Abstract
Mitochondria make functionally relevant contacts with most, if not all, other organelles in the cell. These contacts impact mitochondrial behavior and function as well as a wide variety of cellular functions. Many recent advances have been made in the rapidly growing field of mitochondria contact site biology, and these advances have expanded the known functions of mitochondria contact sites in exciting and unexpected ways.
Introduction
Organelles perform distinct, essential functions in the cell. While once thought to operate independently, it is clear that organelles contact other organelles and these contacts are critical for organelle function and overall cellular homeostasis [1–3], Contact is achieved through proteins that function to directly tether organelles. While some proteins solely serve to physically bridge organelles, others actively participate in interorganelle communication and function. Importantly, the proteins that localize to contact sites establish, maintain, and functionally alter contacts in response to different physiological contexts and impact a wide variety of fundamental cellular functions.
The vast extent to which organelles contact one another has become increasingly evident. While organelle contacts were first visualized by electron microscopy six decades ago [4,5], advances in the ability to visualize organelles and identify the proteins that localize to sites of contact as well as manipulate these contacts have greatly advanced the study of contact site formation and function. Exciting, cutting-edge imaging techniques now allow for the visualization of multiple organelles simultaneously over time with increased spatial and temporal resolution. Thus, the sheer number, extent, and spatiotemporal dynamics of interorganelle contacts within cells can be more fully appreciated. Recent work demonstrates that most organelles make functionally relevant contact, with most, if not all, other organelles in the cell, with functionally relevant contact referring to a tether-mediated contact that impacts the behavior or function of one or both organelles. Here, the focus will be on contacts made between mitochondria and other organelles—specifically, on recent advances that highlight the expanding and unexpected functions of mitochondria-organelle contacts.
The many organelles that mitochondria contact
Contact between mitochondria and the endoplasmic reticulum (ER) has been visually appreciated since the 1950s and functionally appreciated since the 1990s [4–7], Excitingly, over the past decade, contacts between mitochondria and many additional organelles have been identified (Fig. 1). In addition to the ER, mitochondria contact vacuoles/lysosomes, peroxisomes, lipid droplets, endosomes, the Golgi, the plasma membrane (PM), and melanosomes. The number of contacts mitochondria make with a specific organelle can vary dramatically from just a few contacts to hundreds per cell [8]. These contacts are often mediated by multiple, distinct tethering complexes. While these tethering complexes often share some functional overlap, it is clear, as discussed below, that distinct complexes have unique, non-overlapping functions and are differentially regulated in specific biological contexts. In addition, the molecular mechanisms mediating contact and the dynamics and duration of contact can vary dramatically. For example, contacts between mitochondria and lipid droplets can range from tens to hundreds of seconds in Cos7 cells and a mitochondria-PM contact in budding yeast can persist for greater than 45 minutes [8,9]. Thus, not all mitochondria-organelle contacts are created equal; mitochondrial contacts exhibit functional, architectural, and dynamic differences and differentially impact many aspects of mitochondrial behavior and function.
Figure 1. Mitochondria-organelle contacts and functions.
Mitochondria make functionally relevant contact with many organelles in the cell. A subset of mitochondria-organelle contacts and the functions ascribed to these contacts are shown. See text for details.
Mitochondria-organelle contacts and mitochondrial dynamics
Mitochondria form highly elaborate, dynamic, reticular networks in many cell types. The structure and dynamics of the mitochondrial network are, in part, maintained by division and fusion of the organelle [10,11]. Early investigations into the activities of mitochondrial division and fusion were, not surprisingly, approached from a highly mitochondria-centric perspective. However, the finding that sites of mitochondrial division are spatially linked to ER-mitochondria contact sites rapidly changed that view. In both yeast and mammalian cells, the ER is present at the vast majority of mitochondrial division events [12]. The ER-Mitochondria Encounter Structure (ERMES) mediates contact between mitochondria and the ER at sites of mitochondrial division in yeast [13,14], and the identity of the functionally equivalent tether in higher eukaryotes has yet to be determined. ER contact at nascent division sites is associated with the initial constriction of mitochondria and precedes the recruitment of the dynamin related proteins that drive further constriction and scission of the mitochondrial membranes, Dnm1/Drp1 and Dyn2 [11,12,15]. In mammals, the ER-localized formin INF2 has been proposed to stimulate actin polymerization at nascent division sites [16]. Activation of INF2 at sites of ER-mitochondria contact is thought to be mediated by an interaction with the mitochondrial anchored actin nucleating protein, Spire1C [17]. Actin polymerization has been proposed to facilitate mitochondrial division in multiple ways: providing a force generating system to drive the initial constriction of mitochondria, functioning in the direct recruitment of Drp1 to the site of contact, and enhancing ER-mitochondria contact and consequently ER-to-mitochondria calcium transfer leading to constriction of the mitochondrial inner membrane [16,18,19]. Thus, ER-mitochondria contact impacts mechanisms that influence the structure of both the outer and inner mitochondrial membranes during division.
