Abstract
Cell division is critical for development, organ growth, and tissue repair. The later stages of cell division include the formation of the microtubule (MT)-rich central spindle in anaphase, which is required to properly define the cell equator, guide the assembly of the acto-myosin contractile ring and ultimately ensure complete separation and isolation of the two daughter cells via abscission. Much is known about the molecular machinery that forms the central spindle, including proteins needed to generate the antiparallel overlapping interzonal MTs. One critical protein that has garnered great attention is the protein regulator of cytokinesis 1, or Fascetto (Feo) in Drosophila, which forms a homodimer to cross-link interzonal MTs, ensuring proper central spindle formation and cytokinesis. Here, we report on a new direct protein interactor and regulator of Feo we named Feo interacting protein (FIP). Loss of FIP results in a reduction in Feo localization, rapid disassembly of interzonal MTs, and several defects related to cytokinesis failure, including polyploidization of neural stem cells. Simultaneous reduction in Feo and FIP results in very large, tumorlike DNA-filled masses in the brain that contain hundreds of centrosomes. In aggregate, our data show that FIP acts directly on Feo to ensure fully accurate cell division.
INTRODUCTION
Cell division, or mitosis, culminates in the separation of chromatin and cytoplasm into two daughter cells in stages respectively known as anaphase and cytokinesis. These final stages of mitosis require exquisite coordination among three major cellular polymers—actin, microtubules (MTs), and septins. During anaphase, MT-associated proteins (MAPs), molecular motors, and other effector proteins act on interdigitated, antiparallel MTs (referred to here as interzonal MTs) between opposite spindle poles to elongate the mitotic spindle and create the central spindle (D’Avino et al., 2015; Glotzer, 2017). The central spindle helps coordinate the position and assembly of the actin- and myosin-rich cytokinetic apparatus, or contractile ring, which is constructed in conjunction with the actin cross-linking protein Anillin and the septin cytoskeleton (Maddox et al., 2007; Green et al., 2012; Liu et al., 2012). The contractile ring then constricts around the central spindle, which compacts into a midbody, or Flemming body. Finally, the midbody coordinates the abscission process that fully separates the two daughter cells (Mierzwa and Gerlich, 2014). Orchestrating anaphase, cytokinesis, and abscission not only is important for symmetric division but also is critical for asymmetric divisions where two nonidentical daughter cells are produced, as is the case for stem cell divisions. Therefore, it is not surprising that defects in these final stages of mitosis have been linked to defects in cell ploidy, tissue development, and diseases such as cancer (Ganem et al., 2007; Lacroix and Maddox, 2012).
Given the importance of anaphase and cytokinesis, considerable attention has been given to these late mitotic processes, resulting in the identification of well-conserved components that regulate the MT, actin, and septin networks. In search of novel regulators of cytokinesis, we investigated a set of proteins previously found to bind septin 1 and septin 2 in Drosophila using yeast 2 hybrid analysis (Y2H; Shih et al., 2002); they were termed septin-interacting proteins 1, 2, and 3 (Sip1, Sip2, Sip3). Sip2 was of particular interest as we found a striking localization to MTs in interphase and mitosis. However, our work found no evidence of an interaction with septins, or a direct role in regulating septin function. Instead, we discovered that Sip2 binds another conserved cytokinesis molecule called Fascetto (Feo in Drosophila, PRC1 in mammals, Spd-1 in Caenorhabditis elegans) (Jiang et al., 1998; Verbrugghe and White, 2004; Verni et al., 2004), which binds and cross-links MTs to form the central spindle (Mollinari et al., 2002; Subramanian et al., 2010). Thus, we have renamed Sip2 to Feo interacting protein (FIP) and focused our attention on how FIP functions as a Feo regulator.
A lot is known about Feo/PRC1 regulation throughout mitosis. Prior to anaphase, Feo/PRC1 is held inactive through inhibitory phosphorylation by polo-like kinase (Plk1; Neef et al., 2007; Hu et al., 2012) and/or cyclin-dependent kinase 1 (Cdk1) (Zhu et al., 2006; Wang et al., 2015). Following anaphase onset, Feo/PRC1 forms a dimer (D’Avino et al., 2007; Subramanian et al., 2010; Kellogg et al., 2016), binds kinesin-4/Kif4 (Klp3a in Drosophila), and localizes to the MT +ends (Zhu and Jiang, 2005; D’Avino et al., 2007; Bieling et al., 2010; Subramanian et al., 2013; Nguyen et al., 2018). Together, Feo/PRC1 and Kif4/Klp3a function to organize the central spindle in concert with centralspindlin, a heterotetrameric complex comprised of a kinesin-6 motor (MKLP in mammals, Pavarotti in Drosophila, ZEN-4 in C. elegans) and the RhoA GAP (RACGAP1 in mammals, Tumbleweed/RacGAP50C in Drosophila, CYK-4 in C. elegans) (Jantsch-Plunger et al., 2000; Hirose et al., 2001; Mishima et al., 2002; Mishima and Lee, 2015; Kurasawa et al., 2004; White and Glotzer, 2012; Tao et al., 2016). Feo/PRC1 is also involved in recruiting (directly or indirectly) additional proteins to the central spindle necessary for cytokinesis, such as Polo in Drosophila and Plk1 in mammals (D’Avino et al., 2007; Neef et al., 2007), the mammalian MT cross-linker kinesin-5 (Subramanian et al., 2013), the Drosophila kinesin-14 and sticky kinase (Bassi et al., 2013), mammalian CLASP1 (Liu et al., 2009), and mammalian CENP-E (Kurasawa et al., 2004). Thus, Feo/PRC1 is central to the overall organization of the complex protein networks required for daughter cell separation, and its loss-of-function leads to mislocalization of MT-associated proteins, thin/disorganized interzonal MTs, chromosome segregation errors such as chromosome bridge formation, incomplete furrow formation, cytokinesis failure, and ultimately aneuploidy and polyploidy (Li et al., 2018). Uncovering new members of the Feo-dependent cytokinesis pathway, as we have with FIP, is therefore important to fully understand the complex process of daughter cell separation.
RESULTS
FIP is a microtubule and mitotic chromatin-associated protein
To assess the intracellular localization of FIP, Drosophila S2 cells were transiently cotransfected with Ht::FIP (Ht, Halo-tag) and GFP::α-tubulin, fixed, and imaged by confocal microscopy. FIP localization changed dramatically throughout the cell cycle (Figure 1A). In interphase, FIP was enriched on MT +ends, suggesting that it may be involved in regulating MT dynamics. In prometaphase and metaphase, FIP was not detected on MTs, but was clearly enriched around chromosomes, reminiscent of the perichromosomal sheath (Van Hooser et al., 2005; Booth et al., 2016). Following anaphase onset, FIP localized along interzonal MTs, which stimulate and stabilize the site of the cytokinetic furrow. Finally, FIP decorated midbody MTs in late cytokinesis and through abscission, suggesting that FIP might play a role in the very final stages of cell division. To directly monitor FIP dynamics, we performed live 4D imaging of S2 cells expressing GFP::FIP. In agreement with fixed cell analysis, FIP localization rapidly changed from perichromosomal to the spindle midzone at anaphase onset, which then becomes compacted into the midbody MTs as cytokinesis progresses (Figure 1B; Supplemental Video 1).
FIGURE 1:
FIP shows cell cycle–dependent MT and chromatin localization dynamics. (A) Cell cycle stages of S2 cells coexpressing GFP::α-tubulin (red) and Ht::FIP (green; TMR ligand), MeOH fixed, and stained with DAPI (blue). Regions outlined by dashed lines are enlarged in grayscale. Metaphase inset is a single image plane, which best represents FIP perichromosomal localization. All other images are maximum intensity projections. (B) Time series of a S2 cell expressing GFP::FIP (Supplemental Video 1) from metaphase to late telophase. Image stacks of six 0.8-µm sections were collected every 20 s and displayed as projections. Time = min:s relative to anaphase onset at 0:00 (unpublished data). Scale bars: (A) 5 µm, (B) 10 µm.
Video S1.
FIP is perichromosomal in metaphase and at spindle midzone MTs during anaphase and telophase. Corresponds to Figure 1B. Cultured S2 cell transiently expressing GFP::FIP. Video begins in metaphase where FIP is concentrated around mitotic chromatin, and extends through late telophase where FIP concentrates around the cell midbody. Image series is maximum intensity projection of six 0.8μm slices/frame, acquired at 20s/frame and displayed at 10 fps, time is in minutes:seconds relative to NEB, scale bar = 10μm.
The various subcellular localizations of FIP suggested that specific localization motifs or domains might be present, although no known protein domains are computationally predicted (Shih et al., 2002). To determine whether such motifs were present, we truncated FIP into thirds, taking care not to disrupt predicted coiled-coil regions, and generated GFP fusions of each fragment (Figure 2A). The central region of FIP (FIP 220–438) is diffuse throughout the cytoplasm at all cell cycle stages (Figure 2, B–D, center column), the N-terminus (FIP 1–219) conveys the perichomosomal localization in metaphase (Figure 2C, left column), and the C-terminus (FIP439–657) conveys localization to both interphase MT +ends and anaphase/telophase midzone MTs (Figure 2, B–D, right column). Of note, FIP 439–657 precociously localized to MT +ends in mitosis, indicating that upstream regions of FIP are required to prevent mitotic +end tracking (Figure 2C, right column).
