Abstract
Eukaryotic initiation factor 2 (eIF2) is a G protein critical for translation. It is tightly regulated in the integrated stress response (ISR) via phosphorylation of eIF2α and the subsequent control of eukaryotic initiation factor 2B (eIF2B), a multisubunit guanine nucleotide exchange factor. Through studying the localization of eIF2B subunits, we identified cytoplasmic eIF2B bodies in mammalian cells. We highlight a relationship between body size and the eIF2B subunits localizing to them; larger bodies contain all subunits and smaller bodies contain predominantly catalytic subunits. eIF2 localizes to eIF2B bodies and shuttles within these bodies in a manner that correlates with eIF2B activity. On stress, eIF2α-P localizes predominately to larger bodies and results in a decreased shuttling of eIF2. Interestingly, drugs that inhibit the ISR can rescue eIF2 shuttling in a manner correlating to levels of eIF2α-P. In contrast, smaller bodies show increased eIF2 shuttling in response to stress, which is accompanied by the localization of eIF2Bδ to these bodies, suggesting the formation of a novel trimeric complex of eIF2B. This response is mimicked by ISR-inhibiting drugs, providing insight into their potential mechanism of action. This study provides evidence that the composition and function of mammalian eIF2B bodies are regulated by the ISR and the drugs that control it.
INTRODUCTION
The complex process by which ribosomes are first recruited to mRNA and an appropriate start codon for a protein-coding sequence is selected, is defined as translation initiation. In eukaryotes, the highly conserved heterotrimeric G-protein eukaryotic initiation factor 2 (eIF2) is essential for this process. In its active, guanosine triphosphate (GTP) bound form, eIF2 binds to an initiator methionyl-tRNA (Met-tRNAi) molecule to form a ternary complex (TC). The TC is loaded onto the small (40S) ribosomal subunit facilitated by the binding of other eIFs, to form a 43S preinitiation complex (PIC). The PIC is recruited to the 5′-end of a target mRNA molecule and scans the mRNA sequence for a start codon (Hinnebusch and Lorsch, 2012). On start codon recognition, eIF2-GTP is hydrolyzed mediated by the GTPase-activating protein eIF5 (Huang et al., 1997), reducing the affinity of eIF2 for the Met-tRNAi (Erickson and Hannig, 1996; Algire et al., 2005). This stimulates the release of other eIFs allowing the 60S ribosomal subunit to interact with the 40S ribosomal subunit. This initiation process generates a full 80S ribosome with an appropriately positioned Met-tRNAi at the start codon of an mRNA ready to recruit the next amino-acylated tRNA to the subsequent codon in the translation elongation phase (Hinnebusch and Lorsch, 2012).
For subsequent rounds of translation initiation to occur, eIF2-GTP must be replenished within the cell. eIF2-guanosine diphosphate (GDP) is released from the PIC in complex with eIF5, which functions as a GDP dissociation inhibitor (GDI) (Jennings and Pavitt, 2010). eIF2 has a higher affinity for GDP (Erickson and Hannig, 1996), and thus the guanine nucleotide exchange factor (GEF), eukaryotic initiation factor 2B (eIF2B), is required for recycling of eIF2-GDP to the GTP-bound form. Recently, eIF2B was revealed as a dual-functioning protein. In addition to its GEF activity, eIF2B also acts as a GDI displacement factor (GDF) that can release eIF2-GDP from eIF5 (Jennings et al., 2013). Although functionally similar to other GEFs of the Ras superfamily, eIF2B exhibits a greater level of complexity within its quaternary structure, likely due to its dual role as both a GDF and a GEF but also its requirement for translation initiation regulation. Structurally, eIF2B is composed of five nonidentical subunits, termed α through to ε, encoded in human cells by the genes EIF2B1–5, respectively. The γ and ε subunits of eIF2B have been shown to form a catalytic subcomplex, whereas the α, β, and δ subunits are required for the regulation of eIF2B activity in response to various cellular signals (Pavitt et al., 1997, 1998). Structural studies have revealed that eIF2B exists as a decamer in its native form (Gordiyenko et al., 2014; Wortham et al., 2014), and the recently solved crystal structure of Schizosaccharomyces pombe and human eIF2B contains this decamer arranged with a eIF2B(αβδ)2 hexameric regulatory core, bound by two eIF2B(γε) catalytic subcomplexes on opposite sides (Kashiwagi et al., 2016; Tsai et al., 2018; Zyryanova et al., 2018).
Regulation of eIF2B activity provides a critical controlled point in the translation initiation pathway. The most well-studied mechanism of eIF2B regulation forms part of the integrated stress response (ISR), which results in the down-regulation of global protein synthesis and the concomitant expression of stress-specific factors such as ATF4. More specifically, in response to cellular stress, protein kinases phosphorylate the α subunit of eIF2 on Ser 51. In mammalian cells, there are four eIF2α kinases that are activated in response to diverse stress stimuli. These kinases are PKR (protein kinase R), PERK (protein kinase RNA-like endoplasmic reticulum kinase), GCN2 (general control nondepressible 2), and HRI (heme-regulated inhibitor) (Wek et al., 2006). Phosphorylation of eIF2α converts it from a substrate to a competitive inhibitor of eIF2B (Rowlands et al., 1988; Dever et al., 1995) leading to the down-regulation of global protein synthesis. Paradoxically, the translation of a subset of ISR-responsive mRNAs, which support adaptive stress responses, is up-regulated (Young and Wek, 2016). Conditions that promote eIF2α phosphorylation and the inhibition of translation also lead to the formation of stress granules. These cytoplasmic granules contain stalled mRNAs harboring 40S ribosome subunits, various translation initiation factors, and RNA-binding proteins (Buchan and Parker, 2009).
Phosphorylated eIF2α binds specifically to the regulatory eIF2B(αβδ)2 complex that resides in the center of the eIF2B decamer (Pavitt et al., 1997; Kashiwagi et al., 2016, 2017), and the α subunit of eIF2B in particular is critical for this regulation (Hannig et al., 1990; Dever et al., 1993; Fabian et al., 1997; Kimball et al., 1998; Elsby et al., 2011). Recently, the small molecule, integrated stress response inhibitor (ISRIB) was identified in a cell-based screen for inhibitors of PERK activity (Sidrauski et al., 2013). ISRIB reverses phosphorylated eIF2α-induced translational repression (Sidrauski et al., 2013, 2015a; Halliday et al., 2015) through restoration of eIF2B activity (Sekine et al., 2015; Sidrauski et al., 2015b). This restoration of eIF2B activity occurs through stabilization of eIF2B in its decameric form (Sidrauski et al., 2015b). ISRIB binds at the interface between eIF2Bδ and eIF2Bβ promoting the dimerization of two eIF2B(βδγε) tetramers and promoting decamer formation (Tsai et al., 2018; Zyryanova et al., 2018). Recently, the FDA-approved drug dibenzoylmethane (DBM) was found to have similar properties to ISRIB (Halliday et al., 2017) in that it can reverse stress-induced translational depression. However, the mechanism of action of this drug remains unknown, with evidence to suggest that unlike ISRIB, it may act downstream of eIF2B (Halliday et al., 2017).
It has been established in the yeasts Saccharomyces cerevisiae and Candida albicans that eIF2B localizes to large cytoplasmic eIF2B bodies (Campbell et al., 2005; Campbell and Ashe, 2006; Egbe et al., 2015; Moon and Parker, 2018). These bodies represent sites where GEF activity occurs and are potentially associated with eIF2B regulation (Taylor et al., 2010). Overexpression of eIF2B subunits in mammalian cells suggests that eIF2B might form subcomplexes that possess different sensitivities to regulation (Liu et al., 2011; Wortham et al., 2014). The significance of eIF2B localization in terms of the control of protein synthesis has not been investigated in mammalian cells.
The importance of elucidating the mechanisms of eIF2B function in mammalian cells is heightened by the fact that mutations in eIF2B lead to the neurological disorder leukoencephalopathy with vanishing white matter (VWM). Mutations most commonly occur in the ε subunit of eIF2B; however, causative mutations in all five subunits of eIF2B have been identified (Fogli and Boespflug-Tanguy, 2006). Although eIF2B is a global regulator of protein synthesis, VWM primarily manifests as a leukodystrophy characterized by immature astrocytes and an increased number of oligodendrocyte progenitor cells (Dooves et al., 2016). In addition to neurological symptoms, some patients also present with ovarioleukodystrophy and in severe cases, multiple organs are defective, with the severity of the patient symptoms inversely related to age at onset (Bugiani et al., 2010; Hamilton et al., 2018). The cellular mechanisms through which mutations cause VWM pathophysiology remain elusive. A number of VWM mutations have been shown to affect eIF2B complex formation or function; however, some mutations do not impact either of these factors yet cause severe disease (Liu et al., 2011; Wortham and Proud, 2015). Characterizing eIF2B cellular localization may provide a tool to further investigate the mechanisms of VWM pathology.
Here we investigated eIF2B localization in human astrocytic cells and show that eIF2B localizes to heterogeneous populations termed eIF2B bodies. These bodies appear to differ in size and eIF2B subunit makeup. We show that all eIF2B subunits localize to the larger eIF2B bodies, and the shuttling of eIF2 through these bodies appears to be down-regulated by the induction of cellular stress. Interestingly, the ISR inhibitor, ISRIB, attenuates the effects of stress on eIF2 shuttling through these bodies in a manner that correlates to levels of phosphorylated eIF2α. In contrast to the larger eIF2B bodies, the shuttling of eIF2 through small eIF2B bodies is increased in response to cellular stress. This increase in the shuttling of eIF2 appears to be accompanied by an increase in the localization of eIF2Bδ to the small eIF2B bodies. Suprisingly, translation-enhancing drugs, which target the ISR, appear to mimic the effect that stress has on eIF2 shuttling and eIF2Bδ localization to small eIF2B bodies. We propose these two populations of eIF2B bodies, which are differently regulated by the ISR, may be important in mediating the cell’s response to stress. Larger eIF2B bodies provide a source of eIF2B that can be inhibited during stress and promote ISR-specific up-regulation of stress-responsive genes, whereas small eIF2B bodies provide a source of eIF2B that is not inhibited by cellular stress, allowing for low levels of translation to occur so the cell can overcome the stress.
