Abstract
Mitogen-activated protein kinases (MAPKs) mediate numerous eukaryotic signaling responses. They also can modulate their own signaling output via positive or negative feedback loops. In the yeast pheromone response pathway, the MAPK Fus3 triggers negative feedback that dampens its own activity. One target of this feedback is Ste5, a scaffold protein that promotes Fus3 activation. Binding of Fus3 to a docking motif (D motif) in Ste5 causes signal dampening, which was proposed to involve a central cluster of phosphorylation sites in Ste5. Here, we reanalyzed the role of these central sites. Contrary to prior claims, phosphorylation-mimicking mutations at these sites did not impair signaling. Also, the hyperactive signaling previously observed when these sites were mutated to nonphosphorylatable residues arose from their replacement with valine residues and was not observed with other substitutes. Instead, a cluster of N-terminal sites in Ste5, not the central sites, is required for the rapid dampening of initial responses. Further results suggest that the role of the Fus3 D motif is most simply explained by a tethering effect that promotes Ste5 phosphorylation, rather than an allosteric effect proposed to regulate Fus3 activity. These findings substantially revise our understanding of how MAPK feedback attenuates scaffold-mediated signaling in this model pathway.
INTRODUCTION
Protein kinases play a central role in response to extracellular signals. In eukaryotes, the mitogen-activated protein kinase (MAPK) cascade is a common and versatile signaling module consisting of a series of sequentially acting protein kinases. A thoroughly studied example functions in the mating pathway of budding yeast, Saccharomyces cerevisiae, which for decades has served as a model system for investigating eukaryotic signal transduction (Bardwell, 2005; Alvaro and Thorner, 2016). In this pathway (Figure 1A), a MAPK cascade is activated in response to extracellular mating pheromones; these bind a G protein–coupled receptor (GPCR), which then triggers dissociation of a heterotrimeric G protein (Gαβγ). The liberated Gβγ dimer activates the downstream MAP kinase cascade via a crucial intermediary, Ste5, a scaffold protein that has binding sites for Gβγ as well as the pathway kinases (Figure 1B). When activated by pheromone, Gβγ recruits Ste5 to the plasma membrane, and this both initiates and propagates activation of the kinase cascade (Pryciak and Huntress, 1998; Lamson et al., 2006; Maeder et al., 2007; Zalatan et al., 2012). Ultimately, the cascade activates the MAPK Fus3 (plus its semiredundant paralogue, Kss1), which then stimulates downstream mating responses such as cell-cycle arrest, transcriptional induction, and morphological changes.
FIGURE 1:

Pheromone response and negative feedback. (A) The pheromone response pathway. Binding of pheromone to the GPCR triggers dissociation of the G protein heterotrimer (Gαβγ). The free Gβγ dimer then stimulates membrane recruitment of the scaffold protein, Ste5. A cascade of kinase activation ultimately activates the MAPK Fus3, which stimulates both downstream mating responses and negative feedback, which dampens the response. Ste5 is one of several targets of Fus3-mediated negative feedback. (B) Domain structure of Ste5. Names of domains and motifs are shown below the linear structure, and their binding targets are indicated above. SP/TP motifs, which represent minimal phosphorylation sites for MAPKs and CDKs, are marked in red. (C) Ste5 has two clusters of SP/TP sites. The central cluster was originally implicated as a target of Fus3 negative feedback. In those studies, the Thr and Ser sites were mutated to nonphosphorylatable Val and Ala residues, designated 2V2A (or “abcd” in Malleshaiah et al., 2010), as well as to phosphomimetic Glu residues, designated 4E (or “EFGH” in Malleshaiah et al., 2010).
In addition to its positive role in promoting signaling responses, the MAPK Fus3 activates a negative feedback loop that attenuates pathway signaling (Figure 1A). For example, if Fus3 is catalytically inactivated, it becomes more highly phosphorylated by its upstream kinase (Gartner et al., 1992), implying that the Fus3 normally limits its own activation by inhibiting an upstream step. One candidate target of this Fus3-mediated feedback is the scaffold protein Ste5, as its initial pheromone-stimulated membrane localization is subsequently attenuated in a manner that depends on Fus3 kinase activity (Yu et al., 2008; Bush and Colman-Lerner, 2013). Additional candidates include Ste7 (the kinase that activates Fus3) (Zhou et al., 1993; Hao et al., 2012), Msg5 (a phosphatase that dephosphorylates Fus3) (Doi et al., 1994; Nagiec et al., 2015), and Ste18 (the Gγ subunit of the G protein) (Choudhury et al., 2018). Feedback regulation of these multiple targets need not be mutually exclusive and may instead operate in concert. This report will focus on the role of Ste5 as both target and mediator of negative feedback regulation.
The region of Ste5 that binds Fus3 is a short peptide sequence called the docking motif, or D motif (see Figure 1B). Mutating this site to a nondocking variant (ND) causes hyperactive signaling (Bhattacharyya et al., 2006), which indicates that the Fus3-Ste5 interaction is not required for signaling but instead serves to attenuate pathway output. Additional evidence suggested that Fus3 regulates signaling by phosphorylating Ste5 at a central cluster of four SP/TP sites, near the D motif (Figure 1, B and C). A mutation at one of these four sites (T287V) was found to cause hyperactive signaling, similar to (though not as strong as) the ND mutation (Bhattacharyya et al., 2006). A later study mutated all four of these sites to nonphosphorylatable Val and Ala residues (here called 2V2A; see Figure 1C), which also caused hyperactive signaling (Malleshaiah et al., 2010). Conversely, mutation of these sites to phospho-mimicking Glu residues (here called 4E; see Figure 1C) was reported to inhibit signaling (Malleshaiah et al., 2010). Hence, these findings suggested that Fus3 dampened the pheromone response by phosphorylating these four sites in Ste5. It was further suggested that their phosphorylation is antagonized by the phosphatase Ptc1, because cells lacking Ptc1 (ptc1∆) showed poor pheromone response (Malleshaiah et al., 2010).
In addition to the central cluster of phosphorylation sites implicated in negative feedback, Ste5 contains a separate cluster of phosphorylation sites in its N-terminus, flanking a plasma membrane-binding domain (Figure 1, B and C). The sites in this region were originally found to be phosphorylated by a cyclin-dependent kinase (CDK), which inhibits Ste5 membrane localization and pheromone response in cells that have entered the cell division cycle (Strickfaden et al., 2007). CDK phosphorylation of these sites depends on a separate docking sequence (the LP motif, Figure 1B) that is specific for the cyclins Cln1 and Cln2 (Bhaduri and Pryciak, 2011; Bhaduri et al., 2015). Although these two clusters of phosphorylation sites were previously implicated in distinct regulatory circuits, we recently found that CDK and Fus3 activity can each promote phosphorylation of both clusters in vivo (Repetto et al., 2018). This finding is consistent with the fact that CDKs and MAPKs are related kinases that share the same minimal phosphorylation motif (SP or TP). Moreover, the same study revealed that Fus3-dependent control of Ste5 membrane association depended on the N-terminal cluster rather than the central cluster. This surprising finding caused us to revisit the role of the central cluster in regulating pathway output.
Here, we reanalyze how these two clusters of phosphorylation sites relate to the role of Fus3-Ste5 docking, negative feedback, and signaling hyperactivity. In particular, we investigated whether the central cluster is required for signal dampening by Fus3. We observed several conflicts with the previously reported findings. The collective results argue that docking-dependent attenuation of pathway output does not require the ability to modify the central cluster, and instead, depends on the N-terminal cluster. Our findings substantially alter previous interpretations of signaling regulation and feedback control in this well-studied pathway.
RESULTS
Phospho-mimicking mutations at the central sites in Ste5 do not disrupt signaling
To reinvestigate the role of the central sites in controlling downstream signaling outputs, such as MAPK phosphorylation and transcriptional induction, we began by assaying a previously described Ste5 variant with mutations that mimic phosphorylation at all four sites (4E), plus two others that harbored mutations to nonphosphorylatable residues: a previous 2V2A mutant (i.e., Ser/Thr residues replaced with Ala/Val) and a newer 4Q mutant (i.e., replaced with uncharged Gln residues). We observed two conflicts with the earlier results (Malleshaiah et al., 2010), which will be elaborated in turn below: 1) the 4E mutant did not show reduced signaling, and 2) the strong hyperactivity of the 2V2A mutant was not shared by the 4Q mutant.
