Abstract
Integrin α6β4 is an essential, dynamic adhesion receptor for laminin 332 found on epithelial cells, required for formation of strong cell–extracellular matrix (ECM) adhesion and induced migration, and coordinated by regions of the β4C cytoplasmic domain. β4E, a unique splice variant of β4 expressed in normal tissue, contains a cytoplasmic domain of 231 amino acids with a unique sequence of 114 amino acids instead of β4C’s canonical 1089 amino acids. We determined the distribution of α6β4E within normal human glandular epithelium and its regulation and effect on cellular biophysical properties. Canonical α6β4C expressed in all basal cells, as expected, while α6β4E expressed within a subset of luminal cells. α6β4E expression was induced by three-dimensional culture conditions, activated Src, was reversible, and was stabilized by bortezomib, a proteasome inhibitor. α6β4C expressed in all cells during induced migration, whereas α6β4E was restricted to a subset of cells with increased kinetics of cell–cell and cell–ECM resistance properties. Interestingly, α6β4E presented in “ringlike” patterns measuring ∼1.75 × 0.72 microns and containing actin and CD9 at cell–ECM locations. In contrast, α6β4C expressed only within hemidesmosome-like structures containing BP180. Integrin α6β4E is an inducible adhesion isoform in normal epithelial cells that can alter biophysical properties of cell–cell and cell–ECM interactions.
INTRODUCTION
The α6β4 integrin regulates formation of a hemidesmosome (HD) that is essential for normal homeostasis within the stratified epithelium of the skin. The HD remodels and is associated with the response to the physical and chemical microenvironment (Zhang et al., 2011; Osmani et al., 2018). Loss or mutation of the α6β4 integrin or laminin 332 in normal epithelial tissues results in blistering diseases, indicating its essential adhesion and signaling role (Pulkkinen and Uitto, 1998). Integrin α6β4 also has a role in regulating endothelial responses in lung to mechanical stress (Chen et al., 2015).
Within the simplified structure of epithelial glands, α6β4 loss is synonymous with prostatic intraepithelial neoplasia (PIN), which has a high predictive value as a marker of adenocarcinoma (Bonkhoff and Remberger, 1998). The normal prostate gland is composed of two major cell types, basal and secretory luminal cells (Bonkhoff, 1996). The basal cells attach to a basal lamina containing laminin 332 via the α6β4 integrin through the HD interacting with anchoring filaments (laminin 332) that, in turn, interact with anchoring fibers (Collagen VII) (Knox et al., 1994; Davis et al., 2001a). The basal cells of the normal prostate gland contain the stem cell compartment expressing the α6 integrin (Bonkhoff, 1996; Schmelz et al., 2005), similar to other stem cells that require it for maintenance of a stem cell niche in skin, glands, nerves, bone marrow, and developing embryos (Jacques et al., 1998; Emsley and Hagg, 2003; Mueller et al., 2006; Qian et al., 2006, 2007). In the prostate gland, transformed stem cells located in the basal layer lose critical adhesive elements and acquire luminal features (Bonkhoff and Remberger, 1998). Because the α6β4 integrin is critical for normal mechano-transduction, the known loss of the receptor expression in early prostate cancer would predict an alteration of the biophysical properties of normal basal cells of the prostate gland.
Although primary prostate cancer does not express the α6β4 integrin or assemble HDs (Nagle et al., 1995), it was recently reported that β4 integrin is expressed in a subpopulation of human prostate tumor progenitors (Yoshioka et al., 2013). Previous work established that formation of the HD requires the cytoplasmic domain of the β4 integrin (Nievers et al., 2000) to regulate its assembly and disassembly (Wilhelmsen et al., 2007).
Interestingly, an isoform of β4 integrin exists, created by mRNA splicing, that contains a unique cytoplasmic domain without the canonical regions required for HD function (van Leusden et al., 1997; de Melker and Sonnenberg, 1999) and whose function is unknown. This raised the possibility that the β4E variant could be present in early stages of the transition of the normal basal cells to progenitor cells. In this study, we used normal prostate basal cells (RWPE-1)—originally characterized by Bello et al. (1997) and characterized by us in three-dimensional (3D) culture (Wang et al., 2017)—to determine the distribution of α6β4E protein within normal human glandular epithelium, its regulation, and its effect on cellular biophysical properties.
RESULTS
Integrins β4E and β4C are present in human normal prostate basal cells and tissue
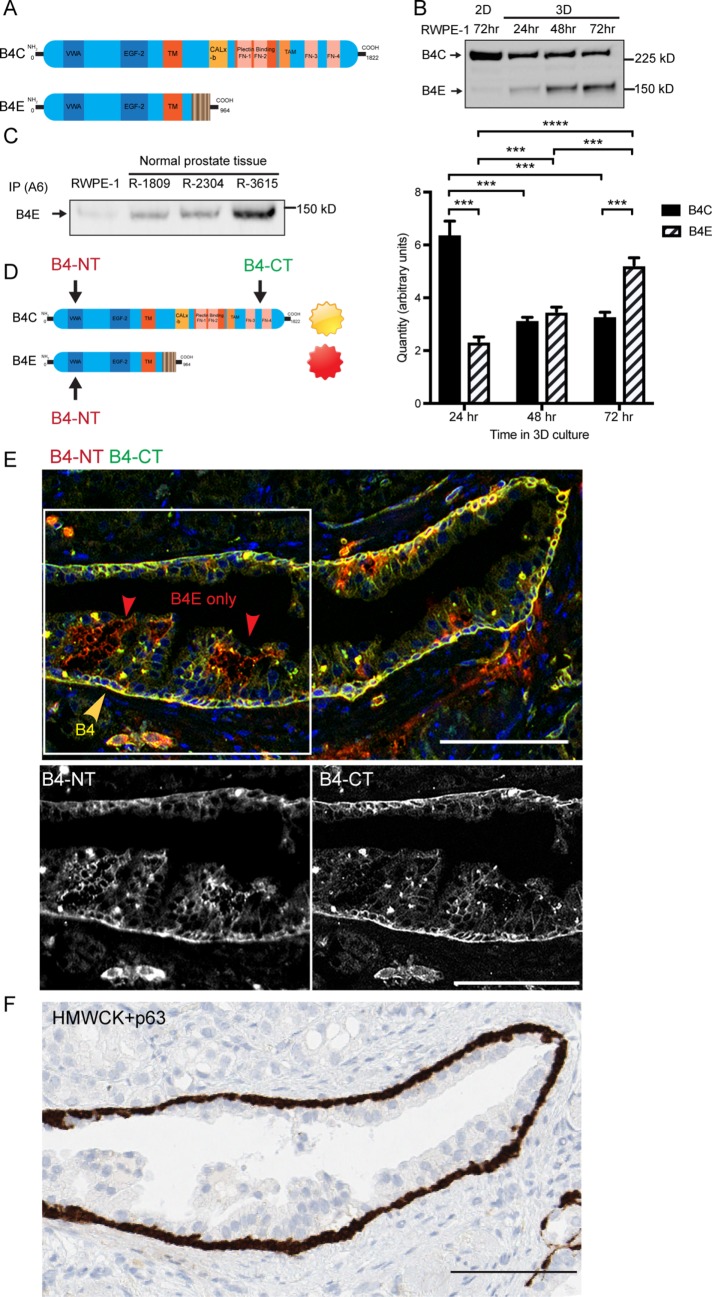
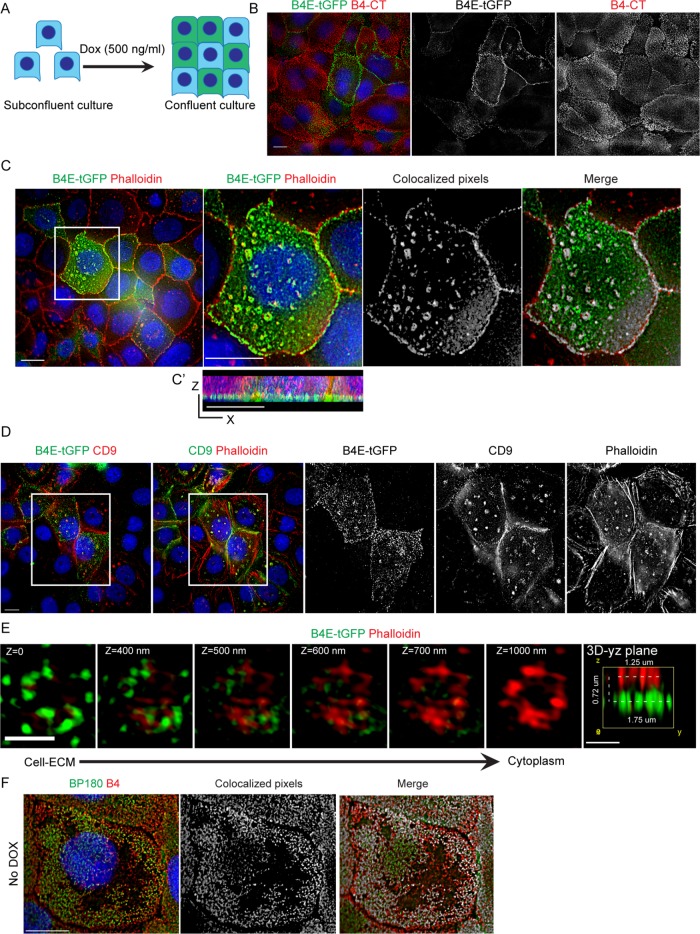
To determine the distribution of α6β4E protein within normal human glandular epithelium, we used the fact that the full-length β4C and the β4E isoform have identical extracellular domains while containing different cytoplasmic sequences (Figure 1A). Using an antibody to the extracellular domain, both forms can be detected by Western blot analysis of the heterodimers (Figure 1B) in the normal basal cell line (RWPE-1) depending on culture conditions. In two-dimensional (2D) culture, only β4C is expressed, whereas under 3D culture conditions, the β4C isoform is expressed with an inducible β4E isoform. The β4C degradation products, inducible with serum-containing media (shown later in Figure 3C) were not observed in 2D or 3D using serum-free media conditions. The induction of β4E over a 24- to 72-h period, detected by Western blot analysis, was verified by a similar increase in the quantity of the mRNA specific for β4E as detected by quantitative reverse transcriptase PCR (qRT-PCR) (Figure 1B). We also tested for the presence of α6β4E heterodimers in human normal prostate tissue samples. Three lysates from normal prostate tissue were immunoprecipitated with an α6-integrin antibody, and we retrieved the β4E integrin from all three samples (Figure 1B). Retrieval of the β4C was not possible, consistent with previous reports that α6β4 in a mature HD complex with the extracellular matrix (ECM) is not readily extractable (Sterk et al., 2000). To distinguish β4C protein and β4E by immunofluorescence microscopy, we used a combination of antibodies that detect the extracellular domain of β4, which would detect both forms, and the cytoplasmic domain–specific antibody to detect only the β4C form (Figure 1D), as previously reported by others (Ni et al., 2005). Using de-identified human prostate tissue, we detected the β4C subunit (Figure 1E, yellow) in the basal cells primarily polarized to the basal lamina. The β4E form (Figure 1E, red) was found within the luminal cell layer in a location between the cells, reminiscent of a previously reported suprabasal location for β4 integrin (Owens et al., 2003). The reference location of the basal cells was verified in a serial section of the same tissue using a basal cell antibody cocktail containing high-molecular-weight cytokeratin (HMWCK) and p63 (Figure 1F, brown).