FIGURE 2:
FIP subcellular localizations rely on specific protein regions. (A) FIP locus and protein structure indicating the promotor region (dashed line); exons (boxes); introns (solid lines); predicted coiled coils (CC, blue); EB1-binding motifs (SxIP, green); the N-terminal, central, and C-terminal fragments used in B–D (amino acid ranges indicated); and guide RNA sites used to generate the CRISPR mutant (magenta arrows; gRNA1 and gRNA2). (B) Interphase S2 cells expressing GFP-tagged FIP1–219, FIP220–438, or FIP439–657; stills from a 60-s time series (2-s intervals, single image plane; grayscale). Dashed lines represent the region of the cell used to create a color-coded “time projection” (rainbow). Only FIP439–657 shows MT +end tracking (rainbow streaks). (C, D) Images of S2 cells in metaphase and cytokinesis expressing GFP-tagged FIP1–219, FIP220–438, and FIP439–657 (green) counterstained for DNA (Hoechst 33342, blue) and MTs (SiR-tubulin, magenta). Regions with dashed lines are shown to the right in grayscale. FIP1-219 is perichromosomal (green arrows) and centrosomal (pink arrow). FIP439–657 localizes to centrosomes (pink arrow) and midzone MTs during cytokinesis (yellow arrow). All metaphase and cytokinesis images are maximum intensity projections through the region of interest. n Values on figure indicate number of cells imaged. Scale bars: 5 µm.
FIP localizes to interphase MT +end through direct EB1-binding
To investigate FIP localization to MT +ends, we performed live two-color imaging of S2 cells coexpressing fluorescently tagged FIP and the highly conserved MT end-binding 1 protein to mark MT +ends (EB1; Vaughan, 2005). In support of our fixed data, FIP colocalized with the characteristic EB1 MT +end-tracking comets in interphase cells (Supplemental Figure S1A and Supplemental Video 2). FIP enrichment at MT +ends dropped from 1.46 ± 0.48-fold (over cytoplasm) in interphase to a nearly undetectable enrichment of 0.36 ± 0.24-fold in metaphase, whereas EB1 enrichment did not appear to change (Supplemental Figure S1, B and C; Supplemental Video 3). This regulated cell-cycle behavior is similar to the direct EB1-binding proteins STIM and CLASP2, which down-regulate tip-tracking behavior in response to mitotic phosphorylation (Kumar et al., 2012; Smyth et al., 2012). Thus, we hypothesized that FIP is also recruited to MT +ends by EB1. To test this hypothesis, we grossly overexpressed EB1 (EB1OE), a condition that drives exogenous EB1 along the MT lattice, and showed that FIP was also mislocalized to the MT lattice (Supplemental Figure S1D). In contrast, overexpression of FIP alone never forced FIP on the MT lattice regardless of expression level (Supplemental Figure S1E), indicating that EB1 is limiting for MT lattice recruitment.
Video S2.
FIP localizes to growing interphase MT +ends. Corresponds to Figure S1A. Interphase S2 cell coexpressingHt::FIP (JF549 ligand, magenta) and EB1::mNG (green) illustrating colocalization of EB1 comets with FIP at MT +ends. Boxed frames in upper right show the EB1 channel in greyscale (top), FIP channel in greyscale (middle) and merge (bottom). Image series is a single slice, acquired at 2 s/frame and displayed at 5fps, time is in minutes:seconds, scale bar = 5μm.
Video S3.
FIP localization to MT +ends approaches background levels in mitosis. Corresponds to Figure S1B. Metaphase S2 cell coexpressingHt::FIP (JF549 ligand, magenta) and EB1::mNG (green) illustrating colocalization of EB1 comets with FIP at MT +ends. Boxed frames on right show the FIP channel in greyscale (bottom), EB1 channel in greyscale (middle), and merge (top). Image series is a single slice, acquired at 2 s/frame and displayed at 5fps, time is in minutes:seconds, scale bar = 5μm.
Using Y2H analysis, we showed that both full-length FIP (FIPFL) and the FIP C-terminus (FIP439–659) directly bind EB1 (Supplemental Figure S1F). Additionally, FIP was coimmunoprecipitated from S2 cells overexpressing GFP::EB1 and Flag::FIP (Figure 7B). Sequence analysis of the FIP C-terminus revealed two consensus Ser-x-Ile-Pro (SxIP) motifs (Figure 2A; Honnappa et al., 2009). To explore the function of these SxIP motifs, we mutated one (fip∆1 or fip∆2) or both (fip∆1-2) SxIP motifs to SNNN (Jiang et al., 2012) and transfected these mutant constructs into S2 cells along with low levels of EB1. FIP∆1 and FIP∆2 exhibited a decreased enrichment at MT +ends, whereas fip∆1-2 was undetectable at +ends (Supplemental Figure S2, A and B; Supplemental Video 4). Furthermore, EB1OE was unable to recruit fip∆1-2 to the MT lattice (Supplemental Figure S1G), confirming that the SxIP motifs mediate EB1–FIP interaction.
FIGURE 7:
FIP directly binds Feo. (A) The indicated FIP protein truncations (horizontal lines) were tested for direct binding to Feo protein truncations (A’) via Y2H. The MT-localization region (439–657) of FIP binds to the N-terminal region (1–346) of Feo, which consists of the dimerization (1–67) and rod (68–351) domains. The blue yeast colony indicates a detectable interaction (growth is on QDOXA plates) between the two minimum fragments (orange lines). Complete interaction data are provided in Supplemental Figure S5. (B) Western blot showing FIP coimmunoprecipitated with both GFP::EB1 (red box) and GFP::Feo (from mitotically enriched cells, blue box). (C) S2 cells coexpressing mNeonGreen::Feo (red) and TagRFP::FIP (green) show identical localization of Feo and FIP beginning at anaphase onset (0:00) through telophase (22:00). Cell fails cytokinesis because it is plated on Con A (Supplemental Video 9). Yellow arrows indicate the approximate position of the cell equator. Scale bar: (C) 5 µm.
Video S4.
FIP localization to interphase MT +ends is EB1-dependent. Corresponds to Figure S2A. Interphase S2 cell coexpressingHt::FIP (top left), or Ht::FIPΔ1-2 (top right), and EB1::mNG (bottom row), illustrating that the FIPΔ1-2 is incapable of MT +end tracking, despite clear EB1 MT +end tracking. In both cases, Halo-tagged FIP constructs are covalently linked to a JF549 Halo Tag ligand. Image series is a single slice, acquired at 2 s/frame and displayed at 5fps, time is in minutes:seconds, scale bar = 5μm.
Given the MT +end localization, we tested whether FIP plays a role in regulating interphase MT dynamics. Live imaging of interphase S2 cells depleted of FIP by double-stranded RNA (dsRNA) and expressing EB1-GFP revealed a slight, but statistically significant, difference in MT growth rate (130 ± 65 nm/s compared with 150 ± 68 nm/s in controls; Supplemental Figure S2, C and D). Thus far, our results suggest that FIP is recruited to MT +ends by a direct interaction with EB1, but does not play a major role in MT growth. While we cannot rule out a role for MT rescue or catastrophe, it is unlikely given the absence of FIP on paused and shrinking MTs.
FIP is required for efficient progression through mitosis
To investigate the role of FIP in a physiologically relevant context, we turned to a detailed analysis of FIP in intact Drosophila melanogaster tissues. We generated transgenic flies expressing GFP::FIP driven by the ubiquitin promoter and imaged larval imaginal wing disc cells. Similar to S2 cells, FIP localized to both midzone MTs during cytokinesis and MT +ends during interphase (Figure 3A). We then used CRISPR to generate fip7, a mutant allele that deletes 153 nucleotides of the promoter sequence, the transcription and translation start sites, and ∼75% (1452/1971) of the coding nucleotides (Figure 2A, gRNAs; Supplemental Figure S3B, PCR confirmation of deleted genomic region; locus was also sequenced). Animals homozygous for fip7/fip7 or fip7/Df(2L)Exel7029 (hereafter both referred to as fip-) are viable and fertile; thus, FIP is a nonessential protein for viability of D. melanogaster reared in typical lab conditions.
FIGURE 3:
FIP is required for efficient cell division. (A) Wing disc cells from transgenic animals expressing FIP::GFP showing FIP localization to interzonal MTs during cytokinesis and MT +end tracking during interphase. (B) Binucleate cell in a fixed fip- wing disc (dashed box, asterisk) stained with phalloidin (green) and anti-lamin (magenta). The region within the dashed box is shown in grayscale on the right. (B’) Percentage of cells that were binucleate; each point represents a single wing disc, ***P < 0.001. (C) Micronuclei in a fixed fip- wing disc (dashed box) stained with phalloidin (green) and anti-lamin (magenta). The region within the dashed box is shown in grayscale on the right. (C’) Percentage of cells with micronuclei; each point represents a single wing disc, **P < 0.01. Scale bars: 5 µm.
Given the localization of FIP, we predicted that loss of FIP would result in cell division defects. Indeed, analysis of fixed fip- wing discs showed binucleate cells (1.08 ± 0.76% of cells; Figure 3, B and B′) and rare incidences of micronuclei (0.54 ± 0.56% of cells; Figure 3, C and C′ ), which suggests a history of cytokinesis failure and possibly chromosome fragmentation or missegregation (Fenech et al., 2011). We next attempted to capture mitotic defects using two-color live imaging of mitosis in fip- wing discs using GFP::Jupiter (marking MTs) and H2AV::mRFP (marking chromosomes). Although we did not capture complete mitotic failure, our live imaging uncovered a slight delay in mitotic progression (nuclear envelope breakdown [NEBD] to anaphase onset of 533 ± 116 s in mutants compared with 493 ± 67 s in controls; Figure 4, A and A’) and a defect in chromosome segregation wherein fip- cells ceased chromosome movement sooner than controls and segregated a shorter distance (Figure 4, B and B’). Parallel experiments using dsRNA knockdown of FIP in S2 cells revealed multinucleate cells (6.2 ± 2.5% vs. 3.7 ± 2.0% in controls) and a decrease in anaphase cell index (0.1 ± 0.0% vs. 0.7 ± 0.8% in controls; Supplemental Figure S2E). Taken together, these results suggest FIP is required for fully accurate chromosome segregation, cytokinesis, and overall mitotic progression.
