Abstract
Chlamydia trachomatis is the most prevalent sexually transmitted bacterial pathogen and the leading cause of preventable blindness in the developing world. C. trachomatis invades the epithelium of the conjunctiva and genital tract and replicates within an intracellular membrane-bound compartment termed the inclusion. To invade and replicate in mammalian cells, Chlamydia remodels epithelial surfaces by reorganizing the cytoskeleton and cell–cell adhesions, reprograms membrane trafficking, and modulates cell signaling to dampen innate immune responses. If the infection ascends to the upper female genital tract, it can result in pelvic inflammatory disease and tissue scarring. C. trachomatis infections are associated with infertility, ectopic pregnancies, the fibrotic disorder endometriosis, and potentially cancers of the cervix and uterus. Unfortunately, the molecular mechanisms by which this clinically important human pathogen subverts host cellular functions and causes disease have remained relatively poorly understood because of the dearth of molecular genetic tools to study Chlamydiae and limitations of both in vivo and in vitro infection models. In this review, we discuss recent advances in the experimental molecular tool kit available to dissect C. trachomatis infections with a special focus on Chlamydia-induced epithelial barrier disruption by regulating the structure, function, and dynamics of epithelial cell–cell junctions.
Keywords: Chlamydia, pathogenesis, genetics, cell-cell junctions, organoids, infection models
Introduction
The order Chlamydiae are obligate intracellular pathogens of eukaryotic cells. These bacteria have reduced genomes and display biphasic developmental stages that alternate between distinct extracellular and intracellular forms 1, 2. Eleven pathogenic Chlamydia species infect vertebrate animals and display tissue-specific tropism 3. The elementary body (EB) is the environmentally stable form of the pathogen that binds and invades target cells. The EB then transitions into the larger intracellular reticulate body (RB) form. RBs replicate and secrete proteins across the parasitophorous membrane-bound vacuole (“inclusion”) to modulate multiple host cellular functions that benefit the bacterium 4.
All Chlamydiae encode a type III secretion (T3S) system to deliver a defined cohort of bacterial proteins (“T3S effectors”) directly into the host cell 5. The Chlamydia trachomatis EB T3S effectors modify the cytoskeleton and stimulate bacterial uptake into a membrane-bound vacuole that is rapidly segregated from degradative trafficking pathways ( Figure 1A) 6, 7. A subset of T3S effectors are inserted into the inclusion membrane. These inclusion membrane proteins (Incs), which are secreted throughout the infectious life cycle, are diverse (~5% of the total coding potential of C. trachomatis) and their molecular function is just beginning to be understood 8. For instance, Incs co-opt the microtubule motor protein dynein to transport nascent inclusions along microtubules toward the centrosome ( Figure 1A) 9. Along the way, the inclusion membrane is modified by lipid kinases, which may be important for evasion of endolysosomal compartments 10. Some Incs contain SNARE (soluble N-ethylmaleimide–sensitive factor attachment protein receptor)-like domains that coordinate fusion between inclusions and other membrane vesicles 11, whereas others promote the recruitment of the endoplasmic reticulum and Golgi complex to the vicinity of the inclusion, possibly to intercept lipid-rich vesicles to support Chlamydia replication ( Figure 1A) 12– 14. As the inclusion matures, it is increasingly encased by a network of F-actin, microtubules, intermediate filaments, and septins, which help confine the bacteria within the inclusion and limit recognition of bacterial products by innate immune sensors 15– 17.
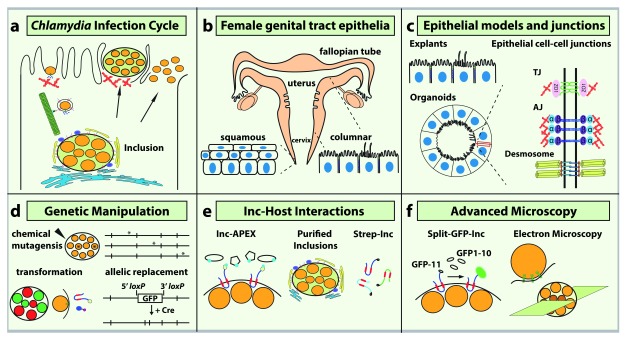
Figure 1. Recent advances in the molecular tool kit and infection models to explore Chlamydia infection.
( a) The Chlamydia trachomatis infection cycle. The elementary body (EB) form of the bacteria remodels actin filaments (red) during entry and traffics along microtubules (green) to the perinuclear region. Inclusion membrane proteins (Incs) recruit the Golgi complex (yellow) and endoplasmic reticulum (blue). At the end of the intracellular cycle, the inclusion exits via actin-dependent extrusion or cell lysis. ( b) Anatomy of the female genital tract and epithelial cell organization in the lower and upper tract. ( c) New epithelial model systems (left) and schematic of polarized columnar epithelial cell–cell junctions (right). Tight junction (TJ) and adherens junction (AJ) complexes recruit adaptor proteins that connect to the actin cytoskeleton (red); desmosomes interact with intermediate filaments (yellow). ( d) New genetic tools for C. trachomatis include chemical mutagenesis and whole-genome sequencing to identify mutations and plasmid transformation to generate fluorescent reporter strains, tagged effectors, and targeted gene disruption via allelic replacement. ( e) New proteomic-based strategies to identify host proteins that interact with Chlamydia Incs. Incs tagged with the enzyme ascorbate peroxidase (APEX) (left) can ligate biotin-phenol on host proteins in close proximity. Purified inclusions (middle) and Strep-tagged Incs (right) were used to identify host proteins recruited to the inclusion and Inc–host protein interactions, respectively. ( f) Summary of advanced microscopy approaches to visualize Chlamydia effector localization using the Split-green fluorescent protein (Split-GFP) system (left), the structure of the T3S apparatus in contact with the plasma membrane (middle), and reticulate body (RB)-to-EB conversion (right). ZO-1, zona occludens 1.
