Summary
Transcription in human mitochondria is driven by a single-subunit, factor-dependent RNA polymerase (mtRNAP). Despite of its critical role in both expression and replication of the mitochondrial genome, transcription initiation by mtRNAP remains poorly understood. Here we report crystal structures of human mitochondrial transcription initiation complexes assembled on both light and heavy strand promoters. The structures reveal how transcription factors TFAM and TFB2M assist mtRNAP to achieve promoter-dependent initiation. TFAM tethers the N-terminal region of mtRNAP to recruit the polymerase to the promoter, whereas TFB2M induces structural changes in mtRNAP to enable promoter opening and trapping of the DNA non-template strand. Structural comparisons demonstrate that the initiation mechanism in mitochondria is distinct from that in the well-studied nuclear, bacterial, or bacteriophage transcription systems, but that similarities are found on the topological and conceptual level. These results provide a framework for studying the regulation of gene expression and DNA replication in mitochondria.
Introduction
Transcription of the human mitochondrial genome is carried out by the single-subunit mitochondrial RNA polymerase (mtRNAP), which initiates at the light strand promoter (LSP) and the divergent heavy strand promoter (HSP). In addition to its pivotal role in producing mitochondrial rRNA, tRNA and mRNA, mtRNAP also generates the RNA primer required for replication of the mitochondrial genome (Agaronyan et al., 2015; Gustafsson et al., 2016). Thus, transcription initiation is a key regulatory step for mitochondrial gene expression and for organelle biogenesis and maintenance.
To achieve promoter specific initiation, mtRNAP requires the mitochondrial transcription factor A (TFAM) and the mitochondrial transcription factor B2 (TFB2M). Previous biochemical studies have established that TFAM functions in promoter recruitment (Gaspari et al., 2004; Morozov et al., 2014), whereas TFB2M is required for DNA opening (Gaspari et al., 2004; Morozov et al., 2014; 2015; Ramachandran et al., 2016). The structures of free mtRNAP (Ringel et al., 2011) and free TFAM (Ngo et al., 2011; Rubio-Cosials et al., 2011) are known, but no structures have been reported for TFB2M or the initiation complex (IC) containing TFAM, TFB2M and mtRNAP. Thus, it remains elusive how TFAM and TFB2M cooperate with mtRNAP to enable transcription initiation.
MtRNAP belongs to the pol A family of single subunit (ss) DNA-dependent RNAPs, which also includes the well-studied RNAP from bacteriophage T7. All ssRNAPs share high sequence homology in their carboxy-terminal domains (CTD) that form a fold resembling a right hand (Jeruzalmi and Steitz, 1998; Kohlstaedt et al., 1992). The CTD forms the catalytic core of these enzymes and comprises the conserved palm and the mobile fingers subdomains. The CTD also contains the ‘specificity loop’, a β-hairpin that binds the major groove of promoter DNA and forms base-specific contacts in T7 RNAP (Cheetham and Steitz, 1999; Gleghorn et al., 2008). Despite the conservation of the CTD, T7 RNAP does not require initiation factors, and accomplishes DNA binding and opening independently (Cheetham et al., 1999), in contrast to mtRNAP.
This functional difference results mainly from distinct amino-terminal regions of ssRNAPs that show very limited homology and differ in size between these enzymes. In phage RNAPs, the amino-terminal regions contain a promoter-binding domain (PBD) (Durniak et al., 2008). In T7 RNAP, the PBD forms a six-helix bundle that includes two DNA-binding elements, the intercalating hairpin and the AT-rich recognition loop (Cheetham et al., 1999; Durniak et al., 2008). The intercalating hairpin separates DNA strands during promoter opening and interacts with the DNA template strand (Cheetham and Steitz, 1999; Gleghorn et al., 2008). The AT-rich recognition loop binds the minor groove of upstream promoter DNA (Cheetham et al., 1999). Whereas the AT-rich recognition loop is reduced and appears to play no role in promoter binding, the specificity loop and intercalating hairpin of mtRNAP were suggested to be functional homologs of their T7 RNAP counterparts (Ringel et al., 2011). However, the positions observed for these elements in the apo structure of mtRNAP appear incompatible with promoter binding and opening, and hence the structural basis for mtRNAP initiation and its dependency on transcription factors remains unknown.
Here we determine crystal structures of human TFB2M and mitochondrial ICs assembled on light and heavy strand promoter DNA. The structures reveal the locations of transcription factors TFAM and TFB2M on the mtRNAP surface and suggest how they enable recruitment of promoter DNA to mtRNAP and DNA opening. We also provide detailed comparisons of the IC structure with structures of functional complexes of T7 RNAP. Our results reveal the distinct nature of mitochondrial transcription initiation and its molecular basis.
Results
Structure of human TFB2M
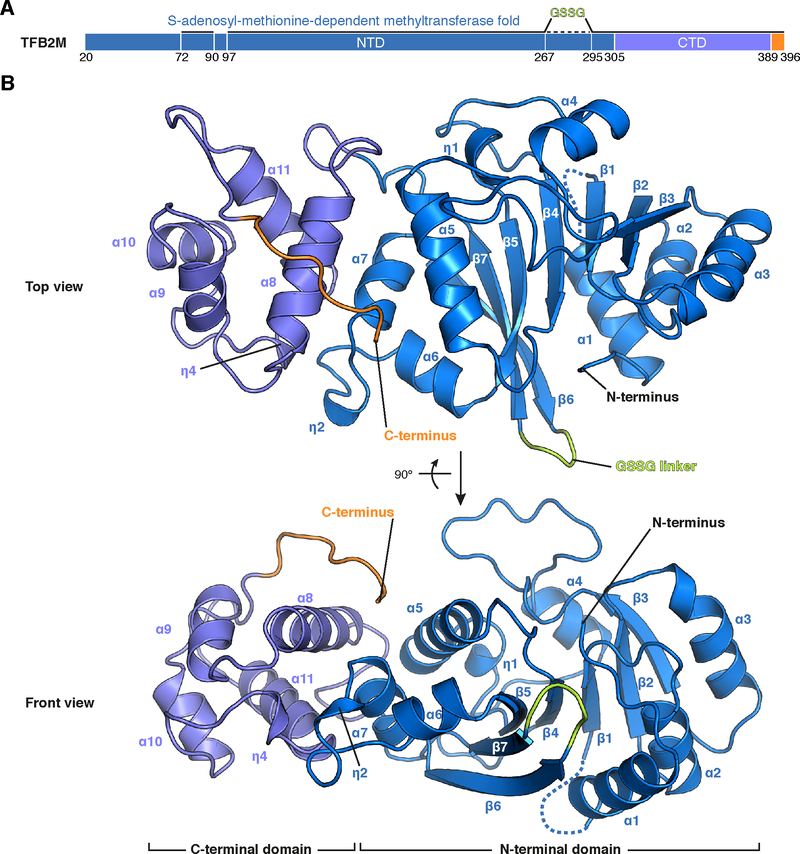
To investigate the mechanism of mitochondrial transcription initiation, we first completed the set of structures for the involved proteins by determining the structure of TFB2M. Extensive crystallization trials using full-length human TFB2M did not yield crystals. We therefore designed a variant lacking apparently flexible regions that may impair crystallization (Methods). This variant, TFB2Mcryst, lacks 62 N-terminal residues and a predicted internal loop (residues 268–294) that was replaced by a short GSSG-linker. Functional characterization of this TFB2M variant shows that replacement of the internal loop does not affect the transcriptional activity (Figure S1C), whereas the N-terminal truncation of TFB2M reduces the activity due to its role in interactions with the priming nucleotide (Figure S1C) (Sologub et al., 2009). TFB2Mcryst yielded crystals that diffracted to 1.75 Å resolution and the structure was solved by molecular replacement (Table S1). The refined model shows very good stereochemistry and contains residues 72–396 of TFB2M with the exception of a short flexible loop (residues 91–96).
The structure (Figure 1) shows that TFB2M resembles the paralogous mitochondrial methyltransferase TFB1M (Guja et al., 2013) and the yeast mitochondrial transcription initiation factor Mtf1 (Schubot et al., 2001) (Figure S1A and B). As predicted from sequence homology, the N-terminal domain (residues 72–305) adopts a fold resembling S-adenosyl-methinonine-dependent methyltransferases with a central seven-stranded β-sheet flanked on either side by three α-helices (Martin and McMillan, 2002). Similar to TFB1M and Mtf1, TFB2M deviates from the canonical methyltransferase fold by an insertion between β6 and β7, which corresponds to the region replaced with the GSSG linker in the crystallization construct (Guja et al., 2013; Schubot et al., 2001). In addition, TFB2M displays a prominent loop insertion between β3 and α4 not found in either of the two other proteins. The C-terminal domain (residues 306–396) consists of four α-helices and an extended C-terminal tail (residues 389–396), which is likely flexible in solution because density for this region was only observed for one of the two copies in asymmetric unit. The structure of TFB2M completes the set of high-resolution structures of proteins involved in mitochondrial transcription initiation.
Figure 1 |. Structure of human TFB2M.
(A) Schematic representation of TFB2M. The N-terminal domain is colored in marine blue, the C-terminal domain in slate and the C-terminal tail in orange. Regions visible in the electron density of the TFB2Mcryst crystals are indicated by a solid black line. The dashed black line represents the internal loop region replaced by a GSSG-linker in order to obtain a crystallizable construct.
(B) Ribbon representation of the human TFB2M structure. Coloring as in (A) with secondary structure elements indicated.
See also Figure S1.
Structure determination of the mitochondrial transcription initiation complex
We then assembled a transcriptionally active IC consisting of TFAM, TFB2M, mtRNAP, and either LSP or HSP DNA containing a pre-melted region spanning register −4 to +3, which corresponds to the DNA region initially unwound around the transcription start site +1 (Ramachandran et al., 2016) (Figure S2A and S2B). After extensive optimization, crystals of the IC were obtained that diffracted to 4.5 Å resolution.
