bundles. Error bars, 95% CI. n ≥ 64. (K) Fold increase of barbed end elongation rates of EnaΔL on fascin (yellow), Singed (blue), α-actinin (green), or fimbrin (purple) bundled filaments. Error bars, SEM. n ≥ 5 barbed ends from at least two movies.
In addition to the leading edge and tips of filopodia, Ena/VASP proteins also localize to focal adhesions and stress fibers (Reinhard et al., 1992; Haffner et al., 1995; Smith et al., 2010; Gateva et al., 2014), which are composed of filaments cross-linked by a Calponin Homology (CH) domain superfamily cross-linker, α-actinin. Bundles formed by α-actinin can contain either parallel or antiparallel filaments with wide spacing (30–36 nm) (Sjöblom et al., 2008). Another CH domain superfamily member, fimbrin/plastin, localizes to the lamellipodia and base of filopodia. In comparison, fimbrin bundles filaments with mixed polarity, but has narrow spacing (10–12 nm), similar to fascin (Hanein et al., 1998).
Ena/VASP has been shown to cluster in cells (Svitkina et al., 2003; Applewhite et al., 2007), and the processive elongation activity of clustered Ena/VASP is enhanced in vitro (Breitsprecher et al., 2008, 2011; Brühmann et al., 2017). Although it was originally thought that clustering of Ena/VASP was required for processive elongation of F-actin and competition with capping protein, individual tetramers are also active processive assembly factors. In solution, human VASP tetramers processively elongate filaments for short ∼1.45 s runs and antagonize capping protein (Hansen and Mullins, 2010) and Drosophila Ena tetramers processively elongate filaments for longer ∼10 s runs and protect against capping (Winkelman et al., 2014). Therefore, it is possible that individual Ena/VASP tetramers also have critical cellular functions, requiring further investigation of the mechanism of Ena/VASP tetramers, particularly on bundled actin filaments such as those found in filopodia.
Within the convergent elongation model for filopodia formation (Svitkina et al., 2003), filaments bundled by fascin within nascent filopodia are likely to have varied lengths as they emerge from the lamellipodium. In these early fascin bundles, in contrast to the shorter actin filaments (trailing filaments), the longest actin filament (leading filament) would have no nearby F-actin at its barbed end. A mechanism must exist so that trailing filament barbed ends can catch up to the leading filament barbed end, resulting in filaments of equal length in mature filopodia. Consistent with this possibility, we previously discovered that Ena makes longer processive runs on the trailing barbed ends of actin filaments bundled by fascin in vitro (Winkelman et al., 2014). Specifically, individual tetramers of a functional version of Drosophila Enabled (EnaΔL), in which a linker in the middle of the sequence is removed (Figure 1A), remain processively associated with trailing barbed ends in fascin bundles threefold longer (Winkelman et al., 2014). However, the underlying molecular mechanisms that facilitate Ena’s enhanced processivity on bundled actin filaments remain unclear. We therefore used a combination of in vitro reconstitution with single-molecule multi-color total internal reflection fluorescence microscopy (TIRFM), kinetic modeling, and analysis of Drosophila culture cells to characterize the dynamics and function of processive elongation of single and bundled filaments by multiple Ena/VASP homologues including Ena, human VASP, and Caenorhabditis elegans UNC-34. Together, our experiments and simulations inform our mechanistic understanding of single Ena/VASP molecules on single and bundled filaments, which demonstrate that avidity of multiple filaments within fascin bundles and multiple Ena arms leads to increased processivity of tetrameric Ena on trailing barbed ends, and reveal a novel regulatory process whereby the particular F-actin bundling protein matters for Ena/VASP processivity.
RESULTS
Ena is more processive on trailing barbed ends of both human and fly fascin (Singed) bundles
To understand what features are important for Drosophila Ena’s enhanced processive run length on trailing barbed ends within human fascin bundles (Figure 1B) (Winkelman et al., 2014), we first tested whether a different fascin homologue also facilitates enhanced residence times. We used two-color TIRFM to directly visualize the assembly of 1.5 μM Mg-ATP-actin monomers (15% Oregon green labeled) with 15 pM fluorescently labeled SNAP(549)-EnaΔL (referred to as EnaΔL) (Figure 1A) and human fascin or fly fascin, Singed. TIRFM allows direct visualization of individual EnaΔL molecule dynamics on single and bundled actin filament barbed ends. EnaΔL processive run lengths were measured for leading and single filament barbed ends (collectively referred to as leading) as well as trailing barbed ends (Figure 1, C and D, and Supplemental Movie S1). Kaplan Meier survival curves were calculated from individual EnaΔL processive runs (Figure 1, E and F), revealing that EnaΔL remains associated with trailing barbed ends (τfascin = 23.7 s, τSinged = 28.1 s) approvimately threefold longer than with leading barbed ends (τfascin = 8.4 s, τSinged = 10.1 s) for both human fascin and fly Singed (Figure 1I and Supplemental Table S1), consistent with our previous findings (Winkelman et al., 2014). Therefore, enhancement of EnaΔL’s processive elongation on trailing barbed ends is not specific to a particular fascin homologue.
Movie S1.
EnaΔL processivity on fascin bundles (corresponds to Figure 1, C, D, I, and K). Spontaneous assembly of 1.5 μM Mg-ATP-actin (15% Oregon green-actin) with 15 pM SNAP(549)-EnaΔL (red) and unlabeled 130 nM human fascin visualized by two-color TIRFM. White arrowheads mark free slow-growing barbed ends, and yellow arrowheads mark fast growing barbed ends associated with EnaΔL. Time interval between frames is 1 s.
Ena’s residence time is not enhanced on trailing barbed ends of fimbrin and α-actinin bundles
To determine whether diverse bundle architectures are similarly sufficient to enhance Ena’s processivity on trailing barbed ends, we tested the effect of bundling proteins with distinct properties. Fascin forms narrow (8–10 nm) bundles consisting of only parallel filaments (Cant et al., 1994; Edwards and Bryan, 1995; Jansen et al., 2011; Yang et al., 2013). In contrast, α-actinin makes more widely spaced bundles (30–36 nm) of both parallel and antiparallel filaments (Sjöblom et al., 2008). Fimbrin has similar narrow spacing to fascin (10–12 nm), but can also make bundles of mixed polarity (Hanein et al., 1998). First, we measured elongation rates of EnaΔL-bound leading and trailing barbed ends of filaments bundled by human fascin, fly fascin Singed, α-actinin, or fimbrin. Two-color TIRFM visualization of control and EnaΔL-bound barbed ends revealed a similar fold increase in EnaΔL-mediated actin elongation for leading (∼2.2- to 3-fold) and trailing (∼2- to 2.5-fold) barbed ends with all four bundling proteins (Figure 1K and Supplemental Tables S2 and S3). Therefore, Ena’s barbed end elongation enhancement is bundling protein independent.
Conversely, Ena’s enhanced processivity on trailing barbed ends is specific to fascin bundles. The average processive run length on leading barbed ends with all four bundling proteins is similar, ∼10 s (Figure 1I). However, there is no enhancement of EnaΔL’s average residence time on trailing barbed ends of α-actinin (τ = 9.4 s) or fimbrin (τ = 8.7 s) bundles (Figure 1, G–I, and Supplemental Table S1). Therefore, F-actin bundling proteins are not universally sufficient to enhance Ena’s processivity on trailing barbed ends. Although fascin exclusively forms parallel bundles, α-actinin and fimbrin form bundles composed of filaments with mixed polarities. We therefore compared EnaΔL’s residence time on trailing barbed ends in parallel and antiparallel two-filament bundles. For both fimbrin and α-actinin bundles, the average residence times for trailing parallel and antiparallel barbed ends are equivalent; thus, neither bundler enhances EnaΔL’s processivity (Figure 1J and Supplemental Table S4). Therefore, neither “fascin-like” filament spacing (8–10 nm) nor polarity (parallel) of actin filaments within bundles is sufficient to facilitate increased processivity on trailing barbed ends. Given that EnaΔL’s approximately threefold enhancement of processivity on trailing barbed ends is specific to fascin, different bundling proteins could regulate Ena’s specific activity for different F-actin networks.