Mid-stage through the infectious cycle, RBs transition back to EBs such that at the end of the cycle the infectious bacteria are released either by an actin-dependent extrusion process whereby the intact inclusion is exocytosed from the cell or by lysis of the host cell which requires the cleavage of cytoskeletal elements and nuclear rupture 18, 19.
Much of our understanding of the cell biology of how Chlamydia interacts with target cells and the molecular mechanisms it uses to manipulate cellular processes is based on observations made in infected cancer cell lines, which in addition to being metabolically and genetically adapted for proliferation, lack positional cues that are available only in the context of tissues. For example, polarized columnar epithelial cells, the in vivo target of C. trachomatis, display a spatial organization of organelles and cell signaling pathways that intimately intertwine stable cell–cell junctions to epithelial function and proliferation ( Figure 1B, C). Such structures and signaling networks are not present—or properly wired—in common cell lines used in Chlamydia research. Fortunately, more sophisticated infection models, coupled with the increasing ability to genetically manipulate Chlamydia, now provide a renewed tool kit to better understand how these pathogens interact with their intact animal hosts.
An expansion of the molecular tool kits available for Chlamydia research
The greatest single advance in Chlamydia biology over the past decade has been the development of methods to perform genetic analysis of C. trachomatis mutants and increasingly robust molecular genetic tools to transform Chlamydia with recombinant DNA ( Figure 1D) 20– 22. As a result, it is now possible to perform targeted gene inactivation and plasmid-based complementation of chromosomal mutations 22– 25. The ability to express exogenous proteins, epitope tags, and fluorescent and other reporter proteins in Chlamydia has also expanded the repertoire of possible technologies that can be applied to study the Chlamydia–host interface 26, 27. For instance, Incs fused to enzymes that enable the biotinylation of proteins identified host factors that are in proximity to the inclusion membrane by affinity capture of biotinylated proteins coupled with tandem mass spectrometry ( Figure 1E) 28, 29.
Two recent studies using quantitative proteomics identified host proteins that interact with Incs and host proteins that are preferentially recruited to the inclusion ( Figure 1E). In one study, intact inclusions were isolated and all associated host proteins were identified and quantified by stable isotope labeling by/with amino acids in cell culture (SILAC)-based mass spectrometry 30. In parallel, a large-scale study identified previously unknown host-binding partners for nearly two thirds of the predicted Incs. Using affinity purification of Strep-tagged Incs coupled to quantitative mass spectroscopy, a second study provided a blueprint for Inc–host interactions 31. The different but complementary approaches presented new clues as to the potential mechanisms used by Chlamydia to subvert various aspects of host cell biology.
The advances in genetic and biochemical approaches have been further complemented with high-resolution microscopy ( Figure 1F). The localization and dynamics of Chlamydia T3S effectors can now be visualized live by using the split-green fluorescent protein (split-GFP) system 32, an approach that relies on fusing the effector with the 16–amino acid GFP-11 β-strand and infecting cells expressing the GFP1-10 β-strands. Upon secretion and complementation, the GFP β-barrel properly folds and fluoresces ( Figure 1F) 33. Furthermore, the application of cryo-electron microscopy revealed with unprecedented resolution that the Chlamydia T3S apparatus changes shape and polarizes toward host cell membrane 34. Similarly, applied serial block-face scanning electron microscopy temporally characterized the process of RB-to-EB conversion during the infection cycle ( Figure 1F) 35.
Recent advances in cell culture infection models
Urogenital serovars of C. trachomatis target the mucosal epithelium. In the female genital tract, the infection typically begins in the endocervix before ascending to the endometrium and fallopian tubes 36. These tissues are comprised largely of polarized columnar epithelial cells with a rich diversity in form and function which is not readily recapitulated in two-dimensional culture settings 37. Thus, new in vitro models that more accurately reconstruct the organization and complexity of the genital tract are essential to better understand the full impact of Chlamydia infection on the epithelial physiology.
Wyrick and colleagues first described key differences in Chlamydia growth in polarized epithelial cells 38. Human endometrial epithelial cancer cells polarized on collagen-coated microcarrier beads significantly enhanced the growth of C. trachomatis serovar E, independent of EB attachment efficiency, compared with non-polarized cells 38. Infections in polarized enterocytes show that the inclusion preferentially captures lipid-rich exocytic vesicles that are specifically trafficked toward the basolateral membrane, suggesting that Chlamydia has adapted to grow in a polarized environment 39. More recently, a human endocervical epithelial cell line (A2EN cells) derived from a healthy patient sample has been shown to polarize, secrete mucin, and express pro-inflammatory cytokines during infection 40, 41.
Meyer and colleagues pioneered the use of ex vivo organotypic cultures and a novel fallopian tube organoid (FTO) model to investigate C. trachomatis infection in primary human epithelia 42, 43. Partially dissected tissue from the ectocervix and fallopian tube—representing the lower and upper genital tracts, respectively—has been cultured ex vivo and infected with Chlamydia 42, 43. These models were recently simplified by culturing isolated tubal epithelial cells in a three-dimensional matrix, generating self-renewing FTOs ( Figure 1C) 44. FTOs consist of secretory and ciliated epithelia, the two most common epithelial cell types in the fallopian tube, and more accurately recapitulate fallopian tube epithelial architecture. Acute infection in FTOs produced a strong inflammatory response while long-term and chronic infection increased epithelial stemness and proliferation and altered the methylation status of genes associated with aging. The latter results provide new insights into the long-term effects of Chlamydia infection on epithelial tissue homeostasis and the correlation between infection and cell proliferation 45.
