Abstract
Background
Colorectal carcinoma (CRC) is one of the most common malignant tumors, and its main cause of death is tumor metastasis. RNA N6-methyladenosine (m6A) is an emerging regulatory mechanism for gene expression and methyltransferase-like 3 (METTL3) participates in tumor progression in several cancer types. However, its role in CRC remains unexplored.
Methods
Western blot, quantitative real-time PCR (RT-qPCR) and immunohistochemical (IHC) were used to detect METTL3 expression in cell lines and patient tissues. Methylated RNA immunoprecipitation sequencing (MeRIP-seq) and transcriptomic RNA sequencing (RNA-seq) were used to screen the target genes of METTL3. The biological functions of METTL3 were investigated in vitro and in vivo. RNA pull-down and RNA immunoprecipitation assays were conducted to explore the specific binding of target genes. RNA stability assay was used to detect the half-lives of the downstream genes of METTL3.
Results
Using TCGA database, higher METTL3 expression was found in CRC metastatic tissues and was associated with a poor prognosis. MeRIP-seq revealed that SRY (sex determining region Y)-box 2 (SOX2) was the downstream gene of METTL3. METTL3 knockdown in CRC cells drastically inhibited cell self-renewal, stem cell frequency and migration in vitro and suppressed CRC tumorigenesis and metastasis in both cell-based models and PDX models. Mechanistically, methylated SOX2 transcripts, specifically the coding sequence (CDS) regions, were subsequently recognized by the specific m6A “reader”, insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), to prevent SOX2 mRNA degradation. Further, SOX2 expression positively correlated with METTL3 and IGF2BP2 in CRC tissues. The combined IHC panel, including “writer”, “reader”, and “target”, exhibited a better prognostic value for CRC patients than any of these components individually.
Conclusions
Overall, our study revealed that METTL3, acting as an oncogene, maintained SOX2 expression through an m6A-IGF2BP2-dependent mechanism in CRC cells, and indicated a potential biomarker panel for prognostic prediction in CRC.
Electronic supplementary material
The online version of this article (10.1186/s12943-019-1038-7) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, N6-methyladenosine (m6A), METTL3, SOX2, IGF2BP2
Background
Colorectal carcinoma (CRC) is a highly lethal cancer with an increasing incidence worldwide [1]. Despite therapeutic advances over the past few decades, the mortality rate of CRC remains high, which is mainly ascribed to recurrence and distant organ metastasis [2]. Recent studies, including ours, have indicated that a small population of cancer cells, called cancer stem-like cells (CSCs), display increased self-renewal ability, and cause chemotherapy resistance, which are possible mechanism for tumor recurrence and metastasis [3–5]. Therefore, exploring the factors that drive tumor initiation and establishing a more accurate model for prognostic prediction in CRC are urgently needed.
Epigenetic regulatory mechanisms, such as DNA methylation, or N6-methyladenosine (m6A), are emerging research frontiers in tumor biology [6–9]. As the most abundant post-transcriptional modification, m6A modification is mainly mediated by m6A WERs (“writers”, “erasers” and “readers”), and is reported to be related to RNA fate control through influencing alternative polyadenylation and pre-mRNA splicing, as well as regulating RNA stability and translation efficiency [10–13]. Our previous study also demonstrated that one of the RNA demethylases, fat-mass and obesity-associated protein (FTO), plays a critical role in cell transformation in leukemia cells [14]. These findings have indicated that m6A has a broad impact on embryonic development, circadian clock control, and the DNA damage response, as well as on tumor progression [10, 14–17]. Furthermore, it is worth noting that these impressive biological functions rely on the target genes of the m6A “writers” or “erasers”, and the fate of the target transcripts generally rely on the specific recognition of m6A “readers” [18]. As a reversible epi-transcriptome modulator, methyltransferase-like 3 (METTL3) is a key member of the m6A methyltransferase complex, and has recently been reported to be essential for tumor progression in leukemia, hepatocellular carcinoma, and malignant glioma via diverse downstream genes [16, 17, 19]. However, the functions of m6A modification and the underlying connection among the m6A “writers”, “readers”, and “targets” are still unexplored in CRC.
Here, we first demonstrated the function of METTL3 in facilitating CRC progression, and identified SRY (sex determining region Y)-box 2 (SOX2) as the downstream target of METTL3. Moreover, insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2) was indicated to prolong the SOX2 life-span. Overall, our study reveals that METTL3 is a promising biomarker for prognostic prediction and a potential therapeutic target in CRC.
Methods
Tissue specimens and patient information
A total of 432 paraffin-embedded, archived CRC specimens and paired adjacent normal tissue samples, including 43 matched liver metastasis tissues and 52 matched lymph node metastasis tissues, were obtained at the SYSUCC (Guangzhou, China) between January 2010 and July 2013 as previously described [20]. The clinical CRC specimens were collected with permission from our Institutional Research Ethics Committee. The clinical characteristics of the samples are summarized in Additional file 1: Table S1.
Methylated RNA immunoprecipitation sequencing (MeRIP-seq)
MeRIP-seq was conducted in accordance with a previously reported protocol with minor modifications [21]. Briefly, 50 μg of total RNA was extracted and purified using RiboMinus™ Eukaryote Kit v2 (A15020, Invitrogen) to deplete the ribosomal RNA from the total RNA. Next, RNA Fragmentation Reagents (AM8740, Invitrogen) were used to shear the RNA into approximately 100-nt fragments. Approximately 1/10 of the fragmented RNA was saved as the input control for further RNA sequencing by RiboBio (Guangzhou, China). The remaining were incubated with an anti-m6A antibody (202,203, Synaptic Systems) for one hour at 4 °C, and then mixed with prewashed Pierce™ Protein A/G Magnetic Beads (88,803, Thermo Scientific) in immunoprecipitation buffer at 4 °C overnight. The m6A antibody was digested with proteinase K digestion buffer and the methylated RNA was purified for further MeRIP sequencing by RiboBio (Guangzhou, China).
MeRIP-qPCR
M6A modifications of individual genes were determined using MeRIP-qPCR assay. Briefly, poly(A) RNA was first purified from 50 μg of total RNA using the Dynabeads™ mRNA Purification Kit (61,006, Invitrogen) and one-tenth of the RNA was saved as the input control. Pierce™ Protein A/G Magnetic Beads (88,803, Thermo Scientific) were prewashed and incubated with 5 μg of anti-m6A antibody (202,003, Synaptic Systems) or rabbit IgG for 2 h at 4 °C with rotation. After 3 washes, the antibody-conjugated beads were mixed with purified poly(A) RNA, and 1 × immunoprecipitation buffer supplemented with RNase inhibitors. Then, the methylated mRNAs were precipitated with 5 mg of glycogen and one-tenth volumes of 3 M sodium acetate in a 2.5 volume of 100% ethanol at − 80 °C overnight after proteinase K digestion. Further enrichment was calculated by qPCR and the corresponding m6A enrichment in each sample was calculated by normalizing to the input.
RNA pull-down assays
RNA was first transcribed by the MEGAscript T7 Transcription Kit (AM1334, Thermo Scientific). Then, the amplified RNA was end-labeled with desthiobiotin by using Pierce RNA 3′ End Desthiobiotinylation Kit (20,163, Thermo Scientific). Finally, RNA pull-down assays were performed using the Pierce Magnetic RNA-Protein Pull-Down Kit (20,164, Thermo Scientific). Up to 50 pmol of biotinylated RNAs was mixed with 2 mg of protein lysates and 50 μl of streptavidin beads. After incubation and three washes, the streptavidin beads were boiled and used for the immunoblotting assay.
RNA immunoprecipitation (RIP) assays
RIP was conducted with the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (17–700, Millipore) according to the manufacturer’s instructions. Briefly, magnetic beads coated with 5 μg of specific antibodies against mouse immunoglobulin G (17–700, Millipore), or IGF2BP2 (ab#128175, Abcam) were incubated with prepared cell lysates overnight at 4 °C. Then, the RNA-protein complexes were washed 6 times and incubated with proteinase K digestion buffer. RNA was finally extracted by phenol-chloroform RNA extraction methods. The relative interaction between IGF2BP2 and SOX2 transcripts was determined by qPCR and normalized to the input.
Vector and m6A mutation assays
The potential m6A sites were predicted using an online tool, SRAMP (http://www.cuilab.cn/sramp/). Full-length SOX2 transcripts, the SOX2 CDS region, the SOX2 three prime untranslated region (3′-UTR), and the m6A motif depleted CDS or 3′-UTR regions were cloned into pcDNA3.1 for the RNA pull down assay. The specific sequences are shown in Additional file 2: Table S2.
