Post-translational phosphorylation is essential to human cellular processes, but the transient, heterogeneous nature of this modification complicates its study in native systems1–3. Here we describe an approach to interrogate phosphorylation and its role in protein-protein interactions on a proteome-wide scale. We genetically encode phosphoserine in recoded E. coli4–6 to generate a peptide-based heterologous representation of the human serine phosphoproteome. We design a single plasmid library encoding >100,000 human phosphopeptides and confirm the site-specific incorporation of phosphoserine in >36,000 of these peptides. We then integrated our phosphopeptide library into an approach called Hi-P to enable proteome-level screens for serine phosphorylation–dependent human protein interactions. Using Hi-P, we find hundreds of known and potentially new phosphoserine-dependent interactors with 14–3-3 proteins and WW domains. These phosphosites retain important binding characteristics of the native human phosphoproteome as shown by motif analysis and pull-downs using full-length phosphoproteins. This technology can be used to interrogate user-defined phosphoproteomes in any organism, tissue or disease of interest.
The interplay between kinases, phosphatases and their substrates results in the presence of dynamic, cell-specific phosphoproteomes. Serine phosphorylation is one of the most common post-translational protein modifications in eukaryotic cells and has an integral role in modulating enzymatic activity and intermolecular interactions1–3. Although human interactome studies have identified tens of thousands of potential protein-protein interactions7, 8, the importance of phosphorylation in these interactions is difficult to ascertain. Recently, the genetically programmed incorporation of phosphoserine (pSer) as a nonstandard amino acid in E. coli has emerged as a powerful tool to unravel the structural and functional importance of protein phosphorylation4–6, 9, 10. However, this technology has been limited to the study of individual proteins and has been unable to uncover insights into the global role of phosphorylation in complex systems without phosphoproteome-level biological techniques.
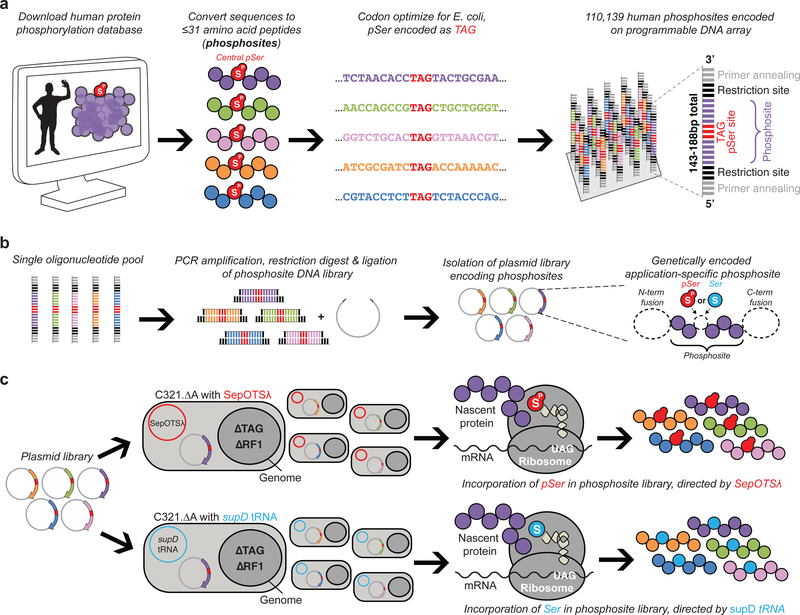
Here we extend this approach to identify proteome-wide phosphoserine-dependent human protein interactions. To encode the pSer component of the human phosphoproteome, 110,139 previously-observed instances of serine phosphorylation11 were designed as singly phosphorylated 16–31 amino acid phosphopeptides, herein referred to as phosphosites (Fig. 1a, Supplementary Data 1). These phosphosites contain a central pSer residue flanked on either side by 15 amino acids from the parent protein, or fewer for sites occurring <15 amino acids from a terminus within the parent protein (Supplementary Fig. 1a,b). Oligonucleotides encoding these phosphosites were synthesized on a programmable DNA microarray and included universal primer annealing and restriction sites, enabling single-pool introduction of the entire phosphosite DNA library into an application-dependent expression vector (Fig. 1b, Supplementary Data 1)12. The central pSer residue in each phosphosite was encoded by a UAG codon. This enabled the flexible, site-specific incorporation of either pSer or Ser in phosphosites by using the pSer orthogonal translation system (SepOTSλ) or the Ser amber suppressor tRNASerCUA (supD tRNA), which respectively incorporate pSer or Ser in response to UAG codons (Supplementary Fig. 1c)5. We also utilized a genomically recoded strain of E. coli (C321.ΔA) that lacks endogenous UAG codons and release factor 1, such that UAG codons which normally cue translational termination can be unambiguously reassigned to pSer or Ser6, 13, 14. Thus, by transforming the phosphosite-encoding plasmid library into C321.ΔA containing either SepOTSλ or supD tRNA, we are able to produce either phosphorylated or non-phosphorylated representations of the human phosphoproteome (Fig. 1c).
Figure 1: Design and display of the synthetic human serine phosphoproteome.
(a) Recombinant human phosphosite DNA sequences were designed based on previously-observed instances of serine phosphorylation from the PhosphoSitePlus database11 and synthesized as oligonucleotides harboring a central TAG codon to direct pSer or Ser incorporation. The 16–31 amino acid phosphosites including the TAG codon were encoded as 48–93 bp oligonucleotides, and additional restriction and primer annealing sites were added to both ends, yielding 143–188 bp sequences. (b) All oligonucleotide sequences encoding phosphosites were liberated from the microarray, PCR-amplified in a single pool, restriction digested, and introduced into an application-dependent expression vector. (c) The phosphosite-encoding plasmid library was then transformed into genomically recoded E. coli (C321.ΔA) lacking all endogenous UAG codons and release factor 1 (RF1), which normally terminates translation at UAG codons. The library was separately transformed into C321.ΔA strains containing either a translation system to insert pSer (SepOTSλ) or Ser (supD tRNA) at UAG codons, enabling the synthesis of either the phosphorylated or unphosphorylated version of the phosphosite library. This workflow was employed for various applications of the phosphosite library, as dictated by the expression vector used for experimentation.
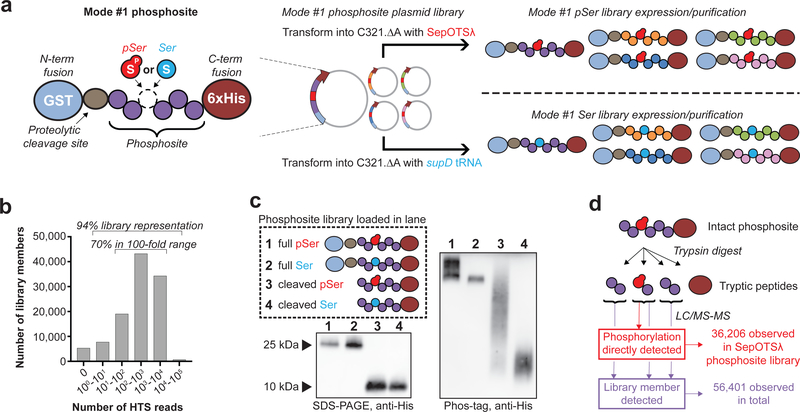
To enable high-level expression of our human phosphosite collection, we first introduced the phosphosite DNA library into a vector encoding an N-terminal GST fusion tag, a proteolytic cleavage site and a C-terminal 6xHis tag, referred to as mode #1 (Fig. 2a). High-throughput sequencing (HTS) analysis confirmed the presence of 94% of the encoded phosphosites in the mode #1 plasmid library, with 70% of sequences falling within a 100-fold range of abundance (Fig. 2b). Immunoblot analysis of full-length and proteolytic cleavage products confirmed production of the mode #1 phosphosite library using either SepOTSλ or supD tRNA, while Phos-tag gel shift analysis demonstrated robust pSer incorporation within the phosphosite library by differential mobility of the pSer library compared to the Ser library (Fig. 2c). Mass spectrometry-based proteomics was used to confirm phosphosite expression and site-specific incorporation of pSer across different mode #1 library preparations (Supplementary Fig. 1d). Evidence for the presence of at least 56,401 phosphosites was obtained across all samples, and pSer was directly observed in 36,206 phosphosites synthesized using SepOTSλ (Fig. 2d, Supplementary Data 2). Comprehensive library validation by proteomics was limited by sample complexity, incomplete DNA representation in the plasmid library, small tryptic peptide fragment lengths, and inclusion of phosphosites not well suited for mass spectrometry (Supplementary Fig. 1e–h, Supplementary Note 1,2).
Figure 2: Expression and LC-MS/MS validation of the synthetic human serine phosphoproteome.
(a) Mode #1 phosphosite library configuration for overexpression in E. coli. The mode #1 plasmid library is introduced into C321.ΔA with either SepOTSλ or supD tRNA to express pSer- or Ser-containing mode #1 phosphosite libraries, respectively. (b) The plasmid library used for mode #1 phosphosite expression encodes ~94% of the 110,139 designed recombinant phosphosites as determined by HTS analysis. (c) Western blot from Phos-tag acrylamide gel illustrates broad mobility shift of recombinant pSer-encoding phosphosite library, either with the GST fusion tag or after enzymatic removal of the GST tag. Similar results were obtained for two other independently purified mode #1 protein libraries. (d) >36,000 unique phosphopeptides generated using SepOTSλ containing pSer at the encoded position were directly observed by LC-MS/MS, and evidence for >56,000 unique phosphosite library members was observed in total across all sample preparations (mode #1 phosphosite libraries made with either SepOTSλ or supD tRNA).
