Extraintestinal pathogenic Escherichia coli (ExPEC) strains are responsible for a majority of human extraintestinal infections globally, resulting in enormous direct medical and social costs. ExPEC strains are comprised of many lineages, but only a subset is responsible for the vast majority of infections. Few systematic surveillance systems exist for ExPEC.
KEYWORDS: Escherichia coli, extraintestinal infections, extraintestinal pathogenic E. coli, molecular epidemiology
SUMMARY
Extraintestinal pathogenic Escherichia coli (ExPEC) strains are responsible for a majority of human extraintestinal infections globally, resulting in enormous direct medical and social costs. ExPEC strains are comprised of many lineages, but only a subset is responsible for the vast majority of infections. Few systematic surveillance systems exist for ExPEC. To address this gap, we systematically reviewed and meta-analyzed 217 studies (1995 to 2018) that performed multilocus sequence typing or whole-genome sequencing to genotype E. coli recovered from extraintestinal infections or the gut. Twenty major ExPEC sequence types (STs) accounted for 85% of E. coli isolates from the included studies. ST131 was the most common ST from 2000 onwards, covering all geographic regions. Antimicrobial resistance-based isolate study inclusion criteria likely led to an overestimation and underestimation of some lineages. European and North American studies showed similar distributions of ExPEC STs, but Asian and African studies diverged. Epidemiology and population dynamics of ExPEC are complex; summary proportion for some STs varied over time (e.g., ST95), while other STs were constant (e.g., ST10). Persistence, adaptation, and predominance in the intestinal reservoir may drive ExPEC success. Systematic, unbiased tracking of predominant ExPEC lineages will direct research toward better treatment and prevention strategies for extraintestinal infections.
INTRODUCTION
Extraintestinal pathogenic Escherichia coli (ExPEC) strains are versatile bacteria that can cause urinary tract, bloodstream, prostate, and other infections at nonintestinal sites. They typically occupy a niche in the intestinal microbiota of humans (and other animals), and it is from this reservoir that they emerge to cause extraintestinal infections (1). ExPEC strains are responsible for an enormous number of human infections globally, both in health care-associated and community settings (2, 3), and a limited set of ExPEC lineages are responsible for most infections (4). ExPEC strains also have the infamous distinction of being associated with the acquisition of new and troubling antibiotic resistance genes (5, 6). The clinical and economic impact of these infections and their optimal management, especially in the face of increasing antibiotic resistance, are challenging and underappreciated (7, 8).
ExPEC strains have been alternatively defined by the number and constellation of virulence genes they possess (“special pathogenicity” definition) and by their identification as predominant lineages in the gut prior to causing extraintestinal infections by mass action (“prevalence” definition) (9–11). Unfortunately, neither definition is truly adequate, nor are the definitions mutually exclusive. Despite the overrepresentation of many classic ExPEC virulence genes (12) in many lineages causing infection, there is still uncertainty about what defines or differentiates commensal E. coli and facultative ExPEC pathogens (13). Important differences among ExPEC strains clearly do exist, as demonstrated by the emergence of pandemic lineages such as E. coli sequence type 131 (ST131), along with its highly drug-resistant ST131 H30Rx (C2) sublineages (4, 14, 15). Genetic determinants of persistence, predominance, and competitiveness within the gut microbiota, which are not clear at present, may help explain the success of some of these lineages (16).
For this review, we define ExPEC based on study identification of E. coli as either the cause of extraintestinal infection or presence in a diagnostic, screening, or surveillance specimen in addition to membership in a major ExPEC lineage. An ExPEC ST is defined as major based on its frequency of detection (in all included studies) and by its estimated summary proportion. Multilocus sequence typing (MLST) is the most common method of identifying ExPEC-associated clonal complexes or lineages. However, whole-genome sequencing (WGS)-based lineage classification is gradually supplanting the 7-locus MLST-based sequence type (ST) and related classification systems. We define the major ExPEC lineages using the Achtman MLST classification scheme (17). Most studies included in this review lack detailed virulence gene characterizations; therefore, strict classification of ExPEC by virulence is not possible. Results will unavoidably include both commensal E. coli and true ExPEC.
The majority of studies investigating ExPEC linages are constrained by their geographic locations, time periods, or retrospectively assembled isolate collections and therefore cannot address questions about the global epidemiology of E. coli causing extraintestinal human infections. MLST databases can be leveraged to investigate the epidemiology of ExPEC lineages (18); however, strain information, location, source, collection time, and other epidemiologic information are often limited or absent. In order to describe global ExPEC lineages and leverage information from a large number of ExPEC studies conducted around the world, we performed a systematic review of the published literature examining ExPEC recovered from humans. Studies of ExPEC from the fields of molecular microbiology, molecular epidemiology, and clinical microbiology were identified. We examine the geographic and temporal distribution of major ExPEC STs. We describe the contribution of major ExPEC STs to extraintestinal infections versus intestinal colonization. We investigate differences in ExPEC ST distribution by study isolate selection or inclusion criteria. Many studies include resistance phenotype as a criterion for isolate selection or inclusion, and inclusion criteria may result in an ST distribution bias. The purpose of this investigation is to reflect on and to harness information from many individual studies conducted to characterize the E. coli strains that cause human extraintestinal infections, to provide context for different ExPEC STs (e.g., temporal or geographic differences), and to help prioritize STs for future study.
METHODS
Search Strategy and Selection Criteria
We conducted a systematic review to characterize the contribution of global ExPEC lineages to human infections. This review is reported in accordance with the PRISMA statement (20). We searched Medline (including in-process and other nonindexed citations) and Embase, using Ovid for both, as well as Scopus from 1 January 1995 to 1 January 2018. All search strings were developed with the assistance of a librarian. Search strings are provided in the supplemental material. Study inclusion criteria were the following: (i) original investigation of extraintestinal pathogenic E. coli (including related search terms such as uropathogenic E. coli) but not other E. coli pathotypes (e.g., E. coli O157:H7 or EHEC); (ii) investigation of E. coli recovered from humans only or from humans and other reservoirs, and (iii) characterization of E. coli strains by MLST or WGS. Genotyping by MLST and/or WGS was required in order to standardize high-resolution lineage definitions that were comparable across the widest array of studies; image-based tools such as pulsed-field gel electrophoresis (PFGE) or other PCR-based molecular fingerprinting tools produce data that are not easily portable or comparable and were excluded. The purpose of the review was to examine the global contribution of all major ExPEC STs to extraintestinal infection; therefore, for consistency, studies that implemented full MLST protocols were included. If study results provided a breakdown of STs or if an alternate MLST method was used but results were mapped back to the Achtman scheme, they were also included. We placed no restrictions on language, year of publication, or study type. We excluded studies that included fewer than 20 unique E. coli isolates (unless they were WGS studies). Reviews, commentaries, or editorials were also excluded from the analysis. Studies had to include humans but could also report results for animals or other sources although MLST results only from human ExPEC isolates are reported in this review. Studies could include E. coli recovered from stool or E. coli recovered from sites of extraintestinal infections; this allowed us to compare the ST distributions in ExPEC infections and the intestinal reservoir. Risk of bias domains was not investigated in this review due to study heterogeneity. Individual patient data were not available in most studies, so ST distributions were analyzed in aggregate. Two investigators (H. M. Geum and A. Guo) independently assessed titles and abstracts for eligible publications. If eligibility could not be determined, the full article was retrieved, and the article methods were screened. We hand searched review articles that we identified for additional references. Discrepancies over study inclusion were adjudicated by consensus.
Data Abstraction
Two reviewers (H. M. Geum and A. Guo) performed data abstraction for included publications. Data were abstracted using a standardized form. The following study-level characteristics were abstracted: number of E. coli isolates investigated (the total and the number characterized by MLST or WGS), geographic region, sample collection dates, antimicrobial susceptibility testing (if available), outpatient or inpatient population, genotyping method (MLST or WGS), MLST method (Achtman, Whittam, or Pasteur) (18, 21, 22), and number of isolates by specific ST. Age and sex were not individually abstracted as age range and sex distribution varied widely by study and were frequently not reported. Some new ST assignments and a small number of rare STs were excluded from our final review data set. For certain studies, it was not clear whether the specific ST or ST complex was being reported. Ultimately, 215 specific STs were selected from the published studies and are reported here. The distribution of STs was investigated by study isolate source, selection criteria, geographic region, and start of sample collection by year.
Study isolate source was classified as follows in our data set: (i) colonization (either isolates from surveillance cultures or isolates from healthy individuals, primarily recovered from stool), (ii) any source (inclusion of ExPEC from multiple infections or sources), (iii) bloodstream infection or blood source (including true bloodstream infections, sepsis or bacteremia, or blood source isolate), (iv) urinary tract infection (UTI) (including cystitis, pyelonephritis, or urine source isolate), (v) respiratory tract infection (including E. coli from lung or sputum cultures), and (vi) other sources, which included studies of E. coli recovered from cases of orthopedic infections, cystic fibrosis, inflammatory bowel disease (IBD), meningitis, and necrotizing enterocolitis.
Many studies included a preselection strategy in their sampling scheme, which was captured during data abstraction. For example, a study might provide MLST data on extended-spectrum β-lactamase (ESBL)-producing E. coli isolates only or on sepsis-associated E. coli only, meaning that the results are representative only of that particular subset of resistant E. coli, infection type, or source. We investigated the distribution of STs in studies that selected isolates for inclusion based on ESBL-producing, carbapenem- or fluoroquinolone-resistant, or multidrug-resistant (MDR) phenotypes versus studies that included isolates based on isolate availability or source without regard to resistance phenotype. To augment conclusions from the systematic review, results from selected WGS studies and larger MLST studies are included in the final part of the Results section as these studies provide a deeper perspective on ExPEC epidemiology, evolution, and pathogenesis.
Statistical Analyses
A meta-analysis was performed including all studies identified in the systematic review. The 20 most common STs, based on frequency of detection and by estimated overall summary proportion, were defined as major ExPEC STs. Calculation of overall proportions, 95% confidence limits, and I2 (study heterogeneity) values were estimated using meta-analysis random-effects models, implemented through the meta package, version 4.9-2, using the metaprop function in R, version 3.5.2. Summary proportions by ST are reported by study characteristics, including study isolate selection criteria, geographic area, source, and isolate collection start date. Estimation of summary proportion by study characteristic was not performed if fewer than three studies were available for analysis. The I2 statistic describes the percentage of variation across studies that is attributable to study heterogeneity rather than random variation. The results for each model include Cochran’s Q (analogous to a chi-square test), which indicates heterogeneity in summary proportions for each ST within a subgroup (e.g., geographic region) analysis. All summary proportions, I2 estimates, confidence limits, and Cochran’s Q are reported for each model in supplemental files.
RESULTS
The search resulted in the identification of 1,964 nonduplicate published articles. The titles and abstracts were reviewed, and 1,508 did not meet the inclusion criteria and were excluded (Fig. 1). Full-text documents were retrieved for 456 articles. After full-text review, 217 articles were included for data abstraction, including 173 articles that reported results for MLST only or MLST plus WGS. A further 44 articles were identified that reported WGS results only. The investigation of ExPEC lineages is based on these 217 studies.
FIG 1.
PRISMA flow diagram for this systematic review. This diagram shows the selection of studies included in the systematic review of ExPEC lineages (20).
Summary of Articles
MLST results were reported in 173 articles (21–194). Almost all of the studies implemented the Achtman MLST scheme. Two studies used the Whittam MLST scheme and were excluded from the review (109, 157). Six studies implemented the Pasteur Institute’s MLST scheme (21). The ST results of four (21, 130, 170, 176) of these six studies were partially, but unambiguously, converted to STs corresponding to the Achtman scheme (195). Therefore, the distribution of human ExPEC STs was derived from 169 studies. The full review data set is provided in Table S1 in the supplemental material. These 169 studies include MLST data for 15,538 isolates and represent a very wide range of scientific disciplines and diverse research questions, sampling methods, study populations, time periods, and regions. Studies including whole-genome sequencing of ExPEC isolates are listed in Table S2.
Studies reported E. coli sample sizes of anywhere between 20 and 3,060 isolates. Multiple studies selected isolates from large national or regional collections of E. coli based either on specimen source (e.g., urine or stool), infection type (e.g., urinary tract or bloodstream infection), or resistance screening, so the initial E. coli sample sizes were larger than the final number of genotyped E. coli isolates. Therefore, we included the number of E. coli isolates from each study that were eligible for MLST or WGS testing. The sample sizes of E. coli characterized by MLST ranged from 5 to 2,166 isolates. A summary of study characteristics is provided in Table 1. Many studies (n = 112, 66%) preselected ExPEC isolates for study inclusion based on a specific antibiotic resistance phenotype or genotype. Seventy-two (64%) of these studies restricted their MLST analyses to ESBL-positive ExPEC. E. coli isolates from a wide range of extraintestinal infections were represented in the studies, including 35 (21%) studies that investigated UTIs and 26 (15%) studies that included isolates recovered from bloodstream or meningitis cases. The patient population was split between inpatient and outpatients, with many (n = 73, 43%) studies including isolates from both inpatients and outpatients. Europe was overrepresented, with 82 (49%) studies, followed by Asia (24%) and North America (11%). This review covers the period since 1995 when MLST was first introduced (196) although the majority of studies were conducted since 2000. Five studies (3%) included ExPEC recovered from the 1990s, 31 (18%) included isolates from 2000 and 2005, 76 (45%) included isolates from 2006 to 2010, and 44 (26%) included isolates from 2011 to 2016. Two studies examined ExPEC collections retrospectively, one examined ExPEC recovered from 1956 to 2000 (45), and another study examined isolates from 1988 to 2003 (28). Eleven studies did not report the date or year in which they started collecting specimens or isolates.
TABLE 1.
Characteristics of included ExPEC studies
| Study characteristic | No. of studies (%) |
|---|---|
| Total studies | 169 (100) |
| Study ExPEC selected based on resistancea | 112 (66) |
| Extended spectrum β-lactamase | 72 (64) |
| Fluoroquinolone/quinolone | 10 (9) |
| Multidrug resistance | 9 (8) |
| Beta-lactamase | 8 (7) |
| Carbapenem | 7 (6) |
| Fosfomycin | 2 (2) |
| Nitrofurantoin | 2 (2) |
| Colistin | 1 (1) |
| Fully susceptible | 1 (1) |
| ExPEC sourcea | |
| Any infection | 72 (43) |
| Urinary tract infections | 49 (29) |
| Bloodstream infection | 26 (15) |
| Colonization | 17 (10) |
| Orthopedic infection | 2 (1) |
| Cystic fibrosis | 1 (0.6) |
| Irritable bowel disease | 1 (0.6) |
| Necrotizing enterocolitis | 1 (0.6) |
| Patient population | |
| Any inpatients | 110 (65) |
| Any outpatients | 103 (61) |
| Both in- and outpatients | 73 (43) |
| Not reported | 28 (17) |
| Geography | |
| Europe | 82 (49) |
| Asia | 41 (24) |
| North America | 18 (11) |
| Africa | 12 (7) |
| Middle East | 5 (3) |
| South America | 3 (2) |
| Central America | 2 (1) |
| Oceania | 2 (1) |
| Global | 3 (2) |
| Not reported | 1 (0.6) |
Some studies selected for more than one type of resistance phenotype or included a combination of infections or sources.
