Abstract
Osteosarcoma is the most frequent primary tumor of bone and is characterized by its high tendency to metastasize in lungs. Although treatment in cases of early diagnosis results in a 5-yr survival rate of nearly 60%, the prognosis for patients with secondary lesions at diagnosis is poor, and their 5-yr survival rate remains below 30%. In the present work, we have used a number of analytical methods to investigate the impact of increased metastatic potential on the biophysical properties and force generation of osteosarcoma cells. With that aim, we used two paired osteosarcoma cell lines, with each one comprising a parental line with low metastatic potential and its experimentally selected, highly metastatic form. Mechanical characterization was performed by means of atomic force microscopy, tensile biaxial deformation, and real-time deformability, and cell traction was measured using two-dimensional and micropost-based traction force microscopy. Our results reveal that the low metastatic osteosarcoma cells display larger spreading sizes and generate higher forces than the experimentally selected, highly malignant variants. In turn, the outcome of cell stiffness measurements strongly depends on the method used and the state of the probed cell, indicating that only a set of phenotyping methods provides the full picture of cell mechanics.
INTRODUCTION
Tumors are classically detected using manual palpation, with malignancies appearing harder to the touch than normal tissues. While it has been demonstrated that the extracellular matrix (ECM) of the majority of cancers does indeed harden (Boyd et al., 2005), cells isolated from these tissues are generally softer than their normal counterparts (Suresh, 2007). For instance, cell softening has been reported among tumor types in bladder cancer (Lekka et al., 1999), breast cancer (Guck et al., 2005), and melanoma (Ochalek et al., 1988). The acquisition of these phenotypic features results from sequential and random adaptions of the cancer cells to their microenvironments, which occur through genetic and epigenetic changes. It is also known that selective pressure is particularly dramatic in the case of metastatic cells, which have to survive in a variety of contrasting environments (Klein, 2013).
Osteosarcoma is the most frequent primary bone cancer and affects mostly children and young adults. Despite the significant progress experienced in surgical and chemotherapeutic treatment of this tumor type, the survival rate of patients with metastatic disease remains below 30% (Picci, 2007). Because of its mesenchymal origin, osteosarcoma does not follow a standard epithelial-to-mesenchymal transition, a process by which carcinoma cells lose their polarity and disassemble their cell–cell junctions, giving rise to mesenchymal-like cells. This difference potentially invalidates many of the observations made in the more frequent carcinomas that originate from epithelial cells (Wolf and Friedl, 2006). In the present work, we have analyzed the biomechanical changes that osteosarcoma cells suffer during metastatic adaptation. With that aim, we have used two paired cell lines composed of a low metastatic osteosarcoma parental cell type and an experimentally derived highly metastatic variant. In both pairs, the low metastatic cells (SaOs-2 and HuO9) were isolated from primary human osteosarcomas, and the corresponding highly metastatic sublines (LM5 and M132) were derived by means of intravenous injection of the parental cell lines in immunodeficient mice and serial retransplantation of the secondary lung tumors that formed. SaOs-2 cells were collected and retransplanted four times to give rise to the highly metastatic LM5 cells (Jia et al., 1999). In turn, the HuO9 cells went through the serial transplantation process three times until the metastatic M132 cell line was established (Kimura et al., 2002). The lasting impact of the adaptations acquired by the neoplastic cells subject to this protocol is confirmed by their significantly enhanced metastatic dissemination. For instance, while intravenously injected HuO9 cells require almost 2 wk to form visible secondary lung tumors, the time needed for the derived M132 cell line to metastasize is almost reduced to half, resulting in a corresponding shortening of the survival rate of the host animals (Kimura et al., 2002; Ren et al., 2015). Similarly, the metastatic tumors formed by the intravenous injection of LM5 cells are more abundant and significantly larger than those of the parental cell line SaOs-2 (Jia et al., 1999).
The biomechanical tests performed in this study comprise indentation-type atomic force microscopy (IT-AFM), real-time deformability (RT-DC), and tensile biaxial stretching, all of which measure the mechanical response of cells to forces applied in different directions (Figure 1). Additionally, we analyzed the morphological differences between the low and the highly metastatic cell lines using fluorescently labeled cells and estimated their force generation capabilities using two-dimensional (2D) traction force microscopy (TFM) and micropillar-based TFM (mTFM). Here, we found that the mechanical properties and behavior of cells strongly depend on the method with which they are tested. We observed a marginally reduced stiffness in free-floating, highly metastatic cells, but an inverse trend when probed in an adhered state through tension. Furthermore, we found a consistent and significant decrease in cellular contractility with increasing metastatic potential. While these results confirm some of the data available in the literature (Indra et al., 2011; Peschetola et al., 2013), they also contradict some of the findings reported by other groups (Mierke et al., 2008; Rosel et al., 2008; Kraning-Rush et al., 2012). This discrepancy reflects the trends reported in literature but shows the importance of different mechanical tests for diagnostic applications and lays a foundation and landmark for future research in the field of cancer biomechanics.
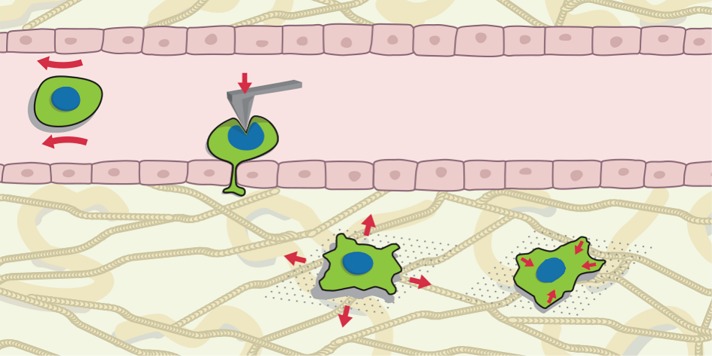
FIGURE 1:
Late stages of the metastatic cascade and biomechanical interrogation. During their metastatic journey, cancer cells are exposed to a number of biophysical challenges. Their adaptation to overcome these threats can be explored using different tools. Each one of the phenotyping techniques relies on the application of a force of known magnitude and tracking of the resulting cell deformation. During blood circulation, shear forces (depicted by red arrows around the cell) and collision are dominant threats. Nuclear size and compressive stiffness (as measured using AFM, red arrow in cell sitting on the endothelium) gain relevance during extravasation, and finally, once malignant cells reach the target organ, tensile stresses caused by tissue deformations (left cell in stroma) are central. Additionally, adhering cells exert contractile forces, which can be decomposed into forces parallel to the surface as well as out-of-plane ones.