FIGURE 4:
FIP is required for interzonal MT stability. (A) 4D time series of control and fip- wing disc expressing GFP::Jupiter (green) and H2Av::mRFP (magenta) progressing through mitosis. (A’) Quantification of mitotic duration (NEBD to anaphase onset) showing a delay in mitotic progression in fip- cells, *P < 0.05. (B) Wing disc cells in telophase showing maximum nuclear separation. Dashed lines represent chromosome separation measurements. (B’) Quantification of maximum chromosome separation (displayed as a percentage of metaphase spindle length to control for cell size); each point represents a single cell, ****P < 0.0001. (C) Stills and kymographs from telophase fip- and control wing discs. Kymographs begin at anaphase (separation of chromosomes, magenta streaks). Wild-type midbody formation in controls is seen as a pair of streaks (green), while the rapid dissolution of MTs (GFP::Jupiter) is seen in the fip- kymograph. Blue arrows indicate the frame of the kymograph used to create the still image at the top. Yellow arrows indicate the average time point, relative to anaphase onset, where the GFP::Jupiter interzonal signal ends. (C’) Interzonal MT duration measured using GFP::Jupiter signal (anaphase onset to the last frame where an intercellular MT bridge was clearly seen); each point represents a single cell, ****P < 0.0001. Horizontal scale bars in A–C = 3 µm; vertical scale bars in C = 480 s.
In searching for the mechanism by which loss of FIP could cause these defects and considering previous work that showed FIP binds septin 1 and septin 2 (Shih et al., 2002), we hypothesized that FIP regulates septins to ensure proper cytokinesis. Indeed, loss of septin family members has been shown to result in cytokinesis failure (Kinoshita et al., 1997; Nagata et al., 2003) and chromosome missegregation (Spiliotis et al., 2005). We were unable, however, to confirm direct FIP–septin interaction by Y2H (Supplemental Figure S3A), and our two-color live imaging of Sep2::GFP and H2AV::mRFP in dividing fip- wing disc cells revealed that the timing of septin 2 recruitment to the cleavage furrow and midbody appeared normal (unpublished data). Most interestingly, we found that septin 2 was less persistent at the midbody (disassembling in 2002 ± 330 s following anaphase onset compared with 2784 ± 623 s in controls; Supplemental Figure S3, C and C’), and high-resolution imaging in fixed fip- cells revealed that septin 2 in the midbody lacked the cylindrical organization characteristic of control midbodies (Supplemental Figure S3D). Given that FIP does not localize to the cell cortex and that we were unable to detect direct FIP–septin interaction, we favor the hypothesis that the defects in septin organization and dynamics were secondary to an upstream defect.
FIP stabilizes interzonal MTs during cytokinesis
On the basis of FIP’s localization to interzonal MTs, we hypothesized that the atypical organization of septin 2 in fip- midbodies was due to defects in MT organization. Our live imaging of GFP::Jupiter (marking MTs) and H2AV::mRFP in fip- wing discs revealed no significant defect in interzonal MT density in fip- wing discs in the first 2 min after anaphase onset (measured by GFP::Jupiter enrichment; unpublished data). Following fip- wing disc cells farther into telophase revealed a prominent defect where interzonal MTs focused into a single point, in contrast to wild-type cells that show a stereotypical organization of two discrete bundles of MTs on either side of the midbody (Figure 4C, parallel vertical streaks in the kymograph). It appears as if the interzonal MTs region is significantly diminished, possibly completely lost. Even more striking, however, was that fip- cells lost all detectable midbody MTs three times faster than controls (Figure 4, C and C’; 350 ± 40 s after anaphase onset compared with 952 ± 130 s in controls).
Our MT analysis suggested that the early stages of cytokinesis, which include specifying the cleavage plane and ingression of the membrane, proceed normally. To show this more directly, we imaged cortical actin using GFP::Moesin and found that 100% of fip- cells formed an actin contractile ring and completed membrane constriction down to the midbody (Supplemental Figure S4, A and B; 154 ± 26 s compared with 153 ± 25 s in controls), no significant difference in actin enrichment in the nascent furrow (Supplemental Figure S4C; 2.62 ± 0.5-fold enrichment compared with 2.44 ± 0.5-fold in controls), and no significant difference in the residence of GFP::Moesin localization to the midbody (882 ± 127 s compared with 986 ± 229 s in controls; Supplemental Figure S4D). Together, these results indicate that FIP is dispensable for early organization of interzonal MTs and actin ring formation but plays an important role in stabilizing the late interzonal MTs required for the very final stages of cell division.
FIP is required for proper neural stem cell ploidy
Previous studies have shown that mutations in mitotic genes can result in variant phenotypes in the wing disc and larval brain (Poulton et al., 2014, 2017). Indeed, unlike wing disc cells, analysis of fip- larval brains revealed striking chromatin masses (Figure 5, A and B; 4.5 ± 4.5 DNA masses per CNS compared with 0/CNS in controls). Expression of GFP::FIP in the fip- background nearly fully rescued the polyploid phenotype (1.1 ± 1.9 polyploid cells per CNS; Figure 5B). High-resolution imaging of these DNA masses showed that they typically reside within single cells, adopting a lobular morphology often surrounded by numerous micronuclei (Figure 5C, red arrows).
FIGURE 5:
Loss of FIP results in failed divisions and polyploidy in brains. (A) Third instar larval brains stained for F-actin (phalloidin) and DNA (DAPI). Dashed boxed region is enlarged to the right, highlighting polyploid neuroblasts (NBs) in the fip- mutant (yellow arrows). (B, D) Quantification of the number of DNA masses in control, mutant, rescue, and RNAi knockdown conditions; each point represents the number of DNA masses per CNS (central brain + VNC), sqh-Gal4 is a ubiquitous RNAi knockdown; Dpn-Gal4 is NB-specific. The numbers on the x-axis (1, 2, 3) refer to the genotype indicated in A; genotype 3 is indicated below the graph. (C) High-resolution image of a polyploid cell adjacent to a healthy NB (top left) stained with anti-lamin (green) and DAPI (magenta) showing many large DNA structures in addition to micronuclei (red arrows). Dashed lines indicate cell boundaries from phalloidin channel (unpublished data). (E) Live images of wild-type and fip- NB expressing GFP::Moesin (magenta) and H2AV::mRFP (green); see Supplemental Videos 5 and 6. Wild-type NB proceed through mitosis with normal timing and an extended version of this movie (unpublished data) confirmed no regression of the cytokinetic furrow. After an extended period in prometaphase and metaphase, the fip- NB initiates asymmetric division (40:45), attempts to complete cytokinesis (46:45), begins massive membrane blebbing at the site of furrow formation (81:15, yellow arrows), and eventually fails cytokinesis/abscission, regressing into a single cell (128:00, blue arrow). Time is in min:s relative to NEB. Scale bars: (A) 150 µm, (C, E) 10 µm.
The distinctive localization of the polyploid cells in the CNS suggested that these cells were derived from central brain neuroblasts (NBs). To explore this possibility, a UAS-FIP RNA interference (RNAi) transgene was driven by either Sqh-GAL4 (a strong and ubiquitous driver) or Dpn-GAL4 (a relatively moderate driver expressed only in NBs). Both gene knockdowns produced identical polyploid cells in the central brain (2.7 ± 3/brain in Sqh-GAL4 and 3.9 ± 2/brain in Dpn-GAL4; Figure 5D). To better understand how these DNA masses arise, we performed live two-color imaging of GFP::Moesin and H2AV::mRFP in fip- brains. We were unsuccessful in capturing the initial event of polyploidization; however, we did capture an early polyploid NB in mitosis, which remained in metaphase for ∼33 min, four times longer than wild-type NB divisions (Figure 5E; Supplemental Videos 5 and 6; Rusan et al., 2008). The fip- NB eventually entered anaphase with chromatin bridges remaining in the division plane and extensive membrane blebbing; ultimately, the NB failed abscission, driving the cell into a greater state of polyploidy. This video also revealed that the micronuclei likely result from lagging chromosomes being independently encased in nuclear membrane. We suspect that once a fip- NB undergoes a mitotic failure, the state of the cell progressively becomes worse with each round of attempted division.
Video S5.
Control NB division. Corresponding to Figure 5E. A wild-type neuroblast undergoes cell division with normal timing and normal chromosome segregation. The division results in the renewal of the NB and the production of a ganglion mother cell (GMC). Not shown is the longer version of the movie that confirms no failure in abscission. Images are single planes acquired at 60 seconds/frame and displayed at 10 fps.
Video S6.
Loss of FIP leads to failed cytokinesis and increased ploidy. Corresponds to Figure 5E. Polyploid neuroblast from a fip- 3rd instar larval brain expressing GFP::Moesin (magenta) and H2AV::mRFP (green). Neuroblast enters anaphase and attempts to complete cytokinesis but fails. The contractile ring regresses and the DNA from the would-be daughter cells collapses into a single DNA mass. Frames were captured at different rates, but displayed at 21fps, time is in minutes, scale bar = 10μm.