RESULTS
eIF2B localizes to a heterogeneous population of cytoplasmic bodies in mammalian glial cells
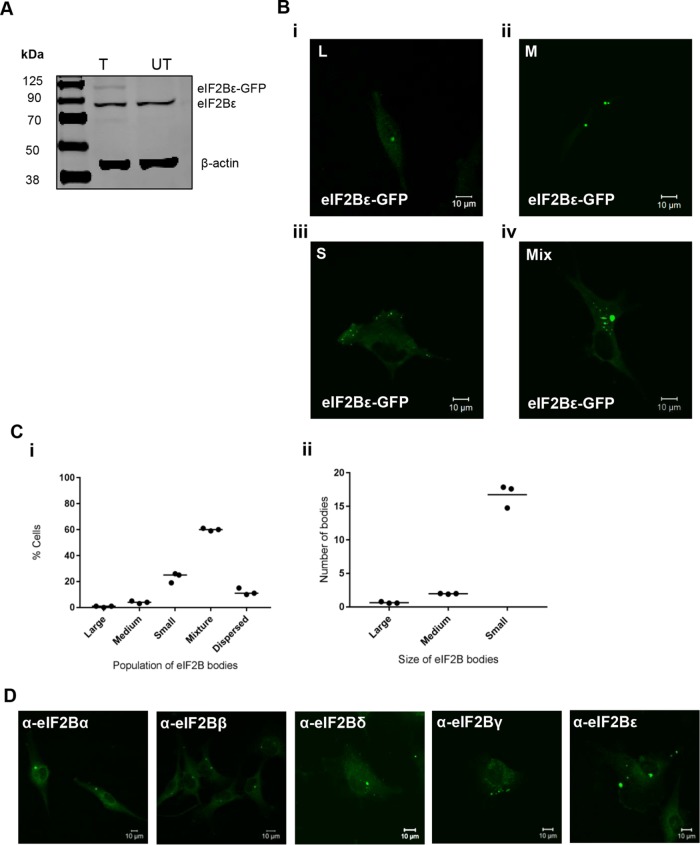
To study the cellular localization of eIF2B in human glial cells, the ε subunit of eIF2B bearing a C-terminal green fluorescent protein (GFP) tag was transiently transfected into the astrocytoma cell line, U373 (Figure 1A). Under normal cellular growth conditions, we observed that eIF2Bε-GFP localized to cytoplasmic foci, which we have termed eIF2B bodies (Figure 1B). Intriguingly, within a population of cells, the number and size of these bodies varied quite dramatically. We initially characterized the bodies by size; classified them into large (≥10 µm2), medium (≥3 µm2 ≤9.99 µm2), and small (≤2.99 µm2); and observed populations of cells containing only large (Figure 1Bi), only medium (Figure 1Bii), only small (Figure 1Biii), and a mixture (Figure 1Biv) of the different-sized bodies. To characterize these different subpopulations, we carried out counts across 100 cells (Figure 1Ci). A minor population of cells (11%) with entirely dispersed localization was also observed. The counts revealed that cells containing a mixture of different-sized bodies were the most frequent. On further investigation of these mixed populations, it appeared that on average, these cells contained one large body, two medium bodies, and >15 small bodies (Figure 1Cii). We have also observed populations of eIF2B bodies in other cell types, demonstrating that this localization is not a specific feature of glial cells (Supplemental Figure S1A). To ensure that this localization was not a result of transient overexpression of eIF2B, immunostaining was carried out for endogenous eIF2B subunits. As shown in Figure 1D, localized foci of each subunit of eIF2B are clearly visible.
FIGURE 1:
eIF2Bε localizes to cytoplasmic bodies in U373 cells. (A) eIF2Bε-GFP was expressed in U373 cells, confirmed by Western blot analysis. (B) Live cell confocal images of U373 cells expressing eIF2Bε-GFP (i) showing localization to only large (L) (≥10 µm2), (ii) only medium (M) (≥3 µm2 ≤9.99 µm2), (iii) only small (S) (≤2.99 µm2), and (iv) a mixture (Mix) of L, M, or S eIF2B bodies. (C) (i) The median percentage of cells, in a population of 100 cells, exhibiting eIF2Bε-GFP localized to only L, only M, only S, a mix of different-sized eIF2B bodies, or dispersed (n = 3); (ii) within the population of cells containing a Mix of eIF2B bodies, the mean number of L, M, and S eIF2B bodies. (D) Confocal images of endogenous eIF2B subunits localizing to cytoplasmic bodies. U373 cells were fixed in methanol and subject to ICC with (left to right) anti-eIF2Bα, anti-eIF2Bβ, anti-eIF2Bδ, anti-eIF2Bγ, or anti-eIF2Bε primary antibodies and visualized using appropriate secondary antibodies conjugated to Alexa Fluor 488.
Owing to the size of the eIF2B bodies, we wanted to ensure that these bodies were not the result of ubiquitin-directed protein aggregation and therefore aggregsomes of misfolded proteins. While ubiquitination can target cells for a number of functions, only the presence of poly-ubiquitin targets specific proteins for degradation by the proteasome (Kleiger and Mayor, 2014). To test this, immunofluorescence was perfomed on cells using the poly-ubiquitin FK1 antibody. As shown in Supplemental Figure S1B, eIF2B bodies do not colocalize with aggregates of poly-ubiquitin, suggesting they are not sites of misfolded proteins.
Another possibility for this localization could be that the GFP tag itself may be responsible for the observed aggregation rather than the eIF2Bε subunit. To address this, cells expressing a GFP-only control plasmid were visualized and no localized foci were observed (Supplemental Figure S2A). Furthermore, we observed cytoplasmic foci with eIF2Bε C-terminally tagged with myc (Supplemental Figure S2B). Quantification of cells harboring eIF2Bε-myc revealed a similar pattern of distribution to the GFP-tagged subunit (Supplemental Figure S2C, i and ii), providing further evidence the eIF2Bε subunit is responsible for the observed localization.
Translation initiation factors are known to aggregate to stress granules during stress (Kedersha and Anderson, 2002). To confirm that these eIF2B bodies are distinct from stress granules, cells were exposed to either ER stress using thapsigargin (Tg) or oxidative stress using sodium arsenite (SA). The stress granule markers G3BP and eIF3b were used to identify stress granules. On exposure to both stresses, G3BP and eIF3b aggregated into distinct stress granules; however, no colocalization of these stress granules with eIF2B bodies was observed in these cells, confirming the eIF2B bodies as distinct cytoplasmic foci (Supplemental Figure S3, A and B).
Hence, we demonstrate that eIF2B can localize to cytoplasmic bodies in human cells. These findings have similarities with our previous work carried out in yeast cells, where cytoplasmic eIF2B bodies were identified (Campbell et al., 2005). However, unlike yeast cells, the human eIF2B bodies are more abundant and diverse in size.
eIF2 localizes with eIF2B bodies and can rapidly shuttle through these sites
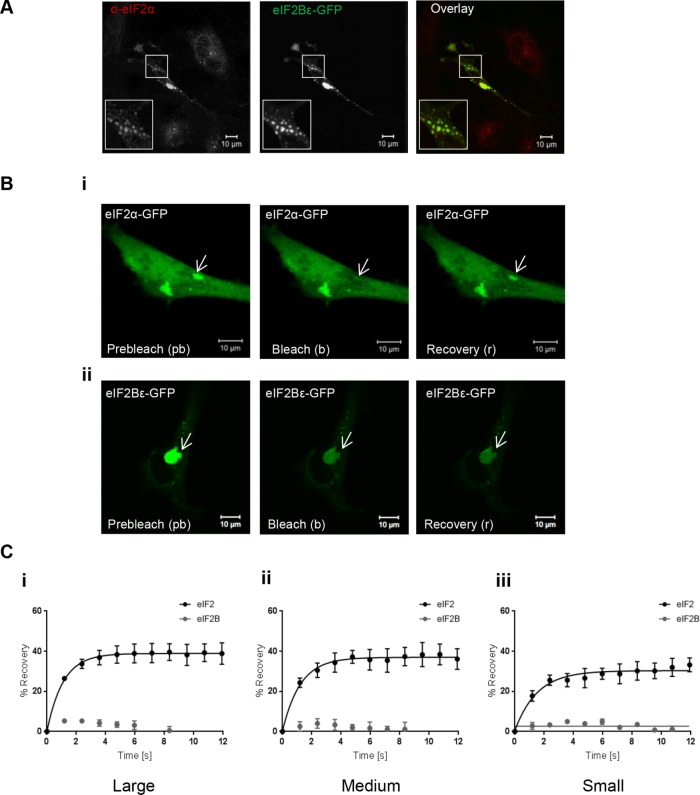
eIF2B is the GEF for eIF2, and a key feature of the yeast eIF2B bodies is the colocalization of eIF2 (Campbell et al., 2005). We next investigated if eIF2 localized to eIF2B bodies. To test this, cells expressing eIF2Bε-GFP were subjected to immunocytochemistry (ICC) with an anti-eIF2α antibody. eIF2α localized to all eIF2B bodies, independent of size (Figure 2A). In yeast, the shuttling of eIF2 through eIF2B bodies in live cells has been shown to correlate with eIF2B GEF activity (Campbell et al., 2005; Singh et al., 2011). To assess whether eIF2 in mammalian eIF2B bodies undergoes similar dynamics, fluorescence recovery after photobleaching (FRAP) analysis was carried out. Prebleach, bleach, and recovery data were collected for each size of eIF2B body (Figure 2B). No recovery of eIF2Bε-GFP was observed after FRAP across any of the different sizes of eIF2B bodies, suggesting that eIF2B is a relatively stable component of the eIF2B bodies. In contrast, recovery of eIF2α-GFP was observed for all categories of eIF2B bodies. Figure 2C shows the percentage of recovery relative to fluorescence in prebleach images for eIF2α-GFP and eIF2Bε-GFP within i) large, ii) medium, and iii) small bodies over a 12-s time period.
FIGURE 2:
eIF2α associates with eIF2B bodies and can shuttle into these sites. (A) Confocal images of U373 cells transfected with eIF2Bε-GFP, fixed in methanol, and subject to ICC with a primary anti-eIF2α antibody, and visualized using an appropriate Alexa Fluor 568–conjugated secondary antibody. (B) FRAP analysis was carried out on (i) eIF2α-GFP localized to eIF2B bodies in U373 cells transfected with eIF2α-GFP and eIF2Bε-RFP to mark the eIF2B body and on (ii) eIF2Bε-GFP in U373 cells transfected with eIF2Bε-GFP. Panels show representative prebleach (pb), bleach (b), and recovery (r) images from FRAP experiments. (C) Normalized FRAP curves for eIF2α-GFP and eIF2Bε-GFP recovery to (i) large, (ii) medium, and (iii) small eIF2B bodies. For large, medium, and small bodies, FRAP analysis was performed on 10 bodies (n = 3). The percentage recovery is presented as mean ± SEM.