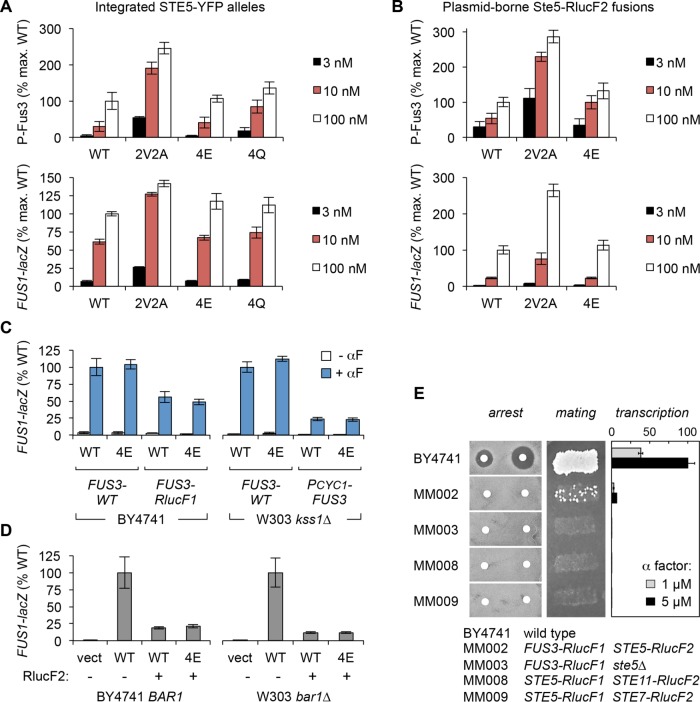
Our experiments did not confirm the claim that the 4E mutation disrupts Ste5 signaling. Instead, we found that the 4E mutant was phenotypically indistinguishable from wild type (WT) in assays of either Fus3 phosphorylation or transcriptional response and regardless of pheromone dose (Figure 2, A and B). Moreover, this was true in the context of either an integrated STE5-YFPx3 allele (Figure 2A) or the same plasmid-borne STE5-RlucF2 alleles used in the prior study (Figure 2B). Testing in several other contexts also failed to reveal a discernable defect for the 4E mutant (Figure 2C); these included the same strain background used previously (BY4741), strains lacking the alternate MAPK Kss1, and strains with reduced Fus3 expression (PCYC1-FUS3, which we hypothesized might increase sensitivity to mild defects). The reason for this stark discrepancy from the previous study is not certain, but one other finding is suggestive. We noticed that split-luciferase gene fusions used in the prior study (RlucF1 and RlucF2) showed substantially reduced signaling activity; for example, signaling was markedly lower for the Ste5-RlucF2 fusion than for unfused Ste5 (Figure 2D), and strains with other luciferase fusions had mild (Figure 2C) to severe (Figure 2E) defects. Hence, because the 4E mutant was previously tested as a Ste5-RlucF2 fusion, it might have appeared to be defective if it was inadvertently compared with an unmatched (e.g., unfused) WT control. Overall, after subjecting the Ste5-4E mutant to numerous tests, we conclude that signaling output is not detectably reduced by phospho-mimicking mutations at these four central sites.
FIGURE 2:
The Ste5-4E mutant is not impaired at signaling. (A) Strains with integrated STE5-YFPx3 alleles were tested for Fus3 phosphorylation (15 min; mean ± SEM, n = 4) and FUS1-lacZ transcription (90 min; mean ± SD, n = 3) in response to varying α-factor doses. Strains: MWY001, MWY060, MWY056, MWY352. Plasmid: pSB231. (B) Plasmid-borne Ste5-RlucF2 fusions were assayed for Fus3 phosphorylation (15 min; mean ± SEM, n = 4) and FUS1-lacZ transcription (90 min; mean ± SD, n = 4). Strain: TCY3106. Plasmids: pPP1044, pSH95-MM100, pSH95-MM115, pSH95-MM130. (C) Transcription was measured for plasmid-borne alleles of Ste5 (WT, 4E) in four contexts. Left, BY4741 strains with FUS3-WT or a FUS3-RlucF1 fusion, treated ± α factor (1 µM, 90 min). Right, W303 kss1∆ strains with FUS3-WT or PCYC1-FUS3, treated ± α factor (100 nM, 90 min). Bars, mean ± SD (n = 4), normalized for each strain background. Strains: PPY2271, MM003, PPY2389, PPY2412. Plasmids: pPP1044, pPP1969, pPP4315. (D) Transcriptional induction by plasmid-borne Ste5 alleles, with (+) or without (–) an RlucF2 fusion, tested in two backgrounds: BY4741 (BAR1; 1 µM α factor, 90 min) and W303 (bar1∆; 10 nM α factor, 90 min). Bars, mean ± SD (n = 4), normalized for each background. The WT plasmid lacking RlucF2 is the direct progenitor of the fusion constructs. Strains: PPY2271, PPY2365. Plasmids: pPP1044, pRS316, pSH95, pSH95-MM100, pSH95-MM130. (E) Strains with RlucF1 and RlucF2 fusions (Malleshaiah et al., 2010) were compared with the parental strain BY4741 in assays of cell-cycle arrest (halo assay using 4 and 20 nM α factor), mating, and transcription (mean ± SD, n = 3). Strains: BY4741, MM002, MM003, MM008, MM009. Mating partner: PT2α. Plasmid: pSB231.
It was also proposed that Ptc1 helps control pheromone signaling by dephosphorylating the four central MAPK sites in Ste5 (Malleshaiah et al., 2010). This suggestion was based on the observation that pheromone response is reduced in ptc1∆ cells. We confirmed this observation, but found that deletion of PTC1 also reduced signaling by the 2V2A and 4E mutant versions of Ste5 (Figure 3, A–C), in which the phosphorylation state of the four central sites could not be altered by the presence or absence of Ptc1. A similar effect of Ptc1 was also seen for the Ste5-ND mutant (Figure 3, A–C), which is defective at Fus3 docking. These results indicate that the signaling defect of ptc1∆ cells cannot be attributed to increased phosphorylation of the four central sites in Ste5 and thus suggest that an indirect cause is more likely. Collectively, our findings with both the ptc1∆ strain and the Ste5-4E mutant do not support the previous claim that signaling is inhibited by hyperphosphorylation at the four central sites of Ste5.
FIGURE 3:

Deletion of PTC1 reduces signaling irrespective of the status of central sites in Ste5. (A) Phospho-MAPK analysis in PTC1 vs. ptc1∆ strains harboring the indicated Ste5 alleles, following treatment with 10 nM α factor (15 min). Strains: TCY3106, MWY393. Plasmids: pPP1044, pPP1969, pPP3044, pPP4314, pPP4315. (B) Quantification of phospho-Fus3 signal from experiments as in (A) (mean ± SD, n = 4). (C) Transcriptional induction was measured after 90 min α-factor treatment, in parallel with the assays above (mean ± SD, n = 4).
Phosphorylation of central sites in Ste5 is not necessary to dampen signaling output
Next, we studied the phenotypes caused by replacing the four central MAPK sites in Ste5 with nonphosphorylatable residues. In agreement with previous observations (Malleshaiah et al., 2010), we found that the 2V2A mutant was strongly hyperactive in signaling assays (Figure 2A). Yet, these strong phenotypes were not shared by the 4Q mutant (Figure 2A), raising questions about whether they can be explained simply by a lack of phosphorylation. To probe the basis for this discrepancy, we made additional Ste5 mutants in which these four phosphorylation sites were replaced with Val, Ala, or Gly residues (4V, 4A, 4G, respectively; see Figure 4A). Remarkably, the hyperactivity of the 2V2A mutant was shared only by the 4V mutant, whereas the 4A and 4G mutants showed signaling that was comparable to (or even slightly lower than) the WT level (Figure 4, B and C). These phenotypic differences were evident in assays of either transcriptional induction or Fus3 phosphorylation and in two different strain backgrounds (Figure 4, B and C). By contrast, Ste5 protein levels were similar for all mutants (Figure 4D). An additional mutant with only one Val and three Ala substitutions, Ste5-3A1V (Figure 4A; Bhaduri and Pryciak, 2011), was also hyperactive (Figure 4E), indicating that a single Val residue could cause signaling behavior to differ sharply from the 4A mutant. We also observed similar results when these mutants were tested in the context of integrated STE5-YFPx3 alleles (Figure 4F); in this case, we also tested two additional alleles in which the four sites were substituted with the uncharged polar residues Gln or Asn (4Q and 4N), for which signaling was elevated slightly but was most similar to the 4E mutant (Figure 4F). Collectively, the results demonstrate that signaling hyperactivity is not a straightforward consequence of removing the phospho-acceptor sites, and instead, it depends on the specific chemical nature of the substitute side chains.
