Abstract
The mitotic spindle is an ensemble of microtubules responsible for the repartition of the chromosomal content between the two daughter cells during division. In metazoans, spindle assembly is a gradual process involving dynamic microtubules and recruitment of numerous associated proteins and motors. During mitosis, centrosomes organize and nucleate the majority of spindle microtubules. In contrast, oocytes lack canonical centrosomes but are still able to form bipolar spindles, starting from an initial ball that self-organizes in several hours. Interfering with early steps of meiotic spindle assembly can lead to erroneous chromosome segregation. Although not fully elucidated, this process is known to rely on antagonistic activities of plus end– and minus end–directed motors. We developed a model of early meiotic spindle assembly in mouse oocytes, including key factors such as microtubule dynamics and chromosome movement. We explored how the balance between plus end– and minus end–directed motors, as well as the influence of microtubule nucleation, impacts spindle morphology. In a refined model, we added spatial regulation of microtubule stability and minus-end clustering. We could reproduce the features of early stages of spindle assembly from 12 different experimental perturbations and predict eight additional perturbations. With its ability to characterize and predict chromosome individualization, this model can help deepen our understanding of spindle assembly.
INTRODUCTION
Oogenesis terminates with meiosis I and II, two successive divisions without intervening DNA replication, leading to the formation of a haploid female gamete, the oocyte. During meiosis I, half of the DNA content is ejected into the first polar body after a reductional division segregating homologous chromosomes. Meiosis II is an equational division resembling mitosis, allowing the separation of sister chromatids in the second polar body. Importantly, oocyte formation is highly prone to chromosome segregation errors, specifically in humans, that can be responsible for spontaneous abortions and chromosomal defects (Nagaoka et al., 2012).
One key characteristic of most oocytes is that they lack centrioles (Szöllösi, 1976; Manandhar et al., 2005), essential elements of centrosomes. Centrosomes are the main microtubule-organizing centers of mitotic cells. They nucleate and organize mitotic spindles, which orchestrate chromosome alignment and segregation. This lack of canonical centrosomes in oocytes imposes peculiar modes of spindle morphogenesis that could contribute to the susceptibility of the female gamete to producing errors in chromosome segregation (Duncan and Wakefield, 2011; Mihajlovic´ and FitzHarris, 2018). Meiosis I, in particular, is more error-prone than meiosis II (Nagaoka et al., 2012), even if the opposite can be true in older mothers (Herbert et al., 2015). Meiosis I spindle morphogenesis relies on an inside-out mode of assembly, first promoting the nucleation of microtubules around chromatin and then defining the spindle poles (Heald et al., 1996; Dumont et al., 2007; Schuh and Ellenberg, 2007; Breuer et al., 2010). Hence, meiotic spindle bipolarity is not predefined by the positions of the two centrosomes on opposite sides of the nucleus, as in mitosis, but instead is progressively established. Bipolarization can take ∼4 h in the mouse; in comparison, the entire process of mitosis lasts approximately 1 h in most cells. Moreover, in the mouse, proper spindle assembly requires the sorting and clustering of multiple acentriolar microtubule-organizing centers (aMTOCs) composed mostly of pericentriolar material at each spindle pole (Schuh and Ellenberg, 2007; Breuer et al., 2010; Kolano et al., 2012). As a result, meiotic spindle poles appear less robust, being anchored not into unique and well-defined centrosomes but into discrete aggregates of aMTOCs, whose final shape varies considerably from one meiotic spindle to the other. In some species, such as Drosophila, nematodes, Xenopus, and even humans, microtubules at spindle poles are not even connected or anchored to detectable aMTOCs (Gard, 1992; Gard et al., 1995; Srayko et al., 2000; Cullen and Ohkura, 2001; Holubcová et al., 2015). However, despite these differences, spindle assembly is overall comparable between oocytes of different species (Bennabi et al., 2016).
In mouse oocytes at meiosis I entry and following nuclear envelope breakdown (NEBD), microtubules are first nucleated and stabilized around chromatin, forming a so-called microtubule ball (Schuh and Ellenberg, 2007; Kitajima et al., 2011). They are then progressively organized into a central array by microtubule motors and microtubule-associated proteins (MAPs), which sort and orient them, leading to a slowly established bipolar structure (Walczak et al., 1998; Dumont et al., 2007; Brunet et al., 2008; FitzHarris, 2009; Breuer et al., 2010; Kolano et al., 2012). Concomitantly, chromosomes undergo a first phase of “individualization” after nuclear envelope fragmentation, during which they are moved toward the periphery of the forming microtubule ball, increasing their relative distances from each other (Kitajima et al., 2011). They are then gathered toward the center of the bipolarized spindle before being separated into two equal halves at anaphase I (Mihajlovic´ and FitzHarris, 2018). Strikingly, skipping the microtubule-ball stage by a slight overexpression of a minus end–directed motor HSET (kinesin 14) results in precocious spindle bipolarization and increased chromosome misalignment, leading to segregation errors (Bennabi et al., 2018). However, overexpression of HSET after spindle bipolarization does not affect spindle morphology nor chromosome alignment (Bennabi et al., 2018), suggesting that errors in chromosome alignment and segregation specifically arise from defects accumulated during the early steps of spindle assembly. Remarkably, the system is very sensitive to the quantity of available HSET: upon inhibition, spindles do not bipolarize or do so with a substantial delay (Matthies, 1996; Bennabi et al., 2018). In contrast, slight overexpression of HSET (1.6-fold) accelerates bipolarization, leading to longer spindles, while higher doses of HSET trigger the formation of monopolar asters. Other perturbations can induce delayed spindle bipolarization, affecting the duration of the microtubule-ball stage, also resulting in an increase in misaligned chromosomes and in aneuploidy (e.g., pericentrin depletion, Baumann et al., 2017; NuMA perturbation, Kolano et al., 2012). However, in these genetic studies, it was not established whether observed aneuploidies were solely due to the perturbation of early stages of spindle assembly. Indeed, depletion or overexpression of proteins involved in meiosis spindle assembly affects not only the timing of spindle bipolarization (accelerated or delayed) but also spindle morphology and thus the repartition of chromosomes along the spindle (for review see Bennabi et al., 2016). Thus, the contribution of early chromosome individualization at the microtubule-ball stage to correcting chromosome alignment and ensuing segregation remains an open question. A role for chromosome individualization could be to separate physically the chromosomes, facilitating their proper capture by microtubules. Alternatively, it could contribute to temporal control to coordinate with the synthesis of components essential for spindle assembly.
Addressing these questions experimentally is challenging, and thus meiotic spindle formation is not yet fully characterized. Previous in silico investigations have described meiotic spindle bipolarization, especially the interplay between minus end– and plus end–directed sliding/clustering of microtubules (MTs; Schaffner and José, 2006; Burbank et al., 2007; Loughlin et al., 2010; Craig et al., 2011). Most models did not consider MTs’ dynamic instability, focusing on “sorting” of MTs into two poles by the different proteins. However, in meiotic spindles, MT growth is often much faster than motor-mediated displacements, also called poleward flux (Lockhart and Cross, 1996; Kapitein et al., 2005; FitzHarris, 2009; Breuer et al., 2010; Needleman et al., 2010; Brugués et al., 2012; Norris et al., 2018). Thus, it is essential to include MT dynamics in models of meiotic spindle formation. Numerical simulations, integrating MT dynamics, reproduced the main features of microtubule movements and dynamics within the Xenopus meiotic spindle, and thus realistic spindle organization along its long axis (Loughlin et al., 2010). However, in this study, MT alignment was imposed at the beginning of the simulation and an oriented and fixed DNA plate was fixed in the middle of the structure. As a result, this model, while very interesting for studying the metaphase steady state, is not informative on the contribution of early steps, in particular the microtubule ball and its transition toward a bipolar structure. Furthermore, the impact of these early stages on the distribution of chromosomes has never been addressed in silico so far.
We hence developed numerical simulations of early stages of spindle assembly integrating the activity of plus end– and minus end–directed motors. We included a more complex description of MT nucleation, considering MT dynamics, chromosome motion, and the contribution of aMTOCs. A first simplified model reproduced different spindle morphologies that were observed by varying HSET levels. It allowed us to explore the sensitivity of spindle assembly and chromosome repartition to the balance of plus end– and minus end–directed motors. To further characterize chromosome motion during spindle assembly and identify perturbations in chromosomal distribution, we extended the first model to include other ingredients necessary to this process. We tested the effect of each component of the model on spindle morphology by varying its concentration and compared the simulated results with corresponding experimental observations whenever these were available. When experimental observations were not available to our knowledge, simulations offered a prediction of the response to the perturbation. Eventually, we quantified chromosome individualization in our simulations and highlighted the conditions for increasing or decreasing it, suggesting experiments that could bring new insights into this process.