To test that our synthetic phosphosites exhibit anticipated binding properties, we generated 12 separate mode #1 phosphosites from our library representing the epitopes of well-established pSer-specific rabbit monoclonal antibodies (Supplementary Data 3). 11 of 12 pSer-encoding mode #1 phosphosites were recognized by their corresponding antibodies, while Ser-encoding phosphosites were not (Supplementary Fig. 2). Similar to previous results from combinatorial arrays of short synthetic phosphopeptides2, 15, we demonstrate that our phosphosites retain sufficient sequence information from their parent proteins to mediate phosphorylation-specific antibody interactions.
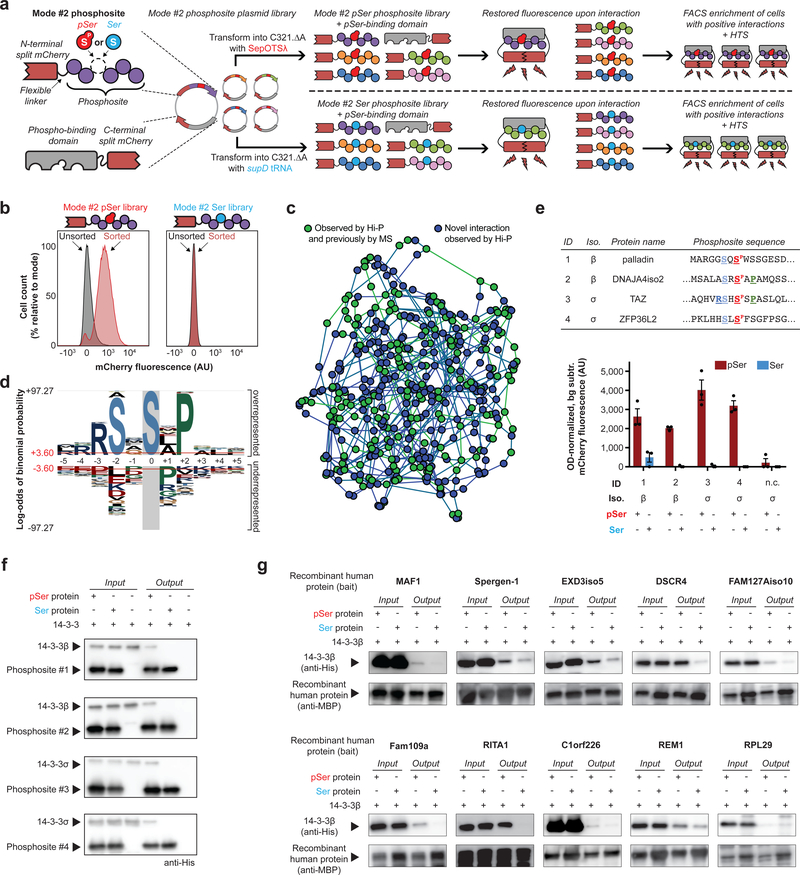
Having characterized the expression of the phosphosite library and examined interactions of a handful of individual phosphosites, we next aimed to identify phosphorylation-dependent interactions on a systems level. Current protein-protein interaction discovery tools, such as yeast two-hybrid systems, do not enable the systematic discovery of interactions coordinated by protein phosphorylation. Recently, bimolecular fluorescence complementation (BiFC) was successfully used to capture a single phosphopeptide-protein interaction using our genetically encoded pSer technology in E. coli16. Briefly, a phosphopeptide fused to a portion of a split mCherry reporter was encoded on the same plasmid as a phosphorylation-binding domain fused to the remaining portion of the split mCherry. The phosphopeptide-protein interaction was then detected by restored intracellular mCherry fluorescence. We decided to integrate our phosphosites into this mCherry-based BiFC system to identify which constituent library members interact with a phosphorylation-binding domain of interest, a platform we named Hi-P. In Hi-P, the phosphosite DNA library is introduced into a vector encoding the N-terminal portion of split mCherry (mode #2). This same vector also encodes a binding domain of interest fused to the C-terminal split mCherry fragment. The mode #2 vector library is then transformed into C321.ΔA containing either SepOTSλ or supD tRNA. Cells harboring productive phosphosite-protein interactions are then isolated by fluorescence-activated cell sorting (FACS) and the implicated phosphosites are identified by HTS (Fig. 3a).
Figure 3: Identification of pSer-dependent protein interactions with 14–3-3 isoforms by Hi-P.
(a) Hi-P experimental workflow. Split mCherry in E. coli enables identification of protein interactions by restored fluorescence signal. Cells expressing mode #2 phosphosites that interact with a phospho-binding protein are isolated by FACS, and implicated phosphosites are identified by HTS. This scheme can be performed with either SepOTSλ or supD tRNA to study pSer- or Ser-containing mode #2 phosphosites, respectively. (b) Hi-P experiments with 14–3-3β yielded increased mean population fluorescence after FACS with SepOTSλ (encoding pSer) but not with supD tRNA (encoding Ser). n=105 cells for flow cytometry observation. (c) A network illustrating the relationships amongst the Hi-P hits using 14–3-3β and SepOTSλ. Each node represents a phosphosite sequence identified by Hi-P and is connected to the two phosphosite nodes closest in sequence space by weighted edges. Green nodes indicate the phosphosite is derived from a protein that has been identified as a 14–3-3 interactor in previous mass spectrometry (MS) experiments17 and blue nodes are novel candidate interactions identified via Hi-P. (d) pLogo analysis19 of 14–3-3β Hi-P results (n = 388 phosphosites above 1,000 HTS read cutoff identified from a single Hi-P experiment). Significance calculated by binomial probability of amino acid frequencies, red lines indicate p = 0.05 significance threshold with Bonferroni correction. (e) Validation of select 14–3-3 Hi-P hits by BiFC shown in bar graph. Top-ranking phosphosite sequences were identified by Hi-P using either 14–3-3β or 14–3-3σ isoforms, as indicated. Amino acids surrounding the central pSer residue (in red) adhering to the RSXSPXP motif are colored and bolded. Background fluorescence for isogenic cells in which mode #2 phosphosite expression was not induced was subtracted, and fluorescence was normalized by OD600. Error bars show s.e.m. centered at mean (n = 3 independent replicates); n.c. = negative control phosphosite not anticipated to interact with 14–3-3 proteins AGPADAPAGAVVGGG[SP/S]PRGRPGPVPAPGLLA. (f) Pull-down analysis of immobilized mode #1 phosphosites confirmed pSer incorporation is necessary for 14–3-3 interaction, representative of two independent replicates. (g) Pull-down analysis of immobilized MBP-fusion full-length recombinant human phosphoproteins confirmed pSer is necessary for or enhances 14–3-3β interaction for 9/10 of the tested proteins, representative of two independent replicates. AU, arbitrary units.
Using Hi-P, we investigated 14–3-3 isoforms β and σ, which are both known to bind pSer/pThr-containing phosphoproteins via a well-defined RSX[SP/TP]XP motif2 and are known to be highly phosphorylation-specific. We first co-expressed the 14–3-3-split mCherry fusion protein with the mode #2 phosphosite library encoding either pSer or Ser. Sequential FACS experiments yielded cell populations with increased mCherry fluorescence only when using the mode #2 phosphosite library containing pSer (Fig. 3b, Supplementary Fig. 3). These results indicate that Hi-P recapitulates the known phosphorylation binding preference of 14–3-3 proteins. HTS results from Hi-P experiments with both 14–3-3 isoforms identified hundreds of interactions previously observed in other experiments17 (Fig. 3c, Supplementary Fig. 4, Supplementary Data 4,5) including the well-characterized in vivo-validated interaction with TAZ18, which was our top-sequenced candidate phosphosite interactor with 14–3-3σ. Given that approximately 60% of Hi-P-identified phosphosite interactors were derived from human proteins never shown to interact with 14–3-3 isoforms, Hi-P can be useful in the identification of both known and candidate novel interactions. We performed network analysis using Levenshtein distances to examine similarity between phosphosite sequences and showed that novel interactions are dispersed amongst known interactors, indicating a lack of systematic bias in 14–3-3 interactions identified by Hi-P (Fig. 3c). Global motif analysis19 of phosphosites identified in 14–3-3 Hi-P experiments revealed a RSXSPXP signature motif, which perfectly matches the canonical 14–3-3 interaction motif (Fig. 3d, Supplementary Fig. 5). Together, these data showed that our phosphosite library was able to recapitulate physiologically relevant interactions. Consistent with previous work, only about half of the identified sequences contained individual elements of the canonical 14–3-3 interaction motif (−3 Arg, −2 Ser or +2 Pro), demonstrating our ability to identify candidate interactors that do not perfectly conform to known motifs (Supplementary Data 6,7)20. To validate our results, we used BiFC to individually examine several mode #2 phosphosites identified as 14–3-3 candidate interactors. Consistent with the Hi-P data, pSer incorporation was necessary to mediate these 14–3-3-phosphosite interactions (Fig. 3e). We also performed in vitro pull-down assays using mode #1 versions of these same phosphosites, confirming the necessity of pSer to coordinate these interactions (Fig. 3f, Supplementary Fig. 6a).
Next, we tested if the interactions identified by Hi-P may apply to the full-length parent proteins from which the phosphosites were derived, thus leading to the discovery of new pSer-mediated protein-protein interactions. Based on phosphosites observed in Hi-P experiments with 14–3-3β, we expressed 10 corresponding full-length recombinant human phosphoproteins that had never previously been observed to interact with 14–3-3 isoforms (Supplementary Data 8). In in vitro pull-down studies, 9 out of 10 of these proteins exhibited interactions with 14–3-3β that were enhanced by or dependent on pSer incorporation (Fig. 3g, Supplementary Fig. 6b). To test a known and a candidate novel interaction site from the same full-length protein, we expressed human FOXO3A containing pSer at well-defined site (pSer253)21 and a new site (pSer413), which were both identified by Hi-P. As with the other tested recombinant phosphoproteins, both forms of FOXO3A showed a pSer-dependent interaction with 14–3-3β, demonstrating that Hi-P can confirm known biology and predict potential new interaction sites in the context of full-length proteins (Supplementary Fig. 6c).