RESULTS
Highly and low metastatic osteosarcoma cells display large morphological differences
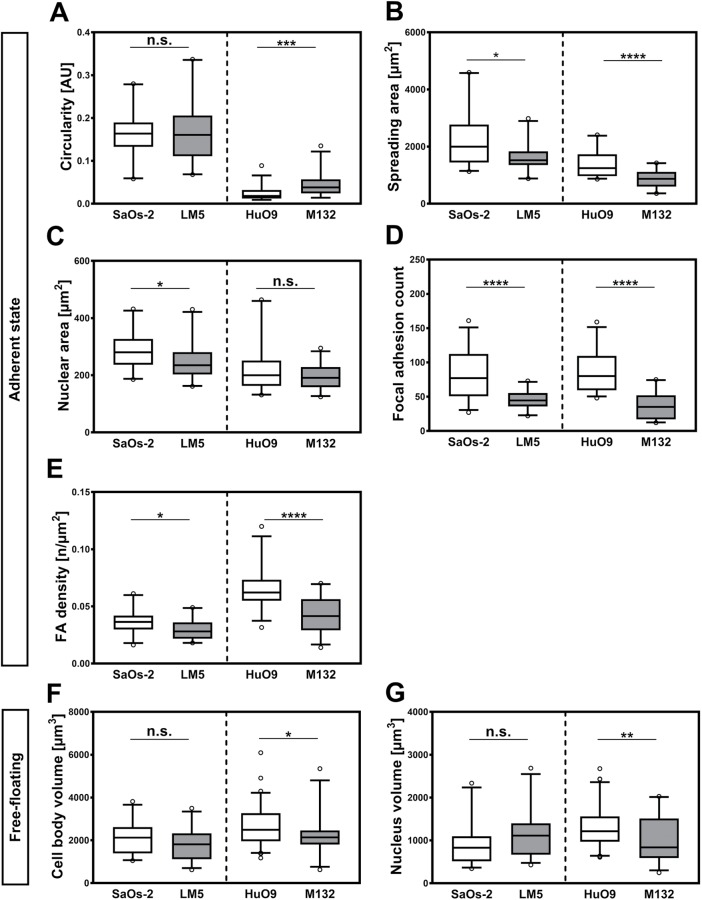
Immunofluorescence staining was used to reveal the morphologic differences between the studied cell model pairs (Figure 2 and Supplemental Figure 1). Labeling of filamentous actin with fluorescent phalloidin shows similar cytoskeletal organization in SaOs-2 and LM5 cells in the adherent state, with cells exhibiting well-defined stress fibers that span the cell body. In turn, adherent M132 and, more evidently, HuO9 cells display thin stress fibers forming abundant filopodial-like structures (Figure 3, top row). These morphological features result in lower cell circularity values for this osteosarcoma pair, with the low metastatic variant displaying the least circular approximation (Figure 4A). Quantitative image analysis of the fluorescently stained samples reveals that, in 2D culture conditions, the highly metastatic variants exhibit significantly smaller spreading areas (Figure 4B). In the SaOs-2/LM5 model, the reduction in spreading area (2254 ± 955 μm2 [n = 24] vs. 1605 ± 479 μm2 [n = 22] for SaOs-2 and LM5, respectively) is accompanied by an equivalent reduction of nuclear projected area (287.9 ± 67.65 μm2 [n = 24] vs. 246.6 ± 64.19 μm2 [n = 22] for SaOs-2 and LM5, respectively). This trend is not found in the HuO9/M132 cell pair, in which the reduction in nuclear projected area (220.9 ± 82.13 μm2 [n = 29] vs. 197.2 ± 43.26 μm2 [n = 27] for HuO9 and M132, respectively; Figure 4C) is less pronounced than the drop in the spreading area of the highly metastatic cell line (1385 ± 453 μm2 [n = 29] vs. 870 ± 304 μm2 [n = 27] for HuO9 and M132, respectively). As a model of cell adhesion and its connection with the ECM in two dimensions, immunofluorescence staining against vinculin, a major component of the focal adhesion complex, was performed (Figure 3, top row, green channel). Analysis of the vinculin signal shows a higher average number of focal adhesions (FAs) in the SaOs-2 and HuO9 cells (80 ± 33 and 87.8 ± 31, respectively) than in their highly metastatic counterparts (45 ± 13 in the case of LM5 and 36.6 ± 19.7 in M132) (Figure 4D). This difference is also evident when the data are normalized by the spreading area, the highly metastatic cells LM5 and M132 having 0.0294 ± 0.0084 FAs/μm2 and 0.0417 ± 0.0154 FAs/μm2, respectively, compared with 0.037 ± 0.0106 FAs/μm2 and 0.0652 ± 0.018 FAs/μm2 for SaOs-2 and HuO9, respectively (Figure 4E).
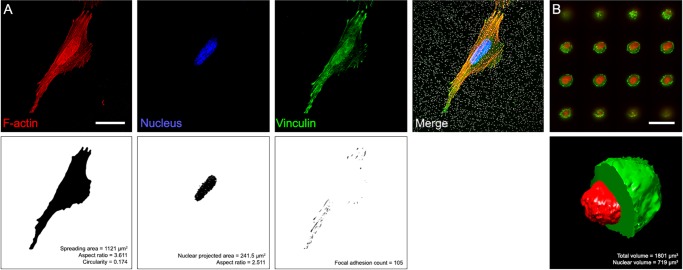
FIGURE 2:
Analysis of immunofluorescence images. (A) Confocal images of the different cell lines stained with NucBlue (blue channel in the top left panel), phalloidin (red channel), and anti-vinculin (green channel) were used to obtain cell spreading area (top right panel), projected area of the nucleus (bottom left panel), and FA number (bottom right panel). (B) For volume estimations, nonadherent cells were stained with phalloidin (green channel) and NucBlue (red channel). In the example, confocal slices of a free-floating SaOs-2 cell (top panel) were reconstructed and segmented to estimate cytoplasmic and nuclear volumes (bottom panel). Scale bars: 25 μm.
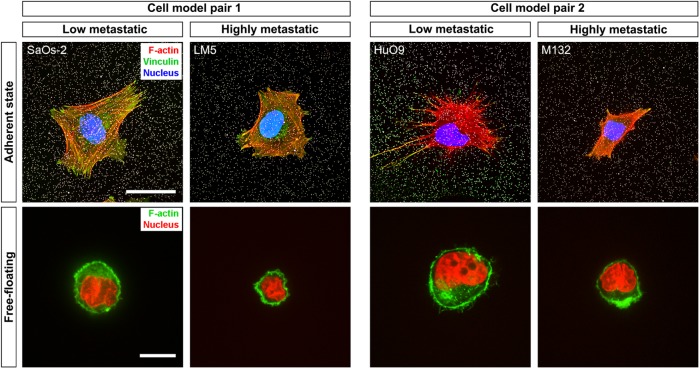
FIGURE 3:
Cell morphology. Cells were stained under two different conditions: cultured on substrates identical to those used in the tensile stiffness and TFM experiments and in the free-floating state. In the images of the cells on 2D substrates (top row), beads on the surface are displayed in white, nuclei in blue, actin cytoskeleton in red, and vinculin in green. The yellow color indicates colocalization of the signal of actin (in the stress fibers) and vinculin. In turn, in the free-floating state (bottom row), the actomyosin cortex, evidenced with phalloidin staining, is shown in green and nuclei in red. Scale bars: 30 μm (images of adherent cells); 15 μm (images of free-floating cells).
FIGURE 4:
Morphometric analysis of cell body, nuclear sizes, and FA counts for highly and low metastatic cell lines. Box plot diagrams showing 5th, 25th, 75th, and 95th percentiles and median values of diverse morphological features of the osteosarcoma models, namely, (A) circularity, (B) spreading area, (C) nuclear projected area, (D) FA count, (E) FA density, (F) free-floating volume, and (G) free-floating nuclear volume. SaOs-2, n = 24; LM5, n = 22; HuO9, n = 29; M132, n = 27. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; n.s., not significant.
