Abstract
MicroRNA (miRNA)-mediated gene regulation contributes to liver pathophysiology, including hepatic stellate cell (HSC) activation and fibrosis progression. Here, we investigated the role of miR-942 in human liver fibrosis. The expression of miR-942, HSC activation markers, transforming growth factor-beta pseudoreceptor BMP and activin membrane-bound inhibitor (BAMBI), as well as collagen deposition, were investigated in 100 liver specimens from patients with varying degree of hepatitis B virus (HBV)-related fibrosis. Human primary HSCs and the immortalized cell line (LX2 cells) were used for functional studies. We found that miR-942 expression was upregulated in activated HSCs and correlated inversely with BAMBI expression in liver fibrosis progression. Transforming growth factor beta (TGF-β) and lipopolyssacharide (LPS), two major drivers of liver fibrosis and inflammation, induce miR-942 expression in HSCs via Smad2/3 respective NF-κB/p50 binding to the miR-942 promoter. Mechanistically, the induced miR-942 degrades BAMBI mRNA in HSCs, thereby sensitizing the cells for fibrogenic TGF-β signaling and also partly mediates LPS-induced proinflammatory HSC fate. In conclusion, the TGF-β and LPS-induced miR-942 mediates HSC activation through downregulation of BAMBI in human liver fibrosis. Our study provides new insights on the molecular mechanism of HSC activation and fibrosis.
Keywords: TGF-β signaling, HSCs, Viral hepatitis, Liver fibrosis
Introduction
Hepatitis B virus (HBV) infection is associated with hepatotoxity and injury, and is a major cause of chronic liver disease in humans. Its persistence is a major cause of liver fibrosis and cirrhosis. Liver fibrosis is generally associated with chronic hepatic inflammation and injury. In this process, hepatic stellate cells (HSCs) are activated by fibrogenic cytokines and mediators, and this is followed by matrix deposition and fibrosis as has been observed extensively in clinical settings (Shi et al. 2017; Yu et al. 2017). Although prevention or reversal of HSC activation has been experimentally shown to attenuate liver fibrosis, there is currently no effective approach to inhibit HSC activation in patients (Koo et al. 2016). One reason for this is that the underlying mechanisms of HSC activation and fate dynamics has not been fully delineated in normal and pathologic conditions of the liver.
MicroRNAs (miRNAs) are small endogenous noncoding RNAs that negatively regulate protein translation through binding to the 3′ untranslated regions (UTRs) of their target messenger RNAs (mRNAs) (Bartel 2004). Recent studies have implicated miRNAs in the pathogenesis of liver fibrosis (Hyun et al. 2016; Lakner et al. 2012; Sombetzki et al. 2015; Zhang et al. 2014) and various other liver disease conditions (Bandiera et al. 2015; Matsuura et al. 2016; Wang et al. 2012; Wu et al. 2016). For example, miR-378a suppresses HSC activation by depleting Gli3 expression, which is associated with smoothened-dependent NF-κB signaling in liver fibrosis (Hyun et al. 2016). miR-22½22 have been reported as new markers for HSC activation and liver fibrosis progression (Ogawa et al. 2012). Further, miR-942 was recently identified as a novel miRNA in hepatocytes and hepatocellular carcinoma cells, specifically as a regulator of apoptosis (Liu et al. 2014b; Yang et al. 2014a). However, the role of miR-942 in HSC activation and human liver fibrosis progression is still unclear.
On the other hand, the transforming growth factor (TGF-β1) is a known driver of liver fibrogenesis (Dooley and ten Dijke 2012). We have previously reported that TGF-β1 signaling is enhanced by TLR4-mediated suppression of BAMBI in HSCs and during hepatic fibrosis (Liu et al. 2014a; Seki et al. 2007). Given that BAMBI is a decoy TGF-β receptor I (TpRI/ALK-5), which interferes with TGF-β1 signaling by capturing its activated ligands, the reduction of BAMBI expression could amplify fibrotic processes by enhancing signaling via ALK-5. Consistently, BAMBI regulation in HSCs appears to play a prominent role in the development and progression of liver inflammation and fibrosis (Friedman 2007). Indeed, we have demonstrated that lipopolysaccharide (LPS)-mediated TLR4 signaling induces an NF-κB p50-HDAC1 repressor complex, which subsequently blunts the transcriptional activity of the BAMBI gene promoter in HSCs (Liu et al. 2014a). In the current study, we investigated the involvement of miR-942 in liver fibrosis progression in patients, and mechanistically explored its role in facilitating HSC activation.
Materials and methods
Liver tissue biopsy specimens and patient information
Liver tissue specimens were obtained from 100 patients with clinically diagnosed liver fibrosis (Stages F0/1 = 16; F2 = 39; F3 = 31; F4 = 14 tissues), who underwent liver biopsy from February 2013 to July 2016. The etiological background of the patients is hepatitis B viral hepatitis, and the human liver specimens are all from Putuo Hospital at Shanghai University of Traditional Chinese Medicine (TCM), Shanghai, China. The stage of liver fibrosis was scored based on the examination of haematoxylin and eosin, Sirius red, Masson trichrome, and reticular fiber staining by three pathologists in the TCM Department of Pathology. Fibrosis stages were defined based on Scheuer criteria (Desmet et al. 1994; Goodman 2007) as follows: grade 0—normal liver; grade 1—an increase of collagen without the formation of septa (small satellite expansion of the portal fields); grade 2—formation of incomplete septa not interconnecting with each other, from the portal tract to the central vein; grade 3—complete but thin septa interconnecting with each other, which divide the parenchyma into separate fragments; and grade 4—complete cirrhosis, similar to grade 3 with thicker septa. Written informed consent was obtained from each patient, and the study was approved by the Clinical Ethics Committee of Putuo Hospital of Shanghai University of TCM within which the work was undertaken. Patients with renal and/or hepatic failure, acute coronary syndromes, valvular heart diseases, autoimmune thyroid diseases, or systematic inflammatory diseases were excluded from our study, and none of the patients had received anti-viral therapies prior to liver biopsy.