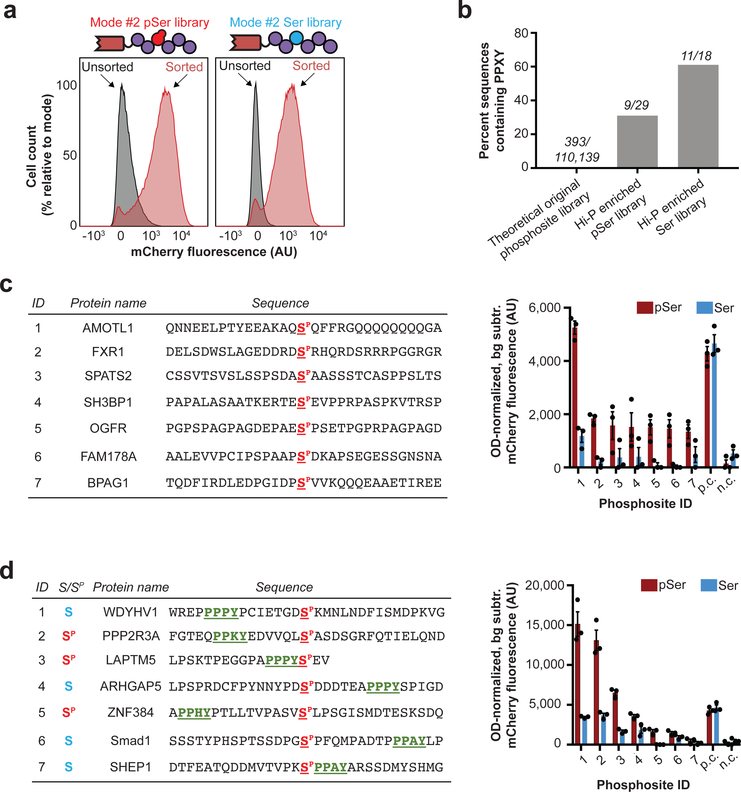
Many protein-protein interactions are driven by small functional domains that recognize protein phosphorylation, imparting phosphorylation-binding properties to diverse classes of proteins. One example is the second WW domain (WW2) of human NEDD4 (E3 ubiquitin ligase neural precursor cell-expressed developmentally downregulated 4) and of the closely related NEDD4–2 (also known as NEDD4L). These domains are of particular interest since both are thought to exhibit mixed modalities of pSer-dependent, pSer-independent, and/or pSer-enhanced ligand binding22–25. We reasoned that Hi-P might be an ideal approach to evaluate the various binding modes of small functional domains, and therefore decided to test the NEDD4 and NEDD4–2 WW2 domains in Hi-P experiments. Consistent with these mixed binding modes and in contrast to 14–3-3 isoforms, Hi-P identified phosphosite interactions from both pSer and Ser mode #2 libraries with the WW2 domains (Fig. 4a, Supplementary Fig. 7a). In all of these Hi-P experiments, we observed a high degree of enrichment for phosphosites containing the well-known pSer-independent WW domain binding motif PPXY (Fig. 4b, Supplementary Fig. 7b, Supplementary Data 9–12)26. As demonstrated in our 14–3-3 experiments, Hi-P can accurately identify interaction motifs. However, further motif analysis of phosphosites identified by Hi-P revealed no sequence element patterns relative to the central pSer residue that were characteristic of NEDD4 or NEDD4–2 WW2 interactors (Supplementary Fig. 8). This was a surprising result, suggesting that non-obvious or highly heterogeneous context cues can drive WW2 interactions. Once again, we validated our Hi-P results by analyzing a targeted set of phosphosites by BiFC. These experiments confirmed the mixed binding modalities of the NEDD4 WW2 domain with phosphosites identified by Hi-P (Fig. 4c,d, Supplementary Fig. 9).
Figure 4: Investigation of NEDD4 WW2 interactions by Hi-P.
(a) Hi-P experiments with cells co-expressing the NEDD4 WW2 domain and the mode #2 phosphosite library yielded increased mean population fluorescence with either SepOTSλ (encoding pSer) or supD tRNA (encoding Ser). n=105 cells for flow cytometry observation. (b) Hi-P experiments using WW2 from NEDD4 resulted in enrichment of PPXY-containing phosphosites in both pSer- and Ser-encoding populations. The raw number of sequences containing PPXY over number of sequences in population are shown above each bar. All data is for a 1,000-read cutoff by HTS. (c) BiFC analysis of select NEDD4 WW2 Hi-P phosphosite hits from SepOTSλ (encoding pSer) experiments and excluding sequences containing the PPXY motif. (d) BiFC analysis of select NEDD4 WW2 Hi-P phosphosite hits, with PPXY motif (green underlined), from experiments with either SepOTSλ (encoding pSer) or supD tRNA (encoding Ser). For BiFC experiments in (c) and (d), background fluorescence for isogenic cells in which mode #2 phosphosite expression was not induced was subtracted, and fluorescence was normalized by OD600. Error bars show s.e.m. centered at mean (n = 3 independent replicates). n.c., negative control mode #2 peptide WFYSPFLE co-expressed with mouse Nedd4 WW2 fused to C-terminal split mCherry22; p.c., positive control mode #2 peptide IPGTPPPNYD co-expressed with mouse Nedd4 WW2 fused to C-terminal split mCherry22; AU, arbitrary units.
We then investigated if the novel phosphosite interactions revealed by Hi-P using the isolated WW domain could be recapitulated with a full-length interaction partner expressed in a human cell line. First, we expressed a targeted library of 20 phosphosites identified from NEDD4 WW2 Hi-P experiments (those from Fig. 4c and Supplementary Fig. 9) with an MBP fusion tag to enhance expression (mode #3, Supplementary Fig. 10a). We excluded phosphosites containing PPXY sequences because this type of WW domain interaction is already well characterized. We then spiked this targeted phosphosite library into mammalian cell lysates expressing full-length human NEDD4 and performed co-immunoprecipitation (co-IP) mass spectrometry experiments (Supplementary Fig. 10b). To address pSer specificity, parallel experiments were performed with pSer and Ser targeted mode #3 phosphosite libraries. We found that several of these phosphosites co-precipitated with NEDD4 in a phosphorylation-dependent manner (Supplementary Data 13). Interestingly, a PPXY-free phosphosite from AMOTL1 was the top candidate interactor with NEDD4 as identified by both BiFC (using just the WW2 domain) and co-IP (using the full-length NEDD4 protein), exhibiting enhanced binding with full-length NEDD4 when pSer was incorporated within the phosphosite. AMOTL1 was previously observed to interact with NEDD4–2 in a PPXY-dependent manner27, but the PPXY-free region identified by Hi-P has never been directly implicated in coordinating an interaction with a NEDD family protein. Overall, results between Hi-P and co-IP may differ because the full-length NEDD4 protein has four total WW domains, while Hi-P only examined the WW2 domain in isolation.
Finally, we evaluated expression-based bias and experimental reproducibility of the platform to further characterize Hi-P. To better understand how phosphosite expression levels may influence identification by Hi-P, we compared the number of Hi-P HTS reads with phosphopeptide ion intensity by mass spectrometry (an indication of phosphosite expression level) for individual phosphosites. We saw no correlation between phosphosite ion intensity and Hi-P reads (Supplementary Fig. 11). To this end, we also note that phosphosite interactors with 14–3-3 isoforms were not identified by Hi-P when using supD tRNA (Fig. 3b, Supplementary Fig. 3), suggesting that differential expression alone of individual phosphosites cannot drive false positive BiFC interactions. We then investigated the reproducibility of Hi-P by performing three independent replicate experiments for 14–3-3β with the pSer-phosphosite library and the WW2 domain of NEDD4 with either the pSer- or Ser-phosphosite libraries (Supplementary Data 14). We observed considerable overlap between data sets for 14–3-3β (Supplementary Fig. 12a,b), but less for the NEDD4 WW2 domain (Supplementary Fig. 12c,d). These results indicate that various “bait” structures may behave differently in Hi-P experiments due to their size, ligand binding kinetics, or binding modalities, which may in turn affect reproducibility. As demonstrated above, low-throughput experiments using full-length proteins or functional domains should be conducted to validate interactions predicted by Hi-P.
In this study, we present a synthetic serine phosphoproteome display technology capable of producing tens of thousands of human phosphosites. Importantly, these phosphosites are identifiable by mass spectrometry and retain important binding characteristics of the native human phosphoproteome. Our synthetic phosphoproteome is encoded by a modular DNA library, enabling diverse applications from phosphopeptide library purification to functional interactome screens, such as Hi-P. Previous work has established the relevance of genetically encoded representations of the human proteome for autoantigen discovery using phage display or as a proteomics standard28, 29. Our technology builds upon this concept, permitting the targeted synthesis of human-derived phosphosites to probe phosphorylation-specific protein interactions. Compared to other high-throughput pull-down and co-immunoprecipitation techniques7, 30, our approach for screening phosphorylation-dependent protein interactions is agnostic to cell type and kinase-independent. Additionally, phosphorylation in our protein library is precisely defined by the genetic code and can be identified by DNA sequencing, thus revealing the amino acid sequence directly responsible for coordinating the interaction. Conversely, our system in its current form cannot address the role of phosphorylation in the context of native eukaryotic systems. Phosphosite expression levels are also difficult to rigorously normalize across the entire library, and the current phosphosite library only addresses single phosphorylation events. Future phosphosite libraries should consider combinatorial phosphorylation to investigate the importance of this modification when incorporated at multiple sites within the same protein. Nevertheless, we have demonstrated that Hi-P can provide proteome-wide surveys of phosphorylation-dependent interactions that mimic full-length proteins, and should be used as a tool to prioritize candidate interactions for further biochemical and cellular validation.