RNA stability assays
CRC cells were seeded in 12-well plates overnight, and then treated with actinomycin D (5 μg/mL, HY-17559, MedChemExpress) at the 0, 3, 6 h. Total RNA was then isolated by TRIzol (15,596,018, Invitrogen) and analyzed by qPCR. The mRNA expression for each group at the indicated time was calculated and normalized by β-Actin. The mRNA half-lives time were estimated according to the linear regression analysis.
Statistical analysis
All data and error bars are presented as the mean ± SDs from at least three independent experiments. All differences between two independent groups were evaluated by a two-tailed Student’s t-test. Survival curves were generated using the Kaplan–Meier method and compared using the log-rank test. Survival data were evaluated by univariate and multivariate Cox regression analyses. To investigate the correlation between two independent groups, the Pearson’s Chi-square test was used. The MedCalc software was used to generate the ROC curve, and the data were analyzed by two-tailed t test. The indicated P values (*P < 0.05 and **P < 0.01) were considered statistically significant.
Additional Materials and Methods are described in Additional file 3.
Results
METTL3 is highly expressed in metastatic CRC and associated with poor prognosis
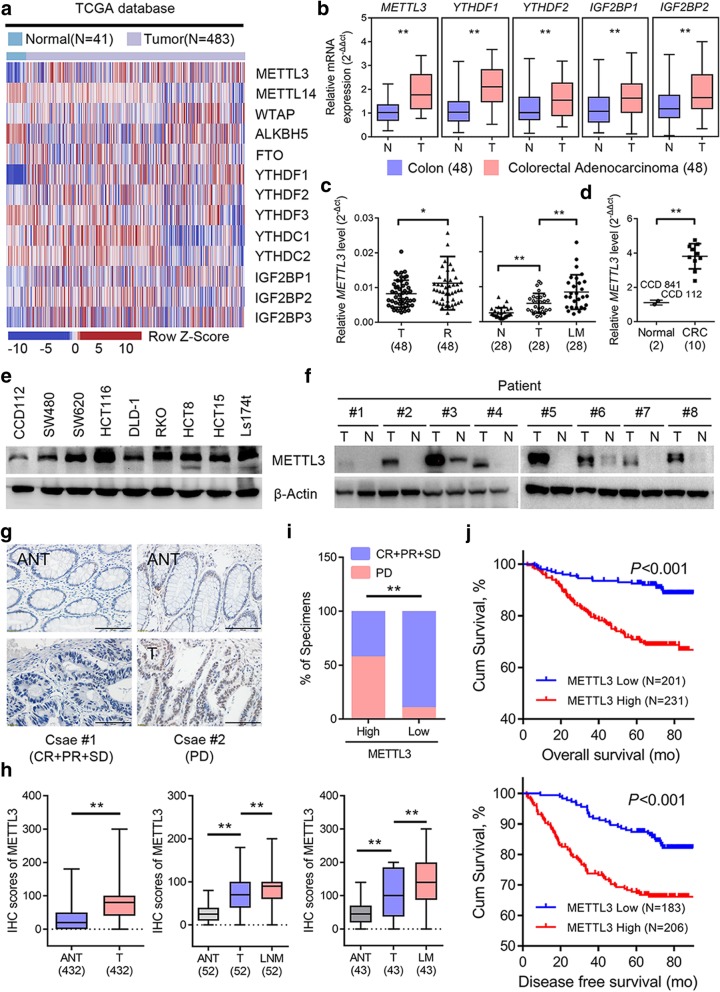
To evaluate the expression profile of m6A WERs in CRC, we analyzed The Cancer Genome Atlas (TCGA) database, and the results showed that several m6A WERs were dysregulated in colon adenocarcinoma (COAD) (Fig. 1a). We next verified that METTL3, YTH N6-methyladenosine RNA binding protein 1 (YTHDF1), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1), and IGF2BP2 were significantly increased in CRC tumors tissues from Sun Yat-sen University Cancer Center (SYSUCC), while the other WERs showed no significant differences (Fig. 1b and Additional file 4: Figure S1a). Additionally, METTL3 was commonly highly expressed in most human cancers from TCGA database (Additional file 4: Figure S1b). These expression differences of METTL3 prompted us to investigate its functional and clinical consequences in CRC. Further validation showed that METTL3 was consistently elevated in recurrent CRC tissues and metastatic liver tissues (Fig. 1c). The METTL3 mRNA and protein level in CRC cell lines were also increased relative to the normal colonic epithelial cell lines (Figs. 1d-e). Moreover, METTL3 protein levels were notably increased in representative CRC patient tissues compared with normal tissue (Fig. 1f). To investigate the clinical implication of METTL3 with CRC, we performed IHC staining for METTL3 in our archived CRC tissue microarray, described previously [20]. Our results indicated that METTL3 staining was increased in primary CRC tissues compared with the adjacent normal tissue. Similarly, the significant elevation of METTL3 was also observed in matched lymph node and liver metastatic foci (Figs. 1g-h). We next explored the correlation between METTL3 with the disease control rate in CRC patients, and found that patients with high METTL3 expression had a poorer benefit from standard chemotherapy (Fig. 1i). Moreover, the CRC patients with high METTL3 expression had both shorter overall survival (OS) and disease-free survival (DFS) (Fig. 1j), which suggests that METTL3 expression might serve as a prognostic marker for OS and DFS in CRC patients.
Fig. 1.
METTL3 is highly expressed in metastatic CRC and associated with poor prognosis. a Heat map profiling the expression of m6A WERs in the TCGA database of COAD. b Real-time PCR analysis of m6A WER expression in 48 paired CRC tumor tissues (T) and adjacent normal tissues (N). c Real-time PCR analysis of METTL3 expression in CRC tissues from patients with recurrence (R, n = 48) and without recurrence (T, n = 48), 28 paired liver metastatic tissues (LM) versus primary tumor tissues (T), and adjacent normal tissues (N). d-e Real-time PCR analysis and Immunoblotting assay of METTL3 expression in normal colonic epithelial cell lines and CRC cell lines. f Immunoblotting assay of METTL3 expression in eight paired CRC primary tumor samples (T) and adjacent normal tissues (N). g Representative images showing METTL3 expression in CRC adjacent normal tissues (ANT) (upper) versus high METTL3 expression in CRC tumor tissues (T) (lower) (scale bar: 100 μm). h METTL3 IHC staining scores in CRC tumor tissues versus ANT (n = 432), paired lymph node metastatic tissues (LNM, n = 52) or paired liver metastatic tissues (LM, n = 43). i Correlation between METTL3 expression with CRC patient response to FOLFOX or XELOX chemotherapy. The data were analyzed by Pearson’s Chi-square test. j Kaplan-Meier analysis of OS time (upper) and DFS time (lower) based on METTL3 expression. CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease. The data in b, c, d, and i, are presented as the mean ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). β-Actin was used as the loading control
SOX2 is regulated by METTL3-mediated m6A modification
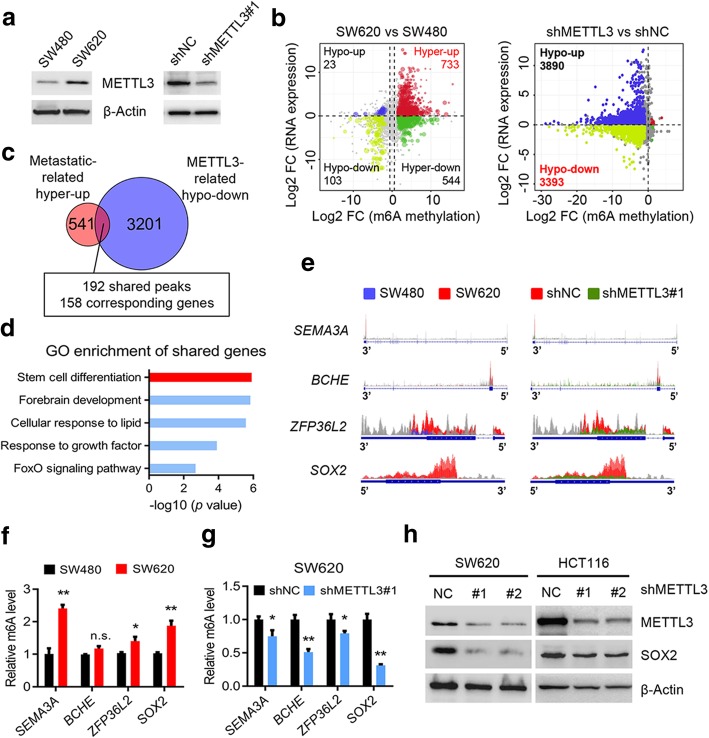
To investigate the potential role of METTL3 in tumor progression, we firstly noted that METTL3 was elevated in SW620 cells compared with SW480 cells (Fig. 2a), a pair of cell lines isolated from abdominal metastatic foci and the primary tumor, respectively, of a single patient. The two cell lines exhibit different metastatic abilities [22], which points to the connection between METTL3 and metastasis. Therefore, we performed MeRIP-seq and RNA-seq in SW480, SW620 and METTL3 knockdown SW620 cells. The results showed that there were generally hyper-methylated peaks in SW620 cells compared with the SW480 cells, and the methylation level of the identified peaks in SW620 cells was downregulated after METTL3 knockdown (Additional file 5: Figures S2a-b). We further investigated the mRNA expression, corresponding to each peak, in our RNA-seq data, and described the distribution of peaks with a significant change in both the RNA level and the m6A level. We found 733 hyper-methylated m6A peaks with higher mRNA expression in SW620 cells versus SW480 cells, and thereafter called these peaks metastatic-related hyper-up peaks. Similarly, we found 3393 hypo-methylated m6A peaks with lower mRNA expression in METTL3-knockdown SW620 cells relative to control SW620 cells, and named these peaks METTL3-related hypo-down peaks (Fig. 2b). Focusing on the peaks in these two groups, we found that 192 specific peaks, corresponding to 158 genes, were shared (Fig. 2c). Interestingly, we found that the shared genes were the most enriched in the stem cell differentiation pathway through GO enrichment analysis on metascape website, indicating that this pathway might be regulated by METTL3 and promoted tumor metastasis via an m6A mechanism (Fig. 2d).
