Abstract
Contractility of the nonmuscle and smooth muscle cells that comprise biological tubing is regulated by the Rho-ROCK (Rho-associated protein kinase) and calcium signaling pathways. Although many molecular details about these signaling pathways are known, less is known about how they are coordinated spatiotemporally in biological tubes. The spermatheca of the Caenorhabditis elegans reproductive system enables study of the signaling pathways regulating actomyosin contractility in live adult animals. The RhoGAP (GTPase-activating protein toward Rho family small GTPases) SPV-1 was previously identified as a negative regulator of RHO-1/Rho and spermathecal contractility. Here, we uncover a role for SPV-1 as a key regulator of calcium signaling. spv-1 mutants expressing the calcium indicator GCaMP in the spermatheca exhibit premature calcium release, elevated calcium levels, and disrupted spatial regulation of calcium signaling during spermathecal contraction. Although RHO-1 is required for spermathecal contractility, RHO-1 does not play a significant role in regulating calcium. In contrast, activation of CDC-42 recapitulates many aspects of spv-1 mutant calcium signaling. Depletion of cdc-42 by RNA interference does not suppress the premature or elevated calcium signal seen in spv-1 mutants, suggesting other targets remain to be identified. Our results suggest that SPV-1 works through both the Rho-ROCK and calcium signaling pathways to coordinate cellular contractility.
INTRODUCTION
Animals are full of biological tubing, including blood and lymphatic vessels, lung airways, salivary glands, digestive canals, and urinary and reproductive tracts. Actomyosin contractility plays a central role in the functioning of these biological tubes, which must dilate and contract with the proper timing and magnitude to generate appropriate responses to changing biological states (reviewed in Sethi et al., 2017). Consequences of misregulated biological tubes can be seen in conditions such as heart disease, hypertension, and asthma (Uehata et al., 1997; Wettschureck and Offermanns, 2002; Seguchi et al., 2007; Lavoie et al., 2009). Previous research has focused on how biochemical factors such as acetylcholine, serotonin, and nitric oxide regulate tissue function in biological tubes and has elucidated many of the downstream signaling mechanisms that regulate cellular and actomyosin contractility. However, mechanical factors such as flow and pressure are also known to regulate tissue function in biological tubes (Smiesko and Johnson, 1993; Gunst et al., 2003; Smith et al., 2003), and the mechanisms by which these mechanical signals regulate cellular contractility are not as well understood. In vivo studies suggest that mechanical cues play a critical role in regulating and coordinating actomyosin contractility (Munjal and Lecuit, 2014).
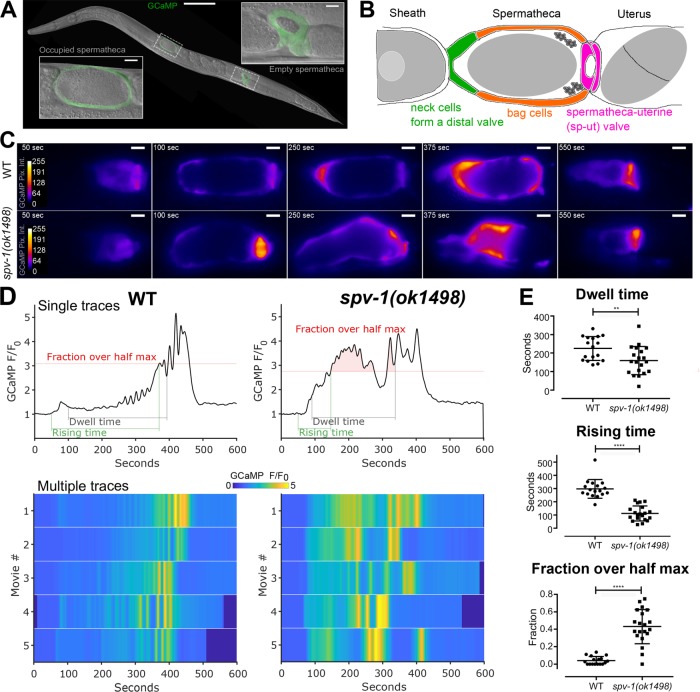
The Caenorhabditis elegans spermatheca (Figure 1A) is a powerful in vivo model system for the study of how cells regulate actomyosin contractility and produce coordinated tissue-level responses to mechanical input. The spermatheca is part of the hermaphroditic gonad, a simple, tubular organ consisting of the oviduct (containing germ cells and oocytes and enclosed by contractile sheath cells), the spermatheca, and the uterus (Kimble and Hirsh, 1979). The spermatheca, consisting of a single layer of 24 myoepithelial cells (Figure 1B), is the site of sperm storage and fertilization. Sheath cell contractions propel the oocyte into the spermatheca, dramatically stretching the cells of the distal neck and spermathecal bag and initiating a process that culminates in spermathecal cell contraction, spermatheca uterine (sp-ut) valve dilation, and expulsion of the fertilized egg into the uterus. This ovulation cycle, including oocyte entry, transit through the spermatheca, and expulsion into the uterus, repeats roughly every 20 min until ∼150 eggs have been produced by each gonad arm. Because the nematode is transparent, and the cells of the spermatheca are clearly visible, the entire ovulatory process can be visualized in live animals using time-lapse microscopy.
FIGURE 1:
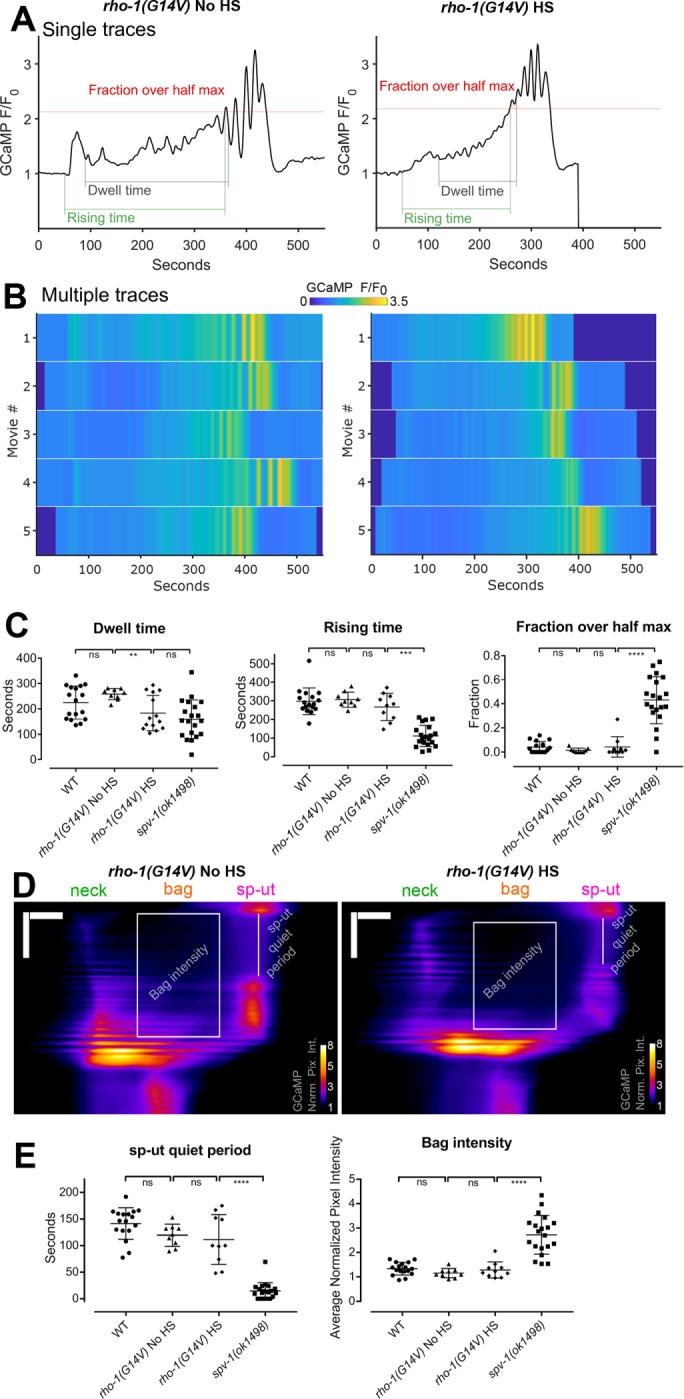
Loss of SPV-1 alters calcium signaling in the spermatheca. (A) An adult nematode with labeled spermathecae, showing one with an embryo inside (left) and one without an embryo inside (right). Scale bars: 100 μm (large image); 10 μm (insets). (B) Tissue-level schematic cartoon of the spermatheca. The spermatheca consists of three distinct regions: the cells closest to the sheath form a neck, or distal valve, that constricts to enclose the newly entered embryo, the central cells form a bag that accommodates the embryo during fertilization and egg shell deposition, and the spermatheca and uterus are connected by the sp-ut valve. (C) Still frames from GCaMP movies of embryo transits in wild-type (WT) (top) and spv-1(ok1498) (bottom) animals. Movies were temporally aligned to the start of oocyte entry at 50 s. (D) GCaMP time series generated from GCaMP movies (Supplemental Movie 1), with metrics highlighted. Dwell time is a tissue function metric calculated as the time from the closing of the distal valve to the opening of the sp-ut valve; rising time is a calcium signaling metric measuring the time from the opening of the distal valve to the first time point where the time series reaches half its maximum; and fraction over half max is a calcium signaling metric measuring how much of the dwell time is spent over the half maximum. Bottom panels show heat maps of five time series for each condition. The top line of the heat map is the time series in the top panel. (E) Quantification of metrics from time series. Error bars display SD, and p values were calculated using Welch’s t test: **, p < 0.01; ****, p < 0.0001.
Contraction of the spermatheca is driven by the well-conserved Rho-ROCK (Rho-associated protein kinase) and calcium signaling pathways, similar to those observed in nonmuscle and smooth muscle cells (Pelaia et al., 2008; Brozovich et al., 2016; Sethi et al., 2017). These two pathways act in concert to regulate the levels of phosphorylated myosin (Somlyo and Somlyo, 2003). The calcium signaling pathway requires activated phospholipase C (PLC-1) (Kariya et al., 2004; Kovacevic et al., 2013), which cleaves the membrane lipid phospatidyl inositol bisphosphate (PIP2) to generate 1,4,5 triphosphate (IP3) and diacylglycerol. IP3 stimulates the release of calcium from the endoplasmic reticulum by binding to the IP3 receptor (Clandinin et al., 1998; Bui and Sternberg, 2002; Yin et al., 2004), and this elevated cytosolic calcium activates myosin light-chain kinase, which phosphorylates the regulatory light chain of myosin (Kelley et al., 2018). The Rho-ROCK pathway acts via RhoA (RHO-1), with active, GTP-bound RHO-1/RhoA activating Rho kinase/ROCK (LET-502) (Wissmann et al., 1997), which phosphorylates and activates myosin and also inhibits myosin phosphatase (Wissmann et al., 1999), resulting in increased phosphorylation and activation of nonmuscle myosin II (Piekny et al., 2003) and contraction of actomyosin fibers. Activation and coordination of both the Rho-ROCK and calcium signaling pathways is required for successful transits of embryos through the spermatheca (Kovacevic et al., 2013; Tan and Zaidel-Bar, 2015). Despite our detailed molecular understanding of these two central pathways, little is known about how they are integrated to regulate actomyosin contractility at the cellular level.