ExPEC Sequence Types
A summary of all 215 MLST STs detected in one or more studies is provided in Table S1. Twenty major ExPEC STs, as defined by the number of positive studies and estimated summary proportions, are shown in Table 2. These include ST131, ST69, ST10, ST405, ST38, ST95, ST648, ST73, ST410, ST393, ST354, ST12, ST127, ST167, ST58, ST617, ST88, ST23, ST117, and ST1193 (by decreasing study positivity). These top STs accounted for 85% of E. coli isolates for which ST results were reported in all included studies. Not surprisingly, ST131 was detected in over 90% of studies, while the next most common STs (ST69 or ST10) were detected in 50% of studies overall. The maximum number of STs identified in any one study was 84, while some studies reported information on a few STs. If a study investigated only a single ST, it was excluded from analysis. As expected, the study heterogeneity estimated by I2, which assesses heterogeneity beyond random variation, was large and ranged from 0.37 to 0.94 (Table 2). An I2 of over 0.75 is generally considered to indicate that study results are very heterogeneous.
TABLE 2.
Summary proportions for the top 20 ExPEC STs: meta-analysis resultsa
| Sequence type | No. of positive studies | Summary proportion (95% CI) | I2 (95% CI) |
|---|---|---|---|
| ST131 | 133 | 0.24 (0.21, 0.27) | 0.94 (0.94, 0.95) |
| ST69 | 87 | 0.05 (0.05, 0.06) | 0.68 (0.60, 0.74) |
| ST10 | 85 | 0.05 (0.04, 0.06) | 0.81 (0.77, 0.84) |
| ST405 | 81 | 0.04 (0.04, 0.05) | 0.74 (0.68, 0.79) |
| ST38 | 72 | 0.06 (0.04, 0.07) | 0.83 (0.79, 0.86) |
| ST95 | 71 | 0.07 (0.06, 0.09) | 0.87 (0.84, 0.89) |
| ST648 | 67 | 0.04 (0.03, 0.05) | 0.71 (0.63, 0.77) |
| ST73 | 62 | 0.08 (0.07, 0.10) | 0.86 (0.83, 0.89) |
| ST410 | 45 | 0.03 (0.03, 0.05) | 0.69 (0.58, 0.77) |
| ST393 | 42 | 0.03 (0.02, 0.03) | 0.75 (0.66, 0.81) |
| ST354 | 40 | 0.02 (0.01, 0.03) | 0.63 (0.49, 0.74) |
| ST12 | 37 | 0.04 (0.03, 0.05) | 0.56 (0.36, 0.70) |
| ST127 | 36 | 0.03 (0.03, 0.04) | 0.46 (0.19, 0.63) |
| ST167 | 35 | 0.03 (0.02, 0.04) | 0.69 (0.57, 0.78) |
| ST58 | 35 | 0.02 (0.01, 0.03) | 0.73 (0.62, 0.80) |
| ST88 | 34 | 0.03 (0.02, 0.04) | 0.84 (0.79, 0.88) |
| ST617 | 34 | 0.02 (0.01, 0.03) | 0.53 (0.30, 0.68) |
| ST23 | 31 | 0.03 (0.02, 0.05) | 0.87 (0.83, 0.90) |
| ST117 | 30 | 0.02 (0.01, 0.02) | 0.37 (0.01, 0.59) |
| ST1193 | 28 | 0.03 (0.02, 0.05) | 0.85 (0.79, 0.89) |
Summary proportions (95% CI) and I2 (95% CI) were estimated using meta-analysis random-effects models, implemented through the meta package, version 4.9-2, using the metaprop function in R, version 3.5.2. The I2 statistic describes the percentage of variation across studies that is attributable to study heterogeneity rather than random variation. Summary proportions were calculated across multiple, diverse studies, including very different study populations, multiple specimen types, and clinical outcomes and should not be interpreted as population-level estimates of an individual ST’s contribution to the global burden of infection.
ExPEC Sequence Types by Isolate Inclusion Criteria
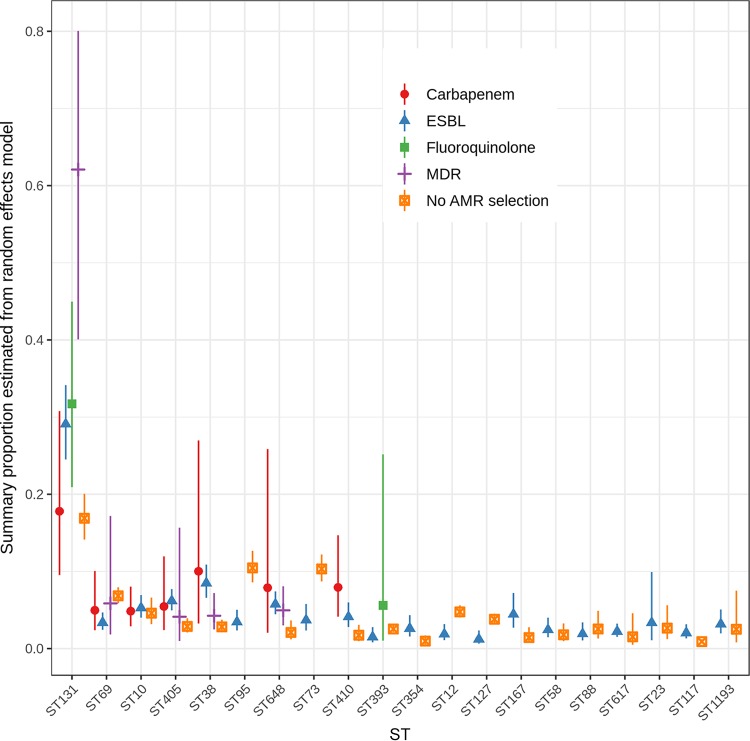
Most studies included ExPEC isolates based on the presence of specific antimicrobial resistance phenotypes, largely ESBL production (Fig. 2; Table S3). This inclusion criterion may influence the identification of important STs in a given study. Other studies, those that did not select based on resistance phenotype, selected isolates based on infection type or specimen source (e.g., UTI isolates) or were convenience samples of ExPEC isolates available to study investigators. Figure 2 shows the estimated overall proportion for the most common 20 ExPEC STs stratified by whether studies selected isolates based on a resistance criterion (n = 112) or not (n = 57). Six studies were excluded from this analysis as they selected for isolates exhibiting resistance to amdinocillin and/or nitrofurantoin (2 studies), colistin (1 study), or fosfomycin (2 studies).
FIG 2.
Summary proportions from random-effects models for the most common ExPEC STs by study isolate inclusion criteria. Six studies were excluded from this analysis as they selected for isolates exhibiting specific drug resistance phenotypes, including amdinocillin and nitrofurantoin (1 study), nitrofurantoin alone (1 study), colistin (1 study), and fosfomycin (2 studies), or only for fully susceptible E. coli isolates (1 study). AMR, antimicrobial resistance.
The primary resistance phenotypes used for isolate selection in these 163 studies included beta-lactam resistance or ESBL production, quinolone or fluoroquinolone resistance, and any multidrug resistance. A small number of studies included carbapenem-resistant ExPEC. Selection based on ESBL production was by far the most common resistance criterion used, and importantly all of the top 20 STs were detected in multiple collections of ESBL-positive isolates (Table 1; Table S3). The estimated overall proportion of ST131 in studies varied by the resistance selection criterion used in the study design. Estimated summary proportions for MDR, ESBL-producing, and carbapenem- and fluoroquinolone-resistant ST131 isolates were 0.62 (95% confidence interval [CI], 0.40, 0.80), 0.29 (95% CI, 0.25, 0.34), 0.18 (95% CI, 0.10, 0.31), and 0.32 (95% CI, 0.21, 0.45), respectively. In contrast, the ST131 prevalence was only 0.17 (95% CI 0.14, 0.20) in studies without resistance selection (Q test P < 0.001). In general, studies selecting on resistance phenotype produced higher estimated summary proportions (in at least one resistance category) than studies that employed no resistance inclusion criterion. This was true except for ST95 (Q test P < 0.001) and ST73 (Q test P < 0.001) and, to a lesser extent, ST69 (Q test P = 0.001), ST12 (Q test P = 0.009), and ST127 (Q test P = 0.003) (Fig. 2; Table S3). Selection for carbapenem-resistant isolates resulted in higher estimated summary proportions for ST38, ST648, and ST410 isolates. Interestingly, these data suggest that summary proportions for ST10, ST58, ST88, ST617, ST23, and ST1193 do not differ between studies employing ESBL-positive selection and studies without resistance selection. In one study of fully susceptible ExPEC isolates, ST131 was not identified at all. Of the 80 isolates characterized by MLST, 35 (44%) belonged to the ST69, ST95, ST73, ST12, and ST127 lineages (80). This suggests that the infection burden of ExPEC lineages such as ST131 (especially MDR ST131) and ST38 may be overestimated, while that of ST95 and ST73 may be underestimated if selection is based on antimicrobial resistance phenotype or genotype. These differences in estimates across studies suggest that inferences about the clinical and public health importance of specific ExPEC STs need to consider study inclusion criteria.
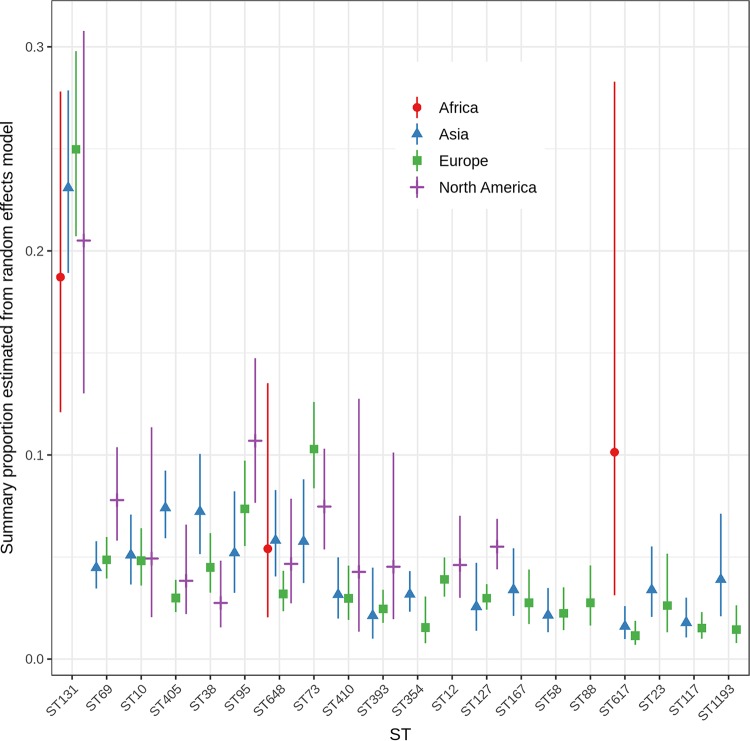
ExPEC Sequence Types by Geographic Area
As expected, the summary proportions for ST131 were elevated for each geographic region compared to those for other major STs. Summary proportion estimates for ST131 were higher for studies from Asia (0.23; 95% CI, 0.19, 0.28) and Europe (0.25; 95% CI, 0.21, 0.30) and lower in African (0.19; 95% CI, 0.12, 0.28) and North American (0.21; 95% CI, 0.13, 0.31) studies. However, these summary proportions were not significantly different (Q test P = 0.57) (Fig. 3; Table S4). ST648 and ST617 were the only other STs for which sufficient data from African studies were available; the summary proportion for ST617 (0.10; 95% CI, 0.03, 0.28) (Q test P = 0.004) was considerably higher than for studies from Asia and Europe. Notably, common STs, including ST95, ST393, and ST354, were absent from, and the summary proportion for ST73 was especially low in, the African studies (Table S1). Underrepresentation of these STs might be the result of isolate selection criteria as a high proportion (83%) of African studies selected isolates based on resistance phenotype. Summary proportions may also underestimate African ExPEC ST diversity due to MLST reference database limitations or due to the scope or sampling methods of included studies.
FIG 3.
Summary proportions from random-effects models for the most common ExPEC STs by geographic region. Six studies were not included: three included global collections of ExPEC, two studies were from Australia only, and one study did not report the geographic source of isolates analyzed.
In 41 studies conducted in Asia, 71% selected isolates based on resistance phenotype. In addition to ST131, Asian studies yielded the highest summary proportions, by region, for ST405 (0.07; 95% CI, 0.06, 0.09) (Q test P < 0.001), ST38 (0.07; 95% CI, 0.05, 0.10) (Q test P =0.01), ST1193 (0.04; 95% CI, 0.02, 0.07) (Q test P = 0.024), and possibly ST648 (0.06; 95% CI, 0.04, 0.08) (Q test P = 0.08), while Asian studies exhibited a lower overall estimate for ST73 (0.06; 95% CI, 0.04, 0.09) (Q test P = 0.03) (Fig. 3; Table S4). ST12 and ST88 were identified in fewer than 3 studies of the 41 studies from Asia.
In 82 studies from Europe, 67% selected isolates based on resistance phenotype. European studies reported the highest overall proportion of ST131 (0.25; 95% CI, 0.21, 0.30) and a higher overall proportion of ST73 (0.25; 95% CI, 0.21, 0.30) (Q test P = 0.03). The estimated proportions for ST405 (0.03; 95% CI, 0.02, 0.04) and ST648 (0.03; 95% CI, 0.02, 0.04) tended to be lower than point estimates from other regions (Fig. 3; Table S4).
In 18 studies set in North America, 44% selected isolates based on resistance phenotype. Reports from North America yielded higher overall proportions of ST95 (0.11; 95% CI, 0.08, 0.15) (Q test P = 0.037), ST69 (0.08; 95% CI, 0.06, 0.10) (Q test P = 0.031), and ST127 (0.06, 95% CI, 0.04, 0.07) (Q test P < 0.001). The proportion of ST38 (0.03; 95% CI, 0.02, 0.05) appears to be lower in North America than elsewhere, and ST354, ST167, ST58, ST88, ST617, ST23, ST117, and ST1193 were identified in fewer than three North American studies (Fig. 3; Table S4). Detection of these STs may be influenced by the limited number of studies undertaken in North America. Interestingly, the summary proportions for ST10 were similar for all regions.
Five studies each recovered isolates from the Middle East and from Central or South America. Studies originating from Central and South America detected ST131, ST69, ST73, and ST62 most commonly, while studies originating from the Middle East detected ST131, ST38, ST410, ST10, and ST617 most commonly. As the sample sizes for these regions are small, they are not included in the data shown in Fig. 3. Two studies included isolates from Australia only, and one study did not report the geographic source of isolates analyzed.