RESULTS
We developed our model with the open source software Cytosim (Nedelec and Foethke, 2007). Cytosim is a very flexible agent-based cytoskeleton simulation engine following Langevin dynamics (see Materials and Methods). We first aimed at identifying the minimal necessary components of the system that would collectively generate the desired behavior in a simulation, given their identified properties. We specifically focused on explaining the tight regulation of spindle assembly by HSET (Bennabi et al., 2018). With this in mind, we identified from the literature (reviewed in Marlow, 2018; Mogessie, Scheffler, and Schuh, 2018) the components that seemed essential to our model (Figure 1; Supplemental Materials): 1) explicit dynamics of all MTs, 2) Ran-mediated MT nucleation around mobile chromosomes, 3) MT-nucleating aMTOCs, 4) the plus end–directed kinesin 5 (Eg5), and 5) the minus end–directed kinesin 14 (HSET). Microtubules are modeled following the two-state dynamic instability model (Mitchison and Kirschner, 1984): they can grow, undergo catastrophes, and then shrink until they disappear (see Materials and Methods and Supplemental Materials). Steric interactions between microtubules were added to account for soft-core repulsion, as well as for the various cross-linkers, steric attraction, and crowding agents (Groen et al., 2011), similarly to previous models (Loughlin et al., 2010; Letort, Politi, et al., 2015). Their effect is to align neighboring microtubules and minimize spatial overlaps. For computational efficiency, we simulated a significantly smaller system than the experimental one (fewer microtubules, chromosomes, aMTOCs, …) and consequently for much shorter times (see Materials and Methods for a discussion of this specific point). This allowed a qualitative study that incurred acceptable computation time and memory. Additional description of the implementation of those components, in particular the list of parameters and their sources, is given in the Supplemental Materials.
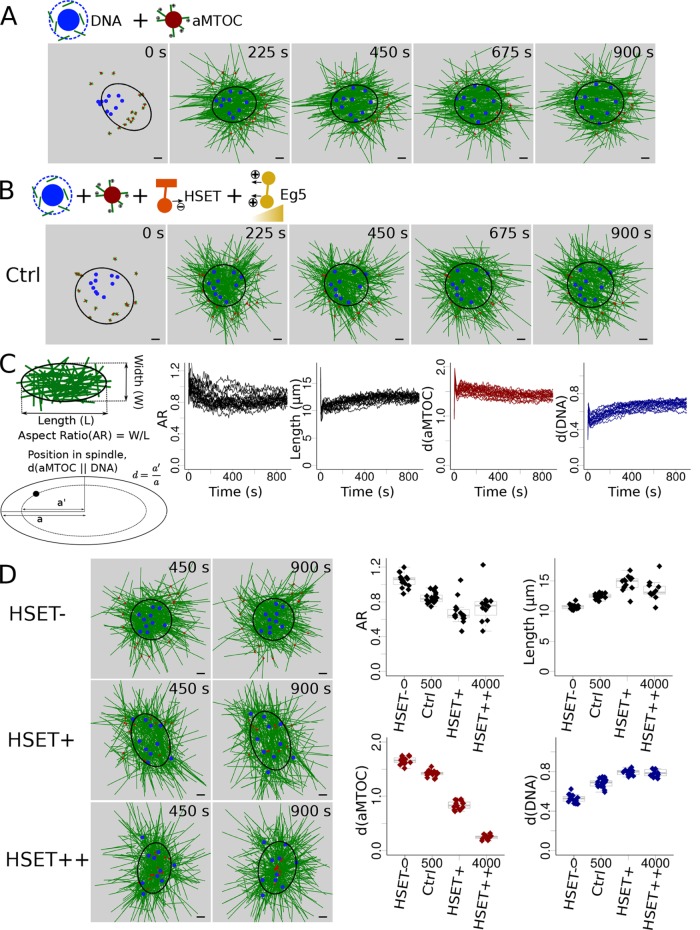
FIGURE 1:
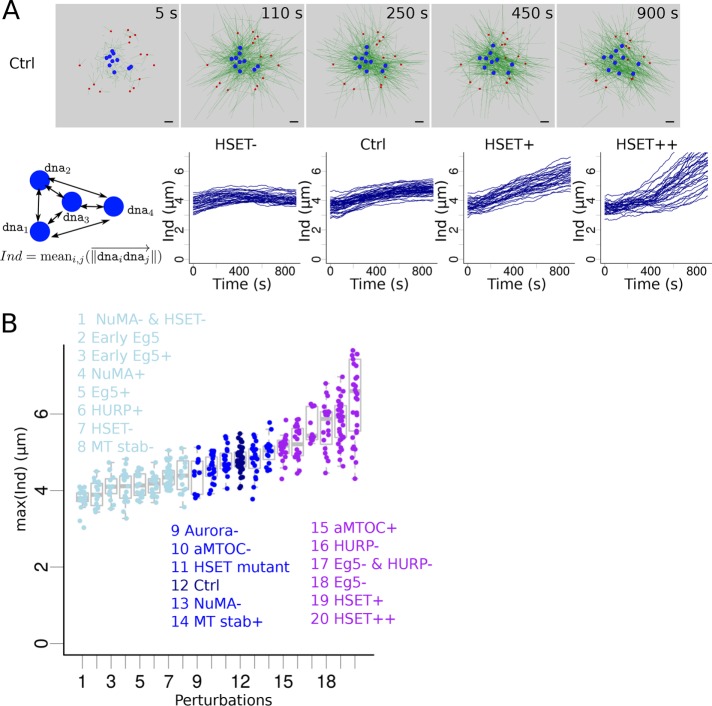
HSET concentration impacts early stages of spindle assembly. (A) Snapshots of a simulation without motors (only MT dynamics and nucleation). (B) Snapshots of a simulation with HSET and Eg5 entities. (C) Analysis of the simulated spindles: schematic description of the measures used (left) to analyze multiple simulations (see Materials and Methods): evolution over time of the spindle aspect ratio (first graph), of spindle length (second graph), of aMTOCs position (third graph), and of DNA position (last graph). Each line represents an individual simulation. (D) Variation of the quantity of simulated HSET and its effect on spindle features. (Left) Snapshots of the simulations when HSET is inhibited (HSET-, top), overexpressed (∼3 times more, HSET+, middle), and highly overexpressed (∼8 times more, HSET++, bottom). (Right) Final values (at t = 900 s) of the spindle features (aspect ratio, top left; spindle length, top right; aMTOC position, bottom left; DNA position, bottom right) in 15 simulations for each of the 4 HSET quantities (HSET-, Ctrl, HSET+, HSET++). Microtubules are green, DNA beads mimicking chromosomes blue, aMTOCs red, HSET orange, and Eg5 dark yellow. Scale bar is 2 µm. For quantification, spindle shapes are fitted with ellipses (see Materials and Methods) represented in black.
Simulations reproduce spindle assembly for a wide range of HSET concentrations
Initially, we placed chromosomes, modeled as beads (Figure 1A, blue), randomly in a disk inside a circle of aMTOCs, modeled as small asters (Figure 1A, red), corresponding to time points right after NEBD when chromosomes are condensed in the cytoplasm, surrounded by aMTOCs. Microtubule nucleation is then triggered around the DNA beads (Supplemental Materials), mimicking the activity of gamma-tubulin and other nucleators activated by the Ran-GTP gradient (Karsenti and Vernos, 2001; Job et al., 2003; Dumont et al., 2007; Kollman et al., 2011). With nucleation only, the forming microtubule mass around the chromosomes pushed them apart and individualized them, while the aMTOCs stayed out of the MT mass (Figure 1A). To trigger correct spindle assembly, we then added the key molecular motors, the kinesin Eg5 and its antagonist kinesin HSET (Supplemental Materials). All the HSET motors were added from the beginning of the simulation, while Eg5 was only progressively activated (see discussion and measurements in Supplemental Materials, Section 4). Indeed, levels of several spindle assembly factors increase progressively during meiosis, including Eg5 (Brunet et al., 2008; Breuer et al., 2010; Bennabi et al., 2018) and early stages have been proposed to be dominated by minus-end motors (Schuh and Ellenberg, 2007; Kitajima et al., 2011). We thus assumed that HSET is efficient immediately after NEBD (Bennabi et al., 2018). With this setup, microtubules assembled into a ball, pushing the chromosome beads apart toward the periphery, and the aMTOCs converged toward the microtubule-ball surface (Figure 1B and Supplemental Movie S1). To characterize the simulated spindle morphologies, we fitted an ellipse around the majority of the microtubule mass and measured its length and aspect ratio (AR: width-to-length ratio, Figure 1C; Materials and Methods). We also characterized the position d of aMTOCs and chromosomes relative to the spindle. For this, we measured the semimajor axis a’ of the ellipse that contained the object of interest (aMTOCs or chromosomes) and had the same aspect ratio as the ellipse that fits the spindle. We further defined the relative position d as the ratio of the semimajor axis of the object’s ellipse a’ to the semimajor axis of the ellipse fitting the spindle a (Figure 1C). If d is close to 0, the aMTOCs/chromosomes are close to the spindle center. This gave a measure of how far inside the spindle the aMTOCs or chromosomes were distributed, and could be compared in spindles of different shapes. The evolution of these measures over time during the simulation reflected the formation of the microtubule ball (Figure 1C), with DNA individualization and aMTOC clustering, consistent with experimental behavior (Schuh and Ellenberg, 2007). Eventually, we altered the quantity of HSET in the simulation to test whether this simple model was sufficient to reproduce the drastic consequences of HSET perturbations on early spindle morphology. As experimentally observed (Bennabi et al., 2018), the microtubule ball could still form upon inhibition of HSET (HSET-), while when HSET was overexpressed (∼3 times the level of the endogenous protein, HSET+), an elongated spindle with spread chromosomes formed (Figure 1D and Supplemental Movie S2). Higher overexpression (HSET++) led to a monopolar spindle (aster) with aMTOCs in the center and DNA at the periphery (Figure 1D). The increase in spindle length under conditions of overexpressed HSET was mainly due to excessive sliding caused by the motor. In addition, greater spreading of DNA beads along the spindle axis would trigger MTs nucleation away from the center, also leading to an increase in length. In conclusion, our first simplified model reproduced different spindle morphologies that were obtained by varying HSET levels experimentally.