The IC crystal structure was determined by a combination of molecular replacement and anomalous diffraction (Methods and Tables S2 and S3), which led to an interpretable electron density map (Figure S2E). The known structures of mtRNAP (Schwinghammer et al., 2013) and TFAM (Ngo et al., 2011) were fitted into the electron density and the newly obtained TFB2M structure could be unambiguously placed. Correct positioning of TFB2M was verified using an anomalous difference Fourier map that revealed selenium peaks for all nine methionine residues (Figure S2C). Most of the DNA could be built, except for parts of the single-stranded region, and the correct sequence register was confirmed using anomalous diffraction from 5-Bromo-Uracil labelled DNA scaffolds (Figure S2D and Table S3). This led to an atomic model for the IC refined to a free R-factor of 31 % (Table S2). We also solved a 4.5 Å resolution crystal structure of the IC assembled on the HSP promoter (Table S4). This structure was essentially identical to the LSP IC (r.m.s.d. = 0.23 Å over 10,136 atoms) (Figure S2F) and in the following we focus our discussion on the LSP IC.
IC structure reveals locations of TFAM, TFB2M, and DNA on mtRNAP
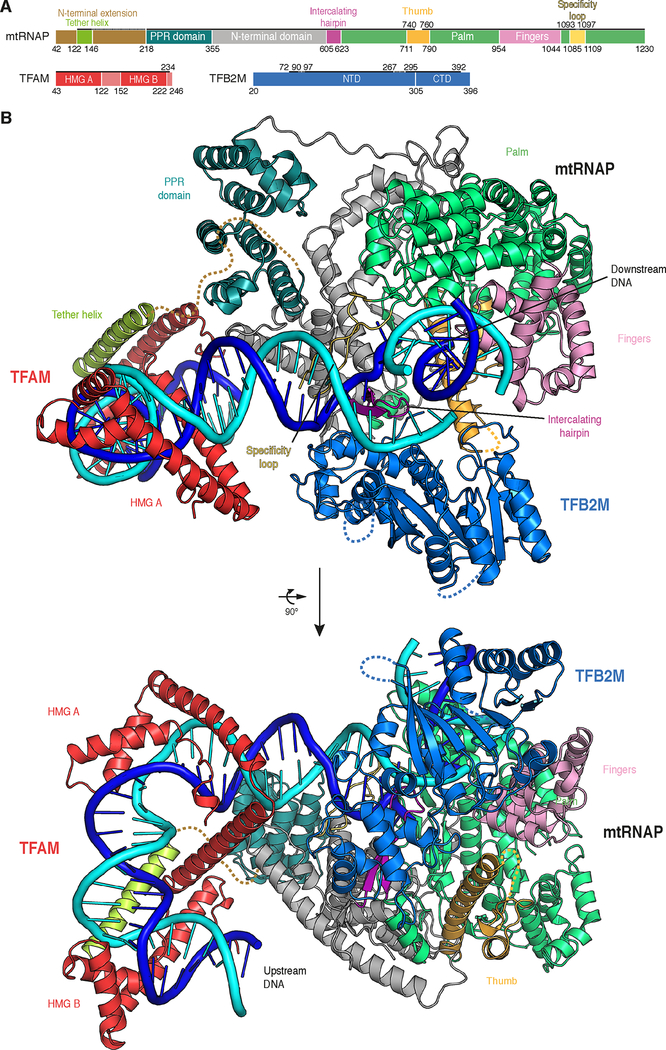
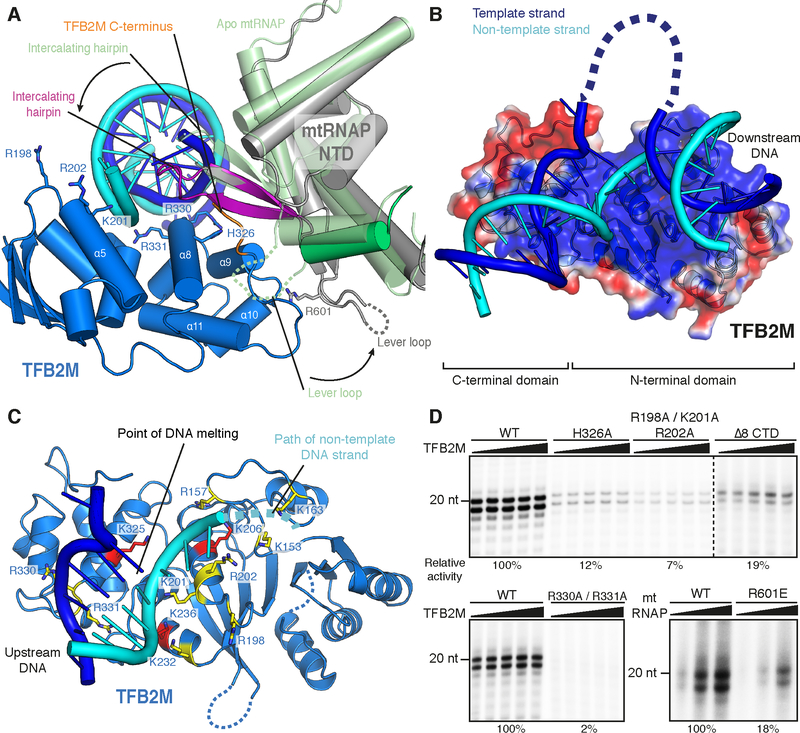
The IC structure (Figure 2B) reveals that mtRNAP is largely unchanged compared to the previously reported EC structure (Schwinghammer et al., 2013), with the exception of the fingers domain, which adopts the ‘clenched’ conformation observed in the apo enzyme (Ringel et al., 2011). Whereas the position of the downstream DNA duplex in the IC is identical to that observed in the EC, the upstream DNA occupies a different location, running along the NTD of mtRNAP. The conserved intercalating hairpin of mtRNAP separates the DNA strands at the upstream edge of the open DNA region observed in the active center cleft of the polymerase (Figure 2B and 3A).
Figure 2 |. Structure of the human mitochondrial transcription initiation complex.
(A) Schematic representation of mtRNAP, TFAM and TFB2M. Important structural elements are indicated with flanking residue numbers. Regions with interpretable electron density in the IC crystal structure are indicated by a solid black line. Regions with density of insufficient quality for model building are indicated by a dashed black line. The colour code is used throughout.
(B) Ribbon representation of the IC structure assembled on LSP DNA. Important structural elements are indicated.
See also Figure S2.
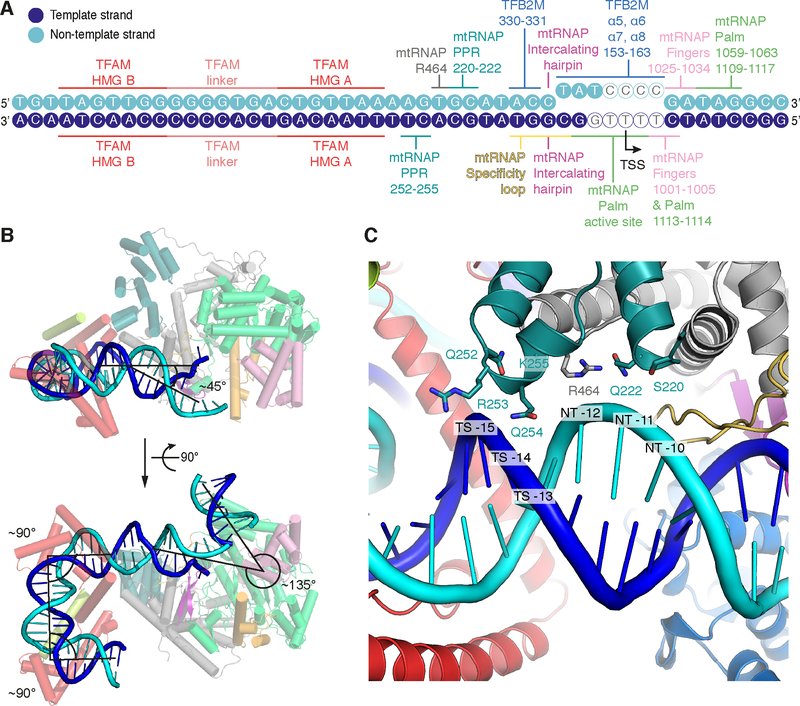
Figure 3 |. Promoter DNA interactions in the IC and DNA bending.
(A) Schematic representation of protein-DNA interactions in the IC. DNA bases of the LSP promoter scaffold used for crystallization are depicted as circles in blue and cyan for the template and non-template strand, respectively. DNA bases lacking density in the IC crystal structure are depicted as hollow circles. Protein regions interacting with the DNA are indicated with coloring as in Figure 2.
(B) DNA bending observed in the IC structure. Ribbon representation of the IC LSP structure with mtRNAP and TFAM in the background and DNA in the foreground. Helices are depicted as cylinders. TFB2M was omitted for clarity. Approximate angles between DNA duplexes are indicated.
(C) Close-up view of the interaction between the PPR domain of mtRNAP and the upstream DNA duplex between the −10 and −15 bases. Several potentially interacting residues in the PPR domain are located close to the DNA backbone. In addition to two regions in the PPR domain (residues 220–222 and 252–255), R454 from the N-terminal domain of mtRNAP is located in proximity to the DNA. Note that the trajectories of sidechains shown are derived from the high-resolution structure of the EC used for molecular replacement (PDB ID 4BOC) (Schwinghammer et al., 2013).
TFB2M contacts the intercalating hairpin and covers the junction between the upstream DNA duplex and the open DNA region (Figure 2B and 3A). TFAM binds DNA 16–39 nt upstream of the transcription start site and induces a ~180° bend into DNA, resembling the free TFAM-DNA complex (Ngo et al., 2011; Rubio-Cosials et al., 2011). In agreement with cross-linking data (Morozov et al., 2014), TFAM does not contact TFB2M, but binds the N-terminal domain (NTD) of mtRNAP at helix D. In addition to the severe upstream bend in the DNA induced by TFAM binding, the trajectory of the DNA is changed by ~45° between mtRNAP and TFAM (Figure 3B). This is apparently caused by interactions of the PPR domain with the DNA backbone at register −10 to −15 (Figure 3B and C), where the DNA minor groove appears widened. In addition, the downstream DNA in the IC duplex encloses an angle of ~135° relative to the upstream duplex at the point of DNA melting (Figure 3B).