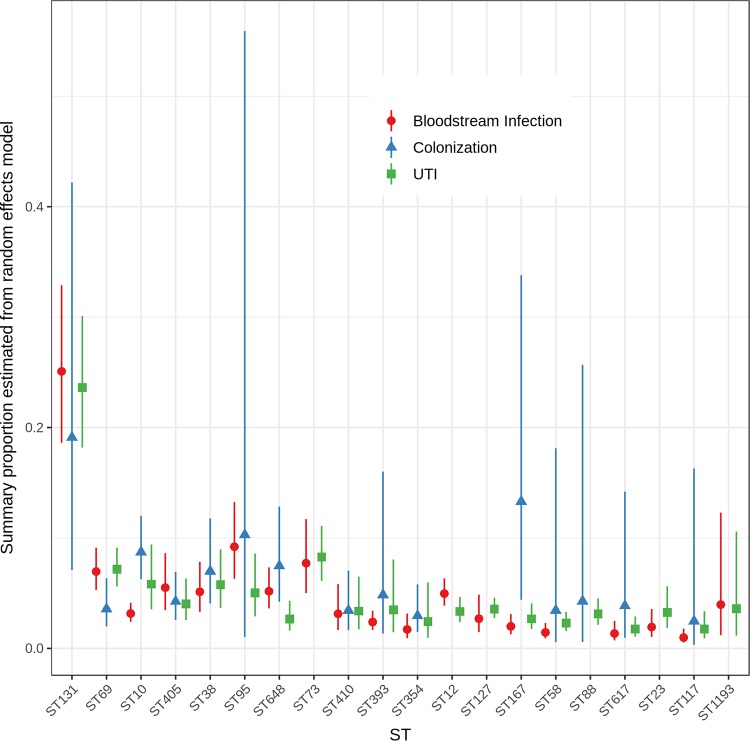
ExPEC Sequence Types by Source
Of the 169 included ExPEC studies, only 79 studies included isolates from a single specimen source or infection type (Table 1). Even though we restricted our analyses to a single reported source, there will be an unavoidable mixture of ExPEC and non-ExPEC in these results. In the 90 studies that included isolates from multiple sources, detailed ST breakdowns by source were not provided in their reports, so these studies could not be included in the meta-analysis. Only 17 studies focused specifically on colonization or surveillance isolates.
The presence of specific STs in the studies varied only modestly by E. coli source (Fig. 4; Table S5). All 20 major STs were identified in three or more studies examining UTI (or urine specimens) and bloodstream infections (or blood specimens). ST73, ST12, ST127, ST23, and ST1193 were each detected in fewer than three studies of colonization or surveillance isolates. ST131 was found at a higher overall proportion in studies that investigated UTI (or urine) (0.24; 95% CI, 0.18, 0.30) and bloodstream infections (or blood isolates) (0.25; 95% CI 0.19, 0.33) but was lower in colonization or surveillance studies (0.19; 95% CI, 0.07, 0.42). These differences were not significant (Q test P = 0.84). ST69 also exhibited this pattern (Q test P = 0.08). Compared to the levels of all of the other STs, ST73 was generally found at elevated levels in UTI (or urine) (0.08; 95% CI, 0.06, 0.11) and bloodstream infections (or blood) (0.08; 95% CI, 0.05, 0.12) (Fig. 4). In contrast, ST10 (0.09; 95% CI 0.06, 0.12) (Q test P < 0.001), ST648 (0.08; 95% CI, 0.04, 0.13) (Q test P = 0.02), and ST167 (0.13; 95% CI, 0.04, 0.34) appeared to be identified at higher proportions in studies looking at colonization or surveillance isolates. Most of the other major STs were identified at similar levels in studies of UTI, bloodstream infections, or stool. Relatively few studies specifically investigated stool isolates, which may explain why a number of STs had low detection rates.
FIG 4.
Summary proportions from random-effects models for the most common ExPEC STs by source. Only those studies that examined isolates recovered exclusively from one source category were included. One study of bloodstream isolates also included isolates from orthopedic infections and was included in the bloodstream/meningitis group.
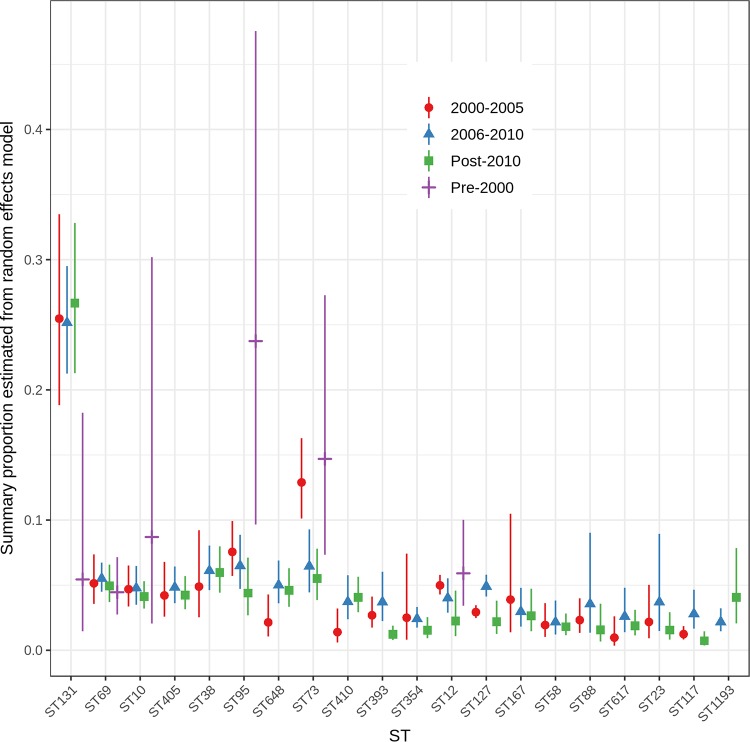
ExPEC Sequence Types by Start Date of Study Isolate Collection
To examine timing of ExPEC ST emergence or persistence, we investigated the distribution of STs by the start date of study isolate collection; time periods were grouped as follows: pre-2000,2001 to 2005, 2006 to 2010, and 2011 or later (Fig. 5; see also Table S6 in the supplemental material). All predominant STs were identified from the inception of MLST testing around 1995; however, there were only 11 pre-2000 studies that were eligible for inclusion in the systematic review and meta-analysis. Aside from the pre-2000 period, virtually all 20 major STs were detected in studies that collected isolates after the year 2000.
FIG 5.
Summary proportions from random-effects models for the most common ExPEC STs by start date of study isolate collection. Dates were determined based on the reported start date for sample collection for each study. In some cases, isolate collection ended in a different period, which makes categories overlap for some studies. Moreover, the number of STs included in databases has grown over time. The absence of some STs might reflect the fact that these STs had not yet been added to the MLST allele database.
The overall estimated proportion of ST131 was low prior to 2000 (0.05; 95% CI, 0.02, 0.18) but increased, and summary proportions were approximately 0.25 in studies from 2000 onwards (Q test P = 0.08) (Table S6). ST73 (Q test P < 0.001), ST95 (Q test P = 0.013), and ST12 (Q test P = 0.1) were all detected more commonly in the pre-2000 period, and the summary proportions gradually declined from 2000 onwards (Fig. 5; Table S6). This suggests that these STs have been long-standing ExPEC lineages and may have ceded ground to newly emerging lineages, such as ST131. Summary proportions for some STs appeared to rise and fall over the time periods (ST88, ST127, ST393, and ST117) although this could just reflect study variability (Fig. 5). There is some evidence that ST1193 increased from 2006 (Q test P = 0.1) although there was only a single study that detected ST1193 in sampling prior to 2006. The summary estimates for ST69, ST58, ST167, ST38, ST405, and ST10 remained remarkably stable over time. Changes in ST detection over time may be influenced by multiple factors, including the following: (i) the emergence and recognition of new ST lineages; (ii) dynamic changes in ST detection due to shifts in ExPEC sources, transmission, or selective pressures; (iii) an expansion in the number of MLST allele profiles contained in the databases (where new STs defined later would be underrepresented at earlier time periods); and (iv) the specific selection of certain STs or lineages in some studies (e.g., ST131).
Whole-Genome Sequencing of ExPEC Lineages
Twenty-eight (16%) of the 173 MLST studies included whole-genome sequencing of E. coli isolates. Forty-four additional studies employed WGS to study ExPEC but did not include information on the distribution of STs and so were not summarized in the systematic review (Table S2). Many of these additional WGS-focused studies examined a small number of genomes or previously deposited genomes or were draft genome announcements. Several WGS studies investigated ST131 specifically (14, 197–201) or examined the evolution, resistance, and virulence gene distributions of ExPEC in general. To complement and augment the meta-analysis results, we provide a narrative summary of results from WGS studies focusing on the molecular epidemiology, evolution, and drivers of the emergence of ExPEC lineages, starting with ST131.
Emergence of ST131 and drivers of success.
The study by Kallonen et al. (166), conducted over an 11-year period, noted a single rapid expansion of ST131 beginning around 1995 in the United Kingdom. Ben Zakour et al. investigated the evolution of 185 previously deposited ST131 genomes, which suggested that the emergence of fluoroquinolone-resistant fimH30 ST131 (clade C) occurred in the 1980s, followed by the rapid expansion of the ST131 lineages, probably originating in North America (197). A split into clades C1 (H30R; fluoroquinolone resistant) and C2 (H30Rx; fluoroquinolone resistance in combination with CTX-M-15 ESBL production) then occurred around 1990. This is generally consistent with other studies that have investigated ST131 genomes and the acquisition of resistance genes and plasmids by the H30R and H30Rx lineages (14, 166, 201).
McNally et al. compared core and accessory ST131 genomes and found that the phylogenetic clustering of ST131 is driven to a large extent by accessory genome content. This provides support for the theory that ST131 has benefited from compensatory mutations that make carriage of multidrug resistance and virulence plasmids possible without a fitness cost. Furthermore, comparison of human and nonhuman source isolates suggests that ST131 has a wide host range (198). Alqasim et al. took a different approach and investigated capsular variation within the ST131 H30Rx C2 clade (13). They show evidence for multiple recombination events at capsular loci in ST131 genomes over time. Capsular variation did not appear to be related to virulence in in vitro testing but may still contribute to the widespread dissemination of this ST131 sublineage (13).
Comparative genome analysis of ST131 versus other ExPEC STs was performed by Kallonen et al. to identify potential drivers of ST131 success in causing a large fraction of total infections. They showed that the SPATE (serine protease autotransporters of Enterobacteriaceae) virulence gene (espC) was strongly associated with ST131, which has been linked to other E. coli pathotypes. This observation was echoed in the study by Shaik et al. that compared ST131 E. coli to other ESBL-producing E. coli lineages (ST38, ST405, and ST648) and found that SPATE was identified exclusively in ST131 isolates (199). In contrast, Clark et al. conducted a WGS-based virulence gene analysis and did not find any genome regions that explained ST131 increased fitness compared to that of non-ST131 ExPEC genomes (202). Kallonen et al. also argue that antimicrobial resistance does not appear to be a major driver in the success of resistant ST131 or other drug-susceptible ExPEC lineages, such as ST73, as these groups were equally maintained in the population over time; this was also shown in the study by Ben Zakour (166, 197). These analyses suggest that the success of ST131 and other major ExPEC lineages may be linked to selection and expansion in E. coli reservoirs, primarily the intestinal tract (166).
Emergence of other major ExPEC STs and drivers of success.
Relatively few studies investigated the genomic basis for non-ST131 ExPEC success. ST73 phylogenies from the Kallonen et al. study exhibited multiple divergent clades, suggesting that ST73 is an older lineage (166). Alhashash et al. investigated 22 isolates of MDR ST73 and also found considerable genome heterogeneity, suggesting a longer period of evolution. The authors reported similar levels of virulence gene carriage and a diverse pool of plasmids containing resistance genes that did not differ between isolates recovered from UTI and bacteremia cases. The authors concluded that the increase in ST73 resistance was not due to the spread of one or more specific clones. So, unlike the highly clonal ST131, ST73 is also highly successful but for reasons that are still unclear (152). A few small studies investigated ST95 (203) and ST69 (166) by WGS but provided few details on the emergence of these STs. Shaik et al. examined ST38, ST405, and ST648 and contrasted these with ST131. ST131 possessed a greater total number of virulence genes, while the non-ST131 isolates shared similar, albeit lower, numbers of virulence genes, but the composition of genes varied (199). Salipante et al. showed no real differences in virulence factors between urine and blood source STs (75). Despite the significant role these major STs have in global infections, we still know fairly little about the apparent differences in performance by the different lineages at different extraintestinal sites and in the gut.
Genomic epidemiology of major ExPEC STs.
Several genomic epidemiology studies were identified that assessed potential ExPEC transmission routes and identified clusters of closely related ExPEC isolates in diverse clinical settings. Salipante et al. investigated the genomic epidemiology of ExPEC recovered from urine and blood at a single health care institution. They observed genetically identical urine and blood source isolates, differing by 0 to 1 single nucleotide polymorphisms (SNPs), collected over short periods. Another pair of isolates exhibiting indistinguishable genomes occurred more than a year apart, without any evidence of an epidemiological link (75). ExPEC genome clusters were also observed in Brodrick et al., where related ESBL-positive E. coli ST131 genomes were identified in 17 patients (112). Clark et al. identified genetically monomorphic ST131 isolates from 10 subjects (202). Finally, Roer et al. studied 552 cephalosporin-resistant ExPEC isolates recovered from bloodstream infections between 2014 and 2015 in Denmark and found evidence of 15 national outbreaks (defined as fewer than 10 SNPs) comprised of seven ST131 outbreaks, with three due to ST12, two due to ST410, and one each due to ST3666, ST58, and ST69 (72).
ExPEC has been associated with many different environmental reservoirs and potential foodborne and other transmission routes (204). Sexual and household transmission has been demonstrated previously using non-WGS molecular methods, but household clustering of ST131 ExPEC has recently been confirmed using WGS (161). Berg et al. identified ST38 human and retail chicken isolates that differed by fewer than 15 SNPs (91). ESBL-positive ST410 isolates from a study by Falgenhauer et al. recovered from human, companion animal, livestock, and farm sources differed by fewer than 70 SNPs (138). Clusters of ESBL-producing E. coli, including ST131, ST2003, ST354, ST38, ST405, and ST410, from clinical and water sources in Thailand also differed by only 3 to 23 SNPs (74). E. coli ST131 has also been shown to have a potentially large host range and has been identified in wild birds, food, and companion animals (198). However, in one study by de Been et al. which examined human and poultry isolates that were considered to be indistinguishable by less discriminatory genotyping methods, genome sequences were more divergent than expected (205). Musicha et al. investigated ExPEC isolates associated with bacteremia and meningitis infections in Malawi and found E. coli to be more diverse than in other regions (34), reinforcing the results from this systematic review. The extent of global differences in ExPEC molecular epidemiology is still not well understood.
A few studies investigated the link between the human intestinal reservoir for ExPEC and development of infection in the same host. This relationship has been known for a long time, but WGS has confirmed the link (41, 121, 161). A study by Nielsen et al. examined fecal and UTI isolates by WGS (41). Multiple E. coli isolates per fecal specimen were investigated, providing information on the predominance of ExPEC ST carriage in the stool. ST95 was associated with fecal predominance in multiple subjects, whereas ST131 was predominant in the gut in only two cases. The authors found no differences in phylogenies of E. coli isolates recovered from feces and those from UTI cases. ST73 and ST12 were overrepresented among the UTI cases; however, no closely related genomes were discovered. The main difference between the fecal and UTI isolates was in their accessory genomes. Moreover, healthy women who had never had a UTI carried fecal E. coli isolates in their gut that were similar to the fecal E. coli isolates recovered from women who went on to develop UTI (41). Intriguingly, one study with detailed assessments of urine and fecal isolates confirmed shared clones between the gut and the urinary tract and showed that the metabolic properties of ExPEC are crucial to fitness in both environments. In fact, isolates that exhibited greater fitness at both sites were most successful (121). These data begin to suggest that the dichotomy between the special pathogenicity and prevalence explanations for ExPEC success be revisited. Identification of genes that contribute to persistence and predominance in the intestinal reservoir, and possibly to host specificity and environmental dissemination, should be the focus of future investigations.
