Abstract
Prostate cancer is one of the most common and heritable human cancers. Our aim was to find germline biomarkers that can predict disease outcome. We previously detected predisposing signals at 2q37, the location of the prostate specific ANO7 gene. To investigate, in detail, the associations between the ANO7 gene and PrCa risk and disease aggressiveness, ANO7 was sequenced in castration resistant tumors together with samples from unselected PrCa patients and unaffected males. Two pathogenic variants were discovered and genotyped in 1769 patients and 1711 unaffected males. Expression of ANO7 vs. PrCa aggressiveness was investigated. Different databases along with Swedish and Norwegian cohorts were used for validation. Case–control and aggressive vs. nonaggressive association analyses were performed against risk and/or cancer aggressiveness. The ANO7 mRNA level and patient survival were analyzed using expression data from databases. Variant rs77559646 showed both risk (OR 1.40; p = 0.009, 95% CI 1.09–1.78) and association with aggressive PrCa (Genotype test p = 0.04). It was found to be an eQTL for ANO7 (Linear model p‐values for Finnish patients p = 0.009; Camcap prostate tumor p = 2.53E‐06; Stockholm prostate tumor cohort p = 1.53E‐13). rs148609049 was not associated with risk, but was related to shorter survival (HR 1.56; 95% CI 1.03–2.36). High ANO7 expression was independently linked to poor survival (HR 18.4; 95% CI 1.43–237). ANO7 genotypes correlate with expression and biochemical relapse, suggesting that ANO7 is a potential PrCa susceptibility gene and that its elevated expression correlates with disease severity and outcome.
Keywords: ANO7, CRPC, expression, prostate cancer, survival
Short abstract
What's new?
The discovery of germline biomarkers to predict outcome in prostate cancer could greatly aid disease management. One such marker of particular interest in this regard is the prostate‐specific gene ANO7, which previous studies have associated with high‐grade prostate cancer. Here, specific germline ANO7 genotypes were associated with increased prostate cancer risk. In patients, ANO7 expression correlated with disease severity, with elevated expression associated with decreased overall survival. The data suggest that ANO7 is a susceptibility marker in prostate cancer and, with further characterization, could be used to inform patient selection strategies and therapeutic approaches.
Introduction
Prostate cancer (PrCa) is one of the most commonly diagnosed cancers and is the sixth most common cause of male cancer‐related death worldwide.1 PrCa is one of the most heritable cancers, with genetic factors estimated to account for 57% of the risk.2, 3 Genome‐wide association studies have discovered over 100 susceptibility loci in the human genome, but only a few have shown an association with disease outcome.
Our earlier linkage studies, together with recent GWAS results, have provided evidence for many significant PrCa risk regions in the genome, such as 2q37.3.4 ANO7 (TMEM16G, NGEP [New Gene Expressed in Prostate])5, 6 resides in this genomic area of interest and was therefore chosen for a detailed analysis. ANO7 is a prostate‐specific member of the anoctamin family of Ca2+‐activated Cl− channels, whose function is yet to be discovered. ANO7 has been reported to be downregulated in metastatic disease,7 and reduced protein expression has been associated with high‐grade PrCa.8
Novel biomarkers are greatly needed as there are many unmet needs in PrCa management. For example, during screening, there are no standards or recommendations as no biomarkers are available to discriminate between indolent cases and aggressive disease except for some rare BRCA2 mutations (NCCN Guidelines for Prostate Early Detection V.1.2018). Germline variants would be particularly attractive biomarkers because they are present at the time of diagnosis and remain static despite treatment, hormonal status, or age.
In this study, the entire ANO7 gene was sequenced. The SNPs (single nucleotide polymorphism) classified as pathogenic were analyzed for association with PrCa risk and outcome. We report that ANO7 genotypes correlate with expression and survival, suggesting that ANO7 is a potential PrCa susceptibility gene and that its elevated expression correlates with disease severity and outcome.