Free-floating size, defined as the volume enclosed by the actomyosin cortex (Figure 2 and Supplemental Figure 2), appears significantly reduced in the highly metastatic cell variants of both models (Figure 3, bottom row), with 2077 ± 47 μm3 (n = 26) versus 1803 ± 713 μm3 (n = 37) for SaOs-2 and LM5, respectively, and 2653 ± 947 μm3 (n = 71) versus 2228 ± 859 μm3 (n = 25) for HuO9 and M132 (Figure 4F). Similar to the morphology of adherent cells, nuclear size follows contrasting tendencies, with the parental cell line of HuO9 and the highly metastatic LM5 cells displaying larger nuclei than their paired cell lines (902 ± 485 μm3 [n = 26] vs. 1132 ± 560 μm3 [n = 37] for SaOs-2 and LM5, respectively, and 1328 ± 493 μm3 [n = 71] vs. 1026 ± 525 μm3 [n = 25] for HuO9 and M132, respectively) (Figure 4G).
Indentation-derived cell stiffness follows divergent trends in both osteosarcoma model pairs
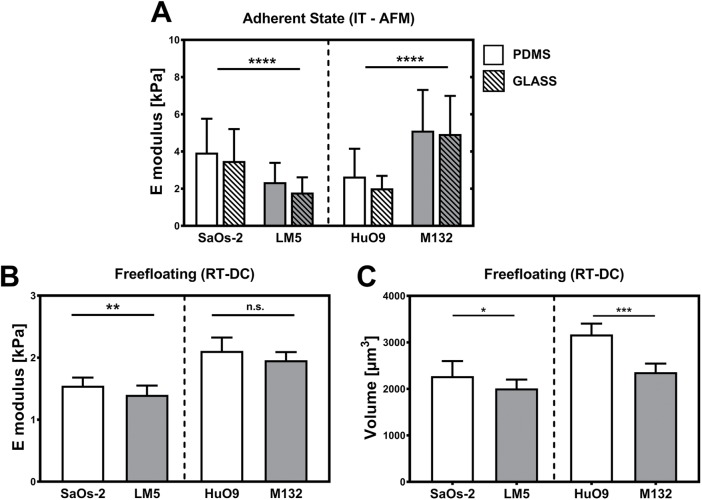
IT-AFM is one of the most widely used methods for probing cells growing on 2D substrates. To exclude effects of the underlying surface on the measurement, we estimated the compressive Young’s modulus (E) for each line in two different experimental conditions: growing on glass and on polydimethylsiloxane (PDMS). AFM indentation results are shown in Figure 5A. SaOs-2 yielded an average compressive modulus of 3.5 ± 1.7 kPa (n = 68 cells) on glass and of 3.9 ± 1.8 kPa (n = 59) on PDMS, while the highly metastatic variant LM5 was significantly softer (p < 0.001) in both conditions (E = 1.8 ± 0.8 kPa [n = 70 cells] on glass, E = 2.4 ± 1.0 kPa [n = 32] on PDMS). In turn, the parental cell line HuO9 was significantly more compliant in compression than its highly metastatic form M132, both on glass (2.0 ± 0.7 kPa [n = 81 cells] vs. 4.9 ± 2.0 kPa [n = 80]) and on PDMS (2.6 ± 1.5 kPa [n = 47 cells] vs. 5.1 ± 2.2 kPa [n = 80]).
FIGURE 5:
IT-AFM and RT-DC. Elastic modulus was estimated on cells attached to (A) PDMS (empty bars) and glass (dashed bars). The analyzed model cell pairs display similar mechanical features on both substrates but different trends for the two model cell lines. (B) In turn, the mechanical properties of free-floating cells were estimated using RT-DC. These experiments reveal higher compliance of the highly metastatic cells compared with the parental cell lines, although this is significant only for the SaOs-2/LM5 model pair. (C) In accordance with the morphological data, the volume of the measured cells was significantly smaller with highly metastatic potential for both model lines. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; n.s., not significant.
In the free-floating state, highly metastatic osteosarcoma cells are softer than their less metastatic counterparts
Deformability of free-floating osteosarcoma cells was estimated using the recently described RT-DC (Otto et al., 2015). In this case, subconfluent cultures of osteosarcoma cells were trypsinized, centrifuged, resuspended in a solution of phosphate-buffered saline (PBS) containing 0.5% (wt/vol) methylcellulose, and immediately analyzed using the RT-DC setup. Results reveal increased deformation in both highly metastatic cell lines (Figure 5B), which is mapped to a bulk stiffness using a fluid dynamics model. Specifically, SaOs-2 (n = 1005, 3 replicates) and HuO9 (n = 903, 3 replicates) display an estimated stiffness of 0.95 ± 0.07 and 1.22 ± 0.09 kPa, respectively, while the highly malignant counterparts LM5 (n = 1550, 3 replicates) and M132 (n = 758, 2 replicates) exhibit a comparable drop in their rigidity (0.82 ± 0.06 and 1.215 ± 0.055 kPa, respectively). However, this is significant only for the SaOs-2/LM5 pair. In turn, the drop in cell body volume estimated from the immunofluorescence images (Figure 4F) was confirmed by the images obtained in the RT-DC setup (Figure 5C).
Highly metastatic cells display slightly higher tensile stiffness
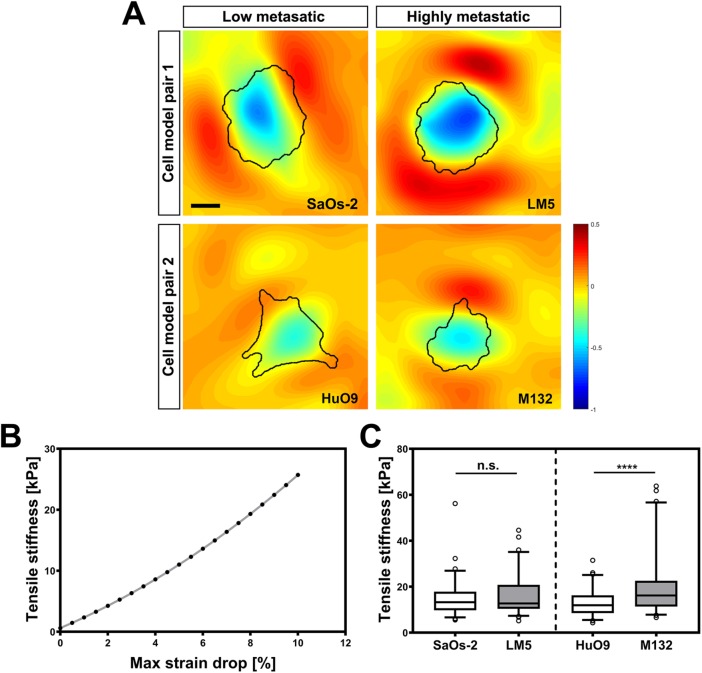
All four osteosarcoma lines were tested in tension using a method in which stiffness is estimated based on the ability to resist biaxial tensile strain applied to a soft PDMS substrate through a regulated vacuum pump (Bartalena et al., 2011). Images of fluorescent beads attached to the substrates in the relaxed and stretched states were used to estimate the deformation map, and the resistance to the deformation of the cell was quantified by calculating the largest strain drop beneath each cell footprint relative to the applied strain (Figure 6A). With the measure called maximal principal strain drop (MPSD) (Bartalena et al., 2011, 2012), the tensile stiffness is estimated by application of an inverse finite-element model of the system (Figure 6B). Both osteosarcoma model lines follow the same trend, with an increased ability to resist the tensile deformation for the highly metastatic cell lines compared with their parental cells (Figure 6C). However, the difference was significant only for the HuO9/M132 cell pair (Supplemental Table 1).