TFAM recruits mtRNAP to promoter DNA
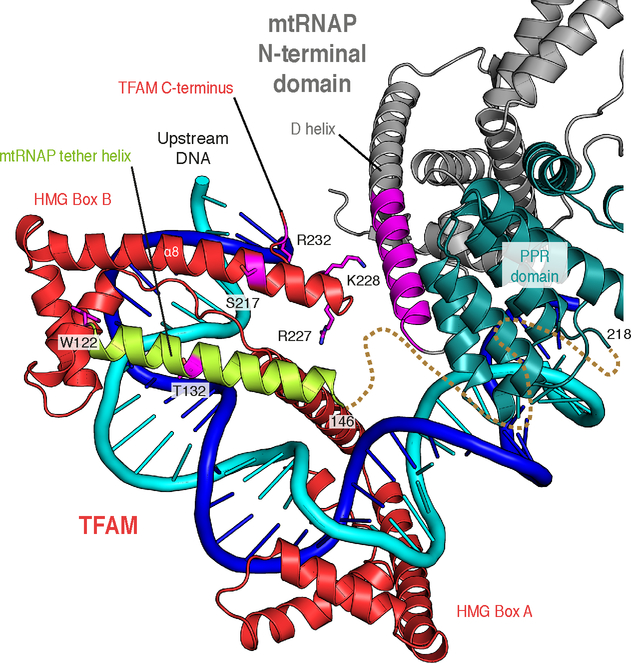
The IC structure explains how TFAM recruits mtRNAP to promoter DNA (Gaspari et al., 2004; Morozov et al., 2014; Posse et al., 2014). The HMG Box B domain of TFAM interacts with a newly observed ‘tether’ helix in the N-terminal extension of mtRNAP, thereby anchoring mtRNAP to the promoter (Figure 4, S3A and Movie S1). The C-terminal tail of TFAM is located close to the PPR domain and residues 444–462 of mtRNAP (D-helix), consistent with published biochemical, genetic and cross-linking data (Dairaghi et al., 1995a; Morozov and Temiakov, 2016; Morozov et al., 2015) (Figure 4). These contacts enable TFAM to recruit mtRNAP and position its active site over the transcription start site for de novo RNA synthesis (Dairaghi et al., 1995b; Morozov et al., 2014). In agreement with cross-linking data (Morozov and Temiakov, 2016), TFAM binding is identical in the structure of the HSP IC (Figure S3B). There, similarly to the LSP IC, TFAM binds to the region that is located 16–39 bp upstream to the HSP transcription start site, in agreement with footprinting data (Fisher et al., 1987). This indicates that the two transcription units in human mitochondria possess similar architecture, in contrast to previous reports that suggested no role of the TFAM C-terminal tail in LSP activation (Uchida et al., 2017) and proposed opposite orientations of TFAM relative to mtRNAP in the IC assembled on HSP DNA (Ngo et al., 2014).
Figure 4 |. TFAM recruits mtRNAP to promoter DNA.
Close-up view of the interaction between mtRNAP and TFAM in the IC. Coloring as in Figure 2. Residues in TFAM and the region in mtRNAP previously shown to be functionally important for initiation or identified as crosslinking-points are shown in magenta (Morozov et al., 2014; 2015). The putative trajectory of the polypeptide connecting the tether helix to the PPR is indicated as a dashed line.
TFB2M assists mtRNAP in DNA opening
The IC structure also reveals how TFB2M assists mtRNAP in promoter opening and stabilization of open DNA (Morozov et al., 2015; Posse and Gustafsson, 2016; Ramachandran et al., 2016). First, TFB2M binds the duplex DNA around base −7 with its conserved arginine residues R330 and R331 (Figure 5A, 5C and Figure S4A and B). Mutation of these residues to alanine severely impairs transcription initiation (Figure 5D). Second, TFB2M induces conformational changes in mtRNAP that stabilize open DNA. Comparison of the IC and the apo mtRNAP structure indicates that TFB2M binding induces a rotation of the PBD of mtRNAP (residues 420–520 and 557–637), which includes the intercalating hairpin. This rotation moves the intercalating hairpin by ~7 Å and positions it between DNA strands (Figure 5A and Movie S1). The intercalating hairpin is further buttressed by TFB2M helix α8, which contains residues that are essential for activity (Morozov et al., 2015), including residue H326, which is critical for transcription initiation (Figure 5A and 5D and Figure S4A).
Figure 5 |. TFB2M stabilizes open DNA and traps the non-template strand.
(A) TFB2M binding induces structural rearrangements in mtRNAP. Ribbon representation of DNA, mtRNAP and TFB2M around the point of DNA melting in the IC. Helices are depicted as cylinders. Functionally important residues in TFB2M are shown as sticks. The palm domain (residues 646–1230) of free mtRNAP (Ringel et al., 2011) (PDB ID 3SPA; transparent pale green) was superimposed on the IC. For clarity, only residues 420–520 and 557–637 of mtRNAP are shown. Grey and green dashed lines indicate the trajectory of unresolved parts of the mtRNAP lever loop in the IC and apo mtRNAP structures, respectively. Arrows indicate the movement induced by TFB2M binding.
(B) Nucleic acid binding by TFB2M. The surface of TFB2M is coloured according to electrostatic potential with positive and negative potential in blue and red, respectively. DNA is shown in cartoon view. Upstream DNA and mtRNAP were omitted for clarity.
(C) Charged residues in TFB2M in proximity to nucleic acid in the IC. TFB2M is shown as ribbon representation in marine blue. DNA is shown in cartoon view with coloring as in (B). Arginine and Lysine residues in the vicinity of the DNA are shown as sticks. Residues conserved as basic (R,K) in human and mouse TFB2M, but not in TFB1M, are shown in yellow. Residues conserved as basic in both TFB2M and TFB1M are shown in red.
(D) Activity of structure-based point mutants of mtRNAP and TFB2M in transcription assays.
The PBD also harbours a ‘lever’ loop (residues 588–604), a structural element that is located adjacent to the intercalating hairpin and found in mtRNAP but not in phage RNAPs. The lever loop is essential for initiation (Morozov et al., 2015) and likely plays a key role in TFB2M-induced rotation of the core NTD. The lever loop would clash with bound TFB2M if it adopted the position observed in free mtRNAP (Figure 5A). In the IC, the lever loop interacts with loop α9-α10 in TFB2M (residues 341–347), and this may stabilize the rotated NTD core. Indeed, mutation of an arginine residue in the lever loop (R601E) results in decreased transcription initiation (Figure 5D). Comparison with the structure of free TFB2M reveals that the C-terminal tail of TFB2M (residues 389–396) has apparently moved to accommodate the intercalating hairpin of mtRNAP in the position observed in the IC (Figure S4A). The C-terminus of TFB2M appears to stabilize the intercalating hairpin, as its shortening by eight amino acids leads to a reduction in activity (Figure 5D).
Finally, TFB2M traps the non-template DNA strand in the open DNA region. This was previously suggested for Mtf1 (Paratkar and Patel, 2010) and is reminiscent, on the topological level, of the bacterial initiation factor sigma (Feklistov and Darst, 2011; Helmann and Chamberlin, 1988; Zhang et al., 2012). The NTD of TFB2M displays a positively charged surface that guides the DNA non-template strand away from the template strand (Figure 5B). Three conserved positively charged residues (R198, K201 and K202) protrude from loop β4-α5 and helix α5 of TFB2M towards the non-template strand and are required for efficient initiation (Figure 5A, 5C and S4A). Further DNA interactions may be formed by the positively charged residues K153, R157, K163 and K206, which line the projected path of the non-template strand, and residues K325, K232 and K236 close to the duplex DNA (Figure 5C). Most of these positively charged residues are conserved in TFB2M from human and mouse, arguing for their functional importance. Consistent with this, these residues are not strongly conserved in the paralog TFB1M, which is not involved in transcription initiation (Figure S4B) (Litonin et al., 2010; Metodiev et al., 2009).
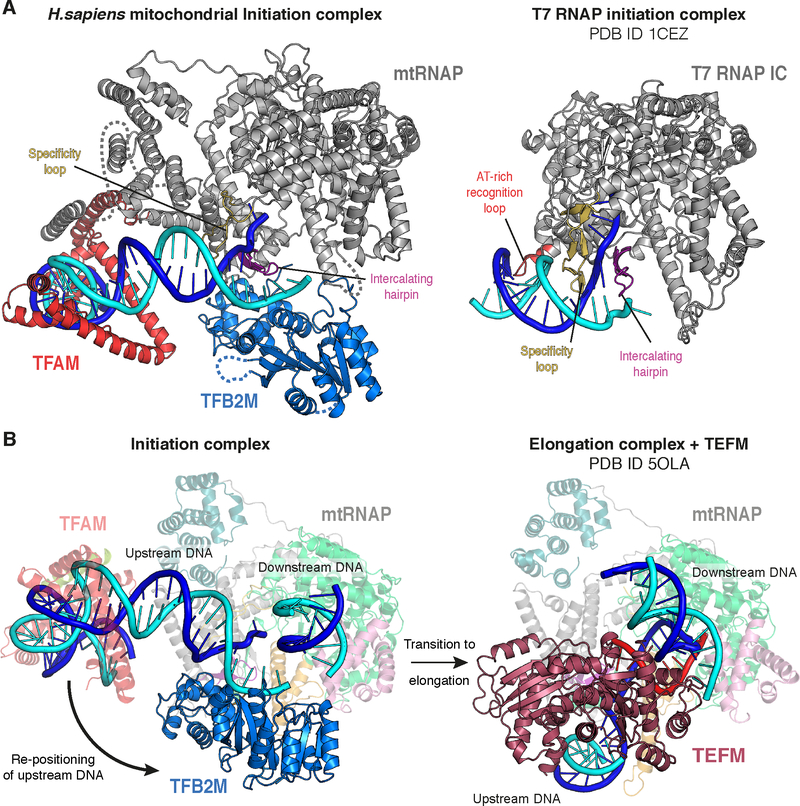
Comparison to T7 RNAP initiation
Comparison of the mitochondrial IC structure to the structure of the T7 RNAP IC (Cheetham et al., 1999) reveals possible reasons for the requirement of initiation factors by mtRNAP. Promoter recognition by T7 RNAP is achieved in part through sequence-specific DNA contacts at registers −5 to −11 formed by the specificity loop (Figure 6A and Figure S5). The specificity loop in mtRNAP shows only fragmented density in the DNA major groove around registers −9 to −7, arguing against a prominent role of this loop in promoter recognition. Consistent with this, LSP and HSP share no sequence homology in this region and DNA base mutations hardly change initiation activity (Gaspari et al., 2004). In addition to the specificity loop, T7 RNAP engages with promoter DNA via the AT-rich recognition loop, which inserts into the upstream DNA minor groove between registers −17 and −13 (Cheetham et al., 1999). (Figure 6A). The structure of the IC demonstrates that mtRNAP does not form sequence-specific contacts with promoter DNA in this region. Instead, only interactions between the PPR domain of mtRNAP and the upstream DNA backbone were detected (Figure 3A and C). Thus, recruitment of mtRNAP to DNA-bound TFAM apparently substitutes for the lack of extensive DNA interactions formed by T7 RNAP with promoter DNA.