Cell culture
The human HSC cell line LX2 (catalog no. SCC640) was purchased from Merck Millipore (Darmstadt, Germany) and cultured in Dulbecco Modified Eagle Medium (DMEM)↱ supplemented with 2% fetal bovine serum (FBS) and 1% penicillin–streptomycin antibiotics. Human (h) HSCs (hHSCs, catalog no. #5300), hepatocytes (catalog no. #5200), hepatic macrophages (hKCs) (catalog no. #5340), and hepatic sinusoidal endothelial cells (hLSECs) (catalog no. #5000) were all purchased from ScienCell Research Laboratories (SanDiego, CA, USA). hHSCs were cultured in DMEM supplemented with 10% FBS, and used between passages 4 and 7. hHepatocytes were cultured in MEM199↱ supplemented with 10% FBS, hKCs were cultured in RMPI1640 supplemented with 10%FBS, hLSEC were cultured in Endothelial Cell Medium (ECM, Cat. #1001) supplemented with 10% FBS.
Immunofluorescent staining
Tri-color immunofluorescence staining was performed on paraffin sections using different combinations of antibodies. After deparaffinization and dehydration, microwave antigen retrieval was performed for 5 min before peroxidase quenching with 3% H2O2 in phosphate-buffered saline (PBS) for 15 min. Subsequently, the sections were blocked with 5% bovine serum albumin (BSA) for 30 min and then were incubated with monoclonal anti-desmin for 1 h at room temperature. Slides were then washed three times with PBS and incubated with the secondary Cy3-conjugated AffiniPure goat anti-mouse antibody (Jackson ImmunoResearch, Cat: 115–165-062, PA, US) for 30 min. The sections were incubated with the BAMBI antibody followed by Cy2-conjugated AffiniPure goat anti-rabbit antibody (Jackson ImmunoResearch, Cat: 111–225-144, PA, US). Imaging was performed using Zeiss (Mannheim, Germany) LSM800 inverted stand and Axio Imager Z2m confocal system.
siRNA and oligonucleotide transfection
Transfection of siRNA targeting NF-κB p50, Smad2/3, BAMBI, as well as non-targeting (scrambled) control was carried out according to manufacturer’s protocol (Santa Cruz Biotech.Inc, CA, USA), using Lipofectamine LTX as transfection reagent. Knockdown efficiency was assessed 48–72 h post-transfection using real-time PCR and western blotting methods. For modulating miRNA, all miRNA mimics, miR-942 inhibitor, and corresponding controls used were synthesized by RiboBio (Guangzhou, China). pEGFPN3-BAMBI and control vector were purchased from Shanghai Generay Biotech Co., Ltd (Shanghai, China). Oligonucleotide transfection was performed with Lipofectamine LTX reagent according to the manufacturer’s protocol, at a final concentration of 120 pmol/L.
Quantitative real-time PCR
Total RNA was extracted using the Trizol method (Takara, Japan). RNA concentration and purity were determined by Colibri Microvolume Spectrometer (Titertek-Berthold, Germany). This was followed by reverse transcription of 1 μg total RNA. PrimeScript RT Reagent Kit were purchased from Takara (Shanghai, China). PCR amplification was conducted in 10 μl total volume consisting of 3 μl cDNA, 5 μl SYBR green mixture, 1.6 μl H2O, and 0.4 μl of primer mix (10 μM). Primers used for quantitative RT-PCR or for cloning are listed in Supporting Table 1. The amplification steps were 40 cycles of denaturation at 94 °C for 15 s, extension at 60 °C for 60 s, using the ABI ViiA 7Dx real-time PCR system (Life Technologies, NY, USA). Gene expression was normalized to 18S rRNA as internal control. For bulge-loop miRNA qRT-PCR, primer set specific for miR-942 was designed by RiboBio (Guangzhou, China). U6 small nucleolar RNA (RiboBio Co., Ltd) was used for miRNA normalization. qRT-PCR values were calculated by relative quantification using the delta-delta CT method.
Immunoblot analysis
Cells were lysed in ice-cold radioimmunoprecipitation assay buffer containing protease inhibitors. The cell lysates were then collected and centrifuged at 11,000×g for 10 min at 4 °C. Protein concentration of the supernatant was measured using Pierce BCA Protein Assay kit according to manufacturer’s instruction (cat no. 23227, Thermo Fisher, IL, USA). 20 μg protein was separated by SDS-PAGE and subsequently transferred onto PVDF membranes (GE Healthcare Life Science). Membranes were blocked for 1 h with 5% nonfat milk, incubated overnight with the respective primary antibodies and for 1 h with secondary antibodies (mouse or rabbit), followed by chemiluminescent detection. Monoclonal antibody against GAPDH was used as loading control. Details of the antibodies used are included in the Supplementary material. Densitometric quantification of immunoblot intensity was assessed using Image J software (https://imagej.nih.gov/ij/).
ChIP assay
Cross-linking was performed by adding 1% formaldehyde directly into plates of cultured cells. Cells were resuspended in sonication buffer and incubated on ice for 10 min to lyse the nuclei. Samples were further sonicated to obtain 200- to 400-bp chromatin fragments. Immunoprecipitation was carried out according to the protocol provided by Upstate Biotechnology and as previously described (Liu et al. 2014a, b). Antibodies for NF-κB p50 and Smad2/3 were incubated with diluted chromatin at 4 °C overnight. Protein A-Sepha-rose blocked with sheared salmon sperm DNA was used to collect Ab-chromatin complexes. Primers used to amplify the − 215 NF-κB binding site of the miR-942 promoter are 5′-AAAGTCATGCCCCACGTGTT-3′ (forward) and 5′-GGCCTGCTCCAAATGTTGTT-3′ (reverse). Primers used to amplify the − 1286 Smad2/3 binding site of the miR-942 promoter were 5′-AGCGCCAATTGCTCTGACTT-3′ (forward) and 5′-ACGGCAGCTTGAGATGCTAA-3′ (reverse).
Statistical analysis
The results were presented as mean ± SD and as representative of three independent experiments, each in biological triplicates. Differences between multiple groups were compared by one-way analysis of variance (ANOVA) with Kruskal–Wallis test using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA, USA). Statistical significance was accepted when P < 0.05. Further details about materials and methods are included in the Supplementary material.