FIGURE 4:

Signaling phenotypes of nonphosphorylatable Ste5 mutants depends on the substitute residue. (A) Diagram of residue substitutions in the Ste5 mutants tested below. (B) Transcriptional induction by plasmid-borne Ste5 alleles was assayed after 90 min α-factor treatment in two strain backgrounds: W303 (bar1∆; 10 nM α factor) and BY4741 (BAR1; 1 µM α factor). Bars, mean ± SD (n = 4). Strains: PPY2271, PPY2365. Plasmids: pRS316, pPP1044, pPP1969, pPP3044, pPP4314, pPP4316, pPP4317, pPP4318. (C) Phospho-MAPK levels in W303 background cells from B (10 nM α factor, 30 min). Top, representative blots. Bottom, quantified results (mean ± SEM, n = 4). (D) Ste5-myc13 levels in the cells from B (top). A nonspecific band on the same blot served as a loading control. (E) The Ste5-3A1V mutant is as hyperactive as the 4V and 2V2A mutants. Fus3 phosphorylation was measured after 15 min treatment with 10 nM α factor. Bars, mean ± SEM (n = 4). Strain: PPY2365. Plasmids: pPP1044, pPP1969, pPP4314, pPP4316, pPP4317, pPP4378. (F) Transcriptional induction in strains with integrated STE5-YFPx3 alleles after treatment with 3 or 10 nM α factor (90 min). Bars, mean ± SD (n = 4). Strains: MWY001, MWY056, MWY060, MWY352, MWY397, MWY399, MWY401, MWY403. Plasmid: pSB231.
To further clarify the role of these phosphorylation sites in signaling attenuation, we assayed the temporal dynamics of Fus3 phosphorylation. With Ste5-WT, Fus3 phosphorylation peaked after 2 min of pheromone stimulation and then declined by 5–15 min (Figure 5A). This rapid dampening of signal output occurred on a timescale similar to that observed previously for Fus3-dependent attenuation of both Ste5 membrane association and Fus3 phosphorylation (Yu et al., 2008; Bush and Colman-Lerner, 2013; Repetto et al., 2018). (Note that they differ from declines observed at later times in other studies that mainly used BAR1 cells [Hao et al., 2008; Choudhury et al., 2018]; see Discussion.) These kinetic patterns were roughly similar at all pheromone doses, although higher doses yielded stronger responses. After the initial dampening, the resumption of phospho-Fus3 signal was accompanied by an increase in total Fus3 levels (Figure 5A), which was similar at all doses and hence was not sufficient to explain the dose-dependent levels of Fus3 phosphorylation. Next, we tested how these temporal dynamics were altered by mutations in Ste5. In contrast to Ste5-WT, the temporal decline was greatly diminished for the Ste5-ND mutant (Figure 5B), though there remained a mild, brief decline (from 2 to 5 min) that promptly reversed (from 5 to 15 min). These behaviors of the Ste5-ND mutant imply that the decline in the 2- to 15-min period for Ste5-WT reflects Fus3 docking-mediated feedback. Notably, the 4A and 4G mutants showed a decline in response that was similar to WT (Figure 5B), indicating that phosphorylation of these sites is not required. In contrast, the 2V2A and 4V mutants showed Fus3 activation that was unusually strong (compared with WT or ND) and declined negligibly during the 2- to 15-min period. Thus, the mutants with Val substitutions show defects in negative feedback that cannot be explained simply by the absence of phosphorylation sites, because the 4A and 4G mutants mimic WT signal-dampening behavior.
FIGURE 5:
Temporal attenuation of signaling requires the Fus3 D motif but not the central phosphorylation sites in Ste5. (A) Temporal dynamics of MAPK phosphorylation was assayed at three different pheromone concentrations. Top, representative example. Bottom, quantified results (mean ± SEM, n = 4), expressed relative to the 2-min peak with 10 nM α factor. Strain: PPY2365. Plasmids: pPP1044 and pPP1969. (B) MAPK phosphorylation dynamics for cells harboring the indicated Ste5 mutants, treated with 10 nM α factor. Top, representative example. Bottom, quantified results (mean ± SEM, n = 4). The dashed region in the left plot is expanded at the right to facilitate comparison of that subset. Strain: PPY2365. Plasmids: pPP1044, pPP1969, pPP3044, pPP4314, pPP4316, pPP4317, pPP4318.
Hyperactivating mutations perturb phosphorylation of distal sites in Ste5
Our findings suggested that the signaling phenotypes of central phosphorylation site mutations are not dictated simply by whether the sites can be phosphorylated, but rather by the chemical nature of the replacement side chains. We speculated that the replacement with nonpolar Val residues might have a side effect of reducing the solvent exposure of the local peptide region and hence impeding access to nearby docking sites. To explore this possibility, we used a mobility-shift assay to monitor Ste5 phosphorylation in vivo. This assay uses an N-terminal fragment of Ste5, Ste5-NT, which includes the D motif plus phosphorylation sites in both the central and N-terminal clusters (Figure 6A). In previous work, we showed that pheromone treatment triggers a shift in mobility of this fragment, which is caused by phosphorylation (i.e., is reversed by phosphatase), and that this depends on both Fus3 kinase activity and Fus3-Ste5 docking (Repetto et al., 2018). We also showed that pheromone can still induce a mobility shift for this fragment even when the central sites cannot be modified (e.g., due to the 4E or 4Q mutation), and this remaining shift depends on the N-terminal sites (Repetto et al., 2018). Thus, here we compared how this remaining shift was affected by the different mutations in the central cluster. We observed two behavioral groups (Figure 6, B and C): mutants in one group (4A, 4G, and 4E) largely retained the WT mobility shift, whereas the shift was strongly disrupted for mutants that incorporate Val residues (4V and 3A1V). It is important to emphasize that the central sites cannot be phosphorylated in any of these mutants, and hence their different phosphorylation patterns must reflect differences in phosphorylation of other sites, elsewhere in the Ste5 fragment. Therefore, these results show that the mutants containing Val substitutions (2V2A and 4V) uniquely perturb phosphorylation of separate, distal sites (Figure 6D), and this phosphorylation defect correlates with the hyperactivity of these same mutants. It is also noteworthy that the mobility shift was disrupted more strongly by the 2V2A and 4V mutations than by the ND mutation (Figure 6C), causing an apparent reduction in both the fraction of molecules phosphorylated and the extent of mobility shift, which could also relate to their stronger hyperactivity.
FIGURE 6:

Valine mutations disrupt pheromone-induced phosphorylation of distal sites in Ste5. (A) Diagram of full-length Ste5 and the Ste5-NT fragment used to detect phosphorylation by mobility shift assays. At bottom are shown the sites of mutations tested in assays below. (B) Mobility shift assays of phosphorylation for Ste5-NT fragments harboring the indicated mutations. Cells were synchronized in G1 (by arresting cdc15-2 cells in mitosis and releasing for 20 min) and then treated briefly ± α factor (1 µM, 5 min) Strain: MWY198. Plasmids: pPP3415, pPP4235, pPP4236, pPP4324, pPP4325, pPP4326. (C) Assays were performed as in B, but here included Ste5-ND (pPP4358) for comparison. (D) Illustration summarizing the results above. Valine substitutions at the central sites have a side effect of disrupting Fus3 phosphorylation of other sites in the Ste5 N-terminus.