These data show that, as in S. cerevisiae (Campbell et al., 2005; Taylor et al., 2010) in human astrocytoma cells, eIF2 can rapidly transit through the eIF2B body, whereas eIF2B is a stable component. In addition, this shuttling of eIF2 was observed in all of the different-sized eIF2B bodies.
eIF2B subunits display unique localization patterns to different-sized eIF2B bodies
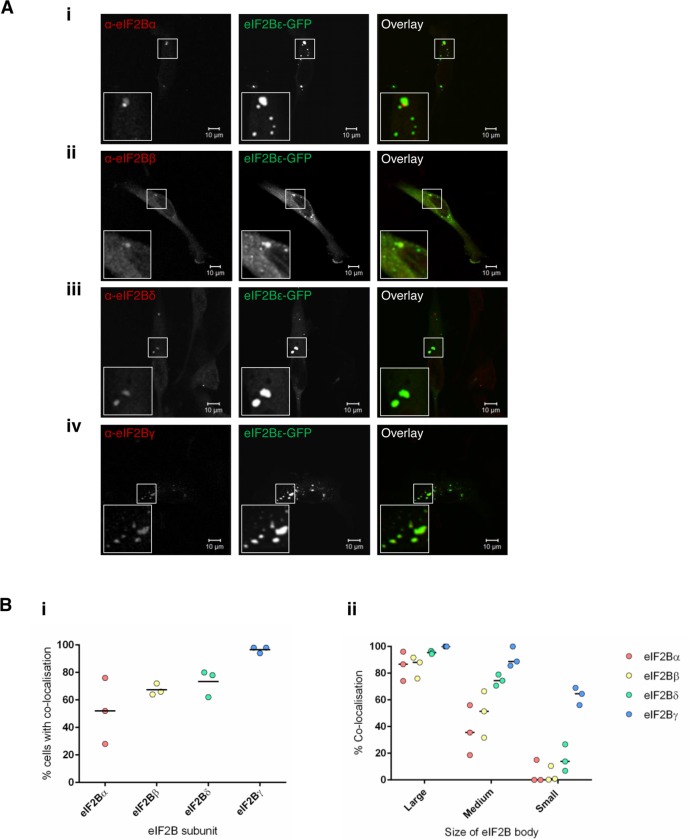
eIF2B exists as a decamer in its native form. Having shown that eIF2Bε localizes to cytoplasmic bodies and that eIF2 can shuttle through these foci, we next investigated whether other eIF2B subunits colocalized with these bodies. U373 cells expressing eIF2Bε-GFP were subjected to ICC with antibodies against the eIF2Bα, β, δ, and γ subunits. Representative images are shown in the panels of Figure 3A. The four eIF2B subunits show discrete patterns of colocalization to the eIF2Bε cytoplasmic bodies in cells containing small, medium, large, and a mixture of different-sized bodies. Intriguingly, 100% colocalization was not observed for all the subunits. For each subunit, we characterized the percentage of cells showing colocalized bodies, and these counts revealed that each eIF2B subunit had a unique pattern of colocalization with the eIF2Bε bodies (Figure 3Bi). Within these populations of cells, we next determined whether the percentage of colocalization observed correlated with the size of the eIF2B body (Figure 3Bii). Remarkably, a correlation was observed between the composition and size of the eIF2B body. For example, for the large eIF2B bodies, all of the subunits colocalized with a median percentage colocalization of greater than 85%. For the small population of eIF2B bodies, eIF2Bγ showed the highest level of colocalization (65%), with the regulatory subunits showing medians of less than 15% for eIF2Bδ and 0% for eIF2Bα and eIF2Bβ (Figure 3Bii). Like the large eIF2B bodies, all subunits colocalized with the medium eIF2B bodies; however, a greater variation among subunits was observed. eIF2Bγ again had the highest level of colocalization (86%); this was followed by eIF2Bδ (74%), eIF2Bβ (51%), and eIF2Bα (36%).
FIGURE 3:
eIF2B (α–γ) subunits colocalize with eIF2B bodies. (A) Confocal images of U373 cells transfected with eIF2Bε-GFP, fixed in methanol, and subject to ICC with primary (i) anti-eIF2Bα, (ii) anti-eIF2Bβ, (iii) anti-eIF2Bδ, and (iv) anti-eIF2Bγ antibodies. All antibodies were visualized using appropriate secondary antibodies conjugated to Alexa Fluor 568. (B) (i) For eIF2Bα, eIF2Bβ, eIF2Bδ, or eIF2Bγ subunits, the percentage of cells that showed colocalization to eIF2Bε-GFP bodies (n = 3 counts of 50 cells). Within the population of cells that showed colocalization, (ii) the median percentage of colocalization for eIF2Bα, β, δ or γ subunits with large, medium and small eIF2Bε-GFP bodies (n = 3 counts of 50 cells).
Here we have demonstrated that mammalian eIF2B bodies differ in size, and that this classification system broadly correlates to the subunits present. The large and medium bodies contain all subunits of eIF2B whereas, intriguingly, the small eIF2B bodies seem to represent a different subcomplex of eIF2B that predominately consists of eIF2Bγ and ε, the catalytic subunits of eIF2B. While these eIF2B subcomplexes have been shown to be formed in vitro (Li et al., 2004), this is one of the first pieces of evidence that suggest that these subcomplexes, lacking regulatory subunits of eIF2B, are formed in vivo.
Phosphorylated eIF2α localizes to large and medium eIF2B bodies during cellular stress and movement of eIF2 through these bodies is impaired
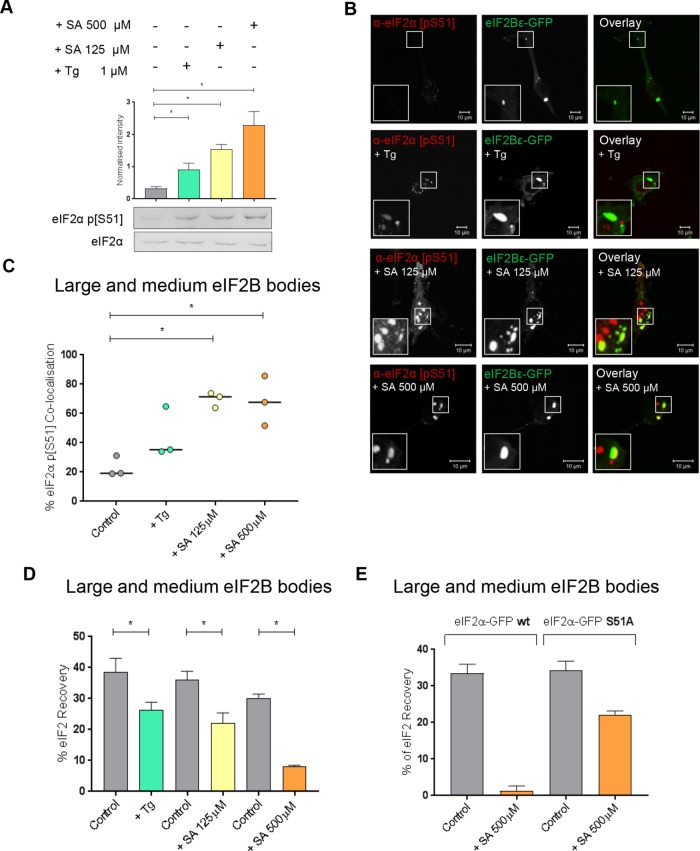
In response to various stress conditions, eIF2α is phosphorylated at serine 51 by an eIF2α kinase, inhibiting eIF2B activity and stimulating the ISR. This regulation is specifically carried out by the regulatory subunits of eIF2B, and in particular eIF2Bα, which recognizes and binds the phosphorylated eIF2α (Kimball et al., 1998). As the large and medium eIF2B bodies (greater than 3 µm2) contained all subunits, we hypothesized that only these bodies would respond to stress and so initially focused our analysis on these eIF2B bodies. SA and Tg were used to induce eIF2α phosphorylation, and the levels of eIF2α phosphorylation induced by each of these stresses were determined using Western blot analysis (Figure 4A). Two concentrations of SA treatment were used, resulting in various levels of eIF2α phosphorylation. The Tg treatment also induced eIF2α phosphorylation to a lesser degree than both SA treatments.
FIGURE 4:
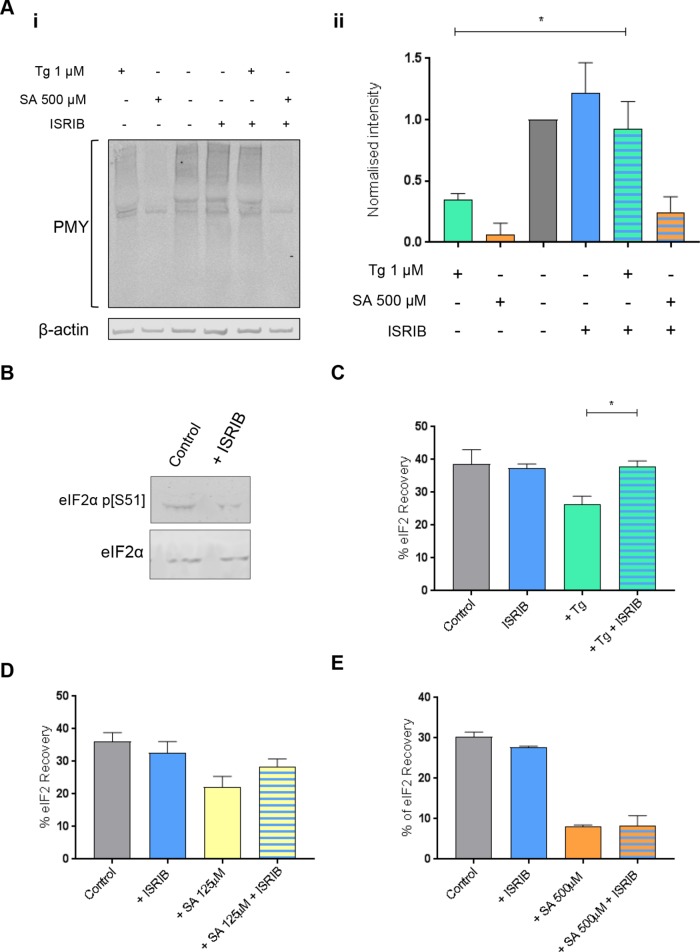
In the presence of Tg- or SA-induced cellular stress, phosphorylated eIF2α localizes to large and medium eIF2B bodies decreasing movement of eIF2 through these bodies. (A) Western blot analysis of the level of eIF2α and eIF2α p[S51] expression in U373 cells treated with 500 μM SA, 125 μM SA, or 1 µM Tg to induce cellular stress. Levels of phosphorylated eIF2α were normalized to levels of total eIF2α and presented as mean ± SD (n = 3). (B) U373 cells were transfected with eIF2Bε-GFP and treated with 500 μM SA, 125 μM SA, or 1 µM Tg. Cells were fixed in methanol and subject to ICC with a primary anti-eIF2α p[S51] antibody. The anti-eIF2α p[S51] antibody was visualized using an appropriate Alexa Fluor 568–conjugated secondary antibody and imaged using confocal microscopy. (C) The median percentage of anti-eIF2α p[S51] colocalized to large and medium eIF2Bε-GFP bodies (≥3 µm2) was determined in a population of 50 cells (n = 3). (D) FRAP analysis was carried out on eIF2α-GFP localized to large and medium eIF2B bodies in U373 cells transfected with eIF2α-GFP in addition to eIF2Bε-RFP to mark the eIF2B bodies. Cells were treated with 1 µM Tg, 125 μM SA, or 500 μM SA to induce cellular stress. FRAP analysis was performed on 10 bodies (n = 3). The mean ± SEM percentage of eIF2α-GFP recovery was determined from normalized FRAP recovery curves. (E) FRAP analysis was carried out on eIF2α-GFP localized to large and medium eIF2B bodies in U373 cells transfected with eIF2α-GFP wt or eIF2α-GFP S51A mutant, in addition to eIF2Bε-RFP to mark the eIF2B bodies. Cells were treated with 500 μM SA to induce cellular stress. FRAP analysis was performed on 10 bodies (n = 2). The mean ± SEM percentage of eIF2α-GFP recovery was determined from normalized FRAP recovery curves. p Values were derived from a Kruskal-Wallis test, followed by a Conover-Inman analysis, *p ≤ 0.05.