Figure 6 |. Comparison to T7 RNAP initiation and transition to elongation.
(A) (Left) Ribbon depiction of the LSP IC. MtRNAP is shown in grey with the intercalating hairpin in purple and the specificity loop in yelloworange. (Right) Ribbon depiction of the T7 RNAP initiation complex (PDB ID 1CEZ) (Cheetham et al., 1999). Coloring as for the mitochondrial IC, with the AT-rich recognition loop in red. The topology around the point of DNA melting is similar in both complexes. See also Figure S5.
(B) (Left) Structure of the LSP IC. TFAM and mtRNAP are depicted transparently for clarity. The movement of the upstream DNA upon transition to the EC is indicated with an arrow. (Right) Structure of the human mitochondrial transcription elongation complex with the elongation factor TEFM bound (PDB ID 5OLA) (Hillen et al., 2017).MtRNAP is depicted transparently for clarity. The position of the downstream DNA duplex is identical in both the IC and the EC. The TFB2M binding site on mtRNAP is occupied by the upstream DNA and TEFM in the EC, demonstrating that a pronounced rearrangement of the upstream DNA must take place during the transition from initiation to elongation and that binding of TFB2M and TEFM to mtRNAP are mutually exclusive.
Opening of the DNA duplex by T7 RNAP is facilitated by the intercalating hairpin, which separates the two DNA strands at the upstream edge of the DNA bubble. In the apo mtRNAP structure, the intercalating hairpin has been observed in a conformation that is incompatible with promoter melting (Ringel et al., 2011). In the mitochondrial IC, however, the intercalating hairpin and specificity loop are arranged as in the T7 RNAP IC (Figure S5). This suggests that binding of TFB2M stabilizes an initiation-competent conformation of mtRNAP that is characterized by properly positioned elements required for DNA opening, including the intercalating hairpin (Figure 6A and Figure S5). In summary, comparison of the mitochondrial IC and the T7 RNAP IC suggests that TFAM compensates for the lack of prominent RNAP-promoter interactions upstream of the point of DNA opening and that TFB2M assists in promoter opening by positioning key structural elements in mtRNAP in a fashion reminiscent of T7 RNAP.
Transition from initiation to elongation
After RNA chain initiation, mtRNAP must lose its interactions with TFAM and TFB2M in order to transition to the elongation phase. In the case of T7 RNAP, this initiation-elongation transition is accompanied by substantial refolding of the polymerase, which destroys the PBD (Tahirov et al., 2002; Yin and Steitz, 2002). In contrast, comparison of the mitochondrial IC with the EC structure (Schwinghammer et al., 2013) demonstrates that the mtRNAP conformation remains largely unchanged. Instead, the DNA rearranges during the initiation-elongation transition. In the EC, upstream DNA is repositioned and occupies the binding site of TFB2M, which must therefore dissociate during the transition (Figure 6B). TFB2M dissociation is also required for binding of the elongation factor TEFM (Figure 6B), which also interacts with the intercalating hairpin of mtRNAP and critically affects processivity of the elongation complex (EC) (Hillen et al., 2017). The transition further creates a channel for RNA exit underneath the intercalating hairpin, which remains in an open conformation and now separates the exiting RNA from the DNA template (Schwinghammer et al., 2013). Thus, TFB2M positions the intercalating hairpin for initiation, and this position of the hairpin is largely maintained during subsequent elongation. These comparisons highlight the dramatic differences between mtRNAP and T7 RNAP with respect to the structural changes that occur during the initiation-elongation transition when the polymerase escapes from the promoter.
Discussion
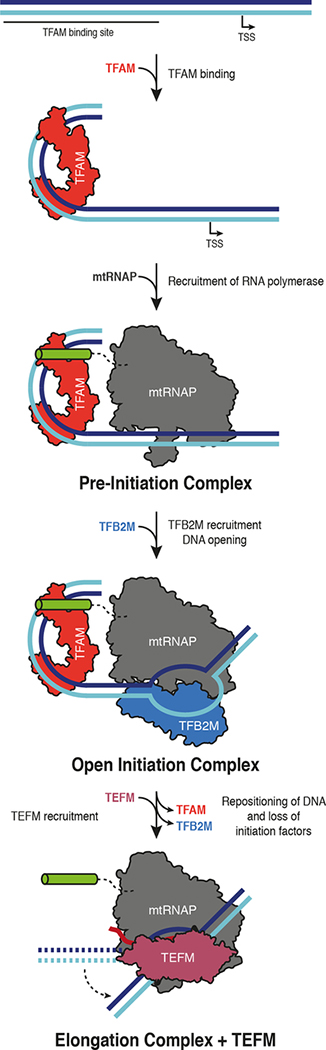
In this study we extend our previous structural work on mitochondrial transcription from elongation (Schwinghammer et al., 2013) to initiation. Our structures of the ICs demonstrate the conserved architecture of the transcription complexes that assemble at divergent human mitochondrial promoters and support the sequential model of transcription initiation (Morozov et al., 2014) (Figure 7 and Movie S1). First, recruitment of mtRNAP to TFAM-bound promoter DNA positions mtRNAP at the transcription start site. This explains the critical role of the distance between the TFAM-binding site and the start site in initiation (Dairaghi et al., 1995b). Subsequent binding of TFB2M induces DNA opening and stabilizes open DNA with the use of conformational changes and binding energy. TFB2M positions the intercalating hairpin of the polymerase for DNA opening. Initial RNA synthesis may then be facilitated by the N-terminal region of TFB2M (residues 21–71), which is mobile in the IC structure but can be cross-linked to the priming nucleotide (Sologub et al., 2009).
Figure 7 |. Model for transcription initiation in human mitochondria.
The TFAM binding site and the transcription start site (TSS) are indicated. TFAM (red) binding bends the DNA upstream of the TSS and leads to recruitment of mtRNAP (gray) to form the closed pre-initiation complex. Binding of TFB2M (blue) leads to melting of the DNA duplex and the bending of the downstream DNA observed in the open initiation complex. Promoter escape and transition to the elongation complex involve repositioning of the upstream DNA duplex (dashed arrow) and binding of TEFM (raspberry), which occupies a similar site on mtRNAP as TFB2M in the IC.
See also Movie S1.
The transition from initiation to elongation is accompanied by a dramatic rearrangement of the upstream DNA, as observed for the related T7 RNAP (Yin and Steitz, 2002). However, in contrast to T7 RNAP (Yin and Steitz, 2002), the conformation of mtRNAP remains largely unchanged during the transition. Instead, the transition involves dissociation of the initiation factors. Comparison of the IC structures with our recent structure of the EC bound by the mitochondrial elongation factor TEFM (Hillen et al., 2017) demonstrates that TFB2M and TEFM binding to mtRNAP are mutually exclusive. Thus, TFB2M must be released before TEFM can bind mtRNAP. These results indicate changes that occur during the initiation-elongation transition of mitochondrial transcription.
Our structural data also show how the initiation mechanism of mtRNAP differs from that used by multisubunit RNAPs. Like mtRNAP, multisubunit RNAPs depend on additional factors for initiation, but these factors are not homologous to TFAM and TFB2M, neither on the sequence level nor at the structural level. There are, however, conceptual similarities between all initiation systems. In particular, TFB2M traps the non-template strand in the open DNA region, in a manner that is topologically similar to the sigma factor required by bacterial RNAP for initiation (Feklistov and Darst, 2011; Murakami and Darst, 2003; Zhang et al., 2012). The eukaryotic RNA polymerase I initiation machinery apparently also uses trapping of the open DNA (Engel et al., 2017; Han et al., 2017; Sadian et al., 2017).
In conclusion, our results provide the structural basis of mitochondrial transcription initiation and suggest structural rearrangements that occur during the transition to transcription elongation. The mitochondrial initiation system employs mechanisms of initiation that are clearly distinct from those observed for nuclear, bacterial or bacteriophage RNAPs. This likely reflects the need for regulating mitochondrial transcription, which is not only required for the expression of essential genes and the synthesis of ribosomal and transfer RNA, but also to generate RNA primers for replication of the mitochondrial genome (Agaronyan et al., 2015; Gustafsson et al., 2016).
STAR METHODS
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Patrick Cramer (patrick.cramer@mpibpc.mpg.de)
Experimental model and subject details
For cloning we used the Escherichia coli strain XL1-Blue (Agilent) and for recombinant expression the Escherichia coli strains BL21CodonPlus (DE3) RIL (Agilent) and Rosetta2 (DE3) pLysS (Merck Millipore). Cells were grown at 37°C as described in the STAR Methods Details section.