Role for N-terminal sites in dampening signal output
The results above show a correlation between the ability to attenuate signal output and the ability to phosphorylate Ste5 at sites away from the central cluster. Moreover, recent work shows that the ability of Fus3 to inhibit Ste5 membrane association (Yu et al., 2008; Bush and Colman-Lerner, 2013) involves phosphorylation sites in the N-terminal cluster rather than the central cluster (Repetto et al., 2018). Together, these findings would seem to predict that these N-terminal sites are involved in dampening signal output. However, our previous work indicated that removing these sites did not cause hyperactive signaling, but only resistance to inhibition by CDK activity (Strickfaden et al., 2007; Bhaduri and Pryciak, 2011). Here, we reexamined this issue by measuring how the N-terminal sites affect Fus3 activation dynamics. We found that a mutant lacking these sites, Ste5-8A, did not display the WT pattern of signal dampening in the first 2–15 min (Figure 7A), and instead it resembled Ste5-ND. Thus, in the early time period (5–30 min), signaling by Ste5-8A was indeed hyperactive. At later times, however, signaling by Ste5-8A reached a plateau, such that it did not keep up with Ste5-ND and instead was eventually matched by Ste5-WT (Figure 7A). This pattern can explain why the Ste5-8A mutant did not appear hyperactive in prior assays, which mostly measured transcriptional readouts after prolonged (≥90 min) pheromone exposure (Strickfaden et al., 2007; Bhaduri and Pryciak, 2011). To understand the difference between the Ste5-8A and Ste5-ND profiles, we tested a variant that combines both mutations, 8A+ND, and found that it behaved like the 8A mutant (Figure 7A); in other words, the 8A mutation reduced the signaling output of the ND mutant. This behavior suggests that the hyperactivity conferred by the 8A mutation is partially counteracted by a mild functional deficiency, perhaps due to slightly reduced Ste5 protein levels (see below and Strickfaden et al., 2007), and that this curbs its signaling output, especially at later times (when Fus3 levels have increased). Overall, these findings indicate that the temporal dampening of signal output depends on both the Fus3 docking site and the N-terminal phosphorylation sites in Ste5, and that the role of the latter was obscured in prior studies because the 8A mutation imparts a separate, mild deficiency.
FIGURE 7:
Role for N-terminal phosphorylation sites in dampening signal output. (A) Fus3 activation dynamics for cells harboring the indicated Ste5-YFPx3 mutants treated with 50 nM α factor. The Ste5-8A mutation, lacking the eight N-terminal phosphorylation sites, was tested alone and in combination with the ND mutation (8A+ND). Top, representative example. Bottom, quantified results (mean ± SEM, n = 8). Strains: MWY001, MWY006, MWY038, MWY042. (B) Representative example of MAPK phosphorylation and Ste5-YFPx3 protein levels for variants that combine phosphorylation site mutations with the ND mutation. Cells were treated with 10 nM α factor for 15 min. Strains: TCY3106, MWY001, MWY006, MWY038, MWY042, MWY056, MWY060, MWY159, MWY397, MWY407, MWY409, MWY412, MWY413. All strains harbored plasmid pSB231. (C) Top, quantification of P-Fus3 levels from experiments as in B (mean ± SEM, n = 6). Middle, FUS1-lacZ levels in the same strains, after treatment with the indicated α-factor concentrations for 90 min (mean ± SEM, n = 6). Bottom, quantification of Ste5-YFP levels in the same strains (mean ± SEM, n = 5).
We extended these analyses further by comparing the effect of combining the ND mutation with mutation of phosphorylation sites at the central cluster, the N-terminal cluster, or both. We reasoned that, if the main role of Fus3 docking in negative feedback is to modify a particular set of sites, then when these sites cannot be modified, there should be no additional effect of disrupting the D motif. We found that, for mutations at the central cluster (4A, 2V2A, and 4E), adding the ND mutation still increased signaling (Figure 7, B and C); while this effect was only slightly detectable for the hyperactive 2V2A mutant, it was clearly evident for the 4A and 4E mutants. Therefore, Fus3-Ste5 docking can lead to a strong attenuation of signaling even when the central sites cannot be modified. In comparison, when the ND mutation was added to the 8A mutant, or to a variant with both 8A and 4E mutations (4E+8A), the increase in signaling was much weaker, though not entirely absent (Figure 7, B and C). These results fit with a model in which Fus3-Ste5 docking regulates signaling primarily via the 8 N-terminal sites, plus residual effects via other mechanisms (see Discussion). Separately, the data also shed light on the complex effects of the 8A mutation. Compared to WT, the 8A mutant showed an increase in Fus3 phosphorylation measured after 15 min, but not in transcriptional induction, which was measured after 90 min (Figure 7C, top vs. middle). This difference in early versus late measures of signaling output is consistent with the temporal plateau in Ste5-8A signaling, noted above (in Figure 7A), and likely reflects a mild (∼ 30–50%) reduction in Ste5 levels for variants harboring the 8A mutation (Figure 7, B and C, bottom). Collectively, these findings confirm a functional link between the Fus3 D motif and the N-terminal phosphorylation sites in Ste5, and they clarify why hyperactive signaling by the Ste5-8A mutant is obscured at late response times.
Comparing the tethering versus allosteric roles of Fus3-Ste5 docking
Docking of Fus3 onto Ste5 can potentially alter signaling output in two distinct ways (Figure 8A). First, by providing a tether that increases kinase-substrate proximity, docking can enhance the ability of Fus3 to phosphorylate sites on Ste5; indeed, this function is evident from gel-shift assays both here (Figure 6C) and in a prior report (Repetto et al., 2018). Second, it can increase Fus3 kinase activity. In particular, a Ste5 peptide spanning the D motif can partially activate Fus3 in vitro, resulting in a monophosphorylated form of the kinase (Bhattacharyya et al., 2006). This D motif peptide binds Fus3 in a bipartite manner, with two distinct contact sequences, sites A and B, separated by a linker (Figure 8B). Shortening or lengthening the linker reduced Fus3 activation in vitro (Bhattacharyya et al., 2006), suggesting that Ste5 activates Fus3 by contacting both lobes of the kinase and deforming them into a favorable orientation. To date, no data distinguish whether the hyperactive signaling by the Ste5-ND mutant reflects a defect in the tethering role, the allosteric role, or both. To clarify this issue, we made Ste5 variants (Figure 8B) in which sites A and B were mutated individually, plus one in which the linker length was increased by three residues, which reduced allosteric activation of Fus3 fourfold in vitro (Bhattacharyya et al., 2006). We found that both the A and B sites were required to dampen signaling, as each single mutant (ND-A and ND-B) was as hyperactive as the full ND mutant (Figure 8, C–E). In contrast, increasing the linker length (ND-L+3) did not alter signaling strength (Figure 8, C–E) or dynamics (Figure 8F). These results imply that the allosteric effect of the D motif might not be necessary for its regulatory function in vivo, and that the tethering role might be sufficient. The need for both A and B sites can be explained because Fus3 does not bind detectably to either site alone (Bhattacharyya et al., 2006). A potential caveat is that the ND-L+3 mutation does not fully disrupt the allosteric effect in vitro (Bhattacharyya et al., 2006), and so the remaining effect might suffice in vivo; future studies might resolve this issue by testing whether the bipartite D motif in Ste5 can be functionally replaced with a monopartite motif from another protein (Remenyi et al., 2005).
FIGURE 8:

Features of the Fus3 D motif required to control signaling output. (A) Docking of Fus3 onto Ste5 can potentially contribute to negative feedback by two routes. It can enhance phosphorylation of sites in Ste5 by increasing their local proximity. It can also partially activate Fus3, whereby contacts with both N- and C-lobes of the kinase induce an allosteric change in conformation that promotes autophosphorylation. (B) The D motif in Ste5 consists of two Fus3-contacting regions (sites A and B) separated by a linker. Lowercase letters show mutations in the original ND mutant, in half-site mutants (ND-A, ND-B) and in a mutant with a lengthened linker (ND-L+3). (C–E) Cells harboring plasmid-borne Ste5 alleles were treated with α factor (10 nM) and assayed for MAPK phosphorylation (15 min) and transcriptional induction (90 min). (C) Representative immunoblots. (D, E) Quantified P-Fus3 and FUS1-lacZ results (mean ± SEM, n = 4). Strain: PPY2365. Plasmids, pRS316, pPP1044, pPP1969, pPP3044, pPP4380, pPP4381, pPP4401. (F) Time course of Fus3 phosphorylation in the strains from C–E harboring WT Ste5 or the ND-L+3 mutant, following treatment with 10 nM α factor (mean ± SEM, n = 4).