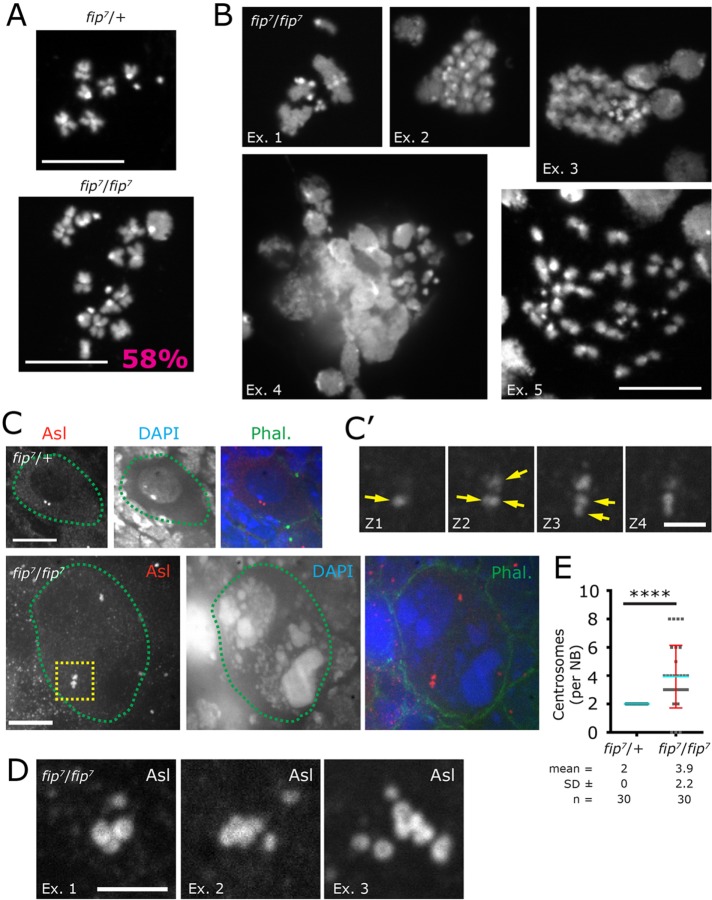
To determine whether the large DNA masses represented an increase in cell ploidy, we performed whole-brain squashes from third instar larvae stained with diamidine-phenylindole dihydrochloride (DAPI). Whereas wild-type NBs contained the normal complement of eight chromosomes (in some cases fewer than eight are detected due to sample preparation), 58% of fip- NBs were polyploid (contained more than eight chromosomes; Figure 6A). Our preparation also revealed many polyploid NBs with other abnormalities such as the formation of synapsed (polytenelike) chromosomes (Figure 6B, Ex.1) and asynchronous chromosome condensation (Figure 6B, Ex. 2–4), which was especially clear when the large DNA masses were captured in a squash. We also analyzed NBs for centrosome number as an additional proxy for cytokinesis failure. We find the normal complement of two centrosomes in wild-type NBs, whereas fip- NBs contained an average of four centrosomes (Figure 6, C–E), which is consistent with our live imaging of NB cytokinesis failure and our analysis of DNA content. Therefore, we can conclude that cells that contain these large DNA masses are polyploid, and that they likely manifest from a subset of the 58% polyploid fip- NBs.
FIGURE 6:
Loss of FIP results in polyploid NBs and centrosome amplification. (A) Third instar larval brain squashes reveal the normal complement of chromosomes in wild-type (heterozygous) NBs, whereas 58% of mutant fip7 NBs are polyploid. (B) Additional representative examples of fip7 NBs showing polytenelike chromosomes (Ex. 1), asynchronous chromosome condensation (Ex. 2, 3), and a large degree of polyploidy (Ex. 5). We also occasionally find the larger DNA aggregates in a state of mixed chromosome condensation (Ex. 4). (C) NBs stained for the centrosome marker Asterless (Asl, red) and DNA (DAPI, blue). Cell outline based on phalloidin staining of cortical actin (Phal., green), and images are maximum intensity projects of all the z-planes that capture the centrosomes. Wild-type NB shows the normal complement of centrosomes, whereas the fip- mutants show elevated centrosome numbers. (C’) Enlarged view of the yellow-boxed centrosome cluster shown in C. Each image in C’ is a different image plane along the z-axis highlighting the supernumerary centrosomes (yellow arrow). (D) Additional examples of supernumerary centrosomes from fip7 mutant NBs. (E) Average centrosome count in control and fip7 NBs, ****P < 0.0001. Scale bars: (A–C) = 10 µm; (C’, D) = 5 µm.
FIP binds the MT lattice via the PRC1 orthologue Feo
To explore the mechanism by which FIP regulates cytokinesis, we sought to identify protein partners of FIP that might function at interzonal MTs. Although we showed that FIP localization did not appear exclusive to MT +ends in the later stages of mitosis, we nevertheless tested whether FIP localization to interzonal MTs was mediated by EB1. However, live imaging of the EB1-binding mutant of FIP (fip∆1-2) revealed normal interzonal MT localization in anaphase and telophase (Supplemental Figure S4E; Supplemental Video 7); thus, interzonal MT recruitment is independent of EB1. We next investigated whether FIP directly interacted with the MT-binding protein abnormal spindle (Asp), which displays a similar anaphase localization to FIP (Asp; Wakefield et al., 2001; Riparbelli et al., 2002) and is known to bind MTs (Saunders et al., 1997; Mollinari et al., 2002; Ito and Goshima, 2015; Schoborg et al., 2015). However, we were unable to confirm a previously reported FIP–Asp interaction (Asp; Giot et al., 2003) using our Y2H system (Supplemental Figure S5). Furthermore, GFP::FIP localization in a asp- genetic background appeared unperturbed (unpublished data). Finally, we hypothesized that FIP is recruited to the central spindle by Feo (Verni et al., 2004), the Drosophila PRC1 orthologue shown to play an interzonal MT stabilization role during anaphase (Wang et al., 2015). We first used Y2H to reveal that full-length FIP and Feo directly interact (Supplemental Figure S5). Y2H analysis using FIP and Feo truncations narrowed the interaction down to FIP220–647 (containing the sequence necessary and sufficient for MT localization in mitosis) and Feo1–346 (containing the dimerization and rod domains) (Figure 7, A and A′). We also confirmed the interaction by coimmunoprecipitation from S2 cells overexpressing GFP::Feo and Flag::FIP (Figure 7B and Supplemental Figure S6A). Importantly, a Feo-FIP IP was successful only when the culture was enriched for mitotic cells using an overnight colchicine treatment, indicating the interaction is likely MT-independent and restricted to mitosis. This is consistent with the localization of Feo and FIP to separate cellular compartments in interphase—Feo is nuclear and FIP is cytoplasmic. As a final confirmation of FIP–Feo interaction in vivo, we performed live imaging of S2 cells coexpressing mNeonGreen::Feo and TagRFP::FIP. In low-expressing interphase cells, FIP tracked MT +ends and Feo was seen along a subset of bundled MTs (Supplemental Figure S6B; Supplemental Video 8), whereas in high-expressing cells, we found that Feo ubiquitously coated and bundled MTs, as expected given Feo’s known role to cross-link MTs in mitosis (Supplemental Figure S6B; Mollinari et al., 2002). In addition, these bundled MTs recruited FIP, which in interphase is found only at the MT +ends regardless of expression level (Supplemental Figure S1E), indicating that Feo can mediate a FIP–MT interaction (Supplemental Figure S6B). At low expression levels, and in the physiologically relevant context of mitosis, Feo and FIP localization were indistinguishable (Figure 7C; Supplemental Video 9). In Drosophila larval wing discs, Feo localization was also identical to FIP, showing translocation from the central spindle in anaphase to the midbody in telophase (Supplemental Figure S7). Together, these results indicate that Feo and FIP likely form a complex and cotarget interzonal MTs at anaphase onset.
Video S7.
FIP localization to interzonal MTs in anaphase and telophase is independent of EB1. Corresponds to Figure S4E. Cultured S2 cell transiently expressing mNG::FIPΔ1-2. Video begins in metaphase, where FIP?1-2 is concentrated as expected around mitotic chromatin, and extends through late telophase, where FIP concentrates as expected around the cell midbody. Image series is maximum intensity projection of six 0.8μm slices, acquired at 20 s/frame and displayed at 10 fps, time is in minutes:seconds relative to NEB, scale bar = 5μm.
Video S8.
Low expression of Feo does not bundle MTs and does not interfere with FIP MT +end tracking in interphase. Corresponds to Figure S6B. Interphase S2 cell coexpressing low quantities of mNeonGreen::Feo (red) and TagRFP::FIP (green) illustrating normal FIP MT +end tracking. Images are single planes acquired at 3 seconds/frame and displayed at 30 fps.
Video S9.
FIP and Feo localization is indistinguishable during anaphase and telophase. Corresponds to Figure 6C. Mitotic S2 cell coexpressing low quantities of mNeonGreen::Feo (red) and TagRFP::FIP (green) illustrating normal FIP MT +end tracking. Images are single planes acquired at 15 seconds/frame and displayed at 15 fps.
Feo requires FIP for proper localization
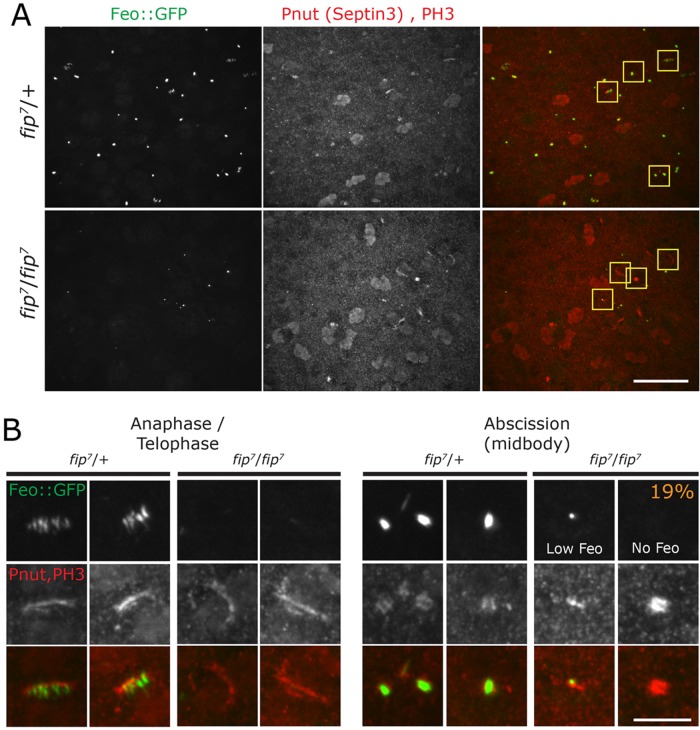
To test how FIP might regulate Feo localization, we performed live imaging of Feo::GFP in wild-type and fip- wing discs dissected from third instar larvae. These videos show a reduction in the number of cells entering mitosis and fewer Feo containing midbodies in the fip- tissue as compared with wild type (Supplemental Videos 10 and 11). To better quantify this observation, we turned to fixed analysis of wing discs from wild-type and fip- animals expressing Feo::GFP and stained for the contractile ring component peanut (Pnut, septin 3).Wild-type wing discs contain a high percentage of mitotic cells, with anaphase/telophase cells showing a clear contractile ring with Feo decorating interzonal MTs that compact down to a barrellike midbody at the end of mitosis. In contrast, fip- wing discs show nearly undetectable Feo on midzone MTs during anaphase and telophase (Figure 8, A and B). At later stages of telophase and midbody formation, Feo levels are extremely low, and in 19% of midbodies (marked by Pnut), no Feo::GFP is detected. Interestingly, the low levels of Feo in fip- cells present as small dots (Figure 8B), mirroring the observation that MTs focus down to a small dot in fip- cells (Figure 4C). Overall, however, Feo::GFP in fixed fip- wing discs was noticeably reduced compared with our live analysis. We suspect that Feo in fip- wing discs does not survive our fixation protocol that uses cold phosphate-buffered saline (PBS) fixation needed to detect Pnut (Figure 8 and Materials and Methods). Nevertheless, there is a consistent trend in both the live and fixed analysis toward a reduced amount of Feo in the absence of FIP. We also note that the effect on Feo localization might be underestimated as these animals are expressing Feo::GFP under the ubiquitin promoter, and thus, likely overexpressing Feo.
