In human pathophysiology, the clash between microbial infection and host immunity contributes to multiple diseases. Cystic fibrosis (CF) is a classical example of this phenomenon, wherein a dysfunctional, hyperinflammatory immune response combined with chronic pulmonary infections wreak havoc upon the airway, leading to a disease course of substantial morbidity and shortened life span.
KEYWORDS: cystic fibrosis, Pseudomonas aeruginosa, airway, antimicrobial peptides, inflammation, innate immunity, lung infection, reactive oxygen species, ROS
SUMMARY
In human pathophysiology, the clash between microbial infection and host immunity contributes to multiple diseases. Cystic fibrosis (CF) is a classical example of this phenomenon, wherein a dysfunctional, hyperinflammatory immune response combined with chronic pulmonary infections wreak havoc upon the airway, leading to a disease course of substantial morbidity and shortened life span. Pseudomonas aeruginosa is an opportunistic pathogen that commonly infects the CF lung, promoting an accelerated decline of pulmonary function. Importantly, P. aeruginosa exhibits significant resistance to innate immune effectors and to antibiotics, in part, by expressing specific virulence factors (e.g., antioxidants and exopolysaccharides) and by acquiring adaptive mutations during chronic infection. In an effort to review our current understanding of the host-pathogen interface driving CF pulmonary disease, we discuss (i) the progression of disease within the primitive CF lung, specifically focusing on the role of host versus bacterial factors; (ii) critical, neutrophil-derived innate immune effectors that are implicated in CF pulmonary disease, including reactive oxygen species (ROS) and antimicrobial peptides (e.g., LL-37); (iii) P. aeruginosa virulence factors and adaptive mutations that enable evasion of the host response; and (iv) ongoing work examining the distribution and colocalization of host and bacterial factors within distinct anatomical niches of the CF lung.
INTRODUCTION
Cystic fibrosis (CF) remains one of the most prevalent, life-shortening genetic diseases in the Caucasian population (1). According to the Cystic Fibrosis Foundation, there are approximately 30,000 people living with CF in the United States and upwards of 70,000 worldwide (2). Incidence varies by country but remains approximately 1 in 3,000 live births in Caucasians in both North America and Europe (3, 4). In the 1950s, CF was an exclusively pediatric disease, as the vast majority of patients did not live beyond infancy (5). However, by the 1990s, life expectancy had improved to approximately 20 years (2). Today, with further advancement in early diagnostic and management strategies, the median life expectancy of a CF patient is 29.6 years in the United States and 31 years in the United Kingdom (2, 6). Moreover, the median predicted survival (i.e., the age beyond which 50% of CF patients born today are expected to live) is 47 years in both the United States and United Kingdom, resulting in an increasingly higher proportion of adult patients (2, 6). Nevertheless, CF patients continue to suffer profound morbidity throughout their lifetimes, necessitating steadfast research efforts to better understand CF pathophysiology and to spur development of novel therapeutics.
CF is caused by a mutation in a gene that encodes the cystic fibrosis transmembrane conductance regulator (CFTR), a chloride and bicarbonate ion transport channel that maintains osmotic balance across multiple epithelial surfaces in the body (7). CFTR mutations are inherited in an autosomal recessive pattern, and approximately 2,000 disease-causing mutations have been identified thus far; these mutations have been subsequently categorized into six classes based on the manner in which CFTR function is perturbed (5, 8). Deletion of a phenylalanine at position 508 (Phe508del or ΔF508) remains the most common mutation and is represented in over two-thirds of CF patients (9). Ultimately, CFTR dysfunction causes an inability to regulate chloride and bicarbonate ion transport across epithelia, resulting in multiple-organ dysfunction, wherein the major clinical manifestations of CF include high chloride content of sweat (which is the basis of the gold-standard diagnostic technique for the disease [i.e., sweat chloride test]), meconium ileus, distal intestinal obstruction, exocrine pancreatic insufficiency, growth disturbance, male infertility, and chronic and recurrent pulmonary infections due to opportunistic pathogens (10–12). Long-term complications of CF also include diabetes and mood disorders, such as depression and anxiety; unfortunately, pulmonary failure remains an inevitable fate for nearly all CF patients (13, 14).
PATHOPHYSIOLOGY OF CF LUNG DISEASE: THE HOST-PATHOGEN INTERFACE
Overview
Obstructive pulmonary disease is the substantial determinant of morbidity and the leading cause of mortality for CF patients (15). There are multiple working hypotheses to explain early events leading to the development of CF lung pathology, and each of these postulates lies at the interface of host-pathogen interactions. Broadly, deficits in the immune response due to CFTR dysfunction result in a predisposition to acute and, ultimately, chronic lung infections with opportunistic pathogens (16). Subsequently, a combination of inflammatory host immune products and bacterial virulence factors drives progressive lung damage throughout a CF patient’s lifetime, resulting in respiratory failure, candidacy for lung transplantation, and, eventually, death (7).
Understanding the early events of CF disease development is essential, as CF patients can display signs of permanent changes to lung architecture within the first 5 years of life (5). By the age of 3 years, CF patients often already show air trapping, mucus obstruction, and bronchiectasis (i.e., enduring enlargement of airways) by computed tomography (CT) scanning; elevated levels of inflammatory markers (e.g., neutrophil elastase [NE], which is described in detail below); as well as polymicrobial infection of the lung with pathogens that persist into late stages of disease (e.g., Staphylococcus aureus and Pseudomonas aeruginosa) (17–20).
In the laboratory, many insights about early disease pathophysiology have been derived from both in vitro and in vivo studies. Wherein the latter have been augmented by the relatively recent development of the CF ferret and pig models (21–23), most animal work within the field has continued, primarily in mouse models (24). Additionally, translational work using bronchoalveolar lavage (BAL) fluid and immune cells obtained directly from CF patients has added significantly to the field’s understanding of the host-microbe interface but, at times, also fueled controversy regarding the role of primary immune dysfunction in CF (25).
Defects in Host Immunity
The most common and generally well-accepted paradigm regarding the development of CF lung disease is the “low-volume hypothesis” (1, 7, 26). CFTR dysfunction results in an inability to secrete chloride and bicarbonate ions into the airway lumen, which normally balances sodium reabsorption via a different channel, the epithelial Na+ channel (ENac); unopposed sodium reabsorption results in net water uptake by the respiratory epithelium, resulting in dehydration of the airway surface liquid (ASL) (27, 28). The ASL has multiple functions, but chief among these roles is hydration of mucus, a key component of the innate immune response as part of the mucociliary ladder (29). Dehydrated mucus ultimately compromises mucociliary and cough clearance of mucus, providing a nidus for colonization and infection by opportunistic pathogens (30).
Relatedly, a second hypothesis for the development of CF lung disease pertains to the altered pH of ASL. Diminished functionality of CFTR reduces bicarbonate secretion into the airway lumen, resulting in decreased pH of the ASL; indeed, some studies have shown that ASL from CF patients is more acidic than that of healthy patients (31–33). The more acidic ASL within the CF lung has multiple consequences. First, CF ASL (derived from the pig model of disease, which recapitulates acidic airway secretions better than the mouse model) exhibits reduced bacterial killing due to compromised function of cationic antimicrobial peptides (AMPs) (34, 35). AMPs are small innate immune proteins, present within epithelial and leukocyte secretions, with broad antimicrobial activity against bacterial and viral pathogens as well as immunomodulatory functions (36). The microbial killing activities of AMPs present within the CF airway, including human β-defensin-3 (hBD-3) as well as LL-37, are reduced under acidic pH conditions (37, 38). AMPs are also further discussed in greater detail below (see “LL-37, an Antimicrobial Peptide with Immunomodulatory Actions”). Second, in the CF pig model, independent of ASL volume, altered pH of ASL also causes mucus tethering and impaired mucus detachment from the lung epithelium (39). This effect also promotes mucus plugging and reduced mucociliary clearance, but the primary mechanism here is the acidic ASL pH rather than ASL dehydration (35, 39). Finally, CF ASL activates proteases, which can directly damage lung tissue and degrade innate immune effectors (40, 41).
Various studies also suggest that primary dysregulation of the immune system (i.e., due to abrogated CFTR function) contributes to CF lung disease, although this theory remains somewhat controversial; much of the disagreement appears to focus upon whether intrinsic immune defects promote a hyperinflammatory microenvironment within the CF lung or if bacterial infection represents the first event that incites early inflammation within the CF lung (42, 43).
Indeed, there are multiple lines of evidence that support a predilection toward hyperinflammation within CF tissues, independent of bacterial infection. Studies have shown elevated concentrations of proinflammatory markers in the cell-free supernatants of CF epithelial cell cultures and in ex vivo CF tissue specimens that are free of infection (compared to healthy controls) (44–47). Research using CF mouse and ferret models demonstrates that newborn animals with CFTR mutations already have inflammation of the lung in the absence of detectable bacteria and fungi; the possibility of early viral infection, however, was not excluded in this work (23, 48, 49). Evidence of inflammation included early neutrophil and macrophage infiltration into the naive mouse lung, whereas elevated concentrations of proinflammatory cytokines (tumor necrosis factor alpha [TNF-α] and interleukin-8 [IL-8]) were observed within BAL fluid of newborn ferrets with CF (compared to healthy animals) (23, 48, 49).
Congruent with these findings in animal models of CF lung disease, BAL fluid taken from 4-week-old CF patients who were culture negative for common CF pathogens contains higher concentrations of relevant proinflammatory cytokines (e.g., IL-6, IL-8, TNF-α, and arachidonic acid derivatives) than in BAL fluid from healthy controls (1, 24, 50–52). CF patients also show reduced concentrations of anti-inflammatory mediators (e.g., IL-10, lipoxin, and docosahexaenoic acid) within BAL fluid compared to healthy patients (1, 24, 50–52). The concentration of glutathione, a critical antioxidant, is reduced within CF airways, whereas CF immune cells overproduce reactive oxygen species (ROS), thereby predisposing patients to greater tissue damage due to oxidative stress (53–55). Given these findings, a critical balance between proinflammatory/anti-inflammatory mediators and oxidant/antioxidant molecules is dysregulated within the CF lung at baseline.
There are multiple proposed mechanisms for how basal inflammation in CF promotes subsequent bacterial infection. One potential explanation is that excess production of proinflammatory cytokines within the airway is responsible for robust, early recruitment of inflammatory cells, specifically neutrophils (further discussed in detail below) (56). Activated neutrophils release multiple products that drive CF lung pathology and inadvertently predispose patients to infection, including serine proteases (e.g., neutrophil elastase) (57). Neutrophil proteases can degrade vital innate immune antimicrobials, including AMPs such as defensins, and even components of the complement system, thus contributing to a secondary vulnerability to bacterial infection (41, 58).
Altered Effector Functions of Immune Cells: the Pathological Role of Neutrophils in the CF Lung
Intrinsically diminished or altered effector functions of both innate and adaptive immune cells in CF patients may also contribute to reduced clearance of opportunistic pathogens and early damage to CF lung tissue (59). Neutrophils normally represent the critical frontline innate immune response to infection: activation of neutrophils via stimulation of their pathogen recognition receptors (PRRs) results in these cells employing versatile effector functions, including phagocytosis, degranulation, and neutrophil extracellular trap (NET) formation (NETosis), a form of programmed cell death that results in the release of DNA and other intracellular contents, to respond to intruders (59, 60). Neutrophil granules contain a wide variety of antimicrobials, including AMPs such as cathelicidins (e.g., LL-37), serine proteases, and enzymes such as myeloperoxidase (MPO), which aid in the synthesis of chlorine-containing ROS (e.g., hypochlorous acid [HOCl]) (61, 62). Neutrophil granular constituents (which can be deployed intracellularly within phagosomes or released extracellularly via degranulation) normally function to clear infection and modulate the immune response (61, 62). However, under chronic inflammatory conditions and when produced in excess, many of these same compounds can have directly toxic effects on host tissues (63).
In CF, neutrophils play a primary role in driving inflammation and tissue damage within the lung (64). Excessive influx of neutrophils into the CF lung (without the concomitant capacity to clear infection) is a histopathological hallmark of the disease; in fact, neutrophils represent the most abundant immune cells within CF lungs (65–67). The presence of neutrophilic inflammation has been noted in a study of CF fetal tissue, suggesting that these immune cells may be recruited early and even influence the prenatal, sterile CF lung environment (68). Under in vitro conditions (in the absence of bacteria), with buffer control or lipopolysaccharide (LPS) stimulation, airway- and blood-derived neutrophils from children with CF secrete significantly more IL-8 than neutrophils from children with non-CF lung disease; given that IL-8 is a potent neutrophil chemokine, CF neutrophils likely contribute to their own abundant recruitment to the lung (69). Moreover, early elevation of IL-8 concentrations within the CF airway likely serves not only to recruit neutrophils but also to promote degranulation and production of ROS that can have deleterious consequences for lung tissue (as discussed below) (59).
Another critical factor which may promote profuse neutrophil migration to the CF lung is high levels of circulating selectins (compared to those in healthy patients); E- and P-selectins mediate the first step of transendothelial migration of neutrophils from circulation into the lung (70). Importantly, although elevated levels of cytokines and circulating selectins may mediate early recruitment of neutrophils to the CF lung, the presence of these cells within the lung throughout the course of disease is likely sustained by acute and chronic bacterial infection (i.e., via the release of pathogen-associated molecular patterns [PAMPs], which can directly stimulate inflammatory responses) (71).
A further complication is that CF neutrophils demonstrate reduced apoptosis (likely secondary to high levels of granulocyte-macrophage colony-stimulating factor [GM-CSF] and reduced levels of IL-10 within the airway) and release more damaging proteolytic compounds than healthy neutrophils, including neutrophil elastase (NE), a highly reliable prognostic indicator in CF patients (72–74). Levels of NE correlate strongly with a more rapid decline of pulmonary function as determined by pulmonary function tests (PFTs) as well as the development of bronchiectasis, the abnormal enlargement of airways that is a key manifestation of obstructive lung disease in CF (17, 18, 75). Intracellular NE breaks down phagocytosed proteins (e.g., bacteria and bacterial products) but, when released into the extracellular milieu, can also directly degrade lung connective tissue (e.g., collagen and elastin) in addition to innate immune effectors, such as complement proteins, as described above (64, 70). NE also directly stimulates the airway epithelium to release IL-8 (76).
Additionally, neutrophils undergo significant NET formation or NETosis within the CF airway, a type of programmed cell death that results in the extracellular release of chromatin (i.e., DNA and histones) as well granular antimicrobial contents (e.g., NE and MPO) (77, 78). NETosis can be stimulated by cytokines (e.g., IL-8 and TNF-α) and/or microbial infection (e.g., bacterial PAMPs such as LPS), and the ultimate manifestation of this process is thought to be a final, “frustrated,” suicidal attempt by neutrophils to clear an infection that has otherwise proven to be resistant to phagocytosis (79). While the primary objective of NETosis in killing bacteria and clearing bacterial infections fails within the context of CF, due to the adaptable and diverse abilities of CF pathogens to evade the host response, secondary, harmful consequences of NETosis on host tissues likely occur (77, 79). For instance, the presence of abundant free DNA within the CF airway is associated with obstruction (i.e., more viscous airway secretions) and decline of pulmonary function (80, 81); accordingly, treatment of CF patients with dornase alpha (DNase) reduces DNA recovery from airway specimens as well as the rate of pulmonary function loss (82, 83). Levels of other NET-associated products, including NE and MPO, have also been associated with poor outcomes (17, 18, 75, 84). A recent study mechanistically linked greater production of NETs by CF neutrophils (than by healthy neutrophils) to reduced apoptosis/longer life spans of these cells (85).
Some have argued that compared to healthy neutrophils, CF neutrophils exhibit other aberrant effector functions, including reduced phagocytosis and degranulation as well as impaired chlorination (i.e., ROS production) of the phagolysosome (86–88). These CFTR-dependent defects in neutrophil function ultimately result in abrogated bacterial killing. While the presence of CFTR in neutrophils was once the subject of debate, the chloride channel has now been localized to phagolysosomes of neutrophils, and its dysfunction has been directly implicated in reduced chlorination of phagocytosed bacterial products (88). In another elegant study, CFTR dysfunction was shown to disrupt sodium and chloride flux within the neutrophil, resulting in reduced degranulation of secondary and tertiary granules (87). Notably, neutrophils derived from patients treated with the novel CFTR potentiator ivacaftor (which improves chloride flux through CFTR in patients with a specific mutation, G551D) showed partial correction of the degranulation defect and improvement in bacterial killing; these data suggest that CFTR dysfunction is directly tied to reduced degranulation of neutrophils (87). In part, this set of work explains why, despite an excessive influx of neutrophils into the CF lung, the primary objectives of surveillance and clearance of pathogens by these cells remain unfulfilled.
Relevant to this discussion, the primary impairment of neutrophil effector functions in CF is not completely established and remains an area of controversy (59). Some studies have directly contradicted the above-mentioned evidence, suggesting instead that CF neutrophils are not deficient in phagocytosis, the production of ROS, or even NET formation (which is an oxidative-burst-dependent process) (59, 89, 90). In other disorders with primary dysfunction of neutrophils, such as chronic granulomatous disease (CGD), there is an expected and observed predilection for invasive, systemic infections; in contrast, in CF, infecting bacterial populations remain localized to the lung, rarely causing disseminated infections (25). This clinical observation suggests that any defects of neutrophil function in CF are likely to be less overt than in other diseases.
Importantly, although neutrophils are understood to be key players in CF lung disease, altered effector functions of CF macrophages and even adaptive immune cells, such as T cells, illustrate that primary immune dysfunction in CF encompasses multiple components of the host defense apparatus (91–93). For instance, autophagy, which is a critical homeostatic and innate immune mechanism, is impaired in epithelial cells and macrophages derived from CF patients (94). In healthy cells, autophagy is induced upon starvation and results in the encapsulation of cytosolic contents within double-membrane compartments known as autophagosomes (95). Autophagosomes subsequently fuse with lysosomes, resulting in the enzymatic degradation of their cargo into substrates for central metabolism (e.g., free amino and fatty acids) (95). Importantly, autophagy also recycles damaged mitochondria (i.e., a source of endogenous oxidative stress), removes aggregated/nonfunctional proteins, and clears phagocytosed and/or intracellular pathogens as well (94–96). Dysfunctional autophagy has been implicated in various diseases in human pathophysiology, including CF (97).
Although the nexus between impairment of CFTR function and abrogated autophagy remains unclear, there are several working hypotheses, which include the following: (i) proteins of the autophagic machinery are sequestered within cytosolic aggregates consisting of misfolded CFTR proteins, and (ii) CF cells exhibit an accumulation of certain microRNAs, which specifically downregulate the expression of genes encoding autophagy proteins (94). Ultimately, dysregulated autophagy in CF results in an increased production of inflammatory cytokines, greater endogenous ROS production, as well as a reduced capacity to clear bacterial infection (as demonstrated in CF macrophages in vitro and CF mouse models in vivo) (94–96). As such, perturbation of autophagy represents another example of a CFTR-intrinsic “hit” to innate immune defenses, promoting both immunopathology and a predisposition to opportunistic infection.
In brief summary, within the hyperinflammatory milieu of the CF lung, the secretion of various neutrophil-derived antimicrobials, including the above-mentioned proteases (e.g., neutrophil elastase), cationic peptides, extracellular DNA (eDNA), and ROS, likely contributes to the progression and exacerbation of disease. Many of these neutrophil effectors are released in an unsuccessful attempt to clear bacterial infection but have secondary, inadvertently damaging effects on host tissues. In the following section, we focus on two neutrophil antimicrobials that are implicated in CF lung disease: LL-37, a cationic AMP, and reactive oxygen species, including hydrogen peroxide (H2O2) and hypochlorous acid (HOCl).
LL-37, an Antimicrobial Peptide with Bactericidal and Immunomodulatory Actions
AMPs are defined as gene-encoded small peptides with antimicrobial activity; this definition specifically excludes innate immune proteins with hydrolytic or enzymatic activity (e.g., lysozyme and chitinases) (98). These small peptides, which have been conserved across eons of natural selection and across diverse taxa of organisms, play a vital role in the innate defense against microbial infection; multiple AMPs exhibit broad killing capacity against an assortment of microbes, including Gram-positive/Gram-negative bacteria, viruses, fungi, and protozoa (99). AMPs have been identified and studied in multiple plant, insect, and mammalian species: in plants and insects especially, AMPs are thought to play an essential role, given the lack of a complex adaptive immune system in these organisms (100). Various forms of AMPs have also been described in animals, in both mammalian and nonmammalian species (99).
Importantly, although bacteria are an often-studied target of AMPs that are produced by more-complex organisms, bacteria are also known to produce their own AMPs, known as bacteriocins (e.g., pyocins of Pseudomonas aeruginosa) (101, 102). Bacteria that express bacteriocins are able to kill and outcompete susceptible members of their own (or very closely related) species; as such, bacteriocins are tools for intraspecies competition within an ecological niche for limiting resources, e.g., space and nutrients, etc. (101, 102). The pyocins of P. aeruginosa may have additional functionality as well. The R/F-pyocin cluster of P. aeruginosa has recently been implicated in bacterial autolysis and eDNA release, which is important for bacterial adherence, biofilm formation, and antibiotic resistance (see below for further discussion of the lys-encoded endolysin) (103).
Human AMPs represent crucial components of the body’s innate immune or rapid response to pathogens. These peptides are largely synthesized and secreted by the epithelia of multiple organ systems (e.g., skin, respiratory, ocular, and gastrointestinal epithelia, etc.) as well as by phagocytic cells, including neutrophils, wherein they are stored within cytoplasmic granules (104). The classification system for AMPs is loosely based on the amino acid composition and secondary structure of these peptides; for example, peptides may be classified by whether they have a linear, alpha-helical structure; the presence of disulfide bonds that maintain beta sheet conformation; or an overrepresentation of a single amino acid (99, 105). Nonetheless, there are some structural and charge commonalities among human AMPs: these peptides are typically cationic (net positive charge) with amphipathic regions (i.e., composed of both hydrophobic and hydrophilic amino acids), which aid in binding or embedding within the negatively charged membranes of pathogens (99). AMPs are also typically synthesized as propeptides, with a highly conserved N-terminal region (often containing a signal peptide for trafficking to the endoplasmic reticulum for secretion) and a variable C-terminal region that exhibits antimicrobial activity; the propeptides are often degraded by endogenous proteases into their final form prior to secretion (106).
Additionally, there are several proposed mechanisms for the antimicrobial activity of AMPs. Two commonly cited models for bacterial killing are the toroidal pore-forming model and the carpet model (107). Both mechanisms rely on the binding of cationic, amphipathic peptides to predominantly negatively charged membranes of bacteria, leading to membrane disruption, loss of osmotic balance, and cell lysis (99). The toroidal pore-forming model is initiated by aggregation of AMPs at the membrane, induction of membrane curvature, and subsequent formation of pores (with the peptides and hydrophilic phosphate heads of lipids within the membranes forming the channels of the pores); the pores promote leakage of intracellular contents and, ultimately, membrane rupture (108, 109). The carpet model manifests as significant local accumulation of AMPs (“carpeting”) on the membrane, followed by disruption of the membrane in a detergent-like manner; the bacterial membrane disintegrates into micellar structures, covered with the peptides (107, 110). There is additional evidence that at sublethal concentrations, AMPs can traverse bacterial membranes and have toxic intracellular effects by disrupting macromolecular synthesis (111, 112).
To date, three main classes of AMPs have been described in humans: defensins, histatins, and LL-37, the only known human cathelicidin (113, 114). Defensins, which exhibit a beta sheet structure, cross-linked by disulfide bridges, are broken up into two subfamilies: α-defensins (including HNP1 to -4, HD5, and HD6) and β-defensins (hBD1 to -4) (114). The α-defensins HNP1 to -4 are predominantly synthesized and stored in neutrophil azurophilic granules, whereas HD5 and HD6 are secreted by the gastrointestinal epithelium (115, 116). β-Defensins (hBD1 to -4) are largely synthesized and secreted by the epithelia of multiple organs (e.g., respiratory, skin, gastrointestinal, and ocular epithelia) in response to infectious or inflammatory stimuli (115, 116). The histatins are histidine-rich peptides present in human saliva with significant antimicrobial activity against fungal species (117).
Although a detailed discussion of the roles of defensins and histatins in human biology is out of scope here, there are a few salient aspects of the regulation and importance of defensins in innate immunity that are worth mentioning in the context of CF. The α-defensins (HNP1 to -4) are constitutively expressed, released from neutrophils upon degranulation, and found in high concentrations under infectious/inflammatory conditions (113); within CF airway specimens, the concentrations of HNP1 to -3 are high enough not only to kill pathogens but also to be cytotoxic to mammalian cells (118). The release of β-defensins from the epithelium (specifically, hBD2 to -4) is induced by infectious and inflammatory (cytokine) stimuli, and the role of hBD3 in the early pathophysiology of CF lung disease, i.e., its presence and reduced antimicrobial efficacy in acidic ASL, has already been discussed above (37). The efficacy and expression of hBD1 and hBD2 are reduced within the CF airway, creating a vulnerability to opportunistic pathogens such as P. aeruginosa (119–121). Deletion or depletion of defensins in mouse models of other diseases (outside the context of CF) has also established the importance of these AMPs in protection from bacterial infection (98). Finally, apart from their antimicrobial functionality, defensins (like other AMPs) also have immunomodulatory actions, implicated in both pro- and anti-inflammatory contexts (122).
The cationic AMP that has been the focus of recent studies by our group is LL-37/hCAP18, the only known human cathelicidin. All cathelicidins, which have been identified in numerous mammalian species, have common structural hallmarks: each has a highly conserved N-terminal domain, which is composed of a signal sequence and a conserved region, termed “cathelin” (cathepsin L inhibitor), and a C-terminal domain, which is released after peptide cleavage, can be of various lengths, and exhibits the antimicrobial activity of the peptide (117). The gene encoding LL-37/hCAP18 is located on chromosome 3, in close proximity to genes encoding MyD88 and Toll-like receptor 9 (TLR9), suggesting potential coregulation of LL-37 expression with genes involved in proinflammatory responses (36). This gene directly encodes the precursor peptide hCAP18, which is subsequently cleaved to release the C-terminal AMP LL-37, so named because it consists of 37 amino acids, beginning with two leucine residues; cleavage occurs immediately before release from neutrophil granules, likely via an endogenous serine protease, proteinase-3 (123). Although LL-37 was first localized within neutrophil-specific granules as well as epithelia (e.g., respiratory and skin epithelia), it has now been found to originate from other immune cells as well, including monocytes, lymphocytes, and NK cells (36, 124).
LL-37 synthesis is induced by proinflammatory cytokines (e.g., TNF-α and IL-6), explaining its abundant presence at sites of infection and inflammatory responses (36, 125). The LL-37-encoding gene also contains vitamin D-responsive elements, theorized to connect the role of LL-37 to autoimmunity (which is correlated with vitamin D deficiency and predisposition to infections) (126); this finding also points to the role of LL-37 in modulation of immune responses (discussed further below).
As an antimicrobial, LL-37 is broadly efficacious against Gram-positive and Gram-negative bacteria, including CF-relevant and antibiotic-resistant pathogens (127); in vitro studies have also demonstrated the efficacy of LL-37 in inhibiting P. aeruginosa biofilm formation (128). Furthermore, LL-37 synergizes with other host-derived airway antimicrobials in killing bacteria (e.g., defensins and lysozyme) (35, 124, 129, 130). There are multiple posited mechanisms by which LL-37 directly kills bacterial cells: there is evidence in the literature for the carpet model of bacterial killing (131) as well as for a modified, toroidal pore-forming model (132, 133), both of which compromise bacterial membrane integrity, resulting in lysis. There is additional evidence that LL-37 preferentially attacks septating (actively growing/dividing) bacterial cells, thereby inhibiting bacterial proliferation (134).
Apart from its role as an antimicrobial (and similar to other AMPs discussed above), LL-37 is also known to modulate the immune response. There are multiple known proinflammatory actions of LL-37. LL-37 treatment of epithelial cells results in the release of IL-8, likely contributing to neutrophil recruitment and subsequent inflammation (135). LL-37 also contributes to the recruitment of immune cells to sites of inflammation by directly binding to a common receptor on neutrophils, monocytes, and T cells called formyl peptide receptor-like 1 (FPRL1) (136). Furthermore, LL-37 directly stimulates neutrophils to release ROS, α-defensins, and IL-8 (137). Conversely, there are multiple anti-inflammatory roles of LL-37 as well. The most well studied is the capacity of LL-37 to bind LPS and thereby reduce LPS-stimulated proinflammatory cytokine secretion and ROS production by macrophages in vitro (138). By extension, the LPS-binding ability of LL-37 reduces mortality in a mouse model of endotoxic shock (i.e., systemic LPS injection coupled with LL-37 delivery) (139).