Fig. 2.
Identification of METTL3 targets via MeRIP-seq and RNA-seq. a, Immunoblotting of METTL3 in SW480 and SW620 cells (left), and in METTL3 knockdown SW620 and control SW620 cells (right). b, Distribution of peaks (fold change > 1.5 or < − 1.5, P < 0.05) with a significant change in both the RNA expression level and m6A level in SW620 cells compared with SW480 cells (left), and in METTL3 knockdown SW620 cells compared to control SW620 cells (right). c, Venn diagram showing the shared peaks between metastatic-related hyper-up peaks and METTL3-related hypo-down peaks. A total of 192 shared peaks corresponding to 158 specific genes were observed. d, GO biological process enrichment analysis of the above shared peaks. e, The m6A abundances in SEMA3A, BCHE, ZFP36L2, and SOX2 transcripts in SW620 cells related to the SW480 cells (left panel), and in METTL3-knockdown SW620 cells (shMETTL3#1) related to the control SW620 cells (shNC) (right panel). f, Gene-specific m6A qPCR analysis of alterations in the m6A level in four representative genes in SW620 and SW480 cells. g, Gene-specific m6A qPCR analysis of alterations in the m6A level in four representative genes in METTL3-knockdown SW620 and control SW620 cells. h, Immunoblotting assay of SOX2 after METTL3-knockdown in SW620 and HCT116 cells. The data in f, and g are presented as the mean ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). β-Actin was used as the loading control. The relative m6A level was normalized by input. The relative expression level was normalized by the β-Actin
We next screened the genes listed in the stem cell differentiation pathway in our m6A-seq data, and found four genes, semaphorin 3A (SEMA3A), butyrylcholinesterase (BCHE), ZFP36 ring finger protein like 2 (ZFP36L2), and SOX2, that exhibited a substantial increase in m6A level in SW620 cells compared with SW480 cells and showed a consistent decreased m6A level in METTL3-knockdown SW620 cells compared with control cells (Fig. 2e). Gene-specific m6A pull down assay and qPCR analysis showed that the m6A levels of SOX2, ZFP36L2, and SEMA3A were increased in SW620 cells compared with SW480 cells (Fig. 2f). However, SOX2 exhibited the most consistent decreased m6A level and mRNA level in METTL3 knockdown CRC cells versus the control cells (Fig. 2g, and Additional file 5: Figures S2c-d). Moreover, the significant decreased protein level of SOX2 was detected after METTL3 inhibition in SW620 and HCT116 cells (Fig. 2h). Considering that SOX2 is considered the important CSC marker to promote tumor initiation and participate in tumor metastasis [23, 24], we presumed that METTL3 promoted CRC stemness and metastasis in an m6A-dependent manner to maintain SOX2 expression.
METTL3 promotes CRC cell stemness in vitro
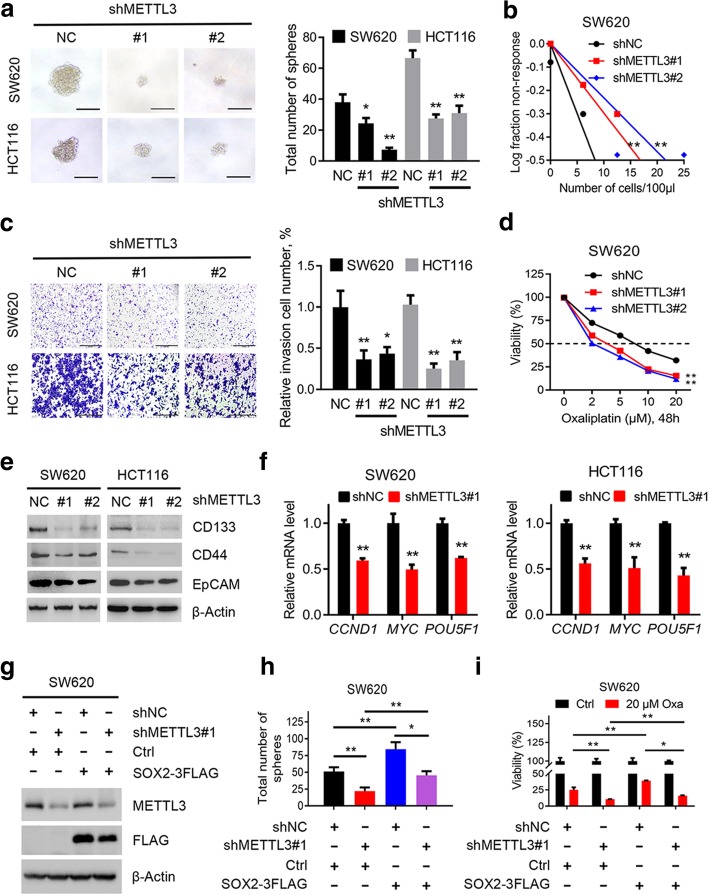
We next performed several experiments to test our hypothesis. Interestingly, a decrease in sphere numbers and sizes as well as a markedly reduced stem cell frequency were observed in METTL3-inhibited SW620 and HCT116 cells compared with the corresponding control cells (Figs. 3a-b and Additional file 6: Figure S3a). The cell colony-formation and invasion abilities of SW620 and HCT116 cells were also impaired after METTL3 inhibition (Fig. 3c and Additional file 6: Figure S3b). As mentioned previously, stemness is thought to be responsible for chemotherapy resistance, and we specifically found that sensitivity to oxaliplatin-based chemotherapy was increased in METTL3-knockdown SW620 and HCT116 cells relative to the control cells (Fig. 3d and Additional file 6: Figure S3c). In addition, the expression of CSC surface antigens such as CD133, CD44, and epithelial cell adhesion molecule (EpCAM), in SW620 and HCT116 cells was remarkably reduced after METTL3 inhibition (Fig. 3e). Moreover, the expression of SOX2 downstream genes, including cyclin D1 (CCND1), MYC proto-oncogene protein (MYC), and POU class 5 homeobox 1 (POU5F1) [25–27], was consistently suppressed in METTL3-knockdown SW620 and HCT116 cells (Fig. 3f). These results revealed the oncogenic role of METTL3, specifically in the promoting of tumor self-renewal, cellular invasion and chemotherapy resistance in CRC cells.
Fig. 3.
METTL3 promotes CRC cell stemness in vitro. a, Representative images and quantification of the in vitro sphere-formation assay of METTL3 knockdown CRC cells and control cells (n = 6). Scale bar: 200 μm. b, In vitro limiting dilution assay of METTL3 knockdown and control SW620 cells. A well not containing spheres (diameter ≥ 50 μm) was defined as a non-response (n = 12). c, Representative images and quantification of invaded METTL3-knockdown and control SW620 and HCT116 cells. Scale bar: 100 μm. d, Cell viability of METTL3-knockdown SW620 cells and control SW620 cells after treatment with oxaliplatin for 48 h. e, Immunoblotting analysis of stem-like cell surface antigen (CD133, CD44, and EpCAM) in METTL3-knockdown and control SW620 and HCT116 cells. f, Real-time PCR analysis of SOX2 targets genes (CCND1, MYC, and POU5F1) in METTL3-knockdown and control SW620 and HCT116 cells. g, Immunoblotting analysis of SOX2 and METTL3 in METTL3-knockdown and control SW620 cells with or without SOX2 overexpression. h, Quantification of the in vitro sphere-formation assay of METTL3 knockdown and control SW620 cells with or without SOX2 overexpression. (n = 6). i, Cell viability of METTL3-knockdown and control SW620 cells with or without SOX2 overexpression after oxaliplatin treatment for 48 h. All data are presented as the mean ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). β-Actin was used as the loading control. The relative expression level was normalized by β-Actin
We questioned whether SOX2 overexpression could rescue the reduction in stemness due to METTL3 inhibition. As expected, SOX2 overexpression in METTL3-knockdown and control CRC cells (Fig. 3g and Additional file 6: Figure S3d) led to the increased sphere formation, and an apparent chemotherapy resistance phenotype (Figs. 3h-i, and Additional file 6: Figures S3e-f). Collectively, the above results indicated the critical role of METTL3 in promoting stemness features through maintaining SOX2 expression in CRC.