To determine whether phosphorylated eIF2α localized to the large and medium eIF2B bodies, ICC using an antibody to phosphorylated eIF2α was performed. Phosphorylated eIF2α localized to these eIF2B bodies; however, cytoplasmic foci distinct from the eIF2B bodies were also observed (Figure 4B). Phosphorylated eIF2α is known to aggregate in stress granule (SG) on the stress conditions used here (Kedersha et al., 1999), and thus the observed foci of phosphorylated eIF2α distinct from eIF2B bodies are likely to be SGs. The percentage colocalization of phosphorylated eIF2α in both large and medium bodies was determined and is shown in Figure 4C. In untreated cells, the majority of eIF2B bodies did not show colocalization with phosphorylated eIF2α localized; however, following induction of stress, phosphorylated eIF2α was observed to colocalize with eIF2B bodies (Figure 4C). Both concentrations of SA induced a significant increase in the percentage colocalization of phosphorylated eIF2α, whereas a smaller increase in the percentage colocalization was observed for Tg-treated cells (Figure 4C). This is likely due to the lower level of phosphorylated eIF2α induced by the Tg treatment (Figure 4A).
The observed increase in the level of phosphorylated eIF2α associated with the large and medium eIF2B bodies after stress (Figure 4C) is consistent with the higher affinity of phosphorylated eIF2α for eIF2B. The tighter binding sequesters eIF2B leading to decreased eIF2B GEF activity (Ramaiah et al., 1994; Pavitt et al., 1998). Based on our studies in yeast (Campbell et al., 2005), the presence of increased phosphorylated eIF2α in eIF2B bodies should reduce the level of eIF2 shuttling. To assess this, eIF2α-GFP shuttling was monitored in the large and medium bodies after treatment with either Tg or SA. In response to all stress treatments, these eIF2B bodies showed a significant decreased recovery of eIF2 (Figure 4D and Supplemental Figure S4A). The greatest decrease in recovery was observed for the higher concentration of SA (500 µM), followed by the lower concentration of SA (125 µM) and then Tg. This decrease in eIF2 recovery directly correlates with the levels of phosphorylated eIF2α observed in Figure 4A and a reduction in total protein synthesis as observed by puromycin incorporation assays (Figure 5A). To confirm that this reduced shuttling of eIF2 was in response to the increased levels of phosphorylated eIF2α, FRAP activity was analyzed in the presence of an eIF2α mutant where Ser51 is replaced by Ala. This mutant cannot be phosphorylated, as confirmed by Western blot (Supplemental Figure S4C) and therefore cannot inhibit eIF2B GEF activity during cellular stress. FRAP analysis following stress treatment with the higher concentration of SA (500 µM) revealed that the eIF2α S51A mutant could still shuttle through the eIF2B body, while the wild-type (WT) eIF2α exhibited severe reduction in shuttling (Figure 4E and Supplemental Figure S4B). These data suggest that the ability of eIF2 to recover within eIF2B bodies is specifically influenced by eIF2α phosphorylation-dependent eIF2B regulation.
FIGURE 5:
ISRIB reverses the effects that the ISR has on eIF2 shuttling in large and medium eIF2B bodies in a manner dependent on levels of phosphorylated eIF2α. (A) (i) Puromycin incorporation assays were carried out on U373 cells treated with 200 nM ISRIB or 1 μM Tg and 500 μM SA alone or in combination with 200 nM ISRIB. (ii) Levels of puromycin where normalized to β-actin and are presented as mean ± SD for each treatment normalized to control cells (n = 3). (B) Western blot analysis of the level of eIF2α and eIF2α p[S51] expression in U373 cells treated with 200 nM ISRIB for 1 h. FRAP analysis was carried out on eIF2α-GFP localized to large and medium eIF2B bodies in U373 cells transfected with eIF2α-GFP and eIF2Bε-RFP to mark the eIF2B bodies. Cells were treated with 200 nM ISRIB alone or in combination with (C) 1 µM Tg, (D) 125 μM SA, or (E) 500 μM SA. FRAP analysis was performed on 10 bodies (n = 3) the mean ± SEM percentage of eIF2α-GFP recovery was determined from normalized FRAP recovery curves; control and stress treatment data from Figure 4D are reshown for relevance. p Values derived from a Kruskal-Wallis test, followed by a Conover-Inman analysis, *p ≤ 0.05.
ISRIB reverses the effects that the ISR has on eIF2 shuttling in large and medium eIF2B bodies dependent on levels of phosphorylated eIF2α
ISRIB has been identified as a small molecule that renders cells insensitive to the inhibitory effects of eIF2α phosphorylation (Sidrauski et al., 2013, 2015a; Halliday et al., 2015) through stabilization of decameric eIF2B (Sidrauski et al., 2015b; Tsai et al., 2018; Zyryanova et al., 2018). We therefore investigated whether ISRIB could attenuate the inhibitory effect of eIF2α phosphorylation on protein synthesis and eIF2 shuttling through the large and medium eIF2B bodies. ISRIB treatment alone did not affect levels of phosphorylated eIF2α (Figure 5B) and had little effect on the rate of protein synthesis or eIF2 recovery to eIF2B bodies (Figure 5, A and C). Puromycin incorporation assays confirmed that ISRIB could overcome the inhibitory effects of Tg on protein synthesis but not at the higher concentration of SA (500 µM) (Figure 5A). These data are consistent with previous studies that highlighted that ISRIB can restore protein synthesis when the ratio of nonphosphorylated eIF2 exceeds that of phosphorylated eIF2 (Sidrauski et al., 2015a).
FRAP analysis was carried out on cells treated with Tg or SA in the presence of ISRIB. A carrier-only control (dimethyl sulfoxide–treated cells) did not show any significant difference in the recovery of eIF2α (Supplemental Figure S5). For Tg-treated cells, ISRIB was found to significantly increase the recovery of eIF2 to untreated levels (Figure 5C and Supplemental Figure S5Ai). A similar trend was observed for the lower concentration of SA (125 µM) (Figure 5D and Supplemental Figure S5Aii), whereas ISRIB could not rescue the recovery of eIF2 at the higher SA concentration (500 µM) (Figure 5E and Supplemental Figure S5Aiii). Thus, our favored interpretation for these observations is that under high levels of phosphorylated eIF2α, and therefore a higher ratio of phosphorylated to unphosphorylated protein, the level of ISRIB used is incapable of reversing the inhibitory effect of eIF2α phosphorylation on the recovery of eIF2.
During stress small eIF2B bodies increase in number and show an increased recovery of eIF2 and a redistribution of eIF2Bδ
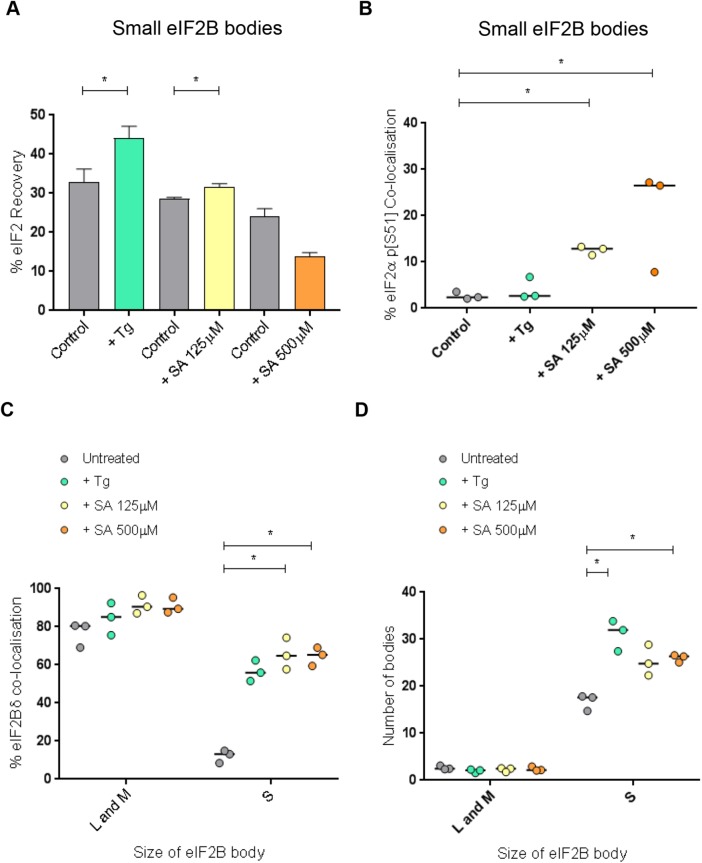
We have shown that in response to cellular stress, phosphorylated eIF2α localizes to large and medium eIF2B bodies and this correlates with a decrease in the rate of eIF2 shuttling, suggesting that these bodies can be regulated via the ISR. We hypothesize that this regulation is due to the presence of regulatory subunits within these large and medium bodies. Interestingly, we have also shown that eIF2 can shuttle through all eIF2B bodies including small bodies (Figure 2Ciii). As these small eIF2B bodies predominantly contained eIF2Bγ and eIF2Bε (Figure 3Bii), we hypothesized that the recovery of eIF2 in small bodies would not be affected during stress. Surprisingly, both Tg and the lower concentration of SA (125 µM) significantly increased the recovery of eIF2 in these small bodies, whereas in contrast, a decrease in the percentage recovery of eIF2 was observed for the higher concentration of SA (500 µM) (Figure 6A and Supplemental Figure S5B, i–iii). As the decrease in shuttling of eIF2 in the large and medium eIF2B bodies correlated with the presence of phosphorylated eIF2α, we hypothesized that this lack of recovery of eIF2 in small bodies at the higher concentration of SA (500 µM) may be due to an increase in localization of phosphorylated eIF2α. In agreement with this when colocalization of phosphorylated eIF2α with small bodies was analyzed, an increase in localization was observed under all stress treatments with the greatest increase observed for the higher concentration of SA (Figure 6B).