Interestingly, ER-mitochondria contacts at sites of division are spatially linked to nucleoids, complexes of mitochondrial DNA (mtDNA) and associated proteins. Studies in yeast first demonstrated a link between ER-associated mitochondrial division and the maintenance and distribution of nucleoids [14], which has more recently been described in mammalian cells [20]. Specifically, the small subset of ER-mitochondria contact sites that go on to divide are spatially and temporally linked to actively replicating nucleoids. The coupling of ER-associated mitochondrial division with mtDNA replication is proposed to serve as a mechanism to distribute newly replicated mtDNA to daughter mitochondria and facilitate the accurate distribution of nucleoids in cells [14,20]. The identity of factors that physically and functionally coordinate mtDNA replication, which occurs in the mitochondrial matrix, with ER-mitochondria contacts and the division machine on the mitochondrial surface are at this point unknown.
A recent study has added yet another unexpected player to the process of mitochondrial division, the lysosome, as sites of mitochondria-lysosome contact have also been spatially linked to sites of mitochondrial division [21]. Lysosomal GTP-bound Rab7 has been shown to be involved in the formation and stabilization of mitochondria-lysosome contacts, which are subsequently destabilized by TBC1D15, a RAB7 GAP that is recruited to mitochondria by the outer mitochondrial membrane protein Fis1. The ER and Drp1 are present at sites of lysosome- marked mitochondrial division raising the question of how contact between mitochondria and multiple organelles at sites of division is regulated and integrated.
The connection between mitochondria-ER contacts and mitochondrial dynamics might extend beyond division. In a recent study using grazing incidence structured illumination microscopy (GI-SIM) to examine ER and mitochondrial dynamics with increased spatial and temporal resolution over standard SIM techniques, the ER was found to be present at over half of the mitochondrial fusion events observed [22]. In comparison to fusion events not associated with the ER, the duration of fusion events from initial contact to completion of fusion were shorter for fusion events associated with the ER. While the molecular basis and functional contributions of ER-mitochondria contact at sites of fusion have yet to be determined, it is clear that ER-mitochondria contacts are intimately connected to the division and fusion dynamics of the mitochondrial network.
Mitochondria-organelle contacts and organelle distribution
In addition to regulating the cellular distribution of mitochondria by impacting mitochondrial dynamics, mitochondria contact sites can also more directly impact the position of mitochondria in cells. In yeast, the Mitochondria-ER-Cortex-Anchor (MECA) tethers mitochondria to the PM and cortical ER, bringing three cellular membranes in close proximity [23], The core protein component of MECA, Num1, interacts directly with the mitochondrial membrane and PM; the molecular basis for the interaction between Num1 and the ER is poorly understood [24–27]. Num1 forms stable contacts between mitochondria and the cell cortex that persist for extended periods of time [9], and the tethering activity of Num1 is required for proper mitochondrial distribution and inheritance during budding yeast mitosis [23,28,29]. Interestingly, the mitochondria-PM contacts sites formed by Num1 also serve as cortical attachment sites for dynein [9], where, once anchored, dynein captures and walks along astral microtubules to help orient the mitotic spindle [30,31], When mitochondrial inheritance is inhibited, Num1-mediated mitochondria-PM contacts are not formed in buds and defects in dynein mediated spindle positioning are observed [9,32]. Thus, Num1-mediated mitochondria-PM tethering not only impacts the spatial distribution of mitochondria within cells but also when and where dynein anchoring occurs and, consequently, the function of dynein in spindle orientation. This surprising connection between a mitochondria-PM contact site and dynein anchoring was recently shown to be conserved in the evolutionarily distant fission yeast S. pombe [33]. The functional and physiological significance of why mitochondria-PM contacts serve as cortical attachment sites for dynein has yet to be determined.
While MECA-mediated tethering is stable in mitotic cells, a recent study indicates that the protein components of MECA, Num1 and Mdm36, are degraded during meiosis, specifically anaphase II, which results in the release of mitochondria from the cell cortex [34]. This programmed release of mitochondria is proposed to be important for subsequent incorporation of mitochondria into spores. Interestingly, after being released from the cell cortex, mitochondria become closely opposed to the nuclear envelope, raising the possibility that a yet to be determined ER-mitochondria tethering complex is upregulated in meiosis to mediate this extensive contact. Thus, the regulation of at least two mitochondria contact sites facilitates mitochondrial repositioning during the meiotic developmental program.