Materials and Methods
Patients and controls
DNA samples from 1769 patients were received from four different sources: the PROSTY study, 9 the Turku Prostate Cancer Consortium, the Tampere University Hospital, and the Auria Biobank (Turku, Finland). These also included germline DNA samples from a total of 664 individuals (from 142 families) with a strong PrCa family history. One index patient (youngest affected) per family was first genotyped, followed by the analysis of his affected and unaffected relatives if proven positive for the variant. A set of females was also genotyped to investigate segregation of other cancers in the families. The mean age of the PrCa patients in the whole sample set was 62.6 years. In all analyses, cases were classified as aggressive if at least one of the following criteria was met: PSA (prostate‐specific antigen) ≥20 ng/mL, Gleason grade ≥ 8, metastatic disease, or PrCa as the cause of death. The classification is based on the ICPCG consortium criteria10 that have been updated by the PRACTICAL consortium (http://practical.icr.ac.uk).
For controls, 1,711 DNA samples from unaffected men were used. Of these, 1,577 originated from the FinRSPC study11 (PSA < 1.4 ng/mL), 12 were from the Finnish Red Cross and 122 were from the Turku University Hospital, Laboratory of Medical Genetics. The details on all cohorts are explained in Supporting Information Table S1a and their clinical characteristics are described in Supporting Information Table S1b. All samples were collected with written and signed informed consent. This research was approved by the Institutional Review Boards of the Tampere and Turku University Hospitals.
To validate the association between germline variants and PrCa risk and prognosis, two Swedish study populations, STHM212 and PROCAP,13 and one Norwegian study population (CONOR)14, 15 were analyzed. STHM2 comprises 3132 PrCa cases and 1429 controls while PROCAP and CONOR are two PrCa cohorts containing 669 and 1455 PrCa patients, respectively.
ANO7 sequencing, variant selection and validation
Eighty‐six samples were NGS (next generation sequencing) sequenced in the initial screening. The ANO7 variants of interest were chosen based on the following criteria: low global allele frequency (minor allele frequency < 0.05) and deleterious impact on protein (CADD [combined annotation dependent depletion] score > 10). This list was compared with a validated set of Sanger sequenced familial PrCa samples, allowing us to ultimately select the overlapping SNPs and validate them in a large validation set (Fig. 1). The selected ANO7 SNPs rs148609049, rs77559646 and rs181722382 are presented in Figure 2. The details on DNA extraction, NGS and Sanger sequencing can be found in the Supporting Information.
Figure 1.

Flow chart of initial ANO7 screening, variant selection and results validation. Blood DNA from 50 PrCa cases, tumor DNA from 22 CRPC tumors and blood DNA from 14 healthy males were sequenced via Illumina Miseq. Libraries were prepared with the Illumina TruSeq Custom amplicon kit (Illumina, San Diego, CA) following the manufacturer's instructions. The TruSeq Custom amplicon kit included all 25 exons (hg19) with 97% coverage of the gene. Variant calling and quality control were conducted using an in‐house pipeline (Supporting Information). The ANO7 variants of interest that were selected were verified from the same samples with Sanger sequencing. The three selected SNPs were further genotyped using TaqMan® SNP Genotyping Assay primers and probes in the validation step. All genotyping rounds were validated in two ways: five positive randomly selected samples were validated with Sanger sequencing and, in addition, 10% of all DNA samples were reanalyzed with TaqMan®. Swedish and Norwegian validation sets are described in the Oncoarray data (http://practical.icr.ac.uk). [Color figure can be viewed at wileyonlinelibrary.com]
Figure 2.
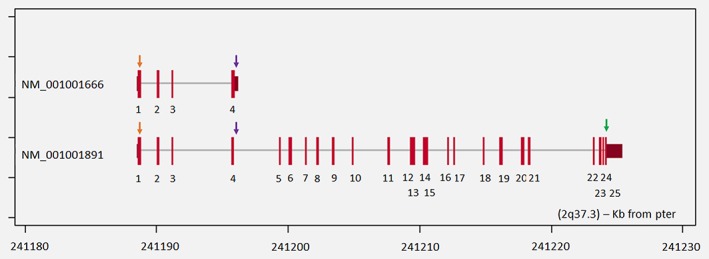
The detected ANO7 mutations. Modified figure from http://atlasgeneticsoncology.org/Genes/GC_ANO7.html. The locations of rs148609049 (orange arrow), rs77559646 (purple arrow) and rs181722382 (green arrow) variants in ANO7 transcripts NM_001001666 and NM_001001891 (hg19).