FIGURE 1:
Integrin β4E and integrin β4C are present within human prostate cell lines and patient-derived tissue, and β4E is distributed within the luminal cell compartment. (A) Schematic of the different functional domains of integrin β4C and integrin β4E variants. (B) Integrin β4 heterodimers (B4C, B4E) can be detected in normal basal cells (RWPE-1) in 2D culture after 72 h or in 3D cultures after 24, 48, or 72 h by immunoprecipitation of α6 integrin, followed by Western blot protein analysis. Quantities of β4C or β4E mRNA specific for each isoform were detected by qRT-PCR using isoform-specific primers. Data are shown as means ± SD, n = 3. Statistical comparison was done by nonparametric, two-tailed Student’s t test (***, p < 0.005). (C) Immunoprecipitation of α6 integrin (IP A6) and retrieval of α6β4E heterodimer from normal prostate tissue. (D) The strategy and location of the epitopes of the β4 N-terminus antibody (B4-NT) and β4C-terminus antibody (B4-CT) were used to detect different β4 variants in human prostate tissue. (E) Epifluorescence images show the basal distribution of the β4C isoform (yellow arrow) and the β4E isoform (red arrow). The color channels for the boxed region were separated to show the distribution of β4 N-terminus antibody (B4-NT) or the β4 C-terminus antibody (B4-CT). The distribution of HMWCK and p63 detects the basal cell layer (F, brown) within a serial section of the same gland shown in E. Scale bars: 100 microns.
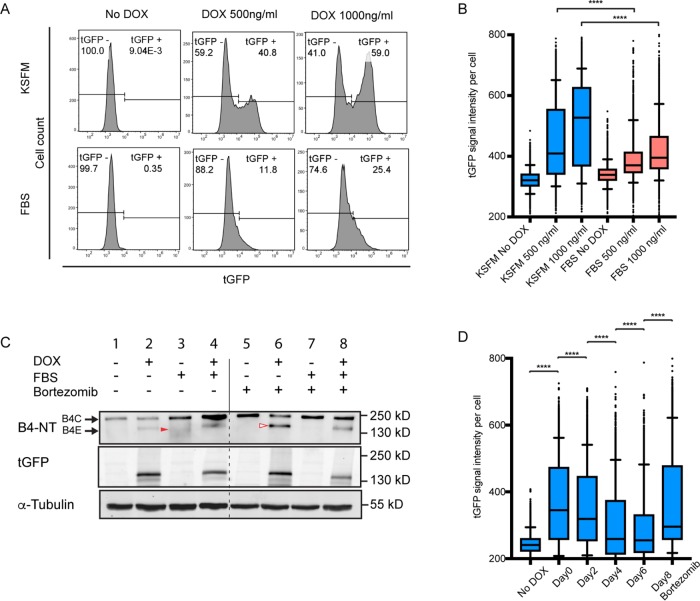
FIGURE 3:
Epithelium-specific regulation of integrin β4E expression and its suppression by serum-induced degradation. (A) Flow-cytometry analysis of the cell population (cell count) containing β4E-tGFP expression (tGFP) in KSFM or in media containing FBS, with or without two concentrations of doxycycline for 24 h. (B) The distribution of the β4E-tGFP (tGFP) intensity per cell is shown under the same conditions as A. In the box-and-whisker plot, whiskers indicate 5–95% percentile range, and dots indicate outliers. Statistical significance was determined by unpaired t test, (****, p < 0.0001). (C) Whole-cell lysate and Western blot analysis of integrin β4C, β4E, and β4E-tGFP (tGFP) expression in cells treated with either 1µg/ml doxycycline in the presence or absence of serum (FBS) or 50 nM bortezomib, using tubulin (α-tubulin) as the loading control. Note that the red filled arrowhead indicates β4C degradation and the red open arrowhead indicates β4E. (D) β4E-tGFP (tGFP) induction intensity per cell in the population either without (No DOX) or with 500 ng/ml doxycycline treatment done immediately after plating (Day 0) or after 2, 4, or 6 d in KSFM, or after 8 d in KSFM and then treated with 50 nM bortezomib for 18 h before analysis. Samples were analyzed by flow cytometry. Data are shown as a box-and-whisker graph. In the box-and-whisker plot, whiskers indicate 5–95% percentile range, and dots indicate outliers. Statistical significance was determined by unpaired t test (****, p > 0.0001).
Integrin β4E-tGFP is inducible in normal RWPE-1 basal cells and results in a functional heterodimer without altering integrin β4C expression
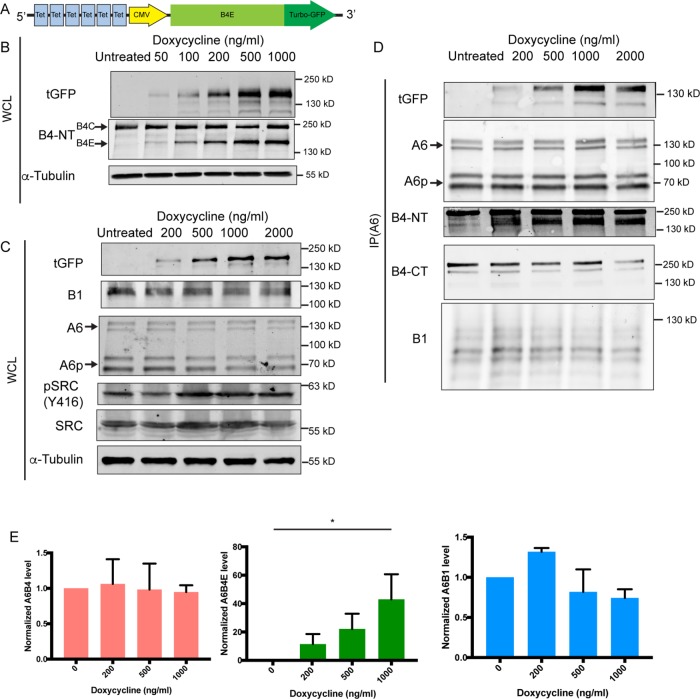
To study the biological characteristics of the β4E integrin in RWPE-1 cells, we constructed a vector for doxycycline-inducible expression of a turbo-GFP (tGFP)-tagged integrin β4E (β4E-tGFP). The construct contains in part a cytomegalovirus (CMV) promoter with the addition of tGFP at the C-terminal end of β4E (Figure 2A) and, as others have shown, GFP tagging does not affect function (Geuijen and Sonnenberg, 2002; Tsuruta et al., 2003). Increasing doxycycline concentrations resulted in increased expression of β4E-tGFP without altering the abundance of the endogenous β4C expression (Figure 2B). Six different concentrations of doxycycline were used, and a dose-dependent and reversible increase of β4E expression was observed by qRT-PCR without changing β4C expression (Supplemental Figure 1). Because we observed a suprabasal expression of β4E in the prostate luminal compartment (Figure 1), and previous work showed that suprabasal expression of β4 integrin was indicative of disrupted TGFβ signaling for clonal expansion of initiated tumor cells (Owens et al., 2003), we determined whether Src signaling was activated in response to β4E induction. Initiated cells have activated Src signaling as an oncogenic event (reviewed in Summy and Gallick, 2003), and c-Src activation is associated with a mouse model of prostate cancer progression (Cai et al., 2011). The increased β4E-tGFP expression resulted in increased activated Src, a marker of initiated tumor cells, as detected by Western blot analysis of the whole-cell lysate (Figure 2C), without affecting the expression of α6 or β1-integrin protein abundance. The β4E-tGFP formed a heterodimer with endogenous α6 integrin and the α6p form (Davis et al., 2001b), as detected by immunoprecipitation of α6 integrin and retrieval of increasing amounts of tGFP and β4E integrin in response to doxycycline as detected by tGFP antibody and the extracellular domain–specific antibody (β4-NT). In addition, α6β1 integrin is barely detectable in the RWPE-1 basal cells, as expected (Sterk et al., 2000) (Figure 2D). Quantitation of the blots is shown in Figure 2E.
