Abstract
The rapid growth of the field of gene editing can largely be attributed to the discovery and optimization of designer endonucleases. These include zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regular interspersed short palindromic repeat (CRISPR) systems including Cas9, Cas12a, and structure-guided nucleases. Zebrafish (Danio rerio) have proven to be a powerful model system for genome engineering testing and applications due to their external development, high fecundity, and ease of housing. As the zebrafish gene editing toolkit continues to grow, it is becoming increasingly important to understand when and how to utilize which of these technologies for maximum efficacy in a particular project. While CRISPR–Cas9 has brought broad attention to the field of genome engineering in recent years, designer endonucleases have been utilized in genome engineering for more than two decades. This chapter provides a brief overview of designer endonuclease and other gene editing technologies in zebrafish as well as some of their known functional benefits and limitations depending on specific project goals. Finally, selected prospects for additional gene editing tools are presented, promising additional options for directed genomic programming of this versatile animal model system.
Keywords: Genome editing, zebrafish, designer nuclease, CRISPR, DNA repair, base editing
Introduction
One of the first applications of programmable designer endonucleases was published in 1996 in which a chimeric zinc finger was fused with the nuclease domain from FokI creating the zinc finger nuclease2. However, it was not until 2008 that the first targeted gene knockouts were reported in zebrafish using ZFNs3,4. Gene knockout and other mutant alleles are generated by the induction of double stranded breaks (DSBs) in the DNA double helix. These breaks are rapidly fixed by endogenous DNA repair pathways that can cause stochastic insertion or deletion (indel) mutations and can lead to disruption of the open reading frame and subsequent protein structure. In the short period since then, the further optimization of ZFNs, TALENs5, CRISPR systems1 including the Cas96,7 as well as Cas12a (formerly known as Cpf1)8,9 nucleases, and the structure guided nuclease10 have facilitated unforeseen opportunities in the zebrafish engineering arena. Though knockdown approaches such as morpholino technology11 provide robust reduction in gene expression, designer endonucleases can enable a gene knockout, which can lead to total ablation of gene products and the generation of mutant lines with relative ease. Even with these new technologies, the challenge to produce fully null mutants is still present. Total gene knockout is often confounded by genetic compensation12,13, induction of unintentional hypomorphism14, and other pathways that are not yet fully elucidated.
The DSBs induced by these technologies are often repaired by the high-efficiency, error-prone non-homologous end joining (NHEJ) pathway15,16,17. Less frequently, DNA breaks are repaired by the low efficiency, homology directed repair (HDR) group of pathways including homologous recombination that utilizes the homologous chromosome or exogenous DNA as a repair template15,18. In recent years, there has been further elucidation of another repair pathway, microhomology mediated end joining (MMEJ). MMEJ utilizes 5–25bp segments on the repair template that have homology around the break point and can assist in aligning the broken strands before joining19,20,21. The alignment conferred by the microhomologous regions can help MMEJ produce a desired and predictable outcome when used with a designer nuclease.
Technologies
Zinc Finger Nucleases – bringing designer nucleases to zebrafish
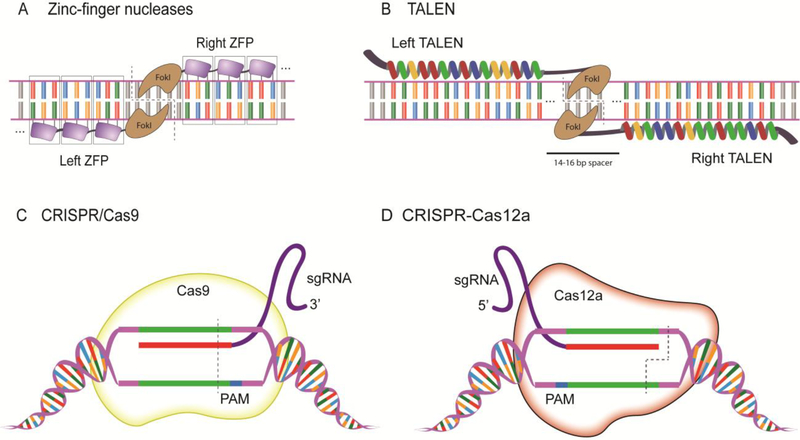
Cys2His2 zinc finger (ZF) sequence-specific DNA binding domains were first discovered in transcription factor IIIA from Xenopus laevis22 and were subsequently engineered for customized nuclease application. ZF nucleases are made by the fusion of the programmable DNA binding domains with the fusion of the FokI endonuclease domain23. The native FokI type IIS restriction enzyme has separable DNA binding and nuclease domains and requires the formation of a dimer to be catalytically active24. ZFNs are currently engineered with three to six zinc finger domains comprised of ~30 amino acids each per domain with two beta strands and one alpha helix, with the alpha helix conferring recognition of 3 nucleotides25. For example, a single ZFN module containing 3 Cys2His2 domains is capable of recognizing 9nt of DNA sequence; a pair of three finger ZFNs are thus capable of recognizing 18nt, which can be sufficient for targeting a unique genomic locus in zebrafish26. FokI functions as a homodimer and requires the specific orientation and spacing of the ZFNs to induce a double stranded break (DSB) in the DNA at a desired gene region (Fig 1a). FokI can be further engineered to function as an obligate heterodimer to increase specificity of cleavage27.
Fig 1. Established guided designer endonucleases used in zebrafish.
(A) Zinc-finger nucleases recognize DNA by the fusion of 3 zinc finger recognition domains on either side of the DNA, specifically binding an 18nt region in this example, and the fused FokI dimerizes and catalyzes cleavage of the DNA. (B) An illustrative 15 repeat TALEN binds on either strand of the DNA separated by a spacer region, and the DSB occurs around the halfway point in the spacer region where the FokI domains dimerize.(C) SpCas9 recognizes the target sequence with the assistance of the sgRNA next to the 3’ PAM sequence and induces a double stranded break 3 base pairs 5’ from the PAM. (D) Cas12a recognizes the target sequence with the assistance of the sgRNA next to the 5’ TTTN PAM sequence and cleaves the DNA at the 18th base on the non-targeted strand and after the 23rd base on the targeted strand to create sticky ends at the DSB. (see text)
Microinjection of ZFN protein into zebrafish embryos has been shown to be well tolerated with as high as 5ng of ZFN mRNA being injected with minimal toxicity and appreciable efficacy28,29. However, the complex nature of ZFN synthesis and design such as unpredicted aberrant interactions between zinc finger subunits in addition to the high cost of construction has largely rendered ZFNs a less accessible technology to zebrafish researchers. However, their pioneering work established gene editing in vertebrate embryos as a viable gene knockout approach that was subsequently deployed in other models such as mice30 and rats31.