A role for mitochondria-PM tethering in development and differentiation, more specifically stem cell fate determination, has been recently reported for mammary stem cells [35]. During the asymmetric division of a mammary stem cell, highly fused mitochondria are tethered to the PM by a complex between Mfn1, an outer mitochondrial membrane fusion protein, and PCKζ, a cortically anchored kinase. The tethered mitochondria, which have enhanced reactive oxygen species scavenging capacity, are asymmetrically inherited by the daughter cell that retains stem cell identity. Even in simple asymmetrically dividing budding yeast cells, Mmr1-mediated ER-mitochondria tethering has been implicated in the asymmetric inheritance and retention of higher functioning mitochondria by daughter cells [36,37]. While we are still in the early stages of understanding the full mechanistic and functional contributions of mitochondria-organelle contacts to the function-dependent distribution of mitochondria in asymmetrically dividing cells, mitochondria contact sites will play pivotal roles in development and differentiation.
Mitochondria-organelle contacts and molecular transport
In addition to influencing the dynamics and distribution of mitochondria, contacts between mitochondria and other organelles have been implicated in the interorganelle exchange of Ca2+, lipid, and various metabolites. The role of ER-mitochondria contacts in Ca2+ transfer has been appreciated and well-studied for some time [6]. Ca2+ transfer between the two organelles impacts mitochondrial bioenergetics, metabolism, and dynamics as well as cell death and autophagy, and perturbations in Ca2+ transfer are associated with many disease states. A discussion of the many players involved in and the functions of Ca2+ transfer between the ER and mitochondria warrants its own reviews; please see [38–42]. The transfer of another metal, iron, has also been associated with mitochondria contacts. Kiss-and-run contacts between mitochondria and endosomes have been implicated in iron transfer between the two organelles [43–45]. While the molecular basis for mitochondria-endosome contacts is unclear, the duration of contact between the two organelles is regulated by intraendosomal iron, suggesting that the contact is influenced by endosomal cargo.
A role for mitochondria contacts in interorganelle lipid transport has also been appreciated for decades [6,7], and several exciting advances have recently been made in our understanding of the molecular bases and mechanisms of transport. Multiple proteins that mediate mitochondria-organelle contact contain lipid binding and transport domains. For example, three of the four proteins in the ER-mitochondria tether ERMES (Mmm1, Mdm12, and Mdm34) contain a synaptotagmin-like-mitochondrial-lipid binding protein (SMP) domain [13,46,47]. SMP domains are conserved lipid binding domains found in proteins that reside at membrane contact sites [48,49]. Mmm1 and Mdm12 have been shown to bind phospholipids in vitro, and the structure of the Mmm1 and Mdm12 complex indicates that the proteins come together to form an extended, continuous hydrophobic channel that likely facilitates phospholipid transport [50–52]. Cells that lack ERMES exhibit defects in phospholipid transport between the ER and mitochondria [13]. Thus, similar to many proteins containing SMP domains, ERMES components serve to tether as well as facilitate transfer of phospholipids between two organelle membranes. While a functional ortholog of the entire ERMES complex has yet to be identified, the SMP domain protein PDZD8 has been recently suggested to be a functional ortholog, or possible paralog, of the ERMES component Mmm1 in metazoans [53,54], PDZD8 mediates ER-mitochondria contact, and PDZD8-mediated contact is proposed to facilitate ER-to-mitochondria Ca2+ transfer. A role for PDZD8 in ER-associated mitochondrial division or lipid transfer between the two organelles has yet to be examined.
Vps13 has been suggested to fulfill some of the functions of the ERMES complex in yeast and metazoans [55–58]. In yeast, Vps13 localizes to multiple interorganelle contact sites, including sites of mitochondria-vacuole contact [56–58], and its closest homolog in metazoans, VPS13A, localizes to ER-mitochondria contacts [55]. Vps13 has phospholipid binding and transfer activities in vitro and, based on the crystal structure of the N-terminal region of Vps13 from Chaetomium thermophilum, the protein has a hydrophobic cavity large enough to accommodate several lipid molecules simultaneously [55]. Consistent with the possibility that Vps13 may serve some ERMES-like functions, Vps13 is required in the absence of ERMES and dominant mutations in Vps13 have been identified as suppressors of ERMES deficiency in yeast [56,58].
The ER-membrane protein complex (EMC) is yet another protein complex in yeast that has been localized to ER-mitochondria contacts and implicated in phospholipid transfer between the two organelles [59]. While the EMC, a complex of 6 proteins (Emc1-6) that has been implicated in ER protein folding [60], is found throughout the ER, a fraction of the complex interacts with Tom5, a component of the translocase of the mitochondrial outer membrane. Cells lacking the EMC exhibit a reduction in ER-mitochondria tethering and in the transfer of phospholipids from the ER to mitochondria [59]. The proportion of the EMC that interacts with Tom5 colocalizes with ERMES; however, data suggest that the EMC and ERMES function independently in their tethering and lipid transport roles. Dissecting the contact site specific functions of the EMC are complicated by the fact that the EMC has been shown to be recently involved in the biogenesis of multipass transmembrane proteins, and thus, could be indirectly affecting contact site formation and function [61–63].