We have developed the C. elegans spermatheca as a model for understanding how cells within a tissue coordinately regulate actomyosin contractility (Kovacevic et al., 2013; Tan and Zaidel-Bar, 2015; Wirshing and Cram, 2017; Kelley et al., 2018). Using the genetically encoded calcium sensor, GCaMP, we have shown that the stretch of incoming oocytes triggers a flash of calcium in the sp-ut valve, followed by dynamic oscillations across the central bag that culminate in contraction and expulsion of the fertilized embryo through the sp-ut valve and into the uterus (Kovacevic et al., 2013). Phospholipid signaling, communication through gap junctions, and the mechanosensor FLN-1/filamin (Kovacevic and Cram, 2010; Kovacevic et al., 2013) are all required for proper spatiotemporal regulation of calcium signaling. Spatiotemporal control of Rho-ROCK signaling is also required for coordinated regulation of contractility in the spermatheca. We have shown that the RhoGAP (GTPase-activating protein toward Rho family small GTPases) SPV-1 controls the spatiotemporal activation of RHO-1 and regulates tissue contractility through LET-502/ROCK (Tan and Zaidel-Bar, 2015). Disruption of these spatiotemporal regulators often results in damage to embryos, reflux of embryos back into the oviduct, or embryos that are trapped in the spermatheca and unable to exit.
In this study, we demonstrate that the RhoGAP SPV-1 is necessary for proper calcium signaling in the C. elegans spermatheca. We further find that SPV-1 is a major regulator of spatiotemporal aspects of spermathecal calcium signaling, controlling the timing and magnitude of calcium signaling in the spermathecal bag and sp-ut cells. Expression of constitutively active alleles of RHO-1 and CDC-42 recapitulate the contractility and calcium-release phenotypes, respectively, of loss of spv-1. This places SPV-1 as a key regulator of both major signaling pathways that modulate actomyosin contractility.
RESULTS
SPV-1 regulates calcium signaling in the spermatheca during embryo transits
To determine whether SPV-1 plays a role in spermathecal calcium signaling, we used a mutant allele of spv-1, ok1498, which causes a 577–base pair frameshift deletion in the RhoGAP domain, resulting in loss of function of the SPV-1 protein (Tan and Zaidel-Bar, 2015). We imaged embryo transits in wild-type and spv-1(ok1498) nematodes expressing the genetically encoded calcium sensor GCaMP3 in the spermatheca (Figure 1C and Supplemental Movie 1). To characterize the spv-1 mutant calcium signaling phenotype, we generated GCaMP time series from the embryo transit movies by calculating the average pixel intensity of each frame (Figure 1, C and D, and Supplemental Movie 1). Plotting time series from many animals as heat maps reveals that spv-1(ok1498) embryo transits consistently show a rapid increase in the onset of calcium activity at the start and elevated calcium levels throughout (Figure 1D).
Movie S1.
GCaMP embryo transit movies and time series display calcium signaling differences in spv-1 mutants. Processed GCaMP embryo transit movies are shown in the bottom panels, while time series are simultaneously displayed in the top panels. A wildtype movie and time series are shown in the left column, while a spv-1 mutant movie and time series are shown in the right column. Metric annotations appear in the time series as they occur in the movies. Both movies and time series were temporally aligned to put oocyte entry at 50 seconds.
To quantify these differences and enable statistical analysis over many embryo transits, we identified one tissue function metric from the movies, dwell time, and two calcium signaling metrics extracted from the GCaMP time series, rising time and fraction over half max (Figure 1D and Supplemental Movie 1). The tissue function metric, dwell time, is defined as the time from distal neck closure to sp-ut valve opening. Dwell time measures the amount of time the embryo remains enclosed by the spermatheca. In spv-1 mutants, faster transit of the embryo through the spermatheca results in a reduced dwell time (Figure 1E). This result is in agreement with a previous observation that embryo transits are faster when SPV-1 is lost (Tan and Zaidel-Bar, 2015). The first calcium signaling metric, rising time, is defined as the time from the start of oocyte entry to the first time point when the GCaMP time series crosses the half maximum. Rising time captures the rate of increase in calcium signal at the start of embryo transit. In spv-1 mutants, a rapid rise in calcium is observed immediately upon entry (Figure 1, D and E). The second calcium signaling metric, fraction over half max, is defined as the duration of the dwell time over the GCaMP half-maximum value divided by the total dwell time. This metric captures the level of calcium throughout embryo transits. In spv-1 mutants, fractions over half max are higher than in wild-type animals (Figure 1, D and E). Taken together, these metrics suggest SPV-1 regulates calcium activity in the spermatheca by dampening calcium signaling, keeping cytosolic calcium at low levels until late in embryo transits.
SPV-1 overexpression results in low calcium signaling and embryo trapping
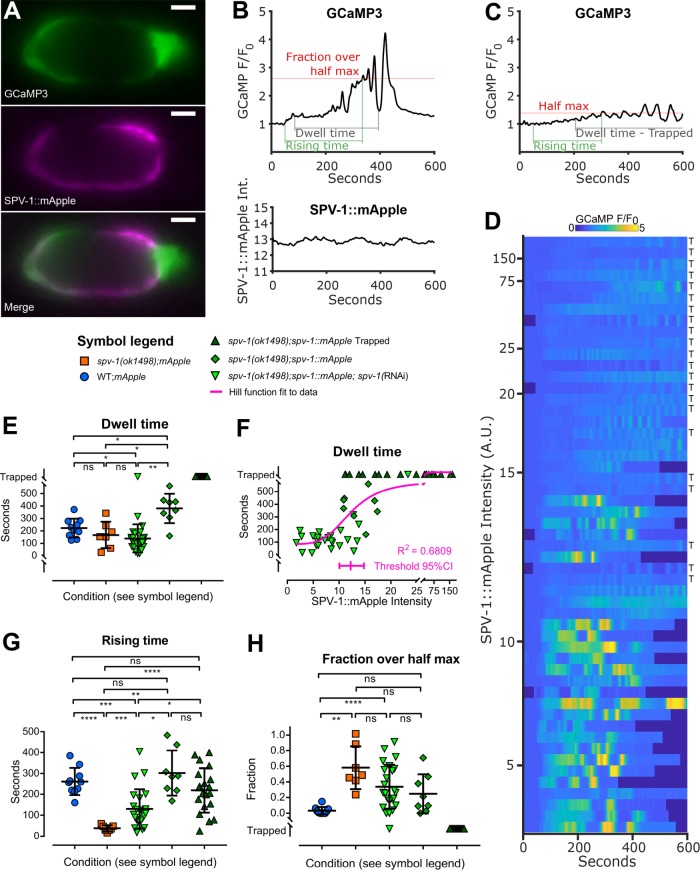
To further understand how SPV-1 regulates calcium signaling, we expressed an SPV-1::mApple translational fusion in the spv-1 mutant background. Animals expressing both the mApple fluorophore and GCaMP were imaged during embryo transits (Figure 2, A and B). In animals expressing high levels of SPV-1::mApple, embryos failed to exit the spermatheca (referred to here as embryo trapping), and spermathecae exhibited low levels of calcium activity (Figure 2, C and D), suggesting that overexpression of SPV-1 results in disrupted tissue function and calcium signaling phenotypes. We next crossed spv-1::mApple into the wild-type background and observed embryo trapping and low calcium signaling in 100% of movies recorded (n = 5; unpublished data). In control experiments, overexpression of mApple alone did not result in embryo trapping or low calcium levels (Figure 2E and Supplemental Figure 1, A–E). These data suggest that high levels of SPV-1 inhibit calcium release and spermathecal tissue contractility.
FIGURE 2:
Spermathecal tissue function and calcium signaling exhibit a threshold response to SPV-1::mApple. (A) Still images from a dual-labeled spermatheca expressing GCaMP and SPV-1::mApple. Scale bars: 10 μm. Brightness is enhanced for presentation. (B) GCaMP time series of normalized average pixel intensity, F/F0 (top), and SPV1::mApple time series of raw average pixel intensity (bottom), from a single embryo transit movie over the same spatial frame and time. (C) A representative GCaMP time series from an SPV-1::mApple–expressing spermatheca containing a trapped embryo, corresponding to the top row of the heat map in D. (D) Heat map showing GCaMP time series from 53 embryo transit movies of varying SPV-1::mApple intensity, ranked with the highest SPV-1::mApple intensity at the top and decreasing with each row. Rows labeled with “T” indicate trapped embryo. (E) Dwell times plotted as a function of condition. (F) Dwell times plotted as a function of SPV-1::mApple intensity. The threshold value from the fitted Hill function is 12.2, with the 95% confidence interval from 10.1 to 14.8. (G) Rising times plotted as a function of condition. (H) Fractions over half max plotted as a function of condition. In E, G, and H, error bars display SD, and p values were calculated using Welch’s ANOVA with Games-Howell multiple comparison: ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Spermathecal tissue function and calcium signaling exhibit a threshold response to SPV-1::mApple levels
Because high levels of SPV-1::mApple resulted in depressed calcium signaling and embryo trapping, we hypothesized that lowering the levels of SPV-1::mApple would result in a more wild-type phenotype in the spv-1 mutant background. To test this in a way that allowed a full sampling of SPV-1::mApple levels, we used spv-1 RNA interference (RNAi) diluted with empty RNAi to four different strengths. These treatments generated spv-1(ok1498); spv-1::mApple animals with a range of SPV-1::mApple levels, measured by mApple fluorescence intensity. Examining the data across all spv-1(ok1498); spv-1::mApple animals, with and without RNAi treatment, shows that spermathecal calcium signaling and embryo transit timing exhibit a threshold response to SPV-1::mApple levels, in which low levels of SPV-1::mApple result in short dwell times and spv-1 mutant-like calcium signaling with a rapid onset of elevated calcium activity, whereas high levels of SPV-1::mApple induce the overexpression phenotypes with embryo trapping and low calcium activity (Figure 2, D and E). Plotting dwell times as a function of SPV-1::mApple intensity clearly displays this threshold behavior (Figure 2F), which can be modeled using a Hill function (Hill, 1910) to determine where, and how rapidly, a system switches from one state to another (Monod et al., 1965). To explore this, we fit a Hill equation to these dwell time data points (Figure 2F) and extracted a threshold value of 12.2 for SPV-1::mApple intensity, with a 95% confidence interval from 10.1 to 14.8. This window of SPV-1::mApple intensity values coincides with the switch from mutant to overexpression behavior in both tissue function (Figure 2, E and F, and Supplemental Figure 2A) and calcium signaling (Figure 2, D, G, and H, and Supplemental Figure 2, B and C). The wild-type phenotype occurs within this window, as well as the spv-1 mutant and overexpression phenotypes.
SPV-1 regulates spatiotemporal aspects of calcium signaling
In wild-type embryo transits, calcium signaling starts with a single pulse in the sp-ut valve, followed by a quiet period after the oocyte enters, then increasing pulses are initiated in the distal neck and travel across the spermathecal bag, culminating in intense calcium pulses and the contractions that expel the embryo into the uterus (Kovacevic et al., 2013). In contrast, in spv-1(ok1498) embryo transits, calcium rises immediately upon oocyte entry, lacks spatial organization, and is elevated compared with wild-type signal, sporadically crashing to a low level and rising again multiple times before the embryo is expelled (Figures 1, B and C, and 3A and Supplemental Movie 1).