Video S1.
Simulation of early meiotic spindle assembly, simplified model. Default simulation with the first model (left). Same simulation, showing also HSET (orange) and Eg5 (yellow) bound to MTs (right). Microtubules are in green, DNA beads in blue, aMTOCs in red, total time is 900 s.
Video S2.
Variation of HSET quantity, simplified model. Simulations with inhibited HSET (HSET-, left), normal level (Ctrl, middle) and over-expressed level (HSET+, right). Microtubules are in green, DNA beads in blue, aMTOCs in red, total time is 900 s.
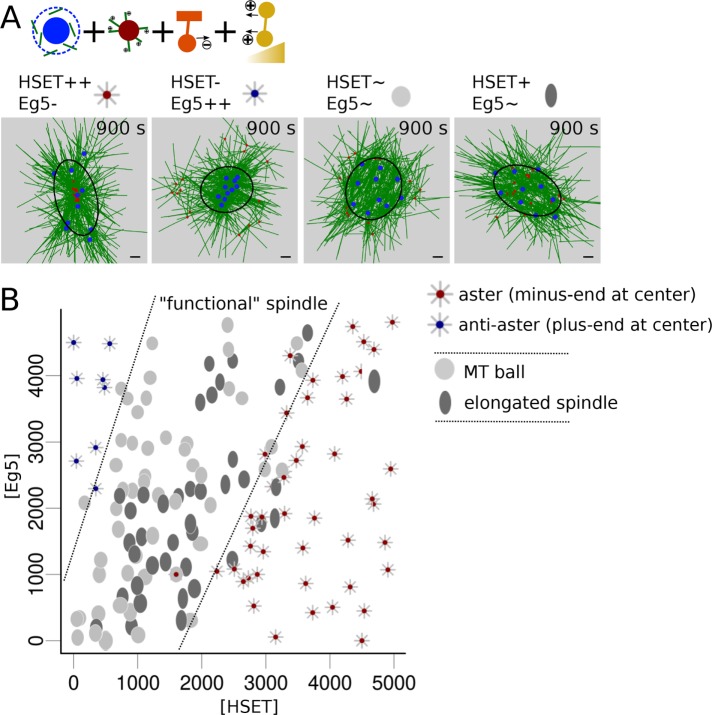
The balance between plus-end and minus-end motors, but also microtubule nucleation, determines early spindle morphologies
Although previous models with static nucleation or static MTs (Surrey, Nedelec, et al., 2001; Schaffner and José, 2006; Burbank et al., 2007; Craig et al., 2011) could not explain this change in spindle length, they highlighted the importance of the balance between plus end– and minus end–directed forces for producing different spindle morphologies (Mountain, 1999; Surrey, Nedelec, et al., 2001; Mitchison et al., 2005; Schaffner and José, 2006; Burbank et al., 2007; Hentrich and Surrey, 2010; Derr, Goodman, et al., 2012). Thus, we next varied the quantity of both HSET and Eg5 in the simulation (Figure 2). Consistent with these previous models and experimental observations (Mountain, 1999; Mitchison et al., 2005; Hentrich and Surrey, 2010; Derr, Goodman, et al., 2012), the system self-organized into one of four different morphologies. When both motors were in low quantities, MTs nucleation and dynamics ruled the network (Mitchison et al., 2005), and the microtubules formed a ball with poor chromosome individualization (Figure 1, A and B). When HSET dominated, a monopolar aster formed, presenting the majority of MT minus ends and aMTOCs clustered in the center (Figure 2, A, first column, and B). When Eg5 dominated, the spindle formed an inverted aster (anti-aster) with the DNA clustered in the center and aMTOCs in the periphery (Figure 2, A, second column, and B). For an intermediate range of Eg5 and HSET concentrations, when there is no clear dominance by any of the motors, a “functional” spindle (not collapsing to an aster) could form (area between dashed lines, Figure 2B). The microtubule ball formed, with chromosomes at the periphery and aMTOCs inside (Figure 2, A, third column, and B). Eventually, a slight increase of HSET activity from these equilibrated conditions provoked the elongation of the system, concomitant with chromosome spreading (Figure 2, A, fourth column, and B).
FIGURE 2:
A window of balanced HSET and Eg5 quantities promotes spindle elongation. (A) Final snapshots of simulations with varying amounts of HSET and Eg5. From left to right: high levels of HSET with low levels of Eg5 (HSET++ Eg5-, left); low levels of HSET with high levels of Eg5 (HSET- Eg5++); normal HSET and Eg5 levels (HSET∼ Eg5∼, Ctrl); high levels of HSET and normal levels of Eg5 (HSET+ Eg5∼, right). Four different morphologies can be identified: 1) aster with aMTOCs clustered in the center, represented by a gray aster with a red dot at the center; 2) inverted aster with DNA clustered toward the center, represented by a gray aster with a blue dot at the center; 3) microtubule ball represented as a gray ellipse (aspect ratio >0.75); and 4) elongated spindle represented as a darker ellipse (aspect ratio <0.75). Ellipses were represented with an aspect ratio and length scaled to their measured values. Microtubules are green, DNA blue, aMTOCs red, HSET orange, and Eg5 dark yellow. Scale bar is 2 µm and the black ellipses represent the fitted spindle ellipse. (B) Spindle morphologies at t = 900 s according to the respective amounts of simulated HSET and Eg5. Dashed lines (arbitrarily drawn) delimit the regime in which the microtubule structures do not collapse, thus leading to a “functional” spindle.
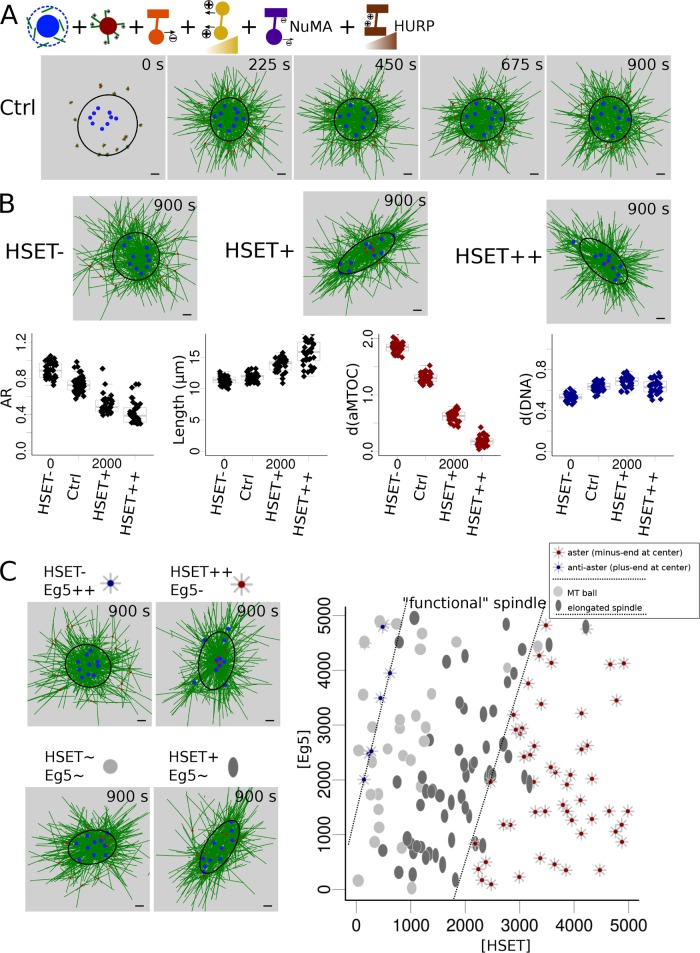
While capturing the main spindle assembly behavior, even the best-organized systems were poorly elongated, having an aspect ratio above 0.6, while experimental ratios were measured around 0.3–0.4 (Bennabi et al., 2018). Simulated microtubule balls were also quite loose networks. Moreover, this initial model did not make it possible to explore a wide range of conditions that would interfere with chromosome individualization. Therefore, we decided to add essential spindle features such as anti-parallel fibers present around chromosomes and stabilized by HURP (or PRC1)-like proteins (Sampath et al., 2004; Bieling et al., 2010; Breuer et al., 2010) and minus-end clustering by NuMA/dynein complexes (Compton, 1998; Kolano et al., 2012). Differences in MT stability along the spindle were indeed shown to have a major effect on mitotic and meiotic spindle assembly (Wollman et al., 2005; Mogilner et al., 2006; Greenan et al., 2010; Loughlin et al., 2010).