FIGURE 6:
Tensile stiffness estimation. (A) Strain distribution around and below the adhering osteosarcoma cells. (B) A finite-element model was developed to convert strain drop values into cell stiffness data. (C) Box plot representation of tensile stiffness of all four osteosarcoma cell lines showing 5th, 25th, 75th, and 95th percentiles and median values. Statistical analysis reveals significant differences between HuO9 and M132 cells and a similar trend in the SaOs-2/LM5. ****, p < 0.0001; n.s., not significant. Scale bar: 15 μm.
Highly metastatic cells show reduced contractility
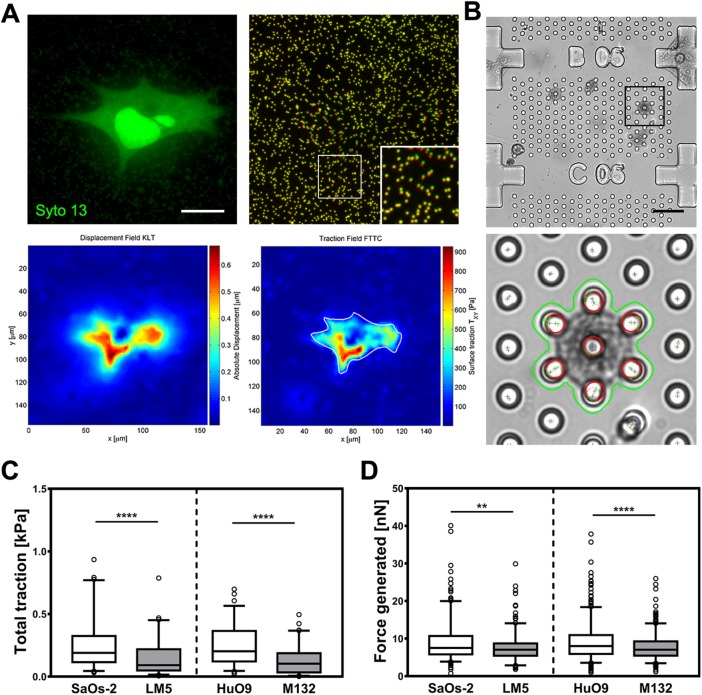
In addition to the morphometric and mechanical analysis, we measured the forces exerted by the osteosarcoma cells using two complementary methods commonly used in the field of TFM (Polacheck and Chen, 2016; Roca-Cusachs et al., 2017), namely, conventional 2D-TFM (Holenstein et al., 2017) and mTFM (Goedecke et al., 2015) (Figure 7, A and B). In 2D-TFM, silicone substrates identical to those used in immunostaining and tensile stiffness estimation were used. In both cell pairs, the total traction exerted by low metastatic cell lines is significantly larger (0.29 ± 0.44 kPa [n = 101] and 0.24 ± 0.16 kPa [n = 71] in the cases of SaOs-2 and HuO9, respectively) than the traction generated by the highly metastatic cells (0.15 ± 0.15 kPa [n = 67] and 0.13 ± 0.11 kPa [n = 80] in the cases of LM5 and M132, respectively; Figure 7C). Similarly, the total force generated by the cells on commercial arrays composed of micropillars followed a similar pattern (Figure 7D). Highly metastatic cell lines show a reduced contractility behavior compared with their parental cell lines, with a mean maximal generated force of 9.18 ± 6.18 nN (n = 304) and 9.13 ± 5.41 nN (n = 482) for the parental cell lines (SaOs-2 and HuO9, respectively) and 7.54 ± 3.73 (n = 245) and 7.69 ± 3.4 nN (n = 387) for the derived cell lines (LM5 and M132, respectively).
FIGURE 7:
Traction force generation. (A) Representative 2D-TFM data set with attached cell (left), pseudo-colored bead images (right, green: before detachment; red: after detachment; inset represents an enlarged view of the boxed region), calculated displacement, and traction field heat maps. (B) Exemplary micropost array (top) with automatically segmented cell and tracked post tip in the deformed state (bottom). (C) Traction per cell measured using 2D-TFM and (D) forces using micropillar force sensors represented as box plot diagrams showing 5th, 25th, 75th, and 95th percentiles and median values. **, p < 0.01; ****, p < 0.0001. Scale bars: 15 μm (A); 40 μm (B).
Cellular mechanisms underlying the acquisition of the metastatic phenotype
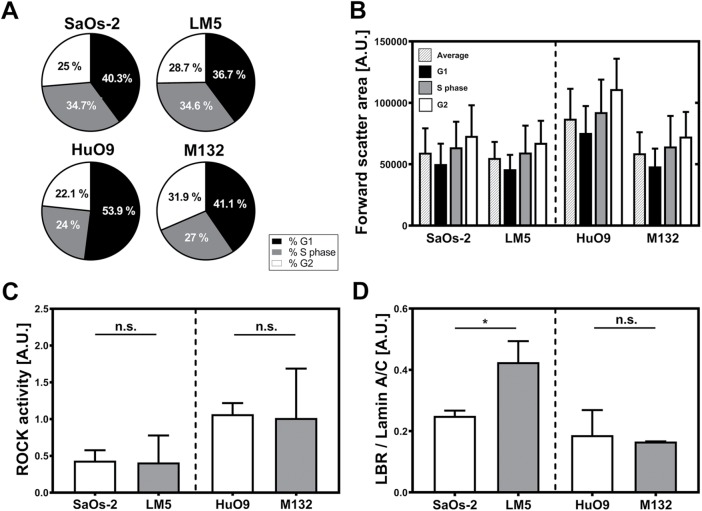
To estimate the impact of differences in cell cycle on the observed mechanical properties of highly and low metastatic cells, we performed flow cytometry. Our results reveal a higher number of cells in G1 phase and a reduced cell count in G2 in the HuO9 cell line compared with the derived M132 cells (Figure 8A). By separately analyzing the forward-scatter area (FSC-A) of osteosarcoma cells in different phases, we observed a trend toward larger values as the cell cycle progresses from G1 to G2 (Figure 8B). Because spreading area (Panagiotakopoulou et al., 2018) and free-floating volume (Figure 8B) in cancer cells are reduced during the G1 phase, the actual size differences in the HuO9/M132 pair might actually be larger when taking the cell cycle into account. Similarly, the total force generated by cancer cells has been reported to be reduced during the G1 phase (Panagiotakopoulou et al., 2018), and consequently, the force generated by the HuO9 cells (Figure 7D) might have been underestimated as well.
FIGURE 8:
(A) Flow cytometry of cells stained with propidium iodide reveals similar cell numbers in G1, S, and G2 phases in the case of the SaOs-2/LM5 cell pair. In turn, HuO9 cell cultures display higher cell counts in G1 phase and less in G2 than the derived cell line M132. (B) Cells in different cell cycle phases display differences in their relative sizes measured as FSC-A. The average cell size (patterned bars) and the size of cells at the different cycle phases followed the same trend observed using other techniques, with the highly metastatic cells displaying smaller sizes. G1 (black bars), S phase (gray bars) and G2 (white bars). (C) ROCK activity shows no statistically significant differences in the osteosarcoma pairs. However, in both highly metastatic cell lines (LM5 and M132), the measured activity was slightly lower than in the parental cell lines (HuO9 and M132) (n = 4). (D) In turn, the ratio between LBR and lamin A/C is significantly higher in LM5 compared with SaOs-2 (p < 0.05), with no statistical differences in the other cell pair (n = 3). *, p < 0.05; n.s., not significant.