Ena’s processive run length increases with bundle size
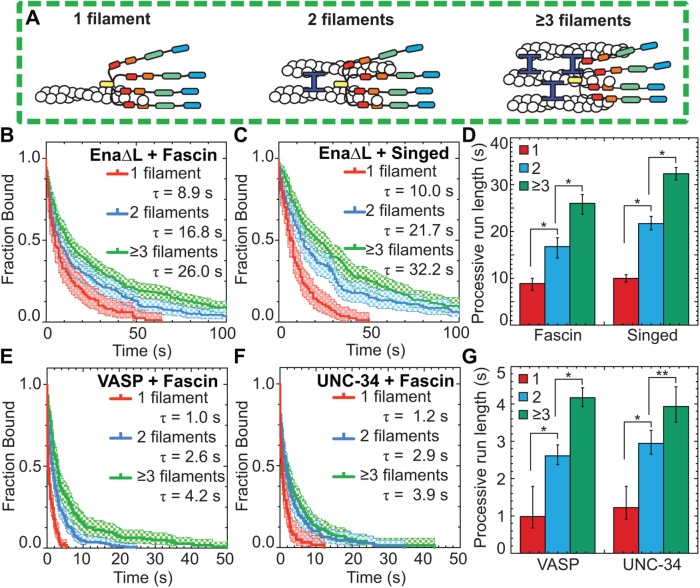
Filopodia are composed of ∼10–30 actin filaments bundled by fascin (Svitkina et al., 2003; Faix and Rottner, 2006), suggesting an avidity mechanism where enhanced processivity depends on Ena/VASP simultaneously associating with a barbed end and sides of neighboring filaments. To test whether the number of filaments in a fascin bundle positively correlates with processive run length, we determined the dependence of EnaΔL’s enhanced processivity on fascin bundle size (Figure 2A). Average run lengths on trailing barbed ends (Figure 1, E and F) were thereby parsed into two-filament bundles or three- or more filament bundles for both human and fly fascin (Figure 2, B–D, and Supplemental Table S1). EnaΔL’s average residence time on trailing barbed ends of a two-filament bundle (τfascin = 16.8 s, τSinged = 21.7 s) is approximately twofold longer than on single filament barbed ends (τfascin = 8.9 s, τSinged = 10.0 s). Furthermore, there is an additional ∼1.5-fold increase in processivity when EnaΔL is bound to trailing barbed ends of three- or more filament bundles (τfascin = 26.0 s, τSinged = 32.2 s) (Figure 2D). Therefore, consistent with an avidity effect, EnaΔL’s processivity increases with the number of fascin-bundled filaments. In contrast, EnaΔL’s enhanced processivity on trailing barbed ends of fascin bundles is not altered by the concentration of fascin (Supplemental Figure S1). This indicates that the mechanism for Ena’s enhanced processivity is independent of fascin concentration and does not likely rely on a direct interaction between fascin and Ena.
FIGURE 2:
Ena/VASP’s processive run length increases with the number of filaments in a fascin bundle. (A) Cartoons of Ena/VASP on a single filament and 2- and 3-filament fascin bundles. (B–G) Two-color TIRFM visualization of 1.5 µM Mg-ATP-actin (15% Oregon green-actin) with fly SNAP(549)-EnaΔL (red), human SNAP(549)-VASP or worm SNAP(549)-UNC-34 and unlabeled 130 nM human fascin or 250 nM Singed as indicated. (B, C) Kaplan–Meier curves representing average processive run lengths (τ) for 15 pM EnaΔL with (B) fascin or (C) Singed on single filaments (red), or bundles with 2 (blue) and ≥3 (green) filaments. Error bars, 95% CI. n ≥ 98. (D) Average processive run lengths for increasing number of filaments in fascin (yellow) or Singed (blue) bundles shown in B and C. Error bars, 95% CI. P values (* <0.0001). (E, F) Kaplan–Meier curves representing run lengths (τ) for (E) 25 pM VASP or (F) 18 pM UNC-34 with fascin on single filaments (red), or bundles with 2 (blue) and ≥3 (green) filaments. Error bars, 95% CI. n ≥ 60. (G) VASP and UNC-34 average processive run lengths for increasing number of filaments in fascin bundles shown in E and F. Error bars, 95% CI. P values (* <0.0001).
Human VASP and worm UNC-34 also have enhanced processive properties on fascin bundles
To determine whether enhanced processivity on fascin-bundled trailing filament barbed ends is conserved among Ena/VASP family members, we extended our analysis to full-length human VASP and full-length worm UNC-34 (Figure 1A). Human VASP is a well-characterized Ena/VASP protein (Bachmann et al., 1999; Chereau and Dominguez, 2006; Breitsprecher et al., 2008; Pasic et al., 2008; Hansen and Mullins, 2010), whereas UNC-34 has not yet been biochemically characterized in vitro despite multiple in vivo studies (Sheffield et al., 2007; Fleming et al., 2010; Havrylenko et al., 2015).
For our initial characterization of the three homologues, we measured the affinity for barbed ends and effect on actin elongation for EnaΔL, VASP, and UNC-34. Initially, the effect of Ena/VASP homologues on actin elongation rates and their apparent affinity (Kd, app) for barbed ends was determined by single-color TIRFM visualization of spontaneous assembly of 1.5 μM Mg-ATP-actin (15% Oregon Green) over a range of concentrations for each unlabeled Ena/VASP homologue (Supplemental Figure S2, A–F). At near saturating concentrations all three Ena/VASP homologues increase actin elongation by a similar amount, ∼1.6- to ∼2.7-fold, but have somewhat varying affinities for actin filament barbed ends ranging from 3.2 nM (EnaΔL) to 6.7 nM (UNC-34) to 12.2 nM (VASP) (Supplemental Figure S2F). Likewise, bulk seeded pyrene actin assembly assays also show that all three Ena/VASP homologues increase actin elongation rates by similar amounts, and fits of assembly rate over a range of Ena/VASP concentrations revealed apparent affinities for barbed ends ranging from 0.7 nM (EnaΔL) to 10.2 nM (UNC-34) to 10.8 nM (VASP) (Supplemental Figure S2, G and H). We then used two-color TIRFM visualizations of red-labeled EnaΔL, VASP, and UNC-34 on fascin bundles to measure actin elongation rates of Ena/VASP-bound leading and trailing barbed ends (Supplemental Figure S2I and Supplemental Movie S2). All three Ena/VASP homologues similarly increase actin elongation approximately two- to threefold on both leading and trailing barbed ends (Supplemental Figure S2J and Supplmental Tables S2 and S5). Enhancement of actin elongation rates by EnaΔL and VASP are similar to previously reported values (Hansen and Mullins, 2010; Winkelman et al., 2014; Brühmann et al., 2017), and the actin elongation properties of UNC-34 are in good agreement with those of the other homologues. Therefore, though Ena, VASP, and UNC-34 vary in their barbed end affinity, they all similarly increase the actin elongation rate of both leading and trailing barbed ends of fascin-bundled filaments.
Movie S2.
UNC-34 processivity on fascin bundles (corresponds to Figure 2, F and G). Spontaneous assembly of 1.5 μM Mg-ATP-actin (15% Oregon green-actin) with 18 pM SNAP(549)-UNC-34 (red) and unlabeled 130 nM human fascin visualized by two-color TIRFM. White arrowheads mark free slow-growing barbed ends, and yellow arrowheads mark fast growing barbed ends associated with UNC-34. Time interval between frames is 0.5 s.
To test whether different Ena/VASP homologues have similarly enhanced processive properties on fascin bundles, two-color TIRFM visualization of 1.5 μM Mg-ATP-actin (15% Oregon Green) was used to quantify the processive run lengths of fluorescently labeled VASP and UNC-34 on fascin bundles (Figure 2, E–G, and Supplemental Movie S2). The average residence time of both VASP (1.0 s) and UNC-34 (1.2 s) on single filament barbed ends is ∼9-fold shorter than EnaΔL (8.9 s), as expected from lower apparent affinities for barbed ends and previously reported values (Hansen and Mullins, 2010). Yet, like EnaΔL, both VASP and UNC-34 have ∼2.5-fold longer processive run lengths on trailing barbed ends of 2-filament fascin bundles (τVASP = 2.6 s, τUNC-34 = 2.9 s), with an additional ∼1.5-fold increase on trailing barbed ends of three- or more filament bundles (τVASP = 4.2 s, τUNC-34 = 3.9 s) (Figure 2, E–G, and Supplemental Table S1). Therefore, enhanced processivity on fascin-bundled trailing barbed ends is conserved from worms to flies to humans, suggesting that enhanced processivity is important for Ena/VASP’s activity in cells.