The epithelium of the female genital tract
Ultrastructural analysis of the lower female genital tract shows squamous ectocervical epithelia and columnar endocervical epithelia, which display differential organization of cell–cell junctions 37. The endometrium resembles columnar epithelia of the endocervix with increased epithelial diversity that includes ciliated and secretory cells and hormonally responsive and crypt-like glandular epithelia ( Figure 1B) 46.
Ultimately, the protective function of the epithelium is accomplished through the formation of intercellular junctions, molecular complexes that link up with the cytoskeleton to reinforce epithelial integrity. However, unlike other epithelial tissues, the endometrium exhibits remarkable changes during hormonal cycles and pregnancy, directly altering the expression profile of cell–cell junction proteins, their organization, dynamics, and barrier function 46, 47. In columnar epithelia, the three main classes of intercellular junctions are the tight junctions (TJs), adherens junctions (AJs), and desmosomes ( Figure 1C) 48. Although their function as molecular linkages has long been appreciated, newer studies have uncovered critical roles in transducing cell signaling pathways during tissue damage, repair, and pathogenic infection 49.
At the apex of the lateral cell membrane, TJs maintain a fence that physically separates the apical and basolateral membranes while gating the flux of ions and solutes through the paracellular space 50. Three families of transmembrane proteins—claudins, occludins, and junctional adhesion molecules (JAMs)—form homotypic interactions between adjacent cells. Adaptor proteins—that is, zona occludens 1–3 (ZO-1–3)—bind to their cytoplasmic tails and scaffold the recruitment of the polarity complex, which specifies apical membrane identity, and actin and microtubules to regulate TJ dynamics and stability 50.
AJs along the basolateral membrane are formed by homotypic interaction between cadherins, calcium-dependent transmembrane proteins 51. The adaptor proteins α- and β-catenin bind to cadherins and the actin cytoskeleton to reinforce AJ stability 52. AJ components, including β-catenin and others such as YES-associated protein (YAP), also function as transcription factors but are excluded from the nucleus through their association with stable AJs 53. AJ assembly is initially mediated by nectins, calcium-independent adhesion molecules, that also bind to AJ and TJ adaptor proteins 54, 55. The related but structurally distinct desmosomes are composed of a second class of cadherins—desmogleins and desmocollins—that connect to intermediate filaments, such as keratins, rigid cytoskeletal elements that provide additional structural support. Indeed, these strong junctions are thought to resist mechanical stress and are essential for the maintenance of epithelial integrity 56.
Many pathogenic viruses and bacteria have evolved diverse strategies to subvert cell–cell adhesions to gain entry into host cells or penetrate further into the underlying tissue 57. For example, Listeria monocytogenes surface protein InlA binds to E-cadherin and the hepatitis C virus binds to claudin-1 to promote their internalization 58, 59. The enteropathogenic Escherichia coli T3S effector EspF disrupts TJs and leads to a loss of epithelial barrier function 60. Importantly, these perturbations to cell–cell junctions can elicit an immune response as lumenal bacteria and toxins leak into the underlying tissue 61.
Chlamydia interactions with the epithelial surface
Initial observations in tumor cell lines indicated that C. trachomatis infection can impact cell–cell junctions. Chlamydia infections disrupt N-cadherin–based AJs in HeLa cells. Gaps were observed between Chlamydia-infected cells and β-catenin relocalized to inclusions based on indirect immunofluorescence 62. These basic observations in two-dimensional cell culture were detailed further in an ex vivo fallopian tube infection model. Infection with a C. trachomatis lymphogranuloma venereum (LGV) biovar disrupted epithelial polarity and cell–cell junction organization through a Wnt-driven paracrine signaling pathway 42. The Wnt pathway consists of secreted glycoproteins that bind to the Frizzled and LRP5/6 receptors, activating downstream signals to regulate tissue organization through cell fate determination, polarity formation, and cell growth 63. Activation of the canonical pathway disrupts the “destruction complex”, stabilizing the Wnt effector β-catenin and allowing its translocation to the nucleus, where it activates target genes 63. In fallopian tube epithelia, β-catenin translocates to the inclusion and a component of the destruction complex, adenomatous polyposis coli (APC), shows more diffuse cytoplasmic localization. Notably, the localization of these Wnt effectors also changed in neighboring uninfected cells but was rescued upon the addition of Wnt inhibitors, suggesting that infection can alter tissue-level Wnt signaling programs 42. More recently, pharmacological inhibition of Wnt signaling in endometrial epithelial cells resulted in smaller and aberrant inclusions and significantly reduced the production of EBs 64.
Chlamydia pneumoniae, the human respiratory pathogen and causative agent of roughly 4 to 6% of community-acquired pneumonia 65, also targets effectors of the Wnt pathway. The C. pneumoniae Inc Cpn1027 interacts with and may recruit the Wnt effectors Caprin2 and glycogen synthesis kinase 3 (GSK3) to the inclusion 66. The C. trachomatis T3S effector TepP regulates GSK3β recruitment to nascent inclusions in endocervical epithelial cells 10, but it is unclear whether these effectors regulate Wnt in the appropriate tissue context. In endothelial cells, C . pneumoniae infection also promotes vascular endothelial (VE)-cadherin phosphorylation on Y658 67, a residue that can control AJ organization and barrier function 68. The actin-binding protein EPS8, which is recruited to nascent inclusions during invasion and interacts with phosphorylated peptides derived from the T3S effector Tarp, also binds to VE-cadherin, alpha-catenin, and TJ components to control junction organization 69– 72. These results suggest that Chlamydia may secrete effectors that can target components of cell–cell junctions and cell signaling pathways that regulate junction organization.