We further measured the Rho-associated protein kinase (ROCK) activity of the osteosarcoma cells lines. We found that the phosphorylation activity of ROCK, which is related to contractility potential, was almost identical in all four cell lines (Figure 8C). Because the composition of the nuclear lamina has been shown to have a central role on the structural integrity of the nucleus, which in turn is key for mechanotransduction signaling (Lammerding et al., 2007; Pajerowski et al., 2007; Harada et al., 2014), we used Western blotting to compare the amounts of lamin B receptor (LBR) and lamin A/C in the analyzed cell lines (Supplemental Figure 3). We observed that, in the highly metastatic LM5 cells, the ratio between LBR and lamin A/C was significantly increased (p < 0.05); however, this difference was not present in the other osteosarcoma pair (Figure 8D).
DISCUSSION
During cancer development and metastatic dissemination, cancer cells acquire genetic and epigenetic modifications that render them more competitive in the neoplastic microenvironment and are transferred to subsequent cell generations (Cairns, 1975; Nowell, 1976; Cameron et al., 2000; Greaves and Maley, 2012). Following this evolutionary concept, changes are not limited to the biochemical pathways of cancer cells, but alter also their biophysical phenotype. For instance, malignant cells show increased migration, reduced adhesion, and higher compliance (Swaminathan et al., 2011; Calzado-Martín et al., 2016; Northey et al., 2017). It therefore has been suggested that these findings can be used to derive diagnostic methods based on these markers to detect metastatic potential (Lekka et al., 1999; Guck et al., 2005; Lopez et al., 2011; Plodinec et al., 2012) or to develop treatment methods that directly target these biophysical characteristics and thus effectively hinder metastasis (Wirtz et al., 2011). However, as many of these results were reported on a single type of cancer, and many of them are contradictory (Indra et al., 2011; Kraning-Rush et al., 2012), no universal molecular or biophysical marker of malignancy has been found to date. Moreover, most of the studies focus on epithelial-derived carcinomas, which during their malignant transformation undergo phenotypic changes commonly referred to as epithelial-to-mesenchymal transition (Sannino et al., 2017). On the other hand, far less is known about these relationships in other types of cancer cells, such as those involved in sarcomas, which are of mesenchymal origin. Among these tumors, osteosarcoma is the most frequent primary bone cancer and affects mostly children and young adults. Its high malignancy and tendency to metastasize in the lungs results in a low survival rate in patients who display secondary lesions. It is therefore of a great interest to create new diagnostic tools to detect the malignancies before the neoplastic cells spread.
In the course of cancer progression, neoplastic cells experience a variety of different mechanical environments (Müller and Silvan, 2019), and conventional techniques for cell stiffness measurements that apply compressive or shear forces to the cell’s upper surface or relatively low-force tensile stretching of cells in suspension might not tell the full story, especially if viewed in isolation. The implications of this limitation have started to emerge, and a number of groups have recently reported multiparametric studies to characterize cancer cells (Wu et al., 2018) and their microenvironments (Pearce et al., 2018). Going a step further, we have combined the best established high- and semi-high throughput techniques to measure the morphological and mechanical properties of two osteosarcoma models. The tools used for biomechanical phenotyping measure cell resistance to forces applied in different directions and consequently challenge different cellular structures (Figure 1). Therefore, the biophysical information extracted by each of these techniques might be relevant to different cellular processes occurring during metastatic dissemination. To gain insight into the adaptations that osteosarcoma cells undergo when acquiring metastatic potential, we have analyzed two paired cell lines in which the parental cells, SaOs-2 and HuO9, were isolated from human primary osteosarcoma tumors and their highly malignant variants (LM5 and M132, respectively) were established by means of serial collection of lung metastases and reinjection in the tail vein (Jia et al., 1999; Kimura et al., 2002). Genomic instability is a limitation of cancer studies involving in vitro culture, especially in case of osteosarcoma-derived cells, which have shown significant genomic differences over longer culturing periods (Muff et al., 2015). Nevertheless, the different metastatic potential of parental and experimentally selected osteosarcoma cell lines seems to remain unaffected by these events (Ren et al., 2015), and such changes might be more relevant in the research of molecular and genetic markers of malignancy (Muff et al., 2012).
The experimental procedure by which the highly metastatic cells were generated repeatedly forces the cells through a process that resembles the late steps of osteosarcoma hematogenous metastasis, namely, blood circulation, adhesion to the blood vessel wall, extravasation, and establishment of secondary lesions. Cells injected through the tail vein have in the first place to subsist in a nonadherent environment in which cells must overcome potentially lethal mechanical threats associated with fluid flow, such as collision, hemodynamic forces, and shear stress (Wirtz et al., 2011; Rejniak, 2012; Massagué and Obenauf, 2016). Under these conditions, the cell cortex, a structural framework with actin filaments and myosin as its main components, acquires a central role in shape maintenance and survival. In both cell pairs, the highly metastatic variants display reduced free-floating stiffness, a trend that has already been described in other tumor types. However, under the same shear conditions, smaller cells deform to a lesser extent than larger ones of identical stiffness, a feature that facilitates the recovery of the original shape after elastic deformations. Therefore, it is possible that, while stiffness reduction of cancer cells favors their success in other steps of the metastatic cascade, the observed size adaptation reduces the risk of membrane rupture during systemic circulation. Because metastasizing cells frequently aggregate in multicellular structures during their systemic circulation, a characteristic that facilitates their arrest in capillaries, a strategy that has been described in a number of tumor types including osteosarcoma (Massagué and Obenauf, 2016), it is not likely that the smaller sizes have a significant impact on the physical arrest of the malignant cells.