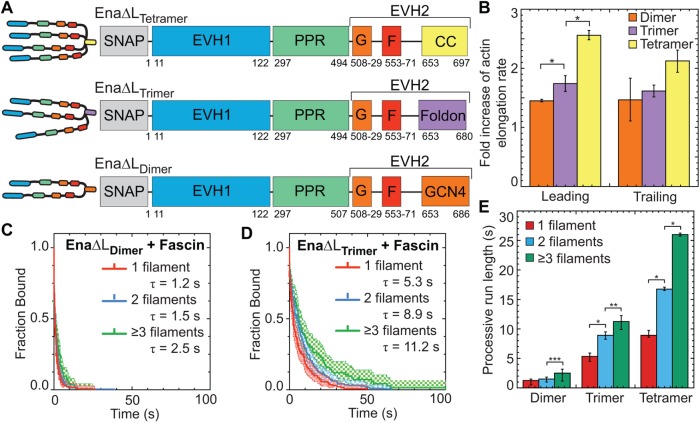
Enhanced elongation and processive run length increases with the number of Ena arms
Wild-type Ena is a tetrameric protein (Kuhnel et al., 2004; Winkelman et al., 2014), with four arms that could facilitate simultaneous associations with a barbed end, neighboring actin filaments, and/or actin monomers for processive elongation. Since we observed that EnaΔL’s average processive run length increases with the number of fascin-bundled filaments (Figure 2), we investigated the importance of Ena’s oligomeric state by measuring actin elongation and processive properties of dimeric and trimeric EnaΔL. Dimer and trimer constructs were formed by replacing EnaΔL’s (referred to in this section as EnaΔLTetramer) coiled-coil tetramerization domain with a GCN4 dimerization domain (Harbury et al., 1993) or a Foldon trimerization domain (Figure 3A) (Güthe et al., 2004; Papanikolopoulou et al., 2004), and the oligomeric state was verified by gel filtration and multi-angle light scattering (Supplemental Figure S3, A–C). Two-color TIRFM was used to visualize 1.5 μM Mg-ATP-actin (15% Alexa-488 labeled) with SNAP(549)-EnaΔLΔCC-GCN4 (referred to as EnaΔLDimer) or SNAP(549)-EnaΔLΔCC-Foldon (referred to as EnaΔLTrimer) on fascin bundles. First, we measured actin elongation rates of EnaΔL-bound leading and trailing barbed ends (Figure 3B and Supplemental Tables S2 and S5). While all constructs increase actin’s elongation rate on both leading and trailing filaments, the fold increase is positively correlated with the number of EnaΔL arms. EnaΔLTetramer has the largest enhancement of actin elongation (2.56-fold leading, 2.1-fold trailing), followed by EnaΔLTrimer (1.74-fold leading, 1.62-fold trailing), and then EnaΔLDimer (1.45-fold leading, 1.46-fold trailing).
FIGURE 3:
The processive run length increases with the number of EnaΔL “arms.” (A) Cartoon and domain organizations of Ena∆LTetramer, Ena∆LTrimer, and Ena∆LDimer. (B–E) Two-color TIRFM visualization of 1.5 µM Mg-ATP-actin (15% Alexa488-actin) with indicated SNAP(549)-EnaΔL construct and 130 nM fascin. (B) Fold increase of barbed end elongation rates of Ena∆LDimer (orange), Ena∆LTrimer (purple), and Ena∆LTetramer (yellow). Error bars, SEM. n ≥ 5 barbed ends from at least two movies. P values (* ≤0.05) (C, D) Kaplan–Meier curves representing average processive run lengths (τ) for (C) 50 pM MBP-SNAP(549)-Ena∆L∆CC-GCN4 or (D) 70 pM MBP-SNAP(549)-Ena∆L∆CC-Foldon with fascin on single filaments (red), or bundles with 2 (blue) and ≥3 (green) filaments. Error bars, 95% CI. n ≥ 93. (E) Average processive run length for increasing number of EnaΔL “arms” on single filaments (red), or fascin bundles with 2 (blue) and ≥3 (green) filaments shown in C and D. Error bars, 95% CI. P values (*** <0.05, ** <0.001, * <0.0001). EnaΔLTetramer data in B and E are also reported in Figures 1K and 2D, respectively.
Similar to actin elongation rates, average processive run length is also positively correlated with number of EnaΔL arms (Figure 3, C–E). Remarkably, although reduced ∼10-fold compared to EnaΔLTetramer, EnaΔLDimer does remain processively associated with single filament (τ = 1.2 s), 2-filament trailing (τ = 1.5 s), and 3- or more filament trailing (τ = 2.5 s) barbed ends (Figure 3, C and E, Supplemental Movie S3, and Supplemental Table S1). EnaΔLTrimer has intermediate processivity on single filament (τ = 5.3 s), 2-filament trailing (τ = 8.9 s), and 3- or more filament trailing (τ = 11.2 s) barbed ends (Figure 3, D and E, and Supplemental Table S1). For each construct, the fluorescence intensity was not correlated with run length (Supplemental Figure S3, D–G), indicating that processive activity is not affected by EnaΔL construct multimerization. EnaΔLTrimer’s processive run lengths are similar to the residence time of EnaΔLTetramer on single filaments but are not comparably enhanced on trailing barbed ends (Figure 3E). Therefore, EnaΔLDimer is sufficient for minimal processive elongation and EnaΔLTrimer is necessary for longer processive runs on single filaments, but EnaΔLTetramer is necessary for the longest processive runs on trailing barbed ends of fascin bundles (Figure 3E). Interestingly, the avidity effect of multiple filaments in a fascin bundle is apparent even with fewer arms than the wild-type tetramer. The positive correlation between processive elongation and Ena arms is consistent with a recent study on chimeric human VASP with Dictyostelium GAB domains on single actin filaments (Brühmann et al., 2017).
Movie S3.
EnaΔLDimer processivity on fascin bundles (corresponds to Figure 3, B, C, and E). Spontaneous assembly of 1.5 μM Mg-ATP-actin (15% Alexa488-actin) with 50 pM MBP-SNAP(549)-EnaΔLΔCC-GCN4 (red) and unlabeled 130 nM human fascin visualized by two-color TIRFM. White arrowheads mark free slow-growing barbed ends, and yellow arrowheads mark fast growing barbed ends associated with EnaΔLDimer. Time interval between frames is 0.5 s.
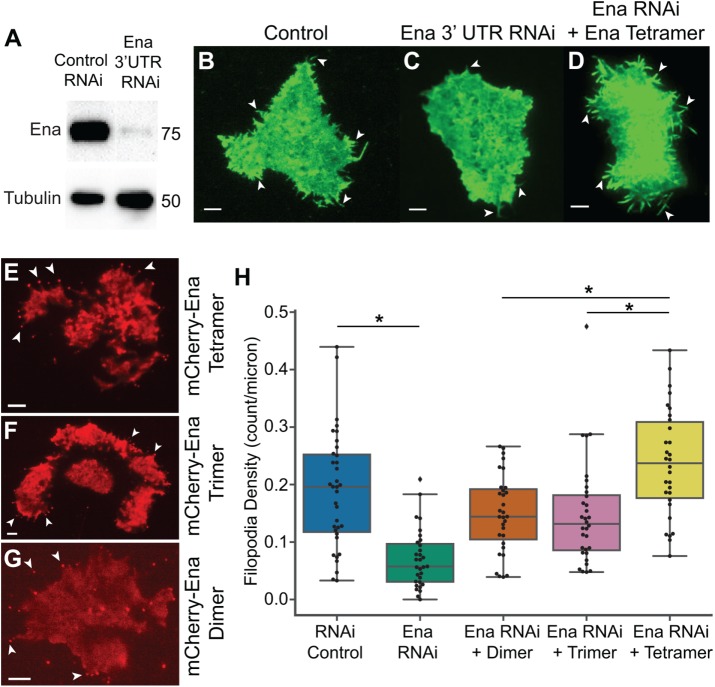
Tetrameric Ena is more effective at forming filopodia in Drosophila culture cells
EnaΔLTetramer is significantly better at processive actin filament assembly than either EnaΔLDimer or EnaΔLTrimer, where EnaΔLTetramer increases the actin elongation rate ∼2- to 2.5-fold and remains processively associated with trailing barbed ends of fascin bundles for ∼25 s (Figure 3, B and E). To determine whether WT EnaTetramer is therefore necessary for proper function in cells, we evaluated the ability of Ena oligomerization constructs to facilitate filopodia in ML-DmD16-c3 Drosophila culture cells, derived from third instar larval wing discs (Figure 4). We knocked down endogenous Ena with dsRNAi against the 3′UTR (Figure 4A), and then expressed GFP-actin (Figure 4, B–D) and full-length Ena constructs labeled with mCherry. A constitutive pIZ plasmid was used to express the Ena constructs: mCherry-Ena (referred to as mCherry-EnaTetramer), mCherry-EnaΔCC-GCN4 (referred to as mCherry-EnaDimer), or mCherry-EnaΔCC-Foldon (referred to as mCherry-EnaTrimer) (Figure 4, E–G, and Supplemental Figure S4A). The activity of the different Ena constructs was analyzed by quantifying filopodia density, through determining the number of filopodia per perimeter of the cell (Figure 4H). Compared to control cells (0.19 ± 0.06 filopodia/micron), RNAi-treated cells without exogenous Ena have a 2.7-fold decrease in filopodia density (0.07 ± 0.03 filopodia/micron). Strikingly, mCherry-EnaTetramer forms significantly more filopodia (0.24 ± 0.05 filopodia/micron) compared to mCherry-EnaTrimer (0.15 ± 0.05 filopodia/micron) and mCherry-EnaDimer (0.15 ± 0.04 filopodia/micron). There was no correlation between filopodia density and GFP-actin fluorescence or mCherry fluorescence (Supplemental Figure S4, B and C). Therefore, Ena tetramers facilitate the production of significantly more filopodia than dimer and trimer constructs following knockdown of endogenous Ena.