In addition to contacts mediating transfer between the ER and mitochondria, mitochondria-lipid droplet and mitochondria-peroxisomes contacts have also been implicated in interorganelle transport. In metazoans, mitochondria-lipid droplet contacts have been shown to participate in both the breakdown and synthesis of fatty acids. During starvation, fatty acids are transferred via direct contact from lipid droplets to mitochondria, where the fatty acids are subsequently metabolized to drive mitochondrial ATP production [64]. Conversely, in brown adipocytes, mitochondria recruited to lipid droplets by Perilipin5 provide ATP to fuel fatty acid synthesis and lipid droplet expansion [65,66]. For mitochondria-peroxisome contacts, Pex34, a peroxisomal protein [67], and Fzo1, a dynamin related GTPase that drives fusion of the mitochondrial outer membrane [68], have recently been identified as mitochondria-peroxisome tethering proteins in yeast [69]. Pex34 is anchored in the peroxisomal membrane, and the molecular basis for the interaction with mitochondria is unknown. While Fzo1 is known to be anchored in the mitochondrial outer membrane, recent evidence suggests that the protein may also localize to the peroxisomal membrane [69]. Thus, Fzo1-Fzo1 interactions may mediate mitochondria-peroxisome tethering similar to the mitochondria-mitochondria tethering role for Fzo1 in outer mitochondrial membrane fusion but without the subsequent membrane fusion event [70]. Interestingly, Pex34-mediated, but not Fzo1-mediated, mitochondria-peroxisome contacts are functionally linked to the transfer of β-oxidation products from peroxisomes to mitochondria [69], highlighting that not all contact sites are created equal.
Mitochondria-organelle contacts and cellular metabolism and homeostasis
The formation and function of many mitochondria-organelle contacts are integrated with cellular metabolism and homeostasis. vCLAMP, for vacuole-mitochondria patch, tethers mitochondria to the vacuole in yeast, and vCLAMP formation is integrated with cellular metabolism as vCLAMPs are decreased in respiratory conditions [71,72]. A core component of vCLAMP is the HOPS (homotypic fusion and vacuole sorting complex) tethering complex subunit Vps39. As a component of HOPS, Vsp39 functions in the endolysosomal fusion pathway [73]. Interestingly, Vps39 is the only component of the HOPS complex that is required for vCLAMP, and it is unclear whether Vps39 localizes to vCLAMP alone or together with the HOPS complex [71,72]. Functional studies on vCLAMP were initially plagued by a problem common to any contact site composed of a protein that also functions outside the context of the contact site; it is difficult to discern whether the phenotypes observed in the absence of the protein are due to loss of contact site activity or the canonical function of the protein. For vCLAMP, this difficulty was overcome by the identification of Vps39 separation of function mutants, which are defective for vCLAMP formation but not for HOPS complex function [74]. Previous work had demonstrated that Vps39 function is critical in the absence of ERMES [71,72]. While the loss of Vps39 function in vCLAMP was thought to underlie the synthetic interaction observed, the vCLAMP-impaired Vps39 mutants were elegantly used to demonstrate that the role of Vps39 in HOPS-dependent trafficking to the vacuole is what becomes critical in the absence of ERMES. This unexpected result, in combination with additional studies using vesicular trafficking mutants, suggests that HOPS function and, more generally, trafficking to the vacuole are critical in ERMES deficient cells [74]. While not critical in the absence of ERMES, the vCLAMP specific functions of Vps39 impact specific cellular stress response pathways and survival during starvation; however, the underlying functional basis for these phenotypes has yet to be determined.
Unlike vCLAMP-mediated mitochondria-vacuole contact, mitochondria-vacuole contact mediated by Vps13 is critical in the absence of ERMES [56–58], highlighting that the mitochondria-vacuole contact mediated by vCLAMP and Vps13 are functionally distinct. Interestingly, however, Vps39 and other components of the vesicular trafficking pathway are required for the ability of Vps13 dominant mutants to suppress defects associated with ERMES deficiency [74]. Thus, transport to the vacuole is required for the Vps13-mediated ERMES bypass. These findings suggest the vacuole is likely acquiring a factor via vesicular transport that is required for the ERMES bypass, and transport of this factor, potentially a phospholipid, from the vacuole to mitochondria may be facilitated by Vps13.