We demonstrated that Hi-P recapitulated known interactions and provided a rapid pipeline for the identification of novel candidate protein-protein interactions for human phosphorylation-binding proteins and domains. Our phosphosite library is derived from the human phosphoproteome, allowing direct interrogation of a vast collection of physiologically relevant binding sites that are not always predictable using motif analysis and bioinformatic approaches, offering distinct advantages over phosphorylation-oriented randomized peptide libraries2, 31. We were able to identify binding partners that do not fully conform to canonical interaction motifs and identify mixed modes of pSer-dependent and pSer-independent binding. One of the advantages of Hi-P is the site-specific identification of protein regions implicated in coordinating protein-protein interactions, which is unknown in most high-throughput interaction discovery workflows. Some phosphosites encoded in our library are also derived from proteins that are generally difficult to isolate or prepare for high-throughput studies, such as transmembrane proteins (e.g. phosphosites derived from Claudin-6 and ZO-1 were identified in Hi-P experiments with 14–3-3 isoforms). This genetically encoded heterologous phosphosite library is amenable to in vivo selections in E. coli, thereby offering a rapid and cost-effective platform capable of interrogating the human serine phosphoproteome (Supplementary Note 3). Our system paves the way for the construction of targeted disease- and tissue-specific phosphoproteomes that can be used to discover new roles of serine phosphorylation, and could be easily modified to investigate diverse post-translational modifications using other genetically encodable nonstandard amino acids with physiological relevance.
Online Methods
Phosphosite DNA library design
15-residue amino acid sequences corresponding to previously observed phosphorylation sites across the human proteome were downloaded from PhosphoSitePlus11 on 11 January 2015. Entries were filtered to include only human sequences containing phosphoserine at the central position, and duplicate entries were removed. The 15-residue sequences were then matched to corresponding full-length human proteins and elongated to contain 31 amino acids (15 on either side of the phosphoserine residue). If phosphoserine occurred within 15 residues of the N- or C-terminus, the peptide sequence was extended to the end of the protein. Using Geneious software (version 8), amino acid sequences were reverse translated and codon optimized for Escherichia coli K12 (high). The central phosphoserine residue was encoded as TAG. Other post-translational modifications were not taken into account. A KpnI site followed by an AAG (Lys) codon were encoded at the 5’ end of the encoded phosphosites, while a HindIII site was included at the 3’ end.
Sequences of primer pairs used to PCR amplify the DNA library were previously described32. Individual primers were blastn searched against the DNA library for entries containing high sequence homology (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The ΔTm between primer-specific and non-specific library sequences was ensured to be ≥15°C to reduce non-specific amplicons (https://www.idtdna.com/calc/analyzer). Ten sets of 20 bp orthogonal primer annealing sequences were encoded in the library to facilitate amplification of DNA subpools, and one set of 20 bp universal primer annealing sequences was encoded in every DNA sequence at the 5’ and 3’ termini (Supplementary Data 1, Supplementary Data 15). This resulted in 110,139 DNA library sequences between 143 and 188 bp in length (Supplementary Data 1).
The DNA library was produced using an oligonucleotide library synthesized by Agilent Technologies having a length of 143–188 nucleotides and sequence complexity of 110,139 with twofold synthesis redundancy12. DNA was provided as a 10 pmol lyophilized pool. Phosphosite DNA was PCR amplified in a single pool using primers End-F and End-R (Supplementary Data 15). The PCR product was then extracted on a 2% agarose gel, digested with KpnI and HindIII restriction enzymes and ligated into either the pNAS1B or pCRT7 vectors modified as described below. The ligation reaction was desalted by drop dialysis (V-Series membrane, Millipore) and then transformed into ElectroMAX DH10B cells by electroporation (1 mm cuvette using Gene Pulser Xcell from Bio-Rad, 1800 V, 200 Ω, 25 mF). and recovered in 700 μL SOC medium for 1 h at 37 °C, 230 rpm. The transformation mixture was then inoculated directly into 50 mL LB with 100 ng/μL ampicillin and grown overnight at 37 °C, 230 rpm, and the plasmid library was isolated by miniprep (Omega Bio-tek). Plasmid libraries will be made available from Addgene (https://www.addgene.org/Jesse_Rinehart).
Strain and plasmid information
The C321.ΔA strain used in all experiments involving protein expression and the SepOTSλ and supD tRNA-encoding plasmids were described previously5 and are available from Addgene (SepOTSλ plasmid: #68292; supD tRNA plasmid: #68307; C321.ΔA strain: #68306). It should be noted that this strain of C321.ΔA has the serB phosphoserine phosphatase locus knocked out, which boosts intracellular availability of phosphoserine for incorporation using the SepOTSλ plasmid4. For all Hi-P and BiFC experiments, a new supD tRNA plasmid was generated by removing the four tRNASep gene copies from the SepOTSγ plasmid (containing SepRS9-EFSep21)5, 33 by NotI restriction digest and replacing them with two gene copies of supD tRNA from the original supD tRNA plasmid so comparisons between pSer- and Ser-encoding proteins were performed in isogenic plasmids/strains except for the tRNASep/supD tRNA locus.
Phosphosite fusion proteins for overexpression and purification (mode #1 and mode #3) were encoded in the pCRT7 Topo tetR pLtetO vector5 with the following modifications: XbaI and HindIII enzymes were used to remove the tetR, pLtetO and recombinant protein expression loci from pCRT7. In parallel, a multiple cloning site containing pBAD, ribosome binding site, and an NdeI site (G1, Supplementary Data 16) were introduced between BamHI and SacI sites in the pNAS1B vector16. The araC and pBAD regions from the modified pNAS1B plasmid were excised using SphI and XhoI enzymes. This insert and the XbaI/HindIII-digested pCRT7 vector were blunted using a Quick Blunting Kit (NEB) and ligated together. Then, primers P1 and P2 (Supplementary Data 17) were used to amplify an N-terminal GST fusion tag and a human rhinovirus 3C proteolytic cleavage site from the pGEX6P-1 vector encoding a GST fusion protein, and adding a multiple cloning site with KpnI and HindIII sites, a 6xHis tag and a TAA stop codon. Primers P3 and P4 (Supplementary Data 17) were used to add NdeI and SacI sites to the P1/P2 PCR product via secondary PCR amplification. This PCR product was introduced into the modified pCRT7 vector via NdeI/SacI sites, and recombinant phosphosite DNA sequences were subsequently inserted via KpnI/HindIII sites (between the proteolytic cleavage site and 6xHis tag). For mode #3 phosphosites (as in Supplementary Fig. 10) or full-length protein studies (as in Fig. 3g), this vector was further modified to replace the C-terminal 6xHis tag with an 8xHis tag, and this modified vector was then used to replace the N-terminal GST tag with and an Avitag (https://www.avidity.com) followed by an MBP tag. Finally, a last pCRT7 derivative vector was generated containing an N-terminal 8xHis tag/Avitag/MBP tag, and with no 8xHis tag on the C-terminus. All modified pCRT7 vectors retained unique KpnI/HindIII cloning sites for the introduction of phosphosite or phosphoprotein DNA.
The split mCherry experiments were performed in the pNAS1B vector16 with the following modifications: The existing KpnI site was removed by A to T substitution. A multiple cloning site containing pBAD and NdeI and PsiI sites (G1, Supplementary Data 16) was introduced between BamHI and SacI sites. The human NEDD4 WW2 domain and the C-terminal split mCherry protein16 with an added 6xHis fusion tag (G2, Supplementary Data 16) were introduced between NdeI and PsiI sites. Primers P5 and P6 (Supplementary Data 17) were used to PCR amplify the region between PsiI and XhoI sites in this vector but with the PsiI-adjacent SacI site removed. This PCR product was then reintroduced into the vector between PsiI and XhoI sites. NdeI and SacI sites 5’ to the C-terminal mCherry cassette allowed the insertion of phosphobinding protein domains of interest (NEDD4–2 WW2, human 14–3-3β and 14–3-3σ from G3, G4 and G5, respectively, Supplementary Data 16). The N-terminal split mCherry cassette (G6, Supplementary Data 16 was introduced between EcoRI and PvuII sites, with internal KpnI/HindIII sites allowing for insertion of a phosphosite cassette. Another HindIII site in the vector had been removed by site-directed mutagenesis using P7 and P8 (Supplementary Data 17).
Phosphosite DNA for targeted clonal BiFC validation experiments (Fig. 3e, Fig. 4c,d, Supplementary Fig. 9) or targeted library experiments (Supplementary Fig. 10) were synthesized by IDT as concatenated <1,000 bp DNA sequences (separated by KpnI/HindIII sites) that were digested and ligated into either the modified pNAS or pCRT7 vector between KpnI and HindIII sites. DNA design and synthesis for full-length recombinant human phosphoproteins used for pull-down experiments with 14–3-3β in Fig. 3g are listed in Supplementary Data 8 and described below. All restriction enzymes and T4 DNA ligase were from NEB, all double-stranded Escherichia coli K12 codon-optimized DNA inserts in Supplementary Data 16 were synthesized by IDT, and all oligonucleotides in Supplementary Data 17 were synthesized by the Keck Biotechnology Resource Laboratory at the Yale School of Medicine. Plasmids used in this work will be made available from Addgene (https://www.addgene.org/Jesse_Rinehart).
High-throughput sequencing information
Plasmid libraries encoding mode #1 phosphosites as in Fig. 2 were grown from an ElectroMAX DH10B glycerol stock containing the previously-electroporated phosphosite DNA library on the modified pCRT7 vector by direct inoculation of the glycerol stock in 100 mL LB with 100 ng/μL ampicillin and grown overnight at 37 °C, 230 rpm. Plasmid library was harvested by maxiprep (Perfectprep, Eppendorf). A KpnI/HindIII digest of approximately 500 μg plasmid library was performed and the phosphosite DNA library insert was extracted on a 2% agarose gel. DNA was used for 75 bp paired-end sequencing.