FIGURE 2:
Dose-dependent induction of integrin β4E-tGFP expression in normal RWPE-1 basal cells results in a functional heterodimer without altering integrin β4C expression. Schematic (A) of the construct used to generate doxycycline-inducible C-terminus tGFP-tagged β4E (β4E-tGFP). (B) Whole-cell lysate (WCL) Western blot analysis of tGFP, integrin β4C (B4C), integrin β4E (B4E), or tubulin (α-tubulin) from cells treated with increasing concentrations of doxycycline for 4 d. Tubulin was used as the loading control. (C) Whole-cell lysate (WCL) Western blot analysis of tGFP, integrin β1 (B1), integrin α6 (A6), integrin α6p (A6p), activated Src (pSRC (Y416)), total Src (SRC), or tubulin (α-tubulin) from cells treated with increasing concentrations of doxycycline for 4 d. Tubulin was used as the loading control. (D) Immunoprecipitation of α6 integrin (IP A6) and retrieval analysis of tGFP, integrin α6 (A6) and the integrin α6p variant (A6p), integrin β4 (B4-NT), integrin β4C (B4-CT), and integrin β1 (B1). (E) Quantitative abundance of α6β4, α6β4E, or α6β1 in response to increasing concentration of doxycycline for 4 d. Data are shown as means ± SD (three independent experiments). Statistical significance determined by one-way analysis of variance (*, p < 0.05).
Epithelial-specific induction of integrin β4E-tGFP expression and suppression by degradation
Because the results indicated that β4E was inducible under RWPE-1 3D growth conditions and was observed in normal epithelial glands in a luminal-type compartment, we next examined whether β4E expression was influenced by tissue culture conditions suitable for epithelial or mesenchymal growth. RWPE-1 cells, as basal stem cells, respond to different media conditions by altering their phenotype (Litvinov et al., 2006; Rodriguez-Teja et al., 2016). Under basal epithelial maintenance conditions, that is, using keratinocyte serum-free media (KSFM), the doxycycline treatment results in a dramatic increase of β4E-tGFP protein expression, from an unexpressed state to expression in 40.8–59% of the population, depending on the concentration of doxycycline, as detected by flow cytometry (Figure 3A). In contrast, under serum-containing media (fetal bovine serum [FBS]) conditions that promote mesenchymal phenotypes, inducible integrin β4E-tGFP protein is strikingly absent from most of the population, because expression values range from 11.8% to 25.4% of the population. Examining the β4E-tGFP protein signal intensity per cell indicates that, under KSFM conditions, there is a statistically significant increase in β4E-tGFP protein as compared with cells in the FBS conditions with both concentrations of doxycycline treatment (Figure 3B). Under both culture conditions, β4E-tGFP mRNA expression is driven by the CMV promoter, so we examined whether the loss of β4E-tGFP protein expression under the FBS condition could be due to increased degradation of β4E-tGFP protein and used the selective and potent proteasome inhibitor bortezomib (Adams and Kauffman, 2004). Previous work showed the degradation of β4 integrin occurs by proteasomal activity in patient-derived keratinocytes (Micheloni et al., 2004). In the absence of bortezomib, under KSFM conditions, no loss of the endogenous β4C was observed (Figure 3C, compare lanes 1 and 2), while under FBS conditions, an increased production of the endogenous β4C and its known degradation products (Giancotti et al., 1992; Potts et al., 1994; von Bredow et al., 1997) (Figure 3C, solid red arrowhead) were observed (Figure 3C, compare lanes 1 and 3). In the presence of bortezomib, under KSFM conditions, both the endogenous β4C and the inducible level of β4E-tGFP (Figure 3C, open red arrowhead) were increased (Figure 3C, compare lanes 2 and 6). In the presence of the proteasome inhibitor bortezomib, under FBS conditions, the endogenous β4C was increased and the degradation products were not observed, as expected (Figure 3C, compare lanes 3 and 7). Under FBS conditions, the induced β4E-tGFP was decreased (Figure 3C, compare lanes 2 and 4), consistent with the flow-cytometry data (Figure 3, A and B). Taken together, the data indicate that, under the influence of FBS, β4E-tGFP protein expression is decreased due to degradation, because it is increased in the presence of the proteasome inhibitor bortezomib (Figure 3C, compare lanes 2 and 6).
The induction of β4E-tGFP could be suppressed by tissue culture conditions, so we next determined the optimal induction time after seeding the cultures. Using flow cytometry, we quantitated β4E-tGFP protein expression in response to doxycycline treatment administered either immediately or after 2,4, or 6 d of growth under KSFM conditions. A summary of the data shows that induction of β4E-tGFP signal intensity per cell (Figure 3D, Day 0) is maximal within 1 d of culture under KSFM conditions, and treatment of cells after 2–6 d (Figure 3D, Days 2–4) of growth, before doxycycline treatment, will dramatically hinder doxycycline’s ability to increase expression of β4E-tGTP. The loss of an “inducible window” of protein expression can be overcome by use of the proteasome inhibitor bortezomib (Figure 3D).
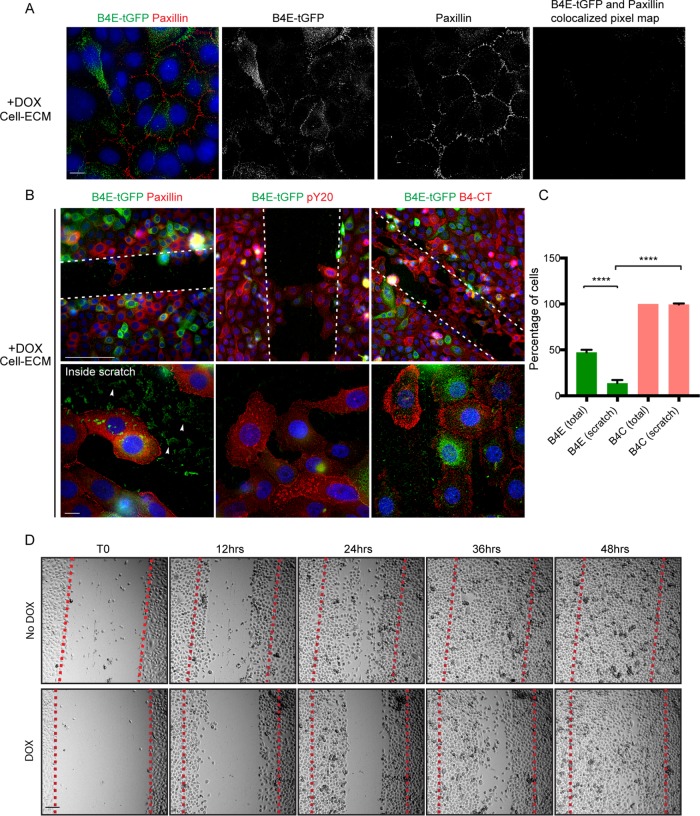
Integrin β4E-tGFP localizes to cell–ECM adhesion sites independent of paxillin or cell migration
Because the induction of β4E-tGFP was optimal during early culture conditions in KSFM and no endogenous β4E was expressed (Figure 1), we next determined whether β4E-tGFP was found within early cell–ECM adhesion sites containing paxillin or expressed in cells induced to migrate. Epifluorescence microscopy analysis of the cell–ECM surface of subconfluent cultures showed that the distribution of β4E-tGFP occurred independent of paxillin adhesion sites (Figure 4A). The induction of migration using a scratch assay resulted in RWPE-1 cells migrating into the scratch and expressing paxillin and phosphotyrosine (pY20) at focal adhesion sites (Figure 4B), as expected. Closer inspection of the β4E-tGFP signal inside the scratch (Figure 4B, inside the scratch, white arrowheads) revealed an ECM surface–associated signal. Approximately 95% of the cells expressed the β4C isoform when migrating into the scratch. Interestingly, while 50% of the cells expressed β4E-tGFP, only 10% entered the scratch (Figure 4C). Taken together, these data suggested that β4E-tGFP expression was associated with nonmigratory cells, and the cellular remnants (“footprints”) prompted us to observe cell migration using time-lapse microscopy. A time-stamped series (Figure 4D) showed an approximate 12-h delay in closure of the scratch in the β4E-tGFP–expressing population compared with the uninduced population (Figure 4, red lines in each panel). Supplemental Movies 1 and 2 are provided to add more detail.