Method Details
Protein expression constructs
A construct of human mtRNAP lacking the N-terminal 104 amino acids (Δ104mtRNAP, E555A natural variant) with an N-terminal 6-His tag and a TEV cleavage site was constructed from the pProEx-based expression plasmid described previously (Sologub et al., 2009). Human TFAM was expressed as a construct lacking the mitochondrial localization sequence (res. 1–42) and two cysteine residues (C49S and without the C-terminal C246), mutations which have been shown to have no effect on activity (Morozov et al., 2014). The construct was cloned from the previously described pET22b-based expression plasmid (Morozov et al., 2014) by inserting a TEV cleavage site between the N-terminal 6-His tag and the coding. Human TFB2M was expressed as a construct lacking either only the predicted mitochondrial localization sequence (res. 1–20; Δ20TFB2M) or lacking the N-terminal 62 residues (Δ62TFB2M). The construct was generated from the pTYB11-based expression vector described previously (Sologub et al., 2009) by removing the intein tag and replacing it with a TEV-cleavable N-terminal 6-His tag.
To obtain a crystallizable variant of human TFB2M, we used limited proteolysis combined with Edman sequencing to map regions sensitive to proteolysis. This approach identified two regions sensitive to cleavage by Trypsin (around residues K63 and R288), indicating potentially less ordered regions of the protein. We then used secondary structure prediction (PSIPRED) (Buchan et al., 2013) together with a computationally generated 3D homology model (Swiss Model) (Biasini et al., 2014), which we compared to S.cerevisiae Mtf1 (PDB ID 1I4W) (Schubot et al., 2001) and M.musculus TFB1M (PDB ID 4GC5) (Guja et al., 2013), to delineate the boundaries of the putative flexible regions. This approach led us to design the crystallization variant, TFB2Mcryst, which lacks 62 amino acids at the N-terminus and a loop region between residues 268 and 294, for which no electron density could be observed in the IC and which was replaced by a short GSSG linker. Expression plasmids for structure determination were generated by round the horn PCR. All TFB2M variants used in transcription assays were generated using QuickChange Mutagenesis kit (Agilent) in N-his Δ20TFB2M background. The R601E mtRNAP variant was generated using Δ119mtRNAP background.
Protein expression and purification
Human mtRNAP was expressed in E.coli BL21 (DE3) RIL cells (Agilent) and purified essentially as described previously (Sologub et al., 2009). Briefly, cells were grown until the OD600 reached 0.6 units and expression was induced by addition of 0.15 mM IPTG and carried out at 16 °C for 18 h. MtRNAP was purified by affinity chromatography using Ni-NTA beads (Qiagen) followed by tag cleavage using TEV protease. The protein was further purified by Heparin affinity chromatography using a HiTrap Heparin HP column (GE Healthcare) followed by size exclusion chromatography using a Superdex 200 Increase 10/300 column (GE Healthcare) equilibrated with mtRNAP Buffer (40 mM Tris/HCl pH 8.0, 300 mM NaCl, 20 mM MgCl2, 5% Glycerol, 10 mM DTT). Peak fractions were pooled, concentrated and stored at −80 °C.
Human TFAM was expressed in E.coli BL21 Rosetta2 pLysS cells (Merck Millipore). Cells were grown until the OD600 reached 1.0 units and expression was induced by addition of 0.8 mM IPTG and carried out at 37 °C for 1.5 h. TFAM was purified by affinity chromatography using a HisTrap HP 5 ml column (GE Healthcare) followed by tag cleavage using TEV protease and reverse Ni-NTA affinity chromatography. The protein was further purified by ion exchange chromatography using a HiTrap SP HP column (GE Healthcare) followed by Heparin affinity chromatography using a HiTrap Heparin HP column (GE Healthcare). Final purification was done by size exclusion chromatography using a HiPrep 16/600 Superdex 75 pg column (GE Healthcare) equilibrated with TFAM buffer (40 mM Tris/HCl pH 8.0, 100 mM NaCl, 5% Glycerol, 10 mM DTT). Peak fractions were pooled, concentrated and stored at −80°C.
Human TFB2M was expressed in E.coli BL21 (DE3) RIL cells (Agilent). Cells were grown in expression media (10 g/l Tryptone, 5 g/l Yeast extract, 50 mM KH2PO4, 50 mM Na2HPO4, 25 mM (NH4)2SO4, 2 mM MgSO4, 0.5 % (v/v) glycerol, 0.05 % (w/v) D-glucose) until the OD600 reached 6.0 units. Expression was induced by addition of 0.1 mM IPTG and carried out at 16°C for 18 h. TFB2M was purified by affinity chromatography using Ni-NTA beads (Qiagen) followed by tag cleavage using TEV protease and reverse Ni-NTA affinity chromatography using a HisTrap HP 5 ml column (GE Healthcare). The protein was further purified by Heparin affinity chromatography using a HiTrap Heparin HP column (GE Healthcare) and size exclusion chromatography using a Superdex 200 Increase 10/300 column (GE Healthcare) equilibrated with TFB2M buffer (40 mM Tris/HCl pH 8.0, 100 mM NaCl, 5% Glycerol, 10 mM DTT). Peak fractions were pooled, concentrated and stored at −80°C.
For expression of selenomethionine-labeled protein, expression cultures were grown in SeMet Base media (Molecular Dimensions) with the addition of L-Selenomethionine (50 μg/ml), L-Lysine (50 μg/ml), L-Threonine (50 μg/ml), L-Phenylalanine (50 μg/ml), L-Leucine (25 μg/ml), L-Isoleucine (25 μg/ml) and L-Valine (25 μg/ml). Selenomethionine-labeled protein was purified as described above.
Crystallization of TFB2M
Crystals of TFB2M could only be obtained using a truncated variant of the protein, TFB2Mcryst (see above for details). TFB2Mcryst was crystallized by the hanging drop vapour diffusion method at 20 °C by mixing 1.5 μl of protein solution (13 mg/ml) with 1.5 μl of reservoir solution containing 100 mM HEPES pH 7.5, 200 mM NaCl, 17% PEG3350 and 0.4 μl of seeding solution produced from previously grown crystals in a similar condition. Crystals were cryo-protected by gradually increasing the Glycerol concentration in the drop to 25% (v/v) final and flash frozen in liquid nitrogen.
Reconstitution and crystallization of the IC
HPLC-purified synthetic DNA oligonucleotides were obtained from IDT. Scaffolds for crystallization were annealed by heating complimentary oligonucleotides (LSP IC: HH NT2 / HH TS1; HSP IC: HH NT6 / HH TS3; LSP Bromine 1: HH NT2 / HH TS1-Br1; LSP Bromine 2: HH NT2-Br2 / HH TS50; LSP Bromine 3: HH NT2 / HH TS1-Br3; LSP Bromine 4: HH NT2-Br4 / HH TS1; LSP Bromine 5: HH NT2 / HH TS1-Br5; LSP Bromine 6: HH NT67 / HH TS1-Br6) (Table S5) to 95°C and step-wise cooling to 4°C (1°/90s) at a final concentration of 0.5 mM in H2O.
The human IC was reconstituted by incubating Δ104 mtRNAP (35 μM) with a 1.1-fold molar excess of scaffold DNA, equimolar amounts of TFAM (43–245 C49S) and a 1.5-fold molar excess of Δ20TFB2M in a buffer containing 30 mM Tris/HCl pH 8.0, 200 mM NaCl, 10 mM MgCl2, 5% glycerol and 10 mM DTT for 20 min at 20°C. The complex was subsequently purified by size exclusion chromatography using a Superdex 200 Increase 3.2/300 column equilibrated with complex crystallization buffer (10 mM Tris/HCl pH 8.0, 100 mM NaCl, 20 mM MgCl2, 10 mM DTT, 1 mM TCEP). Crystals were obtained by the hanging drop vapour diffusion method at 20 °C by mixing equal volumes of protein solution and reservoir solution containing 100 mM BIS-TRIS pH 6.0, 200 mM L-Proline and 5–7% PEG8000. Crystals were cryo-protected by gradually increasing the glycerol concentration in the drop to a final of 25% (v/v) and flash-frozen in liquid nitrogen. For selenomethionine labelling and subsequent crystallization, a construct of mtRNAP lacking the N-terminal 108 amino acids (Δ108mtRNAP) and Δ62TFB2M were used.
Data collection, structure determination and refinement
Diffraction data were collected at beamline X06SA and X10SA at the Swiss Light Source in Villigen, Switzerland, with an EIGER 16M detector (Dectris) or a PILATUS 6M detector (Dectris), respectively, and at beamline P14 operated by EMBL Hamburg at the PETRA III storage ring (DESY Hamburg, Germany) with a Pilatus 6M detector (Dectris). The data was processed using XDS (Kabsch, 2010) and scaled with XSCALE (Kabsch, 2010). For selenomethionine and 5-Bromo-Uracil containing crystals, multiple datasets (from a single crystal or multiple isomorphous crystals) were merged using XSCALE to improve the anomalous signal.
The crystals of TFB2Mcryst belonged to space group P21 and diffracted to a resolution of 1.75 Å with two copies of the protein in the asymmetric unit. The structure of TFB2Mcryst was solved by molecular replacement in Phaser (McCoy et al., 2007) using a partial model of the yeast homolog Mtf1 (PDB ID 1I4W, residues 134–138; 143–198; 242–281) truncated to poly-alanine. Density modification and building of an initial model was done using phenix.autobuild (Adams et al., 2010) and subsequently completed manually in Coot (Emsley et al., 2010). The model was subjected to iterative cycles of refinement in phenix.refine (Adams et al., 2010) and manual model building in Coot until excellent stereochemistry and a free R-factor of 21.4 % was obtained (Table S1). The final model contains residues 72–94, 97–267 and 295–396 of the wild-type TFB2M.