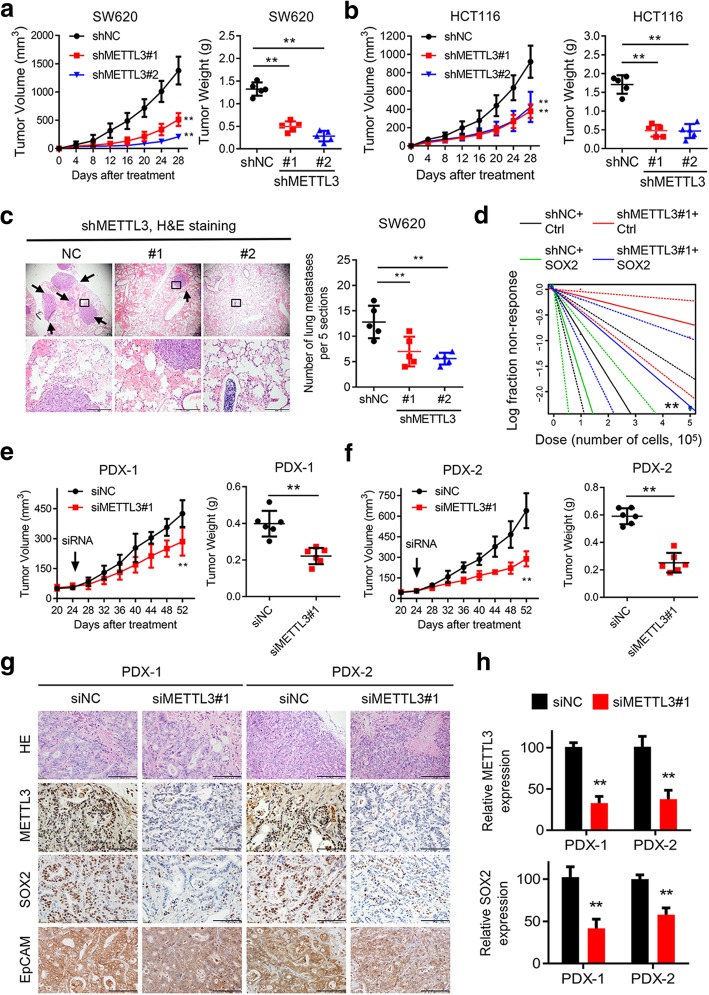
METTL3 drives CRC tumorigenesis and metastasis in vivo
To investigate the function of METTL3 in vivo, we next performed a subcutaneous xenotransplantation assay to determine whether METTL3 contributed to CRC development. The tumor growth rate was slower, and the xenograft tumor weight was reduced, when METTL3-knockdown SW620 and HCT116 cells were implanted, compared with the control cells (Figs. 4a-b, and Additional file 7: Figure S4a). Immunostaining assays indicated that the growth-impaired tumors generated from METTL3-ablated CRC cells had lower expression of SOX2 and EpCAM compared with the control subcutaneous mouse models (Additional file 7: Figure S4b). Moreover, compared with the mice that tail vein injected with METTL3 knockdown cells, the mice injected with control SW620 cells developed more lung metastatic nodules, as observed by histologic examination (Figs. 4c). As demonstrated above, METTL3 maintained self-renewal ability in vitro; therefore, we explored whether a similar effect would exist in vivo. Notably, the frequency of tumorigenic CRC cells was significantly decreased among the METTL3 knockdown SW620 cells (Fig. 4d, and Additional file 7: Figures S4c-d). In addition, SOX2 overexpression subsequently increased the tumor incidence and the frequency of tumorigenic cells among both control and METTL3 knockdown SW620 cells (Fig. 4d and Additional file 7: Figures S4c-d). Alltogether, the xenograft mouse models demonstrated that METTL3 contributed to tumorigenesis and the formation of metastatic foci through maintaining SOX2 expression in CRC.
Fig. 4.
METTL3 drives CRC tumorigenesis and metastasis in vivo. a-b, Subcutaneous tumor models in nude mice showing the tumor growth rate (left) and tumor weights (right) at day 28 after the implantation of METTL3-knockdown and control SW620 and HCT116 cells (n = 5 mice per group). c, Representative H&E staining (scale bar: 100 μm) and quantification of metastatic lung nodules at day 60 after the tail vein injection of METTL3- knockdown or control SW620 cells (n = 5 mice per group). Arrow: metastatic lung nodules. Five sections were evaluated for each lung. d, In vivo limiting dilution assay showing the estimated frequency of CSCs among METTL3-knockdown and control SW620 cells with or without SOX2 overexpression. Response: mice developed subcutaneous tumor (n = 5 mice per group). e-f, Tumor growth rate and tumor weights in two PDX models of intratumoral treatment with siMETTL3 and siNC. g-h, Representative images and quantification of H&E and immunostaining (scale bar: 100 μm) of METTL3, SOX2, and EpCAM in two PDX-based subcutaneous tumor models. All data and error bars are presented as the mean ± SDs. *P < 0.05, **P < 0.01 (Student’s t-test)
PDX tumor models can simulate the physical tumor microenvironment, and the tumor growth corresponds to the treatment evaluations for the original patient [20, 28]. Therefore, we applied two PDX models to evaluate the potential therapeutic effect of METTL3 through intratumoral RNAi injection. In addition, the volumes of tumors treated with METTL3 siRNA were significantly lower than that of tumors in control group (Figs. 4e-f). Moreover, the PDX tumors were isolated and assessed by IHC staining, which showed reduced staining of METTL3, SOX2 and EpCAM expression in the METTL3 siRNA-treated group (Figs. 4g-h). Taken together, the above results further highlighted the crucial roles of METTL3 in CRC tumorigenesis and metastasis in vivo.
IGF2BP2 enhances SOX2 mRNA stability via an m6A-dependent manner
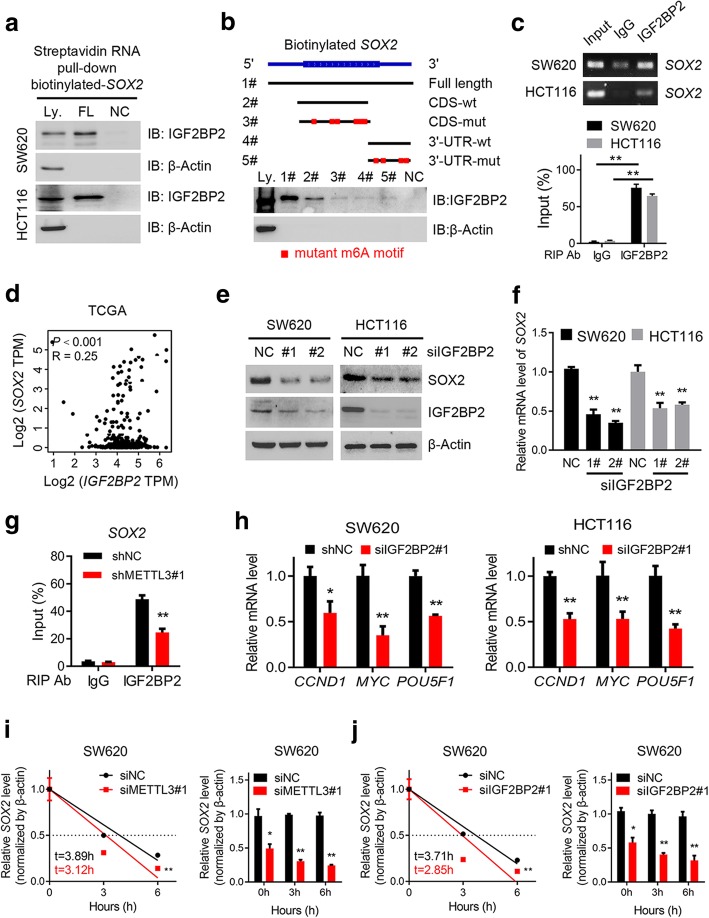
Previous studies had identified two major families of m6A “readers” that might play a specific role in control the fate of the methylated mRNA, such as the YTH family and the IGF2BP family [12, 29, 30]. To elucidate the specific m6A readers of SOX2, and determine the m6A-dependent mechanism of SOX2 regulation, we performed a streptavidin RNA pull-down assay to screen for SOX2-related m6A readers. Interestingly, IGF2BP2, but not other members of the IGF2BP family or the YTH family, specifically bound the SOX2 full-length transcripts in SW620 and HCT116 cells (Fig. 5a and Additional file 8: Figure S5a). As represented in Fig. 2d, our m6A-seq data also provided a clue that the most of the m6A peaks of SOX2 transcripts were located near the stop codon which indicated possible binding sites. Notably, the RNA pull-down assays verified that IGF2BP2 predominantly bound to the SOX2 CDS region, instead of the 3′-UTR in SW620 cells, and the specific binding was significantly impaired after m6A motif depletion (Fig. 5b and Additional file 8: Figure S5b). RIP assays also validated the direct interaction between the IGF2BP2 and SOX2 mRNA in SW620 and HCT116 cells (Fig. 5c). In TCGA database for COAD, a positive correlation between IGF2BP2 and SOX2 expression was observed as shown in Fig. 5d, indicating the potential positive regulatory mechanism. Consistent with our hypothesis, SOX2 protein and mRNA expression were significantly decreased after the siRNA inhibition of IGF2BP2 in SW620 and HCT116 cells (Figs. 5e-f). Additionally, the direct interaction between IGF2BP2 and SOX2 transcripts was impaired in SW620 cells after METTL3 inhibition (Fig. 5g). The expression of SOX2 downstream genes, as shown in Fig. 3f, was also reduced after IGF2BP2 inhibition in SW620 and HCT116 cells (Fig. 5h). Furthermore, we assessed the RNA decay rate in METTL3 or IGF2BP2 inhibited CRC cells and the corresponding control cells. The SOX2 mRNA expression was initially decreased and the SOX2 mRNA half-lives were consistently markedly shortened upon METTL3 or IGF2BP2 inhibition in SW620 and HCT116 cells (Figs. 5i-j, and Additional file 8: Figures S5c-d). Taken together, our data suggested that the methylated SOX2 transcripts were directly recognized by the m6A “reader”, IGF2BP2, which maintained the stability of the transcripts to prevent its degradation and naturally increase its expression via an m6A-IGF2BP2-dependent mechanism.