Chlamydia muridarum, a mouse-adapted Chlamydia, also alters the composition of TJ proteins in mouse oviduct epithelial cells 73. Infection with C. muridarum decreases the expression of ZO-1, claudin-1/2, occludin, and JAM-1 while upregulating the expression of claudin-3 and claudin-4. Accordingly, C. muridarum infection alters transepithelial electrical resistance, indicating that the barrier function of TJs is compromised. These phenotypes were exacerbated by the absence of the Toll-like receptor 3 (TLR3), which initiates double-strand RNA (dsRNA)-dependent type I interferon responses 74. How TLR3 signaling is linked to TJ remodeling is unclear, but type III interferons were recently reported to strengthen epithelial barrier function during Salmonella infection 75.
Mechanistically, it remains to be determined whether reprogramming of cell signaling pathways or the removal of adhesion components and recruitment to the inclusion or both underlie the disruption of cell–cell junctions. Nevertheless, Chlamydia targeting of cell–cell junctions may drive certain aspects of its pathogenesis, including infertility and ectopic pregnancies. During estrus, the endometrium undergoes extensive paracrine-driven remodeling of cell–cell junctions to permit blastocyst adherence and invasion into the epithelial tissue 46. Thus, infection-mediated disruption of epithelial cell–cell junctions may impede implantation. Alternatively, Chlamydia infection is associated with elevated expression of the prokineticins receptor 2 (PROKR2), a G protein–coupled receptor that modulates TJ organization and paracellular permeability, which may prime tubal epithelia for implantation in fallopian tubes of patients with ectopic pregnancies 76, 77.
Chlamydia activates epithelial-to-mesenchymal transitions
The remodeling of epithelial cell–cell junctions is a hallmark of both inflammation and cellular proliferation 78. New studies indicate that multiple Chlamydia biovars can induce the transformation of epithelial cells to a more mesenchymal-like state by activating the epithelial-to-mesenchymal transition (EMT) program. EMT is essential for organogenesis and tissue repair during which epithelial cells adopt features of cells in the mesenchyme like fibroblasts, altering their polarity and the organization of the actin cytoskeleton and cell–cell adhesions 79. Driven by growth factors (for example, transforming growth factor beta [TGF-β], hepatocyte growth factor, and epidermal growth factor) and hormones (estrogen), EMT can be aberrantly activated during tumorigenesis and inflammation-associated fibrosis 78, 80. Most notably, EMT is often coupled to the downregulation of E-cadherin, upregulation of cell–extracellular matrix (cell–ECM) adhesions, deposition of ECM proteins, and enhanced cell motility. This transition is thought to enable tumor cells to disseminate toward the lymph nodes and vasculature 79. Alternatively, inflammation-associated EMT is necessary for tissue repair and ceases at the end of the inflammatory response. However, inflammation during chronic infections can lead to sustained EMT, tissue damage, and organ fibrosis 81, 82.
Chlamydia infection can promote EMT in different epithelial subtypes and through multiple pathways. Oviduct epithelial cells infected with C. trachomatis serovar D increase the expression of microRNAs (miRNAs) that promote EMT 83. By immunofluorescence microscopy, infection downregulates the expression of E-cadherin while upregulating the EMT markers smooth muscle actin (SMA), the matrix-degrading enzyme matrix metalloproteinase-9 (MMP9), fibronectin, and the transcription factors SNAIL1/2 and ZEB1 83, 84. Similar results were observed in conjunctival epithelial cells infected with C. trachomatis serovar B. Infection increases TGF-β expression which signals through SNAIL/ZEB2 and drives the downregulation of E-cadherin and upregulation of fibronectin and SMA. These changes may also involve epigenetic modifications as methylation of E-cadherin, fibronectin, and SMA genes were observed 85.
More recently, a global phosphoproteomic and transcriptomic analysis in ectocervical epithelial explants infected with C. trachomatis serovar L2 indicated that infection activates cell signaling pathways to promote an EMT-like signature 43. The infection initiates at the top of the stratified epithelia and progresses toward the basal layer, suggesting that altering cell–cell junctions during EMT may provide access for subsequent rounds of infection. Indeed, downstream of the MAPK pathway, the transcriptional factors ETS1 and ERF promote EMT in infected cells, leading to the downregulation of E-cadherin and increased cell motility and invasion 43. Collectively, these observations suggest that Chlamydia can promote the transformation of epithelial cells, which may contribute to cancers of the cervix and uterus. However, these results must be confirmed in vivo and in the proper tissue context.
Chlamydia infection in new in vivo models
Extending these observations to a robust animal model of infection has remained challenging. Although the mouse-adapted C. muridarum is used extensively to study the host immune response in mice, this Chlamydia species is less genetically tractable than C. trachomatis. On the other hand, intravaginal inoculations of C57BL/6 mice with C. trachomatis often fail to induce significant pathology because the bacteria do not efficiently ascend to the upper genital tract 86. However, transcervical inoculations 87 that bypass the vaginal vault and the use of more permissive mouse strains (for example, C3H/HeJ) have improved the ability to monitor the infections in the mouse upper genital tract and ensuing pathology and infertility 88. C. trachomatis serovar L2 ascends to the upper genital tract in C3H/HeJ mice when inoculated intravaginally, stimulating a robust immune response and altering epithelial cell height 89. Future studies combining these new infection models and techniques with both host and bacterial genetics will promote better molecular dissection of Chlamydia pathogenesis in live animals.