Previous conclusions were modestly affected by the addition of new components
A second model was simulated, with entities to mimic HURP activity. They are active only around chromosome beads, bind to anti-parallel MTs, and protect the MTs against plus-end depolymerization by decreasing their catastrophe rate (see the Supplemental Materials). As HURP and other plus-end clustering proteins are thought to be brought toward MT plus ends by kinesin 5, they were progressively activated in the simulation, like Eg5 in our model. NuMA-mediated minus-end clustering was modeled by minus-end binding complexes (see the Supplemental Materials) that also had a slight depolymerizing effect on the bound minus end (MCAK-like proteins; Walczak et al., 1996; Desai et al., 1999; Gaetz and Kapoor, 2004; Hueschen et al., 2017). This extended model produced tighter spindle architectures (Figure 3A and Supplemental Movie S3) and more pronounced spindle elongation upon HSET overexpression (Figure 3B and Supplemental Movie S4). Importantly, we still obtained the same four morphologies according to the Eg5/HSET balance, but the spindles were more elongated (Figure 3C and Supplemental Movie S5; aspect ratio could now be below 0.5). Interestingly, anti-asters were less common, even at higher levels of Eg5. This is due to MT stabilization around chromosomes, which prevents tight chromosome grouping in the center (Supplemental Movie S5). Moreover, spindle length was now affected by plus-end stabilization and minus-end depolymerization. Overall, the extended model, like the simpler one, agreed with previous studies on the necessity to balance plus-end and minus-end motors, but also brought new insights into the importance of MT dynamics/nucleation and of chromosome motility in determining spindle morphology.
FIGURE 3:
Refinement of spindle morphologies regimes according to HSET and Eg5 concentrations with the extended model. (A) Snapshots of a simulation with default parameter values (Ctrl) with the new extended model. (B) Variation of HSET amounts. (Top) Final snapshot of simulations with varying HSET concentration: with HSET inhibited (HSET-, left), overexpressed (HSET+, middle), and highly overexpressed (HSET++, right). (Bottom) Measures of spindle features (aspect ratio, first column; spindle length, second column; aMTOCs position, third column; DNA position, last column), defined as explained in Figure 1C, at the end time point (900 s) for repeated simulations for each HSET quantity (inhibition, Ctrl, overexpression, high overexpression). (C) Balance between HSET and Eg5 concentrations. (Left) Final snapshots for varying quantities of HSET and Eg5: HSET inhibition and Eg5 highly overexpressed (HSET- Eg5++, top left); HSET highly overexpressed and Eg5 inhibited (HSET++ Eg5-, top right); HSET and Eg5 default values (HSET∼ Eg5∼, bottom left); HSET overexpressed and Eg5 default value (HSET+ Eg5∼, bottom right). As in Figure 2, four different morphologies are determined: 1) aster with aMTOCs clustered in the center represented by a gray aster with a red dot at the center, 2) inverted aster with DNA clustered toward the center represented by a gray aster with a blue dot at the center, 3) microtubule ball represented as a gray ellipse (aspect ratio > 0.5), and 4) elongated spindle represented as a darker ellipse (aspect ratio < 0.5). Ellipses were plotted with an aspect ratio and length scaled to their measured values. (Right) Final spindle morphologies (at t = 900 s) according to the quantity of simulated HSET and Eg5. Microtubules are green, DNA blue, aMTOCs red, HSET orange, Eg5 dark yellow, NuMA purple, and HURP brown. Scale bar is 2 µm. For quantification, spindle shapes are fitted with ellipses (see Materials and Methods) represented in black.
Video S3.
Simulation of early meiotic spindle assembly, extended model. Default simulation with the second model (left). Same simulation, showing also NuMA (purple) and HURP (brown) bound to MTs (right). Microtubules are in green, DNA beads in blue, aMTOCs in red, total time is 900 s.
Video S4.
HSET over-expression elongates the spindle, extended model. Simulations with normal level of HSET (Ctrl, left) and with HSET over-expressed (HSET+, right).
Video S5.
Different morphologies of the system, extended model. (1st column) inverted aster (Eg5+, HSET-), (2nd) MT-ball (Ctrl values), (3rd) elongated spindle (HSET+), (4th) aster (HSET++). Microtubules are in green, DNA beads in blue, aMTOCs in red, total time is 900 s.
Simulations robustly reproduce experimental perturbations of early stages of meiotic spindle assembly
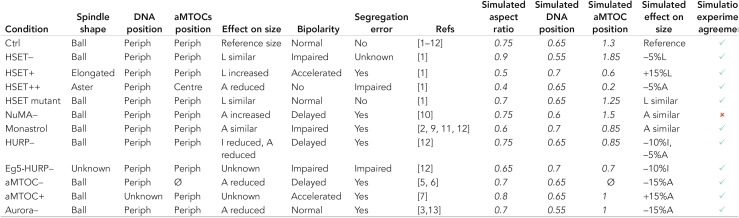
Our second objective was to study the extent of chromosome individualization under different conditions and predict spindle phenotypes. Chromosome individualization is indeed an intriguing step of early meiotic spindle assembly, during which chromosomes are moved toward the periphery of the microtubule ball and separated from each other (Kitajima et al., 2011) before congressing toward the spindle center. We thus confronted the model with available experimental observations (consisting of inhibition or overexpression of various proteins). As we focused on mouse oocyte meiosis I spindle early assembly (between ∼0 and 1.5 h), we limited our comparison to experiments performed in this time frame. Depending on the studies, we collected data from the literature on either spindle shape, aMTOCs position, chromosome position, spindle length, or area or microtubule density. We combined the last three measures into one single measure of “spindle size.” In the majority of cases, the literature provided us with only a qualitative idea of these features. To compare those results, we classified the numerical output of the simulations in global phenotypes, similar to experimental data (Table 1). Specifically, we considered that simulation behavior was in agreement (green check mark) when the measured value of spindle aspect ratio, aMTOC position, or chromosome position was correspondingly above or below our arbitrary fixed threshold of 0.5 (50%). For spindle size, we considered the behavior as correct if the trend of change (increase, decrease, or unchanged) was similar.
TABLE 1:
Perturbation effe5 cts on spindle features.
For each perturbation, experimental observations are given when known (top rows) with below the average values of around 15 simulations (italic text). Refs: [1] Bennabi et al., 2018; [2] Schuh and Ellenberg, 2007; [3] Bury et al., 2017; [4] Yoshida et al., 2015; [5] Baumann et al., 2017; [6] Ma and Viveiros 2014; [7] Manil-Segalen et al., 2018; [8] Kitajima et al., 2011; [9] Clift and Schuh, 2015; [10] Kolano et al., 2012; [11] Mailhes et al., 2004; [12] Breuer et al., 2010; [13] Yu et al., 2016.
A stands for area, L for length, and I for intensity.
Overall, we used experimental data obtained for 12 different perturbations, and predicted the phenotype for eight additional perturbations. The aspect ratio, aMTOC positions, and chromosome positions were correctly simulated under all 12 conditions. The variation in the size of the system agreed with experimental measurements for all tested conditions except one: NuMA inhibition (Table 1, red cross). This difference could be explained by the fact that these experiments were conducted on oocytes expressing a nonfunctional form of NuMA (deleted for its microtubule binding domain; Kolano et al., 2012) and spindle size was already different at NEBD, whereas the simulations were performed in the total absence of NuMA. Nonetheless, this difference might reflect the sensitivity of the system to size: a lot of different factors influence microtubule dynamics, and thus spindle size, and knowledge of those factors and precise effects is still limited. Reassured that the simulations could give a valid prediction of spindle organization, we used them to predict spindle morphologies under other perturbations that have not yet been reported in mouse oocytes (Table 2 and Supplemental Movie S6).
TABLE 2:
Simulation predictions of perturbations.
| Condition | Spindle shape | DNA position | aMTOCs position | Effect on size |
|---|---|---|---|---|
| HURP+ | 0.8 (ball) | 0.5 (center) | 1.65 (periph) | I unchanged |
| NuMA+ | 0.75 (ball) | 0.55 (periph) | 1.35 (periph) | –10%A |
| Eg5+ | 0.8 (ball) | 0.5 (center) | 1.5 (periph) | A unchanged |
| Early Eg5 | 0.9 (inverted aster) | 0.45 (center) | 1.9 (periph) | +5%A |
| MT stab+ | 0.75 (ball) | 0.6 (periph) | 1 (periph) | +25%A |
| MT stab- | 0.7 (ball) | 0.6 (periph) | 1.5 (periph) | –20%A |
| HSET-NuMA- | 0.9 (inverted aster) | 0.4 (center) | 2 (periph) | –15%L |
| Early Eg5+ | 0.95 (inverted aster) | 0.45 (center) | 2.15 (periph) | –10%L |
For each perturbation, average values of around 15 simulations are given.
A stands for area and L for length.
Video S6.
Simulation predictions of perturbations with the extended model. Examples of predicted perturbations : (Left) Early Eg5, (Middle) HURP+ and (Right) Mtstab+. Microtubules are in green, DNA beads in blue, aMTOCs in red, total time is 900 s.