DISCUSSION
In this study, we report several findings that shed light on the mechanisms of negative feedback in the yeast pheromone response pathway. In particular, we probed the feedback effects of the MAPK Fus3 that involve docking to and phosphorylation of the scaffold protein Ste5. Our findings call into question prior suggestions that Fus3 down-regulates Ste5 signaling by phosphorylating the central cluster of sites in Ste5 (Malleshaiah et al., 2010). First, contrary to that report, we found that a mutant variant mimicking phosphorylation at the central sites (Ste5-4E) is functionally indistinguishable from WT. Second, we confirmed that mutating these residues to nonphosphorylatable residues can cause hyperactive signaling, but we found that this depends on the prior choice to replace Thr sites with Val residues, and that it is not observed if these same sites are replaced with Ala or Gly residues. Third, we found that signal output is rapidly dampened in the first few minutes of response, and that this depends on both the Fus3 docking site in Ste5 and the N-terminal phosphorylation sites, but does not require the ability to modify the central sites; indeed, disrupting the Fus3 docking site causes hyperactive signaling even when the central sites are absent. Collectively, our findings argue that Fus3-dependent regulation of signal output involves the N-terminal cluster of phosphorylation sites in Ste5. This view agrees with recent quantitative microscopy data showing that a Fus3-dependent decline of Ste5 membrane localization depends on the N-terminal cluster and not the central cluster (Repetto et al., 2018). Finally, our findings suggest that the Fus3 docking site in Ste5 can promote negative feedback primarily via a tethering function, in which it helps Fus3 phosphorylate sites elsewhere in Ste5, rather than via allosteric effects on Fus3 activity.
Our observations with the Ste5-4A and Ste5-4G mutants show that phosphorylation of the central sites in Ste5 is not required for the WT pattern of signal attenuation. Yet the phenotypes were dramatically different when the substitutions at these positions included Val residues (i.e., 2V2A, 4V, and 3A1V). A basis for these signaling differences was suggested by phosphorylation assays, which revealed that the added Val residues uniquely disrupted docking-dependent phosphorylation of N-terminal sites. We speculate that the Val side chains might be energetically driven to avoid solvent exposure by interacting with other nonpolar residues in adjacent docking motifs, thereby obscuring their access. In accord with this notion, we previously observed a related result in which recognition of the LP motif by Cln2 was reduced by the 3A1V mutation, and not by the 4E mutation (Bhaduri and Pryciak, 2011). We are not aware of a similar case in which the phenotype of nonphosphorylatable substitutions shows such an astonishing dependence on the chosen residue, although this could often go unnoticed as it is common to simply use Ala residues rather than compare multiple different replacement residues. It is also possible that the composition of this region of Ste5 makes it unusually sensitive to the chemical nature of the substitute residues, perhaps due to a propensity for disorder (Oates et al., 2013) and/or an abundance (∼40%) of proline and hydrophobic side chains.
Although our findings (here and in Repetto et al., 2018) show that phosphorylation of the central sites is not essential for the attenuation of either pathway output or Ste5 membrane association, it is worth emphasizing that these sites are indeed phosphorylated in vivo in response to both pheromone addition and cell-cycle entry (Repetto et al., 2018). This provides reason to be cautious about dismissing their possible functions. It remains conceivable that they play a regulatory role that is subtle and requires more sensitive assays to detect, or that operates in only a highly specific signaling context or subpopulation of cells. In this regard we note that the 4A and 4G mutants show some mild differences from WT in the pattern of dampening (see Figure 5B) and slightly reduced signal output (see Figure 4). We have not yet pursued these nuances further, but they might hint at subtle effects worth exploring in future studies.
Several observations conflict with the previous proposal that the phosphatase Ptc1 controls signaling by dephosphorylating the central sites in Ste5 (Malleshaiah et al., 2010). First, we report here that deleting PTC1 reduced signaling even by Ste5-2V2A and Ste5-4E mutants, where the phosphorylation state of these sites could not be changed. Second, the putative Ptc1-binding sequence in Ste5 (residues 277–280, PLLP [Malleshaiah et al., 2010]) overlaps a docking motif for the cyclins Cln1 and Cln2 (residues 278–281, LLPP [Bhaduri and Pryciak, 2011]), and yet, unlike deletion of PTC1, mutation of this motif did not cause reduced signaling in a variety of assays (Bhaduri and Pryciak, 2011; Repetto et al., 2018). Third, based on indirect inference from split-luciferase assays of Ste5-Fus3 interaction (Malleshaiah et al., 2010), it was suggested that pheromone induces Ptc1 to reduce phosphorylation of the central sites in Ste5, but recent mass spectrometry and mobility shift data indicate that pheromone stimulates phosphorylation of these sites (Repetto et al., 2018). While it is conceivable that the actions proposed for Ptc1 affect only a subpopulation of Ste5 molecules (e.g., at mating projections), the model was derived from measurements of the bulk population. We look forward to future studies and additional technologies that can further clarify the dynamic changes in phosphorylation at these sites during the course of pheromone response.
Our findings argue that the N-terminal cluster of phosphorylation sites in Ste5 is the relevant target by which Fus3-Ste5 docking attenuates pathway output. This cluster was originally found to mediate inhibition by CDK activity (Strickfaden et al., 2007), whereas the central cluster was implicated in negative feedback from Fus3 (Bhattacharyya et al., 2006; Malleshaiah et al., 2010). Recent work, however, shows that the effects of CDK and Fus3 are intertwined, as each kinase can drive phosphorylation of both clusters in vivo, and the two kinases can collaborate to fully inhibit Ste5 membrane association (Repetto et al., 2018). Our new findings extend these discoveries by showing that temporal dampening of signal output shows a similar reliance on both the Fus3 docking site and the N-terminal phosphorylation sites in Ste5. They also clarify why mutation of the N-terminal sites (Ste5-8A) did not originally appear to share the hyperactive phenotype seen with mutation of the docking site (Ste5-ND). Namely, the Ste5-8A mutant does display hyperactive signaling at early response times, but this is obscured at later times because the 8A mutations also confer a mild functional deficiency (most likely via slightly reduced protein levels).
We must stress that the hyperactivity and temporal dampening phenotypes observed in our experiments (and in many other studies) reflect the average behavior of asynchronous cell populations, in which responses are not uniform (Oehlen and Cross, 1994; Colman-Lerner et al., 2005). Recent single-cell microscopy studies show that the temporal dampening of both Ste5 membrane recruitment and downstream pathway output is strongest in cells that have recently entered the cell cycle (Durandau et al., 2015; Conlon et al., 2016; Repetto et al., 2018). Similarly, in synchronized cultures, rapid attenuation of signal output depends not only on Fus3 activity but also on cell-cycle stage and Cln1/2-CDK activity (Repetto et al., 2018). Thus, a significant component of the hyperactivity of Ste5-8A and Ste5-ND mutants in asynchronous cultures is likely to reflect the behavior of a subpopulation of cells at the refractory cell-cycle stages. When assayed in G1-synchronized cultures, these mutants did not show strongly increased signal output (Repetto et al., 2018), though mild effects might exist. Thus, in future studies, it will be useful to further interrogate the temporal dynamics of signal output in synchronous cultures, to determine whether mild attenuation from Fus3 feedback can be detected in the absence of the CDK contribution.