FIGURE 3:
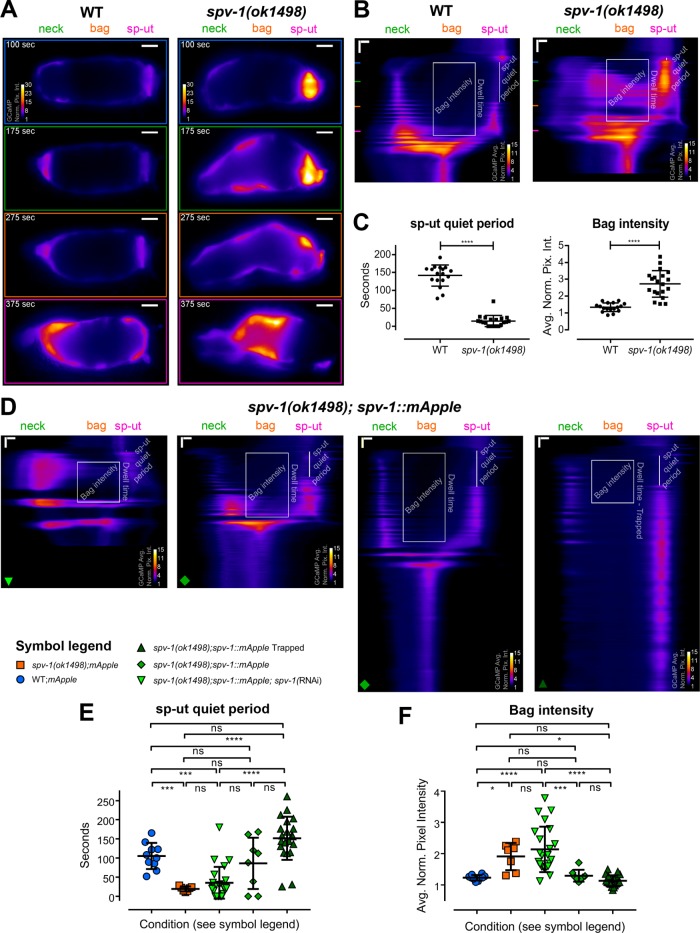
SPV-1 regulates spatiotemporal aspects of calcium signaling. (A) Individual frames from wild-type (left) and spv-1(ok1498) (right) embryo transit movies, the same movies as Figure 1C. All frames follow the color scale indicated in the top frame. Scale bars: 10 μm. (B) Kymograms of the movies in A, generated by averaging over the columns of each movie frame, display the variation in average calcium signaling from the distal valve on the left to the sp-ut valve on the right, with time progressing down (Supplemental Movie 2). Horizontal scale bars: 5 μm; vertical scale bars: 50 s. Colored lines on the left side of the kymograms correspond to the individual frames in A; annotations show the two spatiotemporal calcium signaling metrics used for analysis. The sp-ut quiet period measures the low calcium signaling of the sp-ut valve after oocyte entry, which is lost in spv-1 mutants. Bag intensity measures the average normalized fluorescence intensity of a 25-μm-wide region in the bag section of the spermatheca during the dwell time. (C) Quantification of metrics. Error bars display SD, p values were calculated with Welch’s t test: ****, p < 0.0001. (D) Kymograms from embryo transit movies with SPV-1::mApple intensities of 2.5, 12.6, 16.9, and 78.5, from left to right. Horizontal scale bars: 5 μm; vertical scale bars: 50 s. (E) sp-ut quiet periods plotted as a function of condition. (F) Bag intensities plotted as a function of condition. In E and F, error bars display SD, and p values were calculated using Welch’s ANOVA with Games-Howell multiple comparison: ns, p ≥ 0.05; *, p < 0.05; ***, p < 0.001; ****, p < 0.0001.
To visualize and analyze these spatiotemporal changes, we generated kymograms, in which each frame of the movie is condensed into a horizontal line along the distal to proximal axis of the spermatheca, by averaging over the columns of that frame. Lines representing the individual frames are then stacked vertically, generating kymograms that display the spatial variation of the GCaMP signal from the distal neck to the sp-ut valve over the entire movie (Figure 3, A and B, and Supplemental Movie 2). Wild-type kymograms indicate calcium activity is restricted to the distal (neck) and proximal (sp-ut) ends of the spermatheca during the dwell time. In contrast, spv-1(ok1498) kymograms depict increased activity in the middle of the spermathecal bag as well as in the sp-ut valve (Figure 3B).
Movie S2.
Kymograms display the spatiotemporal differences in spv-1 mutant calcium signaling. Processed GCaMP embryo transit movies, the same shown in Movie 1, are shown in the top panels, while kymograms are simultaneously displayed in the lower panels. A wildtype movie and kymogram are shown in the left column, while a spv-1 mutant movie and kymogram are shown in the right column. Metrics appear on the kymograms as they occur in the movies. Both movies and kymograms were temporally aligned to put oocyte entry at 50 seconds.
To quantify these differences, we identified two prominent spatiotemporal calcium signaling metrics that are consistently altered in spv-1 mutant kymograms (Figure 3, B and C). The first is the sp-ut quiet period. In wild-type kymograms, the sp-ut valve displays a single calcium transient upon oocyte entry, followed by an extended period of low calcium. In spv-1(ok1498) kymograms, the sp-ut valve displays an immediate rise in calcium followed by sustained calcium. The sp-ut quiet period metric quantifies this difference by measuring the time from the end of the initial calcium transient to the next rise in sp-ut calcium. The second spatiotemporal calcium signaling metric is bag intensity. In wild-type kymograms, the central spermathecal cells exhibit low calcium activity that increases and pulses as the tissue expels the egg. In spv-1(ok1498) kymograms, these central cells exhibit rapid, high calcium levels that stay elevated while the spermatheca is occupied. The bag intensity metric quantifies this difference by measuring the average pixel intensity of a 25-µm region of the spermathecal bag over the dwell time. Both spatiotemporal metrics differ significantly between wild-type and spv-1(ok1498) animals (Figure 3C).
We next examined kymograms from the movies with varying SPV-1::mApple intensities (Figure 3D). As expected, low levels of SPV-1::mApple result in short sp-ut quiet periods and high bag intensities, whereas high levels of SPV-1::mApple result in long sp-ut quiet periods and low bag intensities (Figure 3, E and F), with the switch occurring between mApple intensity values of 10.1 and 14.8 (Supplemental Figure 2, D and E). This spatiotemporal analysis indicates that SPV-1 spatially regulates calcium activity in the spermatheca by keeping calcium low in the bag cells and sp-ut valve for a period of time after oocyte entry.
Increasing RHO-1 activity recapitulates transit timing of the spv-1 mutant, but not calcium signaling
Because SPV-1 acts through RHO-1 to regulate contractility of the spermatheca (Tan and Zaidel-Bar, 2015), we speculated that increasing RHO-1 activity might alter calcium signaling in a manner similar to the loss of SPV-1. To test this idea, we obtained nematodes expressing constitutively active RHO-1(G14V) under the control of a heat shock promoter (McMullan and Nurrish, 2011), crossed them with GCaMP-expressing animals, and optimized the heat shock protocol to enable the capture of embryo transit movies (Figure 4, A and B). As expected, increasing RHO-1 activity results in shorter dwell times (Figure 4, A and C). However, increasing RHO-1 activity did not phenocopy spv-1 mutant calcium signaling, showing slow rising times and low fractions over half max (Figure 4, A–C), and long sp-ut quiet periods and low bag intensities (Figure 4, D and E). In all calcium signaling metrics, increased RHO-1 activity did not differ significantly from the non–heat shocked controls. These results suggest that SPV-1 is likely not working through RHO-1 to regulate calcium signaling in the spermatheca.
FIGURE 4:
Increasing RHO-1 activity alters spermathecal contractility but does not recapitulate spv-1(ok1498) mutant calcium signaling. (A) Representative time series from embryo transits with metrics annotated. (B) Heat maps showing time series from multiple embryo transits. The time series in A corresponds to the first row of the heat map. (C) Quantification of time series metrics. (D) Representative kymograms with metrics annotated. Horizontal scale bars: 10 μm; vertical scale bars: 100 s. (E) Quantification of kymogram metrics. In C and E, error bars display SD, and p values were calculated using Welch’s ANOVA with Games-Howell multiple comparison: ns, p ≥ 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. WT and spv-1(ok1498) data are duplicated from Figures 1 and 3.
SPV-1 regulates calcium signaling via its GAP domain
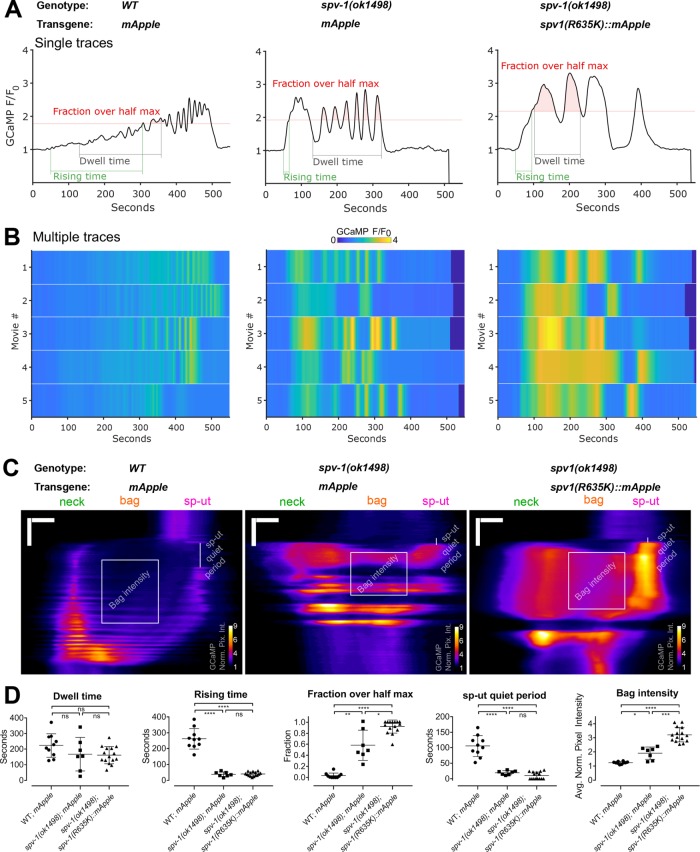
A functional RhoGAP domain is required for SPV-1 to regulate spermathecal contractility and embryo transit timing (Tan and Zaidel-Bar, 2015). To explore whether this activity is also needed for SPV-1 to regulate calcium signaling, we generated transgenic nematodes expressing labeled SPV-1 with a nonfunctional RhoGAP domain, SPV-1(R635K)::mApple, and acquired two-color movies of embryo transits. spv-1(R635K)::mApple was expressed at average mApple fluorescence intensities from 11 to 24, coinciding with mApple fluorescence levels at and above the threshold that induces trapping with RhoGAP functional SPV-1::mApple (Supplemental Figure 1). GCaMP time series, heat maps, and kymograms show that embryo transit calcium levels in spv-1(ok1498); spv-1(R635K)::mApple animals are higher than in the spv-1 mutant controls and well above levels in the wild-type controls (Figure 5, A–C). All of the spv-1(ok1498); spv-1(R635K)::mApple calcium signaling metrics are significantly different from the wild-type controls, and the fractions over half max and bag intensities are significantly increased over the spv-1 mutant controls (Figure 5D). These data suggest that the GAP activity of SPV-1 is required to modulate calcium signaling and indicate that the downstream target of SPV-1 is likely to be a GTPase.
FIGURE 5:
SPV-1 regulates calcium signaling through its GAP domain. (A) Representative time series from embryo transits with metrics annotated. (B) Heat maps showing time series from multiple embryo transits. Time series in A correspond to the first rows of the heat maps. (C) Representative kymograms with metrics annotated. Horizontal scale bars: 10 μm; vertical scale bars: 100 s. (D) Quantification of metrics. Error bars display SD, and p values were calculated using Welch’s ANOVA with Games-Howell multiple comparison: ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. WT;mApple and spv-1(ok1498); mApple data are duplicated from Figures 2 and 3.