FIGURE 4:
Integrin β4E-tGFP at cell–ECM adhesion sites is independent of paxillin or cell migration. (A) Epifluorescence images at the cell–ECM show the distribution of β4E-tGFP (green), paxillin (red), and nuclei (blue) and the black-and-white split channel images and the colocalized pixel map. Scale bars: 10 microns. (B) Scratch-induced (white dotted lines) cell migration reveals the cell–ECM distribution of β4E-tGFP (green) and paxillin (red) (B, top left panel), or β4E-tGFP isoform (green) and phosphotyrosine (pY20, red) (B, top middle panel), or β4E-tGFP (green) and β4C (red) (B, top right panel). All image panels show nuclei (blue). Scale bars: 100 microns. Migrating cells (inside scratch) under each condition (B, bottom panels) are shown. All image panels show nuclei (blue). Scale bars: 10 microns. White arrowheads indicate residual β4E-tGFP (green) in ECM after scratch. (C) Quantification of the percentage of cells in B expressing either β4E-tGFP (total or within the scratch) or β4C (total or within the scratch). Data are shown as means ± SD (three independent experiments). Statistical significance determined by unpaired t test (****, p > 0.0001). (D) Time-lapse microscopy and representative time frames (0, 12, 24, 36, and 48 h) of migrating cells, either with or without doxycycline (DOX, No DOX) into the scratch (red dotted line). Scale bar: 100 microns.
Movie S1.
Live-imaging of induced migration of RWPE-1 cells without doxycycline treatment. RWPE-1 cells were cultured for 4 days and were induced to migrate by a scratch assay procedure. Movie was taken 1 hour after induction for 48 hours. Frames were captured every 10 minutes. Scale bar = 100 microns.
Movie S2.
Live-imaging of induced migration of RWPE-1 cells treated with doxycycline to induce β4E-tGFP. RWPE-1 cells were cultured for 4 days with doxycycline (500 ng/ml) and were induced to migration by a scratch assay procedure. Movie was taken 1 hour after induction for 48 hours. Frames were captured every 10 minutes. Scale bar = 100 microns.
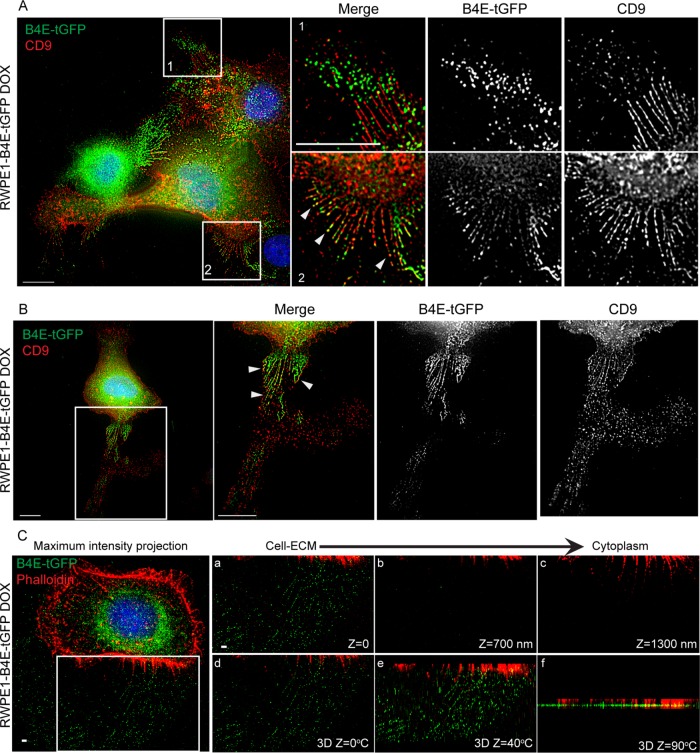
Integrin β4E-tGFP localizes at CD9-positive retraction-fiber regions in migrating cells
Because β4 integrin is an active migration receptor on laminin and can localize to retraction fibers (Geuijen and Sonnenberg, 2002) and CD9 can localize to retraction fibers (Yamada et al., 2013), we investigated the distribution of β4E and CD9 in subconfluent randomly migrating cells under serum-free conditions. Epifluorescence images at the cell–ECM region show that β4E-tGFP localized at the base of CD9-decorated retraction fibers (Figure 5A) and that β4E-tGFP and CD9 codistributed within the retraction fibers (Figure 5B). Interestingly, the distribution of β4E-tGFP relative to CD9 on retraction-fiber regions was dynamic. In migrating cells, β4E-tGFP localized either at the tip of retraction-fiber regions, while CD9 localized at the base of retraction fiber regions (Figure 5A, 1 and 2), or at the base of retraction fiber regions, while CD9 localized at the tip and ECM region (Figure 5B). It is important to note that, under 2D conditions, there is no expression of an endogenous β4E (Figure 1), and only the expressed β4E (β4E-tGFP) was monitored. Three-dimensional superresolution microscopy revealed that β4E-tGFP and F-actin colocalized at the base of the retraction fiber, and the 3D-constructed images using the z-sections showed that β4E-tGFP radiated from the base within the retraction fiber (Figure 5C). Taken together, these data show that actin and β4E-tGFP are colocated at the base of the CD9-containing retraction fiber in migrating cells.
FIGURE 5:
Integrin β4E-tGFP localizes at CD9-positive retraction fiber regions in migrating cells. (A) Epifluorescence images at cell–ECM show the distribution of integrin β4E-tGFP (green, B4E-tGFP), CD9 (red, CD9), and nuclei (blue) in subconfluent randomly migrating cells. Top right panels show merge (first panel), β4E-tGFP only (second panel), and CD9 only (third panel) at higher magnification of the box 1 region. Bottom right panels show merge (first panel), β4E-tGFP only (second panel), and CD9 only (third panel) at higher magnification of box 2 region. Scale bar: 10 microns. White arrowheads indicate colocalized regions of CD9 and β4E-tGFP. (B) Epifluorescence images at cell–ECM show (first panel) the distribution of integrin β4E-tGFP (green, B4E-tGFP), CD9 (red, CD9), and nuclei (blue) in a single migrating cell. The boxed region (B, first panel) is shown at higher magnification (B, second panel, Merge) with corresponding split channels of β4E-tGFP only (third panel), and CD9 only (far-right panel). Arrowheads indicate colocalized regions of CD9 and β4E-tGFP. Scale bar: 10 microns. (C) 3D-SIM superresolution microscopic images show the distribution of integrin β4E-tGFP (green, B4E-tGFP), F-actin (red, Phalloidin), and nuclei (blue) in a single migrating cell. Left, maximum-intensity projected image. The boxed region in the left panel is shown at higher magnification (a–c) as single-plane z-section images to show distributions of β4E-tGFP (green, B4E-tGFP) and F-actin (red, Phalloidin). (d–f) are 3D-constructed images at different z-angles to show the 3D structure of the boxed region. Scale bar: 1 micron.
Integrin β4E-GFP distributes into “ringlike” patterns containing actin and CD9 at the cell–ECM surface
The dramatic distribution of β4E-tGFP associated with retraction fibers during random migration and its cell–cell distribution under nonmigratory conditions prompted us to determine the distribution of the induced integrin in confluent cultures using superresolution microscopy according to the scheme shown in Figure 6A. We analyzed a subpopulation of the RWPE-1 cells that expressed β4E-tGFP (Figure 6B), and the localization of β4E-tGFP was different from that of β4C (Figure 6B). Strikingly, we found a distinctive “ringlike” pattern at the cell–ECM interface (Figure 6C). Because others had reported the interaction of β4C integrin and actin during the early stages of HD formation (Fontao et al., 1999) and basal cells can contain clusters of α6β4C as it becomes incorporated into α3β1 clusters connected to actin filaments (Sterk et al., 2000), we tested whether actin was present. Both β4E-tGFP and F-actin (phalloidin) are colocalized in the ringlike patterns (Figure 6C) and a z-section scan (Figure 6C′) indicates localization of the regions at the cell–ECM surface. The Z-sections can be seen in more detail by inspecting Supplemental Movie 3. Further work showed that CD9 is also localized to these patterns (Figure 6D). In confluent cultures, the use of multicolor superresolution microscopy revealed that the β4E-tGFP signal in the ring region is a 1.75-micron cluster located below the actin signal, which measures ∼1.25 microns (Figure 6E). As expected, the endogenous β4C integrin colocalized with BP180 in RWPE-1 cells (Figure 6F) in a pattern similar to that previously reported by others in keratinocytes (Nahidiazar et al., 2015). Because the increased β4E-tGFP expression resulted in increased activation of Src (Figure 2C), we next investigated whether formation of the ringlike structure was dependent on Src activity. In RWPE1-1 cells treated with PP2, a known Src inhibitor, both β4E-tGFP and F-actin (phalloidin) are colocalized in the ringlike patterns (Supplemental Figure 2, A and B). Thus, inhibition of Src activity did not change the formation of the ringlike patterns or β4E-tGFP localization at the ringlike patterns.