During the next steps of the metastatic progression, adhesion to the endothelial wall and extravasation, cancer cells might benefit from the increased cell body compliance. In vitro time-lapse studies of the extravasation process have shown that, independently of taking place between (paracellular) or through (transcellular) endothelial cells, the area through which tumor cells penetrate is relatively small (Chen et al., 2013), and it is only at the end of the process that the opening seems to enlarge, probably to facilitate the penetration of the nucleus (Chen et al., 2013). Interestingly, the characterized cell model pairs seem to have followed two different adaptation strategies to successfully perform this step. On one hand, the highly metastatic LM5 cells display slightly larger nuclear volumes compared with the low metastatic SaOs-2 cells, the latter being stiffer when measured by IT-AFM, a technique highly influenced by the mechanical properties of the nucleus (Lekka et al., 1999). Analysis of the relative amounts of LBR and lamin A/C points to changes in the composition of the nuclear lamina as responsible for this phenotypic adaptation (Houben et al., 2007). On the other hand, the HuO9/M132 pair shows the opposite trend, with the highly metastatic variant displaying larger nuclei but softer indentation stiffnesses, with the relative amount of LBR and lamin A/C in both cell lines being similar in this case (Figure 8D). Again, biomechanical, molecular, and morphological data need to be analyzed collectively to understand the potential significance of the observed adaptations. During the extravasation step, tumor cells undergo dramatic shape changes, keeping the spherical shape in the luminal side and adopting a final spread morphology upon exiting the lumen (Chen et al., 2013). This is when the stiffness of the stress fibers, spreading area, and force generation might play a relevant role. For adherent cells, our biaxial stretching experiments reveal the opposite trend in the mechanical properties compared to the free-floating state, with highly metastatic cells appearing stiffer than their less metastatic counterparts. Along with spreading area, nuclear area was proportionally reduced for the SaOs-2/LM5 pair but less pronounced for the HuO9/M132 model. It is known that force generation properties of adherent cells are linked to various cell functions such as migration, contraction, mechanosensing, and morphogenesis, with many of these properties also being important during metastasis (Butcher et al., 2009). Nevertheless, previous studies on force magnitudes in cells with different malignancies are inconsistent. Traction stresses are found to increase (Wyckoff et al., 2006; Mierke et al., 2008; Rosel et al., 2008; Kraning-Rush et al., 2011) but also to decrease (Munevar et al., 2001; Indra et al., 2011; Peschetola et al., 2013) in transformed cells or in cells with advanced metastatic potential. We performed TFM with two common but functionally different methods and found significantly decreased traction generation with increased malignancy. As previously mentioned, changes in the composition of the nuclear lamina potentially explain the nuclear softening in LM5 cells and might also be behind the reduced traction forces generated by these cells (Lammerding et al., 2007; Harada et al., 2014; Buxboim et al., 2017). Nevertheless, the activity of ROCK, an enzyme known to promote cell contractility through myosin II phosphorylation, is not affected by these events, and its activity is almost identical within the cell pairs. Although a direct relationship between ROCK activity and cell traction has been reported in other cancer types, it is likely that the lower force generation capability of the highly metastatic cells in our paired cell lines is caused by the cytoskeletal differences and reduced FA counts found in these cells. In fact, taking into account the lower number of FAs per cell in the highly metastatic variants, these lower cell tractions could also indicate a more gradual switch to a less adhesion-dependent migration type, as was suggested previously (Peschetola et al., 2013). Nevertheless, the significance of the reduction in FA count in experimentally selected cells that are directly injected into the bloodstream as single cells, and therefore do not need to detach from the primary malignancy, is not clear. In addition, cells are known to show distinctive migration patterns, adhesion, and traction when cultured in a three-dimensional (3D) ECM-derived matrix such as collagen or fibrin hydrogels. These 3D microenvironments are more physiologically close to the culture conditions found in vivo, as many features such as fibril structure, orientation, and pore size are not available to a cell cultured on an ECM-coated 2D substrate (Hall et al., 2013). However, as the calculation of cell-generated contractility in three dimensions is experimentally and computationally much more expensive, it is currently not possible to perform a high-throughput screening of cellular contractility in three dimensions as was done here in two dimensions. Identifying and elucidating the manifold biochemical and biomechanical events underlying cancer progression represent an enormous research challenge, in particular because the different types of carcinomas and sarcomas all show different paths toward tumorigenesis and metastatic dissemination.
In summary, we performed a thorough biophysical and morphological analysis of two model cell lines for osteosarcoma and compared cells with high metastatic potential with their respective parental, less metastatic cell lines. Through this, we have demonstrated that the definition of “cell stiffness” depends heavily on the means by which it is measured (Wu et al., 2018) and the framework according to which it is analyzed, and therefore a consistent link between cell stiffness and increased metastatic potential can only be identified within the context of the method used to probe stiffness. It therefore appears that the mechanical properties should be probed separately, but analyzed collectively in the study of cancer cell mechanics.
MATERIALs AND METHODS
Cell culture
SaOs-2, LM5, HuO9 and M132 cells were cultured in DMEM/Ham (F12) (Sigma; D8437) medium, supplemented with 10% fetal calf serum (Thermo; 10270106) at 37°C in a humidified 95% air/5% CO2 atmosphere. Cells were cultured between passages 10 and 40, without any noticeable difference in growth rate or morphology over the course of time. For minimization of the impact of osteosarcoma cell variability in in vitro cultures (Muff et al., 2015), experiments were done with at least two independently frozen vials and using different passages. The LM5 cell line was kindly provided by E. S. Kleinerman (M.D. Anderson Cancer Center, Houston, TX), and the HuO9 and M132 cells were kindly provided by M. Tani (National Cancer Center Hospital, Tokyo, Japan).
Immunofluorescence, image acquisition, and segmentation
Cells were fixed with 10% neutral buffered Formalin (Sigma; HT501128) for 20 min at room temperature and permeabilized for 10 min in 0.2% Triton X-100 (Sigma; 93418) in PBS. Samples were then incubated with an anti-vinculin antibody (Sigma; V9131; 1:400). Next, samples were washed several times in PBS and further incubated with an anti-mouse antibody (Thermo; A21202) together with fluorescent phalloidin (Thermo; A22284) and NucBlue (Invitrogen; R37605) for 1 h. Finally, samples were washed with PBS and mounted in Mowiol. Confocal images were acquired using an inverted spinning-disk confocal microscope (iMic; FEI Photonics) using a 60× objective (NA 1.35). For morphological analysis in the free-floating state, cells were trypsinized for 5 min, centrifuged at 250 × g, and resuspended in PBS. F-actin and nuclear staining were performed using the same protocol as in the case of adhering cells. In both conditions, images were preprocessed with ImageJ (National Institutes of Health) (Figure 2) (Schneider et al., 2012), and the analysis of morphological features was performed with CellSegm, a MATLAB (MathWorks, Natick, MA) segmentation toolbox (Hodneland et al., 2013), and represented using the UCSF Chimera package (Pettersen et al., 2004).
IT-AFM
Compressive characterization of osteosarcoma cells adhering to glass or PDMS was performed using a NanoWizard atomic force microscope (JPK Instruments) with silicon, spherical (r = 1 µm) AFM tips (NanoWorld) coated with PLL-PEG to hinder cell attachment, and a spring constant of 0.42 N/m estimated by measuring their resonance frequency in air. The extension/retraction speed was set to 4 µm/s, and indentation depth was limited to ∼1 µm to permit a small strain assumption in the analysis.
Cells were seeded on either glass or PDMS substrates 12–14 h before the experiment and kept at room temperature during the measurement. Force-displacement curves of the central part of at least 40 cells per line and condition were acquired in two separate experiments. JPK Data Processing software that applies the Hertz model for spherical indentation was used to calculate the cell stiffness in terms of Young’s modulus.
RT-DC
RT-DC measurements were performed as previously described (Otto et al., 2015). Briefly, cells were trypsinized, centrifuged, and resuspended at 106 cells/ml in a methylcellulose/PBS solution. Next, cells were pumped at a constant flow rate through the microfluidic chip, placed on an inverted microscope (Axiovert 200M; Carl Zeiss) equipped with a 40× objective and a high-power light source. When the flow profile was stabilized, a high-speed CMOS camera (Mikrotron MC1362) was used to acquire images of the cells being deformed by the flow through the channel. Images of the cells were processed first by background subtraction and image thresholding, then by extracting the cell contour of the binary image (Suzuki and Be, 1985), measuring the perimeter (p) and the cell cross-sectional area (A), and then calculating the cell deformation (D) through the circularity (c), defined as
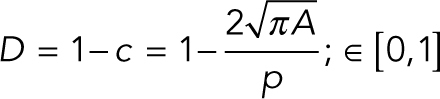 |
Cell stiffness was then estimated using an analytical model that uses the flow profile around a spherical object moving in a cylindrical channel to derive the relationship between cell size–dependent stress, deformation, and elastic properties (Mietke et al., 2015). Measurements of all cell types were repeated at least three times using two different flow rates (0.16 and 0.32 µl/s). As a reference, projected cell area and circularity were additionally extracted for cells passing through a section of the microfluidic chip with a large cross-section, that is, without being deformed.