Prior work makes clear that the normal function of the Fus3 docking site in Ste5 (the D motif) is to attenuate pathway output (Bhattacharyya et al., 2006). Docking interactions between kinases and substrates are widely recognized to enhance the efficiency and specificity of substrate phosphorylation (Remenyi et al., 2006; Ubersax and Ferrell, 2007), generally by providing a tethering interaction that can increase the local concentration of substrate phospho-acceptor sites near the kinase. The D motif in Ste5 also has the unusual property, observed in only a few cases (Remenyi et al., 2006; Ubersax and Ferrell, 2007), of allosterically modulating the activity of its kinase partner, Fus3 (Bhattacharyya et al., 2006). Our results suggest that this allosteric effect might not be essential to the in vivo regulatory role of the Ste5 D site, and that a simple tethering function provides a sufficient explanation. This notion is consistent with the observations that the D motif promotes pheromone-induced phosphorylation of the N-terminal sites in Ste5 and that these sites are important for temporal control of Ste5 membrane localization and pathway output (see Figures 6C and 7 and Repetto et al., 2018). Note that our findings regarding the D motif concern only one of two distinct allosteric effects on Fus3 that have been observed with two separate regions of Ste5. The other effect is mediated by the C-terminal VWA domain of Ste5, which can cause Fus3 to be more readily phosphorylated by its upstream activator, Ste7 (Good et al., 2009); this “coactivation” phenomenon was proposed to increase pathway output and specificity and is not impacted by our results.
It is noteworthy that different dynamics of pheromone response have been observed in different studies. As with our observations, some studies found a rapid initial spike in signaling within the first 1–5 min, followed by a “correction” period (Yu et al., 2008; Bush and Colman-Lerner, 2013; Repetto et al., 2018). These kinetics fit with the speed of Fus3-dependent substrate phosphorylation in vivo, which can be observed within 5 min (Yu et al., 2008; Choudhury et al., 2018; Repetto et al., 2018). Other studies reported a slow gradual rise in signaling, followed by a decline at a considerably later time (e.g., after 30 min) (Hao et al., 2008; Choudhury et al., 2018). We have not observed this “late decline” behavior, though our analyses so far have been restricted to bar1∆ strains in the W303 background; further studies will be necessary to determine whether these qualitative differences are due to differences in strain background, methodological procedures, or degradation of pheromone by the Bar1 peptidase. In some studies that focused mainly on the later decline events, there were indications of an earlier attenuation within the first 5–10 min, declining to roughly 25–65% of initial peak levels (e.g., see Figure 3B in Nagiec et al., 2015 and Supplemental Figure S1B in Choudhury et al., 2018). Thus, it should eventually be possible to harmonize findings and reach a consensus description of the dynamic aspects of this signaling system.
Finally, we emphasize that negative feedback by Fus3 involves several targets. In addition to Ste5, other implicated candidates include the MAPK-activating kinase Ste7, the Gγ subunit Ste18, and the phosphatase Msg5. Our findings suggest that a primary regulatory role of Fus3-Ste5 docking is to promote phosphorylation of the N-terminal sites in Ste5. Nevertheless, there might also be secondary roles, because adding the ND mutation to the Ste5-8A mutant still caused a residual increase in signaling (see Figure 7C); this effect likely does not involve Ste18, because Ste18 phosphorylation by Fus3 does not require the Ste5 D motif (Choudhury et al., 2018). Separately, we note that the Ste5-ND mutant retained a partial reduction in signaling between 2 and 5 min (see Figures 5B and 7A). This might reflect the role of factors other than Ste5, such as the feedback targets mentioned above or ligand-stimulated endocytosis of the pheromone receptor (Jenness and Spatrick, 1986). Because of these complexities, our interpretations here focused on the broader changes in signaling during the period from 2 to 15 min, for which the role of Ste5 is made clear by the stark difference between Ste5-WT and hyperactive variants (e.g., ND, 8A, and 8A+ND mutants). In contrast, Ste5 is likely not the relevant target of Fus3 in controlling stimulus sensitivity (i.e., effective concentration for half-maximal response [EC50]), because blocking Fus3 kinase activity shifts the EC50 for signaling output (Yu et al., 2008) but not the EC50 for Ste5 recruitment (Bush and Colman-Lerner, 2013). We expect that future studies will make it possible to attribute individual regulatory facets to their responsible targets, and thus eventually provide a full understanding of the network of feedback circuits in this system.
MATERIALS AND METHODS
Yeast methods
Standard procedures were used for growth and genetic manipulation of yeast (Rothstein, 1991; Sherman, 2002). Unless described otherwise, cells were grown at 30°C in yeast extract-peptone-dextrose medium with 2% glucose (YPD) or in synthetic complete (SC) medium with 2% glucose. Strains and plasmids are listed in Tables 1 and 2, respectively. Integrated PSTE5-STE5-YFPx3 alleles were introduced at the TRP1 locus of strain TCY3106 using plasmid pPP3379 and its mutant derivatives, which were linearized by digestion with SnaBI (or BstAPI for pPP3542). “Loop-in” integration of these plasmids can introduce single or multiple copies, which were distinguished by PCR as described previously (Repetto et al., 2018); only strains with single copies were used for signaling assays.
TABLE 1:
Yeast strains used in this study.
| Strain bkgda | Name | Relevant genotype | Source |
|---|---|---|---|
| a | TCY3106 | MATa bar1∆ ade2 his3 leu2 trp1 ura3 can1::HO-CAN1 ho::HO-ADE2 ste5∆::natMX4 | Repetto et al., 2018 |
| a | MWY001 | TCY3106 TRP1::PSTE5-STE5-YFPx3_(1x) | Repetto et al., 2018 |
| a | MWY006 | TCY3106 TRP1::PSTE5-ste5(8A)-YFPx3_(1x) | Repetto et al., 2018 |
| a | MWY038 | TCY3106 TRP1::PSTE5-ste5(ND)-YFPx3_(1x) | Repetto et al., 2018 |
| a | MWY042 | TCY3106 TRP1::PSTE5-ste5(8A+ND)-YFPx3_(1x) | This study |
| a | MWY056 | TCY3106 TRP1::PSTE5-ste5(4E)-YFPx3_(1x) | This study |
| a | MWY060 | TCY3106 TRP1::PSTE5-ste5(2V2A)-YFPx3_(1x) | This study |
| a | MWY159 | TCY3106 TRP1::PSTE5-ste5(4E+8A)-YFPx3_(1x) | This study |
| a | MWY198 | MATa ade1 cdc15-2 CLN2-myc13::kanMX6 ste5::ADE2 TRP1::PSTE5-STE5-YFPx3_(3x) | Repetto et al., 2018 |
| a | MWY352 | TCY3106 TRP1::PSTE5-ste5(4Q)-YFPx3_(1x) | This study |
| a | MWY393 | TCY3106 TRP1::PSTE5-STE5-YFPx3_(1x) ptc1∆::hphMX6 | This study |
| a | MWY397 | TCY3106 TRP1::PSTE5-ste5(4A)-YFPx3_(1x) | This study |
| a | MWY399 | TCY3106 TRP1::PSTE5-ste5(4V)-YFPx3_(1x) | This study |
| a | MWY401 | TCY3106 TRP1::PSTE5-ste5(4G)-YFPx3_(1x) | This study |
| a | MWY403 | TCY3106 TRP1::PSTE5-ste5(4N)-YFPx3_(1x) | This study |
| a | MWY407 | TCY3106 TRP1::PSTE5-ste5(4E+ND)-YFPx3_(1x) | This study |
| a | MWY409 | TCY3106 TRP1::PSTE5-ste5(2V2A+ND)-YFPx3_(1x) | This study |
| a | MWY412 | TCY3106 TRP1::PSTE5-ste5(4A+ND)-YFPx3_(1x) | This study |
| a | MWY413 | TCY3106 TRP1::PSTE5-ste5(4E+8A+ND)-YFPx3_(1x) | This study |
| a | PPY2365 | MATa FUS1::FUS1-lacZ::LEU2 ste5::ADE2 bar1∆::hphMX6 | Winters and Pryciak, 2018 |
| a | PPY2389 | MATa FUS1::FUS1-lacZ::LEU2 ste5::ADE2 bar1∆::hphMX6 kss1∆::kanMX6 | Winters and Pryciak, 2018 |
| a | PPY2412 | MATa FUS1::FUS1-lacZ::LEU2 ste5::ADE2 bar1∆::hphMX6 kss1∆::kanMX6 fus3::natMX6::PCYC1-FUS3 | Winters and Pryciak, 2018 |
| b | BY4741 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 | Brachmann et al., 1998 |
| b | MM002 | BY4741 FUS3-RlucF1::natMX6 STE5-RlucF2::hphMX6 | Malleshaiah et al., 2010 |
| b | MM003 | BY4741 FUS3-RlucF1::natMX6 ste5∆ | Malleshaiah et al., 2010 |
| b | MM008 | BY4741 STE5-RlucF1::natMX6 STE11-RlucF2::hphMX6 | Malleshaiah et al., 2010 |
| b | MM009 | BY4741 STE5-RlucF1::natMX6 STE7-RlucF2::hphMX6 | Malleshaiah et al., 2010 |
| b | PPY2271 | BY4741 ste5∆::natMX6 | This study |
| c | PT2α | MATα hom3 ilv1 can1 | Pryciak and Huntress, 1998 |
aStrain background: a, W303 [ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1]; b, BY4741 [his3∆1 leu2∆0 ura3∆0 met15∆0]; c, other.