Fig. 5.
IGF2BP2 enhances SOX2 mRNA stability via an m6A-dependent manner. a, Immunoblotting of IGF2BP2 after RNA pull down assay with cell lysate (Ly.), full-length biotinylated-SOX2 (FL), and beads only (NC) in SW620 and HCT116 cells. b, Immunoblotting of IGF2BP2 with cell lysate (Ly.), full-length biotinylated-SOX2 (#1), the SOX2 CDS region with or without m6A motif mutation (#2, #3), the SOX2 3′-UTR region with or without m6A motif mutation (#4, #5), and beads only (NC) in SW620 cells. c, Agarose electrophoresis and real-time PCR analysis of RIP assays in CRC cells showing the direct binding between the IGF2BP2 protein and SOX2 mRNA. d, Correlation between IGF2BP2 and SOX2 expression in TCGA database for COAD, analyzed with the Gene Expression Profiling Interactive Analysis (GEPIA) online analysis tool (http://gepia.cancer-pku.cn/). e, Immunoblotting of SOX2 after IGF2BP2 inhibition in SW620 and HCT116 cells. f, Real-time PCR analysis of SOX2 after IGF2BP2 inhibition in SW620 and HCT116 cells. g, RIP-qPCR showing the enrichment of SOX2 in SW620 after METTL3 inhibition. h, Real-time PCR analysis of SOX2 downstream genes after IGF2BP2 inhibition in SW620 and HCT116 cells. i-j, The decay rate of mRNA and qPCR analysis of SOX2 at the indicated times after actinomycin D (5 μg/ml) treatment in SW620 cells after METTL3 inhibition (left), and in SW620 cells after IGF2BP2 inhibition (right). The data in c, g, h, i and j are presented as the mean ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). β-Actin and an IgG antibody was used as the negative control. The relative expression level was normalized by β-Actin. The relative SOX2 enrichment in the RIP assay was normalized by input
Clinical correlation between METTL3, SOX2 and IGF2BP2 in CRC
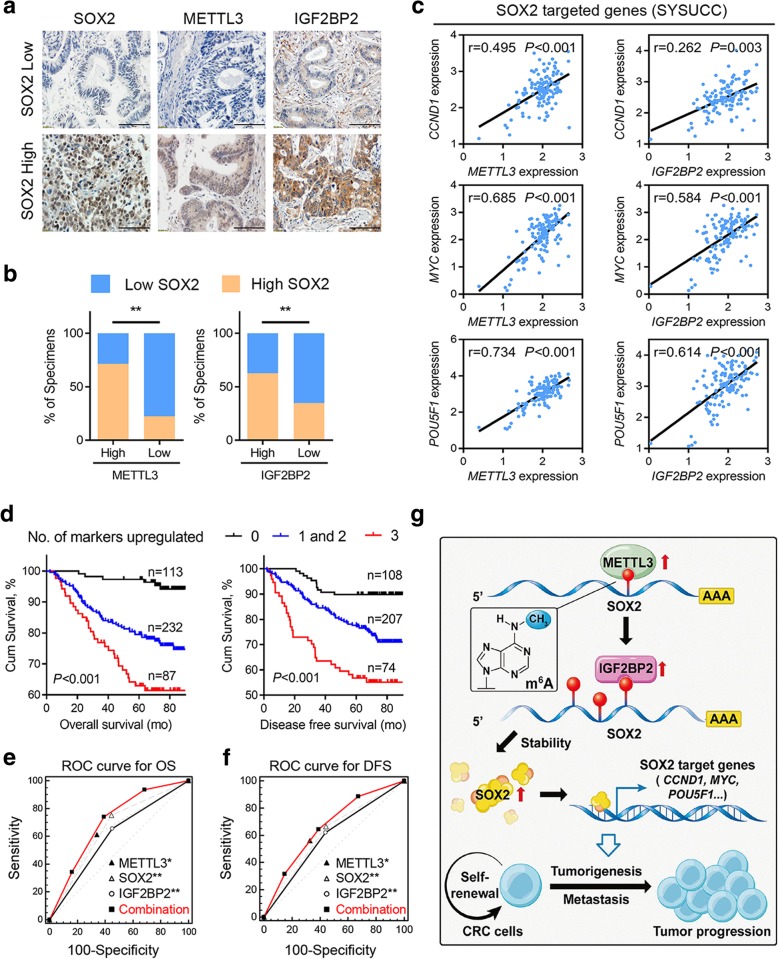
Based on the mechanism we identified above, we proceeded to explore the clinical relevance between METTL3, IGF2BP2, and SOX2 in our study. An IHC assay of SOX2, and IGF2BP2 was performed using the CRC microarray (Fig. 6a). IHC analysis showed that the expression of both SOX2 and IGF2BP2 was significantly increased in CRC tumor tissues compared with that in the paired adjacent normal tissues (Additional file 9: Figure S6a). Consistent with this finding, the Kaplan-Meier survival analysis and log-rank test suggested that high expression of SOX2 and IGF2BP2 notably correlated with shorter overall survival and disease-free survival times (Additional file 9: Figures S6b-c). Notably, SOX2 expression positively correlated with both METTL3 and IGF2BP2 in CRC tissues (Fig. 6b). Moreover, METTL3 or IGF2BP2 expression positively correlated with the SOX2 downstream genes CCND1, MYC, and POU5F1 in our independent cohort of paired CRC tumor and adjacent normal tissues from SYSUCC (Fig. 6c). Similar results were also observed in TCGA database in a COAD cohort (Additional file 9: Figure S6d). Using Cox regression analysis, IHC scores for METTL3, SOX2, and IGF2BP2 expression were analyzed in a CRC patient cohort, and each of these three genes showed a notably increased hazard ratio (HR) for death, indicating that these three genes were independent prognostic factors in our CRC cohorts (Additional file 10: Table S3). Therefore, we attempted to generate a new IHC panel containing METTL3, SOX2, and IGF2BP2 to predict the prognosis of CRC. The Kaplan-Meier survival analysis and log-rank test suggested that the patients with three highly expressed markers had the shortest overall survival and disease-free survival times (Fig. 6d). Moreover, in the receiver operating characteristic (ROC) curve analysis, the combination index of the new IHC panel (METTL3, SOX2, and IGF2BP2) showed an additive predictive value for overall survival compared with any individual marker (Figs. 6e-f). The improved predictive values provided us with a credible IHC panel for evaluating the prognosis of CRC patients. As illustrated in Fig. 6g, METTL3 was highly expressed in CRC patients, and contributed to an increase in the m6A methylation level of SOX2 transcripts. Methylated SOX2 was subsequently recognized by the m6A “reader”, IGF2BP2, to maintain its mRNA stability and expression. Finally, increasing SOX2 expression promoted CRC cell stemness and metastasis through downstream targets of SOX2, leading to CRC progression.