Lam6, also known as Ltc1, localizes to vCLAMP as well as ER-mitochondria and ER-vacuole contacts [75,76]. Lam6 belongs to a StART/VASt domain family of sterol transporting proteins [77]. Indeed, Lam6 has been shown to transport sterols in vitro [76,78]. Lam6 is required for cell viability in the absence of ERMES and is suggested to function in the regulation of local membrane lipid composition perhaps via the regulation of sterols on mitochondria [75,76]. While the function of Lam6 at ER-mitochondria contacts has yet to be fully defined, Lam6 functions at ER-vacuole contacts to create sterol-enriched vacuole membrane domains that regulate TORC1 signaling by spatially segregating TORC1 regulators [79]. Given that Lam6 localizes to multiple contact sites in the cell, it has been proposed that coordinate regulation of the relative levels of Lam6 at each contact may regulate an organelle interaction network that integrates organelle communication and function with cellular metabolism and homeostasis [75,79]. Consistent with this idea, the organelle specific receptors for Lam6 are involved in many aspects of organelle biogenesis and function. Lam6 is recruited to mitochondria by Tom70/71, mitochondrial preprotein import receptors, and to the vacuole by Vac8, which has functions in vacuole transport, autophagy, and nucleus-vacuole junction formation [76,80–82]. If and how Lam6 affects the canonical functions of its organelle receptors and integrates these critical functions with contact site formation and function are outstanding questions.
ER-mitochondria contacts are also intimately tied to cellular homeostasis via their impact on autophagy. Mitophagosomes in yeast and autophagosome in metazoans have been shown to form at ER-mitochondria contacts, which have been suggested to be a source of phospholipid for the forming autophagosome. Disrupting ER-mitochondria contacts, using mutants of ERMES in yeast and by depleting Mfn2 or PACS2 in metazoans, results in decreased formation of autophagic vesicles [83–85]. However, once again highlighting the fact that not all contacts are created equal, decreasing ER-mitochondria contact mediated by the VAPB-PTPIP51 tethering complex in metazoans induces basal autophagy, while increasing VAPB-PTPIP51-mediated contact impairs basal autophagy [86,87]. The effects of VAPB-PTPIP51 on autophagy are likely due to the role the tethering complex plays in the regulation of Ca2+ transfer between the two organelles. Further adding to the complexity, the effects of ER-mitochondria contacts on autophagy vary depending on the autophagic stimulus [87]. Thus, an important yet difficult challenge will be to tease apart the direct and indirect contributions of distinct mitochondria contacts to the mechanisms and regulation of various autophagic pathways that differ in terms of the stimulus received and intended cargo to be cleared.
Shared components of mitochondria contact sites
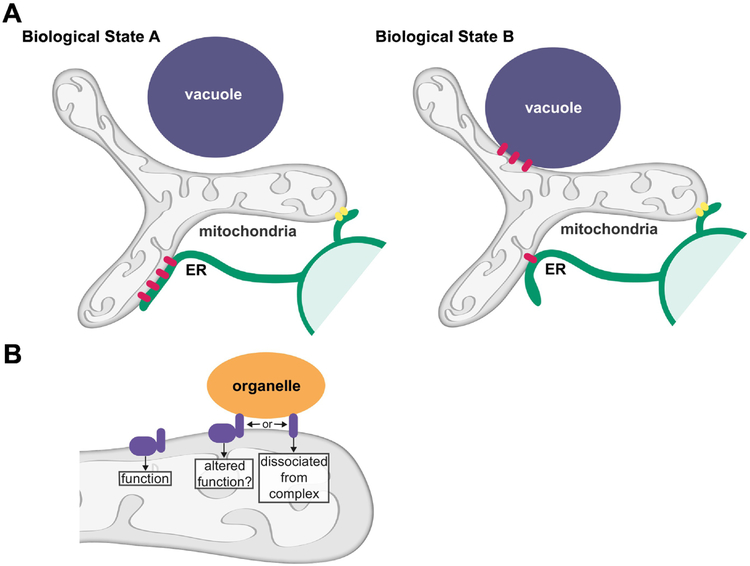
As more proteins that mediate or localize to sites of interorganelle contact are identified, it has become increasingly clear that contact site proteins can localize to more than one site of contact. Thus, a challenge going forward is to understand how the localization of a protein to a specific contact site is determined in specific biological circumstances (Fig. 2A). Changes in nutrient status have been shown to affect the localization of contact site proteins. In glucose conditions, Vps13 localizes to mitochondria-vacuole and mitochondria-endosome contacts, and in respiratory conditions, Vps13 relocalizes to the nucleus-vacuole junction [56,57]. Organelle specific adaptors recruit Vps13 to specific membranes; Vps13 is recruited to mitochondria by Mcp1 and to endosomes and vacuoles by Ypt35 [58,88], These organelle specific adaptors share a related motif, termed the Vps13 adaptor binding (VAB) domain, that mediates the interaction with Vps13 [88]. It has been suggested the adaptors compete for Vps13 binding and changes in the level of a specific adaptor and/or the accessibility or modification state of its VAB domain are used to control Vps13 localization in specific cellular circumstances. Perhaps similar mechanisms are used to regulate the cellular localization of Lam6 and other proteins that localize to multiple sites of contact.
Figure 2. Shared components of mitochondria-contact sites.