Results
High hepatic miR-942 level is associated with HSC activation in human liver fibrosis
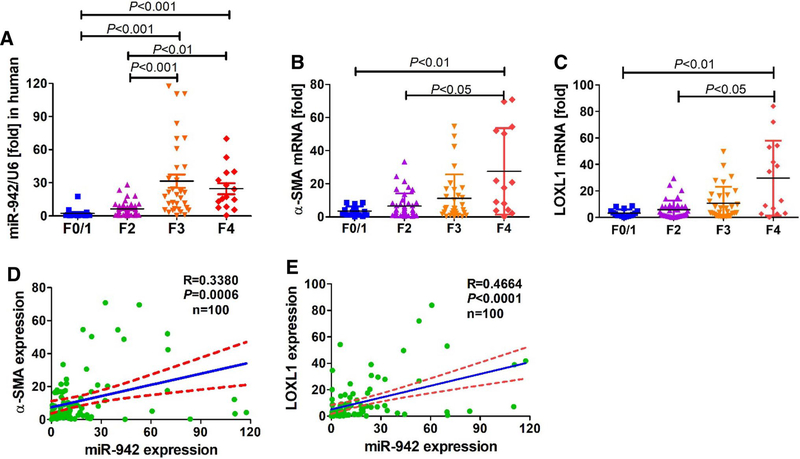
To determine the relevance of miR-942 in human chronic liver disease, we examined 100 HBV-infected human liver tissue specimens at various fibrosis stages. Assessment of liver function parameters showed that patients at advanced fibrosis (F4 stage) showed higher ALT and AST as compared to those with F0/1 fibrosis (P < 0.05, Supporting Table 2). Further, patients at F4 stage presented with significantly higher (P < 0.05) level of fibrosis parameters, e.g. hyaluronic acid, procollagen type III, collagen IV and laminin. We found that miR-942 expression is significantly upregulated in liver tissues at F3 and F4 fibrosis stages compared to F1/0 samples (Fig. 1a).
Fig. 1.
miR-942 expression is induced in activated HSC during human liver fibrogenesis. a mRNA expression of miR-942, b α-SMA, and c LOXL1 in 100 human liver fibrosis specimens (F0/1 = 16; F2 = 39; F3 = 31; F4 = 14) as measured by real-time PCR. Bars show mean ± SD, n = 3 per group. One-way ANOVA (and nonparametric) Kruskal–Wallis test were used for statistical comparison of data. d Pearson correlation between miR-942 and α-SMA or e LOXL1 mRNA levels. r=correlation coefficient. P < 0.05 is statistical significance
Liver fibrosis is known to result from HSC activation. We measured mRNA expression of activation markers α-SMA and lysyl oxidase homolog 1 (LOXL1), and found an increased expression with the severity of fibrosis (Fig. 1b, c). Of note, both α-SMA (r = 0.3380, P = 0.0006, n = 100) and LOXL1 (r=0.4664, P < 0.0001, n = 100) correlated significantly with miR-942 expression (Fig. 1d, e), which also showed a notable similar cell distribution with α-SMA.
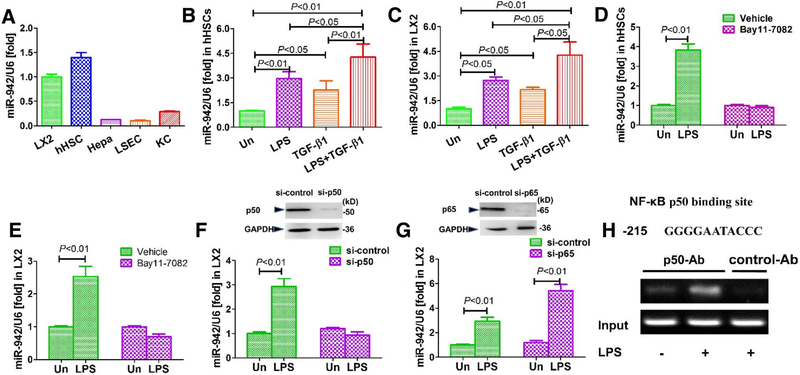
Using commercially available human primary liver cells, we confirmed that HSCs are the predominant liver cell type producing miR-942. Specifically, miR-942 is expressed at a higher level in both LX2 cells and human primary HSCs (hHSC) compared to Kupffer cells (i.e. liver resident macrophages), liver sinusoidal endothelial cells or hepatocytes (Fig. 2a). Importantly, the hepatic stellate cells LX2 cells and hHSC expressed similar levels of miR-942. Based on these results, we hypothesized that miR-942 overexpression contributes to, or results from, HSC activation during liver disease progression.
Fig. 2.
NF-κB p50 mediate miR-942 upregulation in response to LPS. a miR-942 expression in liver cell types after 24 h culture as measured by real-time PCR. Bars show mean ± SD, n = 3 per group. hHSC human primary hepatic stellate cells, Hepa hepatocytes, KCs Kupffer cells, LSECs liver sinusoidal endothelial cells. b miR-942 expression in hHSCs and c LX2 in response to LPS and TGF-β1 stimulation. The cells were stimulated with 5 ng/ml TGF-β1 for 48 h, followed by a 4 h stimulation with 100 ng/ml LPS. Bars show mean ± SD, n = 3 per group of three independent experiments. d miR-942 expression in hHSCs and e LX2 after 4 h stimulation with LPS (100 ng/ml). Where Bay11–7082 is indicated, hHSCs were pre-treated with 1 μM of the inhibitor for 2 h before LPS treatment. Un untreated. Data represent mean ± SD of three independent experiments. f Real-time PCR data following siRNA-mediated knockdown of p50 and (g) p65. On the upper part of the figures: immunoblots showing knockdown efficacy 48 h after transfection. LX2 cells were stimulated with 100 ng/ml LPS for 4 h. Un untreated. Bars show mean ± SD, n = 3 per group of three independent experiments. h Sequence of the NF-κB binding site at position − 215 of the miR-942 promoter as predicted with bioinformatics platforms. Below: a ChIP analysis showing the effect of LPS (100 ng/ml for 1 h) on p50 recruitment to the NF-κB binding site of the human miR-942 promoter. Following treatment with LPS, cells were fixed and soluble chromatin was immunoprecipitated using anti-p50 or control abs (normal mouse IgG), as indicated. The miR-942 promoter fragment containing the – 215-binding site was amplified by PCR. Un untreated
LPS and TGF-β1 are known drivers of liver fibrosis, thus we tested their potential impact on miR-942 expression in HSCs. Indeed, miR-942 expression in hHSCs and LX2 was increased about two-threefold in response to LPS and TGF-β1 stimulation (Fig. 2b, c). Further, miR-942 expression was superinduced by the parallel activation by co-treatment with LPS plus TGF-β1 in hHSCs (Fig. 2b, c), suggesting a synergistic impact upon activating the two signaling pathways. These data together support that miR-942 is crucially increased in HSC-associated liver fibrosis.