Importance of minus end–directed forces in chromosome individualization
We found only three conditions under which chromosomes were not pushed toward the spindle periphery, but remained close to the center (position d ≤ 0.5): expression of Eg5 directly from NEBD (Supplemental Movie S6), double inhibition of HSET and NuMA, and early overexpression of Eg5 (Tables 1 and 2). This strongly suggested, that, as proposed by previous work, early stages of spindle assembly have to be dominated by minus-end forces for efficient chromosome individualization to take place. To further characterize how chromosome individualization was affected by initial conditions, we averaged the distances between chromosome in simulations and followed their evolution over time (Figure 4A). When HSET was inhibited, DNA beads were first pushed apart by microtubule polymerization and then were brought back toward the center when Eg5 activity was high enough (Figure 4A, first graph, and Supplemental Movie S7). When HSET was present at a control concentration, the efficiency of individualization was increased, notably during the first half of simulations, during which most of the individualization happened. During the second half of the simulations, individualization was slowed down by Eg5 and HURP activity (which tend to congress chromosomes to the center; Figure 4A, second graph, and Supplemental Movie S7), consistent with experimental tracking of kinetochore motion (Kitajima et al., 2011). However, overexpression of HSET led to increased individualization specifically during the second half of the simulations, showing that in those conformations, Eg5 and HURP promoted the process of chromosome individualization (Figure 4A, third and fourth graphs, and Supplemental Movie S7). This confirmed that perturbing the balance of activities in the spindle had a drastic effect on chromosome individualization. To identify conditions that had the greatest impact, we compared the maximal individualization values reached in all our previous simulations (Figure 4B). This reflected the maximal separation between chromosomes in the simulation. We ranked the simulation conditions by their median values of this maximal individualization and classified them into three groups: perturbations that modestly perturbed individualization compared with the control case (p value of two-sided Kolmogorov–Smirnov test comparison to control values > 0.01; Figure 4B, dark blue), perturbations that promoted chromosome individualization (p value < 0.01; Figure 4B, purple) and perturbations that decreased chromosome individualization (p value < 0.01; Figure 4B, light blue).
FIGURE 4:
Impact of various perturbations on chromosome individualization. (A) (Top) Snapshots of a simulation with the Ctrl configuration, focusing on early time points. aMTOCs are red, microtubules are green and shown with thin lines for a better visualization of motion of DNA beads (blue). (Bottom) Measure of the DNA beads’ individualization score, Ind: the average distance between the DNA beads (cartoon). Evolution of the individualization score over time for different quantities of simulated HSET: inhibition (HSET-, first column), default (Ctrl, second column), slight overexpression (HSET+, third column), and high overexpression (HSET++, last column). Each line represents an individual simulation. (B) Maximal value of the individualization score reached during a simulation according to the perturbation applied to the system (sorted by maximal score). Perturbations were classified into three groups based on their maximal individualization scores compared with control’s: similar (middle, dark blue), lower (left, light blue), and higher (right, purple) individualization.
Video S7.
Chromosomes individualization, extended model. DNA beads (blue) movement during the simulations for 3 different levels of HSET : (Left) HSET inhibited (HSET-), (Middle) default value (Ctrl) and (Right) HSET over-expressed (HSET+).
In the group comparable to controls (Figure 4B, dark blue), all five experimental conditions consistently underwent spindle assembly through a transient microtubule-ball stage. Hence, potential chromosome segregation errors under such conditions would be caused by later defects (e.g., in chromosome congress). We then examined the conditions in the two other groups to determine whether we could establish correlations between abnormal chromosome individualization (measured from the simulations, Figure 4B) and errors in chromosome segregation (observed experimentally, Table 1). The eight perturbations that resulted in reduced chromosome individualization (Figure 4B, light blue) have not yet been experimentally tested, except for HSET inhibition (HSET-). If the spindle still manages to bipolarize in those conditions, the follow-up of chromosome behavior could tell us whether skipping the chromosome individualization step affects later chromosome repartition.
In the group with greatest chromosome individualization (Figure 4B, purple), three conditions out of six formed asters (HSET++, Monastrol [Eg5-], both Eg5 and HURP inhibited [Eg5- and HURP-]). Under one of these three conditions, the spindle eventually bipolarized, associated with an increase in chromosome segregation errors (Monastrol Eg5- condition; Mailhes et al., 2004). Two conditions out of six (HSET slight overexpression and aMTOC overexpression) led to faster bipolarization and an increase in chromosome segregation errors (Table 1). The last condition, HURP inhibition (HURP-), led to a delayed bipolarization, with errors in chromosome segregation as well (Breuer et al., 2010). Thus, increased chromosome individualization seems to be associated with failure or affected timing of spindle bipolarization and chromosome segregation errors.
DISCUSSION
Female first meiotic spindle assembly is a slow and complex process, involving many different proteins recruited at various rates, and our current knowledge and understanding of the mechanism remains limited. In mouse oocytes, in particular, it is difficult to timely and reversibly inhibit proteins, and as a result we know little about proteins that are rate-limiting or when they become essential. Even if the essential players of spindle assembly have been identified, their relative quantities and timing can drastically affect overall spindle formation, as shown by our simulations. Our model reproduced the effect of varying HSET concentrations on spindle morphology and notably explained the significant spindle elongation observed under conditions of slight HSET overexpression (Figures 1 and 3; Supplemental Movie S4). We explored here how the balance between the quantity of minus end–directed motors, plus end–directed motors, and microtubule dynamics impacted spindle morphology (Figures 2 and 3; Supplemental Movie S5). We observed that different structures can be formed: a microtubule aster (if minus end–motor dominated), an inverted aster (if plus end–motor dominated), a microtubule ball (motors balanced or microtubule nucleation–dominated), or an elongated spindle (slightly more effective minus-end motors than plus-end motors). Strikingly, spindlelike structures (microtubule ball or elongated spindle) can only form for an intermediate range of HSET and Eg5 concentrations (dashed lines, Figures 2 and 3). We also highlighted how changing the timing of “availability” of proteins could affect this outcome (Table 1, Early Eg5 simulation, and Supplemental Materials).
Despite its limitations, our model, based on current knowledge of the proteins involved, reproduced qualitatively nearly all the tested experimental observations on early meiotic spindle assembly in mouse oocytes. We evaluated the simulations based on qualitative experimental spindle descriptions in a wide range of experimental perturbations and found only one disagreement in variation of spindle size (out of 42 measured features, Table 1). As discussed in Results, this difference in behavior is most likely due to our lack of knowledge on the secondary effects of the experimental perturbations. Importantly, these results suggest that a more thorough investigation of the conditions where the model failed would allow us to identify new properties of the corresponding proteins or some of their interactions and partners. Our model focused on early stages of spindle assembly and does not allow long-term simulations. Refining the model to address the evolution of the microtubule ball to a bipolar spindle would be an important following work.
We also tested numerically the response of the system to perturbations for which we do not know yet the experimental outcome (Table 2; Supplemental Movie S6), and thus where the simulation results are predictions. We arbitrarily chose perturbations that seemed interesting to us, but other perturbations (or combination of perturbations) could just as well be simulated. Some of these conditions can be tested experimentally, which would further validate, improve, or refute our working model.
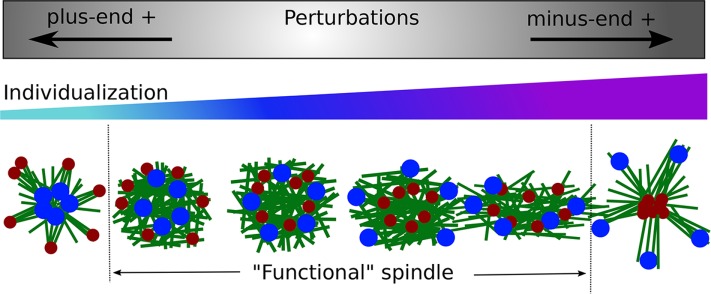
We were interested in testing whether initial chromosome individualization impacted the ensuing chromosome segregation. Using chromosome separation in the simulations, we could sort the perturbations according to their effect on chromosome individualization (Figure 4). We summarized our findings by representing the perturbations and their impact on chromosome individualization and spindle morphologies (Figure 5). Some perturbations do not greatly affect early spindle assembly: the microtubule ball forms with quasi-normal chromosome individualization (Figure 5, middle). In perturbations that favor minus end–directed forces, aMTOCs converge to the center and chromosomes are pushed away, increasing chromosome individualization (Figure 5, right direction). When the minus end–directed forces are strong enough, the system loses its ball shape and elongates or collapses into an aster in the most extreme cases (Figure 5, right). According to our simulations, perturbations that favor plus end–directed forces impair chromosome individualization: instead chromosomes are clustered toward the center and aMTOCs stay at the periphery (Figure 5, left direction). When plus end–directed forces are strongly favored, the system forms an inverted aster with chromosomes loosely clustered in the center (Figure 5, left).
FIGURE 5:
Summary. Spindle morphologies associated with perturbations of chromosome individualization. Schematic representation of the conclusions. Perturbations that favor plus end–directed forces decrease chromosome individualization (left arrow). Perturbations that favors minus end–directed forces increase chromosome individualization (right arrow). In the bottom row, the morphologies of the system were represented, based on the results from Table 1: microtubules are in green, chromosomes in blue, and aMTOCs in red. The regime in which spindlelike structures can form and do not collapse is limited by dashed lines.
The majority of perturbations that have been explored experimentally so far indicate that increased chromosome individualization is usually associated with chromosome segregation errors (Figure 5, right). However, the available literature on this process is too scarce to be conclusive, and we also need more experimental results on perturbations that decrease chromosome individualization (Figure 5, left). Therefore, experimental conditions in which chromosomes congress at the center early on would be informative; our simulations pinpointed experimentally testable conditions that should allow this, in particular early Eg5 expression.