LL-37 is known to be overproduced within the CF airway: LL-37 has been quantitated within CF airway secretions at high, bactericidal levels, with concentrations of up to 30 µg/ml (119, 140). Studies have found that the LL-37 concentration in CF correlates with the level of inflammation (total cell number and total number of neutrophils present within BAL fluid) (119). However, despite the high concentration of LL-37 within CF patient BAL fluid, LL-37 bactericidal activity may be affected by proteolysis (due to neutrophil elastase and cathepsin D) and/or complexation of the peptide with polyanionic glycosaminoglycans (e.g., heparan sulfate and hyaluronic acid) within the airway (140). In addition to inhibiting the activity of hBD3, low pH has been shown to inhibit the antimicrobial activity of LL-37 and the synergism between LL-37 and other airway antimicrobials (37, 129); given that CF ASL pH tends to be acidic, this suggests that the antimicrobial activity of LL-37 may be inhibited by the airway pH in CF patients, compromising a key component of innate immune defense.
On a structural level, this inhibition is thought to be because the alpha-helicity of LL-37, which is directly correlated with its antimicrobial activity, is reduced under acidic conditions (38). However, this effect on LL-37 structure occurs at pH <5, and CF airway pH, based on the above-mentioned pig model and human studies, seldom reaches this threshold (32, 34). Furthermore, other work in the field, including a recent study by Schultz et al. employing a novel fiber-optic pH probe, showed no difference in the pH of ASL derived from healthy and CF children (141, 142). These results suggest that the impact of reduced pH on antimicrobials in CF lungs and perhaps the role of acidic pH in early pathogenesis of CF lung disease, a critical finding from fundamental studies in the pig model, remain a source of some debate (143). These conflicting findings could be explained by differences in the ages of CF patients enrolled in each study. Whereas CF ASL acidity was demonstrated using specimens from human neonates (or newborn pigs), Shultz et al. and others found no pH differences in ASL from older children or adults with CF compared to healthy donors (33, 34, 141, 142). Thus, it is possible that while the very primitive CF airway represents an acidic environment with a corresponding, early compromise of innate immunity (including AMPs such as LL-37), pH perturbation may be abolished with age, wherein other mechanisms would subsequently explain immune dysfunction later in life.
Regardless, the importance of LL-37 in host defense against CF-relevant pathogens has been demonstrated in mouse models of disease. Overexpression of LL-37 within the airway via an adenovirus (AdV) vector reduced the P. aeruginosa burden and the resultant proinflammatory response after bacterial challenge (139). Additionally, in a CF mouse tracheal xenograft model, overexpression of LL-37 within CF xenografts restored killing of P. aeruginosa and S. aureus by ASL derived from the tracheal grafts (compared to ASL derived from healthy grafts) (144). Outside the context of CF, expression of the mouse analog of LL-37, CRAMP (cathelicidin-related antimicrobial peptide), is protective against invasive skin infections due to Streptococcus pyogenes, demonstrating the versatility and relevance of the peptide in innate defense against multiple infectious diseases (145).
Notably, while LL-37 may be present at bactericidal concentrations within the CF airway, bacteria have developed tools to evade AMP stress. AMPs were thought to attack a critical, conserved vulnerability, often called an “Achilles’ heel,” of prokaryotic species (i.e., the negatively charged outer leaflet of their cell membranes), thereby explaining broad efficacy against Gram-positive and Gram-negative species and making it unlikely that bacteria might develop resistance against AMPs (132). This theory has changed over time as numerous bacterial mechanisms of resistance against AMPs have been elucidated (146). These mechanisms include the bacterial capacity to alter the membrane charge (thereby reducing AMP binding) and the expression of AMP efflux systems (146). Moreover, LL-37 at sublethal concentrations has been shown to promote bacterial mutagenesis, thereby directly contributing to bacterial pathoadaptation within the host (147, 148). Here, we further explore bacterial defense against AMPs (and other innate antimicrobials) as well as the contribution of these antimicrobials to bacterial mutagenesis in the context of P. aeruginosa mucoid conversion (see below).
Reactive Oxygen Species
Reactive oxygen species (ROS) are a vital component of the oxygen-dependent microbial killing function of phagocytic cells such as neutrophils. Unfortunately, while the intended action of ROS production is clearance of bacteria (and other microbes), ROS can nonspecifically target host tissues and thus can also play a pathophysiological role in settings of chronic inflammation, including within the CF airway.
Endogenous sources of ROS include aerobic respiration (i.e., oxidative phosphorylation) in mitochondria. Electrons from the electron transport chain (ETC) can directly reduce oxygen (O2) to superoxide (O2·−), which can then undergo dismutation via superoxide dismutase (SOD) to produce hydrogen peroxide (H2O2) (149). Typically, the very low concentration of superoxide and peroxide produced via the ETC can be sufficiently detoxified via intracellular antioxidant enzymes, such as SOD, catalase, and glutathione peroxidase, and therefore do not play a pathophysiological role in most diseases (149). However, a more significant source of ROS within eukaryotic species is the oxidative/respiratory-burst response of phagocytes (150). Neutrophils have been prototypically studied to understand the origins of ROS as host-derived antimicrobials, although eosinophils and monocytes/macrophages also represent important sources of ROS within the innate immune apparatus of the host (29). Although not discussed here, via a mechanism different from ROS production, phagocytes also generate reactive nitrogen species (RNS), which are also potent free-radical antimicrobials (151).
The importance and mechanism of NADPH oxidase-dependent ROS generation within neutrophils are well established. While one isoform of the NADPH oxidase, Nox2, is present in neutrophils (and other phagocytes), other homologs of this system are expressed more broadly across tissues in the body, including Duox1 and Duox2, which are found on the respiratory epithelium (Duox1 and -2 are discussed below in the context of the CF lung) (152–154).
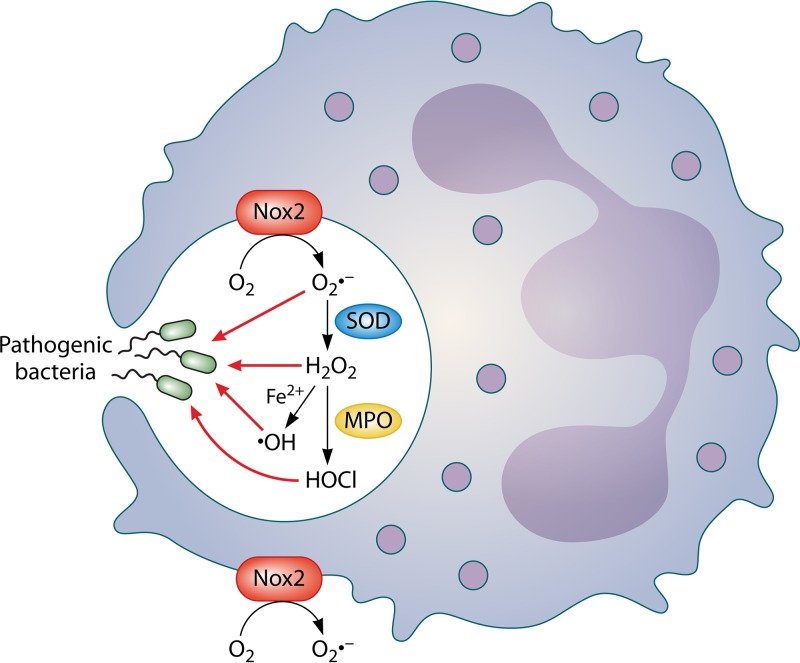
Nox2, a multiprotein complex, includes membrane-associated proteins (gp91phox and p22phox) as well as cytosolic protein components (p67phox, p47phox, p40phox, and RacGTP) (155, 156). In neutrophils at rest, the cytosolic components of Nox2 are present in inactive complexes within secondary granules (152). Neutrophil activation by the engagement of TLRs with microbial products (e.g., LPS and flagellin, etc.) directs opsonophagocytosis of microbes (via complement or Fc receptors), or exposure to proinflammatory cytokines can trigger the ROS burst response in these cells (63, 157). Upon activation of the neutrophil, secondary granules fuse with both the plasma and phagolysosomal membranes; the cytosolic Nox proteins subsequently undergo phosphorylation and translocation to both membranes, where they bind to the membrane-associated proteins of Nox2, making the complex functionally active (152, 155, 156). Functional Nox2 then directly couples the oxidation of NADPH to the reduction of oxygen (O2) to superoxide (O2·−) (Fig. 1): stoichiometrically, the oxidation of one molecule of NADPH liberates two electrons, which can then generate two molecules of O2·− (158).The orientation of the complex (and, as such, the secretion of O2·−) is aimed extracellularly at the plasma membrane and toward the internal contents of a phagolysosome (152). O2·− is then used as a substrate to generate multiple forms of ROS via a stepwise process, as discussed below.
FIG 1.
Simplified model for intracellular and extracellular generation of reactive oxygen species (ROS) by phagocytes. Molecular oxygen is converted by NADPH oxidase 2 (Nox2) to superoxide (O2·−). O2·− is converted to hydrogen peroxide (H2O2) by superoxide dismutase (SOD). H2O2 may then be converted to either hydroxyl free radical (·OH) (via Fenton chemistry in the presence of ferrous iron [Fe2+]) or halogenated ROS, such as hypochlorous acid (HOCl), via myeloperoxidase (MPO). Nox2 is localized within phagosomes as well as on the cell membrane (158).
Importantly, CGD is the inheritance of a nonfunctional NADPH oxidase (impairing ROS production and ROS-dependent processes such as NET formation) that leads to a predisposition for life-threatening, invasive bacterial and fungal infections (e.g., predominant causative agents include S. aureus and Aspergillus fumigatus [63]). As such, CGD indicates the essential role of the Nox complex and intact ROS production for innate immune defense against certain pathogens (159).
Multiple forms of ROS can be synthesized within the phagosome of neutrophils in a stepwise fashion (Fig. 1): O2·− generated by Nox2 can spontaneously dismutate to form H2O2 (via the consumption of 2 hydrogen ions under acidic conditions), or this reaction can be catalyzed by the host enzyme SOD (149). H2O2 formed in this manner can act directly on engulfed bacteria or be converted to other forms of ROS. MPO (which enters the phagosome via fusion with primary granules) catalyzes the conversion of H2O2 to halogenated ROS, represented predominantly by hypochlorite (HOCl) within the phagosome (150). Via Fenton chemistry, in the presence of iron (Fe2+), H2O2 can also be converted to hydroxyl radical (·OH−), a very potent antimicrobial (149).
The primary function of ROS production within neutrophils is killing of engulfed pathogens. There are oxidative and nonoxidative means by which ROS produced within the phagosome contribute to bacterial killing, and here, we primarily focus on H2O2 and HOCl, which were the subjects of our work. H2O2 exhibits a bimodal killing pattern, causing significant bacterial killing in vitro at very low (0.15 mM) and very high (25 mM) concentrations but reduced killing at intermediate concentrations (160). The primary mechanism of H2O2 killing at low concentrations is DNA damage (mode 1 killing): this mode of killing depends on the presence of iron (which generates secondary ROS, likely via Fenton chemistry), promoting the oxidation of guanine bases (i.e., 8-hydroxyguanine production), which is likely to be mutagenic/toxic (161, 162).
In contrast, mode 2 killing by H2O2 (i.e., at high concentrations) can likely target any bacterial macromolecule (161). For instance, ROS can target bacterial proteins, resulting in denaturation and loss of function: iron/sulfur clusters, due to their abundance, rapidly react with ROS (149). Aromatic and sulfur residue-containing amino acids can also react with ROS to produce protein carbonyls, which serve as reactive intermediates that damage other targets (152, 158). Lipid peroxidation of bacterial cell membranes, resulting in membrane damage, has been observed following ingestion by phagocytes, suggesting that the bacterial cell membrane is an important site of ROS action within the phagosome (163). Lipid peroxidation can also produce ketones and aldehyde intermediates that can mediate further oxidative damage to secondary targets as well (149).
Despite being recognized as a potent form of ROS, the exact mechanism of bacterial killing via HOCl remains poorly defined. Proteins are likely to be the main target, as indicated by chlorination of amino acid residues (e.g., chlorination of tyrosine) in the presence of HOCl; sulfur-containing amino acids (e.g., cysteine and methionine) are thought to be other likely targets, based on high in vitro reaction rates (158). In Escherichia coli, HOCl causes lethal DNA damage as well (164).
Importantly, ROS produced within the phagosome facilitate nonoxidative means of bacterial killing as well. When superoxide is generated within the phagosome, there is a compensatory influx of potassium to balance the negative charge of superoxide; this potassium influx activates neutrophil enzymes (e.g., elastase) that are able to proteolytically attack phagocytosed bacteria (165). The ROS burst is also a vital precursor to NET formation, which facilitates trapping and killing of bacteria via extrusion of DNA and associated proteins (e.g., elastase, MPO, histones, and LL-37) (165).
For in vitro bacterial susceptibility testing, the use of ROS concentrations that are most physiologically representative remains a source of some debate within the literature. Although this is a controversial area, most agree that there are differences in ROS concentrations within the extracellular milieu of phagocytes versus those within the phagosome. Some studies have shown that bacteria in vivo are likely to be exposed to H2O2 concentrations ranging from 12 µM within the extracellular environment (released by stimulated neutrophils) to 100 mM within the phagolysosomal compartment (166). This is substantiated by other work that suggests that the rates of ROS generation within the phagosome for both superoxide, the direct precursor of peroxide, and HOCl are several millimoles per minute (167, 168). Indeed, most in vitro investigations of bacterial susceptibility to many forms of ROS, including H2O2, have been performed with ROS concentrations in the millimolar range, mimicking the intraphagosomal setting (161).
As stated above, the overabundance of ROS within the CF airway is attributed to the chronicity of the inflammatory process (i.e., driven by bacterial infection and a continuous influx of neutrophils) as well as an underlying imbalance between ROS and endogenous antioxidants (e.g., glutathione) (54). There is abundant evidence for overproduction of ROS in the CF lung, including multiple studies that have employed specimens derived directly from patients to study markers of oxidative stress (169–176). Studies seldom include the direct measurement of ROS but rather seek to measure oxidized macromolecules as a surrogate for oxidative stress. These markers include hallmarks of lipid peroxidation (e.g., lipid hydroperoxides, 8-isoprostane, 4-hydroxynonenal, and malondialdehyde), protein oxidation (e.g., protein carbonyls, glycophore formation, and chlorinated/halogenated amino acids), DNA oxidation (e.g., 8-hydroxydeoxyguanosine), and neutrophil enzymes that directly participate in ROS formation (e.g., MPO) (169–177).
Many of these studies have shed light on the role of ROS in CF pathogenesis. Compared to healthy patients, concentrations of ROS markers are significantly elevated in lung specimens (i.e., sputum or BAL fluid) from CF patients (169, 170, 173, 175, 176). This elevation in oxidative stress within the airway can be noted in children as young as 2 to 3 years old, suggesting that ROS overproduction is part of the early events of disease development (175, 176). Concentrations of ROS markers are further elevated when CF patients are infected with S. aureus and/or P. aeruginosa, suggesting that bacterial infection is a key determinant of oxidative stress in CF (171, 175, 176). Furthermore, concentrations of systemic markers of oxidative stress (within venous blood) are significantly elevated during pulmonary exacerbations and reduced with treatment, indicating a clear connection with the changing inflammatory/infectious status of CF patients (174). Perhaps most importantly, there is an inverse correlation between markers of oxidative changes within the lung and the pulmonary function of CF patients, drawing a nexus between ROS and lung tissue damage (172).
There are three main sources of ROS production in the CF lung: neutrophils, the airway epithelium, and other immune cells, such as macrophages. Given the predominance of neutrophilic inflammation within the CF lung, neutrophils are thought to be the primary source of ROS, and there are two studies that support this argument. In both studies, the concentrations of markers used as a surrogate for oxidative stress (e.g., halogenated tyrosines and protein carbonyls) directly correlated with the number of neutrophils and neutrophil-associated proteins in young children with CF (i.e., NE and MPO); CF patients also had higher concentrations of ROS markers than non-CF controls (175, 176). While there was a correlation between ROS markers and neutrophils, there was also a strong relationship in both studies between ROS within the airway and infection, particularly with P. aeruginosa (175, 176). Both studies thus conclude that infection is a critical driver of neutrophil-derived ROS production within the CF airway.
The CF airway epithelium is another important source of ROS. Alveolar type II epithelial cells can directly produce ROS via Nox homologs, Duox1 and Duox2 (i.e., dual oxidases) (152, 153). These protein complexes are responsible for continuous H2O2 production (whereas the direct product of Nox is superoxide), even in the absence of infection in CF airways (152). H2O2 generated via Duox complexes can further be used as a substrate (along with thiocyanate [SCN−]) by lipoperoxidase (LPO) to generate hypothiocyanite (OSCN−), another form of antimicrobial ROS (153). In CF, it is theorized that before the establishment of chronic bacterial infection, H2O2 generated by the Duox complexes represents the primary source of ROS; however, following chronic infection, Nox-dependent generation of ROS by phagocytes (primarily polymorphonuclear leukocytes [PMNs]) predominates (54).
Other phagocytic cells within the airway represent the third main source of ROS in CF patients. Nox2 is also expressed in macrophages, which play a role in the phagocytic, innate immune response to bacterial infection within the CF lung (152, 178). Thus, macrophages are likely to be another source of ROS in CF.
A suggested primary defect in the transport of glutathione, a critical endogenous antioxidant, likely contributes to ROS stress within the CF airway. Glutathione exists in a reduced form (GSH) that is oxidized (GSSG) by oxidizing agents such as ROS; GSH is regenerated from GSSG via the pentose phosphate shunt (i.e., through NADPH oxidation) (179). CFTR is responsible for the release of reduced and oxidized forms of glutathione in in vitro studies using a kidney cell line (stably expressing normal CFTR) (180). Furthermore, CFTR mutation in airway epithelial cells (with the homozygous ΔF508 mutation) reduces the extracellular transport of glutathione without affecting the intracellular synthesis of glutathione; conversely, restoration of CFTR in the CFTR−/− cell line (via transfection) increases the extracellular release of GSH, suggesting that aberrant transport of GSH is specific to the CFTR protein (181). Additionally, pancreatic insufficiency in CF patients results in reduced intake/absorption of fat-soluble, dietary antioxidants (e.g., vitamin E and carotenoids), likely contributing to the primary oxidant/antioxidant imbalance in CF patients (54).
Although the primary target of ROS produced within the CF airway is pathogenic microbes, these organisms are adept at evading ROS stress. Specifically, bacteria that infect CF patients have developed multiple strategies to neutralize oxidative species, including expression of antioxidants (e.g., catalase, superoxide dismutase, and peroxidases, etc.), induction of transcriptional stress responses (e.g., via global oxidative stress regulators such as oxyR), and activation of DNA repair machinery (i.e., to correct lesions due to free radicals that would otherwise prevent replication) (162, 182). Additionally, similar to LL-37, sublethal concentrations of ROS can promote bacterial mutagenesis, leading to adaptations that may also facilitate evasion of ROS stress (183). Many of these versatile pathways in bacteria to detoxify ROS are discussed in the context of P. aeruginosa mucoid conversion and evasion of innate immunity (see below).
However, one of the implications of bacteria thriving within an ROS-rich environment in CF is that chronic infection perpetuates the inflammatory response; this leads to an even greater production of ROS, which ultimately overwhelm endogenous host antioxidants (e.g., glutathione), resulting in off-target effects. Indeed, ROS can act as second messengers directly on the airway epithelium to facilitate proinflammatory responses. IL-1β activation of NF-κB depends on intracellular H2O2, which can accumulate within the extracellular milieu of the CF airway and freely diffuse across cell membranes (184). Relatedly, another study shows that CF epithelial cells amass intracellular H2O2 due to reduced expression of Nrf-2, a transcription factor that is central to the antioxidant response. As such, the CF epithelium shows intrinsically reduced expressions of several host antioxidants (e.g., thioredoxin, peroxiredoxins, catalase, and glutathione) but increased expression of superoxide dismutase, thus explaining the intracellular accumulation of H2O2. Increased intracellular H2O2 then directly increases IL-6 and IL-8 secretion by the CF epithelium, thereby linking excess ROS production, dysregulation of antioxidant responses, and the proinflammatory environment of the CF lung (185).
In addition to promoting inflammation within the lung, ROS can directly damage host tissues via the oxidation of macromolecules: lipids, DNA, and proteins (i.e., the same ways in which these oxidants are toxic to pathogens) (54). Prolonged ROS exposure can induce lipid peroxidation, compromising membrane integrity and causing DNA strand breaks, resulting in mutagenesis or cell death and alteration of protein structure/function (186). Different forms of ROS affect the host by distinct mechanisms; for example, H2O2 has been linked to ATP depletion in the airway epithelium, whereas HOCl is implicated in intracellular protein damage via sulfhydryl group oxidation and protein carbonyl formation (187).
Although the collateral effects of ROS illustrate how immune dysregulation (i.e., hyperinflammation) can damage the CF airway, infection is also a critical driver of lung pathology in CF patients. In the following section, we discuss the role of bacterial infection in CF pathogenesis, specifically in the context of early events of disease manifestation.
The “Chicken-and-Egg” Counterargument: Bacterial Infection Precedes Inflammation
The “chicken-and-egg” question of what comes first in the CF lung, inflammation (i.e., primary dysregulation of the immune response) or bacterial infection, remains a source of debate. Thus far, we have discussed studies wherein defects in the immune response, particularly in innate immunity, predispose patients to early inflammation and secondary bacterial infection. However, in direct contrast to the above-mentioned evidence, other elegant work with CF patient specimens as well as the pig model of disease emphasizes the importance of bacterial infection as the first event that drives subsequent inflammation within the CF lung.
Indeed, one landmark study of fetal CF lung tissue suggests that the early CF lung is histopathologically normal, i.e., indistinguishable from non-CF fetal tissue and free of neutrophilic inflammation in the absence of infection (188). The same study also showed that staining for proinflammatory cytokines such as IL-6 and IL-8 is similar between CF and non-CF fetal tissues, indicating the absence of intrinsic inflammation in the prenatal stage of disease (188). Additionally, multiple prospective clinical studies done by Armstrong and colleagues suggest that concentrations of markers of inflammation (e.g., proinflammatory cytokines, total number of neutrophils, and free neutrophil elastase) within BAL fluid from infants diagnosed with CF (aged <1 year) are elevated only directly in response to bacterial infection and resolve with bacterial eradication via antibiotic therapy; infants with CF who are free of infection (either never infected and asymptomatic or cleared infection via antimicrobial treatment) do not show significant elevations in concentrations of inflammatory markers compared to non-CF controls (189–191). Conversely, young patients who become unable to clear their bacterial infection show the highest elevation in concentrations of inflammatory markers. These clinical studies demonstrate that infection not only initiates inflammation within the CF lung but likely also maintains inflammation (189–191).
Similarly, the pig model of CF (CFTR−/−), developed by the Welsh laboratory, substantiates the above-mentioned clinical studies that indicate that infection precedes inflammation within the CF lung. The pig has been validated as a reasonable model of disease, as it exhibits multiple prototypical characteristics of early CF presentation, particularly within the gastrointestinal tract: intestinal obstruction (meconium ileus), pancreatic and hepatobiliary changes, as well as failure to thrive (21, 22). Additionally, there are parallels between early anatomical abnormalities, such as reduced lumen of the trachea, in both human and pig CF lungs (192). In a well-cited publication from the Welsh group, the laboratory demonstrated that in newborn CF pigs free of bacterial infection (although viral infection was not excluded), the CF lung appears to be histopathologically normal compared to healthy swine controls (193). They also found no elevation in levels of proinflammatory cytokines and immune cells within the CF pig lung at this early time point compared to their control animals. However, shortly after birth, the CF pigs become colonized with bacteria, after which there are noticeable inflammatory changes within the airway; the predisposition to bacterial infection in CF pigs but not in non-CF controls is likely due to defects in innate immunity (e.g., acidic pH of ASL or impaired mucociliary clearance), which have been investigated by the Welsh group and are discussed at length above (34, 35, 39). As such, these fundamental, controlled animal studies also suggest that early pathophysiology of CF lung disease is best characterized by lung hyperinflammation that is secondary to bacterial infection.
Progression of Events following Lung Infection and End-Stage Lung Disease in CF
Importantly, CF lung pathobiology is likely to be multifactorial, and each of the above-mentioned chicken-and-egg hypotheses may contribute to the disease process. Based on all studies described above, we speculate that the early events of CF lung disease proceed as follows: (i) CFTR-dependent defects in innate immunity predispose a patient to initial infection; (ii) bacterial infection then incites inflammation, characterized by the production of proinflammatory cytokines and the influx of neutrophils (which produce antimicrobials such as LL-37 and ROS in abundance); and (iii) the inflammatory response is overly exuberant and ineffective in clearing infection (due to bacterial adaptations and the capacity to evade immunity) and inflicts collateral damage on host tissue due to intrinsic deficits in endogenous anti-inflammatory and antioxidant molecules (e.g., IL-10 and glutathione). While these events may initiate the early manifestations of disease, the persistence of infectious and inflammatory factors throughout the disease course drives progression toward chronicity and the ultimate loss of pulmonary function. In other words, CF invariably remains a disease that is borne out of the bacterium-host interface.
Indeed, regardless of the temporal sequence of early events (inflammation and then bacterial infection or vice versa), the later progression, beginning with acute and subsequently chronic bacterial infections of the lung, appears to be well established. Unsuccessful clearance of infection promotes a destructive cycle of inflammation (i.e., neutrophil recruitment, frustrated phagocytosis, and the production of neutrophil-derived antimicrobials) (7). Patients may suffer symptomatic complications of these infectious/inflammatory cycles, often manifested as “pulmonary exacerbations,” characterized by increased coughing, dyspnea, change in sputum volume/color, weight loss, and reduced pulmonary function (194–196). Within a state of chronic pulmonary inflammation, both bacterial and host factors can damage lung tissue, leading to greater inflammation and eventual gross, irreversible changes to airway and parenchymal anatomy: bronchiectasis, cystic changes, mucus plugging, and emphysema (197, 198). The ultimate endpoint of disease is pulmonary failure, leading to candidacy for lung transplantation or death (7). While transplantation remains a palliative life-extending intervention for CF patients who are considered appropriate transplant candidates, it is not curative; there are significant associated complications, and the 5- and 10-year survival rates posttransplantation for children with CF remain at 57% and 45%, respectively (199–201). Notably, expresses andadult CF patients who receive lung transplants have superior outcomes, with 5- and 10-year posttransplantation survival rates of 67% and 55%, respectively (199).
Given the importance of infection as a driver of the disease process, the following discussion turns to the pathogenic microbiome of the CF lung and how the microbial composition changes throughout a CF patient’s lifetime.
THE CF LUNG MICROBIOME
Despite past dogma, it is now a well-accepted paradigm that the healthy lung is not sterile (202–204). The healthy lung microbiome is composed primarily of members of the bacterial phyla Bacteroidetes and Firmicutes, which migrate into the lung from the oropharynx via aspiration of saliva (204). Furthermore, dysbiosis of the healthy lung microbiome is linked to pulmonary as well as systemic diseases (202–204).
Early dysbiosis of the CF lung and colonization with pathogenic bacterial species are attributed to defective or dysregulated immunity, including altered mucociliary clearance (as discussed above) (205). The predominant, “conventional” pathogenic bacterial species that colonize/infect the early CF lung include S. aureus (including methicillin-resistant S. aureus [MRSA]) and Haemophilus influenzae, whereas P. aeruginosa and the Burkholderia cepacia complex dominate in the later stages of disease; whereas early infections are intermittent, separated by periods of clearance and remittance of respiratory symptoms, the CF airways eventually become chronically infected with bacteria (206). Factors that influence the composition of the CF microbiome include patient age, antibiotic treatment, and disease modifier therapy (i.e., ivacaftor) (207–209). In general, as the patient ages, the microbial diversity of the CF lung declines, and chronic infection (i.e., resistant to antibiotic therapy) due to only one or two pathogenic bacterial species, most commonly P. aeruginosa, is established (210).
Other isolated or detected opportunistic pathogens in CF include bacteria such as Stenotrophomonas maltophilia, Achromobacter xylosoxidans, and nontuberculous mycobacteria (i.e., Mycobacterium abscessus); fungi such as Aspergillus and Candida spp.; and viruses (e.g., rhinovirus, influenza virus, and respiratory syncytial virus [RSV]) (211, 212). Although an in-depth discussion of the vast majority of these pathogens is out of scope here, their presence within the CF lung indicates that these species, both individually and via complex polymicrobial interactions, likely influence disease pathobiology.
