Abstract
Ophiopogonin-B (OP-B) has been reported to suppress metastasis and angiogenesis of adenocarcinoma A549 cells in vitro and in vivo. More and more evidences indicate that inflammatory microenvironment facilitates tumor metastasis. Digital Gene Expression (DGE) analysis of non-small cell lung cancer (NSCLC) cell lines showed that OP-B downregulated the expression of linc00668, which promoted progression of cancer. Herein, we simulated the inflammatory microenvironment by co-culturing A549 cells with LPS-treated THP-1 cells and found that the level of linc00668 increased significantly in the mock group, while OP-B treatment inhibited the level of linc00668 and reversed epithelial-mesenchymal transition (EMT) induced by linc00668. In addition, overexpression of linc00668 in A549 cells suppressed the expression of E-cadherin and induced expression of N-cadherin, while OP-B treatment reversed these changes. Bioinformatic prediction and dual-luciferase reporter gene assay validated that linc00668 sponge miR-432-5p and at last acted on EMT to execute the anti-migration function of A549 cells under inflammatory microenvironment. Taken together, OP-B inhibits metastasis of A549 cells via the linc00668/miR-432-5p/EMT axis.
Keywords: NSCLC, metastasis, linc00668, miR-432-5p, epithelial-mesenchymal transition (EMT)
Introduction
Non-small cell lung cancer (NSCLC) accounts for approximately 80~85% of all lung cancer cases, mainly including adenocarcinoma and squamous cell carcinoma. Despite remarkable advances in clinical diagnosis and treatment, the average five-year survival rate of NSCLC patients is only 16% in the USA, and <10% in the UK1, 2. Adenocarcinoma is the most common type of NSCLC, which has high mortality due to its metastasis at an early stage 3, 4. Therefore, preventing metastasis is very important for improving curative effect and prolonging the life of patients.
Increasing evidences indicated that inflammatory microenvironment facilitated tumor metastasis by enhancing epithelial-mesenchymal transition (EMT) 5, 6, while the underlying mechanisms remained to be validated. Long non-coding RNAs (lncRNAs) are a class of RNA transcripts which usually longer than 200 nucleotides in length 7, 8. Recently, lncRNAs have emerged as important regulators of physiological and pathological processes 9, 10. LncRNA-dysregulation has been reported to be correlated with tumor development 11, 12, and recent studies have revealed an essential role of lncRNAs in regulating EMT and tumor metastasis 13. For example, linc00668 has been reported to be up-regulated in gastric cancer tissues and serves as an independent predictor for overall survival of gastric cancer patients 14.
Ophiopogonin-B (OP-B) has been reported to suppress metastasis and angiogenesis of adenocarcinoma A549 cells in vitro and in vivo 15, while the underlying mechanisms remained to be elucidated. In this study, digital gene expression (DGE) analysis and quantitative RT-PCR (qRT-PCR) assay validated that linc00668 is significantly inhibited in A549 cells by OP-B. Herein, we aimed to identify the role of linc00668 in the metastasis of A549 cells under inflammatory microenvironment, and to investigate the regulation effect of OP-B on the process of linc00668-regulated metastasis of A549 cells.
Materials and Methods
Cell culture
A549, 293T and THP-1 cells were obtained from the Cell Bank, Chinese Academy of Sciences (Shanghai, China), and cultured in DMEM-F12 medium (Gibco, Australia) supplemented with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Australia) at 37℃ in a humidified 5% CO2 incubator.
Co-culture system
Human lung adenocarcinoma A549 cells were co-cultured with 10 μg/mL LPS-treated human pro-myelomonocytic cells (THP-1 cells) at a ratio of 1:5 in 6-well tissue culture plates containing transwell inserts (0.4-mm-pore-size polycarbonate membrane, #3412; Corning Costar, Cambridge, MA)6. Next, the co-cultured A549 cells were exposed to 5 μmol/L of OP-B for 24 h.
Cell transfection
A549 cells at exponential stage were used for transfection. Before transfection, 1 × 106 A549 cells were cultured in 6-well plates with 2 mL complete medium for 24 h until they were 90% confluent. The over-expression vector of linc00668 was constructed in pcDNA3.1 (+) by Genscript which purchased from Nanjing Realgene Inc (Nanjing, China). The vectors and microRNAs were transfected, respectively, into A549 cell line by Lipofectamine2000 reagents and cultured with Opti-MEM serum-free medium following the instructions.