A) The biological state of the cell can impact the extent of mitochondria-organelle contacts as well as the localization of contact site proteins. Many mitochondria contact site proteins localize to multiple, distinct contact sites, and the localization of a protein to a specific contact site may differ in specific biological circumstances. In the example shown in A, a mitochondria contact site protein (shown in pink) primarily localizes to and mediates ER-mitochondria contact in Biological State A and primarily localizes to and mediates mitochondria-vacuole contact in Biological State B. The localization of a distinct mitochondria contact site protein (shown in yellow) is not altered between Biological States A and B.
B) Many mitochondrial contact site proteins have well studied functions in other critical cellular processes. The localization of such proteins to a contact site may impact the canonical functions of that protein. For proteins that canonically function within a larger complex, it is not clear if the protein, when present at mitochondria contacts, remains with the complex or dissociates.
Adding to the complexity of mitochondria contact site biology is the increasing number of proteins found at mitochondrial contact sites that have well studied functions in other critical cellular processes (Fig. 2B). For numerous proteins that localize to mitochondria-organelle contacts, the mitochondrial receptor is a member of the outer membrane import machinery; Lam6 interacts with Tom70/71, EMC with Tom5, and Vps39 with Tom40 [59,74,76]. In addition, the ERMES component Mdm10 functions in the biogenesis of mitochondrial outer membrane beta barrel proteins [80]. Other mitochondria contact site proteins have canonical functions in membrane trafficking and remodeling pathways. The vCLAMP component Vps39 functions in endolysosomal trafficking as a member of the HOPS complex [73]. and proteins that drive outer mitochondrial membrane fusion, Fzo1/MFN1 and MFN2 (yeast/mammals) [70]. also mediate mitochondria-organelle contacts. Fzo1 functions in mitochondria-peroxisome tethering [69], while its mammalian homologs have been implicated in contacts between mitochondria and the ER, melanosomes, and lipid droplets in the case of Mfn2 and mitochondria and the PM in the case of Mfn1 [35,89–92]. It is unclear in most, if not all, of these circumstances if and how the canonical functions of the proteins impact or are impacted by contact site formation and function (Fig. 2B). In addition, for proteins that canonically function within a larger complex, it is not clear if the protein, when present at mitochondria contacts, remains with the complex or dissociates. Furthermore, proteins that function in cis within their resident membrane may also act in trans when present at a contact, similar to the resident ER protein Sac1 and its ability to modify PM lipids in trans at contact sites [93]. Going forward, it will be exciting to determine how localization to a contact site impacts the canonical activities of proteins and integrates these activities with contact site function to coordinately regulate organelle biogenesis and behavior.
Concluding Remarks
The contacts mitochondria make with other organelles play critical roles in many aspects of mitochondrial biology and behavior. Furthermore, the functions of mitochondria-organelle contact extend beyond the mitochondrion itself and impact a larger interorganelle communication network and, consequently, a wide variety of cellular functions. While great progress in our understanding of the formation and function of many mitochondria contacts sites has been made, we still have a far way to go before we are able to paint a complete picture of the molecular basis, function, and regulation of all mitochondria-organelle contacts (see Outstanding Questions). Just understanding each contact in isolation, however, will not be enough. It will be critical to understand how distinct mitochondria contacts communicate with one another and with other interorganelle contacts. As our knowledge of interorganelle contact site biology rapidly advances, a complex, dynamic, and interdependent organelle interaction network exists in cells and this network responds to cellular needs. Therefore, the exciting and unexpected knowledge gained about the biology of mitochondria-organelle contacts must be placed into the larger context of the complete organelle interaction network of the cell.
Outstanding questions.
How many distinct mitochondria-organelle contacts exist?
What functions of mitochondrial-organelle contacts have yet to be discovered?
How do distinct mitochondrial contacts communicate with one another and with other interorganelle contacts?
For mitochondrial contact site proteins that have well studied functions in other critical cellular processes, how does the localization of the protein to a contact site impact the canonical activities of that protein and integrate those activities with contact site function?
How can we place the knowledge gained about the biology of mitochondria-organelle contacts into the larger context of the complete organelle interaction network of the cell?
Highlights.
Mitochondria make functionally relevant contacts with most, if not all, other organelles in the cell.
Mitochondria-organelle contacts impact many aspects of mitochondrial behavior and function.
The functions of mitochondria-organelle contact extend beyond the mitochondrion itself and impact an interorganelle communication network and, consequently, a wide variety of cellular functions.
Many mitochondrial contact site proteins localize to multiple contact sites.
Many mitochondrial contact site proteins function in other critical cellular processes, such as protein import, vesicular trafficking, and membrane remodeling pathways. It is unclear how these canonical functions are integrated with contact site function.
Mitochondria-organelle contacts are part of a complex and dynamic organelle interaction network that integrates organelle communication and function with cellular metabolism and homeostasis.