Genotype data analysis
Each assessed SNP was found to be in Hardy–Weinberg equilibrium. The odds ratios (OR) of case–control and case–case analyses were calculated for each variant according to Bland and Altman.16 The databases ExAC03, snp138, ClinVar and 1000 genomes were used to determine the allelic frequencies of the variants. Survival analysis (Kaplan–Meier estimator and Cox proportional hazards analysis) was performed using the “Survival” package and R software (version 3.3.2). Multiple testing correction was not applied, p values <0.05 were considered statistically significant.
eQTL analysis
For the association between rs77559646 genotypes and ANO7 expression levels, the Mann–Whitney U test was applied. We also used Matrix eQTL (expression quantitative trait loci) (version 3.2.2) with the parameters “useModel = modelLINEAR” and “errorCovariance = numeric ()” to test the cis‐eQTL association between rs77559646 genotypes and ANO7 expression levels using the data from Camcap and Stockholm cohorts consisting of 119 and 94 prostate samples, respectively.17 The Finnish cohort included 22 tumor samples in which ANO7 expression was analyzed by RNA‐seq. The sequenced samples were part of Turku Prostate Cancer Consortium treatment cohort and originated from surgically removed prostates. The patients did not receive any prostate cancer therapy before prostatectomy.
Differential gene expression analysis of ANO7 across normal and cancerous prostate tissues
For the differential ANO7 gene expression analysis, the Mann–Whitney U test was performed on normal prostate and PrCa tumor gene expression data from TCGA provisional (downloaded from the cBio Cancer Genomics Portal18) and the Oncomine database.19 R (version 3.2.2) was used to perform the statistical analyses.
Association analysis of ANO7 expression and PrCa outcome
The association between ANO7 expression levels and overall survival was assessed by using a Kaplan–Meier estimator. The expression data on ANO7 and the information on overall survival and clinical outcomes for a collection of 333 prostate adenocarcinoma samples were downloaded from the cBioPortal for Cancer Genomics18 and the Cancer Genome Atlas Research Network.20 The PrCa patients were stratified into two groups based on the deviation of the expression value from the mean expression value of all samples. Thus, scores above zero indicate higher expression than the mean value while scores below zero indicate lower expression. Package “Survival” and R (version 3.2.2) were used to perform the analysis.
Multivariate analysis
For analysis of the association between overall survival, ANO7 expression and clinical variables (Gleason score, PSA, T stage, N stage and age), multivariate Cox proportional hazard analyses were performed on the 333 prostate carcinoma samples described above using R (version 3.2.2). Samples were stratified into high and low expression groups by comparing them to the mean ANO7 expression level as described above. The clinical relevance of overall survival and covariates was performed in several different scenarios. The association was tested against the Gleason score, PSA, age, N stage and clinical T state.
Results
Significant ANO7 variants (Finnish data)
Targeted deep sequencing of the entire ANO7 gene revealed 215 variants (somatic and germline), of which three were selected and validated for further analyses. The variants are listed in the Supporting Information (Supporting Information—Targeted ANO7 sequencing variant list). The same three SNPs were also found in the Sanger screening set.
In the screening set, the variant rs77559646 (Arg158His) was detected in 2/22 (9%) castration resistant FFPE tumor samples and in 2/50 (8%) blood samples of PrCa patients. Variant rs148609049 (Arg30Ter) was found in 4/22 (18%) CRPC (castration‐resistant PrCa) tumors and 2/50 (4%) blood samples. rs181722382 (Leu931Pro) was found in 1 (2%) patient's blood DNA. None of these variants were found among the 14 unaffected controls.