Western blot
The cells were collected and lysed with cell lysis buffer (Beyotime, Shanghai, China). Samples of the lysates were separated on 12% SDS-PAGE gels and transferred to nitrocellulose filter membranes. The membranes were incubated with primary antibodies at 4 °C overnight, including Vimentin, E-cadherin, N-cadherin, ZO-1, ZEB1, Stat3, Stat5 (CST, USA) and β-actin (Santa Cruz, USA), followed by incubation with an HRP-conjugated anti-mouse or anti-rabbit secondary antibody separately. Then immunoreactivity was detected by enhanced chemiluminescence (ECL) (Bio-Rad, USA). Quantitative analysis was performed by Image Lab™ software.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from cells using TRIzol reagent (Invitrogen, USA). For mRNA and lncRNA quantification, the RNA was reverse transcribed into cDNA with a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, USA). EvaGreen 2× qRT-PCR MasterMix-Low ROX (abm, Canada) was used for quantitation with specific primers for the mRNA and lncRNA. GAPDH was used as an internal control. For miRNA quantification, reverse transcription was performed using an miRNA First Strand cDNA Synthesis (Stem-loop Method) Kit (Sangon Biotech, Shanghai, China). A MicroRNA qRT-PCR Kit (SYBR Green Method) (Sangon Biotech, Shanghai, China) was used for quantitation with specific primers for miRNA, and U6 was used as an internal control. The primer sequences were listed in Table S1. All quantitative real-time PCR experiments were performed with an Agilent Technologies Stratagene Mx3000P system (Agilent Technologies, USA). The data was processed with the 2-ΔΔCt method, and the fold changes were normalized to the expression of the internal controls.
Transwell invasion assay
Transwell invasion assay was performed using 8.0 μm Transwell Permeable Supports #3422 (Corning, USA). Cells were harvested 24 h after transfection and 5×104 cells suspended in 100 μl serum-free medium were seeded into the upper chamber pre-coated with Matrigel Matrix (BD Biosciences, USA), and 600 μl medium containing 10% FBS was added to the lower chamber. After incubation for 24 h, cells that did not invaded through the membrane were mechanically removed with a cotton swab. Next, 4% paraformaldehyde was used to fix the cells on the bottom surface of the membrane for 10 min, and then cells were stained with a 0.4% crystal violet solution. The invading cells were imaged using a digital microscopy (Leica, Germany).
Luciferase reporter assay
Cells (293 T) were seeded at 5 × 104 cells/well in 24-well plates and allowed to settle overnight. The pmirGLO Dual-Luciferase miRNA Target Expression Vector was purchased from Promega (USA). After 24h, cells were co-transfected with pmirGLO-Linc00668-WT, pmirGLO-Linc00668-MUT reporter plasmids, negative control (NC) plasmids and NC mimics, miR-432-5p mimics accordingly. 24 h post transfection, cells were lysed using passive lysis buffer (Promega, USA), the luciferase activity was measured by SpectraMax i3x microplate reader (Molecular Devices, US) and normalized to renilla luciferase activity respectively. Experiments were performed in triplicate.
Statistical analyses
Statistical analyses were performed using Graphpad Prism 5.0 software. Data of experiments were expressed as mean ± standard deviation (SD) of at least three independent experiments. Differences between two groups were assessed using Student's t-test. We adopted one-way analysis of variance for multiple comparisons. A p-value < 0.05 was considered statistically significant unless additionally specified.