FIGURE 4:
Tetrameric Ena is necessary for proper filopodia density. (A) Western blot of endogenous Ena expression in control cells and in Ena 3′UTR RNAi-treated cells. β-Tubulin is a loading control. (B–D) Representative fluorescence micrographs of D16 cells with GFP-actin for (B) control treatment, (C) Ena 3′UTR RNAi, and (D) RNAi with transfection of mCherry-EnaTetramer. White arrows indicate representative filopodia. (E–G) Representative fluorescence micrographs of D16 cells transfected with the indicated mCherry-Ena construct. White arrows indicate mCherry-Ena constructs at the tips of filopodia. (H) Boxplot of filopodia density, number of filopodia per micron of cell perimeter, for control cells, Ena 3′UTR RNAi, and RNAi transfected with mCherry-EnaΔCC-GCN4 (mCherry-EnaDimer), mCherry-EnaΔCC-Foldon (mCherry-EnaTrimer), and mCherry-EnaTetramer. n = 3 with at least 10 cells for each experiment. P values (* <0.0005).
Kinetic model of Ena shows a direct correlation between processivity and both bundle size and Ena oligomerization
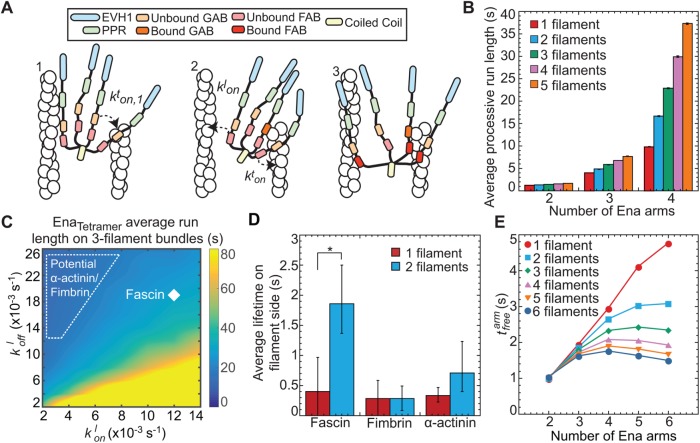
We observed that EnaΔL’s processivity depends on the number of filaments in a fascin bundle (Figure 2D) and number of EnaΔL arms (Figure 3E). Therefore, it is likely that the underlying molecular mechanism for the increased processivity of individual Ena molecules on trailing barbed ends depends on Ena’s ability to simultaneously bind to an elongating barbed end and sides of filaments via its multiple arms (Figure 1B). To investigate this avidity effect, we developed a kinetic model of Ena with varying number of arms, N, binding bundles composed of varying number of actin filaments, n (Figure 5A and Supplemental Figure S5A).
FIGURE 5:
Kinetic model of Ena/VASP on actin bundles shows that processivity positively correlates with both number of Ena arms and bundle size. (A) Modeling schematics showing (from left to right). 1) An Ena arm’s GAB domain binds the trailing barbed end with binding rate kton,1. 2) Once the GAB domain is bound, the FAB domain from the other arms binds to sides of either the trailing filament (kton) or leading filaments (klon). 3) Arms can be bound to the trailing filament, while others bind leading filaments. (B) Bar graph of the average processive run length as a function of number of Ena arms and bundle size. Error bars, SEM. (C) Heat map showing average Ena run length in the case of 3-filament bundles and four Ena arms, with systematic variations of klon and kloff in the simulations. Diamond denotes optimized rates for fascin bundles, and region within dotted line shows potential rates for α-actinin and fimbrin. (D) Average lifetime for SNAP(549)-EnaΔL association to the sides of single filaments (red) and 2-filaments bundled (blue) by fascin, fimbrin, or α-actinin. Error bars, 95% CI. P values (* <0.0001). (E) Average time between binding events (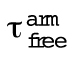 ) for varying arm number and bundle size.
) for varying arm number and bundle size.
Our model considers binding and unbinding kinetics of all N Ena arms on various binding sites of individual actin filaments in a bundle, which together dictate the kinetics of the Ena “molecule” as a whole (Figure 5A). An Ena arm initially binds to the trailing barbed end with an on rate of 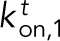 and unbinds with an off rate of
and unbinds with an off rate of 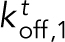 (Figure 5A1). The remaining Ena arms are available to bind and unbind to the side of the trailing filament with a rate
(Figure 5A1). The remaining Ena arms are available to bind and unbind to the side of the trailing filament with a rate 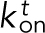 and
and 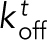 or to the side of other filaments in the bundle with a rate
or to the side of other filaments in the bundle with a rate  and
and 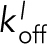 (Figure 5, A2 and A3). A Monte Carlo algorithm was used to integrate rates of binding and unbinding of Ena arms over time. Model parameters were optimized using TIRFM off rates for N ∈ (2,3,4) and n ∈ (1,2, ≥3) (Figure 3E), as described in the Supplemental Materials.
(Figure 5, A2 and A3). A Monte Carlo algorithm was used to integrate rates of binding and unbinding of Ena arms over time. Model parameters were optimized using TIRFM off rates for N ∈ (2,3,4) and n ∈ (1,2, ≥3) (Figure 3E), as described in the Supplemental Materials.
We used the model to characterize Ena’s processive run length at the trailing barbed end. The model shows that increasing both the number of filaments in a bundle and the number of Ena arms increases Ena’s processive run length, which strongly supports the avidity hypothesis. The modeling results are in excellent agreement with the trends observed from our TIRFM data (compare Figure 5B and Figure 3E). Using the model, we tested conditions over a range of both  and
and 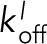 to mimic α-actinin and fimbrin bundles (Figure 1I), where EnaΔL processivity is not enhanced on trailing barbed ends (Figure 5C and Supplemental Figure S5, B–F). The model shows a broad regime that results in the same average processive run length on both leading and trailing barbed ends (Figure 5C, dashed region). This indicates that differences between bundlers could be due to diverse association and dissociation rates caused by differences in how CH domain bundlers and fascin bind F-actin.
to mimic α-actinin and fimbrin bundles (Figure 1I), where EnaΔL processivity is not enhanced on trailing barbed ends (Figure 5C and Supplemental Figure S5, B–F). The model shows a broad regime that results in the same average processive run length on both leading and trailing barbed ends (Figure 5C, dashed region). This indicates that differences between bundlers could be due to diverse association and dissociation rates caused by differences in how CH domain bundlers and fascin bind F-actin.