To that end, there is ample evidence within the clinical literature of the consequences of early and chronic bacterial infection upon CF pulmonary function (and other outcome measures). We have already discussed evidence here that early bacterial colonization of infants with CF promotes inflammation, permanent changes to lung architecture, and decline of pulmonary function (19, 20, 213, 214). In children monitored from infancy to 6 years of age, the loss of diversity of the lung microbiota and the predominance of conventional pathogenic species (e.g., S. aureus and P. aeruginosa) are independently correlated with elevated inflammatory indices within BAL fluid (215). Chronic infection with Gram-negative pathogens of CF, P. aeruginosa and B. cepacia, is associated with a rapid decline in pulmonary function, more frequent exacerbations, and higher rates of mortality (216). Ultimately, 90% of deaths in CF patients are attributed to pulmonary dysfunction directly associated with chronic infection, underscoring the absolute importance of studying the pathogenesis of CF-relevant microbes (212). Given the central role of P. aeruginosa in CF pulmonary disease, the following sections focus on this opportunistic pathogen.
PSEUDOMONAS AERUGINOSA: A CRITICAL PLAYER IN CF LUNG DISEASE
P. aeruginosa is a Gram-negative aerobe/facultative anaerobe that is found ubiquitously in soil and aquatic environments (217). Metabolically, P. aeruginosa is oxidase positive (i.e., preferring to grow in aerobic or microaerobic environments) and non-lactose fermenting but also capable of using nitrite or nitrate as a terminal electron acceptor under anoxic conditions (including within the CF lung) (218). The genomes of multiple strains of P. aeruginosa have been sequenced, demonstrating the versatility and adaptability of this organism: the total genome size is between 5.5 and 7 Mbp and GC rich (i.e., >65% GC content), with an accessory genome of up to 200 kbp, indicating the capacity of P. aeruginosa to acquire genetic elements via horizontal gene transfer: transformation, conjugation, and transduction (219). P. aeruginosa is also capable of three forms of motility, which include flagellum-dependent swimming, flagellum- and pilus-dependent swarming, and pilus-dependent twitching (220).
In the clinical setting, P. aeruginosa was first described by Gessard in 1882 in the article entitled On the Blue and Green Coloration of Bandages, thus documenting the elaboration of a blue-green pigment (pyocyanin) by the bacterium within skin wound pus (221). As an opportunistic pathogen of humans, P. aeruginosa causes both acute and chronic infections in immunocompromised and hospitalized patients. P. aeruginosa has been implicated as a causative agent of endocarditis/bacteremia, skin and soft tissue infections (e.g., burn wounds and surgical-site infections), urinary tract infections, gastrointestinal infections, meningitis, ocular infections, ear infections, and ventilator-associated pneumonia (VAP) (222–224). Among the causative agents of VAP, P. aeruginosa still carries the highest mortality rate (223). Disturbingly, within the hospital environment, P. aeruginosa has been found growing within disinfectants and distilled water and on other inanimate surfaces, including sinks and mops (221, 225–227). Although the nosocomial transmission of P. aeruginosa remains an area of debate, it may be spread via direct contact with contaminated surfaces or hospital equipment (e.g., mechanical ventilators and stethoscopes) or aerosolization or by contaminated health care workers (228–231). In the community, important reservoirs of this bacterium include hot tubs/whirlpools and freshwater lakes, wherein acquisition/infection can cause folliculitis and otitis externa, respectively (232, 233).
P. aeruginosa poses multiple challenges to effective treatment, such as significant antibiotic resistance. Multidrug-resistant (MDR) strains of P. aeruginosa have been documented with greater frequency in the clinical setting and are associated with very poor clinical outcomes; MDR strains employ multiple mechanisms of antibiotic resistance, including overexpression of efflux pumps and beta-lactamases (221, 234, 235). P. aeruginosa has also been studied extensively as a paradigm-defining, biofilm-forming bacterium (236–238). Based on the often-cited Parsek-Singh and Stoodley/Hall-Stoodley criteria, bacterial biofilms are defined as communities of bacteria (i.e., clusters or microcolonies of bacterial cells), adherent to a surface, that elaborate an extracellular matrix composed of bacterial and host materials and exhibit enhanced resistance to antibiotics compared to planktonic bacteria; “planktonic” refers to free-floating, non-surface-adherent bacterial cells (239, 240). Indeed, biofilm formation confers enhanced resistance not only to antibiotics (i.e., up to 1,000-fold greater resistance than for planktonic bacteria) but also to effectors of innate and adaptive immunity (236–238). Additional mechanisms of P. aeruginosa evasion of immune clearance are discussed further below (see P. aeruginosa Virulence Factors, Adaptations, and Host Responses during Chronic Infection).
In the study of human biology and pathophysiology, P. aeruginosa is inextricably linked with CF, wherein it is a prominent causative agent of chronic pulmonary infection. P. aeruginosa is the predominant pathogen infecting adult CF patients; approximately three-quarters of CF adults are infected with this organism (206, 241). Multiple studies also find that P. aeruginosa infection is a determinant of lower/declining lung function and a leading cause of respiratory failure and mortality in CF patients (242–246). In a study examining multiple infecting microbial species as independent correlates of pulmonary outcome measures in CF patients, only P. aeruginosa correlated with reduced pulmonary function, increased concentrations of serum C-reactive protein (CRP) (a systemic marker of inflammation), and higher concentrations of neutrophil elastase in sputum (247). This study, among others mentioned above, suggests the primary importance of studying P. aeruginosa pathogenesis in the context of CF pulmonary disease, despite the diversity of microbial species that may well infect the CF lung throughout a patient’s lifetime.
While the ultimate effects of P. aeruginosa in the chronic/end stages of disease are well established, its impacts on CF patients at an early age are also important. In one study, the median age of P. aeruginosa acquisition was 1.0 year; however, approximately 30% of patients in this cohort were culture positive for P. aeruginosa by as early as 6 months of age (248). In another study, 70% of CF patients were culture positive for P. aeruginosa by 3 years of age, but using a combination of serology and cultures to detect the bacterium, approximately 98% were positive for P. aeruginosa (249). These data suggest that although P. aeruginosa infection may be intermittent at an early age (i.e., prior to the onset of chronic infection), this bacterium is capable of infecting CF patients in infancy, thus influencing the disease course throughout a patient’s lifetime. Infection with P. aeruginosa at infancy is associated with early decline in lung function, elevated concentrations of markers of pulmonary inflammation, and irreversible changes to the airway architecture upon imaging (19, 20, 213, 214). These findings emphasize why many intervention efforts in pediatric CF patients focus upon aggressive antibiotic treatment of P. aeruginosa infections to eradicate the bacterium from the airways (250–252).
Unfortunately, despite improved surveillance and therapeutic regimens, these eradication efforts fail in the long run, as most CF patients become chronically infected with P. aeruginosa (250–252). Moreover, flourishing communities of P. aeruginosa are often isolated from CF patient lungs postmortem or posttransplantation (i.e., within the diseased, explanted lung), suggesting that these bacteria not only are driving the disease process but also are able to outlive the infected tissue and host (253–255). As such, the questions that are central to ongoing investigation in the field pertain to the staying power of P. aeruginosa within the unique, hyperinflammatory environment of the CF lung. What allows P. aeruginosa to endure, proliferate, and profoundly impact all stages of CF pulmonary disease? Importantly, by identifying the tools that P. aeruginosa uses to survive within the CF lung, can we illuminate targets for the development of novel, antimicrobial therapeutics?
Indeed, part of the answer to these questions must include an understanding of P. aeruginosa virulence factors and the remarkable capacity of this bacterium to adapt via genotypic/phenotypic alterations. These pathoadaptive processes are central to the evasion of antimicrobials as well as the immune system and, thus, are essential for establishing chronic infection.
P. AERUGINOSA VIRULENCE FACTORS, ADAPTATIONS, AND HOST RESPONSES DURING CHRONIC INFECTION
P. aeruginosa Evasion of Innate Immunity
The premise that CF is a disease borne out of host-pathogen interactions is well established. Within this paradigm, the survival of P. aeruginosa within the CF lung is predicated upon a clash between bacterial virulence factors (which often arise via genetic/phenotypic adaptations) and the immune response. Unfortunately, the patient is caught in the cross fire between these bacterial/host elements, experiencing all of the deleterious effects on pulmonary function due to chronic P. aeruginosa infection (as described in detail above) and the immunopathological response elicited. In part, P. aeruginosa survival within the lung depends upon successful evasion of the innate immune response (256). Therefore, it is important to understand some of the bacterial factors that shield P. aeruginosa from the hyperinflammatory, neutrophil-rich CF lung microenvironment, thus enabling long-term infection and perpetuation of the disease process.
P. aeruginosa expresses multiple virulence factors during infection that enable evasion of the host response; among these are lipases, proteases, rhamnolipids, pyocyanin, quorum sensing (QS) molecules, catalases, and exopolysaccharides (257). Some of these factors directly damage host tissue and subvert immune cell functions. However, the factors that specifically affect neutrophils are important to consider, as neutrophil recruitment, phagocytosis, and the production of neutrophil-derived antimicrobials (i.e., granular contents) are thought to be essential for P. aeruginosa clearance. Neutropenic mice have been shown to be vulnerable to fatal pulmonary infection with very-low-dose inocula (<100 CFU/mouse) of P. aeruginosa compared to nonneutropenic mice (107 to 108 CFU/mouse) (258). Correspondingly, patients with neutropenia, due to either HIV or cancer chemotherapy treatment, also demonstrate susceptibility to P. aeruginosa pneumonia (259, 260). Furthermore, patients with inherited disorders of neutrophil functions, including leukocyte adhesion deficiency (LAD) and specific granule deficiency, also show a predilection for P. aeruginosa infection (261). Neutrophil-specific granules are known to contain the cathelicidin LL-37; mouse models of LL-37 depletion (in parallel with specific granule deficiency in humans) show reduced clearance of P. aeruginosa, suggesting the importance of LL-37 (and neutrophils) in controlling P. aeruginosa infection in vivo (262, 263).
Despite a profuse influx of neutrophils into the CF lung, CFTR-dependent neutrophil impairment (e.g., reduced chlorination of the phagosome), as discussed above, may contribute to the vulnerability of CF patients to P. aeruginosa (88). However, P. aeruginosa virulence factors also directly interfere with neutrophil-mediated bacterial clearance in multiple ways. For instance, exotoxins produced via the P. aeruginosa type III secretion system directly lyse neutrophils (264); pyocyanin, the blue-green pigment of P. aeruginosa, induces neutrophil apoptosis (265, 266); and rhamnolipids, which are powerful detergents, cause neutrophil necrosis (267). P. aeruginosa alkaline protease has been shown to degrade complement proteins, thus inhibiting complement-mediated opsonophagocytosis by neutrophils (268). The expression of all of the above-mentioned virulence factors is regulated by QS, a mechanism of transcriptional control activated by bacterial density via the production of diffusible signaling molecules (i.e., autoinducers); QS is known to be induced within the CF airway, and QS-deficient P. aeruginosa has been shown to exhibit attenuated virulence (269). In this manner, several P. aeruginosa virulence factors directly eliminate phagocytic cells or impair effector functions that are critical for bacterial clearance in vivo.
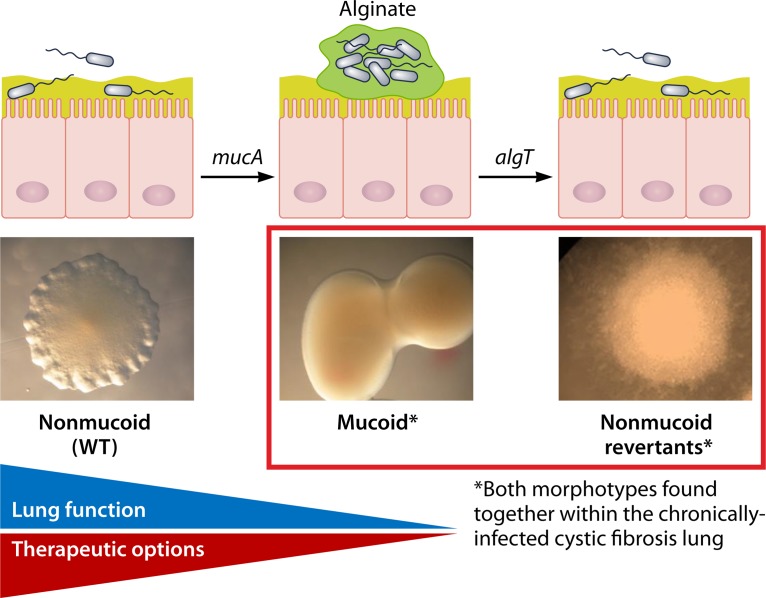
Pathoadaptation during Chronic Infection
Importantly, during chronic infection of the airway, P. aeruginosa is confronted with multiple environmental stresses: intra/interspecies competition for nutrients and space, an anaerobic environment caused by mucus plugging, high concentrations of antibiotics used to aggressively treat infection, and an abundance of neutrophils and neutrophil-derived antimicrobials (e.g., AMPs and ROS) (270). These significant pressures within the airway drive P. aeruginosa microevolution, characterized by the acquisition of spontaneous mutations followed by the selection of genotypically/phenotypically distinct variants better suited for long-term colonization of the airway (i.e., variants exhibiting reduced virulence but greater resistance to antimicrobials and host immunity) (269). This evolutionary process facilitates the transition of P. aeruginosa from an “early/acute” to a “late/chronic” pulmonary pathogen in CF. Some phenotypic characteristics associated with this transition include the downregulation of some virulence factors (i.e., motility appendages [pilus and flagellum] and the above-mentioned proteases, rhamnolipids, and pigments, etc.), increased biofilm formation (i.e., exhibiting characteristics of a sessile, multicellular lifestyle as opposed to planktonic behavior), and upregulation of exopolysaccharide expression (Psl/Pel and alginate) (271).
There are significant benefits conferred by the genotypic/phenotypic conversion of P. aeruginosa into a more chronically adapted CF pathogen. For instance, although P. aeruginosa flagellar motility is necessary for initial colonization of a host, the flagellum is a PAMP (i.e., TLR5 ligand) recognized by the epithelium and phagocytes; as such, downregulation of the flagellum during chronic infection enables evasion of immune cell recognition and phagocytosis while simultaneously diminishing the production of proinflammatory cytokines/chemokines (272–275). The loss of flagellar motility also inhibits P. aeruginosa stimulation of superoxide production and NET formation by neutrophils (276); late isolates of P. aeruginosa from the CF lung, which tend to exhibit reduced motility, are also more resistant to NET-mediated killing (although the precise mechanism of this resistance is not well understood) (90). Similarly, P. aeruginosa LPS, which is also a PAMP (i.e., TLR4 ligand), exhibits lipid A modifications in late CF isolates, causing reduced phagocyte recruitment and lower proinflammatory cytokine production in an animal model of disease (277).
P. aeruginosa within the chronically infected CF lung also demonstrates biofilm characteristics. These bacterial communities appear as microcolonies or aggregates in close association with the airway epithelium and surrounded by neutrophils (253, 254, 278). As stated above, while P. aeruginosa biofilms exhibit significantly enhanced resistance to antibiotics (relative to planktonic bacteria), these communities also confer resistance to innate immune effectors (279). For example, biofilms of P. aeruginosa exhibit reduced activation of the complement system compared to planktonic cultures (280). Furthermore, when neutrophils interact with P. aeruginosa biofilms, the immune cells become immobilized at the upper layers of the microcolonies, engorged with bacteria, and unable to substantively clear the bacterial communities (281). Moreover, while NETs are still induced by P. aeruginosa within biofilms, NET components, including DNA, not only are unable to trap or kill the bacteria but also become incorporated into the extracellular polymeric substances (EPSs) of the biofilm itself (282). As such, P. aeruginosa biofilms can inhibit or coopt the innate immune response to thrive within the CF lung.
Mechanisms of Bacterial Mutagenesis In Vivo and “Insurance Effects”
Pathoadaptation of P. aeruginosa within the CF lung is primarily driven by genetic mutations, often attributed to direct exposure to mutagenic host antimicrobials, in particular, ROS (e.g., H2O2) (269, 283–285). In vitro investigations have supported this premise, as exposure of bacteria to neutrophils or H2O2 promotes an increased frequency of spontaneous mutations (147, 183, 285–287). There is also evidence that AMPs (e.g., LL-37) can induce bacterial mutagenesis at sublethal concentrations, and these data are discussed further in the context of P. aeruginosa mucoid conversion (see below) (147, 148). Host-derived mutagens can damage bacterial DNA, inducing DNA repair mechanisms (e.g., RecA-mediated SOS response, mismatch repair [MMR], translesion repair by polymerase IV [Pol IV] [DinB], and 8-oxo-2-deoxyguanosine [GO] repair by Pol V) (288–290). If these repair mechanisms are overwhelmed, defective due to loss-of-function mutations, or error prone (e.g., translesion repair) or otherwise fail to correct nonlethal DNA lesions, stable mutations may arise in a subpopulation of bacteria (288, 289). Mutational variants may then be selected and maintained within the CF lung if they are better suited to withstand challenging host and antimicrobial pressures. Interestingly, loss-of-function mutations in the MMR apparatus-encoding genes (e.g., mutS or mutL) lead to the emergence of “hypermutator” populations of P. aeruginosa; these hypermutators are isolated in abundance from CF patients in the late stages of disease and exhibit a loss of virulence and significantly heightened resistance to antibiotics (290–293).
Multiple genetic variants of P. aeruginosa, including hypermutators and nonmutators (among others), are often maintained together within the CF lung; indeed, P. aeruginosa isolates from chronically infected CF patients exhibit significant genetic/phenotypic heterogeneity, illustrating the “insurance hypothesis” (269, 294–296). This principle posits that genetic diversity within a group of organisms can confer selective advantages to the entire population (297). As discussed further below, the diversification of P. aeruginosa in vivo likely contributes not only to antibiotic resistance but also to host immunity.
In the following section, three additional P. aeruginosa virulence factors and their roles in combatting host responses are examined in more detail. Some of these factors are overexpressed due to adaptive mutations (e.g., Psl/Pel in the rugose small-colony variants [RSCVs] and alginate in mucoid variants), whereas other factors are either constitutively or inducibly expressed by P. aeruginosa in direct response to host effectors (i.e., catalases and genes regulating autolysis/extracellular DNA [eDNA] release in response to H2O2 exposure).
Bacterial Catalases
Pathogenic bacteria are frequently confronted by oxidative stress in vivo (182). ROS may be generated by bacteria endogenously as a by-product of aerobic respiration or may represent an exogenous pressure from the infected host (162). The genesis of different forms of ROS as part of the innate immune response (i.e., via Nox/Duox complexes) is discussed in detail above. Here, the evolution, mechanism of action, and regulation of the bacterial antioxidant catalase are examined further, illuminating how evasion of ROS stress is critical for P. aeruginosa survival and virulence.
The catalase protein was first biochemically characterized in 1900 and was one of the first enzymes to be crystallized in 1937 (298). Based on functionality and structure, catalases are split into two main groups: heme-containing and nonheme catalases (299). Within the heme-containing catalases, there are two subgroups: monofunctional catalases and bifunctional catalase-peroxidases (300). Nonheme catalases are expressed only by prokaryotes (300). In contrast, monofunctional catalases are widely expressed and demonstrate significant homology across prokaryotes, archaea, and eukaryotes, suggesting that these proteins have been conserved through natural selection and coevolution of bacterial pathogens/humans (298, 300, 301). Both monofunctional catalase and catalase-peroxidases use heme iron as an intermediate to convert H2O2 to nontoxic products, water and oxygen. Heme iron (Fe3+) is first oxidized to Fe4+ by H2O2 and then reduced by a second molecule of H2O2 to recover the original ferric form (299). As such, H2O2 acts first as an oxidizing agent and then as a reducing agent in the process of being detoxified by monofunctional catalases. The stepwise and overall reactions are as follows:
| (reaction 1) |
| (reaction 2) |
| (reaction 3 [overall]) |
where Por is the porphyrin complex of catalase containing heme iron.
Catalase-peroxidases are also ubiquitously expressed across the three domains and subtly differ from monofunctional catalases both structurally and functionally (298, 300, 301). Monofunctional enzymes have only catalase activity and are homotetrameric. In contrast, catalase-peroxidases exhibit both catalase and peroxidase activities and are dimeric (302). Peroxidases catalyze an overall reaction that is similar to that of monofunctional catalase (reaction 3); both types of enzymes are capable of cleaving the peroxidic bond in H2O2 (H-O-O-H) and in organic peroxides such as peroxyacetic acid (R-O-O-H) (302). However, peroxidases can use organic electron donors (instead of a second molecule of H2O2) to recover the ferric form of heme iron in the second reaction. As such, peroxidase activity to detoxify H2O2 would be expressed as follows (298, 300–303):
| (reaction 1) |
| (reaction 2) |
Given that H2O2 is bactericidal (via mechanisms described in detail above), catalase expression protects pathogenic bacteria from the toxic effects of this ROS, whether generated endogenously or by host phagocytes (182, 304). Additionally, catalase also enables circumvention of free radicals that would be formed using H2O2 as a substrate (e.g., HOCl via the enzyme MPO and ·OH via Fenton chemistry), for which bacteria lack specific detoxifying enzymes (289).
P. aeruginosa expresses an impressive arsenal of antioxidants, including three monofunctional catalases (KatA, KatB, and KatC), at least two alkyl hydroperoxide reductases (i.e., peroxidases AhpCF and AhpB), as well as two superoxide dismutases (SodA and SodB) (305, 306). As stated above, superoxide dismutases (in both bacteria and humans) act upstream of catalase by mediating the conversion of superoxide into H2O2. To date, the functionality of KatC is not well understood (307). P. aeruginosa catalase (KatA) expression occurs during infection of the CF lung as patients develop antibodies (within BAL fluid and blood) that are KatA reactive (308). Furthermore, under laboratory conditions, KatA is universally expressed by tested P. aeruginosa clinical isolates, regardless of the site of infection/isolation (i.e., blood isolates for bacteremia infection and sputum isolates for respiratory tract infection); this is not true for all P. aeruginosa antioxidants, as SodA is not expressed in all isolates (305). In total, these findings suggest that catalase likely plays a critical role during P. aeruginosa infection of the CF airway.
KatA is the constitutive, heterotrimeric, stationary-phase/“housekeeping” catalase produced by P. aeruginosa (309, 310). The level of constitutive expression of katA is very high, and exposure to millimolar concentrations of H2O2 does not induce more than a 2-fold change in expression (311). In contrast, katB is inducibly expressed only upon exposure to H2O2 (309, 311). Both catalases are transcriptionally regulated by multiple factors, including OxyR, RpoS, and quorum sensing systems. P. aeruginosa OxyR is a global sensor of H2O2 stress and is homologous to the OxyR protein of E. coli (312). Conserved cysteine residues of OxyR are oxidized in the presence of H2O2, leading to the generation of a disulfide bond, followed by a conformational shift of the protein to its active form; this form of OxyR can then bind to DNA and regulate gene expression (312). Initially, it was thought that only katB expression is regulated by OxyR; in light of the H2O2-inducible transcription of katB, this seemed logical (166, 309). However, further work in the field has now shown that OxyR can promote the transcription of both katA and katB as well as the transcription of other antioxidant-encoding genes (e.g., ahpCF and ahpB) (313–315). Consequently, oxyR mutants exhibit heightened sensitivity to ROS- and neutrophil-mediated killing as well as reduced virulence in animal models of infection (306, 309, 316).
The induction of RpoS (a sigma factor responsive to stress/nutrient limitation) and/or the collaborating stringent response (activated upon amino acid starvation) also upregulates the transcription of both katA and katB (317). This is important because the host presents a nutrient-limiting, stressful environment, wherein we may infer that the expression of catalases is induced via these pathways (318). Indeed, loss-of-function mutations in stress response genes result in increased susceptibility of P. aeruginosa to oxidative stress, decreased catalase expression, and loss of virulence in animal models of disease (317–321). Additionally, two quorum sensing systems in P. aeruginosa also regulate katA, but not katB, transcription; deletion of autoinducer genes of the LasRI/RhlRI systems reduces katA transcription and correspondingly increases the H2O2 susceptibility of the mutants (322). This finding, combined with the role of RpoS in regulating katA transcription, helps to explain why katA expression is maximal at stationary phase (i.e., at a high bacterial cell density).
Although the transcriptional regulation networks of katA and katB appear to be somewhat interconnected, there are notable differences between the two catalase proteins in the degree of protection that each affords against H2O2 exposure, the intracellular localization of both proteins, and extracellular release mechanisms. KatB is not produced during the normal growth cycle (i.e., it exhibits only inducible expression), whereas KatA is the housekeeping catalase and expressed across all phases of growth (323). In the vast majority of studies, KatA has been shown to be essential for protection against bactericidal, millimolar concentrations of H2O2, whereas deletion of katB results in only marginally increased sensitivity of P. aeruginosa to lower doses of H2O2 (323–327). Correspondingly, in a transposon mutant screen seeking to identify P. aeruginosa genes essential for defense against H2O2, both katA and oxyR mutants were identified as highly H2O2 sensitive, although this was not true for katB (316).
In addition, while KatB is localized only within the cytoplasm, KatA is present within both the cytoplasm as well as the periplasm, suggesting that KatA is a released or secreted protein (323). In further investigating the potential extracellular role of KatA, one study showed that cell-free supernatants taken from a P. aeruginosa katA mutant were unable to protect a highly sensitive oxyR mutant from H2O2 stress, whereas supernatants from a katB mutant were able to rescue the oxyR mutant (309). This study speculated that KatA may be released extracellularly via bacterial cell lysis, although direct evidence of this mechanism was not shown. Furthermore, the presence of KatA (not KatB) appears to be indispensable to shield P. aeruginosa biofilms from H2O2 penetration/killing (324, 325). For many of these reasons, it is unsurprising that katA mutants of P. aeruginosa demonstrate significantly reduced virulence in animal models of infection, while this is not true for katB mutants (326, 327). Interestingly, mucosal immunization with KatA increases pulmonary clearance of P. aeruginosa in a rat model of acute infection (328).
Finally, research has identified other important roles for KatA in P. aeruginosa biology. A recent study showed that KatA protects P. aeruginosa from nitric oxide stress under anaerobic conditions, suggesting that KatA is a versatile protein that can detoxify both ROS and RNS (329). Additionally, some studies suggest that katA expression is reduced in P. aeruginosa biofilms compared to planktonic cultures in vitro (although paradoxically, biofilms still exhibit enhanced resistance to H2O2) (330, 331). P. aeruginosa mutability is also increased in biofilms compared to planktonic growth, and indeed, katA mutants demonstrate higher rates of spontaneous mutations than wild-type bacteria (285, 331). As such, the expression of katA (or lack thereof) may influence the capacity of P. aeruginosa to undergo pathoadaptation by detoxifying endogenous or exogenous sources of mutagenic ROS.
Autolysis/eDNA Release
Autolysis, as a form of programmed cell death, is a recognized phenomenon in prokaryotic species and thought to be analogous to apoptosis in eukaryotes (332–334). At first glance, the tendency of bacteria to commit suicide in direct response to certain forms of stress seems counterproductive. However, this behavior, when exhibited by a subset of bacterial cells within a population, is thought to confer significant advantages for the survival of the larger bacterial community during infection (332–334). Using superresolution microscopy and standard bacterial genetics, the mechanism and regulation of P. aeruginosa autolysis were recently elucidated by Turnbull and colleagues in a landmark publication (103).
The stepwise progression of this mechanism, called “explosive cell lysis,” is described as follows (103). Upon exposure of P. aeruginosa to cell wall or genomic stress (i.e., due to antibiotic/ROS treatment), the RecA-mediated SOS response against DNA damage is triggered. As part of the global transcriptional effect of this response, transcription of the R/F-pyocin gene cluster is upregulated (Fig. 2A); this set of genes represents a prophage-derived fraction of the P. aeruginosa accessory genome that is well conserved across multiple clinical and environmental isolates (335, 336). While the role of pyocins in the intraspecies interactions of P. aeruginosa communities is discussed above (in the context of antimicrobial peptides), none of the structural pyocin genes in the R/F-pyocin cluster are required for autolysis. Instead, P. aeruginosa lysis depends upon only one gene within the cluster, lys (PA0629), which encodes a phage-derived endolysin (Fig. 2A). This endolysin traverses the inner membrane and degrades P. aeruginosa peptidoglycan, thereby destabilizing the bacterial cell wall and causing cell lysis (Fig. 2B).
FIG 2.