Both tumor and blood DNA were available from the Auria Biobank patients with castration resistant disease in the validation set. The variants rs148609049 and rs77559646 were detected in 5/64 (7.8%) and 6/64 (9.4%), respectively, of these cases. Importantly, they were found in both tumor and blood DNA. Therefore, in PROSTY study, CRPC cases for which germline DNA was not available, the variants were also considered germline of origin. All variants were also found in their homozygous form in germline.
We analyzed 142 families with a strong PrCa history (3 or more cases per family). The rs148609049 and rs77559646 variants were commonly present in PrCa families but did not completely segregate with the disease (Supporting Information Fig. S2). With rs77559646, there was a trend for affected males to more commonly carry the variant compared to unaffected males. Variant rs181722382 was present only in three families and did not segregate with the disease.
Variant rs181722382 was extremely rare in the data sets. Therefore, in the remaining analyses, we included only rs148609049 and rs77559646. The Kaplan–Meier survival curves of rs148609049 carriers and noncarriers are shown in Figure 3 a and c, indicating that the variant carriers had shorter overall survival than the noncarriers [Overall survival HR = 1.56 (95% CI 1.03–2.37); PrCa‐specific survival HR 1.76 (95% CI 0.91–3.41)]; rs77559646 did not affect outcome (3B, D). Cox regression modeling resulted in an overall significance of p < 10 E −13 (Fig. 3 e). The variables explaining patient survival were Gleason grade [HR 4.7 (95% CI 3.61–6.12)] and rs148609049 [HR 1.56 (1.03–2.37)].
Figure 3.

Kaplan–Meier overall survival plots of rs148609049 (a, c), rs77559646 (b, d) variant carriers (red curve) and noncarriers (blue curve) and Cox regression analysis (e). Figures A and B show PrCa specific survival while C and D represent overall survival. PrCa specific survival does not show a statistically significant trend, i.e., carriers of either variant would have a shortened survival time (a, b). Because of missing cause of death information, the overall survival was also analyzed. Variant rs77559646 AG or A/A genotypes (d) do not have an effect on survival, while the rs148609049 C/T or T/T genotype (c) significantly reduce survival. In the Cox regression analysis, Gleason score and rs148609049 were the most significant factors explaining overall survival (e).
The case–control analysis did not show any differences in genotypic frequencies between PrCa patient groups and healthy males for rs148609049 (OR 1.02; p = 0.86) (Supporting Information Table S4). In contrast, rs77559646 was significantly more frequently observed in patients than in controls (Odds ratio 1.4; p = 0.008) (Supporting Information Table S4) and was strongly associated with PrCa risk. There was no significant difference between the ages in variant carrier and noncarrier groups.
In case–case analysis (Supporting Information Table S5) with aggressive and nonaggressive cases, rs148609049 was not more frequently observed in aggressive cases (Genotype test p = 0.68). Although the Kaplan–Meier and Cox regression analyses did not show any evidence that rs77559646 would associate with more aggressive disease, case–case analysis showed that the variant was significantly more common in aggressive than nonaggressive cases (Genotype test p = 0.04). When examining the clinical characteristics individually (Supporting Information Table S3), rs77559646 carriers were more likely to have an early onset disease (<55 years) and to have high Gleason scores (≥8). In turn, the carriers of variant rs148609049 more often died of PrCa than noncarriers. Although these differences were not statistically significant, they point to the same direction as the survival analyses.
Data validation in other populations
In the Swedish STHM2 population, rs77559646 was associated with PrCa risk (OR 1.36, 95% CI 1.06–1.75) but not aggressive disease in case‐only analysis. Variant rs148609049 showed no association with either risk or aggressive disease in STHM2 (data not shown). Neither rs77559646 nor rs148609049 showed any evidence of association with PrCa‐specific survival in the STHM2, PROCAP or CONOR study populations (data not shown).