NF-κB p50 acts downstream of LPS to upregulate miR-942 expression
Given that LPS is a strong inducer of the transcription factor NF-κB, we hypothesized that NF-κB upon induction may bind to and activate the miR-942 promoter. Accordingly, treatment of both hHSCs and LX2 cells with Bay11–7082, an inhibitor of NF-κB, completely reversed the miR-942 induction caused by LPS (Fig. 2d, e), confirming that NF-κB is required for the elevated expression of miR-942.
To determine the NF-κB subunit responsible for miR-942 upregulation in HSCs, we carried out siRNA-mediated silencing of its p50 or p65 subunits in LX2 cells and subsequently stimulated the cells with LPS. Interestingly, whereas LPS stimulation induced a three-fold increase of miR-942 expression in the siRNA control setting (Fig. 2f), this effect was blunted in the p50-depleted LX2 cells. On the other hand, the knockdown of p65 did not alter the ability of LPS to induce miR-942 (Fig. 2g). These results suggest that NF-κB subunit p50, but not p65, is a mediator of the LPS-dependent upregulation of miR-942 in HSCs. p50 regulates gene expression as a homodimer (Liu et al. 2014a, b) and sequence analysis of the human miR-942 promoter (− 2000 to 0) predicted a p50 binding site (Fig. 2h). Thus, we further performed ChIP analysis for the respective − 215 promoter region of the miR-942 promoter and confirmed that LPS mediates an increase of NF-κB p50 binding (Fig. 2h). These evidences together support an LPS-induced regulation of mrR-942 level via NF-κB signaling.
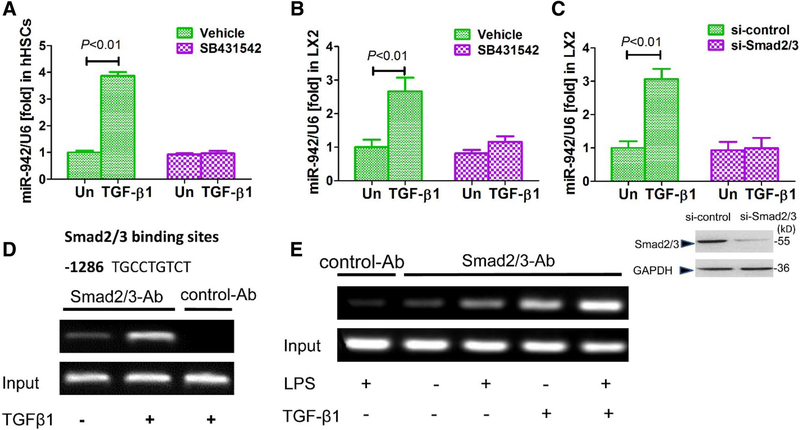
ALK-5 and R-Smad activation mediate miR-942 upregulation in response to TGF-β1
Canonical TGF-β signaling is mediated by its type I receptor ALK-5 and the receptor (R) Smad proteins Smad2 and Smad3 (Massague 2012). Thus, we used SB431542 (a selective inhibitor of ALK-5) to interfere with the TGF-β1 pathway in hHSCs and LX2 cells, and subsequently assessed the impact on miR-942 induction. SB431542 blunted the upregulation of miR-942 mediated by TGF-β1 (Fig. 3a, b), suggesting that ALK-5 activity is required for miR-942 upregulation. Similarly, knockdown of Smad2/3 in LX2 cells suppressed the induction of miR-942 by TGF-β1 (Fig. 3c), further supporting that the canonical Smad2/3 pathway is required for the observed upregulation of miR-942. Bioinformatics analysis identified a Smad2/3 binding site at position − 1286 in the miR-942 promoter (Fig. 3d). Consistently, ChIP assays confirmed that TGF-β1 stimulation induces the binding of Smad2/3 to the predicted miR-promoter site in LX2 cells (Fig. 3d). Of note, stimulation of LX2 cells with LPS also induces R-Smad binding activity to the miR-942 promoter region (Fig. 3e), supporting our prior observation that TGF-β1/LPS cotreatment superinduced miR-942.
Fig. 3.
Smad2/3 mediate the induction of miR-942 expression in response to TGF-β. a Real-time PCR showing miR-942 levels in hHSCs and b in LX2 cells after 24 h stimulation with 5 ng/ml TGF-β1. The cells were pre-treated for 2 h with 10 μM SB431542 (a TGF-β1 receptor inhibitor). Un untreated. Bars show mean ± SD, n = 3 per group, of three independent experiments. c miR-942 levels as measured after 48 h of Smad2/3 knockdown in LX2 cells, and following stimulation with 5 ng/ml TGF-β1 for 24 h. Un untreated. Data represent mean ± SD of three independent experiments, n = 3 per group. One-way ANOVA (and nonparametric) Kruskal–Wallis test was used for statistical comparison of data (a–c). d Predicted Smad2/3 binding site at position − 1286 of the miR-942 promoter as identified by bioinformatics analysis, and ChiP analysis showing the effect of TGF-β stimulation (5 ng/ml). e ChiP assay showing the combined effect of 100 ng/ml LPS and 5 ng/ml TGF-β stimulation (2 h) on the recruitment of Smad2/3 to Smad binding site in the hsa-miR-942 promoter in LX2 cells. Un untreated, Ab antibody. LX2 cells pre-treated with LPS for 2 h in the combination treatment. Then, cells were lysed, nuclei were purified and soluble chromatin was immunoprecipitated using anti-Smad2/3 or normal rabbit IgG as control Ab. Subsequently, the miR-942 promoter fragment containing the – 1286-binding site was amplified by PCR
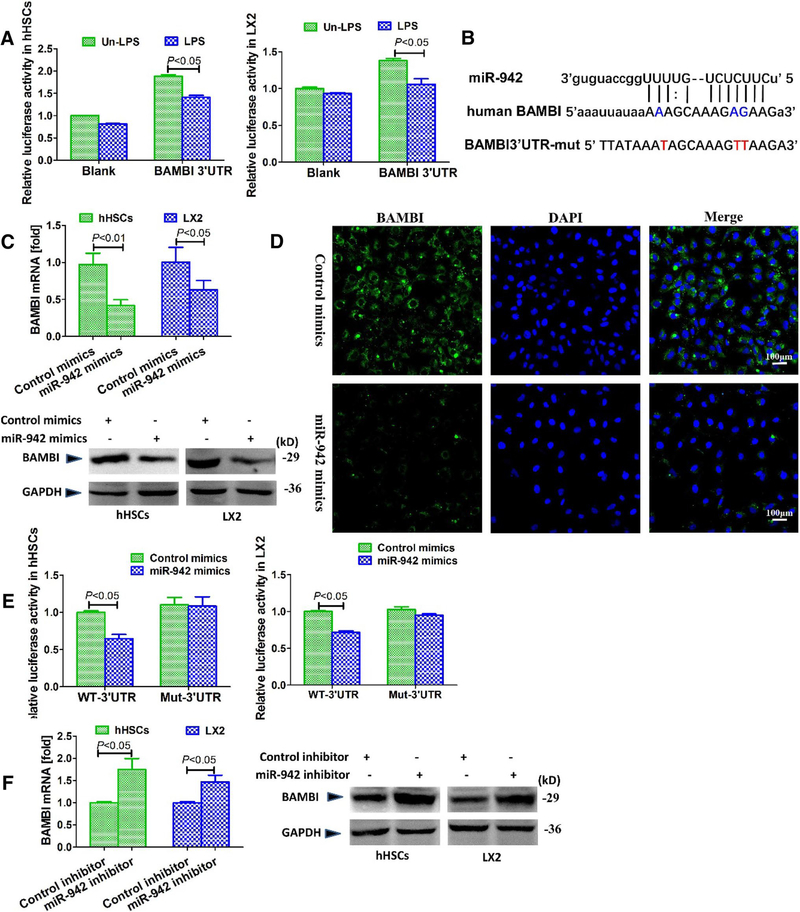
BAMBI is a direct target of miR-942 in HSCs