FIGURE 6:
Integrin β4E-tGFP distributes into ringlike patterns containing F-actin and CD9 at the cell–ECM surface. (A) Schematic shows the use of confluent cultures. (B) Epifluorescence images at the cell–ECM show the distribution of β4E-tGFP (green, B4E-tGTP), β4C (B4-CT, red), nuclei (blue), and the corresponding black-and-white split-channel images. Scale bar: 10 microns. (C) Epifluorescence images at the cell–ECM show the distribution of β4E-tGFP (green, B4E-tGTP), F-actin (Phalloidin, red), and nuclei (blue). Boxed region (C, far-left panel) at higher magnification (C, second panel) (scale bar: 10 microns) shows the distribution of β4E-tGFP (green, B4E-tGTP), F-actin (Phalloidin, red), and nuclei (blue), with the corresponding colocalization of white pixels map (C, third panel) and a merged image (C, far-right panel). x,z-projection (C′) shows β4E-tGFP localized at the cell–ECM plane. (D) Epifluorescence images (D, far-left panel) at the cell–ECM show the distribution of β4E-tGFP (green, B4E-tGTP), CD9 (CD9, red), and nuclei (blue). (D, second panel) shows distribution of CD9 (green), F-actin (phalloidin, red), and nuclei (blue). (E) Representative 3D-SIM superresolution images at different z-sections show the distribution of β4E-tGFP (green, B4E-tGTP) and F-actin (Phalloidin, red) as a ringlike pattern from cell–ECM to cytoplasm. Scale bars: 1 micron. (F) Representative images show β4C localized at HD-like structures at cell–ECM. Left panel shows the distribution of BP180 (Green, BP180) and β4C (red, B4C) at cell–ECM. Middle and right panels show the corresponding colocalization white pixels map between BP180 and β4C. HD-like structures were defined as colocalization of BP180 and β4C. Nuclei are shown in blue. Scale bar: 10 microns.
Movie S3.
Integrin β4E co-localized with actin in "ring-like" patterns at the cell-ECM region. A representative Z-stack image showing β4E-tGFP (green) and actin (red) localized at "ring-like" patterns in RWPE-1 cells treated with doxycycline for 4 days. Z-stacks were captured with 0.2 μm step size. Scale bar = 10 microns.
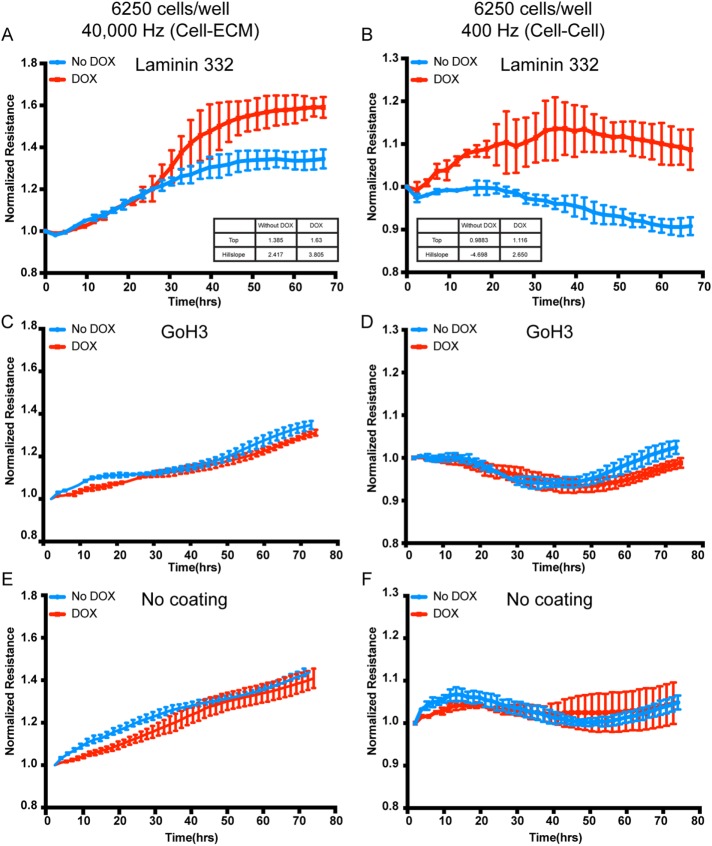
Biophysical properties of cell–cell and cell–ECM are increased by integrin β4E-tGFP expression
The data thus far suggested that the β4E-tGFP integrin was associated with the retraction fiber in randomly migrating cells, and within confluent cultures and tissue, it distributed with a cell–cell location in luminal-type cells. The localization of the laminin-binding integrins, including α6 integrin, in a cell–cell distribution is common in early embryonic cells and early morphogenic or patterning events (reviewed in Bulgakova et al., 2012), providing dynamic biophysical properties. Because our earlier work had shown cell–cell protection from excessive mechanical stretch mediated by the α6β4C (Chen et al., 2015), we tested whether cell–cell and cell–ECM biophysical properties were altered with the induction of β4E-tGFP expression. We used electric cell–substrate impedance sensing (ECIS), because this would provide a sensitive and quantifiable means to test normalized resistance kinetics in both subconfluent and confluent cultures. The induction of β4E-tGFP significantly increased the kinetics of both the cell–ECM and cell–cell normalized resistance measurements (Figure 7, A and B). A comparison of the top and hillslope values indicated that the highest normalized resistance (as indicated by plateau) were increased, with the most dramatic effect seen in altering cell–cell parameters (Figure 7B) after 30 h in the time course. The increase in the cell–ECM parameters that were β4E-tGFP dependent were observed after 24 h in confluent cultures (Figure 7A). The differences in resistance kinetics (cell–ECM or cell–cell) were dependent upon an adhesion-functioning α6 integrin, as detected by the use of a well-characterized function-blocking anti–α6-integrin antibody (GoH3) (Figure 7, C and D). The presence of laminin 332 ligand was also required, because conditions without laminin coating of the wells (no coating) (Figure 7, E and F) resulted in no resistance kinetics difference between the uninduced and doxycycline-induced conditions. In cells that were measured by ECIS at 1 h after plating, there was no detectable difference in cells treated with doxycycline versus untreated cells regardless of cell numbers (6250 cells per well or 12,500 cells per well, respectively) (Supplemental Figure 3).
FIGURE 7:
The kinetics of biophysical properties of cell–ECM and cell–cell are increased by integrin β4E-tGFP expression in confluent cultures. Untreated cells (No DOX) and β4E-tGFP–expressing cells cultured in doxycycline (500 ng/ml) (DOX) were plated on ECIS dishes either precoated with laminin 332 (A,B) or GoH3 function-blocking antibody (C,D) or uncoated (E, F) for 5 d; this was followed by ECIS measurements using 40,000 Hz (A,C,E) or 400 Hz (B,D,F) during β4E-tGFP (DOX) and β4C (No DOX) induction for up to 70 h. (A, B) Inset shows the biophysical parameters determined using a variable slope model, known as a four parameter dose response curve (GraphPad Prism, v8). Equation is: Y = Bottom + (X HillSlope) × (Top-Bottom)/(X HillSlope + EC50 HillSlope). Data are shown as means ± SD (three independent experiments).
DISCUSSION
Previous work reported the existence of a unique β4-integrin isoform produced by partial retention of intronic sequences (van Leusden et al., 1997). The α6β4E protein is expressed in normal prostate glands, distributing within the luminal cells in a cell–cell location. The current report is, to our knowledge, the first description of the reversible induction of the β4E-integrin splice variant in normal basal cells in response to 3D culture conditions. Induction of β4E was context dependent, because epithelial culture conditions were required and inappropriate expression of β4E protein was eliminated by proteasome activity. The induced expression of β4E increased the kinetics of both the cell–cell and cell–ECM biophysical properties in a mature nonmigratory population. Taken together, these data suggest that the splice variant has a specialized function in the homeostasis of the normal epithelium. Many molecular details of the β4E splice variant remain to be determined. We speculate that β6β4E integrin is an ideal candidate to provide strong cell–cell and cell–ECM adhesion during the process of contextual remodeling without committing to HD formation. The future use of animal models will likely add significantly to our understanding of its role in normal tissue homeostasis as well as in pathological processes associated with aberrant β4-integrin function. Understanding the cytoplasmic or lateral protein interactions of this interesting variant will be important, as will knowing whether its expression can be used to halt α6β1-integrin–dependent cancer invasion and metastasis.
The distribution of the β4E form within the luminal cell compartment (Figure 1) is reminiscent of previous reports of suprabasal expression of β4 integrin (as detected only by extracellular domain epitopes) as a high-risk factor for malignant progression in mouse skin carcinogenesis (Tennenbaum et al., 1993). The resulting disrupted TGFβ signaling was permissive for clonal expansion of tumor-initiated cells (Owens et al., 2003). Because initiated tumor cells commonly have activated Src signaling as an oncogenic event (reviewed in Summy and Gallick, 2003) and c-Src activation is associated with a mouse model of invasive prostate cancer progression (Cai et al., 2011), it is intriguing that an increased Src activation accompanies β4E-induced expression in normal basal cells (Figure 2). Given that the prostate basal compartment contains the stem cells for the human prostate gland (Schmelz et al., 2005) and hosts the premalignant changes for the development of prostate cancer (Bonkhoff, 1996), it will be of interest to determine the contribution of β4E integrin to the development of PIN and the abnormal luminal expansion during PIN progression (Montironi et al., 1996).