TALENs
Transcription activator-like effectors (TALEs) are derived from plant pathogen Xanthomonas bacteria, a plant pathogen that hijacks host gene expression to favor its own survival32,33,34. The nuclease activity of transcription activator like effector nucleases (TALENs), like ZFNs, is conferred by the fusion of the FokI endonuclease to the TALE programmable DNA binding cassette32. The fused FokI functions as a homodimer that binds both sides of the target DNA and cleaves upon dimerization with the TALEN pairs35. TALE repeats are comprised of 33–35 amino acid repeats that are identical except for residues 12 and 13. These resides are referred to as the repeat variable diresidue (RVD) domain and are responsible for conferring one-to-one recognition of nucleotides32. TALENs are normally used as dimers with 15–20 TALE repeats binding on one strand separated by a spacer region of 15–20 nucleotides followed by another 15–20 TALE repeats binding the opposite strand; consequently, the DSB occurs at around the halfway point of the spacer where the fused FokI domains have the greatest probability of dimerization and subsequent DNA cleavage (Fig 1b).
Custom TALEN expression vectors are constructed using diverse approaches including Golden Gate36, serial ligation37, and ligation independent38 methods. Conventional TALEN scaffolds function optimally at a locus with a thymidine at the 5’ end of the TALE binding region39,32,40,36. However, later scaffolds have since circumvented this preference without a substantive decrease in efficiency41,42. Due to their high efficacy when delivered in vivo by microinjection and new rapid synthesis protocols such as that developed by Ma et al.43, TALENs remain a useful tool for genome engineering purposes, showing particular efficacy in zebrafish gene knock-in approaches 44 (see also below).
Though TALENs provide many solutions to the problems posed by ZFNs such as unpredicted interaction between the nuclease subunits, the detailed cloning methods involved in TALEN construction has rendered them useful in specific applications such as HR and HDR induction. The specialized employment of TALENs can be largely attributed to the highly accessible nature of CRISPR-Cas9 for NHEJ-based applications.
TALENs have had and continue to have tremendous impacts of the field of genome engineering. TALENs were one of the first technologies to achieve consistent targeted homologous recombination and paved the way for many later targeted integration technologies45. In addition to their pioneering work in integrating exogenous DNA, TALENs were one of the first technologies to remove large spans of DNA approaching nearly 20kb46. TALENs remain a useful tool and in fact have been recently shown to have powerful therapeutic application in chimeric antigen receptor T cell (CAR T) based modalities to treat various cancers47. In addition to therapeutic applications, TALENs continue to be streamlined technologies to investigate specific cell type function48 as well as the expression of poorly understood regulatory pathways49
CRISPR-Cas9
Clustered regular interspersed short palindromic repeats (CRISPR) and CRISPR associated protein-9(Cas9)1 have greatly expanded the genome engineering field. CRISPR systems have been elucidated to protect bacteria from exogenous DNA50. CRISPR RNA (crRNA) and a trans activating CRISPR RNA (tracrRNA) are transcribed from a CRISPR locus and are processed and incorporated into the mature Cas9 complex51,52. The CRISPR locus likewise encodes DNA of previously encountered pathogens in its spacer regions. The invading pathogenic DNA can later be transcribed and used as a guide and direct the binding of the mature Cas9 complex to the intruding virus. Once the complex is guided to the exogenous DNA, the DNA is cleaved and leads to subsequent degradation of the intruding DNA (Fig1c)53,54,55. The CRISPR–Cas9 system utilizes a protospacer adjacent motif (PAM), which is present in the exogenous DNA but absent in the encoded spacer region, to differentiate between the endogenous CRISPR spacer and the pathogenic DNA. In the commonly used CRISPR–Cas9 system derived from the human pathogen Streptococcus pyogenes (SpCas9), a native NGG 3’ PAM sequence is required for efficient cleavage activity56. CRISPR–Cas9 has further been optimized for gene editing by adding a linker region between the tracrRNA and crRNA to form a single hybrid single guide RNA (sgRNA) to simplify molecular construction57.
The SpCas9 system has since been outfitted for zebrafish by codon optimizing the Cas9 protein for zebrafish expression and adding an SV40 large T antigen nuclear localization signal to both the N and C termini58. This codon optimized Cas9 was subsequently cloned into vectors that facilitate the synthesis of capped and polyadenylated Cas9 mRNA via in vitro transcription from either SP6 or T3 promoters58. This method was further optimized by allowing the synthesis of gRNA generated by using two partially overlapping oligonucleotides in a clone-free manner59. One oligonucleotide is specific to the genomic loci of interest whereas the other is a generic oligonucleotide that can be used for all constructs and contains the necessary secondary structure. These oligonucleotides form a double stranded template by annealing at the designed 20nt overlap and are extended via polymerase chain reaction (PCR). The duplex DNA generated from this polymerization reaction can then be used for gRNA synthesis via in vitro transcription reaction60 in a high throughput manner enabling one to synthesize hundreds of gRNA in just a few hours.
Injection of pre-formed gRNA/Cas9 ribonucleoprotein (RNP) complexes has likewise been optimized for zebrafish injections as early as the single cell stage61,62. Other Cas9 systems that have been shown to work well in zebrafish include forms fused to an NLS, and as fusions with several different fluorescent protein domains62,63,64. Other Cas9 mRNA constructs exist such as only C-terminal NLS or only N-terminal NLS tagged Cas9 as well as T7 promoter driven Cas9 for fine-tuned expression and mRNA. Many are now commercially available, reducing the requirement for gene editing to a single custom reagent. This simplification enables both the rapid deployment of this tool for many research studies and also opens the door to active learning classrooms such as an undergraduate development lab (Essner et al., personal communication).