We previously demonstrated that the interaction of NF-κB p50 with HDACi represses the transcriptional activity of BAMBI gene, with subsequent sensitization of HSCs for TGF-β1 signaling (Liu et al. 20i4a, b). Thus, based on the above findings that both TGF-β1/Smad and LPS/NF-κB/p50 regulate miR-942 expression, we reasoned that miR-NAs represent additional regulators of BAMBI expression levels in HSCs. To functionally proof this, we transfected hHSCs and LX2 cells with a human BAMBI 3′UTR luciferase reporter and stimulated the cells with LPS. The luciferase activity decreased significantly (P < 0.05) upon LPS treatment as compared with the control (Fig. 4a), indicating that the same inducer of miR-942 suppressed BAMBI. To further explore potential targets of miR-942, we used online platforms TargetScan (http://www.targetscan.org/) and targets/expression (http://www.microrna.org/microrna/home.do). Interestingly, BAMBI emerged as a potential target of miR-942, as its 3′UTR contained a complementary site for the seed region of miR-942 (Fig. 4b).
Fig. 4.
miR-942 directly binds to and suppress BAMBI. a Relative luciferase activity of BAMBI 3′UTR in hHSCs and LX2 cells. Cells were transfected with BAMBI-3′UTR luciferase reporter plasmids for 18 h and stimulated with 100 ng/ml LPS for 4 h; Renilla luciferase was used for normalization. Bars indicate mean ± SD of three independent experiments, n = 3 per group. b miR-942 binding site predicted in the 3′UTR of human BAMBI mRNA using miRNA database (http://www.miRNA.org). c The effect of miR-942 mimics on BAMBI mRNA and protein levels in HSCs as measured 48 h post-transfection by real-time PCR and immunoblotting, respectively. Bars indicate mean ± SD of three independent experiments, n = 3 per group. d Immunofluorescence showing the repression of BAMBI by miR-942 in LX2 cells. Original magnification × 100. e Relative luciferase activity of BAMBI3′UTR (wt) or BAMBI3′UTR (mut) in hHSCs and LX2 cells, upon co-transfection with miR-942 or control mimics (120 nmol/L) for 18 h. Renilla luciferase was used for normalization. Bars indicate mean ± SD of three independent experiments, n = 3 per group. f BAMBI mRNA and protein expression as measured in HSCs after 72 h treatment with miR-942 inhibitor. Bars indicate mean ± SD of three independent experiments. Where applicable, one-way ANOVA (and nonparametric) Kruskal–Wallis test was used for statistical comparison of data
To determine whether miR-942 regulates BAMBI expression, we transfected hHSCs and LX2 cells with miR-942 mimics. This led to a reduced expression of BAMBI mRNA and protein as observed in real-time PCR, immunoblots and immunofluorescence analyses (Fig. 4c, d). We also performed further validation studies using luciferase reporter plasmids with a mutated (Mut) BAMBI 3′-UTR region. In both hHSCs and LX2 cells, transfection of miR-942 decreases luciferase activity of the reporter construct containing the WT BAMBI 3′-UTR by about 30% compared to the control, whereas luciferase activity of the reporter construct with the Mut-BAMBI 3′-UTR was unaffected (Fig. 4e). These results suggest that the miR-942 binding site of the BAMBI 3′-UTR is required for miR-942-mediated posttranscriptional regulation of BAMBI expression. Consistently, inhibition of miR-942 increases the expression of BAMBI at mRNA and protein levels in hHSCs and LX2 (Fig. 4f). Collectively, these findings demonstrate that BAMBI is post-transcriptionally regulated by miR-942.
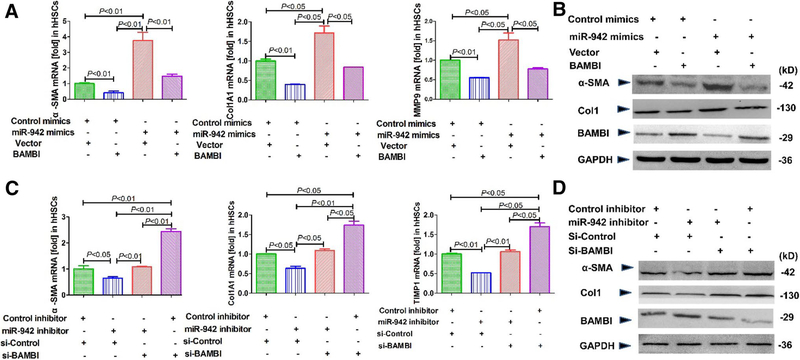
miR-942 mediates HSC activation and fibrogenesis via BAMBI reduction
Given that miR-942 expression levels correlate with HSC activation in fibrosis and decrease BAMBI expression by post-transcriptional regulation; we next investigated whether miR-942 is profibrogenic through BAMBI. BAMBI overexpression reduced the mRNA expression of fibrotic markers α-SMA, collagen 1A and MMP9 in hHSCs (Fig. 5a) and LX2 cells (Supporting Fig.1a), supporting our previous finding that BAMBI interferes with fibrogenic TGF-β1 signaling (Seki et al. 2007). Further, BAMBI overexpression is sufficient to blunt profibrogenic protein expression induced by miR-942 mimics in hHSC and LX2 cells (Fig. 5b). We also found that the knockdown of BAMBI increases α-SMA, Col1A1, TIMP1, and LOXL1 mRNA levels in hHSCs (Fig. 5c, d), and in LX2 cells (Supporting Fig. 1b), but did not further increase the expression of the mentioned profibrogenic genes when the cells were treated with miR-942 inhibitor. Together, these results strongly support that (a) BAMBI is a downstream target of miR-942, and (b) miR-942 mediates HSC activation through BAMBI.