The kinetic model suggests an area of kinetic rates for Ena binding to the side of a leading filament that could account for the specificity of EnaΔL’s enhanced processivity to fascin bundles. Therefore, we further analyzed the TIRFM experiments to determine the dissociation rate of individual EnaΔL molecules from the sides of actin filaments bundled by different bundling proteins at the resolution that we measured its processivity on barbed ends (Figure 1I). We measured the residence time of EnaΔL on sides of single filaments and 2-filament bundles in the presence of fascin, fimbrin, and α-actinin (Figure 5D). EnaΔL associates ∼5-fold longer to the sides of 2-filament bundles formed by fascin, but not on 2-filament bundles formed by fimbrin or α-actinin. This suggests that the specificity for Ena’s enhanced processivity on the barbed end involves an increased affinity of Ena’s F-actin binding domain for actin filaments bundled by fascin.
Finally, we used the model to estimate rates of Ena-mediated filament elongation. While at least one Ena arm associates with the barbed end, its other arms undergo binding and dissociation events. When free, an arm can bind G-actin from solution and transfer it to the barbed end. The elongation rate of the Ena-bound filament should be proportional to the average time that individual arms are free. From the model, the average time that individual arms remain unbound while the Ena molecule is in the bound state,  , increases with N and decreases with n (Figure 5E). This result is consistent with the TIRFM data for the fold increase of actin elongation rate due to EnaΔL on the leading (n = 1 in the model) and trailing barbed ends (n > 1 in the model) (Figures 1K and 3B).
, increases with N and decreases with n (Figure 5E). This result is consistent with the TIRFM data for the fold increase of actin elongation rate due to EnaΔL on the leading (n = 1 in the model) and trailing barbed ends (n > 1 in the model) (Figures 1K and 3B).
DISCUSSION
Ena’s processivity is enhanced specifically on fascin bundles
Ena/VASP proteins are important processive actin elongation factors that are localized to diverse F-actin networks composed of filaments bundled by different cross-linking proteins, including fascin, fimbrin, and α-actinin. Previously, we found that Ena takes ∼3-fold longer processive runs on trailing barbed ends of fascin-bundled F-actin (Winkelman et al., 2014). Here we investigated the mechanism and conservation of Ena/VASP’s processivity at the barbed end of single filaments and filaments bundled by different cross-linking proteins, as well as the physiological relevance of Ena/VASP tetramerization.
We found that although fly EnaΔL’s processivity is enhanced ∼3-fold on trailing barbed ends in fascin bundles, there is no processivity enhancement on trailing barbed ends of α-actinin or fimbrin bundles (Figure 1I). Fimbrin and α-actinin use two CH domains to bundle F-actin, whereas fascin uses β-trefoil domains. We calculated the radius of gyration for the linker between the FAB and CC domains to be ∼4 nm, making the distance between two FAB domains ∼12 nm including the CC domain. Though a stretched linker could allow Ena to reach two α-actinin filaments (∼33 nm), it does not seem likely. However, the distance between fimbrin filaments could very easily be accounted for with Ena’s FAB-to-FAB distance, which further supports that Ena’s specificity for fascin bundles is due to a property beyond simply filament spacing.
Though the exact mechanism for Ena’s specificity for fascin bundles remains unclear, we suggest several non–mutually exclusive hypotheses. First, fascin might hold the trailing filament in a specific register with respect to the leading filament and/or could allow for a particular favorable F-actin conformation (Claessens et al., 2006; Shin et al., 2009) that facilitates easier Ena/VASP binding. Second, fascin could bundle closer to the growing barbed end compared to fimbrin or α-actinin, thereby promoting longer processive Ena runs by keeping trailing barbed ends closer to sides of leading filaments. Third, our kinetic model revealed a broad region of Ena binding kinetics to sides of bundled filaments ( and
and 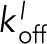 ) that could explain EnaΔL’s lack of enhanced processivity on fimbrin and α-actinin bundles (Figure 5C). Consistent with this possibility, EnaΔL has longer lifetimes on sides of 2-filament fascin bundles compared to 2-filament bundles formed by fimbrin or α-actinin (Figure 5D). This suggests that Ena’s enhanced processivity is facilitated in part by an increased affinity of Ena for F-actin bundled by fascin. Alternatively, Ena’s enhanced association with sides of bundled actin filaments could be merely due to an avidity effect of multiple filaments, which is perturbed by competition between Ena and fimbrin or α-actinin, but not fascin. Competition between Ena and fimbrin or α-actinin, CH domain bundling proteins, could be due to similar binding sites on actin filaments. Many actin binding proteins, including those with CH domains and domains related to FAB domains (WH2), are known to bind in the same target binding cleft of actin (Dominguez, 2004, 2009). Finally, although it remains possible that Ena weakly associates directly with fascin, this is unlikely as EnaΔL’s enhanced processivity remains constant over a range of fascin concentrations (Supplemental Figure S1). Further studies are required to fully understand the mechanisms by which Ena’s processivity is specifically enhanced on actin filaments bundled by fascin. However, this important observation reveals for the first time that bundling proteins and the F-actin networks they form can differentially regulate the activity of processive actin assembly factors, thereby providing a mechanism to allow Ena/VASP proteins to facilitate the assembly of diverse bundled networks with different dynamics in cells. Understanding how different bundling proteins associate with and help form specific F-actin networks in cells will therefore be of critical importance.
) that could explain EnaΔL’s lack of enhanced processivity on fimbrin and α-actinin bundles (Figure 5C). Consistent with this possibility, EnaΔL has longer lifetimes on sides of 2-filament fascin bundles compared to 2-filament bundles formed by fimbrin or α-actinin (Figure 5D). This suggests that Ena’s enhanced processivity is facilitated in part by an increased affinity of Ena for F-actin bundled by fascin. Alternatively, Ena’s enhanced association with sides of bundled actin filaments could be merely due to an avidity effect of multiple filaments, which is perturbed by competition between Ena and fimbrin or α-actinin, but not fascin. Competition between Ena and fimbrin or α-actinin, CH domain bundling proteins, could be due to similar binding sites on actin filaments. Many actin binding proteins, including those with CH domains and domains related to FAB domains (WH2), are known to bind in the same target binding cleft of actin (Dominguez, 2004, 2009). Finally, although it remains possible that Ena weakly associates directly with fascin, this is unlikely as EnaΔL’s enhanced processivity remains constant over a range of fascin concentrations (Supplemental Figure S1). Further studies are required to fully understand the mechanisms by which Ena’s processivity is specifically enhanced on actin filaments bundled by fascin. However, this important observation reveals for the first time that bundling proteins and the F-actin networks they form can differentially regulate the activity of processive actin assembly factors, thereby providing a mechanism to allow Ena/VASP proteins to facilitate the assembly of diverse bundled networks with different dynamics in cells. Understanding how different bundling proteins associate with and help form specific F-actin networks in cells will therefore be of critical importance.
The mechanism of tetrameric Ena acting on fascin bundles for filopodia formation
Given that Ena localizes to filopodia with fascin, lamellipodia with fimbrin, and stress fibers with α-actinin, sensitivity to diverse bundles could play an important role in regulating Ena activity in cells. Filopodia are unique among these networks with long, straight filaments that emerge from a network capped by capping proteins. Lamellipodia have short, branched filaments and stress fibers are contractile, bipolar networks. Thus, filopodia are the ideal network for enhanced Ena/VASP processivity facilitating elongation of longer filaments that requires stronger competition against capping protein to form a protrusive network. It has been established that Ena/VASP proteins have enhanced processive elongation activity when clustered (Breitsprecher et al., 2008, 2011), which is likely critical for filopodia assembly. Ena/VASP can form clusters by associating with lamellipodin (Hansen and Mullins, 2015) or IRSp53 (Krugmann et al., 2001; Disanza et al., 2013). Yet, fluorescence recovery after photobleaching (FRAP) experiments have shown that Ena/VASP clusters at the leading edge turn over rapidly (Applewhite et al., 2007) as individual molecules exchange. We have found that the activity of unclustered Ena/VASP molecules is enhanced severalfold on actin filaments bundled by fascin and is conserved from worms to flies to humans, suggesting that individual Ena/VASP molecules may also have critical cellular roles. For example, the increased residence time on trailing barbed ends of individual Ena/VASP molecules could play a critical role in a feedback mechanism between Ena/VASP and fascin in emerging filopodia (Winkelman et al., 2014). Ena/VASP-associated barbed ends elongate faster, assembling longer actin filaments that contain more fascin binding sites, which subsequently enhance Ena/VASP’s processivity. Within a nascent filopodia, different length filaments could be initially bundled by fascin. Ena/VASP’s rapid turnover in clusters could be partially due to tetramers leaving the clusters to elongate trailing barbed ends once fascin bundles emerge. Our hypothesis is that single tetramers act in the nascent filopodia to help the trailing barbed ends catch up to the leading barbed end. This would allow for all the filaments to reach the same length, resulting in mature filopodia with uniform thickness and aligned barbed ends.