FIGURE 6:
On stress, the number of small eIF2B bodies within the cell increase and these bodies show an increased recovery of eIF2 and a redistribution of eIF2Bδ. (A) FRAP analysis was carried out on eIF2α-GFP localized to small eIF2B bodies in U373 cells transfected with eIF2α-GFP and eIF2Bε-RFP to mark the eIF2B bodies. Cells were treated with 1 µM Tg, 125 μM SA, or 500 μM SA. FRAP analysis was performed on 10 bodies (n = 3) and the mean ± SEM percentage of eIF2α-GFP recovery was determined from normalized FRAP recovery curves. (B) U373 cells were transfected with eIF2Bε-GFP and treated with 1 µM Tg, 125 μM SA, or 500 μM SA. Cells were fixed in methanol and subject to ICC with a primary anti-eIF2α p[S51] antibody. The anti-eIF2α p[S51] antibody was visualized using an appropriate Alexa Fluor 568–conjugated secondary antibody, and the median percentage of anti-eIF2α p[S51] colocalized to small eIF2Bε-GFP bodies was determined in a population of 50 cells (n = 3). (C) U373 cells were transfected with eIF2Bε-GFP and treated with 1 µM Tg, 125 μM SA, or 500 μM SA. Cells were fixed in methanol and subject to ICC with a primary anti-eIF2Bδ antibody and visualized using an appropriate secondary antibody conjugated to Alexa Fluor 568. Within a population of 50 cells, the median percentage of colocalization between anti-eIF2Bδ and small eIF2B bodies was determined (n = 3). (D) U373 cells were transfected with eIF2Bε-GFP, and treated with 1 µM Tg, 125 μM SA, or 500 μM SA. Counts were performed to determine the median number of large and medium (L and M) or small (S) eIF2B bodies within a population of 50 cells (n = 3). p Values derived from a Kruskal-Wallis test, followed by a Conover-Inman analysis, *p ≤ 0.05.
Interestingly, increased GEF activity of eIF2B has been linked to subunit composition, specifically the presence of regulatory subunits (Fabian et al., 1997; Williams et al., 2001; Dev et al., 2010; Liu et al., 2011). To address whether the stress treatments had any impact on the distribution of regulatory subunits within the small eIF2B bodies, which could possibly explain this increased recovery of eIF2, ICC analysis was carried out. Interestingly, the percentage of eIF2Bδ colocalizing to the small eIF2B bodies increased by over 40% for all stresses (Figure 6C).
Having shown under stress the subunit composition of small eIF2B bodies is altered, we were intrigued to determine whether the number of these bodies also changed. We carried out counts across 50 cells and observed no change to the average number of large and medium bodies under stress; however, the number of small bodies increased for all stress treatments (Figure 6D). These data suggest small eIF2B bodies show a different response to stress when compared with the large and medium bodies.
ISRIB treatment mimics the effect of stress on small eIF2B bodies
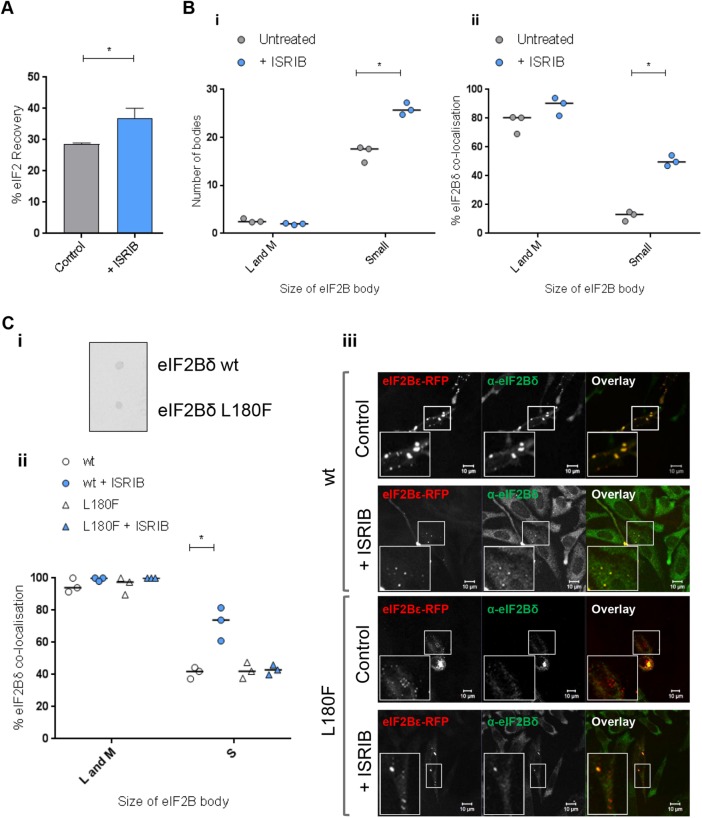
We next investigated whether ISRIB had any impact on small body activity and composition. Rather surprisingly, we identified that ISRIB treatment significantly increased the degree to which eIF2 shuttles through the small eIF2B bodies (Figure 7A and Supplemental Figure S5Biv). The level of increase of eIF2 shutting seen on ISRIB treatment is similar to the increase observed for Tg and SA (125 µM) treatments. Furthermore, an increased number of small bodies and also localization of eIF2Bδ to the small bodies was observed (Figure 7B), a similar trend to all stress conditions (Figure 6, C and D).
FIGURE 7:
ISRIB treatment mimics the effect of stress on small eIF2B bodies. (A) FRAP analysis was carried out on eIF2α-GFP localized to small eIF2B bodies in U373 cells transfected with eIF2α-GFP and eIF2Bε-RFP to mark the eIF2B bodies. Cells were treated with 200 nM ISRIB and FRAP analysis was performed on 10 bodies (n = 3) The percentage of eIF2 that recovered is presented as mean ± SEM. (B) U373 cells were transfected with eIF2Bε-GFP and treated with 200 nM ISRIB. (i) The median number of large and medium (L and M) or small (S) eIF2Bε-GFP bodies was determined within a population of 50 cells (n = 3). (ii) Cells were fixed in methanol and subject to ICC with a primary anti-eIF2Bδ antibody and visualized using an appropriate secondary antibody conjugated to Alexa Fluor 568. Within a population of 50 cells, the median percentage of colocalization between anti-eIF2Bδ and large and medium (L and M) or small (S) eIF2Bε-GFP bodies was determined (n = 3). (C) (i) CHO cells containing eIF2Bδ (WT) or eIF2Bδ (L180F) were subject to dot blot analysis for anti-eIF2Bδ. (ii) The cells were transfected with eIF2Bε-RFP and treated with 200 nM ISRIB. Cells were fixed in methanol and subject to ICC with a primary anti-eIF2Bδ antibody and visualized using an appropriate secondary antibody conjugated to Alexa Fluor 568. Within a population of 25 cells, the median percentage of colocalization between anti-eIF2Bδ and large and medium (L and M) or small (S) eIF2Bε-RFP bodies was determined (n = 3). (iii) Representative images showing the localization of eIF2Bδ to small bodies in eIF2Bδ (WT) and eIF2Bδ (L180F) CHO cells in the presence or absence of ISRIB treatment. p Values derived from a Kruskal-Wallis test, followed by a Conover-Inman analysis, *p ≤ 0.05.
ISRIB has been shown to restore eIF2B activity through stabilization of eIF2B in its decameric form (Tsai et al., 2018; Zyryanova et al., 2018). However, our data suggest that ISRIB may also promote the localization of eIF2Bδ to small eIF2B bodies to which catalytic subunits localize. To confirm that this movement of eIF2Bδ was a specific response to ISRIB exposure, we made use of an ISRIB-resistant mutation in eIF2Bδ (L180F) (Sekine et al., 2015). CHO cells containing wild type (WT) eIF2Bδ or mutant eIF2Bδ (L180F) were transfected with eIF2Bε-red fluorescent protein (RFP), and a similar pattern of eIF2B body localization was observed for both sets of cells (Figure 7Ciii). To ensure that the eIF2Bδ mutant did not affect antibody recognition, dot blot analysis was carried out. Analysis of extracts prepared from both the WT and the mutants cells showed eIF2Bδ was recognized to similar levels in both sets of cells (Figure 7Ci). On ISRIB treatment, as for U373 cells, an increase in eIF2Bδ localization to small bodies was observed for the WT eIF2Bδ. Interestingly, in the mutant cells, no increase in distribution of mutant eIF2Bδ (L180F) to small bodies was observed (Figure 7C, ii and iii). These data suggest that the redistribution of eIF2Bδ to small bodies in response to ISRIB is a direct effect of ISRIB interaction with eIF2Bδ.
DBM treatment has an effect similar to that of ISRIB on small eIF2B body dynamics
We next determined whether this effect of ISRIB on small bodies was unique to ISRIB or whether other drugs, which block the activation of the ISR, also result in similar effects. To test this, we exposed cells to the drug DBM. DBM is a naturally occurring structural analogue of curcumin, with widely reported anti-cancer properties, and was recently identified in a drug screen to identify compounds that decrease the unfolded protein response (UPR) activity in cells and allow a restoration of protein synthesis (Halliday et al., 2017).
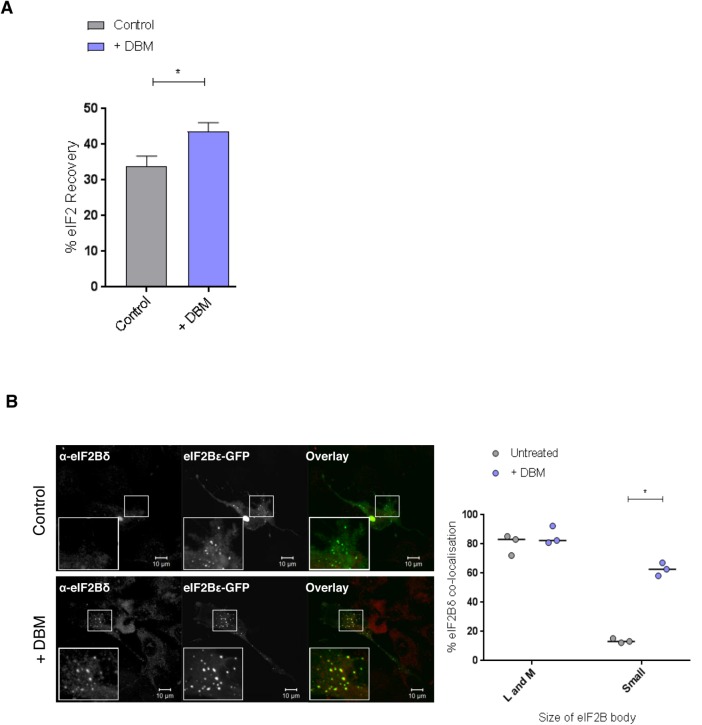
FRAP analysis on DBM-treated cells showed that, like ISRIB, DBM did not affect the recovery of eIF2 in large or medium bodies but increased the degree to which eIF2 shuttles through the small eIF2B bodies (Figure 8A and Supplemental Figure S5Bv). Intriguingly, a similar increase in eIF2Bδ localization was observed in the small bodies on DBM treatment (Figure 8B). These data suggest that DBM may have an effect on small bodies similar to that observed for ISRIB.