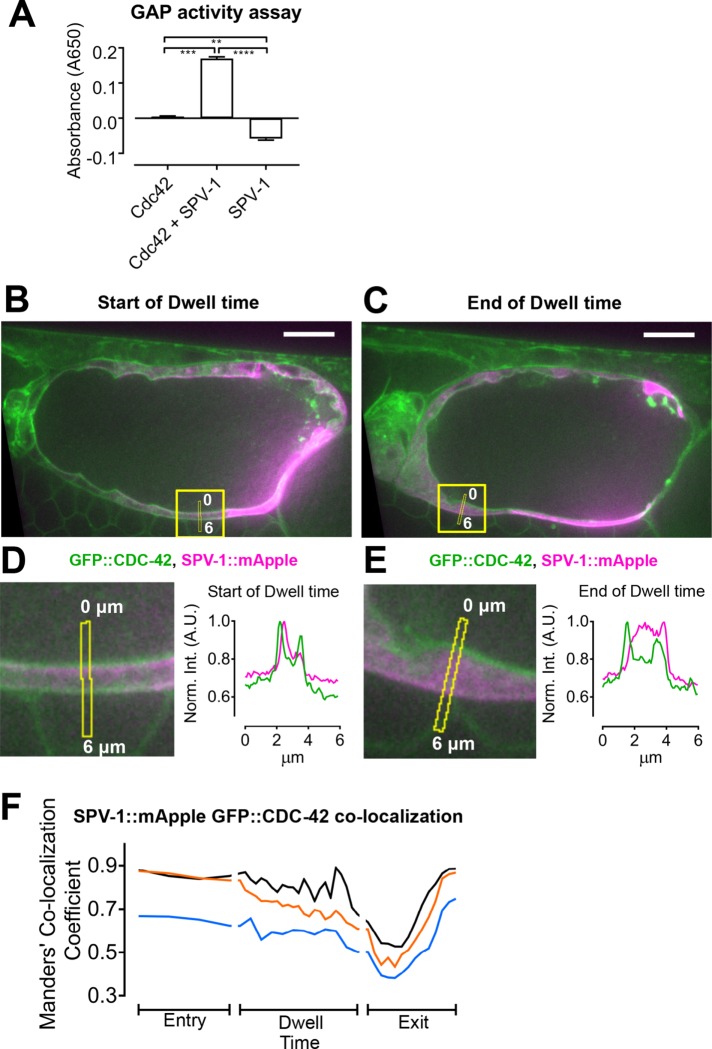
SPV-1 has GAP activity toward Cdc42 and partially colocalizes with CDC-42
In addition to regulating RHO-1, in vitro RhoGAP activity assays indicate SPV-1 has significant GAP activity toward another Rho family GTPase, Cdc42/CDC-42 (Figure 6A; Ouellette et al., 2016). To determine whether CDC-42 is present in the cells of the spermatheca, we obtained and imaged nematodes expressing a green fluorescent protein (GFP)::CDC-42 fusion (Neukomm et al., 2014) and found CDC-42 expressed throughout the spermatheca at the apical and basal membranes (Figure 6, B and C). To investigate the spatial relationship between CDC-42 and SPV-1, we crossed spv-1::mApple into the gfp::cdc-42 animals and acquired two-color movies of embryo transits. We found that SPV-1::mApple partially colocalizes with GFP::CDC-42 at the cell membranes (Figure 6, D and E). To quantify the colocalization of SPV-1::mApple and GFP::CDC-42 in the spermatheca, we calculated the Manders’ colocalization coefficient (Manders et al., 1993; Costes et al., 2004) for each frame of these embryo transit movies (Figure 6F). Manders’ coefficient gives the fraction of SPV-1::mApple pixel intensity in pixels that are also positive for GFP::CDC-42. This analysis shows high colocalization during oocyte entry, with ∼70–90% of the SPV-1::mApple pixel intensity found in GFP::CDC-42–positive pixels. Colocalization decreases during the dwell time and exit, rebounding to high colocalization by the end of embryo exit. These data suggest that SPV-1 may regulate CDC-42 in spermathecal cells.
FIGURE 6:
SPV-1 exhibits GAP activity toward Cdc42 and partially colocalizes with CDC-42 at spermathecal cell membranes. (A) In vitro assay measuring activity of the SPV-1 RhoGAP domain toward recombinant mammalian Cdc42. Error bars display SEM, and p values were calculated using Welch’s ANOVA with Games-Howell multiple comparison: **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. (B, C) The first and last frames of the dwell time from a representative movie, with the quantified region, capturing the same cell, annotated. Scale bars: 10 μm. (D, E) Left, digitally zoomed view of the quantified region, with the line scans annotated; right, average fluorescence intensity along the line scans, with GFP::CDC-42 in green and SPV-1::mApple in magenta. (F) Time series of Manders’ colocalization coefficients for embryo transit movies from three different animals. Manders’ coefficient gives the fraction of SPV-1::mApple pixel intensity in pixels that are also positive for GFP::CDC-42. The black trace is from the same movie displayed in B–E.
Increasing CDC-42 activity recapitulates many aspects of spv-1 mutant calcium signaling
To determine whether increasing CDC-42 activity alters calcium signaling similarly to the loss of SPV-1, we generated nematodes expressing a constitutively active CDC-42(Q61L) (Ziman et al., 1991; Aceto et al., 2006) under the control of a heat shock promoter, crossed this line with our GCaMP sensor line, optimized the heat shock protocol for CDC-42, and acquired embryo transit movies. Increasing CDC-42 activity does not significantly alter dwell times. Comparing animals expressing constitutively active CDC-42(Q61L) with non–heat shocked controls demonstrates that activation of CDC-42 results in faster rising times, increased fractions over half max, shorter sp-ut quiet periods, and higher bag intensities (Figure 7, A–E). Heat shock alone does not significantly alter calcium signaling metrics in control animals (Supplemental Figure 3).
FIGURE 7:
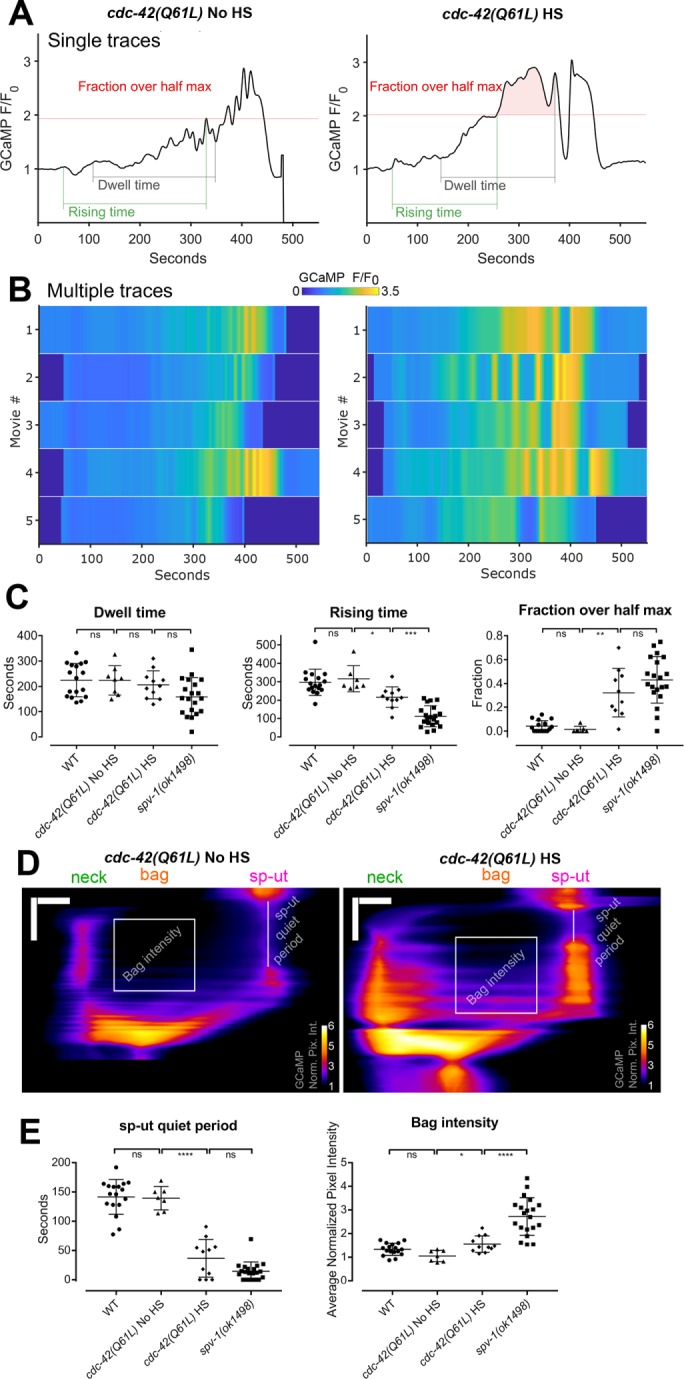
Increasing CDC-42 activity alters spermathecal calcium signaling. (A) Graphs are representative time series from embryo transits with metrics annotated. HS indicates induction of expression by heat shock. (B) Heat maps showing time series from multiple embryo transits. Time series in A corresponds to the first row of the heat map. (C) Quantification of time series metrics. (D) Representative kymograms with metrics annotated. Horizontal scale bars: 10 μm; vertical scale bars: 100 s. (E) Quantification of kymogram metrics. In C and E, error bars display SD, and p values were calculated using Welch’s ANOVA with Games-Howell multiple comparison: ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. WT and spv-1(ok1498) data are duplicated from Figures 1 and 3.
Because both Cdc42 and Rho are known regulators of the actin cytoskeleton (Sit and Manser, 2011), and because rearrangements of the actin cytoskeleton can influence the spatiotemporal regulation of the endoplasmic reticulum and thereby directly affect calcium levels (Wang et al., 2002), we visualized F-actin in dissected spermathecae from animals treated with heat shock conditions. No major alterations in actin cytoskeletal arrangements were observed in response to heat shock (Supplemental Figure 4), suggesting disruption of the actin cytoskeleton is unlikely to underlie the defects in calcium signaling seen in animals expressing constitutively active CDC-42. These data suggest SPV-1 may act through CDC-42 to regulate calcium signaling in the spermatheca during embryo transits, perhaps through effectors that do not directly regulate the actin cytoskeleton.
Decreasing CDC-42 using RNAi does not alter calcium signaling
To further explore CDC-42’s role in calcium signaling, we generated nematodes coexpressing a red calcium sensor, R-GECO, and CDC-42 labeled with GFP. We used RNAi against cdc-42 to deplete CDC-42 levels and recorded embryo transit movies with R-GECO to monitor calcium signaling. Decreasing CDC-42 levels via RNAi does not alter calcium time series in the spv-1 mutant background (Figure 8, A and B) or in the wild-type background (unpublished data). Dwell times and global calcium signaling metrics are unchanged by cdc-42(RNAi) in both wild-type and mutant backgrounds (Figure 8C). Spatiotemporal patterns of calcium signaling are also unchanged by cdc-42(RNAi), indicated by kymograms (Figure 8D) and spatiotemporal calcium signaling metrics (Figure 8E). Measurements of GFP::CDC-42 fluorescence intensity suggest that cdc-42(RNAi) treatments are significantly depleting CDC-42 levels (Figure 8F); however, residual levels of CDC-42 could be sufficient to support the observed calcium dynamics. These results suggest that SPV-1 may regulate calcium signaling through another as yet unidentified effector.