Though efficient editing has been shown in zebrafish as well as other model systems, the CRISPR–Cas9 system is not without its drawbacks. Native SpCas9 has a requirement for an NGG PAM sequence56, a targeting limitation especially when targeting AT rich genomic regions such as introns. This obstacle is reduced as continued engineering of SpCas9 has led to variants with an NGA PAM65,66 and is still in progress to utilize additional PAM targets. Whether these variants are as effective in zebrafish as they are in human DNA editing is yet to be described.
Off-targeting in zebrafish is an important consideration that can be addressed in several ways. First, outcrossing a founder animal results in the natural removal of 98% of DNA harboring any off-target edited (49 out of 50) non-linked mutant chromosomes67. Second, generating more than one allele can reduce the likelihood of a linked, second mutation found on the remaining selected chromosome.
Cas12a
Subsequent to the discovery and utilization of CRISPR–Cas9 as a genome editing tool, a bioinformatic expedition began to identify other bacterial immune systems with desirable qualities such as different PAM requirements and high efficiency nucleases with potential lower off target activity. This work led to the discovery of the CRISPR from Prevotella and Francisella 1 initially described as Cpf1 and now formally called Cas12a68,8,69,70. Cas12a contains a RuvC-like endonuclease domain that is similar to the Cas9 endonuclease domain, suggesting it has a similar role in adaptive immunity as a targeted endonuclease71. Additionally, Cas12a has fewer secondary structure requirements for gRNA, does not require a tracrRNA and utilizes only a sgRNA that can be self-processed8. Cas12a has a 5’ TTTN PAM sequence preference72,8, affording targeting into AT rich gene regions such as zebrafish introns that may be inaccessible with SpCas9. Additionally, Cas12a creates a staggered cut distal from the PAM by cleaving the DNA after the 18th base on the non-targeted strand and after the 23rd base on the targeted strand (Fig 1d). Unlike Cas9, this cleavage does not destroy the original target site and might facilitate greater efficiency of donation of exogenous DNA via single strand annealing53 or homologous recombination approaches. All of these features could lead to simplified delivery in the zebrafish model.
However, initially described Cas12a systems do not work off-the-shelf in zebrafish as either RNP or mRNA73. Further optimization has led to functional systems for zebrafish74, and Cas12a provides a unique opportunity for genome engineering in this model system due to its more compact size, AT rich PAM, and staggered DNA cutting. Cas12a can catalyze higher HDR-based repair in zebrafish relative to SpCas9, possibly due to its repeated cutting that could help drive HDR over nonspecific end joining74.
Further, because Cas12a processes its own sgRNA, this allows for the possibility of Cas12a’s sgRNAto be packaged into a smaller cassette being driven by an RNA Polymerase II promoter. Cas9 sgRNAs are large and require extensive processing, necessitating an RNA Polymerase III promoter such as U6 to drive their expression75,76,77. This added benefit provides a simplified opportunity for tissue-specific expression of Cas12a gRNA in zebrafish, which is challenging with current Cas9 gRNA systems. However, developing technologies are beginning to enable the use of RNA Polymerase II promoters for Cas9 sgRNA with the utilization of ribozymes and other molecular catalysts to process the sgRNA78.
xCas9 – next generation of CRISPR targeting in zebrafish?
To circumvent the issue of the PAM requirement narrowing the target range of CRISPR–Cas9, researchers set out to develop a Cas9 with a broad PAM utilization. Though Staphylococcus aureus Cas9 (SaCas9) has been engineered previously to have an NNNRRT (R=A or G) PAM to increase its targeting range, it suffered a concomitant increase in off-target editing79. Using a variation of phage-assisted continuous evolution (PACE)80, researchers evolved SpCas9 in vitro and selected for variants that bind a greater variety of PAM sequences. This led to the discovery of Cas9 variants, termed xCas9, that were capable of binding NG, NNG, GAA, GAT, and CAA PAMs81. These variants were assessed for genome engineering applications such as transcriptional activation by fusing xCas9 to the VP64 transcriptional activator82, DNA cutting, and cytidine and adenine base editing by fusing xCas9 to third-generation (BE3) base editing architecture containing a nucleobase deaminase83.
In all three parameters tested xCas9 was shown to have a significantly increased activity in each respective application relative to SpCas9. xCas9 provides a tool that has a broad range of PAM targets, greater specificity relative to SpCas9, and less off targets. This suggests that there is not an obligate trade-off between efficiency, PAM requirement, and specificity84. This xCas9 system currently has the largest potential genomic targeting range among known SpCas9 variants Though preliminary data for xCas9 appears promising, overall efficiency has not yet been established. Once cleavage kinetics and gene editing capabilities of xCas9 have been further assessed, this suggests the possibility of highly active and specific Cas9 variants that may be powerful tools in the zebrafish genome engineer’s toolbox in the near future.
Structure guided nuclease – a DNA-guided gene editor
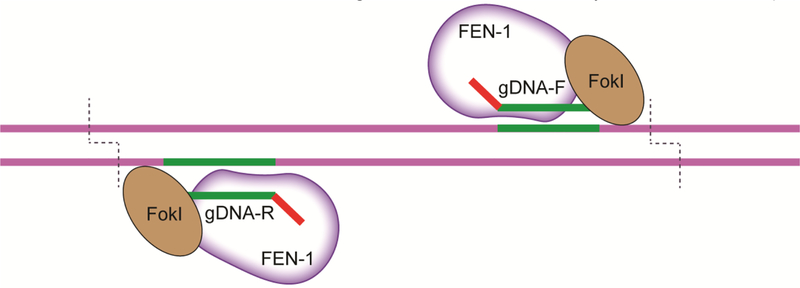
A DNA-guided endonuclease has been recently constructed known as the structure-guided nuclease (SGN)10. This gene editor is a fusion protein of flap-endonuclease-1 (FEN-1) and the cleavage domain of FokI. FEN–1 functions endogenously by recognizing single unpaired 3’ nucleotide overhang “flaps” that occur during DNA replication and repair and removes the lesion for the DNA polymerase to fill in85. This structure-guided nuclease functions in concert with a pair of guide DNA (gDNA) molecules that have a single unpaired 3’ nucleotide and produce a 3’ overhang flap flanking the target site for the FEN-1 domain to recognize and cleave the target (Fig 2). Microinjection of SGN mRNA and gDNA results in the editing of endogenous zebrafish DNA, albeit with modest efficiency in this first generation tool86,10. Interestingly, the structure- guided nuclease preferentially creates large deletions instead of small indels that are observed with other engineered endonucleases10. This unique lack of PAM requirement and creation of large deletions suggests the possibility of exploiting the SGN’s activity by insertion of large repair templates or producing efficient knockouts in zebrafish by removal of large regions of DNA in virtually any genomic locus. The founders of this technology report low overall editing efficacy and this technology has not yet been widely adopted at this time. With further validation and optimization, the SGN could be a powerful tool for the genome engineer in the near future.