Acknowledgments
I would like to thank members of the Lackner Lab for helpful discussions. I would like to apologize to colleagues whose outstanding work on mitochondria-organelle contacts could not be included due to space limitations. This work is supported by the NIH NIGMS grant R01GM120303 to L.L.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eisenberg-Bord M et al. (2016) A Tether Is a Tether Is a Tether: Tethering at Membrane Contact Sites. Dev Cell 39, 395–409 [DOI] [PubMed] [Google Scholar]
- 2.Gatta AT and Levine TP (2017) Piecing Together the Patchwork of Contact Sites. Trends Cell Biol 27, 214–229 [DOI] [PubMed] [Google Scholar]
- 3.Prinz WA (2014) Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J Cell Biol 205, 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard W and Rouiller C (1956) Close topographical relationship between mitochondria and ergastoplasm of liver cells in a definite phase of cellular activity. J Biophys Biochem Cytol 2, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copeland DE and Dalton AJ (1959) An association between mitochondria and the endoplasmic reticulum in cells of the pseudobranch gland of a teleost. J Biophys Biochem Cytol 5, 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizzuto R et al. (1998) Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766 [DOI] [PubMed] [Google Scholar]
- 7.Vance JE (1990) Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem 265, 7248–7256 [PubMed] [Google Scholar]
- 8.Valm AM et al. (2017) Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraft LM and Lackner LL (2017) Mitochondria-driven assembly of a cortical anchor for mitochondria and dynein. J Cell Biol 216, 3061–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labbé K et al. (2014) Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Biol 30, 357–391 [DOI] [PubMed] [Google Scholar]
- 11.Lackner LL (2014) Shaping the dynamic mitochondrial network. BMC Biol 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman JR et al. (2011) ER tubules mark sites of mitochondrial division. Science 334, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornmann B et al. (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murley A et al. (2013) ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife 2, e00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JE et al. (2016) Multiple dynamin family members collaborate to drive mitochondrial division. Nature 540, 139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korobova F et al. (2013) An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manor U et al. (2015) A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife 4, e08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakrabarti R et al. (2018) INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J Cell Biol 217, 251–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji WK et al. (2017) Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J Cell Biol 216, 4123–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis SC et al. (2016) ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong YC et al. (2018) Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y et al. (2018) Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell 175, 1430–1442.e17 [DOI] [PubMed] [Google Scholar]
- 23.Lackner LL et al. (2013) Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci U S A 110, E458–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ping HA et al. (2016) Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J Cell Biol 213, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang X et al. (2009) A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle 8, 3182–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X et al. (2012) A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J Cell Biol 196, 743–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu JW et al. (2004) Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13, 677–688 [DOI] [PubMed] [Google Scholar]
- 28.Cerveny KL et al. (2007) Yeast mitochondrial division and distribution require the cortical Num1 protein. Dev Cell 12, 363–375 [DOI] [PubMed] [Google Scholar]
- 29.Klecker T et al. (2013) The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J Cell Sci 126, 2924–2930 [DOI] [PubMed] [Google Scholar]
- 30.Farkasovsky M and Kuntzel H (2001) Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J Cell Biol 152, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heil-Chapdelaine RA et al. (2000) The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol 151, 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmit HL et al. (2018) The role of mitochondria in anchoring dynein to the cell cortex extends beyond clustering the anchor protein. Cell Cycle 17, 1345–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraft LM and Lackner LL (2019) A conserved mechanism for mitochondria-dependent dynein anchoring. Mol Biol Cell DOI: 10.1091/mbc.E18-07-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawyer EM et al. (2018) Developmental regulation of an organelle tether coordinates mitochondrial remodeling in meiosis. J Cell Biol DOI: 10.1083/jcb.201807097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu MJ et al. (2018) Epithelial-Mesenchymal Transition Directs Stem Cell Polarity via Regulation of Mitofusin. Cell Metab DOI: 10.1016/j.cmet.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 36.McFaline-Figueroa JR et al. (2011) Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 10, 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swayne TC et al. (2011) Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr Biol 21, 1994–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giorgi C et al. (2018) The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol 19, 713–730 [DOI] [PubMed] [Google Scholar]
- 39.Herrera-Cruz MS and Simmen T (2017) Cancer: Untethering Mitochondria from the Endoplasmic Reticulum. Front Oncol 7, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchi S et al. (2017) Endoplasmic Reticulum-Mitochondria Communication Through Ca2+ Signaling: The Importance of Mitochondria-Associated Membranes (MAMs). Adv Exp Med Biol 997, 49–67 [DOI] [PubMed] [Google Scholar]
- 41.Phillips MJ and Voeltz GK (2016) Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 17, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizzuto R et al. (2009) Ca(2+) transfer from the ER to mitochondria: when, how and why. Biochim Biophys Acta 1787, 1342–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das A et al. (2016) Endosome-mitochondria interactions are modulated by iron release from transferrin. J Cell Biol 214, 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamdi A et al. (2016) Erythroid cell mitochondria receive endosomal iron by a “kiss-and-run” mechanism. Biochim Biophys Acta 1863, 2859–2867 [DOI] [PubMed] [Google Scholar]
- 45.Sheftel AD et al. (2007) Direct interorganellar transfer of iron from endosome to mitochondrion. Blood 110, 125–132 [DOI] [PubMed] [Google Scholar]