The IC crystals belonged to space group P21, contained two copies of the complex in the asymmetric unit, and showed diffraction to 4.5 Å resolution. Initial phases were obtained by molecular replacement using Phaser (McCoy et al., 2007) in the PHENIX suite (Adams et al., 2010) and the human mitochondrial transcription EC (PDB ID 4BOC) lacking nucleic acid as search model. The solution was subsequently used as starting phases for molecular replacement combined with single-wavelength anomalous diffraction (MR-SAD) and automated density modification in PHENIX using diffraction data collected from crystals containing selenomethionine-labeled mtRNAP, TFAM and TFB2M and LSP DNA. (Table S2) The resulting electron density map showed clear features of nucleic acids and proteins in addition to the search model used and was phase extended using the higher resolution native dataset in phenix.autobuild. Interpretation of the electron density was facilitated by anomalous scattering from selenium and bromine atoms incorporated into proteins and DNA, respectively (Figure S2C, S2D and Table S2 and S3). Anomalous difference Fourier maps were computed as log-likelihood gradient maps in Phaser (within the PHENIX suite) using phases derived from the refined IC model (Read and McCoy, 2011). Modelling of the IC was done largely using the experimental map and cross-validated with a map generated by MR-SAD with the keyword “phaser_sites_then_phase=True” to obtain a map free of model bias. As one molecule in the asymmetric showed better overall density than the other, model building and analysis was done largely based on the density for this molecule (chains A, C, D, E and F in the final PDB file).
Starting from the model of the EC placed by molecular replacement, individual domains and secondary structure elements of mtRNAP were rigid body fitted in real space manually to fit the experimental electron density. The tip of the thumb domain did not show convincing density and was therefore removed (residues 740–760). The region of mtRNAP corresponding to the specificity loop (residues 1086–1107) showed fragmented density and was modelled based on the structure of the T7 RNAP initiation complex.(Cheetham et al., 1999) The density allowed for modelling the main chain trace of this element lacking only three residues (1094–1096) at the tip, yet the sequence register could not be assigned unambiguously and it was therefore modelled as poly-alanine (residues 1068–1107).
To obtain an initial model of the IC, the crystal structure of TFAM (Ngo et al., 2011) (PDB ID 3TMM) in complex with LSP DNA was fitted into the electron density and the individual domains were rigid body fitted locally in real space using Coot (Emsley et al., 2010). The DNA emerging from TFAM was extended and adjusted as ideal B-DNA to the expected melting point at the beginning of the mismatched region (−4) and rigid body fit locally in real space. The experimental density allowed for modelling of two additional bases of the template and three of the non-template strand, respectively, past the melting point. Although the density clearly showed the trajectory of the phosphate backbone, the conformation of the bases could not be confidently determined at this resolution and was thus modelled based on anomalous peaks from bromine labeled scaffolds with the help of anomalous difference peaks from crystals containing DNA labelled with 5-bromo-uracile at specific sites (Figure S2D). The downstream duplex DNA showed weaker density and was similarly positioned with the help of anomalous difference peaks (Figure S2D). In the crystal form observed, the downstream DNA mediates a crystal contact and may therefore be stabilized in the observed conformation.
To model TFB2M, the crystal structure of TFB2Mcryst was rigid body fit into the experimental IC density. Correct positioning of TFB2M was verified using peaks in an anomalous difference Fourier map calculated from the dataset, which was used for phasing the IC, which included selenomethionine-labelled TFB2M (Figure S2C). Placement of TFB2M led to a single clash between the C-terminal tail of TFB2M with the mtRNAP intercalating hairpin and the non-template DNA (Figure S4A). Since this C-terminal region appears to be flexible, the TFB2M model was truncated to the last residue with clear density in the IC map (residue 392).
After positioning of all known protein structures, a residual unexplained density in the experimental map remained close to the HMG box B of TFAM with three weak peaks in the anomalous difference map for selenium. An anomalous difference map calculated from a dataset obtained from crystals in which only mtRNAP was selenomethionine-labeled indicated that these peaks originate from residues within the polymerase. Based on a unique ‘MRM’ sequence motif found in the thus far not observed N-terminal extension region of mtRNAP and secondary structure prediction using PSIPRED (Buchan et al., 2013), a helix spanning residues 122–146 in mtRNAP was assigned to the unmodeled density, which we termed ‘tether helix’. (Figure S3A).
Refinement of the IC model against the native dataset using phenix.refine with secondary structure restraints, reference model restraints and DNA geometry restraints resulted in a model with good geometry and a free R-factor of 31.0 %. (Table S2) The resulting mFo-DFc map showed difference density for some additional features such as the polypeptide path connecting the mtRNAP tether helix to the PPR domain, which may run along the C-terminal end of TFAM α8, two additional helices of the PPR domain, the missing single-stranded non-template DNA strand, and the loop in TFB2M which was deleted in the TFB2Mcryst variant (residues 268–294) and seems to be positioned close to the N-terminus of TFB2M and the downstream DNA duplex in the IC. However, we refrained from modelling these features due to the limited resolution of the data. Residues 235–237 at the C-terminal tail of TFAM did not show good density and were therefore removed from the model.
The structure of the HSP IC was solved by molecular replacement using the LSP IC as search model and subsequently adjusted to the HSP sequence. The model was refined in phenix.refine using a similar protocol as for the LSP IC and led to a final model with good stereochemistry and a free R of 33.5 %. (Table S4) The identical orientation of TFAM on both LSP and HSP was verified by comparing anomalous difference peaks calculated using structure factors from crystals containing selenomethionine-labeled TFAM and the respective promoter DNA. (Figure S3B)
Figures were prepared using PyMol. Surface charge analysis was performed using the APBS plugin for PyMol (Baker et al., 2001) and displayed with +/− 1kT/e. Figures were prepared using the LSP IC model, unless stated otherwise.
Transcription assays
Standard transcription reactions were carried out using PCR-amplified DNA templates containing the LSP promoter (region −60 to +20) as described previously (Morozov et al., 2015). The reactions contained DNA templates (50 nM), Δ119 mtRNAP (50 nM), Cys-less TFAM (50 nM), Δ20 TFB2M (50 nM) in a transcription buffer containing 40 mM Tris (pH=7.9), 10 mM MgCl2 and 10 mM DTT in the presence of ATP (0.3 mM), GTP (0.3 mM), UTP (0.01 mM) and 0.3 μCi [γ−32P] UTP (800 Ci/mmol). To assay the activity of the IC assembled on pre-melted LSP, the reaction was performed in the presence of ATP (0.3 mM), GTP (0.3 mM) and 0.3 μCi [γ−32P] ATP (800 Ci/mmol) to generate 4–5 nt RNA products. Reactions were carried out at 35°C for 30 min and stopped by addition of an equal volume of 95% formamide/0.05 M EDTA. The products were resolved by 20% PAGE containing 6 M urea and visualized by PhosphorImager (GE Healthcare).
Quantification and Statistical analysis
Data for transcription initiation assays are mean values of at least three technical replicates.
Data and Software availability
Coordinate and structure factor files for the LSP and HSP IC crystal structures and for the human TFB2M crystal structure were deposited with the Protein Data Bank with accession codes 6ERP, 6ERQ and 6ERO, respectively.
Supplementary Material
Figure S1. Related to Figure 1. | Structural comparison of TFB2M homologues and functional characterization of crystallization construct.
(A) Structural comparison of human TFB2M (marine blue) and S.cerevisiae Mtf1(lightpink) (PDB ID 1I4W) (Schubot et al., 2001). The proteins were aligned over 256 residues with an r.m.s.d of 3.9 Å. While both proteins have a loop insertion between β6 and β7, human TFB2M has an additional loop insertion between β3 and α4. Note that the C-terminal tail is not fully resolved in the structure of Mtf1.
(B) Structural comparison of human TFB2M (marine blue) and M.musculus TFB1M (palegreen) (PDB ID 4GC5) (Guja et al., 2013). The proteins were aligned over 264 residues with an r.m.s.d of 3.9 Å. Compared to TFB2M and Mtf1, the insertion between β6 and β7 is small in TFB1M. Furthermore, TFB1M contains a protruding alpha-helix in the N-terminal region which is not predicted to be present in TFB2M. Note that the C-terminal tail is not fully resolved in the structure of TFB1M and the the N-terminal region is not fully resolved in TFB2M.
(C) Functional characterization of truncations used to obtain a crystallizable TFB2M variant. Replacement of the loop between β6 and β7 (residues 269–293) does not affect transcriptional activity, while removal of 62 amino acids from the N-terminus results in a decrease of activity by 40%, consistent with previous reports that the N-terminal region of TFB2M plays a role in priming RNA synthesis (Sologub et al., 2009).
Figure S2. Related to Figure 2. | Structure determination of the human mitochondrial initiation complex.
(A) Activity of the IC used for structure determination. Transcription initiation activity was assayed using radiolabelled ATP and a LSP DNA template resulting in synthesis of 4- and 5-nt RNA.
(B) Reconstitution of the human mitochondrial transcription initiation complex. Shown is a representative size exclusion chromatogram of the final purification step prior to crystallization. The peak containing the IC is indicated. SDS-PAGE analysis of the peak fraction demonstrates stoichiometric presence of mtRNAP, TFAM and TFB2M. The nucleic acid scaffold which was used to assemble the LSP IC is depicted schematically below with each base represented as a circle and the mismatch bubble around the transcription start site indicated. The template strand is colored blue, the non-template strand cyan.
(C) Positioning of the high-resolution TFB2M crystal structure in the IC structure. The TFB2M structure is shown as ribbon representation with colouring as in Figure 1. Peaks in the anomalous difference Fourier map calculated from crystals containing selenomethionine labelled proteins are shown as green mesh at 3.5 σ. All selenium peak positions coincide with methionine residues in the positioned TFB2M structure.