DISCUSSION
In the absence of systematic, large-scale, public health reporting or surveillance systems for ExPEC, reviews of the literature can help capture information on important ExPEC lineages. Leveraging data from a systematic review can depict a more complete picture of the type, evolution, distribution, and characteristics of ExPEC. As expected, ST131 was the undisputed winner. ST131 was detected in 91% of all reviewed studies and exhibited the highest summary proportions, regardless of study isolate selection criteria, geographic region, source, or time period. The other major STs were ST69, ST10, ST405, ST38, ST95, ST648, ST73, ST410, ST393, ST354, ST12, ST127, ST167, ST58, ST617, ST88, ST23, ST117, and ST1193 (in decreasing study positivity) across all 169 studies (Table 2). Other narrative reviews of ExPEC point to a largely overlapping list of important STs (4).
The results of this systematic review are heavily influenced by the selection criteria employed by study authors. The role of certain ExPEC lineages in the global spread of antimicrobial resistance is undisputed; however, the overall contribution of ExPEC to antimicrobial-susceptible extraintestinal infections is also enormous. A large fraction of studies (66%) sampled isolates based on antibiotic resistance phenotype, especially ESBL production. This inclusion criterion influenced the distribution of STs detected and most likely overestimated the importance of certain lineages associated with multidrug resistance and ESBLs, such as ST131, ST405, ST38, ST648, ST410, and ST354, whereas ExPEC lineages exhibiting lower multidrug resistance levels, such as ST95, ST73, ST12, and ST127, may have been underreported.
It is likely that the high estimated proportions of certain MDR lineages globally are due to the large number of studies that included only resistant isolates. Two large retrospective studies that included all isolates irrespective of their susceptibility profiles, conducted in the United Kingdom and Ireland from 2001 until 2010/2011, showed that ST73 was the most common cause of bloodstream infections due to ExPEC (132, 166). This was reinforced in studies from Germany (84) and Canada (187). In contrast, studies that included only MDR isolates (primarily ESBL producers) described ST131 as the most prevalent ST among ExPEC responsible for bloodstream infections (72, 154, 193).
Certain longstanding or “classic” ExPEC STs, such as ST73 and ST95, are successful extraintestinal pathogens but are also persistent intestinal colonizers (4). Interestingly, most of the other major STs were identified at similar proportions in studies of UTI, bloodstream infections, and stool. Such classic STs are often responsible for higher proportions of extraintestinal infections than the MDR lineages, as illustrated in the large studies from the United Kingdom and Ireland (132, 166). The introduction of highly resistant lineages, such as ST131, does not necessarily displace these classic lineages (166). However, constant selection pressures created by antimicrobials can drive the selection and emergence of MDR ExPEC lineages, as noted with ST131. All major STs have now been linked to multidrug resistance gene carriage or ESBL production, and an increasing number of the major STs have been associated with carbapenemase production.
The largest number of ExPEC genomic studies were conducted in Europe and North America and showed similar distributions of ExPEC lineages; however, data from Asia and Africa suggest that different STs may be associated with infections in these regions. This could have resulted from the influence of selection criteria used in these studies or could represent real differences due to distinct selection pressures in these regions. In 2014, a report from the World Health Organization (WHO) showed that when antimicrobial resistance surveillance data are available, appropriate treatment regimens can be selected, resistance trends can be understood, priority areas for interventions can be identified, and the impact of interventions to reduce resistance can be monitored (206). The report revealed the lack of adequate surveillance programs in many parts of the world. This systematic review supports the WHO report conclusions and specifically points to the lack of systematic and unbiased information about ExPEC lineage incidence, especially the absence of this information for many regions of the world, including Africa, Central and South America, the Middle East, and Oceania. Understanding these underrepresented regions will be vital to detecting the emergence of newly successful lineages, especially those harboring extensive drug resistance plasmids.
This review highlights that the epidemiology and population dynamics of ExPEC are complex and variable. Recently, investigators have identified blooms of specific STs over multiple years (207, 208) and have reported an increase in the prominence of other STs, such as MDR ST1193 (209). Other STs appeared to decrease over time (e.g., ST95). This may be due to actual increases in different lineages or could be the result of a heightened focus on MDR ExPEC lineages. Other STs exhibited nearly identical proportions over all periods (e.g., ST10), suggesting that some lineages may be less influenced by selection pressures on ExPEC population dynamics. This is a significant concern for public health, infection prevention, antimicrobial stewardship, and vaccination programs. We need continuous, large-scale global genomic studies that include ExPEC isolates, irrespective of their susceptibility profiles, to fully elucidate the population structure of these successful groups of pathogens.
The results of this review question the absolute distinction between commensal fecal isolates and ExPEC isolates as two separate E. coli pathotypes as isolates from colonization and infection studies share substantial parts of their genetic cores, which for some lineages cannot be distinguished in SNP-based phylogeny. Nielsen et al. make the case that ExPEC bacteria exist equally in the intestinal reservoir and urinary tract, as there were few differences in the core genomes between these pathotypes, except possibly in the accessory genome (41). This has also been supported by data from other non-WGS studies (210, 211). This reinforces the idea that the E. coli in the intestinal reservoir serves as the primary source of the E. coli that gives rise to extraintestinal infections and that there may be no clear distinction between commensal E. coli and ExPEC. Kallonen et al. point to the importance of competition, persistence, and predominance in the intestinal reservoir to the dominance of ST131 and ST73 ExPEC lineages as the cause of infections (166). However, currently there is limited evidence of this distinction in other lineages as most WGS-based studies have focused primarily on ST131. Despite the lack of differences in virulence gene profiles between infection and intestine source isolates, phylogenetic group, as a marker of evolution, clearly distinguishes lineages that contribute most to extraintestinal infections (166). It is also possible that disturbances to the microbiota, as the result of travel or antibiotic use, contribute to transient intestinal acquisition of and colonization with successful ExPEC lineages. This could explain why it could be challenging to differentiate commensal E. coli and ExPEC strains around the time of infection; however, this hypothesis requires testing. The research community still does not fully understand the role of different adaptations in ExPEC that contribute to fitness in the intestinal reservoir and at extraintestinal infection sites.
ExPEC bacteria are transmitted via fecal-oral, household, sexual, and foodborne routes. Local clusters of closely related ExPEC isolates, as shown by WGS, have been detected in multiple settings. Almost all major ExPEC STs have been identified in at least one nonhuman host or environmental source. Understanding the transmission mechanisms for these common and successful STs could go a long way in reducing ExPEC carriage and infection. Multiple factors, from both the host and the environment, shape the genomics of ExPEC. Therefore, the need for a “one-health” (i.e., spanning ExPEC isolates among humans, animals, and their environments) strategy for surveillance and containment across the different sectors will clarify ExPEC transmission routes.
There are multiple limitations to this systematic review. Study populations were very different. The studies included E. coli isolates from a range of sources, including surveillance specimens, UTIs, and bloodstream infections, and from both community-based and health care settings. The values of I2 in the supplemental tables show that study heterogeneity was substantial. Different definitions of “ExPEC” were used in the various studies. In many cases, the patient or isolate sampling procedures and denominators were incompletely described or were not listed. Given the breadth of the literature and the multiple disciplines contributing to ExPEC research, we likely missed some relevant reports and manuscripts. The review adhered to a modified Cochrane review strategy; however, variation in the indexing of studies in publication databases may have influenced our ability to identify relevant studies. It can be a common practice to include the same isolates in more than one study so that different research questions can be addressed. We did our best to exclude studies that used the same or part of the same collection of E. coli isolates although some isolates may have been counted twice in this review. We also acknowledge that there were rare STs that were detected in some of studies but were not captured and abstracted into our review data set. However, we feel confident that the 215 STs included represent the STs contributing most to the burden of human extraintestinal infections around the world. Given the large number of studies that selected their study isolates by resistance phenotype, the ExPEC lineages associated with multidrug resistance (e.g., ST131) will be significantly overrepresented, while lineages not associated with resistance are likely to be underrepresented.
A discrete set of ExPEC lineages contributes to the enormous burden of human extraintestinal infections. Despite the limited number, these STs are highly diverse in genetic background, virulence, and resistance gene carriage. There is mounting evidence that ExPEC success as a pathogen of extraintestinal sites may be mediated by its persistence, adaptation, and predominance or function in the intestinal reservoir. The emphasis on MDR ExPEC ST131 has unfortunately created a gap in our knowledge about other important ExPEC lineages, such as ST73 and ST95. Additional whole-genome sequencing-based studies, using a one-health approach, are urgently needed to understand the phylogeny, function, population dynamics, and molecular epidemiology of these 20 major ExPEC STs. This gap will gradually be filled as genome sequencing becomes part of routine research and public health practice. ExPEC contributes to an enormous infectious disease burden around the world and serves as a reservoir for the development and mobilization of new resistance genes or novel combinations of resistance genes in the gut and at infected extraintestinal body sites. Interventions to tackle the most common ExPEC lineages would go a long way in reducing the burden of disease caused globally by these E. coli strains.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We declare that we have no conflicts of interest.
A.R.M. and J.D.D.P. conceived of and designed the review. A.R.M., H.M.G., and A.G. performed the systematic review. C.D.F. and T.J.E. analyzed the data. A.R.M. and J.D.D.P. wrote the manuscript.
Biographies

Amee R. Manges is an Associate Professor at the University of British Columbia, School of Population and Public Health, and an Adjunct Scientist at the British Columbia Centre for Disease Control, where she supervises a research laboratory. Her primary research interests are in public health and molecular epidemiology, specifically the application of genomics and metagenomics to questions in infectious disease epidemiology and human microbiome research.

Hyun Min Geum received her bachelor of science degree in microbiology and immunology (2016) at the University of British Columbia and is now pursuing her M.D. Her current research interests include infectious diseases, addiction medicine, and public health.

Alice Guo is a public health professional who holds a B.Sc. in microbiology and immunology from the University of British Columbia and an M.Sc. in global population health from the London School of Economics and Political Science. Currently, she is a consultant in the Department of Essential Medicines and Health Products at the World Health Organization. Her interests are in improving population health outcomes globally.

Thaddeus J. Edens is a bioinformatician and scientific computing consultant and holds a Ph.D. in mathematics from the University of California at Davis. His research interests include processing, managing, and analyzing high-dimensional metagenome sequencing data. He also provides consulting services in biostatistics and scientific software development.

Chad D. Fibke is a current M.Sc. student in the University of British Columbia, School of Population and Public Health. His research interests are in molecular and genomic epidemiology and public health bioinformatics.

Johann D. D. Pitout is a Professor at the Cummings Medical School, University of Calgary, and Medical Microbiologist at Calgary Laboratory Services, Alberta, Canada. His main research interests are resistance to antimicrobial agents among Gram-negative bacteria, especially the laboratory detection, characterization, molecular epidemiology, and evolution of bacteria with newer β-lactamases such as AmpC, extended-spectrum β-lactamases, and carbapenemases. He has also been involved in population-based surveillance studies and the role of high-risk clones and mobile genetic elements (such as plasmids) among bacteria producing these newer types of β-lactamases. He serves on the editorial boards of several journals has published over 160 peer-reviewed manuscripts in this field.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CMR.00135-18.