- 46.Kopec KO et al. (2010) Homology of SMP domains to the TULIP superfamily of lipidbinding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics 26, 1927–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee I and Hong W (2006) Diverse membrane-associated proteins contain a novel SMP domain. FASEB J 20, 202–206 [DOI] [PubMed] [Google Scholar]
- 48.Reinisch KM and De Camilli P (2016) SMP-domain proteins at membrane contact sites: Structure and function. Biochim Biophys Acta 1861, 924–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toulmay A and Prinz WA (2012) A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci 125, 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.AhYoung AP et al. (2015) Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci USA 112, E3179–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeong H et al. (2017) Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites. Proc Natl Acad Sci U SA 114, E9502–E9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawano S et al. (2018) Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J Cell Biol 217, 959–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirabayashi Y et al. (2017) ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 358, 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wideman JG et al. (2018) PDZD8 is not the ‘functional ortholog’ of Mmm1, it is a paralog. F1000Res 7, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar N et al. (2018) VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 217, 3625–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lang AB et al. (2015) ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol 210, 883–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park JS et al. (2016) Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol Biol Cell 27, 2435–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.John Peter AT et al. (2017) Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J Cell Biol 216, 3219–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lahiri S et al. (2014) A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol 12, e1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jonikas MC et al. (2009) Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chitwood PJ et al. (2018) EMC Is Required to Initiate Accurate Membrane Protein Topogenesis. Cell 175, 1507–1519.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guna A et al. (2018) The ER membrane protein complex is a transmembrane domain insertase. Science 359, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shurtleff MJ et al. (2018) The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife 7, e37018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rambold AS et al. (2015) Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 32, 678–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benador IY et al. (2018) Mitochondria Bound to Lipid Droplets Have Unique Bioenergetics, Composition, and Dynamics that Support Lipid Droplet Expansion. Cell Metab 27, 869–885.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H et al. (2011) Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res 52, 2159–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tower RJ et al. (2011) The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast. Mol Biol Cell 22, 1727–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hermann GJ et al. (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol 143, 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shai N et al. (2018) Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat Commun 9, 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoppins S and Nunnari J (2009) The molecular mechanism of mitochondrial fusion. Biochim Biophys Acta 1793, 20–26 [DOI] [PubMed] [Google Scholar]
- 71.Elbaz-Alon Y et al. (2014) A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell 30, 95–102 [DOI] [PubMed] [Google Scholar]
- 72.Hönscher C et al. (2014) Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell 30, 86–94 [DOI] [PubMed] [Google Scholar]
- 73.Bröcker C et al. (2012) Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci U S A 109, 1991–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.González Montoro A et al. (2018) Vps39 Interacts with Tom40 to Establish One of Two Functionally Distinct Vacuole-Mitochondria Contact Sites. Dev Cell 45, 621–636.e7 [DOI] [PubMed] [Google Scholar]
- 75.Elbaz-Alon Y et al. (2015) Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep 12, 7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murley A et al. (2015) Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J Cell Biol 209, 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khafif M et al. (2014) Identification and phylogenetic analyses of VASt, an uncharacterized protein domain associated with lipid-binding domains in Eukaryotes. BMC Bioinformatics 15, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gatta AT et al. (2015) A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife 4, e07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murley A et al. (2017) Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling. J Cell Biol 216, 2679–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiedemann N and Pfanner N (2017) Mitochondrial Machineries for Protein Import and Assembly. Annu Rev Biochem 86, 685–714 [DOI] [PubMed] [Google Scholar]
- 81.Tang F et al. (2006) Vac8p, an armadillo repeat protein, coordinates vacuole inheritance with multiple vacuolar processes. Traffic 7, 1368–1377 [DOI] [PubMed] [Google Scholar]
- 82.Wang YX et al. (1998) Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol 140, 1063–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Böckler S and Westermann B (2014) Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev Cell 28, 450–458 [DOI] [PubMed] [Google Scholar]
- 84.Hailey DW et al. (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamasaki M et al. (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 [DOI] [PubMed] [Google Scholar]
- 86.Stoica R et al. (2014) ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun 5, 3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gomez-Suaga P et al. (2017) The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy. Curr Biol 27, 371–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bean BDM et al. (2018) Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J Cell Biol 217, 3593–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boutant M et al. (2017) Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J 36, 1543–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daniele T et al. (2014) Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr Biol 24, 393–403 [DOI] [PubMed] [Google Scholar]
- 91.de Brito OM and Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 [DOI] [PubMed] [Google Scholar]
- 92.Naon D et al. (2016) Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc Natl Acad Sci U S A 113, 11249–11254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stefan CJ et al. (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144, 389–401 [DOI] [PubMed] [Google Scholar]