Fig. 5.
miR-942 promotes HSC activation, fibrogenesis via BAMBI downregulation. a α-SMA, Col1A1, and MMP9 mRNA expression in hHSC, as measured by quantitative real-time PCR after transfection with miR-942 mimics, control mimics or BAMBI expression plasmid (pBAMBI) for 48 h. Data represent mean ± SD of three independent experiments, n = 3 per group. b α-SMA, Col1A1, and BAMBI expression in LX2 cells, as determined by immunoblot after transfection with miR-942 mimics, control mimics or BAMBI expression plasmid (pBAMBI) for 72 h. c Expression of α-SMA, Col1A1, and TIMP1 in hHSCs, as determined by real-time PCR after transfection with miR-942 inhibitor, control inhibitor, si-BAMBI or a si-control for 72 h. Data represent mean ± SD of three independent experiments, n = 3 per group. One-way ANOVA (and nonparametric) Kruskal–Wallis test was used for statistical comparison of data. d α-SMA, Col1A1, and BAMBI expression, as determined by immunoblot in LX2 cells, after transfection with miR-942 inhibitor, control inhibitor, si-BAMBI or si-control for 72 h
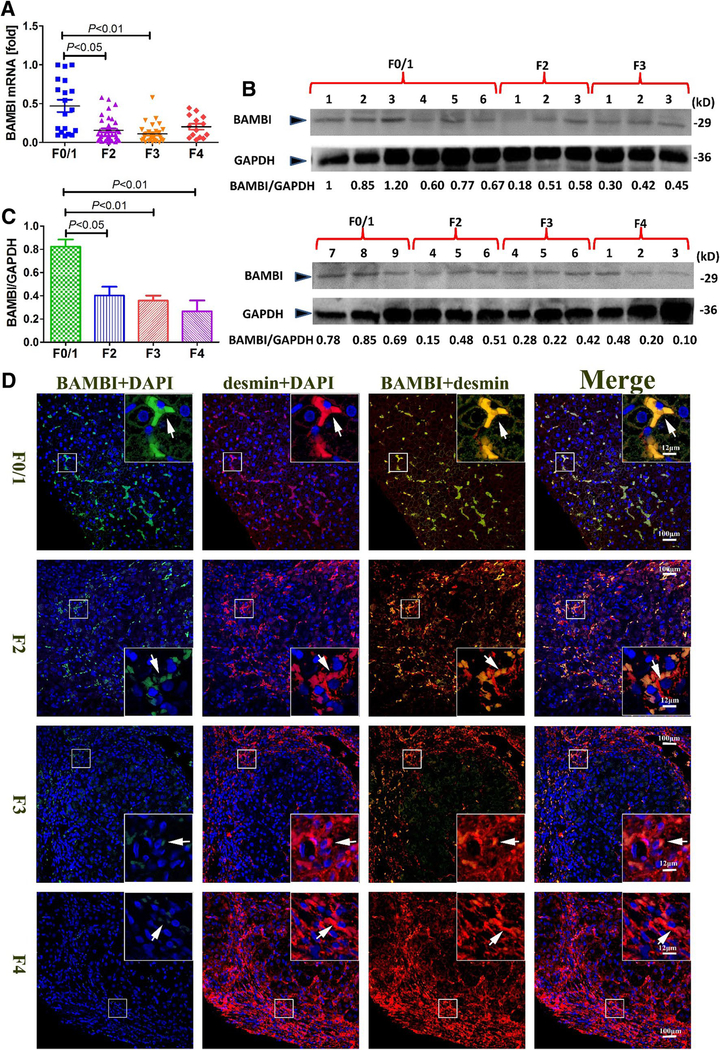
BAMBI is predominantly expressed by HSCs in the liver and is decreased in human hepatic fibrosis
Although we have previously associated BAMBI with HSC activation (Liu et al. 2014a), its implication in human liver fibrosis has not been reported. We found that hepatic BAMBI mRNA and protein levels are significantly low in patients with advanced stage liver fibrosis, when compared to those with mild or no fibrosis, i.e. F0/1 group (Fig. 6a–c). These results were evident when we performed colocalization experiments with desmin staining, which is representative for quiescent as well as activated HSCs. With desmin, we observed a strong overlay in F0/F1 stage liver tissue indicating the expression of BAMBI in desmin-positive quiescent HSC (Fig. 6d). The number of desmin-positive cells in patients with F2–F4 stage is significantly higher in hepatic lobules, ostensibly indicating proliferation and migration of the activated HSCs, whereas BAMBI-positive cells remarkably display weak staining. These findings strongly suggest that BAMBI expression is decreased during HSC activation and fibrosis progression.
Fig. 6.