FIGURE 8:
FIP is required for FEO localization and proper midbody architecture. (A) Peripodial cells in the pouch of control and fip- wing discs expressing Feo::GFP (green) and stained for Pnut (septin 3, red) and PH3 (red). Note that these samples were prepared in cold fixative (Materials and Methods). (B) Enlarged images of peripodial cells show Feo::GFP localization at the anaphase spindle in control but not in fip- mutants. Feo prominently localizes to the midbody in controls, whereas fip- mutant have low levels of Feo that appear focused down to a single dot; in 19% of cells, Feo is completely absent. Scale bars: (A) 20 µm, (B) 5 µm.
Video S10.
Feo localization in wild-typeperipodial wing disc cells. Corresponds to Figure 7.Peripodial cells expressing Feo::GFP indicating the high level of mitotic activity. Feo localizes to interzonal MTs at anaphase and telophase, followed by the focused localization to the midbody. Images acquired at 20s/frame and displayed at 15 fps.
Video S11.
FIP mutants have fewer mitotic divisions in wing disc cells. Corresponds to Figure 7. Peripodial cells expressing Feo::GFP in fip- mutant animals highlighting the low level of mitotic activity. Localization of Feo to anaphase interzonal MTs is rarely observed, while Feo localization to midbodies is minimal. Images acquired at 20s/frame and displayed at 15 fps.
FIP and Feo function in the same pathway to ensure proper cell division
Having established a direct protein interaction, identical intracellular localization, and the partial localization dependence of Feo on FIP, we investigated a genetic interaction between FIP and Feo. For this analysis, we turned to the most prominent phenotype of DNA masses in the central brain (Figure 5). fip- (fip7/fip7 or fip7/fipDf) and feoRNAi (tubulin-Gal4 driving feoRNAi(GL) shown, or feoRNAi(HM) [unpublished data]) brain lobes contain 1.9 ± 2.1 and 0.6 ± .9 DNA masses per lobe, respectively (Figure 9, A and A’; complete data set shown in Supplemental Figure S8), all within the central brain. feoRNAi was used in this experiment because feo null flies die at early larval stages. Importantly, double loss-of-function analysis (fip-; feoRNAi) showed a great enhancement of the phenotype with 8.4 ± 3.6 DNA masses per brain lobe (Figure 9, A and A’), an average that is three times greater than the sum of feoRNAi + fip-. Animals codepleted of FIP and Feo also greatly expand the effected brain tissue beyond the central brain and into the optic lobes (Figure 9A, blue arrows). Animal survival is also affected as double loss of both FIP and Feo result in pupal lethality, whereas fip- or feoRNAi individually gives rise to viable adults. Finally, overexpression of Feo::GFP in the fip- mutant background nearly fully rescued the phenotype with an average of 0.3 ± 0.8 polyploid cells per lobe (Figure 9, A and A’). Results similar to that in the brain lobes were observed when analyzing cells in the ventral nerve cord (VNC; Figure 9, B and B’; complete data set is shown in Supplemental Figure S8). Together, these results indicate that FIP and Feo function together in the same pathway to ensure proper cytokinesis. Consistent with the viability of fip null animals, the genetic interaction data indicate that FIP is a nonessential component of the Feo-cytokinesis pathway because partial loss of Feo using feoRNAi enhanced the null phenotype of fip7/fipDf; if FIP were an essential component, the phenotype would not be enhanced by feoRNAi.
FIGURE 9:
FIP and Feo function in the same pathway to ensure proper ploidy. (A) Third instar larval brains stained for DNA (DAPI) in control (green, 1), single loss-of-function (orange, 2 and 3), and double loss-of-function (blue, 4). Overexpressing Feo in the fip- background (pink, 5) rescues the polyploidy phenotype. Red arrows indicate loss of function; green arrow indicates overexpression. Pink, dashed lines roughly delineate the central brain (right) from the optic lobe (left). Polyploid cells in the central brain (yellow arrows) are present in single loss-of-function, whereas polyploid cells are present in both the central brain (yellow arrows) and optic lobes (blue arrows) in double loss-of-function. (A’) Quantification of polyploid cells per brain lobe. (B) VNC stained for DNA (DAPI); genotypes as indicated at the top. (B’) Quantification of polyploid cells in the VNC. ***P < 0.001, ****P < 0.0001. Scale bar: 150 µm.
Interestingly, double loss-of-function (fip-; feoRNAi) resulted in 64% of brains containing much larger DNA structures in addition to the smaller DNA masses (Figure 10A). These larger structures can be grouped into two categories: “single mass” structures, which are homogenous DNA structures contained within a single huge cell, and “clustered” DNA fragments (Figure 10B). To convey the difference in size, we measured the cross-sectional area of the DNA regions in wild-type, single mutant, and double loss-of-function (Figure 10C) backgrounds. The average cross-sectional area of a wild-type NB nucleus was 100 ± 5 µm2, whereas fip- and feoRNAi nuclei averaged 570 ± 130 µm2 and 550 ± 140 µm2, respectively. The average size of the large single masses in the double loss-of-function was nearly 50 times larger (4742 ± 1821 µm2) than wild type, whereas the clustered nuclei were over 100 times larger (10,147 ± 2815 µm2). Interestingly, these larger DNA structures contain two hallmarks of cancer/tumor cells: a large increase in centrosome numbers, in some cases hundreds of centrosomes (Figure 10, D and D’), and greatly abnormal ploidy (Figure 10E). Taken all of our data in aggregate, we conclude that FIP is indeed a component of the Feo-dependent cytokinesis pathway.
FIGURE 10:
fip- and FeoRNAi codepleted animals form tumorlike masses. (A) fip- mutants show relatively small polyploid cells, whereas double depletion of fip- and FeoRNAi form massive DNA aggregates that span all brain regions. (B) The large DNA aggregates can be generally classified into “single” and “clustered” masses. The numbers 4 and 5 refer to the genotype in C. (C) Quantification showing the cross-sectional area of the DNA masses in the indicated genotypes. ****P < 0.0001. (D, D’) The DNA masses from the fip- and FeoRNAi double-depletion brains show massive centrosome (Asl, red) amplification. (D’) Enlargements of the regions are shown by the yellow arrow in D. (E) Chromosome squash from a large DNA aggregate revealing increased poloidy. Scale bar: (A) 100 µm, (B, D) 50 µm, (D’) 5 µm, (E) 10 µm.
DISCUSSION
We identified a new binding partner and regulator of Feo (PRC1), which we renamed from Sip2 to FIP. FIP localization is highly dynamic throughout the cell cycle, suggesting that it plays several roles. In interphase, FIP binds growing MTs via EB1 and appears to regulate MT growth, whereas in mitosis (prior to anaphase onset), FIP is perichromosomal and plays an unknown role required for fully accurate mitotic progression (Supplemental Figure S9). These first two roles will require further investigation, possibly using other cell types that exhibit more prominent interphase and preanaphase phenotypes in a fip- background.
Our study here focused mainly on investigating FIP’s role during late cytokinesis (Supplemental Figure S9). Using colocalization and double loss-of-function analysis, we were able to place FIP within the Feo-dependent cytokinesis pathway. We found that FIP, while important, is a nonessential component of the Feo pathway. This is supported by the observation that Feo overexpression can nearly fully suppress fip- and because the null phenotype of fip- is much less severe than the early lethality of feo null animals (Perrimon et al., 1989; Verni et al., 2004). Our work on FIP highlights the importance of investigating the role of nonessential modulators of critical cellular processes, because the viable adults that emerge can be defined as animals highly susceptible to disease. In the case of fip- mutants, we do not find catastrophic developmental defects, but the abnormal DNA masses found in ∼5% of NBs likely evolved from a subset of the 58% polyploid NBs. This suggests that much of the neuronal tissue contains cells with abnormal ploidy. Whereas long-term effects of both the larger DNA masses and polypoid NB in fip- flies are unclear, it is well established that polyploidy can give rise to aneuploid cells (Ganem et al., 2007; Davoli and de Lange, 2011) and is a common feature in human cancers (Zack et al., 2013). Therefore, we predict that fip- flies are more prone to develop deficiencies and abnormalities with age as they acquire secondary mutations and are influenced by the environment. Such disadvantaged animals would likely be outcompeted from a population over time.
Our work mainly focused on the most prominent FIP loss-of-function phenotype of large DNA masses in the brain to hypothesize that FIP is required for proper chromosome separation during mitosis. We then show that FIP localizes to, and stabilizes, antiparallel interzonal MTs at the latest stages of cytokinesis. Given that overexpression of FIP did not force its localization onto the MT lattice in interphase or on astral MTs in mitosis, we reasoned that FIP was likely not a MAP and that FIP regulates interzonal MT stability through one of these candidate proteins: EB1, Klp61F (kinesin-5, Eg5 in mammals), Asp, or Feo. Using mutant analysis (fip∆1-2 eliminated EB1 as a candidate), protein localization of FIP::GFP (not localized to the spindle in metaphase eliminated Klp61F as a candidate), and direct protein–protein interaction via Y2H analysis (an interaction with Feo, but not Asp), we determined that FIP binds MTs via the cross-linking protein Feo.