Avidity promotes enhanced Ena processivity on fascin bundles
We hypothesize that avidity between multiple actin filaments in a fascin bundle and multiple Ena arms promotes the formation of long filopodia filaments. We investigated the avidity effect by testing how the number of filaments in a fascin bundle and number of EnaΔL arms affects EnaΔL’s processive run length. Our results strongly indicate that avidity plays a major role, as there is an ∼2-fold increase in EnaΔL’s residence time on trailing barbed ends in 2-filament bundles and an additional ∼1.5-fold increase on bundles with three or more filaments compared to single filament barbed ends (Figure 2, B–D). Similarly, the residence time of both VASP and UNC-34 is longer on trailing barbed ends and is correlated with the number of actin filaments in a fascin bundle (Figure 2, E–G). Furthermore, the residence time of EnaΔLTrimer and EnaΔLTetramer is ∼4.5- and ∼10-fold longer than EnaΔLDimer on fascin bundles with three or more filaments (Figure 3, C–E). A recent study measuring processive elongation using chimeric human VASP with Dictyostelium GAB domains on single filaments (Brühmann et al., 2017) supports our conclusions that enhanced elongation and processive run length are positively correlated with the number of Ena arms. Observing this positive correlation under more “physiological conditions,” a construct using Ena’s unmodified EVH2 domains and on fascin bundles, indicates that these properties are relevant for Ena’s activity in cells and specifically for filopodia.
We further tested the avidity hypothesis by developing a kinetic model that incorporates Ena with differing number of arms binding to single or multiple filaments (Figure 5). Previous models have focused exclusively on modeling the kinetics of Ena/VASP-mediated barbed end elongation of single actin filaments (Hansen and Mullins, 2010; Breitsprecher et al., 2011; Brühmann et al., 2017). VASP-mediated single filament elongation rates were shown to increase linearly with the number of VASP arms in solution as predicted by the model (Breitsprecher et al., 2011). However, this model overlooks the binding kinetics of arms that are not associated with the barbed end. Hence, we developed a kinetic model that explicitly incorporates the binding and unbinding rates of each Ena arm on multiple filaments (Figure 5A). After an Ena arm binds to the barbed end (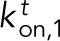 ), the remaining arm(s) are free to bind to the side of the leading filament(s) (
), the remaining arm(s) are free to bind to the side of the leading filament(s) ( ) or the trailing filament (
) or the trailing filament ( ). We quantified the processive run length for various numbers of bundled filaments and Ena arms.
). We quantified the processive run length for various numbers of bundled filaments and Ena arms.
The model demonstrates that the avidity effect of Ena emerges from an effective increase in local concentration of F-actin that allows for more FAB binding sites and from multiple Ena arms with available FAB domains. The avidity effect results in longer residence times near the trailing barbed end. Importantly, if an arm dissociates from the trailing barbed end, Ena will continue to processively elongate the barbed end and not diffuse away given that other arms’ FAB domains are associated with nearby actin filaments. Furthermore, our model that includes multiple arms binding to multiple actin filaments still has a linear correlation of elongation rates with number of Ena arms on single filaments (Figure 5E), as predicted by a previous model (Brühmann et al., 2017). The  is linear with respect to increasing additional Ena arms on single filaments, but with an increasing number of filaments there are diminishing returns by adding more Ena arms.
is linear with respect to increasing additional Ena arms on single filaments, but with an increasing number of filaments there are diminishing returns by adding more Ena arms. 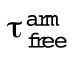 peaks at a tetramer on larger bundles, which we speculate may provide an additional argument for why a tetramer of Ena/VASP is evolutionarily preferred. We also observe that an Ena tetramer is more effective at forming filopodia in Drosophila culture cells compared to dimer and trimer constructs (Figure 4). Since the tetramer has increased residence time on trailing barbed ends and increases actin’s elongation rate above the dimer and trimer, this suggests that the tetramer is necessary for proper actin elongation rates and competition with capping protein to allow for the formation of the correct number of filopodia.
peaks at a tetramer on larger bundles, which we speculate may provide an additional argument for why a tetramer of Ena/VASP is evolutionarily preferred. We also observe that an Ena tetramer is more effective at forming filopodia in Drosophila culture cells compared to dimer and trimer constructs (Figure 4). Since the tetramer has increased residence time on trailing barbed ends and increases actin’s elongation rate above the dimer and trimer, this suggests that the tetramer is necessary for proper actin elongation rates and competition with capping protein to allow for the formation of the correct number of filopodia.
MATERIALS AND METHODS
TIRFM
TIRFM images were collected at 250 ms intervals with a cellTIRF 4Line system (Olympus, Center Valley, PA) fitted to an Olympus IX-71 microscope with through-the-objective TIRF illumination and an iXon EMCCD camera (Andor Technology, Belfast, UK). Mg-ATP-actin (15% Oregon Green or Alexa 488 labeled) was mixed with polymerization TIRF buffer (10 mM imidazole [pH 7.0], 50 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid [EGTA], 50 mM dithiothreitol [DTT], 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, and 0.5% [400 cP] methylcellulose) to induce F-actin assembly and any additional actin binding proteins. This mixture was transferred to a flow cell for imaging at room temperature. For two-color TIRFM, we cyclically imaged labeled actin (1 frame, 488 nm excitation for 50 ms) and SNAP(549)-Ena/VASP (1 frame, 561 nm excitation for 50 ms) (Winkelman et al., 2014).
D16 cell culture
ML-DmD16-c3 (DGRC) cells were cultured in Schneider’s media with 10% fetal bovine serum (Gibco, Waltham, MA), Anti-Anti (Gibco, Waltham, MA), and 10 μg/ml recombinant human insulin (Gibco, Waltham, MA), transfected with FugeneHD (Promega, Madison, WI), and imaged on extracellular matrix (ECM)-coated glass-bottom dishes after 48–72 h. ECM was harvested from ML-DmD17-c3 (DGRC, Bloomington, IN) (Currie and Rogers, 2011). All imaging was performed on a TIRF system mounted on an inverted microscope (Ti-E; Nikon, Tokyo, Japan) using a 100X/1.49NA oil immersion TIRF objective driven by Nikon Elements software unless noted otherwise. Images were captured using an Orca-Flash 4.0 (Hamamatsu, Hamamatsu, Japan) and were processed for brightness and contrast using ImageJ (Schneider et al., 2012) analysis. We quantified >30 cells using CellGeo (Tsygankov et al., 2014). Filopodia were quantified with the criteria of >0.78 μm long and <0.91 μm wide. Protein expression was quantified by Western blot of D16 whole cell lysate. Primary mouse antibodies against Ena (5G2; Drosophila Studies Hybridoma Bank) and tubulin (12G10; Drosophila Studies Hybridoma Bank) were used at 1:200 and 1:500, respectively. Primary rabbit antibody against mCherry (ab167453; Abcam) was used at 1:500. Secondary anti-mouse HRP (Cell Signaling) and anti-rabbit HRP (Sigma) were used at 1:5000.