Because of biological variation, a high-throughput technique like RT-DC can result in a significant difference between two measurements of biological replicates when using a t test or the Mann-Whitney U-test due to the very large sample size. Therefore, to test the significance in the RT-DC experiments, considering the variation between replicates and also the variation of the effect of a treatment, we used an algorithm based on linear mixed models and likelihood ratio test, which was introduced previously (Herbig et al., 2018). For data analysis, ShapeOut, an open-source analysis tool for RT-DC data, was used to perform these tests (Müller and Herbig, 2017).
Tensile testing
The stiffness of cells resulting from their ability to resist applied biaxial tensile strain was calculated using a previously developed system (Bartalena et al., 2011). In brief, cells were stretched using a functional imaging platform coupled with a pressure-actuated biaxial stretcher (StageFlex), and images of the fluorescent beads were acquired in the loaded and relaxed states using a Leica DM5500 microscope equipped with a 40× (NA 0.8) water-immersion objective. On each substrate, between 10 and 20 randomly selected cells were probed three times each in random order. Image pairs were postprocessed with a Matlab (MathWorks, Natick, MA) algorithm using an interface to the ImageJ plug-in bUnwarpJ, which extracts the deformation between the two images by performing an elastic registration using an energy minimization method and represented by B-splines (Arganda-Carreras et al., 2006). The two principal strains (equal to radial and circumferential strains in a 2D equibiaxial strain state) can be calculated using the direct differentiation of the spline-based displacement field. The measured maximal principal strain drop was calculated by normalizing the difference between the applied overall mean strain and minimal substrate strain underneath the cell by the overall mean strain. The Young’s modulus was estimated using an inverse finite-element model (Bartalena et al., 2011).
Additional methods
Additional methods are described in the Supplemental Material.
Supplementary Material
Acknowledgments
We acknowledge the support of the Center for Microscopy and Image Analysis, University of Zurich, in the performance of mTFM experiments and Nino Willi and Denys Sutter for optimizing the image segmentation algorithms. This work was supported by the Swiss National Science Foundation (grant numbers 165670, 138221, 118036 to J.G.S.), by the Commission for Technology and Innovation (grant numbers 14796-PFLS-LSCTI to J.G.S.), by the Kurt and Senta Foundation, and by the Alexander von Humboldt Foundation (AvH Professorship to J.G.).
Abbreviations used:
- 2D
two-dimensional
- 3D
three-dimensional
- AFM
atomic force microscopy
- ECM
extracellular matrix
- FA
focal adhesion
- FSC-A
forward-scatter area
- IT-AFM
indentation-type atomic force microscopy
- LBR
lamin B receptor
- MPSD
maximal principal strain drop
- mTFM
micropillar-based TFM
- NA
numerical aperture
- PBS
phosphate-buffered saline
- PDMS
polydimethylsiloxane
- ROCK
Rho-associated protein kinase
- RT-DC
real-time deformability
- TFM
traction force microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0545) on February 20, 2019.
REFERENCES
- Arganda-Carreras I, Sorzano COS, Marabini R, Carazo JM, Ortiz-de-Solorzano C, Kybic J. (2006). Consistent and elastic registration of histological sections using vector-spline regularization. In: Computer Vision Approaches to Medical Image Analysis, Heidelberg, Germany: Springer, 85–95. [Google Scholar]
- Bartalena G, Grieder R, Sharma RI, Zambelli T, Muff R, Snedeker JG. (2011). A novel method for assessing adherent single-cell stiffness in tension: design and testing of a substrate-based live cell functional imaging device. Biomed Microdevices , 291–301. [DOI] [PubMed] [Google Scholar]
- Bartalena G, Loosli Y, Zambelli T, Snedeker JG. (2012). Biomaterial surface modifications can dominate cell–substrate mechanics: the impact of PDMS plasma treatment on a quantitative assay of cell stiffness. Soft Matter , 673. [Google Scholar]
- Boyd NF, Rommens JM, Vogt K, Lee V, Hopper JL, Yaffe MJ, Paterson AD. (2005). Mammographic breast density as an intermediate phenotype for breast cancer. Lancet Oncol , 798–808. [DOI] [PubMed] [Google Scholar]
- Butcher DT, Alliston T, Weaver VM. (2009). A tense situation: forcing tumour progression. Nat Rev Cancer , 108–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A, Irianto J, Swift J, Athirasala A, Shin J-W, Rehfeldt F, Discher DE. (2017). Coordinated increase of nuclear tension and lamin-A with matrix stiffness outcompetes lamin-B receptor that favors soft tissue phenotypes. Mol Biol Cell , 3333–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. (1975). Mutation selection and the natural history of cancer. Nature , 197–200. [DOI] [PubMed] [Google Scholar]
- Calzado-Martín A, Encinar M, Tamayo J, Calleja M, San Paulo A. (2016). Effect of actin organization on the stiffness of living breast cancer cells revealed by peak-force modulation atomic force microscopy. ACS Nano , 3365–3374. [DOI] [PubMed] [Google Scholar]
- Cameron MD, Schmidt EE, Kerkvliet N, Nadkarni K V, Morris VL, Groom AC, Chambers AF, MacDonald IC. (2000). Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res , 2541–2546. [PubMed] [Google Scholar]
- Chen MB, Whisler JA, Jeon JS, Kamm RD. (2013). Mechanisms of tumor cell extravasation in an in vitro microvascular network platform. Integr Biol , 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedecke N, Bollhalder M, Bernet R, Silvan U, Snedeker J. (2015). Easy and accurate mechano-profiling on micropost arrays. J Vis Exp , 10.3791/53350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC. (2012). Clonal evolution in cancer. Nature , 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M, Lenz D, Erickson HM, Ananthakrishnan R, Mitchell D, Käs J, et al (2005). Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J , 3689–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MS, Long R, Feng X, Huang Y, Hui C-Y, Wu M. (2013). Toward single cell traction microscopy within 3D collagen matrices. Exp Cell Res , 2396–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Swift J, Irianto J, Shin J-W, Spinler KR, Athirasala A, Diegmiller R, Dingal PCDP, Ivanovska IL, Discher DE. (2014). Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol , 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig M, Kräter M, Plak K, Müller P, Guck J, Otto O. (2018). Real-time deformability cytometry: label-free functional characterization of cells. In: Flow Cytometry Protocols, ed. Hawley TS, Hawley RG, New York: Springer, 347–369. [DOI] [PubMed] [Google Scholar]
- Hodneland E, Kögel T, Frei DM, Gerdes H-H, Lundervold A. (2013). CellSegm—a MATLAB toolbox for high-throughput 3D cell segmentation. Source Code Biol Med , 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holenstein CN, Silvan U, Snedeker JG. (2017). High-resolution traction force microscopy on small focal adhesions—improved accuracy through optimal marker distribution and optical flow tracking. Sci Rep , 41633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben F, Ramaekers FCS, Snoeckx LHEH, Broers JLV. (2007). Role of nuclear lamina-cytoskeleton interactions in the maintenance of cellular strength. Biochim Biophys Acta , 675–686. [DOI] [PubMed] [Google Scholar]