Interestingly, our interaction data suggested that FIP enhances Feo’s MT cross-linking role via the N-terminal dimerization domain of Feo (Supplemental Figure S9) and not by acting directly on Feo’s C-terminal MT-binding domain. This allows us to propose several testable models of FIP function. One model is that FIP could directly enhance the dimerization state of Feo, which is required for cross-linking two MTs (Supplemental Figure S9). Loss of FIP in this model would lead to a shift toward a monomeric state of Feo, leading to reduced MT cross-linking and effectively reduced interzonal MT stability. A second model of FIP function is based on previous work showing that Kif4 binds to the N-terminal dimerization and rod domains of PRC1 to recruit MT catastrophe factors and facilitate antiparallel MT “pruning” (Bieling et al., 2010; Nguyen et al., 2018), which maintains proper MT overlap from each spindle pole. Loss of Kif4 leads to increased MT overlap, elongated spindles, and broadening of PRC1 localization in the central spindle (Zhu and Jiang, 2005; Shrestha et al., 2012; Nguyen et al., 2018). Because loss of FIP gives the opposite result where the overlapping interzonal MTs rapidly dissipate, this second model posits that FIP normally works to maintain the correct amount of MT overlap by limiting the amount of Klp3A (Drosophila Kif4) bound to Feo. In this scenario, loss of FIP would result in increased Klp3A levels, increased recruitment of catastrophe factors, and elevated interzonal MT pruning.
Additional models would include mechanisms by which FIP modulates Feo function via competitive or cooperative binding with the many other Feo-binding partners. These proposed models represent exciting future directions and will benefit greatly from in vitro biochemical studies using purified components. Such experiments will also help identify a possible mammalian functional orthologue of FIP.
MATERIALS AND METHODS
Cell culture, transfection, vectors, and dsRNA knockdown
Drosophila S2 cells (Invitrogen) were maintained in SF900 media (Life Technologies) supplemented with Antibiotic-Antimycotic (Life Technologies) at 25°C. FIP (isoform B) and Feo were PCR amplified and cloned into Entry vectors for the Gateway system (Invitrogen) using the following primers: FIP Forward 5′-CACCATGTCTGGCCTCAAGAAATTCC-3′, FIP Reverse 5′-AAACTTTTGGGCATCTCTTATTGT-3′, Feo Forward 5′-CACCATGAACTCGCCGAGCGCCATTG-3′, and Feo Reverse 5′-GAACTGTC-TGCGCGGCTGCACG-3′. We used the manufacturer’s protocol in combination with Gateway destination vectors from the Drosophila Gateway Vector Collection, and our personal collection, to generate fusion constructs tagged with GFP, Flag, mNeonGreen, TagRFP, or Halo under the control of the Actin5c promoter. The following constructs were generated: FIP::GFP, GFP::FIP, TagRFP::FIP, mNeonGreen::Feo, Flag::FIP, GFP::Feo, and GFP::EB1. Transfection was performed as described previously (Schoborg et al., 2015) with the following differences: 1 μg of vector was used for (1–5) million cells. For dsRNA treatment, ∼5 million cells were collected 2–3 d after passaging and resuspended with 2 ml of fresh SF900 media with 20 µg dsRNA. Every 2 d, media were replaced with fresh media and dsRNA. Cells were either fixed and stained on day 6 or transfected on day 4 and allowed to grow for two additional days before imaging. An identically treated well lacking dsRNA was included to control for growth conditions. The following primers sequences were obtained from the Harvard Drosophila RNAi Screening Center and used to generate templates for T7 RNA synthesis reactions (Promega) from FIP cDNA: 5′-ATGGCTGCAAAAAGGCTAC-3′, 5′-CTAAACGTCGACGAATTGTT-3′.
Staining and immunofluorescence
S2 cells were allowed to adhere to concanavalin A (Con A)-coated coverslips for 20–30 min, briefly washed with PBS, and then fixed with either prechilled anhydrous methanol for 20 min at –20°C or 4% paraformaldehyde (PFA; Electron Microscopy Sciences) in PBS for 20 min at room temperature. If necessary, cells were counterstained for 1 h with DAPI and Alexa 568–conjugated phalloidin (Invitrogen), rinsed 3×, and then mounted in Vectashield (Vector Laboratories). Phalloidin was desiccated in a glass well and resuspended in PBS before using to remove methanol. Larval brains and wing discs were obtained from wandering third instar larvae and fixed as inverted carcasses in 4% PFA diluted in PBS with 0.05% Triton X-100 for 30 min at room temperature while rotating. The only exception to this protocol was used to prepare the wing discs in Figure 8, which were fixed in cold PBS and 0.01% Tween-20 (PBST) with 4% PFA, as the anti-Pnut antibody did not work otherwise. All tissues were dissected from the fixed carcasses in Schneider’s Drosophila Medium (Life Technologies) supplemented with Antibiotic-Antimycotic (Life Technologies). Tissues were blocked with 5% normal goat serum (NGS) in PBS with 0.05% or 0.3% Triton-X 100 (PBSTx) for 1 h at room temperature and incubated with primary antibodies diluted in PBSTx + 5% NGS overnight at 4°C. Samples were washed 3× in PBSTx for 20–30 min to 1 h at room temperature and then incubated with secondary antibodies diluted in PBSTx + 5% NGS with DAPI and phalloidin for 2–24 h at room temperature or 4°C. Tissues were washed 3x in PBSTx and then mounted in Vectashield. The following primary antibodies were used: mouse anti-Pnut 1:200 (4C9H4 DSHB), mouse anti-lamin 1:100 (ADL84.12 DSHB), mouse anti-GFP (JL-8) 1:5000 (Clonetech), mouse anti-flag 1:10,000 (M2 Sigma-Aldrich), rabbit anti-GFP 1:1000 (ab 290 AbCam), rabbit anti-phosphorylated histone H3 1:1000 (Millipore), and guinea pig anti-Asl antibodies (1:1500; gift from G. Rogers, University of Arizona, Tucson, AZ). Goat secondary antibodies raised against the appropriate species were Alexa Fluor 488–, 568–, or 647–conjugated 1:500 (Invitrogen). For live imaging of Halo-tagging proteins, cells were stained with the JF549 ligand for >1 h prior to live imaging. A 1-uM stock solution of JF549 in dimethyl sulfoxide was added to culture media at 1:200 for a final working concentration 5 nM of ligand. For fixed imaging of Halo-tagged proteins, S2 cells expressing GFP-Tub and Ht-Sip2 were incubated for 1 h with 100 nM Tetramethylrhodamine Halotag Ligand (TMR-HL), washed several times with PBS, fixed for 10 min in cold MeOH, rehydrated in PBST, and stained for 1 h with DAPI prior to mounting.
Immunoprecipitation and immunoblotting
For immunoprecipitation (IP), transfected Drosophila S2 cells (4 ml) (either treated with 30 µM colchicine for 16 h to increase the mitotic index to ∼20% or treated with ethanol as a control) were spun down, had the supernatant removed, and then resuspended in 1 ml of RIPA buffer (10 mM Tris-HCl, pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid [EGTA], 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and 0.25 μM protease inhibitors [Pierce Protease inhibitor mini EDTA Free; Thermo Fisher Scientific, Waltham, MA]). Fifty microliters of the lysate was removed and saved as the “input” sample, whereas the rest of the lysate was combined with Protein-A–conjugated dynabeads (Life Technologies) prebound to antibody (see below) and incubated for 2 h at 4°C, rotating. The supernatant of the lysate and bead mixture was removed and the beads were washed 3× in lysis buffer. The beads were moved to a fresh tube during the final wash, eluted with 30 μl of Laemmli buffer, and boiled for 5 min. For antibody binding, 50 μl of dynabeads and 1 μl of rabbit anti-gfp antibody (ab 290 AbCam) were added to 1 ml of PBST. The bead mixture was incubated at 4°C, rotating, for 1–2 h. The elute was removed and resolved on SDS–PAGE, along with the input samples, and transferred onto nitrocellulose membrane. Western blots were stained with primary antibodies (mouse anti-GFP [JL-8] 1:5000 [Clontech] or mouse anti-flag 1:10,000 [M2 Sigma-Aldrich]) and anti-mouse peroxidase-conjugated secondary antibodies 1:5000 (Thermo Fisher Scientific). Western blots were visualized using SuperSignal West Dura Extended Substrate (Life Technologies) and imaged using a ChemiDoc MP Imaging System (Bio-Rad).
Fly stocks and genetics
The following fly strains were used: Cyo,P{GFPnls}, Cyo,P{2xTb1-RFP}, TM6B,Tb1, AntpHu, and TM6C,Sb1,Tb1 balancer chromosomes; Tub-GAL-4 (Bloomington stock (BS) #5138); sqh-GAL4 and ubi-Moe::GFP/CyO; TM3/TM6 (gift from D. Kiehart, Duke University); Dpn-GAL4 (gift from M. Sato, Kanazawa University); UAS-EB1-GFP (gift from S. Rogers, University of North Carolina at Chapel Hill); G147 (Morin et al.); H2AV-mRFP (BS #23650); Df(2L)Exel7029 (BS #7802; Sep2::GFP (BS #26257); Ubi-p63E-feo::GFP (BS #59274); Ubi-p63E-feo::mCherry (BS #59278); UAS-FeoRNAi (HM) (BS #28926); and UAS-FeoRNAi (GL) (BS #35467). All fly stocks were maintained on standard cornmeal-agar media at room temperature (19–23°C), and experimental crosses were kept at 25°C until fixation or live imaging. Figure 7, A and B, was obtained from homozygous fip7 larvae, in all other cases, the genotype fip- refers to fip7/Df(2L)Exel7029.