Plasmid construction
Enabled (EnaΔL) constructs were prepared by removing the 6x-His tag from the C-terminus of previously described EnaΔL constructs (MBP-SNAP-EnaΔL or MBP-EnaΔL) (Winkelman et al., 2014) and insertion into a MBP containing plasmid (pet21A) by standard restriction digest and infusion (Clontech, Mountain View, CA) following PCR amplification (iProof; Bio-Rad, Hercules, CA). EnaΔLDimer and EnaΔLTrimer constructs were prepared by removing the coiled-coil domain and adding a Foldon domain (Güthe et al., 2004; Papanikolopoulou et al., 2004) (MBP-SNAP-EnaΔLΔCC-Foldon) or GCN4 domain (Harbury et al., 1993) (MBP-SNAP-EnaΔLΔCC-GCN4) from MBP-SNAP-EnaΔL. UNC-34 was cloned from worm cDNA and inserted into a pet21A vector with MBP-SNAP (New England Biolabs, Ipswich, MA) at XmaI/PacI sites, while also including a flexible linker (GGSGGS) in the forward primer sequence of SNAP constructs. Singed and VASP constructs were cloned from fly and human cDNA libraries, respectively. VASP was inserted into a MBP-SNAP and SNAP containing vector, while Singed was inserted into a pGEX KT Ext plasmid containing GST with a thrombin cleavage site at XbaI/XhoI sites. Plasmids for transfection of mCherry-EnaΔCC-GCN4 and mCherry-EnaΔCC-Foldon were cloned into a pIZ-mCherry-Ena (Bilancia et al., 2014) construct using infusion (Clontech, Mountain View, CA). The RNA interference (RNAi) was designed using Primer3Plus (Untergasser et al., 2012) targeting the 3′ UTR of enabled using forward primer
5′TAATACGACTCACTATAGGGAGACCACGTGATGGCATGTGCATAGGC3′ and reverse primer 5′TAATACGACTCACTATAGGGAGACCACTGCTGAAGACTTGCTGGTTC3′. The 3′UTR was extracted from w1118 strain fly genome, and the DNA region of interest was isolated by PCR amplification and placed in a bluescript SK vector. Double-stranded DNA (dsDNA) was produced using PCR amplification, and dsRNA was produced from the resulting dsDNA using the MEGAscript T7 Transcription kit (Invitrogen, Waltham, MA).
Protein expression and purification
Recombinant Ena/VASP proteins were purified by expressing in Escherichia coli strain BL21-Codon Plus (DE3)-RP (Agilent Technologies, Santa Clara, CA) with 0.25 mM isopropyl β-d-1-thiogalactopyranoside for 16 h at 16°C. Cells were lysed with an Emulsi-Flex-C3 (Avestin, Ottawa, Canada) in extraction buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, 10% glycerol, 0.1 mM DTT) with 0.5 μM phenylmethylsulfonyl fluoride and complete, EDTA-free Protease Inhibitor Cocktail (Roche, Basel, Switzerland) and were clarified. The extract was incubated for 1 h at 4°C with amylose resin (New England Biolabs, Ipswich, MA) and was washed with extraction buffer; then Ena/VASP was batch eluted with elution buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, 10% glycerol, 0.1 mM DTT, 40 mM maltose). Ena/VASP was incubated overnight with and without 1 μM TEV protease to cleave MBP and filtered on a Superdex 200 10/300 GL or Superose 6 Increase 10/300 GL column (GE Healthcare, Little Chalfont, UK) where they eluted as stable oligomers. Ena/VASP constructs were dialyzed against SNAP buffer (20 mM HEPES [pH 7.4], 200 mM KCl, 0.01% NaN3, 10% glycerol, and 0.1 mM DTT). SEC-MALS was performed using DAWN HELEOS II and Optilab T-rEX (Wyatt Technology, Goleta, CA) with a Superdex 200 Increase 10/300 GL column and Akta FPLC (GE Healthcare, Little Chalfont, UK). SEC-MALS data were analyzed using Astra 6.0 (Wyatt Technology, Goleta, CA). SNAP-tagged proteins were labeled with BG-549 (New England Biolabs, Ipswich, MA) following the manufacturers’ protocols. Concentrations of SNAP-tagged proteins were determined by densitometry of Coomassie-stained bands on SDS−PAGE gels compared with standards. Ena/VASP was flash-frozen in liquid nitrogen and stored at −80°C. N-terminal SNAP and MBP tags did not affect Ena/VASP’s activity (Winkelman et al., 2014). Actin was purified from rabbit skeletal muscle acetone powder (Pel-Freez, Rogers, AR) or self-prepared chicken skeletal muscle acetone powder by a cycle of polymerization and depolymerization and gel filtration (Spudich and Watt, 1971). Gel-filtered actin was labeled with Oregon green (Kuhn and Pollard, 2005) on Cys374 or Alexa 488 carboxylic acid succinimidyl ester on lysine residues (Kellogg et al., 1988; Vignjevic et al., 2006b). Human fascin, human α-actinin IV, and Schizosaccharomyces pombe fimbrin were expressed in bacteria and purified as described (Vignjevic et al., 2003; Skau and Kovar, 2010; Li et al., 2016). Singed was purified in the same manner as previously reported for human fascin (Vignjevic et al., 2003).
Glass preparation
Microscope slides and coverslips (#1.5; Fisher Scientific, Waltham, MA) were washed for 30 min with acetone and for 10 min with 95% ethanol, sonicated for 2 h with Helmanex III detergent (Hellma Analytics, Müllheim, Germany), incubated for 2 h with piranha solution (66.6% H2SO4, 33.3% H2O2), washed with deionized water, and dried. Glass then was incubated for 18 h with 1 mg/ml mPeg-Silane (5000 MW) in 95% ethanol, pH 2.0. Parallel strips of double-sided tape were placed on the coverslip to create multiple flow chambers (Zimmermann et al., 2016).
Calculation of residence time and elongation rates
To calculate Ena/VASP’s residence time on barbed ends, SNAP(549)-Ena/VASP fluorescent spots associated with the barbed end were manually tracked using MTrackJ (Meijering et al., 2012) in ImageJ. Spots that did not move were not scored, because they were assumed to be adsorbed to the glass. Events that contained joined barbed ends with no clear leading or trailing barbed bend were not included in the average lifetime calculation. Residence times for single SNAP-549-Ena(ΔL) tetramers were determined by fitting a Kaplan−Meier (Kaplan and Meier, 1958) survival curve with a single exponential equation, f(x) = x0 * exp(−x/T1) to calculate the average lifetime. Kaplan−Meier survival curves were used to account for processive runs that started before imaging began or ends after imaging terminated. Log rank statistical significance tests were done using Prism 7 (GraphPad Software, San Diego, CA). Barbed end elongation rates were calculated by measuring filament lengths over time with ImageJ software. Multiple filament lengths were plotted over time, and the distribution was fitted with a linear equation using KaleidaGraph 4.5 (Synergy Software, Reading, PA). To calculate the number of filaments in a bundle, the TIRFM movie was used to follow the history of the filaments. This could most accurately differentiate between two-filament bundles and three or more filament bundles. Owing to photobleaching of the filaments over time, the actin fluorescence was not used to determine the number of filaments within the bundle.
Fluorescence spectroscopy
Bulk actin assembly was measured from the fluorescence of pyrene-actin with a Safire2 or Infinite M200 Pro (Tecan Systems, Männedorf, Switzerland) fluorescent plate reader (Neidt et al., 2008). Briefly, 5 μM unlabeled Mg-ATP-actin was preassembled into seeds for 1 h by adding 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0. The assay measures the elongation rate of actin by the addition of 20% pyrene-labeled Mg-ATP-actin monomers and actin binding proteins to be assayed. Final protein concentrations are indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank Elena Solomaha of the University of Chicago BioPhysics Core Facility for performing SEC-MALS and Jonathan Winkelman for cloning VASP and UNC-34. We thank Caitlin Anderson, Katie Homa, Cristian Suarez, and other members of the D.R.K. laboratory for helpful discussions. We thank the Drosophila Genomics Resource Center, supported by National Institutes of Health (NIH) grant 2P40OD010949, for Drosophila cells. This material is based on work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1144082 and DGE-1746045 (to A.J.H.), NIH Molecular and Cellular Biology Training Grant T32 GM007183 (to A.J.H.), NIH Grant RO1 GM079265 (to D.R.K.), Department of Defense Army Research Office MURI grant W911NF1410403 (to G.A.V. and D.R.K.), and the University of Chicago Materials Research Science and Engineering Center, funded by National Science Foundation award DMR-1420709 (to G.A.V. and D.R.K.). Acknowledgment is made to the computational resources provided by the Research Computing Center at The University of Chicago.
Abbreviations used:
- CC
coiled coil
- FRAP
fluorescence recovery after photobleaching
- SH3
SRC Homology 3
- TIRFM
total internal reflection fluorescence microscopy
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0500) on January 2, 2019.