FIGURE 8:
DBM shows an effect on small eIF2B bodies similar to that of ISRIB. (A) FRAP analysis was carried out on eIF2α-GFP localized to small eIF2B bodies in U373 cells transfected with eIF2α-GFP and eIF2Bε-RFP to mark the eIF2B bodies. Cells were treated with 20 µM DBM. FRAP analysis was performed on 10 bodies (n = 3), and the percentage of eIF2 that recovered is presented as mean ± SEM. (B) U373 cells were fixed in methanol and subject to ICC with a primary anti-eIF2Bδ antibody and visualized using an appropriate secondary antibody conjugated to Alexa Fluor 568. Within a population of 50 cells, the median percentage of colocalization between anti-eIF2Bδ and large and medium (L and M) or small (S) eIF2Bε-GFP bodies (n = 3) is shown. p Values derived from a Kruskal-Wallis test, followed by a Conover-Inman analysis, *p ≤ 0.05.
DISCUSSION
Previous work in the yeasts S. cerevisiae and C. albicans has shown that eIF2B localizes to cytoplasmic foci, termed eIF2B bodies (Campbell et al., 2005; Egbe et al., 2015), and eIF2 was shown to shuttle through these bodies. Three different strategies to decrease the guanine nucleotide exchange function of eIF2B all inhibited eIF2 shuttling into the foci, showing that in yeast, the measurement of eIF2 shuttling through the eIF2B body correlates precisely with eIF2B GEF activity (Campbell et al., 2005). Here we have extended these studies to look at the functional localization of eIF2B in mammalian cells, which has not previously been evaluated.
We show that eIF2B bodies exist in mammalian cells and eIF2 can shuttle through these bodies. Unlike in yeast where cells exhibited a single eIF2B body, mammalian cells exhibit populations of different-sized bodies. This difference in eIF2B body distribution from yeast to mammalian cells is likely due to differences in organism complexity. Structural analysis of eIF2B has shown that eIF2B forms a heterodecamer in its native form (Gordiyenko et al., 2014; Wortham et al., 2014; Kashiwagi et al., 2016, 2017), but can also form eIF2B subcomplexes that contain varying degrees of the individual eIF2B subunits on overexpression. Biochemical analysis showed that these eIF2B subcomplexes differ in GEF activity in vitro (Liu et al., 2011; Wortham et al., 2014). The eIF2B subunit localization data presented here suggest that the eIF2B subcomplexes, which have been shown to form in vitro, also exist in mammalian cells under normal growth conditions. In this study, we have categorized the different-sized eIF2B bodies based on their eIF2B subunit composition, with large and medium bodies (greater than 3 µm2) containing all subunits and smaller bodies (less than 3 µm2) containing predominantly catalytic subunits, eIF2Bγ and eIF2Bε.
Activation of the ISR by diverse cellular stresses results in the phosphorylation of eIF2α at serine 51. Phosphorylated eIF2 is a competitive inhibitor of eIF2B (Rowlands et al., 1988; Dever et al., 1995) causing the down-regulation of translation initiation. The eIF2B regulatory subunits (α, β, and δ) are essential for mediating the control of eIF2B activity under stress (Pavitt et al., 1997; Krishnamoorthy et al., 2001), and eIF2B complexes containing the regulatory subunits are known to display a higher affinity for eIF2 when present in its phosphorylated form (Pavitt et al., 1998; Kashiwagi et al., 2017). We postulate that having populations of eIF2B subcomplexes, where regulatory subunits are either present or absent, may be an important mechanism in the cellular response to stress. FRAP analysis of eIF2 shuttling through the eIF2B bodies has revealed that during stress, large and medium eIF2B bodies (to which the eIF2B regulatory subunits predominately localize) show attenuated eIF2 shuttling in response to ER and oxidative stress, induced by Tg and SA, respectively. Analysis of the cellular localization of phosphorylated eIF2 in these cells demonstrated that phosphorylated eIF2 localized predominately to these bodies. Additionally, puromycin incorporation assays show a decrease in total protein synthesis under the same stress conditions, demonstrating a correlation among global protein synthesis, the shuttling of eIF2 through, and localization of phosphorylated eIF2α to these eIF2B bodies. In addition, the decrease in shuttling of eIF2 through these bodies during stress is directly related to the phosphorylation of eIF2. eIF2 harboring an S51A mutation that prevents phosphorylation does not show the same decrease in shuttling observed for WT eIF2 during stress. This evidence supports our previous conclusions in yeast where we observed that the rate of recovery of eIF2 in FRAP analysis is relative to the amount of GEF activity in the eIF2B bodies (Campbell et al., 2005; Taylor et al., 2010).
ISRIB has recently been identified as an inhibitor of the ISR, reversing the effects of eIF2α phosphorylation (Sidrauski et al., 2013, 2015a). ISRIB has been shown to stabilize eIF2B in its decameric form, increasing eIF2B GEF activity and restoring translation in stressed cells (Tsai et al., 2018; Zyryanova et al., 2018). As the shuttling of eIF2 in large and medium bodies (which contain all subunits) decreases in a manner correlating with an increased presence of phosphorylated eIF2α, we hypothesized that if ISRIB can restore eIF2B activity, it would rescue eIF2 shuttling within these foci. We found this hypothesis to be true for cells treated with Tg and a low concentration of SA (125 µM). However, ISRIB was unable to rescue eIF2 shuttling through these bodies in cells treated with a higher SA concentration (500 µM). This higher concentration of SA induced greater levels of eIF2α phosphorylation; therefore, our results suggest that ISRIB can restore eIF2 shuttling in a manner that is dependent on the precise levels of eIF2α phosphorylation. This is in agreement with previous studies where ISRIB has been shown to restore eIF2B activity when levels of nonphosphorylated eIF2 are greater than levels of phosphorylated eIF2 (Sidrauski et al. 2015b).
In addition to observing large and medium eIF2B bodies (containing all subunits of eIF2B), we also observed small bodies (less than 3 µm2), which predominately contained eIF2Bγ and eIF2Bε. In vitro GEF assays, in both yeast and mammalian cells, show that eIF2Bγε heterodimers exhibit GEF activity that is unregulatable by phosphorylated eIF2α (Pavitt et al., 1998; Li et al., 2004). We showed that eIF2 localized to these small bodies and was mobile within them. We therefore hypothesized that shuttling of eIF2 would not be down-regulated by stress due to the absence of regulatory subunits in these bodies. Intriguingly, we found eIF2 shuttling through small bodies was in fact significantly increased by cellular stress induced by both Tg and a low concentration of SA (125 µM). Therefore, eIF2 shuttling through these small bodies is not down-regulated by stress and is in fact enhanced. Biochemical assays in yeast have demonstrated that increasing the expression of regulatory eIF2B subunits can enhance GEF activity (Fabian et al., 1997; Williams et al., 2001; Dev et al., 2010; Liu et al., 2011). Interestingly, we have observed an increase in the localization of eIF2Bδ to eIF2B small bodies under stress treatment, which may be responsible for the observed increase in eIF2 shuttling. This is suggestive of the formation of a currently unidentified subcomplex containing eIF2Bδ, γ, and ε, which has a role in the cellular ISR. This complex may not have been previously identified in research that analyzed eIF2B subcomplexes, as such studies did not observe cells under stress. Future studies are required to directly test this hypothesis.
In contrast, when cells were treated with a higher concentration of SA (500 µM), shuttling of eIF2 through small eIF2B bodies was reduced. This higher SA treatment induced a greater increase in the level of eIF2α phosphorylation when compared with the other two stress treatments. Phosphorylated eIF2α is a competitive inhibitor of eIF2B that exhibits greater affinity for eIF2B than unphosphorylated eIF2α (Rowlands et al., 1988). In these cells, we saw a greater induction of phosphorylated eIF2α localizing to these small bodies, and thus our favored explanation for this result is that in the presence of high levels of phosphorylated eIF2α, all eIF2B complexes become inhibited independent of subunit makeup.
In recent years, low levels of cellular stress have been shown to be protective. Preconditioning cells through the induction of low levels of phosphorylated eIF2α appears to prime cells to overcome stress (Lu et al., 2004; Lewerenz and Maher, 2009). Our data suggest that small eIF2B bodies have greater GEF activity under stress, and perhaps these small bodies may be important in providing a source of eIF2B activity that is not inhibited by stress, thus allowing cells to respond and recover from stress. Recent work in S. cerevisiae has observed that induction of stress results in an increase in eIF2B bodies (Moon and Parker, 2018). Interestingly, we did not see an increase in the number of large eIF2B bodies; rather, we observed an increase in the number of small bodies under stress. This further suggests that in mammalian cells, these smaller bodies are important for the cell to respond to stress. Further investigations into the effects that stress has on eIF2B composition may provide a greater insight into how cells respond to stress.
The change in small eIF2B body subunit dynamics in response to stress also appears to be driven by ISRIB treatment, where we observed similar increases in the number of small bodies, eIF2 shuttling, and redistribution of eIF2Bδ to these bodies. Mutational analysis has confirmed that this redistribution of eIF2Bδ is a direct effect of ISRIB. These results appear somewhat self-contradictory, as cellular stress promotes eIF2α phosphorylation, inhibiting eIF2B GEF activity, whereas ISRIB opposes these effects and promotes eIF2B GEF activity. Recent data have shown that ISRIB can stabilize decameric eIF2B and increase the GEF activity in a dose-dependent manner (Wong et al., 2018). In addition to these effects, our data suggest that during stress, ISRIB promotes the formation of small eIF2B bodies containing eIF2Bδ, which appears to have increased GEF activity that is not down-regulated on low levels of stress. It has previously been suggested that in addition to its role in stabilizing the eIF2B decamer, ISRIB may also enhance the basal activity of eIF2B by providing a source of eIF2B that is not inhibited by phosphorylated eIF2α, allowing sufficient levels of ternary complex to sustain protein translation and overcome stress (Sidrauski et al., 2015b). We therefore hypothesize that the effect of ISRIB on the small eIF2B bodies may be responsible for providing a source of eIF2B that is active during stress.