FIGURE 8:
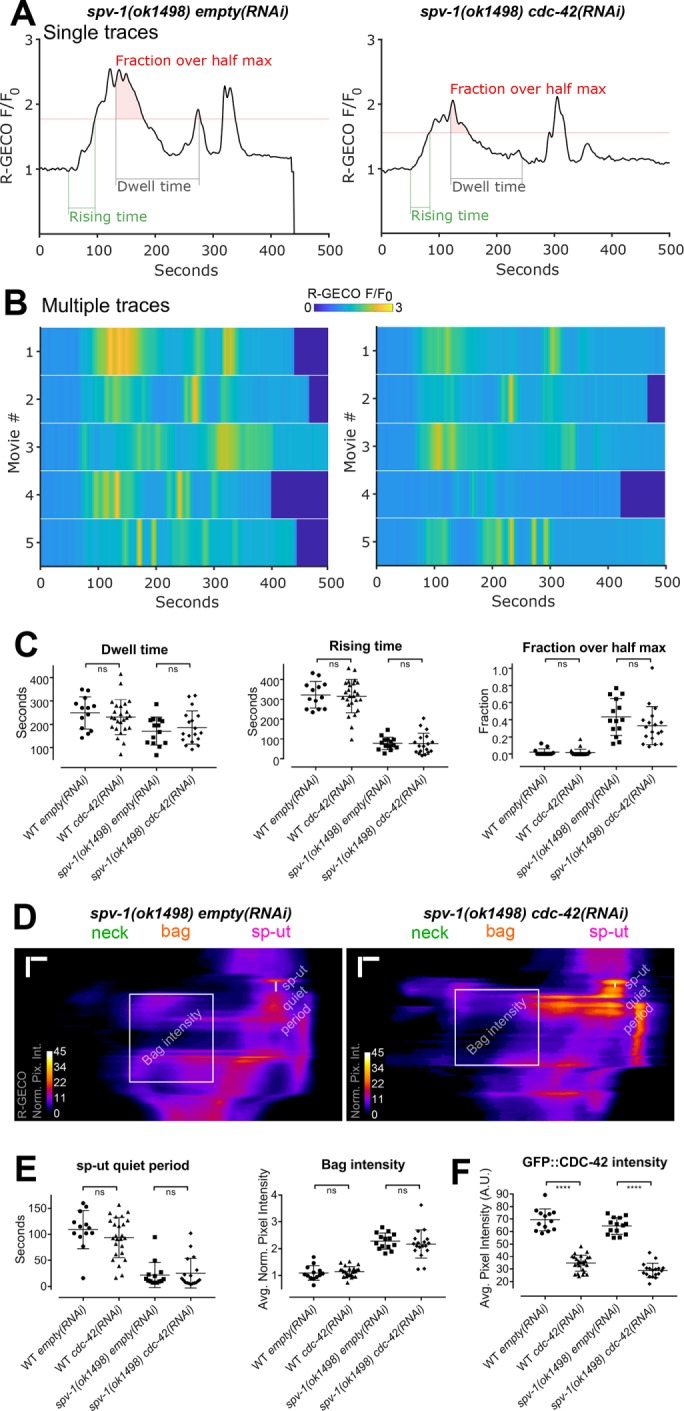
cdc-42(RNAi) does not alter calcium signaling. (A) Representative time series from embryo transits with metrics annotated. (B) Heat maps showing time series from multiple embryo transits. The time series in A corresponds to the first row of the heat map. (C) Quantification of time series metrics. (D) Representative kymograms with metrics annotated. Horizontal scale bars: 5 μm; vertical scale bars: 50 s. (E) Quantification of kymogram metrics. (F) Quantification of GFP::CDC-42 intensity. In C, E, and F, error bars display SD, and p values were calculated using Welch’s t test: ns, p ≥ 0.05; ****, p < 0.0001.
DISCUSSION
In this work, we show that the RhoGAP SPV-1 is a major regulator of calcium signaling in the C. elegans spermatheca during embryo transits. SPV-1 controls spatiotemporal aspects of calcium signaling by keeping calcium low in the spermathecal bag cells and sp-ut valve for a period of time after oocyte entry. This controlled calcium signaling likely contributes to the spatiotemporal patterns of contractility present in wild-type function of the tissue, where contractility must be high in the distal neck to keep the embryo in the spermatheca and yet must also stay low in the central bag cells so that the egg forms with the correct shape. Misregulated calcium signaling, in addition to increased active RHO-1 levels, likely contributes to the misshapen embryos and decreased brood sizes seen in the spv-1(ok1498) mutant (Tan and Zaidel-Bar, 2015).
Spermathecal tissue function and calcium signaling exhibit a threshold response to SPV-1::mApple, suggesting levels of SPV-1 protein must be maintained within a narrow range for wild-type embryo transits to occur. Increases in SPV-1 above wild-type levels lead to overdampening of the signaling pathways regulating contractility, resulting in a tissue that cannot produce productive contractions. Decreases in SPV-1 below wild-type levels lead to insufficient inhibition of the signaling pathways that stimulate contractility, making the tissue hypercontractile (Figure 9, A and B). A threshold response also suggests that feedback mechanisms are involved in controlling spermathecal tissue function and calcium signaling. The molecular mechanisms tightly regulating the expression and activity levels of SPV-1 and characterization of the feedback control system that governs the spermatheca are exciting areas for future investigation.
FIGURE 9:
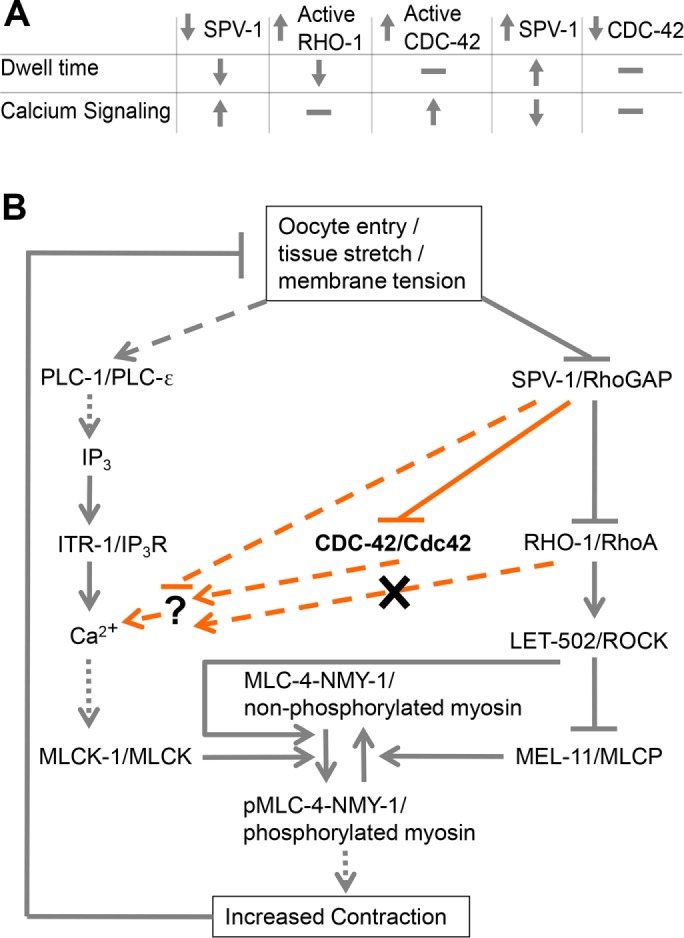
SPV-1 regulates spermathecal contractility via calcium and Rho-ROCK signaling. (A) Summary table of findings. SPV-1 regulates both the Rho-ROCK and calcium signaling pathways, which together are required for activation of myosin and tissue contractility. Increasing RHO-1 activity leads to faster transits (decreased dwell times) similar to decreased SPV-1, but does not alter calcium signaling. Increasing active CDC-42 does not alter dwell times but does alter calcium signaling similar to decreased SPV-1. Increasing SPV-1 increases dwell times, often resulting in transit failures and embryo trapping, and decreases calcium signaling activity and magnitude. Decreasing CDC-42 using RNAi does not alter dwell time or calcium signaling. (B) Proposed model of the network regulating actomyosin contractility in the spermatheca. Gray lines along the outside display previously known interactions. Orange lines display interactions investigated in this study. Solid lines indicate direct interactions, dotted lines indicate resultant interactions with known intermediates not shown, and dashed lines indicate unknown intermediates. SPV-1, CDC-42, and/or an unknown effector symbolized by the question mark could interact with any of the upstream members of the calcium signaling pathway; those arrows were omitted for clarity.
Given previous results suggesting SPV-1 acts through RHO-1 to regulate contractility, it was a surprise to find that constitutively active RHO-1 did not recapitulate the spv-1(ok1498) calcium signaling phenotypes. Although animals expressing constitutively active RHO-1 did exhibit shorter dwell times, suggesting sufficient activation of RHO-1 to stimulate actomyosin contractility, they produced fairly wild-type calcium signaling. RHO-1 is therefore probably working predominantly through LET-502/ROCK and myosin activation and not in the regulation of calcium signaling. Increased phosphorylation of myosin through the Rho-ROCK pathway would lead to calcium sensitization of myosin, a common phenomenon in smooth muscle and nonmuscle systems in which contraction is initiated at lower than normal calcium levels (Somlyo and Somlyo, 2003).
Because altering RHO-1 activity did not affect the calcium signal, we asked whether SPV-1 GAP activity was necessary to regulate calcium during embryo transits. Expression of SPV-1 with a nonfunctional GAP domain (spv-1(R635K)::mApple) resulted in high calcium levels, exceeding those observed in the spv-1 mutant. This suggests that a GTPase acts downstream of SPV-1 to modulate calcium signaling and that SPV-1(R635K) may be able to bind and sequester GTPases, preventing them from being turned off by other GAPs.
Several observations suggest the small GTPase CDC-42 may play a role in the regulation of calcium signaling in the spermatheca. Expressing constitutively active CDC-42 in the spermatheca recapitulates key aspects of the spv-1 mutant calcium signaling phenotypes. In particular, the spv-1 mutant-like increase in calcium signal evident in the increased fractions over half max and the reduced sp-ut quiet periods suggests increased CDC-42 activity may contribute to the stimulation of calcium release downstream of SPV-1. Although calcium levels increase in animals expressing constitutively active CDC-42, the dwell times are unchanged. Therefore, premature and elevated calcium signaling alone does not generate the hyperconstriction of the spermatheca and rapid embryo transits observed in spv-1(ok1498) animals. The hyperconstriction may therefore be driven primarily by RHO-1. We attempted to directly address the question of whether the spv-1 phenotype is the collective result of misregulation of both RHO-1 and CDC-42 by generating animals expressing both of the constitutively active constructs. Unfortunately, developmental and ovulation defects in these animals prevented us from collecting the transit data needed to answer this question.
Despite the phenotypic similarities between loss of spv-1 and increased CDC-42, the calcium intensities in the spermathecal bag and the rising times in animals expressing constitutively active CDC-42 are significantly different from those seen in the spv-1 mutant population. This may be because the levels of CDC-42 activity in a constitutively active mutant activated by heat shock differ from wild-type CDC-42 activity levels in intensity, duration, and/or spatial organization. The uneven recapitulation of the spv-1 mutant calcium signaling phenotype may also suggest that another unidentified protein acts downstream of SPV-1 to regulate calcium signaling.
If SPV-1 works through the inactivation of CDC-42 to regulate calcium release, depletion of CDC-42 might be expected to suppress the spv-1 mutant calcium dynamics. However, depleting CDC-42 using RNAi did not result in altered calcium signaling in the spv-1 mutant or wild-type backgrounds, despite evidence that the treatment is effective at depleting CDC-42. CDC-42 is important for larval development, including proper formation of the gonad (Norman et al., 2005), and depleting it completely results in morphogenetic defects that preclude ovulation. Owing to this, our RNAi treatments were started at later larval stages and were not complete knockdowns. Perhaps residual CDC-42 is sufficient to properly regulate calcium signaling. Another possibility is that SPV-1 regulates calcium through another unknown effector. Our results demonstrate the GAP activity of SPV-1 is required for proper spatiotemporal regulation of calcium signaling, suggesting that a GTPase is a target. Of the known Rho GTPases in C. elegans (CED-10, RAC-2, MIG-2, CDC-42, RHO-1, and CRP-1), SPV-1 has been shown to only have significant GAP activity toward CDC-42 and RHO-1 (Ouellette et al., 2016). Identification of additional SPV-1 effectors is an important avenue for future studies.