Fig. 6.
Clinical correlation between METTL3, SOX2 and IGF2BP2 in CRC. a, Representative images showing high or low expression of METTL3, SOX2 and IGF2BP2 in 432 CRC tumor specimens. b, Correlation between SOX2 and METTL3 or IGF2BP2 in CRC microarray specimens. c, Correlation between METTL3 level (left) or IGF2BP2 level (right) and the levels of SOX2 downstream genes, including CCND1, MYC, and POU5F1, in 63 paired CRC tumor tissues and adjacent normal tissues (SYSUCC cohort). d, Kaplan-Meier analysis of overall survival (OS) for CRC patients (n = 432) based on the number of upregulated molecular markers (Kaplan-Meier analysis with log-rank test). METTL3, SOX2, and IGF2BP2 expression was stratified by the individual medians by IHC analysis, and the patients were divided into three groups as indicated. e, ROC curve analysis for OS for METTL3 [AUC = 0.654, (95% CI, 0.607–0.698)], SOX2 [AUC = 0.635, (95% CI, 0.588–0.681)], and IGF2BP2 [AUC = 0.602, (95% CI, 0.554–0.649)] as individual biomarkers or for the combined panel [AUC = 0.703 (95% CI, 0.658–0.746)]. AUC, area under the curve. *P < 0.05, **P < 0.01 (Student’s t-test). f, ROC curve analysis for DFS for METTL3 [AUC = 0.612, (95% CI, 0.564–0.658)], SOX2 [AUC = 0.615, (95% CI, 0.568–0.661)], and IGF2BP2 [AUC = 0.591, (95% CI, 0.543–0.637)] as individual biomarkers or for the combined panel [AUC = 0.664 (95% CI, 0.618–0.709)]. AUC, area under a curve. g, Proposed working model of the proposed mechanism in this study. *P < 0.05, **P < 0.01 (Student’s t-test)
Discussion
METTL3, acting as the key component of N6-methyltransferase complex, has been reported to play an important role in many tumor types [12, 18, 19, 30–33]. Our results uncover a significant oncogenic role for METTL3 in tumor progression, though there are other studies that had suggested some controversial conclusions. Two independent previous studies stated that both METTL3 and FTO played an oncogenic role in acute myeloid leukemia through the diverse downstream targets [14, 33]. Two other studies stated that either increased or decreased METTL3 expression could respectively promote the self-renewal and tumorigenicity of glioma stem-like cells [32, 34]. Moreover, one study showed that high m6A modification promoted hepatocellular carcinoma progression which was ascribed to high METTL3 expression, while another study believed that was ascribed to low METTL14 expression [19, 35]. Considering the controversial conclusions of m6A and METTL3 in different cancer types, we believe that our current study has uncovered the underlying functions of WERs in CRC and showed the oncogenic role of METTL3 in promoting CRC stemness and metastasis, indicating the broad impact of METTL3 and m6A methylation on cancer development and precision therapy.
Colorectal CSCs are a group of tumor cells with self-renewal ability and multiple differentiation potentials which have strong tumorigenic and metastatic potential [3, 4]. In our previous work, the presence of CSCs in CRC was also suggested to be responsible for chemotherapy resistance [36]. Therefore, the elimination of colorectal CSCs is an important therapeutic strategy to improve the prognosis of CRC patients [37, 38]. Specifically, our study demonstrated that the inhibition of METTL3 could augment the chemotherapy response and decrease the stem cell frequency in CRC both in vitro and in vivo. Moreover, inhibition of METTL3 with siRNA treatment could significantly reduce the tumor size in PDX models. These results suggested that inhibition of METTL3 may be an effective way to diminish CSCs, thereby terminating malignant tumor recurrence and metastasis.
The acknowledged CSCs marker SOX2, was previously reported to be highly expressed and to participate in maintaining the properties of tumor-initiating cells, promoting proliferation in squamous cell carcinoma [23, 39]. CD133, CD166, EpCAM and CD44 are reported to be surface antigen of colorectal CSCs [38]. However, the regulatory mechanism for these CSC markers remains unclear. In this study, we found that inhibition of METTL3 could basically reduce these surface antigens expression, confirmed the oncogenic effect of SOX2 and revealed the m6A-dependent regulatory mechanism to partially explain the common upregulation of SOX2 in CRC. MYC, as one of the SOX2 target gene [27], is reported to be directly controlled by METTL3/IGF2BP2 axis [29]. In our work, we think that MYC can be regulated by both METTL3/IGF2BP2 axis and SOX2 respectively, which might partially explain the elevated expression of MYC in various human cancers. In conclusion, we suggested that METTL3 might be a new CSC marker due to its functions in maintaining the CSC stemness phenotype, providing new ideas and theoretical basis for the diagnosis and treatment of CRC.
M6A readers were reported to be involved in controlling the fate of mRNA, and both the YTHDF2 and IGF2BP1/2/3 were associated with methylated mRNA stability [29, 30]. Our data first identified that only IGF2BP2 directly bound to the specific m6A sites in SOX2 CDS regions and controlled the SOX2 mRNA half-life via an m6A-dependent manner. In fact, before its identification as an m6A reader, IGF2BP2 had already suggested to be associated with tumor progression through preserving the stemness phenotype in glioblastoma and hepatocellular carcinoma [40–42]. Here, we coincidentally verified the high expression of IGF2BP2 in CRC and its regulatory effect on SOX2 mRNA stability to promote CRC stemness. These results might partially account for the roles of IGF2BP2 in preserving the tumor stemness phenotype. However, further molecular mechanisms underlying m6A methylation and mRNA fate deserves extensive study.
Conclusions
In conclusion, our study suggested that METTL3 was essential for CRC progression and provided an attractive m6A-dependent regulatory mechanism. The combined network of “writer” METTL3, “reader” IGF2BP2, and “target” SOX2 highlighted an innovative m6A-dependent gene regulatory mechanism in epigenetics. In addition, the PDX models indicated a promising therapeutic strategy for CRC through the use of the efficient inhibitors of METTL3, which we will focus on developing in the future.