Fig 2. The structure-guided nuclease.
Using a ssDNA guide of at least 20nt, the structure guided nuclease (SGN) dimer cleaves 9–10nt away from the 3’ end of the gDNA on both strands of DNA and creates a large deletion of the intervening genomic locus10,86.
Argonautes – the ups and downs of a genome tool
Argonaute proteins are enzymes involved in the processing and maturation of small RNA molecules involved in the RNA interference (RNAi) system that is implicated in eukaryotic gene regulation87. Promising Argonaute proteins are derived from Rhodobacter sphaeroides and Thermus thermophiles and are implicated in bacteria host defense88. These Argonautes have been shown to utilize DNA guides to induce RNA and DNA target cleavage89. Additionally, the Argonaute from the archaeon Pyrococcus furiosus (PfAgo) has been shown to use small 5’ phosphorylated DNA guides to cleave both single and double stranded DNA at supraphysiological temperatures90.
The search for DNA-guided Argonaute proteins that work at more physiological temperatures is ongoing. Though all claims for DNA editing activity has been retracted91,92,93, the false-start from work with the Natronobacterium gregoryi Argonaute (NgAgo) suggests that NgAgo can be a potentially useful tool in zebrafish gene knockdown94. NgAgo and other Argonaute proteins therefore provide potential tools in the zebrafish engineering toolbox in addition to other gene knockdown technologies such as morpholinos, but lack the possibility of producing total gene knockouts at this time. As a result of the controversial past of NgAgo, researchers should proceed cautiously when investigating claims about potential editing capabilities and applications of this protein.
Oligo Mediated Repair
The previously mentioned gene editing tools can serve to produce gene knock outs or to knock in exogenous genomic sequences into the zebrafish genome. In addition to inserting novel genomic material into the zebrafish genome for analysis, many disease allele variants do not result in completely eliminated gene function, removing the possibility of using a simple knockout as the experimental paradigm95. Therefore it is important to be able to precisely program genetic information to express a foreign gene product or recapitulate a disease state. Designer nucleases introduce DSBs and subsequently activate repair and recombination pathways at the targeted zebrafish genomic locus3,96. Such repair pathways can be exploited using different approaches to produce a desired integration or mutant allele.
One such repair pathway is oligo mediated repair. Oligo mediated repair utilizes single stranded DNA (ssDNA) molecules with homology to the zebrafish genomic locus as a donor sequence to be used as a repair template following the induced DSB97,98. This approach replaces the host locus DNA with a delivered novel insert sequence. Oligo-mediated repair templates are easy to design and provide an efficient way to modify the host zebrafish locus. Using ssDNA that spans the region of a nuclease cut site, typically 1–10% of precise insertions can be detected via an oligo-mediated homology directed repair approach using TALENs and these integrations resulted in germline transmission97,99,100. Further, slightly larger segments such as loxP sites were also successfully inserted via ssDNA coinjection with TALENs to produce a potential conditional knockout allele that was also stably passed on to the germline100. However, donor templates are typically short (<100nt) focusing this approach typically on relatively smaller modifications.
One of the most promising newer technologies in oligo-mediated repair involves the utilization of asymmetric donor DNA in combination with SpCas9 to induce a DSB101. Dissociation of Cas9 from the substrate dsDNA locus is comparatively slow, resulting in Cas9 asymmetrically dissociating the 3’ end of the cleaved DNA on the nontarget strand. By designing ssDNA similar in length to the cleaved DNA strand that is released first in the dissociation of the Cas9 complex, the rate of oligo-mediated repair in human cells when using SpCas9 can be increased by ~10x. With the further elucidation of Cas9 kinetics and ssDNA integration, this technology could be adapted to the zebrafish engineering sphere and produce high efficiency oligo-mediated repair by taking advantage of the asymmetric release of the dsDNA substrate.
Homologous Recombination
To achieve precise genome editing of larger DNA segments in zebrafish, investigators have deployed homologous recombination (HR). HR uses a double-stranded (dsDNA) donor with ~0.5–2kbp regions of homology on either side of the donor template to achieve precise replacement of up to several kb stretches of host sequence102–104. HR enables precise in-frame insertions of an entire protein coding sequence such as a reporter gene to track inheritance. Likewise, loxP sites can be introduced within the region of homology, but flanking the gene of interest, to produce conditional knockout zebrafish lines102. Using a designer nuclease with a dsDNA donor template has yielded up to 15% of injected zygotes with precisely edited genomes transmitted to progeny102,103,45.
The general workflow to drive homologous recombination involves the injection of a designer nuclease and donor dsDNA flanked by ~1kb homology arms on either side of the inserted gene of interest. The 5’ overhangs produced by TALENs as well as the blunt cuts induced by CRISPR–Cas9 both work in principle as substrates for homologous recombination. Donor dsDNA sequences are often designed such that integration destroys the nuclease recognition site and the integrated DNA is effectively protected from subsequent DNA cleavage once properly inserted. How injected DNA is recruited to the cleavage site in the zebrafish genome in not well understood. However, injection of circular donor DNA was shown to preferentially be used as a repair template relative to linear DNA to produce edited alleles using TALENs in zebrafish102,103. Utilizing a well-constructed designer nuclease in combination with a dsDNA donor plasmid are powerful tools to produce knock-in zebrafish that can be used to create stable transgenic lines for further investigation.