In other systems, CDC-42/Cdc42 is known to regulate actin cytoskeletal rearrangements and cell migration as well as protein kinase cascades that can alter transcription of downstream targets (Johnson, 1999; Takai et al., 2001). However, few studies link Cdc42 to the activation of calcium signaling. Perhaps the best-characterized example is found in mast cells, a critical cell in the immune system during inflammatory reactions. Calcium signaling, triggered by antigen binding to immunoglobulin E and FCεRI receptors, controls mast cell exocytosis and degranulation, which releases histamines and other substances. In RBL-2H3 mast cells, dominant active mutants of Cdc42 lead to elevated levels of antigen-stimulated IP3 and cytosolic calcium (Hong-Geller and Cerione, 2000). Further studies have shown that Cdc42 can participate in the regulation of stimulated PIP2 synthesis, the substrate of PLC isozymes, in these cells (Wilkes et al., 2014). This suggests CDC-42 activity in the spermatheca may be controlling calcium activity through the regulation of PIP2 turnover and/or the production of IP3. In addition, in vitro studies have shown that human Cdc42 can stimulate purified PLC-β (Illenberger et al., 1998) and interact with PLC-γ1 (Hong-Geller and Cerione, 2000). It is possible that CDC-42 is affecting the activation of PLC-1/ε or the availability of substrate in the spermathecal cells to control IP3 production and calcium release during embryo transits.
SPV-1 has three human orthologues (Tan and Zaidel-Bar, 2015): ARHGAP29/PARG1 (Saras et al., 1997; Myagmar et al., 2005), HMHA1/ARHGAP45 (de Kreuk et al., 2013; Amado-Azevedo et al., 2018), and GMIP/ARHGAP46 (Aresta et al., 2002). Recently, loss of HMHA1/ARHGAP45 in endothelial cells was found to increase wound healing and cell migration rates, suggesting altered actomyosin contractility, and the cellular response to shear stress, suggesting altered mechanotransduction (Amado-Azevedo et al., 2018). Similar patterns of altered mechanotransduction and contractility are present in the spermatheca when SPV-1 is lost. In mice, ARHGAP29/PARG1 and a binding partner Rasip1 were found to regulate nonmuscle myosin II via RhoA and Cdc42 during blood vessel development, with Rasip1 first activating Cdc42 to break cell–cell adhesions during lumen formation and Rasip1 then activating ARHGAP29/PARG1, inhibiting RhoA, and keeping contractility low so the lumen stays open and the blood vessel can expand as the animal grows (Barry et al., 2016). This behavior of ARHGAP29/PARG1, regulating cellular contractility directly through RhoA and also through Cdc42 in conjunction with a binding partner, further suggests other proteins may interact with SPV-1 to modulate calcium signaling in the spermatheca.
Our results indicate SPV-1 regulates calcium signaling in addition to Rho-ROCK signaling, providing insights into the regulation of calcium signaling, actomyosin contractility, and tissue function in the C. elegans spermatheca. From this evidence, it appears that not only is there interaction between the two central pathways that regulate actomyosin contractility, but a single protein can fine-tune both pathways to generate consistent and robust tissue function. Given conservations of sequence and function, it will be exciting to determine whether SPV-1 orthologues play similar roles in other biological contexts.
MATERIALS AND METHODS
Caenorhabditis elegans strains and culture
Nematodes were grown on nematode growth media (NGM) (0.107 M NaCl, 0.25% wt/vol Peptone [Fisher], 1.7% wt/vol BD Bacto-Agar [Fisher], 0.5% Nystatin [Sigma], 0.1 mM CaCl2, 0.1 mM MgSO4, 0.5% wt/vol cholesterol, 2.5 mM KPO4) and seeded with Escherichia coli OP50 using standard techniques (Myers et al., 1996). Nematodes were cultured at 20°C unless specified otherwise. All lines expressing GCaMP3 were crossed into UN1108 as described in Kovacevic et al. (2013) or UN1417, a separate integration event of the same construct, fln-1p::GCaMP3. GCaMP signal in UN1108 and UN1417 does not differ significantly in any of the described metrics. spv-1(ok1498) animals were crossed with UN1417 animals to generate the line UN1416 (spv-1(ok1498); fln-1p::GCaMP3). Table 1 provides a full list of the C. elegans strains used in this study.
TABLE 1:
Caenorhabditis elegans strains used in this study.
| Strain | Genotype |
|---|---|
| N2 | Wild-type Bristol |
| RZB300 | cdc-42(gk388) spv-1(ok1498) opIs295 II; xbEx1406[spv-1p::spv-1::mApple+pRF4(rol-6(su1006))] |
| UN1108 | xbIs1101[fln-1p::GCaMP3+pRF4(rol-6(su1006))] II |
| UN1416 | spv-1(ok1498) II; xbIs1408[fln-1p::GCaMP3] |
| UN1417 | xbIs1408[fln-1p::GCaMP3] |
| UN1516 | xbIs1408[fln-1p::GCaMP3]; nzIs1[hsp-16.2p::rho-1(G14V)] |
| UN1649 | spv-1(ok1498) II; xbIs1408[fln-1p::GCaMP3]; xbEx1649[spv-1p::spv-1(R635K)::mApple] |
| UN1661 | spv-1(ok1498) II; xbIs1408[fln-1p::GCaMP3]; xbEx1661[spv-1p::spv-1::mApple] |
| UN1662 | spv-1(ok1498) II; xbIs1408[fln-1p::GCaMP3]; xbEx1662[spv-1p::spv-1::mApple] |
| UN1663 | xbIs1408[fln-1p::GCaMP3]; xbEx1661[spv-1p::spv-1::mApple] |
| UN1664 | xbIs1408[fln-1p::GCaMP3]; xbEx1662[spv-1p::spv-1::mApple] |
| UN1665 | spv-1(ok1498) II; xbIs1408[fln-1p::GCaMP3]; xbEx1665[spv-1p::spv-1::mApple] |
| UN1728 | xbIs1101[fln-1p::GCaMP3+pRF4(rol-6(su1006))] II; xbEx1720[hsp-16.2p::cdc-42(Q61L)+myo-3p::mCherry] |
| UN1771 | xbIs1408[fln-1p::GCaMP3]; xbEx1771[fkh-6p::mApple] |
| UN1772 | spv-1(ok1498) II; xbIs1408[fln-1p::GCaMP3]; xbEx1771[fkh-6p::mApple] |
| UN18124 | cdc-42(gk388) opIs295 II; xbEx18124[fln-1p::R-GECO+pRF4(rol-6(su1006))] |
| UN18127 | cdc-42(gk388) spv-1(ok1498) opIs295 II; xbEx18124[fln-1p::R-GECO+pRF4(rol-6(su1006))] |
Construction of spv-1::mApple and SPV-1(R635K)::mApple transgenic animals.
GFP and the spv-1 3′UTR were removed from plasmid pPY1 (spv-1p::spv-1::GFP::spv-1 3′UTR; Tan and Zaidel-Bar, 2015) and replaced with tdTomato and the fln-1 3′UTR from pUN284 (fln-1p::inx-12::tdTomato::fln-1 3′UTR) using restriction digest cloning with the enzymes KpnI and EagI. tdTomato was removed from this plasmid using the enzymes KpnI and NotI, and replaced with mApple amplified from Addgene plasmid 27698 using primers that introduced KpnI and NotI restriction sites to the ends of mApple, generating pUN362. spv-1p::spv-1::mApple::fln-1 3′UTR was then transferred from pUN362 into pUN359, a plasmid modified from pCFJ151 (Frøkjær-Jensen et al., 2008) to facilitate CRISPR insertion at the ttTi5605 transposon site, using restriction digest cloning with enzymes PmeI and SpeI, resulting in pUN427. pUN595 (spv-1p::SPV-1(R635K)::mApple::fln-1 3′UTR) was generated using around-the-horn PCR site-directed mutagenesis on pUN427 with primers designed to introduce the two-nucleotide R635K amino acid mutation and a single-nucleotide silent mutation 30 base pairs downstream to generate a BglII restriction site to facilitate tracking the R635K mutation. Transgenic animals were created by microinjecting N2 nematodes with a DNA solution containing 50 ng/µl of Cas9 plasmid (Chen et al., 2008; Tzur et al., 2013); 30 ng/µl each of two different CRISPR guide plasmids, pUN357 and pUN358; and 45 ng/µl of pUN427 or pUN595. Progeny displaying red fluorescence in the spermatheca were isolated and screened for CRISPR integration. After multiple failed attempts at CRISPR integration, lines expressing the transgenes as extrachromosomal arrays were established and used for experiments. Three spv-1::mApple lines and two SPV-1(R635K)::mApple lines were isolated from independent microinjections. These lines were crossed with UN1416 nematodes to generate animals with dual-labeled spermathecae in the wild-type and spv-1 mutant backgrounds.
Construction of fkh-6p::mApple transgenic animals.
GFP was removed from pUN106 (fkh-6p::GFP::unc-54 3′UTR) and replaced with mApple from pUN427 using restriction digest cloning with enzymes KpnI and EcoRI to create pUN822. Transgenic animals were created by microinjecting N2 nematodes with a DNA solution containing 45 ng/µl of pUN822 and 100 ng/µl of pRF4 (rol-6 injection marker). Progeny displaying red fluorescence in the spermatheca were isolated, and lines expressing the transgene as an extrachromosomal array were established. These lines were crossed with UN1416 nematodes to generate animals with dual-labeled spermathecae in the wild-type and spv-1 mutant backgrounds.
Construction of constitutively active CDC-42 (hsp16.2p::CDC-42(Q61L)::unc-54 3′UTR) animals.
CDC-42(Q61L) was amplified from pJK6 (a gift from Ahna Skop [University of Wisconsin, Madison], made originally by John White [University of Wisconsin, Madison]) using primers engineered with EcoRI 5′ extensions and ligated into pUN597 (hsp16.2::unc-54 3′UTR) between the promoter and 3′UTR to create pUN624. Transgenic animals were created by microinjecting a DNA solution containing 20 ng/μl pUN624 and 25 ng/μl myo-3:mCherry as a co-injection marker into N2 animals. Animals exhibiting mCherry expression in the body wall muscle were segregated to establish the transgenic line UN1720. UN1720 was crossed into UN1108 (fln-1p::GCaMP3), creating the transgenic line UN1728 for calcium studies.
Construction of gfp::cdc-42; spv-1(ok1498); spv-1::mApple animals.
The strain WS5018 (cdc-42(gk388);opIs295 II (gfp::cdc-42)) (Neukomm et al., 2014) was obtained from the Caenorhabditis Genetics Center and crossed with the spv-1(ok1498); spv-1::mApple line generated previously, creating strain RZB300.
Construction of gfp::cdc-42; R-GECO animals.
WS5018 animals were injected with a DNA solution containing 50 ng/μl of pUN526 (fln-1p::R-GECO::fln-1 3′UTR) and 40 ng/μl of pRF4 (rol-6 injection marker). Progeny displaying red fluorescence in the spermatheca were isolated, and a line, UN18124, expressing the transgene as an extrachromosomal array, was established. RZB300 animals lacking red fluorescence were crossed with UN18124 animals to generate the strain UN18127 with gfp::cdc-42 and R-GECO in the spv-1(ok1498) background.
Heat shock protocols for constitutively active rho-1(G14V) and cdc-42(Q61L)
UN1516 (RHO-1(G14V); fln-1p::GCaMP3) animals were synchronized using an “egg prep” in which embryos were released from young gravid adults using an alkaline hypochlorite solution (Hope, 1999). Clean embryos were plated on OP50-seeded NGM and cultured at 20°C until young adulthood (70–72 h post–egg prep). For induction of expression of constitutively active RHO-1(G14V), animals were moved from 20°C to 33°C for 30 min, and then left to recover at 20°C for 1 h before imaging.
UN1728 (CDC-42(Q61L); fln-1p::GCaMP3) animals were synchronized as described earlier. Young adults expressing myo-3::mCherry were segregated and placed at 33°C for 2 h to induce the expression of constitutively active CDC-42(Q61L). Animals were left to recover at 20°C for 1 h before imaging.