REFERENCES
- 1.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol 157:1127–1129. doi: 10.1016/S0022-5347(01)65154-1. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B, Brown P. 2003. Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect Dis Clin North Am 17:227–241. doi: 10.1016/S0891-5520(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 3.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 4.Riley LW. 2014. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 20:380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suskind AM, Saigal CS, Hanley JM, Lai J, Setodji CM, Clemens JQ. 2016. Incidence and management of uncomplicated recurrent urinary tract infections in a national sample of women in the United States. Urology 90:50–55. doi: 10.1016/j.urology.2015.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.François M, Hanslik T, Dervaux B, Le Strat Y, Souty C, Vaux S, Maugat S, Rondet C, Sarazin M, Heym B, Coignard B, Rossignol L. 2016. The economic burden of urinary tract infections in women visiting general practices in France: a cross-sectional survey. BMC Health Serv Res 16:365. doi: 10.1186/s12913-016-1620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plos K, Connell H, Jodal U, Marklund BI, Mårild S, Wettergren B, Svanborg C. 1995. Intestinal carriage of P fimbriated Escherichia coli and the susceptibility to urinary tract infection in young children. J Infect Dis 171:625–631. doi: 10.1093/infdis/171.3.625. [DOI] [PubMed] [Google Scholar]
- 10.Bettelheim KA, Taylor J. 1969. A study of Escherichia coli isolated from chronic urinary infection. J Med Microbiol 2:225–236. doi: 10.1099/00222615-2-3-225. [DOI] [PubMed] [Google Scholar]
- 11.Turck M, Petersdorf RG. 1962. The epidemiology of nonenteric Escherichia coli infections: prevalence of serological groups. J Clin Invest 41:1760–1765. doi: 10.1172/JCI104635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JR, Delavari P, Kuskowski M, Stell AL. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 183:78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- 13.Alqasim A, Scheutz F, Zong Z, McNally A. 2014. Comparative genome analysis identifies few traits unique to the Escherichia coli ST131 H30Rx clade and extensive mosaicism at the capsule locus. BMC Genomics 15:830. doi: 10.1186/1471-2164-15-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TEA, Johnson JR, Didelot X, Walker AS, Crook DW. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162-15. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köhler C-D, Dobrindt U. 2011. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol 301:642–647. doi: 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JPA, Clarke M, Devereaux PJ, Kleijnen J, Moher D. 2009. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. PLoS Med 6:e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560. doi: 10.1186/1471-2164-9-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weihong Q, Lacher DW, Bumbaugh AC, Hyma KE, Ouellette LM, Large TM, Tarr CL, Whittam TS. 2004. EcMLST: an online database for multi locus sequence typing of pathogenic Escherichia coli, p 499–500. In Proceedings of the 2004 IEEE Computational Systems Bioinformatics Conference (CSB 2004). Institute of Electrical and Electronics Engineers, Washington, DC. [Google Scholar]
- 23.Adams-Sapper S, Diep BA, Perdreau-Remington F, Riley LW. 2013. Clonal composition and community clustering of drug-susceptible and -resistant Escherichia coli Isolates from bloodstream infections. Antimicrob Agents Chemother 57:490–497. doi: 10.1128/AAC.01025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adenipekun EO, Jackson CR, Ramadan H, Iwalokun BA, Oyedeji KS, Frye JG, Barrett JB, Hiott LM, Woodley TA, Oluwadun A. 2016. Prevalence and multidrug resistance of Escherichia coli from community-acquired infections in Lagos, Nigeria. J Infect Dev Ctries 10:920. doi: 10.3855/jidc.7997. [DOI] [PubMed] [Google Scholar]
- 25.Alonso N, Miró E, Pascual V, Rivera A, Simó M, Garcia MC, Xercavins M, Morera MA, Espejo E, Gurguí M, Pérez J, Rodríguez-Carballeira M, Garau J, Calbo E, Navarro F, Mirelis B, Coll P. 2016. Molecular characterisation of acquired and overproduced chromosomal blaAmpC in Escherichia coli clinical isolates. Int J Antimicrob Agents 47:62–68. doi: 10.1016/j.ijantimicag.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Miajlovic H, Aogáin MM, Collins CJ, Rogers TR, Smith S. 2016. Characterization of Escherichia coli bloodstream isolates associated with mortality. J Med Microbiol 65:71–79. doi: 10.1099/jmm.0.000200. [DOI] [PubMed] [Google Scholar]
- 27.Mora A, Blanco M, López C, Mamani R, Blanco JE, Alonso MP, García-Garrote F, Dahbi G, Herrera A, Fernández A, Fernández B, Agulla A, Bou G, Blanco J. 2011. Emergence of clonal groups O1:HNM-D-ST59, O15:H1-D-ST393, O20:H34/HNM-D-ST354, O25b:H4-B2-ST131 and ONT:H21,42-B1-ST101 among CTX-M-14-producing Escherichia coli clinical isolates in Galicia, northwest Spain. Int J Antimicrob Agents 37:16–21. doi: 10.1016/j.ijantimicag.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Mora A, López C, Dabhi G, Blanco M, Blanco JE, Alonso M, Herrera A, Mamani R, Bonacorsi S, Moulin-Schouleur M, Blanco J. 2009. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol 9:132. doi: 10.1186/1471-2180-9-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morales-Espinosa R, Hernandez-Castro R, Delgado G, Mendez JL, Navarro A, Manjarrez A, Cravioto A. 2016. UPEC strain characterization isolated from Mexican patients with recurrent urinary infections. J Infect Dev Ctries 10:317. doi: 10.3855/jidc.6652. [DOI] [PubMed] [Google Scholar]
- 30.Moreira da Silva RCR, de Oliveira Martins Júnior P, Gonçalves LF, de Paulo Martins V, de Melo ABF, Pitondo-Silva A, de Campos TA. 2017. Ciprofloxacin resistance in uropathogenic Escherichia coli isolates causing community-acquired urinary infections in Brasília, Brazil. J Glob Antimicrob Resist 9:61–67. doi: 10.1016/j.jgar.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Mshana SE, Imirzalioglu C, Hain T, Domann E, Lyamuya EF, Chakraborty T. 2011. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect 17:1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x. [DOI] [PubMed] [Google Scholar]
- 32.Mshana SE, Falgenhauer L, Mirambo MM, Mushi MF, Moremi N, Julius R, Seni J, Imirzalioglu C, Matee M, Chakraborty T. 2016. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect Dis 16:187. doi: 10.1186/s12879-016-1527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller A, Stephan R, Nüesch-Inderbinen M. 2016. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci Total Environ 541:667–672. doi: 10.1016/j.scitotenv.2015.09.135. [DOI] [PubMed] [Google Scholar]
- 34.Musicha P, Feasey NA, Cain AK, Kallonen T, Chaguza C, Peno C, Khonga M, Thompson S, Gray KJ, Mather AE, Heyderman RS, Everett DB, Thomson NR, Msefula CL. 2017. Genomic landscape of extended-spectrum β-lactamase resistance in Escherichia coli from an urban African setting. J Antimicrob Chemother 72:1602–1609. doi: 10.1093/jac/dkx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura A, Komatsu M, Noguchi N, Ohno Y, Hashimoto E, Matsutani H, Abe N, Fukuda S, Kohno H, Nakamura F, Matsuo S, Kawano S. 2016. Analysis of molecular epidemiologic characteristics of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli colonizing feces in hospital patients and community dwellers in a Japanese city. J Infect Chemother 22:102–107. doi: 10.1016/j.jiac.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Alyamani EJ, Khiyami AM, Booq RY, Majrashi MA, Bahwerth FS, Rechkina E. 2017. The occurrence of ESBL-producing Escherichia coli carrying aminoglycoside resistance genes in urinary tract infections in Saudi Arabia. Ann Clin Microbiol Antimicrob 16:1. doi: 10.1186/s12941-016-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naseer U, Haldorsen B, Simonsen GS, Sundsfjord A. 2010. Sporadic occurrence of CMY-2-producing multidrug-resistant Escherichia coli of ST-complexes 38 and 448, and ST131 in Norway. Clin Microbiol Infect 16:171–178. doi: 10.1111/j.1469-0691.2009.02861.x. [DOI] [PubMed] [Google Scholar]
- 38.Naseer U, Haldorsen B, Tofteland S, Hegstad K, Scheutz F, Simonsen GS, Sundsfjord A. 2009. Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS 117:526–536. doi: 10.1111/j.1600-0463.2009.02465.x. [DOI] [PubMed] [Google Scholar]
- 39.Naseer U, Olsson-Liljequist BE, Woodford N, Dhanji H, Cantón R, Sundsfjord A, Lindstedt B-A. 2012. Multi-locus variable number of tandem repeat analysis for rapid and accurate typing of virulent multidrug resistant Escherichia coli clones. PLoS One 7:e41232. doi: 10.1371/journal.pone.0041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen JB, Albayati A, Jørgensen RL, Hansen KH, Lundgren B, Schønning K. 2013. An abbreviated MLVA identifies Escherichia coli ST131 as the major extended-spectrum β-lactamase-producing lineage in the Copenhagen area. Eur J Clin Microbiol Infect Dis 32:431–436. doi: 10.1007/s10096-012-1764-x. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen KL, Stegger M, Kiil K, Godfrey PA, Feldgarden M, Lilje B, Andersen PS, Frimodt-Møller N. 2017. Whole-genome comparison of urinary pathogenic Escherichia coli and faecal isolates of UTI patients and healthy controls. Int J Med Microbiol 307:497–507. doi: 10.1016/j.ijmm.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novais A, Machado E, Amaral S, Goncalves T, Canton R, Coque T. 2011. Amplification of CTX-M-15-B2-ST131 and emergence of A and D widespread clones producing different ESBLs in Portuguese hospitals. Clin Microbiol Infect 17(Suppl 4):S147. [Google Scholar]
- 43.Nüesch-Inderbinen MT, Baschera M, Zurfluh K, Hächler H, Nüesch H, Stephan R. 2017. Clonal diversity, virulence potential and antimicrobial resistance of Escherichia coli causing community acquired urinary tract infection in Switzerland. Front Microbiol 8:2334. doi: 10.3389/fmicb.2017.02334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ojer-Usoz E, Gonzalez D, Vitas AI. 2017. Clonal diversity of ESBL-producing Escherichia coli isolated from environmental, human and food samples. Int J Environ Res Public Health 14:676. doi: 10.3390/ijerph14070676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olesen B, Scheutz F, Andersen RL, Menard M, Boisen N, Johnston B, Hansen DS, Krogfelt KA, Nataro JP, Johnson JR. 2012. Enteroaggregative Escherichia coli O78:H10, the cause of an outbreak of urinary tract infection. J Clin Microbiol 50:3703–3711. doi: 10.1128/JCM.01909-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortega A, Oteo J, Aranzamendi-Zaldumbide M, Bartolomé RM, Bou G, Cercenado E, Conejo MC, González-López JJ, Marín M, Martínez-Martínez L, Merino M, Navarro F, Oliver A, Pascual Á, Rivera A, Rodríguez-Baño J, Weber I, Aracil B, Campos J. 2012. Spanish multicenter study of the epidemiology and mechanisms of amoxicillin-clavulanate resistance in Escherichia coli. Antimicrob Agents Chemother 56:3576–3581. doi: 10.1128/AAC.06393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aoike N, Saga T, Sakata R, Yoshizumi A, Kimura S, Iwata M, Yoshizawa S, Sugasawa Y, Ishii Y, Yamaguchi K, Tateda K. 2013. Molecular characterization of extraintestinal Escherichia coli isolates in Japan: relationship between sequence types and mutation patterns of quinolone resistance-determining regions analyzed by pyrosequencing. J Clin Microbiol 51:1692–1698. doi: 10.1128/JCM.03049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortega A, Sáez D, Bautista V, Fernández-Romero S, Lara N, Aracil B, Pérez-Vázquez M, Campos J, Oteo J. 2016. Carbapenemase-producing Escherichia coli is becoming more prevalent in Spain mainly because of the polyclonal dissemination of OXA-48. J Antimicrob Chemother 71:2131–2138. doi: 10.1093/jac/dkw148. [DOI] [PubMed] [Google Scholar]
- 49.Oteo J, Cercenado E, Cuevas Ó, Bautista V, Delgado-Iribarren A, Orden B, Pérez-Vázquez M, García-Cobos S, Campos J. 2010. AmpC β-lactamases in Escherichia coli: emergence of CMY-2–producing virulent phylogroup D isolates belonging mainly to STs 57, 115, 354, 393, and 420, and phylogroup B2 isolates belonging to the international clone O25b–ST131. Diagn Microbiol Infect Dis 67:270–276. doi: 10.1016/j.diagmicrobio.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Oteo J, Cercenado E, Fernández-Romero S, Saéz D, Padilla B, Zamora E, Cuevas O, Bautista V, Campos J. 2012. Extended-spectrum-β-lactamase-producing Escherichia coli as a cause of pediatric infections: report of a neonatal intensive care unit outbreak due to a CTX-M-14-producing strain. Antimicrob Agents Chemother 56:54–58. doi: 10.1128/AAC.05103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oteo J, Diestra K, Juan C, Bautista V, Novais Â, Pérez-Vázquez M, Moyá B, Miró E, Coque TM, Oliver A, Cantón R, Navarro F, Campos J. 2009. Extended-spectrum β-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int J Antimicrob Agents 34:173–176. doi: 10.1016/j.ijantimicag.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 52.Oteo J, Orden B, Bautista V, Cuevas O, Arroyo M, Martínez-Ruiz R, Pérez-Vázquez M, Alcaraz M, García-Cobos S, Campos J. 2009. CTX-M-15-producing urinary Escherichia coli O25b-ST131-phylogroup B2 has acquired resistance to fosfomycin. J Antimicrob Chemother 64:712–717. doi: 10.1093/jac/dkp288. [DOI] [PubMed] [Google Scholar]
- 53.Ouédraogo A-S, Sanou S, Kissou A, Poda A, Aberkane S, Bouzinbi N, Nacro B, Ouédraogo R, Van De Perre P, Carriere C, Decré D, Jean-Pierre H, Godreuil S. 2017. Fecal carriage of Enterobacteriaceae producing extended-spectrum beta-lactamases in hospitalized patients and healthy community volunteers in Burkina Faso. Microb Drug Resist 23:63–70. doi: 10.1089/mdr.2015.0356. [DOI] [PubMed] [Google Scholar]
- 54.Paltansing S, Vlot JA, Kraakman MEM, Mesman R, Bruijning ML, Bernards AT, Visser LG, Veldkamp KE. 2013. Extended-spectrum β-lactamase-producing Enterobacteriaceae among travelers from the Netherlands. Emerg Infect Dis 19:1206–1213. doi: 10.3201/eid.1908.130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papagiannitsis CC, Študentová V, Jakubů V, Španělová P, Urbášková P, Žemličková H, Hrabák J. 2015. High prevalence of ST131 among CTX-M-producing Escherichia coli from community-acquired infections, in the Czech Republic. Microb Drug Resist 21:74–84. doi: 10.1089/mdr.2014.0070. [DOI] [PubMed] [Google Scholar]
- 56.Park SH, Byun JH, Choi SM, Lee DG, Kim SH, Kwon JC, Yoo JH, Choi JH. 2012. Characterisation of extended-spectrum beta-lactamase producing Escherichia coli isolated from patients with urinary tract infections in Korea: comparison between community isolates and nosocomial isolates. Clin Microbiol Infect 18(Suppl 3):572. [Google Scholar]