Interestingly, by looking at an important variety of experimental perturbations of early spindle assembly, we noted the robustness of this system. Indeed, challenged by various perturbations, a spindle would still form and bipolarize (8/12 perturbations and partial bipolarization in two of the four other perturbations, Table 1). However, it is also quite sensitive, as these perturbations will often lead to delayed or accelerated spindle assembly, notable variations in spindle morphology, and an increase in chromosome misalignment. The robustness of this system might allow it to resist environmental conditions and still be able to divide, but at the expense of fidelity of chromosome segregation. This relative sensitivity is especially important in aging oocytes, which are prone to higher levels of segregation errors (Nagaoka et al., 2012), where global protein expression and microtubule dynamics are altered (Camlin et al., 2017; Nakagawa and FitzHarris, 2017).
MATERIALS AND METHODS
Numerical simulations
Cytosim (www.cytosim.org) is an open-source program designed to simulate cytoskeletal systems. It calculates the motion of semiflexible filaments according to Langevin dynamics (Nedelec and Foethke, 2007) and can simulate a large number of filaments and associated proteins. It has already been used to study the auto-organization of cytoskeletal systems composed either of actin filaments (Letort, Politi, et al., 2015; Ennomani, Letort, et al., 2016) or of microtubules (Surrey, Nedelec, et al., 2001; Goshima et al., 2005; Loughlin et al., 2010; Letort et al., 2016). Using Cytosim, we developed a model of early meiotic spindle assembly, based on current knowledge. Here, we describe briefly the main components of the model. A more thorough description of these elements and a justification of the model’s assumptions is given in the Supplemental Materials, along with the list of parameter values. We also provide a typical configuration file so that simulations can be directly reproduced.
Microtubule dynamics and nucleation
Microtubule dynamics is modeled following Terrell Hill’s two-state dynamic instability model (Mitchison and Kirschner, 1984): microtubules can grow, undergo catastrophes, and then shrink until they disappear. Because precise dynamics is not known for mouse oocytes spindles, we based our parameters on studies in other meiotic systems, assuming that MTs have a short lifetime on the order of only a few minutes, are only a few microns long, and do not rescue (Needleman et al., 2010).
Steric interactions between microtubules were included to avoid unrealistic overlaps (hard-core repulsion). The model also accounted for numerous cross-linkers present inside the spindle (Loughlin et al., 2010) and steric attractions between neighboring polymers that tend to align them (Groen et al., 2011; Letort, Politi, et al., 2015). In a dense environment, such interactions have indeed been shown to have a strong impact on the cytoskeleton system organization (Letort, Politi, et al., 2015).
Chromosomes were represented individually as motile beads, that is, as spherical entities, with passive binders placed on their surface (Lacroix et al., 2018). These binders can attach to nearby microtubule plus ends, but with a high unbinding rate, as the microtubule–kinetochore interactions were shown to be very unstable during the first hour of meiosis (Kitajima et al., 2011; Głuszek, Cullen, Li, et al., 2015; Yoshida et al., 2015).
Microtubule nucleation can be triggered by aMTOCs and Ran-mediated activation of nucleators as gamma-tubulin (Karsenti and Vernos, 2001; Job et al., 2003; Sampath et al., 2004; Brunet et al., 2008; Kollman et al., 2011). The Ran-mediated nucleation pathway was implemented with diffusible nucleator entities that were activated only close to the chromosome beads (Supplemental Materials). aMTOCs were modeled as small asters able to nucleate a few microtubules each, in all directions equally, whereby the minus ends of these MTs would remain as anchors to the aMTOCs (Letort et al., 2016; Supplemental Materials). The nucleation rate in the simulation was taken to be quite low, as their activity was shown to increase only later during meiosis (Schuh and Ellenberg, 2007; Brunet et al., 2008).
Molecular motors and cross-linkers
Numerous proteins are involved in spindle assembly (Bennabi et al., 2016; Marlow, 2018; Mogessie, Scheffler, and Schuh, 2018), and our knowledge of the system is still too limited to aim for realistic simulations. We integrated only the key identified components.
Kinesin 5, Eg5, present in both mitotic and meiotic spindles, is widely known to play a major role in spindle length regulation and chromosome segregation toward the midzone (Kapoor et al., 2000; Karsenti and Vernos, 2001; Mitchison et al., 2005; Valentine et al., 2006; Dogterom and Surrey, 2013). It is a tetrameric plus end–directed motor, able to slide antiparallel microtubules (Kapitein et al., 2005; Valentine et al., 2006). We modeled Eg5 as diffusing entities (labeled Eg5) composed of two equivalent motors capable of binding nearby microtubules all along their length, linked by a Hookean spring (Supplemental Materials). Minus end–directed motors have been shown to play an important role in balancing Eg5 action in spindle assembly (Mountain, 1999; Surrey, Nedelec, et al., 2001; Mitchison et al., 2005; Schaffner and José, 2006; Burbank et al., 2007; Hentrich and Surrey, 2010; Craig et al., 2011; Derr, Goodman, et al., 2012). In particular, kinesin 14, HSET (Matthies, 1996; Mountain, 1999; Cai et al., 2009; Hepperla et al., 2014; Braun et al., 2017; Norris et al., 2018), an Ncd homologue, strongly impacts meiotic spindle assembly (Bennabi et al., 2018). We thus implemented minus end–directed motors (labeled HSET) as diffusing entities composed of one passive binder, capable of attaching to nearby MTs, linked by a Hookean spring to a minus end–directed motor (Supplemental Materials).
Although these two motors were sufficient to capture the basic features of spindle assembly, other proteins help to shape the spindle by clustering microtubules and affecting their dynamics. Their effect cannot be neglected when the aim is to explore chromosome motion. Indeed, the NuMA/dynein complex was shown to have a strong impact on spindle morphogenesis timings (Kolano et al., 2012) and on pole focusing of mitotic spindle (Walczak et al., 1996) and meiotic acentriolar spindles (Compton, 1998; Khodjakov et al., 2000; Oriola et al., 2018). Moreover, it is thought to have a role in recruiting kinesin 13 family proteins (MCAK, Kif2a/KLP10A) to the microtubule minus ends (Gaetz and Kapoor, 2004), which have a depolymerizing activity (Hunter et al., 2003). To model these effects, we added entities (labeled as NuMA) composed of a domain that could bind and track microtubule minus ends, linked by a Hookean spring to another domain capable of triggering the depolymerization of microtubules it attached to. Finally, microtubules around the spindle central midzone are cross-linked and stabilized in anti-parallel bundles near the chromosomes (due to HURP, PRC1, and other proteins). Thus, we added numerical entities, labeled HURP, composed of two heads that could bind microtubules when close to a DNA bead (Ran-GTPases), move toward its plus end (to simulate kinesin 5–mediated transport toward the MT plus end), but freeze as soon as both heads were bound to anti-parallel MT. When one of the heads was close to the MT plus end, it would also increase MT stability by decreasing its rate of catastrophe.
It is also important to note that we decided not to include branching nucleation, induced notably by the nucleator Augmin, which promotes spindle assembly by triggering nucleation parallel to template microtubules (Goshima et al., 2008; Petry et al., 2011). Indeed, based on observations from other female meiotic spindles, it seemed that Augmins are recruited only at later stages of spindle assembly, to stabilize and favor bipolarization (Meireles et al., 2009; Colombié et al., 2013; Watanabe et al., 2016).
Simulation space
For simplicity, simulations were performed in a two-dimensional setup, and we did not include an implicit description of the oocyte volume in the simulations. Microtubules were not spatially confined, but the other components (nucleators, motors, etc.) were confined into a circle of radius 20 µm. Indeed, initially the spindle was positioned in the cell center without interaction with the cell cortex, so we assumed that any interaction of MTs with the cell cortex could be neglected. Moreover, the confinement of the proteins inside the simulated space makes it possible to control the local availability of proteins around the spindle without having to consider their diffusion in the entire volume of the oocyte. Thus, the effect of increasing oocyte volume on spindle size could be simulated simply by decreasing the number of proteins in the simulation, mimicking the reduction of proteins that are locally active.
Configuration
All simulations were started by placing the DNA beads randomly in a central disk of radius 4 µm, surrounded by a circle of aMTOCS placed 6–9 µm from the center (Figure 1A; t = 0 s). All the entities mimicking kinesin motors or other proteins were added at the start of the simulation, but activation of Eg5 and HURP entities was delayed to account for the rate of their production and recruitment to the spindle. To reduce computational cost, the size of the system was greatly reduced, and the time simulated was also shortened. We simulated only 10 DNA beads and 15 aMTOCs able to generate 800 MTs, with thousands of associated entities, during 900 s. This permits qualitative conclusions on spindle assembly, while not being quantitatively accurate. Those values were varied in the Supplemental Materials to test the sensitivity of the simulations to those parameters.