BAMBI is downregulated in human liver fibrosis. a BAMBI mRNA expression in patients with different liver fibrosis stages (F0/1 = 16; F2 = 39; F3 = 31; F4 = 14 samples), as measured by realtime PCR. b Representative immunoblots of BAMBI protein levels from 24 human liver fibrosis tissues. Samples per group are as follows: F0/1 = 9; F2 = 6; F3 = 6; F4 = 3). c Densitometric quantification of BAMBI immunoblots from b using Image J software. Bars represent mean ± SD. d Representative images of paraffin sections from patients with various liver fibrosis stages indicated. Samples are immunostained for BAMBI (green), desmin (red) and DAPI (blue). Image magnification is ×200, enclosed area shows ×800 magnification; arrows denote co-localization
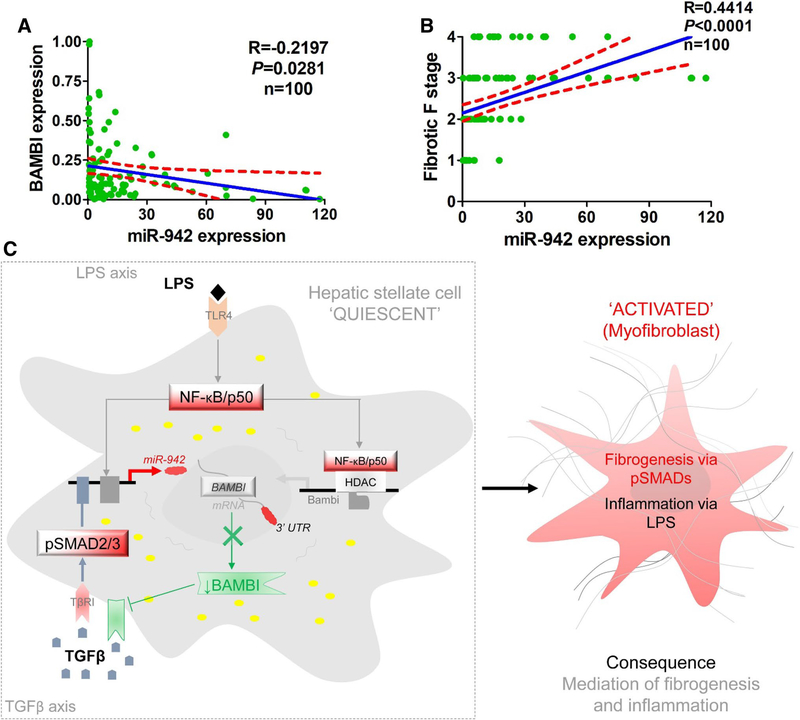
Finally, we found a significant negative correlation between BAMBI mRNA and miR-942 levels (r = − 0.2197, P = 0.0281) (Fig. 7a) in human liver fibrosis. Importantly, miR-942 levels correlated positively with liver fibrosis stages (r = 0.4414, P < 0.0001) (Fig. 7b). Taken together, these data strongly suggest that hepatic miR-942 facilitates liver fibrosis progression by regulating BAMB1 expression and function (Fig. 7c).
Fig. 7.
miR-942 and BAMBI correlation with fibrotic stage and proposed mechanism in HSC activation. a Pearson correlation analysis indicating an inverse relationship between miR-942 and BAMBI mRNA levels. r=correlation coefficient, statistical significance – P < 0.05. b Pearson correlation analysis indicating direct association between high miR-942 expression and fibrotic stage. c Proposed mechanism of miR-942-mediated liver fibrosis. Upon stimulation with TGF-β1 and LPS, Smad2/3 and NF-κB p50 respectively bind to miR-942 promoter. This leads to the induction of miR-942 expression, the subsequent downregulation of BAMBI, and HSC activation. HSC activation then orchestrates fibrosis
Discussion
The quest for antifibrosis therapy has led to several insights on the molecular mechanisms and drivers of liver fibrosis, including HSC activation and the possible role of miRNAs (Nwosu et al. 2016; Moran-Salvador and Mann 2017; Yang et al. 2015). We show that miR-942 is produced by activated HSC, increases significantly in human liver fibrosis, and is positively correlated with fibrosis progression. To the best of our knowledge, this is the first report on miR-942 as a profibrogenic miRNA in human liver fibrosis.
Several miRNAs, including miR-19 (Lakner et al. 2012), miR-200a (Yang et al. 2014b) and miR-29 (Roderburg et al. 2011; Zhang et al. 2014), have recently been implicated in the regulation of HSC activation and liver fibrosis. The microRNA, miR-942, is a novel candidate in liver diseases. Our study reveals that HSCs are the major miR-942-expressing cell type, compared to hepatocytes, KCs, and LSECs, in human liver fibrosis from HBV patients. We identified NF-κB/p50 and Smad2/3 as downstream mediators of the miR-942 promoter via binding to the respective transcription factor binding sites upon LPS and/or TGF-β1 stimulation, and identified BAMBI as a potential target of miR-942. We found this quite interesting, given that we have recently shown a functional link between BAMBI, LPS and TGF-β1 during HSC activation and fibrogenesis in mouse liver (Seki et al. 2007).
Studies have investigated the importance of BAMBI in human diseases (Legg 2016). In this present study, we discovered a miR-942-mediated post-transcriptional mechanism of BAMBI degradation, which is regulated by LPS. Furthermore, TGF-β1/Smad2/3 signaling, which itself is enhanced upon BAMBI depletion, transcriptionally induces miR-942 expression and subsequent BAMBI degradation, therewith representing some kind of feedforward loop. We show that a parallel activation of both TGF-β1 and LPS pathways, which is frequently the case in chronic liver disease situations, leads to superinduction of miR-942 expression, BAMBI depletion and ostensibly HSC-mediated fibrogenesis. Of note, miR-942 expression is increased and BAMBI expression is decreased with progression of HBV-mediated human liver fibrosis. It would be interesting to find out if the effects of miR-942 in cancer and HCV-mediated cell death are as well mechanistically linked to BAMBI/TGF-β1 family signaling.
Clinically, our study suggests that miR-942 could serve as a biomarker that could indicate the degree of liver fibrosis. Further investigations are needed to validate this finding. For example, the presence and level of miR-942 in blood of liver fibrosis patients as compared to controls need to be determined. It will also be interesting to determine if miR-942 can be found in exosomes, using supernatants of cultured and LPS-stimulated HSCs. This could pave way for findings of high diagnostic relevance in liver fibrosis.
Supplementary Material
Acknowledgements
This work was mainly supported by The National Natural Science Foundation of China (No. 81673788 to C. Liu), Peak discipline of Colleges and universities in Shanghai (No. A-U151902 to C Liu), and Research Project of Putuo Hospital Fund (No. 2016204B to C Liu & No. 2017207A to WT M). Federal Ministry of Education and Research grant ‘LiSyM’ (Grant PTJ-FKZ: 031L0043 to S Dooley). National Institutes of Health Grant (No. R01DK085252, R21AA025841 to E. Seki) and Winnick Research award from Cedars-Sinai Medical Center (to E. Seki). Xinglin Scholar of Shanghai University of Traditional Chinese Medicine (to C Liu) and Shanghai Municipal Commission of Health and Family Plan Fund (No. 20144Y0185 to L Wu). We thank Lingzhi Lian and honghui Wen (Department of Pathology, Putuo Hospital) for technical support with fibrotic stage analysis.