Additional files
Table S1. Correlation analysis for clinicopathologic variables in METTL3 expression among 432 colorectal cancer patients. (DOCX 15 kb)
Table S2. The specific sequence of wide-type or m6A motif depletion SOX2 CDS and 3′-UTR. (DOCX 14 kb)
Supplementary materials and methods. (DOCX 35 kb)
Figure S1, related to Fig. 1. METTL3 is highly expressed in human tumors. a, Real-time PCR analysis of m6A WER expression in 48 paired CRC tumor tissues (T) and adjacent normal tissues (N). b, Box plots of METTL3 expression in TCGA database. (TIF 6470 kb)
Figure S2, related to Fig. 2: Identification of METTL3 targets via MeRIP-seq and RNA-seq. a, Volcano Plots showing the numbers of transcripts with significantly increased and decreased m6A peaks (fold change > 1.5 or < − 1.5, P < 0.05) in SW620 cells compared with SW480 cells (left) and in METTL3-knockdown SW620 compared with the control SW620 cells (right). b, Venn diagram showing the shared peaks between metastatic-related hyper-methylated peaks with METTL3-related hypo-methylated peaks. c, Gene-specific m6A qPCR analysis of alterations in the m6A level in four representative genes in METTL3 knockdown HCT116 compared with the control cells. d, Real-time PCR analysis of mRNA expression of four representative genes in METTL3 knockdown and control SW620 and HCT116. The data in c, and d are presented as the means ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). The relative m6A level was normalized by input. The relative expression level was normalized by β-Actin. (TIF 6844 kb)
Figure S3, related to Fig. 3: METTL3 promotes CRC cell stemness in vitro. a, In vitro limiting dilution assay of METTL3-knockdown and control HCT116. A well not containing spheres (diameter ≥ 50 μm) was defined as a non-response (n = 12). b, Total number of colonies formed by METTL3-knockdown versus control SW620 and HCT116 cells. c, Cell viability of METTL3-knockdown HCT116 versus control HCT116 cells after oxaliplatin treatment for 48 h. d, Immunoblotting analysis of SOX2 and METTL3 in METTL3 knockdown and control HCT116 cells with or without SOX2 overexpression. e, Quantification of the in vitro sphere-formation assay of METTL3-knockdown and control HCT116 cells with or without SOX2 overexpression (n = 6). f, Cell viability of METTL3-knockdown and control HCT116 cells with or without SOX2 overexpression when treated with oxaliplatin for 48 h. All data are presented as the mean ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). β-Actin was used as the loading control. (TIF 6517 kb)
Figure S4, related to Fig. 4: METTL3 drives CRC tumorigenesis and metastasis in vivo. a, Subcutaneous tumor models in nude mice showing the tumor size at day 28 after the implantation of METTL3-knockdown and control SW620 and HCT116 cells (n = 5 mice per group). b, Representative and quantification of H&E and immunostaining (scale bar: 100 μm) of METTL3, SOX2, and EpCAM in subcutaneous tumor models of METTL3 knockdown and control SW620 and HCT116 cells. c, Tumor incidence showing the tumorigenesis of the indicated serial of cell numbers of METTL3 knockdown and control SW620 cells with or without SOX2 overexpression. d, Stem cell frequencies of METTL3 knockdown and control SW620 cells with or without SOX2 overexpression. Estimate: 1/ (the estimated stem cell frequency); Lower, Upper: 95% confidence intervals. All data and error bars are presented as the mean ± SDs. *P < 0.05, **P < 0.01 (Student’s t-test). (TIF 9763 kb)
Figure S5 related to Fig. 5: IGF2BP2 enhances SOX2 mRNA stability via an m6A-dependent manner. a, Immunoblotting of IGF2BP1, IGF2BP3, YTHDF1, YTHDF2 after RNA pull down assay with cell lysate (Ly.), full-length biotinylated-SOX2 (FL), and beads only (NC) in SW620 and HCT116 cells. b, Immunoblotting of IGF2BP1, IGF2BP3, YTHDF1, and YTHDF2 with cell lysate (Ly.), full-length biotinylated-SOX2 (#1), the SOX2 CDS region with or without m6A motif mutation (#2, #3), the SOX2 3′-UTR region with or without m6A motif mutation (#4, #5), and beads only (NC) in SW620 cells. c-d, The decay rate of mRNA and qPCR analysis of SOX2 at indicated time after actinomycin D (5 μg/ml) treatment in HCT116 cells after METTL3 inhibition (left), and in HCT116 cells after IGF2BP2 inhibition (right). The date in c, and d are presented as the mean ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). β-Actin was used as the negative control. The relative expression level was normalized by β-Actin. (TIF 7319 kb)
Figure S6 related to Fig. 6: Clinical correlation between METTL3, SOX2 and IGF2BP2 in CRC. a, SOX2 and IGF2BP2 IHC staining scores in primary CRC tumor tissues (T) and adjacent normal tissue (ANT) (n = 432). b-c, Kaplan-Meier analysis of OS and DFS curves based on the expression of SOX2 and IGF2BP2 expression (Kaplan-Meier analysis with the log-rank test). d, Correlation between METTL3 level (left) or IGF2BP2 level (right) with SOX2 target genes, including CCND1, MYC, and POU5F1, in TCGA database for COAD. *P < 0.05, **P < 0.01 (Student’s t-test). (TIF 6918 kb)
Table S3 Univariate and multivariate analyses of prognostic factors for overall survival among 432 colorectal cancer patients. (DOCX 13 kb)
Acknowledgments
We thank all members of the Xu’s laboratory for their advice and technical assistance.
Abbreviations
- 3′-UTR
three prime untranslated region
- BCHE
Butyrylcholinesterase
- CCND1
Cyclin D1
- CDS
Coding sequence
- COAD
Colon adenocarcinoma
- CRC
Colorectal carcinoma
- CSCs
Cancer stem-like cells
- EpCAM
Epithelial cell adhesion molecule
- FTO
Fat-mass and obesity-associated protein
- HR
Hazard ratio
- IGF2BP1
Insulin like growth factor 2 mRNA binding protein 1
- IGF2BP2
Insulin like growth factor 2 mRNA binding protein 2
- IHC
Immunohistochemical
- m6A
N6-methyladenosine
- MeRIP-seq
Methylated RNA immunoprecipitation sequencing
- METTL3
Methyltransferase-like 3
- MYC
MYC proto-oncogene protein
- PDX
Patient-derived xenograft
- POU5F1
POU class 5 homeobox 1
- RIP
RNA immunoprecipitation
- RNA-seq
transcriptomic RNA sequencing
- ROC
Receiver operating characteristic
- RT-qPCR
quantitative real-time PCR
- SEMA3A
Semaphorin 3A
- SOX2
SRY (sex determining region Y)-box 2
- SYSUCC
Sun Yat-sen University Cancer Center
- TCGA
The Cancer Genome Atlas
- YTHDF1
YTH N6-methyladenosine RNA binding protein 1
- YTHDF2
YTH N6-methyladenosine RNA binding protein 2
- ZFP36L2
ZFP36 ring finger protein like 2
Authors’ contributions
Conceptualization, XRH, JHQ, and Li T; Methodology, Li T, HPS, Chen ZH, and ZJ; Bioinformatics Analysis, ZZX, Li XY; Investigation, Lin JF, WQN, WF, and ZZL; Writing -Original Draft, Li T; Writing -Review & Editing, Lin DX, XD, Li B, Chen DM, and KTB; Funding Acquisition, XRH, JHQ, and ZZX; Supervision, XRH, and JHQ. All authors read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (81871951, 81772614, U1611261); Science and Technology Program of Guangzhou (201904020046); National Key R&D Program of China (2018YFC1313300), Natural Science Foundation of Guangdong Province (2018B030306049, 2017A030313485, 2014A030312015, 2018A0303130282); Science and Technology Program of Guangdong (2015B020232008).
Availability of data and materials
All data generated or analyzed during this study are included either in this article or in the additional files. The MeRIP-seq data and RNA-seq data have been deposited in the Genome Sequence Archive (http://gsa.big.ac.cn/) and are accessible under GSA: CRA001257.
Ethics approval and consent to participate
The clinical CRC specimens were conducted with permission from the Institutional Research Ethics Committee of Sun Yat-sen University Cancer Center, China. All animal experiments were performed in accordance with a protocol approved by the ethics committee of the Institutional Animal Care of Sun Yat-sen University Cancer Center, China.
Consent for publication
The content of this manuscript has not been previously published and is not under consideration for publication elsewhere.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ting Li, Pei-Shan Hu and Zhixiang Zuo contributed equally to this work.
Contributor Information
Huai-Qiang Ju, Phone: 86-20-87342280, Email: juhq@sysucc.org.cn.
Rui-Hua Xu, Phone: 86-20-87343333, Email: xurh@sysucc.org.cn.