Working model for Lys-mediated autolysis in P. aeruginosa. (A) Part of the R/F-pyocin gene cluster (PA0628-PA0632), a putative operon containing lys (PA0629), which encodes an endolysin implicated in P. aeruginosa autolysis (“explosive cell lysis”) and eDNA release. As part of the RecA-dependent SOS response, lys transcription is upregulated by cell membrane and genotoxic stresses (caused by exposure to antibiotics or reactive oxygen species [e.g., H2O2]) (103, 336). (B) Lys traverses the inner membrane (IM) via holin proteins (CidB, AplB, and a putative holin, Hol [PA0614]) and degrades peptidoglycan (PG), thereby destabilizing the cell wall and prompting cell lysis (103).
Access of Lys to the periplasmic space is likely mediated by three holin proteins, which are inserted into the inner membrane: CidA, AplB, and a putative holin, Hol (PA0614) (also encoded within the R/F-pyocin cluster) (103, 337, 338) (Fig. 2B). The role of CidA and AlpB in P. aeruginosa autolysis was established in publications that preceded the identification of lys (103, 337, 338). However, Turnbull et al. found that the expression of both lys and hol in trans was necessary to induce cell lysis in E. coli, indicating that the expression of either the endolysin or holins individually is not sufficient to mediate this process; they further posit that in P. aeruginosa, all three holins could potentially partner with Lys, thereby hypothesizing that deletion of one holin-encoding gene may not be sufficient to abrogate autolysis (103). To that end, their laboratory recently showed that deletion of all three holins is required to completely abolish cell lysis or to phenocopy the single deletion of lys (Cynthia Whitchurch, presented at the 16th International Conference on Pseudomonas, Liverpool, United Kingdom, 2017).
There are few additional nuances of P. aeruginosa autolysis that are worth acknowledging. First, P. aeruginosa autolysis was shown to occur in both biofilm and planktonic cultures (103). Autolysis does not occur in every cell within a P. aeruginosa population: Turnbull et. al showed via microscopy that the frequency of cell lysis in different P. aeruginosa strains can vary from 1 in 3,000 to 1 in 100,000 cells (also confirming that the phenotype is conserved across multiple laboratory and clinical strains) (103). Second, other work within the field suggests that the Pseudomonas quinolone signal (PQS) quorum sensing system, which is distinct from the above-mentioned RhlRI/LasRI systems, is required for cell lysis (although Turnbull et al. find that PQS is dispensable for autolysis) (339, 340). One PQS-dependent mechanism occurs via upregulated synthesis of pyocyanin, a phenazine compound that is the blue-green pigment of P. aeruginosa (341). Multiple roles of pyocyanin in virulence and innate immune evasion are discussed above. However, pyocyanin can additionally act as an electron acceptor from molecules such as NADH and as an electron donor to molecular oxygen, thereby participating in the endogenous generation of H2O2 (ROS) by P. aeruginosa (340, 341). H2O2 is then predicted to cause genomic DNA stress, resulting in the recA-dependent autolytic pathway discussed above. Endogenous or exogenous ROS stress has been implicated as a trigger for autolysis in E. coli as well (342).
Additionally, the LasRI system has also been implicated in autolysis: loss-of-function lasR mutants, which exhibit an autolytic phenotype, are isolated in abundance from the chronically infected CF lung (343, 344). While any direct link between LasRI gene regulation and lys-mediated autolysis has not been established, the fact that multiple late-adapted P. aeruginosa isolates exhibit this phenomenon suggests that autolysis likely confers advantages during chronic infection of the host.
Indeed, the “altruistic suicide” of a proportion of P. aeruginosa populations has been shown to be principally responsible for eDNA release (345–347). eDNA is a vital component of the extracellular polymeric substances of P. aeruginosa biofilms and has been shown to have essential roles in P. aeruginosa early attachment/biofilm formation and resistance to antimicrobials such as tobramycin (346, 347). lys mutants are severely compromised in eDNA release as well as microcolony formation (103). Correspondingly, deletion of alpB, which encodes one of the holins involved in autolysis/eDNA release, reduces the capacity of P. aeruginosa to colonize the murine lung (337). Under conditions of host stress (i.e., ROS/antibiotic exposure), autolysis can also facilitate dispersal of P. aeruginosa from established biofilms, enabling colonization of a different niche with more favorable microenvironmental conditions for survival (348, 349). As discussed above, abundant free DNA within the CF lung (likely to include bacterial eDNA as well as host DNA from neutrophils) correlates with viscosity of airway secretions and decline in pulmonary function (80, 81).
Another consequence of bacterial cell lysis is the generation of membrane vesicles (MVs), thought to form during lys-mediated autolysis via the passive enclosure of outer membrane fragments (103). These MVs are distinct from outer membrane vesicles (OMVs), which can form and be released independent of cell lysis/bacterial cell death (350, 351). Similar to OMVs, however, MVs have been shown to colocalize with eDNA and are predicted to encapsulate additional cargo, including “moonlighting” (intracellular) proteins or virulence factors, which may serve as public goods for the benefit of multicellular bacterial populations (103, 269).
Exopolysaccharides: Psl, Pel, and Alginate
The acquisition of adaptive mutations by P. aeruginosa during chronic infection of the CF lung has been well documented (as described above) (269). One set of commonly encountered mutations in chronically adapted variants results in the overproduction of P. aeruginosa exopolysaccharides: Psl, Pel, and alginate (352) (Fig. 3A to C). These polysaccharides form part of the extracellular polymeric substances of P. aeruginosa and represent multifunctional virulence factors, conferring properties of biofilm formation and enhanced resistance to environmental stressors such as antibiotics and host effectors (352–354). The overproduction of alginate, which results in the mucoid colony phenotype of P. aeruginosa, is described in detail below (Fig. 3C). Here, we briefly describe the roles of Psl and Pel in the antibiotic tolerance and immune evasion characteristics of the rugose small-colony variant (RSCV) (Fig. 3B).
FIG 3.
Predominant colony morphologies of P. aeruginosa variants within the cystic fibrosis lung. P. aeruginosa strains were grown and imaged on Vogel-Bonner minimal medium (VBMM) with Congo Red. (A) Wild type (nonmucoid); (B) rugose small-colony variant (RSCV) (nonmucoid); (C) mucoid variant. RSCVs overproduce Psl and Pel exopolysaccharides. Mucoid variants overproduce the alginate exopolysaccharide. The red color of RSCV colonies is due to positive staining with Congo Red. All colonies were imaged at the same magnification. (Adapted from reference 238 with permission.)
The P. aeruginosa RSCV, which describes a colony morphotype of small, wrinkled appearance, is often isolated from the late, chronically infected CF lung (355–357). RSCVs exhibit increased production of Psl and Pel, two of the P. aeruginosa exopolysaccharides (352, 358, 359). Psl and Pel are encoded by 7- and 15-gene operons, respectively, both of which are transcriptionally regulated (in part) by the intracellular second messenger cyclic di-GMP; the acquisition of gene mutations that result in elevated intracellular levels of c-di-GMP promotes the upregulation of Psl/Pel synthesis, thereby producing the RSCV phenotype (352, 358, 359). RSCVs demonstrate hyperaggregation and significantly enhanced biofilm formation compared to isogenic wild-type bacteria; here, “wild type” refers to environmental isolates of the pathogen that are initial or early colonizers of the CF lung but may subsequently undergo adaptive mutagenesis/differentiation (358–360). Given that the expression of either Psl or Pel is required for biofilm formation in P. aeruginosa, the augmented “biofilm phenotype” of RSCVs is attributed to the overproduction of these polysaccharides (352).
Additionally, RSCVs show increased resistance to antipseudomonal antibiotics (e.g., aminoglycosides) and to innate immune effectors, including ROS and AMPs (361, 362). Accumulation of c-di-GMP in these variants contributes to the downregulation of flagella, likely mediating evasion of immune recognition (per the TLR-dependent mechanisms discussed above) (358). Furthermore, in animal models of disease, RSCVs cause more persistent infections than wild-type bacteria and are associated with greater lung histopathology (361). Likely due to a combination of all of these properties, the emergence of RSCVs within the CF lung is associated with poor clinical outcomes, including loss of pulmonary function and prolonged hospitalization (356, 363).
ALGINATE AND THE MUCOID P. AERUGINOSA PHENOTYPE IN CF
There are multiple similarities between the properties of the P. aeruginosa RSCV and the mucoid phenotype. Parallel to the overproduction of Psl/Pel in the RSCV, overproduction of alginate by mucoid variants of P. aeruginosa (Fig. 3C) also confers significant advantages to the bacterium and simultaneously proves disastrous for the CF patient. The mucoid phenotype of P. aeruginosa was first documented by Sonnenschein in 1937, and this phenomenon was subsequently (although imprecisely) characterized as a “capsulated” P. aeruginosa variant based on microscopy (364). Since then, mucoid variants of P. aeruginosa have also been reported in multiple clinical settings of P. aeruginosa infection and across organ systems, including urinary tract infections, non-CF bronchiectasis (e.g., chronic obstructive pulmonary disease [COPD]), and ocular infections (365–367). Nonetheless, within the biomedical literature, the mucoid P. aeruginosa phenotype carries its most significant and common association with the CF lung, leading some to suggest that the phenotype be considered pathognomonic for the disease (367). During intermittent infection of the CF airway, nonmucoid (i.e., environmental, wild-type) P. aeruginosa predominates; at this stage, it is possible to achieve eradication of nonmucoid bacteria with aggressive antibiotic therapy (252, 368). However, the transition to the chronically infected stage of disease, wherein bacteria are recalcitrant to antimicrobials, is associated with the emergence of mucoid P. aeruginosa variants, which profoundly impact the clinical status of the CF patient (369).
The nonmucoid-to-mucoid transition (i.e., mucoid conversion) occurs at a median age of 13 years, although mucoid variants may be noticed by as early as 18 months of age in CF patients; in contrast, the median age of the first (nonmucoid) P. aeruginosa infection is 1 year (248). In the United States, approximately 70% of CF patients become infected with mucoid P. aeruginosa at some point during their disease course (292). As discussed above, there are deleterious effects of early P. aeruginosa colonization on the CF lung (i.e., worsening inflammatory status, architectural changes, and decline of pulmonary function) (19, 20, 213, 214). Unfortunately, mucoid conversion heralds even greater damage to the CF airway: infection with mucoid P. aeruginosa is associated with severe bronchiectasis, accelerated decline in patient lung function, and increased mortality, explaining why mucoid conversion remains a feared complication in the management of CF (242, 248, 252, 370–373).
Alginate Biosynthesis and Regulation
Mucoidy is defined by the overproduction of alginate, which is arguably the most extensively studied and well-characterized exopolysaccharide of P. aeruginosa (352). Alginate, a polyanionic polymer of β-1,4-linked d-mannuronic and l-guluronic acids, is synthesized by both P. aeruginosa and Azotobacter vinelandii, which is a Gram-negative, nonpathogenic, soil-dwelling bacterium (374) (Fig. 4). Various species of brown algae (seaweeds) also produce alginate, which is extracted and sold commercially as alginic acid; the only difference between bacterial and seaweed alginates is that only bacterial alginate is acetylated, although both polysaccharides are composed of the same constituent monosaccharides (374, 375).
FIG 4.
Chemical structure of alginate. Alginate is a polyanionic exopolysaccharide, composed of β-1,4-linked d-mannuronic and l-guluronic acids. (Reproduced from reference 238 with permission.)
Alginate biosynthesis is energetically costly and, as such, is carefully controlled via a complex regulatory network (367) (Fig. 5). The alginate biosynthetic operon consists of 12 genes (algD [PA3540] through algA [PA3551] [algD-algA]) and is regulated via the algD promoter (367, 376, 377). The expression of this operon requires the activity of a sigma factor encoded by algT (also known as algU and σ22); AlgT is encoded in a separate operon (algT-mucA-mucB-mucC-mucD), which also includes the gene products that ultimately regulate AlgT activity (374, 375). Notably, AlgT is a homolog of E. coli σE, which is responsive to multiple forms of envelope stress; as such, not only does AlgT act as a positive regulator of alginate biosynthesis, it also is predicted to regulate more than 290 open reading frames in P. aeruginosa as part of the global stress response (i.e., to heat, osmotic, and oxidative stresses) (378–380).
FIG 5.
Regulation of alginate biosynthesis: mucoid conversion and reversion. In wild-type, nonmucoid P. aeruginosa, the alginate biosynthetic operon (algD-algA) is inactive, as a sigma factor, AlgT, is sequestered at the inner membrane (IM) by its cognate anti-sigma factor, MucA. Acquisition of a mucA mutation results in a truncated MucA protein that is no longer able to bind AlgT. AlgT can then activate alginate biosynthesis in 2 principal ways: (i) by activating the transcription of the alginate biosynthetic operon at the algD promoter (PalgD) and (ii) by activating the transcription of genes encoding three ancillary transcription factors, AlgB, AlgR, and AmrZ, which also bind to PalgD and are essential for alginate production. Mucoid variants of P. aeruginosa can revert back to a nonmucoid phenotype via the acquisition of a secondary (suppressor) mutation in algT, which inactivates alginate biosynthesis (375, 425).
In nonmucoid P. aeruginosa and under conditions of limited environmental stress, AlgT is sequestered by its cognate anti-sigma factor, MucA, which is an inner membrane protein (381–383) (Fig. 5). However, during chronic infection of the CF lung, P. aeruginosa can spontaneously acquire mutations in mucA, resulting in a truncated, nonfunctional MucA protein that is no longer able to sequester AlgT; as such, AlgT is then free to bind to RNA polymerase and direct the transcription of the alginate biosynthesis genes and other downstream targets (381–383). Mutations in mucB and mucD have also been shown to destabilize the MucA-AlgT interaction and result in mucoid conversion; however, 80% of mucoid isolates from CF patients show mutations in mucA, suggesting its primary relevance to the phenomenon of mucoidy in the clinical setting (382–385).
AlgT is also known to act as a positive transcriptional regulator of its own operon and genes encoding ancillary transcription factors that are also necessary for alginate biosynthesis, including algB, algR, and amrZ (377, 386–388) (Fig. 5). AlgB and AlgR are the response regulator proteins of two distinct two-component regulatory systems in P. aeruginosa (389, 390). Although the cognate sensor kinases for both proteins (i.e., KinB and FimS, respectively, and their phosphorylation activities) are not required for alginate biosynthesis, both AlgB and AlgR are required (391). AmrZ (formerly AlgZ) is a ribbon-helix-helix DNA-binding protein that is also essential for alginate biosynthesis (387). Importantly, there are other transcription factors that are also known to bind to the algD promoter and mediate alginate transcription, including integration host factor (IHF), AlgP, and AlgQ; however, to date, AlgT is specifically known to upregulate the transcription of genes encoding the factors AlgB, AlgR, and AmrZ only (377).
Binding sites for AlgB, AlgR, and AmrZ have been demonstrated within the algD promoter region, and similar to AlgT, all of these transcription factors also regulate multiple downstream targets (i.e., virulence factors), independent of alginate biosynthesis (392–394) (Fig. 5). Indeed, AlgR has been shown to be a positive regulator of biofilm formation via quorum sensing and a negative regulator of type III secretion (395, 396). Additionally, AlgR is important for P. aeruginosa defense against some host antimicrobials, and deletion of algR (in a nonmucoid, wild-type strain background) attenuates virulence in a murine model of pulmonary infection (397, 398). Similarly, AlgB is a global regulator of multiple virulence factors, including motility, pyocyanin, and elastase (375). Among its many targets, AmrZ is known to positively regulate twitching motility (via the type IV pilus) and repress Psl synthesis (399).
It is worth noting that although the AlgT sigma factor canonically acts as a positive regulator of transcription, it has been shown to repress genes via its downstream transcription factors (399–402). For instance, in mucoid variants of P. aeruginosa, wherein AlgT and, correspondingly, AmrZ activities are upregulated, Psl and flagellum synthesis is repressed; conversely, mutation of both algT and amrZ results in derepression of Psl/flagellum synthesis, and AmrZ has been shown to directly bind to the promoters of operons encoding Psl/flagella (399, 400). Similarly, repression of type III secretion in mucoid P. aeruginosa is both AlgT and AlgR dependent (401).
Mutagenic Host Immune Effectors Induce Mucoid Conversion
The acquisition of mutations and their contributions to the adaptive evolution of P. aeruginosa are discussed at length above. However, in the context of mucoid conversion, several landmark publications have studied potential host factors (i.e., mutagens) that may promote or increase the frequency of mucA mutations, thereby triggering mucoidy in vivo.
Indeed, the first detailed mechanism for mucoid conversion via host factors was provided by Mathee et al., who showed that exposure of P. aeruginosa biofilms to sublethal concentrations of H2O2 or to activated human neutrophils resulted in the isolation of mucoid variants after 3 to 8 days (183). These mucoid isolates demonstrated the same mucA mutation allele as the one commonly observed in clinical isolates (i.e., mucA22, a deletion of a guanine residue resulting in a frameshift and premature termination of translation) (384). Thus, those authors conclude that exposure to oxidative stress may actually be beneficial to P. aeruginosa, as ROS can promote adaptive mutagenesis of the bacterium. To that end, Mathee et al. propose two possible mechanisms: ROS exposure either directly generates the mucA mutations or selects spontaneous mucA variants more capable of evading oxidative stress.
The nexus between ROS exposure and induced mutagenesis in bacteria has since been better understood; as described above, ROS can directly cause DNA damage, which can either activate error-prone repair mechanisms or overwhelm these systems altogether (147, 183, 285–287). The result is genetic variants, which subsequently undergo selection based on microenvironmental pressures within the host. DNA repair mechanisms implicated in ROS-dependent mutagenesis include MMR (e.g., mutations in mutS-mutL, common in the “hypermutator” phenotype of P. aeruginosa) and translesion repair (i.e., the activity of DinB, an error-prone polymerase) (288–290). Studies have shown that deletion of mutS promotes mucoid conversion of P. aeruginosa upon ROS treatment in vitro, whereas dinB mutation abrogates mucoid conversion (403–405). These findings suggest that the link between exposure to host effectors and the resultant mucoid conversion in P. aeruginosa depends upon DNA damage/repair.
Furthermore, Limoli et al. also elegantly demonstrated that exposure to sublethal amounts of ROS, neutrophils, and LL-37 directly induces mucA mutagenesis in P. aeruginosa, independent of selection (147). This phenomenon of LL-37-induced mutagenesis was also shown to be DinB dependent. The genetic construct used in this study was a chloramphenicol resistance cassette that was placed under the control of an algD promoter in a wild-type, nonmucoid strain that was chloramphenicol sensitive. Thus, mucoid conversion would be detectable upon growth of bacteria on chloramphenicol-containing medium. The nonmucoid strain was then exposed to various host effectors and subsequently plated on both nonselective media (i.e., to quantitate the total number of bacteria) and selective media (i.e., to quantitate spontaneous mucoid variants). There was no statistically significant change in the total number of bacteria across the various treatments, but the frequency of mucoid variants (i.e., number of mucoid colonies/total colonies) increased upon exposure to H2O2, neutrophils, and LL-37, suggesting that mutations were induced and not selected in these P. aeruginosa populations (147).
Fascinatingly, Limoli et al. also demonstrated not only that sublethal concentrations of LL-37 induce mucoid conversion but also that the resulting mucoid variants are more resistant to lethal concentrations of LL-37 (compared to their wild-type, nonmucoid parent) (147). Therefore, a host response mechanism, which is supposed to clear bacteria, instead backfires spectacularly, contributing to bacterial mutation and to resistance.
The Fitness Advantages of Mucoidy (and Associated Controversies)
Indeed, many of the drastic, detrimental clinical manifestations of mucoid conversion are attributed to the phenotypic advantages conferred by alginate overproduction. Although alginate is not required for biofilm formation (406), mucoid variants of P. aeruginosa demonstrate multiple biofilm-like characteristics, including enhanced resistance to antibiotics and to host-derived antimicrobials: several studies show that mucoid variants are more resistant to conventional, antipseudomonal antibiotics than are isogenic, nonmucoid variants (407–410). Additionally, mucoidy confers advantages in evading the immune response. Mucoid variants are more resistant to complement-mediated killing, phagocytic killing by macrophages and neutrophils, and NET-mediated killing (90, 147, 411, 412). Mucoid variants also downregulate the production of flagella, which likely enables evasion of immune detection by the mechanisms specified above (413).
However, despite these observed advantages of mucoidy in vitro and in vivo, which correlate with the clinical observations of chronicity and recalcitrance to therapeutics, there are contradictory findings within the literature as well. Some studies demonstrate that mucoid variants are more susceptible to antibiotics than are paired nonmucoid isolates; these findings suggest that alginate production alone is not a reliable predictor of antibiotic susceptibility in clinical isolates (414, 415). Moreover, whereas some have found that alginate acts as a scavenger of ROS from phagocytic cells (without affecting the viability or oxygen consumption of the phagocytes), others suggest that alginate stimulates a very robust ROS burst response (416–418).
Animal studies with paired mucoid and nonmucoid variants are informative regarding this important topic as well, although they offer similar paradoxes. Mouse models of pulmonary infection with P. aeruginosa generally show that mucoid strains cause more persistent infection than nonmucoid strains (384, 419, 420). However, there are notable exceptions, wherein infection of either a mouse or a seedling model showed an attenuated mucoid phenotype with a significantly higher 50% lethal dose (LD50) (i.e., lower lethality) than for nonmucoid variants (421, 422). The latter studies argue that as part of their chronic adaption, mucoid strains likely stimulate a reduced inflammatory response to escape immune clearance. However, where directly studied, the impact of mucoid versus nonmucoid strains on the immunopathology of the CF lung is still debated. Some have shown no difference between mucoid and nonmucoid strain infections in causing tissue histopathological changes in the animal lung, whereas other studies show greater tissue damage attributed to mucoidy (384, 419, 420, 423). More recent work argues that regardless of colony morphotype, both mucoid and nonmucoid strains that are “late adapted” (i.e., isolated from chronically infected patients) cause reduced mortality and inflammation during murine pulmonary infection compared to “early” nonmucoid isolates (i.e., from initial P. aeruginosa infection) (423, 424). Some of the differences across these studies may be explainable due to variations in infection strategies (i.e., acute versus chronic infection) and how these infections are achieved (e.g., with or without intratracheal inoculation with agarose beads, which are sometimes used to establish chronic P. aeruginosa infection).
Nonetheless, these sometimes conflicting findings point toward an overall, incomplete understanding of the advantages of mucoid conversion. Furthermore, as discussed below, mucoid variants of P. aeruginosa are often found to coexist with other genetic/phenotypic variants within the CF lung, suggesting that intraspecies interactions among these variants may influence the true impact of mucoid conversion on the CF patient.
REVERSION: INSTABILITY OF THE MUCOID PHENOTYPE
P. aeruginosa mucoid variants (mucA mutant and algT+) often revert back to a nonmucoid phenotype both in vitro and in vivo (423, 425–428) (Fig. 5 and 6). The most common pathway for reversion occurs via the acquisition of a second-site, suppressor mutation in algT (mucA and algT mutant) (425–427). A loss-of-function mutation in algT would shut off alginate production, despite a mucA mutation, as AlgT activity is required for transcription of the alginate biosynthesis operon (376). The vast majority (70%) of nonmucoid variants isolated from the chronically infected CF lung have a mucA mutation, suggesting that these populations are predominantly revertants rather than wild-type (mucA+ algT+) bacteria (425). While a second-site mutation in algT is the most commonly identified pathway for reversion in vitro, some revertants isolated both in vitro and in vivo have been shown to lack an algT mutation and thus cannot be complemented back to mucoidy via the expression of algT in trans (425, 426, 428). Although alternative, algT-independent pathways for reversion have not been well characterized, one publication shows that a second-site mutation in PA3257 (algO) can cause reversion in vitro; the function of AlgO is not understood, although the algO mutation was shown to reduce algT transcription, suggesting that either direct or indirect effects on AlgT function may be necessary for reversion (428). In theory, however, secondary mutations in any gene essential for alginate biosynthesis (e.g., algD, algR, algB, and amrZ, etc.) could be found to be responsible for algT-independent phenomena of reversion.
FIG 6.
Paradigm for mucoid conversion and reversion within the cystic fibrosis (CF) lung. CF patients are initially infected by environmental (wild-type [WT]) isolates of P. aeruginosa. During chronic infection of the CF lung, nonmucoid variants of P. aeruginosa commonly acquire a mucA mutation, leading to a phenotypic switch to mucoidy (i.e., alginate overproduction). Mucoid conversion portends a decline in patient lung function, and mucoid variants exhibit increased recalcitrance to antibiotic therapy. However, via the acquisition of second-site, suppressor mutations (e.g., algT), mucoid variants commonly revert back to a nonmucoid phenotype both in vitro and in vivo. Mixed populations of mucoid and nonmucoid revertants of P. aeruginosa are commonly observed within the CF lung, suggesting a selective advantage for mucoid/nonmucoid coinfection. (Adapted from reference 377 with permission.)
There are multiple hypotheses within the literature to explain the instability of the mucoid phenotype. Growth of P. aeruginosa in the absence of in vivo selective pressures is postulated to promote reversion in vitro; in part, this is attributed to the high energetic cost of alginate overproduction, which may not be advantageous under laboratory conditions (427). Additionally, certain growth modalities in vitro demonstrate a propensity for causing reversion, including growth of mucoid strains under static, aerobic conditions in liquid medium or within biofilms (425, 429, 430). Nonetheless, the triggers or microenvironmental cues that promote reversion of mucoid P. aeruginosa within the CF lung remain unknown.
However, in a longitudinal study of nonmucoid and mucoid isolates of P. aeruginosa from a cohort of patients in Denmark, it was found that nonmucoid revertants (mucA and algT mutant) can acquire third-site mutations, which restore mucoidy, and fourth-site mutations that cause reversion once again (431). These findings suggest that with regard to alginate production, P. aeruginosa isolates exhibit phenotypic switching within the CF lung throughout the course of disease. The mechanism of this phenomenon is attributed to sigma factor competition between AlgT and other P. aeruginosa sigma factors, including RpoN and RpoD; briefly, a mutation in algT that results in reversion may not cause a complete loss of function but rather may cause a reduced affinity of AlgT for the RNA polymerase; in this case, transcriptional regulation due to competing transcription factors (e.g., RpoD) may predominate. However, upon the acquisition of a subsequent rpoD mutation, AlgT may be able to more favorably compete for binding to the RNA polymerase and thus upregulate alginate biosynthesis once again (431, 432).
The above-mentioned study and numerous additional reports within the literature indicate that both mucoid and nonmucoid variants coexist within the chronically infected CF lung (253, 254, 269, 425, 431, 433–438) (Fig. 6). If alginate overproduction alone presented a substantial and permanent advantage for P. aeruginosa, we might expect all nonmucoid variants within the lung to be outcompeted. Indeed, the copresence of mucoid strains and nonmucoid revertants within the hyperinflammatory CF lung suggests a broader advantage for mixed-variant P. aeruginosa communities.
To that end, our group recently demonstrated that mixed-variant P. aeruginosa populations exhibit enhanced resistance to innate antimicrobials in vitro (439). Cocultures of mucoid and nonmucoid variants show greater resistance to both LL-37 and H2O2 than monocultures of either variant. In mixed populations, each colony variant contributes to immune evasion via the sharing of public goods: mucoid strains protect nonmucoid strains from LL-37 via the production of alginate. In contrast, nonmucoid revertants (i.e., algT mutants) shield mucoid strains from H2O2 via the overproduction of catalase (KatA). We also demonstrate that the expression of katA is negatively regulated in a mucA background via AlgT and a downstream transcription factor, AlgR. Furthermore, in nonmucoid revertants, KatA release into the extracellular milieu is mediated by the lys-encoded endolysin (i.e., explosive cell lysis) (439).
These findings support the premise that genetically/phenotypically diverse P. aeruginosa populations in vivo may exhibit selective advantages in circumventing host immunity (and other environmental pressures). Further examination of the interactions between different P. aeruginosa colony morphotypes (e.g., mucoid and RSCV) that coinfect the CF airway is ongoing in our laboratory and others. However, one quickly emerging area of interest pertains to studying the host-microbe interface within spatially distinct niches of the CF lung.
CF LUNGS EXHIBIT INTERLOBAR VARIABILITY IN DISEASE: MICROENVIRONMENTAL STUDIES OF THE HOST-PATHOGEN INTERFACE
Clinical imaging studies and histopathological analyses of distinct anatomical regions of CF lungs demonstrate interlobar differences in tissue damage: the upper lobes and right side of CF lungs often show more severe histopathology and signs of permanent architectural damage (i.e., bronchiectasis, bullous emphysema, and air trapping, etc.) upon radiological imaging than the lower lobes and left side of the lungs (197, 198, 440–445). Additionally, to the extent that techniques have measured regional lung function and ventilation/perfusion defects, the upper lobes are more compromised than the lower lobes (446, 447). This interlobar heterogeneity has even prompted surgeons to perform lobectomies to selectively isolate and remove those regions of CF lungs that are most profoundly affected by the disease process (448, 449). Over time, these findings have led both scientists and clinicians to ask whether this interlobar variability is driven by intrinsically anatomical, immune, or pathogen-associated factors.