Results
OP-B inhibits EMT in human lung adenocarcinoma cells in the inflammatory microenvironment
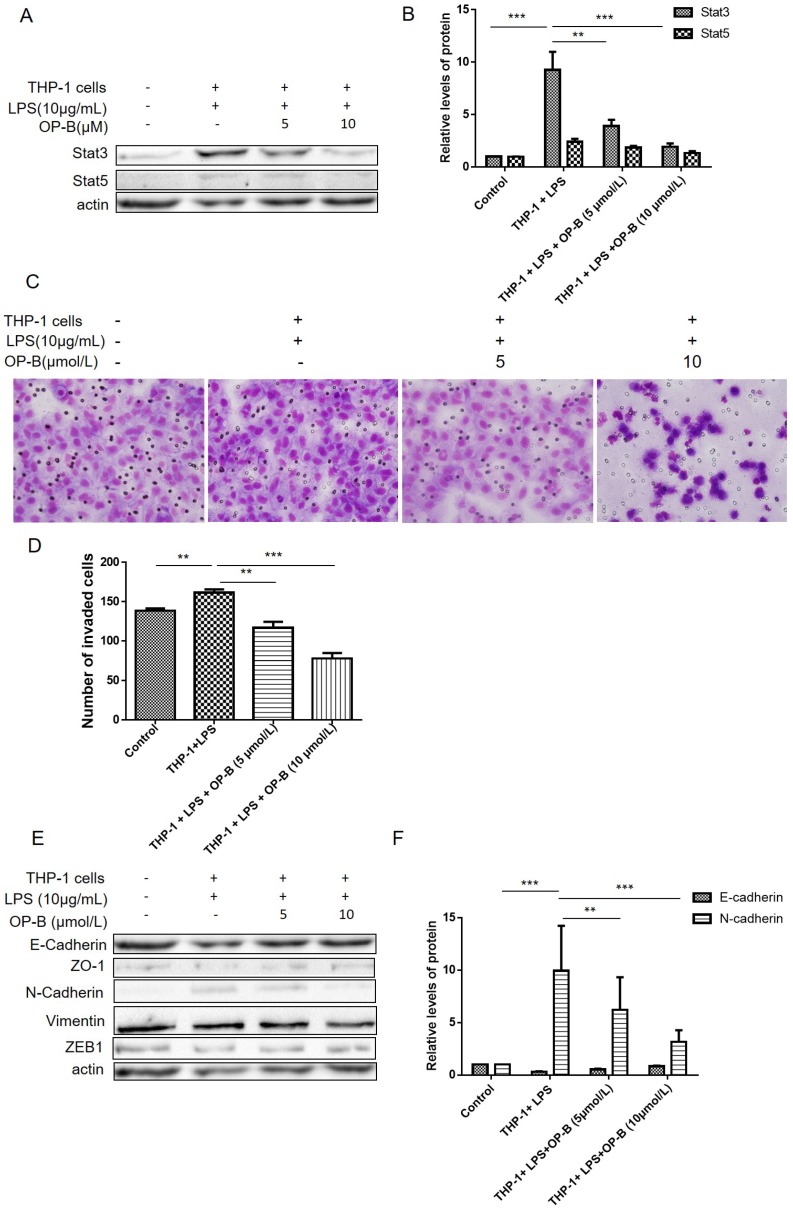
In our previous study, we have reported that OP-B inhibited metastasis of A549 cells in vitro and in vivo. Herein, we established a co-culture system to investigate whether OP-B affected EMT of lung adenocarcinoma cells in the inflammatory microenvironment. Firstly, A549 cells were co-cultured with LPS-stimulated THP-1 cells for 24 h, after that the cells were treated by 5 or 10 μmol/L of OP-B for 24 h. For aberrant Stat3, 5 signaling playing important role in chronic inflammation conditions 16, 17, we then detected the level of Stat3 and Stat5 by western blot, the results showed that the levels of Stat3 and Stat5 were all stimulated under the co-culture condition, while 5 or 10 μmol/L of OP-B treatment significantly decreased the levels of Stat3 and Stat5 (Figure 1A-B).
Figure 1.
OP-B inhibits the migration of A549 cells co-cultured with LPS-challenged THP-1 cells. (A~B), Levels of Stat3 and Stat5 were detected by western blot assay with specific antibodies, β-actin was used as a control. (C~D), Effects of OP-B on the migration of cells across the Transwell chamber in the co-culture system. Images are representative of three independent experiments. (E~F), Effects of OP-B on the levels of EMT-related markers in co-cultured A549 cells. Levels of E-cadherin, ZO-1, N-cadherin, ZEB1, and Vimentin were detected by western blot assay, β-actin was used as a loading control. Error bars represented mean ± SD from three independent experiments. (*), P < 0.05; (**), P < 0.01; (***), P < 0.001.
Then effects of OP-B on the invasion ability of co-cultured A549 cells were evaluated by transwell assay, and the results showed that OP-B prevented the migration of co-cultured A549 cells across the transwell chamber (Figure 1C-D). More importantly, the results of western blot showed that co-culture decreased the expression of epithelial markers E-cadherin and increased the expression of mesenchymal markers N-cadherin and Vimentin. However, OP-B treatment reversed these changes (Figure 1E-F). Meanwhile, the expression of Vimentin, ZO-1 and ZEB1 were not changed by the treatment. These results indicated that OP-B inhibited the migration of A549 cells under the inflammation microenviroment by reversing the EMT markers E-cadherin and N-cadherin.