CRISPR and transgenic animals
Two CRISPR guide RNA sites were determined using http://flycrispr.molbio.wisc.edu/: 5′-GAATACTATTGCCAGAAGGT-3′ and 5′-GCGACGCTGAGGAATACCAG-3′. Each guide was cloned into a separate pU6-Bbsl plasmid, and equimolar amounts were injected into Cas9 embryos by BestGene (Chino Hills, CA). Injected males were mated to yw females and progeny were screened for nonhomologous end joining (NHEJ), excising ∼80% of the FIP locus, by single wing PCR using the primers 5′-GCAAAGGCGCGTCGATCGTTGGC-3′ and 5′-TAGCGGAGCAGTACCAGACTTCTGGG-3′ (Supplemental Figure S3B shows one example). We isolated seven clones and sequenced the FIP locus of each, revealing five different alleles, each of which differed slightly in the exact NHEJ product. We found no significant difference in the penetrance/severity of our primary phenotype (polyploid neuroblasts in the larval brain). Therefore, we randomly selected fip7 for use in this study.
Microscopy and live cell imaging
Live and fixed cell imaging was performed on a Nikon Eclipse Ti inverted microscope equipped with a CSU22 spinning disk unit (Yokogawa) and an ORCA-Flash4.0 CMOS camera (Hamamatsu) with either a 40x/1.30 NA plan Fluor objective, sometimes in conjunction with a 1.5× tube lens, or a 100×/1.49 NA TIRF objective. Solid state laser lines at 405, 491, 561, and 642 nm (VisiTech International). Microscope control and image acquisition were performed using Metamorph (version 7.8.13.0).
Live cell imaging in Supplemental Figures S1, A and B, and S2A and fixed imaging in Supplemental Figure S3C were performed in superresolution mode on a Zeiss LSM 880 equipped with an Airyscan module using a 63×/1.4 NA objective. Figure 4, A–C, and Supplemental Figures S3B and S4A were collected using Airyscan and a Fast module set for 0.5× Nyquist sampling on a 40×/1.4NA Plan-Apochromat objective. All image acquisition was performed using Zen Black software (version 2.3), where Airyscan technology was utilized, raw data were processed and deconvolved within Zen, and processing strength was automatically computed within Zen.
For live imaging of S2 cells, resuspended cells (200 µl) were plated onto a glass-bottom, 35-mm dish (MatTek) coated with 15 µg of Con A and allowed to settle for 20–30 min before imaging. For extended time-lapse imaging, 1.5 ml of conditioned SF900 media was added to the dish, instead of fresh Schneider’s medium, after cells had adequately adhered. When imaging cytokinesis in S2 cells, uncoated dishes were used, as Con A inhibits cytokinesis, and cells were imaged immediately. In Figure 2B, Hoechst 33342 and SiR-tubulin (cytoskeleton) were used at 1.5 and 0.1 µM, respectively.
Larval brains and wing discs were dissected from wandering third instar larvae in Schneider’s Drosophila Medium (Life Technologies) supplemented with Antibiotic-Antimycotic (Life Technologies). Brains and wing discs were briefly washed into PBS and then gently adhered to a poly-l-lysine-coated, glass-bottom 35-mm dish in PBS before replacing PBS with 1.5 ml Schneider’s medium. Wing discs were mounted with the peripodial epithelium facing the coverslip, and brains were mounted with the ventral surface facing the coverslip. In all cases, we used #1.5 coverslips.
Larval brain squashes for ploidy analysis
Dissected third instar larval brains were transferred to a clean drop of 0.7% saline with 0.1 mM Colchicine and incubated for 1.5 h at 25°C. The dissected brains were then transferred to a drop of 0.5% citrate solution and incubated at room temperature for 8 min. The brains were then fixed in acetic acid:methanol:ddH20 (6:6:1) for 20 s, and then individual brains were transferred to 2 μl of 45% acetic acid on separate 20 × 40-mm coverslips and incubated for 2 min. A 22 × 22-mm siliconized coverslip was then placed over the larger coverslip containing the dissected brains, and this assembly was then placed on a glass slide used as a support. The brains were then squashed very hard on the bench top. The complete assembly was then placed on dry ice for 10 min. The smaller coverslip was removed, and the larger coverslip containing the squashed brains was incubated in 95% EtOH at –20°C for 15 min. The larger coverslips were left to air-dry and then incubated in 2x saline sodium citrate (SSC) solution (KD Medical) for 5 min. The coverslips were stained with 1x DAPI for 5 min and then rinsed with fresh 2x SSC for 5 s. The coverslips were then air-dried in a vertical position and mounted onto glass slides with Vectashield.
Image processing and analysis
In all cases where measurements were inherently subjective, images and image series were randomly blinded before analysis to avoid cognitive bias. In all cases where live measurements were compared among multiple conditions, all imaging was performed on the same day to avoid changes in laser level or ambient temperature which could influence fluorescence or cell physiology.
All image processing, quantification, and false coloring was performed using FIJI (ImageJ; National Institutes of Health, Bethesda, MD). Image quantification was performed exclusively on raw data. If rotation was necessary (e.g., to generate kymographs or to reorient tissues), images were rotated only once and bicubic interpolation was used. Unless disclosed here, image manipulation was limited to linear contrast stretching to best represent the main message of each image. Figure 1B and Supplemental Figure S4E were filtered of outlying (hot) pixels using the “remove outliers” command, with a threshold of 75 Gy values (∼50% of the true dynamic range). Images in Supplemental Figure S1B were rotated so that each tip is oriented with the +end facing right.
All image series used for temporal-color coding were first corrected for bleaching using histogram matching so that truly static pixels present as gray in the resulting color-coded image and not a reddish-yellow. Color-coded projections were generated using the Temporal-Color Code feature in FIJI. Images in Figure 5 were filtered with a gamma correction (gamma = 0.65 for phalloidin and 0.75 for DNA) to better represent a mix of bright and dim signals and rotated so that the anterior of the brain faced up.
Kymographs were generated by rotating an image series so that movements of interest occurred predominantly along the x-axis. A rectangular region encompassing the region of interest was resliced without interpolation and maximum intensity projected so that each frame of the original image series represents a single line of pixels on the resulting kymograph.
Enrichment measurements of FIP at MT tips and spindle midzones, and Moesin in the nascent cytokinetic furrow, were calculated by manually defining a region of interest (microtubule tip, nascent furrow, or spindle midzone), and a control region adjacent to the region of interest. Enrichment was calculated using the following equation:
 |
Microtubule growth rates were calculated using a manual tracking plugin for Fiji; each data point represents the average speed of a single MT. Multinucleate, metaphase, and anaphase frequency in FIP dsRNA-treated S2 cells were calculated by dividing the number of multinucleate, metaphase, and anaphase cells by the area of a single field of view (∼37,000 µm2). Micronuclei and binucleate frequency were calculated by dividing the number of aberrant cells in a single wing disc epithelium by the total number of scorable cells (i.e., those easily represented by one or two focal planes) in the region.
NEB to anaphase timing was determined based on a sudden change in the nuclear outline and the first detectable separation of sister chromatids, respectively. Cytokinesis timing was calculated by counting the time between the formation of the nascent furrow and the last frame where a clear separation between furrows could be resolved.
Maximum sister chromosome separation was determined by measuring the space between H2AV masses at their most distant point (153 ± 33 s after anaphase in mutants, 195 ± 17 s in controls). Midbody duration was scored by calculating the time from anaphase onset until the GFP::Jupiter, GFP::Sep2, or GFP::Moesin fluorescence at the midbody was no longer detectable above the background cytoplasmic fluorescence. Only midbodies that persisted within the central slices of the Z stack were used for analysis to avoid midbodies that simply drifted out of the focal plane.
Polyploid cells in the larval CNS were scored based on intense DAPI staining, a larger than average nucleus, abnormal DNA architecture, and multiple nuclei (determined by lamin staining) contained within the same cell. For quantifications, polyploid cells were counted as “one” even if they contained multiple fragmented nuclei or multiple intact nuclei within the same cell as determined by actin staining. For the large DNA aggregates reported for the fip- and Feo codepleted animals, those polyploid cells were also quantified as one polyploid cell, although it is hypothesized that multiple polyploid neuroblasts may have fused together to form the large DNA aggregates based on the dramatically larger cell size and DNA content within those neuroblasts. Polyploid nuclei of the double mutants were considered large single or large clustered masses if the cross-sectional area of the DNA exceeded 1000 µm2.
Yeast-two-hybrid analysis
FIP, Feo, Asp, Sep1 and Sep2 full-length, and FIP and Asp pieces were amplified from cDNA clones by PCR using Phusion (Thermo Fisher Scientific) and with the primers shown in Supplemental Table 1. PCR products were then introduced into Gateway Entry vectors using the pENTr/D-TOPO Kit (Thermo Fisher Scientific). All Y2H experiments were then conducted as described in Galletta and Rusan (2015). In brief, FIP, Feo, Asp, Sep1 and Sep2 full-length, FIP, Feo, and Asp pieces were introduced into pDEST-pGADT7 and pDEST-pGBKT7 using Gateway technology (Thermo Fisher Scientific), transformed into Y187 or Y2HGold yeast strains (Takara Bio USA, Mountain View, CA), and grown in −Leu or −Trp media. After mating bait and prey strains, diplods containing both were selected on −Leu, −Trp (DDO) plates and then replica-plated onto plates of increasing stringency: DDO; −Ade, −Leu, −Trp, −Ura (QDO); −Leu, −Trp plates supplemented with Aureobasidin A (Takara Bio USA) and X-α-Gal (Gold Biotechnology, St. Louis, MO) (DDOXA); and −Ade, −Leu, −Trp, −Ura plates supplemented with Aureobasidin A and X-α-Gal (QDOXA). Interactions were scored based on growth and the development of blue color as appropriate. All plasmids were tested for the ability to drive reporter activity in the presence of an empty vector (autoactivation). Plasmids that conferred autoactivity were omitted from further analysis.