Recent evidence suggests that there are regional (i.e., lobar) differences in the distribution of bacterial and host factors across CF lungs. Multiple publications have examined the diversification/clonality of bacterial variants arising in different parts of the airway using explanted CF lungs. Willner et al. performed 16S sequencing of tissue sections from CF lung explants and found that bacterial populations present, in terms of diversity and abundance of species, were significantly different in each lobe of the lungs (450). These findings were confirmed and extended by seminal work in this field performed by Jorth et al. (451). In examining interlobar variations among P. aeruginosa isolates from CF lung explants, Jorth and colleagues demonstrated the presence of regionally distinct lineages of variants that were compartmentalized by lung lobe; that is, clonal relatedness of bacteria could be stratified spatially within the lungs, and there was limited intermixing among variants from one lobe to another (451).
Furthermore, variants derived from a given lung lobe shared not only genotypic but also phenotypic characteristics, including metabolic profiles, antibiotic resistance, and virulence factor expression. Most strikingly, P. aeruginosa isolates derived from a more damaged (upper lobe) region of the lungs exhibited greater resistance to antibiotics and immune effectors (i.e., to serum and neutrophil killing) and were also more virulent in an animal model of infection than isolates derived from a less damaged (lower lobe) region of the lungs (451). These paradigm-defining data suggest that the microenvironmental factors that select for genotypically/phenotypically distinct bacterial strains within the CF lungs likely vary by lung lobe; furthermore, the bacterial variants ultimately selected may differentially influence focal tissue damage within the organ.
Jorth et al. did not investigate variants of bacteria other than P. aeruginosa, perhaps because previous work by the same group found that in terms of sheer bacterial abundance, tested CF lung explants were often dominated by P. aeruginosa infection (451, 452). Nonetheless, other major coinfecting species also included Burkholderia cenocepacia, S. aureus, S. maltophilia, and A. xylosoxidans (452). Indeed, the influence of these other organisms as well as their genetic/phenotypic diversity across different regions of the CF airway merit further investigation. To that end, a prodigious new approach to 3-dimensionally map the colocalization of multiple bacterial species (e.g., P. aeruginosa and S. aureus) within CF lung explants could yield exciting insights regarding bacterium-bacterium interactions in vivo (453).
Additional work in the field has sought to build on the above-mentioned findings. For instance, Hogan and colleagues examined regional BAL fluid and protected airway brush samples from the lungs of adult CF patients with stable disease (454). This study showed that measures of microbial diversity and abundance, accounting for P. aeruginosa and other bacterial organisms, were generally homogenous across different lung lobes of a given patient; furthermore, lobar tissue damage did not correlate with the regional microbes identified therein. Hogan et al. argue that their data may not agree with those of other studies because their work specifically examined specimens from patients with stable disease, whereas the studies by Jorth et al. and Willner et al. both used tissue from lung explants (i.e., end-stage disease) (450, 451, 454). This argument is partly substantiated by a recent publication that shows lobe-specific diversification of S. maltophilia lineages within a CF lung explant; however, there is also some evidence of global selective pressures within the lung as well, given how some S. maltophilia variants were found across multiple lung lobes (455). Regardless of some incongruencies within the literature, Hogan et al. aptly postulate that interlobar differences in tissue damage within CF lungs may not be the result of bacterial factors alone but rather may be the consequence of bacterium-host interactions.
There has been limited investigation of interlobar variations in host factors within CF lungs in direct relation to bacterial infection. Indeed, two studies using regional BAL fluid from CF patients find that while total bacterial density may differ between lung lobes, it does not correlate with markers of inflammation (e.g., proinflammatory cytokines [IL-8] and number of inflammatory cells) (456, 457). Independent of bacterial infection, however, the upper lobes of the lungs show greater numbers of neutrophils and higher concentrations of neutrophil-derived products (e.g., elastase) than the lower lobes (457).
These results are somewhat perplexing in the context of some above-mentioned studies wherein lavage of whole CF lungs demonstrated correlations between infecting bacterial populations (i.e., the presence of certain bacterial species as well as total bacterial density) and inflammatory indices (189–191). This discrepancy indicates that lobe-specific BAL and whole-lung BAL fluids may provide different answers about host-pathogen interactions. Nonetheless, given that there is some evidence that bacterial infection drives inflammation within CF lungs and conflicting data that suggest that patterns of local inflammation may not correlate with pathogens, more work is needed to uncover regional host-pathogen dynamics within the CF airway. These studies may ultimately enable a better understanding of focal, lobe-specific tissue damage, which remains a hallmark of CF pulmonary disease.
CONCLUSIONS
Scientific advancements have significantly affected the lives of CF patients, including revolutionary work in genetic therapy, but a cure for this devastating disease eludes us. Until a cure is discovered, investments continue in studying the pathogenesis of CF pulmonary disease in both the laboratory and clinical arenas. Indeed, a common thread uniting much of this work is the interaction between a dysfunctional immune response and persistent infection with highly adaptable, opportunistic pathogens, including P. aeruginosa. Although the temporal sequence of early events within the CF lung remains a source of some debate, both host immune effectors and bacterial virulence factors ultimately cause end-organ damage, leading to pulmonary failure and patient death (Fig. 7). Importantly, the genetic plasticity of P. aeruginosa makes it a particularly dangerous pathogen in CF, capable of evading diverse antimicrobial and innate immune stresses. Although P. aeruginosa acquisition is independently associated with poor outcomes, the emergence of specific phenotypic variants (e.g., RSCV or mucoid) further accelerates patient decline. Future work in this area will continue to examine the titanic struggle between bacterial infection and host responses to illuminate how changes in local microenvironments within the CF airway contribute to the overall disease process. In doing so, we continue to strive toward the development of novel antimicrobials, immune modulators, and other disease-modifying therapeutics that could profoundly impact the lives of CF patients.
FIG 7.
The pathogenesis of cystic fibrosis pulmonary disease: a vicious cycle driven by immune dysfunction and bacterial infection. Although the newborn CF lung is histopathologically comparable to a healthy lung, the CFTR mutation causes significant, primary defects in innate immunity (e.g., airway surface liquid [ASL] dehydration and acidification). These gaps in the airway’s basic defenses predispose to early infection with bacterial pathogens, including P. aeruginosa. Bacteria within CF lungs trigger an exuberant, neutrophilic immune response, resulting in excessive inflammation. Bacterial virulence factors enable evasion of host inflammatory products, including reactive oxygen species, elastases, and extracellular DNA, which have collateral, damaging effects on healthy lung tissue. Compromised tissue integrity further contributes to immune dysfunction, initiating a cycle of recurrent and eventually chronic polymicrobial infections within a milieu of pathological inflammation. This disastrous series of events causes permanent destruction of airway architecture, manifesting as bronchiectasis, mucus plugging, and, ultimately, pulmonary failure.
ACKNOWLEDGMENT
We declare no conflict of interest.
Biographies

Sankalp Malhotra, Ph.D., is a medical student at The Ohio State University College of Medicine. He completed his graduate studies under the direction of Daniel J. Wozniak at Ohio State, studying the interaction between mucoid and nonmucoid variants of Pseudomonas aeruginosa and innate immune effectors in cystic fibrosis. His long-term research and clinical interests include host-microbe interactions in immunocompromised patients.

Don Hayes, Jr., M.D., M.S., M.Ed., is a clinician-scientist and Director of the Lung and Heart-Lung Transplant Programs at Nationwide Children’s Hospital and The Ohio State University Wexner Medical Center. His main research focus is tracheobronchial epithelial stem/progenitor cells in cystic fibrosis and lung transplantation. He is specifically interested in the role of these cells in cystic fibrosis pathophysiology and the process of conductive airway reepithelialization post-lung transplant. As of 2018, he has published over 250 research articles.

Daniel J. Wozniak, Ph.D., is a Professor of Microbiology and Microbial Infection and Immunity at The Ohio State University Wexner Medical Center. His chief research interest is Pseudomonas aeruginosa pathogenesis in cystic fibrosis. Specifically, his laboratory studies P. aeruginosa virulence factors, including bacterial exopolysaccharides and biofilm formation, to identify novel therapeutic targets against chronic bacterial infection. As of 2018, he has published over 100 research articles.
REFERENCES
- 1.O’Sullivan BP, Freedman SD. 2009. Cystic fibrosis. Lancet 373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 2.Cystic Fibrosis Foundation. 2017. Cystic Fibrosis Foundation patient registry. 2016 annual data report. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 3.Scotet V, Duguépéroux I, Saliou P, Rault G, Roussey M, Audrézet M-P, Férec C. 2012. Evidence for decline in the incidence of cystic fibrosis: a 35-year observational study in Brittany, France. Orphanet J Rare Dis 7:14. doi: 10.1186/1750-1172-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spoonhower KA, Davis PB. 2016. Epidemiology of cystic fibrosis. Clin Chest Med 37:1–8. doi: 10.1016/j.ccm.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Elborn JS. 2016. Cystic fibrosis. Lancet 388:2519–2531. doi: 10.1016/S0140-6736(16)00576-6. [DOI] [PubMed] [Google Scholar]
- 6.Cystic Fibrosis Trust. 2017. UK Cystic Fibrosis Registry annual data report 2016. Cystic Fibrosis Trust, London, United Kingdom. [Google Scholar]
- 7.Elborn S, Vallieres E. 2014. Cystic fibrosis gene mutations: evaluation and assessment of disease severity. Adv Genomics Genet 4:161–172. [Google Scholar]
- 8.Antoniou S, Elston C. 2016. Cystic fibrosis. Medicine (Baltimore) 44:321–325. doi: 10.1016/j.mpmed.2016.02.016. [DOI] [Google Scholar]
- 9.Davidson DJ, Porteous DJ. 1998. The genetics of cystic fibrosis lung disease. Thorax 53:389–397. doi: 10.1136/thx.53.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutting GR. 2015. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet 16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Boeck K, Vermeulen F, Dupont L. 2017. The diagnosis of cystic fibrosis. Presse Med 46:e97–e108. doi: 10.1016/j.lpm.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Smyth AR, Bell SC, Bojcin S, Bryon M, Duff A, Flume P, Kashirskaya N, Munck A, Ratjen F, Schwarzenberg SJ, Sermet-Gaudelus I, Southern KW, Taccetti G, Ullrich G, Wolfe S. 2014. European cystic fibrosis society standards of care: best practice guidelines. J Cyst Fibros 13:S23–S42. doi: 10.1016/j.jcf.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Marshall BC. 2017. 2016 Cystic Fibrosis Foundation patient registry highlights. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 14.Chin M, Aaron SD, Bell SC. 2017. The treatment of the pulmonary and extrapulmonary manifestations of cystic fibrosis. Presse Med 46:e139–e164. doi: 10.1016/j.lpm.2016.11.030. [DOI] [PubMed] [Google Scholar]
- 15.Strausbaugh SD, Davis PB. 2007. Cystic fibrosis: a review of epidemiology and pathobiology. Clin Chest Med 28:279–288. doi: 10.1016/j.ccm.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Starner TD, McCray PB Jr, American College of Physicians, American Physiological Society. 2005. Pathogenesis of early lung disease in cystic fibrosis: a window of opportunity to eradicate bacteria. Ann Intern Med 143:816–822. doi: 10.7326/0003-4819-143-11-200512060-00010. [DOI] [PubMed] [Google Scholar]
- 17.Rosenow T, Oudraad MCJ, Murray CP, Turkovic L, Kuo W, de Bruijne M, Ranganathan SC, Tiddens HAWM, Stick SM, Australian Respiratory Early Surveillance Team for Cystic Fibrosis. 2015. PRAGMA-CF. A quantitative structural lung disease computed tomography outcome in young children with cystic fibrosis. Am J Respir Crit Care Med 191:1158–1165. doi: 10.1164/rccm.201501-0061OC. [DOI] [PubMed] [Google Scholar]
- 18.Sly PD, Gangell CL, Chen L, Ware RS, Ranganathan S, Mott LS, Murray CP, Stick SM. 2013. Risk factors for bronchiectasis in children with cystic fibrosis. N Engl J Med 368:1963–1970. doi: 10.1056/NEJMoa1301725. [DOI] [PubMed] [Google Scholar]
- 19.Gangell C, Gard S, Douglas T, Park J, De Klerk N, Keil T, Brennan S, Ranganathan S, Robins-Browne R, Sly PD, AREST CF. 2011. Inflammatory responses to individual microorganisms in the lungs of children with cystic fibrosis. Clin Infect Dis 53:425–432. doi: 10.1093/cid/cir399. [DOI] [PubMed] [Google Scholar]
- 20.Ramsey KA, Ranganathan S, Park J, Skoric B, Adams AM, Simpson SJ, Robins-Browne RM, Franklin PJ, De Klerk NH, Sly PD, Stick SM, Hall GL. 2014. Early respiratory infection is associated with reduced spirometry in children with cystic fibrosis. Am J Respir Crit Care Med 190:1111–1116. doi: 10.1164/rccm.201407-1277OC. [DOI] [PubMed] [Google Scholar]
- 21.Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, Rogan MP, Pezzulo AA, Karp PH, Itani OA, Kabel AC, Wohlford-Lenane CL, Davis GJ, Hanfland RA, Smith TL, Samuel M, Wax D, Murphy CN, Rieke A, Whitworth K, Uc A, Starner TD, Brogden KA, Shilyansky J, McCray PB Jr, Zabner J, Prather RS, Welsh MJ. 2008. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers CS, Abraham WM, Brogden KA, Engelhardt JF, Fisher JT, McCray PB Jr, McLennan G, Meyerholz DK, Namati E, Ostedgaard LS, Prather RS, Sabater JR, Stoltz DA, Zabner J, Welsh MJ. 2008. The porcine lung as a potential model for cystic fibrosis. Am J Physiol Lung Cell Mol Physiol 295:L240–L263. doi: 10.1152/ajplung.90203.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keiser NW, Birket SE, Evans IA, Tyler SR, Crooke AK, Sun X, Zhou W, Nellis JR, Stroebele EK, Chu KK, Tearney GJ, Stevens MJ, Harris JK, Rowe SM, Engelhardt JF. 2015. Defective innate immunity and hyperinflammation in newborn cystic fibrosis transmembrane conductance regulator-knockout ferret lungs. Am J Respir Cell Mol Biol 52:683–694. doi: 10.1165/rcmb.2014-0250OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhooghe B, Noël S, Huaux F, Leal T. 2014. Lung inflammation in cystic fibrosis: pathogenesis and novel therapies. Clin Biochem 47:539–546. doi: 10.1016/j.clinbiochem.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 25.Rieber N, Hector A, Carevic M, Hartl D. 2014. Current concepts of immune dysregulation in cystic fibrosis. Int J Biochem Cell Biol 52:108–112. doi: 10.1016/j.biocel.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Clunes MT, Boucher RC. 2007. Cystic fibrosis: the mechanisms of pathogenesis of an inherited lung disorder. Drug Discov Today Dis Mech 4:63–72. doi: 10.1016/j.ddmec.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boucher RC. 2007. Airway surface dehydration in cystic fibrosis: pathogenesis and therapy. Annu Rev Med 58:157–170. doi: 10.1146/annurev.med.58.071905.105316. [DOI] [PubMed] [Google Scholar]
- 28.Garland AL, Walton WG, Coakley RD, Tan CD, Gilmore RC, Hobbs CA, Tripathy A, Clunes LA, Bencharit S, Stutts MJ, Betts L, Redinbo MR, Tarran R. 2013. Molecular basis for pH-dependent mucosal dehydration in cystic fibrosis airways. Proc Natl Acad Sci U S A 110:15973–15978. doi: 10.1073/pnas.1311999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruscia EM, Bonfield TL. 2016. Innate and adaptive immunity in cystic fibrosis. Clin Chest Med 37:17–29. doi: 10.1016/j.ccm.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Matsui H, Grubb BR, Tarran R, Randell SH, Gatzy JT, Davis CW, Boucher RC. 1998. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell 95:1005–1015. doi: 10.1016/S0092-8674(00)81724-9. [DOI] [PubMed] [Google Scholar]
- 31.Cantin AM, Hartl D, Konstan MW, Chmiel JF. 2015. Inflammation in cystic fibrosis lung disease: pathogenesis and therapy. J Cyst Fibros 14:419–430. doi: 10.1016/j.jcf.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Song Y, Salinas D, Nielson DW, Verkman AS. 2006. Hyperacidity of secreted fluid from submucosal glands in early cystic fibrosis. Am J Physiol Cell Physiol 290:C741–C749. doi: 10.1152/ajpcell.00379.2005. [DOI] [PubMed] [Google Scholar]
- 33.Alaiwa MA, Beer AM, Pezzulo AA, Launspach JL, Horan RA, Stoltz DA, Starner TD, Welsh MJ, Zabner J. 2014. Neonates with cystic fibrosis have a reduced nasal liquid pH; a small pilot study. J Cyst Fibros 13:373–377. doi: 10.1016/j.jcf.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pezzulo AA, Tang XX, Hoegger MJ, Abou Alaiwa MH, Ramachandran S, Moninger TO, Karp PH, Wohlford-Lenane CL, Haagsman HP, van Eijk M, Bánfi B, Horswill AR, Stoltz DA, McCray PB Jr, Welsh MJ, Zabner J. 2012. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature 487:109–113. doi: 10.1038/nature11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah VS, Meyerholz DK, Tang XX, Reznikov L, Alaiwa MA, Ernst SE, Karp PH, Wohlford-Lenane CL, Heilmann KP, Leidinger MR, Allen PD, Zabner J, McCray PB Jr, Ostedgaard LS, Stoltz DA, Randak CO, Welsh MJ. 2016. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science 351:503–507. doi: 10.1126/science.aad5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auvynet C, Rosenstein Y. 2009. Multifunctional host defense peptides: antimicrobial peptides, the small yet big players in innate and adaptive immunity. FEBS J 276:6497–6508. doi: 10.1111/j.1742-4658.2009.07360.x. [DOI] [PubMed] [Google Scholar]
- 37.Abou Alaiwa MH, Reznikov LR, Gansemer ND, Sheets KA, Horswill AR, Stoltz DA, Zabner J, Welsh MJ. 2014. pH modulates the activity and synergism of the airway surface liquid antimicrobials β-defensin-3 and LL-37. Proc Natl Acad Sci U S A 111:18703–18708. doi: 10.1073/pnas.1422091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. 1998. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J Biol Chem 273:3718–3724. doi: 10.1074/jbc.273.6.3718. [DOI] [PubMed] [Google Scholar]
- 39.Hoegger MJ, Fischer AJ, McMenimen JD, Ostedgaard LS, Tucker AJ, Awadalla MA, Moninger TO, Michalski AS, Hoffman EA, Zabner J, Stoltz DA, Welsh MJ. 2014. Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science 345:818–822. doi: 10.1126/science.1255825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Britigan BE, Hayek MB, Doebbeling BN, Fick RB. 1993. Transferrin and lactoferrin undergo proteolytic cleavage in the Pseudomonas aeruginosa-infected lungs of patients with cystic fibrosis. Infect Immun 61:5049–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taggart CC, Greene CM, Smith SG, Levine RL, McCray PB Jr, O’Neill S, McElvaney NG. 2003. Inactivation of human beta-defensins 2 and 3 by elastolytic cathepsins. J Immunol 171:931–937. doi: 10.4049/jimmunol.171.2.931. [DOI] [PubMed] [Google Scholar]
- 42.Ratner D, Mueller C. 2012. Immune responses in cystic fibrosis: are they intrinsically defective? Am J Respir Cell Mol Biol 46:715–722. doi: 10.1165/rcmb.2011-0399RT. [DOI] [PubMed] [Google Scholar]
- 43.Stoltz DA, Meyerholz DK, Welsh MJ. 2015. Origins of cystic fibrosis lung disease. N Engl J Med 372:351–362. doi: 10.1056/NEJMra1300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machen TE. 2006. Innate immune response in CF airway epithelia: hyperinflammatory? Am J Physiol Cell Physiol 291:C218–C230. doi: 10.1152/ajpcell.00605.2005. [DOI] [PubMed] [Google Scholar]
- 45.Bonfield TL, Konstan MW, Berger M. 1999. Altered respiratory epithelial cell cytokine production in cystic fibrosis. J Allergy Clin Immunol 104:72–78. doi: 10.1016/S0091-6749(99)70116-8. [DOI] [PubMed] [Google Scholar]
- 46.Tirouvanziam R, de Bentzmann S, Hubeau C, Hinnrasky J, Jacquot J, Péault B, Puchelle E. 2000. Inflammation and infection in naive human cystic fibrosis airway grafts. Am J Respir Cell Mol Biol 23:121–127. doi: 10.1165/ajrcmb.23.2.4214. [DOI] [PubMed] [Google Scholar]
- 47.Tirouvanziam R, Khazaal I, Péault B. 2002. Primary inflammation in human cystic fibrosis small airways. Am J Physiol Lung Cell Mol Physiol 283:L445–L451. doi: 10.1152/ajplung.00419.2001. [DOI] [PubMed] [Google Scholar]
- 48.Legssyer R, Huaux F, Lebacq J, Delos M, Marbaix E, Lebecque P, Lison D, Scholte BJ, Wallemacq P, Leal T. 2006. Azithromycin reduces spontaneous and induced inflammation in ΔF508 cystic fibrosis mice. Respir Res 7:134. doi: 10.1186/1465-9921-7-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer M, Huaux F, Gavilanes X, Van Den Brûle S, Lebecque P, Lo Re S, Lison D, Scholte B, Wallemacq P, Leal T. 2009. Azithromycin reduces exaggerated cytokine production by M1 alveolar macrophages in cystic fibrosis. Am J Respir Cell Mol Biol 41:590–602. doi: 10.1165/rcmb.2008-0155OC. [DOI] [PubMed] [Google Scholar]
- 50.Khan T, Wagener J, Bost T, Martinez J, Accurso F, Riches D. 1995. Early pulmonary inflammation in infants with CF. Am J Respir Crit Care Med 151:1075–1082. doi: 10.1164/ajrccm.151.4.7697234. [DOI] [PubMed] [Google Scholar]
- 51.Rosenfeld M, Gibson RL, McNamara S, Emerson J, Burns JL, Castile R, Hiatt P, McCoy K, Wilson CB, Inglis A, Smith A, Martin TR, Ramsey BW. 2001. Early pulmonary infection, inflammation, and clinical outcomes in infants with cystic fibrosis. Pediatr Pulmonol 32:356–366. doi: 10.1002/ppul.1144. [DOI] [PubMed] [Google Scholar]
- 52.Freedman SD, Blanco PG, Zaman MM, Shea JC, Ollero M, Hopper IK, Weed DA, Gelrud A, Regan MM, Laposata M, Alvarez JG, O’Sullivan BP. 2004. Association of cystic fibrosis with abnormalities in fatty acid metabolism. N Engl J Med 350:560–569. doi: 10.1056/NEJMoa021218. [DOI] [PubMed] [Google Scholar]
- 53.Roum JH, Buhl R, McElvaney NG, Borok Z, Crystal RG. 1993. Systemic deficiency of glutathione in cystic fibrosis. J Appl Physiol 75:2419–2424. doi: 10.1152/jappl.1993.75.6.2419. [DOI] [PubMed] [Google Scholar]
- 54.Galli F, Battistoni A, Gambari R, Pompella A, Bragonzi A, Pilolli F, Iuliano L, Piroddi M, Dechecchi MC, Cabrini G. 2012. Oxidative stress and antioxidant therapy in cystic fibrosis. Biochim Biophys Acta 1822:690–713. doi: 10.1016/j.bbadis.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 55.Velsor LW, Van Heeckeren A, Day BJ. 2001. Antioxidant imbalance in the lungs of cystic fibrosis transmembrane conductance regulator protein mutant mice. Am J Physiol Lung Cell Mol Physiol 281:L31–L38. doi: 10.1152/ajplung.2001.281.1.L31. [DOI] [PubMed] [Google Scholar]
- 56.Conese M, Copreni E, Di Gioia S, De Rinaldis P, Fumarulo R. 2003. Neutrophil recruitment and airway epithelial cell involvement in chronic cystic fibrosis lung disease. J Cyst Fibros 2:129–135. doi: 10.1016/S1569-1993(03)00063-8. [DOI] [PubMed] [Google Scholar]
- 57.Kelly E, Greene CM, McElvaney NG. 2008. Targeting neutrophil elastase in cystic fibrosis. Expert Opin Ther Targets 12:145–157. doi: 10.1517/14728222.12.2.145. [DOI] [PubMed] [Google Scholar]
- 58.van den Berg CW, Tambourgi DV, Clark HW, Hoong SJ, Spiller OB, McGreal EP. 2014. Mechanism of neutrophil dysfunction: neutrophil serine proteases cleave and inactivate the C5a receptor. J Immunol 192:1787–1795. doi: 10.4049/jimmunol.1301920. [DOI] [PubMed] [Google Scholar]
- 59.Hartl D, Gaggar A, Bruscia E, Hector A, Marcos V, Jung A, Greene C, McElvaney G, Mall M, Döring G. 2012. Innate immunity in cystic fibrosis lung disease. J Cyst Fibros 11:363–382. doi: 10.1016/j.jcf.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 60.Witko-Sarsat V, Rieu P, Descamps-Latscha B, Lesavre P, Halbwachs-Mecarelli L. 2000. Neutrophils: molecules, functions and pathophysiological aspects. Lab Invest 80:617–653. doi: 10.1038/labinvest.3780067. [DOI] [PubMed] [Google Scholar]
- 61.Cowland JB, Borregaard N. 2016. Granulopoiesis and granules of human neutrophils. Immunol Rev 273:11–28. doi: 10.1111/imr.12440. [DOI] [PubMed] [Google Scholar]
- 62.Faurschou M, Borregaard N. 2003. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect 5:1317–1327. doi: 10.1016/j.micinf.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Bardoel BW, Kenny EF, Sollberger G, Zychlinsky A. 2014. The balancing act of neutrophils. Cell Host Microbe 15:526–536. doi: 10.1016/j.chom.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 64.Gifford AM, Chalmers JD. 2014. The role of neutrophils in cystic fibrosis. Curr Opin Hematol 21:16–22. doi: 10.1097/MOH.0000000000000009. [DOI] [PubMed] [Google Scholar]
- 65.Danel C, Erzurum SC, McElvaney NC, Crystal RC. 1996. Quantitative assessment of the epithelial and inflammatory cell populations in large airways of normals and individuals with cystic fibrosis. Am J Respir Crit Care Med 153:362–368. doi: 10.1164/ajrccm.153.1.8542144. [DOI] [PubMed] [Google Scholar]
- 66.Hubeau C, Lorenzato M, Couetil JP, Hubert D, Dusser D, Puchelle E, Gaillard D. 2001. Quantitative analysis of inflammatory cells infiltrating the cystic fibrosis airway mucosa. Clin Exp Immunol 124:69–76. doi: 10.1046/j.1365-2249.2001.01456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lammertyn EJ, Vandermeulen E, Bellon H, Everaerts S, Verleden SE, Van Den Eynde K, Bracke KR, Brusselle GG, Goeminne PC, Verbeken EK, Vanaudenaerde BM, Dupont LJ. 2017. End-stage cystic fibrosis lung disease is characterised by a diverse inflammatory pattern: an immunohistochemical analysis. Respir Res 18:10. doi: 10.1186/s12931-016-0489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verhaeghe C, Delbecque K, de Leval L, Oury C, Bours V. 2007. Early inflammation in the airways of a cystic fibrosis foetus. J Cyst Fibros 6:304–308. doi: 10.1016/j.jcf.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 69.Corvol H, Fitting C, Chadelat K, Jacquot J, Tabary O, Boule M, Cavaillon JM, Clement A. 2003. Distinct cytokine production by lung and blood neutrophils from children with cystic fibrosis. Am J Physiol Lung Cell Mol Physiol 284:L997–L1003. doi: 10.1152/ajplung.00156.2002. [DOI] [PubMed] [Google Scholar]
- 70.Downey D, Bell S, Elborn J. 2009. Neutrophils in cystic fibrosis. Thorax 64:81–88. doi: 10.1136/thx.2007.082388. [DOI] [PubMed] [Google Scholar]
- 71.Laval J, Ralhan A, Hartl D. 2016. Neutrophils in cystic fibrosis. Biol Chem 397:485–496. doi: 10.1515/hsz-2015-0271. [DOI] [PubMed] [Google Scholar]
- 72.Bonfield TL, Panuska JR, Konstan MW, Hilliard KA, Hilliard JB, Ghnaim H, Berger M. 1995. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med 152:2111–2118. doi: 10.1164/ajrccm.152.6.8520783. [DOI] [PubMed] [Google Scholar]
- 73.Saba S, Soong G, Greenberg S, Prince A. 2002. Bacterial stimulation of epithelial G-CSF and GM-CSF expression promotes PMN survival in CF airways. Am J Respir Cell Mol Biol 27:561–567. doi: 10.1165/rcmb.2002-0019OC. [DOI] [PubMed] [Google Scholar]
- 74.Taggart C, Coakley RJ, Greally P, Canny G, O’Neill SJ, McElvaney NG. 2000. Increased elastase release by CF neutrophils is mediated by tumor necrosis factor-alpha and interleukin-8. Am J Physiol Lung Cell Mol Physiol 278:L33–L41. doi: 10.1152/ajplung.2000.278.1.L33. [DOI] [PubMed] [Google Scholar]