Effects of linc00668 on lung cancer cell migration and invasion
It's known that Stat3 promotes metastasis by enhancing EMT-associated signaling pathway. In order to find out the underlying mechanisms that OP-B regulating the EMT pathway and whether Stat3 was included in the pathway, we reanalyzed the transcriptome date performed before and carried out bioinformatic analysis.
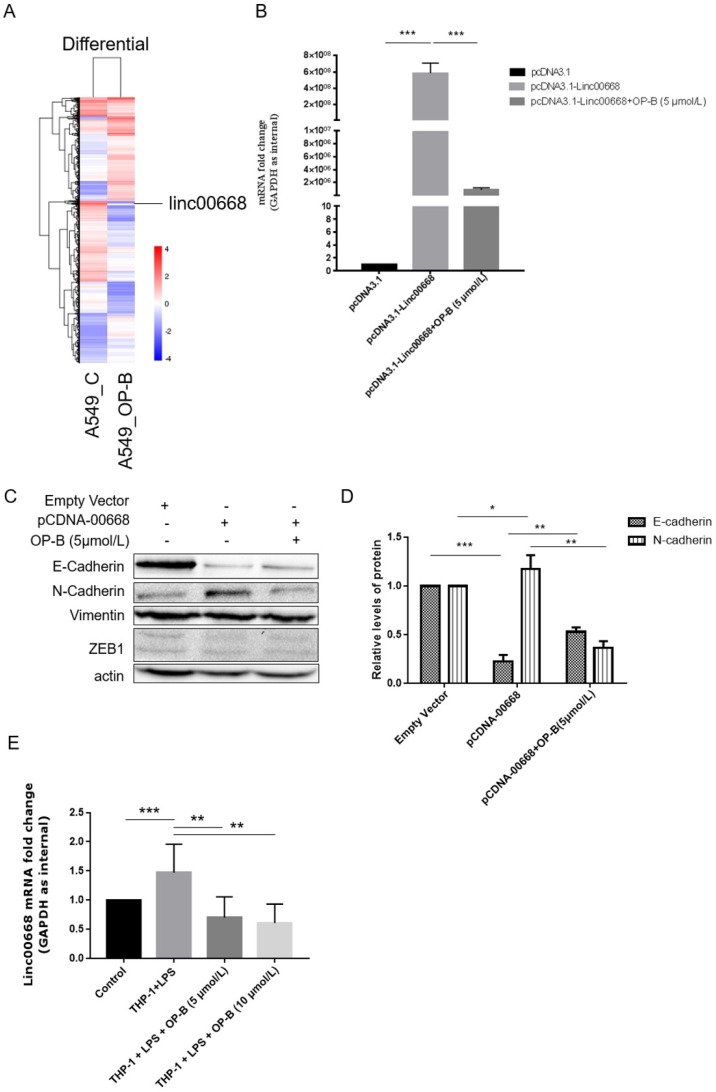
Screening of the genome transcriptome data of RNA-seq showed that OP-B (10 μmol/L) treatment resulted in significant decrease of linc00668 in A549 cells (by 6.672-fold) (Figure 2A). Further results of qRT-PCR validated that in linc00668 overexpressed A549 cell line, 5 μmol/L of OP-B inhibited the expression of linc00668 significantly (P<0.001) (Figure 2B).
Figure 2.
Effects of linc00668 on A549 cell migration and invasion. A, Screening of the genome transcriptome data of RNA-seq in OP-B-treated A549 cells. B, QRT-PCR analysis of linc00668 regulated by OP-B in linc00668 vector-transfected A549 cells. (C~D), Regulation of OP-B on EMT signaling pathway were detected by western blotting in linc00668 vector-transfected A549 cells. Images are representative of three independent experiments. E, QRT-PCR detection of linc00668 in the A549 cells co-cultured with LPS-stimulated THP-1. Error bars represented mean ± SD from three independent experiments. (*), P < 0.05; (**), P < 0.01; (***), P < 0.001.