Though the above mentioned tools provide the opportunity for precise genetic knock-ins into the zebrafish genome there are several caveats to consider. The gene editing approaches in many published manuscripts seem to have a locus-specific effect on efficiency with variation between loci and different research groups. However, elucidation of other DNA repair pathways including MMEJ, single-stranded annealing (SSA), and alternative end joining (alt–EJ) pathways is beginning to indicate that there may be temporal and spatial preference for particular pathways. In fact, it has recently been shown that alt-EJ is favored in early development and this preference may be exploited to generate a desired mutant allele105. Further understanding and exploitation of these and other pathways may begin to yield precise knock-ins with more consistent results between trials and investigators.
Base Editing
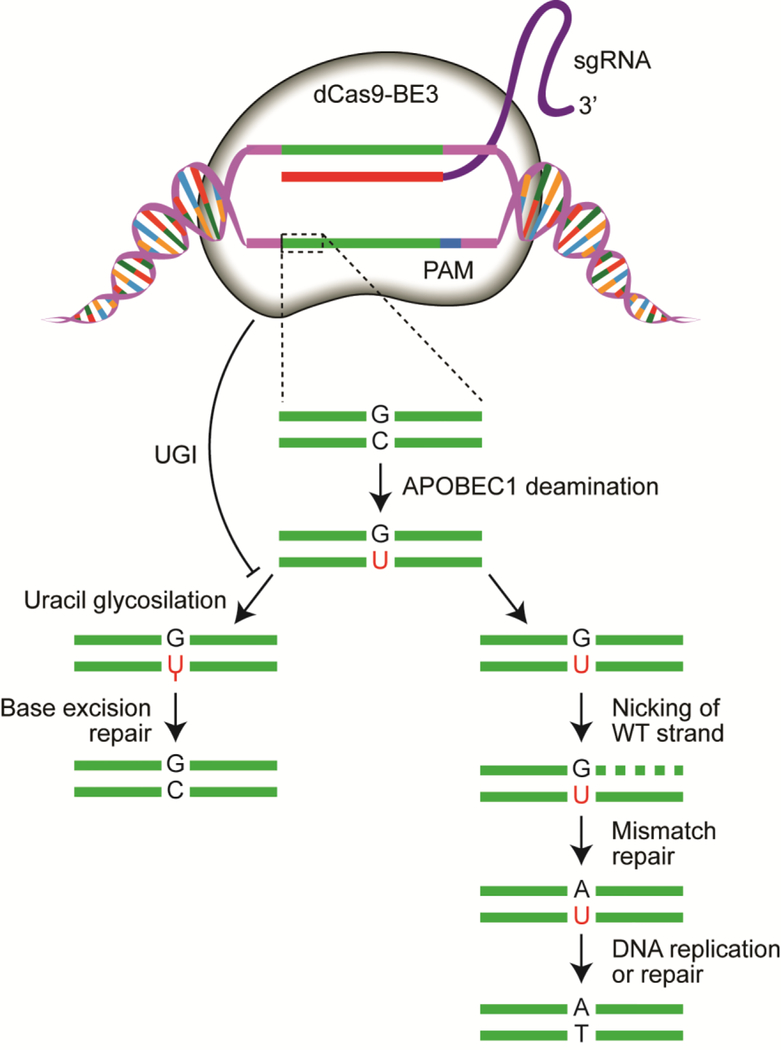
Many genome editing technologies rely on exploiting various endogenous DNA repair pathways to facilitate the integration of exogenous DNA or produce a gene knockout by a frameshift mutation. An alternative editing system would function such to directly edit genomic DNA without relying on exogenous donor DNA. From this notion arose the base editor system (BE)83. The first generation base editor system (BE1) employed a catalytically inert Cas9 (dCas9) fused to a cytidine deaminase (APOBEC1) to induce C:G base pairs to be changed to T:A base pairs, as well as an XTEN linker region to facilitate structural mobility of the BE1 complex. To subvert possible mismatch repair (MMR) at the site of base editing and therefore restore the original C:G base pair, a uracil glycosylase inhibitor (UGI) was fused to the C terminus of BE1 creating the second generation base editor (BE2), which showed a 3-fold increase in vitro editing relative to BE1. Finally, Komor et al. hypothesized that nicking the DNA strand with the unedited G would stimulate newly synthesized DNA and induce MMR to preferentially resolve the U:G base pair into the desired U:A and subsequent T:A products. To facilitate this approach, the catalytic His residue at position 840 in the HNH domain of the dCas9 BE251 was restored to induce DNA nicking activity. This therefore produced the third generation base editor system (BE3) consisting of APOBEC-XTEN-dCas9(A840H)-UGI, which garners an efficient deamination window of ~5nt with preference to act from positions 4 to 8 within the protospacer, counting the end distal to the PAM as position 1 (Fig 3).
Fig 3. The base editor system for non-nuclease editing of the zebrafish genome.
The third generation base editor (BE3) catalyzes G:C to an A:T deamination with the fused APOBEC1 nucleobase deaminase. Base excision repair is prevented by the fused uracil glycosylase inhibitor (UGI). The formation of the AT transition is further biased by introducing a nick on the non-edited strand to drive mismatch repair and subsequent resolution to the A:T basepair106,109.
A BE3:sgRNA complex has been preliminarily tested in zebrafish with promising results106,107. BE3:sgRNA targeting the tyrosinase loci in the zebrafish genome was injected and assessed for base editing 4 days post injection via high throughput sequencing (HTS). BE3 showed minimum indel induction and modest mutations in vivo with as high as 7.7% efficiency of edited zebrafish alleles transmitted to the next generation. The base editing system therefore postulates a possible “DNA-free” editing system that does not rely on a donor DNA template, but can instead directly edit DNA in vivo.
Conclusion
Zebrafish are proving to be a powerful genetic and disease model due to their advantages such as conservation in biological function, high fecundity, rapid embryonic development, and external fertilization that facilitate embryo injection and analysis. The qualitative and quantitative data that can be obtained from the genotype and phenotype of zebrafish allow nearly unmatched high throughput screening relative to other vertebrate genetic models. These benefits also make zebrafish an outstanding system to investigate novel gene editing technologies.
Few current limits are found in the current genome engineering toolbox, but opportunities still remain. The development of a highly active DNA-guided gene editor could be optimized to leverage massively parallel DNA synthesis of oligonucleotides would be well-suited to the high throughput nature of the zebrafish model. Furthermore, perhaps the genome engineer can consider a next generation genome editing tool that involves the direct editing of genomic DNA without having to employ exogenous DNA at all such as an optimized base editor system.