(D) Confirmation of sequence register assignment for DNA by anomalous diffraction from 5-Bromo-Uracil labeled DNA. The refined model of the IC is shown transparently as ribbon representation with a 2mFo-DFc map around the DNA shown as grey mesh at 1.0. Helices are depicted as cylinders. Anomalous difference Fourier maps are depicted as red mesh at 4.0 σ. Shown are the maps from six different 5-Bromo-Uracil labeled DNA scaffolds (Table S3 and Methods).
(E) Comparison of electron densities for the functionally important α8 helix of TFB2M. The 2mFo-DFc map for the high-resolution TFB2M structure is shown at 1.0 σ (top) and the experimental electron density for the IC is shown at 1.0 σ (bottom).
(F) Structure of the IC assembled using HSP promoter DNA. Important structural elements are indicated and colored as for the LSP IC in Figure 2. The structure of the IC is nearly identical on LSP and HSP promoter DNA.
Figure S3. Related to Figure 4. | Identification of the tether helix and conservation of IC architecture.
(A) Identification of the mtRNAP tether helix. (Left) The 2mFo-DFc electron density for the LSP IC is shown as gray mesh at 1.0 σ with the refined model shown as ribbon representation. An anomalous difference Fourier map calculated from the dataset used for phasing is displayed as green mesh at 3 σ. (Right) Secondary structure prediction for the N-terminal extension region of mtRNAP encompassing the tether helix calculated with PSIPRED (Buchan et al., 2013). Modeling of the predicted alpha helix consisting of mtRNAP residues 122–146 leads to methionine residues at the experimentally determined positions close to the helix (M135, M137 and M146). Selenomethionine-labeling of only mtRNAP in the IC confirms that these peaks originate from methionine residues within mtRNAP. (Methods)
(B) Comparison of DNA binding by TFAM in the LSP IC and HSP IC structure. TFAM is shown as ribbon representation in red, template DNA in blue and non-template DNA in cyan. Anomalous difference Fourier maps calculated from crystals containing selenomethionine labelled TFAM in the LSP or HSP IC, respectively, are shown as green mesh at 5 σ. Two strong selenium peaks were observed in both maps, corresponding to M215 and M222 of TFAM. The comparison demonstrates that TFAM binds to both promoter sequences in identical orientations.
Figure S4. Related to Figure 5. | Structural basis of open DNA stabilization and conservation of basic residues between TFB2M and TFB1M.
(A) Close up view of the interaction of the TFB2M C-terminal domain with the intercalating hairpin of mtRNAP and the DNA. Nucleic acid and protein are depicted as ribbon representation with coloring as in Figure 2. The trajectory of the C-terminal tail of TFB2M (res. 393–396) observed in the TFB2M crystal structure but removed from the IC model is indicated as a dashed orange line. The presumably flexible tail would clash with the intercalating hairpin and the non-template DNA in the conformation observed in the free TFB2M structure.
(B) Sequence alignment of human (Homo sapiens) and mouse (Mus musculus) TFB2M and TFB1M. Shown is only the region between residues 114 and 335 of TFB2M, which contains the basic residues that may interact with nucleic acid in the IC. Identical residues are indicated by white character on red background, similar residues by red character and blue boxes (threshold score 0.7). The residues that may be involved in DNA interactions are labeled and indicated by black character on yellow background. With the exception of K206, K236 and K325, all residues are conserved as positively charged in TFB2M, but not in TFB1M. The alignment was computed using Clustal Omega (Goujon et al., 2010; Sievers et al., 2011) and visualized using ESPript 3.0 (Robert and Gouet, 2014).
Figure S5. Related to Figure 6. | Topological similarity between mtRNAP and T7 RNAP around the point of DNA melting.
(A) Ribbon representation of structural elements of human mtRNAP interacting with the DNA around the point of DNA melting. Colouring as in Figure 2 with base numbering indicated. The intercalating hairpin is positioned with the help of TFB2M to melt the DNA duplex and the specificity loop of mtRNAP runs along the DNA groove between bases −7 to −9.
(B) Ribbon representation of the corresponding structural elements of T7 RNAP interacting with the DNA around the point of DNA melting in the T7 RNAP initiation complex (PDB ID 1CEZ) (Cheetham et al., 1999). Coloring as in (A).
Movie S1. Related to Figure 4, 5 and 7. | Animation of the assembly of the human mitochondrial transcription inititation complex.
Animation showing the proposed model for assembly of the mitochondrial IC. Structures are depicted as ribbon representation with coloring as in Figure 2. Following the assembly of the initiation complex, the complex formation is reversed and repeated again from a different perspective in order to highlight the rearrangements in mtRNAP upon TFB2M binding and DNA opening.
Acknowledgements
We thank past and current members of the Cramer and Temiakov laboratories, in particular K. Schwinghammer, S. Neyer, C. Bernecky, A. Parshin and K. Agaronyan. We thank the crystallization facility at the MPIbpc, in particular J. Wawrzinek. Parts of this work were performed at Beamline X06SA and X10SA at the Swiss Light Source at the Paul-Scherrer-Institut, Villigen, Switzerland, and at PETRA-III, DESY Hamburg, Germany. We thank the staff at EMBL beamline P14 of the Deutsches Elektronensynchrotron (DESY) in Hamburg, Germany, in particular I. Bento, and at beamline PXI and PXII at the Swiss Light Source (SLS) in Villigen, Switzerland. H.S.H. was supported by a Boehringer Ingelheim Fonds PhD fellowship. P.C. was supported by the Deutsche Forschungsgemeinschaft (SFB860, SPP1935), the European Research Council Advanced Investigator Grant TRANSREGULON (grant agreement No 693023), and the Volkswagen Foundation. D.T. was supported by NIH RO1 GM104231.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaronyan K, Morozov YI, Anikin M, and Temiakov D (2015). Replication-transcription switch in human mitochondria. Science 347, 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, and McCammon JA (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.a 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research 42, W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DWA, Minneci F, Nugent TCO, Bryson K, and Jones DT (2013). Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Research 41, W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham GM, and Steitz TA (1999). Structure of a transcribing T7 RNA polymerase initiation complex. Science 286, 2305–2309. [DOI] [PubMed] [Google Scholar]
- Cheetham GM, Jeruzalmi D, and Steitz TA (1999). Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature 399, 80–83. [DOI] [PubMed] [Google Scholar]
- Dairaghi DJ, Shadel GS, and Clayton DA (1995a). Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol 249, 11–28. [DOI] [PubMed] [Google Scholar]
- Dairaghi DJ, Shadel GS, and Clayton DA (1995b). Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta 1271, 127–134. [DOI] [PubMed] [Google Scholar]
- Durniak KJ, Bailey S, and Steitz TA (2008). The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science 322, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C, Gubbey T, Neyer S, Sainsbury S, Oberthuer C, Baejen C, Bernecky C, and Cramer P (2017). Structural Basis of RNA Polymerase I Transcription Initiation. Cell 169, 120–131.e122. [DOI] [PubMed] [Google Scholar]
- Feklistov A, and Darst SA (2011). Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell 147, 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP, Topper JN, and Clayton DA (1987). Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell 50, 247–258. [DOI] [PubMed] [Google Scholar]
- Gaspari M, Falkenberg M, Larsson N-G, and Gustafsson CM (2004). The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. Embo J. 23, 4606–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn ML, Davydova EK, Rothman-Denes LB, and Murakami KS (2008). Structural basis for DNA-hairpin promoter recognition by the bacteriophage N4 virion RNA polymerase. Molecular Cell 32, 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, and Lopez R (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Research 38, W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guja KE, Venkataraman K, Yakubovskaya E, Shi H, Mejia E, Hambardjieva E, Karzai AW, and Garcia-Diaz M (2013). Structural basis for S-adenosylmethionine binding and methyltransferase activity by mitochondrial transcription factor B1. Nucleic Acids Research 41, 7947–7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson CM, Falkenberg M, and Larsson N-G (2016). Maintenance and Expression of Mammalian Mitochondrial DNA. Annu. Rev. Biochem 85, 133–160. [DOI] [PubMed] [Google Scholar]
- Han Y, Yan C, Nguyen THD, Jackobel AJ, Ivanov I, Knutson BA, and He Y (2017). Structural mechanism of ATP-independent transcription initiation by RNA polymerase I. Elife 6, e27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD, and Chamberlin MJ (1988). Structure and function of bacterial sigma factors. Annu. Rev. Biochem 57, 839–872. [DOI] [PubMed] [Google Scholar]
- Hillen HS, Parshin AV, Agaronyan K, Morozov YI, Graber JJ, Chernev A, Schwinghammer K, Urlaub H, Anikin M, Cramer P, et al. (2017). Mechanism of Transcription Anti-termination in Human Mitochondria. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeruzalmi D, and Steitz TA (1998). Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. Embo J. 17, 4101–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, and Steitz TA (1992). Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256, 1783–1790. [DOI] [PubMed] [Google Scholar]
- Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, and Temiakov D (2010). Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem 285, 18129–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, and McMillan FM (2002). SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol 12, 783–793. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev MD, Lesko N, Park CB, Cámara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, and Larsson N-G (2009). Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metabolism 9, 386–397. [DOI] [PubMed] [Google Scholar]
- Morozov YI, and Temiakov D (2016). Human Mitochondrial Transcription Initiation Complexes Have Similar Topology on the Light and Heavy Strand Promoters. J. Biol. Chem 291, 13432–13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov YI, Agaronyan K, Cheung ACM, Anikin M, Cramer P, and Temiakov D (2014). A novel intermediate in transcription initiation by human mitochondrial RNA polymerase. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov YI, Parshin AV, Agaronyan K, Cheung ACM, Anikin M, Cramer P, and Temiakov D (2015). A model for transcription initiation in human mitochondria. Nucleic Acids Research 43, 3726–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, and Darst SA (2003). Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol 13, 31–39. [DOI] [PubMed] [Google Scholar]
- Ngo HB, Kaiser JT, and Chan DC (2011). The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nature Publishing Group; 18, 1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo HB, Lovely GA, Phillips R, and Chan DC (2014). Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Comms 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paratkar S, and Patel SS (2010). Mitochondrial transcription factor Mtf1 traps the unwound non-template strand to facilitate open complex formation. J. Biol. Chem 285, 3949–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posse V, and Gustafsson CM (2016). Human Mitochondrial Transcription Factor B2 is Required for Promoter Melting During Initiation of Transcription. J. Biol. Chem jbc.M116.751008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posse V, Hoberg E, Dierckx A, Shahzad S, Koolmeister C, Larsson N-G, Wilhelmsson LM, Hällberg BM, and Gustafsson CM (2014). The amino terminal extension of mammalian mitochondrial RNA polymerase ensures promoter specific transcription initiation. Nucleic Acids Research 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Basu U, Sultana S, Nandakumar D, and Patel SS (2016). Human mitochondrial transcription factors TFAM and TFB2M work synergistically in promoter melting during transcription initiation. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ, and McCoy AJ (2011). Using SAD data in Phaser. Acta Crystallogr. D Biol. Crystallogr 67, 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, and Temiakov D (2011). Structure of human mitochondrial RNA polymerase. Nature 478, 269–273. [DOI] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cosials A, Sidow JF, Jiménez-Menéndez N, Fernández-Millán P, Montoya J, Jacobs HT, Coll M, Bernadó P, and Solà M (2011). Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol 18, 1281–1289. [DOI] [PubMed] [Google Scholar]
- Sadian Y, Tafur L, Kosinski J, Jakobi AJ, Wetzel R, Buczak K, Hagen WJ, Beck M, Sachse C, and Müller CW (2017). Structural insights into transcription initiation by yeast RNA polymerase I. Embo J. 36, 2698–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubot FD, Chen CJ, Rose JP, Dailey TA, Dailey HA, and Wang BC (2001). Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription. Protein Sci. 10, 1980–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinghammer K, Cheung ACM, Morozov YI, Agaronyan K, Temiakov D, and Cramer P (2013). Structure of human mitochondrial RNA polymerase elongation complex. Nat. Struct. Mol. Biol 20, 1298–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sologub M, Litonin D, Anikin M, Mustaev A, and Temiakov D (2009). TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell 139, 934–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, and Yokoyama S (2002). Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature 420, 43–50. [DOI] [PubMed] [Google Scholar]
- Uchida A, Murugesapillai D, Kastner M, Wang Y, Lodeiro MF, Prabhakar S, Oliver GV, Arnold JJ, Maher LJ, Williams MC, et al. (2017). Unexpected sequences and structures of mtDNA required for efficient transcription from the first heavy-strand promoter. Elife 6, e27283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YW, and Steitz TA (2002). Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 298, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, and Ebright RH (2012). Structural Basis of Transcription Initiation. Science 338, 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1. | Structural comparison of TFB2M homologues and functional characterization of crystallization construct.