With the exception of reproducibility studies (as detailed below), PCR amplicons of mode #2 phosphosite DNA libraries in the modified pNAS1B vector for Hi-P experiments were generated using various combinations of primers P9-P16 (Supplementary Data 17), allowing for sample multiplexing and determination of sample of origin from degenerate base ends followed by 2 bp barcodes. DNA was used for 100 bp paired-end sequencing.
DNA samples were end-repaired, A-tailed and adapters were ligated. Indexed libraries that met appropriate cut-offs were quantified by both qRT-PCR (KAPA Biosystems) and insert size distribution was determined with the LabChip GX. Samples with a yield of ≥0.5 ng/μl were used for sequencing.
Sample concentrations were normalized to 350 pM and loaded onto Illumina HiSeq 4000 flow cells at a concentration that yielded 300–350 million passing filter clusters per lane. Each amplicon library was run over 50% of two lanes (multiplexed with 50% exome libraries). The samples were then sequenced using 75 or 100 bp paired-end reads on an Illumina HiSeq 4000 according to Illumina protocols. The 6 bp index was read during an additional sequencing read that automatically followed the completion of read 1. Data generated during sequencing runs was simultaneously transferred to the high-performance computing cluster. A positive control (prepared bacteriophage Phi X library) provided by Illumina was spiked into every lane at a concentration of 0.3% to monitor sequencing quality in real time.
Sequencing reads were first filtered for quality using Trimmomatic34, which applied a sliding window filter of width 2 bp and a Phred score cutoff of 30. If the average quality score over two consecutive bases fell below 30, the read was trimmed to remove the remaining bases. Quality trimmed read pairs were then merged using BBMerge with the stringency set to “strict” (sourceforge.net/projects/bbmap). Using custom scripts, the merged reads were then sorted and assigned to the various input libraries based on barcodes added during the PCR amplification step. The variable sequence region for each amplicon was then extracted, and for each input library the abundance of every unique sequence was calculated.
In order to determine library coverage, sequencing reads were filtered for quality using Trimmomatic with a sliding window filter of 2 bp and Phred score cutoff of 30. Additionally, the first 5 bp were trimmed from the start of the reads. Subsequently, the trimmed read pairs were merged using BBMerge with the stringency set to “strict”. The FASTQ file of merged read pairs was then aligned to a FASTA file containing each of the library member sequences using the BWA-mem algorithm with the –M option. The resultant alignment files were then sorted and indexed using samtools and the mappings to each library member were evaluated using BBMap’s pileup.sh with “secondary=false”. The percent of the total 110,139 possible phosphosites represented in plasmid libraries as determined by HTS are as follows: mode #1 library, 94%; mode #2 library + 14–3-3β, 88%; mode #2 library + 14–3-3σ, 90%, mode #2 library + NEDD4 WW2 domain, 93%; mode #2 library + NEDD4–2 WW2 domain, 98%.
For reproducibility studies, amplicons from FACS-enriched mode #2 phosphosite plasmid libraries were generated using P17 and P18 primers (Supplementary Data 17) and sent to the Massachusetts General Hospital Center for Computational & Integrative Biology DNA Core for 100 bp paired-end amplicon sequencing (approximately 100,000–200,000 reads per sample replicate). Raw read files were similarly processed as detailed above.
Mode #1 phosphosite library expression and purification
20 mL of C321.ΔA cells containing either the SepOTSλ or supD tRNA plasmid were grown to OD600 of 0.4. Cells were then spun down at 4,000 × g for 1 minute, supernatant was decanted, and cells were washed with 20 mL ice cold, deionized water. This was repeated once. Cells were resuspended in 50 μL water, mixed with 1 μL mode #1 library plasmid (approximately 100 ng/μL), and electroporated using parameters stated in the “Phosphosite DNA library design” section. The cells were then resuspended in 1 mL of S.O.C. medium (Thermo) and incubated for 1 h at 30 °C and 230 rpm in a 15 mL culture tube. At least 107 colony-forming units were obtained per electroporation, as calculated by plating serial dilutions. Recovered cells were directly inoculated into 100 mL LB supplemented with 100 ng/μL ampicillin and 25 ng/μL kanamycin and grown overnight at 30 °C, 230 rpm. 4×500 mL of LB with 100 ng/μL ampicillin, 25 ng/μL kanamycin, and 2 mM O-phospho-L-serine was inoculated with overnight culture to an OD600 of 0.15 and grown at 30 °C, 230 rpm until OD600 = 0.6–0.8. Phosphosite library and SepOTSλ expression were then induced with 0.2% arabinose and 1 mM isopropyl β-D-1-thiogalactopyranoside, respectively. Cells were grown for an additional 4 h at 30 °C, 230 rpm. Cells were harvested by centrifugation and frozen at −80 °C.
500 mL cell pellets were resuspended in 6 mL lysis buffer (50 mM Tris pH 7.4, 500 mM NaCl, 500 μM EDTA, 500 μM EGTA, 10% glycerol, 1 mM DTT, 50 mM NaF, 1 mM NaVO4, 1 mg/mL lysozyme, 1 Roche cOmplete protease inhibitor tablet per 50 mL) and sonicated on ice using a QSonica Q500 with 1/8” microtip probe (10 s on, 40 s off, 40% amplitude, on for 3 min total). Combined lysates were then passed over 1 mL equilibrated Ni-NTA resin (Qiagen) in a purification column by gravity. Resin was then washed with 10 mL wash buffer (50 mM Tris pH 7.4, 500 mM NaCl, 500 μM EDTA, 500 μM EGTA, 10% glycerol, 1 mM DTT, 50 mM NaF, 1 mM NaVO4, 20 mM imidazole) and eluted with 5 mL elution buffer (50 mM Tris pH 7.4, 500 mM NaCl, 500 μM EDTA, 500 μM EGTA, 10% glycerol, 1 mM DTT, 50 mM NaF, 1 mM NaVO4, 250 mM imidazole). The eluate was then incubated with 1 mL equilibrated glutathione HiCap resin (Qiagen) mixing end-over-end for 30 min at RT and washed with 10 mL wash buffer by gravity. 4 mL elution buffer (50 mM Tris pH 7.4, 500 mM NaCl, 500 μM EDTA, 500 μM EGTA, 10% glycerol, 1 mM DTT, 50 mM NaF, 1 mM NaVO4, 50 mM reduced L-glutathione) was then passed over the resin. Eluate was buffer exchanged (50 mM Tris pH 7, 150 mM NaCl, 1 mM EDTA, 1mM DTT) and concentrated to ~500 μL using an Amicon Ultra-4 10 kDa molecular weight cutoff spin column (Millipore) and incubated with 20 μL (40 units) PreScission protease (GE Healthcare Life Sciences) end-over-end overnight at 4 °C. The cleaved library was then passed through an Amicon Ultra-0.5 30 kDa molecular weight cutoff to remove the cleaved GST and uncleaved library. The peptide library was then concentrated using Amicon Ultra-0.5 3 kDa molecular weight cutoff and buffer exchanged with 10 mM Tris, pH 8. Concentrated, cleaved phosphosites were quantified by bicinchoninic acid assay and dried by centrifugal vacuum concentration.
Clonal phosphosite expression and evaluation as in Supplementary Fig. 2 was performed by co-transformation of SepOTSλ or supD tRNA-encoding plasmids with the phosphosite DNA on the modified pCRT7 plasmid in chemically competent (standard RbCl method) C321.ΔA cells. Cells were plated on LB agar with 100 ng/μL ampicillin and 25 ng/μL kanamycin and grown for 18 h at 30 °C. Up to 5 colonies were picked and grown in 5 mL 100 ng/μL ampicillin and 25 ng/μL kanamycin and grown overnight at 30 °C, 230 rpm. A glycerol stock was made of each strain, and each stock was restreaked on a selective agar plate and incubated for 18 h at 30 °C. 5 colonies were picked in 5 mL LB 100 ng/μL ampicillin and 25 ng/μL kanamycin and grown in 5 mL LB containing 100 ng/μL ampicillin and 25 ng/μL kanamycin at 30 °C, 230 rpm overnight. Cells were then diluted to OD600 = 0.15 in 20 mL LB with 100 ng/μL ampicillin, 25 ng/μL kanamycin and 2 mM O-phospho-L-serine, grown to mid-log (OD600 = 0.6–0.8), and protein expression was induced with 0.2% arabinose and 1 mM IPTG. Cells were grown for an additional 4 h at 30 °C, 230 rpm. An equivalent number of cells as 1 mL OD600 = 2.5 was spun down for 5 minutes at 4,000 × g, supernatant was aspirated, and cell pellets were frozen at −80 °C overnight, and then lysed for 10 minutes in 40 μL lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1mM DTT, 50 mM NaF, 1 mM NaVO4, 5% glycerol, Roche cOmplete protease inhibitors, 1x Novagen BugBuster). Lysates were then spun down at 21,000 × g for 7 minutes to remove cell debris. 1 μL lysate was run per lane on acrylamide gels. Note that the addition of 2 mM O-phospho-L-serine may be unnecessary to promote phosphoserine incorporation in all preparations, as phosphoserine is already present intracellularly in E. coli as a natural metabolite.
Phos-tag gels and westerns
100 μM Phos-tag acrylamide (Wako) within hand-cast 12% acrylamide gels was used for visualization of phosphosites by western blot. SDS-PAGE gels (4–15% acrylamide, Bio-Rad) and Phos-tag gels were transferred onto PVDF membranes. Anti-His westerns were performed using 1:2,500 diluted rabbit Anti-6xHis antibody (PA1–983B, Thermo Fisher Scientific) in 5% w/v milk in TBST for 1 h and 1:10,000 diluted donkey anti-rabbit HRP (711–035-152, Jackson ImmunoResearch) in 5% w/v milk in TBST for 1 h. HRP-conjugated anti-MBP monoclonal antibody (E80385, New England Biolabs) was used at a 1:500 dilution in 5% w/v milk in TBST for 1 h. Phospho-specific primary antibodies used in Supplementary Fig. 2 are listed in Supplementary Data 3 and were used in 1:1,000 dilutions in 5% w/v milk in TBST for 1 h. Protein bands were then visualized using Clarity ECL substrate (Bio-Rad) and an Amersham Imager 600 (GE Healthcare Life Sciences). All full (uncropped) western blots can be found in Supplementary Note 4–10.