- 57.Park SH, Byun J-H, Choi S-M, Lee D-G, Kim S-H, Kwon J-C, Park C, Choi J-H, Yoo J-H. 2012. Molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in the community and hospital in Korea: emergence of ST131 producing CTX-M-15. BMC Infect Dis 12:149. doi: 10.1186/1471-2334-12-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barillova P, Tchesnokova V, Dübbers A, Küster P, Peters G, Dobrindt U, Sokurenko EV, Kahl BC. 2014. Prevalence and persistence of Escherichia coli in the airways of cystic fibrosis patients—an unrecognized CF pathogen? Int J Med Microbiol 304:415–421. doi: 10.1016/j.ijmm.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 59.Peirano G, Laupland KB, Gregson DB, Pitout J. 2011. Colonization of returning travelers with CTX‐M‐producing Escherichia coli. J Travel Med 18:299–303. doi: 10.1111/j.1708-8305.2011.00548.x. [DOI] [PubMed] [Google Scholar]
- 60.Peirano G, van der Bij AK, Gregson DB, Pitout J. 2012. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum beta-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J Clin Microbiol 50:294–299. doi: 10.1128/JCM.06025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peirano G, van Greune CHJ, Pitout J. 2011. Characteristics of infections caused by extended-spectrum β-lactamase–producing Escherichia coli from community hospitals in South Africa. Diagn Microbiol Infect Dis 69:449–453. doi: 10.1016/j.diagmicrobio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, Krogfelt KA. 2009. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC Microbiol 9:171. doi: 10.1186/1471-2180-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan M-D, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JDD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pietsch M, Eller C, Wendt C, Holfelder M, Falgenhauer L, Fruth A, Grössl T, Leistner R, Valenza G, Werner G, Pfeifer Y. 2017. Molecular characterisation of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates from hospital and ambulatory patients in Germany. Vet Microbiol 200:130–137. doi: 10.1016/j.vetmic.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 65.Pitout JDD, Gregson DB, Campbell L, Laupland KB. 2009. Molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary health region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob Agents Chemother 53:2846–2851. doi: 10.1128/AAC.00247-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, Clabots C, Kuskowski MA, Trott DJ. 2011. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob Agents Chemother 55:3782–3787. doi: 10.1128/AAC.00306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pomba C, López-Cerero L, Bellido M, Serrano L, Belas A, Couto N, Cavaco-Silva P, Rodríguez-Baño J, Pascual A. 2014. Within-lineage variability of ST131 Escherichia coli isolates from humans and companion animals in the south of Europe. J Antimicrob Chemother 69:271–273. doi: 10.1093/jac/dkt343. [DOI] [PubMed] [Google Scholar]
- 68.Poulsen HO, Johansson A, Granholm S, Kahlmeter G, Sundqvist M. 2013. High genetic diversity of nitrofurantoin- or mecillinam-resistant Escherichia coli indicates low propensity for clonal spread. J Antimicrob Chemother 68:1974–1977. doi: 10.1093/jac/dkt159. [DOI] [PubMed] [Google Scholar]
- 69.Ben Sallem R, Ben Slama K, Estepa V, Jouini A, Gharsa H, Klibi N, Sáenz Y, Ruiz-Larrea F, Boudabous A, Torres C. 2012. Prevalence and characterisation of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates in healthy volunteers in Tunisia. Eur J Clin Microbiol Infect Dis 31:1511–1516. doi: 10.1007/s10096-011-1471-z. [DOI] [PubMed] [Google Scholar]
- 70.Qin X, Hu F, Wu S, Ye X, Zhu D, Zhang Y, Wang M. 2013. Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains. PLoS One 8:e61169. doi: 10.1371/journal.pone.0061169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ranjan A, Shaik S, Mondal A, Nandanwar N, Hussain A, Semmler T, Kumar N, Tiwari SK, Jadhav S, Wieler LH, Ahmed N. 2016. Molecular epidemiology and genome dynamics of New Delhi metallo-β-lactamase-producing extraintestinal pathogenic Escherichia coli strains from India. Antimicrob Agents Chemother 60:6795–6805. doi: 10.1128/AAC.01345-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roer L, Hansen F, Thomsen MCF, Knudsen JD, Hansen DS, Wang M, Samulioniené J, Justesen US, Røder BL, Schumacher H, Østergaard C, Andersen LP, Dzajic E, Søndergaard TS, Stegger M, Hammerum AM, Hasman H. 2017. WGS-based surveillance of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. J Antimicrob Chemother 72:1922–1929. doi: 10.1093/jac/dkx092. [DOI] [PubMed] [Google Scholar]
- 73.Rogers BA, Ingram PR, Runnegar N, Pitman MC, Freeman JT, Athan E, Havers S, Sidjabat HE, Gunning E, De Almeida M, Styles K, Paterson DL. 2015. Sequence type 131 fimH30 and fimH41 subclones amongst Escherichia coli isolates in Australia and New Zealand. Int J Antimicrob Agents 45:351–358. doi: 10.1016/j.ijantimicag.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 74.Runcharoen C, Raven KE, Reuter S, Kallonen T, Paksanont S, Thammachote J, Anun S, Blane B, Parkhill J, Peacock SJ, Chantratita N. 2017. Whole genome sequencing of ESBL-producing Escherichia coli isolated from patients, farm waste and canals in Thailand. Genome Med 9:81. doi: 10.1186/s13073-017-0471-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salipante SJ, Roach DJ, Kitzman JO, Snyder MW, Stackhouse B, Butler-Wu SM, Lee C, Cookson BT, Shendure J. 2015. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 25:119–128. doi: 10.1101/gr.180190.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shakir S, Goldbeck J, Robison D, Eckerd A, Chavez-Bueno S. 2014. Genotypic and phenotypic characterization of invasive neonatal Escherichia coli clinical isolates. Am J Perinatol 31:975–982. doi: 10.1055/s-0034-1370341. [DOI] [PubMed] [Google Scholar]
- 77.Shelburne SA, Kim J, Munita JM, Sahasrabhojane P, Shields RK, Press EG, Li X, Arias CA, Cantarel B, Jiang Y, Kim MS, Aitken SL, Greenberg DE. 2017. Whole-genome sequencing accurately identifies resistance to extended-spectrum β-lactams for major Gram-negative bacterial pathogens. Clin Infect Dis 65:738–745. doi: 10.1093/cid/cix417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sherchan JB, Hayakawa K, Miyoshi-Akiyama T, Ohmagari N, Kirikae T, Nagamatsu M, Tojo M, Ohara H, Sherchand JB, Tandukar S. 2015. Clinical epidemiology and molecular analysis of extended-spectrum-β-lactamase-producing Escherichia coli in Nepal: characteristics of sequence types 131 and 648. Antimicrob Agents Chemother 59:3424–3432. doi: 10.1128/AAC.00270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi H, Sun F, Chen J, Ou Q, Feng W, Yong X, Xia P. 2015. Epidemiology of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing nosocomial Escherichia coli infection in China. Ann Clin Microbiol Antimicrob 14:4. doi: 10.1186/s12941-015-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bengtsson S, Naseer U, Sundsfjord A, Kahlmeter G, Sundqvist M. 2012. Sequence types and plasmid carriage of uropathogenic Escherichia coli devoid of phenotypically detectable resistance. J Antimicrob Chemother 67:69–73. doi: 10.1093/jac/dkr421. [DOI] [PubMed] [Google Scholar]
- 81.Shin J, Kim DH, Ko KS. 2011. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J Infect 63:39–47. doi: 10.1016/j.jinf.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 82.Suzuki S, Shibata N, Yamane K, Wachino J.-i, Ito K, Arakawa Y. 2008. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother 63:72–79. doi: 10.1093/jac/dkn463. [DOI] [PubMed] [Google Scholar]
- 83.Totsika M, Beatson SA, Sarkar S, Phan M-D, Petty NK, Bachmann N, Szubert M, Sidjabat HE, Paterson DL, Upton M, Schembri MA. 2011. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS One 6:e26578. doi: 10.1371/journal.pone.0026578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toval F, Köhler C-D, Vogel U, Wagenlehner F, Mellmann A, Fruth A, Schmidt MA, Karch H, Bielaszewska M, Dobrindt U. 2014. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J Clin Microbiol 52:407–418. doi: 10.1128/JCM.02069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Umene YD, Wong LK, Satoh T, Yamane K, Matsui M, Riley LW, Arakawa Y, Suzuki S. 2015. Molecular epidemiological characterization of uropathogenic Escherichia coli from an outpatient urology clinic in rural Japan. J Clin Microbiol 53:681–683. doi: 10.1128/JCM.03068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Usein C-R, Condei M, Cristea D, Ciontea SA, Tatu-Chiţoiu D, Cristea V, Străuţ M. 2014. Escherichia coli ST131 causing invasive infections in Romanian patients—a threat we can no longer ignore. Roum Arch Microbiol Immunol 73:5–8. [PubMed] [Google Scholar]
- 87.Usein C-R, Papagheorghe R, Oprea M, Condei M, Strãuţ M. 2016. Molecular characterization of bacteremic Escherichia coli isolates in Romania. Folia Microbiol (Praha) 61:221–226. doi: 10.1007/s12223-015-0427-6. [DOI] [PubMed] [Google Scholar]
- 88.van der Donk CFM, Schols J, Driessen CJ, Hagenouw RGP, Meulendijks A, Stobberingh EE. 2013. Prevalence and spread of multidrug resistant Escherichia coli Isolates among nursing home residents in the southern part of the Netherlands. J Am Med Dir Assoc 14:199–203. doi: 10.1016/j.jamda.2012.09.026. [DOI] [PubMed] [Google Scholar]
- 89.van der Donk C, van de Bovenkamp J, Bamelis H, Driessen C, Feldhoff K-H, Kalka-Moll W, Magerman K, Stobberingh E. 2013. Prevalence and spread of multidrug-resistant Escherichia coli including ST131 in different patient populations in the Euroregion Meuse–Rhine. Future Microbiol 8:1027–1037. doi: 10.2217/fmb.13.61. [DOI] [PubMed] [Google Scholar]
- 90.Voor In ‘T Holt AF, Wattel AA, Boers SA, Jansen R, Hays JP, Goessens WHF, Vos MC. 2016. Detection of healthcare-related extended-spectrum beta-lactamase-producing Escherichia coli transmission events using combined genetic and phenotypic epidemiology. PLoS One 11:e0160156. doi: 10.1371/journal.pone.0160156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berg ES, Wester AL, Ahrenfeldt J, Mo SS, Slettemeås JS, Steinbakk M, Samuelsen Ø, Grude N, Simonsen GS, Løhr IH, Jørgensen SB, Tofteland S, Lund O, Dahle UR, Sunde M. 2017. Norwegian patients and retail chicken meat share cephalosporin-resistant Escherichia coli and IncK/bla CMY-2 resistance plasmids. Clin Microbiol Infect 23:407.e9–407.e15. doi: 10.1016/j.cmi.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 92.Wang S, Zhao S-Y, Xiao S-Z, Gu F-F, Liu Q-Z, Tang J, Guo X-K, Ni Y-X, Han L-Z. 2016. Antimicrobial resistance and molecular epidemiology of Escherichia coli causing bloodstream infections in three hospitals in Shanghai, China. PLoS One 11:e0147740. doi: 10.1371/journal.pone.0147740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y, Tian G-B, Zhang R, Shen Y, Tyrrell JM, Huang X, Zhou H, Lei L, Li H-Y, Doi Y, Fang Y, Ren H, Zhong L-L, Shen Z, Zeng K-J, Wang S, Liu J-H, Wu C, Walsh TR, Shen J. 2017. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1 -positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis 17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 94.Ward DV, Scholz M, Zolfo M, Taft DH, Schibler KR, Tett A, Segata N, Morrow AL. 2016. Metagenomic sequencing with strain-level resolution implicates uropathogenic E. coli in necrotizing enterocolitis and mortality in preterm infants. Cell Rep 14:2912–2924. doi: 10.1016/j.celrep.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weissman SJ, Hansen NI, Zaterka-Baxter K, Higgins RD, Stoll BJ. 2016. Emergence of antibiotic resistance-associated clones among Escherichia coli recovered from newborns with early-onset sepsis and meningitis in the United States, 2008–2009: table 1. J Ped Infect Dis 5:269–276. doi: 10.1093/jpids/piv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol 78:1353–1360. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wickramasinghe NH, Xu L, Eustace A, Shabir S, Saluja T, Hawkey PM. 2012. High community faecal carriage rates of CTX-M ESBL-producing Escherichia coli in a specific population group in Birmingham, UK. J Antimicrob Chemother 67:1108–1113. doi: 10.1093/jac/dks018. [DOI] [PubMed] [Google Scholar]
- 98.Xia S, Fan X, Huang Z, Xia L, Xiao M, Chen R, Xu Y, Zhuo C. 2014. Dominance of CTX-M-type extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS One 9:e100707. doi: 10.1371/journal.pone.0100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu M, Fan Y, Wang M, Lu X. 2017. Characteristics of extended-spectrum β-lactamases-producing Escherichia coli in fecal samples of inpatients of Beijing Tongren Hospital. Jpn J Infect Dis 70:290–294. doi: 10.7883/yoken.JJID.2016.023. [DOI] [PubMed] [Google Scholar]
- 100.Yaita K, Aoki K, Suzuki T, Nakaharai K, Yoshimura Y, Harada S, Ishii Y, Tachikawa N. 2014. Epidemiology of extended-spectrum β-lactamase producing Escherichia coli in the stools of returning Japanese travelers, and the risk factors for colonization. PLoS One 9:e98000. doi: 10.1371/journal.pone.0098000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yun KW, Kim DS, Kim W, Lim IS. 2015. Molecular typing of uropathogenic Escherichia coli isolated from Korean children with urinary tract infection. Korean J Pediatr 58:20. doi: 10.3345/kjp.2015.58.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bert F, Johnson JR, Ouattara B, Leflon-Guibout V, Johnston B, Marcon E, Valla D, Moreau R, Nicolas-Chanoine M-H. 2010. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J Clin Microbiol 48:2709–2714. doi: 10.1128/JCM.00516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yun KW, Lee M-K, Kim W, Lim IS. 2017. Uropathogenic Escherichia coli ST131 in urinary tract infections in children. Korean J Pediatr 60:221. doi: 10.3345/kjp.2017.60.7.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang J, Zheng B, Zhao L, Wei Z, Ji J, Li L, Xiao Y. 2014. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis 14:659. doi: 10.1186/s12879-014-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao S-Y, Wang Y-C, Xiao S-Z, Jiang X-F, Guo X-K, Ni Y-X, Han L-Z. 2015. Drug susceptibility and molecular epidemiology of Escherichia coli in bloodstream infections in Shanghai, China, 2011–2013. Infect Dis (Auckl) 47:310–318. doi: 10.3109/00365548.2014.990509. [DOI] [PubMed] [Google Scholar]
- 106.Beyrouthy R, Robin F, Dabboussi F, Mallat H, Hamzé M, Bonnet R. 2014. Carbapenemase and virulence factors of Enterobacteriaceae in North Lebanon between 2008 and 2012: evolution via endemic spread of OXA-48. J Antimicrob Chemother 69:2699–2705. doi: 10.1093/jac/dku181. [DOI] [PubMed] [Google Scholar]
- 107.Bi W, Li B, Song J, Hong Y, Zhang X, Liu H, Lu H, Zhou T, Cao J. 2017. Antimicrobial susceptibility and mechanisms of fosfomycin resistance in extended-spectrum β-lactamase-producing Escherichia coli strains from urinary tract infections in Wenzhou, China. Int J Antimicrob Agents 50:29–34. doi: 10.1016/j.ijantimicag.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 108.Adler A, Miller-Roll T, Assous MV, Geffen Y, Paikin S, Schwartz D, Weiner-Well Y, Hussein K, Cohen R, Carmeli Y. 2015. A multicenter study of the clonal structure and resistance mechanism of KPC-producing Escherichia coli isolates in Israel. Clin Microbiol Infect 21:230–235. doi: 10.1016/j.cmi.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 109.Bidet P, Mahjoub‐Messai F, Blanco J, Blanco J, Dehem M, Aujard Y, Bingen E, Bonacorsi S. 2007. Combined multilocus sequence typing and O serogrouping distinguishes Escherichia coli subtypes associated with infant urosepsis and/or meningitis. J Infect Dis 196:297–303. doi: 10.1086/518897. [DOI] [PubMed] [Google Scholar]