- 75.Mayer-Hamblett N, Aitken ML, Accurso FJ, Kronmal RA, Konstan MW, Burns JL, Sagel SD, Ramsey BW. 2007. Association between pulmonary function and sputum biomarkers in cystic fibrosis. Am J Respir Crit Care Med 175:822–828. doi: 10.1164/rccm.200609-1354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakamura H, Yoshimura K, McElvaney NG, Crystal RG. 1992. Neutrophil elastase in respiratory epithelial lining fluid of individuals with cystic fibrosis induces interleukin-8 gene expression in a human bronchial epithelial cell line. J Clin Invest 89:1478–1484. doi: 10.1172/JCI115738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brinkmann V, Zychlinsky A. 2012. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol 198:773–783. doi: 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papayannopoulos V, Staab D, Zychlinsky A. 2011. Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving DNase therapy. PLoS One 6:e28526. doi: 10.1371/journal.pone.0028526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Law SM, Gray RD. 2017. Neutrophil extracellular traps and the dysfunctional innate immune response of cystic fibrosis lung disease: a review. J Inflamm (Lond) 14:29. doi: 10.1186/s12950-017-0176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marcos V, Zhou-Suckow Z, Onder Yildirim A, Bohla A, Hector A, Vitkov L, Krautgartner WD, Stoiber W, Griese M, Eickelberg O, Mall MA, Hartl D. 2015. Free DNA in cystic fibrosis airway fluids correlates with airflow obstruction. Mediators Inflamm 2015:408935. doi: 10.1155/2015/408935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kirchner KK, Wagener JS, Khan TZ, Copenhaver SC, Accurso FJ. 1996. Increased DNA levels in bronchoalveolar lavage fluid obtained from infants with cystic fibrosis. Am J Respir Crit Care Med 154:1426–1429. doi: 10.1164/ajrccm.154.5.8912759. [DOI] [PubMed] [Google Scholar]
- 82.Ratjen F, Paul K, Van Koningsbruggen S, Breitenstein S, Rietschel E, Nikolaizik W. 2005. DNA concentrations in BAL fluid of cystic fibrosis patients with early lung disease: influence of treatment with dornase alpha. Pediatr Pulmonol 39:1–4. doi: 10.1002/ppul.20134. [DOI] [PubMed] [Google Scholar]
- 83.Konstan MW, Wagener JS, Pasta DJ, Millar SJ, Jacobs JR, Yegin A, Morgan WJ. 2011. Clinical use of dornase alfa is associated with a slower rate of FEV1decline in cystic fibrosis. Pediatr Pulmonol 46:545–553. doi: 10.1002/ppul.21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meyer KC. 2004. Neutrophils, myeloperoxidase, and bronchiectasis in cystic fibrosis: green is not good. J Lab Clin Med 144:124–126. doi: 10.1016/j.lab.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 85.Gray RD, Hardisty G, Regan KH, Smith M, Robb CT, Duffin R, Mackellar A, Felton JM, Paemka L, McCullagh BN, Lucas CD, Dorward DA, McKone EF, Cooke G, Donnelly SC, Singh PK, Stoltz DA, Haslett C, McCray PB, Whyte MKB, Rossi AG, Davidson DJ. 2017. Delayed neutrophil apoptosis enhances NET formation in cystic fibrosis. Thorax 73:134–144. doi: 10.1136/thoraxjnl-2017-210134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alexis NE, Muhlebach MS, Peden DB, Noah TL. 2006. Attenuation of host defense function of lung phagocytes in young cystic fibrosis patients. J Cyst Fibros 5:17–25. doi: 10.1016/j.jcf.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pohl K, Hayes E, Keenan J, Henry M, Meleady P, Molloy K, Jundi B, Bergin DA, McCarthy C, McElvaney OJ, White MM, Clynes M, Reeves EP, McElvaney NG. 2014. A neutrophil intrinsic impairment affecting Rab27a and degranulation in cystic fibrosis is corrected by CFTR potentiator therapy. Blood 124:999–1009. doi: 10.1182/blood-2014-02-555268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Painter RG, Valentine VG, Lanson NA, Leidal K, Zhang Q, Lombard G, Thompson C, Viswanathan A, Nauseef WM, Wang G, Wang G. 2006. CFTR expression in human neutrophils and the phagolysosomal chlorination defect in cystic fibrosis. Biochemistry 45:10260–10269. doi: 10.1021/bi060490t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McKeon DJ, Cadwallader KA, Idris S, Cowburn AS, Pasteur MC, Barker H, Haworth CS, Bilton D, Chilvers ER, Condliffe AM. 2010. Cystic fibrosis neutrophils have normal intrinsic reactive oxygen species generation. Eur Respir J 35:1264–1272. doi: 10.1183/09031936.00089709. [DOI] [PubMed] [Google Scholar]
- 90.Young RL, Malcolm KC, Kret JE, Caceres SM, Poch KR, Nichols DP, Taylor-Cousar JL, Saavedra MT, Randell SH, Vasil ML, Burns JL, Moskowitz SM, Nick JA. 2011. Neutrophil extracellular trap (NET)-mediated killing of Pseudomonas aeruginosa: evidence of acquired resistance within the CF airway, independent of CFTR. PLoS One 6:e23637. doi: 10.1371/journal.pone.0023637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang P-X, Murray TS, Villella VR, Ferrari E, Esposito S, D’Souza A, Raia V, Maiuri L, Krause DS, Egan ME, Bruscia EM. 2013. Reduced caveolin-1 promotes hyperinflammation due to abnormal heme oxygenase-1 localization in lipopolysaccharide-challenged macrophages with dysfunctional cystic fibrosis transmembrane conductance regulator. J Immunol 190:5196–5206. doi: 10.4049/jimmunol.1201607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lamothe J, Valvano MA. 2008. Burkholderia cenocepacia-induced delay of acidification and phagolysosomal fusion in cystic fibrosis transmembrane conductance regulator (CFTR)-defective macrophages. Microbiology 154:3825–3834. doi: 10.1099/mic.0.2008/023200-0. [DOI] [PubMed] [Google Scholar]
- 93.Kushwah R, Gagnon S, Sweezey NB. 2013. Intrinsic predisposition of naive cystic fibrosis T cells to differentiate towards a Th17 phenotype. Respir Res 14:138. doi: 10.1186/1465-9921-14-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cormet-Boyaka E, Caution K, Amer AO. 2016. Autophagy in cystic fibrosis pathogenesis and treatment, p 245–265. In Gorbunov N. (ed), Autophagy in current trends in cellular physiology and pathology. IntechOpen, London, United Kingdom. [Google Scholar]
- 95.Villella VR, Esposito S, Bruscia EM, Maiuri MC, Raia V, Kroemer G, Maiuri L. 2013. Targeting the intracellular environment in cystic fibrosis: restoring autophagy as a novel strategy to circumvent the CFTR defect. Front Pharmacol 4:1. doi: 10.3389/fphar.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ryter SW, Choi AMK. 2015. Autophagy in lung disease pathogenesis and therapeutics. Redox Biol 4:215–225. doi: 10.1016/j.redox.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dikic I, Elazar Z. 2018. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 98.Maróti G, Kereszt A, Kondorosi É, Mergaert P. 2011. Natural roles of antimicrobial peptides in microbes, plants and animals. Res Microbiol 162:363–374. doi: 10.1016/j.resmic.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 99.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 100.Tam JP, Wang S, Wong KH, Tan WL. 2015. Antimicrobial peptides from plants. Pharmaceuticals (Basel) 8:711–757. doi: 10.3390/ph8040711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parret AHA, De Mot R. 2002. Bacteria killing their own kind: novel bacteriocins of Pseudomonas and other γ-proteobacteria. Trends Microbiol 10:107–112. doi: 10.1016/S0966-842X(02)02307-7. [DOI] [PubMed] [Google Scholar]
- 102.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. doi: 10.1016/S0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 103.Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rivas-Santiago B, Serrano CJ, Enciso-Moreno JA. 2009. Susceptibility to infectious diseases based on antimicrobial peptide production. Infect Immun 77:4690–4695. doi: 10.1128/IAI.01515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zanetti M, Gennaro R, Romeo D. 1995. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett 374:1–5. doi: 10.1016/0014-5793(95)01050-O. [DOI] [PubMed] [Google Scholar]
- 106.Zanetti M. 2005. The role of cathelicidins in the innate host defenses of mammals. Curr Issues Mol Biol 7:179–196. [PubMed] [Google Scholar]
- 107.Li Y, Xiang Q, Zhang Q, Huang Y, Su Z. 2012. Overview on the recent study of antimicrobial peptides: origins, functions, relative mechanisms and application. Peptides 37:207–215. doi: 10.1016/j.peptides.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 108.Mihajlovic M, Lazaridis T. 2010. Antimicrobial peptides bind more strongly to membrane pores. Biochim Biophys Acta 1798:1494–1502. doi: 10.1016/j.bbamem.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sengupta D, Leontiadou H, Mark AE, Marrink SJ. 2008. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim Biophys Acta 1778:2308–2317. doi: 10.1016/j.bbamem.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 110.Wimley WC. 2010. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol 5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patrzykat A, Friedrich CL, Zhang L, Mendoza V, Hancock REW. 2002. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob Agents Chemother 46:605–614. doi: 10.1128/AAC.46.3.605-614.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ulvatne H, Samuelsen Ø, Haukland HH, Krämer M, Vorland LH. 2004. Lactoferricin B inhibits bacterial macromolecular synthesis in Escherichia coli and Bacillus subtilis. FEMS Microbiol Lett 237:377–384. doi: 10.1016/j.femsle.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 113.Tecle T, Tripathi S, Hartshorn KL. 2010. Defensins and cathelicidins in lung immunity. Innate Immun 16:151–159. doi: 10.1177/1753425910365734. [DOI] [PubMed] [Google Scholar]
- 114.Cole AM, Waring AJ. 2002. The role of defensins in lung biology and therapy. Am J Respir Med 1:249–259. doi: 10.1007/BF03256616. [DOI] [PubMed] [Google Scholar]
- 115.Steinstraesser L, Kraneburg U, Jacobsen F, Al-Benna S. 2011. Host defense peptides and their antimicrobial-immunomodulatory duality. Immunobiology 216:322–333. doi: 10.1016/j.imbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 116.Yang D, Chertov O, Oppenheim JJ. 2001. Participation of mammalian defensins and cathelicidins in anti-microbial immunity: receptors and activities of human defensins and cathelicidin (LL-37). J Leukoc Biol 69:691–697. [PubMed] [Google Scholar]
- 117.Bals R. 2000. Epithelial antimicrobial peptides in host defense against infection. Respir Res 1:141–150. doi: 10.1186/rr25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soong LB, Ganz T, Ellison A, Caughey GH. 1997. Purification and characterization of defensins from cystic fibrosis sputum. Inflamm Res 46:98–102. doi: 10.1007/s000110050114. [DOI] [PubMed] [Google Scholar]
- 119.Chen CI-U, Schaller-Bals S, Paul KP, Wahn U, Bals R. 2004. Beta-defensins and LL-37 in bronchoalveolar lavage fluid of patients with cystic fibrosis. J Cyst Fibros 3:45–50. doi: 10.1016/j.jcf.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 120.Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. 1997. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553–560. doi: 10.1016/S0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 121.Dalcin D, Ulanova M. 2013. The role of human beta-defensin-2 in Pseudomonas aeruginosa pulmonary infection in cystic fibrosis patients. Infect Dis Ther 2:159–166. doi: 10.1007/s40121-013-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hancock REW, Haney EF, Gill EE. 2016. The immunology of host defence peptides: beyond antimicrobial activity. Nat Rev Immunol 16:321–334. doi: 10.1038/nri.2016.29. [DOI] [PubMed] [Google Scholar]
- 123.Sørensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N. 2001. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97:3951–3959. doi: 10.1182/blood.V97.12.3951. [DOI] [PubMed] [Google Scholar]
- 124.Bals R, Wang X, Zasloff M, Wilson J. 1998. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc Natl Acad Sci U S A 95:9541–9546. doi: 10.1073/pnas.95.16.9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Frohm M, Agerberth B, Ahangari G, Stahle-Backdahl M, Liden S, Wigzell H, Gudmundsson GH. 1997. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J Biol Chem 272:15258–15263. doi: 10.1074/jbc.272.24.15258. [DOI] [PubMed] [Google Scholar]
- 126.Wang T-T, Nestel FP, Bourdeau V, Nagai Y, Wang Q, Liao J, Tavera-Mendoza L, Lin R, Hanrahan JW, Mader S, White JH, Hanrahan JH. 2004. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J Immunol 173:2909–2912. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- 127.Saiman L, Tabibi S, Starner TD, San Gabriel P, Winokur PL, Jia HP, McCray PB Jr, Tack BF. 2001. Cathelicidin peptides inhibit multiply antibiotic-resistant pathogens from patients with cystic fibrosis. Antimicrob Agents Chemother 45:2838–2844. doi: 10.1128/AAC.45.10.2838-2844.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Overhage J, Campisano A, Bains M, Torfs ECW, Rehm BHA, Hancock REW. 2008. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect Immun 76:4176–4182. doi: 10.1128/IAI.00318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen X, Niyonsaba F, Ushio H, Okuda D, Nagaoka I, Ikeda S, Okumura K, Ogawa H. 2005. Synergistic effect of antibacterial agents human β-defensins, cathelicidin LL-37 and lysozyme against Staphylococcus aureus and Escherichia coli. J Dermatol Sci 40:123–132. doi: 10.1016/j.jdermsci.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 130.Singh PK, Tack BF, McCray PB Jr, Welsh MJ. 2000. Synergistic and additive killing by antimicrobial factors found in human airway surface liquid. Am J Physiol Lung Cell Mol Physiol 279:L799–L805. doi: 10.1152/ajplung.2000.279.5.L799. [DOI] [PubMed] [Google Scholar]
- 131.Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y. 1999. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J 341:501–513. doi: 10.1042/0264-6021:3410501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zeth K, Sancho-Vaello E. 2017. The human antimicrobial peptides dermcidin and LL-37 show novel distinct pathways in membrane interactions. Front Chem 5:86. doi: 10.3389/fchem.2017.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Y, Qian Z, Ma L, Hu S, Nong D, Xu C, Ye F, Lu Y, Wei G, Li M. 2016. Single-molecule visualization of dynamic transitions of pore-forming peptides among multiple transmembrane positions. Nat Commun 7:12906. doi: 10.1038/ncomms12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. 2011. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc Natl Acad Sci U S A 108:E77–E81. doi: 10.1073/pnas.1101130108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van Wetering S, Tjabringa GS, Hiemstra PS. 2005. Interactions between neutrophil-derived antimicrobial peptides and airway epithelial cells. J Leukoc Biol 77:444–450. doi: 10.1189/jlb.0604367. [DOI] [PubMed] [Google Scholar]
- 136.Yang D, Chen Q, Schmidt AP, Anderson GM, Wang JM, Wooters J, Oppenheim JJ, Chertov O. 2000. LL-37, the neutrophil granule- and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J Exp Med 192:1069–1074. doi: 10.1084/jem.192.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zheng Y, Niyonsaba F, Ushio H, Nagaoka I, Ikeda S, Okumura K, Ogawa H. 2007. Cathelicidin LL-37 induces the generation of reactive oxygen species and release of human α-defensins from neutrophils. Br J Dermatol 157:1124–1131. doi: 10.1111/j.1365-2133.2007.08196.x. [DOI] [PubMed] [Google Scholar]
- 138.Dürr UHN, Sudheendra US, Ramamoorthy A. 2006. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta 1758:1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 139.Bals R, Weiner DJ, Moscioni AD, Meegalla RL, Wilson JM. 1999. Augmentation of innate host defense by expression of a cathelicidin antimicrobial peptide. Infect Immun 67:6084–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bergsson G, Reeves EP, McNally P, Chotirmall SH, Greene CM, Greally P, Murphy P, O’Neill SJ, McElvaney NG. 2009. LL-37 complexation with glycosaminoglycans in cystic fibrosis lungs inhibits antimicrobial activity, which can be restored by hypertonic saline. J Immunol 183:543–551. doi: 10.4049/jimmunol.0803959. [DOI] [PubMed] [Google Scholar]
- 141.McShane D, Davies JC, Davies MG, Bush A, Geddes DM, Alton EWFW. 2003. Airway surface pH in subjects with cystic fibrosis. Eur Respir J 21:37–42. doi: 10.1183/09031936.03.00027603. [DOI] [PubMed] [Google Scholar]
- 142.Schultz A, Puvvadi R, Borisov SM, Shaw NC, Klimant I, Berry LJ, Montgomery ST, Nguyen T, Kreda SM, Kicic A, Noble PB, Button B, Stick SM. 2017. Airway surface liquid pH is not acidic in children with cystic fibrosis. Nat Commun 8:1409. doi: 10.1038/s41467-017-00532-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Figueira MF, Webster MJ, Tarran R. 2018. Rebuttal from Miriam F. Figueira, Megan J. Webster and Robert Tarran. J Physiol 596:3443–3444. doi: 10.1113/JP276146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bals R, Weiner DJ, Meegalla RL, Wilson JM. 1999. Transfer of a cathelicidin peptide antibiotic gene restores bacterial killing in a cystic fibrosis xenograft model. J Clin Invest 103:1113–1117. doi: 10.1172/JCI6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 146.Nizet V. 2006. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr Issues Mol Biol 8:11–26. [PubMed] [Google Scholar]
- 147.Limoli DH, Rockel AB, Host KM, Jha A, Kopp BT, Hollis T, Wozniak DJ. 2014. Cationic antimicrobial peptides promote microbial mutagenesis and pathoadaptation in chronic infections. PLoS Pathog 10:e1004083. doi: 10.1371/journal.ppat.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rodríguez-Rojas A, Makarova O, Müller U, Rolff J. 2015. Cationic peptides facilitate iron-induced mutagenesis in bacteria. PLoS Genet 11:e1005546. doi: 10.1371/journal.pgen.1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vatansever F, de Melo WCMA, Avci P, Vecchio D, Sadasivam M, Gupta A, Chandran R, Karimi M, Parizotto NA, Yin R, Tegos GP, Hamblin MR. 2013. Antimicrobial strategies centered around reactive oxygen species—bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol Rev 37:955–989. doi: 10.1111/1574-6976.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Robinson JM. 2008. Reactive oxygen species in phagocytic leukocytes. Histochem Cell Biol 130:281–297. doi: 10.1007/s00418-008-0461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol 2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 152.Rada B, Leto TL. 2008. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib Microbiol 15:164–187. doi: 10.1159/000136357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fischer H. 2009. Mechanisms and function of DUOX in epithelia of the lung. Antioxid Redox Signal 11:2453–2465. doi: 10.1089/ars.2009.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pongnimitprasert N, El-Benna J, Foglietti M, Gougerot-Pocidalo M, Bernard M, Braut-Boucher F. 2008. Potential role of the “NADPH oxidases” (NOX/DUOX) family in cystic fibrosis. Ann Biol Clin (Paris) 66:621–629. doi: 10.1684/abc.2008.0285. [DOI] [PubMed] [Google Scholar]
- 155.Vignais PV. 2002. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell Mol Life Sci 59:1428–1459. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nauseef WM. 2004. Assembly of the phagocyte NADPH oxidase. Histochem Cell Biol 122:277–291. doi: 10.1007/s00418-004-0679-8. [DOI] [PubMed] [Google Scholar]
- 157.Laroux FS, Romero X, Wetzler L, Engel P, Terhorst C. 2005. Cutting edge: MyD88 controls phagocyte NADPH oxidase function and killing of Gram-negative bacteria. J Immunol 175:5596–5600. doi: 10.4049/jimmunol.175.9.5596. [DOI] [PubMed] [Google Scholar]
- 158.Winterbourn CC, Kettle AJ, Hampton MB. 2016. Reactive oxygen species and neutrophil function. Annu Rev Biochem 85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 159.Holland SM. 2013. Chronic granulomatous disease. Hematol Oncol Clin North Am 27:89–99. doi: 10.1016/j.hoc.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Imlay JA, Linn S. 1986. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol 166:519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fang FC. 2011. Antimicrobial actions of reactive oxygen species. mBio 2:e00141-11. doi: 10.1128/mBio.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Demple B. 1991. Regulation of bacterial oxidative stress genes. Annu Rev Genet 25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 163.Shohet SB, Pitt J, Baehner RL, Poplack DG. 1974. Lipid peroxidation in the killing of phagocytized pneumococci. Infect Immun 10:1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dukan S, Touati D. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol 178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Paiva CN, Bozza MT. 2014. Are reactive oxygen species always detrimental to pathogens? Antioxid Redox Signal 20:1000–1037. doi: 10.1089/ars.2013.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Panmanee W, Hassett DJ. 2009. Differential roles of OxyR-controlled antioxidant enzymes alkyl hydroperoxide reductase (AhpCF) and catalase (KatB) in the protection of Pseudomonas aeruginosa against hydrogen peroxide in biofilm vs. planktonic culture. FEMS Microbiol Lett 295:238–244. doi: 10.1111/j.1574-6968.2009.01605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bonvillain RW, Painter RG, Ledet EM, Wang G. 2011. Comparisons of resistance of CF and non-CF pathogens to hydrogen peroxide and hypochlorous acid oxidants in vitro. BMC Microbiol 11:112. doi: 10.1186/1471-2180-11-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. 2006. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J Biol Chem 281:39860–39869. doi: 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- 169.Antus B. 2016. Oxidative stress markers in sputum. Oxid Med Cell Longev 2016:2930434. doi: 10.1155/2016/2930434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hull J, Vervaart P, Grimwood K, Phelan P. 1997. Pulmonary oxidative stress response in young children with cystic fibrosis. Thorax 52:557–560. doi: 10.1136/thx.52.6.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sadowska-Bartosz I, Galiniak S, Bartosz G, Rachel M. 2014. Oxidative modification of proteins in pediatric cystic fibrosis with bacterial infections. Oxid Med Cell Longev 2014:389629. doi: 10.1155/2014/389629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Starosta V, Rietschel E, Paul K, Baumann U, Griese M. 2006. Oxidative changes of bronchoalveolar proteins in cystic fibrosis. Chest 129:431–437. doi: 10.1378/chest.129.2.431. [DOI] [PubMed] [Google Scholar]
- 173.Witko-Sarsat V, Delacourt C, Rabier D, Bardet J, Nguyen AT, Descamps-Latscha B. 1995. Neutrophil-derived long-lived oxidants in cystic fibrosis sputum. Am J Respir Crit Care Med 152:1910–1916. doi: 10.1164/ajrccm.152.6.8520754. [DOI] [PubMed] [Google Scholar]
- 174.McGrath LT, Mallon P, Dowey L, Silke B, McClean E, McDonnell M, Devine A, Copeland S, Elborn S. 1999. Oxidative stress during acute respiratory exacerbations in cystic fibrosis. Thorax 54:518–523. doi: 10.1136/thx.54.6.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thomson E, Brennan S, Senthilmohan R, Gangell CL, Chapman AL, Sly PD, Kettle AJ, Australian Respiratory Early Surveillance Team for Cystic Fibrosis. 2010. Identifying peroxidases and their oxidants in the early pathology of cystic fibrosis. Free Radic Biol Med 49:1354–1360. doi: 10.1016/j.freeradbiomed.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 176.Kettle AJ, Chan T, Osberg I, Senthilmohan R, Chapman ALP, Mocatta TJ, Wagener JS. 2004. Myeloperoxidase and protein oxidation in the airways of young children with cystic fibrosis. Am J Respir Crit Care Med 170:1317–1323. doi: 10.1164/rccm.200311-1516OC. [DOI] [PubMed] [Google Scholar]
- 177.Brown RK, Kelly FJ. 1994. Role of free radicals in the pathogenesis of cystic fibrosis. Thorax 49:738–742. doi: 10.1136/thx.49.8.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ziady AG, Hansen J. 2014. Redox balance in cystic fibrosis. Int J Biochem Cell Biol 52:113–123. doi: 10.1016/j.biocel.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Aquilano K, Baldelli S, Ciriolo MR. 2014. Glutathione: new roles in redox signalling for an old antioxidant. Front Pharmacol 5:196. doi: 10.3389/fphar.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Linsdell P, Hanrahan JW. 1998. Glutathione permeability of CFTR. Am J Physiol 275:C323–C326. doi: 10.1152/ajpcell.1998.275.1.C323. [DOI] [PubMed] [Google Scholar]
- 181.Gao L, Kim KJ, Yankaskas JR, Forman HJ. 1999. Abnormal glutathione transport in cystic fibrosis airway epithelia. Am J Physiol Lung Cell Mol Physiol 277:L113–L118. doi: 10.1152/ajplung.1999.277.1.L113. [DOI] [PubMed] [Google Scholar]
- 182.Hassett DJ, Cohen MS. 1989. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J 3:2574–2582. doi: 10.1096/fasebj.3.14.2556311. [DOI] [PubMed] [Google Scholar]
- 183.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JIA, Jensen P, Johnsen AH, Givskov M, Ohman DE, Soren M, Hoiby N, Kharazmi A. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 184.Li Q, Engelhardt JF. 2006. Interleukin-1β induction of NFκB is partially regulated by H2O2-mediated activation of NFκB-inducing kinase. J Biol Chem 281:1495–1505. doi: 10.1074/jbc.M511153200. [DOI] [PubMed] [Google Scholar]
- 185.Chen J, Kinter M, Shank S, Cotton C, Kelley TJ, Ziady AG. 2008. Dysfunction of Nrf-2 in CF epithelia leads to excess intracellular H2O2 and inflammatory cytokine production. PLoS One 3:e3367. doi: 10.1371/journal.pone.0003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wright DT, Cohn LA, Li H, Fischer B, Li CM, Adler KB. 1994. Interactions of oxygen radicals with airway epithelium. Environ Health Perspect 102:85–90. doi: 10.2307/3432221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cochrane CG. 1991. Cellular injury by oxidants. Am J Med 91:23S–30S. doi: 10.1016/0002-9343(91)90280-B. [DOI] [PubMed] [Google Scholar]
- 188.Hubeau C, Puchelle E, Gaillard D. 2001. Distinct pattern of immune cell population in the lung of human fetuses with cystic fibrosis. J Allergy Clin Immunol 108:524–529. doi: 10.1067/mai.2001.118516. [DOI] [PubMed] [Google Scholar]
- 189.Armstrong DS, Grimwood K, Carzino R, Carlin JB, Olinsky A, Phelan PD. 1995. Lower respiratory infection and inflammation in infants with newly diagnosed cystic fibrosis. BMJ 310:1571–1572. doi: 10.1136/bmj.310.6994.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Gutierrez JP, Hull J, Olinsky A, Phelan EM, Robertson CF, Phelan PD. 1997. Lower airway inflammation in infants and young children with cystic fibrosis. Am J Respir Crit Care Med 156:1197–1204. doi: 10.1164/ajrccm.156.4.96-11058. [DOI] [PubMed] [Google Scholar]
- 191.Armstrong DS, Hook SM, Jamsen KM, Nixon GM, Carzino R, Carlin JB, Robertson CF, Grimwood K. 2005. Lower airway inflammation in infants with cystic fibrosis detected by newborn screening. Pediatr Pulmonol 40:500–510. doi: 10.1002/ppul.20294. [DOI] [PubMed] [Google Scholar]
- 192.Meyerholz DK, Stoltz DA, Namati E, Ramachandran S, Pezzulo AA, Smith AR, Rector MV, Suter MJ, Kao S, McLennan G, Tearney GJ, Zabner J, McCray PB Jr, Welsh MJ. 2010. Loss of cystic fibrosis transmembrane conductance regulator function produces abnormalities in tracheal development in neonatal pigs and young children. Am J Respir Crit Care Med 182:1251–1261. doi: 10.1164/rccm.201004-0643OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Stoltz DA, Meyerholz DK, Pezzulo AA, Ramachandran S, Rogan MP, Davis GJ, Hanfland RA, Wohlford-Lenane C, Dohrn CL, Bartlett JA, Nelson GA IV, Eugene C, Taft PJ, Ludwig PS, Estin M, Hornick EE, Launspach JL, Samuel M, Rokhlina T, Karp PH, Ostedgaard LS, Uc A, Starner TD, Horswill AR, Brogden KA, Prather RS, Richter SS, Shilyansky J, McCray PB Jr, Zabner J, Welsh MJ. 2010. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci Transl Med 2:29ra31. doi: 10.1126/scitranslmed.3000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Waters V, Ratjen F. 2015. Pulmonary exacerbations in children with cystic fibrosis. Ann Am Thorac Soc 12:S200–S206. doi: 10.1513/AnnalsATS.201502-098AW. [DOI] [PubMed] [Google Scholar]
- 195.Bhatt JM. 2013. Treatment of pulmonary exacerbations in cystic fibrosis. Eur Respir Rev 22:205–216. doi: 10.1183/09059180.00006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Abbott J, Holt A, Hart A, Morton AM, MacDougall L, Pogson M, Milne G, Rodgers HC, Conway SP. 2009. What defines a pulmonary exacerbation? The perceptions of adults with cystic fibrosis. J Cyst Fibros 8:356–359. doi: 10.1016/j.jcf.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 197.Mets OM, Roothaan SM, Bronsveld I, Luijk B, Van De Graaf EA, Vink A, De Jong PA. 2015. Emphysema is common in lungs of cystic fibrosis lung transplantation patients: a histopathological and computed tomography study. PLoS One 10:e0128062. doi: 10.1371/journal.pone.0128062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Perera PL, Screaton NJ. 2011. Radiological features of bronchiectasis. Eur Respir Mon 52:44–67. [Google Scholar]