Trypsin digestion of phosphosite libraries
In-solution trypsin digest of ~200 μg of the GST-cleaved phosphosite peptide library was performed as in Pirman, et. al.5 Briefly, 200 μg dried peptides were resuspended in 100 μL 10 mM Tris pH 8.5 and 25 μL of solubilization buffer (1 mM EDTA, 10 mM DTT, 0.5% acid labile surfactant II from Protea, 10 mM Tris pH 8.5) and heated at 55 °C for 35 min to reduce cysteines. Samples were then placed on ice for 1 min, and then 20 μL of 1 M Tris pH 8.5 and 47 μL of 100 mM iodoacetamide were added to the samples, and left in the dark at room temperature for 30 min for cysteine alkylation. 7 μL of 200 mM DTT was then added to quench the reaction, and then 1.1 mL water, 6.7 μL CaCl2, 133 μL 1M Tris pH 8.5 and 60 μL 0.5 mg/mL sequence-grade trypsin (Promega) were added to each sample. Trypsin digest was performed for 16 h at 37 °C. Surfactant was then cleaved using 125 μL 20% trifluoroacetic acid and incubating samples at room temperature for 15 min. Peptides were desalted using two C18 silica MicroSpin Columns (The Nest Group) in sequence, and eluted in 300 μL 80% acetonitrile 0.1% trifluoroacetic acid and dried by centrifugal vacuum.
Phosphopeptide ERLIC fractionation
ERLIC fractionation was performed as in Ferdaus, et. al.35, with the following modifications. For both Ser- and pSer-encoding samples, two independently-generated (the same HTS-confirmed mode #1 vector library was electroporated in separate experiments into the expression strain) GST-cleaved peptide libraries of approximately 10 μg and 100 μg trypsin-digested peptides were dissolved in 10 μL or 100 μL 85% ACN/0.1% FA, respectively. Samples were injected on a PolyWAX LP column (150 × 1.0 mm, 5 mg particle diameter, 300 Å pore, PolyLC), and mixtures of 85% acetonitrile/0.1% formic acid (solvent A) and 30% acetonitrile/0.1% formic acid (solvent B) were flowed at 50 μL/min using an Agilent 1100 Series HPLC and a 70-minute method: (min/%B, linear ramping between steps) 0/0, 5/0, 22/8, 47/45, 57/100, 62/100, 70/0. One fraction was collected every 2 minutes for 60 minutes followed by one fraction every 5 minutes for 10 minutes. Fractions were dried by centrifugal vacuum concentration. Fractions were resuspended in 0.6 μL 70% formic acid, 0.4 μL 50% acetonitile/0.1% formic acid, 7 μL 0.1% trifluoroacetic acid. 5 μL of each fraction was used for mass spectrometry analysis.
TiO2 enrichment
~100 μg of GST-cleaved trypsin-digested library peptides were subjected to tip-based TiO2 phosphopeptide enrichment, based on the technique by Kettenbach, et. al.36 400 μg TiO2 (Titansphere, GL Science) placed over a 0.6 mm diameter 3M Empore C18 disk in a pipette tip were equilibrated with 50% ACN/0.5% TFA. Peptides were solubilized in 40 μL 50% ACN/0.5% TFA and flowed through the tip by centrifugation. The tip was then washed with 2 × 45 μL 50% ACN/0.5% TFA and 40 μL 80% ACN/0.1% FA. Phosphopeptides were eluted using 40 μL 1% NH4OH into a tube containing 2.5 μL 70% FA followed by 40 μL ACN 0.1% FA. Peptides were then dried by centrifugal vacuum concentration, resuspended in 50 μL water, dried again, then resolubilized in 0.9 μL 70% FA, 0.6 μL 50% ACN/0.1% FA, 11.1 μL 0.1% TFA. Peptides concentration was estimated by A280 and 2 μg in a 5 μL volume of each sample were used for mass spectrometry analysis.
Mass spectrometry
LC-MS/MS was performed using an EASY-nLC 1000 UPLC (Thermo) paired with a Q Exactive Plus (Thermo), except for the second ERLIC replicate samples (100 μg tryptic peptide preparation) and co-IP experiments which were run using an ACQUITY UPLC M-Class (Waters) and Q Exactive Plus. The analytical column employed was a 65 cm long, 75 μm internal diameter PicoFrit column (New Objective) packed in-house to a length of 50 cm with 1.9 μm ReproSil-Pur 120 Å C18-AQ (Dr. Maisch) using methanol as the packing solvent. Peptide separation was achieved using mixtures of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) with either a 90-minute gradient (used for all samples except TiO2-enriched samples): (min/%B, linear ramping between steps) 0/1, 2/7, 60/24, 65/48, 70/80, 75/80, 80/1, 90/1; or a 290-minute gradient (only used for TiO2-enriched samples): (min/%B, linear ramping between steps) 0/1, 2/7, 265/30, 270/60, 275/99, 280/99, 285/1, 290/1. All gradients were performed with a flowrate of 250 nL/min. At least one blank injection (5 μL 2% B) was preformed between samples to eliminate peptide carryover on the analytical column. 100 fmol of trypsin-digested BSA or 100 ng trypsin-digested wildtype K-12 MG1655 E. coli proteins were run periodically between samples as quality control standards.
The mass spectrometer was operated with the following parameters: (MS1) 70,000 resolution, 3e6 AGC target, 300–1700 m/z scan range; (data dependent-MS2) 17,500 resolution, 1e6 AGC target, top 10 mode, 1.6 m/z isolation window, 27 normalized collision energy, 90 s dynamic exclusion, unassigned and +1 charge exclusion. Mass spectra were searched with MaxQuant37 v1.5.1.2 using a custom database containing all possible 110,139 synthetic phosphosites encoded on the original oligonucleotide array (≤ 31 amino acid phosphosites plus the encoded lysine residue on the C-terminus, Supplementary Data 1) in addition to the E. coli proteome (EcoCyc K-12 MG1655 v17, downloaded 24 Jun 2015). The searches treated carbamidomethyl (Cys) as a fixed modification, and acetyl (N-terminal), oxidation (Met), deamidation (Asn, Gln), and phosphorylation (Ser/Thr/Tyr) as variable modifications. Up to 3 missed trypsin cleavage events were allowed, and peptides identified have a minimum length of 5 amino acids. The false discovery rate was set at 1%.
Phosphorylated tryptic peptides as identified by MaxQuant were matched to the anticipated phosphosite sequences. If phosphorylation was identified by MaxQuant with >50% certainty at a genetically encoded position, the corresponding phosphosite was counted in our tally. For calculations of the number of phosphosites present in the library regardless of phosphorylation status, the number of “leading razor” phosphosites identified in MaxQuant was tallied for all observed tryptic peptides. A list of the number of identified phosphosites and pSer-containing phosphosites for each sample preparation can be found in Supplementary Data 2. A discussion of alternative phosphosite identification and pSer localization parameters using relaxed stringencies can be found in Supplementary Note 1, and these results are also reported in Supplementary Data 2.
Fluorescence-activated cell sorting for Hi-P
20 mL of C321.ΔA cells containing either the SepOTSλ or supD tRNA (on plasmid modified to include SepRS9-EFSep21)5,33 plasmid were grown to an OD600 of 0.4. Cells were then spun down at 4,000 × g for 1 minute, supernatant was decanted, and cells were washed with 20 mL ice cold, deionized water. This was repeated once. Cells were resuspended in 50 μL water, mixed with 1 μL mode #2 library plasmid (approximately 100 ng/μL), and electroporated using the parameters stated above. The cells were then resuspended in 1 mL of S.O.C. medium (Thermo) and incubated for 1 h at 30 °C and 230 rpm in a 15 mL culture tube. At least 107 colony-forming units were obtained per electroporation, as calculated by plating serial dilutions. Recovered cells were directly inoculated in 50 mL of LB with 100 ng/μL ampicillin and 25 ng/μL kanamycin and grown overnight at 30 °C and 230 rpm.
The next morning, cultures were diluted to an OD600 of 0.15 in 5 mL of LB containing 100 ng/μL ampicillin, 25 ng/μL kanamycin, and 2 mM O-phospho-L-serine and grown at 30 °C and 230 rpm. The cells were grown until OD600 reached mid-log (0.6–0.8), then protein expression was induced using 1 mM IPTG, 0.2% arabinose, and 100 ng/μL anhydrotetracycline, and grown at 20 °C and 230 rpm for 20–24 h. 100 μL of cells were spun down at 4,000 × g and supernatant was removed. Cells were then resuspended in 3 mL ice cold M9 minimal media in a 5 mL polystyrene tube (Falcon).