Recently, the drug DBM was identified to have similar properties to ISRIB in decreasing UPR activity in cells and allowing a restoration of protein synthesis (Halliday et al., 2017). However, unlike ISRIB, DBM was shown not to stabilize the eIF2B decameric complex, and its mechanism of action remains largely unknown. DBM-treated cells also showed an increased localization of eIF2Bδ to small bodies, correlating with an increase in eIF2 shuttling through these bodies, and these data suggest that eIF2Bδ, γ, and ε subcomplexes may be involved in DBM’s mechanism of action, again providing a source of eIF2B that can remain active under stress conditions.
eIF2B consolidates into large cytoplasmic bodies in both yeasts and mammalian cells. The situation in mammalian cells is more intricate, as eIF2B bodies of various sizes exist with various subunit compositions. The large and medium eIF2B bodies harbor regulatory subunits and likely represent sites of GEF activity vulnerable to stress-induced repression. The small eIF2B bodies are mainly composed of catalytic eIF2B subunits and appear to be differentially regulated by the ISR, likely providing a level of eIF2B GEF activity during stress that allows cells to survive and respond. The presence of these diverse bodies in different abundances may allow different cell types to harbor unique stress responses and perhaps contribute to the tissue specificity of VWM, a key area of future research.
MATERIALS AND METHODS
Cell culture
U373 astrocytoma cells were cultured in MEM, supplemented with 10% (vol/vol) fetal bovine serum (FBS), 1% (wt/vol) nonessential amino acids, 1% (wt/vol) sodium pyruvate, 1% (wt/vol) glutamine, and 1% (wt/vol) penicillin/streptomycin, all purchased from Life Technologies. MG-63 cells were cultured in RPMI supplemented with 10% (vol/vol) FBS and 1% (wt/vol) penicillin/streptomycin. HEK293 and HepG2 cells were cultured in DMEM supplemented with 10% FBS (vol/vol) and 1% (wt/vol) penicillin/streptomycin. CHO-C30 cells and CHO-C30 cells harboring the L180F mutation within the eIF2B4 gene (Sekine et al., 2015) were a kind gift from D. Ron (Cambridge Institute for Medical Research). Cells were cultured in Nutrient Mixture F12 Ham (Sigma) supplemented with 10% fetal calf serum (vol/vol) (FetalClone II; Thermo) and 1% (wt/vol) penicillin/streptomycin, (Life Technologies). All cells were maintained at 37°C under 5% CO2 and were routinely tested for contamination with MycoAlert Mycoplasma Detection Kit purchased from Lonza (Slough, UK).
Plasmids
pCMV6-AC-GFP plasmid vectors encoding EIF2B5 and EIF2S1 were purchased from OriGene (Rockville, MD). The coding open reading frame (ORF) of EIF2B5 from the pCMV6-AC-GFP vector was cloned into a pCMV6-AC-RFP vector (OriGene). The construct was verified by sequencing. Site-directed mutagenesis was performed on the EIF2S1 plasmid to generate an S51A mutation using a QuikChange II Site-Directed Mutagenesis Kit (Agilent, Stockport, UK) in accordance with manufacturer’s instructions. The construct was verified by sequencing. pHM2Bε was a kind gift from N. Wortham (University of Southampton, Southhampton, UK).
Transient transfections
One day prior to transfection, CHO cells were seeded at a density of 1 × 105 cells, and all other cells were seeded at a density of 8 × 104 cells in a six-well plate or fluorodish. Transfections were performed by chemical transfection with 1 mg/ml polyethylenimine (PEI; Sigma-Aldrich, Dorset, UK). Plasmid DNA (1–3 µg) was diluted in 100 µl of serum and antibiotic-free cell culture medium. Diluted DNA was mixed with PEI and incubated at room temperature (RT) for 10 min. The volume of PEI used was based on a 3:1 ratio of PEI (µg):plasmid DNA (µg). Antibiotic-free cell culture media (600 µl) was added to the transfection mix, and the total volume was added to the cells. Cells were incubated at 37°C for 23 h, with an additional 2 ml of antibiotic-free media added at the 2.5-h point. Cell culture media was changed to complete media and cells were incubated at 37°C for 24–48 h prior to imaging.
Protein analysis
Cells were washed with phosphate-buffered saline (PBS) (Sigma-Aldrich) and lysed in CelLytic M (Sigma-Aldrich) containing protease inhibitor cocktail (Sigma-Aldrich), as per manufacturer’s instructions. Cell lysates were subject to a bicinchoninic acid (BCA) protein assay to determine the protein concentration. Bovine serum albumin (BSA; Sigma-Aldrich) was diluted in CelLytic M to make appropriate standards to generate a standard curve. Ten microliters of each sample/standard was incubated with 200 µl of BCA reagent (0.4% [wt/vol] copper sulfate in BCA) for 30 min and the absorbance at 570 nm determined. Protein samples were concentrated, where appropriate, using Viva spin 2 columns (Fisher Scientific) in accordance with the manufacturer’s instructions. For dot blot analysis, 1 μl of protein samples was applied to a nitrocellulose membrane and allowed to air dry. Membranes were blocked in Tris-buffered saline supplemented with 0.1% (vol/vol) Tween 20 (TBST) and 5% (wt/vol) nonfat milk (Premier Foods, London, UK) for 1 h. Primary antibodies were diluted in block solution and incubated with membranes overnight. Membranes were then washed in TBST and incubated for 1 h with appropriate LiCor (Cambridge, UK) secondary antibodies diluted 1:10,000 in 5% (wt/vol) nonfat milk in TBST. Dot blots were visualized on a LiCor Odyssey Scanner with Image Studio Lite software. For Western blot analysis, protein samples were diluted in 4 × SDS–PAGE sample buffer (Expedeon, Swavesey, UK) and incubated at 95°C for 4 min. Total protein (60 μg) was resolved on a 10% polyacrylamide gel and transferred onto nitrocellulose membrane. Membranes were blocked in 5% (wt/vol) nonfat milk or BSA in TBST and probed with primary antibodies diluted in TBST supplemented with 5% (wt/vol) nonfat milk. The following antibodies were used: eIF2Bε (1:500, ARP61329_P050; Aviva Systems Biology, San Diego, CA), eIF2Bγ (1:500, sc-137248; Santa Cruz Biotechnology, Dallas, TX), eIF2α (1:100, sc-11386; Santa Cruz Biotechnology), and phospho-eIF2α (ser51) (1:1000, 44728G, Invitrogen, Fisher Scientific); and β-actin (1:1000, ab8224; Abcam, Cambridge, UK). Membranes were then washed with TBST, probed with LiCor secondary antibodies (1:10,000, goat anti-rabbit IRDye 680RD P/N 925-68071 and 1:10,000 goat anti-mouse IRDye 800CW P/N 925-32210; LiCor), and visualized on a LiCor Odyssey Scanner with Image Studio Lite software.
ICC
Cells were grown on coverslips in six-well plates and transfected as described above. Cells were fixed in ice-cold methanol (Fisher Scientific) at –20°C for 15 min. Following fixation, cells were washed with PBS supplemented with 0.5% (vol/vol) Tween 20 (PBST), three times for 3 min, and then blocked in PBS supplemented with 1% (wt/vol) BSA. Cells were then washed with PBST, three times for 3 min, and probed with primary antibodies diluted in PBS supplemented with 1% (wt/vol) BSA overnight at 4°C. The following antibodies were used: eIF2Bα (1:25, 18010-1-AP; Proteintech, Manchester, UK), eIF2Bβ (1:25, 11034-1-AP; Proteintech), eIF2Bδ (1:50, sc-271332; Santa Cruz Biotechnology), eIF2Bγ (1:50, sc-137248; Santa Cruz Biotechnology), eIF2Bε (1:500, ARP61329_P050; Aviva Systems Biology), eIF2α (1:20, FL-315 sc-11386; Santa Cruz Biotechnology), and phospho-eIF2α (ser51) (1:100, ab32157; Abcam), eIF2Bγ (1:100, 11296-2-AP; Proteintech), myc (1:100, ab18185; Abcam), G3BP (1:100, ab56574; Abcam), eIF3b (1:100, ab40799; Abcam), poly-ubiquitinylated conjugates, and FK1 (1:100, BML-PW8805; Enzo). Following primary antibody incubation, cells were washed with PBST, four times for 3 min, probed with an appropriate Alexa Fluor–conjugated secondary antibody (Thermo Fisher Scientific), and diluted in PBS supplemented with 1% (wt/vol) BSA for 1 h at RT. Cells were then washed with PBST four times for 3 min and mounted using VECTASHIELD HardSet Antifade Mounting Medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Cells were viewed on a Zeiss LSM 510 or Zeiss LSM 800 confocal microscope.
Confocal Imaging and FRAP analysis
Imaging was performed using either a Zeiss LSM 510 or a Zeiss LSM 800 confocal microscope. The LSM 510 confocal was used with Zeiss 2009 software, a 40 X plan-apochromat oil objective, and an argon laser with a maximum output of 25 mW at 55% laser transmission. The LSM 800 confocal was used with Zen Blue software, a 40X plan-apochromat oil objective, and a diode laser with a maximum output of 10 mW at 0.2% laser transmission. For FRAP analysis, the LSM 510 confocal microscope was used. Bleaching was carried out with 23 iterations and 100% laser power (488-nm argon laser). An image was captured before bleaching, and then after bleaching, 12 images were captured for 589.82 ms, with a 600-ms interval. After generation of prebleach, bleach, and recovery images, the images from each experiment were analyzed in accordance to the methodology by Campbell and Ashe (2007). FRAP curves were fitted using a one-phase association equation. The percentage of eIF2 recovery was determined as the mobile phase of the recovery curve – the percentage of unbleached proteins that recovered.
Cell treatments
To induce cellular stress, cells were treated with 1 µM Tg for 1 h, or 125 µM SA for 30 min, or 500 µM SA for 1 h, with or without the addition of 200 nM ISRIB (Sigma-Aldrich). For DBM treatment, cells were incubated with 20 µM DBM for 1 h.
Puromycin incorporation assays
The 6.7 × 105 cells were seeded in T75 flasks. One day later, culture media were changed to fresh media and cells were either untreated or treated with 1 µM Tg or 500 µM SA with or without the addition of 200 nM ISRIB, as described above. For puromycin labeling, 91 µM puromycin (Thermo Fisher) and 208 µM emetine (Sigma-Aldrich) were added during the last 5 min before harvest. Cells were washed twice in ice-cold PBS containing 355 µM cycloheximide (Calbiochem), immunoblot analysis was carried out as described earlier using a primary antibody for puromycinylated protein (1:500, clone 12D10, MABE343; Merck), and a primary antibody for β-actin (1:2000, ab8227; Abcam) was used as a loading control.
Measurements and statistical analysis
To determine the statistical significance of the difference among groups of data, the data were first subject to a Shapiro-Wilk test for normality. Data were considered parametric when p < 0.05. All data were found to be nonparametric. For the comparison of three or more groups of data, a Kruskal-Wallis test was performed followed by a Conover-Inman post-hoc test. Differences in data were considered significant when p < 0.05.