Association analysis of ANO7 expression with SNP genotype and PrCa severity
The status of ANO7 across different types of normal human tissues and cancers was analyzed by querying the data from the Oncomine database.19 The results showed a strikingly higher expression of ANO7 in prostate glands or tumor samples compared to other normal tissues (Fig. 4 a) and cancer samples (Fig. 4 b). Our eQTL analysis using a Finnish cohort of 22 prostate tumor samples revealed a significant association between the rs77559646 G/A risk genotype and elevated ANO7 gene expression (Fig. 5 a). Notably, this association was also observed in two independent collections of 119 and 94 PrCa samples (Fig. 5 b and c, respectively). This result suggests that the rs77559646 G/A genotype may contribute to the elevated risk of having PrCa via upregulation of ANO7 expression. Consistent with this, we observed a striking upregulation of ANO7 expression in PrCa in two independent clinical PrCa datasets (Fig. 6). To further assess the clinical relevance of our observations, we performed a Kaplan–Meier analysis and examined the association of ANO7 expression and overall survival in a large cohort of PrCa patients20 (N = 289). We found that patients with tumors expressing higher levels of ANO7 had decreased overall survival (Fig. 6). Together, these analyses suggest that ANO7 is a potential PrCa susceptibility gene and that its elevated expression correlates with disease severity.
Figure 4.

Prostate‐lineage‐specific expression of ANO7. The mRNA level of ANO7 is significantly upregulated in the prostate gland (a) and prostate tumors (b) compared to other organs and cancers, respectively. The p values were examined using Student's t‐test. ANO7 expression intensity is shown as log2 median‐centred as downloaded from the licensed Oncomine database19 and displayed by boxplots in R (version 3.2.2). [Color figure can be viewed at wileyonlinelibrary.com]
Figure 5.

eQTL analysis of the association between rs77559646 and ANO7 gene expression. The risk genotype G/A of rs77559646 was significantly associated with increased mRNA expression of ANO7. The ANO7 mRNA levels were examined with RNA‐seq and displayed as RPKM values in a collection of 22 prostate cancer samples (Turku Prostate Cancer Consortium treatment cohort) (a), and by Illumina Expression BeadChip‐based transcriptional profiling in a collection of 119 (b) and 94 (c) human prostate tissue samples17 (levels are log2 transformed and quantile normalized). The ANO7 mRNA expression data in prostate tumors tissues were displayed as a five‐number distribution (minimum, first quartile, median, third quartile, and maximum) according to the rs77559646 genotypes. The p values were assessed via linear regression model. [Color figure can be viewed at wileyonlinelibrary.com]
Figure 6.

ANO7 is highly expressed in prostate cancer and is associated with poor overall survival in patients with prostate cancer. The ANO7 transcript levels were strikingly increased in prostate cancer tissues compared to normal tissues in a collection of cohorts consisting of 550 (a) and 21 (b) samples, respectively.18, 24 The p values were calculated with the Mann–Whitney U test. In (a), ANO7 expression was determined by RNA sequencing; in (b) ANO7 expression intensity is shown as log2 median‐centred intensity as reported in the Oncomine database 19. In (c) the Kaplan–Meier curve and the estimate of the risk for overall survival in a large cohort of PrCa patients with higher (n = 115) or lower (n = 174) expression levels of ANO7 are shown. Patients with tumors expressing higher levels of ANO7 show decreased overall survival. The number of patients in each group at 20‐month intervals is indicated. The p values were calculated by the Log‐rank test and Cox regression model.
The variant rs148609049 was only present in the Finnish cohort, and the variant was not eQTL for ANO7 (data not shown).
Discussion
In this study, the ANO7 gene was analyzed in the context of PrCa susceptibility for the first time. Three validated germline ANO7 variants were evaluated against clinical data. The stop‐gained variant in exon 1 (rs148609049) was clearly correlated with poor survival following a PrCa diagnosis according to Kaplan–Meier, Cox regression and Hazard ratio analyses of the Finnish PrCa patients. Significant decrease in survival was only seen in overall but not in PrCa specific survival, which likely results from fewer cause‐specific events and thus limited statistical power of the analysis.