Future directions
The recent advances in the Chlamydia experimental tool kit have brought about a new era in Chlamydia research. With the ability to specifically disrupt genes and express genes in trans, new mechanisms by which Chlamydia subverts its host will be identified. Applying these molecular tools in model systems that better mimic the in vivo physiology will significantly accelerate our understanding of the infection process, especially as these tools are applied to other C. trachomatis serovars with distinct tissue tropisms. For example, using more sophisticated infection models and defined C. trachomatis mutants, we now test how specific virulence factors promote infection and affect epithelial cell growth, function, organization, and potentially transformation. However, more work is necessary to decode the genetics of the urogenital and ocular biovars and generate the relevant in vivo models of infection that best recapitulate the host responses observed in humans. Collectively, these new tools and models can be prioritized for the identification of new therapeutic targets or harnessed for the rational design of vaccines for this clinically important pathogen.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Ming Tan, Departments of Microbiology and Molecular Genetics, and Medicine, University of California, Irvine, California, USA
Kenneth Fields, Department of Microbiology, Immunology & Molecular Genetics, University of Kentucky College of Medicine, Kentucky, USA
Peter Timms, Faculty of Science, Health, Education and Engineering, University of the Sunshine Coast, Queensland, Australia
Funding Statement
This work received funding support from the National Institute of Allergy and Infectious Diseases (grants R01AI134891 and F32AI138372).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 3 approved]
References
- 1. Horn M: Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol. 2008;62:113–31. 10.1146/annurev.micro.62.081307.162818 [DOI] [PubMed] [Google Scholar]
- 2. Schachter J: Chlamydial infections (first of three parts). N Engl J Med. 1978;298(8):428–35. 10.1056/NEJM197802232980805 [DOI] [PubMed] [Google Scholar]
- 3. Bachmann NL, Polkinghorne A, Timms P: Chlamydia genomics: Providing novel insights into chlamydial biology. Trends Microbiol. 2014;22(8):464–72. 10.1016/j.tim.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 4. Elwell C, Mirrashidi K, Engel J: Chlamydia cell biology and pathogenesis. Nat Rev Microbiol. 2016;14(6):385–400. 10.1038/nrmicro.2016.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mueller KE, Plano GV, Fields KA: New frontiers in type III secretion biology: The Chlamydia perspective. Infect Immun. 2014;82(1):2–9. 10.1128/IAI.00917-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Betts HJ, Wolf K, Fields KA: Effector protein modulation of host cells: Examples in the Chlamydia spp. arsenal. Curr Opin Microbiol. 2009;12(1):81–7. 10.1016/j.mib.2008.11.009 [DOI] [PubMed] [Google Scholar]
- 7. Carabeo RA, Dooley CA, Grieshaber SS, et al. : Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cell Microbiol. 2007;9(9):2278–88. 10.1111/j.1462-5822.2007.00958.x [DOI] [PubMed] [Google Scholar]
- 8. Rockey DD, Rosquist JL: Protein antigens of Chlamydia psittaci present in infected cells but not detected in the infectious elementary body. Infect Immun. 1994;62(1):106–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mital J, Lutter EI, Barger AC, et al. : Chlamydia trachomatis inclusion membrane protein CT850 interacts with the dynein light chain DYNLT1 (Tctex1). Biochem Biophys Res Commun. 2015;462(2):165–70. 10.1016/j.bbrc.2015.04.116 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Carpenter V, Chen YS, Dolat L, et al. : The Effector TepP Mediates Recruitment and Activation of Phosphoinositide 3-Kinase on Early Chlamydia trachomatis Vacuoles. mSphere. 2017;2(4): pii: e00207-17. 10.1128/mSphere.00207-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hackstadt T, Scidmore-Carlson MA, Shaw EI, et al. : The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1999;1(2):119–30. 10.1046/j.1462-5822.1999.00012.x [DOI] [PubMed] [Google Scholar]
- 12. Derré I, Swiss R, Agaisse H: The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER- Chlamydia inclusion membrane contact sites. PLoS Pathog. 2011;7(6):e1002092. 10.1371/journal.ppat.1002092 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Agaisse H, Derré I: Expression of the Effector Protein IncD in Chlamydia trachomatis Mediates Recruitment of the Lipid Transfer Protein CERT and the Endoplasmic Reticulum-Resident Protein VAPB to the Inclusion Membrane. Infect Immun. 2014;82(5):2037–47. 10.1128/IAI.01530-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stanhope R, Flora E, Bayne C, et al. : IncV, a FFAT motif-containing Chlamydia protein, tethers the endoplasmic reticulum to the pathogen-containing vacuole. Proc Natl Acad Sci U S A. 2017;114(45):12039–12044. 10.1073/pnas.1709060114 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Kumar Y, Valdivia RH: Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe. 2008;4(2):159–69. 10.1016/j.chom.2008.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Volceanov L, Herbst K, Biniossek M, et al. : Septins Arrange F-Actin-Containing Fibers on the Chlamydia trachomatis Inclusion and Are Required for Normal Release of the Inclusion by Extrusion. mBio. 2014;5:e460. 10.1128/mBio.01802-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wesolowski J, Weber MM, Nawrotek A, et al. : Chlamydia Hijacks ARF GTPases To Coordinate Microtubule Posttranslational Modifications and Golgi Complex Positioning. mBio. 2017;8(3): pii: e02280-16. 