TABLE 2:
Plasmids used in this study.
| Name | Description | Source |
|---|---|---|
| pPP1044 | CEN ARS HIS3 FUS1-lacZ | Lamson et al., 2006 |
| pPP1969 | CEN ARS URA3 PSTE5-STE5-myc13 TCYC1 | Winters et al., 2005 |
| pPP3044 | CEN ARS URA3 PSTE5-ste5(ND)-myc13 TCYC1 | Takahashi and Pryciak, 2008 |
| pPP3379 | Integrating TRP1 PSTE5-STE5(WT)-YFPx3 TSTE5 | Repetto et al., 2018 |
| pPP3380 | Integrating TRP1 PSTE5-ste5(8A)-YFPx3 TSTE5 | Repetto et al., 2018 |
| pPP3415 | CEN ARS URA3 PSTE5-ste5_1-370(WT)-HA3 TCYC1 | Bhaduri and Pryciak, 2011 |
| pPP3475 | Integrating TRP1 PSTE5-ste5(ND)-YFPx3 TSTE5 | Repetto et al., 2018 |
| pPP3476 | Integrating TRP1 PSTE5-ste5(8A+ND)-YFPx3 TSTE5 | This study |
| pPP3532 | Integrating TRP1 PSTE5-ste5(4E)-YFPx3 TSTE5 | Repetto et al., 2018 |
| pPP3542 | Integrating TRP1 PSTE5-ste5(2V2A)-YFPx3 TSTE5 | This study |
| pPP4046 | Integrating TRP1 PSTE5-ste5(4E+8A)-YFPx3 TSTE5 | This study |
| pPP4235 | CEN ARS URA3 PSTE5-ste5_1-370(3A1V)-HA3 TCYC1 | This study |
| pPP4236 | CEN ARS URA3 PSTE5-ste5_1-370(4E)-HA3 TCYC1 | Repetto et al., 2018 |
| pPP4259 | Integrating TRP1 PSTE5-ste5(4Q)-YFPx3 TSTE5 | Repetto et al., 2018 |
| pPP4270 | Integrating TRP1 PSTE5-ste5(4E+ND)-YFPx3 TSTE5 | This study |
| pPP4272 | Integrating TRP1 PSTE5-ste5(2V2A+ND)-YFPx3 TSTE5 | This study |
| pPP4301 | Integrating TRP1 PSTE5-ste5(4E+8A+ND)-YFPx3 TSTE5 | This study |
| pPP4314 | CEN ARS URA3 PSTE5-ste5(2V2A)-myc13 TCYC1 | This study |
| pPP4315 | CEN ARS URA3 PSTE5-ste5(4E)-myc13 TCYC1 | This study |
| pPP4316 | CEN ARS URA3 PSTE5-ste5(4A)-myc13 TCYC1 | This study |
| pPP4317 | CEN ARS URA3 PSTE5-ste5(4V)-myc13 TCYC1 | This study |
| pPP4318 | CEN ARS URA3 PSTE5-ste5(4G)-myc13 TCYC1 | This study |
| pPP4320 | Integrating TRP1 PSTE5-ste5(4A)-YFPx3 TSTE5 | This study |
| pPP4321 | Integrating TRP1 PSTE5-ste5(4V)-YFPx3 TSTE5 | This study |
| pPP4322 | Integrating TRP1 PSTE5-ste5(4G)-YFPx3 TSTE5 | This study |
| pPP4323 | Integrating TRP1 PSTE5-ste5(4N)-YFPx3 TSTE5 | This study |
| pPP4324 | CEN ARS URA3 PSTE5-ste5_1-370(4A)-HA3 TCYC1 | This study |
| pPP4325 | CEN ARS URA3 PSTE5-ste5_1-370(4V)-HA3 TCYC1 | This study |
| pPP4326 | CEN ARS URA3 PSTE5-ste5_1-370(4G)-HA3 TCYC1 | This study |
| pPP4358 | CEN ARS URA3 PSTE5-ste5_1-370(ND)-HA3 TCYC1 | This study |
| pPP4378 | CEN ARS URA3 PSTE5-ste5(3A1V)-myc13 TCYC1 | This study |
| pPP4380 | CEN ARS URA3 PSTE5-ste5(ND-A)-myc13 TCYC1 | This study |
| pPP4381 | CEN ARS URA3 PSTE5-ste5(ND-B)-myc13 TCYC1 | This study |
| pPP4401 | CEN ARS URA3 PSTE5-ste5(ND-L+3)-myc13 TCYC1 | This study |
| pSB231 | CEN ARS URA3 FUS1-lacZ | Trueheart et al., 1987 |
| pSH95 | CEN ARS URA3 PSTE5-STE5 | Bhattacharyya et al., 2006 |
| pSH95-MM100 | CEN ARS URA3 PSTE5-STE5-RlucF2 | Malleshaiah et al., 2010 |
| pSH95-MM115 | CEN ARS URA3 PSTE5-ste5(2V2A)-RlucF2 | Malleshaiah et al., 2010 |
| pSH95-MM130 | CEN ARS URA3 PSTE5-ste5(4E)-RlucF2 | Malleshaiah et al., 2010 |
| pRS316 | CEN ARS URA3 vector | Sikorski and Hieter, 1989 |
Ste5 mutations
Mutations denoted by abbreviated names in the text and figures are specified as follows:
Ste5-2V2A: T267V S276A T287V S329A
Ste5-4E: T267E S276E T287E S329E
Ste5-4V: T267V S276V T287V S329V
Ste5-4A: T267A S276A T287A S329A
Ste5-4G: T267G S276G T287G S329G
Ste5-3A1V: T267A S276A T287V S329A
Ste5-4Q: T267Q S276Q T287Q S329Q
Ste5-4N: T267N S276N T287N S329N
Ste5-ND: Q292A I294A Y295A L307A P310A N315A
Ste5-8A: T4A S11A T29A S43A S69A S71A S81A T102A
Pheromone response assays
Asynchronous cultures were treated with α factor using concentrations and durations indicated in each figure. To measure transcriptional responses, cells harboring an integrated or plasmid-borne FUS1-lacZ reporter were treated with α factor for 90 min and then were collected and assayed for β-galactosidase activity by colorimetric assay as described previously (Winters and Pryciak, 2018).
To measure MAPK phosphorylation, cells were treated ± α factor as indicated, and then 2-ml samples were harvested by centrifugation. Cell pellets were immediately frozen in liquid nitrogen and stored at –80˚C before preparing cell extracts, as described below. To measure the kinetics of Fus3 activation, we used a procedure described recently (Repetto et al., 2018). The first sample (0 min) was collected immediately before adding α factor. After α-factor treatment, cells were harvested by centrifugation for 30 s, supernatants were aspirated, and cell pellets were frozen by placing the tubes in a liquid nitrogen bath at the stop time indicated (i.e., 2, 5, 15, 30, 45, 60 min). To accomplish this, samples were transferred to 2-ml microcentrifuge tubes 90 s. before the designated stop time to allow sufficient time to collect and freeze cell pellets.
Ste5-NT phosphorylation assays
Mobility-shift assays were used to monitor pheromone-induced phosphorylation of a Ste5-NT fragment in vivo following methods described previously (Repetto et al., 2018). Plasmids expressing HA-tagged Ste5 fragments were transformed into MWY198, a temperature-sensitive cdc15-2 strain. To synchronize the cells in G1 phase, the cdc15-2 cultures were grown at 25°C, arrested at 37°C for 3 h, and then released by transfer back to 25°C (using shaking water baths). Then, 20 min after release, aliquots were collected and incubated ± α factor (1 µM, 5 min). Two-milliliter samples were collected by centrifugation, and cell pellets were frozen before preparation of whole-cell extracts.