References
- 1.Bray Freddie, Ferlay Jacques, Soerjomataram Isabelle, Siegel Rebecca L., Torre Lindsey A., Jemal Ahmedin. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Xu RH, Muro K, Morita S, Iwasa S, Han SW, Wang W, Kotaka M, Nakamura M, Ahn JB, Deng YH, et al. Modified XELIRI (capecitabine plus irinotecan) versus FOLFIRI (leucovorin, fluorouracil, and irinotecan), both either with or without bevacizumab, as second-line therapy for metastatic colorectal cancer (AXEPT): a multicentre, open-label, randomised, non-inferiority, phase 3 trial. Lancet Oncol. 2018;19:660–671. doi: 10.1016/S1470-2045(18)30140-2. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 4.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 5.Hu PS, Xia QS, Wu F, Li DK, Qi YJ, Hu Y, Wei ZZ, Li SS, Tian NY, Wei QF, et al. NSPc1 promotes cancer stem cell self-renewal by repressing the synthesis of all-trans retinoic acid via targeting RDH16 in malignant glioma. Oncogene. 2017;36:4706–4718. doi: 10.1038/onc.2017.34. [DOI] [PubMed] [Google Scholar]
- 6.Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, Flagg K, Hou J, Zhang H, Yi S, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A. 2017;114:7414–7419. doi: 10.1073/pnas.1703577114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, Yi S, Shi W, Quan Q, Li K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16:1155–1161. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 8.Flavahan William A., Gaskell Elizabeth, Bernstein Bradley E. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357(6348):eaal2380. doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng X, Su R, Feng X, Wei M, Chen J. Role of N (6)-methyladenosine modification in cancer. Curr Opin Genet Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)a methyltransferase METTL3 promotes translation in human Cancer cells. Mol Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, Hao Y-J, Zhang Y, Li M-M, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. m6A RNA methylation is regulated by MicroRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-Methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. RNA m(6)a methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes Leukemogenesis via mRNA m(6)a modification. Cell Stem Cell. 2018;22:191–205 e199. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, et al. m(6)a demethylase ALKBH5 maintains Tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 20.Ju Huai-Qiang, Lu Yun-Xin, Chen Dong-Liang, Zuo Zhi-Xiang, Liu Ze-Xian, Wu Qi-Nian, Mo Hai-Yu, Wang Zi-Xian, Wang De-Shen, Pu Heng-Ying, Zeng Zhao-Lei, Li Bo, Xie Dan, Huang Peng, Hung Mien-Chie, Chiao Paul J, Xu Rui-Hua. Modulation of Redox Homeostasis by Inhibition of MTHFD2 in Colorectal Cancer: Mechanisms and Therapeutic Implications. JNCI: Journal of the National Cancer Institute. 2018;111(6):584–596. doi: 10.1093/jnci/djy160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 22.Witty JP, McDonnell S, Newell KJ, Cannon P, Navre M, Tressler RJ, Matrisian LM. Modulation of matrilysin levels in colon carcinoma cell lines affects tumorigenicity in vivo. Cancer Res. 1994;54:4805–4812. [PubMed] [Google Scholar]
- 23.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 24.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Gomez EC, Wang J, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z, Zeng X, Xu J, Xu D, Li J, Jin H, Jiang G, Han X, Huang C. Isorhapontigenin suppresses growth of patient-derived glioblastoma spheres through regulating miR-145/SOX2/cyclin D1 axis. Neuro-Oncology. 2016;18:830–839. doi: 10.1093/neuonc/nov298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SB, Seo KW, So AY, Seo MS, Yu KR, Kang SK, Kang KS. SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ. 2012;19:534–545. doi: 10.1038/cdd.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu YX, Ju HQ, Liu ZX, Chen DL, Wang Y, Zhao Q, Wu QN, Zeng ZL, Qiu HB, Hu PS, et al. ME1 regulates NADPH homeostasis to promote gastric Cancer growth and metastasis. Cancer Res. 2018;78:1972–1985. doi: 10.1158/0008-5472.CAN-17-3155. [DOI] [PubMed] [Google Scholar]
- 29.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, Liu Y, Zhang X, Zhang W, Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. M(6)a RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. The N(6)-methyladenosine (m(6)a)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m(6)a modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 35.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 36.Ju HQ, Lu YX, Chen DL, Tian T, Mo HY, Wei XL, Liao JW, Wang F, Zeng ZL, Pelicano H, et al. Redox regulation of stem-like cells though the CD44v-xCT Axis in colorectal Cancer: mechanisms and therapeutic implications. Theranostics. 2016;6:1160–1175. doi: 10.7150/thno.14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 38.Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138:2151–2162. doi: 10.1053/j.gastro.2009.12.063. [DOI] [PubMed] [Google Scholar]
- 39.Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate hedgehog signaling in lung squamous cell carcinoma. Cancer Cell. 2014;25:139–151. doi: 10.1016/j.ccr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao J, Mu Q, Huang H. The roles of insulin-like growth factor 2 mRNA-binding protein 2 in Cancer and Cancer stem cells. Stem Cells Int. 2018;2018:4217259. doi: 10.1155/2018/4217259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Degrauwe N, Schlumpf TB, Janiszewska M, Martin P, Cauderay A, Provero P, Riggi N, Suva ML, Paro R, Stamenkovic I. The RNA binding protein IMP2 preserves glioblastoma stem cells by preventing let-7 target gene silencing. Cell Rep. 2016;15:1634–1647. doi: 10.1016/j.celrep.2016.04.086. [DOI] [PubMed] [Google Scholar]
- 42.Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, Radovanovic I, Rheinbay E, Provero P, Stamenkovic I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Correlation analysis for clinicopathologic variables in METTL3 expression among 432 colorectal cancer patients. (DOCX 15 kb)
Table S2. The specific sequence of wide-type or m6A motif depletion SOX2 CDS and 3′-UTR. (DOCX 14 kb)
Supplementary materials and methods. (DOCX 35 kb)
Figure S1, related to Fig. 1. METTL3 is highly expressed in human tumors. a, Real-time PCR analysis of m6A WER expression in 48 paired CRC tumor tissues (T) and adjacent normal tissues (N). b, Box plots of METTL3 expression in TCGA database. (TIF 6470 kb)
Figure S2, related to Fig. 2: Identification of METTL3 targets via MeRIP-seq and RNA-seq. a, Volcano Plots showing the numbers of transcripts with significantly increased and decreased m6A peaks (fold change > 1.5 or < − 1.5, P < 0.05) in SW620 cells compared with SW480 cells (left) and in METTL3-knockdown SW620 compared with the control SW620 cells (right). b, Venn diagram showing the shared peaks between metastatic-related hyper-methylated peaks with METTL3-related hypo-methylated peaks. c, Gene-specific m6A qPCR analysis of alterations in the m6A level in four representative genes in METTL3 knockdown HCT116 compared with the control cells. d, Real-time PCR analysis of mRNA expression of four representative genes in METTL3 knockdown and control SW620 and HCT116. The data in c, and d are presented as the means ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). The relative m6A level was normalized by input. The relative expression level was normalized by β-Actin. (TIF 6844 kb)
Figure S3, related to Fig. 3: METTL3 promotes CRC cell stemness in vitro. a, In vitro limiting dilution assay of METTL3-knockdown and control HCT116. A well not containing spheres (diameter ≥ 50 μm) was defined as a non-response (n = 12). b, Total number of colonies formed by METTL3-knockdown versus control SW620 and HCT116 cells. c, Cell viability of METTL3-knockdown HCT116 versus control HCT116 cells after oxaliplatin treatment for 48 h. d, Immunoblotting analysis of SOX2 and METTL3 in METTL3 knockdown and control HCT116 cells with or without SOX2 overexpression. e, Quantification of the in vitro sphere-formation assay of METTL3-knockdown and control HCT116 cells with or without SOX2 overexpression (n = 6). f, Cell viability of METTL3-knockdown and control HCT116 cells with or without SOX2 overexpression when treated with oxaliplatin for 48 h. All data are presented as the mean ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). β-Actin was used as the loading control. (TIF 6517 kb)
Figure S4, related to Fig. 4: METTL3 drives CRC tumorigenesis and metastasis in vivo. a, Subcutaneous tumor models in nude mice showing the tumor size at day 28 after the implantation of METTL3-knockdown and control SW620 and HCT116 cells (n = 5 mice per group). b, Representative and quantification of H&E and immunostaining (scale bar: 100 μm) of METTL3, SOX2, and EpCAM in subcutaneous tumor models of METTL3 knockdown and control SW620 and HCT116 cells. c, Tumor incidence showing the tumorigenesis of the indicated serial of cell numbers of METTL3 knockdown and control SW620 cells with or without SOX2 overexpression. d, Stem cell frequencies of METTL3 knockdown and control SW620 cells with or without SOX2 overexpression. Estimate: 1/ (the estimated stem cell frequency); Lower, Upper: 95% confidence intervals. All data and error bars are presented as the mean ± SDs. *P < 0.05, **P < 0.01 (Student’s t-test). (TIF 9763 kb)
Figure S5 related to Fig. 5: IGF2BP2 enhances SOX2 mRNA stability via an m6A-dependent manner. a, Immunoblotting of IGF2BP1, IGF2BP3, YTHDF1, YTHDF2 after RNA pull down assay with cell lysate (Ly.), full-length biotinylated-SOX2 (FL), and beads only (NC) in SW620 and HCT116 cells. b, Immunoblotting of IGF2BP1, IGF2BP3, YTHDF1, and YTHDF2 with cell lysate (Ly.), full-length biotinylated-SOX2 (#1), the SOX2 CDS region with or without m6A motif mutation (#2, #3), the SOX2 3′-UTR region with or without m6A motif mutation (#4, #5), and beads only (NC) in SW620 cells. c-d, The decay rate of mRNA and qPCR analysis of SOX2 at indicated time after actinomycin D (5 μg/ml) treatment in HCT116 cells after METTL3 inhibition (left), and in HCT116 cells after IGF2BP2 inhibition (right). The date in c, and d are presented as the mean ± SDs (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test). β-Actin was used as the negative control. The relative expression level was normalized by β-Actin. (TIF 7319 kb)
Figure S6 related to Fig. 6: Clinical correlation between METTL3, SOX2 and IGF2BP2 in CRC. a, SOX2 and IGF2BP2 IHC staining scores in primary CRC tumor tissues (T) and adjacent normal tissue (ANT) (n = 432). b-c, Kaplan-Meier analysis of OS and DFS curves based on the expression of SOX2 and IGF2BP2 expression (Kaplan-Meier analysis with the log-rank test). d, Correlation between METTL3 level (left) or IGF2BP2 level (right) with SOX2 target genes, including CCND1, MYC, and POU5F1, in TCGA database for COAD. *P < 0.05, **P < 0.01 (Student’s t-test). (TIF 6918 kb)
Table S3 Univariate and multivariate analyses of prognostic factors for overall survival among 432 colorectal cancer patients. (DOCX 13 kb)
Data Availability Statement
All data generated or analyzed during this study are included either in this article or in the additional files. The MeRIP-seq data and RNA-seq data have been deposited in the Genome Sequence Archive (http://gsa.big.ac.cn/) and are accessible under GSA: CRA001257.