10.1128/mBio.02280-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hybiske K, Stephens RS: Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A. 2007;104(27):11430–5. 10.1073/pnas.0703218104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Snavely EA, Kokes M, Dunn JD, et al. : Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis. 2014;71(3):336–51. 10.1111/2049-632X.12179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Kahane S, Cutcliffe LT, et al. : Development of a Transformation System for Chlamydia trachomatis: Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector. PLoS Pathog. 2011;7(9):e1002258. 10.1371/journal.ppat.1002258 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Kari L, Goheen MM, Randall LB, et al. : Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A. 2011;108(17):7189–93. 10.1073/pnas.1102229108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kokes M, Dunn JD, Granek JA, et al. : Integrating Chemical Mutagenesis and Whole-Genome Sequencing as a Platform for Forward and Reverse Genetic Analysis of Chlamydia. Cell Host Microbe. 2015;17(5):716–25. 10.1016/j.chom.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Mueller KE, Wolf K, Fields KA: Gene Deletion by Fluorescence-Reported Allelic Exchange Mutagenesis in Chlamydia trachomatis. mBio. 2016;7(1):e01817–15. 10.1128/mBio.01817-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keb G, Hayman R, Fields KA: Floxed-Cassette Allelic Exchange Mutagenesis Enables Markerless Gene Deletion in Chlamydia trachomatis and Can Reverse Cassette-Induced Polar Effects. J Bacteriol. 2018;200(24): pii: e00479-18. 10.1128/JB.00479-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sixt BS, Bastidas RJ, Finethy R, et al. : The Chlamydia trachomatis Inclusion Membrane Protein CpoS Counteracts STING-Mediated Cellular Surveillance and Suicide Programs. Cell Host Microbe. 2017;21(1):113–121. 10.1016/j.chom.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agaisse H, Derré I: A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One. 2013;8(2):e57090. 10.1371/journal.pone.0057090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bastidas RJ, Valdivia RH: Emancipating Chlamydia: Advances in the Genetic Manipulation of a Recalcitrant Intracellular Pathogen. Microbiol Mol Biol Rev. 2016;80(2):411–27. 10.1128/MMBR.00071-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rucks EA, Olson MG, Jorgenson LM, et al. : Development of a Proximity Labeling System to Map the Chlamydia trachomatis Inclusion Membrane. Front Cell Infect Microbiol. 2017;7:40. 10.3389/fcimb.2017.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dickinson MS, Anderson LN, Webb-Robertson BJM, et al. : Proximity-dependent proteomics of the Chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites. PLoS Pathog. 2019;15(4):e1007698. 10.1371/journal.ppat.1007698 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Aeberhard L, Banhart S, Fischer M, et al. : The Proteome of the Isolated Chlamydia trachomatis Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components. PLoS Pathog. 2015;11(6):e1004883. 10.1371/journal.ppat.1004883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mirrashidi KM, Elwell CA, Verschueren E, et al. : Global Mapping of the Inc-Human Interactome Reveals that Retromer Restricts Chlamydia Infection. Cell Host Microbe. 2015;18(1):109–21. 10.1016/j.chom.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaddoum L, Magdeleine E, Waldo GS, et al. : One-step split GFP staining for sensitive protein detection and localization in mammalian cells. BioTechniques. 2010;49(4):727–8, 730, 732 passim. 10.2144/000113512 [DOI] [PubMed] [Google Scholar]
- 33. Wang X, Hybiske K, Stephens RS: Direct visualization of the expression and localization of chlamydial effector proteins within infected host cells. Pathog Dis. 2018;76(2). 10.1093/femspd/fty011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nans A, Saibil HR, Hayward RD: Pathogen-host reorganization during Chlamydia invasion revealed by cryo-electron tomography. Cell Microbiol. 2014;16(10):1457–72. 10.1111/cmi.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee JK, Enciso GA, Boassa D, et al. : Replication-dependent size reduction precedes differentiation in Chlamydia trachomatis. Nat Commun. 2018;9(1):45. 10.1038/s41467-017-02432-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor BD, Haggerty CL: Management of Chlamydia trachomatis genital tract infection: Screening and treatment challenges. Infect Drug Resist. 2011;4:19–29. 10.2147/IDR.S12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blaskewicz CD, Pudney J, Anderson DJ: Structure and function of intercellular junctions in human cervical and vaginal mucosal epithelia. Biol Reprod. 2011;85(1):97–104. 10.1095/biolreprod.110.090423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guseva NV, Dessus-Babus S, Moore CG, et al. : Differences in Chlamydia trachomatis Serovar E Growth Rate in Polarized Endometrial and Endocervical Epithelial Cells Grown in Three-Dimensional Culture. Infect Immun. 2007;75(2):553–64. 10.1128/IAI.01517-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moore ER, Fischer ER, Mead DJ, et al. : The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic. 2008;9(12):2130–40. 10.1111/j.1600-0854.2008.00828.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herbst-Kralovetz MM, Quayle AJ, Ficarra M, et al. : Quantification and comparison of toll-like receptor expression and responsiveness in primary and immortalized human female lower genital tract epithelia. Am J Reprod Immunol. 2008;59(3):212–24. 10.1111/j.1600-0897.2007.00566.x [DOI] [PubMed] [Google Scholar]