Supplementary Material
Acknowledgments
We thank Brian Galletta and Matthew Hannaford for critical reading and discussion of the manuscript and Ryan O’Neill for significant help with fly genetics. We thank Xufeng Wu and Chris Combs at the NHLBI Light Microscopy core facility for training, guidance, and discussion. This work is supported by the Division of Intramural Research at the National Institutes of Health/National Heart, Lung, and Blood Institute (1ZIAHL006126 to N.M.R.).
Abbreviations used:
- Asp
abnormal spindle
- BS
Bloomington stock
- Cdk1
cyclin-dependent kinase 1
- Con A
concanavalin A
- DAPI
diamidine-phenylindole dihydrochloride
- EB1
end-binding 1
- EB1 OE
overexpressed EB1
- Feo
Fascetto
- FIP
Fascetto interacting protein
- FIP FL
full-length FIP
- Ht
Halo-tag
- IP
immunoprecipitation
- MAPs
MT-associated proteins
- MT
microtubule
- NB
neuroblast
- NEB
nuclear envelope breakdown
- NGS
normal goat serum
- NHEJ
nonhomologous end joining
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- Plk1
polo-like kinase
- Pnut
peanut
- PRC1
protein regulator of cytokinesis 1
- Sip
septin-interacting proteins
- SSC
saline sodium citrate
- SxIP
Ser-x-Ile-Pro
- TMR-HL
Tetramethylrhodamine Halotag Ligand
- VNC
ventral nerve cord
- Y2H
yeast 2 hybrid.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-09-0573) on February 6, 2019.
REFERENCES
- Bassi ZI, Audusseau M, Riparbelli MG, Callaini G, D’Avino PP. (2013). Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc Natl Acad Sci USA , 9782–9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. (2010). A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell , 420–432. [DOI] [PubMed] [Google Scholar]
- Booth DG, Beckett AJ, Molina O, Samejima I, Masumoto H, Kouprina N, Larionov V, Prior IA, Earnshaw WC. (2016). 3D-CLEM reveals that a major portion of mitotic chromosomes is not chromatin. Mol Cell , 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Archambault V, Przewloka MR, Zhang W, Lilley KS, Laue E, Glover DM. (2007). Recruitment of Polo kinase to the spindle midzone during cytokinesis requires the Feo/Klp3A complex. PLoS One , e572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Giansanti MG, Petronczki M. (2015). Cytokinesis in animal cells. Cold Spring Harb Perspect Biol , a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, de Lange T. (2011). The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. :585–610. [DOI] [PubMed] [Google Scholar]
- Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P. (2011). Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis , 125–132. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Rusan NM. (2015). A yeast two-hybrid approach for probing protein-protein interactions at the centrosome. Methods Cell Biol , 251–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. (2007). Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev , 157–162. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. (2003). A protein interaction map of Drosophila melanogaster. Science , 1727–1736. [DOI] [PubMed] [Google Scholar]
- Glotzer M. (2017). Cytokinesis in metazoa and fungi. Cold Spring Harb Perspect Biol 9, a022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. (2012). Cytokinesis in animal cells. Annu Rev Cell Dev Biol , 29–58. [DOI] [PubMed] [Google Scholar]
- Hirose K, Kawashima T, Iwamoto I, Nosaka T, Kitamura T. (2001). MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J Biol Chem , 5821–5828. [DOI] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell , 366–376. [DOI] [PubMed] [Google Scholar]
- Hu CK, Ozlu N, Coughlin M, Steen JJ, Mitchison TJ. (2012). Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol Biol Cell , 2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Goshima G. (2015). Microcephaly protein Asp focuses the minus ends of spindle microtubules at the pole and within the spindle. J Cell Biol , 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. (2000). CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol , 1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R. (1998). PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell , 877–885. [DOI] [PubMed] [Google Scholar]
- Jiang K, Toedt G, Montenegro Gouveia S, Davey NE, Hua S, van der Vaart B, Grigoriev I, Larsen J, Pedersen LB, Bezstarosti K, et al (2012). A proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol , 1800–1807. [DOI] [PubMed] [Google Scholar]
- Kellogg EH, Howes S, Ti SC, Ramirez-Aportela E, Kapoor TM, Chacon P, Nogales E. (2016). Near-atomic cryo-EM structure of PRC1 bound to the microtubule. Proc Natl Acad Sci USA , 9430–9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Kumar S, Mizoguchi A, Ide C, Kinoshita A, Haraguchi T, Hiraoka Y, Noda M. (1997). Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev , 1535–1547. [DOI] [PubMed] [Google Scholar]
- Kumar P, Chimenti MS, Pemble H, Schonichen A, Thompson O, Jacobson MP, Wittmann T. (2012). Multisite phosphorylation disrupts arginine-glutamate salt bridge networks required for binding of cytoplasmic linker-associated protein 2 (CLASP2) to end-binding protein 1 (EB1). J Biol Chem , 17050–17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. (2004). Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J , 3237–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Maddox AS. (2012). Cytokinesis, ploidy and aneuploidy. J Pathol , 338–351. [DOI] [PubMed] [Google Scholar]
- Li J, Dallmayer M, Kirchner T, Musa J, Grunewald TGP. (2018). PRC1: linking cytokinesis, chromosomal instability, and cancer evolution. Trends Cancer , 59–73. [DOI] [PubMed] [Google Scholar]
- Liu J, Fairn GD, Ceccarelli DF, Sicheri F, Wilde A. (2012). Cleavage furrow organization requires PIP(2)-mediated recruitment of anillin. Curr Biol , 64–69. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang Z, Jiang K, Zhang L, Zhao L, Hua S, Yan F, Yang Y, Wang D, Fu C, et al (2009). PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis. J Biol Chem , 23059–23071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox AS, Lewellyn L, Desai A, Oegema K. (2007). Anillin and the septins promote asymmetric ingression of the cytokinetic furrow. Dev Cell , 827–835. [DOI] [PubMed] [Google Scholar]
- Mierzwa B, Gerlich DW. (2014). Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell , 525–538. [DOI] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M. (2002). Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell , 41–54. [DOI] [PubMed] [Google Scholar]
- Mishima M, Lee KY. (2015). Central spindle robustness by PRC1-centralspindlin interaction. Cell Cycle , 3515–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol , 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Kawajiri A, Matsui S, Takagishi M, Shiromizu T, Saitoh N, Izawa I, Kiyono T, Itoh TJ, Hotani H, Inagaki M. (2003). Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules. J Biol Chem , 18538–18543. [DOI] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA. (2007). Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol , 436–444. [DOI] [PubMed] [Google Scholar]
- Nguyen PA, Field CM, Mitchison TJ. (2018). Prc1E and Kif4A control microtubule organization within and between large Xenopus egg asters. Mol Biol Cell , 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Engstrom L, Mahowald AP. (1989). Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on the X chromosome. Genetics , 333–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton JS, Cuningham JC, Peifer M. (2014). Acentrosomal Drosophila epithelial cells exhibit abnormal cell division, leading to cell death and compensatory proliferation. Dev Cell , 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton JS, Cuningham JC, Peifer M. (2017). Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size. J Cell Biol , 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G, Glover DM, Avides Mdo C. (2002). A requirement for the Abnormal Spindle protein to organise microtubules of the central spindle for cytokinesis in Drosophila. J Cell Sci , 913–922. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Akong K, Peifer M. (2008). Putting the model to the test: are APC proteins essential for neuronal polarity, axon outgrowth, and axon targeting? J Cell Biol , 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM. (1997). The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J Cell Biol , 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg T, Zajac AL, Fagerstrom CJ, Guillen RX, Rusan NM. (2015). An Asp-CaM complex is required for centrosome-pole cohesion and centrosome inheritance in neural stem cells. J Cell Biol , 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Hales KG, Pringle JR, Peifer M. (2002). Identification of septin-interacting proteins and characterization of the Smt3/SUMO-conjugation system in Drosophila. J Cell Sci , 1259–1271. [DOI] [PubMed] [Google Scholar]
- Shrestha S, Wilmeth LJ, Eyer J, Shuster CB. (2012). PRC1 controls spindle polarization and recruitment of cytokinetic factors during monopolar cytokinesis. Mol Biol Cell , 1196–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth JT, Beg AM, Wu S, Putney JW, Jr, Rusan NM. (2012). Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr Biol , 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis ET, Kinoshita M, Nelson WJ. (2005). A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science , 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Ti SC, Tan L, Darst SA, Kapoor TM. (2013). Marking and measuring single microtubules by PRC1 and kinesin-4. Cell , 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. (2010). Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell , 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Fasulo B, Warecki B, Sullivan W. (2016). Tum/RacGAP functions as a switch activating the Pav/kinesin-6 motor. Nat Commun , 11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Yuh P, Heald R. (2005). The perichromosomal layer. Chromosoma , 377–388. [DOI] [PubMed] [Google Scholar]
- Vaughan KT. (2005). TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J Cell Biol , 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbrugghe KJ, White JG. (2004). SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr Biol , 1755–1760. [DOI] [PubMed] [Google Scholar]
- Verni F, Somma MP, Gunsalus KC, Bonaccorsi S, Belloni G, Goldberg ML, Gatti M. (2004). Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr Biol , 1569–1575. [DOI] [PubMed] [Google Scholar]
- Wakefield JG, Bonaccorsi S, Gatti M. (2001). The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J Cell Biol , 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Brust-Mascher I, Scholey JM. (2015). The microtubule cross-linker Feo controls the midzone stability, motor composition, and elongation of the anaphase B spindle in Drosophila embryos. Mol Biol Cell , 1452–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EA, Glotzer M. (2012). Centralspindlin: at the heart of cytokinesis. Cytoskeleton , 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, et al (2013). Pan-cancer patterns of somatic copy number alteration. Nat Genet , 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Jiang W. (2005). Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci USA , 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Lau E, Schwarzenbacher R, Bossy-Wetzel E, Jiang W. (2006). Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc Natl Acad Sci USA , 6196–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.