Cell extracts and immunoblotting
Whole-cell extracts were prepared by lysis in trichloroacetic acid as described previously (Pope et al., 2014) using frozen cell pellets from 2-ml cultures. Protein concentrations were measured by bicinchoninic acid (BCA) assay (Pierce #23225), and equal amounts (10 µg) were loaded per lane. Proteins were resolved by SDS–PAGE (10% acrylamide) and transferred to polyvinylidene difluoride (PVDF) membranes in a submerged tank. Membranes were blocked (1 h, room temperature) in TTBS (0.2% Tween-20, 20 mM Tris-HCl, 500 mM NaCl, pH 7.5) containing 5% nonfat milk and then probed with antibodies in the equivalent solution. Primary antibodies were rabbit anti–phospho-p44/42 (1:1000; Cell Signaling Technology #9101), rabbit anti-myc (1:200; Santa Cruz Biotechnologies #sc-789), rabbit anti-G6PDH (1:100,000; Sigma #A9521), goat anti-Fus3 (1:5000; Santa Cruz Biotechnologies #sc-6773), mouse anti-HA (1:1000; Covance #MMS101R), and mouse anti-GFP (1:2000; Takara #632381). HRP-conjugated secondary antibodies were goat anti-rabbit (1:3000; Jackson ImmunoResearch #111-035-144), goat anti-mouse (1:3000; Bio-Rad #170-6516), or donkey anti-goat (1:3000; Santa Cruz #sc-2020). Enhanced chemilluminescent detection used a Bio-Rad Clarity substrate (#170-5060). Exposures were captured on x-ray film, and densitometry was performed using ImageJ (https://imagej.nih.gov/ij/). Before probing with another antibody, blots were stripped (2 h, 37°C) using Restore stripping buffer (ThermoFisher #21059), washed three times with TTBS (5 min each), and reblocked in TTBS + 5% milk.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 GM-057769). We thank Wendell Lim, Mohan Malleshaiah, and Stephen Michnick for sharing yeast strains and plasmids. We also thank Alejandro Colman-Lerner for many helpful discussions during the course of this work and for comments on the manuscript.
Abbreviations used:
- CDK
cyclin-dependent kinase
- D motif
docking motif
- EC50
effective concentration for half-maximal response
- GPCR
G protein–coupled receptor
- MAPK
mitogen-activated protein kinase
- ND
nondocking mutant
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-12-0793) on February 6, 2019.
REFERENCES
- Alvaro CG, Thorner J. (2016). Heterotrimeric G protein-coupled receptor signaling in yeast mating pheromone response. J Biol Chem , 7788–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L. (2005). A walk-through of the yeast mating pheromone response pathway. Peptides , 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri S, Pryciak PM. (2011). Cyclin-specific docking motifs promote phosphorylation of yeast signaling proteins by G1/S Cdk complexes. Curr Biol , 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri S, Valk E, Winters MJ, Gruessner B, Loog M, Pryciak PM. (2015). A docking interface in the cyclin Cln2 promotes multi-site phosphorylation of substrates and timely cell-cycle entry. Curr Biol , 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. (2006). The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science , 822–826. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast , 115–132. [DOI] [PubMed] [Google Scholar]
- Bush A, Colman-Lerner A. (2013). Quantitative measurement of protein relocalization in live cells. Biophys J , 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S, Baradaran-Mashinchi P, Torres MP. (2018). Negative feedback phosphorylation of Ggamma subunit Ste18 and the Ste5 scaffold synergistically regulates MAPK activation in yeast. Cell Rep , 1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. (2005). Regulated cell-to-cell variation in a cell-fate decision system. Nature , 699–706. [DOI] [PubMed] [Google Scholar]
- Conlon P, Gelin-Licht R, Ganesan A, Zhang J, Levchenko A. (2016). Single-cell dynamics and variability of MAPK activity in a yeast differentiation pathway. Proc Natl Acad Sci USA , E5896–E5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K. (1994). MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J , 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durandau E, Aymoz D, Pelet S. (2015). Dynamic single cell measurements of kinase activity by synthetic kinase activity relocation sensors. BMC Biol , 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Nasmyth K, Ammerer G. (1992). Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev , 1280–1292. [DOI] [PubMed] [Google Scholar]
- Good M, Tang G, Singleton J, Remenyi A, Lim WA. (2009). The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell , 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG. (2008). Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell , 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Yildirim N, Nagiec MJ, Parnell SC, Errede B, Dohlman HG, Elston TC. (2012). Combined computational and experimental analysis reveals mitogen-activated protein kinase-mediated feedback phosphorylation as a mechanism for signaling specificity. Mol Biol Cell , 3899–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness DD, Spatrick P. (1986). Down regulation of the alpha-factor pheromone receptor in S. cerevisiae. Cell , 345–353. [DOI] [PubMed] [Google Scholar]
- Lamson RE, Takahashi S, Winters MJ, Pryciak PM. (2006). Dual role for membrane localization in yeast MAP kinase cascade activation and its contribution to signaling fidelity. Curr Biol , 618–623. [DOI] [PubMed] [Google Scholar]
- Maeder CI, Hink MA, Kinkhabwala A, Mayr R, Bastiaens PI, Knop M. (2007). Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat Cell Biol , 1319–1326. [DOI] [PubMed] [Google Scholar]
- Malleshaiah MK, Shahrezaei V, Swain PS, Michnick SW. (2010). The scaffold protein Ste5 directly controls a switch-like mating decision in yeast. Nature , 101–105. [DOI] [PubMed] [Google Scholar]
- Nagiec MJ, McCarter PC, Kelley JB, Dixit G, Elston TC, Dohlman HG. (2015). Signal inhibition by a dynamically regulated pool of monophosphorylated MAPK. Mol Biol Cell , 3359–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi Z, Uversky VN, Obradovic Z, Kurgan L, et al. (2013). D(2)P(2): database of disordered protein predictions. Nucleic Acids Res , D508–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR. (1994). G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev , 1058–1070. [DOI] [PubMed] [Google Scholar]
- Pope PA, Bhaduri S, Pryciak PM. (2014). Regulation of cyclin-substrate docking by a G1 arrest signaling pathway and the Cdk inhibitor Far1. Curr Biol , 1390–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak PM, Huntress FA. (1998). Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev , 2684–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Bhattacharyya RP, Lim WA. (2005). The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol Cell , 951–962. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Lim WA. (2006). Docking interactions in protein kinase and phosphatase networks. Curr Opin Struct Biol , 676–685. [DOI] [PubMed] [Google Scholar]
- Repetto MV, Winters MJ, Bush A, Reiter W, Hollenstein DM, Ammerer G, Pryciak PM, Colman-Lerner A. (2018). CDK and MAPK synergistically regulate signaling dynamics via a shared multi-site phosphorylation region on the scaffold protein Ste5. Mol Cell , 938–952 e936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. (1991). Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol , 281–301. [DOI] [PubMed] [Google Scholar]
- Sherman F. (2002). Getting started with yeast. Methods Enzymol , 3–41. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics , 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. (2007). A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell , 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Pryciak PM. (2008). Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr Biol , 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J, Boeke JD, Fink GR. (1987). Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol , 2316–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE., Jr (2007). Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol , 530–541. [DOI] [PubMed] [Google Scholar]
- Winters MJ, Lamson RE, Nakanishi H, Neiman AM, Pryciak PM. (2005). A membrane binding domain in the ste5 scaffold synergizes with gbetagamma binding to control localization and signaling in pheromone response. Mol Cell , 21–32. [DOI] [PubMed] [Google Scholar]
- Winters MJ, Pryciak PM. (2018). Analysis of the thresholds for transcriptional activation by the yeast MAP kinases Fus3 and Kss1. Mol Biol Cell , 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RC, Pesce CG, Colman-Lerner A, Lok L, Pincus D, Serra E, Holl M, Benjamin K, Gordon A, Brent R. (2008). Negative feedback that improves information transmission in yeast signalling. Nature , 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. (2012). Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science , 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Gartner A, Cade R, Ammerer G, Errede B. (1993). Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol Cell Biol , 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]