- 41. Buckner LR, Lewis ME, Greene SJ, et al. : Chlamydia trachomatis infection results in a modest pro-inflammatory cytokine response and a decrease in T cell chemokine secretion in human polarized endocervical epithelial cells. Cytokine. 2013;63(2):151–65. 10.1016/j.cyto.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kessler M, Zielecki J, Thieck O, et al. : Chlamydia trachomatis Disturbs Epithelial Tissue Homeostasis in Fallopian Tubes via Paracrine Wnt Signaling. Am J Pathol. 2012;180(1):186–98. 10.1016/j.ajpath.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 43. Zadora PK, Chumduri C, Imami K, et al. : Integrated Phosphoproteome and Transcriptome Analysis Reveals Chlamydia-Induced Epithelial-to-Mesenchymal Transition in Host Cells. Cell Rep. 2019;26(5):1286–1302.e8. 10.1016/j.celrep.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 44. Kessler M, Hoffmann K, Brinkmann V, et al. : The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun. 2015;6:8989. 10.1038/ncomms9989 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Kessler M, Hoffmann K, Fritsche K, et al. : Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat Commun. 2019;10(1):1194. 10.1038/s41467-019-09144-7 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Grund S, Grümmer R: Direct Cell-Cell Interactions in the Endometrium and in Endometrial Pathophysiology. Int J Mol Sci. 2018;19(8): pii: E2227. 10.3390/ijms19082227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Someya M, Kojima T, Ogawa M, et al. : Regulation of tight junctions by sex hormones in normal human endometrial epithelial cells and uterus cancer cell line Sawano. Cell Tissue Res. 2013;354(2):481–94. 10.1007/s00441-013-1676-9 [DOI] [PubMed] [Google Scholar]
- 48. Garcia MA, Nelson WJ, Chavez N: Cell-Cell Junctions Organize Structural and Signaling Networks. Cold Spring Harb Perspect Biol. 2018;10(4): pii: a029181. 10.1101/cshperspect.a029181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zihni C, Balda MS, Matter K: Signalling at tight junctions during epithelial differentiation and microbial pathogenesis. J Cell Sci. 2014;127(Pt 16):3401–13. 10.1242/jcs.145029 [DOI] [PubMed] [Google Scholar]
- 50. Zihni C, Mills C, Matter K, et al. : Tight junctions: From simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17(9):564–80. 10.1038/nrm.2016.80 [DOI] [PubMed] [Google Scholar]
- 51. Nelson WJ: Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochem Soc Trans. 2008;36(Pt 2):149–55. 10.1042/BST0360149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Buckley CD, Tan J, Anderson KL, et al. : Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346(6209):1254211. 10.1126/science.1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Benham-Pyle BW, Pruitt BL, Nelson WJ: Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science. 2015;348(6238):1024–7. 10.1126/science.aaa4559 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Takai Y, Nakanishi H: Nectin and afadin: novel organizers of intercellular junctions. J Cell Sci. 2003;116(Pt 1):17–27. 10.1242/jcs.00167 [DOI] [PubMed] [Google Scholar]
- 55. Rikitake Y, Mandai K, Takai Y: The role of nectins in different types of cell-cell adhesion. J Cell Sci. 2012;125(Pt 16):3713–22. 10.1242/jcs.099572 [DOI] [PubMed] [Google Scholar]
- 56. Nekrasova O, Green KJ: Desmosome assembly and dynamics. Trends Cell Biol. 2013;23(11):537–46. 10.1016/j.tcb.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Bonazzi M, Cossart P: Impenetrable barriers or entry portals? The role of cell-cell adhesion during infection. J Cell Biol. 2011;195(3):349–58. 10.1083/jcb.201106011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lecuit M, Dramsi S, Gottardi C, et al. : A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 1999;18(14):3956–63. 10.1093/emboj/18.14.3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Evans MJ, von Hahn T, Tscherne DM, et al. : Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446(7137):801–5. 10.1038/nature05654 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. McNamara BP, Koutsouris A, O’Connell CB, et al. : Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J Clin Invest. 2001;107(5):621–9. 10.1172/JCI11138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. König J, Wells J, Cani PD, et al. : Human Intestinal Barrier Function in Health and Disease. Clin Transl Gastroenterol. 2016;7(10):e196. 10.1038/ctg.2016.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Prozialeck WC, Fay MJ, Lamar PC, et al. : Chlamydia trachomatis disrupts N-cadherin-dependent cell-cell junctions and sequesters beta-catenin in human cervical epithelial cells. Infect Immun. 2002;70(5):2605–13. 10.1128/iai.70.5.2605-2613.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cadigan KM, Peifer M: Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol. 2009;1(2):a002881. 10.1101/cshperspect.a002881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kintner J, Moore CG, Whittimore JD, et al. : Inhibition of Wnt Signaling Pathways Impairs Chlamydia trachomatis Infection in Endometrial Epithelial Cells. Front Cell Infect Microbiol. 2017;7:501. 10.3389/fcimb.2017.00501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bartlett JG: Diagnostic tests for agents of community-acquired pneumonia. Clin Infect Dis. 2011;52 Suppl 4:S296–304. 10.1093/cid/cir045 [DOI] [PubMed] [Google Scholar]
- 66. Flores R, Zhong G: The Chlamydia pneumoniae Inclusion Membrane Protein Cpn1027 Interacts with Host Cell Wnt Signaling Pathway Regulator Cytoplasmic Activation/Proliferation-Associated Protein 2 (Caprin2). PLoS One. 2015;10(5):e0127909. 10.1371/journal.pone.0127909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu J, Miao G, Wang B, et al. : Chlamydia pneumoniae infection promotes monocyte transendothelial migration by increasing vascular endothelial cell permeability via the tyrosine phosphorylation of VE-cadherin. Biochem Biophys Res Commun. 2018;497(2):742–8. 10.1016/j.bbrc.2018.02.145 [DOI] [PubMed] [Google Scholar]