To investigate whether the regulation of OP-B on EMT pathway was correlated with linc00668, we overexpressed linc00668 in A549 cells and detected the expression of E-cadherin and N-cadherin. The results showed that overexpression of linc00668 suppressed the expression of E-cadherin (P<0.001) and induced the expression of N-cadherin (P<0.05). However, treatment with OP-B reversed these changes (P<0.01) (Figure 2C-D). Similar to the above results, the level of Vimentin and ZEB1 were not changed under this treatment.
Similarly, through co-culture with LPS-stimulated THP-1, we set up an inflammation microenvironment for A549 cells, detection of linc00668 by qRT-PCR showed that the level of it increased significantly in the mock group (P<0.001), while OP-B treatment decreased the level of linc00668 (P<0.01) (Figure 2E).
Linc00668 targeted miR-432-5p and repressed its expression
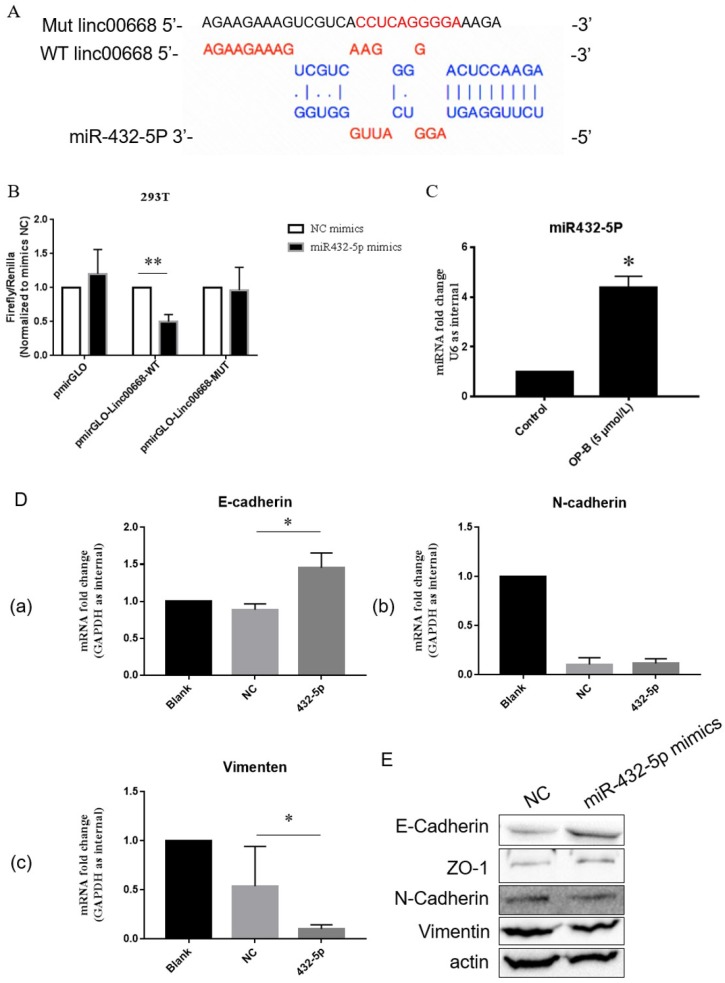
Further bioinformatics prediction showed that linc00668 could potentially bind to miR-432-5p (Figure 3A). To verify that, we constructed linc00668 3' UTR wild type or mutated (predicted miR-432-5p binding sites) luciferase plasmids in the pmirGLO dual luciferase reporter vector, and luciferase activity was evaluated after co-transfection of miRNA and luciferase plasmids.
Figure 3.
The oncogenic role of linc00668 is partly dependent on its regulation of miR-432-5p. A, Diagrammatic sketch of the binding sites for miR-432-5p in linc00668. B, Binding detection of miR-432-5p with linc00668 by luciferase reporter assay in 293T cells. C, QRT-PCR detection of the level of miR-432-5p in A549 cells treated by OP-B. D, After transfected with miR-432-5p mimics, the expression of E-Cadherin (a), N-Cadherin (b) and Vimentin (c) in A549 cells were detected by QRT-PCR. E, Detection of E-Cadherin, ZO-1, N-Cadherin and Vimentin in A549 cells transfected with miR-432-5p mimics. Error bars represented mean ± SD from three independent experiments. (*), P < 0.05; (**), P < 0.01.