RNAi
The RNAi protocol was performed essentially as described in Timmons and Fire (1998). HT115(DE3) bacteria transformed with the double-stranded RNA (dsRNA) construct of interest were grown overnight in Luria broth supplemented with 40 mg/ml ampicillin. The following day, 300 μl of the cultured bacteria was seeded on NGM/RNAi plates supplemented with 25 μg/ml carbenicillin and disopropylthio-β-galactoside and left for 24–72 h to induce dsRNA expression. empty RNAi, that is, HT115(DE3) bacteria transformed with L4440 without a gene-specific insert, is used as a control in RNAi experiments to account for the different bacterial food source as well as nonspecific activation of the RNAi machinery. The diluted spv-1 RNAi experiment was conducted by mixing overnight cultures of spv-1 and empty RNAi bacteria in volume ratios of 1:1, 1:3, 1:9, or 1:19 for seeding NGM/RNAi plates. Embryos were collected using an egg prep, as described earlier. Clean embryos were transferred to the seeded NGM/RNAi plates and incubated at 20°C for 65–70 h before imaging. For cdc-42 RNAi clean embryos were deposited on OP50 NGM plates and incubated at 20°C for 36–48 h; this was followed by transfer to cdc-42 or empty RNAi plates and incubation at 20°C for another 24–36 h before imaging.
Image acquisition
For all GCaMP and R-GECO studies, animals were immobilized with 0.05-micron polystyrene beads (Polysciences, Warrington, PA), mounted on slides with 5–10% agarose pads (Kim et al., 2013) and imaged using a 60×/1.40 NA oil-immersion objective on a Nikon Eclipse 80i microscope equipped with a SPOT RT3 CCD camera (Diagnostic Instruments, Sterling Heights, MI) controlled by SPOT Advanced imaging software (v. 5.0) with Peripheral Devices and Quantitative Imaging modules. Images were acquired at 1600 × 1200 pixels, using the full camera chip, and saved as 8-bit TIFF files. Fluorescence excitation was provided by a Nikon Intensilight C-HGFI 130-W mercury lamp and shuttered with a Lambda 10-B SmartShutter (Sutter Instruments, Novato, CA), also controlled through the SPOT software. Single-channel GCaMP time-lapse movies were acquired using a GFP filter set (470/40× 495lpxr 525/50m) (Chroma Technologies, Bellows Falls VT) at 1 frame per second, with an exposure time of 75 ms, gain of 8, and neutral density of 16. Two-channel GCaMP/mApple and GFP/R-GECO time-lapse movies were acquired using an EGFP/mCherry filter set, with excitation filters 470/40× and 572/35× housed in the SmartShutter filter wheel to enable rapid switching, and emissions passing through a single filter cube containing beamsplitter 59022bs and emission filter 59022m (520/20m and 640/40m) (Chroma Technologies). For GCaMP/mApple movies, one GCaMP frame and one mApple frame were acquired every 2 s, with exposure time of 40 ms and gain of 16 for the GCaMP channel, exposure time of 75 ms and gain of 32 for the mApple channel, and neutral density of 32 for both channels. For GFP/R-GECO movies, one GFP frame and one R-GECO frame were acquired every 2 s, with exposure time of 50 ms, gain of 16, and neutral density of 16 for both channels.
For GFP::CDC-42/SPV-1::mApple studies, animals were mounted with 2 µl of M9 buffer on slides with 10% agarose pads and imaged using a 100×/1.4 NA oil-immersion objective on a Nikon Ti confocal microscope equipped with a spinning-disk head (CSU-X1; Yokogawa, Tokyo, Japan) and Prime95b camera (Photometrics, Tucson, AZ) controlled using Metamorph software (Molecular Devices, Sunnyvale, CA), and saved as 16-bit TIFF files. Z-stacks were acquired with 1-µm spacing between slices, with Z-stacks acquired every 10 s. GFP was excited using the 481-nm laser, and mApple was excited with the 561-nm laser, both at 35% laser power and 200-ms exposure time.
Image processing
Only successful embryo transits, meaning the embryo exited through the sp-ut valve, or whole embryos trapped in the spermatheca were analyzed in this work. GCaMP and R-GECO movies were processed using a custom Fiji script. First, the movie was registered using the Fiji plug-in StackReg (Thévenaz et al., 1998) to correct for any movement of the animal during image acquisition. The movie was then rotated to a standard orientation with the occupied spermatheca horizontal and the sp-ut valve on the right side of the movie. Finally, the movie was cropped to 800 × 400 pixels, annotated as processed, and saved as an 8-bit TIFF file. For GCaMP/mApple and GFP/R-GECO movies, the two channels were registered separately and then recombined before rotation and cropping. The angle of rotation and positioning of the cropping box were determined by manual user input.
GFP::CDC-42/SPV-1::mApple movies were not processed beyond selecting a single z-plane for analysis. Fiji was used to manually track and annotate the cells and to manually draw and quantify line scans.
Generation of GCaMP and R-GECO time series
GCaMP and R-GECO time series were generated by calculating the average pixel intensity for each frame of the processed movie. Normalized average pixel intensity time series were generated by normalizing the entire time series to the baseline value, F0, calculated as the average of the 30 frames before the start of oocyte entry (Kovacevic et al., 2013).
Extraction of metrics from time series
Custom Fiji and Matlab code was used to document and archive manually annotated time points, computationally identified metrics, and additional data for each movie. The global calcium signaling metrics rising time and fraction over half max were adapted from Christo et al. (2015).
Manual annotation of time points and calculation of dwell time.
Time points were determined by visual inspection. Four significant time points were annotated for each movie: 1) distal neck open, when the oocyte starts to enter the spermatheca; 2) distal neck close, when the embryo is completely enclosed; 3) sp-ut valve open, when the embryo starts to exit; and 4) sp-ut valve close, when the embryo completely exits the spermatheca. In ambiguous cases, three individuals scored each time point, and the average of the three values was used. Dwell time is calculated as the frame when the sp-ut valve opens minus the frame when the distal neck closes.
Calculation of max, half max, and rising time.
Each GCaMP and R-GECO time series was smoothed using a moving average filter, Matlab function filter, with a window size of 5. The max value was determined using the built-in Matlab function, and the baseline value, previously used to normalize the time series, was recalled. The half-max value was calculated by taking half the difference between the max and the baseline, and adding the baseline again. The first time point where the time series was above the half-max value was recorded as the end of the rising time, with distal neck open as the start of the rising time. Rising time was calculated as first time point above half max minus distal neck open.
Calculation of fraction over half max.
For the dwell time of each time series, the time points above the half-max value were counted, and this number was divided by the total dwell time to give the fraction over half max.
Calculation of mApple and GFP::CDC-42 average pixel intensity.
For GCaMP/mApple and GFP::CDC-42/R-GECO movies, mApple and GFP::CDC-42 time series were generated by calculating the average pixel intensity of the mApple or GFP channel for each frame of the processed movie. The average of the 30 frames before oocyte entry, analogous to the GCaMP/R-GECO baseline, was recorded as the value for the mApple or GFP::CDC-42 average pixel intensity. For presentation in Figure 2 the mApple time series was smoothed using a moving average filter with a window size of 50, and a constant 2 was added to every value of the time series to accommodate the decreased intensity of the higher averaging. GCaMP/mApple movies were acquired in three batches over 15 mo, with a microscope deep cleaning and fluorescence lamp replacement between batches. SPV-1(R635K)::mApple average pixel intensity values were found to be consistent within batches, so the average SPV-1(R635K)::mApple average pixel intensity value for each batch was used to establish ratios to bring all the mApple values from the first two batches into agreement with the third batch.
Adjustment of metrics for trapped movies.
Trapped dwell times were assigned a value of 575, slightly above the highest value in the data set, for plotting purposes and fitting of the Hill function. These trapped dwell times were excluded from statistical analysis. Trapped fractions over half max were assigned a value of −0.10 for plotting purposes and excluded from statistical analysis.
Fit to Hill function and determination of SPV-1::mApple threshold.
Using GraphPad Prism 7.04, dwell times were arranged in an x,y table with mApple values as x and dwell times as y, with all spv-1(ok1498); spv-1::mApple movies (i.e., trapped, untreated, and RNAi) in a single column. Nonlinear regression curve fit was used, with [agonist] versus response − variable slope (four parameters). The EC50 in the resulting table is presented here as the threshold.
Generation of kymograms
For every frame of the processed movie, the pixel intensities of each column were averaged to generate a single pixel representing that column. This operation was performed for each column of the frame, condensing the frame into a single pixel line. This operation was carried out for every frame of the movie, with the single pixel lines stacking to generate a two-dimensional image that represents the spatiotemporal dynamics of the entire movie (Figure 3, A and B, and Supplemental Movie 2). This was implemented in custom Fiji code using the commands Image>Stacks>Reslice followed by Image>Stacks>Z Project (Average Intensity).
Extraction of spatiotemporal calcium signaling metrics from kymograms
The sp-ut quiet period was visually determined and indicated as a line drawn from the end of the first calcium transient to the start of the next rise in calcium level. The sp-ut quiet period was then calculated as the vertical distance between the endpoints of the line, measured in seconds.
Before calculation of the bag intensity, the kymogram was normalized by dividing the 32-bit kymogram image by the GCaMP or R-GECO time-series baseline value, calculated earlier as the average of the 30 frames before the start of oocyte entry. A left bound at the distal neck and a right bound at the sp-ut valve were then identified. The bag intensity was calculated within a rectangle computationally selected to represent the lowest average calcium signal in a 25-μm region of the spermathecal bag during the dwell time. For trapped SPV-1::mApple kymograms, the average dwell time of the WT;mApple controls was used.
In vitro RhoGAP activity assay
The RhoGAP activity assay was conducted as previously described (Tan and Zaidel-Bar, 2015).
Calculation of Manders’ colocalization coefficient
Manders’ colocalization coefficients (Manders et al., 1993) were calculated using the Costes algorithm (Costes et al., 2004), implemented in the built-in Fiji function Colocalization Threshold. Custom Fiji code was written to allow the Colocalization Threshold function to analyze movies frame by frame. Background subtraction was applied to the GFP::CDC-42 SPV-1::mApple movies before running the Colocalization Threshold function, using the built-in Fiji function Subtract Background. Values for Manders’ coefficients were found to vary with the degree of background subtraction. For determination of the proper level of background subtraction in an unbiased manner, custom Fiji code was written to vary the rolling ball radius from 10 to 500 pixels, at 10 pixel increments. Manders’ coefficients and the R below threshold value from the Costes algorithm were calculated and collected for each frame of the movie at each background-subtraction level. The R below threshold values for each frame of the movie were summed, resulting in a single R below threshold value for the entire movie for each background subtraction level. Given that the Costes algorithm ideally returns an R below threshold value of 0, the background subtraction level resulting in the movie R below threshold value closest to 0 was used as the proper level of background subtraction. The thresholded Manders’ coefficient, giving the amount of mApple intensity in pixels above the mApple intensity threshold that are also above the GFP intensity threshold, divided by the total mApple intensity in pixels above the mApple intensity threshold, is reported here.
For facilitating comparison, the time series of Manders’ coefficients over the embryo transit movies were segmented into entry time, dwell time, and exit time through the identification of time points as noted earlier. The segments for each movie were then normalized to the average entry time, dwell time, or exit time for all GFP::CDC-42 SPV-1::mApple movies, using the Matlab function interp1 to resample each segment to the same length.
Statistical analysis
Statistical tests were conducted at a significance level of 0.05. When two groups were compared, p values were calculated using Welch’s t test in GraphPad Prism. When more than two groups were compared, p values were calculated using Welch’s analysis of variance (ANOVA) with Games-Howell multiple comparisons in R (R Core Team, 2017), using the oneway function in the package userfriendlyscience (Peters, 2017).