Other reports show that, in the developing and small intestine, α6β4 is found ubiquitously at the base of the epithelium, but its crypt form is immunologically distinct and appears to be functionally inactive (Beaulieu, 1997; Teller and Beaulieu, 2001). We note that, in these previous studies, the functional activity of β4C was defined by the canonical cytoplasmic domain, which is missing in the β4E variant. The β4E variant increases the biophysical properties of cell–cell and cell–ECM interactions (Figure 7), which may be important in normal homeostasis. Recent modeling of likely stress requirements in tissues such as the lung, in which α6β4 responds to mechanical stretching (Chen et al., 2015), predicts that airway peristalsis rearranges cells and stimulates a mechano-sensitive process to reform cell–cell adhesions (Bokka et al., 2016). We note that both normal lung and duodenal (small intestine) tissues express β4E, as indicated by RNA analysis (van Leusden et al., 1997). It remains to be determined whether the β4E protein is present in these tissues. Considering the current study, a cell–cell location of β4E would provide a testable molecular explanation for maintaining strength of adhesion without the accompanying adhesion complex formation dictated by the canonical β4C cytoplasmic domain.
The results reported here showed that α6β4C was expressed in all cells during induced migration, consistent with previous reports (reviewed in Mercurio et al., 2001). However, the α6β4E expression was distinct from α6β4C and restricted to a subset of migratory cells with increased cell–cell and cell–ECM resistance properties. Surprisingly, superresolution microscopy revealed a ringlike pattern of β4E-tGFP at the cell–ECM surface in confluent cultures, containing both actin and CD9. In the plasma membrane, ringlike structures called flat clathrin lattices (FCLs) exist on dorsal or ventral surfaces (Grove et al., 2014) and contain specific receptors, including the αvβ5 integrin (Zuidema et al., 2018). The content of the FCL is dynamic, because lowering actomyosin contractility will increase the redistribution of integrin β5 to FCLs (Zuidema et al., 2018). FCLs are F-actin–controlled structures that oppose migration (Leyton-Puig et al., 2017) or in other systems are contractility–independent structures formed as a result of frustrated endocytosis (Baschieri et al., 2018). Because the work reported here shows that β4E integrin is associated with F-actin in ringlike structures during nonmigratory conditions on the basal cell surface, it will be of interest to determine whether β4E redistributes or resides in FCLs during the transition from subconfluent random migration to confluent, nonmigratory cultures. Recent advances in new tools for the quantitative analysis of clathrin-mediated endocytosis during differentiation would likely assist these studies (Dambournet et al., 2018). A limitation of the current work is that the model uses an induced β4E-tGFP without the expression of an endogenous β4E. Future work will likely determine the dynamic of the endogenously produced β4E in 3D culture and uncover other regulatory features.
Finally, we note that, in both subconfluent migratory and confluent nonmigratory cultures, β4E-tGFP was found codistributed with CD9, either in retraction fibers (Figure 5) or in F-actin–containing ringlike structures (Figure 6), respectively. Previous work reported that CD9 interacts with α6β4 (Baudoux et al., 2000), is found in retraction fibers (Yamada et al., 2013), and is diffusely distributed during the assembly of (pre)-HD structures (Yamada et al., 2013). We speculate that α6β4E may associate with CD9 to serve as a temporary and dispensable cell–ECM adhesion site for the retraction fiber during migration and/or may associate with CD9 to provide increased biophysical properties during cell–cell closure events. In either scenario, we speculate that the β4E integrin containing the unique cytoplasmic domain may serve as an intermediary form that could provide a laminin adhesion function before committing to HD assembly. We also note that β4E residency in the specialized FCL endocytic platform may explain, in part, the different internalization rates of laminin-binding integrins, because α6 pairs with β4, β4E, and/or β1 as compared with α3, which pairs only with β1 (Das et al., 2017).
MATERIALS AND METHODS
Cell culture
RWPE-1 cells were obtained from the American Type Culture Collection (ATCC CRL-11609TM) and cultured in KSFM (GIBCO, kit cat. no. 17005-042), supplemented with 0.05 mg/ml bovine pituitary extract (BPE) (provided with the KSFM kit), 5 ng/ml EGF (provided with the KSFM kit), 100 IU penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin (MP Biomedicals, cat. no. 1674049). RWPE-1 identity was verified by allelic signature of 15 different genetic markers.
Three-dimensional. cell culture
RWPE-1 cells (80,000 cells in 45 μl) were placed into the top well chamber of a 96-well hanging-drop plate (Biomatrix 96, cat. no. HDP1096) according to the manufacturer’s suggestions and cultured for up to 72 h. The cells were collected by centrifugation of the collected drops. Approximately one-third of the plate was required to obtain 10–20 µg of protein for analysis.
Overexpression vector constructs
For generation of inducible β4E-tGFP expression constructs, the coding region for β4E-tGFP gene was inserted between AgeI-MluI sites of the pTRIPZ/shRNA lentiviral vector downstream of the Tet/ON promoter (Open Biosystems). Others have shown GFP tagging of β4 does not impair its function (Geuijen and Sonnenberg, 2002; Tsuruta et al., 2003). Lentivirus packaging vectors pMD2.G (Addgene plasmid #12259) and psPAX2 (Addgene plasmid #12260) were gifts from Didier Trono (Addgene, www.addgene.org). pTRIPZ-β4E-tGFP plasmid and lentivirus packaging vectors were cotransfected into HEK-293T cells using Lipofectamine 3000 (Thermo Fisher Scientific, cat. no. L3000008). Virus was harvested 24 and 48 h after transfection, filtered through a 0.45-µm filter, and added to the growth media of cells supplemented with 8 μg/ml polybrene (Sigma-Aldrich, cat. no. H9268). Stable cells were selected with 500 ng/ml puromycin for at least 2 wk. Cells were then induced with doxycycline (500 ng/ml) for 5 d and FACS sorted for GFP-positive cells. Sorted cells were maintained in normal growth medium without doxycycline.
qRT-PCR
qRT-PCR was performed using TaqMan RNA-to-Ct 1-Step Kit (Thermo Fisher, cat. no. 4392653) and Applied Biosystem StepOnePlus Real-Time PCR System (Thermo Fisher, cat. no. 4376600). β4C and β4E specific primers and probes were obtained using TaqMan Gene Expression Assays. Hs01103167_g1 was used to specifically detect integrin β4C mRNA, and Hs01108014_g1 was used to specifically detect integrin β4E mRNA.
Immunofluorescence microscopy
Cells were cultured on coverslips, washed in phosphate-buffered saline (PBS), and then fixed in 2% paraformaldehyde for 20 min at room temperature. After fixation, cells were rehydrated in PBS for 5 min, permeabilized in PBS-0.5% Triton X-100 for 5 min, blocked in blocking buffer (PBS, 5% normal goat serum [Sigma], 0.1% Triton X-100, and 2 mM NaN3) for 30 min, and then incubated with primary antibodies in blocking buffer for 1 h at room temperature. Cells were washed three times, 5 min for each wash, in PBS-0.1% Triton X-100 and then incubated with secondary antibodies and Hoechst 33342 (1:1000; Invitrogen) for 1 h at room temperature. Cells were washed three times, 5 min for each wash, in PBS-0.1% Triton X-100 and mounted in 0.1M n-propyl gallate, 90% (by volume) glycerol, and 10% PBS solution.
Immunohistochemistry
For immunofluorescence microscopy of Formalin-fixed paraffin-embedded (FFPE) tissue samples, FFPE blocks were sectioned at 5-µm thickness and mounted on slides. Slides were baked at 65°C overnight; deparaffined through three 7-min washes in xylene; and then passed through 100, 75, and 50% isopropanol and ddH2O for rehydration. Antigen retrieval was performed using EnVision FLEX Target Retrieval Solution, High pH (DAKO, DM828) buffer and heated at 97°C using a decloaking chamber for 20 min. Slides were washed in washing buffer (0.1 M Tris-HCl, 0.3 M NaCl, 0.1% Tween 20, and 7.7 mM NaN3, pH 7.6, at 25°C) followed by blocking buffer (5% normal goat serum, 0.1 M Tris-HCl, and 0.15 M NaCl, pH 7.6, at 25°C) for 30 min at room temperature. Primary antibodies were diluted in blocking buffer and incubated at 4°C overnight in a humidified chamber. Slides were washed three times in wash buffer and incubated with secondary antibody and Hoechst 33342 (1:1000; Invitrogen) for 30 min to 1 h at room temperature. Slides were washed three times in washing buffer and then mounted using ProLong Diamond Antifade Mountant (Thermo Fisher Scientific, P36970) and stored in the dark at room temperature overnight to cure the mountant. Slides were imaged or stored at −20°C for future analysis.
HMWCK and p63 staining was performed on the Ventana BenchMark ULTRA instrument, and antigen retrieval was 64 min with 16-min antibody incubation at 36°C; the detection was accomplished with Ventana OptiView IHC DAB Detection Kit.
Antibodies
We used antibodies and stains for immunofluorescence microscopy at the following working concentrations: anti-β4 N-terminus antibody (1:20 for FFPE tissue samples and 1:100 for fixed cell lines; Abcam ab110167, 439-9B), anti-β4 C-terminus antibody (1:50 for FFPE tissue samples and 1:100 for fixed cell lines; Thermo Fisher Scientific MA5-17104, 10B10D5), anti-HMWCK and p63 (Ventana Basal Cell Cocktail), anti-paxillin antibody (1:100; BD Transduction Laboratories 610052, clone 349), anti-phosphotyrosine antibody (1:100; BD Transduction Laboratories, clone PY20), anti-BP180 (1:100; clone 1D1, R.B. Nagle, University of Arizona), anti-CD9 (1:100; GeneTex GTX19715, clone ALB 6), and phalloidin (Alexa Fluor 647 Phalloidin; 1:40; Thermo Fisher Scientific A22287). Secondary antibodies were purchased from Jackson Immunoresearch and used at 1:1500.