Perturbation simulations
To assay the effect of inhibition or overexpression of some proteins, we modified the quantities of simulated corresponding entities (Table 1). To test the inhibition of a protein, we removed the corresponding entity entirely (total inhibition), and for overexpression, we doubled its quantity compared with that in the default configuration. Monastrol was simulated as total removal of Eg5 and a twofold reduction of HURP quantity, as HURP is recruited by kinesins to the central spindle (Breuer et al., 2010). The HSET mutant N593K was modeled by blocking the activity of the motor head (velocity set to 0), similarly to the experimental situation (Bennabi et al., 2018). Aurora inhibition was simulated by decreasing the number of nucleators (both aMTOCs and gamma-tubulin; Bury et al., 2017).
We also did not include perturbations of the Ran gradient (inhibition or overexpression; Brunet et al., 1998; Dumont et al., 2007; Cesario and McKim, 2011; Bury et al., 2017), as it affects many proteins in a direct or indirect manner. We did verify, however, that when MTs were only nucleated from aMTOCs, they were clustered in an aster at the center (Schuh and Ellenberg, 2007), arguing for the dominance of minus end–directed motors at this stage.
Analysis
To characterize spindle morphologies, we fitted an ellipse around the microtubules, first by determining its center as the isobarycenter of microtubule ends, and then by selecting the main axis as the direction with the maximal microtubule density projection. An ellipse length was then determined such that 80% of microtubule end projections on the main axis were inside the ellipse. The width was determined similarly so that 80% of microtubule end projections on the perpendicular axis were inside the ellipse. This construction of the ellipse made it possible to estimate the shape of the spindle, while not being too sensitive to the position of individual MTs. To quantify the position of one aMTOC or DNA bead relative to the spindle, that is, how far it was from the center, we defined a new ellipse, with the same aspect ratio and center point as the spindle ellipse, that contained the object of interest (aMTOC or DNA bead). We then calculated the relative position, d, as the ratio of the semimajor axis of this ellipse to the semimajor axis of the spindle ellipse (Figure 1C). For each spindle, the measured position d of aMTOCs or DNA beads was then the average ratio of all aMTOCs or DNA beads. With this definition, a measure that was close to 0 indicated that all objects were close to the center, while higher measures indicated that they were toward the periphery or outside of the spindle.
To differentiate between MT ball–shaped spindles, elongated spindles, and asters, we also measured the aspect ratio of the fitted ellipses as being the ratio between its width and its length. An aspect ratio of 1 indicates a circular spindle (MT ball), while smaller values describe more elongated spindles. In the simplified model, we fixed the threshold between an elongated spindle and a microtubule ball at 0.75, as elongation was quite low. In the more extended model, we could use a more adequate ratio of 0.5, reflecting that the length of the spindle was at least twice its width. Microtubule asters could have a wide range of aspect ratios (circular or elongated asters), so we termed as asters spindles in which all aMTOCs (and so the majority of MTs minus ends) were clustered in the center (aMTOCs position measures less than 0.5). Similarly, we termed as inverted asters the opposite kind of asters in which the DNA beads (and so the majority of MTs plus ends) were clustered in the center (DNA position measures less than 0.5).
To quantify the individualization of the chromosomes in the simulations, we took the average of the distances between all the beads at each time point (Figure 4A). This allowed us to quantify and compare whether DNA beads were together or instead spread apart at different regions of the spindle, regardless of the size of the spindle.
Code availability
Cytosim source code (C++) is available at https://github.com/nedelec/cytosim. We provide a typical configuration file in the Supplemental Materials to make it possible to reproduce the simulations.
Oocyte collection and culture
Oocytes were collected and cultured as described in Bennabi et al. (2018).
Immunofluorescence
After in vitro culture of oocytes, their zona pellucida was removed by incubation in Pronase for Prophase I oocytes or by incubation in acid Tyrode’s medium (pH 2.3) after NEBD. Oocytes were fixed for 10 min at –20°C in 100% methanol on coverslips treated with gelatin and polylysine (as described in Manil-Ségalen et al., 2018). Oocytes were left in phosphate-buffered saline (PBS) overnight at 4°C. After 30 min of blocking in 0.5% Triton X-100, 3% bovine serum albumin (BSA), antibody staining was performed in PBS, 0.5% Triton X-100, 3% BSA. As primary antibody, we used rabbit anti-Eg5 (Novus Biologicals; 1:500). As secondary antibody, we used Alexa-594–labeled anti-rabbit (Invitrogen; 1:500). DNA was stained with Prolong-DAPI (6-diamidino-2-phenylindole) (10 µg/ml final DAPI).
Imaging
Spinning disk images were acquired using a 60×/1.4NA objective on a Zeiss Axioobserver Z1 microscope equipped with a cMOS camera coupled to a Yokogawa CSU-W1 spinning disk. Metamorph Software (Universal Imaging) was used to collect data.
Supplementary Material
Acknowledgments
We thank the members of the Verlhac/Terret team for helpful discussions and Renata Basto for sharing Kinesin 5 antibody. This work was funded by the Agence Nationale de la Recherche (ANR) (ANR-16-CE13 to MET) and by Inca (PLBIO 2016-270-TRAN). This work has received support from the Fondation Bettencourt Schueller and under the program “Investissements d’Avenir” launched by the French Government and implemented by the ANR, with the references ANR-10-LABX-54 MEMO LIFE, ANR-11-IDEX-0001-02 PSL* Research University. F.N. was supported by the Centre for Modelling in the Biosciences ( https://www.bioms.de) and the European Molecular Biology Laboratory (EMBL).
Abbreviations used:
- aMTOC
acentriolar microtubule-organizing center
- AR
aspect ratio
- MAPs
microtubule-associated proteins
- MT
microtubule
- NEBD
nuclear envelope breakdown
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0644) on January 16, 2019.
REFERENCES
Boldface names denote co–first authors.
- Baumann C, Wang X, Yang L, Viveiros MM. (2017). Error-prone meiotic division and subfertility in mice with oocyte-conditional knockdown of pericentrin. J Cell Sci , 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennabi I, Terret M-E, Verlhac M-H. (2016). Meiotic spindle assembly and chromosome segregation in oocytes. J Cell Biol , 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennabi I, Quéguiner I, Kolano A, Boudier T, Mailly P, Verlhac M, Terret M. (2018). Shifting meiotic to mitotic spindle assembly in oocytes disrupts chromosome alignment. EMBO Rep e45225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. (2010). A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell , 420–432. [DOI] [PubMed] [Google Scholar]
- Braun M, Lansky Z, Szuba A, Schwarz FW, Mitra A, Gao M, Ludecke A, Rein ten Wolde P, Diez S. (2017). Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding. Nat Chem Biol , 1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer M, Kolano A, Kwon M, Li CC, Tsai TF, Pellman D, Brunet S, Verlhac MH. (2010). HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J Cell Biol , 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugués J, Nuzzo V, Mazur E, Needleman DJ. (2012). Nucleation and transport organize microtubules in metaphase spindles. Cell , 554–564. [DOI] [PubMed] [Google Scholar]
- Brunet S, Polanski Z, Verlhac MH, Kubiak JZ, Maro B. (1998). Bipolar meiotic spindle formation without chromatin. Curr Biol , 1231–1234. [DOI] [PubMed] [Google Scholar]
- Brunet S, Dumont J, Lee KW, Kinoshita K, Hikal P, Gruss OJ, Maro B, Verlhac M-H. (2008). Meiotic regulation of TPX2 protein levels governs cell cycle progression in mouse oocytes. PLoS One , e3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank KS, Mitchison TJ, Fisher DS. (2007). Slide-and-cluster models for spindle assembly. Curr Biol , 1373–1383. [DOI] [PubMed] [Google Scholar]
- Bury L, Coelho PA, Simeone A, Ferries S, Eyers CE, Eyers PA, Zernicka-Goetz M, Glover DM. (2017). Plk4 and Aurora A cooperate in the initiation of acentriolar spindle assembly in mammalian oocytes. J Cell Biol , 3571–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Weaver LN, Ems-McClung SC, Walczak CE. (2009). Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell , 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camlin NJ, McLaughlin EA, Holt JE. (2017). Kif4 is essential for mouse oocyte meiosis. PLoS One , e0170650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario J, McKim KS. (2011). RanGTP is required for meiotic spindle organization and the initiation of embryonic development in Drosophila. J Cell Sci , 3797–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, Schuh M. (2015). A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat Commun , 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombié N, Głuszek AA, Meireles AM, Ohkura H. (2013). Meiosis-specific stable binding of augmin to acentrosomal spindle poles promotes biased microtubule assembly in oocytes. PLoS Genet , e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA. (1998). Focusing on spindle poles. J Cell Sci , 1477–1481. [DOI] [PubMed] [Google Scholar]
- Craig EM, Dey S, Mogilner A. (2011). The emergence of sarcomeric, graded-polarity and spindle-like patterns in bundles of short cytoskeletal polymers and two opposite molecular motors. J Phys Condens Matter , 374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Ohkura H. (2001). Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat Cell Biol , 637–642. [DOI] [PubMed] [Google Scholar]
- Derr ND, Goodman BS, Jungmann R, Leschziner AE, Shih WM, Reck-Peterson SL. (2012). Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science , 662–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell , 69–78. [DOI] [PubMed] [Google Scholar]
- Dogterom M, Surrey T. (2013). Microtubule organization in vitro. Curr Opin Cell Biol , 23–29. [DOI] [PubMed] [Google Scholar]
- Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac MH. (2007). A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J Cell Biol , 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan T, Wakefield JG. (2011). 50 ways to build a spindle: the complexity of microtubule generation during mitosis. Chromosome Res , 321. [DOI] [PubMed] [Google Scholar]