(A) Structural comparison of human TFB2M (marine blue) and S.cerevisiae Mtf1(lightpink) (PDB ID 1I4W) (Schubot et al., 2001). The proteins were aligned over 256 residues with an r.m.s.d of 3.9 Å. While both proteins have a loop insertion between β6 and β7, human TFB2M has an additional loop insertion between β3 and α4. Note that the C-terminal tail is not fully resolved in the structure of Mtf1.
(B) Structural comparison of human TFB2M (marine blue) and M.musculus TFB1M (palegreen) (PDB ID 4GC5) (Guja et al., 2013). The proteins were aligned over 264 residues with an r.m.s.d of 3.9 Å. Compared to TFB2M and Mtf1, the insertion between β6 and β7 is small in TFB1M. Furthermore, TFB1M contains a protruding alpha-helix in the N-terminal region which is not predicted to be present in TFB2M. Note that the C-terminal tail is not fully resolved in the structure of TFB1M and the the N-terminal region is not fully resolved in TFB2M.
(C) Functional characterization of truncations used to obtain a crystallizable TFB2M variant. Replacement of the loop between β6 and β7 (residues 269–293) does not affect transcriptional activity, while removal of 62 amino acids from the N-terminus results in a decrease of activity by 40%, consistent with previous reports that the N-terminal region of TFB2M plays a role in priming RNA synthesis (Sologub et al., 2009).
Figure S2. Related to Figure 2. | Structure determination of the human mitochondrial initiation complex.
(A) Activity of the IC used for structure determination. Transcription initiation activity was assayed using radiolabelled ATP and a LSP DNA template resulting in synthesis of 4- and 5-nt RNA.
(B) Reconstitution of the human mitochondrial transcription initiation complex. Shown is a representative size exclusion chromatogram of the final purification step prior to crystallization. The peak containing the IC is indicated. SDS-PAGE analysis of the peak fraction demonstrates stoichiometric presence of mtRNAP, TFAM and TFB2M. The nucleic acid scaffold which was used to assemble the LSP IC is depicted schematically below with each base represented as a circle and the mismatch bubble around the transcription start site indicated. The template strand is colored blue, the non-template strand cyan.
(C) Positioning of the high-resolution TFB2M crystal structure in the IC structure. The TFB2M structure is shown as ribbon representation with colouring as in Figure 1. Peaks in the anomalous difference Fourier map calculated from crystals containing selenomethionine labelled proteins are shown as green mesh at 3.5 σ. All selenium peak positions coincide with methionine residues in the positioned TFB2M structure.
(D) Confirmation of sequence register assignment for DNA by anomalous diffraction from 5-Bromo-Uracil labeled DNA. The refined model of the IC is shown transparently as ribbon representation with a 2mFo-DFc map around the DNA shown as grey mesh at 1.0. Helices are depicted as cylinders. Anomalous difference Fourier maps are depicted as red mesh at 4.0 σ. Shown are the maps from six different 5-Bromo-Uracil labeled DNA scaffolds (Table S3 and Methods).
(E) Comparison of electron densities for the functionally important α8 helix of TFB2M. The 2mFo-DFc map for the high-resolution TFB2M structure is shown at 1.0 σ (top) and the experimental electron density for the IC is shown at 1.0 σ (bottom).
(F) Structure of the IC assembled using HSP promoter DNA. Important structural elements are indicated and colored as for the LSP IC in Figure 2. The structure of the IC is nearly identical on LSP and HSP promoter DNA.
Figure S3. Related to Figure 4. | Identification of the tether helix and conservation of IC architecture.
(A) Identification of the mtRNAP tether helix. (Left) The 2mFo-DFc electron density for the LSP IC is shown as gray mesh at 1.0 σ with the refined model shown as ribbon representation. An anomalous difference Fourier map calculated from the dataset used for phasing is displayed as green mesh at 3 σ. (Right) Secondary structure prediction for the N-terminal extension region of mtRNAP encompassing the tether helix calculated with PSIPRED (Buchan et al., 2013). Modeling of the predicted alpha helix consisting of mtRNAP residues 122–146 leads to methionine residues at the experimentally determined positions close to the helix (M135, M137 and M146). Selenomethionine-labeling of only mtRNAP in the IC confirms that these peaks originate from methionine residues within mtRNAP. (Methods)
(B) Comparison of DNA binding by TFAM in the LSP IC and HSP IC structure. TFAM is shown as ribbon representation in red, template DNA in blue and non-template DNA in cyan. Anomalous difference Fourier maps calculated from crystals containing selenomethionine labelled TFAM in the LSP or HSP IC, respectively, are shown as green mesh at 5 σ. Two strong selenium peaks were observed in both maps, corresponding to M215 and M222 of TFAM. The comparison demonstrates that TFAM binds to both promoter sequences in identical orientations.
Figure S4. Related to Figure 5. | Structural basis of open DNA stabilization and conservation of basic residues between TFB2M and TFB1M.
(A) Close up view of the interaction of the TFB2M C-terminal domain with the intercalating hairpin of mtRNAP and the DNA. Nucleic acid and protein are depicted as ribbon representation with coloring as in Figure 2. The trajectory of the C-terminal tail of TFB2M (res. 393–396) observed in the TFB2M crystal structure but removed from the IC model is indicated as a dashed orange line. The presumably flexible tail would clash with the intercalating hairpin and the non-template DNA in the conformation observed in the free TFB2M structure.
(B) Sequence alignment of human (Homo sapiens) and mouse (Mus musculus) TFB2M and TFB1M. Shown is only the region between residues 114 and 335 of TFB2M, which contains the basic residues that may interact with nucleic acid in the IC. Identical residues are indicated by white character on red background, similar residues by red character and blue boxes (threshold score 0.7). The residues that may be involved in DNA interactions are labeled and indicated by black character on yellow background. With the exception of K206, K236 and K325, all residues are conserved as positively charged in TFB2M, but not in TFB1M. The alignment was computed using Clustal Omega (Goujon et al., 2010; Sievers et al., 2011) and visualized using ESPript 3.0 (Robert and Gouet, 2014).
Figure S5. Related to Figure 6. | Topological similarity between mtRNAP and T7 RNAP around the point of DNA melting.
(A) Ribbon representation of structural elements of human mtRNAP interacting with the DNA around the point of DNA melting. Colouring as in Figure 2 with base numbering indicated. The intercalating hairpin is positioned with the help of TFB2M to melt the DNA duplex and the specificity loop of mtRNAP runs along the DNA groove between bases −7 to −9.
(B) Ribbon representation of the corresponding structural elements of T7 RNAP interacting with the DNA around the point of DNA melting in the T7 RNAP initiation complex (PDB ID 1CEZ) (Cheetham et al., 1999). Coloring as in (A).
Movie S1. Related to Figure 4, 5 and 7. | Animation of the assembly of the human mitochondrial transcription inititation complex.
Animation showing the proposed model for assembly of the mitochondrial IC. Structures are depicted as ribbon representation with coloring as in Figure 2. Following the assembly of the initiation complex, the complex formation is reversed and repeated again from a different perspective in order to highlight the rearrangements in mtRNAP upon TFB2M binding and DNA opening.
Data Availability Statement
Coordinate and structure factor files for the LSP and HSP IC crystal structures and for the human TFB2M crystal structure were deposited with the Protein Data Bank with accession codes 6ERP, 6ERQ and 6ERO, respectively.