- 110.Blanco J, Mora A, Mamani R, Lopez C, Blanco M, Dahbi G, Herrera A, Blanco JE, Alonso MP, Garcia-Garrote F, Chaves F, Orellana MA, Martinez-Martinez L, Calvo J, Prats G, Larrosa MN, Gonzalez-Lopez JJ, Lopez-Cerero L, Rodriguez-Bano J, Pascual A. 2011. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother 66:2011–2021. doi: 10.1093/jac/dkr235. [DOI] [PubMed] [Google Scholar]
- 111.Brisse S, Diancourt L, Laouenan C, Vigan M, Caro V, Arlet G, Drieux L, Leflon-Guibout V, Mentre F, Jarlier V, Nicolas-Chanoine M-H. 2012. Phylogenetic distribution of CTX-M- and non-extended-spectrum-lactamase-producing Escherichia coli isolates: group B2 isolates, except clone ST131, rarely produce CTX-M enzymes. J Clin Microbiol 50:2974–2981. doi: 10.1128/JCM.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brodrick HJ, Raven KE, Kallonen T, Jamrozy D, Blane B, Brown NM, Martin V, Török ME, Parkhill J, Peacock SJ. 2017. Longitudinal genomic surveillance of multidrug-resistant Escherichia coli carriage in a long-term care facility in the United Kingdom. Genome Med 9:70. doi: 10.1186/s13073-017-0457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cagnacci S, Gualco L, Debbia E, Schito GC, Marchese A. 2008. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST 131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J Clin Microbiol 46:2605–2612. doi: 10.1128/JCM.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cai JC, Zhang R, Hu YY, Zhou HW, Chen G-X. 2014. Emergence of Escherichia coli sequence type 131 isolates producing KPC-2 carbapenemase in China. Antimicrob Agents Chemother 58:1146–1152. doi: 10.1128/AAC.00912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Calhau V, Ribeiro G, Mendonça N, Da Silva GJ. 2013. Prevalent combination of virulence and plasmidic-encoded resistance in ST 131 Escherichia coli strains. Virulence 4:726–729. doi: 10.4161/viru.26552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cao X, Cavaco LM, Lv Y, Li Y, Zheng B, Wang P, Hasman H, Liu Y, Aarestrup FM. 2011. Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J Clin Microbiol 49:2496–2501. doi: 10.1128/JCM.02503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cao XL, Shen H, Xu YY, Xu XJ, Zhang ZF, Cheng L, Chen JH, Arakawa Y. 2017. High prevalence of fosfomycin resistance gene fosA3 in bla CTX-M-harbouring Escherichia coli from urine in a Chinese tertiary hospital during 2010–2014. Epidemiol Infect 145:818–824. doi: 10.1017/S0950268816002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chang J, Yu J, Lee H, Ryu H, Park K, Park Y-J. 2014. Prevalence and characteristics of lactose non-fermenting Escherichia coli in urinary isolates. J Infect Chemother 20:738–740. doi: 10.1016/j.jiac.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 119.Adler A, Gniadkowski M, Baraniak A, Izdebski R, Fiett J, Hryniewicz W, Malhotra-Kumar S, Goossens H, Lammens C, Lerman Y, Kazma M, Kotlovsky T, Carmeli Y, MOSAR. 2012. Transmission dynamics of ESBL-producing Escherichia coli clones in rehabilitation wards at a tertiary care centre. Clin Microbiol Infect 18:E497–E505. doi: 10.1111/j.1469-0691.2012.03999.x. [DOI] [PubMed] [Google Scholar]
- 120.Chen C-M, Ke S-C, Li C-R, Chiou C-S, Chang C-C. 2014. Prolonged clonal spreading and dynamic changes in antimicrobial resistance of Escherichia coli ST68 among patients who stayed in a respiratory care ward. J Med Microbiol 63:1531–1541. doi: 10.1099/jmm.0.075937-0. [DOI] [PubMed] [Google Scholar]
- 121.Chen SL, Wu M, Henderson JP, Hooton TM, Hibbing ME, Hultgren SJ, Gordon JI. 2013. Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Sci Transl Med 5:184ra60. doi: 10.1126/scitranslmed.3005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chmielarczyk A, Pobiega M, Wójkowska-Mach J, Romaniszyn D, Heczko PB, Bulanda M. 2015. Bloodstream infections due to Enterobacteriaceae among neonates in Poland—molecular analysis of the isolates. Pol J Microbiol 64:217–225. doi: 10.5604/01.3001.0009.2117. [DOI] [PubMed] [Google Scholar]
- 123.Chung H-C, Lai C-H, Lin J-N, Huang C-K, Liang S-H, Chen W-F, Shih Y-C, Lin H-H, Wang J-L. 2012. Bacteremia caused by extended-spectrum-β-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob Agents Chemother 56:618–622. doi: 10.1128/AAC.05753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ciesielczuk H, Jenkins C, Chattaway M, Doumith M, Hope R, Woodford N, Wareham DW. 2016. Trends in ExPEC serogroups in the UK and their significance. Eur J Clin Microbiol Infect Dis 35:1661–1666. doi: 10.1007/s10096-016-2707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Coelho A, Mora A, Mamani R, Lopez C, Gonzalez-Lopez JJ, Larrosa MN, Quintero-Zarate JN, Dahbi G, Herrera A, Blanco JE, Blanco M, Alonso MP, Prats G, Blanco J. 2011. Spread of Escherichia coli O25b:H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain). J Antimicrob Chemother 66:517–526. doi: 10.1093/jac/dkq491. [DOI] [PubMed] [Google Scholar]
- 126.Crémet L, Broquet A, Jacqueline C, Chaillou C, Asehnoune K, Corvec S, Caroff N. 2016. Innate immune evasion of Escherichia coli clinical strains from orthopedic implant infections. Eur J Clin Microbiol Infect Dis 35:993–999. doi: 10.1007/s10096-016-2628-6. [DOI] [PubMed] [Google Scholar]
- 127.Croxall G, Hale J, Weston V, Manning G, Cheetham P, Achtman M, McNally A. 2011. Molecular epidemiology of extraintestinal pathogenic Escherichia coli isolates from a regional cohort of elderly patients highlights the prevalence of ST131 strains with increased antimicrobial resistance in both community and hospital care settings. J Antimicrob Chemother 66:2501–2508. doi: 10.1093/jac/dkr349. [DOI] [PubMed] [Google Scholar]
- 128.Cullik A, Pfeifer Y, Prager R, von Baum H, Witte W. 2010. A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J Med Microbiol 59:580–587. doi: 10.1099/jmm.0.016188-0. [DOI] [PubMed] [Google Scholar]
- 129.Dan M, Yair Y, Samosav A, Gottesman T, Yossepowitch O, Harari-Schwartz O, Tsivian A, Schreiber R, Gophna U. 2015. Escherichia coli isolates from patients with bacteremic urinary tract infection are genetically distinct from those derived from sepsis following prostate transrectal biopsy. Int J Med Microbiol 305:464–468. doi: 10.1016/j.ijmm.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 130.Agabou A, Lezzar N, Ouchenane Z, Khemissi S, Satta D, Sotto A, Lavigne J-P, Pantel A. 2016. Clonal relationship between human and avian ciprofloxacin-resistant Escherichia coli isolates in North-Eastern Algeria. Eur J Clin Microbiol Infect Dis 35:227–234. doi: 10.1007/s10096-015-2534-3. [DOI] [PubMed] [Google Scholar]
- 131.Danzeisen JL, Wannemuehler Y, Nolan LK, Johnson TJ. 2013. Comparison of multilocus sequence analysis and virulence genotyping of Escherichia coli from live birds, retail poultry meat, and human extraintestinal infection. Avian Dis 57:104–108. doi: 10.1637/10218-042812-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 132.Day MJ, Doumith M, Abernethy J, Hope R, Reynolds R, Wain J, Livermore DM, Woodford N. 2016. Population structure of Escherichia coli causing bacteraemia in the UK and Ireland between 2001 and 2010. J Antimicrob Chemother 71:2139–2142. doi: 10.1093/jac/dkw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Day MJ, Rodríguez I, van Essen-Zandbergen A, Dierikx C, Kadlec K, Schink A-K, Wu G, Chattaway MA, DoNascimento V, Wain J, Helmuth R, Guerra B, Schwarz S, Threlfall J, Woodward MJ, Coldham N, Mevius D, Woodford N. 2016. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J Antimicrob Chemother 71:1178–1182. doi: 10.1093/jac/dkv485. [DOI] [PubMed] [Google Scholar]
- 134.den Reijer PM, van Burgh S, Burggraaf A, Ossewaarde JM, van der Zee A. 2016. The widespread presence of a multidrug-resistant Escherichia coli ST131 clade among community-associated and hospitalized patients. PLoS One 11:e0150420. doi: 10.1371/journal.pone.0150420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dias RCS, Marangoni DV, Smith SP, Alves EM, Pellegrino F, Riley LW, Moreira BM. 2009. Clonal composition of Escherichia coli causing community-acquired urinary tract infections in the State of Rio de Janeiro, Brazil. Microb Drug Resist 15:303–308. doi: 10.1089/mdr.2009.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Díaz MA, Hernández-Bello JR, Rodríguez-Baño J, Martínez-Martínez L, Calvo J, Blanco J, Pascual A. 2010. Diversity of Escherichia coli strains producing extended-spectrum beta-lactamases in Spain: second nationwide study. J Clin Microbiol 48:2840–2845. doi: 10.1128/JCM.02147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dohmen W, Van Gompel L, Schmitt H, Liakopoulos A, Heres L, Urlings BA, Mevius D, Bonten MJM, Heederik D. 2017. ESBL carriage in pig slaughterhouse workers is associated with occupational exposure. Epidemiol Infect 145:2003–2010. doi: 10.1017/S0950268817000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Falgenhauer L, Imirzalioglu C, Ghosh H, Gwozdzinski K, Schmiedel J, Gentil K, Bauerfeind R, Kämpfer P, Seifert H, Michael GB, Schwarz S, Pfeifer Y, Werner G, Pietsch M, Roesler U, Guerra B, Fischer J, Sharp H, Käsbohrer A, Goesmann A, Hille K, Kreienbrock L, Chakraborty T. 2016. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int J Antimicrob Agents 47:457–465. doi: 10.1016/j.ijantimicag.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 139.Fam N, Leflon-Guibout V, Fouad S, Aboul-Fadl L, Marcon E, Desouky D, El-Defrawy I, Abou-Aitta A, Klena J, Nicolas-Chanoine M-H. 2011. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb Drug Resist 17:67–73. doi: 10.1089/mdr.2010.0063. [DOI] [PubMed] [Google Scholar]
- 140.Feglo P, Adu-Sarkodie Y, Ayisi L, Jain R, Spurbeck RR, Springman AC, Engleberg NC, Newton DW, Xi C, Walk ST. 2013. Emergence of a novel extended-spectrum-beta-lactamase (ESBL)-producing, fluoroquinolone-resistant clone of extraintestinal pathogenic Escherichia coli in Kumasi, Ghana. J Clin Microbiol 51:728–730. doi: 10.1128/JCM.03006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Aibinu I, Odugbemi T, Koenig W, Ghebremedhin B. 2012. Sequence Type ST131 and ST10 Complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin Microbiol Infect 18:E49–E51. doi: 10.1111/j.1469-0691.2011.03730.x. [DOI] [PubMed] [Google Scholar]
- 142.Francois P, Fankhauser-Rodriguez C, Baud D, Von Dach E, Schrenzel J. 2017. Meeting abstracts from International Conference on Prevention and Infection Control (ICPIC 2017). Antimicrob Resist Infect Control 6(Suppl 3):52. doi: 10.1186/s13756-017-0201-4. [DOI] [Google Scholar]
- 143.Gerhold G, Schulze MH, Gross U, Bohne W. 2016. Multilocus sequence typing and CTX-M characterization of ESBL-producing E. coli: a prospective single-centre study in Lower Saxony, Germany. Epidemiol Infect 144:3300–3304. doi: 10.1017/S0950268816001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 145.Giufre M, Graziani C, Accogli M, Luzzi I, Busani L, Cerquetti M. 2012. Escherichia coli of human and avian origin: detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J Antimicrob Chemother 67:860–867. doi: 10.1093/jac/dkr565. [DOI] [PubMed] [Google Scholar]
- 146.Golding GR, Persaud N, Levett PN, McDonald RR, Irvine J, Nsungu M, Woods S, Khan M, Mataseje LF, Mulvey MR. 2012. Characterization of Escherichia coli urinary tract infection isolates in remote northern Saskatchewan communities: the Northern Antibiotic Resistance Partnership. Diagn Microbiol Infect Dis 74:242–247. doi: 10.1016/j.diagmicrobio.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 147.Ha YE, Kang C-I, Cha MK, Park SY, Wi YM, Chung DR, Peck KR, Lee NY, Song J-H. 2013. Epidemiology and clinical outcomes of bloodstream infections caused by extended-spectrum β-lactamase-producing Escherichia coli in patients with cancer. Int J Antimicrob Agents 42:403–409. doi: 10.1016/j.ijantimicag.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 148.Hammerum AM, Larsen J, Andersen VD, Lester CH, Skovgaard Skytte TS, Hansen F, Olsen SS, Mordhorst H, Skov RL, Aarestrup FM, Agersø Y. 2014. Characterization of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. J Antimicrob Chemother 69:2650–2657. doi: 10.1093/jac/dku180. [DOI] [PubMed] [Google Scholar]
- 149.Hansen F, Olsen SS, Heltberg O, Justesen US, Fuglsang-Damgaard D, Knudsen JD, Hammerum AM. 2014. Characterization of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. Microb Drug Resist 20:316–324. doi: 10.1089/mdr.2013.0157. [DOI] [PubMed] [Google Scholar]
- 150.Hasan B, Laurell K, Rakib MM, Ahlstedt E, Hernandez J, Caceres M, Järhult JD. 2016. Fecal carriage of extended-spectrum β-lactamases in healthy humans, poultry, and wild birds in León, Nicaragua—a shared pool of bla CTX-M genes and possible interspecies clonal spread of extended-spectrum β-lactamases-producing Escherichia coli. Microb Drug Resist 22:682–687. doi: 10.1089/mdr.2015.0323. [DOI] [PubMed] [Google Scholar]
- 151.Hefzy EM, Hassuna NA. 2017. Fluoroquinolone-resistant sequence type 131 subgroups O25b and O16 among extraintestinal Escherichia coli isolates from community-acquired urinary tract infections. Microb Drug Resist 23:224–229. doi: 10.1089/mdr.2016.0040. [DOI] [PubMed] [Google Scholar]
- 152.Alhashash F, Wang X, Paszkiewicz K, Diggle M, Zong Z, McNally A. 2016. Increase in bacteraemia cases in the East Midlands region of the UK due to MDR Escherichia coli ST73: high levels of genomic and plasmid diversity in causative isolates. J Antimicrob Chemother 71:339–343. doi: 10.1093/jac/dkv365. [DOI] [PubMed] [Google Scholar]
- 153.Helldal L, Karami N, Welinder-Olsson C, Moore ERB, Åhren C. 2017. Evaluation of MLVA for epidemiological typing and outbreak detection of ESBL-producing Escherichia coli in Sweden. BMC Microbiol 17:8. doi: 10.1186/s12866-016-0922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hertz FB, Nielsen JB, Schønning K, Littauer P, Knudsen JD, Løbner-Olesen A, Frimodt-Møller N. 2016. Population structure of drug-susceptible, -resistant and ESBL-producing Escherichia coli from community-acquired urinary tract infections. BMC Microbiol 16:63. doi: 10.1186/s12866-016-0681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ho P-L, Ng K-Y, Lo W-U, Law PY, Lai E-Y, Wang Y, Chow K-H. 2016. Plasmid-mediated OqxAB is an important mechanism for nitrofurantoin resistance in Escherichia coli. Antimicrob Agents Chemother 60:537–543. doi: 10.1128/AAC.02156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Horner C, Fawley W, Morris K, Parnell P, Denton M, Wilcox M. 2014. Escherichia coli bacteraemia: 2 years of prospective regional surveillance (2010–12). J Antimicrob Chemother 69:91–100. doi: 10.1093/jac/dkt333. [DOI] [PubMed] [Google Scholar]
- 157.Houdouin V, Bonacorsi S, Bidet P, Blanco J, De La Rocque F, Cohen R, Aujard Y, Bingen E. 2008. Association between mortality of Escherichia coli meningitis in young infants and non-virulent clonal groups of strains. Clin Microbiol Infect 14:685–690. doi: 10.1111/j.1469-0691.2008.02019.x. [DOI] [PubMed] [Google Scholar]