Using a BD FACSAria III, cells were interrogated for mCherry-based fluorescence using a 561-nm laser. Cells were sorted using a gate empirically determined to yield substantially enriched fluorescent signal in regrown cell populations, which differed for each phospho-binding domain (see Supplementary Note 11). Cells were sorted directly into 1 mL LB without antibiotic, recovered at 30 °C and 230 rpm for 3 h, and then supplemented with 2 mL LB with a final concentration of 100 ng/μL ampicillin and 25 ng/μL kanamycin. After 24 h, sorted cell populations were then further supplemented with 2 mL LB with 100 ng/μL ampicillin and 25 ng/μL kanamycin and grown at 30 °C and 230 rpm for an additional 16 h. The procedure for protein expression, preparation for FACS, and cell sorting was repeated, using the same sorting and gating parameters as the first round of sorting. Cells were then recovered, regrown, induced and prepared for FACS as above. Cellular mCherry fluorescence was then observed using the FACSAria III. Plasmid libraries isolated by miniprep of twice-sorted cell populations were prepared for HTS as described above. The Hi-P (FACS/HTS) workflow was repeated in independent duplicate experiments (the same HTS-verified mode #2 vector library was separately electroporated in the expression strain) for the NEDD4 and NEDD4–2 WW2 domains and in a single experiment for 14–3-3 isoforms with SepOTSλ. For reproducibility studies (mode #2 phosphosite library with 14–3-3β with SepOTSλ, NEDD4 WW2 with SepOTSλ, or NEDD4 WW2 with supD tRNA), this workflow was performed in independent triplicate experiments. An example control experiment is shown in the Supplementary Note 11, where cells harboring a known domain-peptide interaction pair (mouse Nedd4 WW2 and peptide IPGTPPPNYD)22 were mixed at known ratios with cells encoding the Nedd4 WW2 and the N-terminal split mCherry with no fusion peptide; iterative sorting rounds enabled enrichment cells encoding the known interacting proteins at every tested dilution. Note that in all experiments, the addition of 2 mM O-phospho-L-serine may be unnecessary to promote phosphoserine incorporation, as phosphoserine is already present intracellularly in E. coli as a natural metabolite.
Plate reader BiFC assays
C321.ΔA cells harboring either the SepOTSλ or supD tRNA (on plasmid including SepRS9-EFSep21)5,33 plasmids were electroporated as detailed above with clonal vectors encoding the phosphorylation-binding domain fused to C-terminal split mCherry and the UAG-containing phosphosite fused to N-terminal split mCherry. Recovered cells were plated on LB agar containing 100 ng/μL ampicillin and 25 ng/μL kanamycin and grown for 18 h at 30 °C. Five colonies were then inoculated in 200 μL LB with 100 ng/μL ampicillin and 25 ng/μL kanamycin in a 96-well plate; this was performed in independent triplicate. Cultures were grown for 16–18 h at 30 °C, 530 rpm in a Jitterbug microplate shaker (Boekel). Cultures were then diluted to OD600 of about 0.15 in a total of 200 μL LB supplemented with 100 ng/μL ampicillin, 25 ng/μL kanamycin, 2 mM O-phospho-L-serine, 1 mM IPTG, 0.2% arabinose and 100 ng/μL anhydrotetracycline, and grown at 30 °C, 530 rpm in the microplate shaker for 24 h. These conditions were found to be sufficient to detect appreciable fluorescent signal with the BiFC system via time-course assay. 100 μL cells were then diluted in 100 μL LB and OD600 and fluorescence (580 nm excitation, 610 nm emission) readings were taken on a Synergy H1 microplate reader (BioTek). In parallel, the same strains were grown under identical conditions except without anhydrotetracycline (no mode #2 phosphosite expression) to establish baseline strain fluorescence values for background subtraction. Cells were diluted to ensure fluorescence and OD600 measurements fell within the linear range of the plate reader. For data analysis, background-subtracted fluorescence values were normalized by OD600 measurements, and negative values were treated as values of zero (below limit of detection). Positive and negative control interactions without UAG codons for the mouse Nedd4 WW2 domain were previously described22. The negative control for the 14–3-3 experiment was an arbitrary phosphosite from our library not detected by Hi-P experiments with 14–3-3 isoforms and not containing any 14–3-3 interaction motif elements with sequence AGPADAPAGAVVGGG[SP/S]PRGRPGPVPAPGLLA.
DNA design for full-length phosphoprotein pulldowns
To select novel phosphoproteins for interaction validation, phosphosite sequences identified by Hi-P using the 14–3-3β protein with SepOTSλ and read more than 1,000 times by HTS (Supplementary Data 4) were sorted by corresponding full-length protein length. Only proteins <300 amino acids in length and without annotated disulfide bonds or transmembrane domains were considered, as they were considered to be more likely to be able to be expressed heterologously in E. coli. All proteins that had been previously identified by high-throughput or low-throughput studies as candidate interactors with 14–3-3 proteins in the ANIA database were excluded17. Of the remaining candidate protein sequences for synthesis, proteins were sorted based on the distance of the location of phosphorylation from the protein termini, and the ten proteins with the most internal phosphorylation sites were selected in order to increase the likelihood that the phosphorylation site is given enough sequence context to participate in protein folding. Protein sequences were reverse translated and codon optimized for expression in K-12 E. coli using Geneious software v8 (and avoiding introduction of new KpnI/HindIII sites). Sequence complexity for DNA synthesis was minimized using an online tool from IDT (https://www.idtdna.com/site/Order/gblockentry). The desired position for phosphoserine incorporation was designated by a TAG codon. 5’-GGTACCAAG (KpnI plus the additional encoded Lys residue found in the phosphosite library) and AAGCTT-3’ were appended to all sequences to allow for restriction digest and ligation into expression vectors. Sequence-verified dsDNA was synthesized by Twist Bioscience. All full-length recombinant human phosphoprotein sequence information can be found in Supplementary Data 8. All of these genes were introduced into the modified pCRT7 vector encoding the N-terminal 8xHis tag/Avitag/MBP tag, except for FAM127A and RPL9 which were introduced into the same vector with N-terminal Avitag/MBP tag and the C-terminal 8xHis tag for reasons of poor expression. Note that for unknown reasons, the N-terminal 8xHis tag/Avitag/MBP tag is not strongly detectable by anti-His antibody.
Pull-downs
GST-fusion phosphosites with C-terminal 6xHis tags (mode #1) or MBP-fusion full-length phosphoproteins (mode #3) with either N- or C-terminal 8xHis tag were expressed clonally with either the SepOTSλ or supD tRNA plasmids in C321.ΔA in 500 mL cultures and purified using Ni-NTA resin as detailed in the library preparation section. 14–3-3β and 14–3-3σ proteins fused to C-terminal split mCherry with a 6xHis tag were expressed using the same vector as for Hi-P experiments, but instead transformed into BL21, and purified with Ni-NTA in the same manner as detailed in the library preparation section. Purified proteins were buffer exchanged using Amicon Ultra-0.5 10 kDa MWCO columns in storage buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 500 μM EDTA, 500 μM EGTA, 20% glycerol, 1 mM DTT, 50 mM NaF, and 1 mM NaVO4. 10 μg mode #1 phosphosite calculated by Coomassie-stained SDS-PAGE was immobilized on 10 μL pre-equilibrated glutathione HiCap resin (Qiagen) in a total of 100 μL binding buffer (50 mM Tris pH 7.4, 500 mM NaCl, 500 μM EDTA, and 1 mM DTT) and incubated end-over-end at 4 °C for 1 h. The resin was then washed twice with 100 μL binding buffer, resin was spun at 100 × g for 1 minute and supernatant was removed. 2 μg 14–3-3 proteins (as estimated by Coomassie stain) was then added to the resin in 10 μL total binding buffer, and 10 μL slurry was removed for SDS-PAGE analysis (“input”). 95 μL binding buffer was then added, and sample was incubated end-over-end for 14–16 h at 4 °C. Resin was then washed twice with 100 μL binding buffer, buffer was removed after spin, and finally 5 μL binding buffer was added to the resin. This final 10 μL slurry was used for SDS-PAGE analysis (“output”). Input and output samples were incubated at 95 °C for 5 minutes in 10 μL 2x Laemmli buffer, and 0.5 μL of each sample was run per lane in 10 μL total sample volume on 4–15% acrylamide gels (Bio-Rad). The exact same protocol was used for pull-downs using the MBP-fusion full-length phosphoproteins (mode #3 configuration), but using amylose resin (NEB) instead of glutathione HiCap resin.
Network analysis
Network analysis was performed by representing each Hi-P hit with the 7 amino acids before and after the encoded phosphoserine site. Custom Python scripts and the NetworkX package (https://networkx.github.io) were used to calculate the Levenshtein distance between each of the sequences, generate the network, and export it as a .gexf file. Gephi (https://gephi.org) was then used to visualize the network using ForceAtlas 2 graph layout algorithm with the following settings: gravity = 1.0; scaling =1.2; edge weight influence = 1; and LinLog mode enabled.