As shown in Figure 3B, co-transfection with pmirGLO-linc00668-WT vector and miR-432-5p mimic reduced luciferase reporter activity significantly in 293 T compared with the control group (P<0.01). Results of qRT-PCR showed that OP-B treatment enhanced the level of miR-432-5p in A549 cells (P<0.05) (Figure 3C). Following results of qRT-PCR showed that overexpression of miR-432-5p in A549 cells resulted in the increasement of E-cadherin or reduction of Vimentin significantly (P<0.05) (Figure 3D). Meanwhile, the results of western blot also validated that miR-432-5p could inhibit the EMT signaling pathway (Figure 3E).
Discussion
More and more evidence showed that increasement of inflammatory cells and soluble inflammatory factors like chemokines and cytokines in primary cancer sites are strongly associated with poor prognosis due to metastasis 18, 19. As an important member of inflammation signal pathway, Stat3 plays a significant role in the occurrence, development, and invasion of tumors 17. Activated Stat3 can destroy to varying degree extracellular matrix and lead to the degradation and destruction of basement membranes, providing suitable environment for the early metastasis of tumor cells 17, 20. In addition, Stat3 can also promote EMT process and propel the transformation of chronic inflammation to cancer 20, 21.
Recent studies on Chinese lung cancer patients found that circulating inflammation markers were elevated in the lung cancer patients several years before the diagnosis. Shiels et al. found that inflammatory microenvironments formed earlier than the occurrence of EGFR mutations 22.
Natural compounds from traditional Chinese medicine have potential in inhibiting metastasis of cancer cells. Our previous study found that OP-B inhibited the migration and metastasis of A549 cells in vitro and in vivo 15, whether it inhibits cancer cell-metastasis in the inflammatory microenvironment is still unclear.
In the simulated inflammatory microenvironment, we found that OP-B treatment remarkably suppressed the invasion of A549 cells, and significantly reversed the changes in the expression of EMT markers in A549 cells, especially the expression of N-cadherin. In addition, OP-B treatment significantly decreased the levels of Stat3, the key master regulator that control cancer proliferation and invasion (Figure 1A). The results suggested that OP-B suppressed the invasion of human lung adenocarcinoma cells in the inflammatory microenvironment. However, the underlying mechanism warrants further elucidation.
Recently, a large number of lncRNAs have been reported to be implicated in regulating NSCLC proliferation, migration and invasion. Clinical data analysis showed that linc00668 was highly expressed in solid tumor, and correlated with the malignancy of tumor patients. While, whether linc00668 affects invasion and whether OP-B inhibits migration through acting on linc00668 warrants further investigation.
Analysis of the transcriptome date showed that linc00668 was significantly downregulated by OP-B in A549 cells, which was validated by qRT-PCR (Figure 2A and B). Furthermore, investigation found that the level of linc00668 increased significantly in THP-1 and A549 co-cultured system compared with that in A549 cells, while OP-B treatment suppressed the expression of linc00668 in the co-cultured system (Figure 2E). Meanwhile results of western blot showed that overexpression of linc00668 induced the reduction of E-cadherin, while promoted the level of N-cadherin (Figure 2C-D).
Bioinformatics prediction showed that linc00668 could potentially bind to miR-432-5p. That was validated by double luciferase reporter gene experiment (Figure 3A-B). Results of qRT-PCR showed that OP-B treatment increased the level of miR-432-5p in A549 cells (Figure 3C). Further experiments by qRT-PCR and western blot showed that overexpression of miR-432-5p resulted in inhibition of EMT signaling pathway (Figure 3D-E).
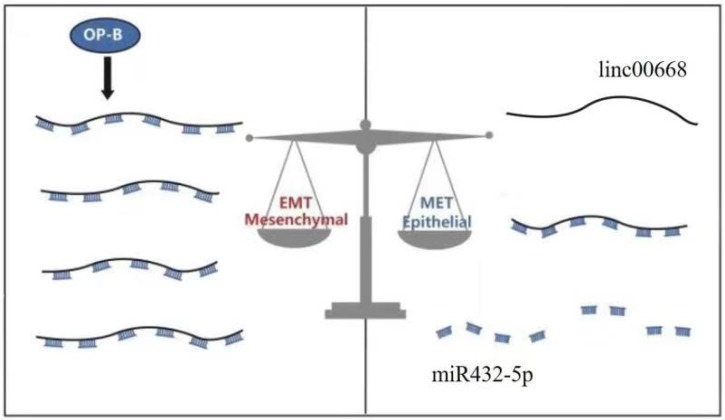
Taken together, Ophiopogonin-B inhibits metastasis in lung adenocarcinoma A549 cells via the Linc00668/miR-432-5p/EMT axis (Figure 4).