- 158.Hu Y-Y, Cai J-C, Zhou H-W, Chi D, Zhang X-F, Chen W-L, Zhang R, Chen G-X. 2013. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl Environ Microbiol 79:5988–5996. doi: 10.1128/AEM.01740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hussain A, Ranjan A, Nandanwar N, Babbar A, Jadhav S, Ahmed N. 2014. Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob Agents Chemother 58:7240–7249. doi: 10.1128/AAC.03320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Johnson JK, Robinson GL, Pineles LL, Ajao AO, Zhao L, Albrecht JS, Harris AD, Thom KA, Furuno JP. 2017. Carbapenem MICs in Escherichia coli and Klebsiella species producing extended-spectrum β-lactamases in critical care patients from 2001 to 2009. Antimicrob Agents Chemother 61:e01718-16. doi: 10.1128/AAC.01718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Johnson JR, Davis G, Clabots C, Johnston BD, Porter S, DebRoy C, Pomputius W, Ender PT, Cooperstock M, Slater BS, Banerjee R, Miller S, Kisiela D, Sokurenko EV, Aziz M, Price LB. 2016. Household clustering of Escherichia coli sequence type 131 clinical and fecal isolates according to whole genome sequence analysis. Open Forum Infect Dis 3:ofw129. doi: 10.1093/ofid/ofw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Alghoribi MF, Gibreel TM, Farnham G, Al Johani SM, Balkhy HH, Upton M. 2015. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J Antimicrob Chemother 70:2757–2762. doi: 10.1093/jac/dkv188. [DOI] [PubMed] [Google Scholar]
- 163.Jørgensen SB, Søraas AV, Arnesen LS, Leegaard TM, Sundsfjord A, Jenum PA. 2017. A comparison of extended spectrum β-lactamase producing Escherichia coli from clinical, recreational water and wastewater samples associated in time and location. PLoS One 12:e0186576. doi: 10.1371/journal.pone.0186576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jørgensen SB, Søraas A, Sundsfjord A, Liestøl K, Leegaard TM, Jenum PA. 2017. Fecal carriage of extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae after urinary tract infection—a three year prospective cohort study. PLoS One 12:e0173510. doi: 10.1371/journal.pone.0173510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kaase M, Pfennigwerth N, Lange F, Anders A, Gatermann SG. 2015. Molecular epidemiology of VIM-1 producing Escherichia coli from Germany referred to the National Reference Laboratory. Int J Med Microbiol 305:784–789. doi: 10.1016/j.ijmm.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 166.Kallonen T, Brodrick HJ, Harris SR, Corander J, Brown NM, Martin V, Peacock SJ, Parkhill J. 2017. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res 27:1437–1449. doi: 10.1101/gr.216606.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kanamori H, Parobek CM, Juliano JJ, Johnson JR, Johnston BD, Johnson TJ, Weber DJ, Rutala WA, Anderson DJ. 2017. Genomic analysis of multidrug-resistant Escherichia coli from North Carolina community hospitals: ongoing circulation of CTX-M-producing ST131- H 30Rx and ST131- H 30R1 strains. Antimicrob Agents Chemother 61:e00912-17. doi: 10.1128/AAC.00912-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kang C-I, Cha MK, Kim SH, Ko KS, Wi YM, Chung DR, Peck KR, Lee NY, Song J-H. 2013. Clinical and molecular epidemiology of community-onset bacteremia caused by extended-spectrum β-lactamase-producing Escherichia coli over a 6-year period. J Korean Med Sci 28:998. doi: 10.3346/jkms.2013.28.7.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Karami N, Helldal L, Welinder-Olsson C, Åhrén C, Moore E. 2013. Sub-typing of extended-spectrum-β-lactamase-producing isolates from a nosocomial outbreak: application of a 10-loci generic Escherichia coli multi-locus variable number tandem repeat analysis. PLoS One 8:e83030. doi: 10.1371/journal.pone.0083030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Karfunkel D, Carmeli Y, Chmelnitsky I, Kotlovsky T, Navon-Venezia S. 2013. The emergence and dissemination of CTX-M-producing Escherichia coli sequence type 131 causing community-onset bacteremia in Israel. Eur J Clin Microbiol Infect Dis 32:513–521. doi: 10.1007/s10096-012-1765-9. [DOI] [PubMed] [Google Scholar]
- 171.Kim J, Bae IK, Jeong SH, Chang CL, Lee CH, Lee K. 2011. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J Antimicrob Chemother 66:1263–1268. doi: 10.1093/jac/dkr106. [DOI] [PubMed] [Google Scholar]
- 172.Kim S, Sung JY, Cho HH, Kwon KC, Koo SH. 2016. Characteristics of the molecular epidemiology of CTX-M-producing Escherichia coli isolated from a tertiary hospital in Daejeon, Korea. J Microbiol Biotechnol 26:1643–1649. doi: 10.4014/jmb.1603.03063. [DOI] [PubMed] [Google Scholar]
- 173.Alhashash F, Weston V, Diggle M, McNally A. 2013. Multidrug-resistant Escherichia coli bacteremia. Emerg Infect Dis 19:1699–1701. doi: 10.3201/eid1910.130309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kim Y, Oh T, Nam Y, Cho S, Lee H. 2017. Prevalence of ST131 and ST1193 among bloodstream isolates of Escherichia coli not susceptible to ciprofloxacin in a tertiary care university hospital in Korea, 2013–2014. Clin Lab 63:1541–1543. doi: 10.7754/Clin.Lab.2017.170319. [DOI] [PubMed] [Google Scholar]
- 175.Lafolie J, Nicolas-Chanoine M-H, Grenouillet F, Hocquet D, Bertrand X. 2014. Prevalence of Escherichia coli sequence type 131 and its H30 subclone among E. coli isolates in a French hospital. Int J Antimicrob Agents 44:466–468. doi: 10.1016/j.ijantimicag.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 176.Lara FBM, Nery DR, de Oliveira PM, Araujo ML, Carvalho FRQ, Messias-Silva LCF, Ferreira LB, Faria-Junior C, Pereira AL. 2017. Virulence markers and phylogenetic analysis of Escherichia coli strains with hybrid EAEC/UPEC genotypes recovered from sporadic cases of extraintestinal infections. Front Microbiol 8:146. doi: 10.3389/fmicb.2017.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lau SH, Kaufmann ME, Livermore DM, Woodford N, Willshaw GA, Cheasty T, Stamper K, Reddy S, Cheesbrough J, Bolton FJ, Fox AJ, Upton M. 2008. UK epidemic Escherichia coli strains A-E, with CTX-M-15 beta-lactamase, all belong to the international O25:H4-ST131 clone. J Antimicrob Chemother 62:1241–1244. doi: 10.1093/jac/dkn380. [DOI] [PubMed] [Google Scholar]
- 178.Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 46:1076–1080. doi: 10.1128/JCM.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lee MY, Choi HJ, Choi JY, Song M, Song Y, Kim S-W, Chang H-H, Jung S-I, Kim Y-S, Ki HK, Son JS, Kwon KT, Heo ST, Yeom J-S, Shin SY, Chung DR, Peck KR, Song J-H, Ko KS. 2010. Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J Infect 60:146–153. doi: 10.1016/j.jinf.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 180.Leflon-Guibout V, Blanco J, Amaqdouf K, Mora A, Guize L, Nicolas-Chanoine M-H. 2008. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J Clin Microbiol 46:3900–3905. doi: 10.1128/JCM.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lemaître C, Mahjoub-Messai F, Dupont D, Caro V, Diancourt L, Bingen E, Bidet P, Bonacorsi S. 2013. A conserved virulence plasmidic region contributes to the virulence of the multiresistant Escherichia coli meningitis strain S286 belonging to phylogenetic group C. PLoS One 8:e74423. doi: 10.1371/journal.pone.0074423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lewis JA, Moore PCL, Arnold DL, Lawrance LM. 2016. Multi-locus sequence typing of Escherichia coli isolates with acquired ampC genes and ampC promoter mutations. Diagn Microbiol Infect Dis 86:265–267. doi: 10.1016/j.diagmicrobio.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 183.Ma L, Siu LK, Lin J-C, Wu T-L, Fung C-P, Wang J-T, Lu P-L, Chuang Y-C. 2013. Updated molecular epidemiology of carbapenem-non-susceptible Escherichia coli in Taiwan: first identification of KPC-2 or NDM-1-producing E. coli in Taiwan. BMC Infect Dis 13:599. doi: 10.1186/1471-2334-13-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Alkeskas A, Ogrodzki P, Saad M, Masood N, Rhoma NR, Moore K, Farbos A, Paszkiewicz K, Forsythe S. 2015. The molecular characterisation of Escherichia coli K1 isolated from neonatal nasogastric feeding tubes. BMC Infect Dis 15:449. doi: 10.1186/s12879-015-1210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Maciuca IE, Williams NJ, Tuchilus C, Dorneanu O, Guguianu E, Carp-Carare C, Rimbu C, Timofte D. 2015. High prevalence of Escherichia coli-producing CTX-M-15 extended-spectrum beta-lactamases in poultry and human clinical isolates in Romania. Microb Drug Resist 21:651–662. doi: 10.1089/mdr.2014.0248. [DOI] [PubMed] [Google Scholar]
- 186.Mahjoub-Messai F, Bidet P, Caro V, Diancourt L, Biran V, Aujard Y, Bingen E, Bonacorsi S. 2011. Escherichia coli isolates causing bacteremia via gut translocation and urinary tract infection in young infants exhibit different virulence genotypes. J Infect Dis 203:1844–1849. doi: 10.1093/infdis/jir189. [DOI] [PubMed] [Google Scholar]
- 187.Manges AR, Harel J, Masson L, Edens TJ, Portt A, Reid-Smith RJ, Zhanel GG, Kropinski AM, Boerlin P. 2015. Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog Dis 12:302–310. doi: 10.1089/fpd.2014.1860. [DOI] [PubMed] [Google Scholar]
- 188.Manges AR, Mende K, Murray CK, Johnston BD, Sokurenko EV, Tchesnokova V, Johnson JR. 2017. Clonal distribution and associated characteristics of Escherichia coli clinical and surveillance isolates from a military medical center. Diagn Microbiol Infect Dis 87:382–385. doi: 10.1016/j.diagmicrobio.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 189.Manges AR, Tabor H, Tellis P, Vincent C, Tellier P-P. 2008. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg Infect Dis 14:1575–1583. doi: 10.3201/eid1410.080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Matsumura Y, Noguchi T, Tanaka M, Kanahashi T, Yamamoto M, Nagao M, Takakura S, Ichiyama S. 2017. Population structure of Japanese extraintestinal pathogenic Escherichia coli and its relationship with antimicrobial resistance. J Antimicrob Chemother 72:1040–1049. doi: 10.1093/jac/dkw530. [DOI] [PubMed] [Google Scholar]
- 191.Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S. 2012. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-beta-lactamase-producing Escherichia coli in Japan. J Antimicrob Chemother 67:2612–2620. doi: 10.1093/jac/dks278. [DOI] [PubMed] [Google Scholar]
- 192.Mavroidi A, Miriagou V, Liakopoulos A, Tzelepi Ε, Stefos A, Dalekos GN, Petinaki E. 2012. Ciprofloxacin-resistant Escherichia coli in central Greece: mechanisms of resistance and molecular identification. BMC Infect Dis 12:371. doi: 10.1186/1471-2334-12-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mellmann A, Bletz S, Böking T, Kipp F, Becker K, Schultes A, Prior K, Harmsen D. 2016. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 54:2874–2881. doi: 10.1128/JCM.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Merino I, Shaw E, Horcajada JP, Cercenado E, Mirelis B, Pallarés MA, Gómez J, Xercavins M, Martínez-Martínez L, De Cueto M, Cantón R, Ruiz-Garbajosa P. 2016. CTX-M-15- H 30Rx-ST131 subclone is one of the main causes of healthcare-associated ESBL-producing Escherichia coli bacteraemia of urinary origin in Spain. J Antimicrob Chemother 71:2125–2130. doi: 10.1093/jac/dkw133. [DOI] [PubMed] [Google Scholar]
- 195.Denamur E, Clermont O, Gordon D. 2015. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 161:980–988. doi: 10.1099/mic.0.000063. [DOI] [PubMed] [Google Scholar]
- 196.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.McNally A, Oren Y, Kelly D, Pascoe B, Dunn S, Sreecharan T, Vehkala M, Välimäki N, Prentice MB, Ashour A, Avram O, Pupko T, Dobrindt U, Literak I, Guenther S, Schaufler K, Wieler LH, Zhiyong Z, Sheppard SK, McInerney JO, Corander J. 2016. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet 12:e1006280. doi: 10.1371/journal.pgen.1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shaik S, Ranjan A, Tiwari SK, Hussain A, Nandanwar N, Kumar N, Jadhav S, Semmler T, Baddam R, Islam MA, Alam M, Wieler LH, Watanabe H, Ahmed N. 2017. Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal Pathogenic Escherichia coli (ExPEC) lineages. mBio 8:e01596-17. doi: 10.1128/mBio.01596-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TEA, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TEA, Johnson JR, Didelot X, Walker AS, Crook DW. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Clark G, Paszkiewicz K, Hale J, Weston V, Constantinidou C, Penn C, Achtman M, McNally A. 2012. Genomic analysis uncovers a phenotypically diverse but genetically homogeneous Escherichia coli ST131 clone circulating in unrelated urinary tract infections. J Antimicrob Chemother 67:868–877. doi: 10.1093/jac/dkr585. [DOI] [PubMed] [Google Scholar]
- 203.Stephens CM, Skerker JM, Sekhon MS, Arkin AP, Riley LW. 2015. Complete genome sequences of four Escherichia coli ST95 isolates from bloodstream infections. Genome Announc 3:e01241-15. doi: 10.1128/genomeA.01241-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Johnson JR, Manges AR. 2015. Reservoirs of extraintestinal pathogenic Escherichia coli. Microbiol Spectr 3:UTI-0006-2012. doi: 10.1128/microbiolspec.UTI-0006-2012. [DOI] [PubMed] [Google Scholar]
- 205.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, Du Y, Hu J, Lei Y, Li N, Tooming-Klunderud A, Heederik DJJ, Fluit AC, Bonten MJM, Willems RJL, de la Cruz F, van Schaik W. 2014. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet 10:e1004776. doi: 10.1371/journal.pgen.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland: https://www.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1. [Google Scholar]
- 207.Yamaji R, Rubin J, Thys E, Friedman CR, Riley LW. 2018. Persistent pandemic lineages of uropathogenic Escherichia coli in a college community from 1999 to 2017. J Clin Microbiol 56:e01834-17. doi: 10.1128/JCM.01834-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Roer L, Overballe-Petersen S, Hansen F, Schønning K, Wang M, Røder BL, Hansen DS, Justesen US, Andersen LP, Fulgsang-Damgaard D, Hopkins KL, Woodford N, Falgenhauer L, Chakraborty T, Samuelsen Ø, Sjöström K, Johannesen TB, Ng K, Nielsen J, Ethelberg S, Stegger M, Hammerum AM, Hasman H. 2018. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e00337-18. doi: 10.1128/mSphere.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tchesnokova VL, Rechkina E, Larson L, Ferrier K, Weaver JL, Schroeder DW, She R, Butler-Wu SM, Aguero-Rosenfeld ME, Zerr D, Fang FC, Ralston J, Riddell K, Scholes D, Weissman S, Parker K, Spellberg B, Johnson JR, Sokurenko EV. 2019. Rapid and extensive expansion in the United States of a new multidrug-resistant Escherichia coli clonal group, sequence type 1193. Clin Infect Dis 68:334–337. doi: 10.1093/cid/ciy525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. 2008. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol 46:2529–2534. doi: 10.1128/JCM.00813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Srivastava R, Agarwal J, Srivastava S, Mishra B. 2014. Role of special pathogenicity versus prevalence theory in pathogenesis of acute cystitis caused by Escherichia coli. J Med Microbiol 63:1038–1043. doi: 10.1099/jmm.0.073270-0. [DOI] [PubMed] [Google Scholar]