Supplementary Material
Acknowledgments
We thank Graham Pavitt (University of Manchester) for helpful discussions and providing critical comments. We thank David Ron for kindly gifting us the CHO cells. We thank Noel Wortham (University of Southampton) and Nancy Kedersha (Harvard Medical School) for suggestions regarding antibodies. We thank our colleagues Christine Le Maitre and Lucy Crooks for their advice on statistical analysis. We gratefully acknowledge the support for this work through a graduate teaching assistant PhD studentship to R.E.H. from the Biomolecular Sciences Research Centre and Department of Biosciences and Chemistry at Sheffield Hallam University, Sheffield, UK.
Abbreviations used:
- BCA
bicinchoninic acid
- BSA
bovine serum albumin
- DBM
dibenzyolmethane
- eIF2
eukaryotic initiation factor 2
- eIF2B
eukaryotic initiation factor 2B
- FBS
fetal bovine serum
- FRAP
fluorescence recovery after photobleaching
- GCN2
general control nondepressible 2
- GDF
GDI displacement factor
- GDI
GDP dissociation inhibitor
- GDP
guanosine diphosphate
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GTP
guanosine triphosphate
- ICC
immunocytochemistry
- ISR
integrated stress response
- ISRIB
integrated stress response inhibitor
- Met-tRNAi
initiator methionyl-tRNA
- ORF
open reading frame
- PBS
phosphate-buffered saline
- PEI
polyethylenimine
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- PIC
preinitiation complex
- RFP
red fluorescent protein
- RT
room temperature
- SA
sodium arsenite
- SG
stress granule
- TBS
Tris-buffered saline
- TBST
TBS and Tween 20
- TC
ternary complex
- Tg
thapsigargin
- UPR
unfolded protein response
- VWM
vanishing white matter
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0538) on February 6, 2019.
REFERENCES
- Algire MA, Maag D, Lorsch JR. (2005). Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell , 251–262. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol Cell , 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugiani M, Boor I, Powers JM, Scheper GC, Van der Knaap MS. (2010). Leukoencephalopathy with vanishing white matter: a review. J Neuropathol Exp Neurol , 987–996. [DOI] [PubMed] [Google Scholar]
- Campbell SG, Ashe MP. (2006). Localization of the translational guanine nucleotide exchange factor eIF2B: a common theme for GEFs? Cell Cycle , 678–680. [DOI] [PubMed] [Google Scholar]
- Campbell SG, Ashe MP. (2007). An approach to studying the localization and dynamics of eukaryotic translation factors in live yeast cells. Methods Enzymol , 33–45. [DOI] [PubMed] [Google Scholar]
- Campbell SG, Hoyle NP, Ashe MP. (2005). Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: implications for translation control. J Cell Biol , 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev K, Qiu H, Dong J, Zhang F, Barthlme D, Hinnebusch AG. (2010). The beta/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol Cell Biol , 5218–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Chent JJ, Barbert GN, Cigan AM, Feng L, Donahue TF, Londontl IM, Katzet MG, Hinnebusch AG. (1993). Mammalian eukaryotic initiation factor 2a kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci USA , 4616–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Yang W, Aström S, Byström AS, Hinnebusch AG. (1995). Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol Cell Biol , 6351–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooves S, Bugiani M, Postma NL, Polder E, Land N, Horan ST, van Deijk AL, van de Kreeke A, Jacobs G, Vuong C, et al (2016). Astrocytes are central in the pathomechanisms of vanishing white matter. J Clin Invest , 1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egbe NE, Paget CM, Wang H, Ashe MP. (2015). Alcohols inhibit translation to regulate morphogenesis in C. albicans. Fungal Genet Biol , 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsby R, Heiber JF, Reid P, Kimball SR, Pavitt GD, Barber GN. (2011). The alpha subunit of eukaryotic initiation factor 2B (eIF2B) is required for eif2-mediated translational suppression of vesicular stomatitis virus. J Virol , 9716–9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson FL, Hannig EM. (1996). Ligand interactions with eukaryotic translation initiation factor 2: role of the gamma-subunit. EMBO J , 6311–6320. [PMC free article] [PubMed] [Google Scholar]
- Fabian JR, Kimball SR, Heinzinger NK, Jefferson LS. (1997). Subunit assembly and guanine nucleotide exchange activity of eukaryotic initiation factor-2B expressed in Sf9 cells. J Biol Chem , 12359–12365. [DOI] [PubMed] [Google Scholar]
- Fogli A, Boespflug-Tanguy O. (2006). The large spectrum of eIF2B-related diseases. Biochem Soc Trans , 22–29. [DOI] [PubMed] [Google Scholar]
- Gordiyenko Y, Schmidt C, Jennings MD, Matak-Vinkovic D, Pavitt GD, Robinson CV. (2014). eIF2B is a decameric guanine nucleotide exchange factor with a γ2ε2 tetrameric core. Nat Commun , 3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday M, Radford H, Serkine Y, Moreno J, Verity N, Quesne J, Ortori CA, Barrett DA, Formont C, Fischer PM, et al (2015). Partial restoration of protein synthesis rates by the small moecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis , e1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday M, Radford H, Zents KAM, Molloy C, Moreno JA, Verity NC, Smith E, Ortori CA, Barrett DA, Bushell M, Mallucci GR. (2017). Repurposed drugs targeting eIF2α-P-mediated translational repression prevent neurodegeneration in mice. Brain , 1768–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EMC, van der Lei HDW, Vermeulen G, Gerver JAM, Lourenço CM, Naidu S, Mierzewska H, Gemke RJBJ, de Vet HCW, Uitdehaag BMJ, et al (2018). Natural history of vanishing white matter. Ann Neurol , 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig EM, Williams NP, Wek RC, Hinnebusch AG. (1990). The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics , 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. (2012). The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol , a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HK, Yoon H, Hannig EM, Donahue TF. (1997). GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev , 2396–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Pavitt GD. (2010). eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature , 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Zhou Y, Mohammad-Qureshi SS, Bennett D, Pavitt GD. (2013). eIF2B promotes eIF5 dissociation from eIF2*GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev , 2696–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K, Takahashi M, Nishimoto M, Hiyama TB, Higo T, Umehara T, Sakamoto K, Ito T, Yokoyama S. (2016). Crystal structure of eukaryotic translation initiation factor 2B. Nature , 122–125. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K, Ito T, Yokoyama S. (2017). Crystal structure of eIF2B and insights into eIF2-eIF2B interactions. FEBS J , 868–874. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. (2002). Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans , 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol , 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Fabian JR, Pavitt GD, Hinnebusch AG, Jefferson LS. (1998). Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2alpha. Role of the alpha- and delta-subunits of eIF2B. J Biol Chem , 12841–12845. [DOI] [PubMed] [Google Scholar]
- Kleiger G, Mayor T. (2014). Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol , 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG. (2001). Tight binding of the phosphorylated subunit of initiation factor 2 (eif2) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol Cell Biol , 5018–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewerenz J, Maher P. (2009). Basal levels of eIF2alpha phosphorylation determine cellular antioxidant status by regulating ATF4 and xCT expression. J Biol Chem , 1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang X, Van Der Knaap MS, Proud CG. (2004). Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol Cell Biol , 3295–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, van der Lei HDW, Wang X, Wortham NC, Tang H, van Berkel CGM, Mufunde TA, Huang W, van der Knaap MS, Scheper GC, et al (2011). Severity of vanishing white matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Hum Mutat , 1036–1045. [DOI] [PubMed] [Google Scholar]
- Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP. (2004). Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J , 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Parker R. (2018). Analysis of eIF2B bodies and their relationships with stress granules and P-bodies. Sci Rep , 12264–12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG. (1998). eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev , 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Yang W, Hinnebusch AG. (1997). Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol Cell Biol , 1298–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiah KVA, Davies MV, Chen J-J, Kaufman RJ, Davies V, Scherer JB, Choi SY, Hershey JWB, Kaufman RJ. (1994). Expression of mutant eukaryotic initiation factor 2 alpha subunit (eIF-2alpha) reduces inhibition of guanine nucleotide exchange activity of eIF-2B mediated by eIF-2 alpha phosphorylation. Mol Cell Biol , 4546–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands AG, Panniers R, Henshaw EC. (1988). The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem , 5526–5533. [PubMed] [Google Scholar]
- Sekine Y, Zyryanova A, Crespillo-Casado A, Fischer PM, Harding HP, Ron D. (2015). Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science , 1027–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK-H, Wilson C, et al (2013). Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife , e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P, Placzek A, Stoica L, Zhou H, Bell J, Friedlander M, Krnjević K, et al (2015a). The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife , 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Tsai JC, Kampmann M, Hearn BR, Vedantham P, Jaishankar P, Sokabe M, Mendez AS, Newton BW, Tang EL, et al (2015b). Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. Elife , e07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Zhou D, Jennings MD, Fukao A, Lee B, Ikeda Y, Chiorini JA, Campbell SG, Ashe MP, et al (2011). Mechanisms of translational regulation by a human eIF5-mimic protein. Nucleic Acids Res , 8314–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EJ, Campbell SG, Griffiths CD, Reid PJ, Slaven JW, Harrison RJ, Sims PFG, Pavitt GD, Delneri D, Ashe MP. (2010). Fusel alcohols regulate translation initiation by inhibiting eIF2B to reduce ternary complex in a mechanism that may involve altering the integrity and dynamics of the eIF2B body. Mol Biol Cell , 2202–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JC, Miller-Vedam LE, Anand AA, Jaishankar P, Nguyen HC, Renslo AR, Frost A, Walter P. (2018). Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science , 6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang H-Y, Anthony TG. (2006). Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans , 7–11. [DOI] [PubMed] [Google Scholar]
- Williams DD, Price NT, Loughlin AJ, Proud CG. (2001). Characterization of the mammalian initiation factor eIF2B complex as a GDP dissociation stimulator protein. J Biol Chem , 24697–24703. [DOI] [PubMed] [Google Scholar]
- Wong YL, LeBon L, Edalji R, Lim H, Sun S, Sidrauski C. (2018). The small molecule ISRIB rescues the stability and activity of Vanishing White Matter Disease eIF2B mutant complexes. ELife , 32733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortham NC, Martinez M, Gordiyenko Y, Robinson CV, Proud CG. (2014). Analysis of the subunit organization of the eIF2B complex reveals new insights into its structure and regulation. FASEB J , 2225–2237. [DOI] [PubMed] [Google Scholar]
- Wortham NC, Proud CG. (2015). eIF2B: recent structural and functional insights into a key regulator of translation. Biochem Soc Trans , 1234–1240. [DOI] [PubMed] [Google Scholar]
- Young SK, Wek RC. (2016). Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem , 16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyryanova AF, Weis F, Faille A, Alard AA, Crespillo-Casado A, Sekine Y, Harding HP, Allen F, Fromont C, Fischer PM, et al (2018). Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B. Science , 1533–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.