- 199.Moreno P, Alvarez A, Carrasco G, Redel J, Guaman HD, Baamonde C, Algar FJ, Cerezo F, Salvatierra A. 2016. Lung transplantation for cystic fibrosis: differential characteristics and outcomes between children and adults. Eur J Cardiothorac Surg 49:1334–1343. doi: 10.1093/ejcts/ezv377. [DOI] [PubMed] [Google Scholar]
- 200.Rosenblatt RL. 2009. Lung transplantation for cystic fibrosis. Respir Care 54:777–787. doi: 10.4187/002013209790983197. [DOI] [PubMed] [Google Scholar]
- 201.Adler FR, Aurora P, Barker DH, Barr ML, Blackwell LS, Bosma OH, Brown S, Cox DR, Jensen JL, Kurland G, Nossent GD, Quittner AL, Robinson WM, Romero SL, Spencer H, Sweet SC, van der Bij W, Vermeulen J, Verschuuren EAM, Vrijlandt EJLE, Walsh W, Woo MS, Liou TG. 2009. Lung transplantation for cystic fibrosis. Proc Am Thorac Soc 6:619–633. doi: 10.1513/pats.2009008-088TL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.O’Dwyer DN, Dickson RP, Moore BB. 2016. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J Immunol 196:4839–4847. doi: 10.4049/jimmunol.1600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shukla SD, Budden KF, Neal R, Hansbro PM. 2017. Microbiome effects on immunity, health and disease in the lung. Clin Transl Immunol 6:e133. doi: 10.1038/cti.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dickson RP, Huffnagle GB. 2015. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog 11:e1004923. doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.LiPuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Surette MG. 2014. The cystic fibrosis lung microbiome. Ann Am Thorac Soc 11:S61–S65. doi: 10.1513/AnnalsATS.201306-159MG. [DOI] [PubMed] [Google Scholar]
- 207.Pittman JE, Wylie KM, Akers K, Storch GA, Hatch J, Quante J, Frayman KB, Clarke N, Davis M, Stick SM, Hall GL, Montgomery G, Ranganathan S, Davis SD, Ferkol TW, Australian Respiratory Early Surveillance Team for Cystic Fibrosis. 2017. Association of antibiotics, airway microbiome and inflammation in infants with cystic fibrosis. Ann Am Thorac Soc 14:1548–1555. doi: 10.1513/AnnalsATS.201702-121OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bernarde C, Keravec M, Mounier J, Gouriou S, Rault G, Férec C, Barbier G, Héry-Arnaud G. 2015. Impact of the CFTR-potentiator ivacaftor on airway microbiota in cystic fibrosis patients carrying a G551D mutation. PLoS One 10:e0124124. doi: 10.1371/journal.pone.0124124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hisert KB, Heltshe SL, Pope C, Jorth P, Wu X, Edwards RM, Radey M, Accurso FJ, Wolter DJ, Cooke G, Adam RJ, Carter S, Grogan B, Launspach JL, Donnelly SC, Gallagher CG, Bruce JE, Stoltz DA, Welsh MJ, Hoffman LR, McKone EF, Singh PK. 2017. Restoring cystic fibrosis transmembrane conductance regulator function reduces airway bacteria and inflammation in people with cystic fibrosis and chronic lung infections. Am J Respir Crit Care Med 195:1617–1628. doi: 10.1164/rccm.201609-1954OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Caverly LJ, Zhao J, LiPuma JJ. 2015. Cystic fibrosis lung microbiome: opportunities to reconsider management of airway infection. Pediatr Pulmonol 50:S31–S38. doi: 10.1002/ppul.23243. [DOI] [PubMed] [Google Scholar]
- 211.Huang YJ, LiPuma JJ. 2016. The microbiome in cystic fibrosis. Clin Chest Med 37:59–67. doi: 10.1016/j.ccm.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gilligan PH. 1991. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev 4:35–51. doi: 10.1128/CMR.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K, Wainwright C. 2002. Early airway infection, inflammation, and lung function in cystic fibrosis. Arch Dis Child 87:306–311. doi: 10.1136/adc.87.4.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Pillarisetti N, Williamson E, Linnane B, Skoric B, Robertson CF, Robinson P, Massie J, Hall GL, Sly P, Stick S, Ranganathan S. 2011. Infection, inflammation, and lung function decline in infants with cystic fibrosis. Am J Respir Crit Care Med 184:75–81. doi: 10.1164/rccm.201011-1892OC. [DOI] [PubMed] [Google Scholar]
- 215.Frayman KB, Armstrong DS, Grimwood K, Ranganathan SC. 2017. The airway microbiota in early cystic fibrosis lung disease. Pediatr Pulmonol 52:1384–1404. doi: 10.1002/ppul.23782. [DOI] [PubMed] [Google Scholar]
- 216.Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ, Ranganathan SC, Waters VJ, Ratjen FA. 2014. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, Gram-negative bacteria, and multiple infections. Ann Am Thorac Soc 11:1120–1129. doi: 10.1513/AnnalsATS.201402-050AS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tummler B, Bosshammer J, Breitnestein S, Brockhausen I, Gudowius P, Herrmann C, Herrman S, Heuer T, Kubesch P, Mekus F, Romling U, Schmidt KD, Spangenberg C, Walter S. 1997. Vaccination against Pseudomonas aeruginosa. Behring Inst Mitt 98:249–255. [PubMed] [Google Scholar]
- 218.Schobert M, Jahn D. 2010. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol 300:549–556. doi: 10.1016/j.ijmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 219.Klockgether J, Cramer N, Wiehlmann L, Davenport CF, Tümmler B. 2011. Pseudomonas aeruginosa genomic structure and diversity. Front Microbiol 2:150. doi: 10.3389/fmicb.2011.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Meliani A, Bensoltane A. 2017. Pseudomonas chemotaxis, motility and host-pathogen interactions. MOJ Immunol 5:00167. [Google Scholar]
- 221.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011-2014. Infect Control Hosp Epidemiol 37:1288–1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Crabtree TD, Gleason TG, Pruett TL, Sawyer RG. 1999. Trends in nosocomial pneumonia in surgical patients as we approach the 21st century: a prospective analysis. Am Surg 65:706–710. [PubMed] [Google Scholar]
- 224.Gaynes R, Edwards JR, National Nosocomial Infections Surveillance System. 2005. Overview of nosocomial infections caused by Gram-negative bacilli. Clin Infect Dis 41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 225.Favero MS, Carson LA, Bond WW, Petersen NJ. 1971. Pseudomonas aeruginosa: growth in distilled water from hospitals. Science 173:836–838. doi: 10.1126/science.173.3999.836. [DOI] [PubMed] [Google Scholar]
- 226.Vasil ML. 1986. Pseudomonas aeruginosa: biology, mechanisms of virulence, epidemiology. J Pediatr 108:800–805. doi: 10.1016/S0022-3476(86)80748-X. [DOI] [PubMed] [Google Scholar]
- 227.Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mayank D, Anshuman M, Singh RK, Afzal A, Baronia AK, Prasad KN. 2009. Nosocomial cross-transmission of Pseudomonas aeruginosa between patients in a tertiary intensive care unit. Indian J Pathol Microbiol 52:509–513. doi: 10.4103/0377-4929.56143. [DOI] [PubMed] [Google Scholar]
- 229.Quick J, Cumley N, Wearn CM, Niebel M, Constantinidou C, Thomas CM, Pallen MJ, Moiemen NS, Bamford A, Oppenheim B, Loman NJ. 2014. Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: an observational study using whole-genome sequencing. BMJ Open 4:e006278. doi: 10.1136/bmjopen-2014-006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yakupogullari Y, Otlu B, Dogukan M, Gursoy C, Korkmaz E, Kizirgil A, Ozden M, Durmaz R. 2008. Investigation of a nosocomial outbreak by alginate-producing pan-antibiotic-resistant Pseudomonas aeruginosa. Am J Infect Control 36:e13–e18. doi: 10.1016/j.ajic.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 231.Tsutsui A, Suzuki S, Yamane K, Matsui M, Konda T, Marui E, Takahashi K, Arakawa Y. 2011. Genotypes and infection sites in an outbreak of multidrug-resistant Pseudomonas aeruginosa. J Hosp Infect 78:317–322. doi: 10.1016/j.jhin.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 232.Ratnam S, Hogan K, March SB, Butler RW. 1986. Whirlpool-associated follicilitis caused by Pseudomonas aeruginosa: report of an outbreak and review. J Clin Microbiol 23:655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.van Asperen IA, de Rover CM, Schijven JF, Oetomo SB, Schellekens JFP, van Leeuwen NJ, Colle C, Havelaar AH, Kromhout D, Sprenger MWJ. 1995. Risk of otitis externa after swimming in recreational fresh water lakes containing Pseudomonas aeruginosa. BMJ 311:1407–1410. doi: 10.1136/bmj.311.7017.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hirsch EB, Tam VH. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res 10:441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. 2006. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother 50:43–48. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. 2010. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol 59:253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 237.Rasamiravaka T, Labtani Q, Duez P, El Jaziri M. 2015. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int 2015:759348. doi: 10.1155/2015/759348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Parsek MR, Singh PK. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 240.Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell Microbiol 11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 241.Sanders DB, Fink A. 2016. Background and epidemiology. Pediatr Clin North Am 63:567–584. doi: 10.1016/j.pcl.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Henry RL, Mellis CM, Petrovic L. 1992. Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr Pulmonol 12:158–161. doi: 10.1002/ppul.1950120306. [DOI] [PubMed] [Google Scholar]
- 243.Kerem E, Corey M, Gold R, Levison H. 1990. Pulmonary function and clinical course in patients with cystic fibrosis after pulmonary colonization with Pseudomonas aeruginosa. J Pediatr 116:714–719. doi: 10.1016/S0022-3476(05)82653-8. [DOI] [PubMed] [Google Scholar]
- 244.Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, Laxova A, Green CG, Collins J, Farrell PM. 2001. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol 32:277–287. doi: 10.1002/ppul.2009.abs. [DOI] [PubMed] [Google Scholar]
- 245.Coburn B, Wang PW, Diaz Caballero J, Clark ST, Brahma V, Donaldson S, Zhang Y, Surendra A, Gong Y, Elizabeth Tullis D, Yau YCW, Waters VJ, Hwang DM, Guttman DS. 2015. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 5:10241. doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Pamukcu A, Bush A, Buchdahl R. 1995. Effects of Pseudomonas aeruginosa colonization on lung function and anthropometric variables in children with cystic fibrosis. Pediatr Pulmonol 19:10–15. doi: 10.1002/ppul.1950190103. [DOI] [PubMed] [Google Scholar]
- 247.Zemanick ET, Harris JK, Wagner BD, Robertson CE, Sagel SD, Stevens MJ, Accurso FJ, Laguna TA. 2013. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS One 8:e62917. doi: 10.1371/journal.pone.0062917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Li Z, Kosorok MR, Farrell PM, Laxova A, West SEH, Green CG, Rock MJ, Splaingard ML. 2005. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293:581–588. doi: 10.1001/jama.293.5.581. [DOI] [PubMed] [Google Scholar]
- 249.Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith AL, Ramsey BW. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis 183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 250.Mayer-Hamblett N, Kloster M, Rosenfeld M, Gibson RL, Retsch-Bogart GZ, Emerson J, Thompson V, Ramsey BW. 2015. Impact of sustained eradication of new Pseudomonas aeruginosa infection on long-term outcomes in cystic fibrosis. Clin Infect Dis 61:707–715. doi: 10.1093/cid/civ377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hansen CR, Pressler T, Høiby N. 2008. Early aggressive eradication therapy for intermittent Pseudomonas aeruginosa airway colonization in cystic fibrosis patients: 15 years experience. J Cyst Fibros 7:523–530. doi: 10.1016/j.jcf.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 252.Langan KM, Kotsimbos T, Peleg AY. 2015. Managing Pseudomonas aeruginosa respiratory infections in cystic fibrosis. Curr Opin Infect Dis 28:547–556. doi: 10.1097/QCO.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 253.Bjarnsholt T, Jensen PO, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Hoiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol 44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 254.Hoiby N, Ciofu O, Bjarnsholt T. 2010. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 255.Rudkjøbing VB, Thomsen TR, Alhede M, Kragh KN, Nielsen PH, Johansen UR, Givskov M, Høiby N, Bjarnsholt T. 2012. The microorganisms in chronically infected end-stage and non-end-stage cystic fibrosis patients. FEMS Immunol Med Microbiol 65:236–244. doi: 10.1111/j.1574-695X.2011.00925.x. [DOI] [PubMed] [Google Scholar]
- 256.Brennan S. 2008. Innate immune activation and cystic fibrosis. Paediatr Respir Rev 9:271–280. doi: 10.1016/j.prrv.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 257.Gonçalves-de-Albuquerque CF, Silva AR, Burth P, Rocco PRM, Castro-Faria MV, Castro-Faria-Neto HC. 2016. Possible mechanisms of Pseudomonas aeruginosa-associated lung disease. Int J Med Microbiol 306:20–28. doi: 10.1016/j.ijmm.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 258.Koh AY, Priebe GP, Ray C, Van Rooijen N, Pier GB. 2009. Inescapable need for neutrophils as mediators of cellular innate immunity to acute Pseudomonas aeruginosa pneumonia. Infect Immun 77:5300–5310. doi: 10.1128/IAI.00501-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Fujitani S, Sun HY, Yu VL, Weingarten JA. 2011. Pneumonia due to Pseudomonas aeruginosa. Part I: epidemiology, clinical diagnosis, and source. Chest 139:909–919. doi: 10.1378/chest.10-0166. [DOI] [PubMed] [Google Scholar]
- 260.Carratala J, Roson B, Fernandez-Sevilla A, Alcaide F, Gudiol F. 1998. Bacteremic pneumonia in neutropenic patients with cancer. Arch Intern Med 158:868–872. doi: 10.1001/archinte.158.8.868. [DOI] [PubMed] [Google Scholar]
- 261.Andrews T, Sullivan KE. 2003. Infections in patients with inherited defects in phagocytic function. Clin Microbiol Rev 16:597–621. doi: 10.1128/CMR.16.4.597-621.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Huang LC, Reins RY, Gallo RL, McDermott AM. 2007. Cathelicidin-deficient (Cnlp−/−) mice show increased susceptibility to Pseudomonas aeruginosa keratitis. Invest Ophthalmol Vis Sci 48:4498–4508. doi: 10.1167/iovs.07-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Beaumont PE, McHugh B, Gwyer Findlay E, Mackellar A, Mackenzie KJ, Gallo RL, Govan JRW, Simpson AJ, Davidson DJ. 2014. Cathelicidin host defence peptide augments clearance of pulmonary Pseudomonas aeruginosa infection by its influence on neutrophil function in vivo. PLoS One 9:e99029. doi: 10.1371/journal.pone.0099029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Diaz MH, Shaver CM, King JD, Musunuri S, Kazzaz JA, Hauser AR. 2008. Pseudomonas aeruginosa induces localized immunosuppression during pneumonia. Infect Immun 76:4414–4421. doi: 10.1128/IAI.00012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Allen L, Dockrell DH, Pattery T, Lee DG, Cornelis P, Hellewell PG, Whyte MK. 2005. Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. J Immunol 174:3643–3649. doi: 10.4049/jimmunol.174.6.3643. [DOI] [PubMed] [Google Scholar]
- 266.Managò A, Becker KA, Carpinteiro A, Wilker B, Soddemann M, Seitz AP, Edwards MJ, Grassmé H, Szabò I, Gulbins E. 2015. Pseudomonas aeruginosa pyocyanin induces neutrophil and mitochondrial acid sphingomyelinase. Antioxid Redox Signal 22:1097–1110. doi: 10.1089/ars.2014.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Alhede M, Bjarnsholt T, Jensen PØ, Phipps RK, Moser C, Christophersen L, Christensen LD, van Gennip M, Parsek M, Høiby N, Rasmussen TB, Givskov M. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155:3500–3508. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- 268.Laarman AJ, Bardoel BW, Ruyken M, Fernie J, Milder FJ, Van Strijp JAG, Rooijakkers SH. 2012. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J Immunol 188:386–393. doi: 10.4049/jimmunol.1102162. [DOI] [PubMed] [Google Scholar]
- 269.Dobrindt U, Hacker JH, Svanborg C. 2013. Preface. Between pathogenicity and commensalism. Curr Top Microbiol Immunol 358:v–vii. [DOI] [PubMed] [Google Scholar]
- 270.Hogardt M, Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol 300:557–562. doi: 10.1016/j.ijmm.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 271.Bhagirath AY, Li Y, Somayajula D, Dadashi M, Badr S, Duan K. 2016. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med 16:174. doi: 10.1186/s12890-016-0339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Cobb LM, Mychaleckyj JC, Wozniak DJ, Lopez-Boado YS. 2004. Pseudomonas aeruginosa flagellin and alginate elicit very distinct gene expression patterns in airway epithelial cells: implications for cystic fibrosis disease. J Immunol 173:5659–5670. doi: 10.4049/jimmunol.173.9.5659. [DOI] [PubMed] [Google Scholar]
- 273.Lovewell RR, Hayes SM, O’Toole GA, Berwin B. 2014. Pseudomonas aeruginosa flagellar motility activates the phagocyte PI3K/Akt pathway to induce phagocytic engulfment. Am J Physiol Lung Cell Mol Physiol 306:L698–L707. doi: 10.1152/ajplung.00319.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lovewell RR, Patankar YR, Berwin B. 2014. Mechanisms of phagocytosis and host clearance of Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol 306:L591–L603. doi: 10.1152/ajplung.00335.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Patankar YR, Lovewell RR, Poynter ME, Jyot J, Kazmierczak BI, Berwin B. 2013. Flagellar motility is a key determinant of the magnitude of the inflammasome response to Pseudomonas aeruginosa. Infect Immun 81:2043–2052. doi: 10.1128/IAI.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Floyd M, Winn M, Cullen C, Sil P, Chassaing B, Yoo D, Gewirtz A, Goldberg J, McCarter L, Rada B. 2016. Swimming motility mediates the formation of neutrophil extracellular traps induced by flagellated Pseudomonas aeruginosa. PLoS Pathog 12:e1005987. doi: 10.1371/journal.ppat.1005987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Cigana C, Curcuru L, Leone MR, Ierano T, Lore NI, Bianconi I, Silipo A, Cozzolino F, Lanzetta R, Molinaro A, Bernardini ML, Bragonzi A. 2009. Pseudomonas aeruginosa exploits lipid A and muropeptides modification as a strategy to lower innate immunity during cystic fibrosis lung infection. PLoS One 4:e8439. doi: 10.1371/journal.pone.0008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kragh KN, Alhede M, Jensen PØ, Moser C, Scheike T, Jacobsen CS, Seier Poulsen S, Eickhardt-Sørensen SR, Trøstrup H, Christoffersen L, Hougen H-P, Rickelt LF, Kühl M, Høiby N, Bjarnsholt T. 2014. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun 82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Jensen PØ, Givskov M, Bjarnsholt T, Moser C. 2010. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol 59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- 280.Jensen ET, Kharazmi A, Garred P, Kronborg G, Fomsgaard A, Mollnes TE, Hoiby N. 1993. Complement activation by Pseudomonas aeruginosa biofilms. Microb Pathog 15:377–388. doi: 10.1006/mpat.1993.1087. [DOI] [PubMed] [Google Scholar]
- 281.Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, James E, Beyenal H, Lewandowski Z, Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, Duffy JE, Beyenal H, Lewandowski Z. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol 171:4329–4339. doi: 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- 282.Caceres SM, Malcolm KC, Taylor-Cousar JL, Nichols DP, Saavedra MT, Bratton DL, Moskowitz SM, Burns JL, Nick A. 2014. Enhanced in vitro formation and antibiotic resistance of nonattached Pseudomonas aeruginosa aggregates through incorporation of neutrophil products. Antimicrob Agents Chemother 58:6851–6860. doi: 10.1128/AAC.03514-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ciofu O, Tolker-Nielsen T, Østrup P, Wang H, Høiby N. 2015. Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv Drug Deliv Rev 85:7–23. doi: 10.1016/j.addr.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 284.Dettman JR, Rodrigue N, Aaron SD, Kassen R. 2013. Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110:21065–21070. doi: 10.1073/pnas.1307862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Boles BR, Singh PK. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A 105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Sakai A, Nakanishi M, Yoshiyama K, Maki H. 2006. Impact of reactive oxygen species on spontaneous mutagenesis in Escherichia coli. Genes Cells 11:767–778. doi: 10.1111/j.1365-2443.2006.00982.x. [DOI] [PubMed] [Google Scholar]
- 287.Lin S, Daniela I, Christoph S, Chua SL, Ding Y, Liu Y, Cai Z, Zhou J, Swarup S, Drautz-Moses DI, Schuster SC, Kjelleberg S, Givskov M, Yang L. 2016. Reactive oxygen species drive evolution of pro-biofilm variants in pathogens by modulating cyclic-di-GMP levels. Open Biol 6:160162. doi: 10.1098/rsob.160162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Cirz RT, Romesberg FE. 2007. Controlling mutation: intervening in evolution as a therapeutic strategy. Crit Rev Biochem Mol Biol 42:341–354. doi: 10.1080/10409230701597741. [DOI] [PubMed] [Google Scholar]
- 289.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Oliver A, Mena A. 2010. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect 16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- 291.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol 190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Hauser AR, Jain M, Bar-Meir M, McColley SA. 2011. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev 24:29–70. doi: 10.1128/CMR.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Høiby N. 2010. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 156:1108–1119. doi: 10.1099/mic.0.033993-0. [DOI] [PubMed] [Google Scholar]
- 294.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Workentine ML, Sibley CD, Glezerson B, Purighalla S, Norgaard-Gron JC, Parkins MD, Rabin HR, Surette MG. 2013. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One 8:e60225. doi: 10.1371/journal.pone.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, Brockhurst MA, Winstanley C. 2011. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med 183:1674–1679. doi: 10.1164/rccm.201009-1430OC. [DOI] [PubMed] [Google Scholar]
- 297.Boles BR, Thoendel M, Singh PK. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci U S A 101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Klotz MG, Loewen PC. 2003. The molecular evolution of catalatic hydroperoxidases: evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol Biol Evol 20:1098–1112. doi: 10.1093/molbev/msg129. [DOI] [PubMed] [Google Scholar]
- 299.Eason MM, Fan X. 2014. The role and regulation of catalase in respiratory tract opportunistic bacterial pathogens. Microb Pathog 74:50–58. doi: 10.1016/j.micpath.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 300.Zamocky M, Furtmüller PG, Obinger C. 2008. Evolution of catalases from bacteria to humans. Antioxid Redox Signal 10:1527–1548. doi: 10.1089/ars.2008.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Zámocký M, Gasselhuber B, Furtmüller PG, Obinger C. 2012. Molecular evolution of hydrogen peroxide degrading enzymes. Arch Biochem Biophys 525:131–144. doi: 10.1016/j.abb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Vidossich P, Alfonso-Prieto M, Rovira C. 2012. Catalases versus peroxidases: DFT investigation of H2O2 oxidation in models systems and implications for heme protein engineering. J Inorg Biochem 117:292–297. doi: 10.1016/j.jinorgbio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 303.Jones P, Dunford HB. 1977. On the mechanism of compound I formation from peroxidases and catalases. J Theor Biol 69:457–470. doi: 10.1016/0022-5193(77)90152-7. [DOI] [PubMed] [Google Scholar]
- 304.Poole K. 2014. Stress responses as determinants of antimicrobial resistance in Pseudomonas aeruginosa: multidrug efflux and more. Can J Microbiol 60:783–791. doi: 10.1139/cjm-2014-0666. [DOI] [PubMed] [Google Scholar]
- 305.Britigan BE, Miller RA, Hassett DJ, Pfaller MA, McCormick ML, Rasmussen GT. 2001. Antioxidant enzyme expression in clinical isolates of Pseudomonas aeruginosa: identification of an atypical form of manganese superoxide dismutase. Infect Immun 69:7396–7401. doi: 10.1128/IAI.69.12.7396-7401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Lau GW, Britigan BE, Hassett DJ. 2005. Pseudomonas aeruginosa OxyR is required for full virulence in rodent and insect models of infection and for resistance to human neutrophils. Infect Immun 73:2550–2553. doi: 10.1128/IAI.73.4.2550-2553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Mossialos D, Tavankar GR, Zlosnik JEA, Williams HD. 2006. Defects in a quinol oxidase lead to loss of KatC catalase activity in Pseudomonas aeruginosa: KatC activity is temperature dependent and it requires an intact disulphide bond formation system. Biochem Biophys Res Commun 341:697–702. doi: 10.1016/j.bbrc.2005.12.225. [DOI] [PubMed] [Google Scholar]
- 308.Moore R, Kyd JM, Carzino R, Armstrong D, Grimwood K, Otczyk DC, Cripps AW. 2013. Mucosal and systemic antibody responses to potential Pseudomonas aeruginosa vaccine protein antigens in young children with cystic fibrosis following colonization and infection. Hum Vaccin Immunother 9:506–514. doi: 10.4161/hv.23226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Hassett DJ, Alsabbagh E, Parvatiyar K, Howell ML, Wilmott RW, Ochsner UA. 2000. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J Bacteriol 182:4557–4563. doi: 10.1128/JB.182.16.4557-4563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Ma JF, Ochsner UA, Klotz MG, Nanayakkara VK, Howell ML, Johnson Z, Posey JE, Vasil ML, Monaco JJ, Hassett DJ. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J Bacteriol 181:3730–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol 182:4533–4544. doi: 10.1128/JB.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Jo I, Chung I-Y, Bae H-W, Kim J-S, Song S, Cho Y-H, Ha N-C. 2015. Structural details of the OxyR peroxide-sensing mechanism. Proc Natl Acad Sci U S A 112:6443–6448. doi: 10.1073/pnas.1424495112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Heo Y-J, Chung I-Y, Cho W-J, Lee B-Y, Kim J-H, Choi K-H, Lee J-W, Hassett DJ, Cho Y-H. 2010. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J Bacteriol 192:381–390. doi: 10.1128/JB.00980-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Palma M, DeLuca D, Worgall S, Quadri LEN. 2004. Transcriptome analysis of the response of Pseudomonas aeruginosa to hydrogen peroxide. J Bacteriol 186:248–252. doi: 10.1128/JB.186.1.248-252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Wei Q, Minh PNL, Dötsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, Plaisance S, Charlier D, Hassett D, Häussler S, Cornelis P. 2012. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 40:4320–4333. doi: 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Choi YS, Shin DH, Chung IY, Kim SH, Heo YJ, Cho YH. 2007. Identification of Pseudomonas aeruginosa genes crucial for hydrogen peroxide resistance. J Microbiol Biotechnol 17:1344–1352. [PubMed] [Google Scholar]
- 317.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. 2013. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol 195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Vogt SL, Green C, Stevens KM, Day B, Erickson DL, Woods DE, Storey DG. 2011. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun 79:4094–4104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Cochran WL, Suh SJ, McFeters GA, Stewart PS. 2000. Role of RpoS and AlgT in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide and monochloramine. J Appl Microbiol 88:546–553. doi: 10.1046/j.1365-2672.2000.00995.x. [DOI] [PubMed] [Google Scholar]
- 320.Jargensen F, Bally M, Chapon-Herve V, Michel G, Lazdunski A, Williams P, Stewart AB. 2016. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145:835–844. doi: 10.1099/13500872-145-4-835. [DOI] [PubMed] [Google Scholar]