The variant rs77559646 located in exon 4 constitutes a missense mutation in the short ANO7 isoform and a splice region change in the long isoform (Fig. 2). Our results showed that this variant increases PrCa risk. The eQTL analysis indicated that ANO7 expression was significantly increased with the G/A genotype. Splicing prediction programs suggested that rs77559646 might influence mRNA splicing and could thereby disturb the balance between the short and the long transcript.21
The study was carried out only using Caucasian patients from Northern Europe. Finland has a founder population known to have genetic enrichments, like the recently identified PrCa risk variant G84E in HOXB1322 but Sweden and Norway have allele frequencies similar to other European populations. As shown, ANO7 was statistically significant in all patient cohorts studied but significance of ANO7 variants in other ethnic backgrounds warrant additional analyses.
Although variants rs148609049 and rs77559646 are both rare, a total of 11 PrCa families carried them both. Six other families carried only rs148609049, while two families only carried rs77559646. Although the segregation was not complete, the variants were enriched in the families compared to the general population. This is not surprising since the original linkage analyses that detected the chromosomal location 2q374 were performed on familial PrCa patients. Variant rs181722382, located in exon 25, was too rare to be analyzed.
We carefully analyzed ANO7 expression as prior results on ANO7 expression in PrCa are somewhat conflicting. Earlier studies using antibodies against ANO7 suggested no correlation between ANO7 protein expression and Gleason grade (C‐terminal antibody), or that ANO7 protein expression is downregulated in high grade prostatic tumors (N‐terminal antibody).8, 23 Chandran et al.7 using a probe‐based microrarray analysis, found that ANO7 mRNA expression was downregulated in metastases compared to the primary tumor, while there was no difference between normal and cancerous prostate on the group level. Kiessling et al.6 on the other hand, reported high individual variability of ANO7 expression between individual samples, both in terms of absolute expression levels and of expression level ratios between matched normal and tumorous prostate.
In our study, higher ANO7 mRNA expression strongly correlated with poor overall survival in PrCa patients. Multivariate analysis compared high and low ANO7 expression and showed an even stronger negative survival effect than the model with the three selected SNPs, indicating that there might be additional variants that regulate expression. Importantly, our survival study does not address the question of whether ANO7 expression was upregulated or downregulated in the tumor compared to the normal tissue.
ANO7 belongs to the anoctamin family of membrane proteins which function as ion channels and/or phospholipid scramblases. The function of ANO7 in PrCa is poorly understood and warrants further investigation. However, our results suggest that this gene plays a central role in PrCa progression. The two ANO7 SNPs reported herein could be suitable for clinical PrCa testing.
Conclusions
In this study, we show that ANO7, a gene specifically expressed in the prostate, is a promising biomarker to predict PrCa aggressiveness. We identified two pathogenic variants that could possibly be utilized in genetic testing of PrCa patients. The stop‐gain variant rs148609049 in exon 1 contributes to decreased survival time. The splice site/missense variant rs77559646 in exon 4 increases the risk of PrCa and is also associated with aggressive PrCa. The same variant is an eQTL for ANO7, and high mRNA levels of ANO7 strongly correlate with poor survival. ANO7 gene expression is highly elevated in PrCa tumors versus normal prostate tissue, and high expression strongly increases the PrCa patient mortality rate. These findings should spur further research to elucidate the regulatory networks of ANO7.
Supporting information
Appendix S1: Supplementary Material
Appendix S2: Supplementary Material
Appendix S3: Supplementary Material
Appendix S4: Supplementary Material
Acknowledgments
The authors wish to thank all the patients and families who participated in this study. The authors also thank Ms. Pauliina Toivonen and Mr. Jukka Karhu for their technical assistance.
Members from the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium are provided in the Supplement/footnotes. Information of the consortium can be found at http://practical.icr.ac.uk
Conflict of interest: The authors declare no potential conflicts of interest.
[Correction added on June 20, 2019 after first online publication: copyright updated.]