Chemicals
Doxycycline was purchased from Sigma-Aldrich (D9871) and was used as indicated in each experiment. Bortezomib was purchased from Selleck Chemicals (cat. no. 1013) and was used at 50 ng/ml overnight.
Immunoblotting
Cell extracts were produced by lysing cells in cold RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1% vol/vol Triton X-100, 1% wt/vol Na deoxycholate, 0.1% SDS, pH7.4). Samples of equal total protein were resolved by SDS-PAGE, blotted, probed with primary and secondary antibodies, and scanned on an Odyssey imager (Li-Cor Biosciences). Care was taken to avoid saturating the scans. Antibodies used for immunoblots included: anti-β4 N-terminus antibody (1:1000; Abcam EPR8558), anti–β4C-terminus antibody (1:1000; Thermo Fisher Scientific MA5-17104, 10B10D5), anti-tGFP antibody (1:1000; ORIGENE TA183081, clone OTI2H8), anti-α6 antibody (1:1000; A6NT, Cress lab), anti-β1 antibody (1:1000; Cell Signaling Technology 9699, clone D2E5), anti-Src antibody (1:1000; Cell Signaling Technology 2108), anti-phospho-Src family (Tyr416) (1:1000; Cell Signaling Technology 2101), and anti–α-tubulin DM1A (1:1000; Sigma-Aldrich). IRDye 800CW secondary antibodies (Li-Cor Biosciences) were prepared according to the manufacturer’s instructions and used at 1:3000 dilutions.
Flow cytometry
Cells were trypsinized, washed with PBS, and analyzed using a BD Accuri C6 flow cytometer (BD Biosciences).
ECIS measurements
Electrical properties of confluent or wounded epithelium were measured using electric ECIS as described previously (Wegener et al., 2000). Cell adhesion measurements were based on changes in resistance/capacitance to current flow applied at different frequencies (Applied Biophysics, Troy, NY). A 96-well plate (Applied Biophysics, 96W10idf PET) was coated with laminin at 4°C overnight, and cells were inoculated at 6250 or 12,500 cells per well in 200 ml in triplicate, and resistance/capacitance was measured at 400 and 40,000 Hz. Biophysical parameters were determined using a variable slope model, known as a four parameter dose response curve (GraphPad Prism, v8): Y = Bottom + (X HillSlope) × (Top-Bottom)/(X HillSlope + EC50 HillSlope). EC50 is the concentration of agonist that gives a response halfway between Bottom and Top. HillSlope describes the steepness of the family of curves. Top and Bottom are plateaus in the units of the y-axis.
Structured illumination superresolution microscopy
Superresolution 3D-structured illumination imaging was performed on a Zeiss ELYRA S1 system equipped with an Alpha Plan-APO 100×/1.46 oil-immersion objective (Zeiss), and 405-, 488-, 561-, and 642-nm lasers and pco.edge camera (PCO-Tech). Image stacks with three rotations per plane, 18 z-steps, and a z-distance of 0.1 µm were acquired and computationally reconstructed to generate superresolution optical serial sections to yield resolutions in x,y of 110 nm and in z of 300 nm. Color channels were aligned using an alignment parameter. SI reconstruction and image processing was performed with the Zen v. 2.3 black-imaging software package (Zeiss).
Statistical methods
The statistical significance of differences in average measurements was evaluated using GraphPad Prism v. 8.0. Means are taken to be significantly different if p < 0.05. In figures, an asterisk (*) indicates 0.05 > p ≥ 0.01; a double asterisk (**) indicates 0.01 > p ≥ 0.001; a triple asterisk (***) indicates 0.001 > p ≥ 0.0001, a quadruple asterisk (****) indicates 0.0001 > p, and not significant (ns) indicates p ≥ 0.05 for the indicated pairwise comparison. Error bars in all figures indicate SD.
Supplementary Material
Acknowledgments
We thank the staff of the University of Arizona Cancer Center core support services (supported by P30 CA23074), Tissue Acquisition and Molecular Analysis Shared Resource for the tissue processing, Flow Cytometry Shared Resource for cell-sorting expertise, and the University of Arizona Microscopy Core service for the superresolution microscopy; Dan Buster for assistance with image analysis and Joseph Mascarenhas for assistance with the ECIS instrument; and William L. Harryman for editing assistance and electronic submission of the article. We acknowledge all the funding sources that made the work possible: National Institutes of Health, National Cancer Institute (NIH-NCI) RO1CA159406 (to A.E.C.), NIH-NCI T32CA009213 (to A.E.C.), NIH, National Heart, Lung, and Blood Institute P01 HL126609 (to J.G.N.G., Project 3 [A.E.C]), and support from the Tim and Diane Bowden Fellowship in Cancer Biology (to M.W.).
Abbreviations used:
- 2D
two-dimensional
- 3D
three-dimensional
- β4E-tGFP
tGFP-tagged integrin β4E
- CMV
cytomegalovirus
- ECIS
electric cell–substrate impedance sensing
- ECM
extracellular matrix
- FBS
fetal bovine serum
- FCL
flat clathrin lattice
- FFPE
Formalin-fixed paraffin-embedded
- HD
hemidesmosome
- HMWCK
high-molecular-weight cytokeratin
- KFSM
keratinocyte serum-free media
- PBS
phosphate-buffered saline
- PIN
prostatic intraepithelial neoplasia
- qRT-PCR
quantitative reverse transcriptase PCR
- tGFP
turbo-GFP.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0652) on March 13, 2019.
REFERENCES
- Adams J, Kauffman M. (2004). Development of the proteasome inhibitor Velcade (bortezomib). Cancer Invest , 304–311. [DOI] [PubMed] [Google Scholar]
- Baschieri F, Dayot S, Elkhatib N, Ly N, Capmany A, Schauer K, Betz T, Vignjevic DM, Poincloux R, Montagnac G. (2018). Frustrated endocytosis controls contractility-independent mechanotransduction at clathrin-coated structures. Nat Commun , 3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudoux B, Castanares-Zapatero D, Leclercq-Smekens M, Berna N, Poumay Y. (2000). The tetraspanin CD9 associates with the integrin alpha6beta4 in cultured human epidermal keratinocytes and is involved in cell motility. Eur J Cell Biol , 41–51. [DOI] [PubMed] [Google Scholar]
- Beaulieu JF. (1997). Extracellular matrix components and integrins in relationship to human intestinal epithelial cell differentiation. Prog Histochem Cytochem , 1–78. [DOI] [PubMed] [Google Scholar]
- Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. (1997). Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis , 1215–1223. [DOI] [PubMed] [Google Scholar]
- Bokka KK, Jesudason EC, Warburton D, Lubkin SR. (2016). Quantifying cellular and subcellular stretches in embryonic lung epithelia under peristalsis: where to look for mechanosensing. Interface Focus , 20160031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkhoff H. (1996). Role of the basal cells in premalignant changes of the human prostate: a stem cell concept for the development of prostate cancer. Eur Urol , 201–205. [DOI] [PubMed] [Google Scholar]
- Bonkhoff H, Remberger K. (1998). Morphogenetic concepts of normal and abnormal growth in the human prostate. Virchows Arch , 195–202. [DOI] [PubMed] [Google Scholar]
- Bulgakova NA, Klapholz B, Brown NH. (2012). Cell adhesion in Drosophila: versatility of cadherin and integrin complexes during development. Curr Opin Cell Biol , 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Babic I, Wei X, Huang J, Witte ON. (2011). Invasive prostate carcinoma driven by c-Src and androgen receptor synergy. Cancer Res , 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Epshtein Y, Ni X, Dull RO, Cress AE, Garcia JG, Jacobson JR. (2015). Role of integrin beta4 in lung endothelial cell inflammatory responses to mechanical stress. Sci Rep , 16529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambournet D, Sochacki KA, Cheng AT, Akamatsu M, Taraska JW, Hockemeyer D, Drubin DG. (2018). Genome-edited human stem cells expressing fluorescently labeled endocytic markers allow quantitative analysis of clathrin-mediated endocytosis during differentiation. J Cell Biol , 3301–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das L, Anderson TA, Gard JM, Sroka IC, Strautman SR, Nagle RB, Morrissey C, Knudsen BS, Cress AE. (2017). Characterization of laminin binding integrin internalization in prostate cancer cells. J Cell Biochem , 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Cress AE, Dalkin BL, Nagle RB. (2001a). Unique expression pattern of the alpha6beta4 integrin and laminin-5 in human prostate carcinoma. Prostate , 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Rabinovitz I, Futscher BW, Schnolzer M, Burger F, Liu Y, Kulesz-Martin M, Cress AE. (2001b). Identification of a novel structural variant of the alpha 6 integrin. J Biol Chem , 26099–26106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melker AA, Sonnenberg A. (1999). Integrins: alternative splicing as a mechanism to regulate ligand binding and integrin signaling events. Bioessays , 499–509. [DOI] [PubMed] [Google Scholar]
- Emsley JG, Hagg T. (2003). Alpha6beta1 integrin directs migration of neuronal precursors in adult mouse forebrain. Exp Neurol , 273–285. [DOI] [PubMed] [Google Scholar]