Figure 4.
A schematic model of the molecular mechanisms associated with OP-B- regulated metastasis in A549 cells. OP-B inhibited the process that linc00668 sponge miR-432-5p and at last acted on EMT to execute the anti-migration function of A549 cells under inflammatory microenvironment.
Supplementary Material
Supplementary table.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (No. 81503374), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Research Foundation of Education Bureau of Jiangsu Province (No. 16KJA360001). China and Europe taking care of healthcare solutions (No. PIRSES-GA-2013-612589). Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. KYCX18_1566).
Author Contributions
Dr Meijuan Chen and Xu Zhang designed the study. Cheng Hu and Rilei Jiang performed the experiment. Ziyu Cheng, Yueyang Lu, Ling Gu, Hongxiao Li, Liqiu Li and Qian Gao provided help to the experiment. Meijuan Chen and Cheng Hu wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T. et al. Non-small-cell lung cancer. Nature reviews Disease primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 3.Collins LG, Haines C, Perkel R, Enck RE. Lung cancer: diagnosis and management. American family physician. 2007;75:56–63. [PubMed] [Google Scholar]
- 4.Riihimaki M, Hemminki A, Fallah M, Thomsen H, Sundquist K, Sundquist J. et al. Metastatic sites and survival in lung cancer. Lung cancer. 2014;86:78–84. doi: 10.1016/j.lungcan.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Yang D, Cao X, Wang F, Jiang H, Feng D, Guo H. et al. LFG-500, a novel synthetic flavonoid, suppresses epithelial-mesenchymal transition in human lung adenocarcinoma cells by inhibiting NLRP3 in inflammatory microenvironment. Cancer letters. 2017;400:137–48. doi: 10.1016/j.canlet.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 7.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome research. 2015;25:1456–65. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature reviews Genetics. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 10.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nature reviews Molecular cell biology. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer research. 2017;77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q, Deng F, Qin Y, Zhao Z, Wu Z, Xing Z. et al. Long non-coding RNA regulation of epithelial-mesenchymal transition in cancer metastasis. Cell death & disease. 2016;7:e2254. doi: 10.1038/cddis.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang E, Yin D, Han L, He X, Si X, Chen W. et al. E2F1-induced upregulation of long noncoding RNA LINC00668 predicts a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically silencing of CKIs. Oncotarget. 2016;7:23212–26. doi: 10.18632/oncotarget.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Hu C, Guo Y, Jiang R, Jiang H, Zhou Y. et al. Ophiopogonin B suppresses the metastasis and angiogenesis of A549 cells in vitro and in vivo by inhibiting the EphA2/Akt signaling pathway. Oncology reports. 2018;40:1339–47. doi: 10.3892/or.2018.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naugler WE, Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends in molecular medicine. 2008;14:109–19. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature reviews Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grugan KD, McCabe FL, Kinder M, Greenplate AR, Harman BC, Ekert JE. et al. Tumor-associated macrophages promote invasion while retaining Fc-dependent anti-tumor function. Journal of immunology. 2012;189:5457–66. doi: 10.4049/jimmunol.1201889. [DOI] [PubMed] [Google Scholar]
- 19.Matsui A, Yokoo H, Negishi Y, Endo-Takahashi Y, Chun NA, Kadouchi I. et al. CXCL17 expression by tumor cells recruits CD11b+Gr1 high F4/80- cells and promotes tumor progression. PloS one. 2012;7:e44080. doi: 10.1371/journal.pone.0044080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nature reviews Cancer. 2014;14:736–46. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 21.Zhang CH, Guo FL, Xu GL, Jia WD, Ge YS. STAT3 activation mediates epithelial-to-mesenchymal transition in human hepatocellular carcinoma cells. Hepato-gastroenterology. 2014;61:1082–9. [PubMed] [Google Scholar]
- 22.Shiels MS, Shu XO, Chaturvedi AK, Gao YT, Xiang YB, Cai Q. et al. A prospective study of immune and inflammation markers and risk of lung cancer among female never smokers in Shanghai. Carcinogenesis. 2017;38:1004–10. doi: 10.1093/carcin/bgx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table.