Supplementary Material
Acknowledgments
We thank Ismar Kovacevic, Pei Yi Tan, Anand Asthagiri, Javier Apfeld, and members of the Cram and Apfeld labs for helpful feedback and discussions. This work was supported by a grant from the National Institutes of Health, National Institute of General Medical Sciences (GM110268) to E.J.C., a grant from the Israel Science Foundation (grant no. 1293/17) to R.Z.-B., a National Science Foundation/Molecular and Cellular Biosciences–U.S. Israel Binational Science Foundation award (1816640) to E.J.C. and R.Z.-B., and a National Science Foundation East Asia and Pacific Summer Institutes fellowship (1414889) to J.B. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
Abbreviations used:
- ANOVA
analysis of variance
- dsRNA
double-stranded RNA
- GFP
green fluorescent protein
- IP 3
1,4,5 triphosphate
- LET
lethal
- NGM
nematode growth media
- PIP 2
phospatidyl inositol bisphosphate
- PLC-1
phospholipase C
- RhoGAP
GTPase-activating protein toward Rho family small GTPases
- RNAi
RNA interference
- ROCK
Rho-associated protein kinase
- sp-ut
spermatheca uterine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0633) on February 6, 2019.
REFERENCES
- Aceto D, Beers M, Kemphues KJ. (2006). Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev Biol , 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado-Azevedo J, Reinhard NR, van Bezu J, van Nieuw Amerongen GP, van Hinsbergh VWM, Hordijk PL. (2018). The minor histocompatibility antigen 1 (HMHA1)/ArhGAP45 is a RacGAP and a novel regulator of endothelial integrity. Vascul Pharmacol , 38–47. [DOI] [PubMed] [Google Scholar]
- Aresta S, de Tand-Heim M-F, Béranger F, de Gunzburg J. (2002). A novel Rho GTPase-activating-protein interacts with Gem, a member of the Ras superfamily of GTPases. Biochem J , 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry DM, Koo Y, Norden PR, Wylie LA, Xu K, Wichaidit C, Azizoglu DB, Zheng Y, Cobb MH, Davis GE, et al (2016). Rasip1-mediated Rho GTPase signaling regulates blood vessel tubulogenesis via non-muscle myosin II. Circ Res , 810–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozovich FV, Nicholson CJ, Degen CV, Gao YZ, Aggarwal M, Morgan KG. (2016). Mechanisms of vascular smooth muscle contraction and the basis for pharmacologic treatment of smooth muscle disorders. Pharmacol Rev Pharmacol Rev , 476–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui YK, Sternberg PW. (2002). Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol Biol Cell , 1641–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Vega VB, Ng H-H. (2008). Transcriptional regulatory networks in embryonic stem cells. Cold Spring Harb Symp Quant Biol , 203–209. [DOI] [PubMed] [Google Scholar]
- Christo SN, Diener KR, Hayball JD. (2015). The functional contribution of calcium ion flux heterogeneity in T cells. Immunol Cell Biol , 694–704. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW. (1998). Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell , 523–533. [DOI] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. (2004). Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J , 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kreuk B-J, Schaefer A, Anthony EC, Tol S, Fernandez-Borja M, Geerts D, Pool J, Hambach L, Goulmy E, Hordijk PL. (2013). The human minor histocompatibility antigen1 is a RhoGAP. PLoS One , e73962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Hopkins CE, Newman B, Thummel JM, Olesen S, Grunnet M, Jorgensen EM. (2008). Single copy insertion of transgenes in C. elegans. , 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunst SJ, Tang DD, Opazo Saez A. (2003). Cytoskeletal remodeling of the airway smooth muscle cell: a mechanism for adaptation to mechanical forces in the lung. Respir Physiol Neurobiol , 151–168. [DOI] [PubMed] [Google Scholar]
- Hill AV. (1910). The possible effects of the aggregation of the molecules of hæmoglobin on its dissociation curves. J Physiol , iv–vii. [Google Scholar]
- Hong-Geller E, Cerione RA. (2000). Cdc42 and Rac stimulate exocytosis of secretory granules by activating the IP 3/calcium pathway in RBL-2H3 mast cells. J Cell Biol , 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. (1999). C. elegans: A Practical Approach, New York: Oxford University Press. [Google Scholar]
- Illenberger D, Schwald F, Pimmer D, Binder W, Maier G, Dietrich A, Gierschik P. (1998). Stimulation of phospholipase C-β 2 by the Rho GTPases Cdc42Hs and Rac1. EMBO J , 6241–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI. (1999). Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev , 54–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya K-I, Bui YK, Gao X, Sternberg PW, Kataoka T. (2004). Phospholipase C epsilon regulates ovulation in Caenorhabditis elegans. Dev Biol , 201–210. [DOI] [PubMed] [Google Scholar]
- Kelley CA, Wirshing ACE, Zaidel-Bar R, Cram EJ. (2018). The myosin light-chain kinase MLCK-1 relocalizes during Caenorhabditis elegans ovulation to promote actomyosin bundle assembly and drive contraction. Mol Biol Cell , 1975–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sun L, Gabel CV, Fang-Yen C. (2013). Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One , 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. (1979). The postembryonic cell lineages of the hermaphrodite and male gonad in Caenorhabditis elegans. Dev Biol , 396–417. [DOI] [PubMed] [Google Scholar]
- Kovacevic I, Cram EJ. (2010). FLN-1/filamin is required for maintenance of actin and exit of fertilized oocytes from the spermatheca in C. elegans. Dev Biol , 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic I, Orozco JM, Cram EJ. (2013). Filamin and phospholipase C-ε are required for calcium signaling in the Caenorhabditis elegans spermatheca. PLoS Genet , e1003510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie TL, Dowell ML, Lakser OJ, Gerthoffer WT, Fredberg JJ, Seow CY, Mitchell RW, Solway J. (2009). Disrupting actin-myosin-actin connectivity in airway smooth muscle as a treatment for asthma? Proc Am Thorac Soc , 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ, Aten JA. (1993). Measurement of co-localization of objects in dual-colour confocal images. J Microsc , 375–382. [DOI] [PubMed] [Google Scholar]
- McMullan R, Nurrish SJ. (2011). The RHO-1 RhoGTPase modulates fertility and multiple behaviors in adult C. elegans. PLoS One , e17265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux J. (1965). On the nature of allosteric transitions: a plausible model. J Mol Biol , 88–118. [DOI] [PubMed] [Google Scholar]
- Munjal A, Lecuit T. (2014). Actomyosin networks and tissue morphogenesis. Development , 1789–1793. [DOI] [PubMed] [Google Scholar]
- Myagmar B-E, Umikawa M, Asato T, Taira K, Oshiro M, Hino A, Takei K, Uezato H, Kariya K. (2005). PARG1, a protein-tyrosine phosphatase-associated RhoGAP, as a putative Rap2 effector. Biochem Biophys Res Commun , 1046–1052. [DOI] [PubMed] [Google Scholar]
- Myers CD, Goh P-Y, Allen TS, Bucher EA, Bogaert T. (1996). Developmental genetic analysis of troponin T mutations in striated and non-striated muscle cells of Caenorhabditis elegans. J Cell Biol , 1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm LJ, Zeng S, Frei AP, Huegli PA, Hengartner MO. (2014). Small GTPase CDC-42 promotes apoptotic cell corpse clearance in response to PAT-2 and CED-1 in C. elegans. Cell Death Differ , 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman KR, Fazzio RT, Mellem JE, Espelt MV, Strange K, Beckerle MC, Maricq AV. (2005). The Rho/Rac-family guanine nucleotide exchange factor VAV-1 regulates rhythmic behaviors in C. elegans. Cell , 119–132. [DOI] [PubMed] [Google Scholar]
- Ouellette MH, Martin E, Lacoste-Caron G, Hamiche K, Jenna S. (2016). Spatial control of active CDC-42 during collective migration of hypodermal cells in Caenorhabditis elegans. J Mol Cell Biol , 313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaia G, Renda T, Gallelli L, Vatrella A, Busceti MT, Agati S, Caputi M, Cazzola M, Maselli R, Marsico SA. (2008). Molecular mechanisms underlying airway smooth muscle contraction and proliferation: implications for asthma. Respir Med , 1173–1181. [DOI] [PubMed] [Google Scholar]
- Peters G-JY. (2017). Userfriendlyscience: quantitative analysis made accessible. Available at https://userfriendlyscience.com/(accessed 15 February 2018).
- Piekny AJ, Johnson J-LF, Cham GD, Mains PE. (2003). The Caenorhabditis elegans nonmuscle myosin genes nmy-1 and nmy-2 function as redundant components of the let-502/Rho-binding kinase and mel-11/myosin phosphatase pathway during embryonic morphogenesis. Development , 5695–5704. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A Language and Environment for Statistical Computing, Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Saras J, Franzén P, Aspenström P, Hellman U, Gonez LJ, Heldin CH. (1997). A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. J Biol Chem , 24333–24338. [DOI] [PubMed] [Google Scholar]
- Seguchi O, Takashima S, Yamazaki S, Asakura M, Asano Y, Shintani Y, Wakeno M, Minamino T, Kondo H, Furukawa H, et al (2007). A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J Clin Invest , 2812–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi K, Cram EJ, Zaidel-Bar R. (2017). Stretch-induced actomyosin contraction in epithelial tubes: mechanotransduction pathways for tubular homeostasis. Semin Cell Dev Biol , 146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit S, Manser E. (2011). Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci , 679–683. [DOI] [PubMed] [Google Scholar]
- Smiesko V, Johnson PC. (1993). The arterial lumen is controlled by flow-related shear stress. Physiology , 34–38. [Google Scholar]
- Smith PG, Roy C, Zhang YN, Chauduri S. (2003). Mechanical stress increases RhoA activation in airway smooth muscle cells. Am J Respir Cell Mol Biol , 436–442. [DOI] [PubMed] [Google Scholar]
- Somlyo AP, Somlyo AV. (2003). Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol Rev , 1325–1358. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. (2001). Small GTP-binding proteins. Physiol Rev , 153–208. [DOI] [PubMed] [Google Scholar]
- Tan PY, Zaidel-Bar R. (2015). Transient membrane localization of SPV-1 drives cyclical actomyosin contractions in the C. elegans spermatheca. Curr Biol , 141–151. [DOI] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann UE, Unser M. (1998). A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process , 27–41. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. (1998). Specific interference by ingested dsRNA. Nature , 854. [DOI] [PubMed] [Google Scholar]
- Tzur YB, Friedland AE, Nadarajan S, Church GM, Calarco JA, Colaiácovo MP. (2013). Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics , 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, Tamakawa H, Yamagami K, Inui J, Maekawa M, et al (1997). Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature , 990–994. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mattson MP, Furukawa K. (2002). Endoplasmic reticulum calcium release is modulated by actin polymerization. J Neurochem , 945–952. [DOI] [PubMed] [Google Scholar]
- Wettschureck N, Offermanns S. (2002). Rho/Rho-kinase mediated signaling in physiology and pathophysiology. J Mol Med , 629–638. [DOI] [PubMed] [Google Scholar]
- Wilkes MM, Wilson JD, Baird B, Holowka D. (2014). Activation of Cdc42 is necessary for sustained oscillations of Ca2+ and PIP2 stimulated by antigen in RBL mast cells. Biol Open , 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirshing ACE, Cram EJ. (2017). Myosin activity drives actomyosin bundle formation and organization in contractile cells of the Caenorhabditis elegans spermatheca. Mol Biol Cell , 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A, Ingles J, Mains PE. (1999). The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the rac signaling pathway. Dev Biol , 111–127. [DOI] [PubMed] [Google Scholar]
- Wissmann A, Ingles J, Mcghee JD, Mains PE. (1997). Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev , 409–422. [DOI] [PubMed] [Google Scholar]
- Yin X, Gower NJD, Baylis HA, Strange K. (2004). Inositol 1,4,5,-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans . Mol Biol Cell , 3938–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Brien JMO, Ouellette LA, Church WR, Johnson DI. (1991). Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol , 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.