The zebrafish genome is net AT rich with open reading frames (ORFs) being largely GC rich108. Therefore, optimization of tools such as Cas12a that can take advantage of its unique PAM requirement or the structure guided nuclease that lacks a sequence requirement entirely will potentially facilitate improved targeting into introns and allow for insertion into these regions. Though CRISPR–Cas9 has brought the field of genome engineering into the mainstream view in recent years, novel genome editing technologies are improving at an unforeseen rate. With the assistance of the powerful zebrafish model, the genome engineer’s toolbox continues to grow with increasingly specific and diverse activities for most any application the investigator may choose.
Highlights.
The zebrafish is a powerful system for developing new genome engineering tools
Designer nucleases enable precise editing of genomic DNA
Gene editing tools from diverse origins from bacteria to artificially constructed tools provide unique advantages and limitations
Exploiting endogenous DNA repair mechanisms can produce favored outcomes depending on gene editing tool application
DNA can now be directly edited without the use of an exogenous repair template
Acknowledgements
This work was supported by U.S. National Institutes of Health (NIH) OD020166 and GM63904 and the Mayo Foundation. B.W.S is supported in part by NIH R25 GM 55252, “Initiative for Maximizing Student Development”. B.W.S. and G.M.G. are supported by Mayo Clinic Graduate School of Biomedical Sciences. We would like to thank H.A. and W.A.G. for insightful discussions that informed this chapter.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Richardson CD, K. K, Feng SJ, Bray NL, Schaefer AJ, Floor S, Corn J CRISPR-Cas9 genome editing in human cells works via the Fanconi anemia pathway. bioRxiv. 2017;136028 CRISPR-Cas9 genome editing in human cells works via the Fanconi anemia pathway. bioRxiv (2017). [DOI] [PubMed] [Google Scholar]
- 2.Kim YG, Cha J & Chandrasegaran S Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences 93, 1156–1160, doi: 10.1073/pnas.93.3.1156 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyon Y et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature Biotechnology 26, 702–708, doi: 10.1038/nbt1409 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urnov FD, Rebar EJ, Holmes MC, Zhang HS & Gregory PD Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics 11, 636–646, doi: 10.1038/nrg2842 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Christian M et al. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 186, 757–U476, doi: 10.1534/genetics.110.120717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823, doi: 10.1126/science.1231143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS et al. Evolution and classification of the CRISPR-Cas systems. Nature Reviews Microbiology 9, 467–477, doi: 10.1038/nrmicro2577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771, doi: 10.1016/j.cell.2015.09.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shmakov S et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nature reviews. Microbiology 15, 169–182, doi: 10.1038/nrmicro.2016.184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu S et al. An alternative novel tool for DNA editing without target sequence limitation: the structure-guided nuclease. Genome biology 17, 186, doi: 10.1186/s13059-016-1038-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasevicius A & Ekker SC Effective targeted gene ‘knockdown’ in zebrafish. Nature genetics 26, 216–220, doi: 10.1038/79951 (2000). [DOI] [PubMed] [Google Scholar]
- 12.El-Brolosy MA & Stainier DYR Genetic compensation: A phenomenon in search of mechanisms. PLoS Genetics 13, doi: 10.1371/journal.pgen.1006780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi A et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230, doi:10.1038/nature1458010.1038/nature14580https://www.nature.com/articles/nature14580#supplementary-informationhttps://www.nature.com/articles/nature14580#supplementary-information (2015). [DOI] [PubMed] [Google Scholar]
- 14.Jokela H et al. Deleting the mouse Hsd17b1 gene results in a hypomorphic Naglu allele and a phenotype mimicking a lysosomal storage disease. Scientific Reports 7, 16406, doi: 10.1038/s41598-017-16618-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyman C & Kanaar R in Annual Review of Genetics Vol. 40 Annual Review of Genetics 363–383 (2006). [DOI] [PubMed]
- 16.Burma S, Chen BP & Chen DJ Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA repair 5, 1042–1048, doi: 10.1016/j.dnarep.2006.05.026 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Lieber MR in Annual Review of Biochemistry, Vol 79 Vol. 79 Annual Review of Biochemistry (eds Kornberg RD, Raetz CRH, Rothman JE, & Thorner JW) 181–211 (2010). [Google Scholar]
- 18.Cannan WJ & Pederson DS Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. Journal of Cellular Physiology 231, 3–14, doi: 10.1002/jcp.25048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennardo N, Cheng A, Huang N & Stark JM Alternative-NHEJ Is a Mechanistically Distinct Pathway of Mammalian Chromosome Break Repair. Plos Genetics 4, doi: 10.1371/journal.pgen.1000110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ata H et al. Toward Precision Molecular Surgery: Robust, Selective Induction of Microhomology-mediated End Joining in vivo. bioRxiv, doi: 10.1101/291187 (2018). [DOI] [Google Scholar]
- 21.McVey M & Lee SE MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends in genetics : TIG 24, 529–538, doi: 10.1016/j.tig.2008.08.007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J, McLachlan AD & Klug A Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. The EMBO journal 4, 1609–1614 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bibikova M et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Molecular and Cellular Biology 21, 289–297, doi: 10.1128/mcb.21.1.289-297.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bitinaite J, Wah DA, Aggarwal AK & Schildkraut I FokI dimerization is required for DNA cleavage. Proceedings of the National Academy of Sciences of the United States of America 95, 10570–10575, doi: 10.1073/pnas.95.18.10570 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavletich NP & Pabo CO ZINC FINGER DNA RECOGNITION - CRYSTAL-STRUCTURE OF A ZIF268-DNA COMPLEX AT 2.1-A. Science (New York, N.Y.) 252, 809–817, doi: 10.1126/science.2028256 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Doyon Y et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26, 702–708, doi: 10.1038/nbt1409 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo J, Gaj T & Barbas CF 3rd. Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. Journal of molecular biology 400, 96–107, doi: 10.1016/j.jmb.2010.04.060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyon Y et al. Heritable Targeted Gene Disruption in Zebrafish Using Designed Zinc Finger Nucleases. Nature biotechnology 26, 702–708, doi: 10.1038/nbt1409 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng X, Noyes MB, Zhu LJ, Lawson ND & Wolfe SA Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nature Biotechnology 26, 695–701, doi: 10.1038/nbt1398 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carbery ID et al. Targeted Genome Modification in Mice Using Zinc-Finger Nucleases. Genetics 186, 451–459, doi: 10.1534/genetics.110.117002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geurts AM et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science (New York, N.Y.) 325, 433, doi: 10.1126/science.1172447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogdanove AJ & Voytas DF TAL effectors: customizable proteins for DNA targeting. Science (New York, N.Y.) 333, 1843–1846, doi: 10.1126/science.1204094 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Moscou MJ & Bogdanove AJ A simple cipher governs DNA recognition by TAL effectors. Science (New York, N.Y.) 326, 1501, doi: 10.1126/science.1178817 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Boch J & Bonas U Xanthomonas AvrBs3 Family-Type III Effectors: Discovery and Function. Annual Review of Phytopathology 48, 419–436, doi: 10.1146/annurev-phyto-080508-081936 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Cade L et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Research 40, 8001–8010, doi: 10.1093/nar/gks518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cermak T et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research 39, e82, doi: 10.1093/nar/gkr218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reyon D et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30, 460–465, doi: 10.1038/nbt.2170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K & Hornung V A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol 31, 76–81, doi: 10.1038/nbt.2460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ & Stoddard BL The crystal structure of TAL effector PthXo1 bound to its DNA target. Science (New York, N.Y.) 335, 716–719, doi: 10.1126/science.1216211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boch J et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science (New York, N.Y.) 326, 1509–1512, doi: 10.1126/science.1178811 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Campbell JM, Hartjes KA, Nelson TJ, Xu X & Ekker SC The New and TALENted Genome Engineering Toolbox. Circulation research 113, 571–587, doi: 10.1161/circresaha.113.301765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamb BM, Mercer AC & Barbas CF Directed evolution of the TALE N-terminal domain for recognition of all 5′ bases. Nucleic Acids Research 41, 9779–9785, doi: 10.1093/nar/gkt754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma AC et al. FusX: A Rapid One-Step Transcription Activator-Like Effector Assembly System for Genome Science. Human Gene Therapy 27, 451–463, doi: 10.1089/hum.2015.172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng Y et al. Making designer mutants in model organisms. Development (Cambridge, England) 141, 4042–4054, doi: 10.1242/dev.102186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin J, Chen J & Solnica-Krezel L Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development (Cambridge, England) 141, 3807– 3818, doi: 10.1242/dev.108019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma AC High Efficiency In Vivo Genome Engineering with a Simplified 15-RVD GoldyTALEN Design. 8, doi: 10.1371/journal.pone.0065259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qasim W et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene- edited CAR T cells. Science translational medicine 9, doi: 10.1126/scitranslmed.aaj2013 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Moreno RL, Williams K, Jones KL & Ribera AB Investigation of Islet2a function in zebrafish embryos: Mutants and morphants differ in morphologic phenotypes and gene expression. PloS one 13, e0199233, doi: 10.1371/journal.pone.0199233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz LE et al. Epigenetic regulators Rbbp4 and Hdac1 are overexpressed in a zebrafish model of RB1 embryonal brain tumor, and are required for neural progenitor survival and proliferation. Disease models & mechanisms 11, doi: 10.1242/dmm.034124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, N.Y.) 315, 1709–1712, doi: 10.1126/science.1138140 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 337, 816–821, doi: 10.1126/science.1225829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishimasu H et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949, doi: 10.1016/j.cell.2014.02.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, N.Y.) 315, 1709–1712, doi: 10.1126/science.1138140 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Brouns SJJ et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science (New York, N.Y.) 321, 960–964, doi: 10.1126/science.1159689 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hale CR et al. RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex. Cell 139, 945–956, doi: 10.1016/j.cell.2009.07.040 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cong L et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (New York, N.Y.) 339, 819–823, doi: 10.1126/science.1231143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Briner AE et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Molecular cell 56, 333–339, doi: 10.1016/j.molcel.2014.09.019 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Jao LE, Wente SR & Chen W Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America 110, 13904–13909, doi: 10.1073/pnas.1308335110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bassett AR, Tibbit C, Ponting CP & Liu JL Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell reports 4, 220–228, doi: 10.1016/j.celrep.2013.06.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varshney GK et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome research 25, 1030–1042, doi: 10.1101/gr.186379.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xin Y & Duan C Microinjection of Antisense Morpholinos, CRISPR/Cas9 RNP, and RNA/DNA into Zebrafish Embryos. Methods in molecular biology (Clifton, N.J.) 1742, 205–211, doi: 10.1007/978-1-4939-7665-2_18 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Burger A et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development (Cambridge, England) 143, 2025–2037, doi: 10.1242/dev.134809 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Hu P, Zhao X, Zhang Q, Li W & Zu Y Comparison of Various Nuclear Localization Signal-Fused Cas9 Proteins and Cas9 mRNA for Genome Editing in Zebrafish. G3: Genes|Genomes|Genetics 8, 823–831, doi: 10.1534/g3.117.300359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang N et al. Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Cell research 23, 465, doi:10.1038/cr.2013.4510.1038/cr.2013.45https://www.nature.com/articles/cr201345#supplementary-informationhttps://www.nature.com/articles/cr201345#supplementary-information (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirano S, Nishimasu H, Ishitani R & Nureki O Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9. Molecular cell 61, 886–894, doi: 10.1016/j.molcel.2016.02.018 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Kleinstiver BP et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485, doi: 10.1038/nature14592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kondrychyn I, Garcia-Lecea M, Emelyanov A, Parinov S & Korzh V Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics 10, 418, doi: 10.1186/1471-2164-10-418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schunder E, Rydzewski K, Grunow R & Heuner K First indication for a functional CRISPR/Cas system in Francisella tularensis. International journal of medical microbiology : IJMM 303, 51–60, doi: 10.1016/j.ijmm.2012.11.004 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Vestergaard G, Garrett RA & Shah SA CRISPR adaptive immune systems of Archaea. RNA biology 11, 156–167, doi: 10.4161/rna.27990 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Makarova KS et al. An updated evolutionary classification of CRISPR-Cas systems. Nature reviews. Microbiology 13, 722–736, doi: 10.1038/nrmicro3569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a Class 2 CRISPR-Cas system. Cell 163, 759–771, doi: 10.1016/j.cell.2015.09.