- Fontao L, Stutzmann J, Gendry P, Launay JF. (1999). Regulation of the type II hemidesmosomal plaque assembly in intestinal epithelial cells. Exp Cell Res , 298–312. [DOI] [PubMed] [Google Scholar]
- Geuijen CA, Sonnenberg A. (2002). Dynamics of the alpha6beta4 integrin in keratinocytes. Mol Biol Cell , 3845–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti FG, Stepp MA, Suzuki S, Engvall E, Ruoslahti E. (1992). Proteolytic processing of endogenous and recombinant beta 4 integrin subunit. J Cell Biol , 951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Metcalf DJ, Knight AE, Wavre-Shapton ST, Sun T, Protonotarios ED, Griffin LD, Lippincott-Schwartz J, Marsh M. (2014). Flat clathrin lattices: stable features of the plasma membrane. Mol Biol Cell , 3581–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques TS, Relvas JB, Nishimura S, Pytela R, Edwards GM, Streuli CH, ffrench-Constant C. (1998). Neural precursor cell chain migration and division are regulated through different beta1 integrins. Development , 3167–3177. [DOI] [PubMed] [Google Scholar]
- Knox JD, Cress AE, Clark V, Manriquez L, Affinito KS, Dalkin BL, Nagle RB. (1994). Differential expression of extracellular matrix molecules and the alpha 6-integrins in the normal and neoplastic prostate. Am J Pathol , 167–174. [PMC free article] [PubMed] [Google Scholar]
- Leyton-Puig D, Isogai T, Argenzio E, van den Broek B, Klarenbeek J, Janssen H, Jalink K, Innocenti M. (2017). Flat clathrin lattices are dynamic actin-controlled hubs for clathrin-mediated endocytosis and signalling of specific receptors. Nat Commun , 16068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov IV, Vander Griend DJ, Xu Y, Antony L, Dalrymple SL, Isaacs JT. (2006). Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res , 8598–8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio AM, Rabinovitz I, Shaw LM. (2001). The alpha 6 beta 4 integrin and epithelial cell migration. Curr Opin Cell Biol , 541–545. [DOI] [PubMed] [Google Scholar]
- Micheloni A, De Luca N, Tadini G, Zambruno G, D’Alessio M. (2004). Intracellular degradation of beta4 integrin in lethal junctional epidermolysis bullosa with pyloric atresia. Br J Dermatol , 796–802. [DOI] [PubMed] [Google Scholar]
- Montironi R, Bostwick DG, Bonkhoff H, Cockett AT, Helpap B, Troncoso P, Waters D. (1996). Origins of prostate cancer. Cancer , 362–365. [DOI] [PubMed] [Google Scholar]
- Mueller FJ, Serobyan N, Schraufstatter IU, DiScipio R, Wakeman D, Loring JF, Snyder EY, Khaldoyanidi SK. (2006). Adhesive interactions between human neural stem cells and inflamed human vascular endothelium are mediated by integrins. Stem Cells , 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle RB, Hao J, Knox JD, Dalkin BL, Clark V, Cress AE. (1995). Expression of hemidesmosomal and extracellular matrix proteins by normal and malignant human prostate tissue. Am J Pathol , 1498–1507. [PMC free article] [PubMed] [Google Scholar]
- Nahidiazar L, Kreft M, van den Broek B, Secades P, Manders EM, Sonnenberg A, Jalink K. (2015). The molecular architecture of hemidesmosomes, as revealed with super-resolution microscopy. J Cell Sci , 3714–3719. [DOI] [PubMed] [Google Scholar]
- Ni H, Dydensborg AB, Herring FE, Basora N, Gagne D, Vachon PH, Beaulieu JF. (2005). Upregulation of a functional form of the beta4 integrin subunit in colorectal cancers correlates with c-Myc expression. Oncogene , 6820–6829. [DOI] [PubMed] [Google Scholar]
- Nievers MG, Kuikman I, Geerts D, Leigh IM, Sonnenberg A. (2000). Formation of hemidesmosome-like structures in the absence of ligand binding by the (alpha)6(beta)4 integrin requires binding of HD1/plectin to the cytoplasmic domain of the (beta)4 integrin subunit. J Cell Sci , 963–973. [DOI] [PubMed] [Google Scholar]
- Osmani N, Pontabry J, Comelles J, Fekonja N, Goetz JG, Riveline D, Georges-Labouesse E, Labouesse M. (2018). An Arf6- and caveolae-dependent pathway links hemidesmosome remodeling and mechanoresponse. Mol Biol Cell , 435–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DM, Romero MR, Gardner C, Watt FM. (2003). Suprabasal alpha6beta4 integrin expression in epidermis results in enhanced tumourigenesis and disruption of TGFbeta signalling. J Cell Sci , 3783–3791. [DOI] [PubMed] [Google Scholar]
- Potts AJ, Croall DE, Hemler ME. (1994). Proteolytic cleavage of the integrin beta 4 subunit. Exp Cell Res , 2–9. [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Uitto J. (1998). Hemidesmosomal variants of epidermolysis bullosa. Mutations in the alpha6beta4 integrin and the 180-kD bullous pemphigoid antigen/type XVII collagen genes. Exp Dermatol , 46–64. [DOI] [PubMed] [Google Scholar]
- Qian H, Georges-Labouesse E, Nystrom A, Domogatskaya A, Tryggvason K, Jacobsen SE, Ekblom M. (2007). Distinct roles of integrins alpha6 and alpha4 in homing of fetal liver hematopoietic stem and progenitor cells. Blood , 2399–2407. [DOI] [PubMed] [Google Scholar]
- Qian H, Tryggvason K, Jacobsen SE, Ekblom M. (2006). Contribution of alpha6 integrins to hematopoietic stem and progenitor cell homing to bone marrow and collaboration with alpha4 integrins. Blood , 3503–3510. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Teja M, Breit C, Clarke M, Talar K, Wang K, Mohammad MA, Pickwell S, Etchandy G, Stasiuk GJ, Sturge J. (2016). How to study basement membrane stiffness as a biophysical trigger in prostate cancer and other age-related pathologies or metabolic diseases. J Vis Exp, PMID: 27684203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz M, Moll R, Hesse U, Prasad AR, Gandolfi JA, Hasan SR, Bartholdi M, Cress AE. (2005). Identification of a stem cell candidate in the normal human prostate gland. Eur J Cell Biol , 341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk LM, Geuijen CA, Oomen LC, Calafat J, Janssen H, Sonnenberg A. (2000). The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin alpha6beta4 and may regulate the spatial organization of hemidesmosomes. J Cell Biol , 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summy JM, Gallick GE. (2003). Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev , 337–358. [DOI] [PubMed] [Google Scholar]
- Teller IC, Beaulieu JF. (2001). Interactions between laminin and epithelial cells in intestinal health and disease. Expert Rev Mol Med , 1–18. [DOI] [PubMed] [Google Scholar]
- Tennenbaum T, Weiner AK, Belanger AJ, Glick AB, Hennings H, Yuspa SH. (1993). The suprabasal expression of alpha 6 beta 4 integrin is associated with a high risk for malignant progression in mouse skin carcinogenesis. Cancer Res , 4803–4810. [PubMed] [Google Scholar]
- Tsuruta D, Hopkinson SB, Lane KD, Werner ME, Cryns VL, Jones JC. (2003). Crucial role of the specificity-determining loop of the integrin beta4 subunit in the binding of cells to laminin-5 and outside-in signal transduction. J Biol Chem , 38707–38714. [DOI] [PubMed] [Google Scholar]
- van Leusden MR, Kuikman I, Sonnenberg A. (1997). The unique cytoplasmic domain of the human integrin variant beta4E is produced by partial retention of intronic sequences. Biochem Biophys Res Commun , 826–830. [DOI] [PubMed] [Google Scholar]
- von Bredow DC, Nagle RB, Bowden GT, Cress AE. (1997). Cleavage of beta 4 integrin by matrilysin. Exp Cell Res , 341–345. [DOI] [PubMed] [Google Scholar]
- Wang M, Nagle RB, Knudsen BS, Rogers GC, Cress AE. (2017). A basal cell defect promotes budding of prostatic intraepithelial neoplasia. J Cell Sci , 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener J, Keese CR, Giaever I. (2000). Electric cell-substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exp Cell Res , 158–166. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Kuikman I, Margadant C, van Rheenen J, Sonnenberg A. (2007). Serine phosphorylation of the integrin beta4 subunit is necessary for epidermal growth factor receptor induced hemidesmosome disruption. Mol Biol Cell , 3512–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Mugnai G, Serada S, Yagi Y, Naka T, Sekiguchi K. (2013). Substrate-attached materials are enriched with tetraspanins and are analogous to the structures associated with rear-end retraction in migrating cells. Cell Adh Migr , 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T, Otero J, Chen Y, Kim YM, Koutcher JA, Satagopan J, Reuter V, Carver B, de Stanchina E, Enomoto K, et al (2013). Beta4 integrin signaling induces expansion of prostate tumor progenitors. J Clin Invest , 682–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. (2011). A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature , 99–103. [DOI] [PubMed] [Google Scholar]
- Zuidema A, Wang W, Kreft M, Te Molder L, Hoekman L, Bleijerveld OB, Nahidiazar L, Janssen H, Sonnenberg A. (2018). Mechanisms of integrin alphaVbeta5 clustering in flat clathrin lattices. J Cell Sci , PMID: 30301780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.