- 321.Suh S, Silo-Suh L, Woods DE, Daniel J, West SEH, Ohman DE, Hassett DJ. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol 181:3890–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Hassett DJ, Ma JF, Elkins JG, McDermott TR, Ochsner UA, West SE, Huang CT, Fredericks J, Burnett S, Stewart PS, McFeters G, Passador L, Iglewski BH. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol 34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 323.Brown SM, Howell ML, Vasil ML, Anderson AJ, Hassett DJ. 1995. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol 177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol 65:4594–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Stewart PS, Roe F, Rayner J, Elkins JG, Lewandowski Z, Ochsner UA, Hassett DJ. 2000. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 66:836–838. doi: 10.1128/AEM.66.2.836-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Lee JS, Heo YJ, Lee JK, Cho YH. 2005. KatA, the major catalase, is critical for osmoprotection and virulence in Pseudomonas aeruginosa PA14. Infect Immun 73:4399–4403. doi: 10.1128/IAI.73.7.4399-4403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Shin DH, Choi YS, Cho YH. 2008. Unusual properties of catalase A (KatA) of Pseudomonas aeruginosa PA14 are associated with its biofilm peroxide resistance. J Bacteriol 190:2663–2670. doi: 10.1128/JB.01580-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Thomas LD, Dunkley ML, Moore R, Reynolds S, Bastin DA, Kyd JM, Cripps AW. 2000. Catalase immunization from Pseudomonas aeruginosa enhances bacterial clearance in the rat lung. Vaccine 19:348–357. doi: 10.1016/S0264-410X(00)00146-8. [DOI] [PubMed] [Google Scholar]
- 329.Su S, Panmanee W, Wilson JJ, Mahtani HK, Li Q, VanderWielen BD, Makris TM, Rogers M, McDaniel C, Lipscomb JD, Irvin RT, Schurr MJ, Lancaster JR, Kovall RA, Hassett DJ. 2014. Catalase (KatA) plays a role in protection against anaerobic nitric oxide in Pseudomonas aeruginosa. PLoS One 9:e91813. doi: 10.1371/journal.pone.0091813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Frederick JR, Elkins JG, Bollinger N, Hassett DJ, McDermott TR. 2001. Factors affecting catalase expression in Pseudomonas aeruginosa biofilms and planktonic cells. Appl Environ Microbiol 67:1375–1379. doi: 10.1128/AEM.67.3.1375-1379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Driffield K, Miller K, Bostock JM, O’Neill AJ, Chopra I. 2008. Increased mutability of Pseudomonas aeruginosa in biofilms. J Antimicrob Chemother 61:1053–1056. doi: 10.1093/jac/dkn044. [DOI] [PubMed] [Google Scholar]
- 332.Lewis K. 2000. Programmed death in bacteria. Microbiol Mol Biol Rev 64:503–514. doi: 10.1128/MMBR.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Bayles KW. 2014. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 12:63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Rice KC, Bayles KW. 2008. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev 72:85–109. doi: 10.1128/MMBR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 336.Chang W, Small DA, Toghrol F, Bentley WE. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. doi: 10.1186/1471-2164-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.McFarland KA, Dolben EL, LeRoux M, Kambara TK, Ramsey KM, Kirkpatrick RL, Mougous JD, Hogan DA, Dove SL. 2015. A self-lysis pathway that enhances the virulence of a pathogenic bacterium. Proc Natl Acad Sci U S A 112:8433–8438. doi: 10.1073/pnas.1506299112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.D’Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol 184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Häussler S, Becker T. 2008. The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 4:e1000166. doi: 10.1371/journal.ppat.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Das T, Ibugo AI, Klare W, Manefield M. 2016. Role of pyocyanin and extracellular DNA in facilitating Pseudomonas aeruginosa biofilm formation, p 23–42. In Dhanasekaran D. (ed), Microbial biofilms: importance and applications. IntechOpen, London, United Kingdom. [Google Scholar]
- 342.Noor R, Murata M, Yamada M. 2009. Oxidative stress as a trigger for growth phase-specific σ-dependent cell lysis in Escherichia coli. J Mol Microbiol Biotechnol 17:177–187. doi: 10.1159/000236029. [DOI] [PubMed] [Google Scholar]
- 343.D’Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Smith EE, Nguyen H, Ernst RK, Freeman TJL, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, Miller SI. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros 8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Nedelcu AM, Driscoll WW, Durand PM, Herron MD, Rashidi A. 2011. On the paradigm of altruistic suicide in the unicellular world. Evolution 65:3–20. doi: 10.1111/j.1558-5646.2010.01103.x. [DOI] [PubMed] [Google Scholar]
- 346.Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. 2011. Extracellular DNA in biofilms. Int J Artif Organs 34:824–831. doi: 10.5301/ijao.5000051. [DOI] [PubMed] [Google Scholar]
- 347.Okshevsky M, Meyer RL. 2015. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol 41:341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 348.Kirov SM, Webb JS, O’May CY, Reid DW, Woo JKK, Rice SA, Kjelleberg S. 2007. Biofilm differentiation and dispersal in mucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 153:3264–3274. doi: 10.1099/mic.0.2007/009092-0. [DOI] [PubMed] [Google Scholar]
- 349.Webb JS, Thompson LS, James S, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S, Charlton T. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol 185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Metruccio MME, Evans DJ, Gabriel MM, Kadurugamuwa JL, Fleiszig SMJ. 2016. Pseudomonas aeruginosa outer membrane vesicles triggered by human mucosal fluid and lysozyme can prime host tissue surfaces for bacterial adhesion. Front Microbiol 7:871. doi: 10.3389/fmicb.2016.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Bonnington KE, Kuehn MJ. 2014. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta 1843:1612–1619. doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Limoli DH, Jones CJ, Wozniak DJ. 2015. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr 3(3):MB-0011-2014. doi: 10.1128/microbiolspec.MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Sousa A, Pereira M. 2014. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens 3:680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Cullen L, McClean S. 2015. Bacterial adaptation during chronic respiratory infections. Pathogens 4:66–89. doi: 10.3390/pathogens4010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Malone JG. 2015. Role of small colony variants in persistence of Pseudomonas aeruginosa infections in cystic fibrosis lungs. Infect Drug Resist 8:237–247. doi: 10.2147/IDR.S68214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Schneider M, Mühlemann K, Droz S, Couzinet S, Casaulta C, Zimmerli S. 2008. Clinical characteristics associated with isolation of small-colony variants of Staphylococcus aureus and Pseudomonas aeruginosa from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol 46:1832–1834. doi: 10.1128/JCM.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Häussler S, Tümmler B, Weissbrodt H, Rohde M, Steinmetz I. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis 29:621–625. [DOI] [PubMed] [Google Scholar]
- 358.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Kirisits MJ, Prost L, Starkey M, Parsek R, Parsek MR. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Häußler S, Ziegler I, Löttel A, Götz FV, Rohde M, Wehmhöhner D, Saravanamuthu S, Tümmler B, Steinmetz I. 2003. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J Med Microbiol 52:295–301. doi: 10.1099/jmm.0.05069-0. [DOI] [PubMed] [Google Scholar]
- 361.Pestrak MJ, Chaney SB, Eggleston HC, Dellos-Nolan S, Dixit S, Mathew-Steiner SS, Roy S, Parsek MR, Sen CK, Wozniak DJ. 2018. Pseudomonas aeruginosa rugose small-colony variants evade host clearance, are hyper-inflammatory, and persist in multiple host environments. PLoS Pathog 14:e1006842. doi: 10.1371/journal.ppat.1006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- 363.Evans TJ. 2015. Small colony variants of Pseudomonas aeruginosa in chronic bacterial infection of the lung in cystic fibrosis. Future Microbiol 10:231–239. doi: 10.2217/fmb.14.107. [DOI] [PubMed] [Google Scholar]
- 364.Cetin E, Toreci K, Ang O. 1965. Encapsulated Pseudomonas aeruginosa (Pseudomonas aeruginosa Mucosus) strains. J Bacteriol 89:1432–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Pugashetti BK, Metzger HM, Vadas L, David S. 1982. Phenotypic differences among clinically isolated mucoid Pseudomonas aeruginosa strains. J Clin Microbiol 16:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Murphy TF, Brauer AL, Eschberger K, Lobbins P, Grove L, Cai X, Sethi S. 2008. Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 177:853–860. doi: 10.1164/rccm.200709-1413OC. [DOI] [PubMed] [Google Scholar]
- 367.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Talwalkar JS, Murray TS. 2016. The approach to Pseudomonas aeruginosa in cystic fibrosis. Clin Chest Med 37:69–81. doi: 10.1016/j.ccm.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 369.Wilson R, Dowling RB. 1998. Lung infections. 3. Pseudomonas aeruginosa and other related species. Thorax 53:213–219. doi: 10.1136/thx.53.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Farrell PM, Collins J, Broderick LS, Rock MJ, Li Z, Kosorok MR, Laxova A, Gershan WM, Brody AS. 2009. Association between mucoid Pseudomonas infection and bronchiectasis in children with cystic fibrosis. Radiology 252:534–543. doi: 10.1148/radiol.2522081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Konig B, Friedl P, Pederson SS, Konig W. 1992. Alginate—its role in neutrophil responses and signal transduction toward mucoid Pseudomonas aeruginosa bacteria. Int Arch Allergy Immunol 99:98–106. doi: 10.1159/000236341. [DOI] [PubMed] [Google Scholar]
- 372.Demko CA, Byard PJ, Davis PB. 1995. Gender differences in cystic fibrosis: Pseudomonas aeruginosa infection. J Clin Epidemiol 48:1041–1049. doi: 10.1016/0895-4356(94)00230-N. [DOI] [PubMed] [Google Scholar]
- 373.Parad RB, Gerard CJ, Zurakowski D, Nichols DP, Pier GB. 1999. Pulmonary outcome in cystic fibrosis is influenced primarily by mucoid Pseudomonas aeruginosa infection and immune status and only modestly by genotype. Infect Immun 67:4744–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Hay ID, Rehman ZU, Moradali MF, Wang Y, Rehm BHA. 2013. Microbial alginate production, modification and its applications. Microb Biotechnol 6:637–650. doi: 10.1111/1751-7915.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Hay ID, Wang Y, Moradali MF, Rehman ZU, Rehm BHA. 2014. Genetics and regulation of bacterial alginate production. Environ Microbiol 16:2997–3011. doi: 10.1111/1462-2920.12389. [DOI] [PubMed] [Google Scholar]
- 376.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A 90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ramsey DM, Wozniak DJ. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol 56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 378.Martin DW, Schurr MJ, Yu H, Deretic V. 1994. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to sigma E and stress response. J Bacteriol 176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize sigma22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 380.Wood LF, Ohman DE. 2012. Identification of genes in the sigma22 regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth. mBio 3:e00094-12. doi: 10.1128/mBio.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Xie ZD, Hershberger CD, Shankar S, Ye RW, Chakrabarty AM. 1996. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol 178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. 1996. Control of AlgU, a member of the σE-like family of stress sigma factors, by the negative regulators mucA and mucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol 178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Mathee K, McPherson CJ, Ohman DE. 1997. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Boucher JC, Yu H, Mudd MH, Deretic V. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun 65:3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol 178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Wozniak DJ, Ohman DE. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol 176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Baynham PJ, Wozniak DJ. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol 22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 388.Wozniak DJ, Sprinkle AB, Baynham PJ. 2003. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J Bacteriol 185:7297–7300. doi: 10.1128/JB.185.24.7297-7300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Yu H, Mudd M, Boucher JC, Schurr MJ, Deretic V. 1997. Identification of the algZ gene upstream of the response regulator algR and its participation in control of alginate production in Pseudomonas aeruginosa. J Bacteriol 179:187–193. doi: 10.1128/jb.179.1.187-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Ma S, Wozniak DJ, Ohman DE. 1997. Identification of the histidine protein kinase KinB in Pseudomonas aeruginosa and its phosphorylation of the alginate regulator AlgB. J Biol Chem 272:17952–17960. doi: 10.1074/jbc.272.29.17952. [DOI] [PubMed] [Google Scholar]
- 391.Ma S, Selvaraj U, Ohman DE, Quarless R, Hassett DJ, Wozniak DJ. 1998. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol 180:956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Xu B, Soukup RJ, Jones CJ, Fishel R, Wozniak DJ. 2016. Pseudomonas aeruginosa AmrZ binds to four sites in the algD promoter, inducing DNA-AmrZ complex formation and transcriptional activation. J Bacteriol 198:2673–2681. doi: 10.1128/JB.00259-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Leech AJ, Sprinkle A, Wood L, Wozniak DJ, Ohman DE. 2008. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J Bacteriol 190:581–589. doi: 10.1128/JB.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Mohr CD, Leveau JHJ, Krieg DP, Hibler NS, Deretic V. 1992. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J Bacteriol 174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Okkotsu Y, Little AS, Schurr MJ. 2014. The Pseudomonas aeruginosa AlgZR two-component system coordinates multiple phenotypes. Front Cell Infect Microbiol 4:82. doi: 10.3389/fcimb.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Pritchett CL, Little AS, Okkotsu Y, Frisk A, Cody WL, Covey CR, Schurr MJ. 2015. Expression analysis of the Pseudomonas aeruginosa AlgZR two-component regulatory system. J Bacteriol 197:736–748. doi: 10.1128/JB.02290-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Lizewski SE, Schurr JR, Jackson DW, Frisk A, Carterson AJ, Schurr MJ. 2004. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J Bacteriol 186:5672–5684. doi: 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Lizewski SE, Lundberg DS, Michael J, Schurr MJ. 2002. The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect Immun 70:6083–6093. doi: 10.1128/IAI.70.11.6083-6093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Jones CJ, Ryder CR, Mann EE, Wozniak DJ. 2013. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J Bacteriol 195:1637–1644. doi: 10.1128/JB.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Tart AH, Blanks MJ, Wozniak DJ. 2006. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J Bacteriol 188:6483–6489. doi: 10.1128/JB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Wu W, Badrane H, Arora S, Baker HV, Jin S. 2004. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J Bacteriol 186:7575–7585. doi: 10.1128/JB.186.22.7575-7585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Jones AK, Fulcher NB, Balzer GJ, Urbanowski ML, Pritchett CL, Schurr MJ, Yahr TL, Wolfgang MC. 2010. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J Bacteriol 192:5709–5717. doi: 10.1128/JB.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Sanders LH, Rockel A, Lu H, Wozniak DJ, Sutton MD. 2006. Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. J Bacteriol 188:8573–8585. doi: 10.1128/JB.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Moyano AJ, Luján AM, Argaraña CE, Smania AM. 2007. MutS deficiency and activity of the error-prone DNA polymerase IV are crucial for determining mucA as the main target for mucoid conversion in Pseudomonas aeruginosa. Mol Microbiol 64:547–559. doi: 10.1111/j.1365-2958.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- 405.Sanders LH, Devadoss B, Raja GV, O’Connor J, Su S, Wozniak DJ, Hassett DJ, Berdis AJ, Sutton MD. 2011. Epistatic roles for Pseudomonas aeruginosa mutS and dinB (DNA pol IV) in coping with reactive oxygen species-induced DNA damage. PLoS One 6:e18824. doi: 10.1371/journal.pone.0018824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O’Toole GA, Parsek MR. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A 100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Hodges NA, Gordon CA. 1991. Protection of Pseudomonas aeruginosa against ciprofloxacin and β-lactams by homologous alginate. Antimicrob Agents Chemother 35:2450–2452. doi: 10.1128/AAC.35.11.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Hentzer M, Teitzel GM, Balzer GJ, Molin S, Givskov M, Matthew R, Heydorn A, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Goltermann L, Tolker-Nielsen T. 2017. Importance of the exopolysaccharide matrix in antimicrobial tolerance of Pseudomonas aeruginosa aggregates. Antimicrob Agents Chemother 61:e02696-16. doi: 10.1128/AAC.02696-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. 2011. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 55:4469–4474. doi: 10.1128/AAC.00126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. 2001. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun 69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. 2005. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol 175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 413.Garrett ES, Perlegas D, Wozniak DJ. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J Bacteriol 181:7401–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Owlia P, Nosrati R, Alaghehbandan R, Lari AR. 2014. Antimicrobial susceptibility differences among mucoid and non-mucoid Pseudomonas aeruginosa isolates. GMS Hyg Infect Control 9:Doc13. doi: 10.3205/dgkh000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Ciofu O, Fussing V, Bagge N, Koch C, Høiby N. 2001. Characterization of paired mucoid/non-mucoid Pseudomonas aeruginosa isolates from Danish cystic fibrosis patients: antibiotic resistance, beta-lactamase activity and RiboPrinting. J Antimicrob Chemother 48:391–396. doi: 10.1093/jac/48.3.391. [DOI] [PubMed] [Google Scholar]
- 416.Simpson JA, Smith SE, Dean RT. 1989. Scavenging by alginate of free radicals released by macrophages. Free Radic Biol Med 6:347–353. doi: 10.1016/0891-5849(89)90078-6. [DOI] [PubMed] [Google Scholar]
- 417.Learn DB, Brestel EP, Seetharama S. 1987. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect Immun 55:1813–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Pedersen SS, Kharazmi A, Espersen F, Hoiby N. 1990. Pseudomonas aeruginosa alginate in cystic fibrosis sputum and the inflammatory response. Infect Immun 58:3363–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Hoffmann N, Rasmussen TBT, Jensen PP, Stub C, Hentzer M, Molin S, Ciofu O, Givskov M, Johansen HK, Høiby N. 2005. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect Immun 73:2504–2514. doi: 10.1128/IAI.73.4.2504-2514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Yu H, Hanes M, Chrisp CE, Boucher JC, Deretic V. 1998. Microbial pathogenesis in cystic fibrosis: pulmonary clearance of mucoid Pseudomonas aeruginosa and inflammation in a mouse model of repeated respiratory challenge. Infect Immun 66:280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Yu H, Boucher JC, Hibler NS, Deretic V. 1996. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma factor AlgU (sigmaE). Infect Immun 64:2774–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Silo-Suh L, Suh S-J, Sokol PA, Ohman DE. 2002. A simple alfalfa seedling infection model for Pseudomonas aeruginosa strains associated with cystic fibrosis shows AlgT (sigma-22) and RhlR contribute to pathogenesis. Proc Natl Acad Sci U S A 99:15699–15704. doi: 10.1073/pnas.242343999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Bragonzi A, Paroni M, Nonis A, Cramer N, Montanari S, Rejman J, Di Serio C, Döring G, Tümmler B. 2009. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am J Respir Crit Care Med 180:138–145. doi: 10.1164/rccm.200812-1943OC. [DOI] [PubMed] [Google Scholar]
- 424.Cigana C, Lorè NI, Riva C, De Fino I, Spagnuolo L, Sipione B, Rossi G, Nonis A, Cabrini G, Bragonzi A. 2016. Tracking the immunopathological response to Pseudomonas aeruginosa during respiratory infections. Sci Rep 6:21465. doi: 10.1038/srep21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Ciofu O, Lee B, Johannesson M, Hermansen NO, Meyer P, Hoiby N. 2008. Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology 154:103–113. doi: 10.1099/mic.0.2007/010421-0. [DOI] [PubMed] [Google Scholar]
- 426.Schurr MJ, Martin DW, Mudd MH, Deretic V. 1994. Gene cluster controlling conversion to alginate-overproducing phenotype in Pseudomonas aeruginosa: functional analysis in a heterologous host and role in the instability of mucoidy. J Bacteriol 176:3375–3382. doi: 10.1128/jb.176.11.3375-3382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.DeVries CA, Ohman DE. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol 176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Sautter R, Ramos D, Schneper L, Ciofu O, Wasserman T, Heydorn A, Hentzer M, Hoiby N, Kharazmi A, Molin S, DeVries CA, Ohman DE, Mathee K. 2012. A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and AlgT/U activity results in loss of alginate production. Gene 498:242–253. doi: 10.1016/j.gene.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Govan JRW, Fyfe JAM, McMillan C. 1979. The instability of mucoid Pseudomonas aeruginosa: fluctuation test and improved stability of the mucoid form in shaken culture. J Gen Microbiol 110:229–232. doi: 10.1099/00221287-110-1-229. [DOI] [PubMed] [Google Scholar]
- 430.Wyckoff TJO, Thomas B, Hassett DJ, Wozniak DJ. 2002. Static growth of mucoid Pseudomonas aeruginosa selects for non-mucoid variants that have acquired flagellum-dependent motility. Microbiology 148:3423–3430. doi: 10.1099/00221287-148-11-3423. [DOI] [PubMed] [Google Scholar]
- 431.Damkiær S, Yang L, Molin S, Jelsbak L. 2013. Evolutionary remodeling of global regulatory networks during long-term bacterial adaptation to human hosts. Proc Natl Acad Sci U S A 110:7766–7771. doi: 10.1073/pnas.1221466110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Yin Y, Withers TR, Wang X, Yu HD. 2013. Evidence for sigma factor competition in the regulation of alginate production by Pseudomonas aeruginosa. PLoS One 8:e72329. doi: 10.1371/journal.pone.0072329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Clark ST, Diaz Caballero J, Cheang M, Coburn B, Wang PW, Donaldson SL, Zhang Y, Liu M, Keshavjee S, Yau YCW, Waters VJ, Elizabeth Tullis D, Guttman DS, Hwang DM. 2015. Phenotypic diversity within a Pseudomonas aeruginosa population infecting an adult with cystic fibrosis. Sci Rep 5:10932. doi: 10.1038/srep10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Yang L, Haagensen JAJ, Jelsbak L, Johansen HK, Sternberg C, Hoiby N, Molin S. 2008. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J Bacteriol 190:2767–2776. doi: 10.1128/JB.01581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Bragonzi A, Wiehlmann L, Klockgether J, Cramer N, Worlitzsch D, Döring G, Tümmler B. 2006. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152:3261–3269. doi: 10.1099/mic.0.29175-0. [DOI] [PubMed] [Google Scholar]
- 436.Seale TW, Thirkill H, Tarpay M, Flux M, Rennert OM. 1979. Serotypes and antibiotic susceptibilities of Pseudomonas aeruginosa isolates from single sputa of cystic fibrosis patients. J Clin Microbiol 9:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Tai AS, Sherrard LJ, Kidd TJ, Ramsay KA, Buckley C, Syrmis M, Grimwood K, Bell SC, Whiley DM. 2017. Antibiotic perturbation of mixed-strain Pseudomonas aeruginosa infection in patients with cystic fibrosis. BMC Pulm Med 17:138. doi: 10.1186/s12890-017-0482-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Troxler BR, Hoover WC, Britton LJ, Gerwin AM, Rowe SM. 2012. Pseudomonas aeruginosa in patients with cystic fibrosis. Pediatr Pulmonol 47:1113–1122. doi: 10.1002/ppul.22543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Malhotra S, Limoli DH, English AE, Parsek MR, Wozniak DJ. 2018. Mixed communities of mucoid and nonmucoid Pseudomonas aeruginosa exhibit enhanced resistance to host antimicrobials. mBio 9:e00275-18. doi: 10.1128/mBio.00275-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Goddard M. 2011. Histopathology of bronchiectasis. Eur Respir Mon 52:22–31. [Google Scholar]
- 441.Dasenbrook EC, Lu L, Donnola S, Weaver DE, Gulani V, Jakob PM, Konstan MW, Flask CA. 2013. Normalized T1 magnetic resonance imaging for assessment of regional lung function in adult cystic fibrosis patients—a cross-sectional study. PLoS One 8:e73286. doi: 10.1371/journal.pone.0073286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Li Z, Sanders DB, Rock MJ, Kosorok MR, Collins J, Green CG, Brody AS, Farrell PM. 2012. Regional differences in the evolution of lung disease in children with cystic fibrosis. Pediatr Pulmonol 47:635–640. doi: 10.1002/ppul.21604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Maffessanti M, Candusso M, Brizzi F, Piovesana F. 1996. Cystic fibrosis in children: HRCT findings and distribution of disease. J Thorac Imaging 11:27–38. doi: 10.1097/00005382-199601110-00002. [DOI] [PubMed] [Google Scholar]
- 444.Mott LS, Park J, Gangell CL, De Klerk NH, Sly PD, Murray CP, Stick SM. 2013. Distribution of early structural lung changes due to cystic fibrosis detected with chest computed tomography. J Pediatr 163:243.e3–248.e3. doi: 10.1016/j.jpeds.2012.12.042. [DOI] [PubMed] [Google Scholar]
- 445.Nemec SF, Bankier AA, Eisenberg RL. 2013. Upper lobe-predominant diseases of the lung. AJR Am J Roentgenol 200:W222–W237. doi: 10.2214/AJR.12.8961. [DOI] [PubMed] [Google Scholar]
- 446.Gyepes MT, Bennett LR, Hassakis PC. 1969. Regional pulmonary blood flow in cystic fibrosis. Am J Roentgenol Radium Ther Nucl Med 106:567–575. doi: 10.2214/ajr.106.3.567. [DOI] [PubMed] [Google Scholar]
- 447.Kaireit TF, Sorrentino SA, Renne J, Schoenfeld C, Voskrebenzev A, Gutberlet M, Schulz A, Jakob PM, Hansen G, Wacker F, Welte T, Tümmler B, Vogel-Claussen J. 2017. Functional lung MRI for regional monitoring of patients with cystic fibrosis. PLoS One 12:e0187483. doi: 10.1371/journal.pone.0187483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Rolla M, D’Andrilli A, Rendina EA, Diso D, Venuta F. 2011. Cystic fibrosis and the thoracic surgeon. Eur J Cardiothorac Surg 39:716–725. doi: 10.1016/j.ejcts.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 449.Sheikh SI, McCoy K, Ryan-Wenger NA, Patel A, Kirkby S. 2014. Lobectomy in patients with cystic fibrosis. Can Respir J 21:e63–e66. doi: 10.1155/2014/709671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, Conrad D. 2012. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J 6:471–474. doi: 10.1038/ismej.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, Harding CL, Radey MC, Rezayat A, Bautista G, Berrington WR, Goddard AF, Zheng C, Angermeyer A, Brittnacher MJ, Kitzman J, Shendure J, Fligner CL, Mittler J, Aitken ML, Manoil C, Bruce JE, Yahr TL, Singh PK. 2015. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, Fligner CL, Singh PK. 2012. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A 109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Garg N, Wang M, Hyde E, da Silva RR, Melnik AV, Protsyuk I, Bouslimani A, Lim YW, Wong R, Humphrey G, Ackermann G, Spivey T, Brouha SS, Bandeira N, Lin GY, Rohwer F, Conrad DJ, Alexandrov T, Knight R, Dorrestein PC. 2017. Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe 22:705.e4–716.e4. doi: 10.1016/j.chom.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Hogan DA, Willger SD, Dolben EL, Hampton TH, Stanton B, Morrison HG, Sogin ML, Czum J, Ashare A. 2016. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One 11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Chung H, Lieberman TD, Vargas SO, Flett KB, McAdam AJ, Priebe GP, Kishony R. 2017. Global and local selection acting on the pathogen Stenotrophomonas maltophilia in the human lung. Nat Commun 8:14078. doi: 10.1038/ncomms14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Gutierrez JP, Grimwood K, Armstrong DS, Carlin JB, Carzino R, Olinsky A, Robertson CF, Phelan PD. 2001. Interlobar differences in bronchoalveolar lavage fluid from children with cystic fibrosis. Eur Respir J 17:281–286. doi: 10.1183/09031936.01.17202810. [DOI] [PubMed] [Google Scholar]
- 457.Meyer KC, Sharma A. 1997. Regional variability of lung inflammation in cystic fibrosis. Am J Respir Crit Care Med 156:1536–1540. doi: 10.1164/ajrccm.156.5.9701098. [DOI] [PubMed] [Google Scholar]