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim HK et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nature methods 14, 153–159, doi: 10.1038/nmeth.4104 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Moreno-Mateos MA et al. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nature communications 8, 2024, doi: 10.1038/s41467-017-01836-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moreno-Mateos MA et al. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nature communications 8, 2024, doi: 10.1038/s41467-017-01836-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ablain J, Durand EM, Yang S, Zhou Y & Zon LI A CRISPR/Cas9 vector system for tissue- specific gene disruption in zebrafish. Developmental cell 32, 756–764, doi: 10.1016/j.devcel.2015.01.032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin L et al. Multiplex Conditional Mutagenesis Using Transgenic Expression of Cas9 and sgRNAs. Genetics 200, 431–441, doi: 10.1534/genetics.115.176917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ding D, Chen K, Chen Y, Li H & Xie K Engineering Introns to Express RNA Guides for Cas9- and Cpf1-Mediated Multiplex Genome Editing. Molecular plant 11, 542–552, doi: 10.1016/j.molp.2018.02.005 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Lee RT, Ng AS & Ingham PW Ribozyme Mediated gRNA Generation for In Vitro and In Vivo CRISPR/Cas9 Mutagenesis. PloS one 11, e0166020, doi: 10.1371/journal.pone.0166020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kleinstiver BP et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 33, 1293–1298, doi: 10.1038/nbt.3404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Badran AH & Liu DR Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nature communications 6, 8425, doi:10.1038/ncomms942510.1038/ncomms9425https://www.nature.com/articles/ncomms9425#supplementary-informationhttps://www.nature.com/articles/ncomms9425#supplementary-information (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57, doi:10.1038/nature2615510.1038/nature26155https://www.nature.com/articles/nature26155#supplementary-informationhttps://www.nature.com/articles/nature26155#supplementary-information (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chavez A et al. Highly efficient Cas9-mediated transcriptional programming. Nature methods 12, 326–328, doi: 10.1038/nmeth.3312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424, doi: 10.1038/nature17946 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63, doi: 10.1038/nature26155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harrington JJ & Lieber MR The characterization of a mammalian DNA structure-specific endonuclease. The EMBO journal 13, 1235–1246 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Varshney GK & Burgess SM DNA-guided genome editing using structure-guided endonucleases. Genome biology 17, doi: 10.1186/s13059-016-1055-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hutvagner G & Simard MJ Argonaute proteins: key players in RNA silencing. Nature Reviews Molecular Cell Biology 9, 22, doi: 10.1038/nrm2321 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Swarts DC et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Research 43, 5120–5129, doi: 10.1093/nar/gkv415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan YR et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Molecular cell 19, 405– 419, doi: 10.1016/j.molcel.2005.07.011 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Swarts DC et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Research 43, 5120–5129, doi: 10.1093/nar/gkv415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cai M, Si Y, Zhang J, Tian Z & Du S Zebrafish Embryonic Slow Muscle Is a Rapid System for Genetic Analysis of Sarcomere Organization by CRISPR/Cas9, but Not NgAgo. Marine biotechnology (New York, N.Y.) 20, 168–181, doi: 10.1007/s10126-018-9794-8 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Khin NC, Lowe JL, Jensen LM & Burgio G No evidence for genome editing in mouse zygotes and HEK293T human cell line using the DNA-guided Natronobacterium gregoryi Argonaute (NgAgo). PloS one 12, e0178768, doi: 10.1371/journal.pone.0178768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Javidi-Parsijani P et al. No evidence of genome editing activity from Natronobacterium gregoryi Argonaute (NgAgo) in human cells. PloS one 12, e0177444, doi: 10.1371/journal.pone.0177444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qi J et al. NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. Cell research 26, 1349–1352, doi: 10.1038/cr.2016.134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wray GA The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics 8, 206, doi: 10.1038/nrg2063 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Auer TO & Del Bene F CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods (San Diego, Calif.) 69, 142–150, doi: 10.1016/j.ymeth.2014.03.027 (2014). [DOI] [PubMed] [Google Scholar]
- 97.Bedell VM et al. In vivo Genome Editing Using High Efficiency TALENs. Nature 491, 114–118, doi: 10.1038/nature11537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hruscha A et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development (Cambridge, England) 140, 4982–4987, doi: 10.1242/dev.099085 (2013). [DOI] [PubMed] [Google Scholar]
- 99.Ma ACH TALEN-Mediated Mutagenesis and Genome Editing. 1451, 17–30, doi: 10.1007/978-1-4939-3771-4_2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bedell VM et al. In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118, doi: 10.1038/nature11537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Richardson CD, Ray GJ, DeWitt MA, Curie GL & Corn JE Enhancing homology- directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34, 339–344, doi: 10.1038/nbt.3481 (2016). [DOI] [PubMed] [Google Scholar]
- 102.Hoshijima K, Jurynec MJ & Grunwald DJ Precise Editing of the Zebrafish Genome Made Simple and Efficient. Developmental cell 36, 654–667, doi: 10.1016/j.devcel.2016.02.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Irion U, Krauss J & Nusslein-Volhard C Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development (Cambridge, England) 141, 4827–4830, doi: 10.1242/dev.115584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zu Y et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nature methods 10, 329–331, doi: 10.1038/nmeth.2374 (2013). [DOI] [PubMed] [Google Scholar]
- 105.Thyme SB & Schier AF Polq-Mediated End Joining Is Essential for Surviving DNA Double-Strand Breaks during Early Zebrafish Development. Cell reports 15, 707–714, doi: 10.1016/j.celrep.2016.03.072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rees HA et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nature communications 8, 15790, doi:10.1038/ncomms1579010.1038/ncomms15790https://www.nature.com/articles/ncomms15790#supplementary-informationhttps://www.nature.com/articles/ncomms15790#supplementary-information (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang Y et al. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nature communications 8, 118, doi: 10.1038/s41467-017-00175-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Phillips RB & Reed KM Localization of repetitive DNAs to zebrafish (Danio rerio) chromosomes by fluorescence in situ hybridization (FISH). Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 8, 27–35 (2000). [DOI] [PubMed] [Google Scholar]
- 109.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471, doi: 10.1038/nature24644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]