REFERENCES
- Applewhite DA, Barzik M, Kojima S-I, Svitkina TM, Gertler FB, Borisy GG. (2007). Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell , 2579–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M. (1999). The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem , 23549–23557. [DOI] [PubMed] [Google Scholar]
- Ball LJ, Jarchau T, Oschkinat H, Walter U. (2001). EVH1 domains: structure, function and interactions. FEBS Lett , 45–52. [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. (2005). Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem , 28653–28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. (2009). Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci , 1947–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell , 509–521. [DOI] [PubMed] [Google Scholar]
- Bilancia CG, Winkelman JD, Tsygankov D, Nowotarski SH, Sees JA, Comber K, Evans I, Lakhani V, Wood W, Elston TC, et al. (2014). Enabled negatively regulates Diaphanous-driven actin dynamics in vitro and in vivo. Dev Cell , 394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J. (2008). Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J , 2943–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TEB, Small JV, Curth U, Dickinson RB, Faix J. (2011). Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J , 456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brühmann S, Ushakov DS, Winterhoff M, Dickinson RB, Curth U, Faix J. (2017). Distinct VASP tetramers synergize in the processive elongation of individual actin filaments from clustered arrays. Proc Natl Acad Sci USA , E5815–E5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. (2010). A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol , 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K, Knowles BA, Mooseker MS, Cooley L. (1994). Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J Cell Biol , 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Dominguez R. (2006). Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J Struct Biol , 195–201. [DOI] [PubMed] [Google Scholar]
- Claessens MMAE, Bathe M, Frey E, Bausch AR. (2006). Actin-binding proteins sensitively mediate F-actin bundle stiffness. Nat Mater , 748–753. [DOI] [PubMed] [Google Scholar]
- Currie JD, Rogers SL. (2011). Using the Drosophila melanogaster D17-c3 cell culture system to study cell motility. Nat Protoc , 1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A, Bisi S, Winterhoff M, Milanesi F, Ushakov DS, Kast D, Marighetti P, Romet-Lemonne G, Müller HM, Nickel W, et al. (2013). CDC42 switches IRSp53 from inhibition of actin growth to elongation by clustering of VASP. EMBO J , 2735–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. (2004). Actin-binding proteins–-a unifying hypothesis. Trends Biochem Sci , 572–578. [DOI] [PubMed] [Google Scholar]
- Dominguez R. (2009). Actin filament nucleation and elongation factors—structure–function relationships. Crit Rev Biochem Mol Biol , 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R, Holmes KC. (2011). Actin structure and function. Annu Rev Biophys , 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, Bryan J. (1995). Fascins, a family of actin bundling proteins. Cell Motil Cytoskeleton , 1–9. [DOI] [PubMed] [Google Scholar]
- Faix J, Rottner K. (2006). The making of filopodia. Curr Opin Cell Biol , 18–25. [DOI] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R. (2007). Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J , 4597–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming T, Chien S-C, Vanderzalm PJ, Dell M, Gavin MK, Forrester WC, Garriga G. (2010). The role of C. elegans Ena/VASP homolog UNC-34 in neuronal polarity and motility. Dev Biol , 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateva G, Tojkander S, Koho S, Carpen O, Lappalainen P. (2014). Palladin promotes assembly of non-contractile dorsal stress fibers through VASP recruitment. J Cell Sci , 1887–1898. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB. (2007). Filopodia: the fingers that do the walking. Sci STKE , re5. [DOI] [PubMed] [Google Scholar]
- Güthe S, Kapinos L, Möglich A, Meier S, Grzesiek S, Kiefhaber T. (2004). Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J Mol Biol , 905–915. [DOI] [PubMed] [Google Scholar]
- Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann SM, Walter U. (1995). Molecular cloning, structural analysis and functional expression of the proline-rich focal adhesion and microfilament-associated protein VASP. EMBO J , 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D, Volkmann N, Goldsmith S, Michon A-M, Lehman W, Craig R, DeRosier D, Almo S, Matsudaira P. (1998). An atomic model of fimbrin binding to F-actin and its implications for filament crosslinking and regulation. Nat Struct Mol Biol , 787–792. [DOI] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. (2010). VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol , 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. (2015). Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments. Elife 2015, , 10.7554/eLife.06585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T. (1993). A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science , 1401–1407. [DOI] [PubMed] [Google Scholar]
- Havrylenko S, Noguera P, Abou-Ghali M, Manzi J, Faqir F, Lamora A, Guérin C, Blanchoin L, Plastino J. (2015). WAVE binds Ena/VASP for enhanced Arp2/3 complex–based actin assembly. Mol Biol Cell , 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S, Collins A, Yang C, Rebowski G, Svitkina T, Dominguez R. (2011). Mechanism of actin filament bundling by fascin. J Biol Chem , 30087–30096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. (1958). Nonparametric estimation from incomplete observations. J Am Stat Assoc , 457–481. [Google Scholar]
- Kellogg DR, Mitchison TJ, Alberts BM. (1988). Behaviour of microtubules and actin filaments in living Drosophila embryos. Development , 675–686. [DOI] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. (2001). Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol , 1645–1655. [DOI] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. (2005). Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J , 1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnel K, Jarchau T, Wolf E, Schlichting I, Walter U, Wittinghofer A, Strelkov SV. (2004). The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc Natl Acad Sci USA , 17027–17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Christensen JR, Homa KE, Hocky GM, Fok A, Sees JA, Voth GA, Kovar DR. (2016). The F-actin bundler α-actinin Ain1 is tailored for ring assembly and constriction during cytokinesis in fission yeast. Mol Biol Cell , 1821–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. (2008). Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol , 446–454. [DOI] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. (2012). Methods for cell and particle tracking. Methods Enzymol , 183–200. [DOI] [PubMed] [Google Scholar]
- Neidt EM, Skau CT, Kovar DR. (2008). The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem , 23872–23883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolopoulou K, Forge V, Goeltz P, Mitraki A. (2004). Formation of highly stable chimeric trimers by fusion of an adenovirus fiber shaft fragment with the foldon domain of bacteriophage T4 fibritin. J Biol Chem , 8991–8998. [DOI] [PubMed] [Google Scholar]
- Pasic L, Kotova T, Schafer DA. (2008). Ena/VASP proteins capture actin filament barbed ends. J Biol Chem , 9814–9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell , 453–465. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. (2009). Actin, a central player in cell shape and movement. Science , 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. (1992). The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J , 2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods. Available at www.nature.com/articles/nmeth.2089 (accessed July 30, 2018). [DOI] [PMC free article] [PubMed]
- Sebé-Pedrós A, Burkhardt P, Sánchez-Pons N, Fairclough SR, Lang BF, King N, Ruiz-Trillo I. (2013). Insights into the origin of metazoan filopodia and microvilli. Mol Biol Evol , 2013–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield M, Loveless T, Hardin J, Pettitt J. (2007). C. elegans Enabled exhibits novel interactions with N-WASP, Abl, and cell-cell junctions. Curr Biol , 1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Drew KRP, Bartles JR, Wong GCL, Grason GM. (2009). Cooperativity and frustration in protein-mediated parallel actin bundles. Phys Rev Lett , 238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom B, Salmazo A, Djinovic´-Carugo K. (2008). Alpha-actinin structure and regulation. Cell Mol Life Sci , 2688–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau CT, Kovar DR. (2010). Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr Biol , 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Blankman E, Gardel ML, Luettjohann L, Waterman CM, Beckerle MC. (2010). A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Dev Cell , 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem , 4866–4871. [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol , 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsygankov D, Bilancia CG, Vitriol EA, Hahn KM, Peifer M, Elston TC. (2014). CellGeo: a computational platform for the analysis of shape changes in cells with complex geometries. J Cell Biol , 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res , e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. (2006a). Role of fascin in filopodial protrusion. J Cell Biol , 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Peloquin J, Borisy GG. (2006b). In vitro assembly of filopodia-like bundles. Methods Enzymol , 727–739. [DOI] [PubMed] [Google Scholar]
- Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. (2003). Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol , 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, Kovar DR. (2014). Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc Natl Acad Sci USA , 4121–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Huang F-K, Huang J, Chen S, Jakoncic J, Leo-Macias A, Diaz-Avalos R, Chen L, Zhang JJ, Huang X-Y. (2013). Molecular mechanism of fascin function in filopodial formation. J Biol Chem , 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann D, Morganthaler AN, Kovar DR, Suarez C. (2016). In vitro biochemical characterization of cytokinesis actin-binding proteins. In: Yeast Cytokinesis, New York: Humana Press, 151–179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.