References
- 1. Fitzmaurice C, Dicker D, Pain A, et al. The Global Burden of Cancer 2013. JAMA Oncol 2015;1:505–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mucci LA, Hjelmborg JB, Harris JR, et al. Familial risk and heritability of cancer among twins in Nordic countries. JAMA 2016;315:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hjelmborg JB, Scheike T, Holst K, et al. The heritability of prostate cancer in the Nordic Twin Study of Cancer. Cancer Epidemiol Biomarkers Prev 2014;23:2303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cropp CD, Simpson CL, Wahlfors T, et al. Genome‐wide linkage scan for prostate cancer susceptibility in Finland: evidence for a novel locus on 2q37.3 and confirmation of signal on 17q21‐q22. Int J Cancer 2011;129:2400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bera TK, Das S, Maeda H, et al. NGEP, a gene encoding a membrane protein detected only in prostate cancer and normal prostate. Proc Natl Acad Sci U S A 2004;101:3059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kiessling A, Weigle B, Fuessel S, et al. D‐TMPP: a novel androgen‐regulated gene preferentially expressed in prostate and prostate cancer that is the first characterized member of an eukaryotic gene family. Prostate 2005;64:387–400. [DOI] [PubMed] [Google Scholar]
- 7. Chandran UR, Ma C, Dhir R, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007;7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mohsenzadegan M, Madjd Z, Asgari M, et al. Reduced expression of NGEP is associated with high‐grade prostate cancers: a tissue microarray analysis. Cancer Immunol Immunother 2013;62:1609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kellokumpu‐Lehtinen PL, Harmenberg U, Joensuu T, et al. 2‐Weekly versus 3‐weekly docetaxel to treat castration‐resistant advanced prostate cancer: a randomised, phase 3 trial. Lancet Oncol 2013;14:117–24. [DOI] [PubMed] [Google Scholar]
- 10. Schaid DJ, McDonnell SK, Zarfas KE, et al. Pooled genome linkage scan of aggressive prostate cancer: results from the International Consortium for Prostate Cancer Genetics. Hum Genet 2006;120:471–85. [DOI] [PubMed] [Google Scholar]
- 11. Finne P, Stenman UH, Määttänen L, et al. The Finnish trial of prostate cancer screening: where are we now? BJU Int 2003;92(Suppl 2):22–6. [DOI] [PubMed] [Google Scholar]
- 12. Nordström T, Aly M, Eklund M, et al. A genetic score can identify men at high risk for prostate cancer among men with prostate‐specific antigen of 1‐3 ng/ml. Eur Urol 2014;65:1184–90. [DOI] [PubMed] [Google Scholar]
- 13. Szulkin R, Holmberg E, Stattin P, et al. Prostate cancer risk variants are not associated with disease progression. Prostate 2012;72:30–9. [DOI] [PubMed] [Google Scholar]
- 14. Szulkin R, Karlsson R, Whitington T, et al. Genome‐wide association study of prostate cancer‐specific survival. Cancer Epidemiol Biomarkers Prev 2015;24:1796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naess O, Søgaard AJ, Arnesen E, et al. Cohort profile: cohort of Norway (CONOR). Int J Epidemiol 2008;37:481–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bland JM, Altman DG. Statistics notes. The odds ratio. BMJ 2000;320:1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whitington T, Gao P, Song W, et al. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat Genet 2016;48:387–97. [DOI] [PubMed] [Google Scholar]
- 18. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data‐mining platform. Neoplasia 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Network CGAR. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paronetto MP, Passacantilli I, Sette C. Alternative splicing and cell survival: from tissue homeostasis to disease. Cell Death Differ 2016;23:1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laitinen VH, Wahlfors T, Saaristo L, et al. HOXB13 G84E mutation in Finland: population‐based analysis of prostate, breast, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 2013;22:452–60. [DOI] [PubMed] [Google Scholar]
- 23. Das S, Hahn Y, Walker DA, et al. Topology of NGEP, a prostate‐specific cell: cell junction protein widely expressed in many cancers of different grade level. Cancer Res 2008;68:6306–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arredouani MS, Lu B, Bhasin M, et al. Identification of the transcription factor single‐minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res 2009;15:5794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supplementary Material
Appendix S2: Supplementary Material
Appendix S3: Supplementary Material
Appendix S4: Supplementary Material