- Ennomani H, Letort G, Guérin C, Martiel J-L, Cao W, Nédélec F, De La Cruz EM, Théry M, Blanchoin L. (2016). Architecture and connectivity govern actin network contractility. Curr Biol , 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzHarris G. (2009). A shift from kinesin 5-dependent metaphase spindle function during preimplantation development in mouse. Development , 2111–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz J, Kapoor TM. (2004). Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol , 465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL. (1992). Microtubule organization during maturation of Xenopus oocytes: assembly and rotation of the meiotic spindles. Dev Biol , 516–530. [DOI] [PubMed] [Google Scholar]
- Gard DL, Cha BJ, Roeder AD. (1995). F-actin is required for spindle anchoring and rotation in Xenopus oocytes: a re-examination of the effects of cytochalasin B on oocyte maturation. Zygote , 17–26. [DOI] [PubMed] [Google Scholar]
- Głuszek AA, Cullen CF, Li W, Battaglia RA, Radford SJ, Costa MF, McKim KS, Goshima G, Ohkura H. (2015). The microtubule catastrophe promoter Sentin delays stable kinetochore–microtubule attachment in oocytes. J Cell Biol , 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Nédélec F, Vale RD. (2005). Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol , 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. (2008). Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol , 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan G, Brangwynne CP, Jaensch S, Gharakhani J, Jülicher F, Hyman AA. (2010). Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr Biol , 353–358. [DOI] [PubMed] [Google Scholar]
- Groen AC, Coughlin M, Mitchison TJ. (2011). Microtubule assembly in meiotic extract requires glycogen. Mol Biol Cell , 3139–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. (1996). Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature , 420. [DOI] [PubMed] [Google Scholar]
- Hentrich C, Surrey T. (2010). Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J Cell Biol , 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepperla AJ, Willey PT, Coombes CE, Schuster BM, Gerami-Nejad M, McClellan M, Mukherjee S, Fox J, Winey M, Odde DJ, et al (2014). Minus-end directed kinesin-14 motors align anti-parallel microtubules to control metaphase spindle length. Dev Cell , 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert M, Kalleas D, Cooney D, Lamb M, Lister L. (2015). Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harb Perspect Biol , a017970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holubcová Z, Blayney M, Elder K, Schuh M. (2015). Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science , 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueschen CL, Kenny SJ, Xu K, Dumont S. (2017). NuMA recruits dynein activity to microtubule minus-ends at mitosis. ELife , 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. (2003). The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell , 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job D, Valiron O, Oakley B. (2003). Microtubule nucleation. Curr Opin Cell Biol , 111–117. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature , 114–118. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol , 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. (2001). The mitotic spindle: a self-made machine. Science , 543–548. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. (2000). Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol , 59–67. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. (2011). Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell , 568–581. [DOI] [PubMed] [Google Scholar]
- Kolano A, Brunet S, Silk AD, Cleveland DW, Verlhac M-H. (2012). Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc Natl Acad Sci USA , E1858–E1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. (2011). Microtubule nucleation by γ-tubulin complexes. Nat Rev Mol Cell Biol , 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Letort G, Pitayu L, Sallé J, Stefanutti M, Maton G, Ladouceur A-M, Canman JC, Maddox PS, Maddox AS, et al. (2018). Microtubule dynamics scale with cell size to set spindle length and assembly timing. Dev Cell , 496–511.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort G, Politi AZ, Ennomani H, Théry M, Nedelec F, Blanchoin L. (2015). Geometrical and mechanical properties control actin filament organization. PLoS Comput Biol , e1004245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort G, Nedelec F, Blanchoin L, Théry M. (2016). Centrosome centering and decentering by microtubule network rearrangement. Mol Biol Cell , 2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart A, Cross RA. (1996). Kinetics and motility of the Eg5 microtubule motor. Biochemistry , 2365–2373. [DOI] [PubMed] [Google Scholar]
- Loughlin R, Heald R, Nédélec F. (2010). A computational model predicts Xenopus meiotic spindle organization. J Cell Biol , 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Viveiros MM. (2014). Depletion of pericentrin in mouse oocytes disrupts microtubule organizing center function and meiotic spindle organization. Mol Reprod Dev , 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailhes JB, Mastromatteo C, Fuseler JW. (2004). Transient exposure to the Eg5 kinesin inhibitor monastrol leads to syntelic orientation of chromosomes and aneuploidy in mouse oocytes. Mutat Res , 153–167. [DOI] [PubMed] [Google Scholar]
- Manandhar G, Schatten H, Sutovsky P. (2005). Centrosome reduction during gametogenesis and its significance. Biol Reprod , 2–13. [DOI] [PubMed] [Google Scholar]
- Manil-Ségalen M, Łuksza M, Kanaan J, Marthiens V, Lane SIR, Jones KT, Terret M-E, Basto R, Verlhac M-H. (2018). Chromosome structural anomalies due to aberrant spindle forces exerted at gene editing sites in meiosis. J Cell Biol , 3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow FL. (2018). Recent advances in understanding oogenesis: interactions with the cytoskeleton, microtubule organization, and meiotic spindle assembly in oocytes. F1000Research , 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies HJ. (1996). Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J Cell Biol , 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles AM, Fisher KH, Colombié N, Wakefield JG, Ohkura H. (2009). Wac: a new augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol , 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihajlović AI, FitzHarris G. (2018). Segregating chromosomes in the mammalian oocyte. Curr Biol , R895–R907. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. (1984). Dynamic instability of microtubule growth. Nature , 237–242. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. (2005). Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell , 3064–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogessie B, Scheffler K, Schuh M. (2018). Assembly and positioning of the oocyte meiotic spindle. Annu Rev Cell Dev Biol , 6.1-6.23. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Wollman R, Civelekoglu-Scholey G, Scholey J. (2006). Modeling mitosis. Trends Cell Biol , 88–96. [DOI] [PubMed] [Google Scholar]
- Mountain V. (1999). The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol , 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. (2012). Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet , 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, FitzHarris G. (2017). Intrinsically defective microtubule dynamics contribute to age-related chromosome segregation errors in mouse oocyte meiosis-I. Curr Biol , 1040–1047. [DOI] [PubMed] [Google Scholar]
- Nedelec F, Foethke D. (2007). Collective Langevin dynamics of flexible cytoskeletal fibers. New J Phys , 427–427. [Google Scholar]
- Needleman DJ, Groen A, Ohi R, Maresca T, Mirny L, Mitchison T. (2010). Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: evidence that the spindle environment does not stabilize microtubules. Mol Biol Cell , 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Jung S, Singh P, Strothman CE, Erwin AL, Ohi MD, Zanic M, Ohi R. (2018). Microtubule minus-end aster organization is driven by processive HSET-tubulin clusters. Nat Commun , 2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriola D, Needleman DJ, Brugués J. (2018). The physics of the metaphase spindle. Annu Rev Biophys , 655–673. [DOI] [PubMed] [Google Scholar]
- Petry S, Pugieux C, Nedelec FJ, Vale RD. (2011). Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc Natl Acad Sci USA , 14473–14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. (2004). The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell , 187–202. [DOI] [PubMed] [Google Scholar]
- Schaffner SC, José JV. (2006). Biophysical model of self-organized spindle formation patterns without centrosomes and kinetochores. Proc Natl Acad Sci USA , 11166–11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. (2007). Self-Organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell , 484–498. [DOI] [PubMed] [Google Scholar]
- Srayko M, Buster DW, Bazirgan OA, McNally FJ, Mains PE. (2000). MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev , 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Surrey T, Nedelec F, Leibler S, Karsenti E. (2001). Physical properties determining self-organization of motors and microtubules. Science , 1167–1171. [DOI] [PubMed] [Google Scholar]
- Szöllösi D. (1976). Oocyte maturation and paternal contribution to the embryo in mammals. In: Developmental Biology and Pathology, ed. Gropp A, Benirschke K, Berlin/Heidelberg: Springer, 9–27. [DOI] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Block SM. (2006). Eg5 steps it up! Cell Div , 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. (1996). XKCM1: A Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell , 37–47. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol , 903–913. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Shioi G, Furuta Y, Goshima G. (2016). Intra-spindle microtubule assembly regulates clustering of microtubule-organizing centers during early mouse development. Cell Rep , 54–60. [DOI] [PubMed] [Google Scholar]
- Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A. (2005). Efficient chromosome capture requires a bias in the “search-and-capture” process during mitotic-spindle assembly. Curr Biol , 828–832. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kaido M, Kitajima TS. (2015). Inherent instability of correct kinetochore-microtubule attachments during meiosis I in oocytes. Dev Cell , 589–602. [DOI] [PubMed] [Google Scholar]
- Yu C, Ji S-Y, Sha Q-Q, Dang Y, Zhou J-J, Zhang Y-L, Liu Y, Wang Z-W, Hu B, Sun Q-Y, et al. (2016). BTG4 is a meiotic cell cycle–coupled maternal-zygotic-transition licensing factor in oocytes. Nat Struct Mol Biol , 387–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.