Co-immunoprecipitation experiments
DNA encoding candidate phosphosite interactors with the NEDD4 WW2 domain (PPXY-free phosphosites identified via Hi-P with SepOTSλ shown in Fig. 4c and Supplementary Fig. 9; PPXY-containing sequences removed to reduce likelihood of pSer-independent interactions) was first ligated in a mixed pool into the modified pCRT7 expression vector encoding an N-terminal 8xHis-Avitag-MBP fusion (mode #3, Supplementary Fig. 10a). This ligation, performed as described for other phosphosite plasmid libraries in “Phosphosite DNA library design”, was then transformed into chemically competent (standard RbCl method) DH10B cells to amplify the plasmid library, which was then isolated by miniprep. >10,000 colony-forming units were obtained in these mode #3 plasmid library preparative steps. The purified vector library was then introduced into the C321.ΔA cells containing either the SepOTSλ or supD tRNA-encoding plasmids, and protein expression on a 500 mL scale followed by Ni-NTA purification was performed as above. HEK293-T cells (ATCC CRL-11268) were cultured in Dulbecco’s Modified Eagle Medium (Life Technologies) with 10% fetal bovine serum (Atlanta Biologicals), 1x GlutaMAX (Gibco) and 1x penicillin-streptomycin (Gibco) at 37 °C and 5% CO2. HEK cells from a 100 mm plate were transiently transfected with a 15 μg vector encoding HA-tagged human NEDD4 isoform 4 (Addgene #27002) using Lipofectamine 3000 (Invitrogen) and the manufacturer’s protocol. After 48 h, cells were washed twice with 10 mL 1x PBS, and then 500 μL HEK lysis buffer was used to lyse cells (final concentration: 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 2.5 mM EDTA, 2.5 mM EGTA, 5% glycerol, 1 mM DTT, 1% Triton X-100, 1x phosphatase inhibitor cocktail 1 [Sigma P2850], 1x protease inhibitor cocktail [Roche cOmplete]). 95 μL of HEK lysate for either cells overexpressing HA-NEDD4 or without HA-NEDD4 (negative control) was mixed with 20 μL of agarose-conjugated monoclonal anti-HA antibody (Santa Cruz F-7) and 100 μg of pSer-containing or Ser-containing MBP-fusion targeted mode #3 phosphosite library (estimated by Coomassie-stained SDS-PAGE gel). Independent duplicate experiments were performed for each sample (the same HEK lysate and the same mode #3 libraries were used in separate replicate tubes). 5 μL of sample was removed for western analysis after mixing these components. Samples were incubated mixing end-over-end at 4 °C for 5 h, then washed twice with HEK lysis buffer, spinning down at 100 × g for 1 minute after washes. After removing the wash supernatant, 5 μL of resin was then removed for western analysis (Supplementary Fig. 10b). The remaining resin was used for a trypsin digest (performed on-resin without eluting bound proteins), using the same trypsin digest protocol as above scaled for 50 μg of protein. After desalting samples (see “Trypsin digestion of phosphosite libraries”), each sample was run in technical duplicate by LC-MS/MS, injecting 2 μg of peptides as determined by A280 over a 90-minute gradient (see “Mass spectrometry”). Mass spectra were searched using MaxQuant as above using a search database including all PPXY-free phosphosites from Supplementary Data 9. Phosphosite intensities and total spectral counts for observed phosphosites are reported in Supplementary Data 13. Phosphosites that were not observed in negative control experiments and had a higher average intensity in pSer-library experiments compared to Ser-library experiments were considered to be candidate pSer-dependent interactions via co-IP. NiNTA-purified targeted mode #3 phosphosite libraries (i.e. the reagent libraries used for co-IP experiments) were also analyzed by mass spectrometry (2 μg peptides, 90-minute gradient), and phosphosites that were identified are listed in Supplementary Data 13. All identified phosphosites from the purified targeted library contained pSer at the encoded position when expressed using SepOTSλ and not when using supD tRNA.
Comparison of LC-MS/MS and Hi-P datasets to evaluate system bias due to expression
For each phosphosite identified unambiguously by LC-MS/MS, the maximum intensity of the corresponding tryptic phosphopeptide across both ERLIC and TiO2 enrichment datasets was compared to the number of HTS reads for the phosphosite from deep sequencing datasets (i.e. from all HTS samples excluding Hi-P reproducibility studies), as presented in Supplementary Fig. 11.
Reproducibility studies
Hi-P was repeated in independent triplicate experiments for mode #2 phosphosite plasmid libraries (the same HTS-verified mode #2 plasmid library was separately electorporated into the expression strain) with either 14–3-3β + SepOTSλ, the NEDD4 WW2 domain + SepOTSλ, or the NEDD4 WW2 domain + supD tRNA. FACS-enriched candidate interacting phosphosite DNA was sequenced by HTS at the Massachusetts General Hospital Center for Computational & Integrative Biology DNA Core and sequence reads were processed as described in the “High-throughput sequencing information” section (Supplementary Data 14). Phosphosite-mapped DNA sequences were compared across replicate experiments, and Venn diagrams were constructed detailing the overlap between the replicate experiments (Supplementary Fig. 12). A read number cutoff was not imposed for this analysis.
Statistics
For pLogo analysis, significance was calculated by binomial probability of amino acid frequencies, and red lines indicate p = 0.05 significance threshold with Bonferroni correction19. All pLogo analysis was performed using a background of all possible theoretical phosphosites (listed in Supplementary Data 1) that are as long as or longer than the queried sequence length, aligned to the central pSer residue. Sample sizes and error bars are defined in figure legends. Types and number of replicates are defined for each experiment throughout the online methods. Additional details can be found in the Life Sciences Reporting Summary.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository38 with the dataset identifier PXD008707. High-throughput sequencing raw data files have been uploaded to the Sequence Read Archive (SUB3834588).
Code availability
Custom scripts used for mass spectrometry data analysis, HTS sequencing analysis, and network analysis are available online at https://github.com/rinehartlab.
Life Sciences Reporting Summary
A Life Sciences Reporting Summary is available.
Supplementary Material
Acknowledgements
We thank J. Niemann for his assistance with selecting phospho-specific antibodies to probe phosphosite synthesis, P. Anderson for assistance with oligonucleotide library synthesis, the Yale Center for Genome Analysis for high-throughput sequencing and K. Mohler for helpful comments on the manuscript. K.W.B. is supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1122492. R.S. received support from the Raymond and Beverly Sackler Institute for Biological, Physical and Engineering Sciences at Yale University and from Southern Connecticut State University School of Graduate Studies, Research, and Innovation. J.R. is supported by the NIH (GM117230, GM125951, DK0174334, CA209992). F.J.I. acknowledges support from the NIH (GM117230, GM125951, CA209992), DARPA (HR0011–15-C-0091), DuPont Inc., and the Arnold and Mabel Beckman Foundation. M.G. is supported by the NIH (HG008126).
Footnotes
Competing interests
J.R., F.J.I., K.W.B., and J.R.S. have filed a provisional patent application with the US Patent and Trademark Office (US Patent Application No. 62/639,279) concerning phosphopeptide-encoding oligonucleotide libraries and methods for detecting phosphorylation-dependent molecular interactions detailed in this work.
References
- 1.Ubersax JA & Ferrell JE Mechanisms of specificity in protein phosphorylation. Nature Reviews Molecular Cell Biology 8, 530–541 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Yaffe MB et al. The Structural Basis for 14–3-3:Phosphopeptide Binding Specificity. Cell 91, 961–971 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Johnson GL & Lapadat R Mitogen-Activated Protein Kinase Pathways Mediated by ERK, JNK, and p38 Protein Kinases. Science 298, 1911–1912 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Park H-S et al. Expanding the Genetic Code of Escherichia coli with Phosphoserine. Science 333, 1151–1154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirman NL et al. A flexible codon in genomically recoded Escherichia coli permits programmable protein phosphorylation. Nature communications 6, 8130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lajoie MJ et al. Genomically Recoded Organisms Expand Biological Functions. Science 342, 357–360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huttlin EL et al. Architecture of the human interactome defines protein communities and disease networks. Nature (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hein MY et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 163, 712–723 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Heo J-M, Ordureau A, Paulo JA, Rinehart J & Harper WJ The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Molecular Cell 60, 7–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ordureau A et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proceedings of the National Academy of Sciences 112, 6637–6642 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornbeck PV et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Research 43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeProust EM et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic acids research 38, 2522–2540 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amiram M et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nature Biotechnology 33, 1272–1279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isaacs FJ et al. Precise Manipulation of Chromosomes in Vivo Enables Genome-Wide Codon Replacement. Science 333, 348–353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou S et al. SH2 domains recognize specific phosphopeptide sequences. Cell 72, 767–778 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Sawyer N et al. Designed Phosphoprotein Recognition in Escherichia coli. ACS Chemical Biology 9, 2502–2507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tinti M et al. ANIA: ANnotation and Integrated Analysis of the 14–3-3 interactome. Database 2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanai F et al. TAZ: a novel transcriptional co‐activator regulated by interactions with 14‐3‐3 and PDZ domain proteins. The EMBO Journal 19, 6778–6791 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Shea JP et al. pLogo: a probabilistic approach to visualizing sequence motifs. Nature Methods 10 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Johnson C et al. Bioinformatic and experimental survey of 14–3-3-binding sites. Biochemical Journal 427, 69–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzivion G, Dobson M & Ramakrishnan G FoxO transcription factors; Regulation by AKT and 14–3-3 proteins. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1813, 1938–1945 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Lu P-J, Zhou X, Shen M & Lu K Function of WW Domains as Phosphoserine- or Phosphothreonine-Binding Modules. Science 283, 1325–1328 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Edwin F, Anderson K & Patel TB HECT Domain-containing E3 Ubiquitin Ligase Nedd4 Interacts with and Ubiquitinates Sprouty2. Journal of Biological Chemistry 285, 255–264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spagnol G et al. Structural Studies of the Nedd4 WW Domains and Their Selectivity for the Connexin43 (Cx43) Carboxyl Terminus. Journal of Biological Chemistry 291, 7637–7650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao S et al. Ubiquitin Ligase Nedd4L Targets Activated Smad2/3 to Limit TGF-β Signaling. Molecular Cell 36, 457–468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang B & Kumar S Nedd4 and Nedd4–2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death & Differentiation 17, 68–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skouloudaki K & Walz G YAP1 Recruits c-Abl to Protect Angiomotin-Like 1 from Nedd4-Mediated Degradation. PLoS ONE 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larman BH et al. Autoantigen discovery with a synthetic human peptidome. Nature Biotechnology 29, 535–541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto M et al. A large-scale targeted proteomics assay resource based on an in vitro human proteome. Nature Methods 14, 251–258 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Collins BC et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14–3-3 system. Nature Methods 10, 1246–1253 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Marx H et al. A large synthetic peptide and phosphopeptide reference library for mass spectrometry-based proteomics. Nature Biotechnology 31, 557–564 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Kosuri S et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nature Biotechnology 28, 1295–1299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S et al. A Facile Strategy for Selective Incorporation of Phosphoserine into Histones. Angewandte Chemie 125, 5883–5887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferdaus MZ et al. SPAK and OSR1 play essential roles in potassium homeostasis through actions on the distal convoluted tubule. The Journal of physiology (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kettenbach AN & Gerber SA Rapid and Reproducible Single-Stage Phosphopeptide Enrichment of Complex Peptide Mixtures: Application to General and Phosphotyrosine-Specific Phosphoproteomics Experiments. Analytical Chemistry 83, 7635–7644 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Vizcaíno JA et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature Biotechnology 32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository38 with the dataset identifier PXD008707. High-throughput sequencing raw data files have been uploaded to the Sequence Read Archive (SUB3834588).