- Indra I, Undyala V, Kandow C, Thirumurthi U, Dembo M, Beningo KA. (2011). An in vitro correlation of mechanical forces and metastatic capacity. Phys Biol , 15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia SF, Worth LL, Kleinerman ES. (1999). A nude mouse model of human osteosarcoma lung metastases for evaluating new therapeutic strategies. Clin Exp Metastasis , 501–506. [DOI] [PubMed] [Google Scholar]
- Kimura K, Nakano T, Park Y-B, Tani M, Tsuda H, Beppu Y, Moriya H, Yokota J. (2002). Establishment of human osteosarcoma cell lines with high metastatic potential to lungs and their utilities for therapeutic studies on metastatic osteosarcoma. Clin Exp Metastasis , 477–485. [DOI] [PubMed] [Google Scholar]
- Klein CA. (2013). Selection and adaptation during metastatic cancer progression. Nature , 365–372. [DOI] [PubMed] [Google Scholar]
- Kraning-Rush CM, Califano JP, Reinhart-King CA. (2012). Cellular traction stresses increase with increasing metastatic potential. PLoS One , e32572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraning-Rush CM, Carey SP, Califano JP, Smith BN, Reinhart-King CA. (2011). The role of the cytoskeleton in cellular force generation in 2D and 3D environments. Phys Biol , 15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Dahl KN, Discher DE, Kamm RD. (2007). Nuclear mechanics and methods. Methods Cell Biol , 269–294. [DOI] [PubMed] [Google Scholar]
- Lekka M, Laidler P, Gil D, Lekki J, Stachura Z, Hrynkiewicz AZ. (1999). Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. Eur Biophys J , 312–316. [DOI] [PubMed] [Google Scholar]
- Lopez JI, Kang I, You W-K, McDonald DM, Weaver VM. (2011). In situ force mapping of mammary gland transformation. Integr Biol , 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Obenauf AC. (2016). Metastatic colonization by circulating tumour cells. Nature , 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke CT, Rösel D, Fabry B, Brábek J. (2008). Contractile forces in tumor cell migration. Eur J Cell Biol , 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietke A, Otto O, Girardo S, Rosendahl P, Taubenberger A, Golfier S, Ulbricht E, Aland S, Guck J, Fischer-Friedrich E. (2015). Extracting cell stiffness from real-time deformability cytometry: theory and experiment. Biophys J , 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muff R, Ram Kumar RM, Botter SM, Born W, Fuchs B. (2012). Genes regulated in metastatic osteosarcoma: evaluation by microarray analysis in four human and two mouse cell line systems. Sarcoma , 937506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muff R, Rath P, Ram Kumar RM, Husmann K, Born W, Baudis M, Fuchs B. (2015). Genomic instability of osteosarcoma cell lines in culture: impact on the prediction of metastasis relevant genes. PLoS One , e0125611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DA, Silvan U. (2019). On the biomechanical properties of osteosarcoma cells and their microenvironment. Int J Dev Biol (in press). [DOI] [PubMed] [Google Scholar]
- Müller P, Herbig M. (2017). ShapeOut. https://github.com/ZELLMECHANIK-DRESDEN/ShapeOut (accessed March 5, 2019).
- Munevar S, Wang Y, Dembo M. (2001). Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J , 1744–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northey JJ, Przybyla L, Weaver VM. (2017). Tissue force programs cell fate and tumor aggression. Cancer Discov , 1224–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell P. (1976). The clonal evolution of tumor cell populations. Science , 23–28. [DOI] [PubMed] [Google Scholar]
- Ochalek T, Nordt FJ, Tullberg K, Variants MC, Burger MM. (1988). Correlation between cell deformability and metastatic potential in B16-F1 melanoma cell variants. Cancer Res , 5124–5128. [PubMed] [Google Scholar]
- Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, Girardo S, Pagliara S, Ekpenyong A, Jacobi A, Wobus M, et al (2015). Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nat Methods , 199–202. [DOI] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. (2007). Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA , 15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotakopoulou M, Lendenmann T, Pramotton FM, Giampietro C, Stefopoulos G, Poulikakos D, Ferrari A. (2018). Cell cycle-dependent force transmission in cancer cells. Mol Biol Cell , 2528–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce OMT, Delaine-Smith RM, Maniati E, Nichols S, Wang J, Böhm S, Rajeeve V, Ullah D, Chakravarty P, Jones RR, Montfort A, et al (2018). Deconstruction of a metastatic tumor microenvironment reveals a common matrix response in human cancers. Cancer Discov , 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschetola V, Laurent VM, Duperray A, Michel R, Ambrosi D, Preziosi L, Verdier C. (2013). Time-dependent traction force microscopy for cancer cells as a measure of invasiveness. Cytoskeleton (Hoboken) , 201–214. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem , 1605–1612. [DOI] [PubMed] [Google Scholar]
- Picci P. (2007). Osteosarcoma (osteogenic sarcoma). Orphanet J Rare Dis , 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, Hyotyla JT, Aebi U, Bentires-Alj M, Lim RYH, Schoenenberger CA. (2012). The nanomechanical signature of breast cancer. Nat Nanotechnol , 757–765. [DOI] [PubMed] [Google Scholar]
- Polacheck WJ, Chen CS. (2016). Measuring cell-generated forces: a guide to the available tools. Nat Methods , 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejniak KA. (2012). Investigating dynamical deformations of tumor cells in circulation: predictions from a theoretical model. Front Oncol , 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Mendoza A, Zhu J, Briggs JW, Halsey C, Hong ES, Burkett SS, Morrow J, Lizardo MM, Osborne T, Li SQ, et al (2015). Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget , 29469–29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Conte V, Trepat X. (2017). Quantifying forces in cell biology. Nat Cell Biol , 742–751. [DOI] [PubMed] [Google Scholar]
- Rosel D, Brabek J, Tolde O, Mierke CT, Zitterbart DP, Raupach C, Bicanova K, Kollmannsberger P, Pankova D, Vesely P, Folk P, et al (2008). Up-regulation of Rho/ROCK signaling in sarcoma cells drives invasion and increased generation of protrusive forces. Mol Cancer Res , 1410–1420. [DOI] [PubMed] [Google Scholar]
- Sannino G, Marchetto A, Kirchner T, Grünewald TGP. (2017). Epithelial-to-mesenchymal and mesenchymal-to-epithelial transition in mesenchymal tumors: a paradox in sarcomas? Cancer Res , 4556–4561. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh S. (2007). Biomechanics and biophysics of cancer cells. Acta Biomater , 413–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Be K. (1985). Topological structural analysis of digitized binary images by border following. Comput Vis Graph Image Process , 32–46. [Google Scholar]
- Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC, Superfine R. (2011). Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res , 5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz D, Konstantopoulos K, Searson PC. (2011). The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer , 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Friedl P. (2006). Molecular mechanisms of cancer cell invasion and plasticity. Br J Dermatol , 11–15. [DOI] [PubMed] [Google Scholar]
- Wu P-H, Aroush DR-B, Asnacios A, Chen W-C, Dokukin ME, Doss BL, Durand-Smet P, Ekpenyong A, Guck J, Guz NV, Janmey PA, et al (2018). A comparison of methods to assess cell mechanical properties. Nat Methods , 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. (2006). ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol , 1515–1523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.