Abbreviations
- α-SMA
α-Smooth muscle actin
- BAMBI
BMP and activin membrane-bound inhibitor
- BSA
Bovine serum albumin
- ChIP
Chromatin immunoprecipitation
- ECM
Extracellular matrix
- HSC
Hepatic stellate cell
- hHSC
Primary human HSC
- KCs
Kupffer cells
- LPS
Lipopolysaccharide
- LSECs
Liver sinusoidal endothelial cells
- mRNA
Messenger RNA
- PBS
Phosphate-buffered saline
- TGFβ
Transforming growth factor β
- UTR
Untranslated regions
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00204-018-2278-9) contains supplementary material, which is available to authorized users.
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297 [DOI] [PubMed] [Google Scholar]
- Bandiera S, Pfeffer S, Baumert TF, Zeisel MB (2015) miR-122—a key factor and therapeutic target in liver disease. J Hepatol 62(2):448–457. 10.1016/j.jhep.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Desmet V, Gerber M, Hoofnagle J, Manns M, Scheuer P (1994) Classification of chronic hepatitis: diagnosis, rading and staging. Hepatology 6(19):1513–1520 [PubMed] [Google Scholar]
- Dooley S, ten Dijke P (2012) TGF-beta in progression of liver disease. Cell Tissue Res 347(1):245–256. 10.1007/s00441-011-1246-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL (2007) A deer in the headlights: BAMBI meets liver fibrosis. Nat Med 13(11):1281–1282. 10.1038/nm1107-1281 [DOI] [PubMed] [Google Scholar]
- Goodman ZD (2007) Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J Hepatol 47(4):598–607. 10.1016/j.jhep.2007.07.006 [DOI] [PubMed] [Google Scholar]
- Hyun J, Wang S, Kim J et al. (2016) MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun 7:10993 10.1038/ncomms10993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JH, Lee HJ, Kim W, Kim SG (2016) Endoplasmic reticulum stress in hepatic stellate cells promotes liver fibrosis via PERK-mediated degradation of hnrnpa1 and up-regulation of SMAD2. Gastroenterology 150(1):181–193. 10.1053/j.gastro.2015.09.039 [DOI] [PubMed] [Google Scholar]
- Lakner AM, Steuerwald NM, Walling TL et al. (2012) Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology 56(1):300–310. 10.1002/hep.25613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg K (2016) Autoimmunity: a controlled performance by BAMBI. Nat Rev Rheumatol 12(2):72 10.1038/nrrheum.2016.5 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen X, Yang L, Kisseleva T, Brenner DA, Seki E (2014a) Transcriptional repression of the transforming growth factor beta (TGF-beta) Pseudoreceptor BMP and activin membrane-bound inhibitor (BAMBI) by nuclear factor kappaB (NF-kappaB) p50 enhances TGF-beta signaling in hepatic stellate cells. J Biol Chem 289(10):7082–7091. 10.1074/jbc.M113.543769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zuo C, Wang X et al. (2014b) miR-942 decreases TRAIL-induced apoptosis through ISG12a downregulation and is regulated by AKT. Oncotarget 5(13):4959–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J (2012) TGFbeta signalling in context. Nat Rev Mol Cell Biol 13(10):616–630. 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K, De Giorgi V, Schechterly C et al. (2016) Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis C. Hepatology 64(3):732–745. 10.1002/hep.28660/suppinfo [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Salvador E, Mann J (2017) Epigenetics and liver fibrosis. Cell Mol Gastroenterol Hepatol 4(1):125–134. 10.1016/j.jcmgh.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwosu ZC, Alborzinia H, Wolfl S, Dooley S, Liu Y (2016) Evolving insights on metabolism, autophagy, and epigenetics in liver myofibroblasts. Front Physiol 7:191 10.3389/fphys.2016.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Enomoto M, Fujii H et al. (2012) MicroRNA-22½22 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut 61(11):1600–1609. 10.1136/gutjnl-2011-300717 [DOI] [PubMed] [Google Scholar]
- Roderburg C, Urban GW, Bettermann K et al. (2011) Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 53(1):209–218. 10.1002/hep.23922 [DOI] [PubMed] [Google Scholar]
- Seki E, De Minicis S, Osterreicher CH et al. (2007) TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med 13(11):1324–1332. 10.1038/nm1663 [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao J, Zhang X et al. (2017) Activated hepatic stellate cells impair NK cell anti-fibrosis capacity through a TGF-beta-dependent emperipolesis in HBV cirrhotic patients. Sci Rep 7:44544 10.1038/srep44544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sombetzki M, Loebermann M, EC R (2015) Vector-mediated micro-RNA-21 silencing ameliorates granulomatous liver fibrosis in Schistosoma japonicum infection. Hepatology 61(6):1787–1789 [DOI] [PubMed] [Google Scholar]
- Wang XW, Heegaard NH, Orum H (2012) MicroRNAs in liver disease. Gastroenterology 142(7):1431–1443. 10.1053/j.gastro.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ng R, Chen X, Steer CJ, Song G (2016) MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut 65(11):1850–1860. 10.1136/gutjnl-2014308430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Meng X, Xue B, Liu N, Wang X, Zhu H (2014a) MiR-942 mediates hepatitis C virus-induced apoptosis via regulation of ISG12a. PLoS One 9(4):e94501 10.1371/journal.pone.0094501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Tao H, Hu W et al. (2014b) MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis. Cell Signal 26(11):2381–2389. 10.1016/j.cellsig.2014.07.016 [DOI] [PubMed] [Google Scholar]
- Yang JJ, Tao H, Deng ZY, Lu C, Li J (2015) Non-coding RNA-mediated epigenetic regulation of liver fibrosis. Metab Clin Exp 64(11):1386–1394. 10.1016/j.metabol.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Yu X, Lan P, Hou X et al. (2017) HBV inhibits LPS-induced NLRP3 inflammasome activation and IL-1beta production via suppressing the NF-kappaB pathway and ROS production. J Hepatol 66(4):693–702. 10.1016/j.jhep.2016.12.018 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ghazwani M, Li J et al. (2014) MiR-29b inhibits collagen maturation in hepatic stellate cells through down-regulating the expression of HSP47 and lysyl oxidase. Biochem Biophys Res Commun 446(4):940–944. 10.1016/j.bbrc.2014.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.