- 68. Potter MD, Barbero S, Cheresh DA: Tyrosine phosphorylation of VE-cadherin prevents binding of p120- and beta-catenin and maintains the cellular mesenchymal state. J Biol Chem. 2005;280(36):31906–12. 10.1074/jbc.M505568200 [DOI] [PubMed] [Google Scholar]
- 69. Lane BJ, Mutchler C, Al Khodor S, et al. : Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 2008;4(3):e1000014. 10.1371/journal.ppat.1000014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Giampietro C, Disanza A, Bravi L, et al. : The actin-binding protein EPS8 binds VE-cadherin and modulates YAP localization and signaling. J Cell Biol. 2015;211(6):1177–92. 10.1083/jcb.201501089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fredriksson K, van Itallie CM, Aponte A, et al. : Proteomic analysis of proteins surrounding occludin and claudin-4 reveals their proximity to signaling and trafficking networks. PLoS One. 2015;10(3):e0117074. 10.1371/journal.pone.0117074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lie PP, Mruk DD, Lee WM, et al. : Epidermal growth factor receptor pathway substrate 8 (Eps8) is a novel regulator of cell adhesion and the blood-testis barrier integrity in the seminiferous epithelium. FASEB J. 2009;23(8):2555–67. 10.1096/fj.06-070573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kumar R, Gong H, Liu L, et al. : TLR3 deficiency exacerbates the loss of epithelial barrier function during genital tract Chlamydia muridarum infection. PLoS One. 2019;14(1):e0207422. 10.1371/journal.pone.0207422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Medzhitov R: Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135–45. 10.1038/35100529 [DOI] [PubMed] [Google Scholar]
- 75. Odendall C, Voak AA, Kagan JC: Type III IFNs Are Commonly Induced by Bacteria-Sensing TLRs and Reinforce Epithelial Barriers during Infection. J Immunol. 2017;199(9):3270–9. 10.4049/jimmunol.1700250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shaw JL, Wills GS, Lee KF, et al. : Chlamydia trachomatis infection increases fallopian tube PROKR2 via TLR2 and NFκB activation resulting in a microenvironment predisposed to ectopic pregnancy. Am J Pathol. 2011;178(1):253–60. 10.1016/j.ajpath.2010.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guilini C, Urayama K, Turkeri G, et al. : Divergent roles of prokineticin receptors in the endothelial cells: angiogenesis and fenestration. Am J Physiol Heart Circ Physiol. 2010;298(3):H844–52. 10.1152/ajpheart.00898.2009 [DOI] [PubMed] [Google Scholar]
- 78. Kalluri R, Weinberg RA: The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–8. 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thiery JP, Acloque H, Huang RY, et al. : Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 80. Eger A, Stockinger A, Schaffhauser B, et al. : Epithelial mesenchymal transition by c-Fos estrogen receptor activation involves nuclear translocation of beta-catenin and upregulation of beta-catenin/lymphoid enhancer binding factor-1 transcriptional activity. J Cell Biol. 2000;148(1):173–88. 10.1083/jcb.148.1.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. López-Novoa JM, Nieto MA: Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 2009;1(6–7):303–14. 10.1002/emmm.200900043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hofman P, Vouret-Craviari V: Microbes-induced EMT at the crossroad of inflammation and cancer. Gut Microbes. 2012;3(3):176–85. 10.4161/gmic.20288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Igietseme JU, Omosun Y, Stuchlik O, et al. : Role of Epithelial-Mesenchyme Transition in Chlamydia Pathogenesis. PLoS One. 2015;10(12):e0145198. 10.1371/journal.pone.0145198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Igietseme JU, Omosun Y, Nagy T, et al. : Molecular Pathogenesis of Chlamydia Disease Complications: Epithelial-Mesenchymal Transition and Fibrosis. Infect Immun. 2018;86(1): pii: e00585-17. 10.1128/IAI.00585-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rajić J, Inic-Kanada A, Stein E, et al. : Chlamydia trachomatis Infection Is Associated with E-Cadherin Promoter Methylation, Downregulation of E-Cadherin Expression, and Increased Expression of Fibronectin and α-SMA-Implications for Epithelial-Mesenchymal Transition. Front Cell Infect Microbiol. 2017;7:253. 10.3389/fcimb.2017.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sturdevant GL, Caldwell HD: Innate immunity is sufficient for the clearance of Chlamydia trachomatis from the female mouse genital tract. Pathog Dis. 2014;72(1):70–3. 10.1111/2049-632X.12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Coers J, Gondek DC, Olive AJ, et al. : Compensatory T cell responses in IRG-deficient mice prevent sustained Chlamydia trachomatis infections. PLoS Pathog. 2011;7(6):e1001346. 10.1371/journal.ppat.1001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pal S, Tifrea DF, Zhong G, et al. : Transcervical Inoculation with Chlamydia trachomatis Induces Infertility in HLA-DR4 Transgenic and Wild-Type Mice. Infect Immun. 2018;86(1): pii: e00722-17. 10.1128/IAI.00722-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shaw JH, Behar AR, Snider TA, et al. : Comparison of Murine Cervicovaginal Infection by Chlamydial Strains: Identification of Extrusions Shed In vivo. Front Cell Infect Microbiol. 2017;7:18. 10.3389/fcimb.2017.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]